Abstract
Mammalian reovirus virions undergo partial disassembly of the outer capsid upon exposure to proteases in vitro, producing infectious subvirion particles (ISVPs) that lack protein ς3 and contain protein μ1/μ1C as endoprotease-generated fragments μ1δ/δ and φ. ISVPs are thought to be required for two early steps in reovirus infection: membrane penetration and activation of the particle-bound viral transcriptase complexes. Genetic and biochemical evidence implicates outer-capsid protein μ1 in both these steps. To determine whether the cleavage of μ1/μ1C is relevant to the unique properties of ISVPs, we analyzed the properties of novel subvirion particles that lacked ς3 yet retained μ1/μ1C in an uncleaved but cleavable form. These detergent-plus-protease subvirion particles (dpSVPs) were produced by treating virions with chymotrypsin in the presence of micelle-forming concentrations of alkyl sulfate detergents. Infections with dpSVPs in murine L or canine MDCK cells provided evidence that the cleavage of μ1/μ1C during viral entry into these cells is dispensable for reovirus infection. Additionally, dpSVPs behaved like ISVPs in their capacity to permeabilize lipid bilayers and to undergo transcriptase activation in vitro, supporting the conclusion that cleavage of μ1/μ1C to μ1δ/δ and φ during viral entry is not required for either membrane penetration or transcriptase activation in cells. The capacity of alkyl sulfate detergents to inhibit the cleavage of μ1/μ1C in a reversible fashion suggests a specific association between virus particle and detergent micelles that may mimic virus particle-phospholipid membrane interactions during reovirus entry into cells.
All viruses must cross a membrane barrier to gain access to the cytoplasm of their host cells. In enveloped animal viruses, entry is achieved by a viral surface protein(s) that mediates fusion between viral and cellular membranes (27). Viruses that lack a lipid envelope, however, cannot use membrane fusion as a generalized mechanism to penetrate host membranes. Instead, entry by nonenveloped viruses is conceived to occur via either the local disruption of a host membrane (3) or the formation of a membrane-spanning pore (48), as mediated by a viral surface protein analogous to the fusion proteins of enveloped viruses.
A common theme that has emerged from studies of the fusion machinery of different enveloped viruses (e.g., togaviruses [36], orthomyxoviruses [32], and paramyxoviruses [49]) is the requirement for posttranslational proteolytic processing of fusion and/or accessory proteins prior to membrane penetration. In many instances, these cleavages prime the fusion proteins for membrane interaction by removing constraints on conformational changes needed for membrane insertion or by generating a new protein terminus that is hydrophobic and can insert into the membrane as a result of these conformational changes. Several groups of nonenveloped viruses, including the rotaviruses (2, 11, 31), adenoviruses (23), picornaviruses (35), and reoviruses (20, 30, 55, 59), also require proteolytic processing for efficient viral entry. To expand current knowledge of the requirement for proteolysis in membrane penetration by nonenveloped viruses, we performed studies to identify which proteolytic events make mammalian reoviruses competent for entry.
The maximally stable infectious form of reovirus is the virion, composed of eight proteins ranging from 12 to 600 in copy number (see references 28 and 45 for reviews). These eight structural proteins are organized into two concentric icosahedral capsids (19). The inner capsid encloses the viral genome, which consists of 10 double-stranded RNA segments. The structural framework of the outer capsid is formed by proteins μ1 (600 copies primarily as cleaved fragments μ1N and μ1C, but also some uncleaved μ1) and λ2 (60 copies). Several lines of evidence indicate that μ1 and its fragments effect the membrane penetration step during infection (24–26, 37, 41, 43, 44, 57). λ2 forms pentameric spikes at the fivefold axes of the outer capsid (19, 61) and is involved in the capping of mRNAs extruded through these spikes in the transcribing particle (12, 50). The other two outer-capsid proteins, ς1 (36 to 48 copies) and ς3 (600 copies), decorate the primary lattice formed by μ1 and λ2. ς1 is found at the fivefold axes and mediates viral attachment to host cell receptors preceding endocytic uptake of the virus (34). ς3 engages in close interactions with the underlying μ1 molecules, suggesting that its primary role may be to stabilize virions (16, 17), probably by regulating the exposure and conformational status of μ1 (19, 37, 43).
When virions are treated in vitro with exogenous proteases (such as chymotrypsin [CHT]), stepwise disassembly of the outer capsid is observed (5, 29, 51). ς3 is degraded, and μ1/μ1C is endoproteolytically cleaved to yield two particle-bound fragments (N-terminal μ1δ/δ and C-terminal φ [41]), producing infectious subvirion particles (ISVPs) (5, 29, 51). The cleavage of μ1/μ1C is initiated shortly after ς3 degradation commences (4, 7, 41). Although both virions and ISVPs are infectious, only ISVPs can lead to interactions with lipid bilayers in vitro (25, 26, 37, 43, 57). This fact, along with observations that ISVP-like particles are generated from infecting virions at early times postinfection (7, 10, 52, 53, 55), has been taken as evidence that the ISVP is a necessary intermediate in reovirus infection. Indeed, the processing of virion proteins by one or more lysosomal cysteine proteases appears to be required for the infectivity of virions, but not ISVPs, in murine L-cell fibroblasts (1, 30). These results suggest that ς3 plays a key regulatory role in infection by reovirus virions: it stabilizes the outer capsid until the particle enters a suitable hydrolytic compartment along the endosomal-lysosomal pathway, where ς3 is degraded, and μ1/μ1C is cleaved to μ1δ/δ and φ (43, 55). Membrane penetration, likely mediated by the cleavage products of μ1 (24–26, 37, 41, 43, 44, 57), can then ensue.
While it seems nearly certain that ς3 removal from reovirus virions is an obligatory step in entry into cells, the importance of the μ1/μ1C cleavage is less clear because it has not been experimentally separable from ς3 degradation (4, 7, 41). μ1, like the fusion proteins of enveloped viruses, probably undergoes structural rearrangements in order to interact with its target membrane during penetration (9, 39). It thus seems reasonable to hypothesize that the cleavage of μ1/μ1C into fragments μ1δ/δ and φ is analogous to maturational cleavages that activate the fusion proteins of many enveloped viruses for membrane interaction, such as the cleavage of influenza virus hemagglutinin HA0 into fragments HA1 and HA2 (32). One possibility is that the cleavage of μ1/μ1C provides protein sequences on either side of the cleavage site with the conformational mobility needed to insert into the membrane bilayer during membrane disruption (41). In this report, we describe an investigation of the role played by the μ1/μ1C cleavage in reovirus infection.
MATERIALS AND METHODS
Reagents.
All enzymes and chemicals were from Sigma Chemical Co. (St. Louis, Mo.) unless otherwise stated. Highly pure stocks of alkyl sulfate detergents were kindly provided by R. R. Rueckert (University of Madison—Wisconsin). Additional stocks of the alkyl sulfates sodium tetradecyl sulfate (14 carbons [14SO4]; Aldrich, Milwaukee, Wis.), sodium dodecyl sulfate (SDS) (12 carbons [12SO4], and sodium octyl sulfate (8 carbons [8SO4]) were also tested. Detergents from both sources gave identical results. Nucleotides were obtained from Pharmacia (Piscataway, N.J.), and [α-32P]GTP was obtained from Dupont NEN (Wilmington, Del.). RNasin was obtained from Promega (Madison, Wis.).
Cells and viruses.
Spinner-adapted murine L cells were grown in suspension in Joklik’s modified minimal essential medium (Irvine Scientific, Irvine, Calif.) supplemented to contain 2% fetal bovine serum, 2% neonatal bovine serum (HyClone Laboratories, Logan, Utah), 2 mM glutamine, and penicillin (1 U/ml)-streptomycin (1 μg/ml) (Irvine Scientific). Plaque assays to determine infectious titers were performed as described previously (22).
Preparation of purified virions.
Purified virions of strain type 1 Lang (T1L) were obtained as described previously (41). Virion buffer contains 150 mM NaCl, 10 mM MgCl2, and 10 mM Tris (pH 7.5). Particle concentrations of purified virion preparations were measured as described previously (54). To generate purified virions containing [35S]methionine/cysteine-labeled proteins, Tran35S-label (12.5 μCi/ml; ICN Biochemicals, Costa Mesa, Calif.) was added to the virus-cell suspension at the initiation of infection. Specific activities of the labeled virions were ≈2 × 106 particles/cpm for Fig. 6B and 106 particles/cpm for all other experiments.
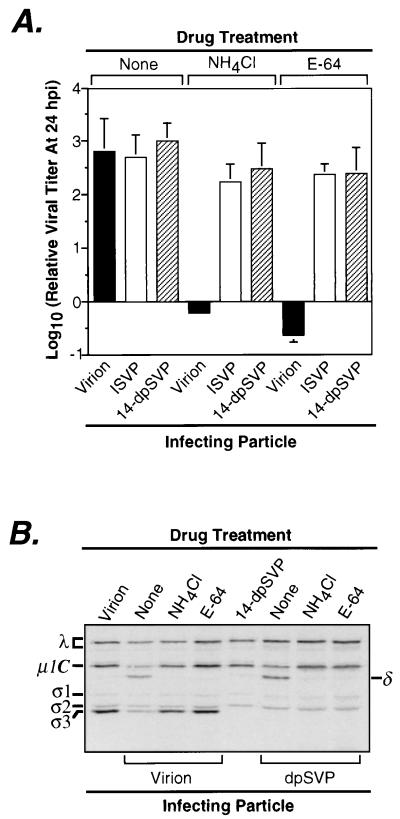
FIG. 6.
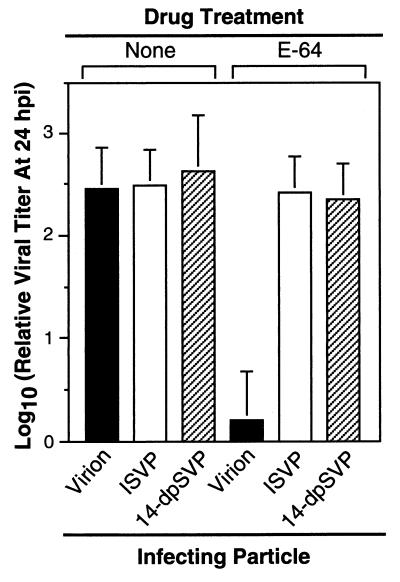
Infectivity of 14-dpSVPs in L cells in the absence and presence of NH4Cl or E-64. (A) T1L virions, ISVPs, or 14-dpSVPs were used to infect L-cell monolayers at an MOI of 3 PFU/cell in the absence and presence of NH4Cl (20 mM) or E-64 (300 μM) in the cell growth medium. Samples were harvested at 24 h postinfection (hpi), and infectious titers were measured by plaque assay. Each bar is the mean log10 PFU/milliliter derived from three independent experiments. (B) Intracellular proteolysis of reovirus particles during infection in L cells. L-cell monolayers were infected with purified 35S-labeled T1L virions or 14-dpSVPs at an MOI of 8 × 104 particles/cell, with NH4Cl (20 mM) or E-64 (300 μM) absent or present in the cell growth medium, and incubated at 37°C for 4 h. Proteins present in cytoplasmic extracts prepared from these samples were resolved by SDS-PAGE, and viral proteins were visualized by phosphorimaging. Reference lanes loaded with untreated virions and 14-dpSVPs are also shown.
ISVPs and dpSVPs.
ISVPs were prepared by digestion of virions with CHT as described previously (41). Detergent-plus-protease subvirion particles (dpSVPs) were also generated as described above except that detergents were included in the CHT digestion reactions. Inclusion of 2 mM SDS (12SO4) or 1 mM 14SO4 resulted in virus particles designated 12-dpSVPs and 14-dpSVPs, respectively. Terminated reactions were often prepared directly for SDS-polyacrylamide gel electrophoresis (PAGE). Alternatively, reactions were diluted into phosphate-buffered saline (PBS) at room temperature and analyzed for infectious titers by plaque assay.
dpSVPs were purified from reaction mixtures as follows. CHT-detergent treatment mixtures originally containing 5 × 1012 T1L virions/ml were treated with phenylmethylsulfonyl fluoride, incubated at 4°C for 20 min, and centrifuged at 16,000 × g for 2 min to remove precipitated detergent. Supernatants were loaded atop 11-ml step gradients containing the following layers (from the bottom up): CsCl at 1.50 g/cm3 (3 ml), CsCl at 1.30 g/cm3 (2 ml), 20% (wt/vol) sucrose (3 ml), and 5% sucrose (3 ml). After centrifugation in a Beckman SW41 rotor at 25,000 rpm and 5°C for 2 h, dpSVPs could be recovered as an optically homogeneous band near the junction of the two CsCl layers. Particles were dialyzed into virion buffer and stored at 4°C. Particle concentrations were measured in the manner described previously for measuring ISVP concentrations (54).
Measurement of CMCs.
To determine detergent critical micellar concentrations (CMCs) for different detergents in virion buffer at 37°C, we used the dye solubilization method of Vuillez-Le Normand and Eiselé (60). Assays were performed as described previously (60) except that (i) the dye was dried in microtubes by lyophilization and (ii) 200 μl of detergent was added to each tube to solubilize the dye. Solubilization was allowed to proceed for 16 to 24 h before 150-μl aliquots of each dilution were removed to a microplate and used to measure A595 in a microplate reader (Bio-Rad, Hercules, Calif.).
SDS-PAGE.
Samples were prepared for SDS-PAGE as described previously (41). SDS-PAGE was carried out on 10% acrylamide gels, and proteins were visualized by staining with Coomassie brilliant blue. Gels loaded with radiolabeled proteins were dried onto filter paper and visualized with the PhosphorImager system (Molecular Dynamics, Sunnyvale, Calif.).
Single-step growth curves.
T1L virions, ISVPs, or 14-dpSVPs were allowed to attach to L cells (2 × 107 cells/ml) suspended in growth medium at a multiplicity of infection (MOI) of 3 PFU/cell. Attachment was carried out with periodic mixing at 4°C for 1 h. The cell-virus suspension was sedimented at 400 × g at 4°C, and the cell pellet was washed with ice-cold PBS to remove unbound virus. Cells were resuspended in growth medium and dispensed into 2-dram (7.4-ml) glass vials at a density of 4 × 105 cells/vial. Vials were transferred to a 37°C incubator, and time points were taken by freezing individual vials at specified times. Duplicate vials were set up for each time point. Vials were subjected to freeze-thawing to generate cell lysates that were analyzed for infectious titers by plaque assay. Growth curves of reovirus particles in the presence of NH4Cl were generated in the same fashion except that 20 mM NH4Cl was included in the growth medium.
Endpoint experiments for infectivity.
Growth of T1L virions, ISVPs, and 14-dpSVPs over a 24-h period in L cells was monitored in the absence and presence of inhibitors of virion infectivity. Experiments were set up as described for single-step growth curves, but only 0- and 24-h time points were collected for each particle type and drug treatment. NH4Cl (20 mM in virion buffer), and trans-epoxysuccinyl-l-leucylamido-(4-guanidino)butane (E-64; 300 μM in dimethyl sulfoxide) were the inhibitors used. Cells were incubated with 300 μM E-64 at 37°C for 1 h prior to virus attachment to maximize the inhibitory effect of this agent (29). Dimethyl sulfoxide alone did not significantly inhibit viral growth at the concentration (1%, vol/vol) used in these experiments (data not shown).
Determination of the extent of proteolysis of T1L virus particles within L cells.
L-cell monolayers in 60-mm-diameter dishes were incubated at 4°C for 20 min, washed with ice-cold PBS, and then infected with purified [35S]methionine/cysteine-labeled virions or 14-dpSVPs at an MOI of 8 × 104 particles/cell. Virus attachment to cells was allowed to proceed for 2 h, after which unbound inoculum was removed by washing with PBS. Cells were overlaid with growth medium containing no drug, 20 mM NH4Cl, or 300 μM E-64 and incubated at 37°C for 4 h. A mock sample (no drug, 0-h incubation at 37°C) was also included for each particle type. At 0 or 4 h postinfection, cells were chilled, scraped into ice-cold lysis buffer (0.5% [vol/vol] Triton X-100, 140 mM NaCl, 10 mM Tris [pH 7.4]), and incubated on ice for 30 min. The cell lysates were then centrifuged at 940 × g for 10 min, and the resulting postnuclear supernatants were diluted into 9 volumes of ice-cold acetone. Precipitated protein was sedimented at 9,000 × g for 15 min, supernatants were decanted, and pellets were dried overnight under reduced pressure. The lyophilized protein was suspended in sample buffer and boiled briefly. Equal counts of radioactivity per lane were loaded onto a 10% acrylamide gel for SDS-PAGE.
Hemolysis experiments.
The capacity of T1L virions, ISVPs, 14-dpSVPs, and cores to lyse erythrocytes (RBCs) was determined. Citrated bovine calf RBCs (Colorado Serum Co., Denver, Colo.) were washed with ice-cold PBS and suspended in PBS at a stock concentration of 30% just prior to use. Hemolysis reactions contained Tris-Cl (10 mM, pH 7.5), NaCl or CsCl (200 mM), RBCs (3%), and either purified virus particles (3 × 1012 particles/ml) or Triton X-100 (1%) in a total volume of 30 μl. Reactions were initiated by transfer to 37°C and terminated by removal onto ice after 40 min. Samples were centrifuged at 300 × g for 5 min, and 15 μl of the supernatant was diluted into 185 μl of virion buffer in a microplate (Costar, Cambridge, Mass.). The extent of hemoglobin release from RBCs was determined by measuring A415 with a microplate reader and expressed as a percentage (taking hemolysis induced by Triton X-100 to be 100%).
Transcriptase activation experiments.
The transcriptase activity of T1L virions, ISVPs, 14-dpSVPs, and cores in the presence of NaCl or CsCl was measured in vitro as described previously (38) except that reaction mixtures included NaCl or CsCl (200 mM) and purified virus particles (3 × 1012 particles/ml) in a total volume of 30 μl. Reactions were initiated by transfer to 37°C and terminated by removal onto ice after 1 h. Each reaction volume was spotted onto a 3-mm-diameter cellulose paper circle (Whatman, Maidstone, England), and 32P-labeled transcripts were detected by trichloroacetic acid precipitation onto the filter followed by liquid scintillation counting (38).
RESULTS
SDS inhibits cleavage of μ1/μ1C by CHT.
When virions of reovirus strain T1L were treated with CHT under defined conditions in vitro, the particles underwent processing to generate ISVPs (Fig. 1A), in that the ς3 protein was rapidly degraded into small peptides and the μ1/μ1C protein was cleaved in a more limited fashion to yield the stable fragments μ1δ/δ (63/59 kDa; N-terminal portions of μ1/μ1C) and φ (13 kDa; C-terminal portion of μ1/μ1C, not resolved from the dye front in Fig. 1A but seen in other gels) (41). As expected, the T1L ς1 protein and proteins associated with the viral core were not cleaved.
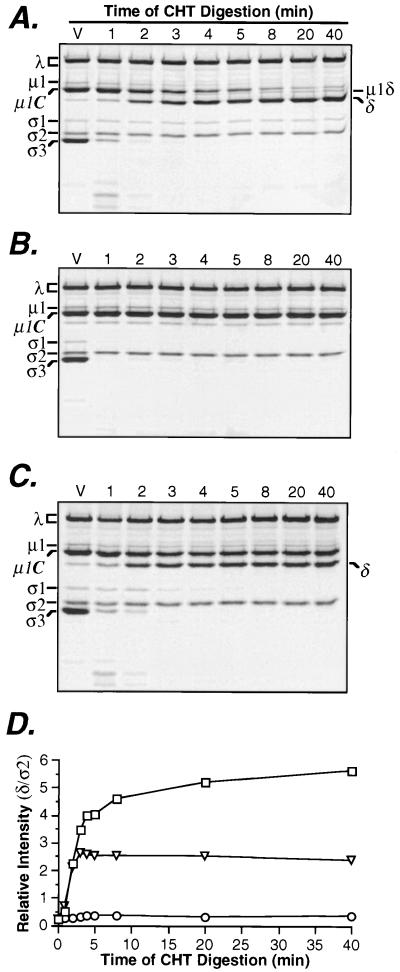
FIG. 1.
Effect of SDS on cleavage of μ1/μ1C protein by CHT. (A) Purified 35S-labeled T1L virions (5 × 1012 particles/ml) were treated with CHT at 37°C for specified times. Viral proteins were resolved by SDS-PAGE and visualized by phosphorimaging to monitor cleavages of proteins ς3 and μ1/μ1C. (B) The experiment was performed exactly as for panel A except that SDS (2 mM) was included in the CHT digestion reactions. (C) To illustrate the rapid inhibitory action of SDS on μ1/μ1C cleavage, SDS (2 mM) was added to a single CHT digest of virions, 2 min after initiation of the reaction. Time points from 3 to 40 min were collected from this single digest, whereas 1- and 2-min time points were obtained from separate digests. (D) The gels in panels A to C were captured as bitmap files, and protein bands corresponding to δ and ς2 were quantitated for each lane, using the ImageQuant program (Molecular Dynamics). The band intensity of δ was divided by the corresponding intensity of ς2 in the same lane, to correct for variations in gel loading. The relative band intensity of δ band is plotted against time of CHT digestion for gels in panels A (□), B (○), and (▿). In panels A to C, a reference lane was loaded with untreated virions (V).
Different results were obtained when the treatment was performed in the presence of 2 mM SDS (Fig. 1B). In that case, degradation of ς3 was accelerated, as indicated by the failure to detect intact ς3 or ς3-containing peptides after 1 min of CHT digestion. In addition, the T1L ς1 protein was made newly sensitive to CHT cleavage by the presence of SDS. In contrast to the SDS-enhanced degradation of ς3 and ς1, the CHT-mediated cleavage of μ1/μ1C was blocked by SDS. Even after 60 min of treatment, the amount of uncleaved μ1/μ1C remained unchanged and the amount of δ fragment was no greater than the small amount normally found in purified virions. The reovirus core proteins remained resistant to CHT cleavage in the presence of SDS (Fig. 1B). Similar results were obtained for strains type 2 Jones, type 3 Dearing, and type 3 clone 9 (data not shown) except that the ς1 protein of type 3 Dearing was sensitive to CHT cleavage in the absence of SDS, as previously reported (40). Thus, the capacity of SDS to block μ1/μ1C cleavage by CHT during the generation of ISVPs is common to strains representing the three known serotypes of mammalian reoviruses.
Inhibition of μ1/μ1C cleavage is not explained by a gradual reduction in CHT activity.
The observation that 2 mM SDS enhanced cleavage of ς3 and ς1 (Fig. 1B) suggested that its capacity to block cleavage of μ1/μ1C could not be explained by a general inhibition of CHT activity. Nonetheless, since μ1/μ1C cleavage begins later in the time course and proceeds at a slower pace (Fig. 1A), it was possible that its inhibition by SDS reflected a gradual reduction in CHT activity in the presence of detergent. To address this possibility, we performed experiments in which CHT treatment of T1L virions was begun without SDS. SDS (2 mM) was then added to the reaction after 2 min of digestion, when ς3 had been degraded into small peptides but only about 40% of μ1/μ1C had been cleaved. In this case, it was observed that cleavage of μ1/μ1C ceased immediately upon addition of SDS, at the same time that ς1 was made newly sensitive to degradation (Fig. 1C and D). These findings provide evidence that the inhibition of μ1/μ1C cleavage by SDS is not a consequence of gradual reduction in CHT activity.
Micelles of alkyl sulfate detergents are needed for inhibiting μ1/μ1C cleavage by CHT.
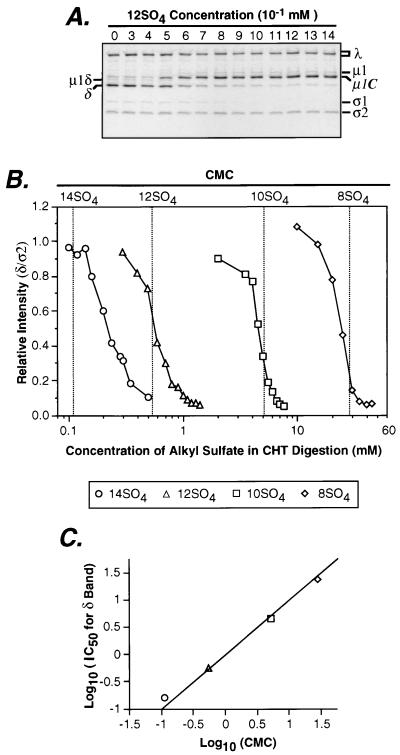
Reasoning that chemical relatives of SDS might also inhibit μ1/μ1C cleavage, we examined the capacities of alkyl sulfate detergents with different carbon chain lengths to block μ1/μ1C cleavage. Like SDS (12SO4), 14SO4, decyl sulfate (10 carbons [10SO4]), and 8SO4 were found to block μ1/μ1C cleavage by CHT without slowing the cleavage of ς3 (data not shown). However, dose-response curves indicated that the inhibition of cleavage titrated at a different concentration range for each alkyl sulfate. Specifically, the concentrations of alkyl sulfate required for half-maximal inhibition of the μ1/μ1C cleavage (IC50) were found to be 0.16 (±0.03 [standard deviation {SD}]) mM for 14SO4, 0.58 (±0.03) mM for 12SO4, 4.5 (±0.03) mM for 10SO4, and 24 (±0.9) mM for 8SO4 (Fig. 2B). Each of these concentrations approximated the CMC of the respective detergent (54) at conditions similar to those used for CHT treatment, suggesting that detergent micelles are required for blocking μ1/μ1C cleavage.
FIG. 2.
Concentrations of alkyl sulfate detergents required to inhibit μ1/μ1C cleavage by CHT. (A) Purified 35S-labeled reovirus T1L virions (5 × 1011 particles/ml) were digested with CHT at 37°C for 20 min in the presence of different concentrations of 12SO4. Viral proteins were resolved by SDS-PAGE, and visualized by phosphorimaging. CHT digestion reactions of virions in the presence of 8SO4, 10SO4, or 14SO4 were performed in a similar fashion (data not shown). (B) From the preceding gels, the intensity of the δ band divided by that of the ς2 band was calculated for each concentration of detergent and used as a measure of the extent of μ1/μ1C cleavage. δ/ς2 ratios were normalized to unity and plotted against detergent concentration. Representative detergent dose-response curves are shown for 8SO4, 10SO4, 12SO4, and 14SO4. CMC values independently measured for each of these detergents in virion buffer at 37°C are displayed as dotted vertical lines. (C) The dose-response curves in panel B were generated in triplicate, and each curve was separately fit to a logistic equation by using SigmaPlot software (SPSS, Chicago, Ill.) to calculate the IC50. The average of the log10 IC50 is plotted against the average of the log10 CMC for each detergent. Error bars for both x and y axes are shown but are embedded within the markers. The line corresponding to exact correlation between detergent CMC and IC50 is shown.
To confirm the involvement of detergent micelles in the inhibition of μ1/μ1C cleavage, we measured the CMC of each detergent at the same conditions used for CHT treatment (in virion buffer at 37°C) (58). The values were determined to be 0.11 (±0.02) mM for 14SO4, 0.54 (±0.03) mM for 12SO4, 5.1 (±0.1) mM for 10SO4, and 28 (±4) mM for 8SO4 (Fig. 2B), which are very similar to the IC50s for inhibition of μ1/μ1C cleavage by each detergent (Fig. 2B and C). These findings strongly suggest that the alkyl sulfates must form micelles before blocking the cleavage of μ1/μ1C at the δ-φ junction. The results also indicate that, within the tested limits, the length of its carbon chain does not affect the capacity of an alkyl sulfate to block μ1/μ1C cleavage, except insofar as chain length affects CMC. The same concentrations of 12SO4, 10SO4, and 8SO4 that blocked μ1/μ1C cleavage also made the T1L ς1 protein sensitive to CHT cleavage. Thus, micelles of these detergents appear to be required for enhancing ς1 cleavage as well. 14SO4 was the only alkyl sulfate tested that did not render the T1L ς1 protein sensitive to CHT cleavage (see below).
Purification of dpSVPs.
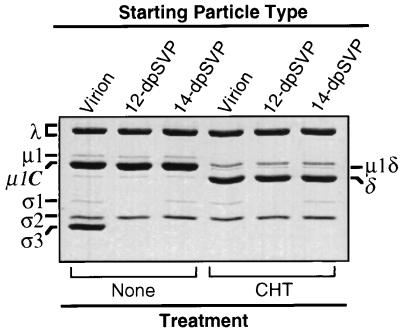
To characterize the particles generated by detergent and CHT treatments (dpSVPs), we isolated them by centrifugation through sucrose-CsCl gradients. We then examined the protein contents of purified dpSVPs by SDS-PAGE and found them to be as expected from previous analyses of raw digests. Thus, 12-dpSVPs contained degraded ς3 and ς1 but uncleaved μ1/μ1C, while 14-dpSVPs contained degraded ς3 but uncleaved ς1 and μ1/μ1C (Fig. 3). When these particles were used as substrates for a second round of CHT treatment, the μ1/μ1C protein was cleaved to yield the fragments μ1δ/δ (Fig. 3) and φ (data not shown), providing evidence that each detergent was removed from the particles during purification and that the inhibition of μ1/μ1C cleavage by each detergent is reversible.
FIG. 3.
In vitro treatment of purified dpSVPs with CHT. Purified aliquots of T1L virions, 12-dpSVPs, and 14-dpSVPs at 5 × 1012 particles/ml were left untreated or digested with CHT for 10 min at 37°C. Viral proteins were resolved by SDS-PAGE and visualized by Coomassie brilliant blue staining.
Infectivity of dpSVPs.
To determine whether dpSVPs might be useful for studies of reovirus entry into cells, we measured the infectivities of different purified preparations of virions and their CHT-generated dpSVPs. Comparing the plaque titer with the particle concentration of each purified preparation yielded a ratio for the number of particles per PFU of each (Table 1). In summary, these experiments showed that the purified 14-dpSVPs retained full infectivity relative to virions. In contrast, the infectivity of purified 12-dpSVPs was greatly reduced. The loss of infectivity observed upon CHT and 12SO4 treatment correlated with the cleavage of ς1.
TABLE 1.
Infectivities of purified dpSVPs
| Particle type | Prepn no. | P/PFUa | Relative P/PFUb |
|---|---|---|---|
| Virion | 1 | 140 | 1.0 |
| 2 | 250 | 1.0 | |
| 14-dpSVP | 1 | 200 | 1.2 |
| 2 | 200 | 1.2 | |
| 3 | 110 | 1.2 | |
| 12-dpSVP | 1 | 4.0 × 105 | 3.4 × 10−4 |
| 2 | 3.7 × 105 | 3.7 × 10−4 | |
| 3 | 3.2 × 105 | 4.2 × 10−4 |
Particle (P)/PFU values for each virus were obtained by dividing P/milliliter by PFU/milliliter and are presented as averages for each particle type (n = 3). Each replicate measurement was derived from a different preparation of particles. Particle concentrations were determined by measuring A260 and multiplying by the appropriate conversion factor (54). Viral infectivities were measured by plaque assay (22).
Defined as the ratio of P/PFU for a preparation of virions to P/PFU of a given particle type (virion or dpSVP) derived from that preparation of virions.
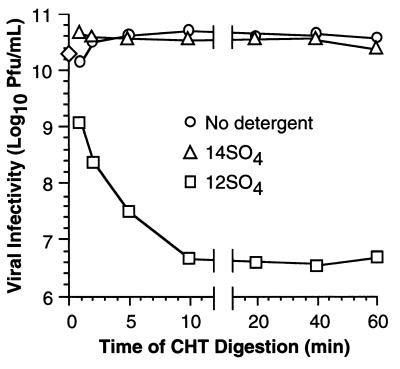
We also measured the infectivities of dpSVPs directly from raw digests. T1L virions in the presence of no detergent, 2 mM 12SO4, or 1 mM 14SO4 were incubated with CHT over a time course, and aliquots were removed at intervals for determining infectivity (Fig. 4). To confirm that the protein contents of the particles were as expected, namely, ISVPs or dpSVPs according to the digestion conditions, samples from each series were also analyzed by SDS-PAGE (data not shown). T1L ISVPs and 14-dpSVPs exhibited little change in infectivity over the time course (Fig. 4). In contrast, 12-dpSVPs rapidly lost infectivity, dropping to about 0.001 times the starting titer by 10 min. As noted above, the maintenance of infectivity in 14SO4 and its loss in 12SO4 correlated with the sensitivity of protein ς1 to cleavage by CHT (Fig. 1B and C). Since treatments with 14SO4 and CHT had no adverse effects on infectivity, we used 14-dpSVPs for further studies of reovirus entry.
FIG. 4.
Infectivity of dpSVPs in raw digests measured as a function of time. Purified T1L virions (5 × 1012 particles/ml) were incubated with CHT (○), CHT plus 14SO4 (1 mM) (▵), or CHT plus 12SO4 (2 mM) (□) at 37°C over a time course. Reactions were terminated at specified times, and infectious titers were determined by plaque assay. Each point represents an average of two independent log10 PFU/milliliter determinations (SD < 0.30 log10 PFU/ml). The infectious titer of untreated virions was obtained from three independent determinations and is depicted as the mean log10 PFU/milliliter value at 0 min (◊).
Growth curves with 14-dpSVPs.
ISVPs differ from virions in several biochemical characteristics, including the absence of ς3, the cleavage of μ1/μ1C into fragments μ1δ/δ and φ, and an apparent change in ς1 protein conformation (19). In addition, ISVPs differ from virions in several properties relating to entry into cells, which must be attributable to one or more of these biochemical differences (reviewed in reference 42). One such difference is that ISVPs exhibit a shortened lag phase in their single-cycle growth curve relative to virions (5, 13, 55). A likely basis for this difference is the requirement for ς3 to be removed from virions by lysosomal proteases (30, 55, 59). ISVPs, having already lost ς3 in vitro, can bypass this proteolytic step and presumably gain access to the cytoplasm of the host cell earlier than virions. In addition to ς3 degradation, virions also undergo μ1/μ1C cleavage at early times of infection, whereas this cleavage has already been effected in vitro during generation of ISVPs (41). A requirement for μ1/μ1C cleavage during viral entry may also contribute to virions having a longer lag phase in their growth curve than ISVPs.
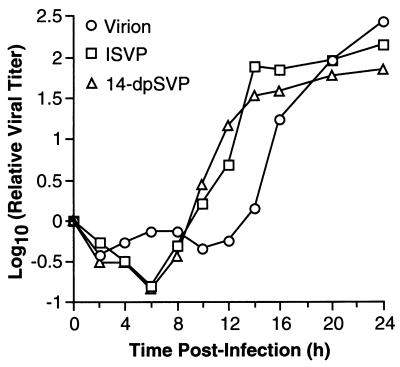
To determine the contribution of μ1/μ1C cleavage to the more rapid growth of ISVPs, we compared single-cycle growth curves generated in parallel for T1L virions, ISVPs, and 14-dpSVPs. The results confirmed the difference in growth kinetics between virions and ISVPs and also demonstrated that the growth kinetics of ISVPs and 14-dpSVPs are very similar, despite their difference in μ1/μ1C cleavage (Fig. 5). These findings suggest that the cleavage of μ1/μ1C into fragments μ1δ/δ and φ during viral entry into host cells contributes little or nothing to the shortened lag phase of ISVPs.
FIG. 5.
Single-step growth curves of 14-dpSVPs in L cells. L-cell monolayers were infected with purified T1L virions, ISVPs, or 14-dpSVPs at an MOI of 3 PFU/cell and harvested at different times postinfection. The infectious titer in each sample was measured by plaque assay. Each point represents the average of two independent log10 PFU/milliliter determinations (SD < 0.50 log10 PFU/ml for all time points).
Infectivity of 14-dpSVPs in the presence of NH4Cl or E-64.
Another property in which virions and ISVPs differ is that ISVPs can replicate in the presence of NH4Cl or E-64, while virions cannot (30, 55). The evidence indicates that these agents block infectivity by blocking the cleavage of outer-capsid proteins during infection with virions, and that they in turn have no effect on the infectivity of ISVPs because cleavage of the outer-capsid proteins has already been effected in vitro with those particles. The capacity of NH4Cl and E-64 to block removal of ς3 from virions during entry almost certainly contributes to the sensitivity of virions to these compounds (42, 43, 55). However, because ς3 removal is prerequisite for μ1/μ1C cleavage to occur, experiments to date (4, 37) have been unable to assess conclusively whether the latter cleavage makes any significant contribution to the capacity of ISVPs to replicate in the presence of NH4Cl or E-64.
To test the contribution of μ1/μ1C cleavage to the capacity of ISVPs to replicate in the presence of either NH4Cl or E-64, we measured the infectivities of T1L virions, ISVPs, and 14-dpSVPs in the presence of these agents. The results confirmed the effects of NH4Cl and E-64 on infections by virions or ISVPs and also showed that 14-dpSVPs, like ISVPs, retain full infectivity in the presence of these compounds (Fig. 6A). In addition, single-cycle growth curves determined for ISVPs and 14-dpSVPs in the presence of NH4Cl (data not shown) were virtually identical, allowing us to rule out the possibility that these compounds can affect the time course of infection by 14-dpSVPs without reducing the final yield of viral progeny. Since not even a minor difference in the responses of 14-dpSVPs and ISVPs to NH4Cl was observed, the findings suggest that the cleavage of μ1/μ1C into fragments μ1δ/δ and φ during viral entry contributes little or nothing to the capacity of ISVPs to replicate in the presence of NH4Cl.
Cleavage of μ1/μ1C protein during infection of L cells with 14-dpSVPs.
The capacity of 14-dpSVPs to initiate infection of L cells in the presence of either NH4Cl or E-64 suggested that cleavage of μ1/μ1C at the δ-φ junction during entry is not required for reovirus infection of these cells. It remained possible, however, that μ1/μ1C cleavage continues to occur during infections with 14-dpSVPs even in the presence of NH4Cl or E-64. For example, μ1/μ1C cleavage might be mediated by a cellular protease that does not require low pH and is not inhibited by E-64. If so, then μ1/μ1C cleavage within cells might yet be required for infection. To test whether μ1/μ1C cleavage occurs during infections with 14-dpSVPs, with or without NH4Cl or E-64 being present, we used radiolabeled virions and 14-dpSVPs to initiate infection in the presence or absence of each inhibitor, recovered the radiolabeled particles at 4 h postinfection, and used SDS-PAGE to monitor the extent of μ1/μ1C cleavage (Fig. 6B). The gels showed that with either virions or 14-dpSVPs, cleavage of μ1/μ1C to generate the μ1δ/δ fragment occurred normally in the absence of inhibitors but was substantially reduced by either NH4Cl or E-64 (Fig. 6B). In fact, little or no additional cleavage of μ1/μ1C was observed even at 10 h postinfection in the presence of these agents (data not shown). Since either NH4Cl or E-64 was capable of greatly reducing the extent of μ1/μ1C cleavage within cells, but 14-dpSVPs remained fully infectious in the presence of each inhibitor, these findings strongly suggest that the cleavage of μ1/μ1C at the δ-φ junction during viral entry is dispensable for reovirus infection of L cells.
Infectivity of 14-dpSVPs in MDCK cells.
To determine whether the infective properties of 14-dpSVPs were unique to L cells, we performed an experiment similar to that shown in Fig. 6A, but this time using MDCK cells. The behaviors of T1L virions, ISVPs, and 14-dpSVPs were determined in parallel for comparison. The results showed that E-64 was as effective at blocking infection with virions in MDCK cells as in L cells (Fig. 7). In addition, again as in L cells, E-64 was incapable of blocking infection with either ISVPs or 14-dpSVPs in MDCK cells (Fig. 7). These findings strongly suggest that the cleavage of μ1/μ1C at the δ-φ junction during viral entry is dispensable for infection of MDCK cells as well and raise the possibility that this cleavage is dispensable for infection of all cells. Similar studies were attempted with NH4Cl but could not be adequately completed due to the high concentrations of this compound that were needed to block infection of MDCK cells with virions (data not shown).
FIG. 7.
Infectivity of 14-dpSVPs in MDCK cells in the absence and presence of NH4Cl or E-64. The infectivities of purified T1L virions, ISVPs, or 14-dpSVPs in MDCK cells in the absence or presence of E-64 (300 μM) were determined exactly as for Fig. 6A except that MDCK cells were substituted for the L cells. Each bar represents an average log10 PFU/milliliter derived from three independent experiments. hpi, hours postinfection.
Capacity of 14-dpSVPs to mediate hemolysis of RBCs.
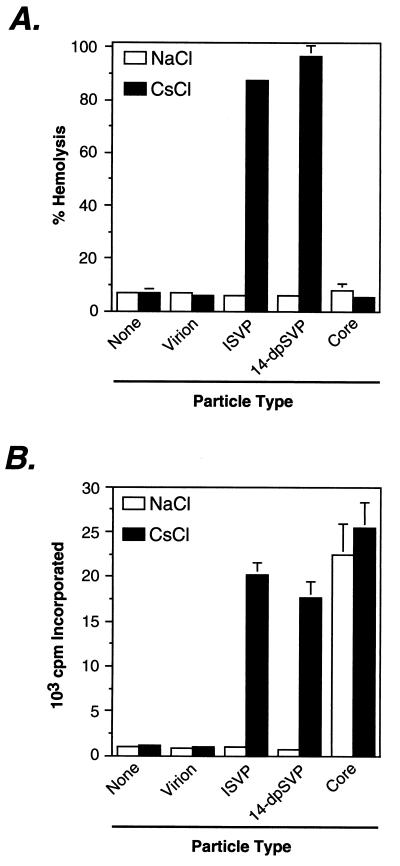
To initiate infection, reovirus particles must penetrate a cellular membrane in a process involving μ1 and/or one or more of its fragments, thereby gaining access to the host cell’s cytoplasm (24–26, 37, 41, 43, 44, 57). The capacity of 14-dpSVPs to replicate normally in L or MDCK cells at conditions that resulted in substantial blockade of μ1/μ1C cleavage provided indirect evidence that cleavage of μ1/μ1C during entry is dispensable for membrane penetration. To test the role of μ1/μ1C cleavage in membrane penetration more directly, we examined the capacity of virions, ISVPs, 14-dpSVPs, and cores to permeabilize lipid bilayers in vitro, using a hemolysis assay. In the presence of NaCl, none of the reovirus particle types demonstrated lysis of RBCs (Fig. 8A). However, when NaCl was replaced by CsCl, which is known to activate reovirus ISVPs for interaction with lipid bilayers (39, 57), ISVPs but not virions or cores induced hemolysis as expected (Fig. 8A). 14-dpSVPs induced hemolysis under the same conditions and to a similar extent as ISVPs (Fig. 8A), providing additional evidence that μ1/μ1C cleavage at the δ-φ junction during viral entry is dispensable for membrane penetration by mammalian reoviruses.
FIG. 8.
Capacity of 14-dpSVPs to effect hemolysis and undergo activation of the virus-bound transcriptase. (A) Bovine calf RBCs were incubated with purified T1L virions, ISVPs, 14-dpSVPs, or cores (3 × 1012 particles/ml) at 37°C for 1 h in the presence of NaCl or CsCl (200 mM). The extent of hemolysis was determined by measuring A415 and expressed as a percentage (taking hemolysis induced by Triton X-100 to be 100%). Each bar represents the average ± SD for three trials. (B) Purified T1L virions, ISVPs, 14-dpSVPs, or cores (3 × 1012 particles/ml) were incubated with a transcription reaction mixture including ribonucleoside triphosphates (2 mM each) and [α-32P]GTP (2.25 μCi) at 37°C for 1 h in the presence of NaCl or CsCl (200 mM). 32P incorporation into reovirus transcripts was quantified by trichloroacetic acid precipitation onto filter paper followed by liquid scintillation counting. Each bar represents the average ± SD for three trials.
Capacity of 14-dpSVPs to undergo transcriptase activation.
In conjunction with viral entry, the particle-bound viral transcriptase activity must be switched on, allowing viral mRNAs to be synthesized in the cytoplasm of the infected cell (7, 43). The infectivity of 14-dpSVPs in L or MDCK cells in the presence of NH4Cl or E-64 suggested that cleavage of μ1/μ1C during viral entry is not necessary for infecting reovirus particles to undergo transcriptase activation. To test the role of μ1/μ1C cleavage in transcriptase activation more directly, we examined the capacity of T1L virions, ISVPs, 14-dpSVPs, and cores to undergo transcriptase activation in vitro. As expected, cores, which possess a constitutively active transcriptase, displayed transcriptase activity in the presence of either NaCl or CsCl (Fig. 8B). ISVPs exhibited transcriptase activity upon addition of CsCl but not NaCl, Cs+ being an activating monovalent cation (8), while virions were inactive in the presence of either NaCl or CsCl (Fig. 8B). 14-dpSVPs underwent switch-on of transcriptase activity at the same conditions and to a similar extent as ISVPs (Fig. 8B), providing additional evidence that cleavage of μ1/μ1C to μ1δ/δ and φ during viral entry is dispensable for transcriptase activation.
DISCUSSION
Is μ1/μ1C cleavage required for membrane penetration by mammalian reoviruses?
Genetic and biochemical evidence indicates that the reovirus μ1 protein is involved in membrane penetration during viral entry into cells, perhaps via direct interactions with the membrane bilayer. For example, only ISVPs (which have μ1 and its fragments as the major surface proteins) can permeabilize lipid bilayers in in vitro assays, and they do so in a manner genetically controlled by the M2 gene, which encodes μ1 (25, 26, 37, 39, 43, 57). Since proteolytic processing of certain proteins is required for penetration into their host cells by a number of enveloped and nonenveloped viruses (see the introduction), the cleavage of μ1/μ1C at the δ-φ junction, which normally accompanies the degradation of ς3 during viral entry, was thought to be necessary for membrane penetration by these viruses (41). In this study, however, we demonstrated that μ1/μ1C cleavage during entry is dispensable for reovirus infection and that particles with uncleaved μ1/μ1C (14-dpSVPs) can permeabilize lipid bilayers in vitro with a similar efficacy as particles with cleaved μ1/μ1C (ISVPs), suggesting that this cleavage may not be required for membrane penetration.
Structural features of μ1 consistent with a role in membrane interaction include the modification of its N terminus with a myristoyl group (44) and the presence of several predicted amphipathic α-helices, including one pair that flanks the cleavage junction between fragments μ1δ/δ and φ in ISVPs (41). Such findings have engendered a model for membrane penetration in which μ1 is analogous to the fusion proteins of enveloped viruses (41). Features of that model include the interaction of the N-myristoyl group and the amphipathic α-helices described above with cellular membranes during entry. The model also predicted that cleavage of protein μ1/μ1C to μ1δ/δ and φ is required to remove existing conformational restraints on μ1, as seen with the influenza virus protein hemagglutinin (27), allowing one or both of the helices that flank the δ-φ cleavage junction to partition into the target membrane. Our current results suggest that the cleavage aspect of the preceding model is incorrect.
While it is possible that the amphipathic α-helices predicted to flank the δ-φ junction do not really exist or are not involved in membrane penetration, findings with the bacterial toxin colicin A (14) suggest another model in which these helices may interact with membrane in a cleavage-independent manner. In the case of colicin A, two amphipathic α helices joined by a short connector loop (47) constitute the initial membrane insertion domain (33). This helix-loop-helix structure is inserted into the target membrane upon an acid-triggered conformational change in the pore-forming domain of the colicin protein (58), without necessity for a proteolytic cleavage within the loop to free the helices for membrane insertion. A similar mechanism is used by diphtheria toxin (46). More detailed biochemical studies of membrane interaction by μ1 are warranted to determine whether this or some other model for membrane penetration is most applicable to reovirus entry into cells.
Is μ1/μ1C cleavage required for transcriptase activation?
The reovirus transcriptase enzymes, which are responsible for the synthesis of mRNAs during viral replication, are located within the inner capsid of the reovirus particle (18, 50). The capacity of these enzymes to synthesize full-length mRNAs is latent in virions and ISVPs but active in core particles derived by in vitro protease treatment of virions or ISVPs (see references 45 and 50 for reviews). A strain difference in the treatment conditions needed to generate cores and activate transcript elongation was mapped to the M2 gene, suggesting that μ1 plays a regulatory role in the switch-on of transcriptase activity (15). According to a current model, virions undergo transcriptase activation via two steps (7, 21, 43). In the first step, virions undergo proteolysis to ISVPs. Then, protease-independent structural changes in μ1, which we believe are linked to membrane penetration (45), yield transcriptionally active particles. Since cleavage of the μ1/μ1C protein at the δ-φ junction normally accompanies the first, protease-dependent step, it has been speculated (6, 21, 43) that this cleavage may be necessary for the conformational changes in μ1 that yield transcriptase activation. However, our results indicating that cleavage at the δ-φ junction during entry is dispensable for infection and that 14-dpSVPs can undergo transcriptase activation in vitro suggest that this cleavage is not necessary for the switch-on of reovirus transcription.
Why is μ1/μ1C cleavage conserved among reoviruses?
Despite the evidence presented in this study, cleavage of μ1/μ1C to generate the μ1δ/δ fragment is highly conserved among all three serotypes of mammalian reoviruses. This conservation also extends to the genetically distinct avian reoviruses, which exhibit similar entry-related cleavages in μ2, the polypeptide analogous to μ1 in those viruses (20). It is thus possible that cleavage at the δ-φ junction during entry is required for infection of host mammals in nature. Alternatively, this cleavage may simply reflect some structural aspect of μ1 that is essential for one or more of its functions. For example, the δ-φ cleavage site may lie within a solvent-exposed loop whose exposure and flexibility is vital for μ1 function. Sites within this loop that are susceptible to protease cleavage during reovirus entry may not be subject to negative selection because, as we showed in this study, cleavage in this region of μ1/μ1C has a neutral effect on viral infectivity.
How do alkyl sulfate detergents inhibit μ1/μ1C cleavage?
Earlier studies of the effects of alkyl sulfate detergents on reovirus virions concluded that SDS (1%) elutes ς3 from particles without observable effects on other polypeptides (16, 17). The effect of detergent treatment on the proteolytic digestion of virion proteins was not investigated. We now report that the simultaneous treatment of virions with micelle-forming concentrations of alkyl sulfate detergent and protease affects not only ς3 but also ς1 and μ1/μ1C, two other components of the outer capsid; however, the nature of these effects is not the same for all three polypeptides. In particular, ς3 is rendered hypersensitive to protease digestion, and ς1, normally resistant to protease treatment in the case of T1L, is newly sensitized to protease cleavage by all but one of the alkyl sulfates tested. The capacity of alkyl sulfates to enhance the protease sensitivity of ς3 and ς1 is as one might expect for these known protein denaturants. In contrast, the cleavage of μ1/μ1C to μ1δ/δ and φ, which normally occurs upon addition of protease, is completely but reversibly blocked by alkyl sulfates. Denaturation of viral proteins by detergent is unlikely to account for the reversible blockade of a specific proteolytic cleavage.
To account for the blockade of μ1/μ1C cleavage by alkyl sulfates, we propose a model in which detergent micelles bind to the viral outer capsid without causing global protein denaturation. Bound micelles may block the μ1/μ1C cleavage either by directly excluding protease molecules from their recognition sites on the viral surface or by eliciting conformational changes in the outer capsid that render these sites inaccessible to protease. The postulated virus-detergent interactions appear to represent a special case of intrinsic binding cooperativity as defined by Tanford (56), where a protein has a hydrophobic loop or tail that can serve either as a nucleus for the formation of an amphiphilic micelle or as a target for the insertion of preformed micelles. Because protein μ1 is a major component of the outer capsid (600 copies) and suffers blockade of cleavage by the detergent, we consider it likely that the regions of the viral particle involved in detergent interaction are located in μ1. Virus-micelle binding may mimic early phases of the μ1-phospholipid membrane interaction postulated to occur during viral entry. For example, a reversible surface-seeking interaction between reovirus ISVPs and phospholipid membranes, mediated by a constitutively exposed region of the μ1 protein, may occur during the initial phases of membrane penetration before extensive conformational changes in the ISVP have taken place. We are currently designing experiments to address these hypotheses.
ACKNOWLEDGMENTS
We thank Rebecca Margraf and Stephan Harrison for technical assistance; Michael Parsons, Rebecca Leidner, and Xiu-Hua Zhou for laboratory support; and the other members of our laboratory for helpful discussions. We are also grateful to Simon Noble, Roland Rueckert, and Leslie Schiff for critical reviews of a preliminary version of the manuscript.
This work was supported by NIH grant R29 AI39533 and by a grant to the Institute for Molecular Virology from the Lucille P. Markey Charitable Trust. M.L.N. received additional support as a Shaw Scientist from the Milwaukee Foundation. K.C. was additionally supported by predoctoral fellowships from the Wisconsin Alumni Research Foundation and the Howard Hughes Medical Institute.
REFERENCES
- 1.Baer G S, Dermody T S. Mutations in reovirus outer-capsid protein ς3 selected during persistent infections of L cells confer resistance to protease inhibitor E64. J Virol. 1997;71:4921–4928. doi: 10.1128/jvi.71.7.4921-4928.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bass D, Baylor M R, Chen C, Mackow E M, Bremont M, Greenberg H B. Liposome-mediated transfection of intact viral particles reveals that membrane penetration determines permissivity of tissue culture cells to rotavirus. J Clin Invest. 1992;90:2313–2320. doi: 10.1172/JCI116119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumenthal R, Seth P, Willingham M C, Pastan I. pH dependent lysis of liposomes by adenovirus. Biochemistry. 1986;25:2231–2237. doi: 10.1021/bi00356a057. [DOI] [PubMed] [Google Scholar]
- 4.Bodkin D K, Nibert M L, Fields B N. Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J Virol. 1989;63:4676–4681. doi: 10.1128/jvi.63.11.4676-4681.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borsa J, Copps T P, Sargent M D, Long D G, Chapman J D. New intermediate subviral particles in the in vitro uncoating of reovirus virions by chymotrypsin. J Virol. 1973;11:552–564. doi: 10.1128/jvi.11.4.552-564.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borsa J, Long D G, Sargent M D, Copps T P, Chapman J D. Reovirus transcriptase activation in vitro: involvement of an endogenous uncoating activity in the second stage of the process. Intervirology. 1974;4:171–188. doi: 10.1159/000149856. [DOI] [PubMed] [Google Scholar]
- 7.Borsa J, Sargent M D, Lievaart P A, Copps T P. Reovirus: evidence for a second step in the intracellular uncoating and transcriptase activation process. Virology. 1981;111:191–200. doi: 10.1016/0042-6822(81)90664-4. [DOI] [PubMed] [Google Scholar]
- 8.Borsa J, Sargent M D, Long D G, Chapman J D. Extraordinary effects of specific monovalent cations on activation of reovirus transcriptase by chymotrypsin in vitro. J Virol. 1973;11:207–217. doi: 10.1128/jvi.11.2.207-217.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandran, K., and M. L. Nibert. Unpublished data.
- 10.Chang C T, Zweerink H J. Fate of parental reovirus in infected cell. Virology. 1971;46:544–555. doi: 10.1016/0042-6822(71)90058-4. [DOI] [PubMed] [Google Scholar]
- 11.Clark S M, Roth J R, Clark M L, Barnett B B, Spendlove E S. Trypsin enhancement of rotavirus infectivity. J Virol. 1981;39:816–822. doi: 10.1128/jvi.39.3.816-822.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cleveland D R, Zarbl H, Millward S. Reovirus guanylyltransferase is L2 gene product λ2. J Virol. 1986;60:307–311. doi: 10.1128/jvi.60.1.307-311.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox D C, Clinkscales W. Infectious reovirus subviral particles: virus replication, cellular cytopathology, and DNA synthesis. Virology. 1976;74:259–261. doi: 10.1016/0042-6822(76)90152-5. [DOI] [PubMed] [Google Scholar]
- 14.Cramer W A, Cohen F S, Merrill A R, Song H Y. Structure and dynamics of the colicin E1 channel. Mol Microbiol. 1990;4:519–526. doi: 10.1111/j.1365-2958.1990.tb00619.x. [DOI] [PubMed] [Google Scholar]
- 15.Drayna D, Fields B N. Activation and characterization of the reovirus transcriptase: genetic analysis. J Virol. 1982;41:110–118. doi: 10.1128/jvi.41.1.110-118.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drayna D, Fields B N. Biochemical studies on the mechanism of chemical and physical inactivation of reovirus. J Gen Virol. 1982;63:161–170. doi: 10.1099/0022-1317-63-1-161. [DOI] [PubMed] [Google Scholar]
- 17.Drayna D, Fields B N. Genetic studies on the mechanism of chemical and physical inactivation of reovirus. J Gen Virol. 1982;63:149–159. doi: 10.1099/0022-1317-63-1-149. [DOI] [PubMed] [Google Scholar]
- 18.Dryden, K. A., D. L. Farsetta, G.-J. Wang, J. M. Keegan, B. N. Fields, T. S. Baker, and M. L. Nibert. Submitted for publication.
- 19.Dryden K A, Wang G, Yeager M, Nibert M L, Coombs K M, Furlong D B, Fields B N, Baker T S. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duncan R. The low pH-dependent entry of avian reovirus is accomplished by two specific cleavages of the major outer capsid protein μ2C. Virology. 1996;29:179–189. doi: 10.1006/viro.1996.0235. [DOI] [PubMed] [Google Scholar]
- 21.Ewing D D, Sargent M D, Borsa J. Switch-on of transcriptase function in reovirus: analysis of polypeptide changes using 2-D gels. Virology. 1985;144:448–456. doi: 10.1016/0042-6822(85)90285-5. [DOI] [PubMed] [Google Scholar]
- 22.Furlong D B, Nibert M L, Fields B N. ς1 protein of mammalian reoviruses extends from the surfaces of viral particles. J Virol. 1988;62:246–256. doi: 10.1128/jvi.62.1.246-256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greber U F, Webster P, Weber J, Helenius A. The role of the adenovirus protease on virus entry into cells. EMBO J. 1996;15:1766–1777. [PMC free article] [PubMed] [Google Scholar]
- 24.Hazelton P R, Coombs K C. The reovirus mutant tsA279 has temperature-sensitive lesions in the L2 and M2 genes: the M2 gene is associated with decreased viral protein production and blockade in transmembrane transport. Virology. 1995;207:46–58. doi: 10.1006/viro.1995.1050. [DOI] [PubMed] [Google Scholar]
- 25.Hooper J W, Fields B N. Monoclonal antibodies to reovirus ς1 and μ1 proteins inhibit chromium release from mouse L cells. J Virol. 1996;70:672–677. doi: 10.1128/jvi.70.1.672-677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hooper J W, Fields B N. Role of the μ1 protein in reovirus stability and capacity to cause chromium release from host cells. J Virol. 1996;70:459–467. doi: 10.1128/jvi.70.1.459-467.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughson F M. Structural characterization of viral fusion proteins. Curr Biol. 1995;5:265–274. doi: 10.1016/s0960-9822(95)00057-1. [DOI] [PubMed] [Google Scholar]
- 28.Joklik W K. Recent progress in reovirus research. Annu Rev Genet. 1985;19:537–575. doi: 10.1146/annurev.ge.19.120185.002541. [DOI] [PubMed] [Google Scholar]
- 29.Joklik W K. Studies on the effect of chymotrypsin on reovirions. Virology. 1972;49:700–801. doi: 10.1016/0042-6822(72)90527-2. [DOI] [PubMed] [Google Scholar]
- 30.Jones, L., K. Subramanian, R. Margraf, B. N. Fields, and M. L. Nibert. Submitted for publication.
- 31.Kaljot K T, Shaw R D, Rubin D H, Greenberg H B. Infectious rotavirus enters cells by direct cell membrane penetration, not by endocytosis. J Virol. 1988;62:1136–1144. doi: 10.1128/jvi.62.4.1136-1144.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klenk H D, Rott R, Orlich M, Blodorn J. Activation of influenza A viruses by trypsin treatment. Virology. 1975;68:426–439. doi: 10.1016/0042-6822(75)90284-6. [DOI] [PubMed] [Google Scholar]
- 33.Lakey J H, Massotte D, Heitz F, Dasseux J L, Faucon J F, Parker M W, Pattus F. Membrane insertion of the pore-forming domain of colicin A. A spectroscopic study. Eur J Biochem. 1991;196:599–607. doi: 10.1111/j.1432-1033.1991.tb15855.x. [DOI] [PubMed] [Google Scholar]
- 34.Lee P W, Hayes E C, Joklik W K. Protein ς1 is the reovirus cell attachment protein. Virology. 1981;108:156–163. doi: 10.1016/0042-6822(81)90535-3. [DOI] [PubMed] [Google Scholar]
- 35.Lee W-M, Monroe S S, Rueckert R R. Role of maturation cleavage in infectivity of picornaviruses: activation of an infectosome. J Virol. 1993;67:2110–2122. doi: 10.1128/jvi.67.4.2110-2122.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lobigs M, Garoff H. Fusion function of the Semliki Forest virus spike is activated by proteolytic cleavage of the envelope glycoprotein precursor p62. J Virol. 1990;64:1233–1240. doi: 10.1128/jvi.64.3.1233-1240.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lucia-Jandris P, Hooper J W, Fields B N. Reovirus M2 gene is associated with chromium release from mouse L cells. J Virol. 1993;67:5339–5345. doi: 10.1128/jvi.67.9.5339-5345.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luongo C L, Dryden K A, Farsetta D L, Margraf R L, Severson T F, Olson N H, Fields B N, Baker T S, Nibert M L. Localization of a C-terminal region of λ2 protein in reovirus cores. J Virol. 1997;71:8035–8040. doi: 10.1128/jvi.71.10.8035-8040.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nibert M L. Ph.D. thesis. Cambridge, Mass: Harvard University; 1993. [Google Scholar]
- 40.Nibert M L, Chappell J D, Dermody T S. Infectious subvirion particles of reovirus type 3 Dearing exhibit a loss in infectivity and contain a cleaved ς1 protein. J Virol. 1995;69:5057–5067. doi: 10.1128/jvi.69.8.5057-5067.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nibert M L, Fields B N. A carboxy-terminal fragment of protein μ1/μ1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J Virol. 1992;66:6408–6418. doi: 10.1128/jvi.66.11.6408-6418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nibert M L, Fields B N. Early steps in reovirus infection of cells. In: Wimmer E, editor. Cellular receptors for animal viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 341–364. [Google Scholar]
- 43.Nibert M L, Furlong D B, Fields B N. Mechanisms of viral pathogenesis. Distinct forms of reoviruses and their roles during replication in cells and host. J Clin Invest. 1991;88:727–734. doi: 10.1172/JCI115369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nibert M L, Schiff L A, Fields B N. Mammalian reoviruses contain a myristoylated structural protein. J Virol. 1991;65:1960–1967. doi: 10.1128/jvi.65.4.1960-1967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nibert M L, Schiff L A, Fields B N. Reoviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1557–1596. [Google Scholar]
- 46.Oh K J, Zhan H, Cui C, Hideg K, Collier R J, Hubbell W L. Organization of diphtheria toxin T domain in bilayers: a site-directed spin labeling study. Science. 1996;273:810–812. doi: 10.1126/science.273.5276.810. [DOI] [PubMed] [Google Scholar]
- 47.Parker M W, Pattus F, Tucker A D, Tsernglou D. Structure of the membrane pore-forming fragment of colicin A. Nature. 1989;337:93–96. doi: 10.1038/337093a0. [DOI] [PubMed] [Google Scholar]
- 48.Rueckert R R. Picornaviridae. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 477–519. [Google Scholar]
- 49.Scheid A, Choppin P W. Identification of biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis, and infectivity of proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology. 1974;57:475–490. doi: 10.1016/0042-6822(74)90187-1. [DOI] [PubMed] [Google Scholar]
- 50.Shatkin A J, Kozak M. Biochemical aspects of reovirus transcription and translation. In: Joklik W K, editor. The Reoviridae. New York, N.Y: Plenum Press; 1983. pp. 79–106. [Google Scholar]
- 51.Shatkin A J, LaFiandra A J. Transcription by infectious subviral particles of reovirus. J Virol. 1972;10:698–706. doi: 10.1128/jvi.10.4.698-706.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silverstein S C, Astell C, Levin D H, Schonberg M, Acs G. The mechanisms of reovirus uncoating and gene activation in vivo. Virology. 1972;47:797–806. doi: 10.1016/0042-6822(72)90571-5. [DOI] [PubMed] [Google Scholar]
- 53.Silverstein S C, Schonberg M, Levin D H, Acs G. The reovirus replicative cycle: conservation of parental RNA and protein. Proc Natl Acad Sci USA. 1970;67:275–281. doi: 10.1073/pnas.67.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith R E, Zweerink H J, Joklik W K. Polypeptide components of virions, top component and cores of reovirus type 3. Virology. 1969;39:791–810. doi: 10.1016/0042-6822(69)90017-8. [DOI] [PubMed] [Google Scholar]
- 55.Sturzenbecker L J, Nibert M, Furlong D, Fields B N. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J Virol. 1987;61:2351–2361. doi: 10.1128/jvi.61.8.2351-2361.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanford C. The hydrophobic effect. 2nd ed. New York, N.Y: John Wiley & Sons; 1980. [Google Scholar]
- 57.Tosteson M T, Nibert M L, Fields B N. Ion channels induced in lipid bilayers by subvirion particles of the nonenveloped mammalian reoviruses. Proc Natl Acad Sci USA. 1993;90:10549–10552. doi: 10.1073/pnas.90.22.10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van der Goot F G, Gonzalez-Manas J M, Lakey J H, Pattus F. A “molten globule” membrane insertion intermediate of the pore-forming domain of colicin A. Nature. 1991;354:408–410. doi: 10.1038/354408a0. [DOI] [PubMed] [Google Scholar]
- 59.Virgin H W IV, Mann M A, Tyler K L. Protective antibodies inhibit reovirus internalization and uncoating by intracellular proteases. J Virol. 1994;68:6719–6729. doi: 10.1128/jvi.68.10.6719-6729.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vuillez-Le Normand B, Eiselé J L. Determination of detergent critical micellar concentration by solubilization of a colored dye. Anal Biochem. 1993;208:241–243. doi: 10.1006/abio.1993.1039. [DOI] [PubMed] [Google Scholar]
- 61.White C K, Zweerink H J. Studies on the structure of reovirus cores: selective removal of polypeptide λ2. Virology. 1976;70:171–180. doi: 10.1016/0042-6822(76)90247-6. [DOI] [PubMed] [Google Scholar]