Abstract
To harness radiometals in clinical settings, a chelator forming a stable complex with the metal of interest and targets the desired pathological site is needed. Toward this goal, we previously reported a unique set of chelators that can stably bind to both large and small metal ions, via a conformational switch. Within this chelator class, py-macrodipa is particularly promising based on its ability to stably bind several medicinally valuable radiometals including large 132/135La3+, 213Bi3+, and small 44Sc3+. Here, we report a 10-step organic synthesis of its bifunctional analogue py-macrodipa-NCS, which contains an amine-reactive –NCS group that is amenable for bioconjugation reactions to targeting vectors. The hydrolytic stability of py-macordipa-NCS was assessed, revealing a half-life of 6.0 d in pH 9.0 aqueous buffer. This bifunctional chelator was then conjugated to a prostate-specific membrane antigen (PSMA)-binding moiety, yielding the bioconjugate py-macrodipa-PSMA, which was subsequently radiolabeled with large 132/135La3+ and small 47Sc3+, revealing efficient and quantitative complex formation. The resulting radiocomplexes were injected into mice bearing both PSMA-expressing and PSMA-non-expressing tumor xenografts to determine their biodistribution patterns, revealing delivery of both 132/135La3+ and 47Sc3+ to PSMA+ tumor sites. However, partial radiometal dissociation was observed, suggesting that py-macrodipa-PSMA needs further structural optimization.
Keywords: Anticancer agents, Chelate, PSMA, Radiopharmaceuticals, Rare earth
Graphical Abstract

A bifunctional chelator py-macrodiap-NCS was prepared over 10 steps. Py-macrodipa-NCS shows adequate hydrolytic stability and was conjugated to a PSMA-targeting vector, yielding a bioconjuate py-macrodipa-PSMA. Py-macrodipa-PSMA delivered both large radiometal 132/135La3+ and small 47Sc3+ to PSMA+ tumors, but further structural optimization is needed to improve its in vivo radiocomplex stability.
INTRODUCTION
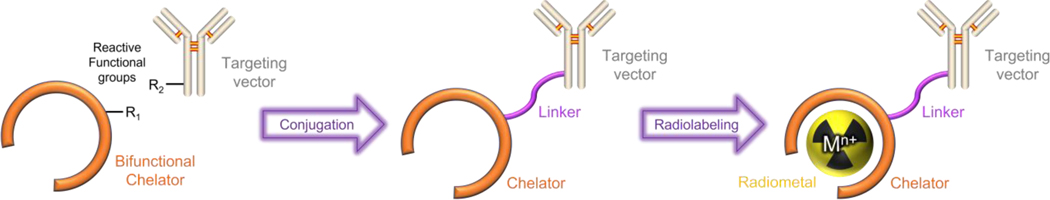
The radioisotopes of metallic elements, or radiometal ions, are important for the development of new radiopharmaceutical agents.[1–3] For example, over 30 radiometal-containing drugs have been approved for various types of therapy and diagnosis to date.[4] In many cases, the use of radiometals in nuclear medicine requires that they be attached to a targeting vector that preferentially accumulates at the site of disease. Attachment of the radiometal to the targeting vector is afforded by a bifunctional chelator, which needs to rapidly form radiometal complexes that are stable in vivo.[5–7] This type of radiopharmaceutical construct, which contains these required components, is depicted in Figure 1, where the radiometal–chelator complex and the biological targeting vector are covalently linked together. Radiometals located at different areas in the periodic table can have significantly disparate chemical properties, and there have been substantial efforts in the field to develop and optimize chelating agents to accommodate them for different radiopharmaceutical applications.[7–9]
Figure 1.
Schematic representation of a metal-chelate-based bioconjugate used for nuclear medicine. Reproduced from ref [22]. The targeting vector can be a small-molecule, peptide, antibody, polysaccharide, lipid, or nanoparticle.
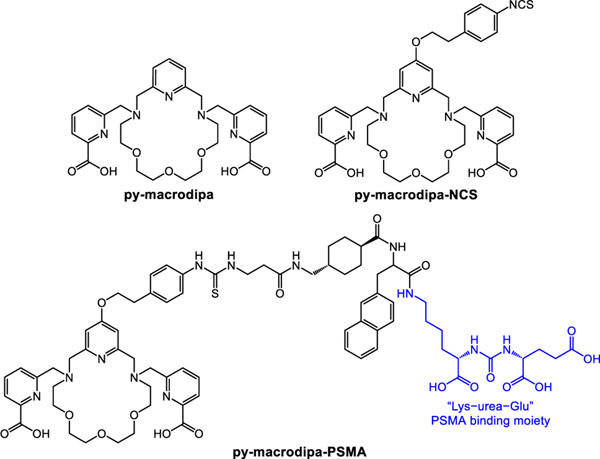
Typically, efforts in the development of new bifunctional chelators have focused on optimizing their coordination chemistry for a specific radiometal ion. However, researchers have gradually recognized the value of chelating agents that can stably bind to radiometal ions with substantially disparate chemical properties, which can accelerate the clinical translation of corresponding radiopharmaceuticals. Recent studies have revealed that a bispidine ligand can rapidly and stably bind to both 225Ac3+ and 177Lu3+.[10] Likewise, some acyclic picolinate ligands were also found to mutually stabilize 225Ac3+and 111In3+.[11,12] In our contribution to this area, we recently reported a unique class of chelators that possess dual size selectivity, a property that confers good affinity for both the large and small metal ions. This unusual selectivity pattern arises from a significant change in ligand binding mode to afford two distinct complex conformations that maximally stabilize both large and small metal ions.[13–15] Within this class, py-macrodipa (Figure 2) forms inert complexes with three medically relevant radionuclides, the large 135La3+ and 213Bi3+ ions, as well as the small 44Sc3+ ion.[14,16] These three radionuclides have suitable emissions for Auger electron therapy, alpha therapy, and positron emission tomography, respectively.[17–19] The ability to radiolabel and stabilize both 135La3+ and 44Sc3+, whose six-coordinate ionic radii are 1.032 and 0.745 Å, respectively,[20] is notable because the state-of-the-art chelator 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA)[21] has a limited ability to bind and stabilize larger radiometals. Thus, for chelators like py-macrodipa, one could envision them to be useful for assembling theranostic radiopharmaceutical agents that can employ the therapeutic properties of large radiometal ions, like 135La3+, with diagnostic properties of smaller radiometal ions, like 44Sc3+, within the same chemical construct. Although the fundamental coordination chemistry and radiolabeling properties of py-macrodipa revealed its promise for these applications, its use within nuclear medicine would most likely require it to be conjugated to a biological targeting vector.
Figure 2.
Structures of Ligands Discussed in This Work.
In order to furnish a targeting vector onto py-macrodipa via a covalent linkage, a reactive moiety that readily reacts with functional groups present on targeting vectors has to be installed on the chelator.[5–7] Toward this goal, we synthesized the novel chelator py-macrodipa-NCS (Figure 2), a bifunctional analogue of py-macrodipa with an amine-reactive isothiocyanate group (–NCS) that reacts readily with primary amines (–NH2) to form a thiourea linkage.[23] Following the workflow shown in Figure 1, this bifunctional py-macrodipa-NCS was conjugated to the prostate-specific membrane antigen (PSMA)-targeting small molecule Glu-urea-Lys-nap-trans-β-Ala, yielding the bioconjugate py-macrodipa-PSMA (Figure 2). As a proof-of-principle demonstration, this bioconjugate was further radiolabeled with 132/135La3+ and 47Sc3+, two radiometals useful for radiopharmaceutical purposes, and studied in vivo to evaluate its potential for nuclear medicine that will serve the basis for more comprehensive studies of this type in the future.
Results and Discussion
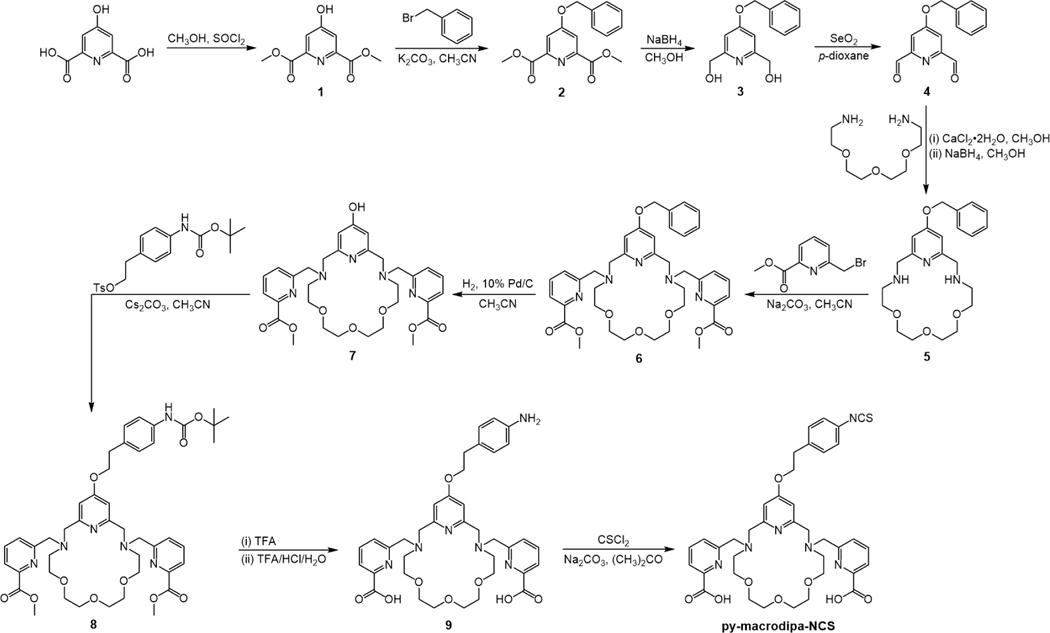
The bifunctional chelator py-macrodipa-NCS was synthesized over 10 steps following the pathway shown in Scheme 1. Briefly, the pyridyl-containing macrocycle was constructed first, followed by the attachment of picolinate pendent donors, and the introduction of the isothiocyanate (–NCS) functional group. The identity and purity of the intermediates and final product were determined by NMR spectroscopy, mass spectrometry, and analytical HPLC, as shown in Figures S1−S15 within the Supporting Information (SI).
Scheme 1.
Synthetic Route to py-macrodipa-NCS.
This synthesis was initiated via the esterification of chelidamic acid in MeOH to yield 1, which was subsequently benzyl-protected and then reduced with NaBH4 to afford the glycol 3.[24] Compound 3 was oxidized to the dialdehyde 4 by SeO2. In general, the construction of the macrocyclic core is the critical step in synthesizing a macrocyclic ligand. As inspired from prior literature,[25] we leveraged Ca2+ as a template in the cyclization of the macrocycle 5. Dialdehyde 4 was allowed to react with 1,11-diamino-3,6,9-trioxaundecane in the presence of CaCl2 and then treated with NaBH4 to form the 18-membered macrocycle 5 via reductive amination. The picolinate pendent arms were installed under standard nucleophilic substitution conditions to yield 6, and the benzyl protecting group was then removed by palladium-catalyzed hydrogenation to afford 7. Subsequently, 8 was prepared by a SN2 reaction between 7 and 4-((tert-butoxycarbonyl)amino)phenethyl 4-methylbenzenesulfonate.[24] The Boc-deprotection and methyl ester hydrolysis were conducted under highly acidic conditions to yield 9. In the final step, the primary amine was converted to an isothiocyanate group with thiophosgene, affording the desired bifunctional chelator py-macrodipa-NCS after purification by preparative HPLC. The overall yield from 3 to py-macrodipa-NCS is 16% over seven steps.
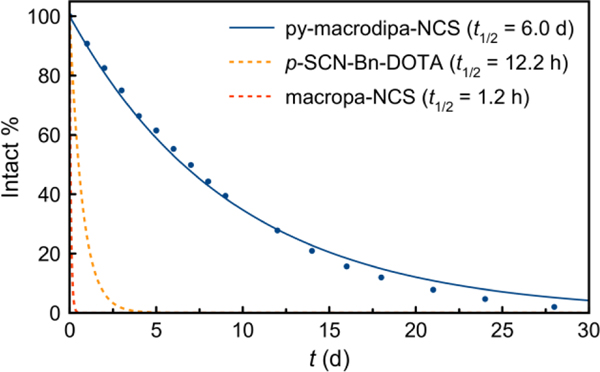
As an electrophilic moiety, the –NCS group is susceptible to hydrolysis, which would revert it back to the non-conjugatable –NH2 form. As such, the reaction on the –NCS group of py-macrodipa-NCS with the desired biological targeting vector is in competition with its hydrolysis. If the hydrolysis proceeds too rapidly, this bifunctional chelator will be ineffective. Furthermore, rapid hydrolysis would also make ambient humidity a problem for its long-term storage or shipment to other research facilities. In this regard, we evaluated the aqueous stability of py-macrodipa-NCS. It was incubated in a pH 9.0 NaHCO3/Na2CO3 buffer, and the solution composition was monitored by HPLC. Over time, the peak corresponding to py-macrodipa-NCS (tR = 18.2 min) decayed with the concomitant formation of new species. A plot of the peak integration of the signal corresponding to py-macrodipa-NCS versus time and an exponential fit to these data revealed the hydrolysis half-life (t1/2) for this compound to be 6.0 ± 0.8 d (Figures 2 and S16, SI). Importantly, this t1/2 is substantially longer than the typical reaction timescale used for successful bioconjugations (< 2 d) involving the –NCS group, suggesting that hydrolysis should not negatively affect its use for this application. Furthermore, the aqueous stability of py-macrodipa-NCS is greater than those of p-SCN-Bn-DOTA and macropa-NCS (Figure 3), the bifunctional analogues of two well-known chelators DOTA and macropa. Under identical conditions, the t1/2 for hydrolysis of p-SCN-Bn-DOTA and macropa-NCS were measured to be 12.2 h and 1.2 h, respectively.[26] The more rapid hydrolysis rate of macropa-NCS in comparison to py-macrodipa-NCS is most likely a consequence of the location of the –NCS functional group on the electron-deficient picolinate heterocycle in the former. Thus, the relatively remote placement of the –NCS group on py-macrodipa-NCS is beneficial with respect to its stability, as well as its lower likelihood of influencing metal binding.
Figure 3.
The hydrolysis of py-macrodipa-NCS at pH 9.0 and RT. The intact compound percentage was plotted versus reaction time and fitted into an exponential decay model.
Having validated sufficient aqueous stability of py-macrodipa-NCS, we next sought to conjugate this compound to a small-molecule targeting vector to obtain the bioconjugate py-macrodipa-PSMA (Figure 2), which was characterized by mass spectrometry and analytical HPLC (Figures S19–S20). Specifically, the well-known “Lys−urea−Glu” moiety was used. This targeting vector, which was prepared by solid-phase peptide synthesis,[27] has high affinity for the PSMA, a membrane protein that is overexpressed on the surface of various subtypes of prostate cancer cells.[28] The naphthyl and tranexamic acid moieties present in py-macrodipa-PSMA, are also employed by the recently FDA-approved radiotherapeutic [177Lu]Lu3+–PSMA-617 (Pluvicto™)[29] and confer additional enhancement to the PSMA target.[30]
With this bioconjugate on hand, we next evaluated its potential use as a radiopharmaceutical agent, via several initial proof-of-principle studies. Medicinally relevant radioisotopes of two rare-earth metals, 132/135La3+ and 47Sc3+ (t1/2 = 80.4 h), were produced as previously described[31,32] and assessed with py-macrodipa-PSMA. 132La3+ (t1/2 = 4.6 h, 58% electron capture and 42% β+ emission), 135La3+ (t1/2 = 18.9 h, 100% electron capture), and 47Sc3+ (t1/2 = 80.4 h, 100% β– emission) have properties suitable for positron emission tomography, Auger electron therapy, and beta therapy, respectively.[17,19,33] Importantly, La3+ and Sc3+ are the largest and smallest ions, respectively, within the rare-earth series.[20] Thus, they provide a good platform for evaluating the ability of py-macrodipa-PSMA to bind and deliver disparately-sized radiometal ions to the target in vivo. Radiolabeling of py-macrodipa-PSMA with 132/135La3+ and 47Sc3+ was efficient. Consistent with the results obtained for the unconjugated py-macrodipa chelator,[14] py-macrodipa-PSMA was quantitatively radiolabeled by 132/135La3+ and 47Sc3+ within 20 min at room temperature (Figure S21, SI), producing molar activities of 3.2 × 10–7 nmol/Bq and 1.3 × 10–6 nmol/Bq, respectively. The difference in molar activity arose due to the comparatively large reaction volume and low molar activity of the parent 47ScCl3 solution; this limitation is a consequence of the production methodology that can be improved with further optimization of isotope separation approaches.
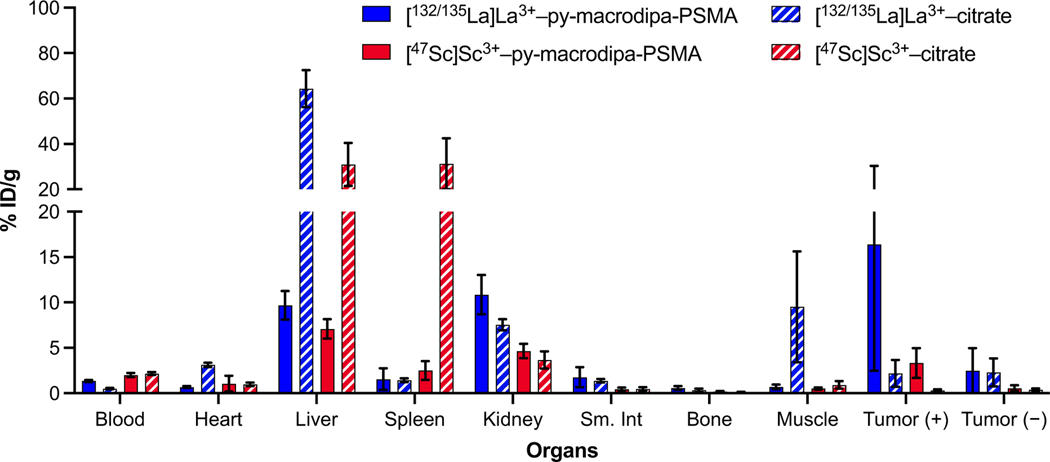
The [132/135La]La3+– and [47Sc]Sc3+–py-macrodipa-PSMA complexes were then injected into mice (n = 5) with both PC3-PIP (PSMA-expressing, PSMA+) and PC3-flu (PSMA-non-expressing, PSMA–) tumor xenografts. [132/135La]La3+– or [47Sc]Sc3+–citrate was injected into another mice cohort (n = 4) to determine distribution of the free ions. All mice were sacrificed 2 h post injection, and biodistribution studies were carried out by counting the 132/135La3+ or 47Sc3+ radioactivity in different organs. These results are summarized in Figure 4 and Tables S1–S2, SI. Remarkably, both the activities of 132/135La3+ or 47Sc3+ accumulated significantly within the PSMA+, but not PSMA– tumor xenografts, indicating that py-macrodipa-PSMA delivered both radiometals ions to the desired tumor sites. The accumulated activity of 132/135La3+ in the PSMA+ tumor (11.0 %ID·g−1) is higher than that of 47Sc3+ (3.3 %ID·g−1), indicating a more effective delivery of the former radioisotope. This difference may be a consequence of different pharmacokinetic properties that arise from the distinct conformations that py-macrodipa attains in binding large and small ions,[14] highlighting that chelator conformations may have an important role in biological properties. As an alternative explanation, the significant difference in molar activity of the two radiometal constructs could account for the lower tumor uptake of the 47Sc3+, which, by virtue of its lower molar activity, contains more free, unlabeled ligand in solution that could block uptake. In addition to the PSMA+ tumor uptake, the biodistribution also reveals the accumulation of activity in non-target tissues. For example, both radiometals accumulated within the liver (9.7 %ID·g−1 for [132/135La]La3+–py-macrodipa-PSMA and 7.1 %ID·g−1 for [47Sc]Sc3+–py-macrodipa-PSMA), at levels that are higher than those for some recently reported examples that employ a similar PSMA-targeting vector (0.1 %ID·g−1 for [132/135La]La3+–macrodipa-DUPA 4 h post injection and 1.2 %ID·g−1 [47Sc]Sc3+–picaga-DUPA 2 h post injection).[34,35] Because the free radiometal ions in their citrate salt forms also accumulate in this organ, the biodistribution of their py-macrodipa-PSMA complexes could signify, albeit not conclusively, partial dissociation of radiometals from the chelator. Analysis of the urine metabolites of these mice by radio-HPLC reveals peaks with the same retention times as those of free radiometal ions (Figure S22, SI), further affirming possible dechelation of the py-macrodipa complexes in vivo. Although the origin of this in vivo instability is uncertain, it does highlight challenges and concerns that arise when chelators are conjugated to biological targeting vectors, as similarly observed in other published work.[36,37] Thus, even though py-macrodipa-PSMA was effective at transporting both 132/135La3+ and small 47Sc3+ to PSMA+ tumors, further tuning of its structure is still needed to optimize the in vivo stability of its radiocomplexes. Furthermore, we highlight that these initial in vivo studies are limited based on the time points assessed, but serve as an initial demonstration of the potential use of the bifunctional analogue of py-macrodipa, the synthesis of which is the focus of this manuscript.
Figure 4.
Bar graph of biodistribution analysis 2 hours post injection of [132/135La]La3+–py-macrodipa-PSMA (n = 5), [132/135La]La3+–citrate (n = 4), [47Sc]Sc3+–py-macrodipa-PSMA (n = 5) and [47Sc]Sc3+–citrate (n = 4) in mice bearing both the PSMA+ and PSMA– tumor xenografts.
CONCLUSION
In this work, a synthetic route for py-macrodipa-NCS, a bifunctional analogue of py-macrodipa, has been developed. This chemical modification has enabled this unique dual-size-selective chelator to be applied as a targeted radiopharmaceutical agent via bioconjugation to the amine-reactive –NCS functional group. Because –NCS functional groups are typically electrophilic and susceptible to hydrolysis, their stability is a concern for the use of bifunctional chelators with this moiety. Importantly, py-macrodipa-NCS is sufficiently stable at pH 9.0 and under ambient conditions in air, which allows it to be easily transported between laboratories and also ensures that it efficiently reacts with amines in aqueous solutions. As a proof-of-principle, the prostate-cancer-targeting bioconjugate py-macrodipa-PSMA was prepared from this bifunctional chelator. Subsequently, py-macrodipa-PSMA was radiolabeled with both 132/135La3+ and 47Sc3+. Consistent with our earlier studies with py-macrodipa, the radiolabeling of both these large and small radiometal ions proceeded effectively, demonstrating that the dual-size-selective property of the parent ligand is retained upon conjugation. Furthermore, the resulting radiocomplexes were evaluated in mice bearing both PSMA+ and PSMA– tumor xenografts. These in vivo studies revealed that py-macrodipa-PSMA was able to deliver radioactivity to the desired PSMA+ tumor site, albeit with different efficiencies for 132/135La3+ and 47Sc3+. This latter observation implies that the conformational differences afforded by the py-macrodipa chelator can affect pharmacokinetics[38] or differences in employed molar activity were large enough to impact target accumulation. Analysis of metabolites of these radiolabeled complexes, however, showed that loss of the radiometal ions occurred in vivo. Because we had previously observed excellent stability of both complexes of unfunctionalized py-macrodipa in human serum, this result demonstrates how radiometal complex stability can be substantially different when incorporated into targeted agents, consistent with similar observations in the literature.[36,37] In any case, this study provides another recent example of leveraging the same construct for targeted nuclear medicine applications with two chemically disparate radiometals[10–12], like 132/135La3+ and 47Sc3+ in this case. We further note that these initial in vivo studies are limited by the lack of additional time points and the lower molar activity of the 47Sc used. They serve to highlight the potential use of the bifunctional analogue of py-macrodipa, the synthesis and characterization of which was the focus of the present study. In addition, we acknowledge that the use of 132/135La3+ and 47Sc3+ as a theranostic pair for downstream clinical applications is not feasible because of their sufficiently different half-lives as well as the fact that 47Sc3+ is a therapeutic β-emitter. However, the choice of these radionuclides is by a desire to demonstrate the proof-of-principle of chelating and delivering radiometal ions with substantially different ionic radii. Ongoing work in this area may lead to multifunctional radiopharmaceutical agents that can be modified for different therapeutic and diagnostic applications by simply altering the radionuclide used. Further efforts will be directed to tuning the bioconjugate structures to optimize the tumor selectivity and minimize radiometal release, as well as gaining an understanding of how bioconjugation affects the stability profiles of the chelator.
Experimental Section
Methods and Materials for Synthesis.
Deionized H2O (≥18 MΩ·cm) was obtained from an Elga Purelab Flex 2 water purification system. Organic solvents were of ACS grade or higher. All other reagents were purchased from commercial sources and used without further purification. The bifunctional chelator py-macrodipa-NCS was synthesized as demonstrated in Scheme 1. Compounds 1, 2, 3, 6-bromomethylpyridine-2-carboxylic methyl ester, and 4-((tert-butoxycarbonyl)amino)phenethyl 4-methylbenzenesulfonate were prepared as previously described.[14,24]
High-performance liquid chromatography (HPLC) for the synthesis of py-macrodipa-NCS and its intermediates consisted of a CBM-20A communications bus module, an LC-20AP (preparative) or LC-20AT (analytical) pump, and an SPD-20AV UV−Vis detector monitoring at 270 nm (Shimadzu, Japan). Semipreparative purification was performed using an Epic Polar preparative column, 120 Å, 10 μm, 25 cm × 20 mm (ES Industries, West Berlin, NJ) at a flow rate of 14 mL/min. Analytical chromatography was carried out using an Ultra Aqueous C18 column, 100 Å, 5 μm, 250 mm × 4.6 mm (Restek, Bellefonte, PA) at a flow rate of 1.0 mL/min. All HPLC methods employed use a binary mobile phase (H2O/CH3OH or H2O/CH3CN, 0.1% TFA added for all solvents) gradient. NMR spectra were acquired on a 500 MHz Bruker AVIII HD spectrometer equipped with a 5 mm, broadband Prodigy cryoprobe operating at 499.76 and 125.68 MHz for 1H and 13C observations, respectively. All NMR experiments were carried out at 25 °C. High-resolution mass spectra (HRMS) were acquired on a Thermo Scientific Exactive Orbitrap mass spectrometer with a heated electrospray (HESI) ion source.
Synthesis of 4.
To a solution of 3 (1.29 g, 5.26 mmol) in dry p-dioxane (30 mL), SeO2 (1.28 g, 11.54 mmol) was added, and this suspension was heated at 95 °C for 2.5 h. Afterwards, the reaction mixture was allowed to cool to RT and then filtered. This filtrate was concentrated to dryness under reduced pressure, giving a pink solid. This material was then rinsed with 3 × 5 mL of CH2Cl2, and these three portions of CH2Cl2 suspension were filtered and combined. The combined filtrate was concentrated under reduced pressure and dried under vacuum overnight, affording 4 as a pale-orange solid (1.15 g, 91%). 1H NMR (500 MHz, CDCl3) δ: 10.11 (s, 2H, –CHO), 7.72 (s, 2H, py), 7.45–7.36 (m, 5H, Ph), 5.25 (s, 2H, –OCH2Ph–). 13C{1H} NMR (126 MHz, CDCl3) δ: 192.3, 166.7, 154.8, 134.5, 128.9, 128.8, 127.7, 111.8, 71.0. ESI-HRMS m/z: 242.0815; calcd for [C14H11NO3 + H]+: 242.0812.
Synthesis of 5.
Dry CH3OH (10 mL) was added to a flask containing 4 (0.56 g, 2.32 mmol) and CaCl2·2H2O (0.36 g, 2.45 mmol), and a solution of 1,11-diamino-3,6,9-trioxaundecane (0.46 g, 2.39 mmol) in dry CH3OH (3 mL) was then dropwise added with stirring. This mixture was stirred at room temperature for 15 min, and then heated at 60 °C for 2.5 h, with a drying tube equipped on the condenser. Afterwards, the reaction mixture was cooled to 0 °C with an ice bath, and NaBH4 (0.44 g, 11.63 mmol) was slowly added with vigorous stirring. The reaction mixture was stirred at RT overnight. H2O (12 mL) was then added, and the mixture was stirred for another 1.5 h. The CH3OH of this mixture was removed under reduced pressure, and the leftover liquid was extracted with 8 × 30 mL of CH2Cl2. The combined extractants were dried over Na2SO4, concentrated to dryness under reduced pressure, and dried under vacuum overnight to afford 5 (0.80 g, 89%) as a pale-yellow oil. This product had a >95% purity and was used in the next step without further purification. 1H NMR (500 MHz, CDCl3) δ: 7.41–7.32 (m, 5H, Ph), 6.66 (s, 2H, py), 5.08 (s, 2H, –OCH2Ph–), 3.81 (s, 4H, –NCH2py–), 3.67 (m, 8H, –CH2–), 3.62 (m, 4H, –CH2–), 2.83 (t, 4H, J = 4.8 Hz, –CH2–). 13C{1H} NMR (126 MHz, CDCl3) δ: 165.5, 160.2, 135.8, 128.7, 128.3, 127.5, 107.5, 70.6, 70.4, 70.3, 69.8, 55.0, 49.1. ESI-HRMS m/z: 402.2389; calcd for [C22H31N3O4 + H]+: 402.2387.
Synthesis of 6.
To a mixture of 5 (0.76 g, 1.89 mmol), 6-bromomethylpyridine-2-carboxylic methyl ester (0.83 g, 3.61 mmol), and Na2CO3 (0.77 g, 7.26 mmol), dry CH3CN (30 mL) was added. This suspension was then heated at 45 °C for 44 hours with a drying tube equipped on the condenser. Afterwards, the suspension was allowed to cool to RT and filtered. The resulting filtrate (~30 mL), which contains 6, was directly used in the next step. ESI-HRMS m/z: 402.2389; calcd for [C22H31N3O4 + H]+: 402.2387.
Synthesis of 7.
To the above-mentioned filtrate containing 6, 10% Pd/C (0.25 g) was added, and the mixture was stirred under H2 atmosphere at RT. The reaction progress was monitored by analytical HPLC, and sometimes more 10% Pd/C was added when the reaction became slow. After the reaction was complete (which usually took about 5 d), the suspension was filtered, and the filtrate was concentrated to dryness under reduced pressure, giving a yellow oil. This crude material was then dissolved in H2O containing 10% CH3OH and 0.1% TFA (3 mL). Following filtration, this solution was injected into the preparative HPLC system to purify the product (Method: 0–10 min, 90% H2O/CH3OH; 10–45 min, 90% → 0% H2O/CH3OH). Pure fractions were combined, concentrated under reduced pressure, and lyophilized to yield the 3.5TFA salt of 7 as a white solid (0.67 g, 35% from 5). 1H NMR (500 MHz, D2O, pD ≈ 2) δ: 8.07 (dd, 2H, J = 7.8, 1.0 Hz, py), 7.48 (t, 2H, J = 7.8 Hz, py), 7.27 (dd, 2H, J = 7.8, 1.0 Hz, py), 6.85 (s, 2H, py), 4.65 (m, 8H, –NCH2py–), 3.98 (t, 4H, J = 4.9 Hz, –CH2–), 3.91 (s, 6H, –CH3), 3.75 (m, 8H, –CH2–), 3.67 (s, 4H, –CH2–). 13C{1H} NMR (126 MHz, D2O, pD ≈ 2) δ: 166.7, 166.6, 152.0, 150.5, 147.3, 140.2, 129.4, 126.4, 113.2, 70.2, 70.1, 64.4, 58.5, 58.4, 56.2, 53.9. ESI-HRMS m/z: 610.2874; calcd for [C31H39N5O8 + H]+: 610.2871. Elemental analysis: found %: C 45.64, H 4.54, N 7.20; calcd % for C31H39N5O8·3.5CF3COOH: C 45.25, H 4.25, N 6.94. Analytical HPLC: tR = 18.89 min (Method: 0–5 min, 90% H2O/CH3OH; 5–25 min, 90% → 0% H2O/CH3OH).
Synthesis of 8.
To a mixture of 7·3.5TFA (0.20 g, 0.20 mmol) and Cs2CO3 (0.39 g, 1.20 mmol), dry CH3CN (8 mL) was added. With a drying tube equipped on the condenser, this mixture was heated at 70 °C for 30 min, after which 4-((tert-butoxycarbonyl)amino)phenethyl 4-methylbenzenesulfonate (0.12 g, 0.31 mmol) was added. This reaction mixture was then heated at 70 °C for 48 h. Afterwards, the suspension was filtered, and the filtrate was concentrated to dryness under reduced pressure, giving a yellow solid. This crude material, which contains the desired compound 8, was directly used in the next step. ESI-HRMS m/z: 867.3696; calcd for [C44H56N6O10 + K]+: 867.3690.
Synthesis of 9.
The entirety of the crude material from the last step was dissolved in TFA (2 mL), and this mixture was heated at 60 °C for 22 h, during which the tert-butyloxycarbonyl (Boc) protecting group was removed. Afterwards, 6 M HCl (2 mL) was added, and the reaction mixture was heated at 60 °C for another 22 h, during which the two methyl esters on picolinate arms were hydrolyzed. The TFA/HCl/H2O was then removed under reduced pressure, and the resulting residue was dissolved in H2O containing 20% CH3OH and 0.1% TFA (3.0 mL). Following filtration, this solution was injected into the preparative HPLC system to purify the product (Method: 0–5 min, 90% H2O/CH3OH; 5–35 min, 90% → 0% H2O/CH3OH). Pure fractions were combined, concentrated under reduced pressure, and lyophilized to yield the 4TFA salt of 9 as a white solid (0.16 g, 77% from 7). 1H NMR (500 MHz, D2O, pD ≈ 2) δ: 7.86 (m, 4H, py), 7.58 (dd, 2H, J = 6.5, 2.3 Hz, py), 7.50 (m, 2H, py), 7.42 (m, 2H, py), 6.77 (s, 2H, py), 4.66 (s, 4H, –NCH2py–), 4.60 (s, 4H, –NCH2py–), 4.22 (t, 2H, J = 6.1 Hz, –CH2–), 4.00 (t, 4H, J = 4.9 Hz, –CH2–), 3.76 (m, 8H, –CH2–), 3.70 (t, 4H, J = 4.9 Hz, –CH2–), 3.14 (t, 2H, J = 6.1 Hz, –CH2–). 13C{1H} NMR (126 MHz, D2O, pD ≈ 2) δ: 167.9, 167.2, 152.3, 150.4, 147.8, 140.3, 140.2, 131.3, 129.0, 128.9, 125.9, 123.7, 111.8. ESI-HRMS m/z: 701.3269; calcd for [C37H44N6O8 + H]+: 701.3293. Elemental analysis: found %: C 46.53, H 4.55, N 7.36; calcd % for C37H44N6O8·4CF3COOH: C 46.72, H 4.18, N 7.26. Analytical HPLC: tR = 17.12 min. (Method: 0–5 min, 90% H2O/CH3OH; 5–25 min, 90% → 0% H2O/CH3OH).
Synthesis of py-macrodipa-NCS.
To a mixture of 9·4TFA (0.094 g, 0.081 mmol) and Na2CO3 (0.107 g, 1.01 mmol), dry acetone (8 mL) was added. This suspension was heated at 50 °C for 30 min, after which CSCl2 (90 μL, ACROS Organics, 85%) was slowly added. The reaction mixture was then heated at reflux for 2 h and then concentrated under reduced pressure. This crude material solid was dissolved in H2O containing 10% CH3CN and 0.1% TFA (1.5 mL). Following filtration, this solution was injected into the preparative HPLC system to purify the product (Method: 0–10 min, 90% H2O/CH3CN; 10–40 min, 90% → 0% H2O/CH3CN). Pure fractions were combined, concentrated under reduced pressure, and lyophilized to yield the 3TFA salt of py-macrodipa-NCS as a nearly-white/pale-yellow solid (0.065 g, 74%). 1H NMR (500 MHz, D2O, pD ≈ 2) δ: 7.88 (m, 4H, py), 7.27 (dd, 2H, J = 7.3, 1.5 Hz, py), 7.29 (m, 2H, Ph), 7.14 (m, 2H, Ph), 6.78 (s, 2H, py), 4.58 (m, 8H, –NCH2py–), 4.25 (t, 2H, J = 5.7 Hz, –CH2–), 3.98 (t, 4H, J = 4.6 Hz, –CH2–), 3.75 (m, 8H, –CH2–), 3.66 (t, 4H, J = 4.6 Hz, –CH2–), 3.05 (t, 2H, J = 5.7 Hz, –CH2–). 13C{1H} NMR (126 MHz, D2O, pD ≈ 2) δ: 168.0, 167.4, 152.3, 150.2, 148.0, 140.2, 138.7, 134.5, 131.0, 129.5, 129.1, 126.2, 126.0, 112.0, 70.2, 70.1, 69.7, 64.4, 58.8, 58.7, 56.5, 34.8. ESI-HRMS m/z: 743.2831; calcd for [C38H42N6O8S + H]+: 743.2858. Elemental analysis: found %: C 48.43, H 4.35, N 7.78; calcd % for C38H42N6O8S·3CF3COOH: C 48.71, H 4.18, N 7.75. Analytical HPLC: tR = 18.22 min (Method: 0–5 min, 90% H2O/CH3CN; 5–25 min, 90% → 0% H2O/CH3CN).
Aqueous Stability of py-macrodipa-NCS.
The hydrolytic stability of py-macrodipa-NCS was evaluated at pH 9.0 and RT (22 °C). Py-macrodipa-NCS and caffeine were dissolved 0.1 M NaHCO3/Na2CO3 buffer (pH 9.0), and the hydrolysis reaction of py-macrodipa-NCS was monitored quantitatively by analytical HPLC (Method: 0–5 min, 90% H2O/CH3CN; 5–25 min, 90% → 0% H2O/CH3CN). Caffeine was used as an internal reference for the peak integration of the HPLC chromatograms. The intact percentage of py-macrodipa-NCS over time was fit to an exponential decay model, affording first order rate constants and half-lives. Three independent replicates were performed.
Synthesis of Glu-urea-Lys-nap-trans-β-Ala.
For the purification and characterization of Glu-urea-Lys-nap-trans-β-Ala the following instruments and conditions were used. High-resolution ESI mass spectrometry was carried out at the Stony Brook University Center for Advanced Study of Drug Action (CASDA) mass spectrometry facility with an Agilent LC–UV–TOF spectrometer. Semi-preparative HPLC was carried out using a Shimadzu HPLC-20AR equipped with a binary gradient, pump, UV–Vis detector, and manual injector on a Phenomenex Luna C18 column (250 mm × 21.2 mm, 100 Å, AXIA packed). Method A (Preparative purification method). A = 0.1% TFA in water, B = 0.1% TFA in MeCN. Gradient: 0–5 min: 5% B; 5–24 min: 5–95% B. Analytical HPLC analysis was carried out using a Shimadzu HPLC-20AR equipped with a binary gradient, pump, UV–Vis detector, autoinjector, and Laura radio-detector on a Phenomenex Luna C18 column (150 mm × 3 mm, 100 Å). Method B (Analytical HPLC analysis). A = 0.1% TFA in water, B = 0.1% TFA in MeCN with a flow rate of 0.8 mL/min, UV detection at 220 and 270 nm. Gradient 0–2 min: 5% B; 2–14 min 5–95% B; 14–16 min 95% B; 16–16.5 min 95–5% B; 16.5–20 min 5% B.
The Fmoc-trans-nap-Lys-urea-Glu-Wang resin (200 mg, 0.16 mmol) was prepared via established solid-phase chemistry.[27] Fmoc-β-L-alanine (199 mg, 0.64 mmol) was coupled to the pre-loaded resin using PyBOP (167 mg, 0.32 mmole) in the presence of DIPEA (112 μL, 0.64 mmol) over a 12 h time period. The resin was then treated with a mixture of 20% piperidine in DMF to remove Fmoc group. After deprotection, all resin was washed with DMF, DCM, Et2O, then dried and treated with a mixture of TFA/TIS/H2O (95%/2.5%/2.5%) to cleave Glu-urea-Lys-nap-trans-β-Ala from the resin. After cleavage from the resin the compound was lyophilized. The crude product was purified by reverse-phase semi-preparative HPLC (Method A) and characterized by analytical HPLC (Method B) and mass spectrometry. Yield: (0.0241 g, 21 %). ESI-HRMS m/z: 727.3655; calcd for [C36H50N6O10 + H]+: 727.3661.
Conjugation of py-macrodipa-NCS to Glu-urea-Lys-nap-trans-β-Ala.
The bifunctional chelator py-macrodipa-NCS·3TFA (0.012 g, 0.011 mmol) and Glu-urea-Lys-nap-trans-β-Ala (0.008 g, 0.011 mmol) were dissolved in 200 μL and 800 μL of 0.1 M NaHCO3/Na2CO3 buffer (pH 9.0), respectively. These two solutions were combined and stirred for 3 d at RT. This reaction mixture was then filtered and injected into the preparative HPLC system to purify the product (Method: 0–10 min, 90% H2O/CH3CN; 10–40 min, 90% → 0% H2O/CH3CN). Pure fractions were combined, concentrated under reduced pressure, and lyophilized to yield py-macrodipa-PSMA as a white fluffy solid (0.114 g). ESI-HRMS m/z: 1469.6419; calcd for [C74H92N12O18S + H]+: 1469.6446. Analytical HPLC: tR = 16.15 min (Method: 0–5 min, 90% H2O/CH3CN; 5–25 min, 90% → 0% H2O/CH3CN).
Methods and Materials for Radiochemistry.
Caution! Work with radioactive 135La and 44Sc should only be carried out by trained personnel at facilities equipped to safely handle and store these radionuclides. [132/135La]LaCl3 was obtained from the Engle Lab at University of Wisconsin—Madison. [47Sc]ScCl3 was obtained from the Lapi lab at University of Alabama at Birmingham. Radio-HPLC analysis was carried out on the Shimadzu HPLC-20AR equipped with a binary gradient, pump, UV–Vis detector, autoinjector and a Laura radiodetector on a Phenomenex Luna (5 μm, 150 mm x 3 mm, 100 Å). Solvent A = 10 mM NaOAc (pH 5.5); solvent B = MeCN. Gradient: 0–2 min, 5% B; 2–14 min, 5–95% B; 14–16 min, 95% B; 16–16.5 min, 95–5% B; 16.5–20 min, 5% B.
132/135La Radiolabeling.
Py-macrodipa-PSMA (6 μL, 1 mM, 20% DMSO in 0.5 M NaOAc buffer, pH 5.5) was combined with 132/135La3+ (537 μCi, 66 μL) and additional NaOAc buffer (320 μL, 20% DMSO in NaOAc). Quantitative radiolabeling was achieved in 20 min at room temperature and studies were performed without purification.
47Sc Radiolabeling.
Py-macrodipa-PSMA (24 μL, 1 mM, 20% DMSO in 0.5 M NaOAc buffer, pH 5.5) was combined with 47Sc (529 μCi, 93 μL, 0.1 M HCl) and additional NaOAc buffer (283 μL, 0.5 M NaOAc buffer, pH 5.5). Due to the propensity of free 47Sc3+ to be retained on the HPLC column, radiolabeling was confirmed using TLC (50 mM EDTA mobile phase). Under these conditions, free 47Sc3+ moves with the solvent front (Rf > 0.9) and radiolabeled complex remains at the baseline (Rf < 0.1). Quantitative radiolabeling was achieved in 20 min at room temperature and studies were performed without purification.
Ex Vivo Biodistribution and Metabolite Analysis.
All animal experiments were conducted according to the guidelines of the Institutional Animal Care and Use Committee (IACUC) at Stony Brook Medicine operating under federal assurance number #A3011–01. Male Ncr mice (Taconic Biosciences, Rensselaer, NY) were inoculated subcutaneously with PC3-PIP (PSMA+) cells and PC3-flu (PSMA−) cells (1.0 × 106 for PC3-PIP and 0.5 × 106 for PC3-flu) suspended in Matrigel (1:2 DPBS:Matrigel), on the right and left shoulders respectively. When the tumors reached a suitable size, either [132/135La]La3+–py-macrodipa-PSMA (25–34 μCi/0.93–1.26 MBq) and [132/135La]La3+–citrate (21–33 μCi/0.78–1.22 MBq) or [47Sc]Sc3+–py-macrodipa-PSMA (38–80 μCi/1.41–2.96 MBq) and [47Sc]Sc3+–citrate (61–84 μCi/2.26–3.10 MBq) were injected via tail vein catheter. After 2 h, animals were sacrificed by cervical dislocation, and selected tissues were collected and weighed. Activity of tissues was counted using a γ-counter (1470 PerkinElmer Wizard) and the radioactivity associated with each organ was expressed as %ID·g−1. To assess the amount of intact complex remaining, urine was directly injected to the radio-HPLC, and eluate was collected in 30 s increments from 0–20 min. Activity in each tube was quantified using a γ-counter and the counts were used to reconstruct the metabolite trace, which was then compared to the original radiolabeling trace.
Supplementary Material
Acknowledgement
This research was supported by the National Institutes of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Numbers R21EB027282 and R01EB029259. This research made use of the NMR Facility at Cornell University, which was supported, in part, by the U.S. National Science Foundation under award number CHE-1531632. This work was also supported by U.S. Department of Energy, Isotope Program under grant award DESC0020197, and the Horizon-broadening Isotope Production Pipeline Opportunities (HIPPO) program under grant DESC0022550. K.E.M. acknowledges the National Institutes of Health T32 Chemical Biology Interface Training Program (T32 GM136572).
Footnotes
Conflict of Interest
J.J.W. holds equity in Ratio Therapeutics (Boston, MA), which has licensed parts of this technology.
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.
REFERENCES
- [1].Kostelnik TI, Orvig C, Chem. Rev 2019, 119, 902–956. [DOI] [PubMed] [Google Scholar]
- [2].Boros E, Packard AB, Chem. Rev 2019, 119, 870–901. [DOI] [PubMed] [Google Scholar]
- [3].Notni J, Wester H-J, J. Label. Compd. Radiopharm 2018, 61, 141–153. [DOI] [PubMed] [Google Scholar]
- [4].Brugarolas P, Comstock J, Dick DW, Ellmer T, Engle JW, Lapi SE, Liang SH, Parent EE, Kishore Pillarsetty NV, Selivanova S, Sun X, Vavere A, Scott PJH, J. Nucl. Med. Technol 2020, 48, 34S–39S. [PubMed] [Google Scholar]
- [5].Price EW, Orvig C, Chem. Soc. Rev 2014, 43, 260–290. [DOI] [PubMed] [Google Scholar]
- [6].Zeglis BM, Lewis JS, Dalton Trans. 2011, 40, 6168–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hu A, Wilson JJ, Acc. Chem. Res 2022, 55, 904–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sneddon D, Cornelissen B, Curr. Opin. Chem. Biol 2021, 63, 152–162. [DOI] [PubMed] [Google Scholar]
- [9].Holik HA, Ibrahim FM, Elaine AA, Putra BD, Achmad A, Kartamihardja AHS, Molecules 2022, 27, 3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cieslik P, Kubeil M, Zarschler K, Ullrich M, Brandt F, Anger K, Wadepohl H, Kopka K, Bachmann M, Pietzsch J, Stephan H, Comba P, J. Am. Chem. Soc 2022, 144, 21555–21567. [DOI] [PubMed] [Google Scholar]
- [11].Wharton L, de G M, Zhang C, Zeisler J, Rodríguez-Rodríguez C, Osooly M, Radchenko V, Yang H, Lin K-S, Bénard F, Schaffer P, Orvig C, Bioconjugate Chem. 2022, 33, 1900–1921. [DOI] [PubMed] [Google Scholar]
- [12].Wharton L, Zhang C, Zeisler J, Rodríguez-Rodríguez C, Osooly M, Radchenko V, Yang H, Lin K-S, Bénard F, Schaffer P, Orvig C, Bioconjugate Chem. 2022, 33, 2381–2397. [DOI] [PubMed] [Google Scholar]
- [13].Hu A, MacMillan SN, Wilson JJ, J. Am. Chem. Soc 2020, 142, 13500–13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hu A, Aluicio-Sarduy E, Brown V, MacMillan SN, V Becker K, Barnhart TE, Radchenko V, Ramogida CF, Engle JW, Wilson JJ, J. Am. Chem. Soc 2021, 143, 10429–10440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hu A, Simms ME, Kertesz V, Wilson JJ, Thiele NA, Inorg. Chem 2022, 61, 12847–12855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hu A, Brown V, MacMillan SN, Radchenko V, Yang H, Wharton L, Ramogida CF, Wilson JJ, Inorg. Chem 2022, 61, 801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fonslet J, Lee BQ, Tran TA, Siragusa M, Jensen M, Kibédi T, Stuchbery AE, Severin GW, Phys. Med. Biol 2018, 63, 015026. [DOI] [PubMed] [Google Scholar]
- [18].Ahenkorah S, Cassells I, Deroose CM, Cardinaels T, Burgoyne AR, Bormans G, Ooms M, Cleeren F, Pharmaceutics 2021, 13, 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Müller C, Domnanich KA, Umbricht CA, van der Meulen NP, Br. J. Radiol 2018, 91, 20180074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shannon RD, Acta Crystallogr., Sect. A: Found. Adv 1976, 32, 751–767. [Google Scholar]
- [21].Stasiuk GJ, Long NJ, Chem. Commun 2013, 49, 2732–2746. [DOI] [PubMed] [Google Scholar]
- [22].Hu A, Wilson JJ, in Radiopharmaceutical Therapy. (Eds.: Lewis JS, Zeglis BM, Bodei L), Springer Science+Business Media, 2023, 123–144. [Google Scholar]
- [23].Lattuada L, Barge A, Cravotto G, Giovenzana GB, Tei L, Chem. Soc. Rev 2011, 40, 3019–3049. [DOI] [PubMed] [Google Scholar]
- [24].Li L, de G. Jaraquemada-Peláez M, Aluicio-Sarduy E, Wang X, Barnhart TE, Cai W, Radchenko V, Schaffer P, Engle JW, Orvig C, Dalton Trans. 2020, 49, 5547–5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lüning U, Baumstark R, Peters K, von Schnering HG, Liebigs Ann. der Chemie 1990, 129–143. [Google Scholar]
- [26].Thiele NA, Brown V, Kelly JM, Amor-Coarasa A, Jermilova U, MacMillan SN, Nikolopoulou A, Ponnala S, Ramogida CF, Robertson AKH, Rodríguez-Rodríguez C, Schaffer P, Williams C, Babich JW, Radchenko V, Wilson JJ, Angew. Chem., Int. Ed 2017, 56, 14712–14717. [DOI] [PubMed] [Google Scholar]
- [27].Śmiłowicz D, Schlyer D, Boros E, Meimetis L, Mol. Pharm 2022, 19, 3217–3227. [DOI] [PubMed] [Google Scholar]
- [28].Kwon H, Son S-H, Byun Y, Asian J Org. Chem 2019, 8, 1588–1600. [Google Scholar]
- [29].Sartor O, Herrmann K, J. Nucl. Med 2022, 63, 823–829. [DOI] [PubMed] [Google Scholar]
- [30].Benešová M, Bauder-Wüst U, Schäfer M, Klika KD, Mier W, Haberkorn U, Kopka K, Eder M, J. Med. Chem 2016, 59, 1761–1775. [DOI] [PubMed] [Google Scholar]
- [31].Aluicio-Sarduy E, Hernandez R, Olson AP, Barnhart TE, Cai W, Ellison PA, Engle JW, Sci. Rep 2019, 9, 10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Loveless CS, Blanco JR, Diehl III GL, Elbahrawi RT, Carzaniga TS, Braccini S, Lapi SE, J. Nucl. Med 2021, 62, 131LP–136. [DOI] [PubMed] [Google Scholar]
- [33].Aluicio-Sarduy E, Barnhart TE, Weichert J, Hernandez R, Engle JW, J. Nucl. Med 2021, 62, 1012–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Aluicio-Sarduy E, Thiele NA, Martin KE, Vaughn BA, Devaraj J, Olson AP, Barnhart TE, Wilson JJ, Boros E, Engle JW, Chem. - Eur. J 2020, 26, 1238–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Vaughn BA, Koller AJ, Chen Z, Ahn SH, Loveless CS, Cingoranelli SJ, Yang Y, Cirri A, Johnson CJ, Lapi SE, Chapman KW, Boros E, Bioconjugate Chem. 2021, 32, 1232–1241. [DOI] [PubMed] [Google Scholar]
- [36].Abou DS, Thiele NA, Gutsche NT, Villmer A, Zhang H, Woods JJ, Baidoo KE, Escorcia FE, Wilson JJ, Thorek DLJ, Chem. Sci 2021, 12, 3733–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kadassery KJ, King AP, Fayn S, Baidoo KE, MacMillan SN, Escorcia FE, Wilson JJ, Bioconjugate Chem. 2022, 33, 1222–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wu C, Kobayashi H, Sun B, Yoo TM, Paik CH, Gansow OA, Carrasquillo JA, Pastan I, Brechbiel MW, Bioorg. Med. Chem 1997, 5, 1925–1934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.