Abstract
Interferons (IFNs) play crucial roles in antiviral defenses. Despite using the same Janus-activated kinase (JAK)–signal transducer and activator of transcription (STAT) signaling cascade, type I and III IFN receptors differ in the magnitude and dynamics of their signaling in terms of STAT phosphorylation, gene transcription, and antiviral responses. These differences are not due to ligand-binding affinity and receptor abundance. Here, we investigated the ability of the intracellular domains (ICDs) of IFN receptors to differentiate between type I and III IFN signaling. We engineered synthetic, heterodimeric type I and III IFN receptors that were stably expressed at similar amounts in human cells and responded to a common ligand. We found that our synthetic type I IFN receptors stimulated STAT phosphorylation and gene expression to greater extents than did the corresponding type III IFN receptors. Furthermore, we identified short, “box motifs” within ICDs that bind to JAK1 that were sufficient to encode differences between the type I and III IFN receptors. Together, our results indicate that specific regions within the ICDs of IFN receptor subunits encode different downstream signaling strengths that enable type I and III IFN receptors to produce distinct signaling outcomes.
INTRODUCTION
Interferons (IFNs) are a diverse family of cytokines that are critical for the innate immune response to pathogen infection (1–3). Classified into types I, II, and III, IFNs antagonize many viruses, such as influenza virus, vesicular stomatitis virus, West Nile virus, yellow fever virus, hepatitis viruses, and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (4–11). Given their crucial role in regulating viral infection, type I IFNs (including IFN-α and IFN-β) have long been used as therapeutics against many viral infections, such as hepatitis B virus (HBV) and hepatitis C virus (HCV). In addition, the type III IFNs, including IFN-λ, are being examined for their therapeutic potential against viruses that target epithelial barrier surfaces, such as HBV, HCV, and SARS-CoV-2 (12–15).
Type I and III IFNs signal through cognate cell surface receptors to induce the transcription of many IFN-stimulated genes (ISGs) that subsequently control cellular behaviors and innate immune responses, including pathogen clearance (16). When bound to ligand, the type I and III IFN receptors each assemble as heterodimers. Type I IFN receptors consist of IFNAR1 and IFNAR2, whereas type III IFN receptors consist of IFNLR1 and interleukin-10 receptor beta (IL-10RB). The intracellular domains of their two subunits are associated with either Janus kinase 1 (JAK1) or tyrosine kinase 2 (TYK2) (17, 18). These kinases phosphorylate residues on the IFN receptors that provide docking sites for signal transducer and activator of transcription (STAT) proteins (19), which are themselves phosphorylated by JAK1 and TYK2 before translocating to the nucleus to induce the transcription of ISGs.
Despite their reliance on similar signaling components, type I and III IFNs produce substantially different transcriptional and antiviral outputs (20, 21). Type I IFNs generally induce ISG transcription to a greater extent than do their type III counterparts, and they may activate a subset of genes that type III IFNs do not (22, 23). Their signaling dynamics also differ; signaling by type III IFNs results in more sustained STAT phosphorylation and ISG expression profiles, whereas type I IFN signaling is more transient (24–26). These differences in signaling magnitude and dynamics are thought to enable type I and III IFNs to work in tandem against the same virus at different times of infection (27); however, it is still not fully understood how these signaling differences arise. Although some of these signaling differences can be explained by differences in IFN receptor abundance and ligand-binding affinities, studies suggest that there may be additional factors at play (20). Over-expressing IFNLR1 does not enable type III IFNs to induce ISG transcription similarly to type I IFNs (28). Similarly, although receptor-binding affinities vary greatly across type I IFNs, these binding affinities do not exhibit a clear relationship with signaling strength (29), and engineered high-affinity variants of type III IFNs still do not fully mimic the transcriptional or antiviral potency of a type I IFN (30). Therefore, the question remains of how these two classes of IFNs produce diverse signaling outcomes despite using the same JAK-STAT pathway.
Because there is relatively little sequence homology between the type I and III IFN receptors (31), we hypothesized that sequence differences between the intracellular domains (ICDs) of the receptors might contribute to downstream signaling differences between type I and III IFNs. However, comparing the signaling capabilities of these ICDs within their native context has been challenging. Here, we engineered synthetic IFN receptors that could be expressed at similar amounts and activated by the same ligand, by fusing the transmembrane domain (TMD) and ICD of each type I and III IFN receptor subunit to the extracellular domain (ECD) of the erythropoietin receptor (EPOR). We then mutated the EPOR extracellular domain to ensure heterodimeric binding to erythropoietin (EPO), so the chimeric IFN receptors would mimic the heterodimeric architecture of endogenous IFN receptors. This system enabled us to bypass differences in ligand affinity, because our chimeric receptors would respond to the same ligand, and control for differences in receptor abundance by sorting cells for similar amounts of each receptor subunit. Here, we showed that sequence differences within the IFNAR ICD enable type I IFN receptors to signal more strongly than type III IFN receptors with respect to STAT phosphorylation and ISG expression.
RESULTS
Establishing synthetic type I and III IFN receptors
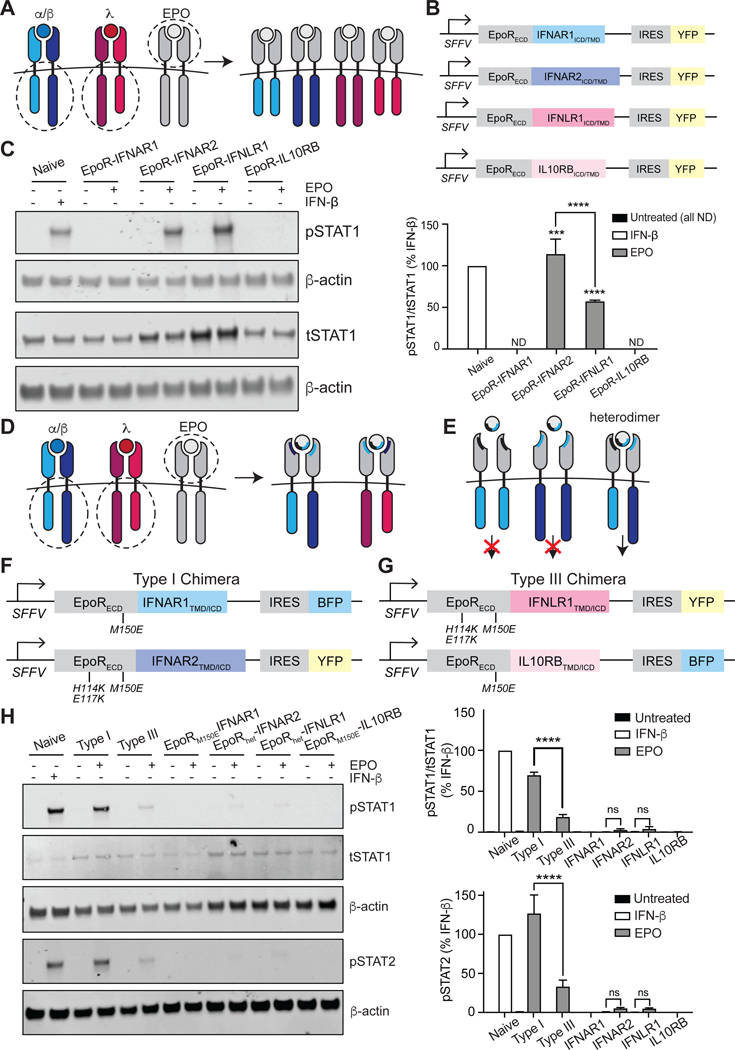
Because receptor abundances and ligand-binding affinities cannot fully explain the differences in signaling between type I and III IFN receptors, we set out to test whether the ICDs of the IFN receptors influenced signaling. The ICDs of cognate IFN receptor subunits are highly divergent, with only 18.57% identity between hIFNAR2 and hIFNLR1 and 27.04% identity between hIFNLR1 and hIL10RB. We thus sought to establish a system in which the ICDs of the type I and III IFN receptors could be compared at identical amounts and upon dimerization by the same ligand (Fig. 1A). Drawing on a precedent set by other groups (32, 33), we decided to engineer receptor chimeras wherein the extracellular, ligand-binding domain of the erythropoietin receptor (EPOR) was fused to the transmembrane and ICDs of each IFN receptor subunit (Fig. 1A).
Fig. 1. Synthetic IFN receptors reveal signaling differences between type I and III IFN receptor ICDs.
(A) Schematic of the homodimeric receptor chimeras generated from the EPOR ECD and the TMDs and ICDs of IFNAR1, IFNAR2, IFNLR1, or IL-10RB. (B) The chimeric IFN receptors were expressed under the control of an SFFV promoter and contained downstream internal ribosome entry sites (IRES) enabling expression of YFP so that cells could be sorted for equivalent expression levels. (C) Left: U2OS cells expressing the indicated homodimeric IFN receptor chimeras were stimulated with EPO (100 ng/ml) for 30 min and then analyzed by Western blotting with antibodies specific for phosphorylated STAT1(pSTAT1) and total STAT1 (tSTAT1). Naïve cells treated with IFN-β (1000 IU/ml) for 30 min were included as a positive control; β-actin was used as a loading control. Right: The relative abundances of pSTAT1 to tSTAT1 in the indicated samples from three independent experiments were determined by densitometry analysis and are expressed as a percentage of the pSTAT1 abundance in IFN-β–treated naive cells, which was set at 100%. (D) Schematic of the heterodimeric IFN receptor chimeras generated by mutating the ECDs from the chimeras shown in (A) so that only one side of EPO can bind (indicated by blue/black shading). (E) Schematic illustrating that signaling only occurs when both subunits of a given heterodimer are present in the cell. (F and G) To facilitate heterodimeric ligand binding, the ECDs of EPOR-IFNAR1 and EPOR-IL10RB contain the point mutation M150E, and the ECDs of EPOR-IFNAR2 and EPOR-IFNLR1 contain the three point mutations H114K, E117K, and M150E. All chimeras were expressed under an SFFV promoter and contained an IRES for downstream expression of either BFP or YFP. (H) Left: U2OS cells expressing chimeric type I or III IFN receptor pairs or individual subunits were treated with EPO (100 ng/ml) for 30 min and then analyzed by Western blotting with antibodies specific for phosphorylated (p) and total (t) STAT1 and pSTAT2. Naïve U2OS cells treated with IFN-β (1000 IU/ml) for 30 min were included as a positive control; β-actin was used as a loading control. Top right: The relative abundances of pSTAT1 to tSTAT1 in the indicated samples from three independent experiments were determined by densitometry analysis and are expressed as a percentage of the pSTAT1 abundance in IFN-β–treated naïve cells, which was set at 100%. Bottom right: The relative abundance of pSTAT2 in the indicated samples was determined by densitometry analysis and is expressed as a percentage of pSTAT2 abundance in IFN-β–treated naïve cells, which was set at 100%. Western blots in (C) and (H) are representative of three independent experiments. ***P < 0.001, ****P < 0.0001; ns, not significant; ND, not detected.
Unlike IFN receptors, EPOR binds to its ligand EPO as a homodimer (34), meaning that EPOR fusions would enable us to engineer homodimers as well as heterodimers of each chimeric IFN receptor. Indeed, previous studies with fusions of the EPOR with type I and II IFN receptors have demonstrated that certain subunits are fully competent to signal as nonphysiological homodimers (IFNAR2), whereas others can only signal when recruited into heterodimeric complexes (IFNAR1) (32, 33). To extend this EPOR fusion strategy to the type III IFN receptor, we first engineered four sets of chimeras that would homodimerize upon EPO ligand binding: EPOR-IFNAR1, EPOR-IFNAR2, EPOR-IFNLR1, and EPOR-IL10RB (Fig. 1A). The TMD of each IFN receptor subunit was also included in the chimera to preserve the correct orientation of the ICDs. The ECDs of IFNAR (35) and IFNLR (30) complexes were overlayed with that of EPOR (36). Computationally aligning the structures (37) revealed that there is only modest structural similarity between the receptor-ligand complexes (TM scores from 0.3 to 0.5), results that reflect the presence of similar domains in somewhat different arrangements, such as the fibronectin type III domains present in each ECD (fig. S1). Each construct was linked to a yellow fluorescent protein (YFP) expressed with an internal ribosome entry sequence (IRES), enabling us to sort cells for similar receptor expression (Fig. 1B). We used lentiviral transduction to stably express one construct at a time in U2OS human bone osteosarcoma cells, which lack endogenous EPOR (38). We stimulated the four resulting cell lines with a saturating concentration (100 ng/ml) of EPO and measured the abundance of phosphorylated STAT1 (pSTAT1) at 30 min. Our results showed that stimulation of homodimers of the longer, JAK1-associated subunits of both type I and III IFN receptors (EPOR-IFNAR2 and EPOR-IFNLR1, respectively) with EPO resulted in increased pSTAT1 abundance, whereas stimulation of the shorter, TYK2-associated subunits (EPOR-IFNAR1 and EPOR-IL10RB) did not (Fig. 1C). We also observed a two-fold greater abundance of pSTAT1 in cells expressing the IFNAR2 homodimer compared with that in cells expressing IFNLR1, providing initial evidence that the IFNAR2 and IFNLR1 ICDs may indeed possess different intrinsic signaling capabilities, despite both binding to JAK1.
Given that IFN receptors are canonically thought to signal only as heterodimers (39), we next set out to engineer obligate heterodimeric versions of our synthetic receptors (Fig. 1, D and E). To do this, we sought to mutate regions within the ECD of the EPOR that would constrain it to bind to EPO asymmetrically. Each subunit of EPOR binds to EPO at distinct binding sites (termed sites 1 and 2) within the ECD, generating an asymmetric complex around the ligand (40). A previous study used a rational protein design strategy to engineer two EPOR variants with reduced binding affinity to one of the two sites on EPO (site 1 null: H114K/E117K; site 2 null: M150E), with binding to the other site unaffected (40). According to this study, dimerization should only occur when cells co-express both a “site 1 null” and “site 2 null” EPOR mutant subunit. To test whether such mutations would convert our homodimer receptor fusions into heterodimers, we first engineered four U2OS cell lines with subunits that each expressed a single binding-site mutation in the EPOR ECD: EPORM150E-IFNAR1TMD-ICD, EPORH114K/E117K-IFNAR2TMD-ICD, EPORH114K/E117K-IFNLR1TMD-ICD, and EPORM150E-IL10RBTMD-ICD. We then treated all four cell lines with EPO (100 ng/ml) to induce homodimerization and measured the abundance of pSTAT1 after 30 min. As expected, the “site 2” null versions of the EPOR-IFNAR1 and EPOR-IL10RB fusions did not produce any pSTAT1 in response to EPO; however, stimulation of the “site 1” null EPOR-IFNAR2 and EPOR-IFNLR1 fusions with EPO resulted in detectable pSTAT1 (fig. S2). These data suggest that EPOR ECDs harboring “site 1” mutations were still sufficient to drive some residual, undesirable receptor homodimerization.
The ICDs of type I and III IFN chimeras encode signal strength
We hypothesized that the residual homodimerization-induced signaling observed from the “site 1” null mutant receptors could be further weakened by engineering EPOR-IFNAR2 and EPOR-IFNLR1 receptors to contain both “site 1” and “site 2” mutations. To test this hypothesis, we constructed the following receptor variants: EPORH114K/E117K/M150E-IFNAR2TMD-ICD and EPORH114K/E117K/M150E-IFNLR1TMD-ICD. For brevity, these site 1/site 2–null receptor subunits were termed EPORhet-IFNAR2 and EPORhet-IFNLR1, whereas the site 2–null cognate subunits were termed EPORM150E-IL10RB and EPORM150E-IFNAR1. We established cell lines that expressed each individual subunit, as well as cell lines that co-expressed a site 1 mutant short subunit with a site 1/site 2 double-mutant long subunit. The coding sequence of each chimera was placed upstream of an IRES-fluorophore (Fig. 1, F and G), enabling us to sort cell lines with comparable amounts of both receptor subunits between the type I and III systems (fig. S3A). After sorting, the abundances of the chimeric IFN receptor subunits were presumably greater than those of their endogenous counterparts, as exemplified by EPORM150E-IFNAR1 which was ~five-fold more abundant than endogenous IFNAR1 (fig. S3B). However, sorting cells to express similar fluorescence intensities of our chimeric IFN receptor subunits enabled us to directly compare the signaling capabilities between our type I and III IFN receptor chimeras without signaling variability arising from differences in abundance.
We next set out to assess signaling from our candidate heterodimeric chimeric IFN receptors. Cells expressing either individual subunits or both subunits (for our heterodimeric type I or type III IFN receptor chimeras) were stimulated with EPO (100 ng/ml) for 30 min. As expected, cells expressing only one subunit failed to induce STAT1 or STAT2 phosphorylation (Fig. 1H), indicating that the combined site1/site 2 mutations in the EPOR ECD were indeed sufficient to abolish homodimer signaling. In contrast, our heterodimeric IFN receptor chimeras each stimulated STAT1 and STAT2 phosphorylation to different extents. Specifically, stimulation of the type I IFN receptor chimera resulted in a three-fold increase in pSTAT1 abundance relative to that caused by stimulation of the type III chimera and comparable amounts of pSTAT1 and pSTAT2 as were induced by IFN-β signaling in naïve U2OS cells (Fig. 1H). Indeed, the difference in pSTAT1 abundance between cells expressing the type I chimera and the type III chimera was comparable to that between IFN-β– and IFN-λ–stimulated naïve (untransduced) cells (Fig. 2A), suggesting that our chimeric IFN receptors recapitulate the signaling patterns observed for endogenous IFN receptors. Together, these data establish a synthetic set of heterodimerizing type I and III IFN receptors and indicate that major differences in type I and III IFN signaling persist even when differences in receptor abundance and ligand binding-affinity are eliminated.
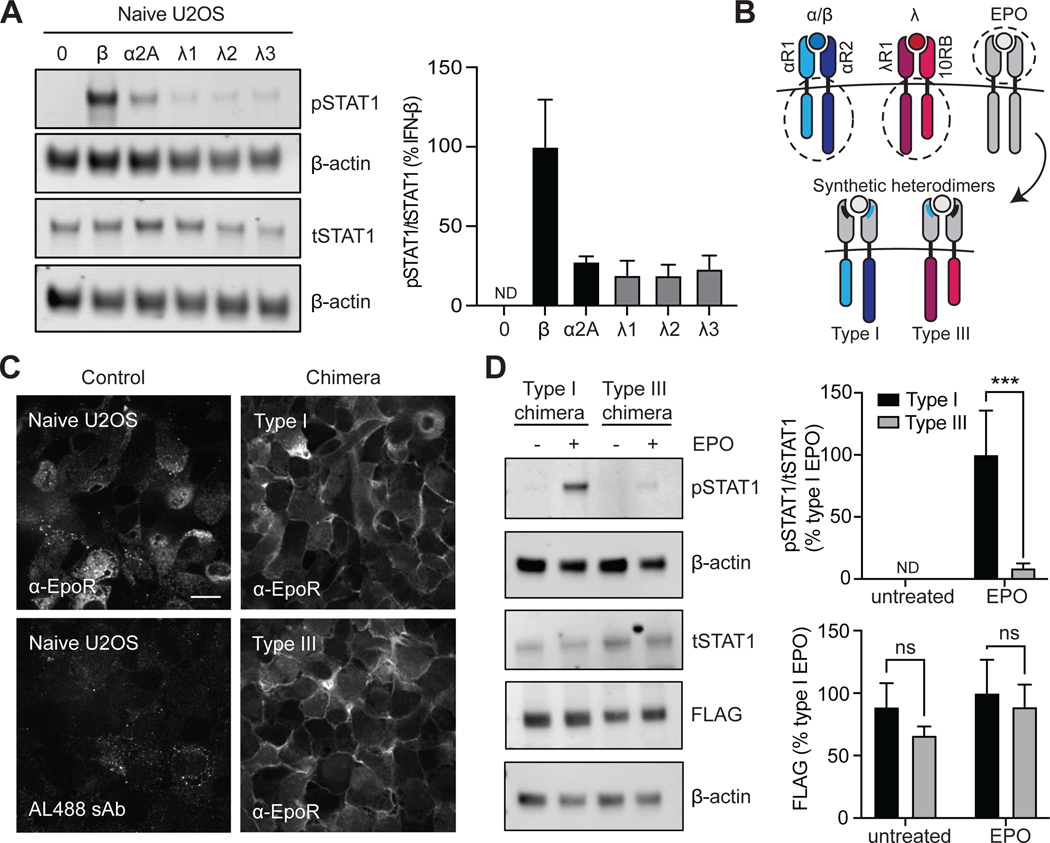
Fig. 2. Signaling differences between synthetic type I and III IFN receptors are independent of receptor abundance.
(A) Left: Naïve U2OS cells were stimulated with recombinant IFN-β (1000 IU/ml), IFN-α2A (1000 IU/ml), IFN-λ1 (100 ng/ml), IFN-λ2 (100 ng/ml), or IFN-λ3 (100 ng/ml) for 30 min and then analyzed by Western blotting with antibodies specific for pSTAT1 and tSTAT1. β-actin was used as a loading control. Right: The relative abundances of pSTAT1 to tSTAT1 in the indicated samples of three independent experiments were determined by densitometry analysis and are expressed as a percentage of the pSTAT1 abundance in IFN-β–treated naïve cells, which was set at 100%. (B) Schematic showing type I and III IFN receptor chimeras. The type I chimera consists of EPORhet-IFNAR2 paired with EPORM150E-IFNAR1, whereas the type III chimera consists of EPORhet-IFNLR1 paired with EPORM150E-IL10RB. (C) U2OS cells expressing equivalent amounts of either the chimeric type I or type III IFN receptor pairs were subjected to immunofluorescence staining and imaged by confocal microscopy to confirm the cell surface expression of EPOR. Scale bar, 10 μm. (D) Left: U2OS cells expressing the indicated FLAG-tagged chimeric IFN receptors were treated with EPO (100 ng/ml) for 30 min and then analyzed by Western blotting with antibodies against the indicated proteins. β-actin was used as a loading control. Top right: The relative abundances of pSTAT1 to tSTAT1 in the indicated samples from three independent experiments were determined by densitometry analysis and are expressed as a percentage of the pSTAT1 abundance in EPO-treated “Type I” cells, which was set at 100%. Bottom right: The relative abundances of FLAG-tagged proteins were determined by densitometry analysis. Western blots in (A) and (D) are representative of three independent experiments. ***P < 0.001; ns, not significant; ND, not detected.
We further confirmed that the type I and III IFN receptor chimeras (Fig. 2B) were similarly expressed using two methods. First, we observed similar receptor amounts on the cell surface by immunofluorescence microscopy with an anti-EPOR antibody without cell permeabilization (Fig. 2C). Second, we established cell lines expressing FLAG-tagged variants of the EPORhet-IFNAR2 and EPORhet-IFNLR1 constructs together with either EPORM150E-IFNAR1 or EPORM150E-IL10RB, respectively, and sorted cells for dual YFP and BFP expression, as with the previous heterodimers. FLAG-tagged protein abundances appeared consistent between the type I and type III IFN receptor chimeras, whereas EPO-stimulated pSTAT1 abundance was still significantly greater in cells expressing the chimeric type I IFN receptors than that in cells expressing the chimeric type III receptors (Fig. 2D). Together, these data confirm that type I and III receptors possess distinct signaling capabilities even when receptor protein amounts are equivalent.
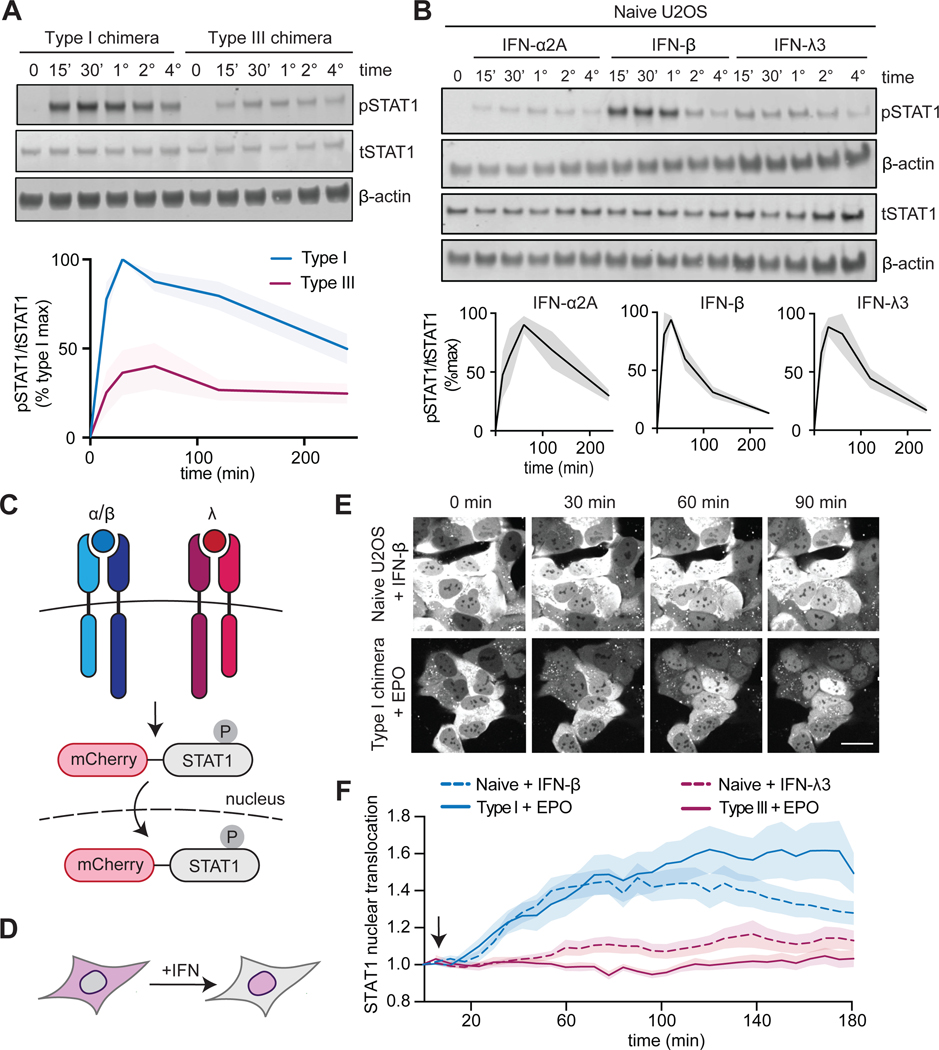
IFN signaling is dynamic, with differences in transient and sustained signaling driven by particular ligands and receptors (24–26); thus, we sought to compare the dynamics of our type I and type III IFN receptor chimeras with respect to IFN stimulation of naïve U2OS cells. We found that signaling was sustained for both the type I and the type III IFN receptor chimeras after EPO treatment, with high amounts of pSTAT1 observed for at least 4 hours (Fig. 3A). In contrast, signaling through the endogenous type I and III IFN receptors appeared to be more transient, with maximal pSTAT1 abundance at 1 hour, which was reduced to basal amounts in 2 to 3 hours (Fig. 3B). We next monitored the nuclear translocation of pSTAT1, which is required for downstream signaling. We generated an mCherry-STAT1 fusion protein that enabled us to visualize re-localization of STAT1 in response to IFN (Fig. 3, C and D) (41). We observed the nuclear localization of mCherry-STAT1 by time-lapse confocal microscopy within 30 min of treatment of naïve cells with IFN-β or of cells expressing the type I IFN receptor chimera with EPO (Fig. 3E). In contrast, we observed substantially less pSTAT1 translocation in response to IFN-λ stimulation of naïve cells or EPO stimulation of cells expressing the type III IFN receptor chimera (Fig. 3F), consistent with the substantially weaker STAT phosphorylation responses observed under these conditions. Together, our results indicate that signaling magnitude is an intrinsic property to IFN receptor ICDs, with type I IFN receptors driving stronger responses than type III IFN receptors, even when differences in ligand-binding affinity and receptor abundance are eliminated. In contrast, regulation of the duration of activation appears to be ICD-independent and is possibly influenced by receptor abundance ligand-binding affinity, or other factors that distinguish our synthetic receptors from endogenous IFN receptors.
Fig. 3. STAT activation dynamics are sustained after activation of chimeric type I and III IFN receptors.
(A) Top: U2OS cells expressing chimeric type I or III IFN receptors were treated with EPO (100 ng/ml) for the indicated times and analyzed by Western blotting with antibodies against the indicated proteins. β-actin was used as a loading control. Bottom: The relative abundances of pSTAT1 to tSTAT1 in the indicated samples from three independent experiments were determined by densitometry analysis and are expressed as a percentage of the maximal pSTAT1 abundance in EPO-treated “Type I chimera” cells, which was set at 100%. The shaded area indicates the SEM. (B) Top: Naïve U2OS cells were treated with recombinant IFN-β (1000 IU/ml), IFN-α2A (1000 IU/ml), IFN-λ1 (100 ng/ml), IFN-λ2 (100 ng/ml), or IFN-λ3 (100 ng/ml) for the indicated times and analyzed by Western blotting for pSTAT1 and tSTAT1. β-actin was used as a loading control. Bottom: The relative abundances of pSTAT1 to tSTAT1 in the indicated samples from three independent experiments were determined by densitometry analysis and are expressed as a percentage of the maximal pSTAT1 abundance in IFN-β–treated naïve cells, which was set at 100%. The shaded area indicates the SEM. Data are representative of three independent experiments. (C and D) Schematics illustrating the nuclear localization of the mCherry-STAT1 live cell reporter in cells undergoing type I or III IFN signaling. (E) Representative images of mCherry-STAT1 fluorescence in naïve U2OS cells treated with IFN-β (1000 IU/ml) or in cells expressing the chimeric type I IFN receptor and treated with EPO (100 ng/ml). Treatments were started after the first set of images was taken (4 min). Scale bar, 10 μm. (F) Cells expressing mCherry-STAT1 and either type I or III IFN receptor chimeras were treated with EPO (100 ng/ml) and analyzed by confocal microscopy to determine nuclear fluorescence. Naïve U2OS cells treated with IFN-β (1000 IU/ml) or IFN-λ3 (100 ng/ml) were imaged as positive controls. Images were collected every 4 min for 3 hours. The arrow indicates the addition of ligand (immediately after the first round of imaging). Each condition included n>20 replicates; the shaded region shows the SEM. (A to F) The type I chimera consists of EPORhet-IFNAR2 paired with EPORM150E-IFNAR1, whereas the type III chimera consists of EPORhet-IFNLR1 paired with EPORM150E-IL10RB.
Signaling through chimeric IFN receptors potently stimulates gene expression
We next sought to establish whether signaling from our chimeric type I and III IFN receptors could stimulate downstream cellular responses. IFN signaling is usually characterized by a robust and rapid gene expression response in which the expression of a number of ISGs is induced with distinct dynamics over the hours after ligand stimulation (42). The products of many ISGs are involved in inhibiting viral replication, thereby providing some protection against the viral infection of cell populations (43). We thus set out to test whether our type I and type III IFN receptor chimeras stimulated robust ISG expression and to quantify differences in the magnitude and dynamics of ISG expression in each case.
We performed bulk RNA-barcoding and -sequencing (44) of RNA extracted from naïve U2OS cells treated with IFN-β (1000 IU/ml) or IFN-λ3 (100 ng/ml), and of cells expressing type I or III IFN receptor chimeras and treated with EPO (100 ng/ml) for 0, 1, 3, or 8 hours. We applied stringent filtering criteria to identify a subset of genes whose changes in expression could be accurately quantified across all conditions. Principal component analysis (PCA) revealed that the directions of highest variance in the dataset were reflective of condition-dependent changes in gene expression, showing responses that increased in magnitude over time for each stimulus (fig. S4A). We kept all high-scoring genes on either PC1 or PC2 (fig. S4B; 120 genes). As a second method to identify hits, we captured genes with high variance between conditions relative to the six untreated replicates (fig. S4C; 100 genes). The final 216 hits were manually inspected, and responses dominated by noise between replicates were excluded; the remaining 96 genes exhibited clearly defined responses between conditions (fig. S4D). Of these, 32 genes exhibited differences in expression between cell lines, but failed to show an IFN-stimulated response. We thus analyzed the remaining 64 genes to quantify the dynamics of IFN-stimulated transcription in each condition.
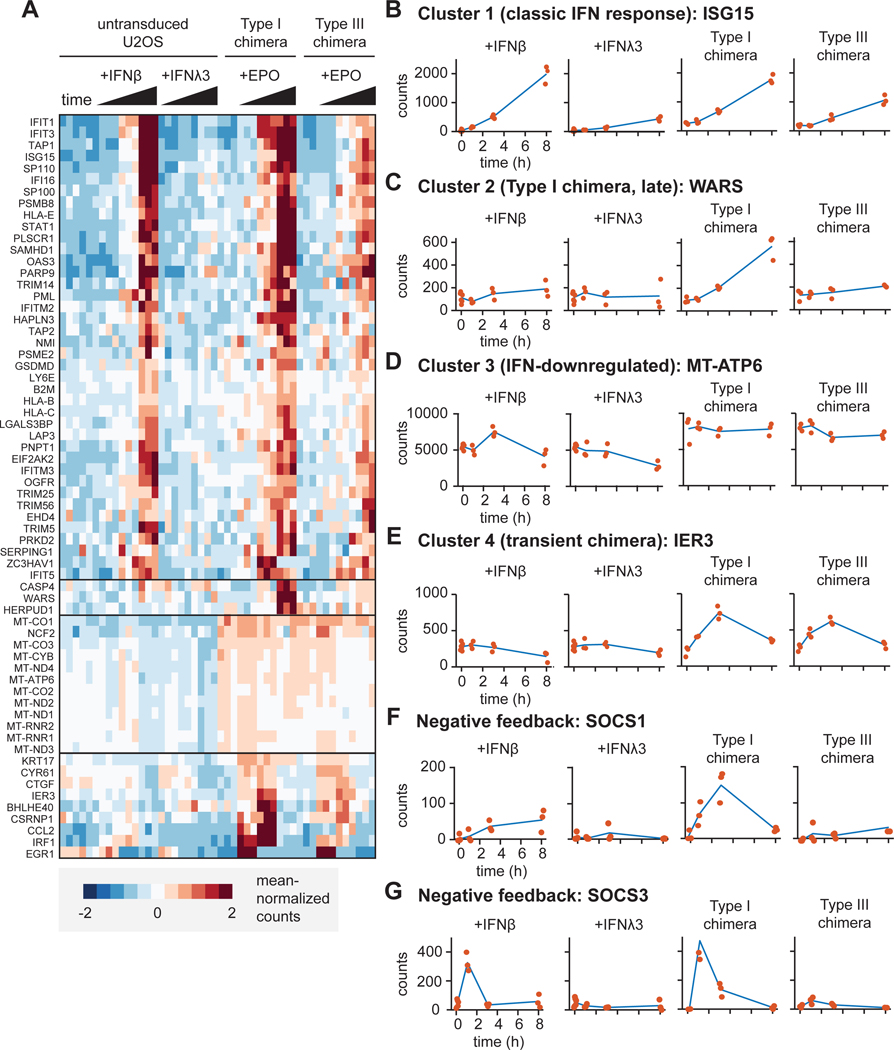
Hierarchical clustering revealed distinct classes of responses from these 64 genes (Fig. 4A). The first cluster was heavily enriched for classical ISGs whose expression was induced by the type I IFN receptor chimera in a similar pattern as that observed in naïve U2OS cells treated with IFN-β (Fig. 4, A and B). As expected, the type I IFN receptor chimera exhibited a stronger ISG response compared to that of the type III IFN receptor chimera (Fig. 4B). These data demonstrate that chimeric IFN receptors mount potent ISG transcriptional responses, and that the extent of ISG expression remains greater in response to type I IFN receptor chimeras than in response to type III IFN receptor chimeras, mirroring differences in the abilities of the endogenous receptors to stimulate STAT1 phosphorylation. The magnitude of this difference, however, was smaller between type I and type III IFN receptor chimeras treated with EPO compared to the magnitude of the difference between naïve cells treated with IFN-β or IFN-λ3 (Fig. 4B), suggesting that receptor abundance and ligand-binding affinity likely do play some role in defining the magnitude of the transcriptional response.
Fig. 4. RNA expression profiles differ between cells expressing chimeric type I and III IFN receptors.
(A to G) Bulk RNA-seq analysis was performed on RNA extracted from naïve U2OS cells treated with IFN-β (1000 IU/ml) or IFN-λ3 (100 ng/ml) and cells expressing chimeric type I or III IFN receptors treated with EPO (100 ng/ml) for the indicated times. Data includes three replicates for each condition. (A) Heat-map showing clustered gene expression patterns for 64 differentially expressed genes. Solid black horizontal lines indicate different clusters. (B) Cluster 1 genes were defined by their time-dependent induction in response to endogenous and chimeric IFN receptor signaling. (C) Cluster 2 genes were defined by being induced at 8 hours by the chimeric type I IFN receptor only. (D) Cluster 3 genes were defined by their time-dependent decreased expression in response to endogenous IFN receptors, but not chimeric IFN receptors. (E) Cluster 4 genes were defined by their transient induction by chimeric IFN receptors, but not endogenous IFN receptors. (B to E) The expression patterns of representative genes are shown for each cluster. Red dots indicate individual data points. (F) SOCS1 expression over time. (G) SOCS3 expression over time. The type I chimera consists of EPORhet-IFNAR2 paired with EPORM150E-IFNAR1, whereas the type III chimera consists of EPORhet-IFNLR1 paired with EPORM150E-IL10RB. ISG15, interferon-stimulated gene 15; WARS, tryptophanyl-tRNA synthetase; MT-ATP6, mitochondrially encoded ATP synthase 6; IER3, immediate early response 3; SOCS1, suppressor of cytokine signaling 1; SOCS3, suppressor of cytokine signaling 3.
Our analysis further revealed three additional clusters of gene expression patterns (Fig. 4, C to E). In cluster 2, expression of a small group of genes was increased at 8 hours in response to stimulation of the type I IFN receptor chimeras (Fig. 4C). We speculate that the expression of these genes may be the result of a sustained, high-amplitude signaling that is unique to the type I IFN receptor chimera (Fig. 3, A and B). Note that all three genes in this cluster have been implicated in promoting apoptosis. Expression of another set of genes, cluster 3, was decreased in response to IFN-λ3 but was unaffected by simulation of type I or type III IFN receptor chimeras (Fig. 4D). The products of many of these genes appear to be involved in cellular respiration, and in particular the electron transport chain, such as MT-ATP6, which forms a subunit of ATP synthase. These signaling differences may be the result of different signaling magnitudes between our type III IFN receptor chimera and endogenous IFN-λ signaling. Lastly, we identified a fourth cluster of genes whose expression was transiently decreased by both the type I and type III IFN receptor chimeras but not by endogenous receptors (Fig. 4E). These genes likely reflect gene expression caused by differences between the chimeras and endogenous receptors (for example, differences in receptor abundance or ECD orientation during signaling). Nevertheless, despite some cell-specific effects of chimeric receptor expression, together, our findings suggest that differences in signal strength between type I and III IFN receptor chimeras can be observed at the level of gene expression as well as at the level of STAT phosphorylation.
We previously observed that our type I and III IFN receptor chimeras produced more sustained STAT1 phosphorylation than did endogenous IFN receptors (Fig. 3, A and B), leading us to test whether we might observe differences between endogenous and chimeric IFN receptors in terms of expression patterns for ISGs involved in the negative regulation of IFN responses. Expression of genes encoding suppressor of cytokine signaling (SOCS) proteins is induced upon IFN signaling, and SOCS proteins subsequently limit IFN signaling by targeting the kinases associated with the type I and III IFN receptors. SOCS3 binds to and directly inhibits JAK1, JAK2 and TYK2 (45), whereas SOCS1 interacts with phosphotyrosine residues on TYK2 (46). We found increased transcription of SOCS1 (Fig. 4F) and SOCS3 (Fig. 4G) in naïve U2OS cells treated with IFN-β, as well as in EPO-treated cells expressing type I IFN receptor chimeras. These data suggest that endogenous and chimeric type I IFN receptor signaling are both capable of inducing negative feedback mechanisms through SOCS proteins. However, whereas SOCS3 expression was transient for both endogenous and chimeric type I IFN receptor activation, SOCS1 expression was transient only for activation of the chimeric type I IFN receptor. It is therefore conceivable that the altered dynamics of negative regulation contribute to the differences that we observed in STAT1 phosphorylation dynamics between endogenous and chimeric IFN receptors; however, this does not exclude the possibility that one or several other factors are also responsible.
Type I and III chimeric IFN receptors inhibit the replication of an RNA virus
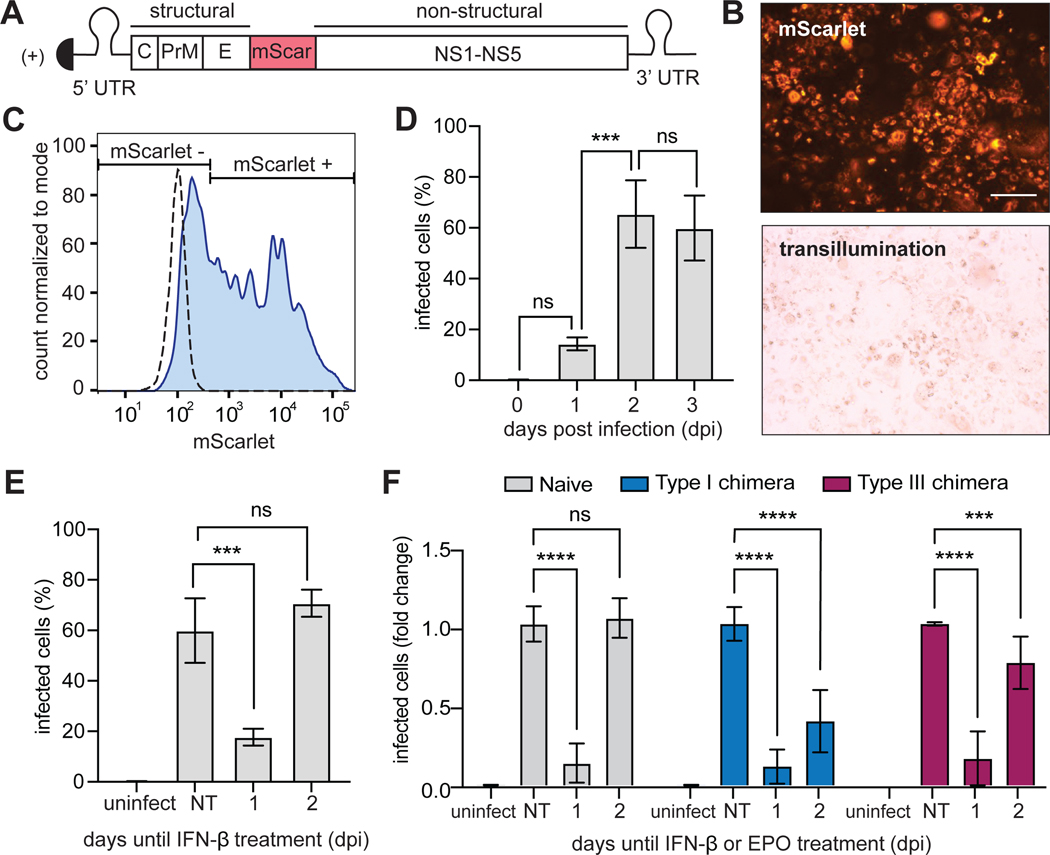
To assess the antiviral capabilities of our chimeric IFN receptors, we generated a fluorescent version of the yellow fever virus (YFV) vaccine strain (YFV-17D) (Fig. 5A). YFV-17D, a positive sense, single-stranded RNA virus, is the prototype of the Flavivirus genus within the Flaviviridae family (47), which can infect U2OS cells. Because a previous study showed that type I and III IFN signaling can antagonize the replication of YFV (8), we rationalized that this reporter virus would enable us to measure differences in the abilities of our chimeric IFN receptors to induce an antiviral response. Expression of mScarlet within the viral polyproteins enabled us to visualize infected cells by microscopy (Fig. 5B) and to quantify them by flow cytometry. To establish the kinetics of infection, U2OS cells were infected at an MOI of 1 and analyzed by flow cytometry to quantify the frequency of mScarlet-expressing cells (Fig. 5C). We found that YFV-17D-mScarlet infection peaked by two days post-infection (dpi) in naïve U2OS cells and was sustained through 3 dpi, with ~60% of cells expressing mScarlet (Fig. 5D). Thus, for subsequent experiments, we used 3 dpi as the final readout. We then sought to determine the effect that IFN signaling had on the replication of YFV-17D-mScarlet. When infected U2OS cells were treated with IFN-β (1000 IU/ml) at 1 dpi, the frequency of infected cells was reduced from 60 to 20% at 3 dpi (Fig. 5E). In contrast, when cells were treated with IFN-β at 2 dpi, the percentage of virally infected cells was not affected (Fig. 5E), suggesting that IFN signaling disrupted YFV-17D-mScarlet replication at early times (at 1 dpi) but had limited effects once maximal viral replication was established (at 2 dpi).
Fig. 5. YFV-17D-mScarlet replication is disrupted by signaling through endogenous or chimeric IFN receptors.
(A) The yellow fever virus (YFV)-17D viral genome was generated with an in-frame insertion of the mScarlet fluorescent protein between the regions encoding the E and NS1 proteins. (B) U2OS cells infected with YFV-17D-mScarlet were imaged in the RFP and transillumination channels. Scale bars, 200 μm. (C) Gating strategy for quantifying the frequency of YFV-17D-mScarlet–infected cells by flow cytometry. Dashed line, uninfected cells; solid line, infected cells (at an MOI of 1) 3 dpi. (D) Naïve (untransduced) U2OS cells were infected with YFV-17D-mScarlet at an MOI of 1 and fixed at the indicated times. The percentage of infected cells at each time was quantified by flow cytometry. (E) Naïve (untransduced) U2OS cells infected with YFV-17D-mScarlet (at an MOI of 1) were treated with IFN-β (1000 IU/ml) at either 1 or 2 dpi. Cells under all conditions were then fixed at 3 dpi and analyzed by flow cytometry to detect mScarlet as a measure of the percentage of cells that were infected. Uninfected and untreated groups were included as controls. (F) U2OS cells expressing chimeric type I or III IFN receptors were treated with EPO (100 ng/ml) at either 1 or 2 dpi and fixed at 3 dpi for analysis by flow cytometry to detect mScarlet-expressing cells. Naïve cells treated with IFN-β (1000 IU/ml) were included as a positive control. The type I chimera consists of EPORhet-IFNAR2 paired with EPORM150E-IFNAR1, whereas the type III chimera consists of EPORhet-IFNLR1 paired with EPORM150E-IL10RB. Infected cells are shown as a proportion of untreated naïve cells, which was set at 1. Data in (D) to (F) are means ± SD. ***P < 0.001, ****P < 0.0001; ns, not significant; uninfect, uninfected; NT, no treatment.
Similarly, when U2OS cells expressing the chimeric IFN receptors were treated with EPO (100 ng/ml) at 1 dpi, the frequency of mScarlet+ cells remained ~5- to 10-fold less at 3 dpi compared with the untreated group, indicating that YFV-17D-mScarlet replication was suppressed by activation of the chimeric IFN receptors at 1 dpi, similarly to the effect of IFN-β (Fig. 5F). However, and in contrast to IFN-β, EPO treatment at 2 dpi also partially reduced the size of the mScarlet+ cell population (type I: a 60% reduction in maximal infection; type III: a 25% reduction of maximal infection) (Fig. 5F). These data suggest that the chimeric IFN receptors both have antiviral capabilities when stimulated at either 1 or 2 dpi; however, given that the fold-reduction in viral replication was greater in cells expressing the chimeric type I IFN receptor, it appears that this receptor may exert a greater antiviral response overall. Note that we also found that the basal extent of infection was different between cells expressing type I and III IFN receptor chimeras. By 3dpi, only 26% of cells with the type I IFN receptor chimera expressed mScarlet, compared to 98% of cells expressing the type III IFN receptor chimera (fig. S5A). Further analysis of early time points post-infection revealed that differences in the extent of infection between naïve and chimera-expressing cells did not appear until 2 dpi, when naïve cells displayed the highest percentage of infected cells (fig. S5B). These data may indicate differences in tonic signaling due to overexpression of individual chimeric IFN receptor components, even when the heterodimeric IFN receptor complexes are not formed. Together, our data suggest that both type I and III IFN receptor chimeras are capable of inducing antiviral states, although the chimeric type I IFN receptor may be more efficient.
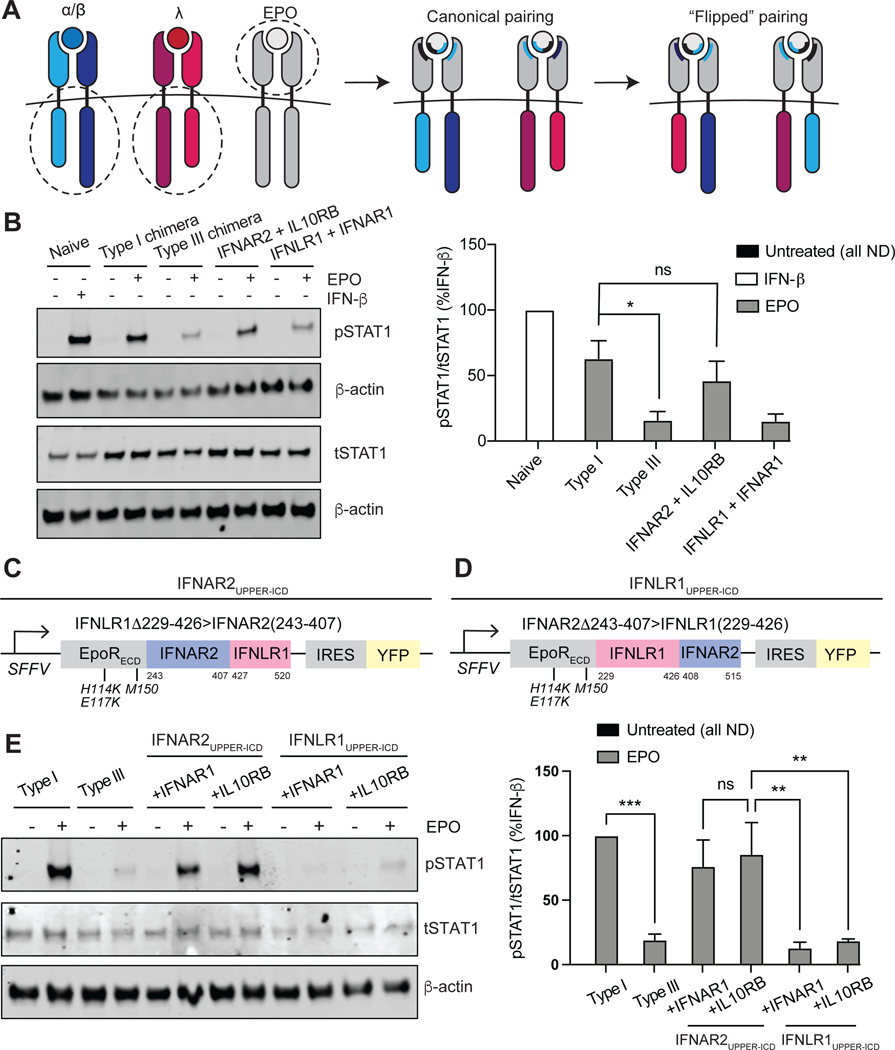
Type I IFN signaling strength requires the membrane-proximal region of IFNAR2
We next sought to determine which subunit of the type I and III IFN receptor ICDs was responsible for defining signaling strength. To this end, we generated cell lines expressing noncanonical combinations of long and short chimeric subunits: EPORhet-IFNAR2 paired with EPORM150E-IL10RB, and EPORhet-IFNLR1 paired with EPORM150E-IFNAR1 at comparable amounts to those of the original type I and III IFN receptor chimera lines (Fig. 6A). EPO treatment of cells expressing EPORhet-IFNAR2 resulted in the greatest increase in pSTAT1 abundance, as detected by Western blotting analysis, regardless of the short subunit that it was paired with; conversely, cells expressing EPORhet-IFNLR1 exhibited reduced amounts of pSTAT1 (Fig. 6B). These data suggest that IFNAR2 determines the strength of signaling by the type I IFN receptor, and that both IFNAR1 and IL10RB can stimulate JAK-STAT signaling to similar extents when paired with a JAK1-associated subunit.
Fig. 6. The membrane-proximal half of the IFNAR2 ICD is necessary for chimeric type I IFN receptor signaling strength.
(A) Schematic illustrating noncanonical, “flipped” chimeric pairings: EPORhet-IFNAR2 paired with EPORM150E-IL10RB; EPORhet-IFNLR1 paired with EPORM150E-IFNAR1. The ECDs of EPORM150E-IFNAR1 and EPORM150E-IL10RB contain the point mutation M150E, whereas the ECDs of EPORhet-IFNAR2 and EPORhet-IFNLR1 contain the three point mutations H114K, E117K, and M150E. All chimeras were expressed under an SFFV promoter and contained an IRES for concurrent expression of either BFP or YFP. (B) Left: U2OS cells expressing the canonical and noncanonical chimeric subunit pairings at similar abundances were treated with EPO (100 ng/ml) for 30 min and analyzed by Western blotting for phosphorylated and total STAT1. β-actin was used as a loading control. Naïve U2OS cells treated with IFN-β (1000 IU/ml) for 30 min were included as a positive control. Western blots are representative of three independent experiments. Right: The relative abundances of pSTAT1 to tSTAT1 in the indicated samples from three independent experiments were determined by densitometry analysis and are expressed as a percentage of the pSTAT1 abundance in IFN-β–treated naïve cells, which was set at 100%. (C) The “IFNAR2UPPER-ICD” triple chimera was generated by fusing the EPOR ECD (containing the mutations H114K, E117K, and M150E) to amino acid residues 243 to 407 of IFNAR2 and 427 to 520 of IFNLR1. (D) The “IFNLR1UPPER-ICD” triple chimera was generated by fusing the EPOR ECD (containing the mutations H114K, E117K, and M150E) to amino acid residues 229 to 426 of IFNLR1 and 408 to 515 of IFNAR2. (E) Left: U2OS cells were transduced to express equivalent amounts of the canonical IFN receptor chimeras (type I or III) or the “UPPER-ICD” chimeras paired with either EPORM150E-IFNAR1 or EPORM150E-IL10RB (each with the mutation M150E in EPOR). Cells were treated with EPO (100 ng/ml) for 30 min before being analyzed by Western blotting for pSTAT1 and tSTAT1. β-actin was used as a loading control. Western blots are representative of three independent experiments. Right: The relative abundances of pSTAT1 to tSTAT1 in the indicated samples of three independent experiments were determined by densitometry analysis and are expressed as a percentage of the pSTAT1 abundance in EPO-treated “type I” cells, which was set at 100%. *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant; ND, not detected.
To identify a minimal region of the long subunit responsible for determining signaling strength, we swapped regions of EPORhet-IFNAR2 into EPORhet-IFNLR1, and vice versa, to generate additional chimeras. The ICDs of IFN receptors typically contain linear amino acid motifs that serve as docking and phosphorylation sites for JAKs and STATs (fig. S6). We first swapped the C-terminal regions between EPORhet-IFNAR2 and EPORhet-IFNLR1 that have been identified as important for STAT binding and activation (Fig. 6, C and D) (48–50). Treatment of cells with EPO revealed that the chimeric subunit containing the “upper ICD” region of IFNAR2 (amino acid residues 243 to 407) attached to the presumed STAT-binding region of IFNLR1 (residues 427 to 520) stimulated potent, type I IFN–like STAT1 phosphorylation when it was paired with either EPORM150E-IFNAR1 or EPORM150E-IL10RB (Fig. 6E). Conversely, fusing the upper ICD of IFNLR1 (residues 229 to 426) to the presumed STAT-binding region of IFNAR2 (residues 408 to 515) induced only a low level of STAT1 phosphorylation comparable to that induced by the chimeric type III IFN receptor (Fig. 6E). Thus, it appears that type I and type III IFN receptor chimeras differ in signaling potency solely due to sequence differences within the upper two thirds of the ICDS of IFNAR2 and IFNLR1, and not within the regions responsible for STAT binding.
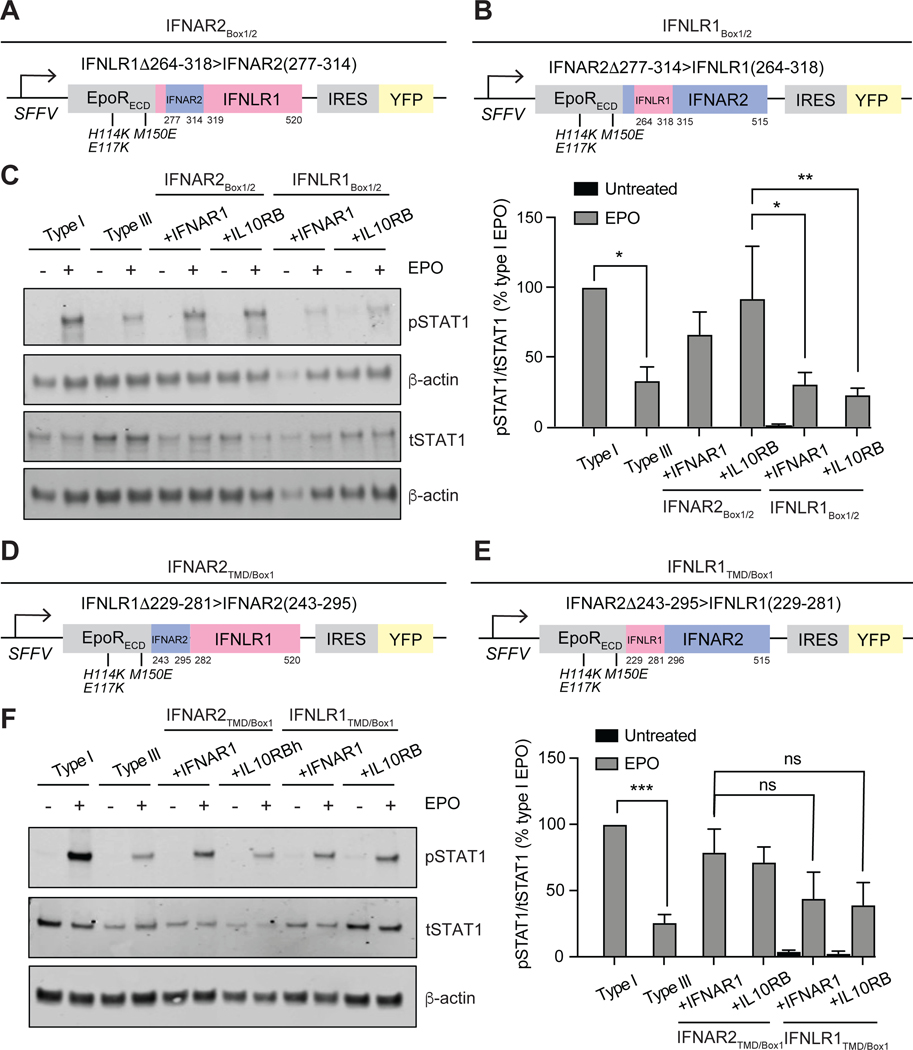
To further narrow down the sequences responsible for encoding signaling strength, we made additional swaps between EPORhet-IFNAR2 and EPORhet-IFNLR1 within their upper-ICD sequences, using alignments between IFNAR2 and IFNLR1 to identify regions of local conservation (fig. S6). One particularly informative set of chimeras involved swapping the JAK1-binding box 1 and 2 motifs between EPORhet-IFNAR2 and EPORhet-IFNLR1 (51, 52). One chimera (“IFNAR2Box1/2”) contained only a 38-residue sequence from IFNAR2 (residues 277 to 314) inserted in place of residues 264 to 318 of IFNLR1, leaving most of the TMD and ICD of IFNLR1 intact (Fig. 7A). Another chimera (“IFNLR1Box1/2”) contained the converse swap, incorporating the JAK1-binding box motifs of IFNLR1, but leaving the TMD and most of the ICD of IFNAR2 intact (Fig. 7B). These chimeras were expressed in cells together with either EPORM150-IFNAR1 or EPORM150E-IL10RB and sorted for similar amounts of cell surface receptor, as for all previous experiments. Treatment of the cells with EPO revealed that only the 38-residue JAK1-binding box motif region from IFNAR2 was sufficient for EPORhet-IFNLR1 to confer strong STAT1 phosphorylation when paired with either EPORM150E-IFNAR1 or EPORM150E-IL10RB (Fig. 7C), whereas the 55-resiude “box” motifs from IFNLR1 converted EPORhet-IFNAR2 to a weaker, type III-like receptor. These results further suggest that the TMDs of IFNAR2 and IFNLR1, similarly to their STAT-binding sequences, are interchangeable in terms of signal strength, and that signal strength differences lay primarily in a short region within their ICDs, corresponding to their JAK-binding box motifs.
Fig. 7. The IFNAR2 box motif region confers type I signaling strength to the chimeric type III IFN receptor.
(A) The “IFNAR2BOX1/2” triple chimera was generated by fusing the EPOR ECD (containing the mutations H114K, E117K, and M150E) to the TMD-ICD of IFNLR1, where amino acid residues 264 to 318 were replaced with residues 277 to 314 of IFNAR2. (B) The “IFNLR1BOX1/2” triple chimera was generated by fusing the EPOR ECD (containing the mutations H114K, E117K, and M150E) to the TMD-ICD of IFNAR2, where amino acid residues 277 to 314 were replaced with residues 264 to 318 of IFNRL1. All of the chimeras were expressed under an SFFV promoter and contained an IRES for concurrent expression of YFP. (C) Left: U2OS cells were transduced to express equivalent amounts of the canonical IFN receptor chimeras (type I or III) or “BOX1/2” chimeras paired with either EPORM150E-IFNAR1 or EPORM150E-IL10RB (each with the mutation M150E in EPOR). Cells were treated with EPO (100 ng/ml) for 30 min before being analyzed by Western blotting for phosphorylated (p) or total (t) STAT1. β-actin was used as a loading control. Western blots are representative of three independent experiments. Right: The relative abundances of pSTAT1 to tSTAT1 in the indicated samples from three independent experiments were determined by densitometry analysis and are expressed as a percentage of the pSTAT1 abundance in EPO-treated “type I” cells, which was set at 100%. (D) The “IFNAR2TMD/BOX1” triple chimera was generated by fusing the EPOR ECD (containing the mutations H114K, E117K, and M150E) to amino acid residues 243 to 295 of IFNAR2 and 282 to 520 of IFNLR1. (E) The “IFNLR1TMD/BOX1” triple chimera was generated by fusing the EPOR ECD (containing the mutations H114K, E117K, and M150E) to amino acid residues 229 to 281 of IFNLR1 and 296 to 515 of IFNAR2. All of the IFN receptor chimeras were expressed under an SFFV promoter and contained an IRES for concurrent expression of YFP. (F) Left: U2OS cells were transduced to express equivalent amounts of the canonical IFN receptor chimeras (type I or III) or the “TMD/BOX1” chimeras paired with either EPORM150E-IFNAR1 or EPOM150E-IL10RB. Cells were treated with EPO (100 ng/ml) for 30 min before being analyzed by Western blotting for pSTAT1 and tSTAT1. β-actin was used as a loading control. Western blots are representative of three independent experiments. Right: The relative abundances of pSTAT1 to tSTAT1 in the indicated samples from three independent experiments were determined by densitometry analysis and are expressed as a percentage of the pSTAT1 abundance in EPO-treated “type I” cells, which was set at 100%. *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant.
The JAK1-binding sequence of IFNAR2 has been studied in detail and subdivided further into two sub-motifs termed “box 1” and “box 2,” which engage distinct surface patches on JAK1 (51). To test whether signaling strength could be further assigned to just a single box motif, we generated chimeras that split the two box motifs from one another, and again assessed the extent of STAT1 phosphorylation in response to EPO. One chimera (“IFNAR2TMD/Box1”) contained a portion of IFNAR2 ranging from the TMD to the end of box 1 (residues 243 to 295) fused to residues 282 to 520 of IFNLR1 (Fig. 7D), whereas the other chimera (“IFNLR1TMD/Box1”) contained the converse sequences of IFNLR1 fused to IFNAR2 (Fig. 7E). Unlike in previous experiments, this swap produced chimeric IFN receptors that both signaled at similar, intermediate strengths that could not be statistically distinguished from one another (Fig. 7F). These data suggest that both box 1 and 2 motifs partially contribute to the overall strength of an IFN receptor and provide opportunities to further tune signaling strength to intermediate levels.
Data from experiments with our chimeric IFN receptors suggest that the JAK1-binding box motifs are the primary determinants of IFN receptor signaling strength. We next sought to test whether these motifs were sufficient to alter the strength of full-length IFN receptors. We reasoned that replacing the JAK1-binding motif of IFNLR1 with that of IFNAR2 might be sufficient to increase the magnitude of IFN-λ signaling. We generated full-length IFNLR1 with either its natural JAK1-binding box motifs [IFNLR1(full length)] or the box motifs of IFNAR2 [IFNLR1(IFNAR2 box motifs)]. We expressed these bicistronic constructs (each expressing an IRES-YFP) in mouse embryonic fibroblasts (MEFs) that were also transfected with plasmid expressing full-length IL-10RB (fig. S7A). Note that MEFs lack endogenous type III IFN signaling, thus providing a clean background on which to conduct these experiments. We subsequently treated cells with hIFN-λ3 (100 ng/ml) of 30 min and measured pSTAT1 abundance by Western blotting. Cells expressing IFNLR1(IFNAR2 box motifs) had three- to four-fold more normalized pSTAT1 abundance compared to that in cells expressing IFNLR1(full length) (fig. S7B). These findings support the results from our EPOR chimera experiments, demonstrating that JAK1-binding box motifs can alter IFN signaling even in the context of the full-length IFN receptor. Note, however, that we also observed differences in the amounts of total STAT1 protein between naïve MEFs and MEFs over-expressing full-length IFNLR1 constructs (fig. S7B). This may be due to enhanced tonic IFN signaling caused by the overexpression of IFN receptor components, consistent with previous studies suggesting that IFN signaling regulates STAT1 abundance (53, 54). Future studies will provide a more comprehensive understanding of how box motifs regulate signaling in full-length and endogenous type III IFN receptors.
DISCUSSION
Here, we engineered heterodimeric IFN receptor chimeras to directly compare the signaling strengths of type I and III IFN receptor ICDs when receptor abundance and ligand-binding affinity were held constant. We showed that the chimeric type I IFN receptor stimulated STAT1/2 phosphorylation, ISG expression, and an antiviral response to greater extents than did the chimeric type III IFN receptor. This greater signal strength appeared to be mediated by the ICD of IFNAR2, regardless of whether it was associated with IFNAR1 or IL-10RB. Furthermore, we found that a single, membrane-proximal sequence encompassing the JAK1-binding box motifs (51) was sufficient to encode signaling strength. Swapping this region into EPORhet-IFNLR1 conferred greater signal strength to the chimeric type III IFN receptor, whereas swapping the converse region from IFNLR1 into EPORhet-IFNAR2 was sufficient to reduce signaling strength in the chimeric type I IFN receptor.
Similar box motifs appear in many other cytokine receptors, with box 1 tending to be proline-rich and box 2 containing many hydrophobic residues (51, 55). Box motifs have also been identified for IFNLR1, which appear to be crucial for its interaction with JAK1 (52, 56); however, there is little sequence homology between this region and the box motifs of IFNAR2. It is possible that these sequence differences affect how IFNAR2 and IFNLR1 interact with JAK1, which may explain the signaling differences that we observed between their ICDs. JAK family members rely on precise structural rearrangements upon receptor binding to facilitate kinase activity (57–60). In the case of JAK1 specifically, activation appears to depend on structural rearrangement of the pseudokinase domain upon receptor binding, leading to release of self-inhibition and promotion of kinase activity within the catalytic region (61–63). Future structural analysis may help to determine whether residue differences in the IFNAR2 and IFNLR1 box motifs lead to differential JAK1 binding, activation, or both.
In contrast, our study found no discernable differences in chimeric type I IFN receptor signal strength when the remainder of IFNAR2 was replaced with IFNLR1, including the TMD and the STAT-binding and activation domains. It is not surprising that the TMDs of IFNAR2 and IFNLR1 appear to be interchangeable, because previous studies have observed that the TMD of IFNAR2 can be replaced with that of IFNAR1 with no discernable effect on signaling (64). On the other hand, it is surprising that the STAT-binding domains of IFNAR2 and IFNLR1 appear to be interchangeable. Despite some similar STAT activation residues between IFNLR1 and IFNAR2 (49, 50, 65, 66), their overall STAT-binding regions are markedly different, including a long stretch of negatively charged amino acid residues that appear only in IFNLR1 (fig. S6). Nevertheless, swapping these membrane-distal regions did not affect the signaling strength of the resulting receptors. It would be interesting to test whether these findings extend beyond the type I and III IFN receptors to other members of the class 2 cytokine receptor family. It is possible that the box motifs tune receptor strength, whereas distinct STAT-binding motifs direct output to different combinations of STAT proteins, enabling evolution to independently tune signaling strength and downstream specificity within this class of receptors.
Independent of signaling strength, our study also found differential signaling dynamics between our chimeric IFN receptors and endogenous IFN receptors. Both type I and III IFN receptor chimeras produced sustained STAT1 phosphorylation over at least 4 hours, in contrast to endogenous type I IFN signaling which was much more transient. This suggests that signaling dynamics are encoded elsewhere, perhaps in receptor abundance, receptor internalization, regulation of the endogenous genomic locus, or the affinity between IFNs and their cognate receptors. Both ligand-binding affinity and receptor abundance influence signaling dynamics for other receptors, such as epidermal growth factor receptor (EGFR) (67). In future studies, it would be exciting to further explore the roles played by receptor abundance and ligand-binding affinity, for example by engineering EPO/EPOR variants with different affinities or preparing cell lines with a range of chimeric receptor amounts or insertion into the endogenous locus. These systems could also address how signaling differences are encoded for IFNs of the same type. For example, a system with more temporal control may be able to directly measure how the activation time of IFN receptors directly contributes to downstream signaling differences within the type I or III IFN subfamily (68).
We described the use of synthetic heterodimeric IFN receptors in which ligand-induced dimerization was mediated by engineered variants of the EPO receptor. Although EPOR fusions have been widely used to induce receptor homodimerization for decades, our system extends this classic approach by adapting EPOR to stimulate signaling solely through IFN receptor heterodimers, mimicking endogenous signaling. We expect similar approaches to transfer to other chimeric receptors for which EPOR fusions have been successfully applied, including receptor tyrosine kinases (69). Nevertheless, one possible limitation of our system is that it only accounts for IFN receptor heterodimers, whereas it has been suggested that cytokine receptors may also assemble into higher-order signaling clusters at the plasma membrane (70, 71). It is tempting to speculate that this might occur for the type III IFN receptor, given data showing that IFN-λ, in contrast to IFN-α, does not require TYK2 for signaling (72), which is thought to bind to and stabilize the IL-10RB subunit. Without TYK2 present on IL-10RB, it is unclear how IFNLR1 becomes phosphorylated in response to IFN-λ. One intriguing possibility is that type III receptor dimers may cluster to enable JAK1 on other IFNLR1 chains to phosphorylate all ICD residues required for STAT docking. More work is needed to understand the differential requirement for TYK2 between type I and III IFN receptors and whether IFN receptors form alternative structures beyond the canonical heterodimer model.
Our study identified specific contributions of the IFN receptor ICDs to signaling strength by our ability to hold other parameters (for example, ligand-binding affinity and receptor abundance) constant. However, differences in these other parameters are certainly present in the endogenous IFN receptor signaling systems of cells and are likely to play important roles. Follow-up work could explore how additional features of IFN signaling depend on these parameters. For example, our study sought to equalize receptor abundance between the chimeric type I and III IFN receptors, but in doing so, likely produced chimeric receptors that were more abundant than their endogenous counterparts. It will be interesting for future studies to explore how IFN receptor abundance affects signaling. Further investigation could also shed light on how the roles played by receptor endocytosis and negative feedback, which our RNA-seq data suggests may differ between type I and III IFN receptors. Finally, we designed identical EPOR fusions for both the type I and III chimeric receptors, but it remains possible that the ECDs of endogenous IFN receptors adopt different conformations (for example. producing differences in the relative location of the membrane-spanning domains) that are not equally matched by EPOR fusion. Future efforts could characterize the involvement of the ECD in signaling differences between endogenous type I and III IFN receptors. Thus, whereas our study points to one important mechanism that appears to regulate type I and III IFN signaling, there are still many other possible factors that should continue to be explored.
Our work also raises implications for IFN-λ therapies in the future. Efforts to engineer IFN-λ variants that bind to their receptor more strongly have only partially increased signaling strength and have not achieved the same signaling magnitude as that of type I IFNs (30); however, this could be seen as a benefit. Type I IFN therapies produce an array of unwanted side effects, possibly due to their robust signaling across all nucleated cell types. Signaling by IFN-λ, on the other hand, is restricted in cell type (73) and signaling outputs, with no reason to suspect that this makes it less effective against viral infection. As we have shown, even the reduced JAK-STAT signaling strength of the type III IFN receptor chimera conferred cells with a substantial antiviral response against YFV-17D. Thus, as next-generation IFN therapies are developed, it will be important to continue elucidating exactly how type I and III signaling is regulated. The inherent mechanisms within the type III IFN receptor that encode its reduced signal strength may affect the way in which IFN-λ can be used for therapy.
MATERIALS AND METHODS
Antibodies and reagents
Recombinant human EPO (E5546), recombinant human IFN-β (IF014), and recombinant human IFN-α2A (SRP4594) were purchased from Sigma-Aldrich. Recombinant human IFN-λ1 (1598–1L), IFN-λ2 (1587-IL), and IFN-λ3 (5259-IL) were purchased from R&D Systems. Primary antibodies for Western blotting were purchased from Cell Signaling Technologies: pSTAT1 (Tyr701) (9167 CST; 1:1000 dilution), pSTAT2 (Tyr690) (88410 CST; 1:1000 dilution), tSTAT1 (9H2) (9176 CST; 1:1000 dilution), tSTAT1 (D1K9Y) (14994 CST; 1:1000 dilution), and β-actin (3700 CST; 1:2500 dilution). Primary antibody against the FLAG tag was purchased from Sigma-Aldrich (F7425; 1:1000 dilution). Primary antibody against IFNAR1 was purchased from Abcam (ab45172; 1:1000 dilution). DyLight800-conjugated goat anti-mouse IgG (SA5–35521; 1:10,000 dilution) and DyLight680-conjugated goat anti-rabbit IgG (35568; 1:10,000 dilution) secondary antibodies were purchased from Thermo Fisher Scientific.
Cell culture
Unless otherwise stated, all cells (U2OS, 293T-lentiX, and Huh7.5 cells) were cultured under standard conditions at 37°C with 5% (v/v) CO2 and in DMEM (Thermo Fisher Scientific) containing 10% (v/v) heat-inactivated fetal bovine serum (FBS) and 1% v/v penicillin/streptomycin (Corning Inc.). Upon reaching confluency, cells were trypsinized with 0.05% trypsin/EDTA and re-plated onto cell culture dishes.
Generation of MEFs
All mouse experiments were registered and approved by the Institutional Animal Care and Use Committee of Princeton University (#3063). C57BL/6 mice were obtained from the Jackson Laboratory. A protocol for isolating MEFs was detailed previously (74). Briefly, mouse embryos were collected from the uterine horns of pregnant C57BL/6 mice (embryonic day 13 to14), rinsed briefly in 70% ethanol, and kept in PBS without Ca2+and Mg2+ until the time of dissection. Embryos were then dissected in a petri dish, first to separate the placenta and embryonic sac and then to remove the head and red organs. The remaining parts of embryos were washed in PBS and finely minced with a razor blade. When the parts of the embryo were small enough to pipette, 1 ml of 0.05% trypsin/EDTA containing 100 Kunitz units of DNase1 were added per embryo, and the tissue was then incubated in a 50-ml tube for 15 min at 37°C, with manual dissociation by pipetting occurring every 5 min. Trypsin was inactivated with an equal volume of freshly prepared MEF medium (450 ml of DMEM, 50 ml of FBS, 5 ml of 200 mM L-glutamine, 5 ml of penicillin/streptomycin). Cells were centrifuged at 300g for 5 min to remove the supernatant and then were resuspended in fresh MEF medium. A volume of cells equivalent to 3 or 4 embryos was plated in T150 flasks that had been precoated for 2 hours with 0.2% gelatin. Adherent cells were either expanded for immediate use or were plated in one well of a 6-well plate and transduced with 2 ml of lentivirus containing SV40 large T antigen (Addgene #1780). After 12 to 16 hours, the cell culture medium was replaced and the cells were expanded for experiments.
Plasmid construction
The plasmid encoding human EPOR was a gift from J. Tavernier (Ghent University) and was subcloned into a pHR backbone before site-directed mutagenesis was conducted to introduce the mutations described in the study. The plasmid encoding human STAT1 was a gift from A. Perantoni (NCI, Addgene #12301) and was subcloned into a pHR backbone encoding mCherry. H2B was ordered as a gblock gene fragment from Integrated DNA Technologies (IDT) and inserted into a pHR backbone encoding mIRFP. All plasmid backbones were either digested with the appropriate restriction enzymes or linearized by PCR amplification. Inserts were cloned by PCR amplification or synthesized (IDT). Assembly reactions were performed with the inFusion kit (Clontech), and stellar competent E. coli cells (Clontech) were transformed according to manufacturer’s instructions. All final plasmids were validated by sequencing (Eton Bioscience or Genewiz).
Generation of the YFV-17D fluorescent reporter
To generate the YFV-17D-mScarlet plasmid, the first 27 nucleotides of the sequence encoding YFV-17D NS1, the Gaussia Luciferase (GLuc) gene, and a dengue virus E linker coding sequence (E Stem and transmembrane domain) were amplified by PCR from pBSC-YFV-GLuc (75) (provided by Laura Gil, Oswaldo Cruz Foundation). The amplified gene cassette was then inserted between the E and NS1 coding sequences of pACNR-FLYF-17D-RL (provided by C. Rice, The Rockefeller University), through Infusion-based molecular cloning, yielding pACNR-FLYF-17D-Gluc. The pACNR-FLYF-17D-Gluc construct was then digested with Nar I because two Nar I sites were introduced to flank the Gluc-coding sequence. The m-Scarlet-I gene was amplified by PCR and subcloned in place of the Gluc-coding sequence into Nar I–digested pACNR-FLYF-17D-Gluc through Infusion-based molecular cloning.
YFV-17D-mScarlet virus production
Plasmid DNA was propagated in JM109 cells (Promega) at 30°C. Twenty micrograms of maxi-prepped DNA (Macherey-Nagel) were digested with Afl II (NEB) for 5 hours at 37°C and purified with a 1:1 ratio of phenol:chloroform:isoamyl alcohol (Acros Organics). The aqueous layer was collected in a total volume of 150 μl, and DNA was precipitated overnight at −20°C through the addition of 15 μl of 3 M sodium acetate (Sigma) and 165 μl of 100% ethanol. The DNA was spun down, washed with 70% ethanol, dried, and resuspended in 20 μl of RNA-free DDI water. The DNA was subjected to in vitro transcription with the mMessage mMachine SP6 transcription kit (Invitrogen, Waltham, MA), and RNA was purified with the MEGAclear transcription clean-up kit (Invitrogen). A 50% confluent, 10-cm plate of human Huh7.5 hepatoma cells was transfected with 10 μg of purified RNA with the TransIT-mRNA transfection kit (Mirus). Culture medium was collected once a day for 4 days. The collected virus was concentrated ~two-fold with an Amicon xcentrifugal filter (Millipore). Viral titers were calculated with a plaque-forming unit (PFU) assay.
Antiviral response assay
Naïve or receptor chimera–expressing U2OS cells were plated on a flat-bottom 96-well plate (at 10,000 cells/well) and left to adhere for 24 hours. The cells were then infected with YFV-17D-mScarlet at a multiplicity of infection (MOI) of 1 in fresh growth medium. For treatment with IFN-β or EPO, an additional 50 μl of treatment-containing medium was supplemented into the appropriate wells and mixed by pipetting. At the end of the infection period, the medium was aspirated from the wells, and the cells were resuspended and fixed with CytoFix solution (BD Biosciences) before being stored at 4°C in PBS. The frequencies of infected cells were determined by quantifying mScarlet expression with a BD LSRII Multi-Laser Analyzer w/ HTS flow-cytometer (BD Biosciences).
Lentiviral production and transduction
To produce lentivirus, HEK293T cells were grown to 90% confluency in 6-well or 10-cm plates pre-coated with poly-L-lysine. Cells were then co-transfected with the plasmid of interest (0.67 or 4 μg for either 6-well or 10-cm plates respectively) and the lentiviral packaging plasmids VSV-G (0.095 or 0.57 μg) and HIV Gal-Pol (0.67 or 4 μg) with the X-tremeGENE9 transfection reagent (Sigma-Aldrich). Culture medium containing lentiviral pseudoparticles was collected after 24 and 48 hours and filtered with a 0.45-μm filter. Stocks of lentiviral pseudoparticles were supplemented with 1:1000 polybrene (from a stock concentration of 4 mg/ml) and 1:50 HEPES (from a stock of 1 M) before storage at −80°C. For transduction, U2OS cells were grown in 6-well plates to ~40% confluency before the addition of 200 to 500 μl of pseudoparticles in 2 ml of growth medium. Pseudoparticle-containing medium was replaced by normal growth medium at 18 to 24 hours after transduction, and all experiments were performed at least 48 hours after transduction to ensure stable integration of the plasmid. In cases where receptor amounts needed to be controlled, cells were trypsinized as described earlier, resuspended at a density of 4 × 106 cells/ml, and sorted for high abundance on a BD FACSAria Fusion (BD Biosciences). Cell surface protein amounts were subsequently confirmed using a BD LSRII Multi-Laser Analyzer w/ HTS (BD Biosciences).
Western blotting
Cells were lysed in ice-cold RPPA buffer [1% Triton X-100, 50 mM HEPES buffer, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 100 mM NaF, 10 mM sodium pyrophosphate, 1 mM Na3VO4, 10% (v/v) glycerol] supplemented with freshly prepared protease and phosphatase inhibitors. Protein amounts were quantified with a Pierce BCA kit (Thermo Fisher Scientific), before samples were mixed with 6x Laemmli buffer/2-mercaptoethanol, heated at 95°C for 5 min, and loaded into a 4 to 12% Bis-Tris gel (Invitrogen) for electrophoresis. Gels were transferred to a nitrocellulose membrane with the iBlot dry transfer system (Thermo Fisher Scientific), blocked in Tris-buffered saline with 0.1% (v/v) Tween 20 (TBST) with 5% (v/v) milk for 30 min at room temperature, and incubated with primary antibody overnight at 4°C. Before imaging, membranes were washed three times for 5 min in TBST and incubated for 1 hour in either IRDye 680CW or 800CW secondary antibodies (Licor) (1:10,000). Imaging was performed with the Li-Cor Odyssey Infrared Imaging System (Licor).
Immunofluorescence
Cells were allowed to grow to 70 to 90% confluency in a 96-well plate and then fixed at 4°C for 10 min in CytoFix solution (BD Biosciences). After fixation, cells were washed with PBS and then permeabilized at −20°C for 10 min in ice-cold 90% (v/v) methanol. Cells were washed again in PBS before being blocked at room temperature for 1 hour with IF buffer [PBS supplemented with 10 % (v/v) FBS and 2 mM EDTA]. Cells were incubated overnight at 4°C in primary antibody diluted in IF-T buffer (IF buffer with 0.3 % Triton X-100). The following day, cells were washed three times in IF-T buffer, incubated at room temperature for 1 hour in secondary antibody diluted 1:100 in IF-T buffer, washed three times again, and then imaged with a confocal microscope.
Confocal microscopy
Cells were plated on collagen-treated, 0.17-mm glass-bottomed, black-walled 96-well plates (In Vitro Scientific). Cells were plated 24 hours before imaging and allowed to adhere to the plate. Immediately before imaging, 50 μl of mineral oil was added on top of the medium to prevent evaporation. Cells were kept at 37°C with 5% (v/v) CO2 while being imaged. All imaging was performed with a Nikon Eclipse Ti microscope with a Prior linear motorized stage, a Yokogawa CSU-X1 spinning disk, an Agilent laser line module containing 405-, 488-, 561-, and 650-nm lasers, an iXon DU897 EMCCD camera, and a x60 oil immersion objective lens. For all IFN treatments, 50 μl of recombinant IFN mixed with growth medium was pipetted onto cells after the imaging had begun. Quantification of nuclear intensities was determined with the TrackMate (76) plugin with ImageJ software (NIH).
RNA-seq
Cells were plated at 200,000 cells/well in 12-well plates and the following day were treated with either IFN-β (1000 IU/ml) or IFN-λ3 (100 ng/ml, in the case of naïve cells) or EPO (100 ng/ml, in the case of chimera-expressing cells) for 0, 1, 3, or 8 hours. Cells were then washed with PBS and lysed for RNA extraction with an RNeasy mini kit (Qiagen). RNA quality was assessed with an Agilent Bioanalyzer (Agilent) or Agilent TapeStation 4200 (Agilent). To profile gene expression from each sample, bulk RNA barcoding and sequencing (BRB-Seq) (44) was performed with some modifications. Reverse transcription (RT) was performed with 50 ng of total RNA per sample with SuperScript IV (Invitrogen) and a distinctly barcoded oligo(dT)30 primer for each sample (modified with the Illumina TruSeq Read 1 priming site instead of Nextera Read 1) with the following thermocycling program: 50°C, 15 min; 55°C, 15 min; 60°C, 15 min; 80°C, 15 min. After RT, up to 24 barcoded first-strand cDNA samples were pooled into a single tube. For each cDNA pool, second-strand cDNA was synthesized by the Gubler-Hoffman nick translation approach per the BRB-Seq protocol. Then, instead of Tn5 tagmentation, each cDNA pool was fragmented and end-repaired with the NEBNext Ultra II FS DNA Module (NEB) at 37°C for 6 min and then at 65°C for 30 min. DNA adaptor (37.5 pmol) was then duplexed and ligated with Illumina Read 2 using optimized ligation conditions (77) at 20°C for 2 hours before ligation products were purified with sequential 1.0X AMPure XP SPRI cleanups (Beckman Coulter). Each cDNA pool was amplified with 12 cycles of PCR using a P5 containing primer and a distinct multiplexed i7 indexing primer (Chromium i7 Multiplex Kit, 10X Genomics), followed by 0.55/0.75X double-sided size-selection with SPRIselect (Beckman Coulter). Libraries were amplified for another 6 cycles of PCR with P5 and P7 primers, and a final 0.7X SPRIselect cleanup was performed. All cDNA pools were sequenced on a NovaSeq SP v1.5 flowcell (Illumina) with 28 cycles Read 1 [covering the within-pool sample barcode and unique molecular index (UMI)], 8 cycles Index Read 1 (pool barcode), and 101 cycles Read 2 (cDNA template). The RNA-seq oligonucleotides used were as follows: R1T_RTP,CTACACGACGCTCTTCCGATCTNNNNN [8 bp sample barcode ]NNNNNNNNNNVVVVVTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTVN, 4nm, ultramer plate *TruSeq Read 1, within-pool sample barcode, UMI, anchored (dT)30; R2T_T,/5phos/GATCGGAAGAGCACACGTCTGAACTCCAGTCA, 100 nm, HPLC; R2T_B,GCTCTTCCGATC*T, 100 nm, HPLC; P5_R1T,AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTC, 100 nm, HPLC.
RNA-seq read processing
Reads were demultiplexed with Picard v2.21.1 (from within viral-ngs v.1.25.0) with the read structure ‘5S8B15M8B101T’ and Q10M2 mismatch tolerance, which simultaneously processes both the within-pool sample barcode (from the RT primer) and the pool barcode (from the i7 indexing primer). Reads were then mapped to the human genome hg38 with STAR v2.7.10a, and reads mapping to the comprehensive gene annotation on the primary assembly were counted with htseq-count v1.99.2. To analyze the resulting data set, we restricted our analysis to genes with a high confidence in detection, so genes with exactly zero reads in any condition were excluded, resulting in a dataset of 6199 genes across 48 replicates and conditions. We then normalized reads for each replicate by the total number of reads detected in that replicate and mean-normalized rows to measure fold-changes in responding genes. We performed principal component analysis (PCA) to identify high-scoring genes on either PC1 or PC2, and then identified additional genes that displayed a high degree of variance between conditions relative to the untreated naïve condition. The remaining genes were manually inspected for noise between replicates.
Statistical analysis
Data are expressed as means ± SEM unless otherwise stated in the figure legends. Statistical analyses between groups were calculated by one-way or two-way analysis of variance (ANOVA) with a Tukey-Kramer post test (multiple comparisons) or Student’s unpaired t test with GraphPad Prism 8 software (GraphPad Software). P < 0.05 was considered a statistically significant difference between means.
Supplementary Material
Acknowledgments:
We are grateful to S. Kotenko (Rutgers New Jersey Medical School) for thoughtful discussions throughout the duration of this project, as well as J. Tavernier (Ghent University) for sharing the EPOR-IFNAR1 and EPOR-IFNAR2 expression plasmids, and A. Perantoni (NCI) for the STAT1 expression plasmid. Huh7.5 cells and the pACNR-FLYF-17D-RL plasmid were kindly provided by C. Rice (The Rockefeller University). Within the Department of Molecular Biology, we thank C. DeCoste and K. Rittenbach at the Flow Cytometry Facility, G. Laevsky and S. Wang at the Confocal Imaging Facility, a Nikon Center of Excellence, and A. Biswas for their assistance. We also thank W. Wang at the Genomics Core Facility in the Lewis Sigler Institute for Integrative Genomics. We also thank all members of the Ploss and Toettcher labs for providing feedback on this manuscript.
Funding:
This study was funded by National Institutes of Health (NIH) grants R01AI138797, R01AI107301, R01AI146917, R01AI153236, and R01AI168048 (to A.P.); a Burroughs Wellcome Fund Award for Investigators in Pathogenesis (to A.P.); American Cancer Society Research Scholar Award RSG-15-048-01-MPC (to A.P.); a Vallee Scholars Award (to J.E.T.); NSF CAREER Award 1750663 (to J.E.T.); NIH grant DP2EB04247 (to J.E.T.); NSF CAREER Award 1750663 (to J.E.T.); NIGMS, NIH grant T32GM007388 (to E.V.M.); and Damon Runyon Postdoctoral Fellowship DRG-2432-21 (to A.E.L.). The Princeton University Flow Cytometry Resource Facility is supported, in part, with funding from NCI-CCSG P30CA072720-5921.
Footnotes
Competing interests: B.A. is a member of the scientific advisory board of Arbor Biotechnologies, holds equity in Celsius Therapeutics, and has received research funding from Genmab that is not related to this work. B.A. has filed patent applications related to RNA-seq technologies. J.E.T. is a member of the scientific advisory boards of Nereid Therapeutics and Prolific Machines. The remaining authors declare that they have no competing interests.
Data and materials availability:
The full RNA-seq data set can be accessed at the NCBI Gene Expression Omnibus, accession number GSE218585. All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. Raw data for the figures can be found in data file S1. All materials can be shared through a material transfer agreement.
References and Notes
- 1.McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A, Type I interferons in infectious disease. Nat Rev Immunol. 15 (2015), pp. 87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotenko S. v., Rivera A, Parker D, Durbin JE, Type III IFNs: Beyond antiviral protection. Semin Immunol. 43 (2019), doi: 10.1016/j.smim.2019.101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teijaro JR, Type i interferons in viral control and immune regulation. Curr Opin Virol. 16 (2016), doi: 10.1016/j.coviro.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazear HM, Daniels BP, Pinto AK, Huang AC, Vick SC, Doyle SE, Gale M, Klein RS, Diamond MS, Interferon-λ restricts West Nile virus neuroinvasion by tightening the blood-brain barrier. Sci Transl Med. 7 (2015), doi: 10.1126/scitranslmed.aaa4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fink K, Lang KS, Manjarrez-Orduno N, Junt T, Senn BM, Holdener M, Akira S, Zinkernagel RM, Hengartner H, Early type I interferon-mediated signals on B cells specifically enhance antiviral humoral responses. Eur J Immunol. 36 (2006), doi: 10.1002/eji.200635993. [DOI] [PubMed] [Google Scholar]
- 6.Coro ES, Chang WLW, Baumgarth N, Type I IFN Receptor Signals Directly Stimulate Local B Cells Early following Influenza Virus Infection. The Journal of Immunology. 176 (2006), doi: 10.4049/jimmunol.176.7.4343. [DOI] [PubMed] [Google Scholar]
- 7.Pinto AK, Daffis S, Brien JD, Gainey MD, Yokoyama WM, Sheehan KCF, Murphy KM, Schreiber RD, Diamond MS, A temporal role of type I interferon signaling in CD8 + T cell maturation during acute West Nile virus infection. PLoS Pathog. 7 (2011), doi: 10.1371/journal.ppat.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Douam F, Soto Albrecht YE, Hrebikova G, Sadimin E, Davidson C, Kotenko S. v., Ploss A, Type III interferon-mediated signaling is critical for controlling live attenuated yellow fever virus infection in vivo. mBio. 8 (2017), doi: 10.1128/mBio.00819-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felgenhauer U, Schoen A, Gad HH, Hartmann R, Schaubmar AR, Failing K, Drosten C, Weber F, Inhibition of SARS–CoV-2 by type I and type III interferons. Journal of Biological Chemistry. 295 (2020), doi: 10.1074/jbc.AC120.013788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klinkhammer J, Schnepf D, Ye L, Schwaderlapp M, Gad HH, Hartmann R, Garcin D, Mahlakõiv T, Staeheli P, IFN-λ prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. Elife. 7 (2018), doi: 10.7554/eLife.33354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anggakusuma A. Frentzen E. Gürlevik Q. Yuan, Steinmann E, Ott M, Staeheli P, Schmid-Burgk J, Schmidt T, Hornung V, Kuehnel F, Pietschmann T, Control of Hepatitis C Virus Replication in Mouse Liver-Derived Cells by MAVS-Dependent Production of Type I and Type III Interferons. J Virol. 89 (2015), doi: 10.1128/jvi.03129-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sohn SY, Hearing J, Mugavero JA, Kirillov V, Gorbunova E, Helminiak L, Mishra S, Mackow E, Hearing P, Reich NC, Kim HK, Interferon-Lambda Intranasal Protection and Differential Sex Pathology in a Murine Model of SARS-CoV-2 Infection. mBio. 12 (2021), doi: 10.1128/mBio.02756-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phillips S, Mistry S, Riva A, Cooksley H, Hadzhiolova-Lebeau T, Plavova S, Katzarov K, Simonova M, Zeuzem S, Woffendin C, Chen PJ, Peng CY, Chang TT, Lueth S, de Knegt R, Choi MS, Wedemeyer H, Dao M, Kim CW, Chu HC, Wind-Rotolo M, Williams R, Cooney E, Chokshi S, Peg-interferon lambda treatment induces robust innate and adaptive immunity in chronic hepatitis B patients. Front Immunol. 8 (2017), doi: 10.3389/fimmu.2017.00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flisiak R, Kawazoe S, Znoyko O, Assy N, Gadano A, Kao JH, Lee KS, Zwirtes R, Portsmouth S, Dong Y, Xu D, Kumada H, Srinivasan S, Peginterferon Lambda-1a/ribavirin with daclatasvir or peginterferon alfa-2a/ribavirin with telaprevir for chronic hepatitis c genotype 1b. Journal of Interferon and Cytokine Research. 36 (2016), doi: 10.1089/jir.2015.0173. [DOI] [PubMed] [Google Scholar]
- 15.Reis G, Moreira Silva EAS, Medeiros Silva DC, Thabane L, Campos VHS, Ferreira TS, Santos CVQ, Nogueira AMR, Almeida APFG, Savassi LCM, Figueiredo-Neto AD, Dias ACF, Freire Júnior AM, Bitarães C, Milagres AC, Callegari ED, Simplicio MIC, Ribeiro LB, Oliveira R, Harari O, Wilson LA, Forrest JI, Ruton H, Sprague S, McKay P, Guo CM, Limbrick-Oldfield EH, Kanters S, Guyatt GH, Rayner CR, Kandel C, Biondi MJ, Kozak R, Hansen B, Zahoor MA, Arora P, Hislop C, Choong I, Feld JJ, Mills EJ, Glenn JS, Early Treatment with Pegylated Interferon Lambda for Covid-19. New England Journal of Medicine. 388, 518–528 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haller O, Kochs G, Weber F, The interferon response circuit: Induction and suppression by pathogenic viruses. Virology. 344 (2006), pp. 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Velazquez L, Fellous M, Stark GR, Pellegrini S, A protein tyrosine kinase in the interferon α β signaling pathway. Cell. 70 (1992), doi: 10.1016/0092-8674(92)90105-L. [DOI] [PubMed] [Google Scholar]
- 18.Müller M, Briscoe J, Laxton C, Guschin D, Ziemiecki A, Silvennoinen O, Harpur AG, Barbieri G, Witthuhn BA, Schindler C, Pellegrini S, Wilks AF, Ihle JN, Stark GR, Kerr LM, The protein tyrosine kinase JAK1 complements defects in interferon-α/β and -γ Signal transduction. Nature. 366 (1993), doi: 10.1038/366129a0. [DOI] [PubMed] [Google Scholar]
- 19.Nadeau OW, Domanski P, Usacheva A, Uddin S, Platanias LC, Pitha P, Raz R, Levy D, Majchrzak B, Fish E, Colamonici OR, The Proximal Tyrosines of the Cytoplasmic Domain of the β Chain of the Type I Interferon Receptor Are Essential for Signal Transducer and Activator of Transcription (Stat) 2 Activation. Journal of Biological Chemistry. 274 (1999), doi: 10.1074/jbc.274.7.4045. [DOI] [PubMed] [Google Scholar]
- 20.Mesev E. v., LeDesma RA, Ploss A, Decoding type I and III interferon signalling during viral infection. Nat Microbiol. 4 (2019), pp. 914–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazear HM, Schoggins JW, Diamond MS, Shared and Distinct Functions of Type I and Type III Interferons. Immunity. 50 (2019), pp. 907–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Z, Hamming OJ, Ank N, Paludan SR, Nielsen AL, Hartmann R, Type III Interferon (IFN) Induces a Type I IFN-Like Response in a Restricted Subset of Cells through Signaling Pathways Involving both the Jak-STAT Pathway and the Mitogen-Activated Protein Kinases. J Virol. 81, 7749–7758 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolen CR, Ding S, Robek MD, Kleinstein SH, Dynamic expression profiling of type I and type III interferon-stimulated hepatocytes reveals a stable hierarchy of gene expression. Hepatology. 59 (2014), doi: 10.1002/hep.26657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maher SG, Sheikh F, Scarzello AJ, Romero-Weaver AL, Baker DP, Donnelly RP, Gamero AM, IFN-α and IFN-λ differ in their antiproliferative effects and duration of JAK/STAT signaling activity. Cancer Biol Ther. 7, 1109–1115 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcello T, Grakoui A, Barba–Spaeth G, Machlin ES, v Kotenko S, Macdonald MR, Rice CM, Interferons α and λ Inhibit Hepatitis C Virus Replication With Distinct Signal Transduction and Gene Regulation Kinetics. Gastroenterology. 131, 1887–1898 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Jilg N, Lin W, Hong J, Schaefer EA, Wolski D, Meixong J, Goto K, Brisac C, Chusri P, Fusco DN, Chevaliez S, Luther J, Kumthip K, Urban TJ, Peng LF, Lauer GM, Chung RT, Kinetic differences in the induction of interferon stimulated genes by interferon-α and interleukin 28B are altered by infection with hepatitis C virus. Hepatology. 59 (2014), doi: 10.1002/hep.26653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galani IE, Triantafyllia V, Eleminiadou EE, Koltsida O, Stavropoulos A, Manioudaki M, Thanos D, Doyle SE, S. v. Kotenko, K. Thanopoulou, E. Andreakos, Interferon-λ Mediates Non-redundant Front-Line Antiviral Protection against Influenza Virus Infection without Compromising Host Fitness. Immunity. 46 (2017), doi: 10.1016/j.immuni.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 28.Pervolaraki K, Rastgou Talemi S, Albrecht D, Bormann F, Bamford C, Mendoza JL, Garcia KC, McLauchlan J, Höfer T, Stanifer ML, Boulant S, Differential induction of interferon stimulated genes between type I and type III interferons is independent of interferon receptor abundance. PLoS Pathog. 14 (2018), doi: 10.1371/journal.ppat.1007420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavoie TB, Kalie E, Crisafulli-Cabatu S, Abramovich R, DiGioia G, Moolchan K, Pestka S, Schreiber G, Binding and activity of all human alpha interferon subtypes. Cytokine. 56, 282–289 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Mendoza JL, Schneider WM, Hoffmann H-H, Vercauteren K, Jude KM, Xiong A, Moraga I, Horton TM, Glenn JS, De Jong YP, Rice CM, Christopher Garcia K, The IFN-λ-IFN-λR1-IL-10Rβ Complex Reveals Structural Features Underlying Type III IFN Functional Plasticity. Immunity. 46, 379–392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Renauld JC, Class II cytokine receptors and their ligands: Key antiviral and inflammatory modulators. Nat Rev Immunol. 3 (2003), doi: 10.1038/nri1153. [DOI] [PubMed] [Google Scholar]
- 32.Pattyn E, van Ostade X, Schauvliege L, Verhee A, Kalai M, Vandekerckhove J, Tavernier J, Dimerization of the interferon type I receptor IFNaR2–2 is sufficient for induction of interferon effector genes but not for full antiviral activity. Journal of Biological Chemistry. 274, 34838–34845 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Muthukumaran G, Kotenko S, Donnelly R, Ihle JN, Pestka S, “Chimeric Erythropoietin-Interferon Receptors Reveal Differences in Functional Architecture of Intracellular Domains for Signal Transduction*” (1997), (available at http://www-jbc.stanford.edu/jbc/). [DOI] [PubMed] [Google Scholar]
- 34.Watowich SS, Yoshimura A, Longmore GD, Hilton DJ, Yoshimura Y, Lodish HF, “Homodimerization and constitutive activation of the erythropoietin receptor” (1992). [DOI] [PMC free article] [PubMed]
- 35.Thomas C, Moraga I, Levin D, Krutzik PO, Podoplelova Y, Trejo A, Lee C, Yarden G, Vleck SE, Glenn JS, Nolan GP, Piehler J, Schreiber G, Garcia KC, Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell. 146, 621–632 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Syed RS, Reid SW, Li C, Cheetham JC, Aoki KH, Liu B, Zhan H, Osslund TD, Chirino AJ, Zhang J, Finer-Moore J, Elliott S, Sitney K, Katz BA, Matthews DJ, Wendoloski JJ, Egrie J, Stroud RM, Efficiency of signalling through cytokine receptors depends critically on receptor orientation. Nature. 395, 511–516 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Mukherjee S, Zhang Y, MM-align: a quick algorithm for aligning multiple-chain protein complex structures using iterative dynamic programming. Nucleic Acids Res. 37, e83–e83 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laugsch M, Metzen E, Svensson T, Depping R, Jelkmann W, Lack of functional erythropoietin receptors of cancer cell lines. Int J Cancer. 122 (2008), doi: 10.1002/ijc.23201. [DOI] [PubMed] [Google Scholar]
- 39.Urin V, Shemesh M, Schreiber G, CRISPR/Cas9-based Knockout Strategy Elucidates Components Essential for Type 1 Interferon Signaling in Human HeLa Cells. J Mol Biol. 431 (2019), doi: 10.1016/j.jmb.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 40.Zhang YL, Radhakrishnan ML, Lu X, Gross AW, Tidor B, Lodish HF, Symmetric Signaling by an Asymmetric 1 Erythropoietin: 2 Erythropoietin Receptor Complex. Mol Cell. 33, 266–274 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Köster M, Hauser H, Dynamic redistribution of STAT1 protein in IFN signaling visualized by GFP fusion proteins. Eur J Biochem. 260 (1999), doi: 10.1046/j.1432-1327.1999.00149.x. [DOI] [PubMed] [Google Scholar]
- 42.de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR, Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 69 (2001). [PubMed] [Google Scholar]
- 43.Schoggins JW, Interferon-Stimulated Genes: What Do They All Do? Annu Rev Virol. 6 (2019), doi: 10.1146/annurev-virology-092818-015756. [DOI] [PubMed] [Google Scholar]
- 44.Alpern D, Gardeux V, Russeil J, Mangeat B, Meireles-Filho ACA, Breysse R, Hacker D, Deplancke B, BRB-seq: Ultra-affordable high-throughput transcriptomics enabled by bulk RNA barcoding and sequencing. Genome Biol. 20 (2019), doi: 10.1186/s13059-019-1671-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Babon JJ, Kershaw NJ, Murphy JM, Varghese LN, Laktyushin A, Young SN, Lucet IS, Norton RS, Nicola NA, Suppression of Cytokine Signaling by SOCS3: Characterization of the Mode of Inhibition and the Basis of Its Specificity. Immunity. 36, 239–250 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piganis RAR, De Weerd NA, Gould JA, Schindler CW, Mansell A, Nicholson SE, Hertzog PJ, Suppressor of cytokine signaling (SOCS) 1 inhibits type I interferon (IFN) signaling via the interferon alpha receptor (IFNAR1)-associated tyrosine kinase Tyk2. J Biol Chem. 286, 33811–33818 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Douam F, Ploss A, Yellow Fever Virus: Knowledge Gaps Impeding the Fight Against an Old Foe. Trends Microbiol. 26 (2018), doi: 10.1016/j.tim.2018.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stanifer ML, Pervolaraki K, Boulant S, Differential regulation of type I and type III interferon signaling. Int J Mol Sci. 20 (2019), doi: 10.3390/ijms20061445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Charis Wagner T, Velichko S, Vogel D, Sandhya Rani MR, Leung S, Ransohoff RM, Stark GR, Daniel Perez H, Croze E, Interferon signaling is dependent on specific tyrosines located within the intracellular domain of IFNAR2c: Expression of IFNAR2c tyrosine mutants in U5A cells. Journal of Biological Chemistry. 277 (2002), doi: 10.1074/jbc.M108928200. [DOI] [PubMed] [Google Scholar]
- 50.Nguyen VP, Saleh AZM, Arch AE, Yan H, Piazza F, Kim J, Krolewski JJ, Stat2 binding to the interferon-α receptor 2 subunit is not required for interferon-α signaling. Journal of Biological Chemistry. 277 (2002), doi: 10.1074/jbc.M111161200. [DOI] [PubMed] [Google Scholar]
- 51.Usacheva A, Sandoval R, Domanski P, Kotenko S. v., Nelms K, Goldsmith MA, Colamonici OR, Contribution of the Box 1 and Box 2 motifs of cytokine receptors to Jak1 association and activation. Journal of Biological Chemistry. 277 (2002), doi: 10.1074/jbc.M205757200. [DOI] [PubMed] [Google Scholar]
- 52.Ferrao R, Wallweber HJA, Ho H, Tam C, Franke Y, Quinn J, Lupardus PJ, The Structural Basis for Class II Cytokine Receptor Recognition by JAK1. Structure. 24 (2016), doi: 10.1016/j.str.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gough DJ, Messina NL, Hii L, Gould JA, Sabapathy K, Robertson APS, Trapani JA, Levy DE, Hertzog PJ, Clarke CJP, Johnstone RW, Functional Crosstalk between Type I and II Interferon through the Regulated Expression of STAT1. PLoS Biol. 8, e1000361 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gough DJ, Messina NL, Clarke CJP, Johnstone RW, Levy DE, Constitutive Type I Interferon Modulates Homeostatic Balance through Tonic Signaling. Immunity. 36, 166–174 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murakami M, Narazaki M, Hibi M, Yawata H, Yasukawa K, Hamaguchi M, Taga T, Kishimoto T, Critical cytoplasmic region of the interleukin 6 signal transducer gp130 is conserved in the cytokine receptor family. Proc Natl Acad Sci U S A. 88 (1991), doi: 10.1073/pnas.88.24.11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang D, Wlodawer A, Lubkowski J, Crystal Structure of a Complex of the Intracellular Domain of Interferon λ Receptor 1 (IFNLR1) and the FERM/SH2 Domains of Human JAK1. J Mol Biol. 428 (2016), doi: 10.1016/j.jmb.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Babon JJ, Lucet IS, Murphy JM, Nicola NA, Varghese LN, The molecular regulation of Janus kinase (JAK) activation. Biochemical Journal. 462 (2014), doi: 10.1042/BJ20140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee TS, On the regulation and activation of JAK2: A novel hypothetical model. Molecular Cancer Research. 11 (2013), doi: 10.1158/1541-7786.MCR-12-0555. [DOI] [PubMed] [Google Scholar]
- 59.Brooks AJ, Dai W, O’Mara ML, Abankwa D, Chhabra Y, Pelekanos RA, Gardon O, Tunny KA, Blucher KM, Morton CJ, Parker MW, Sierecki E, Gambin Y, Gomez GA, Alexandrov K, Wilson IA, Doxastakis M, Mark AE, Waters MJ, Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science (1979). 344 (2014), doi: 10.1126/science.1249783. [DOI] [PubMed] [Google Scholar]
- 60.Lupardus PJ, Ultsch M, Wallweber H, Kohli PB, Johnson AR, Eigenbrot C, Structure of the pseudokinase-kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition. Proc Natl Acad Sci U S A. 111 (2014), doi: 10.1073/pnas.1401180111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toms A. v., Deshpande A, McNally R, Jeong Y, Rogers JM, Kim CU, Gruner SM, Ficarro SB, Marto JA, Sattler M, Griffin JD, Eck MJ, Structure of a pseudokinase-domain switch that controls oncogenic activation of Jak kinases. Nat Struct Mol Biol. 20 (2013), doi: 10.1038/nsmb.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lupardus PJ, Skiniotis G, Rice AJ, Thomas C, Fischer S, Walz T, Garcia KC, Structural snapshots of full-length Jak1, a transmembrane gp130/IL-6/IL-6Rα cytokine receptor complex, and the receptor-Jak1 holocomplex. Structure. 19 (2011), doi: 10.1016/j.str.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Glassman CR, Tsutsumi N, Saxton RA, Lupardus PJ, Jude KM, Christopher Garcia K, Structure of a Janus kinase cytokine receptor complex reveals the basis for dimeric activation. Science (1979). 376 (2022), doi: 10.1126/science.abn8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sharma N, Longjam G, Schreiber G, Type I interferon signaling is decoupled from specific receptor orientation through lenient requirements of the transmembrane domain. Journal of Biological Chemistry. 291 (2016), doi: 10.1074/jbc.M115.686071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dumoutier L, Tounsi A, Michiels T, Sommereyns C, Kotenko S. v., Renauld JC, Role of the interleukin (IL)-28 receptor tyrosine residues for antiviral and antiproliferative activity of IL-29/interferon-λ1. Similarities with type 1 interferon signaling. Journal of Biological Chemistry. 279 (2004), doi: 10.1074/jbc.M404789200. [DOI] [PubMed] [Google Scholar]
- 66.Shemesh M, Lochte S, Piehler J, Schreiber G, IFNAR1 and IFNAR2 play distinct roles in initiating type I interferon-induced JAK-STAT signaling and activating STATs. Sci Signal. 14 (2021), doi: 10.1126/scisignal.abe4627. [DOI] [PubMed] [Google Scholar]
- 67.Freed DM, Bessman NJ, Kiyatkin A, Salazar-Cavazos E, Byrne PO, Moore JO, Valley CC, Ferguson KM, Leahy DJ, Lidke DS, Lemmon MA, EGFR Ligands Differentially Stabilize Receptor Dimers to Specify Signaling Kinetics. Cell. 171, 683–695.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tischer DK, Weiner OD, Light-based tuning of ligand half-life supports kinetic proofreading model of T cell signaling. Elife. 8 (2019), doi: 10.7554/eLife.42498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ohashi H, Maruyama K, Liu YC, Yoshimura A, Ligand-induced activation of chimeric receptors between the erythropoietin receptor and receptor tyrosine kinases. Proc Natl Acad Sci U S A. 91 (1994), doi: 10.1073/pnas.91.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hansen G, Hercus TR, McClure BJ, Stomski FC, Dottore M, Powell J, Ramshaw H, Woodcock JM, Xu Y, Guthridge M, McKinstry WJ, Lopez AF, Parker MW, The Structure of the GM-CSF Receptor Complex Reveals a Distinct Mode of Cytokine Receptor Activation. Cell. 134 (2008), doi: 10.1016/j.cell.2008.05.053. [DOI] [PubMed] [Google Scholar]
- 71.Boulanger MJ, Chow D, Brevnova EE, Christopher Garcia K, Hexameric Structure and Assembly of the Interleukin-6/ IL-6-Receptor/gp130 Complex. Mol. Cell. Biol. 7 (1993). [Google Scholar]
- 72.Fuchs S, Kaiser-Labusch P, Bank J, Ammann S, Kolb-Kokocinski A, Edelbusch C, Omran H, Ehl S, Tyrosine kinase 2 is not limiting human antiviral type III interferon responses. Eur J Immunol. 46, 2639–2649 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Sommereyns C, Paul S, Staeheli P, Michiels T, IFN-lambda (IFN-λ) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 4 (2008), doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jozefczuk J, Drews K, Adjaye J, Preparation of Mouse Embryonic Fibroblast Cells Suitable for Culturing Human Embryonic and Induced Pluripotent Stem Cells. JoVE (Journal of Visualized Experiments), e3854 (2012). [DOI] [PMC free article] [PubMed]
- 75.Kassar TC, Magalhães T, Júnior JVJS, Carvalho AGO, da Silva ANMR, Queiroz SRA, Bertani GR, Gil LHVG, Construction and characterization of a recombinant yellow fever virus stably expressing Gaussia luciferase. An Acad Bras Cienc. 89 (2017), doi: 10.1590/0001-3765201720160196. [DOI] [PubMed] [Google Scholar]
- 76.Tinevez J-Y, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW, TrackMate: An open and extensible platform for single-particle tracking. Methods. 115 (2017), doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 77.Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, Lai MM, Shishkin AA, Bhat P, Takei Y, Trinh V, Aznauryan E, Russell P, Cheng C, Jovanovic M, Chow A, Cai L, McDonel P, Garber M, Guttman M, Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell. 174 (2018), doi: 10.1016/j.cell.2018.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full RNA-seq data set can be accessed at the NCBI Gene Expression Omnibus, accession number GSE218585. All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. Raw data for the figures can be found in data file S1. All materials can be shared through a material transfer agreement.