Abstract
Metastasis is responsible for most breast cancer–related deaths; however, identifying the cellular determinants of metastasis has remained challenging. Here, we identified a minority population of immature THY1+/VEGFA+ tumor epithelial cells in human breast tumor biopsies that display angiogenic features and are marked by the expression of the oncogene, LMO2. Higher abundance of LMO2+ basal cells correlated with tumor endothelial content and predicted poor distant recurrence–free survival in patients. Using MMTV-PyMT/Lmo2CreERT2 mice, we demonstrated that Lmo2 lineage–traced cells integrate into the vasculature and have a higher propensity to metastasize. LMO2 knockdown in human breast tumors reduced lung metastasis by impairing intravasation, leading to a reduced frequency of circulating tumor cells. Mechanistically, we find that LMO2 binds to STAT3 and is required for STAT3 activation by tumor necrosis factor–α and interleukin-6. Collectively, our study identifies a population of metastasis-initiating cells with angiogenic features and establishes the LMO2-STAT3 signaling axis as a therapeutic target in breast cancer metastasis.
LMO2 modulates STAT3 signaling to promote breast cancer metastasis.
INTRODUCTION
While notable progress has been made to treat early-stage breast cancer, treatment options and outcomes for metastatic breast cancer have been largely unchanged in a decade (1–3). To improve outcomes for patients with breast cancer, it is critical to identify and elucidate signaling pathways active in metastatic cells. However, it has been difficult to pinpoint cancer cell populations involved in metastasis as they represent a transient state (4). Previous studies using lineage tracing and cell surface marker profiling have implicated distinct subsets of tumor epithelial cells in breast cancer metastasis, primarily using lineage markers such as E-cadherin (5), N-cadherin (6), and S100A4 (7). Recent studies have also suggested that metastatic cells display hybrid features of both epithelial and mesenchymal lineages (8), but the precise molecular identity of these cells remains unknown.
Our previous work has demonstrated that in breast cancer, minority populations of phenotypically immature cells in the tumor are enriched in tumor-initiating potential and metastasis (9–11). Recent advances in single-cell technologies have revealed complex transcriptional landscapes in human tumors and enabled precise molecular characterization of these minority cell populations (12). However, the functional and clinical significance of these populations remains unclear. To better understand the transcriptional heterogeneity in breast cancer, we recently performed single-cell RNA sequencing (scRNA-seq) in primary patient samples and developed a novel computational method that can predict immature cell populations in silico (13). Using our scRNA-seq data, bulk tumor expression deconvolution, lineage tracing, and functional assays, we have now identified a clinically relevant population of metastasis-initiating cells that express the hematopoietic transcription factor and T cell oncogene, LMO2. Here, we mechanistically define the role of LMO2 in breast cancer metastasis by its association with tumor vasculature and identify LMO2 as a previously unknown regulator of signal transducers and activators of transcription 3 (STAT3) signaling in breast cancer.
RESULTS
LMO2 is expressed in a minority population of immature THY1+/VEGFA+ human breast cancer cells
To dissect the substructure of the epithelial cell populations in breast cancer, we started by analyzing scRNA-seq profiles (13) of triple-negative (n = 5) or estrogen receptor–positive breast cancer (n = 13) from patient tumor specimens. We identified a minority population of THY1+ cells that were largely restricted to the basal compartment, comprising 11% of all basal cells (fig. S1, A and B, and table S1). Moreover, within this subset, 33% of cells expressed VEGFA (fig. S1, A and B). We were struck by this combination since THY1+ cells are enriched in reconstitution potential in the normal mammary gland (14) and tumorigenic potential in mouse tumors (15), and VEGFA is a proangiogenic factor linked to tumor growth and distant metastasis (16, 17). To determine whether THY1+/VEGFA+ cells represent a potential immature cell population, we applied CytoTRACE, a computational framework for predicting cellular differentiation status on the basis of single-cell transcriptional diversity (13). We found that relative to other basal cells, THY1+/VEGFA+ cells are predicted to be significantly less differentiated (Fig. 1A), suggesting a role for this population in tumor growth or metastasis. To understand the distribution of THY1+/VEGFA+ basal cells across breast cancer clinical subtypes, we performed deconvolution analysis using CIBERSORTx of three different clinical cohorts (18–20). We found that THY1+/VEGFA+ basal cells were more abundant among human epidermal growth factor receptor 2 (HER2)–enriched and basal subtypes of breast cancer compared to the luminal and normal subtypes (fig. S1C). Moreover, THY1+/VEGFA+ basal cells were significantly enriched in higher-grade tumors (fig. S1D).
Fig. 1. Identification of an immature basal epithelial population associated with proangiogenic signaling and poor survival in human breast cancer.
(A) Differentiation scores of basal epithelial cells from 17 human breast tumors profiled by scRNA-seq. Differentiation scores (0, more differentiated; 1, less differentiated) were determined by CytoTRACE (13). *P < 0.1; **P < 0.05; ***P < 0.01, unpaired two-tailed t test. (B) Plot showing protein-coding genes ordered by their enrichment in THY1+/ VEGFA+ basal cells from (A). (C) Paired bar plots showing fraction of LMO2+ cells in THY1+/VEGFA+ cells (red) and other cells (blue) in two human breast cancer datasets, tumor cells only, (n = 659) (23), and the basal cells (n = 910) from this study. Individual and combined P values by Fisher’s method. *P < 0.1; **P < 0.05; ***P < 0.01. (D) Heatmap depicting the top 30 differentially expressed genes, with selected lineage markers, in LMO2+ (n = 7 cells) versus LMO2− (n = 903 cells) basal epithelial cells from (A). A random subsample of 50 LMO2− basal cell transcriptomes is shown. Color scale (above) represents z score–normalized expression per gene. (E) Differential enrichment of the HALLMARK_ANGIOGENESIS pathway in LMO2+ versus LMO2− human breast cancer datasets described in (C). An empirical P value was calculated by Monte Carlo Approach (Materials and Methods). Combined P value by Fisher’s method *P < 0.1; **P < 0.05; ***P < 0.01. (F and G) Cell-type and survival association of LMO2+ basal cells across 508 bulk human breast tumor transcriptomes (18) deconvolved using CIBERSORTx. ER, estrogen receptor. (F) Coassociation patterns among cell type abundance profiles in bulk breast tumors, quantified by Pearson correlation. (G) Kaplan Meier curves showing differences in distant recurrence–free survival (DRFS) stratified by the median abundance of LMO2+ basal epithelial cells. DRFS was modeled as a function of LMO2+ basal cell status and ESR1 status (Materials and Methods). Adjusted log-rank P value and hazard ratio (HR) with 95% confidence interval for LMO2+ basal cell status is shown. ER, estrogen receptor.
To identify potential molecular regulators within this population, we next searched for genes with expression patterns that overlap THY1 and VEGFA expression in our dataset. Intriguingly, we found that LMO2, a hematopoietic stem cell regulator (21) and T cell oncogene (22), was among the top five hits (Fig. 1B and table S2). LMO2 also marked THY1+/VEGFA+ cells in an independent scRNA-seq atlas of triple-negative human breast tumors (23), corroborating this result (Fig. 1C). Analysis of the LMO2+ basal epithelial subset showed that these cells not only express THY1 and epithelial cytokeratins (Fig. 1D) but also display a coherent gene expression program significantly enriched in angiogenesis genes, including VEGFA and S100A4 (Fig. 1E and table S3).
We next measured the relative abundance of distinct endothelial, immune, stromal, and epithelial populations in human breast tumors with respect to LMO2+ basal cells. As LMO2 is expressed in multiple cell types, including immune, stromal, and endothelial cells (ECs) (21, 24, 25), the expression of this gene is insufficient to distinguish cell types. Therefore, we defined unique transcriptional signatures for various niche and breast epithelial cells from our scRNA-seq data and used CIBERSORTx to calculate the cellular composition of bulk RNA admixtures from breast cancer clinical cohorts (26) (Materials and Methods). In line with our previous results, we observed a notable correlation between the abundance of LMO2+ basal cells and EC content imputed in 508 breast tumors (r = 0.45; P < 2 × 10−16; Fig. 1F) (18).
Human LMO2+ basal cells are associated with poor outcomes in patients with breast cancer
Deconvolution of an additional 3024 human breast tumors from three clinical cohorts (18–20) found a significant increase in basal LMO2+ cells with worsening clinical grade and stage of the tumor (fig. S2, A and B), suggesting that LMO2+ cell number increase with tumor progression. Furthermore, our analysis revealed that basal LMO2+ cells are more abundant in “basal” breast cancer subtypes, which correlate with more aggressive breast cancers as compared to other Prediction Analysis of Microarray 50 (PAM50) classes (fig. S2C) (27). Importantly, higher levels of LMO2+ basal cells were significantly associated with inferior distant recurrence–free survival (DRFS) (Fig. 1G and fig. S2D), independent of estrogen receptor status. These data link the abundance of LMO2+ basal epithelial cells with more aggressive breast tumors and distant metastasis.
Lmo2 lineage–traced cells have a higher propensity to metastasize
To experimentally verify our in silico findings, we began by using the CreERT2 system to delineate the fate of epithelial cells that have expressed LMO2 in breast tumors. We obtained Lmo2CreERT2 mice and crossed them to Rosa26mTmG reporter and MMTV-PyMT tumor mice to generate triple-transgenic Lmo2CreERT2/Rosa26mTmG/MMTV-PyMT mice, which we termed Lmo2-PyMT (Fig. 2A). MMTV-PyMT tumors are an aggressive luminal subtype of breast cancer (28) that metastasize to the lungs (29) and have been extensively used to explore the cellular underpinnings of breast cancer metastasis (5, 7, 30). Recent studies have also demonstrated that MMTV-PyMT tumors are heterogeneous and contain populations of cells with basal characteristics (31, 32), allowing us to explore the role of Lmo2 in these tumor cells. As Lmo2 is expressed in other cells such as stromal and ECs (33), we orthotopically transplanted lineage-depleted (CD45−/CD31−/Ter119−) tumor cells from TdTomato-fluorescent Lmo2-PyMT into nonfluorescent BL6 mice to clearly assess the contribution of Lmo2 lineage–traced breast cancer cells from the tumor. After tumor formation, we pulsed the mice with tamoxifen to induce expression of green fluorescent protein (GFP) in Lmo2-expressing cells (Fig. 2B). At 48 hours after pulse, we verified that expression of Lmo2 was enriched in the transplanted GFP+ cancer cells (Fig. 2C and figs. S3 and S4A). Flow cytometry analysis and quantification demonstrated that GFP+ cells represented a minor fraction of all tumor cells and expressed epithelial cellular adhesion molecule (EpCAM), confirming their epithelial identity (Fig. 2C and fig. S3A). Moreover, as PyMT tumors are luminal in nature with a minority population of basal cells (31, 32), we found that at 48 hours after pulse, GFP+ cells expressed both luminal and basal cytokeratin with a slight reduction in luminal cytokeratins, specifically in cytokeratin-18 (fig. S4, A to D).
Fig. 2. Lmo2 lineage–traced tumor epithelial cells integrate into the vasculature and can form metastasis in PyMT tumors.
(A) Schematic diagram showing generation of the triple transgenic Rosa26mTmG reporter with MMTV-PyMT and Lmo2-CreERT2 mice (referred to as Lmo2-PyMT). (B) Schematic diagram showing the experimental scheme for Lmo2-PyMT tumors treated with tamoxifen. (C) Left Panel: FACS analysis of Lmo2-PyMT tumors 48 hours after tamoxifen pulse. Cells are gated on lineage− (CD45−, CD31−, and Ter119−) and DAPI− cells (see fig. S3) and analyzed using TdTomato+ and GFP+. Middle and Right Panels: EpCAM and CD49f expression status in GFP+ and TdTomato+ cells. (D) Quantification of GFP+ cells from Lmo2-PyMT tumors (n = 5 mice). (E) Representative immunofluorescence image of Lmo2 lineage–traced cells (GFP+ green) colocalizing and integrating with endomucin (magenta)–stained tumor vasculature. High-resolution magnification of insets 1 and 2 are presented. Scale bars, 50 μm. (F) Schematic diagram showing the experimental scheme for Lmo2-PyMT tumors treated with tamoxifen to trace metastatic cells. (G) Left Panel: FACS analysis of Lmo2-PyMT tumors at tumor end point from (F). Cells are gated on lineage− (CD45−, CD31−, and Ter119−) and DAPI− cells (see fig. S3) and analyzed using TdTomato+ and GFP+. Middle and Right Panel: EpCAM and CD49f expression status in GFP+ and TdTomato+ cells. Panel 4: Quantification of TdTomato+ and GFP+ cells from Lmo2-PyMT tumors (n = 4 mice). (H) Panel 1: Representative image of metastasis shown. Scale bar, 100 μm. Panel 2: Quantification of total number and area of GFP+ and TdTomato+ lung metastasis in Lmo2-PyMT tumors. Each dot represents a single metastatic focus (n = 4 mice). Data are shown as means ± SD, and statistical analysis was performed by unpaired, two-sided Wilcoxon rank sum test, *P < 0.05.
To assess the population dynamics of Lmo2 lineage–traced cells, we plated TdTomato+ tumor cells from Lmo2-PyMT mice in three-dimensional (3D) organoid assays and pulsed the organoids with 4-hydroxytamoxifen. Consistent with the in vivo model, lineage-traced GFP+ cells comprised a minority of tumor organoids (~2%) 7 days after pulse (fig. S4E). This percentage was unchanged even after 4 weeks in culture (fig. S4E), suggesting similar proliferative capacity between GFP+ and TdTomato+ cells. We confirmed this by plating sorted GFP+ and TdTomato+ cells in 3D organoid cultures and showing that both populations formed organoids at similar frequencies (fig. S4F).
To determine whether Lmo2+ cells coassociate with ECs, as predicted in silico (Fig. 1F), we stained the vasculature with endomucin and visualized their colocalization with 3D imaging. Consistent with our in silico analysis, we found that GFP+ cells are significantly enriched in areas with higher endothelial content and vice versa (fig. S4G), with ~64% Lmo2 lineage–traced cells residing near tumor blood vessels (fig. S4H). Unexpectedly, ~14% showed colocalization with tumor vasculature and appeared to be incorporated into the tumor vasculature (Fig. 2E and figs. S4H and S5, A and B). We also found that Lmo2 lineage–traced cells that are closer to vasculature have an elongated cellular morphology or long projections, suggesting that they are in the process of incorporating into the vasculature (fig. S5, C and D).
Given that the abundance of LMO2+ cells in patients with breast cancer predicts DRFS (Fig. 1G) and Lmo2 lineage–traced cells reside closer to and incorporate into tumor vasculature, we next tested whether Lmo2+ cells have metastatic capabilities. As dissemination of metastatic cells occurs continuously during tumor growth, to lineage trace tumor cells expressing Lmo2, we pulsed Lmo2-PyMT mice with tamoxifen two to three times per week once the tumors were palpable and continued until the tumor size endpoint (see Materials and Methods; Fig. 2F). At the end of the experiment, we found that in the primary tumor, only 10 to 15% of tumor cells were GFP+ (Fig. 2G). Unexpectedly, although the tumor was mostly TdTomato+, the lungs had a disproportionately higher number of GFP+ metastases, several of which were larger than the TdTomato+ metastases (P = 0.034, Wilcoxon signed-rank unpaired test) (Fig. 2H). These data suggest that Lmo2 lineage–traced cells have a higher propensity to form metastases in the PyMT mice, although we cannot exclude that some TdTomato+ cells may have switched to GFP+ at the metastatic site. Our results are consistent with our findings in human breast cancer patients (Fig. 1G). Furthermore, a subset of GFP tumor cells did not remain Lmo2 positive (fig. S4I), suggesting that expression of Lmo2 in some cells represents a transient state, in agreement with previous studies linking transient cell states to metastasis (8).
LMO2 knockdown abrogates lung metastasis in human breast cancer models
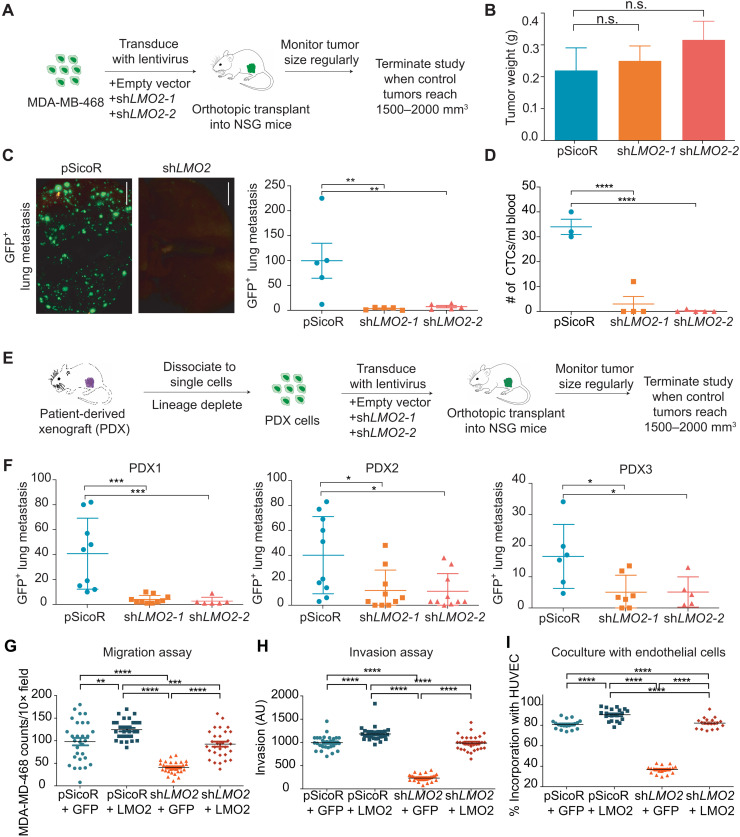
To understand the functional role of LMO2 in human breast cancer, we knocked down LMO2 expression in MDA-MB-468 cells using two independent short hairpin RNA (shRNA) vectors tagged with a GFP reporter (fig. S6, A to C). We then implanted the cells orthotopically in immunodeficient mice (Fig. 3A). In contrast to a previous report (34), knockdown of LMO2 did not significantly affect primary tumor growth (Fig. 3B and fig. S7, A and B) or proliferation in vitro (fig. S7C). Nevertheless, LMO2 knockdown tumors had significantly fewer lung metastases relative to control [P = 0.003, analysis of variance (ANOVA); Fig. 3C]. Moreover, LMO2 knockdown in tumor-bearing mice led to a significantly reduced number of circulating tumor cells compared to control mice (P < 0.0001, ANOVA; Fig. 3D), implicating LMO2 in cancer cell shedding, a key step in metastasis initiation. To extend our findings to more clinically relevant models, we used patient-derived xenograft (PDX) models previously generated in our laboratory (10). Consistent with our MDA-MB-468 studies, knockdown of LMO2 markedly decreased metastasis to the lung in three different PDX models of breast cancer (Fig. 3, E and F) but did not significantly affect tumor growth (fig. S7, D to I).
Fig. 3. Knockdown of LMO2 reduces lung metastasis in human breast cancer.
(A) Schematic of LMO2 knockdown in MDA-MB-468 cells followed by orthotopic transplant in NSG mice to evaluate tumor burden and metastases. (B) Tumor weight in control (pSicoR) and LMO2 knockdown tumors generated from MDA-MB-468 cell xenografts as shown in (A) (n = 5 mice per group). not significant (n.s.), P > 0.05, ANOVA. (C) Spontaneous GFP+ lung metastases from mice with control and LMO2 knockdown tumors in (B) (n = 5 mice per group). Left: Representative immunofluorescence image. Scale bar, 5 mm. Right: Quantification. **P < 0.01, ANOVA. (D) Number of circulating tumor cells in control and LMO2 knockdown tumors (n = 3 mice in pSicoR, 4 in shLMO2-1, and 5 in shLMO2-2). ****P < 0.0001, ANOVA. (E) Schematic of LMO2 knockdown in patient-derived xenografts (PDXs) followed by orthotopic transplant in NSG mice to evaluate tumor burden and metastases. (F) Number of spontaneous GFP+ lung metastases in control and LMO2 knockdown tumors using PDXs. Data are combined from three independent experiments for PDX1 and PDX3 and from two independent experiment for PDX2 (n = 9 mice per group for PDX1, n = 6 mice per group for PDX2, n = 10mice per group for PDX3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ANOVA. (G) MDA-MB-468 cells infected with shRNA targeting 3′UTR of LMO2 or a control shRNA pSicoR were infected with an empty vector control “GFP” or a LMO2 overexpression vector “+LMO2” to generate pSicoR+GFP, pSicoR + LMO2, shLMO2 + GFP, and shLMO2 + LMO2. Transwell migration quantified at 24 hours. (H) Spheroid invasion assay was quantified at day 5 using the breast cancer cells from (G). (I) MDA-MB-468 cells from (G) were cocultured with HUVEC cells, and the percentage of breast cancer cells colocalizing with HUVEC tubes was quantified using ImageJ. For all experiments in (G) to (I), n = 3 and 10 images were analyzed per condition per n. **P < 0.01, ***P < 0.001, ****P < 0.0001, ANOVA (A to I) All data are means ± SD. AU, arbitrary units.
To better understand how LMO2 affects metastasis, we rigorously studied the effects of LMO2 knockdown in vitro in MDA-MB-468 cells. Knockdown of LMO2 showed significant impairment in the ability of cancer cells to migrate across transwells and invade through a 3D hydrogel matrix (fig. S8, A and B). Since LMO2+ epithelial cells associated with ECs in patient samples and MMTV-PyMT tumors, we tested whether knockdown of LMO2 decreased this association in coculture assays. We found that in 3D coculture assays with human vascular ECs (HUVECs), LMO2 knockdown significantly affected incorporation of cancer cells into HUVEC tubes (fig. S8C). To confirm that the effects of knockdown were specific to LMO2, we overexpressed LMO2 in cells with shRNA targeting the 3′ untranslated region (3′UTR). We found that all phenotypes of migration (Fig. 3G and fig. S8D), invasion (Fig. 3H and fig. S8E), and incorporation into the vasculature in vitro (Fig. 3I and fig. S8F) could be rescued by overexpression of LMO2 in LMO2-deficient cells. Lastly, to test whether LMO2 is required after metastatic cells enter circulation, we injected control and LMO2 knockdown cells into the tail vein. We found that LMO2 knockdown did not significantly affect the formation of lung metastases when cells were directly injected in the tail vein (fig. S8G). This suggests that LMO2 is critical for the initial dissemination of cancer cells from the tumor but not for extravasation and formation of metastatic foci.
RNA-seq identifies LMO2 as a regulator of IL6-JAK-STAT3 signaling
To elucidate the molecular function of LMO2 in breast cancer cells, we performed bulk RNA-seq of MDA-MB-468 cells after transfection with control and LMO2 shRNA vectors (Fig. 4A). Among the top 50 genes down-regulated after LMO2 knockdown were genes previously implicated in metastasis, such as BMP2 (35), LGR6 (36), EGR4 (37), TDO2 (38), and S100A4 (Fig. 4A and table S4) (39). Using gene set enrichment analysis (GSEA) (40), we found that inflammatory pathways, such as tumor necrosis factor–α (TNF-α) via nuclear factor κB (NF-κB) signaling, interleukin-6 (IL-6)–Janus kinase (JAK)–STAT3 signaling, and interferon-γ (IFN-γ) response, were significantly down-regulated in LMO2 knockdown as compared to control conditions (Fig. 4B). To confirm our findings in primary patient samples, we performed single-sample GSEA (ssGSEA) in our scRNA-seq dataset and a larger published dataset of primary human breast cancer cells (23). We found that IL6-JAK-STAT3 signaling was significantly enriched in LMO2+ versus LMO2− single cells (Fig. 4C) compared to other pathways (fig. S9, A and B). Moreover, IL6-JAK-STAT3 signaling was also significantly enriched in Lmo2+ PyMT cancer cells in two different scRNA-seq datasets (31, 32), further confirming our findings in a mouse tumor model (fig. S9C).
Fig. 4. LMO2 regulates the IL6-JAK-STAT3 pathway and binds to STAT3.
(A) Top: Schematic of bulk RNA-seq analysis in MDA-MB-468 cells infected with shRNAs targeting LMO2 or a control pSicoR. Bottom: Heatmap showing top and bottom 50 genes differentially expressed between control and LMO2 knockdown conditions, ordered by P adjusted value. (B) Left: Hallmark gene sets found to be significantly enriched by GSEA analysis. Normalized enrichment scores (corresponding to control pSicoR versus LMO2 knockdown) and false discovery rate (FDR) Q values are determined by the GSEA software. An FDR Q value cutoff of <0.25 was used to select significant gene sets. Right: Enrichment plots for HALLMARK_TNFα_SIGNALING_VIA_NFκB and HALLMARK_IL6_JAK_STAT3_SIGNALING are depicted. NES, normalized enrichment score. (C) Differential enrichment of the HALLMARK_IL6_JAK_STAT3_SIGNALING pathway in LMO2+ versus LMO2− cells from two independent human breast cancer datasets as described in Fig. 1C. **P < 0.05. (D) Proximity-mediated ligation assay showed that LMO2 had a stronger interaction with STAT3 compared to NF-κB in vitro (n = 3, 10 images were analyzed per condition per n). Statistical analysis was performed by ANOVA with Dunnett’s adjustment. ****P < 0.0001.
In the hematopoietic system, LMO2 is an adaptor protein that facilitates formation of functional protein complexes, which then activate transcription of downstream targets (25). Hence, we asked whether LMO2 may similarly behave as a bridging molecule to drive downstream signaling in breast epithelial cells. Using proximity ligation assays (PLA), we found that LMO2 had a significantly high binding affinity to STAT3 but not to NF-κB, further confirming our pathway analysis (Fig. 4D).
As epithelial-to-mesenchymal transition (EMT) is linked to increased metastatic capability, we specifically tested whether LMO2 knockdown affects EMT. Our comparison of classical EMT genes between control and LMO2 knockdown in MDA-MB-468 cells showed divergent expression of various EMT-related genes in the two populations (fig. S10, A and B). This suggested to us that the EMT pathway is not primarily affected after LMO2 knockdown, although we did observe minor changes in genes implicated in EMT, such as the epithelial marker EpCAM (41) and the mesenchymal marker S100A4 (39), which define the end points of each state (fig. S10A). We also analyzed our scRNA-seq data for canonical EMT genes and found that, consistent with our knockdown experiments, EMT gene sets and specific cytokeratin, epithelial, and mesenchymal genes were expressed at similar levels in both LMO2+ and LMO2− basal cells (fig. S10, C and D).
LMO2 is required for STAT3 activation by IL-6 and TNF-α
To demonstrate specificity and functional significance of the LMO2-STAT3 interaction, we first showed that LMO2 knockdown significantly reduced LMO2-STAT3 binding (P < 0.0001, ANOVA; Fig. 5A). We also confirmed the LMO2-STAT3 interaction using coimmunoprecipitation (co-IP) assays of LMO2 with STAT3 (Fig. 5B) and a reverse co-IP of STAT3 with LMO2 (Fig. 5C). In breast cancer, STAT3 is activated by cytokines, such as IL-6 (42), TNF-α (43), IFN-α (44), and IFN-γ (45) as well as growth factors such as epidermal growth factor (EGF) (42), leading to phosphorylation of STAT3. Dimerization of pSTAT3 and translocation to the nucleus activates transcription of downstream target genes involved in several processes, including metastasis (38, 46). To understand whether the STAT3-LMO2 interaction influences downstream STAT3 signaling, we used a STAT3-luciferase reporter assay. We stimulated control or LMO2 knockdown cells with IL-6, TNF-α, IFN-γ, IFN-α, and EGF. We found that cells with knockdown of LMO2 were unable to induce transcription of the STAT3-luciferase reporter when treated with IL-6 and TNF-α as compared to control (Fig. 5D), but STAT3-luciferase was activated by IFN-γ, IFN-α, and EGF treatment (Fig. 5D). On a molecular level, we found that LMO2 knockdown significantly reduced STAT3 phosphorylation at Tyr705 when treated with IL-6 and TNF-α (Fig. 5E and fig. S11A). However, LMO2 knockdown did not affect STAT3 phosphorylation on treatment with other STAT3 activators such as EGF, IFN-α, and IFN-γ (fig. S11, B and C), confirming our luciferase reporter assay. This suggests that LMO2 function in breast cancer cells is specific to activation of STAT3 signaling through IL-6 and TNF-α.
Fig. 5. LMO2 stabilizes STAT3 signaling in breast cancer cells.
(A) Left: Proximity-mediated ligation assay between LMO2 and STAT3 in control and LMO2 knockdown cells. Right: Quantification of n = 3 experiments, 10 images per condition per n. Scale bar, 60 μm. ****P < 0.0001, ANOVA. (B) Western blot of the input, immunoprecipitated beads (control), IgG (control), and LMO2 to pull down STAT3. One representative blot (n = 3). (C) Western blot of the input, immunoprecipitated beads (control), IgG (control), and STAT3 to pull down LMO2. One representative blot (n = 3). (D) STAT3-luciferase reporter activity in control and LMO2 knockdown cells treated with IL-6, TNF-α, IFN-α, IFN-γ, and EGF. n = 3. *P < 0.05, ***P < 0.001 and ****P < 0.0001, n.s. P > 0.05, two-way ANOVA. (E) Immunoblotting (left) and quantification (right) of phosphorylated STAT3 in control and LMO2 knockdown cells treated with IL-6 and TNF-α. n = 3. ***P < 0.001, ****P < 0.0001, two-way ANOVA. (F) Proximity ligation assays between STAT3 and JAK2 in control and LMO2 knockdown cells. n = 3. ****P < 0.0001, ANOVA. (G) Proximity ligation assays between STAT3 and PIAS3 in control and LMO2 knockdown cells. n = 3. ****P < 0.0001, ANOVA. (H) Schematic of proposed mechanism of LMO2 in breast cancer metastasis. Cancer cells that express LMO2 have stabilized STAT3 signaling in response to IL-6 and TNF-α from the microenvironment, allowing these cells to intravasate into the circulation by incorporating into the vasculature.
To understand how LMO2 regulates phosphorylation of STAT3, we examined the interaction of STAT3 with its upstream activator JAK2 and its cytoplasmic inhibitor PIAS3. Knockdown of LMO2 decreased the interaction of STAT3 with JAK2 (Fig. 5F) and allowed for increased interaction with its inhibitor, PIAS3 (Fig. 5G). This suggests that LMO2 works as an adaptor protein in the cytoplasm to stabilize the STAT3-JAK2 interaction, thereby allowing efficient phosphorylation and activation of STAT3 while simultaneously preventing its negative regulation by PIAS3 (Fig. 5H). Indeed, directly inhibiting STAT3 with the small molecule, Stattic (47), reduced the binding of MDA-MB-468 cells to HUVEC tubes in vitro (fig. S12). In addition, direct inhibition of STAT3 has been previously demonstrated to reduce metastasis in experimental models, but STAT3 inhibitors have had limited therapeutic success in human breast cancer (47). This LMO2-mediated control of a core inflammatory response pathway likely enables cancer cells to rapidly transition between cellular phenotypes required for metastasis and represents a therapeutic vulnerability that could be targeted.
DISCUSSION
Here, we have identified a population of THY1+/VEGFA+ human basal epithelial cells with higher transcriptional diversity that are marked by expression of LMO2. LMO2+ basal cells were associated with inferior DRFS and enriched in the aggressive basal-like PAM50 subtype (27). We demonstrated that Lmo2 lineage–traced epithelial cells have a higher propensity to form lung metastases. LMO2 knockdown decreased lung metastasis in multiple tumor models of human breast cancer by affecting multiple steps during intravasation, resulting in fewer circulating tumor cells.
Efficient metastasis of tumor cells requires transition from a proliferative state to an invasive state and back to a proliferative state at a distant site (5). Previous studies using mouse tumor models have demonstrated the requirement of a basal epithelial program in metastasis and acquisition of mesenchymal features (6, 48). We identified LMO2+ cells as a minority population of basal cells in human primary patient samples; however, its expression is not exclusive to the basal compartment. This is consistent with previous studies showing that luminal progenitors in triple-negative breast cancer and breast cancer gene (BRCA) mutated tumors acquire basal characteristics, express basal cytokeratins, and are prone for metastasis (49). Our current data do not allow us to exclude the influence of EMT on metastasis downstream of the STAT3/LMO2 axis but does suggest that LMO2+ cells do not represent a fully mesenchymal state. Given recent studies showing that hybrid epithelial-mesenchymal states harbor higher plasticity (50), we speculate that LMO2+ cells could represent a hybrid cell population that has higher levels of activated STAT3 signaling and ability to bind to tumor vasculature. Indeed, it has been shown that hybrid epithelial-mesenchymal cells in metastasis express angiogenic factors (8).
Our results highlight a role for LMO2 in early metastatic dissemination by affecting intravasation but not extravasation. This difference is possibly because intravasation mostly occurs at an abnormal leaky tumor vasculature site, whereas extravasation of the migrated cancer cell involves normal blood vessels (51). In addition, previous studies have shown that creating a tumor microenvironment at the primary tumor is critical for metastasis (52). Thus, based on our data, we speculate that a minority population of cells is induced to express LMO2 in such an environment, thereby stabilizing STAT3 signaling to promote metastasis.
Although LMO2 knockdown did not significantly affect tumor weight, there was a trend toward slightly smaller tumors. Previously, it has been demonstrated that aggressive metastatic cancer cells can reseed the primary tumor and increase tumor growth in a process called tumor self-seeding (53). A previous study demostrated that LMO2 is among the top 50 genes overexpressed in self-seeding metastatic cells (53). Thus, reduction in self-seeding of the tumor can likely explain the nonsignificant decrease in tumor weight observed in LMO2 knockdown tumors.
It is also important to note that only a subset of Lmo2 lineage–traced cells showed vascular phenotypes, suggesting specific epigenetic regulation that is activated in the presence of TNF-α and IL-6 from the microenvironment. Our observations highlight a heterogeneous, cancer cell–intrinsic response to the microenvironment, while previous studies have demonstrated that there is a reciprocal effect of cancer cells on the tumor microenvironment with recruitment of macrophages and cross-talk with tumor ECs during metastasis (54). As our functional studies were done in immunocompromised mice, which have only some components of the innate immune system, future studies will explore the interaction of Lmo2+ cells with immune cells using syngeneic models of breast cancer.
LMO2 has been extensively studied in hematological malignancies (22, 39) and ECs (24, 33) and is well established as a transcriptional adaptor protein (25). Recent studies have attempted to understand the role of LMO2 in breast cancer (34, 55) but have suffered from contradictory results, were limited to cell lines, and did not attribute LMO2 to any particular tumor cell population. We demonstrate that LMO2 is a previously unidentified binding partner of STAT3 in breast cancer cells and modulates STAT3 signaling in response to IL-6 and TNF-α. We speculate that the expression of LMO2 provides the necessary threshold to stabilize STAT3 signaling, which, in turn, enables the tumor cells to enter a transient metastatic state (4) and escape the primary tumor. STAT3 signaling is involved in several processes, and its targets may be defined in unison with other contextual signals such as inflammation. Several studies have linked chronic inflammation in cancer to metastasis (56). By serving as a critical molecular link between these processes, our results define a previously unidentified function for LMO2 in breast cancer metastasis. The development of new methods targeting adaptor proteins (57) and small molecules that disrupt the LMO2-STAT3 axis (58, 59) could provide novel therapeutic strategies to modulate STAT3 signaling and inhibit metastatic colonization in breast cancer.
MATERIALS AND METHODS
Preparation of single-cell suspensions for human and mouse tissues
For human samples, informed consent was obtained after the approval of protocols by the Stanford University and City of Hope Institutional Review Boards (IRB no. 4344). Tumor biopsies from human breast cancer patients (n = 18) were obtained from the primary site (n = 16), lymph nodes (n = 1; paired primary), or brain metastasis (n = 2) during surgical resection of breast tumors at Stanford Hospital and City of Hope National Medical Center (table S1). Samples were mechanically dissociated into <1- to 2-mm3 pieces with a razor blade and then digested at 37oC with 1500 U of collagenase and 500 U of hyaluronidase in advanced Dulbecco’s modified Eagle’s medium (DMEM)/F-12 (Thermo Fisher Scientific), 2 mM glutamax (Invitrogen), and an antibiotic/antimycotic mix containing penicillin (120 μg/ml), streptomycin (100 μg/ml), and amphotericin-B (0.25 μg/ml) (PSA) for 4 to 6 hours with hourly pipetting for 5 min. After digestion, cells were treated with ammonium-chloride-potassium (ACK) lysis buffer to deplete red blood cells and then incubated with 10 U of dispase to further dissociate the tissue into single cells and 1000 U of deoxyribonuclease I to prevent cell clumping. Cells were filtered through a 70-μm nylon mesh and washed with staining buffer containing 2% fetal bovine serum (FBS) and PSA in Hank’s balanced salt solution. Single-cell suspensions of fresh breast tissue were then stained with fluorescent antibodies to prepare for fluorescence-activated cell sorting (FACS).
Flow cytometry
To reduce nonspecific antibody binding, single cells were blocked with rat immunoglobulin G (IgG; 10 μg/ml) on ice for 10 min. Cells were then stained, in the dark, on ice for 30 min. FACS was performed with a 130-μm nozzle on a BD FACSAria II with BD FACSDiva software. Side scatter and forward scatter profiles (area and width) were used to eliminate debris and cell doublets. Dead cells were eliminated by excluding 4′,6-diamidino-2-phenylindole (DAPI)–positive cells. For scRNA-seq, human breast epithelia and niche cells were isolated as described in the section below (“Single-cell RNA sequencing”). For mouse Lmo2-PyMT tumor studies, tumor epithelial cells were enriched by negative gating of lineage markers CD45, Ter119, and CD31. For xenograft studies, human tumor epithelial cells were enriched by negative gating of the major histocompatibility protein H-2kd, expressed on the plasma membrane of all nucleated mouse cells. A complete list of antibodies is provided in table S5.
Single-cell RNA sequencing
A total of 1902 scRNA-seq profiles of tumor and adjacent-normal human breast basal (n = 660), luminal progenitors (n = 532), and mature luminal (n = 710) cells were acquired from a previous study [accession code: GSE138536 (13)]. 250 additional basal epithelia and 207 stromal cells were collected from human breast tumors using the same strategies for cell sorting, library construction, and data processing as previously described (13). Briefly, by negative gating of lineage cells expressing CD45, CD31, CD3, CD16, and CD64, cancer-associated fibroblasts (CAFs; n = 6) and ECs (n = 126) were sorted as lineage−CD49f−EpCAM−. Although ECs are generally CD31+, the enzymatic digestion during tumor cell isolation cleaves surface CD31. Thus, these cells were detected on flow cytometry as CD31−. CAFs and ECs were then distinguished on the basis of transcriptional expression of FAP and PDGFRA for CAFs and CDH5, EMCN, and PECAM1 for ECs. Hematopoietic populations were sorted separately based on surface expression of CD45 followed by CD14+ for macrophages (n = 21), CD3+CD4+ for CD4 T cells (n = 22), CD3+CD8+ for CD8 T cells (n = 21), and CD19+ for B cells (n = 11). A matrix containing gene-level transcripts per million (TPM) for all single cells (n = 2359) has been deposited in Gene Expression Omnibus under the accession GSE159285. Metadata for each single cell is also available in GSE159285 and in table S1.
Predicted ordering of single cells by differentiation status
Single-cell level prediction of differentiation states in human breast tumor basal epithelial cells (n = 910) was performed in R using the CytoTRACE package publicly available at https://cytotrace.stanford.edu (13). In brief, CytoTRACE (13) is a computational tool that assigns a ranked differentiation score to each single cell in scRNA-seq data based on the number of genes expressed. Less differentiated cells are assigned a higher score, while more differentiated cells are assigned a lower score per the algorithm.
Deconvolution of bulk breast tumors
CIBERSORTx was used to deconvolve cell type abundances from (i) microarray gene expression data of 508 bulk breast tumors from the Investigation of Serial Studies to Predict Your Therapeutic Response with Imaging and Molecular Analysis (I-SPY1) clinical trial (18); (ii) microarray gene expression data of 1981 bulk breast tumors from Metabric Discovery (n = 995) and Validation (n = 986) cohorts (19); (iii) RNA-seq data from 1033 bulk breast tumors from The Cancer Genome Atlas (20). Default parameters as described in the “Tutorial” page at http://cibersortx.stanford.edu/ were used to generate a signature matrix from scRNA-seq data. Quantile normalization was run on microarray (but not RNA-seq) data, and bulk-mode batch correction (B-mode) was applied for cross-platform deconvolution.
ssGSEA analysis
Single-sample enrichment of the “HALLMARK_ANGIOGENESIS,” “HALLMARK_E2F_TARGETS,” “HALLMARK_TNFα_SIGNALING_VIA_NFκB,” “HALLMARK_IL6_JAK_STAT3_SIGNALING,” and the dbEMT (60) gene sets was calculated using ssGSEA as implemented in the R GSVA package (v1.30.0) (61). To ensure a fair comparison between LMO2-positive and LMO2-negative populations, an empirical P value was calculated by comparing the mean enrichment in LMO2+ basal cells versus a size-matched collection of LMO2– basal cells randomly sampled 10,000 times. Enrichment was defined as the number of cells expressing a given gene (TPM > 0) divided by the total number of cells expressing that gene.
Survival analysis of deconvolved cell populations
The survival v3.1.12 R package was used to analyze the association of LMO2+ basal cells and other deconvolved cell populations with DRFS in the I-SPY1 cohort (n = 508 human breast cancer samples). Samples were evenly stratified into “high” and “low” groups based on whether tumors had greater than or less than the median abundance of the deconvolved cell population. A Cox proportional hazard model was then used to calculate the effect of the abundance of the deconvolved cell population on DRFS, adjusting for ESR1 status as a possible confounder.
Mice
Rosa26mTmG (stock #007576), C57BL6 (stock #000664), NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) (stock #005557), and MMTV-PyMT (stock #022974) mice were purchased from The Jackson Laboratory. Lmo2CreERT2 transgenic mice were generated by pronuclear injection in C57Bl/6/CBA (gift from T.R.). All tumor xenotransplantation was done in NSG mice. All mice used for this study were maintained at the Stanford Animal Facility in accordance with the guidelines of the animal care use committee [Administrative Panel on Laboratory Animal Care (APLAC) #10868].
Cell lines
MDA-MB-231, MDA-MB-468, and 293T cells were obtained from American Type Culture Collection. These cells were certified by the vendors to be mycoplasma free. None of the cell lines used are listed in the database of commonly misidentified cell lines maintained by International Cell Line Authentication Committee (ICLAC). Cell lines have not been authenticated, but all cell lines used were passaged less than 10 times from when the original cells from the vendors were thawed. Cell lines are routinely tested for mycoplasma contamination and were mycoplasma free. MDA-MB-231, MDA-MB-468, and 293T cells were grown in DMEM (Invitrogen) supplemented with PSA, 10% FBS (Hyclone), Glutamax (Thermo Fisher Scientific), and sodium pyruvate (Life Technologies).
Immunofluorescence staining in paraffin sections
Tumors were fixed in formalin and embedded in paraffin for immunostaining. Sections were deparaffinized, dehydrated, and microwaved for 20 min at 95°C in sodium citrate buffer [10 mM sodium citrate and 0.05% Tween 20 (pH 6.0)] for antigen retrieval. Tissue sections were incubated overnight at 4°C with primary antibodies diluted in phosphate-buffered saline (PBS) + 5% bovine serum albumin (BSA) (antibodies are listed in table S5). Samples were subsequently washed with PBS and were incubated with anti-GFP in Alexa Fluor 488 (1:500), anti-mouse Alexa Fluor in either 488 or 594 (1:500), anti-rat Alexa Flour 594 (1:500), and anti-rabbit in either Alexa Flour 488 or 647 (1:500) conjugated secondary antibodies (Invitrogen) at 1:500 in PBS + 5% BSA for 1 hour at room temperature (RT). All the immunofluorescence sections and cells were mounted in ProLong Gold with DAPI. Images were acquired by a Carl Zeiss LSM 710 Meta confocal microscope. Images were processed using ImageJ.
Real-time PCR
A total of 10,000 GFP+ or TdTomato+ PyMT tumor cells were sorted into 1.5 microfuge tubes and spun down, or lineage-depleted PDX cells were resuspended in RLT buffer. RNA was extracted using the RNeasy Micro Kit (Qiagen, #74034). RNA was reverse-transcribed to cDNA using a SuperScript III First Strand Synthesis kit (Life Technologies, #11752-050) according to the manufacturer’s instructions. cDNA was preamplified 15 cycles according to the cell number using TaqMan pre-amp mastermix (Applied Biosystems, #4391128) and target gene Taqman primer pool. Preamplified cDNA was then subjected to the real-time polymerase chain reaction (PCR) for specific gene target according to the manufacturer’s instruction using 7900HT Real-Time PCR system (Applied Biosystems). All expression data are normalized to Actnb, Gapdh, ACTNB, and GAPDH. Data were analyzed by SDS2.4 software and Excel.
Whole-mount immunostaining in tumors
Procedure was performed as previously described (62). Tumors were dissected in PBS, immediately fixed in 4% paraformaldehyde at 4°C for 1 hour, followed by two 15-min washes with PBS at 4°C. The tumors were then chopped to 3- to 4-mm3 chunks and incubated in anti-endomucin prepared in at least five volumes of PBS containing 0.5% Triton (0.5% PBT) for 6 hours at RT followed by incubating them at 4°C overnight. The tumor chunks were then washed in 20 volumes of 0.5% PBT for 6 hours with a change in wash buffer every hour at RT followed by a wash overnight at 4°C. The tumors were then incubated in 1:250 dilution of secondary antibodies prepared in five volumes of 0.5% PBT for 2 hours at RT followed by overnight at 4°C. The tumors were then washed in 20 volumes of 0.5% PBT for six consecutive days, for 6 hours at RT followed by overnight at 4°C each day. All steps were performed with gentle but continuous shaking. The tumors were lastly cleared with 2 volumes of Vectashield (Vector, catalog no. H-1000) for 2 hours at RT, after which they were imaged immediately or stored in −20°C. Steps involving antibody or Vectashield incubations were performed in 1.5- to 2-ml tubes, and washes were performed in 50-ml tubes.
Confocal imaging for whole mounts
Tumor chunks were flattened between a 1.5-mm-thick microscope coverslip (Fisherbrand, catalog no. 22266858) and a double-concave microscope slide (Sail brand, catalog no. 7104) with their anterior walls (watershed regions) facing the coverslip. The tumors were then imaged using an inverted Zeiss LSM-700 confocal microscope. Digital images of multiple z-stacks for each scanned area were captured with Zeiss Zen software and complied together using the ImageJ software. Threshold limits were set to 80% saturation, and boundary of endomucin-stained channel was used to demarcate vasculature. Within demarcated boundary, GFP+ cells were counted. Total GFP+ cells were also counted. %Integrated into vasculature was determined as (GFP+ cells inside vasculature × 100/total GFP+ cells in field) and as an average across all z planes per image. Stacks were analyzed individually to assess vascular integration in each plane and to reduce errors of maximum intensity projections. For GFP and endomucin area calculations, threshold limits were set using Otsu dark with a minimum value of 10,000 in FIJI for GFP and endomucin-stained channels and used to generate binary masks. The area of each masked image was measured with FIJI and then divided by the total area of the original image to calculate the area fraction of GFP and endomucin for each image. For cell length calculations, the scale is set using the scale bar associated with the image (Analyze>set scale). In the region of interest (ROI) manager, the line tool is selected, the diameter of the cell is drawn and recorded using the record function, and the measurements are documented in the ROI manager.
Plasmids and lentivirus
For knockdown experiments, the lentiviral vector used was pSicoR-GFP. The sequences for the LMO2 shRNA are 5′-GACGCATTTCGGTTGAGAA-3′ and 5′-GCATCCTGTGACAAGCGGATT-3′. For overexpression of LMO2, the cDNA was purchased from Genescript and cloned into the pHIV-Zsgreen vector (Addgene, #18121) for lentiviral expression. Ectopic expression was verified using immunoblotting. For rescue experiments, the LMO2 shRNA targeting the 3′UTR (5′-GACGCATTTCGGTTGAGAA-3′) was cloned into the pRSI12-U6-(sh)-HTS4-UbiC-TagRFP-2A-Puro shRNA expression vector (Cellecta). Viruses were produced in 293T cells using the second-generation lentiviral system and transfection using Lipofectamine 2000 (Life Technologies). Supernatants were collected at 48 and 72 hours, filtered with a 0.45-μm filter, and precipitated with lentivirus precipitation solution (Alstem LLC) per the manufacturer’s instructions or concentrated by ultracentrifugation. Viral titers were determined by flow cytometry analyses of 293T cells infected with serial dilutions of concentrated virus.
Xenograft tumor cell infection and engraftment
Dissociated single cells from xenografts were stained with biotin anti-mouse H-2Kd microbeads and depleted of mouse cells by using AUTO MACS (Miltenyi BioTec). Tumor cells were infected with pSicoR, shLMO2-1, and shLMO2-2, at a multiplicity of infection (MOI) = 25 for knockdown experiments for xenograft tumor. For MDA-MB-468 xenografts, cells in culture were infected at an MOI = 5 and sorted for GFP-expressing cells before xenotransplantation. MOI is calculated on the basis of infection in 293T cells. The infected cells were washed and resuspended in staining media containing 50% Matrigel and injected in the fourth abdominal fat pad by subcutaneous injection at the base of the nipple of female NSG mice (20,000 cells per mouse), except for circulating tumor cell experiment, where infected MDA-MB-468 cells were injected contralaterally in the second/third and fourth mammary fat pads. Mice were monitored every week for tumor growth. All mice in the experiment (control and LMO2 knockdown) were euthanized if tumor growth reached end point (1500 to 2000 mm3) in any of the mice in the experiment, the tumors were ulcerated, or mice showed signs of distress. Tumor size was measured using a caliper. Tumors were harvested, weighed, and dissociated to determine percentage of infected cells using FACS analysis. Tumor weight is plotted as “weight × fraction infected.” The number of GFP+ lung metastases was counted using the ImageJ software. In the case of PDX2, lung metastases were not discrete, so the lungs were dissociated into single cells, and the percent of GFP+ cells was calculated. For the tail vein injection experiment, 20,000 PDX1 tumor cells were resuspended in PBS and injected via the tail vein. The mice were closely monitored to assess any signs of distress. All the mice were euthanized 6 weeks after injection, and the lungs were harvested. GFP+ lung metastases were counted using the ImageJ software.
For mouse tumors, lineage-depleted (CD45−/CD31−/Ter119−) tumor cells from TdTomato-fluorescent Lmo2-PyMT were orthotopically transplanted into nonfluorescent BL6 mice. Mice were injected intraperitoneally with tamoxifen. Tamoxifen was dissolved in corn oil at a concentration of 10 mg/ml. Each mouse received a dose of 1.5 mg, as previously described (63). The mice were analyzed either 48 hours after a single pulse or pulsed with tamoxifen two to three times per week once the tumors were palpable until tumor end point at 2 cm3. At the end of the experiment, tumors were harvested, divided for histology, and FACS-analyzed as described above. Lungs were evaluated for both the number and area of GFP+ or red fluorescent protein–positive (RFP+) metastases and quantified using ImageJ.
Circulating tumor cell enumeration
Quantification of circulating tumor cells was performed in mice xenotransplanted with MDA-MB-468 cells infected with either pSicoR, shLMO2-1, or shLMO2-2. Blood (400 to 900 μl) was collected via cardiac puncture using a 25-gauge needle attached to a 1-ml syringe. The blood was collected directly into K3-EDTA tubes. ACK lysis was performed to remove red blood cells. The cells were washed twice with FACS buffer and fixed with 2% paraformaldehyde in PBS for 10 min. The cells were again washed twice in PBS and resuspended in 100 μl of PBS. The cells were then spread on charged glass slides and allowed to air dry. Subsequently, the slides were counterstained with DAPI, and the number of GFP+ circulating tumor cells was manually counted under the microscope. The numbers were normalized to represent 1 ml of blood collected for each mouse.
Migration and invasion assays
For migration assays, MDA-MB-468 and MDA-MB-231 cells were infected with either empty vector control or shRNA against LMO2. Subsequently, the cells were serum-starved for 24 to 48 hours, and 100,000 cells were plated in a Transwell dish in a 24-well plate containing 5% serum. After 36 hours of incubation, the cells in the upper chamber were removed with a cotton swab, and the cells attached to the underside of the membrane were fixed in 4% paraformaldehyde. The membrane was subsequently cut and stained with 0.1% crystal violet and mounted for imaging. For 3D spheroid invasion assays, the kit was purchased from Trevigen (#3500-096-K), and the protocol was performed as per manufacturer’s instructions.
HUVEC integration
Sixty microliters of growth factor–reduced Matrigel was plated per well in a 96-well plate and allowed to gel for 30 to 60 min at 37°C. HUVEC cells were trypsinized, neutralized, and resuspended in media. Calcein (0.5 μl) was added per 5 ml of media and incubated at 37°C. Cells were washed once to remove calcein. A total of 15,000 cells were plated per well onto the Matrigel coating. Once HUVEC tubes were formed at 4 hours, 5000 MDA-MD-468 cells transduced with either pSicoR, shLMO2-1, or shLMO2-2 or treated with 5 μM of Stattic were added. Images were acquired between 6 and 8 hours of tube formation.
Bulk RNA-seq
MDA-MB-468 cells (200,000) infected with either pSicoR, shLMO2-1, or shLMO2-2 were harvested, and RNA extraction was performed using the RNeasy Micro Kit (Qiagen, #74034). RNA samples were then submitted to Novogene Co. for library construction and sequencing. Briefly, following RNA quality check, mRNA was enriched using the NEBNext Poly(A) mRNA Magnetic Isolation Module (#E7490), and cDNA libraries were constructed using the NEBNext Ultra II RNA Library Prep Kit for Illumina (#E7770 and #E7775). Libraries were fragment-analyzed by LabChip, quantified by quantitative PCR using the KAPA Library Quantification kit (#KR0405), and then sequenced on the Illumina NovaSeq 6000 platform to obtain 2× 150 base pair paired-end reads.
For data processing, raw FASTQ reads were aligned to the GENCODE v29 reference transcripts (GRCh38.p12) using Salmon (64) v0.12 with flags -l IU, --seqBias, --gcBias, --posBias, --useVBOpt, --rangeFactorizationBins 4, and --validateMapping. To calculate differentially expressed genes between control and knockdown samples, we used the R package DESeq2 (65) (version 1.22.2) following the authors’ instructions. Briefly, the gene-level count matrices were created by importing the quantification data from Salmon using tximport (66) (version 1.10.1). The DESeqDataSet was constructed from the resulting tximport processed object along with sample information using the function DESeqDataSetFromTximport. The differentially expressed genes were calculated using the DESeq() function, and results were summarized with the results() function.
GSEA was performed on a preranked list of genes differentially expressed (Q value < 0.1) between control and knockdown conditions (n = 1963 genes) ordered by log2 fold change using the Broad Institute’s software (40). The top and bottom 50 genes from the GSEA input list were mean-centered and scaled before presentation as a heatmap.
Co-IP assay
Co-IP experiments were carried out using the Pierce Co-IP Kit (#26149, Thermo Fisher Scientific) as per the manufacturer’s protocol. For LMO2 and STAT3 interaction, MDA-MB-468 were grown in complete medium in a 10-cm dish to reach 90% confluence. These were lysed with 0.6 ml of ice-cold IP lysis buffer containing protease and phosphatase inhibitors for 1 hour at 4°C upon gentle agitation. For the antibody immobilization step, 20 μg of rabbit anti-LMO2 (Abcam, #ab91652) or 20 μg of rabbit anti-STAT3 [Cell signaling technologies (CST), #4904] or, as a control, 20 μg of rabbit IgG, was diluted onto the AminoLink Plus Coupling Resin. The cell lysates were precleared with control agarose resin, and co-IP was carried out by adding 1 mg of the precleared cell lysate to the antibody immobilized resin, with end over end mixing at 4°C overnight. After elution into 50 μl, the sample was analyzed by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gel and followed by immunoblotting to detect protein-protein interaction.
Western blotting
MDA-MB-468 cells were seeded and serum-starved for 24 hours. They were subsequently treated with TNF-α (20 ng/ml), IL-6 (20 ng/ml), EGF (10 ng/ml), IFN-α (20 ng/ml), or IFN-γ (40 ng/ml) for 30 min. Whole-cell lysates were generated by lysis with radioimmunoprecipitation assay buffer, along with protease and phosphatase inhibitor cocktails (Thermo Fisher Scientific). For cell fraction, the cells were lysed using the Pierce subcellular fractionation kit (PI78840). SDS-PAGE gels were run at 120 V for 75 min, transferred onto polyvinylidene difluoride membranes (#IPFL00010, Millipore, Billerica, MA) at 70 V for 90 min. Membranes were blocked with LI-COR blocking buffer (#927-40000) for 1 hour at RT and then subsequently probed with primary antibodies diluted at 1:1000 in 5% BSA/tris-buffered saline–Tween 0.1% (TBST) overnight at 4°C. Incubation with secondary antibodies for 1 hour at RT containing fluorophores at 1:20,000 dilution (IRDye 800CW conjugated goat anti-rabbit; #926-32211, LI-COR Biosciences, Lincoln, NE) enabled visualization on the Odyssey Infrared Imaging System from LI-COR Biosciences. Washes in between incubations were done for 10 min × 3 using TBST.
Luciferase reporter assay
STAT3 firefly luciferase reporter lentivirus (PLV-10065-50) was purchased from Cellomics Technologies. A stable cell line with MDA-MB-468 was generated using puromycin selection. The cells were then infected with either pSicoR, shLMO2-1, or shLMO2-2. For the reporter assay, 10,000 MDA-MB-468 cells were seeded in full serum media. After attachment, they were treated with TNF-α (20 ng/ml), IL-6 (20 ng/ml), EGF (10 ng/ml), IFN-α (20 ng/ml), or IFN-γ (40 ng/ml) in full serum for 4 hours. The cells were then lysed, and luciferase activity was measured using Dual-Luciferase Reporter Assay System (Promega, #E1960). All fold changes were calculated based on untreated cells in the same group, i.e., pSicoR, shLMO2-1, or shLMO2-2 that are untreated. All experiments were performed in triplicate, and the experiment was repeated three times.
DUOLink proximity ligation assay
For the proximity ligation (PLA) assay (DUOLink, OLink Biosciences, Sigma-Aldrich, #DUO92102), MDA-MB-468 were seeded on 13-mm glass coverslips. The cells were fixed with ice-cold 100% methanol for 5 min at −20°C and then rehydrated thrice in PBS for 5 min each. Coverslips were blocked for 30 min with 3% BSA/PBS and then incubated with appropriate dilution of primary antibodies in 1% BSA/PBS for 1 hour in a moist environment at RT. Primary antibodies were used at 1:200 dilutions to characterize the interaction between LMO2, STAT3, JAK2, and PIAS3. All antibodies are listed in table S5. As a negative control, rabbit and mouse anti-IgGs were used in 1:200 dilutions. Subsequently, the manufacturer’s instructions were followed to complete the PLA assay.
Statistical analysis
All graphs show the average as central values, and error bars indicate ± SD unless otherwise indicated. P values are calculated using paired or unpaired t test, ANOVA, Extreme Limiting Dilution Analysis (ELDA), Wilcoxon rank-sum test, Fisher’s exact test, Monte Carlo Approach, Mann-Whitney, and log-rank test as indicated in the figure legends. All P and Q values were calculated using GraphPad prism or R version ≥3.5.2, unless otherwise stated. For animal studies, sample size was not predetermined to ensure adequate power to detect a prespecified effect size, no animals were excluded from analyses, experiments were not randomized, and investigators were not blinded to group allocation during experiments.
Acknowledgments
We thank P. Lovelace, C. Carswell Crumpton, and other flow cytometry staff for help and animal facility core members. The Wolverine Aria instrument was funded by NIH grant S10-1S10RR02933801. We thank D. Heiser and B. Nazario for technical assistance and M. Cuadro for administrative assistance. We thank the Stanford Neuroscience Microscopy Service, supported by NIH grant NS069375. We thank B. Abrams, UCSC Life Sciences Microscopy Center, RRID: SCR_021135, for technical support during image acquisition and processing. The Lmo2CreERT2 mice were a gift from T.R.
Funding: This work was supported by NIH/NCI (U01CA154209-01 and P01 CA139490-05), the Breast Cancer Research Foundation (to M.F.C.), the U.S. Department of Defense (W81XWH-11-1-0287 and W81XWH-13-1-0281 to M.F.C. and W81XWH-12-1-0020 to S.S.S.), the National Cancer Institute (R0490CA187192-03 and R01CA255450 A.M.N., 5R01CA100225-09 to M.F.C., and PHS grant no. CA09302 to G.S.G.), the Stanford Bio-X Interdisciplinary Initiatives Seed Grants Program (IIP) (A.M.N. and M.F.C.), the Virginia and D.K. Ludwig Fund for Cancer Research (A.M.N. and M.F.C), the Stinehart-Reed Foundation (A.M.N.), the Donald E. and Delia B. Baxter Foundation (A.M.N.), the Stanford School of Medicine Dean’s Fellowship (J.A.), Stanford Bio-X Bowes Graduate Student Fellowship (G.S.G.), and the Stanford Medical Science Training Program (G.S.G.). K.R.-H. is supported by the NIH/NHLBI (R01-HL128503) and is an HHMI Investigator.
Author contributions: S.S.S. and M.F.C. conceived and designed the study. S.S. and J.A. performed experiments and analyzed data with supervision from M.F.C. G.S.G. analyzed single-cell and bulk RNA sequencing data with assistance from C.B.S. and supervision from A.M.N. I.F. and V.H.-A. performed experiments and analyzed data during the revisions with supervision from S.S. and assistance from A.M.N. A.H.K. assisted with the PDX studies. W.H.D.H. assisted with the metastasis experiments. S.D. performed staining for visualization of tumor vasculature under the supervision of K.R.-H. T.A.P. assisted with the circulating cells experiment under the supervision of P.A.B. D.Q. provided technical support. F.M.D. assisted with the collection of patient specimens. J.P.T. assessed the enrichment of genes in LMO2+ cells and provided guidance in the project. T.H.R. provided the Lmo2CreERT2 mice. S.S., J.A., G.S.G., A.M.N., and M.F.C., wrote the manuscript. All authors commented on the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data generated or analyzed during this study are included in this published article (and its supplementary information files). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All single-cell and bulk RNA-seq data generated in this study have been deposited in the Gene Expression Omnibus with the primary accession code GSE159285 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159285). All bioinformatic tools used in this study are published and publicly available. Custom scripts and source data are available on Dyrad at doi:10.5061/dryad.stqjq2c6j. PDXs generated in the manuscript will be available upon request with an appropriate material transfer agreement with Stanford University. Requests for the Lmo2CreERT2 mice should be addressed to T.R. (terry.rabbitts@icr.ac.uk).
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S5
REFERENCES AND NOTES
- 1.Siegel R. L., Miller K. D., Fuchs H. E., Jemal A., Cancer statistics, 2022. CA Cancer J. Clin. 72, 7–33 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Siegel R., Ward E., Brawley O., Jemal A., Cancer statistics, 2011. CA Cancer J. Clin. 61, 212–236 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Esposito M., Ganesan S., Kang Y., Emerging strategies for treating metastasis. Nat. Cancer 2, 258–270 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu W., Kang Y., Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev. Cell 49, 361–374 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Padmanaban V., Krol I., Suhail Y., Szczerba B. M., Aceto N., Bader J. S., Ewald A. J., E-cadherin is required for metastasis in multiple models of breast cancer. Nature 573, 439–444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y., Lv Z., Zhang S., Wang Z., He L., Tang M., Pu W., Zhao H., Zhang Z., Shi Q., Cai D., Wu M., Hu G., Lui K. O., Feng J., Nieto M. A., Zhou B., Genetic fate mapping of transient cell fate reveals N-cadherin activity and function in tumor metastasis. Dev. Cell. 54, 593–607.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Fischer K. R., Durrans A., Lee S., Sheng J., Li F., Wong S. T. C., Choi H., El Rayes T., Ryu S., Troeger J., Schwabe R. F., Vahdat L. T., Altorki N. K., Mittal V., Gao D., Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527, 472–476 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pastushenko I., Brisebarre A., Sifrim A., Fioramonti M., Revenco T., Boumahdi S., Van Keymeulen A., Brown D., Moers V., Lemaire S., De Clercq S., Minguijón E., Balsat C., Sokolow Y., Dubois C., De Cock F., Scozzaro S., Sopena F., Lanas A., D’Haene N., Salmon I., Marine J.-C., Voet T., Sotiropoulou P. A., Blanpain C., Identification of the tumour transition states occurring during EMT. Nature 556, 463–468 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Al-Hajj M., Wicha M. S., Benito-Hernandez A., Morrison S. J., Clarke M. F., Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. 100, 3983–3988 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sikandar S. S., Kuo A. H., Kalisky T., Cai S., Zabala M., Hsieh R. W., Lobo N. A., Scheeren F. A., Sim S., Qian D., Dirbas F. M., Somlo G., Quake S. R., Clarke M. F., Role of epithelial to mesenchymal transition associated genes in mammary gland regeneration and breast tumorigenesis. Nat. Commun. 8, 1669 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H., Patel M. R., Prescher J. A., Patsialou A., Qian D., Lin J., Wen S., Chang Y.-F., Bachmann M. H., Shimono Y., Dalerba P., Adorno M., Lobo N., Bueno J., Dirbas F. M., Goswami S., Somlo G., Condeelis J., Contag C. H., Gambhir S. S., Clarke M. F., Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc. Natl. Acad. Sci. U.S.A. 107, 18115–18120 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawson D. A., Kessenbrock K., Davis R. T., Pervolarakis N., Werb Z., Tumour heterogeneity and metastasis at single-cell resolution. Nat. Cell Biol. 20, 1349–1360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gulati G. S., Sikandar S. S., Wesche D. J., Manjunath A., Bharadwaj A., Berger M. J., Ilagan F., Kuo A. H., Hsieh R. W., Cai S., Zabala M., Scheeren F. A., Lobo N. A., Qian D., Yu F. B., Dirbas F. M., Clarke M. F., Newman A. M., Single-cell transcriptional diversity is a hallmark of developmental potential. Science 367, 405–411 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lobo N. A., Zabala M., Qian D., Clarke M. F., Serially transplantable mammary epithelial cells express the Thy-1 antigen. Breast Cancer Res. 20, 121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho R. W., Wang X., Diehn M., Shedden K., Chen G. Y., Sherlock G., Gurney A., Lewicki J., Clarke M. F., Isolation and molecular characterization of cancer stem cells in MMTV-Wnt-1Murine breast tumors. Stem Cells 26, 364–371 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Mercurio A. M., Lipscomb E. A., Bachelder R. E., Non-angiogenic functions of VEGF in breast cancer. J. Mammary Gland Biol. Neoplasia 10, 283–290 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Zhao D., Pan C., Sun J., Gilbert C., Drews-Elger K., Azzam D. J., Picon-Ruiz M., Kim M., Ullmer W., El-Ashry D., Creighton C. J., Slingerland J. M., VEGF drives cancer-initiating stem cells through VEGFR-2/Stat3 signaling to upregulate Myc and Sox2. Oncogene 34, 3107–3119 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Esserman L. J., Berry D. A., Cheang M. C. U., Yau C., Perou C. M., Carey L., DeMichele A., Gray J. W., Conway-Dorsey K., Lenburg M. E., Buxton M. B., Davis S. E., van’t Veer L. J., Hudis C., Chin K., Wolf D., Krontiras H., Montgomery L., Tripathy D., Lehman C., Liu M. C., Olopade O. I., Rugo H. S., Carpenter J. T., Livasy C., Dressler L., Chhieng D., Singh B., Mies C., Rabban J., Chen Y.-Y., Giri D., Au A., Hylton N.; I-SPY 1 TRIAL Investigators , Chemotherapy response and recurrence-free survival in neoadjuvant breast cancer depends on biomarker profiles: Results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657). Breast Cancer Res. Treat. 132, 1049–1062 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curtis C., Shah S. P., Chin S.-F., Turashvili G., Rueda O. M., Dunning M. J., Speed D., Lynch A. G., Samarajiwa S., Yuan Y., Gräf S., Ha G., Haffari G., Bashashati A., Russell R., McKinney S., Langerød A., Green A., Provenzano E., Wishart G., Pinder S., Watson P., Markowetz F., Murphy L., Ellis I., Purushotham A., Børresen-Dale A.-L., Brenton J. D., Tavaré S., Caldas C., Aparicio S., The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Network , Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamada Y., Warren A. J., Dobson C., Forster A., Pannell R., Rabbitts T. H., The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc. Natl. Acad. Sci. U.S.A. 95, 3890–3895 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larson R. C., Osada H., Larson T. A., Lavenir I., Rabbitts T. H., The oncogenic LIM protein Rbtn2 causes thymic developmental aberrations that precede malignancy in transgenic mice. Oncogene 11, 853–862 (1995). [PubMed] [Google Scholar]
- 23.Kim C., Gao R., Sei E., Brandt R., Hartman J., Hatschek T., Crosetto N., Foukakis T., Navin N. E., Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell 173, 879–893.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamada Y., Zhong Y., Miki S., Taura A., Rabbitts T. H., The transcription factor complex LMO2/TAL1 regulates branching and endothelial cell migration in sprouting angiogenesis. Sci. Rep. 12, 7226 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chambers J., Rabbitts T. H., LMO2 at 25 years: A paradigm of chromosomal translocation proteins. Open Biol. 5, 150062 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newman A. M., Steen C. B., Liu C. L., Gentles A. J., Chaudhuri A. A., Scherer F., Khodadoust M. S., Esfahani M. S., Luca B. A., Steiner D., Diehn M., Alizadeh A. A., Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 37, 773–782 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perou C. M., Sørlie T., Eisen M. B., van de Rijn M., Jeffrey S. S., Rees C. A., Pollack J. R., Ross D. T., Johnsen H., Akslen L. A., Fluge Ø., Pergamenschikov A., Williams C., Zhu S. X., Lønning P. E., Børresen-Dale A.-L., Brown P. O., Botstein D., Molecular portraits of human breast tumours. Nature 406, 747–752 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Guy C. T., Cardiff R. D., Muller W. J., Induction of mammary tumors by expression of polyomavirus middle T oncogene: A transgenic mouse model for metastatic disease. Mol. Cell. Biol. 12, 954–961 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fantozzi A., Christofori G., Mouse models of breast cancer metastasis. Breast Cancer Res. 8, 212 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beerling E., Seinstra D., de Wit E., Kester L., van der Velden D., Maynard C., Schäfer R., van Diest P., Voest E., van Oudenaarden A., Vrisekoop N., van Rheenen J., Plasticity between epithelial and mesenchymal states unlinks EMT from metastasis enhancing stem cell capacity. Cell Rep. 14, 2281–2288 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeo S. K., Zhu X., Okamoto T., Hao M., Wang C., Lu P., Lu L. J., Guan J.-L., Single-cell RNA-sequencing reveals distinct patterns of cell state heterogeneity in mouse models of breast cancer. eLife 9, e58810 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valdés-Mora F., Salomon R., Gloss B. S., Law A. M. K., Venhuizen J., Castillo L., Murphy K. J., Magenau A., Papanicolaou M., Rodriguez de la Fuente L., Roden D. L., Colino-Sanguino Y., Kikhtyak Z., Farbehi N., Conway J. R. W., Sikta N., Oakes S. R., Cox T. R., O’Donoghue S. I., Timpson P., Ormandy C. J., Gallego-Ortega D., Single-cell transcriptomics reveals involution mimicry during the specification of the basal breast cancer subtype. Cell Rep. 35, 108945 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Gratzinger D., Zhao S., West R., Rouse R. V., Vogel H., Gil E. C., Levy R., Lossos I. S., Natkunam Y., The transcription factor LMO2 is a robust marker of vascular endothelium and vascular neoplasms and selected other entities. Am. J. Clin. Pathol. 131, 264–278 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y., Huang D., Wang Z., Wu C., Zhang Z., Wang D., Li Z., Zhu T., Yang S., Sun W., LMO2 attenuates tumor growth by targeting the Wnt signaling pathway in breast and colorectal cancer. Sci. Rep. 6, 36050 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang P., Chen A., He W., Li Z., Zhang G., Liu Z., Liu G., Liu X., He S., Xiao G., Huang F., Stenvang J., Brünner N., Hong A., Wang J., BMP-2 induces EMT and breast cancer stemness through Rb and CD44. Cell Death Discov. 3, 17039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leushacke M., Barker N., Lgr5 and Lgr6 as markers to study adult stem cell roles in self-renewal and cancer. Oncogene 31, 3009–3022 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Matsuo T., Dat L. T., Komatsu M., Yoshimaru T., Daizumoto K., Sone S., Nishioka Y., Katagiri T., Early growth response 4 is involved in cell proliferation of small cell lung cancer through transcriptional activation of its downstream genes. PLOS ONE 9, e113606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boye K., Maelandsmo G. M., S100A4 and metastasis: A small actor playing many roles. Am. J. Pathol. 176, 528–535 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nieto M. A., Huang R. Y.-J., Jackson R. A., Thiery J. P., EMT: 2016. Cell 166, 21–45 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Zhong Z., Wen Z., Darnell Jr. J. E., Stat3: A STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 264, 95–98 (1994). [DOI] [PubMed] [Google Scholar]
- 42.De Simone V., Franzè E., Ronchetti G., Colantoni A., Fantini M. C., Di Fusco D., Sica G. S., Sileri P., MacDonald T. T., Pallone F., Monteleone G., Stolfi C., Th17-type cytokines, IL-6 and TNF-α synergistically activate STAT3 and NF-kB to promote colorectal cancer cell growth. Oncogene 34, 3493–3503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beadling C., Guschin D., Witthuhn B. A., Ziemiecki A., Ihle J. N., Kerr I. M., Cantrell D. A., Activation of JAK kinases and STAT proteins by interleukin-2 and interferon alpha, but not the T cell antigen receptor, in human T lymphocytes. EMBO J. 13, 5605–5615 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Will A., Hemmann U., Horn F., Röllinghoff M., Gessner A., Intracellular murine IFN-gamma mediates virus resistance, expression of oligoadenylate synthetase, and activation of STAT transcription factors. J. Immunol. 157, 4576–4583 (1996). [PubMed] [Google Scholar]
- 45.Azare J., Leslie K., Al-Ahmadie H., Gerald W., Weinreb P. H., Violette S. M., Bromberg J., Constitutively activated Stat3 induces tumorigenesis and enhances cell motility of prostate epithelial cells through integrin β6. Mol. Cell. Biol. 27, 4444–4453 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barbieri I., Pensa S., Pannellini T., Quaglino E., Maritano D., Demaria M., Voster A., Turkson J., Cavallo F., Watson C. J., Provero P., Musiani P., Poli V., Constitutively active Stat3 enhances Neu-mediated migration and metastasis in mammary tumors via upregulation of Cten. Cancer Res. 70, 2558–2567 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Schust J., Sperl B., Hollis A., Mayer T. U., Berg T., Stattic: A small-molecule inhibitor of STAT3 activation and dimerization. Chem. Biol. 13, 1235–1242 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Ye X., Brabletz T., Kang Y., Longmore G. D., Nieto M. A., Stanger B. Z., Yang J., Weinberg R. A., Upholding a role for EMT in breast cancer metastasis. Nature 547, E1–E3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lim E., Vaillant F., Wu D., Forrest N. C., Pal B., Hart A. H., Asselin-Labat M.-L., Gyorki D. E., Ward T., Partanen A., Feleppa F., Huschtscha L. I., Thorne H. J., kConFab, Fox S. B., Yan M., French J. D., Brown M. A., Smyth G. K., Visvader J. E., Lindeman G. J., Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 15, 907–913 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Yang J., Antin P., Berx G., Blanpain C., Brabletz T., Bronner M., Campbell K., Cano A., Casanova J., Christofori G., Dedhar S., Derynck R., Ford H. L., Fuxe J., García de Herreros A., Goodall G. J., Hadjantonakis A.-K., Huang R. Y. J., Kalcheim C., Kalluri R., Kang Y., Khew-Goodall Y., Levine H., Liu J., Longmore G. D., Mani S. A., Massagué J., Mayor R., McClay D., Mostov K. E., Newgreen D. F., Nieto M. A., Puisieux A., Runyan R., Savagner P., Stanger B., Stemmler M. P., Takahashi Y., Takeichi M., Theveneau E., Thiery J. P., Thompson E. W., Weinberg R. A., Williams E. D., Xing J., Zhou B. P., Sheng G.; EMT International Association (TEMTIA) , Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 21, 341–352 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shenoy A. K., Lu J., Cancer cells remodel themselves and vasculature to overcome the endothelial barrier. Cancer Lett. 380, 534–544 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harney A. S., Arwert E. N., Entenberg D., Wang Y., Guo P., Qian B.-Z., Oktay M. H., Pollard J. W., Jones J. G., Condeelis J. S., Real-time imaging reveals local, transient vascular permeability, and tumor cell intravasation stimulated by TIE2hi macrophage–derived VEGFA. Cancer Discov. 5, 932–943 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim M.-Y., Oskarsson T., Acharyya S., Nguyen D. X., Zhang X. H.-F., Norton L., Massagué J., Tumor self-seeding by circulating cancer cells. Cell 139, 1315–1326 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Borriello L., Karagiannis G. S., Duran C. L., Coste A., Oktay M. H., Entenberg D., Condeelis J. S., The role of the tumor microenvironment in tumor cell intravasation and dissemination. Eur. J. Cell Biol. 99, 151098 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y., Wang Z., Huang D., Wu C., Li H., Zhang X., Meng B., Li Z., Zhu T., Yang S., Sun W., LMO2 promotes tumor cell invasion and metastasis in basal-type breast cancer by altering actin cytoskeleton remodeling. Oncotarget 8, 9513–9524 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu J., Lin P., Zhou B., Inflammation fuels tumor progress and metastasis. Curr. Pharm. Des. 21, 3032–3040 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y., Jiang X., Feng F., Liu W., Sun H., Degradation of proteins by PROTACs and other strategies. Acta Pharm. Sin. B 10, 207–238 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Milton-Harris L., Jeeves M., Walker S. A., Ward S. E., Mancini E. J., Small molecule inhibits T-cell acute lymphoblastic leukaemia oncogenic interaction through conformational modulation of LMO2. Oncotarget 11, 1737–1748 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.A cell-based screening method using an intracellular antibody for discovering small molecules targeting the translocation protein LMO2, (available at https://science.org/doi/10.1126/sciadv.abg1950?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%20%200pubmed). [DOI] [PMC free article] [PubMed]
- 60.Zhao M., Kong L., Liu Y., Qu H., dbEMT: An epithelial-mesenchymal transition associated gene resource. Sci. Rep. 5, 11459 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hänzelmann S., Castelo R., Guinney J., GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Das S., Goldstone A. B., Wang H., Farry J., D’Amato G., Paulsen M. J., Eskandari A., Hironaka C. E., Phansalkar R., Sharma B., Rhee S., Shamskhou E. A., Agalliu D., de Jesus Perez V., Woo Y. J., Red-Horse K., A unique collateral artery development program promotes neonatal heart regeneration. Cell 176, 1128–1142.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rios A. C., Fu N. Y., Lindeman G. J., Visvader J. E., In situ identification of bipotent stem cells in the mammary gland. Nature 506, 322–327 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soneson C., Love M. I., Robinson M. D., Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Res. 4, 1521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Tables S1 to S5