Abstract
A basic question in adeno-associated virus (AAV) biology has been whether adenovirus (Ad) infection provided any function which directly promoted replication of AAV DNA. Previously in vitro assays for AAV DNA replication, using linear duplex AAV DNA as the template, uninfected or Ad-infected HeLa cell extracts, and exogenous AAV Rep protein, demonstrated that Ad infection provides a direct helper effect for AAV DNA replication. It was shown that the nature of this helper effect was to increase the processivity of AAV DNA replication. Left unanswered was the question of whether this effect was the result of cellular factors whose activity was enhanced by Ad infection or was the result of direct participation of Ad proteins in AAV DNA replication. In this report, we show that in the in vitro assay, enhancement of processivity occurs with the addition of either the Ad DNA-binding protein (Ad-DBP) or the human single-stranded DNA-binding protein (replication protein A [RPA]). Clearly Ad-DBP is present after Ad infection but not before, whereas the cellular level of RPA is not apparently affected by Ad infection. However, we have not measured possible modifications of RPA which might occur after Ad infection and affect AAV DNA replication. When the substrate for replication was an AAV genome inserted into a plasmid vector, RPA was not an effective substitute for Ad-DBP. Extracts supplemented with Ad-DBP preferentially replicated AAV sequences rather than adjacent vector sequences; in contrast, extracts supplemented with RPA preferentially replicated vector sequences.
A central feature of adeno-associated virus (AAV) biology is that productive infection in cell culture requires coinfection by a helper virus (either adenovirus [Ad] or herpesvirus) (reviewed in reference 3). The requirement for Ad or herpesvirus is not absolute, however, since treatment of cells with genotoxic agents will render the cells permissive for the production of small amounts of AAV (51–53). Presumably, therefore, Ad and herpesvirus do not provide any unique functions which cannot be provided, under some conditions, by the cell infected with AAV alone. AAV gene expression is enhanced by Ad infection (39), and open reading frame 6 of the E4 region of Ad is important for the conversion of the single-stranded AAV genome into a double-stranded form which is the substrate for subsequent steps in DNA replication (12a, 12b). It has been unknown whether Ad infection makes a further, direct contribution to AAV DNA replication.
Ni et al. (34) developed an in vitro AAV DNA replication assay in which a double-stranded AAV substrate with both ends in a closed hairpin configuration replicated in an extract from Ad-infected cells which had been supplemented with the AAV Rep protein. They saw little or no replication in an extract from uninfected HeLa cells. We have reported an assay using open-ended linear duplex DNA in which an extract from uninfected HeLa cells supplemented with Rep protein did replicate AAV DNA (47). However, if an extract from Ad-infected cells was substituted for the uninfected cell extract, full-length replication was substantially enhanced and there was significantly less defective product (46). There are two conclusions from these reports. The first is that if the need for AAV gene expression and the conversion of single-stranded to double-stranded DNA are bypassed, AAV DNA replication can occur in vitro. The second conclusion is that Ad infection makes an additional direct contribution to AAV DNA replication which substantially increases the production of full-length AAV DNA. We have further demonstrated that the difference between the two extracts was that the Ad-infected extract provided a helper function related to elongation during replication (46).
This enhanced elongation could be from either stimulation of cellular factors or the direct contribution of an Ad-encoded protein. With respect to the latter possibility, it has been shown previously, in vivo, that among the Ad proteins required for Ad DNA replication, the DNA polymerase and the terminal protein make no contribution to AAV DNA replication. The data on the Ad DNA-binding protein (Ad-DBP) have been ambiguous (reviewed in reference 5). AAV DNA replication is thought to occur by a single-stranded displacement mechanism. The AAV genome contains an inverted terminal repeat which can form a hairpin and thereby serve as the primer to initiate synthesis. At the end of each round of replication, the newly made strand hairpins on itself to initiate a subsequent round of replication (reviewed in reference 3). Since the viral genome is single stranded, the first round of replication produces a matching second strand but all subsequent rounds involve the genome-length displacement of the nontemplate strand. This is a feature not shared by the replication mechanism of the host cell, which is thought to involve simultaneous replication of both strands. In our investigation of the failure of processivity in in vitro AAV DNA replication in extracts from uninfected cells, it appeared that the elongating strands were dissociating prematurely from the template followed by template strand switch to the displaced strand (46). Ad DNA replication also occurs by a single-stranded displacement mechanism (reviewed in reference 41). Thus, in its requirement to maintain an extensive length of displaced single-stranded DNA in solution, AAV DNA replication shares a basic similarity with Ad DNA replication. It seems possible that the direct helper effect of Ad may be related to this common feature.
In this report, we show that the component of the Ad-infected cell extract which supports processive replication of AAV DNA is Ad-DBP, which has been shown to be necessary for the processive replication of Ad DNA (25). Additionally we show that addition of excessive amounts of the human single-stranded DNA-binding protein (replication protein A [RPA]) to an extract from uninfected cells can also support processive replication. Presumably it is RPA which supports AAV DNA replication in the absence of Ad-DBP.
MATERIALS AND METHODS
Cell extracts.
Replication extracts from uninfected HeLa cells and from HeLa cells infected with Ad were prepared as described previously (16, 45) in a modification of a procedure originally described by Wobbe et al. (50). Whole-cell lysates were made by spinning down cells and freezing, thawing, then mixing them with an equal volume of buffer L (100 mM Tris [pH 6.8], 200 mM dithiothreitol, 4% sodium dodecyl sulfate, 0.2% bromophenol blue, 20% glycerol, 200 μg of phenylmethylsulfonyl fluoride per ml). The mixture was boiled for 5 min and spun for 10 min at 12,000 × g, and the supernatant was analyzed by polyacrylamide gel electrophoresis and Western blotting.
Proteins.
Ad-DBP purified from infected cells was a kind gift of G. Droguette and M. Horwitz. Ad-DBP made in a baculovirus expression system was a generous gift of R. Hay (29). Escherichia coli single-stranded DNA-binding protein (SSB) (48) and human proliferating cell nuclear antigen (3,000 U/mg of protein [17]) were purified as described previously. RPA was purified as described previously (19) both from HeLa cells and from E. coli expressing p11d-tRPA (15). DNA polymerase delta (phosphocellulose pool, 40 U/mg of protein; heparin-Sepharose pool, 850 U/mg of protein) and DNA replication factor C (phosphocellulose pool, 320 U/mg of protein; final fraction, 5,400 U/mg of protein) were purified by a modification of the method of Lee et al. (23).
In vitro DNA replication.
In vitro DNA replication and analysis of radioactively labeled replication products by gel electrophoresis were performed as described previously (47). Reactions were performed with approximately 100 μg of cellular protein except where noted otherwise. Substrate was a BglII digest of plasmid pAV2, except in one experiment where it was undigested pAV2, as noted. pAV2 has been described elsewhere (22). It consists of the entire genome of AAV2 inserted into a pBR322 derivative by means of BglII linkers. AAV Rep68 was expressed in and purified from E. coli either as a maltose-binding protein–Rep fusion protein (8) or as a His-tagged Rep68 fusion (consisting of the entire Rep68 peptide sequence with six histidine residues fused to the amino terminus [23a]). PhosphorImager (Molecular Dynamics) scanning of dried gels was performed with ImageQuant version 3.0 software.
Antibodies.
Monoclonal antibody (MAb) 37-3, a MAb to Ad-DBP, was a generous gift of Doug Brough (9). MAb RPA34-19 was purchased from Oncogene Research Products (catalog no. NA18).
Immunodepletions.
Equal volumes of cellular extract (20 mg/ml) and MAb 37-3 (0.9 mg/ml in 10 mM NaPO4 with 0.02% sodium azide) were mixed. The mixture was made 50 mM in NaCl and shaken at 0°C for 1 h. Protein G-agarose beads (2.5 mg of protein per ml; Calbiochem catalog no. 539207) were added to 10% of the volume. The mixture was shaken at 0°C for an additional hour and spun for 3 min at 12,000 × g, and the supernatant was brought to 10% glycerol and stored at −80°C.
Western blots.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 12% gels and transferred to nitrocellulose. Filters were blocked by incubation in 50 mM Tris (pH 7.6)–0.2 M NaCl–5% Carnation nonfat dry milk–0.05% Tween 20 and then incubated with 10 μg of MAb RPA34-19 in 10 ml of the same buffer. The second antibody was anti-mouse immunoglobulin G (IgG) conjugated to alkaline phosphatase (Sigma A-4312) used at 1:10,000 dilution.
RESULTS
Supplementation of AAV DNA replication in uninfected cell extracts with extracts from Ad-infected cells.
Previously we had shown that extracts from Ad-infected cells were able to replicate AAV DNA with much greater processivity than extracts from uninfected cells. To identify the components of the Ad extract responsible for this helper activity, extracts from Ad-infected cells were mixed with extracts from uninfected cells. As the ratio of uninfected to infected extract was increased, there was an increase in full-length replication (Fig. 1). Addition of increasing amounts of the extract from Ad-infected cells led to a geometric increase in synthesis of full-length DNA, which was suggestive of either an inhibitor in the uninfected extract or a positive factor(s) in the extract from Ad-infected cells which acts cooperatively. An inhibitor seemed less likely because addition of uninfected cell extract to a final fraction of 0.25 did not decrease the level of replication below that seen using extracts from Ad-infected cells (data not shown). This result suggested it would be reasonable to search for helper functions by adding purified known replication components or fractionated Ad extract to the uninfected cell extract and assaying for an increase in full-length replication.
FIG. 1.
Mixing extracts from uninfected and Ad-infected cells. Reactions were performed and analyzed as described in Materials and Methods. DNA replications were performed with various amounts of extract protein from either Ad-infected or uninfected cells. Shown are relative amounts of protein from each extract versus relative amounts of incorporation into full-length DNA.
Supplementation of AAV DNA replication in uninfected cell extracts with purified human DNA replication proteins.
Since the only nonstructural proteins supplied by AAV are the Rep proteins, AAV DNA replication must involve cellular and/or adenovirus replication proteins. AAV DNA replication is thought to proceed by a single-stranded displacement mechanism (reviewed in reference 3). The major cellular proteins which participate in the leading-strand component of cellular DNA synthesis (which may be similar to replication by single-stranded displacement) have been identified and cloned. Purified RPA, replication factor C, proliferating cell nuclear antigen and polymerase delta were added to an extract from uninfected HeLa cells in the replication assay (Fig. 2A). Reactions were performed as described in Materials and Methods, using linear duplex AAV DNA as the template and 20 μg of extract from uninfected cells. A substantial enhancement was seen only in reactions to which RPA (also known as human single-stranded DNA-binding protein) had been added (lanes 6 and 7). The enhancement with the protein which had been produced in an E. coli expression system was indistinguishable from the enhancement using RPA which had been purified from HeLa cells. Interestingly, E. coli single-stranded binding protein (SSB) (lane 5) gave only a very slight enhancement of full-length replication. Assays performed with suboptimal amounts of added RPA which were then supplemented with added E. coli SSB also demonstrated the inability of E. coli SSB to enhance replication in this system (data not shown).
FIG. 2.
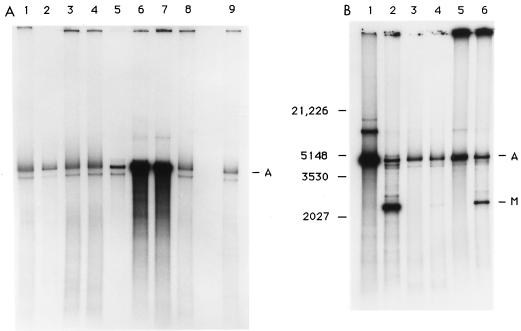
(A) Replication of AAV DNA in an uninfected extract to which purified human DNA replication proteins were added. Reactions were performed as described in Materials and Methods, using an extract from uninfected HeLa cells except that only 20 μg of cellular protein was included in the reaction. Reactions were supplemented with various replication proteins: lane 1, 0.04 μg of replication factor C (final pool); lane 2, 1.0 μg of replication factor C (phosphocellulose pool); lane 3, 0.2 μg of polymerase delta (heparin-Sepharose pool); lane 4, 1.25 μg of polymerase delta (phosphocellulose pool); lane 5, 0.8 μg of E. coli SSB; lane 6, 1.0 μg of RPA expresseed in E. coli; lane 7, 0.25 μg of RPA from HeLa cells; lane 8, 0.1 μg of proliferating cell nuclear antigen; lane 9, no added protein. (B) MboI susceptibility of replication products. Lanes 1 and 2, ad-infected cell extract; lanes 3 and 4, uninfected cell extract; lanes 5 and 6, uninfected cell extract supplemented with RPA; lanes 1, 3, and 5, undigested replication products; lanes 2, 4, and 6, reaction products digested with MboI. A, position of full-length duplex AAV; M, position of the largest MboI digestion product of AAV. Size markers shown on the left in nucleotides are from a HindIII and EcoRI digestion of lambda DNA.
Previously we had shown that the fundamental block in replication with extracts from uninfected cells involved processivity and that replication was much more processive when an Ad-infected extract was used. If replication can proceed completely through two rounds, the duplex AAV molecule will contain two unmethylated strands and therefore be digestible by MboI. As shown previously, replication with an extract from Ad-infected cells leads to the production of labeled DNA which is largely MboI susceptible, whereas replication with an extract from uninfected cells leads to the production of only a small amount of labeled DNA which is MboI susceptible (Fig. 2B). Replication in an extract from uninfected cells supplemented with RPA leads to an increase in MboI-susceptible full-length DNA compared to the unsupplemented, uninfected extract, demonstrating an increase in processivity with the addition of RPA.
In contrast, when RPA was added to an extract from Ad-infected cells, no enhancement of replication was seen (data not shown). Rather, there was a slight decrease (30%) in full-length replication.
Supplementation of AAV DNA replication in uninfected extracts with Ad-DBP.
Ad codes for a single-stranded DNA-binding protein (Ad-DBP) which has been shown to play an essential role in the processivity of Ad DNA replication (25). Because both Ad and AAV apparently replicate by a single-stranded displacement mechanism, it seemed possible that Ad-DBP might be playing a similar role in AAV replication in Ad-infected cells. To test this possibility, the standard replication reaction was performed with and without the addition of Ad-DBP purified from Ad-infected cells. Addition of Ad-DBP led to an increase in full-length product (Fig. 3A; compare lanes 1 and 2 with lane 3). To ensure that the increased replication was due to the Ad-DBP and not to another protein found in Ad-infected cells which might be copurified with Ad-DBP, we repeated the assay using purified Ad-DBP made in a baculovirus expression system. The baculovirus protein also gave increased replication of full-length AAV substrate (Fig. 3B). The effect was significantly better when 1.5 μg was used rather than 0.75 μg (a 7.2-fold increase over the unsupplemented extract, compared with a 2.3-fold increase). This may reflect the cooperative nature of the enhancement suggested by the extract mixing experiment shown in Fig. 1. In addition, Fig. 3B shows a decrease in the intensity of the smear of labeled DNA of approximately 300 to 1,000 nucleotides with the addition of AD-DBP. This is also the case when an Ad-infected cell extract is used in place of an uninfected cell extract. We previously characterized this heterogeneous collection of DNA as short inverted repeats which resulted from the lack of processivity in the in vitro AAV DNA replication system using extracts from uninfected cells (46). AAV DNA replicated in an Ad-DBP-supplemented extract from uninfected cells was also more susceptible to MboI than AAV DNA replicated in the unsupplemented extract (Fig. 3C).
FIG. 3.
Replication of AAV DNA in uninfected cell extracts supplemented with Ad-DBP. (A) Replication in an extract from uninfected cells supplemented with Ad-DBP purified from Ad-infected cells. Lane 1, 2.7 μg of Ad-DBP; lane 2, 1.3 μg of Ad-DBP, lane 3, unsupplemented. (B) Replication in an extract from uninfected cells supplemented with Ad-DBP made in a baculovirus expression system. Lane 1, 1.5 μg of Ad-DBP; lane 2, 0.75 μg of Ad-DBP; lane 3, unsupplemented. (C) MboI digest of replication products from assays performed in an extract from uninfected cells supplemented either with RPA or Ad-DBP. Lane 1, unsupplemented; lane 2, 1.0 μg of RPA; lane 3, 1.5 μg of Ad-DBP from a baculovirus expression system. M designates the largest MboI digestion product of AAV. Bands labeled A and B are observed if protein is not extracted from the reaction; both represent full-length AAV DNA, but presumably of alternative structures, as described previously (46).
Immunodepletion of extracts from Ad-infected cells with an antibody to the Ad-DBP.
We have demonstrated three ways in which the replication in vitro of full-length duplex AAV DNA in an extract from uninfected HeLa cells can be enhanced: supplementation with an extract from Ad-infected cells, supplementation with RPA, and supplementation with Ad-DBP. Next we wanted to determine whether the enhanced replication seen with the Ad-infected extract was at least partly attributable to one or both of the two DNA-binding proteins.
Initially we immunodepleted Ad-DBP from the Ad-infected extract by using a MAb to the N-terminal portion of Ad-DBP (MAb 37-3). The results are shown in Fig. 4A. Lanes 1 and 2 show the effects of depleting and mock depleting an extract from uninfected cells, respectively. The MAb actually enhanced replication in the uninfected extract. In the extract from Ad-infected cells, however, the immunodepletion (lane 3) substantially reduced replication of the AAV substrate compared with mock depletion (lane 4) (70% reduction). To ensure that MAb 37-3 was not depleting or in some other way interfering with the activity of RPA, we repeated the experiment with an extract from uninfected cells to which RPA had been added before the immunodepletion. A comparison of the effect of depletion on the supplemented extract with the effect of depletion on the extract from Ad-infected cells is shown in Fig. 4B. Lanes 1 and 2 show replication in the mock-depleted and the immunodepleted supplemented extracts from uninfected cells, respectively. Immunodepletion with MAb 37-3 did not decrease replication with the supplemented extract, just as it did not decrease replication with the unsupplemented extract. In fact, replication is 16% higher in the immunodepleted extract. This result shows that immunodepletion with MAb 37-3 does not interfere with the activity of RPA and also does not interfere with some component of the replication machinery which is functional only when activity is more substantial than is seen with unsupplemented uninfected cell extract. Immunodepletion of the extract from Ad-infected cells (lane 4) again leads to a substantial reduction in AAV DNA replication (84% reduction) compared to the mock-depleted extract (lane 3).
FIG. 4.
(A) Replication performed with extracts immunodepleted with MAb 37-3. Lanes 1 and 2, extracts from uninfected cells; lanes 3 and 4, extracts from Ad-infected cells; lanes 1 and 3, extracts which had been immunodepleted with MAb 37-3; lanes 2 and 4, extracts which had been mock depleted. (B) Replications performed with extracts from Ad-infected cells and extracts from uninfected cells supplemented with RPA. Lanes 1 and 2, extracts from uninfected cells which had been supplemented with RPA prior to immunodepletion; lanes 3 and 4, extracts from Ad-infected cells; lanes 1 and 3, mock depletion; lanes 2 and 4, immunodepletion with MAb 37-3. All reactions were performed with approximately 50 μg of cellular protein. A and B designate two forms of full-length AAV.
We saw an increase in replication whenever we added the antibody to assays in which the extract from uninfected cells was used and believe that it is due to a slight nonspecific enhancement from some component of the antibody buffer.
Western blots of the extracts performed with MAb 37-3 showed the presence of substantial amounts of the Ad-DBP in the extract from Ad-infected cells but no detectable Ad-DBP in the extract from the uninfected cells (data not shown).
Supplementation of immunodepleted extracts.
To ensure that the reduced replication seen with immunodepleted extract was due to a depletion only of a DNA-binding protein activity and to further ensure that the DNA-binding protein which has been depleted was Ad-DBP and not RPA, the following was done. Figure 5A shows one series of assays performed with the mock-depleted (lane 1) and depleted extracts from Ad-infected cells (lanes 2 to 4). Lane 2 shows the results of replication with the depleted extract alone. Lanes 3 and 4 show the results of replication with the depleted extract which had been supplemented with 1.5 μg of Ad-DBP made in the baculovirus expression system and 1.0 μg of RPA made in the E. coli expression system, respectively. Immunodepletion reduced replication to 25% of the level of the undepleted extract. Addition of the Ad-DBP and RPA restored replication to 51 and 90%, respectively, of the undepleted level. In the case of RPA, replication was restored to almost the same level as before immunodepletion, but restoration was not as complete with Ad-DBP. We have noted that in this assay, if the amounts of Rep protein and substrate DNA are kept constant, but the amount of cellular protein is reduced as was the case in this assay, the ability of Ad-DBP to supplement AAV replication is substantially reduced for unknown reasons (data not shown). We think that this is why added Ad-DBP does not restore replication as completely as does added RPA. In contrast, Fig. 5B shows a replication assay performed with the mock-depleted extract from Ad-infected cells alone and with the mock-depleted extract supplemented with 1.0 μg of RPA. In the case of the mock-depleted extract, the addition of the DNA-binding protein gave no enhancement of replication. These results demonstrate that it was the depletion of a DNA-binding protein activity which gave reduced replication in the immunodepletion experiment.
FIG. 5.
(A) Supplementation of immunodepleted extracts. Replication assays of AAV were performed as described in Materials and Methods in a mock-depleted extract from Ad-infected cells (lane 1) and an MAb 37-3-depleted extract from Ad-infected cells (lanes 2 to 4). After depletion, extracts were unsupplemented (lanes 1 and 2) or supplemented with 1.5 μg of Ad-DBP (lane 3) or 1.0 μg of RPA (lane 4). (B) Supplementation of a mock-depleted extract. Replication of AAV was performed as described in Materials and Methods in a mock-depleted extract from Ad-infected cells. Lane 1, extract supplemented with RPA after mock depletion; lane 2, extract not supplemented. (C) Western blot of depleted and mock-depleted extracts from Ad-infected cells with an antibody to the 34-kDa subunit of RPA. Lane 1, depleted extract; lane 2, mock-depleted extract. Size markers, shown on the right, are in kilodaltons. Additional bands in lane 1 are residual antibody from the immunodepletion, detected by the secondary antibody in the detection system (goat anti-mouse IgG).
To further ensure that immunodepletion by MAb 37-3 did not inadvertently bring down RPA, a Western blot was performed with the mock-depleted and immunodepleted extract, using MAb RPA34-19, a MAb which recognizes the 34-kDa subunit of RPA. From the Western blot shown in Fig. 5C, it is apparent that the 34-kDa subunit of RPA has not been decreased in quantity by immunodepletion with MAb 37-3. The two additional bands seen in the depleted lane are presumably from MAb 37-3, which failed to bind the protein G beads in the immunodepletion. Since these are mouse antibodies, they were detected by the detection system which made use of an alkaline phosphatase-conjugated goat antibody to mouse IgG.
Ad infection does not induce higher levels of RPA in infected cells.
The data that we have presented show that Ad-DBP is responsible for at least a significant portion of the higher levels of replication seen in extracts from Ad-infected cells. However, increased levels of RPA might still be partly responsible for the higher levels of replication. Either Ad infection could induce expression of RPA or Ad infection could alter the nuclear membrane, leading to the extraction of higher percentages of RPA in infected cells than in an extract from uninfected cells. To test these possibilities, we performed Western blotting on the replication extracts made from infected and uninfected cells and used in all of the above-described assays (Fig. 6A) and on a whole-cell lysate made from infected and uninfected cells. In comparing equal amounts of protein, MAb RPA34-19 detected no increase in RPA in infected cells. Apparently Ad infection neither induces higher levels of RPA nor alters the physiology of the cell such that RPA becomes more easily extractable at the 28-h time point at which these extracts were made.
FIG. 6.
Western blots comparing levels of RPA from Ad-infected and uninfected cells. (A) Western blot of extracts used for replication assays. Lane 1, extract from uninfected cells; lane 2, extract from Ad-infected cells. (B) Western blot of whole-cell lysate. Lane 1, lysate of Ad-infected cells; lane 2, lysate of uninfected cells. Size markers, shown on the right, are in kilodaltons.
Ad-DBP enhances AAV DNA replication in the context of a plasmid substrate more efficiently than does RPA.
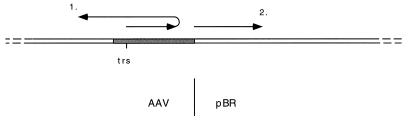
In vitro AAV DNA replication using either RPA or Ad-DBP was substantially equivalent in an assay measuring replication of linear duplex AAV. We have previously reported a replication assay in which the substrate is a plasmid in which the AAV genome is inserted into a plasmid vector (47). Rep-dependent replication in this assay involves rescue of the AAV sequences from the plasmid backbone as well as replication, and we think that this combined rescue-replication is a useful model for rescue-replication of the chromosomally integrated AAV genome. It was noted that in this assay in which replication initiated at the AAV origin in the context of a circular plasmid, replication of pBR sequences rather than AAV sequences often occurred. This is not so surprising since the initial direction of replication from a nick at the AAV origin is necessarily outward through the inverted terminal repeat toward the contiguous vector sequences. In order for AAV sequences, rather than pBR sequences, to be replicated, the replication complex must reverse direction at the AAV/pBR boundary, perhaps by template strand switching, for which we previously proposed a model (47). Figure 7 is a schematic illustrating the two possible directions for replication initiating at the AAV terminal resolution site when the AAV genome is inserted into a plasmid vector.
FIG. 7.
Schematic showing initiation of replication at the terminal resolution site (trs) of AAV when the AAV genome has been inserted into a plasmid vector. Replication can be either back into AAV sequences (1) or forward into vector sequences (2). The shaded region denotes the 145-base inverted terminal repeat of AAV DNA.
When the intact plasmid pAV2 was used as the substrate for in vitro AAV DNA replication, there was a fundamental difference between extracts from uninfected cells and extracts from Ad-infected cells. Whereas extracts from uninfected cells replicated pBR about as frequently as AAV, extracts from Ad-infected cells preferentially replicated AAV. We compared the effects of adding Ad-DBP or RPA to an uninfected extract to those obtained with unsupplemented uninfected and Ad-infected extracts when the substrate was the intact plasmid pAV2 (Table 1). As expected, the extract from Ad-infected cells gave a higher ratio of AAV to pBR replication than did the extract from uninfected cells. As was found with the assay using duplex AAV DNA as the substrate, both Ad-DBP and RPA induced a substantial increase in replication. With the addition of Ad-DBP to an assay using an intact plasmid as the substrate, there was an AAV-to-pBR ratio which was similar to that from the Ad-infected cell extract; however, the addition of RPA gave the opposite result. Addition of RPA to the extract from uninfected cells gave an AAV-to-pBR ratio which was substantially lower than the ratio for the uninfected extract alone.
TABLE 1.
Relative incorporation into AAV and pBR sequences in replication reactions using intact pAV2 as the substrate
| Sequence | Incorporation into full-length rescued AAV and pBRa
|
|||
|---|---|---|---|---|
| Uninfected | Uninfected + RPA | Uninfected + Ad-DBP | Ad infected | |
| AAV | 1.0 | 1.0 | 1.0 | 1.0 |
| pBR | 0.90 | 2.40 | 0.52 | 0.57 |
Measured with a PhosphorImager, with incorporation into AAV normalized to 1.0 in each case. Reactions were performed and analyzed as described in Materials and Methods except that undigested pAV2 was used as the substrate.
DISCUSSION
A basic question in AAV biology has been the nature of the helper effect provided by Ad in productive AAV replication, and a component of this question has been whether Ad provided any function which directly promoted replication of AAV DNA. Previously we demonstrated that full-length replication of an open-ended, duplex AAV DNA substrate was 50-fold better in extracts made from Ad-infected cells supplemented with the AAV Rep protein than in Rep-supplemented extracts made from uninfected cells. A closer examination of replication in the two extracts demonstrated that initiation in each was approximately equivalent but that in the extract from uninfected cells, most replication events led to the dissociation of the elongating strand from the template strand after a few hundred bases. The goal of the studies described in this report was to determine whether the enhancement of processivity was the result of cellular factors whose activity was stimulated by Ad infection or was the result of an Ad protein(s) participating directly in AAV DNA replication.
The data presented demonstrate that the addition of either the human RPA or the Ad-DBP to an extract from uninfected cells supports enhanced AAV DNA replication. The enhancement in both cases was associated with an increase in processivity as demonstrated by MboI susceptibility and the disappearance of short replication products characteristic of displacement of the elongating strand from the template. Previously we had shown that replication of short substrate molecules in extracts from uninfected cells gave almost as much full-length product as replication of short substrates in extracts from Ad-infected cells, which was consistent with the hypothesis that the difference between the two extracts reflected a difference in processivity. In agreement with these results, while RPA and Ad-DBP each increased full-length replication of the short substrates, they did so to a much lesser extent than for longer wild-type-length substrates (data not shown). To show that the abundant single-stranded binding activity which promotes more efficient replication was already present in the extract from Ad-infected cells, we added RPA to this extract and saw no enhancement. The finding that Ad DNA replication takes place in an environment of increased single-stranded binding activity and that AAV replication can be enhanced by supplying an excess of this activity is not an unexpected result. In normal cellular replication, the displaced strand is believed to be incorporated immediately into the lagging-strand replication complex, while in both Ad and AAV replication there presumably are significant lengths of displaced, single-stranded DNA.
One possible mechanism for enhancement of single-stranded binding activity upon Ad infection is an enhancement of cellular single-stranded binding activity. We looked for an increase in absolute amounts of RPA both in the replication extract and in whole-cell lysates of infected cells. By Western blot analysis, we saw no increase after Ad infection. Also, the relative amount of RPA compared to total protein was approximately the same in the replication extracts as in the total-cell lysate. Recently it has been shown (31) that the assembly of the intact RPA from its three components is a cell cycle-dependent phenomenon. Assembly of the complete RPA is necessary for RPA support of DNA replication (12, 15, 18). In addition, phosphorylation of RPA has been shown to be a cell cycle-dependent phenomenon (10, 11). The phosphorylation state of RPA may play a role in the modulation of DNA replication in cells (13). We have not measured what fraction of the components is assembled into the complete RPA or the phosphorylation status of the RPA in our extracts. We do not think, however, that an Ad-induced assembly or phosphorylation of RPA plays a significant role in AAV DNA replication in our assay, because the amount of added RPA which achieved stimulation comparable to maximally effective amounts of Ad-DBP or that seen in the infected cell extract was 10-fold that of RPA naturally present in the extracts. Another possibility for enhancement of single-stranded binding activity is Ad recruitment of RPA into replication centers within the nucleus. It has been shown that during cellular DNA replication, RPA is found concentrated in replication foci (1, 4, 21, 31). It has also been shown that during Ad DNA replication, Ad-DBP is concentrated at replication foci (30, 37, 44). This of course raises the available concentrations of these components in a way which is hard to mimic in a soluble replication system. Additionally, it is possible the Ad infection affects the level of activity of a single-stranded DNA-binding protein other than RPA. It has been noted that a second cellular single-stranded DNA-binding protein (PC4) is capable of playing a role in the simian virus 40 in vitro replication system (35).
The second possible mechanism for enhancement is Ad-DBP, which is synthesized early in infection and is present in large amounts. This protein has been shown to play several roles in Ad DNA replication (reviewed in references 14 and 43) and, in particular, is essential for the elongation step (25). When it was added to an extract from uninfected cells, there was a significant enhancement of AAV DNA replication in our assay. As with RPA, both the Ad-DBP purified from infected cells and that made in an expression system enhanced replication, making it unlikely that the effect is due to a second factor copurifying with Ad-DBP.
Immunodepletion of the infected cell extract with an antibody to Ad-DBP substantially reduced the ability of these extracts to support AAV DNA replication. The restoration of replication capacity by the addition of either Ad-DBP or RPA (both produced in expression systems) to the immunodepleted extract demonstrated that only a single-stranded binding activity had been depleted. The controls showing that RPA was unaffected by the immunodepletion demonstrated that whatever effect Ad infection might have on endogenous RPA activity, the primary single-stranded binding protein support for AAV DNA replication in the Ad-infected extract is supplied by Ad-DBP. It is interesting that Weitzman et al. (49) have shown that in Ad-AAV-coinfected cells, AAV DNA is recruited into Ad replication foci. In particular, they have shown colocalization between Ad-DBP and AAV DNA and between Ad-DBP and the AAV Rep protein. Although our data do not absolutely exclude a role for Ad-induced enhancement of endogenous single-stranded binding protein activity, the presence of large amounts of Ad-DBP in the infected cell (concentrated at the foci containing replicating AAV DNA) and its ability to support AAV DNA replication make it likely that this protein plays an important role in AAV DNA replication in the cell as we have shown that it does in the extract made from these cells.
We noted one substantial difference between the two proteins with regard to the ability to support in vitro AAV DNA replication. When the substrate for replication was not the isolated duplex form of the AAV genome but rather the AAV genome inserted into a plasmid vector, extracts from uninfected cells supplemented with RPA preferentially replicated adjacent vector sequences rather than AAV sequences. In contrast, when the same extract was supplemented instead with Ad-DBP, replication was now preferentially of AAV sequences. In this respect, supplementation of the uninfected extract with Ad-DBP gave replication quite similar to that seen with the extract from Ad-infected cells, while supplementation of the uninfected extract with RPA was quite dissimilar. This last result supports the tentative conclusion that Ad-DBP plays a primary role in AAV DNA replication and suggests that Ad-DBP may play an important role in the excision and replication of an integrated genome upon Ad infection of latently infected cells.
It is interesting to speculate on what might be the source of this difference. Previously (47) we suggested a model to explain how AAV replication turns back on itself in the plasmid context. In that model, when the replication complex passes through the inverted terminal repeat, the now separated DNA strands of the original template may fold up into a hairpin conformation, thereby displacing the newly synthesized strand and its associated replication complex. The newly made strand is now free to fold on itself and replicate back into AAV. This model suggests that replication back into AAV is dependent on self-base pairing within each strand and that replication with Ad-DBP allows self-pairing more readily than does replication with RPA.
There is an extensive and somewhat ambiguous literature on the possible role of Ad-DBP on AAV replication (reviewed in reference 5). It was reported that temperature-sensitive mutations in the E2A gene (which codes for Ad-DBP) caused two- to fivefold-reduced synthesis of AAV DNA when this Ad was used as the helper for AAV at the nonpermissive temperature (28). In contrast, other reports characterized the same mutants as efficient helpers (26, 42). Further investigation of the best-characterized temperature-sensitive E2A mutant (Adts125) seemed to show that synthesis of replicative-form AAV DNA was normal at the nonpermissive temperature, and the decrease in production of infectious particles was due only to other effects of the E2A mutation (27).
Two sets of more definitive experiments have been done more recently. Kitchingman and colleagues (33, 38) constructed a series of point mutants in Ad-DBP, including several targeted to the putative single-stranded DNA binding domain. When transfected into AAV-infected Cos cells, several of these mutants, especially those targeted to the putative DNA binding site, supported almost no synthesis of AAV DNA. In the second set of experiments, Carter et al., making use of a cell line which expresses the E2a gene (20), produced Ad which makes no detectable Ad-DBP. With this mutant used as a helper, AAV DNA replication was reduced severalfold (6). Both sets of experiments imply a role for Ad-DBP in AAV DNA replication. The much greater effect seen with several of the mutants in the first set of experiments can probably best be explained by hypothesizing that these mutants were demonstrating a dominant negative effect. There would of course be no such effect in the second set of experiments, since there was no Ad-DBP; in these experiments, the AAV DNA replication which was seen was most likely supported by the cell’s endogenous RPA. We think that our results are not inconsistent with the last two sets of experiments.
The requirement for single-stranded DNA-binding protein stimulation in this assay seems specific since E. coli SSB gave no enhancement. If the requirement is specific, it is unexpected that two proteins as dissimilar in sequence as the human and Ad single-stranded binding proteins both support replication. Earlier experiments with mutants in the Ad pol gene (reviewed in reference 5) demonstrated that a cellular polymerase replicated AAV DNA, and in this work the supplemented extracts were from uninfected cells; therefore, the replication machinery must have been cellular. It is consequently not surprising that the human protein RPA supported replication. What is somewhat unexpected is that Ad-DBP supports elongation in what is essentially a cellular replication system. Since Ad has its own polymerase and apparently does not utilize a helicase, there would be no selection for Ad-DBP to cooperate with the cellular replication components. NF I and NF III are cellular components involved in Ad DNA replication but only in initiation (32, 36), while the enhancement described in this report is due to elongation. Stimulation of polymerase delta replication by Ad-DBP has been observed previously, but in an assay in which all single-stranded DNA-binding proteins tested gave stimulation (18). Presumably, therefore, in that assay there was no specific protein-protein interaction.
A possibility for further investigation, therefore, is that with respect to the enhanced elongation that we have described, both Ad-DBP and RPA interact with an AAV protein which remains a component of the replication complex. The Rep protein might remain a continuous component of the replication complex, in its role as a helicase. It is reasonable that the AAV Rep protein might have become adapted to both cellular and Ad replication components. The possible role of the Rep protein as a helicase for AAV DNA replication is consistent with previous characterization of Rep as a helicase (16b) and with the data of this study which suggest that one of the components of the replication complex must be able to interact specifically with both Ad-DBP and RPA.
The conclusions of this work with regard to the role of single-stranded DNA-binding proteins in AAV DNA replication may help to reconcile two apparently paradoxical observations. Exposure of cells to ionizing radiation and other genotoxic agents renders them permissive for AAV DNA replication (without helper virus) (51–53). Similar treatment also enhances AAV vector transgene expression (2, 40). While this may in part be caused by an increased amount of double-stranded transcriptional template, it has also been suggested that prolonged transgene expression may be a consequence of enhanced vector integration (40). Perhaps DNA replication induced by radiation is, at least initially, less processive than that induced by Ad coinfection. As mentioned above, changes in the replicative capacity of cells have been associated with changes in the phosphorylation and assembly of RPA, and such changes can be induced by DNA-damaging agents. For example, it has been shown that UV light-induced DNA synthesis arrest is mediated at least in part through transient phosphorylation-related alterations in RPA which also render the protein temporarily nonfunctional in the simian virus 40 in vitro DNA replication assay (7). Replication in the context of an inactive RPA might involve frequent dissociation of the elongating strand, which could foster integration. It might be informative to investigate whether there is a correlation between those DNA-damaging and synthesis-inhibiting agents which give higher transduction efficiencies and those which change the functioning of RPA.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant GM50032.
We are greatly indebted to Douglas Brough for his kind gift of MAb 37-3 and to Gustavo Droguette, Marshall Horwitz, and Ron Hay for their kind gifts of the Ad-DBP. We also thank A. LeGall and A. Muesch for advice on the immunodepletion experiments; D. Brough, D. Klessig, J. Hurwitz, C. H. Young, E. Falck-Pedersen, R. M. Linden, and E. Winocur for helpful discussions; and N. Cortez for excellent technical assistance.
REFERENCES
- 1.Adachi Y, Laemmli U K. Identification of nuclear pre-replication centers for DNA synthesis in Xenopus egg extracts: immunolocalization study of replication protein A. J Cell Biol. 1992;119:1–15. doi: 10.1083/jcb.119.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander I E, Russell D W, Miller A D. DNA-damaging agents greatly increase the transduction of nondividing cells by adeno-associated virus vectors. J Virol. 1994;68:8282–8287. doi: 10.1128/jvi.68.12.8282-8287.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berns K I. Parvovirus replication. Microbiol Rev. 1990;54:316–329. doi: 10.1128/mr.54.3.316-329.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardoso M C, Leonhardt H, Nadal-Ginard B. Reversal of terminal differentiation and control of DNA replication: cyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell. 1993;74:979–992. doi: 10.1016/0092-8674(93)90721-2. [DOI] [PubMed] [Google Scholar]
- 5.Carter B J. Adeno associated helper functions. In: Tijssen P, editor. CRC handbook of parvoviruses. Boca Raton, Fla: CRC; 1990. pp. 255–282. [Google Scholar]
- 6.Carter B J, Antoni B A, Klessig D F. Adenovirus containing a deletion of the early region 2A gene allows growth of adeno-associated virus with decreased efficiency. Virology. 1992;191:473–476. doi: 10.1016/0042-6822(92)90213-9. [DOI] [PubMed] [Google Scholar]
- 7.Carty M P, Zernik-Kobak M, McGrath S, Dixon K. UV light induced DNA synthesis arrest in HeLa cells is associated with changes in phosphorylation of human single-stranded DNA-binding protein. EMBO J. 1994;13:2114–2123. doi: 10.1002/j.1460-2075.1994.tb06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiorini J A, Weitzman M D, Owens R A, Urcelay E, Safer B, Kotin R M. Biologically active Rep proteins of adeno-associated virus type 2 produced as fusion proteins in Escherichia coli. J Virol. 1994;68:797–804. doi: 10.1128/jvi.68.2.797-804.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cleghon V, Piderit A, Brough D E, Klessig D E. Phosphorylation of the adenovirus DNA-binding protein and epitope mapping of monoclonal antibodies against it. Virology. 1993;197:564–575. doi: 10.1006/viro.1993.1630. [DOI] [PubMed] [Google Scholar]
- 10.Din S-U, Brill S J, Fairman M P, Stillman B. Cell-cycle-regulated phosphorylation of DNA replication factor A from human and yeast cells. Genes Dev. 1990;4:968–977. doi: 10.1101/gad.4.6.968. [DOI] [PubMed] [Google Scholar]
- 11.Dutta A, Stillman B. cdc2 family kinases phosphorylate a human cell DNA replication factor, RPA, and activate DNA replication. EMBO J. 1992;11:2189–2199. doi: 10.1002/j.1460-2075.1992.tb05278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erdile L F, Heyer W-D, Kolodner R, Kelly T J. Characterization of a cDNA encoding the 70-kDa single stranded DNA-binding subunit of human replication protein A and the role of the protein in DNA replication. J Biol Chem. 1991;266:12090–12098. [PubMed] [Google Scholar]
- 12a.Ferrari F K, Samulski T, Shenk T, Samulski R J. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J Virol. 1996;70:3227–3234. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12b.Fisher K J, Gao G-P, Weitzman M D, DeMatteo R, Burda J F, Wilson J M. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading strand synthesis. J Virol. 1996;70:520–532. doi: 10.1128/jvi.70.1.520-532.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fotedar R, Roberts J M. Cell cycle regulated phosphorylation of RPA-32 occurs within the replication initiation complex. EMBO J. 1992;11:2177–2187. doi: 10.1002/j.1460-2075.1992.tb05277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hay R T, Freeman A, Leith I, Monaghan A, Webster A. Molecular interactions during adenovirus DNA replication. In: Doerfler W, Bohm P, editors. The molecular repertoire of adenoviruses II. Berlin, Germany: Springer-Verlag; 1995. pp. 31–48. [DOI] [PubMed] [Google Scholar]
- 15.Henricksen L A, Umbricht C B, Wold M S. Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 16.Hong G, Ward P, Berns K I. In vitro replication of adeno-associated virus DNA. Proc Natl Acad Sci USA. 1992;89:4673–4677. doi: 10.1073/pnas.89.10.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16b.Im D S, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 17.Kelman Z. Interactions between DNA polymerase and other cellular proteins. Ph.D. thesis. New York, N.Y: Cornell University Medical College; 1996. [Google Scholar]
- 18.Kenny M K, Lee S-H, Hurwitz J. Multiple functions of human single-stranded-DNA binding protein in simian virus 40 DNA replication: single-strand stabilization and stimulation of DNA polymerase alpha and delta. Proc Natl Acad Sci USA. 1989;86:9757–9761. doi: 10.1073/pnas.86.24.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenny M K, Schlegel U, Furneaux H, Hurwitz J. The role of human single-stranded DNA binding protein and its individual subunits in simian virus 40 DNA replication. J Biol Chem. 1990;265:7693–7700. [PubMed] [Google Scholar]
- 20.Klessig D F, Brough D E, Cleghon V. Introduction, stable integration, and controlled expression of a chimeric adenovirus gene whose product is toxic to the recipient human cell. Mol Cell Biol. 1984;4:1354–1362. doi: 10.1128/mcb.4.7.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krude T. Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp Cell Res. 1995;220:304–311. doi: 10.1006/excr.1995.1320. [DOI] [PubMed] [Google Scholar]
- 22.Laughlin C A, Tratschin J D, Coon H, Carter B J. Cloning of infectious adeno-associated virus genomes in bacterial plasmids. Gene. 1983;23:65–73. doi: 10.1016/0378-1119(83)90217-2. [DOI] [PubMed] [Google Scholar]
- 23.Lee S H, Kwong A D, Pan Z Q, Hurwitz J. Studies on the activator 1 protein complex, an accessory for proliferating cell nuclear antigen-dependent DNA polymerase delta. J Biol Chem. 1991;266:594–602. [PubMed] [Google Scholar]
- 23a.Linden, R. M., and K. I. Berns. Unpublished data.
- 24.Linden R M, Winocur E, Berns K I. The recombination signals for adeno-associated virus site-specific integration. Proc Natl Acad Sci USA. 1996;93:7966–7972. doi: 10.1073/pnas.93.15.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindenbaum J O, Field J, Hurwitz J. The adenovirus DNA binding protein and adenovirus DNA polymerase interact to catalyze elongation of primed DNA templates. J Biol Chem. 1986;261:10218–10277. [PubMed] [Google Scholar]
- 26.McPherson R A, Ginsberg H S, Rose J A. Adeno-associated virus helper activity of adenovirus DNA binding protein. J Virol. 1982;44:666–673. doi: 10.1128/jvi.44.2.666-673.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyers M, Carter B J. Adenoassociated virus replication: the effect of 1-canavanine or a helper mutation on accumulation of viral capsids and progeny single stranded DNA. J Biol Chem. 1981;256:567–570. [PubMed] [Google Scholar]
- 28.Meyers M, Laughlin C A, Jay F T, Carter B J. Adeno helper virus functions for growth of adeno-associated virus: effects of temperature sensitive mutations in adenovirus early region 2. J Virol. 1980;35:65–75. doi: 10.1128/jvi.35.1.65-75.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monaghan A, Webster A, Hay R T. Adenovirus DNA binding protein: helix destabilising properties. Nucleic Acids Res. 1994;22:742–748. doi: 10.1093/nar/22.5.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murti K G, Davis D S, Kitchingman G R. Localization of adenovirus-encoded DNA replication proteins in the nucleus by immunogold electron microscopy. J Gen Virol. 1990;71:2847–2857. doi: 10.1099/0022-1317-71-12-2847. [DOI] [PubMed] [Google Scholar]
- 31.Murti K G, He D C, Brinkley B R, Scott R, Lee S H. Dynamics of human replication protein subunit distribution and partitioning in the cell cycle. Exp Cell Res. 1996;223:279–289. doi: 10.1006/excr.1996.0083. [DOI] [PubMed] [Google Scholar]
- 32.Nagata K, Guggenheimer R A, Enomoto T, Lichy J H, Hurwitz J. Adenovirus DNA replication in vitro: identification of a host factor that stimulates synthesis of the preterminal protein-dCMP complex. Proc Natl Acad Sci USA. 1982;79:6438–6442. doi: 10.1073/pnas.79.21.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neale G A M, Kitchingman G R. Conserved region 3 of the adenovirus type 5 DNA-binding protein is important for interaction with single-stranded DNA. J Virol. 1990;64:630–638. doi: 10.1128/jvi.64.2.630-638.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ni T H, Zhou X, McCarty D M, Zolotukhin I, Muzyczka N. In vitro replication of adeno-associated virus DNA. J Virol. 1994;68:1128–1138. doi: 10.1128/jvi.68.2.1128-1138.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan Z Q, Ge H, Amin A A, Hurwitz J. Transcription-positive cofactor 4 forms complexes with HSSB (RPA) on single-stranded DNA and influences HSSB-dependent enzymatic synthesis of simian virus 40 DNA. J Biol Chem. 1996;271:22111–22116. doi: 10.1074/jbc.271.36.22111. [DOI] [PubMed] [Google Scholar]
- 36.Pruijn G J M, van Driel W, van der Vliet P C. Nuclear factor III, a novel sequence specific DNA-binding protein from HeLa cells stimulating adenovirus DNA replication. Nature. 1986;322:656–659. doi: 10.1038/322656a0. [DOI] [PubMed] [Google Scholar]
- 37.Puvion-Dutilleul F, Puvion E. Replicating single-stranded adenovirus type 5 DNA molecules accumulate within well delimited intra nuclear areas of lytically infected HeLa cells. Eur J Cell Biol. 1990;52:379–388. [PubMed] [Google Scholar]
- 38.Quinn C O, Kitchingman G R. Functional analysis of the adenovirus type 5 DNA-binding protein: site-directed mutants which are defective for adeno-associated virus helper activity. J Virol. 1986;60:653–661. doi: 10.1128/jvi.60.2.653-661.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richardson W D, Westphal H. A cascade of adenovirus early functions is required for expression of adeno-associated virus. Cell. 1981;27:133–141. doi: 10.1016/0092-8674(81)90367-6. [DOI] [PubMed] [Google Scholar]
- 40.Russell D W, Alexander I A, Miller A D. DNA synthesis and topoisomerase inhibitors increase transduction by adeno-associated virus vectors. Proc Natl Acad Sci USA. 1995;92:5719–5723. doi: 10.1073/pnas.92.12.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shenk T. Adenoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2111–2148. [Google Scholar]
- 42.Strauss S E, Ginsberg H S, Rose J A. DNA minus temperature-sensitive mutants of adeno-associated virus replication. J Virol. 1976;17:140–148. doi: 10.1128/jvi.17.1.140-148.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van der Vliet P C. Adenovirus DNA replication. In: Doerfler W, Bohm P, editors. The molecular repertoire of adenoviruses II. Berlin, Germany: Springer-Verlag; 1995. pp. 1–30. [Google Scholar]
- 44.Voelkerding K, Klessig D F. Identification of two nuclear subclasses of the adenovirus type 5-encoded DNA binding protein. J Virol. 1986;60:353–362. doi: 10.1128/jvi.60.2.353-362.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward P, Berns K I. In vitro rescue of an integrated hybrid adeno-associated/simian virus 40 genome. J Mol Biol. 1991;218:791–804. doi: 10.1016/0022-2836(91)90267-a. [DOI] [PubMed] [Google Scholar]
- 46.Ward P, Berns K I. In vitro replication of adeno associated virus DNA: enhancement by extracts from adenovirus infected cells. J Virol. 1996;70:4495–4501. doi: 10.1128/jvi.70.7.4495-4501.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward P, Urcelay E, Kotin R, Safer B, Berns K I. Adeno-associated virus DNA replication in vitro: activation by a maltose binding protein/Rep 68 fusion protein. J Virol. 1994;68:6029–6037. doi: 10.1128/jvi.68.9.6029-6037.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiner J H, Bertsch L L, Kornberg A. The deoxyribonucleic acid unwinding protein of Escherichia coli: properties and functions in replication. J Biol Chem. 1975;250:1972–1980. [PubMed] [Google Scholar]
- 49.Weitzman M D, Fisher K J, Wilson J M. Recruitment of wild type and recombinant adeno-associated virus into adenovirus replication centers. J Virol. 1996;70:1845–1854. doi: 10.1128/jvi.70.3.1845-1854.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wobbe C R, Dean F, Weissbach L, Hurwitz J. In vitro replication of duplex circular DNA containing the Simian virus 40 DNA origin site. Proc Natl Acad Sci USA. 1985;81:5710–5714. doi: 10.1073/pnas.82.17.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yakinoglu A O, Heilbronn R, Burkle A, Schlehofer J R, zur Hausen H. DNA amplification of adeno-associated virus as a response to cellular genotoxic stress. Cancer Res. 1988;48:3123–3129. [PubMed] [Google Scholar]
- 52.Yakobson B, Koch T, Winocur E. Replication of adeno-associated virus in synchronized cells without the addition of helper virus. J Virol. 1987;61:972–981. doi: 10.1128/jvi.61.4.972-981.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yakobson B, Hrynko T A, Peak M J, Winocur E. Replication of adeno-associated virus in cells irradiated with UV light at 254 nm. J Virol. 1989;63:1023–1030. doi: 10.1128/jvi.63.3.1023-1030.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]