Abstract
This study significantly expands both the scope and method of identification for construction of a previously reported tandem mass spectral library of 74 human milk oligosaccharides (HMOs) derived from results of combined LC-MS/MS experiments and comprehensive structural analysis of HMOs. In the present work, a hybrid search “bootstrap” identification method was employed that substantially broadens the coverage of milk oligosaccharides and thereby increases utility use of a spectrum library-based method for the rapid tentative identification of all distinguishable glycans in milk. This involved hybrid searching of the previous library, which was itself constructed using the hybrid search of oligosaccharide spectra in the NIST 17 Tandem MS Library. The general approach appears applicable to library construction of other classes of compounds. The coverage of oligosaccharides was significantly extended using milks from a variety of mammals, including bovine, Asian buffalo, African lion, and goat. This new method led to the identification of another 145 oligosaccharides, including an additional 80 HMOs from reanalysis of human milk. The newly identified compounds were added to a freely available mass spectral reference database of 219 milk oligosaccharides. We also provide suggestions to overcome several limitations and pitfalls in the interpretation of spectra of unknown oligosaccharides.
Graphical Abstract
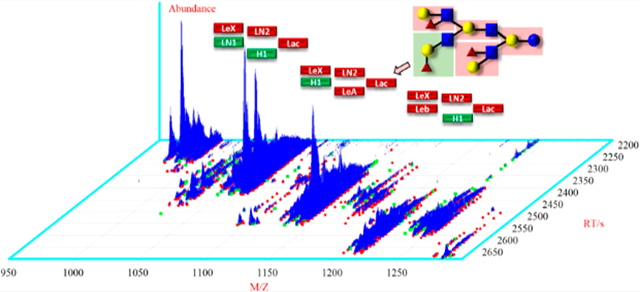
Mammalian milk oligosaccharides (MMOs) from humans and animals have been of great interest to researchers because of their functional role in food nutrition. Milk contains nutrients, immunoglobulins, and other bioactive factors that help neonates grow and protect them against infection and inflammation.1 To date, chemical structures of 162 different human milk oligosaccharides (HMOs) have been reported by mass spectrometry.2 Mass spectrometry coupled to liquid chromatography has become a routine technique for the identification of oligosaccharides in biological mixtures. In particular, hydrophilic interactions liquid chromatography (HILIC) has been shown by Albrecht et al.3 to be well suited to the characterization of individual glycans. This method enables the reliable use of retention time as a means of confirming the identity of glycans. Using these ideas, in a previous report,4 we developed a high-mass-accuracy, highresolution tandem mass spectral library of annotated oligosaccharides in human milk. This was derived from a Standard Reference Material (SRM) using chromatographyretention time-based experiments, MS fragmentation analysis, and library matching of spectra against a reference library with the newly developed “hybrid search”.5 Once interpreted, spectra were assigned and merged to form consensus spectra, and a reference library was created to enable the efficient tentative identification of milk glycans from their spectra or assist in their interpretation.6 The chemical structure of milk oligosaccharides is distinct from other glycans and their conjugates. Lactose or N-acetyl-lactosamine are the typical building blocks attached at the reducing end of milk oligosaccharides.7 Known neutral oligosaccharides are composed of 3 to 12 monosaccharides such as glucose (Glc), fucose (Fuc), galactose (Gal), and N-acetyl-glucosamine (GlcNAc) including N-acetyl-galactosamine (GalNAc) linked by alpha or beta glycosidic bonds. Milks of lactating animals such as cattle, goat, and water buffalo contain oligosaccharides not found in human milk that have been shown to benefit human health. Previous studies have reported that some animal milks contain acidic oligosaccharides with N-acetylneuraminic acid (Neu5Ac), resulting in an oligosaccharide profile similar to human milk.3,8 Additionally, N-glycolylneuraminic acid (Neu5Gc) is present in nonhuman milks,3 adding to the complexity of analysis.
Mature goat milk contains 0.3 g/L oligosaccharides, approximately 10 times more than the concentration in the milks of cow, water buffalo, or sheep.3 Interestingly, while the colostrum and mature milk of African lion contain significant amounts of oligosaccharides,9 prior to this work, other mammals were reported to have oligosaccharides amounts lower by a factor of 20 compared with humans (~12 g/L),10 necessitating additional analytical steps prior to characterization.
The present study characterizes and compares chemical structures derived from tandem mass spectra of oligosaccharides in bovine milk (SRMs 1549a and 1849a) and human milk (SRM 1954) to expand the current NIST reference database of milk oligosaccharides. These studies were extended using samples from water buffalo, Saanen goat, and African lion. Analysis involved the use of HILIC-MS/MS and mass spectral library search methods. The library searching includes both direct and hybrid search functions of the NIST MS search program against reference MS databases of the NIST 17 Tandem MS Full Library and the Milk Oligosaccharide libraries along with other data analysis tools. We also describe a new method of the newly developed “hybrid” search. This, in effect, uses results of prior hybrid identifications to make new hybrid identifications, amounting to a “bootstrapping” means of identification that may prove to be of general utility for the tentative identification of various classes of yet unidentified compounds.
MATERIALS AND METHODS
Human Milk Standard Reference Material (SRM) 195411 was purchased from the National Institute of Standards and Technology (NIST, Gaithersburg, MD). Two commercial bovine milk SRMs, 1549a and 1849a, were also obtained at NIST. The SRM 1549a12 is a commercial bovine whole milk powder while SRM 1849a13 is a hybrid mixture of commercial bovine milk infant and adult formula. Freeze-dried mature milk of Saanen goat and Asian water buffalo were kindly donated by the Philippine Carabao Center, (Nueva Ecija, Philippines). A sample of mature milk from wild African lionesses was kindly provided by the Animal Nutrition Science, Smithsonian’s National Zoo (Washington, DC, USA). Milk samples were stored in a sterile container and kept frozen (−80 °C) until use. A list of all commercially available milk oligosaccharides of high purity (>90%) used in the study is presented in the Supporting Information, Table S1. Water used in the sample preparation was LC-MS grade. All other chemicals used were of analytical grade.
Tandem MS Library Building of Oligosaccharides.
Extraction of Oligosaccharides.
Isolation and purification of oligosaccharides were performed by solid phase extraction (SPE) as described previously for human milk.4 Powdered milk samples (1.0 g) were first dissolved and homogenized in 5 mL water, and then, 0.5 mL aliquots were centrifuged at 14000 × g, 4 °C for 30 min. The liquid layer was transferred by pipet to an Eppendorf tube and mixed with four volumes of Foch solution (2:1 volume fraction of chloroform−methanol) and centrifuged at 14000 ×g, 4 °C for 30 min. The upper layer was transferred by pipet and deproteinated overnight at −20 °C by adding two volumes of ethanol into the mixture, then centrifuged at 14000 × g, 4 °C for 30 min. The decanted liquid was evaporated to dryness under the stream of nitrogen gas under a fume hood prior HILIC-MS analyses.
Analysis of Oligosaccharides by HILIC-MS/MS.
The chromatographic separations were performed on an ACQUITY Glycoprotein BEH Amide column, 300 Å (1.7 μm, 2.1 mm × 150 mm, Waters Corporation, Milford, MA, USA) using the Ultimate 3000 UHPLC system (Thermo Scientific) coupled to an Orbitrap mass spectrometer (Thermo Scientific Orbitrap Fusion Lumos) as previously described.4 The composition of the two mobile phases was 10 mmol/L ammonium formate with 0.1% (volume fraction) formic acid (A) and 99.9% (volume fraction) ACN with 0.1% (volume fraction) formic acid (B). The acquisition time was 65 min, and the mobile phase had a flow rate of 400 μL/min, pH 4.5 with a column oven temperature of 35 °C. The injection volume was 10 μL. Mass spectra were acquired over a range of collisional energies in an HCD cell at NCE (normalized collision energies) values of 10, 15, 20, 40, and 50, and in an ion trap (FT-IT) at 35% NCE. Each sample was analyzed in triplicate. Commercial milk oligosaccharides standards, including nonfucosylated, fucosylated, and sialylated isomers were analyzed and used as standards.
Experimental Flowchart.
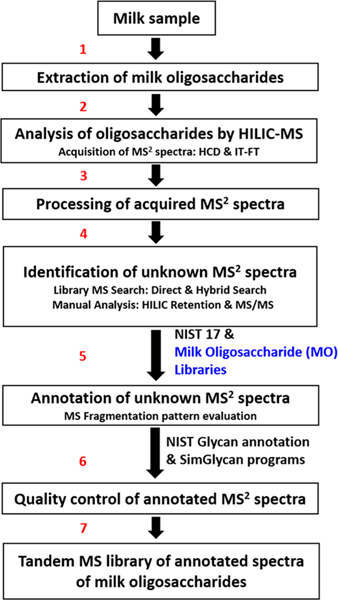
The previous database search method4 used only the NIST 17 Tandem MS Library14 for oligosaccharide identification. The workflow is illustrated in Figure 1. The current method employed Milk Oligosaccharide (MO) libraries (blue text), both the HMO library from that earlier work and an intermediate library derived from nonhuman milks. The first stage in this analysis is illustrated by steps 1−7, with step 5 using the previously published HMO library in addition to the NIST 17 Tandem MS Library, leading to an intermediate library (180 HILIC-MS runs) in the last step. All samples including human milk were reanalyzed to ensure consistent retention times and the highest quality spectra. This involved analysis of 216 HILIC-MS runs using both the original HMO library and intermediate library cited above. This final stage proceeded again through steps 1−7 with the addition of the intermediate oligosaccharide library in step 5.
Figure 1.
Schematic diagram of the steps employed for building a comprehensive tandem mass spectral library of milk oligosaccharides.
Library Search and Data Analysis.
In an earlier study, a database search was applied to a set of unknown spectra using NIST MS Search 2.315 with both direct and hybrid search functions against the NIST 17 Tandem MS Library as diagrammed in Figure 1. Library searching finds the spectra that most closely match to the experimentally acquired unknown spectra and assigns each a “goodness of fit” score that serves to create a ranked “hit list”.6 The search parameters for precursor m/z (400 to 2000) values and product ion masses allow an error tolerance of 10 ppm (10 × 10−6 mg/kg). The search program uses a modified vector dot product to generate a match factor (MF) score that ranges from 0 (no peaks in common) to 999 (identical spectra).16 In terms of spectral matching, searches with MF scores greater than 900 were considered near perfect, while MF scores greater than 850 and greater than 800 were very good and good, respectively, as reported.5 The spectra with lower scores (400−799) were examined manually and processed by SimGlycan and an automated in-house glycan annotation program as previously reported4,17 (Supporting Information). Because of possible changes in peak intensities with structural details, the hybrid search, which could not compensate for such changes, requires manual examination for confirmation.
The key to the tentative identification of compounds not in the library was the DeltaMass (DM) value generated by the hybrid search, which is the difference in mass between the query and library compounds. When scores were sufficiently high (>800), and DM values corresponded to multiples of common glycan components, spectra were manually examined and assigned putative structures. The glycan components considered are galactose or glucose (m/z 162.053), N-acetylglucosamine (m/z 203.079), fucose (m/z 146.058), N-acetylneuraminic acid (m/z 291.095), and N-glycolyneuraminic acid (m/z 307.090) or a combination of two or more monosaccharides. As in the previous study,4 theoretical m/z and experimental m/z values must match within 10 ppm. When more than one structure was possible, MS fragmentation patterns, negative and positive mode spectra, MF scores, collision energy, and retention times of candidate compounds were examined as described in the Supporting Information.
RESULTS AND DISCUSSION
Analysis of Human and Other Mammalian Milks.
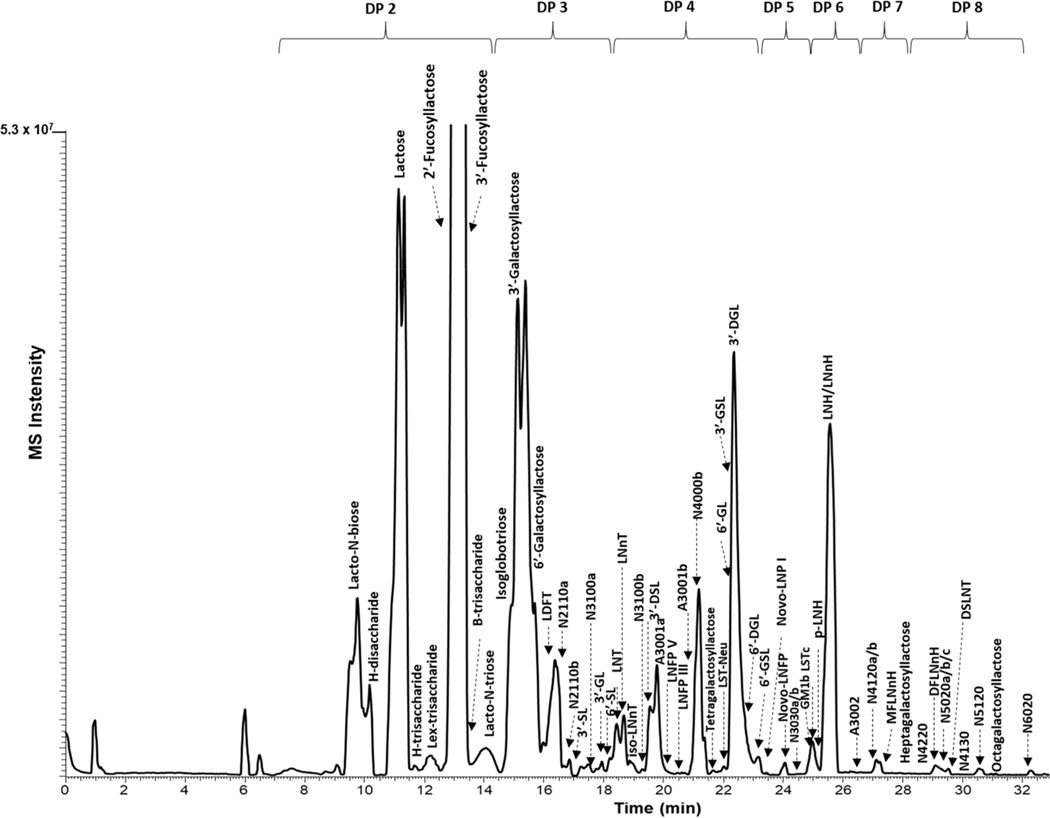
The initial HILIC-MS analysis of oligosaccharides was adapted from previous work for human milk4 utilizing MS/MS analysis of commercial oligosaccharide nonfucosylated, fucosylated, and sialylated oligosaccharide isomers (Figures S11.1−S11.4), as well as HILIC chromatograms (Figures S13.1−S13.5) and NIST 17 MS library. This method was similarly performed for the milks of African lion, Asian buffalo, bovine, and goat. The HILIC-MS analysis of mature milk of the African lion resulted to several sugar units in an oligomer, denoted as the degree of polymerization (DP), which in this work varies between 2 and 8. In some cases, the relative elution on the HILIC column between acidic and neutral oligosaccharides was controlled by polarity rather than size as previously observed.4 For example, the fucosylated oligosaccharide difucosyllactose (LDFT) (DP 4, m/z 657.22) eluted earlier than an acidic oligosaccharide, 3′-sialyllactose (SL) (DP 3, m/z 656.19). Among the oligosaccharides identified for the African lion, 19 are fucosylated, 27 nonfucosylated, and 16 are sialylated oligosaccharides (Table 1). A previous study of African lion milk18 reported four oligosaccharides–all of which were identified in the present analysis. Base peak chromatograms of other mammalian milks in this study are presented in Figure S2.
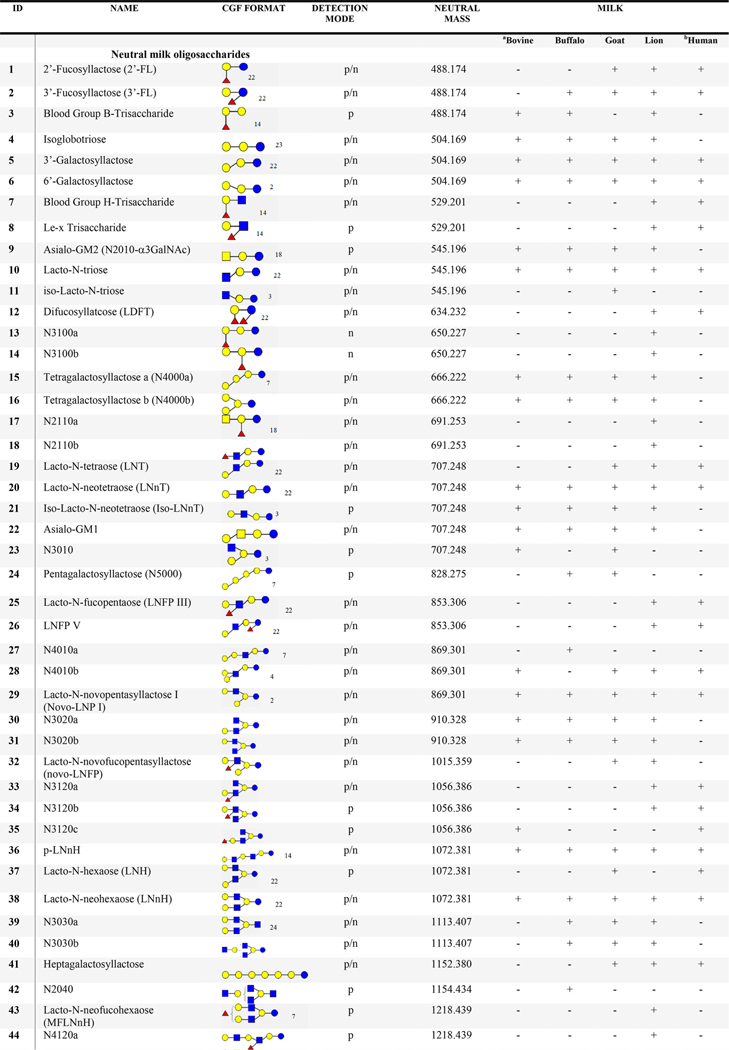
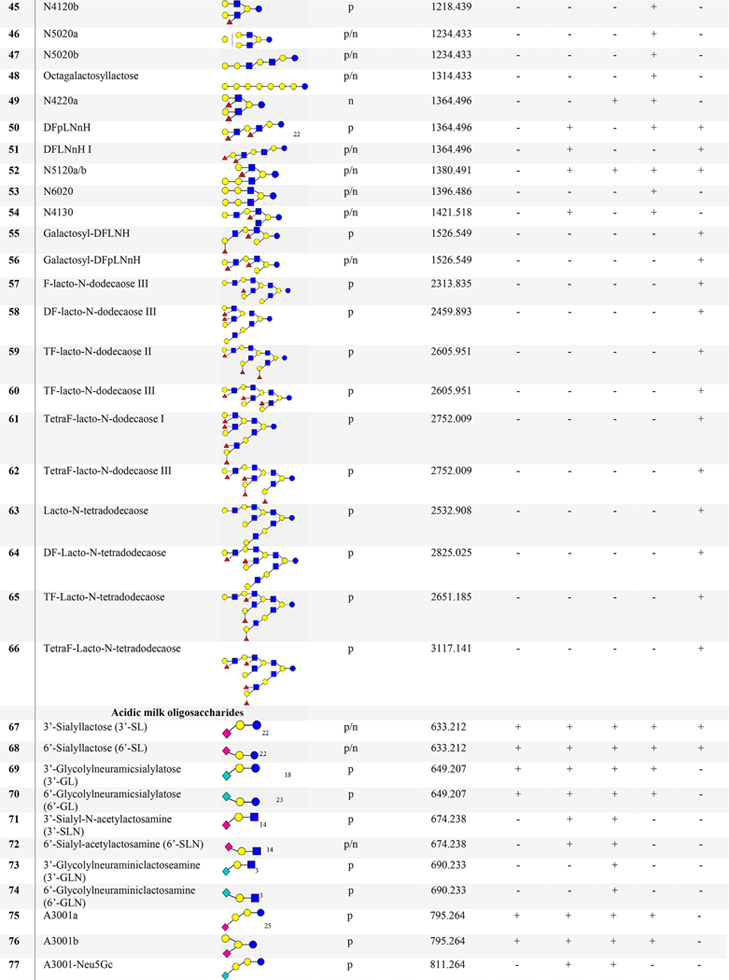
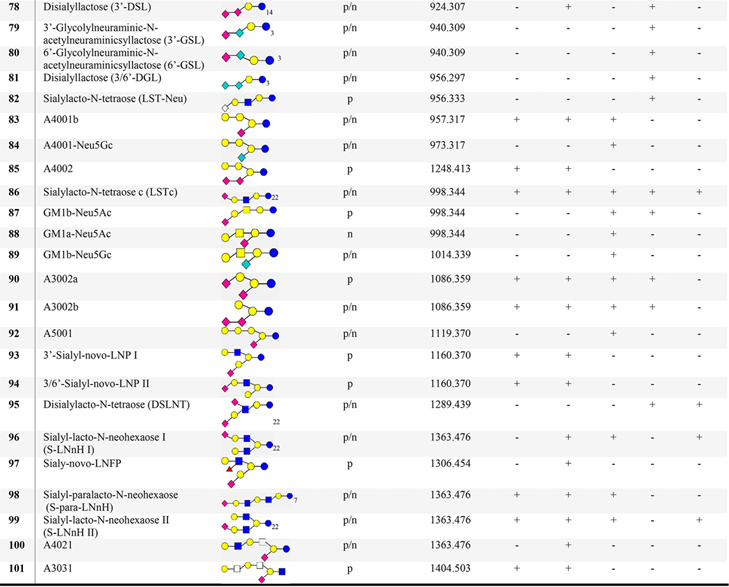
Table 1.
List of Proposed Structures Identified in Milks from Humans and Bovine Reference Materials, Water Buffalo, Saanen Goat, and African Lionc
|
|
|
Bovine Milk Standard Reference Material (SRM) 1549a.
Human Milk SRM 1954.
N4120: N = neutral; 4 = no. of Glc/Gal; 1 = no. of Fuc; 2 = no. of GlcNAc/GalNAc; 0 = no. of Neu5Ac/Neu5Gc. Blue circle = glucose (Glc); yellow circle = galactose (Gal); blue square = N-acetylglucosamine (GlcNAc), white square = HexNAc; yellow square = N-acetylgalactosamine (GalNAc); red triangle = fucose (Fuc); pink diamond = N-acetylneuraminic acid (Neu5Ac); aqua blue diamond = N-glycolylneuraminic acid (Neu5Gc), s = sialic acid; A = acidic; p = positive; n = negative; + detected; - not detected.
Identification of Unknown Spectra.
As described above, spectral libraries of milk oligosaccharides were searched along with spectra from the NIST 17 Tandem MS Library for the chemical identification of other mammalian milks. Using the final oligosaccharide library described above, both direct and hybrid database searchings were performed for each MS2 spectra, and the following information was recorded: hybrid and direct scores, retention times, spectra, DM, precursor types, charge states, and collision energy. All spectra were clustered according the precursor m/z (<10 ppm), energy, and retention time (±0.3 min) to generate consensus spectra.
Direct Search Identification.
As an illustration of a direct search, sialylated oligomers, 3′-sialyllactose and 6′-sialyllactose eluting at 16 and 18 min (Figure 2), generated scores of 987 and 841, respectively. In addition, 2′- and 3′-FL (Table 1, 1 and 2), LNT (Table 1, 19), and LNnT (Table 1, 20) were also directly identified with scores of 907, 882, 854, and 885, respectively. These oligosaccharides are well-known primary components of human milk.
Figure 2.
Base peak chromatogram of abundant oligosaccharides derived from the HILIC-MS analysis of African lion milk. Degree of polymerization (DP) varies from 2 to 8. Refer to Table 1 for the description and annotation of peaks. Annotation number N5120 means 5 hexose, 1 fucose, 2 GlcNAc/GalNAc, 0 Neu5Ac/Neu5Gc. A (acidic) and N (neutral) oligosaccharides.
Hybrid Search Identification.
Hybrid search identifications were considered when the direct search scores were less than 800, and DM values were consistent with the gain or loss of monosaccharide components, as described in the method section. Hybrid match factors (hMF) are generally high (>800) for two spectra that differ by a single moiety that did not significantly affect the fragmentation mechanism.4
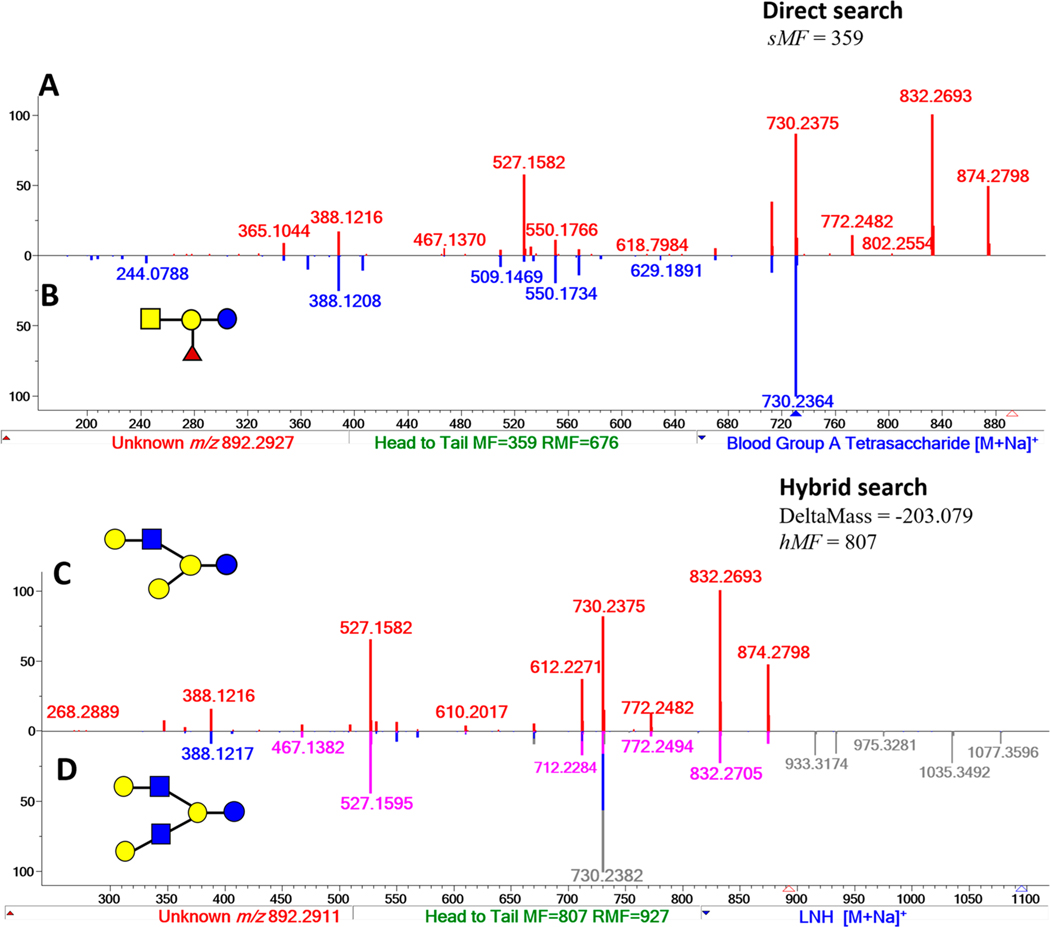
For example, a MS2 spectrum with precursor m/z 892.2927 in the FT-IT data of African lion milk was searched against the NIST 17 Tandem MS library and the intermediate Milk Oligosaccharide Library described earlier (Figure 3). The highest score from a direct search of this spectrum was 359 for a match with Blood Group A-Tetrasaccharide (Figure 3A and B) in the NIST library, indicating the overlap of several peaks with spectra in the NIST 17 library. In contrast, a hybrid search of the same spectrum generated hMF of 807 with a DM of −203.079 Da against a library spectrum of lacto-N-hexaose (LNH, m/z 1095.3709). For illustration, the MS2 peaks (gray line) are shifted by −203.078 Da (pink line) in Figure 3C and D. This mass difference is equal to GlcNAc suggesting a structure for novo-LNP I (Table 1, 29). This compound is a new HMO oligosaccharide2 known to predominate in bovine milk.7
Figure 3.
Mass spectral matching of a query spectrum (A) m/z 892.2927 of an unknown oligosaccharide against NIST 17 and the intermediate Milk Oligosaccharide MS Library using the direct and hybrid search functions using the NIST MS Search v2.3 software.15 The query spectrum, novo-LNP I (C), matched the library spectrum of (B) Blood Group A-Tetrasaccharide by direct search with a match factor of 359 and (D) lacto-N-hexaose, LNH by the hybrid search with a match factor of 807.
Reanalysis of Human Milk Oligosaccharides (HMO).
In the previous work,4 a library of 74 HMO spectra was derived from a comprehensive chemical structure analysis of oligosaccharides in NIST human milk reference material.
Adding the Human Milk Oligosaccharide MS Library to the search, as described above, led to the identification of an additional 80 HMOs. Many of the newly identified HMO structures contain core structures of lacto-N-decaose (LND), para-lacto-N-decaose (para-LND), and lacto-N-octaose (LNO) with a single, double, or combination of both fucose and sialic acid units as previously reported.2 As an illustration, the Trifuco lacto-N-decaose (TriF-LND) isomers have retentions from 38 to 43 min (Figure S1) and were identified by the hybrid search in this study (Figure S1a). The TriF-LND oligosaccharides have six different structures based on the acquired MS2 spectra and MS/MS annotation (Figures S1.1−S1.5). Although complete linkage information such as position and stereochemistry of the glycosidic bonds may not be fully established by tandem MS/MS, the assumption made for the chemical structures of TriF-LND isomers was based in the previously reported papers for human milk.2,19 In addition, galactosyl-lacto-N-hexaoses (N5020), lacto-N-dodecaose (N7050), and lacto-N-tetradodecaose (N8060) carrying fucose residues (Table 1, 55−66) were also identified by the hybrid search DeltaMass, corresponding to three or more HMO sugar residues. Fucosylated-lacto-N-dodecaoses (Table 1, 59 and 60) were proposed according to the core structures of HMOs.2 The latter proposed oligosaccharides contain 15 monosaccharide units and have not been reported to be present in human milk (Figures S1.6−S1.9). Newly identified HMOs including their annotated MS2 spectra20 are given in the Milk Oligosaccharide MS Library (https://chemdata.nist.gov/dokuwiki/doku.php?id=peptidew:lib:nonhumanmilk) and Table S4.
Analysis of Nonhuman Milk Oligosaccharides (Non-HMO).
Table 1 summarizes a total of 90 oligosaccharides found in nonhuman milks, of which 25 were also found in human milk. Furthermore, the remaining 65 oligosaccharides were found unique to nonhuman milks. The identified oligosaccharides have three classifications discussed below: neutral nonfucosylated, neutral fucosylated, and acidic sialylated oligosaccharides. HILIC chromatograms (Figures S13.1−S13.3) and tandem MS spectra (Figures S11.1−S11.5) of various commercial oligosaccharide and nonfucosylated, fucosylated, and sialylated isomeric compounds were also analyzed to confirm identification.
Nonfucosylated Oligosaccharides.
The following neutral oligosaccharides (Table 1), not represented in the existing library, were identified in milks of bovine, buffalo, goat, and lion: isoglobotriose, tetragalactosyllactose, N5020, and N6020. Moreover, chemical structures with varying numbers of sugar units were observed among neutral oligosaccharides.
Tetrasaccharide isomers, N4000a (Table 1, 15) was reported to be present in wallaby milk,7 while N4000b (Table 1, 16) is an epitope of glycolipids. Furthermore, linear (RT 21.0 min) and branched (RT 22.1 min, 23.2 min) structures for three galactosyl pentasaccharide isomers were observed eluting at different retention times. The assignment of putative isomeric structures for galactosyl pentasaccharides was confirmed based on their chromatographic separation and MS/MS annotation. The pentasaccharides such as N4010a (Table 1, 27) was observed in buffalo milk, while N4010b (Table 1, 28) was found in the milks of bovine, goat, and lion. Moreover, the oligosaccharide, novo-LNP I (Table 1, 29) was identified in all mammalian milks and was previously reported detected in the milks of herbivores.3,7
Fucosylated Oligosaccharides.
Twenty-three fucosylated oligosaccharides were added to the library. Oligosaccharides with 3−8 sugar units (Table 1), such as 2′/3′-FL, LDFT, and fucosylated LNnH oligosaccharides (Table 1, 49−51) were detected.
For nonhuman fucosylated tetrasaccharides, fucosylated lacto-N-triose and a branched sequence (N2100b) were observed (Table 1, 17 and 18). The latter oligosaccharide was reported in African lion milk.18 Furthermore, fucosylated LNnH was also identified in lion milk, while the difucosyloligomer, DFpLNnH I (Table 1, 51) was found in buffalo milk. Multiple adducts, protonated (NCE 10%) and sodiated (NCE 40%), of fucosylated oligosaccharides were re-examined for possible rearrangement of ions involving the migration of fucose residues. Protonated ion B3 (m/z 658.255) and sodiated ion Y4 (m/z 730.237) are diagnostic ions for two fucose residues present at the terminal GlcNAc-Gal linkage (Figure S4). As a result, HCD [M+Na]+ MS2 spectra of fucosylated oligosaccharides appeared to be stable and consistent with the product ions of HCD [M+H]+ MS2 spectra which are in contrast to previously reported CID experiments.21 Note that 2′-FL was found in goat and lion milks but not in the milk of buffalo as previously reported.18 Interestingly, most of the identified fucosylated compounds were found in the milk of the African lion.
Sialylated Oligosaccharides.
Library searching led to the assignment of 36 sialylated compounds (Table 1) containing N-acetylneuraminic acid (Neu5Ac) or N-glycolyneuraminic (Neu5Gc) oligosaccharides. Oligomers containing one or two Neu5Ac/Neu5Gc units such as GSL and DGL (Table 1, 79−81) were detected in the milk of lion, while DSL (Table 1, 78) was observed in lion and buffalo milks. Aside from Neu5Ac-lactoses, Neu5Gc oligosaccharides were identified in goat milk. Fragmentation patterns of GSL and DGL are described in Figure S5. Oligosaccharide A4001 is a conjugate sugar of a glycolipid, which predominates in nonhuman mammalian milks.7
When available, commercial sialylated oligosaccharides were used to confirm their identity through their retention and their positive and negative polarity spectra. This included the sialylated pentasaccharide isomers LSTc, GM1b, and GM1a which eluted at 25.3, 24.9, and 22.6 min, respectively. Their negative MS2 spectra were compared with the MS2 spectra of oligosaccharides found in the milks of lion and goat. In depth analysis of the product ions of GM1a shows C-type fragment ions, m/z 282.13 and m/z 833.30, and a set of A-type ions, m/z 424.15, m/z 586.19, and m/z 628.21, suggesting a terminal Gal-GalNAc-Gal sequence. Relatively high intensity B-type ions, m/z 290.09 and m/z 364.12, and a set of X-type ions, m/z 877.29 and m/z 937.32, were observed, which were caused by the cleavage of sialyl glycosidic and cross-ring linkages. Moreover, C-type ions of precursor m/z 997.34 signals for GM1b. Linkages were also reinforced by the cross-ring cleavages, m/z 406.14, m/z 859.28, and m/z 937.31. The analysis of the negative ion MS2 spectra of coeluting oligosaccharides, LSTd and GM1b found A-type ions, enable the determination of Gal-GlcNAc and Gal-GalNAc sequences. Also, differences in product ion intensities of the terminal sialic acid cleavages were apparent between the two isomeric compounds. These findings discriminate the isomeric sialylated pentasaccharides (Figure S6). Overall, 24 Neu5Ac-oligomers and nine Neu5Gc-oligomers have been identified. Other sialylated oligomers are a combination of both Neu5Gc and Neu5Ac. Interestingly, sialylated isomers (Table 1, 75 and 76, 90 and 91), A3001a/b and A3002a/b were detected in all nonhuman milk samples. The 3′-glycolylneuraminic acid (3′Neu5Gc) is abundant in African lion milk, while the 3′-acetylneuraminic acid (3′-Nue5Ac) is dominant among goat, bovine, and buffalo milks (Figures S5 and S7).
Mass Spectral Library of Mammalian Milk Oligosaccharides.
Oligosaccharide spectra led to the assignment of 90 compounds (Table 1) from the milks of bovine (35), buffalo (49), goat (52), and lion (62). Only 25 of these identified oligosaccharides in animal milks were found in previous work.4 Moreover, an additional of 80 oligosaccharides from the reanalysis of human milk SRM were identified and added to the library (Milk Oligosaccharide MS Library, https://chemdata.nist.gov/dokuwiki/doku.php?id=peptidew:lib:nonhumanmilk).
In this study, the milk oligosaccharides from humans were compared with animal species. Table 2 shows the number of unique oligosaccharides identified in each mammalian milk. Next to humans, African lion has the greatest number of unique oligosaccharides. These were primarily fucosylated Table 2. Number of Identified Oligosaccharides in Milks from Humans, Bovine Reference Materials, Asian Buffalo, Saanen Goat, and African Lion oligosaccharides and a mixture of N-glycolylneuraminic and N-acetylneuraminic acid-lactoses.
Table 2.
Number of Identified Oligosaccharides in Milks from Humans, Bovine Reference Materials, Asian Buffalo, Saanen Goat, and African Lion
| Source | Unique | Human | Bovine | Asian buffalo | Goat | African lion |
|---|---|---|---|---|---|---|
| Human | 123 | 154 | 13 | 16 | 19 | 24 |
| Bovine | 0 | 13 | 35 | 32 | 29 | 25 |
| Asian buffalo | 3 | 16 | 32 | 49 | 36 | 31 |
| Goat | 9 | 19 | 29 | 36 | 52 | 34 |
| African lion | 13 | 24 | 25 | 31 | 34 | 62 |
A library of consensus spectra of all identified milk oligosaccharides were generated for FT-IT (615 spectra) and HCD (1990 spectra) data sets. This represents 2605 annotated consensus MS2 spectra available online20 of different precursor ions (Table S3) in positive or negative polarity. This newly constructed mass spectral reference database has a total of 219 milk oligosaccharides. For nonhuman neutral oligosaccharides, 31 are nonfucosylated and 23 fucosylated, while acidic oligosaccharides consist of 36 sialylated (Neu5Ac/Neu5Gc). In contrast to human milk, the number of sialylated oligosaccharides is higher than the number of fucosylated oligosaccharides observed in animal milks. These findings are consistent with previous animal milk studies.3,23
Illustration of Library Use.
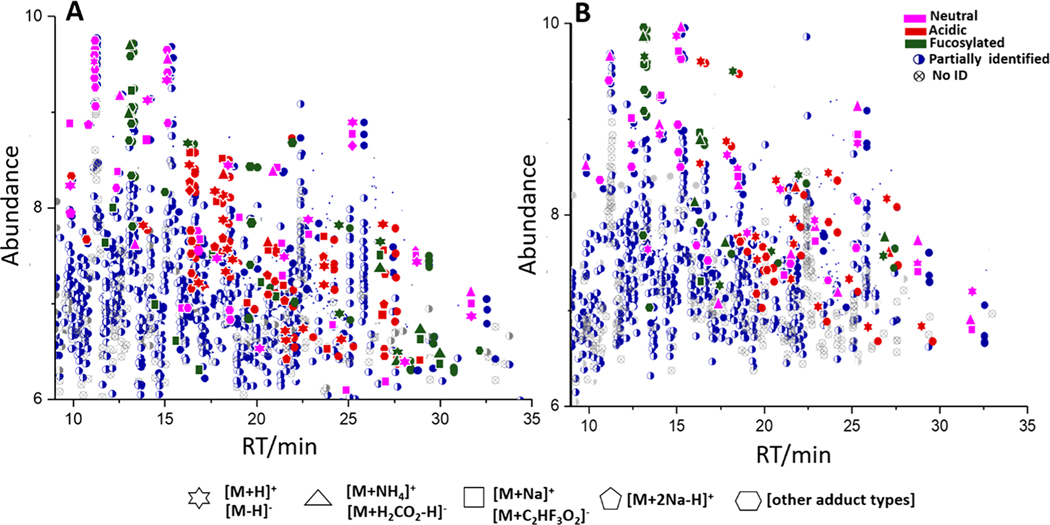
Using the newly created library, oligosaccharides in lion milk having scores above 800 are shown in Figure 4. In this analysis, 70% of the total extracted MS2 spectra in a single HILIC-MS run were assigned to oligosaccharides. Note that abundant ions generated multiple adduct types. The presence of multiple precursor ions and charge states26 for a single analyte is an unavoidable consequence of the electrospray ionization. Additionally, there is a higher degree of potential spectrum variability due to the energy dependence of the fragmentation process. For example, signals at 13.3 min were derived from 3′-FL that have positive [M+H]+, [M+NH4]+, [M+Na]+ (Figure 4A) and negative [M-H]−, [M+H2CO2H]−, [M+C2HF3O2]− (Figure 4B) adduct types. Also, disialylacto-N-tetraose (Table 1, 95) generates multiple adducts and charge states (Figures S10.1− 10.3) and often produces similar confirmatory fragmentation patterns. Nonfucosylated oligosaccharides (N5020 and N6020) at higher retention times were identified as both positive and negative precursor ions. In addition, sialylated oligosaccharides have clear signals (red) in both analyses. The overview of positive and negative ions of lion milk oligosaccharides discriminates the identified nonfucosylated (pink), fucosylated (green), and sialylated (red) oligosaccharides including the partially identified and unidentified compounds. Equivalent plots for other mammals are given in Figures S8 and S9.
Figure 4.
Ion abundance versus retention of oligosaccharides in African lion milk. Neutral (pink), acidic (red), and fucosylated (green) oligosaccharides were identified in (A) positive and (B) negative ion polarity. The extracted MS spectra of African lion milk were hybrid searched against the Milk Oligosaccharide MS libraries. The precursor ions with a threshold score above 800 and DeltaMass of zero were selected for this identification.
Concerns Regarding Building an Oligosaccharide Spectral Library.
In the interpretation of MS2 spectra of unknown oligosaccharides, one must be aware of certain limitations and pitfalls in searching for similar compounds in a reference library. Accurate information on masses, the nature and abundance of the precursor/product ions, possible isomeric structures, low abundance, and lack of related spectra (Supporting Information) are important factors in the identification process.
Accurate information on product masses and peak intensities must be available from high-quality, reliable reference spectral libraries, and all spectra should be measured on high mass accuracy instruments. This accuracy lessens the chance of making false identifications in the analysis of complex mixtures.
Multiple fragmentation types of the same oligosaccharide can provide more confident identification than from a single method alone. Various instrument settings (FT-IT/HCD) in negative and positive polarities and multiple collision energies create fragment-rich and reproducible spectra. For library searching, inclusion of spectra from varying individual voltages, e.g, 10, 20, 35, 40, 50 eV, is preferred because these spectra allow for more fine-grained library matching.27 ESI-MS mobile phase solvents and modifiers may generate different adducts for the same compound that may be useful for interpreting spectra.
Isomers whose product ions contain insufficient information for full structural identification must rely on external information. For example, discrimination of oligosaccharides with GalNAc or GlcNAc in the sequence of N4130 and A3031 (Table 1) by mass spectral information alone is ambiguous. The use of commercially available oligosaccharide standards or specific carbohydrate enzymes can lead to more confident assignment of these structures; however, authentic standards were often unavailable for these compounds. Moreover, any tentative identification by the hybrid search necessarily reduces the confidence in precise structural features.
Lastly, it is important to note that library search scores represent the degree of matching submitted spectra with library spectra of defined compounds. However, unlike peptide identification in proteomics analysis, depending on the class of compound, a spectrum may match multiple compounds. While matching spectra generally means the underlying compounds have considerable structural similarity, structural features may differ in cases where those features are not distinguishable from their mass spectra.6 For example, electron ionization spectra for terpenes are commonly indistinguishable, as unfortunately are many glycans in tandem spectra. Consequently, it is the responsibility of the analyst to make final structural determinations, which often involve use of prior information. Ion mobility and other methods show promise for making such structural distinctions.
CONCLUSIONS
A previous HMO library of 74 oligosaccharides was expanded by 145 oligosaccharides using both new data from four nonhuman mammals and an enhanced identification strategy involving the recently developed hybrid search spectral library method. These four mammals were African lion, Asian water buffalo, bovine, and goat. The new identification strategy utilized a two-stage process that employed libraries constructed using reference libraries derived from previous hybrid-search-based identifications. The first stage used the previous HMO library, and the second stage used an intermediate library that included spectra derived from the first stage. The final library contained 80 more HMOs than the original library. Such a “bootstrap” approach appears to provide a general means of expanding libraries for other classes of compounds.
We report overlaps of oligosaccharides between different mammals and note that oligosaccharides from the African lion differed substantially among nonhuman species. It contains a significant number of fucosylated or Neu5Gc-linked oligosaccharides.
We also illustrate methods for making structural assignments including a description of some of the problem areas in this process–especially regarding the assignment of glycan structures, including unresolved isomers. It is known that the current fragmentation methods are insufficient to reveal all structural details. Various other methods, such as reduction/permethylation combined with multistage mass spectrometry (MSn), have been proposed to provide more structural detail. Future extensions of spectral libraries may also assist the alternative strategies. While spectral libraries can significantly increase the efficiency and scope of compound identification, as for any library identification the final decision concerning the identity, including assignment of detailed connectivity or stereochemistry of a given compound, must be made by a skilled analyst. Hence, we wish to emphasize that library “identifications” especially regarding glycans, for which isomer distinction is an endemic problem, must always be considered tentative. For the most definitive identifications, when available, a reference standard should always be employed.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Oleg Toropov, Dr. Sanford Markey, and Dr. Xinjian Yan for technical support and fruitful discussions. We also thank our collaborators, Mike Maslanka and Michael Jakubasz of the Nutrition Laboratory, Smithsonian National Zoological Park and Conservation Biology, and Penelope Merced and Jessica Asuncion for your technical contributions.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.0c00342.
Comprehensive annotation of fragment ions for glycans (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.analchem.0c00342
The Milk Oligosaccharide MS Library can be accessed at https://chemdata.nist.gov/dokuwiki/doku.php?id=peptidew:lib:nonhumanmilk.
All commercial instruments and materials used in the study are for the experimental purpose only. Such identification does not intend recommendation or endorsement by the National Institute of Standards and Technology, nor does it intend that the materials or instruments used are necessarily the best available for the purpose. The animal specimens used were approved by Animal Care and Use Coordinator Research protection Office.
The authors declare no competing financial interest.
Contributor Information
Concepcion Africano Remoroza, National Institute of Standards and Technology, Gaithersburg, Maryland 20899, United States.
Yuxue Liang, National Institute of Standards and Technology, Gaithersburg, Maryland 20899, United States.
Tytus D. Mak, National Institute of Standards and Technology, Gaithersburg, Maryland 20899, United States
Yuri Mirokhin, National Institute of Standards and Technology, Gaithersburg, Maryland 20899, United States.
Sergey L. Sheetlin, National Institute of Standards and Technology, Gaithersburg, Maryland 20899, United States
Xiaoyu Yang, National Institute of Standards and Technology, Gaithersburg, Maryland 20899, United States.
Joice V. San Andres, Department of Animal Science,University of Nebraska-Lincoln, Lincoln, Nebraska 68583-0908, United States Department of Animal Science, Central Luzon State University, Nueva Ecija 3120, Philippines.
Michael L. Power, Nutrition Laboratory, Smithsonian Conservation Biology Institute, National Zoological Park, Washington, DC 20008, United States
Stephen E. Stein, National Institute of Standards and Technology, Gaithersburg, Maryland 20899, United States
REFERENCES
- (1).Ballard O; Morrow AL Pediatr. Clin. North Am. 2013, 60 (1), 49–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Urashima T; Hirabayashi J; Sato S; Kobata A. Trends Glycosci. Glycotechnol. 2018, 30 (172), SE51–SE65. [Google Scholar]
- (3).Albrecht S; Lane JA; Marino K; Al Busadah KA; Carrington SD; Hickey RM; Rudd PM Br. J. Nutr. 2014, 111 (07), 1313–1328. [DOI] [PubMed] [Google Scholar]
- (4).Remoroza CA; Mak TD; De Leoz MLA; Mirokhin YA; Stein SE Anal. Chem. 2018, 90 (15), 8977–8988. [DOI] [PubMed] [Google Scholar]
- (5).Cooper BT; Yan X; Simón-Manso Y; Tchekhovskoi DV; Mirokhin YA; Stein SE Anal. Chem. 2019, 91 (21), 13924–13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Stein S. Anal. Chem. 2012, 84 (17), 7274–7282. [DOI] [PubMed] [Google Scholar]
- (7).Urashima T; Saito T; Nakamura T; Messer M. Glycoconjugate J. 2001, 18 (5), 357–371. [DOI] [PubMed] [Google Scholar]
- (8).Claps S; Di Napoli MA; Sepe L; Caputo AR; Rufrano D; Di Trana A; Annicchiarico G; Fedele V. Small Ruminant Research 2014, 121 (1), 116–119. [Google Scholar]
- (9).De Waal HO; Osthoff G; Hugo A; Myburgh J; Botes P. Mammalian Biology 2004, 69 (6), 375–383. [Google Scholar]
- (10).Mineguchi Y; Miyoshi M; Taufik E; Kawamura A; Asakawa T; Suzuki I; Souma K; Okubo M; Saito T; Fukuda K; Asakuma S; Urashima T. Glycoconjugate J. 2018, 35 (6), 561–574. [DOI] [PubMed] [Google Scholar]
- (11).SRM 1954 - Organic Contaminants in Fortified Human Milk. NIST. https://www-s.nist.gov/srmors/view_detail.cfm?srm=1954 (accessed July 2020).
- (12).SRM1549a - Whole Milk Powder. NIST. https://www-s.nist.gov/srmors/view_cert.cfm?srm=1549a (accessed July 2020).
- (13).SRM1849a - Infant/Adult Nutritional Formula I (milk-based). NIST.https://www-s.nist.gov/srmors/view_cert.cfm?srm=1849a (accessed July 2020).
- (14).NIST/EPA/NIH Mass Spectral Library with Search Program, June 6, 2017. NIST. https://www.nist.gov/srd/nist-standardreference-database-1a-v17 (accessed July 2020). [Google Scholar]
- (15).NIST Peptide Library Browser and BSA Library, June 6, 2017. NIST. http://chemdata.nist.gov/dokuwiki/doku.php?id=peptidew:nistmssearch (accessed July 2020). [Google Scholar]
- (16).Wallace WE; Ji W; Tchekhovskoi DV; Phinney KW; Stein SE J. Am. Soc. Mass Spectrom. 2017, 28 (4), 733–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Klein J; Zaia JJ Proteome Res. 2019, 18 (9), 3532–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Senda A; Hatakeyama E; Kobayashi R; Fukuda K; Uemura Y; Saito T; Packer C; Oftedal OT; Urashima T. Animal Science Journal 2010, 81 (6), 687–693. [DOI] [PubMed] [Google Scholar]
- (19).Blank D; Geyer H; Maass K; Yamashita K; Geyer R. Anal. Biochem. 2012, 421 (2), 680–690. [DOI] [PubMed] [Google Scholar]
- (20).MS Library of Free Oligosaccharides in Milk from a Variety of Mammals, 2019. NIST. https://chemdata.nist.gov/dokuwiki/doku.php?id=peptidew:lib:nonhumanmilk (accessed July 2020). [Google Scholar]
- (21).Wuhrer M; Deelder AM; van der Burgt YE M. Mass Spectrom. Rev. 2011, 30 (4), 664–680. [DOI] [PubMed] [Google Scholar]
- (22).Remoroza CA; Mak T; Stein SE Milk SRM 1953 Glycan MS Library, 2018. https://chemdata.nist.gov/srmd-1953-glycan/ (accessed July 2020). [Google Scholar]
- (23).Oliveira DL; Wilbey RA; Grandison AS; Roseiro LB Int. J. Dairy Technol. 2015, 68 (3), 305–321. [Google Scholar]
- (24).SimGlycan Database; Premier Biosoft, 2017. [Google Scholar]
- (25).Urashima T; Taufik E; Fukuda K; Asakuma S. Biosci., Biotechnol., Biochem. 2013, 77 (3), 455–466. [DOI] [PubMed] [Google Scholar]
- (26).Yang X; Neta P; Stein SE Anal. Chem. 2014, 86 (13), 6393–6400. [DOI] [PubMed] [Google Scholar]
- (27).Kind T; Tsugawa H; Cajka T; Ma Y; Lai Z; Mehta SS; Wohlgemuth G; Barupal DK; Showalter MR; Arita M; Fiehn O. Mass Spectrom. Rev. 2018, 37 (4), 513–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.