Abstract
Stress granules and P-bodies are conserved cytoplasmic biomolecular condensates whose assembly and composition are well documented, but whose clearance mechanisms remain controversial or poorly described. Such understanding could provide new insight into how cells regulate biomolecular condensate formation and function, and identify therapeutic strategies in disease states where aberrant persistence of stress granules in particular is implicated. Here, I review and compare the contributions of chaperones, the cytoskeleton, post-translational modifications, RNA helicases, granulophagy and the proteasome to stress granule and P-body clearance. Additionally, I highlight the potentially vital role of RNA regulation, cellular energy, and changes in the interaction networks of stress granules and P-bodies as means of eliciting clearance. Finally, I discuss evidence for interplay of distinct clearance mechanisms, suggest future experimental directions, and suggest a simple working model of stress granule clearance.
Keywords: Stress Granules, P-bodies, Chaperones, Cytoskeleton, Proteasome, Granulophagy, RNA helicases, RNA modification, Ubiquitination, Phosphorylation, G3BP, VCP, mRNA, translation, mRNA decay
INTRODUCTION
Stress granules (SGs) and P-bodies (PBs) are paradigm biomolecular condensates, also referred to as membraneless organelles1. They consist largely of non-translating mRNA-protein complexes (mRNPs), and their assembly, composition, dynamics, and function has been the focus of considerable research effort and speculation for almost 25 years2–6. In contrast, how SGs and particularly PBs undergo clearance in cells, either via disassembly or degradative means is less well understood, though many pathways and factors have been implicated. SG and PB clearance mechanisms help determine SG and PB abundance, understanding of which may reveal new functional insights, and be pertinent to understanding the dynamics and function of other biomolecular condensates. Additionally, SG clearance defects are linked to the pathogenesis of Amyotrophic Lateral Sclerosis (ALS) and other neurodegenerative diseases7–9. Thus, understanding SG and PB clearance may identify future therapeutic targets.
In this review, I will discuss reported and putative mechanisms of SG and PB clearance, assess the importance and integration of distinct clearance pathways, identify gaps in knowledge, and suggest future experimental priorities. A quantitative summary of reported SG and PB clearance effects is presented in Table S1 for additional reference. Finally, a simple working model of SG clearance will be presented.
SG AND PB CLEARANCE - ASSEMBLY IN REVERSE, OR A DISTINCT PROCESS?
Several reversible mechanisms exist that oppositely impact assembly and clearance of both SGs and PBs. One example is the entry and exit of mRNAs to and from polysomes. SG and PB assembly is stimulated by translation repression, whereas their clearance correlates with translational recovery10,11, though sometimes translation recovers with SGs still partly evident12,13. Cycloheximide, and other elongation inhibitors, prevent assembly of SGs and PBs by trapping mRNPs in polysomes, but also hasten clearance of already formed SGs14,15 and non-stress induced PBs16,17 by presumably the same mechanism. Stress-induced mammalian PBs may clear significantly more slowly than SGs in the presence of cycloheximide14, unlike in yeast18, though extended cycloheximide treatment still results in full mammalian PB clearance19. Finally, microscopy-based studies20–25 have visualized mRNP entry and exit to/from SGs and PBs during and following stress. Collectively, these data mostly support a model of constant mRNA exchange between polysomes, SGs and PBs.
However, SG (and perhaps PB) clearance is not just a reversal of assembly events. First, whereas SG assembly in human cells is typically described as coalescence of multiple small, spherical foci into fewer, larger, and more irregular condensates, SG clearance is often morphologically distinct in that SGs either dissolve or undergo fracturing with rough-edged SGs sometimes harboring filament-like structures protruding from their surface15,26–28. Second, clearance can occur significantly more slowly11,29 or rapidly30 than assembly following distinct stresses5, though this may also depend on where one draws the line on “fully assembled”. Third, SGs exhibit an evolving proteomic composition during stress progression31, with uniquely enriched factors during stress recovery32. Finally, some factors impact only clearance, but not assembly (e.g., Hsp7013,33, MCM, RVB34, DYRK335, p6236) or vice versa (e.g., Chaperonin-containing T complex34). Different mechanisms of clearance are also utilized depending on stress and cellular context (e.g.,31,37; discussed later). Thus, SG and PB clearance likely involves a mix of reversing assembly-driving processes, and unique clearance mechanisms whose use depends on cellular and stress context (Figure 1).
Figure 1: Reported and putative means of SG clearance.
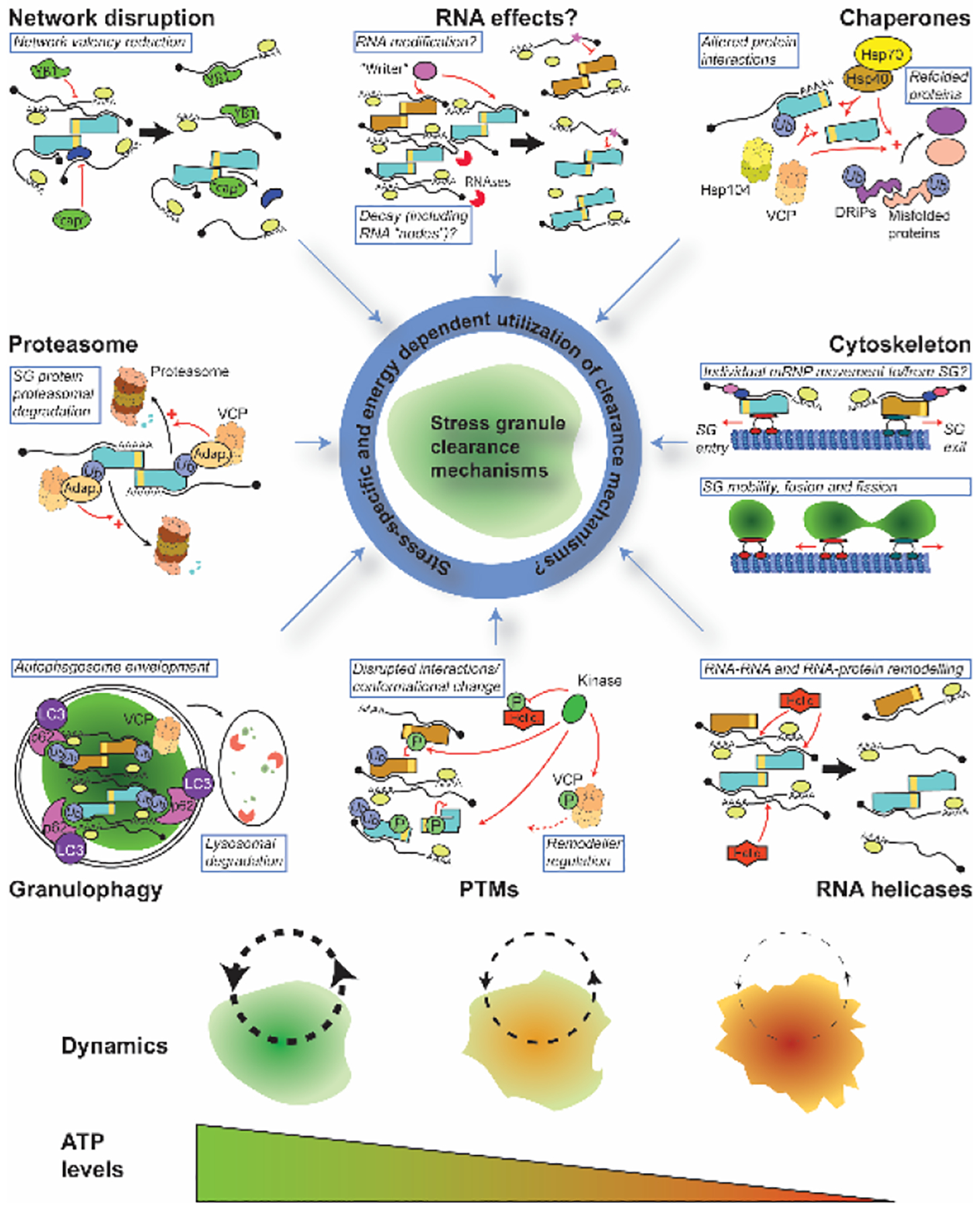
SG clearance is not fully understood, but multiple mechanisms have been reported including chaperone activity, cytoskeletal transport, RNA helicases, post-translational modifications (PTMs), granulophagy, proteasomal activity, and SG network disruption. Direct effects on SG-resident mRNA molecules have not been reported but are feasible given the multivalent, SG-scaffolding nature of mRNA and reported impacts of mRNA modification. A selection of examples of each clearance mechanism are depicted and identified in blue boxes; those with question marks are putative and/or not unambiguously demonstrated. Teal and orange objects with a yellow band indicate RNA-binding proteins with self-interacting domains (analogous to SG scaffolding proteins like G3BP1/2). “Helic.”, “Adap.” and “cap” refer to RNA helicases, VCP adaptor proteins, and network valency reducing, or “capping” proteins197, respectively. Black arrows indicate a transition from one state to another, while red arrows/inhibitory lines indicate direct action of a protein or enzyme on a target with functional consequences. Dynamics, meaning the rate of mRNP entry and exit to/from SGs, and their fluidity, likely depends on ATP levels, which is required by almost all of the SG clearance mechanisms proposed. See main text for more details.
CHAPERONE BASED MECHANISMS OF SG AND PB CLEARANCE
Several chaperones localize in and impact SG assembly and clearance38, including heat shock proteins (Hsps), which generally bind hydrophobic regions of misfolded proteins, and are categorized based on molecular weight, ATPase activity, and function. Hsps and their regulators/co-chaperones can limit accumulation of misfolded proteins in SGs, which can facilitate conversion of SGs to aberrant non-dynamic states, particular under heat shock stress (HS)18,31,33,38. Roles for key chaperones in SG (and PB) regulation are discussed below.
Hsp70s:
Hsp70s hydrolyze ATP to bind cyclically to substrates, promote protein disaggregation and refolding, and play integral roles in regulating nascent protein folding during translation39. In human cells, Hsp70, and an Hsp70 nucleotide exchange factor BAG3, promote clearance of HS or proteasome inhibition (MG132)-induced SGs, in correlation with translational recovery10,31,33,40. Overexpression or prior stress-mediated accumulation of Hsp70 also prevents SG assembly via MG132 or arsenite stress10. HS-induced SGs co-localize with misfolded proteins and Hsp7031,41, whereas under MG132 or arsenite stress, Hsp70 does not localize in SGs10,33. However, in all three stresses, Hsp70 inhibition exacerbates accumulation of ubiquitin signal and misfolded proteins in SGs, including ALS-associated aggregation prone proteins, and misfolded nascent translation products (“Defective ribosomal products”; DRiPs). SGs harboring such misfolded proteins clear more slowly than those that do not31,33.
Similarly, in yeast and Drosophila, Hsp70 and Hsp110 - an Hsp70 subclass that stimulates Hsp70 nucleotide exchange, and has its own ATPase chaperone activity - facilitate SG clearance following HS and sodium azide stress. This again correlates with translational recovery11,13,33. Unlike glucose deprivation SGs18, HS and sodium azide-induced SGs co-localize with Hsp70 and misfolded protein aggregates, again suggesting stress-specific links between SGs and misfolded proteins. Proteins in HS-induced SGs exchange more rapidly with the cytoplasm than aggregate-prone reporter proteins, and thus unsurprisingly, SGs also clear faster than protein aggregates11,18. This mirrors in vitro work demonstrating Pab1 (SG marker) biomolecular condensates are more rapidly dispersed by Hsp70 and other chaperones (particularly Hsp104 and 110) than aggregate-prone protein reporters42. Inhibiting Hsp70 function genetically or chemically does not induce SG assembly in yeast13 or human cells33, or affect assembly under various stresses examined, suggesting a role for Hsp70 primarily in clearance.
Collectively, these findings suggest Hsp70 aids SG clearance by preventing accumulation of misfolded, ubiquitinated proteins in SGs, the importance of which likely depends on the protein misfolding burden caused by a given stress. No SG-localized substrates are known to specifically recruit Hsp70, though many interactions with misfolded proteins, aided by Hsp40s and small Hsps (see below), seem likely. Additionally, Hsp70s and Hsp110 can robustly bind U-rich RNA, and even impact mRNA stability43,44, suggesting an unexplored recruitment and effector mechanism in clearance.
Hsp40s:
Hsp40 proteins work with Hsp70s, increasing Hsp70 ATPase activity via their J-domains, and conferring Hsp70 substrate specificity via C-terminal client domains45. In human cells, several Hsp40s localize in SGs34,46,47, though no specific substrates are known. However, Hdj1 and Hdj2 rely on a G/F-rich intrinsically disordered region (IDR) sequence present in most Hsp40s to both phase separate and localize in arsenite-induced SGs, suggesting possibly promiscuous interactions. Phase separation or substrate binding via their C-terminal domain may relieve an autoinhibitory interaction within Hsp40s that sequesters the J-domain, thus recruiting and stimulating Hsp70 activity46,48. In yeast, the homologs of Hdj1 and 2, Sis1 and Ydj1, localize in sodium azide-induced SGs and regulate specific SG clearance mechanisms. Ydj1 promotes SG disassembly and translation recovery, whereas Sis1 targets SGs to vacuolar compartments, presumably via “granulophagy” (see later)13.
Hsp104:
In yeast, this AAA+ ATPase “disaggregase” chaperone, absent in metazoa, forms a hexameric ring-like structure through which misfolded substrates are threaded through and unfolded49. Hsp104 also localizes18 in and facilitates clearance of HS-induced SGs, and the resumption of translation post-HS, based on genetic and inhibitor studies11,18 Intriguingly, Hsp104 (and Hsp70) are recruited to SGs following HS in part by a glycolysis metabolite mediating allosteric modulation, and solubilization, of the SG-localizing amyloid form of pyruvate kinase (Cdc19)50. Pyruvate kinase is key for ATP production, and though specific Hsp104 and Hsp70 recruitment mechanisms in this context are unclear, solubilization of pyruvate kinase in SGs, aided by Hsp104 activity, promotes efficient SG clearance. Thus, SG clearance and ATP production are coupled processes50, which may act in a positive feedback loop to further aid SG clearance via other ATP-dependent mechanisms (see below and Box 1).
BOX 1 -. THE IMPORTANCE OF ENERGY IN SG CLEARANCE.
ATP levels are a key regulator of both SG assembly and clearance. In human cells and yeast, strongly reducing ATP levels by 50–80% by inhibiting glycolysis (e.g., Glucose depletion, 2-DG, CP91149), or oxidative phosphorylation (e.g., FCCP, CCCP, oligomycin) induces SGs without initially increasing eIF2α phosphorylation29,97. In contrast, a combinatorial block to glycolysis and mitochondrial function in human cells, which reduces ATP levels even more, blocks arsenite-induced SG induction and induces cell death34,97, suggesting SGs are induced by low ATP levels, but still require a minimal level of ATP to form. ATP depletion can also drive SG assembly in G3BP1/2ΔΔ (key SG assembly mutant) cells, and greater RNA partitioning in SGs than eIF4A inhibition alone94. Following arsenite-induced SG assembly, ATP depletion blocks SG movement, fusion and reduces SG dynamics34. Finally, reducing ATP depletion below 50% of normal in human cells causes a near-complete block of arsenite and HS-induced SG clearance over 1.5hrs37, indicating the importance of ATP-based clearance processes.
Various ATP-dependent mechanisms (e.g., helicases, chaperones) may counteract the inherent tendency of non-translating mRNA and associated RNA-binding proteins to phase separate and form condensates. In vitro studies suggest that ATP itself may also play a role as a biological hydrotrope (i.e., solubilizer of hydrophobic proteins) to antagonize condensate formation132. Subsequent in vitro work with reconstituted SGs, induced by specific RNA molecules and using yeast lysates, suggests both roles of ATP may limit SG assembly, though hydrolysis of ATP (and GTP) is required to facilitate clearance of already formed SGs, suggesting involvement of ATP and GTP-dependent machineries133. What GTP-dependent machineries may be involved is unclear, but the translational apparatus seems a natural candidate.
Most SG clearance mechanisms discussed in this review (Figure 1) require cellular energy to function. The relative energy requirements of each pathway in SG clearance are unclear but could have a decisive impact on how SGs clear under stress conditions that significantly deplete cellular energy reserves.
Small Hsps/”Holdases”:
Small Hsps, which lack ATPase activity, generally maintain substrates in a conformation for processing by other chaperones51. Hspb8 aids clearance of MG132-induced SGs by recruiting Bag3 and Hsp70 to SGs harboring misfolded proteins, particularly DRiPs33. Hspb1 (Hsp27) also progressively localizes in HS and Diethyl maleate-induced SGs, but not arsenite, UV or MG132-induced SGs31,33,41,52. However, Hspb1 depletion still mildly slows SG clearance during proteasome inhibition33. Like Hsp70, SG recruiting mechanisms for small Hsps are unclear, but likely involve multiple promiscuous interactions with misfolded protein substrates, though Hspb1 also can bind RNA41.
Cdc48/VCP:
Like Hsp104, Cdc48(yeast)/VCP(human) is another hexameric AAA+ ATPase chaperone, though it is conserved throughout eukaryotes and acts more specifically on ubiquitinated substrates53,54. Diverse roles including protein refolding, endolysosomal trafficking, and protein degradation (via both proteasomal and autophagic means) depend largely on Cdc48/VCP cofactors that confer substrate specificity.
In yeast, Cdc48 inactivation causes SG accumulation during entry into quiescence, and impairs vacuolar targeting of SG material, implying a role in granulophagy55. In human cells, VCP (Valosin-containing protein) localizes in SGs following multiple stresses, and inhibition, either by knockdown or several different inhibitors, slows SG clearance following HS37 and arsenite stress, though not other stresses56. Expression of neurodegenerative disease associated VCP mutants induces constitutive SG assembly55, and slow SG clearance following HS37 and arsenite stress in various models57,58. VCP localization in HS-induced SGs depends on the VCP adaptor FAF2, and ubiquitination of the key mammalian SG assembly protein G3BP137, whereas another VCP adaptor, ZFAND1, aids VCP recruitment and clearance of SGs following arsenite stress58. ZFAND1 depleted cells also accumulate DRiPs in SGs, suggesting VCP may complement Hsp70 functions in SG clearance. As discussed later, VCP is linked to SG clearance via autophagy dependent, independent, and proteasomal-dependent means (Figure 1), implying a multi-faceted role in regulation of SG dynamics.
PBs and chaperones:
Only a few cases of Hsps impacting PB dynamics are known, most of which differ from SG effects. In yeast, similar to SGs, Hsp104 deletion slows PB clearance, leading to formation of aggregate-like structures harboring SG, PB and misfolded proteins18. Unlike SGs, following sodium azide stress, PB clearance is unaffected by Hsp70 inhibition13. Ydj1 aids localization of Dhh1 and Lsm1 (core PB proteins) to foci under acute or chronic glucose deprivation, though Edc3 (another core PB protein) is unaffected59. Finally, in human cells, inhibition of Hsp90, a homodimeric ATPase chaperone considered more selective than Hsp70 in stabilizing unstable substrates and regulating misfolded proteins60, aids PB assembly by an unclear mechanism61,62. In contrast, Hsp90 inhibition slows SG clearance, primarily due to destabilization of the kinase substrate DYRK3 (see later)63.
CYTOSKELETAL BASED MECHANISMS OF SG AND PB CLEARANCE
Though understudied of late, the cytoskeleton and associated motor proteins impact SG and PB clearance, assembly, mobility, and cellular localization26,64,65.
Microtubule-based mechanisms:
MT depolymerization reduces the size and mobility of SGs during assembly in many human cell models12,27,66–70. However, the effects of MT stabilization on SG assembly are controversial66,69. While not required to sustain SG assembly27, MTs promote SG mobility and coalescence into larger foci65, and perhaps SG-PB interactions71. Conversely, PB assembly in yeast and human cells is stimulated by MT depolymerization, though PBs also become largely immobile and exhibit compositional differences72–74. SG clearance following arsenite stress, facilitated by cycloheximide treatment, is strongly impaired by MT depolymerization27. The role of MTs in PB clearance is unknown.
Several MT motor and MT-binding proteins localize in SGs and PBs12,34,47,75–77, but consensus on their role is lacking. Inhibition of dynein motor proteins, which drive retrograde movement on MTs, impairs SG and PB assembly following arsenite and thapsigargin stress12,67,76, whereas non-stressed PBs are unaffected by MT motor perturbations12. Dynein inhibition also weakly inhibits SG clearance in P19 cells76, though no qualitative effects on SG dynamics were observed in other studies68,69. Kinesin, which drives anterograde movement, facilitates SG clearance in NIH3T cells12. Finally, the p50 isoform of Nesprin-1 is primarily a PB localizing, MT-binding protein whose expression promotes PB assembly, mobility, SG-PB interactions and SG clearance following H2O2 but not arsenite stress77.
Actin-based mechanisms:
SGs do not localize with, nor are obviously affected by actin-disrupting drugs67,69, whereas PBs do show some evidence of actin/myosin-based regulation. Specifically, many myosin motor proteins localize in PBs78,79, and immobile PBs associate with actin bundles in U2-OS cells; mobile PBs, in contrast, associate with MTs71 (except in plants80). In yeast, conditional inactivation of the Myosin protein Myo2 slows PB clearance induced by chronic nutrient deprivation81. In HeLa cells, the ortholog Myo5a localizes in PBs, and knockdown reduces PB numbers, whilst a dominant negative Myo5a impairs PB mobility79. Though not clarified, this could solely reflect an actin-based process, or involve microtubule-based function, by virtue of known Myo5-kinesin-MT interactions82.
Although actin filaments do not bind SGs, lamellar actin retrograde flow during stress in U2-OS cells may complement directed transport to “push” small, nascent SGs towards perinuclear regions. Here, subsequent non-specific capillary-based interactions of SGs with MTs are proposed to facilitate granule fusion independent of motor functions, reduce their mobility and lead to deformation of spherical SGs as they conform around the MT network83,84.
RNA HELICASE BASED MECHANISMS OF SG AND PB CLEARANCE
RNA helicases are strong candidates for regulating SG and PB clearance, given that they can prevent, remodel, or disrupt RNA-RNA and mRNP interactions85. Many members of both the non-processive DEAD and processive DExH class of RNA helicases localize in SG and PBs32,34,47,75,86,87. Several RNA helicases also harbor IDRs, which often facilitate condensate formation, particularly in the presence of ATP and RNA88. Typically, RNA helicases bind ATP and RNA co-operatively, and exhibit low affinity for RNA after ATP-hydrolysis. Thus, regulation of RNA helicase ATPase activity is key to RNP remodeling, and successive rounds of RNA binding. ATPase activity can be activated by helicase-interacting proteins harboring MIF4G domains, such as Not1 (Dhh1)89and eIF4G (eIF4A90 and Ded188), which in vitro can drive dissolution of helicase/RNA condensates. ATPase mutant versions of Dhh189, Ded191 and DDX3X (human Ded1 homolog)92 induce formation of PBs (Dhh1) and SGs (Ded1/DDX3), consistent with possible ATPase-reliant clearance functions.
eIF4A:
eIF4A is a SG-localizing helicase best known for its role in unwinding of 5’UTR structure during translation initiation. Inhibition of eIF4A is also a strong eIF2α-phosphorylation independent inducer of SG assembly93–96. While eIF4A’s translation-enhancing role likely antagonizes SG assembly and may facilitate clearance, a distinct role for eIF4A in limiting RNA recruitment to SGs is suggested by arsenite stress and eIF4A inhibition exhibiting additive effects on SG assembly, without additive impacts on translation repression94. ATP-dependent eIF4A binding to RNA also inhibits SG assembly and SG-PB interactions94. In contrast, eIF4A inhibition correlates with specific RNAs and G3BP1 concentrating more strongly in arsenite-induced SGs during both assembly and clearance post-stress94,97. These findings are consistent with eIF4A limiting SG assembly, and/or aiding clearance, though the latter point remains to be directly examined.
eIF4A is a curiously abundant protein in cells (Top 1%; ~10–100 fold excess of other eIF4F factors)98,99, with a 3–5 fold excess over other SG-localized helicases, and 5–50 copies per mRNA in human cell models94. Thus, it appears well suited to an “RNA chaperone” role94. While eIF4A binds and is activated by eIF4G and eIF4B100, indicating a targeting to mRNAs, the scope of eIF4A specificity and regulation remains unclear, as eIF4A lacks substantial helicase-domain flanking sequence that typically dictate interactions, localization, and regulation of other RNA helicases101. Thus, eIF4A may also act somewhat non-specifically as an “RNA disaggregase”102.
Ded1/DDX3:
Ded1 (yeast)/DDX3 (human) is another SG-localizing DEAD-box helicase best known for unwinding 5’UTR structure during translation initiation. Several studies (with one exception103) suggest that Ded1/DDX3 significantly impacts both SG assembly and clearance, albeit via distinct activities. Regarding assembly, overexpression of WT Ded191,104 and DDX392 drives SG assembly in the absence of stress, correlating with reductions in translation rate at the single cell level. This depends not on Ded1/DDX3’s helicase or ATPase activity, but rather interactions with eIF4F factors, resulting in formation of translationally stalled mRNPs91,105,106. However, Ded1/DDX3 ATPase mutants, or DDX3 depletion, also induce SGs in the absence of stress91,105, possibly reflecting a combination of impaired translation and reduced SG clearance (see below). Ded1/DDX3’s IDR domain is also required to form condensates, localize in, and induce SGs in the absence of stress when overexpressed92,104,105.
Regarding clearance, ATPase-deficient Ded1 exhibits slower clearance of sodium azide-induced SGs in yeast following cycloheximide treatment88, suggesting DDX3 ATPase/helicase activities facilitate SG clearance. Finally, several DDX3 helicase domain mutations are associated with medulloblastoma and intellectual disability. These mutations inhibit helicase activity, scanning and translation of structured 5’UTR mRNAs, and induce SG-like foci in various models92,107,108.
Dhh1/DDX6:
Dhh1 (yeast)/DDX6 (human) is a highly studied helicase which promotes translation repression and mRNA decapping109,110, and is a key marker and assembly protein for PBs89,111–113. Dhh1 mutants defective in RNA or ATP binding impair PB assembly89,114. In contrast, Dhh1 ATPase mutants, or mutations disrupting Dhh1 binding with Not1 (Dhh1/DDX6 ATPase activator)115, exhibit constitutive PBs, increased RNA binding, and strongly impair PB clearance89,114. Dhh1 ATPase mutants additionally become trapped in PBs unlike WT Dhh1, which cycles in and out of PBs with a half-life of ~30s110. Curiously, in human iPSC cells, catalytically dead DDX6 expression blocks PBs116; why similar DDX6 and Dhh1 mutations should oppositely affect PB assembly is unclear but may reflect differences in model systems and approach. Properties of Dhh1/DDX6 that may aid PB assembly include its high RNA binding affinity (Kd 1–2 nM)114,117, its significant stoichiometric excess to mRNA (~7-fold117; Top 10% protein by abundance98) and ability to oligomerize117. Thus, besides promoting translation repression, Dhh1 may drive PB assembly by interacting with RNA and forming oligomeric scaffolds, whereas ATP hydrolysis likely facilitates PB clearance by reducing Dhh1 RNA binding.
Altered Dhh1-PB protein interactions may also occur due to ATP and RNA binding and ATPase-driven conformational changes that impact PB clearance. For instance, DDX6 binding to Edc3 or Pat1 (other PB assembly proteins) is disrupted upon CNOT1 binding118. An ATPase mutant Xp54 (Xenopus Dhh1) also interacts with distinct PB-associated proteins versus WT119. Active disruption by Dhh1/DDX6 of RNA-RNA and RNA-protein interactions within PBs is another possible clearance mechanism, albeit direct detection of Dhh1/DDX6 helicase activities remain somewhat controversial114,117,120.
DDX6 progressively re-localizes from PBs to SGs during arsenite stress and may facilitate a PB-SG maturation process121–123. Indeed, DDX6 KO cells, or cells rescued with a DDX6 ATPase mutant, or depleted of CNOT1, all exhibit unusual PB-SG hybrid granules, suggest DDX6 promotes both PB clearance and separation of SGs from PBs123, perhaps by facilitating release or remodeling of PB mRNPs into SG mRNP-like states.
DHX36/RHAU:
This human helicase binds G-quadruplex containing mRNAs (rG4s) which are enriched in SGs124, and promotes translation and decay of such mRNAs125. DHX36 localizes to SGs following many stresses47,86,87, and it’s depletion induces SG assembly in the absence of stress87, and increases SG assembly following arsenite. This may reflect an accumulation of non-translating rG4 RNAs as SGs “seeds”, an observed increase in eIF2α phosphorylation, or both125. A role for DHX36 in promoting rG4 mRNA exit from SGs, and thus clearance, is possible but currently unknown.
“DNA” helicases in SGs:
Three DNA helicase complexes that facilitate DNA unwinding and chromatin remodeling for DNA replication, repair and transcription purposes can also regulate SG dynamics. Minichromosome maintenance helicase (MCM)126 and RuvB-like helicase (RVB)127 localize in yeast and human SGs34, and inhibit SG clearance following sodium azide (yeast) or arsenite stress (human cells). MCM and RVB are not known to exert RNA helicase activity, and with DNA not known to be a SG component, their mode of action remains mysterious. Finally, Bloom’s syndrome protein128 can bind and unwind both DNA quadruplex (dG4) and rG4 sequences, localize in human SGs under many stresses, and inhibit SG formation via a proposed mechanism similar to that of DHX3687,129.
RNA Helicase function and regulation:
Despite similar functionality, eIF4A antagonizes SG assembly, DDX6 promotes PB assembly, and Ded1/DDX3 can promote both assembly and clearance of SG assembly depending on expression levels. These distinct behaviors may reflect a combination of protein interactors, RNA binding affinity, abundance, and regulation of ATPase activity, some or all of which may vary within and outside of SGs and PBs. For example, Dhh1 ATPase activity is significantly lower than Ded1 or eIF4A in vitro, due in part to inhibitory intramolecular interactions114. Thus, Dhh1 may dissociate from mRNA less often than eIF4A or Ded1, thus predisposing it to maintaining PB assembly. Ded1 condensate formation limits its helicase activity and may aid sequestration of translationally repressed mRNAs with structured 5’UTRs in SG130. Finally, all the aforementioned helicases undergo many post-translational modifications (PTMs; see Biogrid/Uniprot), though very few are characterized. One exception is eIF4A Thr164 phosphorylation by cyclin dependent kinase A, which blocks RNA helicase activity, possibly by perturbing RNA binding131. Further characterization of helicase regulatory mechanisms, and examination of helicase properties within and outside of SGs and PBs is an important area of future study.
POST TRANSLATIONAL MODIFICATIONS AS REGULATORS OF SG AND PB CLEARANCE
Numerous PTMs on proteins besides helicases impact clearance and assembly of both SGs and PBs. During stress, the speed and reversibility of most PTMs offers obvious benefits to altering SGs and PBs in such a way that may aid cell survival134. Below, I focus on phosphorylation and ubiquitination events that impact SG and PB clearance, though other modifications (summarized in Table S1) and discussed elsewhere135,136 also play important roles.
PHOSPHORYLATION
eIF2α phosphorylation and SG-associated translation:
Following many stresses, eIF2α phosphorylation by one of four stress-induced kinases in human cells (GCN4, HRI, PERK, PKR) limits translation initiation as part of the integrated stress response. This reduces Met-tRNA ternary complex levels, and thus drives SG and PB assembly by increasing the non-translating mRNP pool137. Recently, other eIF2α stress-inducible kinases have been proposed including MARK2, which phosphorylates eIF2α in response to cytoplasmic protein misfolding sensed by a PKC-Hsp90-dependent mechanism138, and FAM69C, which responds to both HS and arsenite stress, and drives SG assembly in microglia139.
SGs were originally proposed to exclusively harbor translationally repressed mRNPs, but single molecule mRNA translation studies have modified this view. While most SG-localized mRNAs are non-translating, mRNA reporters with 5’UTRs of genes that are both translationally stimulated (ATF4) and repressed (RPL32) during stress can accumulate and translate in SGs140. For the ATF4 reporter, translation frequency of SG-associated transcripts approached 30%, with elongation rates similar to non-SG-localized ATF4 transcripts. While most mRNA reporter transcripts localized close to SG outer edges, consistent with other studies22, no significant effect of an mRNA’s localization on or deep within a SG impacted the likelihood of translation140. PBs showed no interaction with the ATF4 reporter, and thus may be more translationally silent than SGs, though studies with other mRNAs, or sensitive spatial translation assays, are required for more certainty.
Translation of mRNAs localized in SGs or PBs could rapidly facilitate their extraction from either granule, and aid subsequent SG/PB clearance. Despite contrary initial data29, phosphorylated eIF2α (and perhaps eIF2B) can accumulate in SGs during stress and stress recovery34,68,141,142. Once eIF2α is dephosphorylated, translation on SG-associated mRNAs might proceed rapidly given the high local concentration of eIFs and 40S subunits. Specific RNA helicases are involved in all steps of translation143,144, particularly initiation (e.g., eIF4A, Ded1/DDX3), which could disrupt mRNA-mRNA and mRNA-protein interactions sustaining SG assembly. Elongating ribosomes also display potent helicase activity145,146. However, whether translation itself actively disassembles SGs, or occurs after SG clearance remains unclear. Consistent with the second possibility, single molecule translation studies with an mRNA reporter (KDM5B) in U2-OS cells suggest that SGs largely undergo clearance a few minutes prior to detection of distinct translation activity. However, transiently SG-localized mRNAs undergoing low levels of translation may also have simply escaped detection22.
Phosphatases:
If translation itself is a key SG disassembling force, and not a downstream consequence, then eIF2α de-phosphorylation could be key to SG clearance. Notably, PP1 phosphatase acts on phospho-eIF2α147, and treatment with PP1 inhibitors slows trehalose-stimulated SG clearance following arsenite stress148. Additionally, chronic MG132 treatment increases PP1 subunit levels, which limits phospho-eIF2α accumulation during subsequent stress. This in turn inhibits SG assembly, and speeds SG clearance149. Such preconditioning is distinct from that involving Hsp70 accumulation (also following MG132 treatment)10, and thus may occur simultaneously to limit SG assembly. PP1 subunits also localize in arsenite-induced SGs150. Generally, though, little is known about whether PP1, and the activity of other SG-localizing phosphatases34,47,86 impacts SG clearance via eIF2α de-phosphorylation, and/or by targeting other phosphorylation substrates. Given the many phosphorylation events that impact SGs, this is an important area of exploration.
DYRK3:
Inhibitors, knockdown and catalytic mutants of dual specificity tyrosine-phosphorylation-regulated kinase 3 (DYRK3) impair SG clearance in various stress and cell line contexts35. An IDR in DYRK3’s N-terminus drives SG localization under stress. While phosphotargets and DYRK3 interaction partners were identified, some of which localize in SGs35,151, it remains unclear which DYRK3 phosphorylation events aid SG clearance. DYRK3-facilitated SG clearance involves regulation by the chaperone Hsp90, inhibition of which delays SG clearance in various stress and cell line contexts. Hsp90 binds to and stabilizes DYRK3, and the absence of Hsp90 leads to proteasomal-mediated degradation of DYRK363. Finally, DYRK3 activity also limits formation of SGs, but not PBs, and some nuclear biomolecular condensates (e.g., splicing speckles) but not others (e.g., Cajal bodies). All of these condensates clear during mitosis when DYRK3 abundance relative to substrates is maximal151.
CDK:
In yeast, Cdc28, the cyclin dependent kinase (CDK) that governs cell cycle regulation, localizes in SGs, and aids SG clearance following release from a joint glucose deprivation and HS stress152. CDKs localize in SGs in other systems (including human CDK1, CDK2, CDK434,152, and CDKA1 in plants153), and CDK2 and CDK4 inhibitors strongly impair SG clearance following arsenite stress in HeLa cells35. Additionally, yeast and human SG clearance rates are slower in G1 phase, when CDK activity is lower, than in S, G2 or early M-phase when CDK activity is higher152. Many SG-localized proteins are Cdc28 phosphorylation targets, but no specific target is known that explains Cdc28/CDK-mediated SG clearance effects.
Syk:
Expression, localization within SGs and the catalytic activity of the tyrosine kinase Syk facilitates clearance of MG132-induced SGs in MCF7 cells154. Syk-dependent phospho-tyrosine modified proteins also accumulate within SGs under these conditions. SG clearance correlates with a decrease in eIF2α phosphorylation levels, and an apparent increase in autophagosome levels, suggesting Syk could aid SG clearance both by restoring translation and aiding granulophagy. Indeed, blocking autophagy suppresses Syk-mediated effects on SG clearance154.
Focal Adhesion Kinase (FAK):
P19 carcinoma cells show some reliance on FAK kinase activity for HS-induced SG clearance155, which localizes in SGs along with an mRNA-binding protein Grb7, a FAK substrate. Blocking Grb7 phosphorylation by FAK impairs SG clearance following HS. Based on in vivo and in vitro binding assays, it was proposed that Grb7 direct binding to HuR, another SG-localizing protein, is disrupted by FAK-mediated phosphorylation, thus underpinning FAK’s SG clearance effect155.
UNC-51 like autophagy activating kinase 1/2 (ULK1/2):
ULK1 and 2 regulate macroautophagy initiation, but also localize within HS-induced SGs, and interact with several SG localized proteins, including VCP156. Inhibiting ULK1/2 slows SG clearance following transient HS and arsenite stress in various cell lines, whereas ULK1/2 stimulation strongly increases HS-induced SG clearance. These effects are autophagy independent, and instead rely on ULK1/2 stress-induced phosphorylation and activation of VCP ATPase activity, which enhances SG clearance156.
PB regulatory kinases:
Fewer examples of regulation of PB clearance and assembly by kinases are known. In yeast, enhanced PKA kinase activity, using a Ras2 constitutive allele or PKA overexpression, promotes clearance of PBs induced by glucose deprivation, in a manner dependent on phosphorylation of Pat1 (a PB assembly and decapping factor) at S456 and S457157. Elevated PKA activity also limits PB assembly under many other PB-inducing stresses but does not impact SGs157.
In human cells, JNK kinase has been linked to contrasting effects on both PB assembly and clearance that depend on the substrate and PB-inducing stimulus. Following arsenite stress, but not other stresses, the human PB assembly factor and eIF4E binding protein, 4E-T, is phosphorylated at 6 serine residues by JNK, which also localizes in arsenite-induced PBs. Such phosphorylation facilitates assembly of larger PBs and possibly 4E-T self-association158. JNK also binds and phosphorylates Dcp1a at S315, with expression of phosphomimetic or phosphonull S315 alleles of Dcp1a strongly reducing or increasing PB levels respectively159. Dcp1a is hyperphosphorylated during mitosis, including at the S315 site, which coincides with PB clearance160. Finally, in various cancer cell models, based on inhibitor and phosphomimetic/null mutations, Pim kinase 1 and 3 phosphorylate Edc3 at S161, which prevents its localization in PBs and limits PB assembly, possibly via sequestration of other PB-assembly factors161.
UBIQUITINATION
Ubiquitination regulates SG clearance:
Most studies, primarily in human cells, indicate an important role for ubiquitination in SG clearance56,162. Ubiquitin (Ub) can localize in SGs to varying degrees dependent on the stress applied31,37,40,56,58,67,163–166. Free mono-Ub and non-conjugated Ub chains163,165, as well as Ub-conjugated SG proteins, featuring various types of linkage-specific forms of Ub, have been reported56,163. Since distinct Ub-linkage types often specify different outcomes (e.g., K48-linked chains favor proteasomal degradation, K63-linked chains favor autophagy167,168), it is noteworthy that different stresses also lead to varying degrees and specificities of Ub-linkages in SGs. HS generally results in a stronger Ub signal within SGs than arsenite stress. Variable levels of K48 and K63 SG-localized Ub signals have been reported in HS, whereas K63 is generally more abundant in arsenite-induced SGs31,37,56,163,164.
A role for ubiquitination in SG clearance is further suggested by inhibition of the E1 enzyme UBA1, which blocks all ubiquitination events in cells. Specifically, three studies using HeLa, HEK293T and iPSC-derived neuronal cell models exhibited significantly impaired SG clearance following HS56,164 or arsenite stress56, though another study using similar methods reported no significant effects165. Differences here could partly reflect the timing of E1 inhibition and the duration of effective inhibitor action56. Finally, chemical inhibition of all deubiquitination events in cells also impairs clearance of arsenite-induced SGs, with a lesser effect on HS SGs56, suggesting SG ubiquitination alone is not always pro-clearance, and instead plays a complex and stress-specific role.
SG ubiquitination substrates:
A key step in deciphering how ubiquitination impacts SG clearance is identifying ubiquitinated SG substrates, and the E3 ligases and deubiquitinase enzymes that regulate such modifications. An unbiased screen of ubiquitination changes in HEK293T cells following different stresses revealed significant ubiquitination of many mRNP proteins and known SG components, particularly following HS stress (less so arsenite). One SG-localized protein that accumulates Ub modifications is the well described SG assembly protein G3BP1 (see later, and Box 2). Specifically, ubiquitination sites in the NTF2L dimerization domain, and the RNA-binding RRM1 domain of G3BP1, were identified through K-R mutagenesis as being ubiquitination sites that aid SG clearance following HS stress37. NTF2L ubiquitination also aided G3BP1 interaction with and recruitment of VCP to SGs, suggesting that VCP may remove G3BP1 from SGs. Alternatively, ubiquitination of G3BP1’s NTF2L and RRM domains could simply impair interactions (e.g., dimerization, Caprin1 binding, RNA binding) that sustain SG assembly.
BOX 2: G3BP1/2 AND SGs: MORE THAN JUST A SCAFFOLD PROTEIN?
G3BP1/2 RNA binding, protein binding and dimerization activities facilitate SG assembly217. However, other reported G3BP1 functions could impact SG clearance, but have not yet been investigated in this context. G3BP1 seemingly harbors an as yet unmapped endonuclease activity that degrades the c-myc 3’UTR in vitro218,219. G3BP1 also binds and promotes degradation of circular RNAs and mRNAs with highly structured 3’UTRs, in combination with the RNA helicase and nonsense-mediated decay factor, Upf1220. G3BP1 RNA binding domains and S149 phosphorylation are required for these activities. Naturally, an endonuclease activity could clear SGs by targeting SG transcripts for decay. Conversely, G3BP1/2 mRNA binding is also linked to increased mRNA stability221,222. Whether S149 phosphostatus, or another means of regulation govern these opposite outcomes remains unclear.
G3BP1 interaction with USP10 reportedly stabilizes levels of both proteins172,178, but also may inhibit USP10 deubiquitinase activity223,224. Thus, beyond USP10 limiting effects on SG network valency172,197,198, G3BP1-USP10 interaction, or USP10 inhibition by other means, could preserve the ubiquitinated status of several USP10 SG substrates, thus impacting VCP, proteasomal or granulophagy clearance mechanisms. Conversely, interaction of the yeast homologs of G3BP and USP10, namely Bre5 and Ubp3, stimulates Ubp3 deubiquitinase activity225. Absence of Ubp3 deubiquitinase activity, or deletion of either Bre5 or Ubp3, blocks SG assembly in yeast under chronic nutrient deprivation226. Why USP10 and Ubp3 regulation by G3BP and Bre5 appear opposite, whether USP10 deubiquitinase activity and levels are coordinated by G3BP1/2 or otherwise227, and the importance of USP10 deubiquitinase to SG clearance remains unclear.
A DNA and RNA helicase activity for G3BP1 has been reported in vitro228 which could impact SG clearance if functional in vivo. Specifically, a helicase isolated from HeLa cell nuclear fractionation that required ATP and Mg2+, bound ATP, and which could unwind DNA, RNA or RNA/DNA duplexes was identified as G3BP1. No subsequent study has validated this property in vivo, nor identified a putative helicase domain.
Finally, as partly detailed elsewhere217, G3BP1 has been implicated in RasGAP signaling (though this appears discredited229,230), mTOR signaling231, ribosome quality control178, regulation of mRNA translation232 and stability220–222, all of which could directly or indirectly impact SG dynamics. While roles in SG assembly seem generally similar for G3BP1 and its paralog G3BP2, the latter does bind distinct RNAs, and exhibits tissue-specific expression. However, a clear G3BP2-specific function remains generally elusive217,233.
Super-resolution microscopy indicates that K63 and K48 ubiquitination on HS-induced SGs primarily localize on the surface of SGs in the “shell” region, or in cavities directly adjacent to G3BP1 signal. VCP and the proteasome co-localize strongly with SG-associated K48 Ub signal, suggesting a possible role in proteasomal degradation of specific SG substrates56 (see later). Indeed, inhibition of VCP or the proteasome increases G3BP1 ubiquitination during HS stress recovery37.
E3 Ub ligases:
Several E3 Ub ligases localize in SGs based on IP-MS150 and proximity ligation data32,47,86, though none have been clearly shown to promote SG clearance. One candidate is TRIM21, which localizes in arsenite-induced SGs, and at least inhibits SG assembly169. TRIM21 ligates K63-linked Ub chains to G3BP1 during arsenite stress, without inducing obvious changes in G3BP1 abundance, suggesting a possible antagonism to SG-sustaining G3BP1 interactions. Consistent with this, K63-ubiquitinated G3BP1 undergoes LLPS less readily than non-ubiquitinated G3BP1 in vitro. However, TRIM21 and G3BP1 also show increased physical interaction with p62 and NDP52 selective autophagy receptors under arsenite stress. TRIM21 also interacts with core autophagic components (e.g., LC3B, ULK1, BECN1) under these same conditions. While suggestive, whether autophagic function is required for TRIM21-mediated effects on SG assembly or clearance remains unclear169.
Deubiquitinases:
Several deubiquitinase enzymes also localize in SGs. USP5 and USP13 localize to HS-induced SGs, but not SGs induced by other stresses163. Super resolution microscopy indicates localization to SG shells that only partially overlaps with core SG maker proteins (PABP1, G3BP1). Knockdown of either USP5 or USP13 increases Ub accumulation within HS-induced SGs, accelerates their assembly, and slows clearance in a manner dependent on USP5/13 catalytic activity163. USP5 preferentially cleaves free Ub-chains, whereas USP13 acts on protein-conjugated Ub-chains, suggesting that turnover of both types of Ub chain facilitate SG clearance, possibly by disrupting SG protein interactions that rely on Ub-binding.
Ubp3 (S. cerevisiae and S. pombe)/USP10 (human) is another deubiquitinase that localizes to SGs under many stress conditions170–174, though it’s impacts on SGs apparently differ between species. Specifically, Ubp3 has no impact on SG formation in S. pombe173, but is required, with its cofactor Bre5 (G3BP1 homolog), for SG assembly in S. cerevisiae in a manner reliant on its deubiquitinase activity171. In human cells, USP10 has been reported to both stimulate174, in a deubiquitinase-independent manner, and inhibit172 SG assembly, with reduced G3BP1 dimerization and/or Caprin1 interactions suggested as a possible SG-inhibitory mechanism. Specific Ubp3/USP10 targets in SGs and impacts on SG clearance remain unclear. However, amongst several known substrates and functions175, ribosomal protein ubiquitination is regulated by Bre5/G3BP1 and Ubp3/USP10 in yeast and human cells under conditions of translational stalling and starvation, which regulates ribophagy176–178. Autophagy itself is also stimulated by USP10-mediated deubiquitination and subsequent maintenance of Beclin levels, and thus activity of the Vps34 PI3K complex179. Thus, G3BP1’s role as deubiquitinase co-factor may play unappreciated roles in regulating SG clearance (see Box 2).
PBs and Ubiquitination:
Little is known about PBs and Ubiquitination. However, Ub knockdown or blocking K63 Ub chain formation prevents cytokine-induced Dcp1a phosphorylation by JNK and subsequent PB assembly180. The E3 Ub ligase TRAF6 was identified as a Dcp1a binder whose expression maintains levels of other PB proteins (EDC3, XRN1 and DCP2); indeed, TRAF6 KO cells lack PBs entirely. Dcp1a is heavily ubiquitinated by K63 and K29 modifications. However, despite binding Dcp1a, TRAF6 KD does not impact Dcp1a ubiquitination180, suggesting another E3 ligase is involved, possibly regulated by TRAF6.
CLEARANCE OF SGs AND PBs BY GRANULOPHAGY
Autophagy, or more precisely “macroautophagy”, involves sequestration of often large, insoluble substrates (e.g., organelles, protein aggregates) in autophagosomes, which traffic to and fuse with acidic degradative organelles (lysosomes in metazoa, vacuoles in yeast/plants). Here, autophagosome contents are degraded and recycled. Autophagy can act selectively or non-selectively, with the former generally defined and regulated by specific autophagic “receptor” proteins binding to specific “cargos” (i.e., substrate proteins). Autophagic receptor proteins then recruit Atg8, a key autophagosome assembly protein, and additional core autophagy machinery. Several selective autophagic pathways are known that clear damaged or deleterious substrates181.
“Granulophagy” refers to the selective autophagic clearance of SGs and PBs in eukaryotic cells55, though SGs are more studied. Granulophagy’s role in SG clearance seems to vary depending on stress and cellular context36,37,40,55,57,148,154,182,183. In yeast, constitutive SGs accumulate at low levels following autophagy inhibition, with detection of SG protein degradation products in vacuoles under chronic but not transient stress (55 and our unpublished data). In human cells, SGs also accumulate at low levels in the absence of stress following autophagy blocks55. Bafilomycin treatment (lysosomal inhibitor) significantly increases SG accumulation following coxsackievirus A16 (CA16) infection183, and slows SG clearance following prolonged (90min) periods of HS stress37. In some studies, impairing autophagy significantly slows SG clearance following proteasomal inhibition154 or arsenite stress57,184, but not others33,156. Such differences could reflect distinct cell models, inhibitor usage (drug and/or dose), or distinct SG quantification approaches33,57,154,156.
Granulophagy receptor candidates:
Various autophagic receptors may aid SG and PB clearance in a manner dependent on the composition and dynamic state of SGs. p62 and NDP52 both localize with SGs under arsenite stress in a manner dependent on their Ub-binding domains, and physically interact with G3BP1169. p62 also increasingly localizes over time in SGs chronically induced via an optogenetic-based G3BP1-driven assembly mechanism in U2-OS and iPSC-derived neurons; such co-localization correlates with SGs becoming less dynamic185. Knockdown of p62 and NDP52 impairs SG clearance following arsenite stress169,182. p62 knockdown also increases SG levels following CA16 viral infection, with p62 Ub-binding domain mutants exhibiting a similar effect183.
NDP52 may be a preferential PB granulophagy receptor as PB levels under non-stress conditions are significantly increased following NDP52 knockdown. NDP52 also localizes in PBs to a greater extent than p62182. NDP52 binds Pat1 and facilitates autophagy-dependent clearance of PBs following Kaposi’s sarcoma associated herpesvirus (KSHV) infection186,187.
Recently, the chaperonin subunit CCT2 was identified as a non-Ub binding autophagy receptor that preferentially acts on protein aggregates with very low dynamics188. CCT2 localizes in SGs34, and thus may harbor a SG clearance role involving granulophagy189.
Granulophagy cargo candidates:
Several SG proteins have emerged as putative granulophagy cargos, whose utilization and importance may also be context specific. Histone Deacetylase 6 (HDAC6) interacts with p62 during CA16 viral infection in a manner stimulated by HDAC6’s Ub binding domain (UBD), which also drives SG poly-Ub enrichment in CA16 infected cells183. HDAC6 UBDΔ cells also exhibit higher SG levels following CA16 infection.
C9orf72 is another reported p62 interactor and SG and PB-localizing protein36,190, repeat expansion mutations in which are associated with ALS191. KD of C9ORF72 impairs SG clearance following arsenite stress36 to a similar degree as p62 KD, which could reflect a p62 SG recruitment function, a role for C9ORF72 in autophagic flux192–195, or other novel functions.
G3BP1 is another putative granulophagy cargo given its critical SG assembly role, K63-ubiquitination status under HS stress37, p62-interaction, and ubiquitination by TRIM21169. However, no study has clearly shown that G3BP1-p62 interaction, or G3BP ubiquitination events are required for granulophagy.
SG/PB physical juxtaposition with autophagic machinery:
It is unclear if SGs and PBs are enveloped whole by autophagosomes, or whether partial fragmentation of the granule is required; both processes may also occur. Autophagosomes can be large enough (0.5–1.5 μm)196 for engulfment of most SG and PBs (0.1–2 μm)1. Indeed, LC3 (Atg8) foci co-localization with SGs has been commonly observed36,182–184, with super resolution microscopy suggesting p62, NDP52 and LC3 appear on the surface of some arsenite induced SGs36,169. Furthermore, FUS (Fused in Sarcoma; an ALS-associated SG-localizing protein) and p62 labelled structures presumed to be SGs show evidence by electron microscopy of autophagosome engulfment36.
PROTEASOMAL-BASED CLEARANCE OF SGs
The proteasome, which localizes in both human58 and yeast SGs150, is implicated in regulation of SG and PB dynamics. Specifically, in human cells, proteasomal activity aids SG clearance in a stress-dependent manner (e.g., arsenite-induced SGs strongly reliant; HS-induced SGs moderately reliant; sorbitol-induced SGs not reliant)56,58. A key factor in proteasome-based SG clearance is ZFAND1, which drives proteasome localization to SGs and their clearance following arsenite stress58. Proteasomal inhibition also induces SGs, likely due to accumulation of misfolded proteins with SG-seeding potential10,33,38. Unlike SGs, proteasomal inhibition reduces PB levels by an unknown mechanism10. Currently, no specific substrates of proteasomal activity in SGs critical for clearance are known.
CLEARANCE VIA ALTERATIONS IN THE NETWORK STRENGTH OF SGs AND PBs
SGs and PBs are sustained by a network of protein-protein, protein-RNA and RNA-RNA interactions. Given this, certain proteins or RNAs may serve roles as core interaction “nodes” that sustain part or all of the condensate network via a combination of possessing a high valency, high affinity interactions, high local concentration, and favorable entry/exit dynamics. Molecules that “bridge” nodes may connect subnetworks together, helping sustain a larger network, or underpin interactions of distinct condensates such as SGs and PBs. Targeting nodes and bridges for degradation or inhibiting their interactions could therefore efficiently promote SG and PB clearance. Disrupting SG and PB networks with incorporation of molecules that “cap” and thus reduce the network interconnectivity is another possible clearance mechanism. Recent publications focusing on G3BP1 and 2197–199 have driven a focus on these concepts and are excellently reviewed here5. Below, only known, and putative examples of SG network disruption that may promote clearance are discussed, as knowledge for PBs is lacking, though similar concepts likely apply.
G3BP1/2 – a regulated central node sustaining SG networks:
Characterized SG and PB assembly proteins often exhibit high valency and harbor some or all of the following5: dimerization or oligomerization domains, IDRs, and RNA binding domains. Possessing all of these features, G3BP1 and 2 represent the most important known human SG assembly proteins, being essential for SG assembly under many (e.g., arsenite, thapsigargin, eIF4A inhibition172,200) but not all tested stresses (e.g., HS, sorbitol)198. G3BP1/2 exhibit domain homology and redundancy in rescuing SG assembly in G3BP1/2ΔΔ backgrounds when expressed ectopically, with each harboring an NTF2L dimerization domain, an RRM RNA binding domain, and 3 IDRs, the 3rd of which also harbors RNA binding activity. G3BP1 NTF2L and RNA binding domains are essential to SG assembly197–199.
G3BP1 exists in either a closed or open state, the latter of which promotes SG assembly. In the closed state, IDR1-IDR3 bind via electrostatic interactions that limit RNA binding. In response to long relatively unstructured RNAs, which accumulate during stress-induced polysome collapse, G3BP1 adopts an open state, allowing the IDR3 and RRM domains to bind RNA and promote SG assembly198,199. In vitro and in vivo data with phosphomutant alleles suggests IDR1 phosphorylation, particularly at S149, favors IDR1-IDR3 interaction, and thus impedes SG assembly by reducing RNA binding and RNA-induced condensate formation198.
Whether G3BP S149 undergoes stress-induced changes in phosphorylation is controversial200–202. One study demonstrated that Casein Kinase 2 localizes in SGs following arsenite stress and phosphorylates G3BP1 at S149 in vitro and in vivo. G3BP1 S149 exhibited lower phosphorylation levels during arsenite stress versus unstressed cells, with phosphorylation rebounding during recovery202. This supports a proposed “tunable” switch model of RNA-mediated condensation of G3BP1, and subsequent SG assembly198,199.
Regardless, PTM of G3BP1 (also including acetylation203, methylation204,205 and ubiquitination37) offers many means to facilitate SG clearance by altering G3BP1 RNA binding and RNA-induced condensation, disrupting interactions with other proteins (including itself), degrading, or extracting G3BP1 from SGs. All of these possibilities could reduce the strength of the SG interaction network, thus promoting clearance.
G3BP1/2 binding partners that disrupt SG networks:
Distinct protein interactions with G3BP1/2 may also alter SG network strength and favor assembly or clearance. The NTF2L domain not only aids G3BP1/2 dimerization, but also binds proteins that positively (e.g., Caprin1, UBAP2L) or negatively (e.g., USP10) impact SG formation, potentially by adding additional RNA or protein binding valency to G3BP1/2 complexes, thus potentially connecting (Caprin1, UBAP2L) or blocking (USP10) G3BP1/2 interactions with other SG subnetworks5,197–199. Caprin1 and USP10 bind competitively to the NTF2L domain172,206, without disrupting G3BP1 dimerization207,208. USP10 overexpression blocks SG assembly172,208, and is hypothesized to act as a capping protein that limits G3BP1/2 interaction valency in the SG network5,197.
The viral nonstructural protein 3 (nsP3) of Semliki Forest virus binds at the same site as USP10 via a pair of FGDF motifs in its C-terminus, sequestering G3BP1/2 into viral replication centers and aiding viral replication by disrupting SG assembly209. USP10 harbors its own single FGDF motif, and over-expression of either USP10 or nsP3 blocks SG assembly following multiple stresses in an FGDF-dependent manner208, likely by blocking G3BP1 interactions with other SG-promoting proteins (e.g., Caprin1, UBAP2L). Recently, small molecule FGDF-peptide mimics have been developed that prevent in vitro condensation of G3BP1, RNA and Caprin1, block assembly of arsenite and HS-induced SGs, and readily clear already formed SGs induced by various stress and mutant stimuli210.
General protein-mediated disruption of SG networks:
YB-1 may disrupt SG networks by targeting SG interactions involving RNA. YB-1 is a highly abundant multimer-forming protein that strongly localizes in SGs211, and for which contrasting effects on SG assembly have been reported166,212–214. Regardless, YB-1 preferentially binds non-translating mRNA, can disrupt TIA1-mRNA aggregates in vitro, and at endogenous levels, YB-1 facilitates translation and SG clearance following arsenite stress211,215. YB-1 SG clearance effects rely at least partly on it’s cold-shock domain that disrupts RNA structure in an ATP-independent manner215,216. Thus, like eIF4A, it has been described as an RNA chaperone211. SG network strength may also be disrupted by broader compositional changes during recovery. Following arsenite stress, SG recovery enriched proteins generally harbor fewer IDR domains, and exhibit lower phase separation potential than SG proteins localized during assembly32.
RNA-BASED REGULATION OF SG AND PB CLEARANCE
RNA is an essential component of the networks underlying SG and PB formation. This reflects a combinatorial effect of RNA-protein and intermolecular RNA-RNA interactions. Supporting this, trapping mRNA in polysomes with cycloheximide blocks assembly and facilitates clearance of both granule types (see earlier), whereas introducing an excess of non-translating mRNA (or ssDNA) induces SGs166. Interestingly, expression of RNAse L, a viral-induced ssRNA endonuclease, strongly reduces SGs in vivo, but does not impact PBs234. Whether this indicates RNAse L accessibility issues to PB RNAs, or that PBs, once assembled, are not reliant on RNA to sustain them, is unclear. Paradoxically, in vitro, isolated PBs are fully cleared by RNAse A treatment16, whereas SGs are RNAse resistant34. Regardless, known, and putative examples of how RNAs impact SG and PB assembly and thus potentially clearance are discussed below.
RNA nodes in SG and PB networks?
Whilst the lncRNA NEAT1 provides a clear example of an RNA scaffold driving formation of a biomolecular condensate (paraspeckles)235,236, there is no known singular RNA that drives SG assembly. This is perhaps unsurprising given the number of RNA molecules that localize in and presumably drive SG assembly redundantly (e.g., approximately 42,000 SG-localized RNAs in each U2-OS cell under arsenite stress124). However, certain RNA molecules could be more important than others, as variables such as length, lack of structure, low levels of translation, and binding sites for SG-enriched proteins all predispose RNAs to SG enrichment124. Recently, snoRNAs have been proposed to help bridge G3BP1 and UBAP2L interactions and aid SG assembly, based on UBAP2L RNA-IP and sequencing, snoRNA KDs, and a reduced UBAP2L-G3BP1 IP interaction with RNAse present237. This contrasts with an absence of detectable snoRNAs in SGs124, and a robust RNase-insensitive G3BP1-UBAP2L IP interaction described elsewhere197.
In yeast, under non-stress conditions, the RPS28B mRNA is required for PB assembly, and facilities PB assembly under stress238. This reflects a role for the long 3’UTR of RPS28B which binds Edc3 (enhancer of decapping 3; a yeast PB assembly factor) and Dhh1, possibly acting as a nucleating PB scaffold, and a 3’UTR-mediated establishment of an Edc3-Rsp28b protein interaction that also aids PB assembly by an unknown mechanism238.
Impact of mRNA modifications.
mRNAs can undergo numerous modifications due to the activity of “writer” enzymes. “Readers” bind these modified bases and may confer altered regulation of modified mRNAs, while “erasers” remove mRNA modifications. Many writer, reader and eraser enzymes localize in SGs and PBs based on compositional and microscopy datasets34,75,239–242, though in most cases the significance of this is unknown.
m6A, which is enriched in 3’UTRs and near stop codons, and occurs on 0.1–1.8% of A bases (dependent on context, study)243, may impact SG and PB composition and dynamics, though findings remain controversial. First, several labs using FISH and SG RNA-seq approaches sensitive to methylation observe that m6A-modified RNAs are enriched in SGs in various cell and stress contexts239,240,242; analogous findings have been made for m7G244 and m1A245-modifeid RNA. In contrast, another study argued mRNAs with multiple m6A modification sites show no SG enrichment246. Second, KO or depletion of the m6A writer (METTL3/14) enzyme does not impact SG assembly239,240. Third, KD of three m6A reader proteins (YTHDF1, 2 and 3) singly or in combination impairs arsenite-induced SG assembly242, though another study saw no effect with YTHDF3 KD239, possibly reflective of use of a G3BP1 over-expression cell line model. How might m6A readers impact SG assembly, but the m6A modification be dispersible? One possibility is that YTHDF1, 2 and 3, perhaps via their IDRs, stimulate interactions aiding SG assembly246, though this remains untested. In contrast, triple KD of YTHDF1, 2 and 3 induces PBs, possibly related to effects of YTHDF proteins on mRNA decay, though preventing m6A modification itself again has no PB impacts247.
IP-MS approaches to identify RNA binding proteins influenced by m6A modification discovered that in certain sequence contexts, G3BP1/2 RNA binding is repelled by m6A, whereas other SG proteins (e.g., FMR, FXR1/2) and the PB-localized 5’–3’ Exonuclease XRN1, preferentially bind m6A221,248. >1000 G3BP1/2 RNA binding sites in 3’UTRs overlap with m6A sites, thus m6A-driven dissociation of G3BP1/2 mRNA binding could theoretically weaken SG networks and facilitate SG clearance.
Impact of RNA structure.
Intermolecular RNA-RNA interactions promote SG assembly, and extensively structured RNAs (e.g., tRNAs) are generally excluded from SGs124,249. However, specific intramolecular RNA secondary structures, particularly G-quadruplexes (rG4s), can impact SG mRNA targeting and SG dynamics. Transfection of rG4 RNAs promotes stress-independent SG assembly in a small fraction of U2-OS cells in an RNA length and eIF2α phosphorylation-dependent manner250. rG4 RNAs accumulate and are enriched within SGs251. rG4 RNA derived from a C9ORF72 repeat expansion is notably static in SGs250, in contrast to G3BP1/2, which along with other SG proteins (DDX3X, DHX36, FMRP) directly binds rG4 RNAs via their RRM and RGG domains222. Finally, pre-incubation of small molecules that bind rG4 structures slow arsenite-induced SG assembly, possibly due to impaired rG4 binding by G3BP1/2 and other SG proteins251. Thus, disrupting rG4 structures and interactions could theoretically be a SG clearance promoting mechanism.
RNA degradation as a clearance mechanism?
Since mRNA is an abundant, high-valency molecule sustaining SG and PB formation, a simple way to clear granules, besides mRNA exit, would be to promote mRNA decay. Given enrichment of mRNA decay factors in PBs, this seems at first glance a highly probable as a PB clearance mechanism, though whether mRNA decay occurs in PBs remains controversial.
Supporting PBs as mRNA decay sites, PB numbers and volume in yeast increase significantly following mutations in mRNA decapping factors (Dcp1/2) or Xrn1 (5’–3’ major cytoplasmic exonuclease)252. Similarly, Dcp2 KD in U2-OS cells increases PB numbers and increases PB accumulation of an mRNA reporter19. mRNAs stalled in decay due to strong secondary structures hindering Xrn1 progress also accumulate in PBs252. However, these results do not preclude decay having initiated outside of the PB. Finally, in vitro data suggests Dcp1/2 mRNA decapping activities are facilitated in condensates in the presence of Edc3253.
Other evidence argues against PBs as a site of mRNA decay. First, normal mRNA decay rates for various reporters are typically observed in models where visible PBs are genetically blocked254,255. Second, mRNA decay intermediates are not detected in isolated PB transcriptomes in cells with functioning mRNA decay75. Third, PB-localized mRNAs can return to translation during stress recovery21,256,257. Fourth, some mRNAs undergo co-translational decay in yeast258. Finally, single molecule analyses for individual reporters indicates mRNA decay does not occur in PBs257,259.
It remains possible that a subset of mRNAs degrade in PBs. Whether a sufficient fraction of the PB mRNA network is degraded to facilitate PB clearance, versus simply returned to translation, is unknown. In contrast, preferential targeting of mRNAs with a key PB scaffolding role (e.g., RPS28B in yeast238) could be an efficient means to stimulate PB clearance.
SGs also harbor both RNA exonucleolytic and endonucleolytic enzymes (e.g., Xrn1, Angiogenin, Eri1, and perhaps G3BP1/2; see Box 2), and the ribonuclease/angiogenin inhibitor RNH132,34,260, though no role for these factors in facilitating SG clearance via mRNA decay, targeted or otherwise, has been described.
RNA dynamics during SG and PB clearance.
The extent, rate and specificity with which mRNAs exit SGs and PBs during clearance, particularly following distinct stresses, remains largely unclear. Based on specific mRNA reporter studies, a significant fraction of SG and PB-localized transcripts can exit during stress recovery14,20–22,261. The dynamics of all poly(A) SG-localized mRNA in live cells has also been examined under arsenite stress, using injection of fluorescently-labelled poly(U) oligos. Interestingly, 1/3rd of mRNAs diffuse rapidly (half-life in SGs of 40s), 1/3rd diffuse slowly (half-life of 275s) and the remaining 1/3rd do not exchange at all25. Unfortunately, this method was not applied to the study of mRNA exit during SG or PB clearance, but it would be informative to do so. Recently, a study utilized RNA-seq of transient arsenite-induced SGs, and polysome-associated RNA following stress recovery to suggest that >95% of SG-associated mRNA re-enter translation, particularly if subject to m6A modification262. However, this work did not determine whether mRNAs entering translation post stress previously resided in SGs or were simply repressed elsewhere in the cytosol (or derived from nuclear export). Finally, single molecule mRNA reporter imaging combined with FRAP indicates that mRNA exchange dynamics in PBs decreases with stress (chronic nutrient deprivation), with a larger fraction becoming nearly immobile in PBs under stress versus non-stress conditions19.
Collectively, these data highlight a fundamental gap in understanding with clearance implications; namely, to what extent SG and PB mRNA populations exit and re-enter translation, versus other possible fates including targeted mRNA decay or degradation via granulophagy. It is often assumed that mRNAs exit SGs and PBs en masse, at least following transient stresses. As stated earlier, this usually correlates with translational recovery. However, given that the fraction of bulk cytoplasmic mRNA localization in SG ranges from 10–15% with RNA-seq124 or 5–50% with poly(A)-FISH studies263, translation recovery is likely driven mostly by non-SG/PB mRNAs being relieved of their repression. Closer study of this issue is warranted.
INDEPENDENCE AND INTERPLAY OF SG AND PB CLEARANCE MECHANISMS
Given the many SG and PB clearance mechanisms identified to date, it is useful to discern under what conditions one clearance mechanism is particularly favored over others, or whether several clearance pathways work redundantly or in combination. Evidence supporting all these scenarios currently exists.
Several preferential SG clearance mechanisms following a given stress are known. HS-induced SGs in human cells that do not accumulate significant quantities of misfolded proteins are preferentially cleared by Hsp70 during recovery. In contrast, more persistent aberrant SGs enriched in misfolded proteins, which also exhibit reduced dynamics based on FRAP data, increasingly undergo MT-based transport to the aggresome, followed by autophagic (granulophagy) clearance31,33. Similarly, transient (30 min) HS stress induces SG clearance that is insensitive to bafilomycin, implying autophagy independence, whereas SGs induced by longer HS (90 min) are bafilomycin sensitive, implying autophagic dependance37. Clearance of arsenite and HS-induced SGs are sensitive to VCP and proteasomal inhibition, whereas SGs induced by osmotic stress are insensitive. Clearance of arsenite-induced SGs is also more sensitive to deubiquitinase inhibition than HS-induced SG clearance56. Distinct VCP adaptor proteins (ZFAND1 and FAF2) aid clearance of arsenite and HS-induced SGs respectively37,58. In ZFAND1 KD cells, or proteasome-inhibited cells, arsenite also leads to the formation of aberrant SGs in which misfolded, nascently translated proteins and autophagic proteins accumulate58. In ZFAND1 KD cells, treatment with bafilomycin following arsenite causes a 5-fold accumulation in aberrant SGs 3hrs after arsenite removal. Collectively, these data demonstrate stress-specific SG clearance mechanisms, and suggest that granulophagy may compensate for the inability to clear SGs via Hsp chaperones31,33 or proteasomal/ZFAND1/VCP-dependent mechanisms58.
Other studies suggest that multiple SG clearance mechanisms may act together under specific conditions. In yeast, Hsp40 proteins Ydj1 and Sis1, in tandem with Hsp70, simultaneously promote clearance of sodium azide-induced SGs, albeit Ydj1 facilitates SG disassembly and translational recovery, whereas Sis1 aids granulophagy13. Similarly, in human cells, both Hsp70 and autophagy are implicated via knockdown studies in weakly aiding SG clearance in proliferating cells subject to daily low-dose arsenite stress. Notably, combinatorial Hsp70 and autophagy inhibition via KDs does not enhance SG clearance defects seen with single blocks alone, possibly suggesting an epistatic relationship (or incomplete KDs)264. More strikingly, senescent cells show upregulation of both Hsp70 and autophagic activity and are strongly inhibited in SG assembly. Knockdown of either Hsp70 or Atg5 (autophagy block) equally rescues SG assembly in senescent cells following arsenite stress, again suggesting epistasis. However, neither autophagy nor Hsp70 inhibition impacts SG clearance in senescent cells264. Thus, cellular growth state, and the nature of the stress, impact SG clearance mechanisms.
Given the compositional and structural complexity of SGs, it is likely that under some conditions, >1 clearance mechanism may facilitate complete SG clearance. Specific clearance mechanisms (e.g., VCP activity and the proteasome58; MTs and autophagy31, Syk kinase, VCP and autophagy154, ULK kinase and VCP156) may also function together and thus be epistatic, though with rare exceptions264, combinatorial inhibition studies of distinct SG clearance mechanisms to assess this are lacking. Alternatively, heterogeneous populations of SGs may exist in most stress contexts, each of which has their specific preferred clearance mechanism. Importantly, no single clearance mechanism has been identified that completely blocks SG clearance under any given stress (Table S1). Thus, in the absence of a known homogenous SG population, careful analysis of the physical state, composition, and dynamics of individual SGs may be necessary to distinguish whether multiple clearance pathways indeed function simultaneously31,38 or in a compensatory manner. Such questions equally apply to PBs, though better foundational knowledge of PB clearance pathways is required in the first instance.
GAPS IN KNOWLEDGE AND FUTURE EXPERIMENTAL DIRECTIONS
Many processes are known that impact SG clearance in specific contexts, with only a handful well described for PBs. This may reflect in part the greater connection in the literature of aberrant SG dynamics and clearance to disease states, though altered PB dynamics have also recently been linked to cancer161, Parkinson’s disease265 and forms of intellectual disability266. Thus, an obvious area for future progress is to better characterize PB clearance. Initial comparisons of whether PB clearance is acted upon by similar mechanism that underly SG clearance may yield general insights for other biomolecular condensates. As discussed throughout the review, several observations suggest that PBs may be surprisingly distinct in their clearance (Table S2), though more work is necessary to understand the significance of these differences, and their underlying mechanisms. Regardless, many key questions remain regarding the mechanisms and context under which specific processes impact clearance of both SGs and PBs (Box 3).
BOX 3: KEY UNANSWERED QUESTIONS REGARDING SG AND PB CLEARANCE.
Do Hsp chaperones bind and act on specific protein (or RNA?) substrates in SGs to facilitate clearance?
What factors dictate VCP-mediated clearance of SGs via autophagy, proteasomal or other means?
How do cytoskeletal elements and associated motor proteins interface with SG and PB components and regulate clearance?
Is eIF4A a non-specific RNA disaggregase that limits assembly, and facilitates clearance of SGs?
What are the regulatory mechanisms affecting specificity and activity of SG/PB-resident helicases, and how does this impact SG and PB clearance rates?
Does mRNA translation within SGs facilitate their clearance via disruption of RNA-RNA and RNA-protein contacts?
Many protein (and RNA?) modifications impact SG and PB dynamics; what are the roles and regulatory mechanisms of erasers of such modifications in SG and PB clearance?
What E3 Ub ligases impact SG clearance, and what are their substrates?
What are the key receptor and cargo interactions that underpin granulophagy?
What are the key substrates of proteasome-mediated SG clearance?
Is G3BP1’s role in SG dynamics limited to its multivalent RNA-protein scaffolding function, or do other reported activities contribute?
Does RNA decay play any role in SG or PB clearance? If so, do specific RNA molecules exist that preferentially scaffold SGs and PBs, and whose targeted decay would have significant impact?
What are the dynamics of bulk mRNA exit (or decay) within SGs and PBs during clearance?
What factors (e.g., stress, condensate composition, physical state) dictate the use of specific clearance pathways either singly or in combination?
Do cellular energy levels impact the use of specific SG and PB clearance mechanisms?
What experimental approaches are preferable to better understand SG and PB clearance? Whilst strong progress has been made in identifying SG clearance mechanisms in particular, most studies have focused on candidate-based analyses rather than utilizing unbiased genome-wide genetic screening approaches to identify key mechanisms. Thus, unbiased genetic and chemical screening methods remain one obvious area to pursue. However, given issues of redundancy, adaptation, or incomplete knockdown, complementing screening methods with biochemical purification studies to assess changes in condensate composition and modification status during and following stress are especially welcome32,267. A notable study of this type examined SG composition during and following arsenite stress, over a 2hr recovery time course, using APEX2-based proximity labelling32. This approach revealed over 200 “disassembly-engaged proteins” (DEPs) that preferentially enrich in SGs during clearance. These included autophagy proteins, ubiquitination factors, chaperones, RNA helicases, SUMOlyation enzymes, cytoskeletal proteins and interestingly mitochondrial proteins, the significance of which remains unclear. Another recent temporally resolved compositional analysis of HS-induced SGs also revealed that the proteasome, VCP and select translation initiation components only enrich within SGs following >1hr of HS stress267, further suggesting temporally and composition-specific clearance mechanisms may operate. Super-resolution and live cell microscopy methods, including single molecule and FRAP studies will remain pivotal in revealing the underlining mechanisms of SG and PB clearance under a given set of experimental conditions. Finally, increased testing of putative SG and PB clearance mechanisms singly and in combination, over a range of stress types, doses and durations would greatly help elucidate key clearance mechanisms, redundancies and epistatic relationships.
Several factors complicate the interpretation and comparison of SG and PB clearance data in prior studies that could be improved in future work. First, clearly distinguishing assembly versus clearance effects is often difficult when using KOs or treatments (e.g., inhibitors) administered prior or coincident with the addition of the inducing stress. Such approaches can significantly alter the levels of SGs or PBs induced between control and experimental cells, thus distorting interpretation of clearance phenotypes. Second, related to this, rates of SG or PB clearance are rarely calculated, and well resolved clearance time courses are often lacking. Finally, reported metrics of SG or PB abundance often do not fully convey potential differences in datasets. For example, % cells with foci does not reveal differences in the average number condensates/cell, their intensity, size, or area relative to cell volume. While experimental practicality often underlies these issues, ideally, quick inactivation strategies for a process of interest (e.g., rapidly acting drugs, conditional inactivation alleles) should be utilized at the point of stress recovery commencing, with detailed time course data being collected. No single metric is perfect for measuring SG and PB levels, and accurate unbiased quantification can be challenging268. Nonetheless, use of multiple metrics and automated quantification methods should be pursued whenever reasonably possible.
WORKING MODEL OF SG CLEARANCE AND SUMMARY
Although not all SG clearance observations are consistent (Table S1), a general working model (Figure 1) is that stress-specific variations in SG composition and physical state (e.g., the extent of misfolded or modified proteins/mRNAs) likely elicits stress-specific clearance mechanisms; this would mirror the existence of stress-specific assembly mechanisms269,270. SG clearance mechanisms likely vary in importance and redundancy based on stress duration, dose, and cellular context. Granulophagy may become more critical as SGs transition into aberrant, less dynamic states38. Maintenance of SG dynamics34,97, and indeed most reported SG clearance mechanisms also require cellular energy, but the relative energy demands of each pathway, and how this impacts their usage, remain unclear. Finally, RNA itself represents a potentially appealing target for directly effecting SG clearance, possibly through the act of SG-localized translation, modification, structural alteration, or degradation, though these areas remain poorly explored for now.
Understanding of PB clearance remains relatively nascent, but this is an important area for future study which may also impact SGs, given that PBs and SGs physically interact, may exchange components134 and share fundamental assembly mechanisms and compositional elements (i.e., non-translating mRNPs). Despite this, differences in PB versus SG clearance (Table S2) suggests we may be missing important insight into condensate clearance mechanisms. Thus, to achieve conceptual coherence, in SG and PB clearance, the field needs continued perseverance.
Supplementary Material
Acknowledgements
I acknowledge the great support and patience of Professor John Davey and Kara Raymond, which enabled the completion of this manuscript. Thanks to Telsa Mittelmeier for help with proof reading. This article was supported by the National Institute of General Medical Sciences (R01-GM114564).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest
I declare that I have no competing interests with regards to publication of the review “Stress granule and P-body clearance: seeking coherence in acts of disappearance”.
References
- 1.Hirose T, Ninomiya K, Nakagawa S & Yamazaki T A guide to membraneless organelles and their various roles in gene regulation. Nature Reviews Molecular Cell Biology 2022 24:4 24, 288–304 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Luo Y, Na Z & Slavoff SA P-Bodies: Composition, Properties, and Functions. Biochemistry 57, 2424–2431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glauninger H, Wong Hickernell CJ, Bard JAM & Drummond DA Stressful steps: Progress and challenges in understanding stress-induced mRNA condensation and accumulation in stress granules. Mol Cell 82, 2544–2556 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Decker CJ & Parker R P-bodies and stress granules: Possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 4, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofmann S, Kedersha N, Anderson P & Ivanov P Molecular mechanisms of stress granule assembly and disassembly. Biochim Biophys Acta Mol Cell Res 1868, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Protter DSW & Parker R Principles and Properties of Stress Granules. Trends Cell Biol 26, 668–679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramaswami M, Taylor JP & Parker R Altered ribostasis: RNA-protein granules in degenerative disorders. Cell 154, 727–36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li YR, King OD, Shorter J & Gitler AD Stress granules as crucibles of ALS pathogenesis. J Cell Biol 201, 361–72 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Gu J & Sun Q Aberrant Stress Granule Dynamics and Aggrephagy in ALS Pathogenesis. Cells 2021, Vol. 10, Page 2247 10, 2247 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazroui R, Di Marco S, Kaufman RJ & Gallouzi I-E Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol Biol Cell 18, 2603–18 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherkasov V et al. Coordination of Translational Control and Protein Homeostasis during Severe Heat Stress. Curr Biol 23, 2452–62 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Loschi M, Leishman CC, Berardone N & Boccaccio GL Dynein and kinesin regulate stress-granule and P-body dynamics. J Cell Sci 122, 3973–82 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walters RW, Muhlrad D, Garcia J & Parker R Differential effects of Ydj1 and Sis1 on Hsp70-mediated clearance of stress granules in Saccharomyces cerevisiae. RNA 21, 1660–71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mollet S et al. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol Biol Cell 19, 4469–79 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kedersha N et al. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 151, 1257–68 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M & Parker R Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11, 371–82 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cougot N, Babajko S & Séraphin B Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol 165, 31–40 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kroschwald S et al. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aizer A et al. Quantifying mRNA targeting to P-bodies in living human cells reveals their dual role in mRNA decay and storage. J Cell Sci 127, 4443–4456 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Bhattacharyya SN, Habermacher R, Martine U, Closs EI & Filipowicz W Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb Symp Quant Biol 71, 513–21 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Brengues M, Teixeira D & Parker R Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310, 486–9 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moon SL et al. Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nat Cell Biol 21, 162–168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Souquere S et al. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J Cell Sci 122, 3619–3626 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Wilbertz JH et al. Single-Molecule Imaging of mRNA Localization and Regulation during the Integrated Stress Response. Mol Cell 73, 946–958.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Okabe K, Tani T & Funatsu T Dynamic association-dissociation and harboring of endogenous mRNAs in stress granules. J Cell Sci 124, 4087–95 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Bartoli KM, Bishop DL & Saunders WS The role of molecular microtubule motors and the microtubule cytoskeleton in stress granule dynamics. Int J Cell Biol 2011, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nadezhdina ES, Lomakin AJ, Shpilman AA, Chudinova EM & Ivanov PA Microtubules govern stress granule mobility and dynamics. Biochim Biophys Acta Mol Cell Res 1803, 361–371 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Wheeler JR, Matheny T, Jain S, Abrisch R & Parker R Distinct stages in stress granule assembly and disassembly. Elife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kedersha N et al. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell 13, 195–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofmann S, Cherkasova V, Bankhead P, Bukau B & Stoecklin G Translation suppression promotes stress granule formation and cell survival in response to cold shock. Mol Biol Cell 23, 3786–800 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mateju D et al. An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J 36, 1669–1687 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marmor-Kollet H et al. Spatiotemporal Proteomic Analysis of Stress Granule Disassembly Using APEX Reveals Regulation by SUMOylation and Links to ALS Pathogenesis. Mol Cell 80, 876–891.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganassi M et al. A Surveillance Function of the HSPB8-BAG3-HSP70 Chaperone Complex Ensures Stress Granule Integrity and Dynamism. Mol Cell 63, 796–810 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Jain S et al. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell (2016) doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wippich F et al. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 152, 791–805 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Chitiprolu M et al. A complex of C9ORF72 and p62 uses arginine methylation to eliminate stress granules by autophagy. Nat Commun 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gwon Y et al. Ubiquitination of G3BP1 mediates stress granule disassembly in a context-specific manner. Science 372, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alberti S, Mateju D, Mediani L & Carra S Granulostasis: Protein Quality Control of RNP Granules. Front Mol Neurosci 10, 84 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walters RW & Parker R Coupling of Ribostasis and Proteostasis: Hsp70 Proteins in mRNA Metabolism. Trends Biochem Sci 40, 552–559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang A et al. Cancer Cells Evade Stress-Induced Apoptosis by Promoting HSP70-Dependent Clearance of Stress Granules. Cancers (Basel) 14, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahboubi H, Moujaber O, Kodiha M & Stochaj U The Co-Chaperone HspBP1 Is a Novel Component of Stress Granules that Regulates Their Formation. Cells 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoo H, Bard JAM, Pilipenko EV & Drummond DA Chaperones directly and efficiently disperse stress-triggered biomolecular condensates. Mol Cell 82, 741–755.e11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henics T et al. Mammalian Hsp70 and Hsp110 proteins bind to RNA motifs involved in mRNA stability. Journal of Biological Chemistry 274, 17318–17324 (1999). [DOI] [PubMed] [Google Scholar]
- 44.Kishor A et al. Hsp70’s RNA-binding and mRNA-stabilizing activities are independent of its protein chaperone functions. J Biol Chem 292, 14122–14133 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kampinga HH & Craig EA The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11, 579–592 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu J et al. Hsp40 proteins phase separate to chaperone the assembly and maintenance of membraneless organelles. Proc Natl Acad Sci U S A 117, 31123–31133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Markmiller S et al. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 172, 590–604.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karamanos TK, Tugarinov V & Clore GM Unraveling the structure and dynamics of the human DNAJB6b chaperone by NMR reveals insights into Hsp40-mediated proteostasis. Proc Natl Acad Sci U S A 116, 21529–21538 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shorter J & Southworth DR Spiraling in Control: Structures and Mechanisms of the Hsp104 Disaggregase. Cold Spring Harb Perspect Biol 11, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cereghetti G et al. Reversible amyloids of pyruvate kinase couple cell metabolism and stress granule disassembly. Nat Cell Biol 23, 1085–1094 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bakthisaran R, Tangirala R & Rao CM Small heat shock proteins: Role in cellular functions and pathology. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 1854, 291–319 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Kedersha NL, Gupta M, Li W, Miller I & Anderson P RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol 147, 1431–42 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyer H, Bug M & Bremer S Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol 14, 117–123 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Meyer H & Weihl CC The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J Cell Sci 127, 3877–83 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buchan JR, Kolaitis R-M, Taylor JP & Parker R Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153, 1461–74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tolay N & Buchberger A Comparative profiling of stress granule clearance reveals differential contributions of the ubiquitin system. Life Sci Alliance 4, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodriguez-Ortiz CJ et al. The Myoblast C2C12 Transfected with Mutant Valosin-Containing Protein Exhibits Delayed Stress Granule Resolution on Oxidative Stress. Am J Pathol 186, 1623–1634 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turakhiya A et al. ZFAND1 Recruits p97 and the 26S Proteasome to Promote the Clearance of Arsenite-Induced Stress Granules. Mol Cell 70, 906–919.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Cary GA, Vinh DBN, May P, Kuestner R & Dudley AM Proteomic analysis of Dhh1 complexes reveals a role for Hsp40 chaperone Ydj1 in yeast P-body assembly. G3: Genes, Genomes, Genetics 5, 2497–2511 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schopf FH, Biebl MM & Buchner J The HSP90 chaperone machinery. Nat Rev Mol Cell Biol 18, 345–360 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Pare JM et al. Hsp90 regulates the function of argonaute 2 and its recruitment to stress granules and P-bodies. Mol Biol Cell 20, 3273–3284 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suzuki Y et al. The Hsp90 inhibitor geldanamycin abrogates colocalization of eIF4E and eIF4E-transporter into stress granules and association of eIF4E with eIF4G. J Biol Chem 284, 35597–35604 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mediani L et al. Hsp90-mediated regulation of DYRK3 couples stress granule disassembly and growth via mTORC1 signaling. EMBO Rep 22, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buchan JR mRNP granules: Assembly, function, and connections with disease. RNA Biol 11, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chudinova EM & Nadezhdina ES Interactions between the Translation Machinery and Microtubules. Biochemistry (Mosc) 83, (2018). [DOI] [PubMed] [Google Scholar]
- 66.Ivanov PA, Chudinova EM & Nadezhdina ES Disruption of microtubules inhibits cytoplasmic ribonucleoprotein stress granule formation. Exp Cell Res 290, 227–33 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Kwon S, Zhang Y & Matthias P The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev 21, 3381–94 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fujimura K, Katahira J, Kano F, Yoneda Y & Murata M Microscopic dissection of the process of stress granule assembly. Biochim Biophys Acta 1793, 1728–37 (2009). [DOI] [PubMed] [Google Scholar]
- 69.Chernov KG et al. Role of Microtubules in Stress Granule Assembly. Journal of Biological Chemistry 284, 36569–36580 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kolobova E et al. Microtubule-dependent association of AKAP350A and CCAR1 with RNA stress granules. Exp Cell Res 315, 542–555 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aizer A et al. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol Biol Cell 19, 4154–66 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sweet TJ, Boyer B, Hu W, Baker KE & Coller J Microtubule disruption stimulates P-body formation. RNA 13, 493–502 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aizer A & Shav-Tal Y Intracellular trafficking and dynamics of P bodies. Prion 2, 131 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hurst Z, Liu W, Shi Q & Herman PK A distinct P-body-like granule is induced in response to the disruption of microtubule integrity in Saccharomyces cerevisiae. Genetics 222, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hubstenberger A et al. P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol Cell 68, 144–157.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Tsai NP, Tsui YC & Wei LN Dynein motor contributes to stress granule dynamics in primary neurons. Neuroscience 159, 647–656 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rajgor D et al. Mammalian microtubule P-body dynamics are mediated by nesprin-1. J Cell Biol 205, 457–475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hubstenberger A, Noble SL, Cameron C & Evans TC Translation repressors, an RNA helicase, and developmental cues control RNP phase transitions during early development. Dev Cell 27, 161–173 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lindsay AJ & McCaffrey MW Myosin Va is required for P body but not stress granule formation. Journal of Biological Chemistry 286, 11519–11528 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Steffens A, Jaegle B, Tresch A, Hülskamp M & Jakoby M Processing-body movement in Arabidopsis depends on an interaction between myosins and DECAPPING PROTEIN1. Plant Physiol 164, 1879–1892 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chang W et al. Myo2p, a class V myosin in budding yeast, associates with a large ribonucleic acid-protein complex that contains mRNAs and subunits of the RNA-processing body. RNA 14, 491–502 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang JD et al. Direct interaction of microtubule- and actin-based transport motors. Nature 397, 267–270 (1999). [DOI] [PubMed] [Google Scholar]
- 83.Böddeker TJ, Rusch A, Leeners K, Murrell MP & Dufresne ER Actin and microtubules position stress granules. bioRxiv 2023.06.04.543599 (2023) doi: 10.1101/2023.06.04.543599. [DOI] [Google Scholar]
- 84.Böddeker TJ et al. Non-specific adhesive forces between filaments and membraneless organelles. Nat Phys 18, 571–578 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weis K & Hondele M The Role of DEAD-Box ATPases in Gene Expression and the Regulation of RNA-Protein Condensates. Annu Rev Biochem 91, 197–219 (2022). [DOI] [PubMed] [Google Scholar]
- 86.Youn JY et al. High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Mol Cell 69, 517–532.e11 (2018). [DOI] [PubMed] [Google Scholar]
- 87.Chalupníková K et al. Recruitment of the RNA Helicase RHAU to Stress Granules via a Unique RNA-binding Domain. Journal of Biological Chemistry 283, 35186–35198 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hondele M et al. DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573, 144–148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mugler CF et al. ATPase activity of the DEAD-box protein Dhh1 controls processing body formation. Elife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schütz P et al. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci U S A 105, 9564–9569 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hilliker A, Gao Z, Jankowsky E & Parker R The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol Cell 43, 962–72 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Valentin-Vega YA et al. Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation. Sci Rep 6, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mazroui R et al. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2α phosphorylation. Mol Biol Cell 17, 4212–4219 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tauber D et al. Modulation of RNA Condensation by the DEAD-Box Protein eIF4A. Cell 180, 411–426.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mokas S et al. Uncoupling stress granule assembly and translation initiation inhibition. Mol Biol Cell 20, 2673–83 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dang Y et al. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J Biol Chem 281, 32870–8 (2006). [DOI] [PubMed] [Google Scholar]
- 97.Wang T et al. Intracellular energy controls dynamics of stress-induced ribonucleoprotein granules. Nat Commun 13, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Itzhak DN, Tyanova S, Cox J & Borner GHH Global, quantitative and dynamic mapping of protein subcellular localization. Elife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Beck M et al. The quantitative proteome of a human cell line. Mol Syst Biol 7, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Harms U, Andreou AZ, Gubaev A & Klostermeier D eIF4B, eIF4G and RNA regulate eIF4A activity in translation initiation by modulating the eIF4A conformational cycle. Nucleic Acids Res 42, 7911–7922 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Putnam AA & Jankowsky E DEAD-box helicases as integrators of RNA, nucleotide and protein binding. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 1829, 884–893 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ripin N & Parker R Are stress granules the RNA analogs of misfolded protein aggregates? RNA 28, 67–75 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cui BC et al. Pharmacological inhibition of DEAD-Box RNA Helicase 3 attenuates stress granule assembly. Biochem Pharmacol 182, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aryanpur PP, Mittelmeier TM & Bolger TA The RNA Helicase Ded1 Regulates Translation and Granule Formation during Multiple Phases of Cellular Stress Responses. Mol Cell Biol 42, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shih JW et al. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem J 441, 119–129 (2012). [DOI] [PubMed] [Google Scholar]
- 106.Shih JW, Tsai TY, Chao CH & Wu Lee YH Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene 27, 700–714 (2008). [DOI] [PubMed] [Google Scholar]
- 107.Brown NP, Vergara AM, Whelan AB, Guerra P & Bolger TA Medulloblastoma-associated mutations in the DEAD-box RNA helicase DDX3X/DED1 cause specific defects in translation. J Biol Chem 296, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lennox AL et al. Pathogenic DDX3X Mutations Impair RNA Metabolism and Neurogenesis during Fetal Cortical Development. Neuron 106, 404–420.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Coller J & Parker R General translational repression by activators of mRNA decapping. Cell 122, 875–886 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Carroll JS, Munchel SE & Weis K The DExD/H box ATPase Dhh1 functions in translational repression, mRNA decay, and processing body dynamics. Journal of Cell Biology 194, 527–537 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ayache J et al. P-body assembly requires DDX6 repression complexes rather than decay or Ataxin2/2L complexes. Mol Biol Cell 26, 2579–2595 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rao BS & Parker R Numerous interactions act redundantly to assemble a tunable size of P bodies in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences 114, E9569–E9578 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Teixeira D & Parker R Analysis of P-body assembly in Saccharomyces cerevisiae. Mol Biol Cell 18, 2274–87 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dutta A, Zheng S, Jain D, Cameron CE & Reese JC Intermolecular interactions within the abundant DEAD-box protein Dhh1 regulate its activity in Vivo. Journal of Biological Chemistry 286, 27454–27470 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mathys H et al. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol Cell 54, 751–765 (2014). [DOI] [PubMed] [Google Scholar]
- 116.Di Stefano B et al. The RNA Helicase DDX6 Controls Cellular Plasticity by Modulating P-Body Homeostasis. Cell Stem Cell 25, 622–638.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ernoult-Lange M et al. Multiple binding of repressed mRNAs by the P-body protein Rck/p54. RNA 18, 1702–1715 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ozgur S et al. Structure of a Human 4E-T/DDX6/CNOT1 Complex Reveals the Different Interplay of DDX6-Binding Proteins with the CCR4-NOT Complex. Cell Rep 13, 703–711 (2015). [DOI] [PubMed] [Google Scholar]
- 119.Minshall N, Kress M, Weil D & Standart N Role of p54 RNA helicase activity and its C-terminal domain in translational repression, P-body localization and assembly. Mol Biol Cell 20, 2464–2472 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Weston A & Sommerville J Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res 34, 3082–3094 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wilczynska A, Aigueperse C, Kress M, Dautry F & Weil D The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J Cell Sci 118, 981–992 (2005). [DOI] [PubMed] [Google Scholar]
- 122.Buchan JR, Muhlrad D & Parker R P bodies promote stress granule assembly in Saccharomyces cerevisiae. Journal of Cell Biology 183, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Majerciak V, Zhou T, Kruhlak M & Zheng Z RNA helicase DDX6 and scaffold protein GW182 in P-bodies promote biogenesis of stress granules. Nucleic Acids Res 1, 13–14 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Khong A et al. The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Mol Cell 68, 808–820.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sauer M et al. DHX36 prevents the accumulation of translationally inactive mRNAs with G4-structures in untranslated regions. Nat Commun 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bell SD & Botchan MR The minichromosome maintenance replicative helicase. Cold Spring Harb Perspect Biol 5, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jha S & Dutta A RVB1/RVB2: running rings around molecular biology. Mol Cell 34, 521 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kaur E, Agrawal R & Sengupta S Functions of BLM Helicase in Cells: Is It Acting Like a Double-Edged Sword? Front Genet 12, 634789 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Danino Y et al. BLM helicase protein negatively regulates stress granule formation through unwinding RNA G-quadruplex structures. Nucleic Acids Res 1, 13–14 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Iserman C et al. Condensation of Ded1p Promotes a Translational Switch from Housekeeping to Stress Protein Production. Cell 181, 818–831.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bush MS et al. eIF4A RNA Helicase Associates with Cyclin-Dependent Protein Kinase A in Proliferating Cells and Is Modulated by Phosphorylation. Plant Physiol 172, 128–140 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Patel A et al. Biochemistry: ATP as a biological hydrotrope. Science (1979) 356, 753–756 (2017). [DOI] [PubMed] [Google Scholar]
- 133.Begovich K & Wilhelm JE An In Vitro Assembly System Identifies Roles for RNA Nucleation and ATP in Yeast Stress Granule Formation. Mol Cell 79, 991–1007.e4 (2020). [DOI] [PubMed] [Google Scholar]
- 134.Buchan JR & Parker R Eukaryotic stress granules: the ins and outs of translation. Mol Cell 36, 932–41 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ohn T & Anderson P The role of posttranslational modifications in the assembly of stress granules. Wiley Interdiscip Rev RNA 1, 486–493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hofweber M & Dormann D Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics. J Biol Chem 294, 7137–7150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wek RC Role of eIF2α Kinases in Translational Control and Adaptation to Cellular Stress. Cold Spring Harb Perspect Biol 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lu YN et al. MARK2 phosphorylates eIF2α in response to proteotoxic stress. PLoS Biol 19, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wu Z et al. FAM69C functions as a kinase for eIF2α and promotes stress granule assembly. EMBO Rep 24, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mateju D et al. Single-Molecule Imaging Reveals Translation of mRNAs Localized to Stress Granules. Cell 183, 1801–1812.e13 (2020). [DOI] [PubMed] [Google Scholar]
- 141.Kimball SR, Horetsky RL, Ron D, Jefferson LS & Harding HP Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am J Physiol Cell Physiol 284, C273–84 (2003). [DOI] [PubMed] [Google Scholar]
- 142.Kedersha N et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 169, 871–84 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mikhailova T et al. RNA helicase DDX19 stabilizes ribosomal elongation and termination complexes. Nucleic Acids Res 45, 1307–1318 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gross T et al. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science 315, 646–649 (2007). [DOI] [PubMed] [Google Scholar]
- 145.Amiri H & Noller HF A tandem active site model for the ribosomal helicase. FEBS Lett 593, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Takyar S, Hickerson RP & Noller HF mRNA helicase activity of the ribosome. Cell 120, 49–58 (2005). [DOI] [PubMed] [Google Scholar]
- 147.Rojas M, Vasconcelos G & Dever TE An eIF2α-binding motif in protein phosphatase 1 subunit GADD34 and its viral orthologs is required to promote dephosphorylation of eIF2α. Proc Natl Acad Sci U S A 112, E3466–E3475 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dimasi P, Quintiero A, Shelkovnikova TA & Buchman VL Modulation of p-eIF2α cellular levels and stress granule assembly/disassembly by trehalose. Sci Rep 7, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shelkovnikova TA et al. Chronically stressed or stress-preconditioned neurons fail to maintain stress granule assembly. Cell Death Dis 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jain S et al. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 164, 487–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rai AK, Chen JX, Selbach M & Pelkmans L Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 559, 211–216 (2018). [DOI] [PubMed] [Google Scholar]
- 152.Yahya G et al. Stress granules display bistable dynamics modulated by Cdk. J Cell Biol 220, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kosmacz M et al. Protein and metabolite composition of Arabidopsis stress granules. New Phytologist 222, 1420–1433 (2019). [DOI] [PubMed] [Google Scholar]
- 154.Krisenko MO et al. Syk Is Recruited to Stress Granules and Promotes Their Clearance through Autophagy. Journal of Biological Chemistry 290, 27803–27815 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tsai N-P, Ho P-C & Wei L-N Regulation of stress granule dynamics by Grb7 and FAK signalling pathway. EMBO J 27, 715–26 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang B et al. ULK1 and ULK2 Regulate Stress Granule Disassembly Through Phosphorylation and Activation of VCP/p97. Mol Cell 74, 742–757.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Shah KH, Zhang B, Ramachandran V & Herman PK Processing body and stress granule assembly occur by independent and differentially regulated pathways in Saccharomyces cerevisiae. Genetics 193, 109–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cargnello M et al. Phosphorylation of the Eukaryotic Translation Initiation Factor 4E-Transporter (4E-T) by c-Jun N-Terminal Kinase Promotes Stress-Dependent P-Body Assembly. Mol Cell Biol 32, 4572–4584 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rzeczkowski K et al. c-Jun N-terminal kinase phosphorylates DCP1a to control formation of P bodies. J Cell Biol 194, 581–596 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Aizer A, Kafri P, Kalo A & Shav-Tal Y The P Body Protein Dcp1a Is Hyper-phosphorylated during Mitosis. PLoS One 8, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bearss JJ et al. EDC3 phosphorylation regulates growth and invasion through controlling P-body formation and dynamics. EMBO Rep 22, e50835 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Krause LJ, Herrera MG & Winklhofer KF The Role of Ubiquitin in Regulating Stress Granule Dynamics. Front Physiol 13, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xie X et al. Deubiquitylases USP5 and USP13 are recruited to and regulate heat-induced stress granules through their deubiquitylating activities. J Cell Sci 131, (2018). [DOI] [PubMed] [Google Scholar]
- 164.Maxwell BA et al. Ubiquitination is essential for recovery of cellular activities after heat shock. Science 372, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Markmiller S et al. Active Protein Neddylation or Ubiquitylation Is Dispensable for Stress Granule Dynamics. Cell Rep 27, 1356–1363.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bounedjah O et al. Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules. Nucleic Acids Res 42, 8678–8691 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yau R & Rape M The increasing complexity of the ubiquitin code. Nature Cell Biology 2016 18:6 18, 579–586 (2016). [DOI] [PubMed] [Google Scholar]
- 168.Komander D & Rape M The Ubiquitin Code. 10.1146/annurev-biochem-060310-170328 81, 203–229 (2012). [DOI] [PubMed] [Google Scholar]
- 169.Yang C et al. Stress granule homeostasis is modulated by TRIM21-mediated ubiquitination of G3BP1 and autophagy-dependent elimination of stress granules. Autophagy 1–18 (2023) doi: 10.1080/15548627.2022.2164427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ohn T, Kedersha N, Hickman T, Tisdale S & Anderson P A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol 10, 1224–31 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nostramo R, Varia SN, Zhang B, Emerson MM & Herman PK The catalytic activity of the Ubp3 deubiquitinating protease is required for efficient stress granule assembly in S. cerevisiae. Mol Cell Biol 36, MCB.00609–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kedersha N et al. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol 212, 845–60 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang CY et al. Analysis of stress granule assembly in Schizosaccharomyces pombe. RNA 18, 694–703 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Takahashi M et al. Stress Granules Inhibit Apoptosis by Reducing Reactive Oxygen Species Production. Mol Cell Biol 33, 815–829 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bhattacharya U, Neizer-Ashun F, Mukherjee P & Bhattacharya R When the chains do not break: the role of USP10 in physiology and pathology. Cell Death Dis 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kraft C, Deplazes A, Sohrmann M & Peter M Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol 10, 602–610 (2008). [DOI] [PubMed] [Google Scholar]
- 177.Ossareh-Nazari B et al. Ubiquitylation by the Ltn1 E3 ligase protects 60S ribosomes from starvation-induced selective autophagy. J Cell Biol 204, 909–917 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Meyer C, Garzia A, Morozov P, Molina H & Tuschl T The G3BP1-Family-USP10 Deubiquitinase Complex Rescues Ubiquitinated 40S Subunits of Ribosomes Stalled in Translation from Lysosomal Degradation. Mol Cell 77, 1193–1205.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 179.Liu J et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 147, 223–234 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tenekeci U et al. K63-Ubiquitylation and TRAF6 Pathways Regulate Mammalian P-Body Formation and mRNA Decapping. Mol Cell 62, 943–957 (2016). [DOI] [PubMed] [Google Scholar]
- 181.Zaffagnini G & Martens S Mechanisms of Selective Autophagy. J Mol Biol 428, 1714 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Guo H et al. Autophagy supports genomic stability by degrading retrotransposon RNA. Nat Commun 5, 5276 (2014). [DOI] [PubMed] [Google Scholar]
- 183.Zheng Y et al. HDAC6, A Novel Cargo for Autophagic Clearance of Stress Granules, Mediates the Repression of the Type I Interferon Response During Coxsackievirus A16 Infection. Front Microbiol 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Seguin SJ et al. Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly. Cell Death Differ 21, 1838–51 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhang P et al. Chronic optogenetic induction of stress granules is cytotoxic and reveals the evolution of ALS-FTD pathology. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Corcoran JA & McCormick C Viral activation of stress-regulated Rho-GTPase signaling pathway disrupts sites of mRNA degradation to influence cellular gene expression. Small GTPases 6, 178–85 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Robinson C-A et al. Kaposi’s sarcoma-associated herpesvirus (KSHV) utilizes the NDP52/CALCOCO2 selective autophagy receptor to disassemble processing bodies. PLoS Pathog 19, e1011080 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ma X et al. CCT2 is an aggrephagy receptor for clearance of solid protein aggregates. Cell 185, 1325–1345.e22 (2022). [DOI] [PubMed] [Google Scholar]
- 189.Hu R, Qian B, Li A & Fang Y Role of Proteostasis Regulation in the Turnover of Stress Granules. Int J Mol Sci 23, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Maharjan N, Künzli C, Buthey K & Saxena S C9ORF72 Regulates Stress Granule Formation and Its Deficiency Impairs Stress Granule Assembly, Hypersensitizing Cells to Stress. Mol Neurobiol 54, 3062–3077 (2017). [DOI] [PubMed] [Google Scholar]
- 191.Balendra R & Isaacs AM C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat Rev Neurol 14, 544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Nassif M, Woehlbier U & Manque PA The Enigmatic Role of C9ORF72 in Autophagy. Front Neurosci 11, 442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sellier C et al. Loss of C9ORF72 impairs autophagy and synergizes with polyQ Ataxin-2 to induce motor neuron dysfunction and cell death. EMBO J 35, 1276–97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Farg MA et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet 23, 3579–95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Webster CP et al. The C9orf72 protein interacts with Rab1a and the ULK1 complex to regulate initiation of autophagy. EMBO J 35, 1656–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Parzych KR & Klionsky DJ An Overview of Autophagy: Morphology, Mechanism, and Regulation. Antioxid Redox Signal 20, 460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sanders DW et al. Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization. Cell 181, 306–324.e28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yang P et al. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell 181, 325–345.e28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Guillén-Boixet J et al. RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 181, 346–361.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Tourrière H et al. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol 160, 823–31 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 201.Panas MD et al. Phosphorylation of G3BP1-S149 does not influence stress granule assembly. J Cell Biol 218, 2425–2432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Reineke LC et al. Casein Kinase 2 Is Linked to Stress Granule Dynamics through Phosphorylation of the Stress Granule Nucleating Protein G3BP1. Mol Cell Biol 37, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Gal J et al. The Acetylation of Lysine-376 of G3BP1 Regulates RNA Binding and Stress Granule Dynamics. Mol Cell Biol 39, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Tsai WC et al. Arginine Demethylation of G3BP1 Promotes Stress Granule Assembly. J Biol Chem 291, 22671–22685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Tsai WC, Reineke LC, Jain A, Jung SY & Lloyd RE Histone arginine demethylase JMJD6 is linked to stress granule assembly through demethylation of the stress granule-nucleating protein G3BP1. J Biol Chem 292, 18886–18896 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Song D et al. Yin and yang regulation of stress granules by Caprin-1. Proc Natl Acad Sci U S A 119, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Vognsen T, Møller IR & Kristensen O Crystal Structures of the Human G3BP1 NTF2-Like Domain Visualize FxFG Nup Repeat Specificity. PLoS One 8, e80947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Panas MD et al. Viral and cellular proteins containing FGDF motifs bind G3BP to block stress granule formation. PLoS Pathog 11, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Panas MD et al. Sequestration of G3BP coupled with efficient translation inhibits stress granules in Semliki Forest virus infection. Mol Biol Cell 23, 4701–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Freibaum BD et al. Identification of small molecule inhibitors of G3BP-driven stress granule formation. bioRxiv 2023.06.27.546770 (2023) doi: 10.1101/2023.06.27.546770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Hamon L, Budkina K & Pastré D YB-1 Structure/Function Relationship in the Packaging of mRNPs and Consequences for Translation Regulation and Stress Granule Assembly in Cells. Biochemistry (Moscow) 87, S20–S31 (2022). [DOI] [PubMed] [Google Scholar]
- 212.Lyons SM, Achorn C, Kedersha NL, Anderson PJ & Ivanov P YB-1 regulates tiRNA-induced Stress Granule formation but not translational repression. Nucleic Acids Res 44, 6949–6960 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Somasekharan SP et al. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J Cell Biol 208, 913–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Bley N et al. Stress granules are dispensable for mRNA stabilization during cellular stress. Nucleic Acids Res 43, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Budkina K et al. YB-1 unwinds mRNA secondary structures in vitro and negatively regulates stress granule assembly in HeLa cells. Nucleic Acids Res 49, 10061–10081 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Evdokimova VM et al. The major protein of messenger ribonucleoprotein particles in somatic cells is a member of the Y-box binding transcription factor family. J Biol Chem 270, 3186–3192 (1995). [DOI] [PubMed] [Google Scholar]
- 217.Sidibé H, Dubinski A & Vande Velde C The multi‐functional RNA‐binding protein G3BP1 and its potential implication in neurodegenerative disease. J Neurochem 157, 944 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Gallouzi I et al. A novel phosphorylation-dependent RNase activity of GAP-SH3 binding protein: a potential link between signal transduction and RNA stability. Mol Cell Biol 18, 3956–3965 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Tourrière H et al. RasGAP-associated endoribonuclease G3Bp: selective RNA degradation and phosphorylation-dependent localization. Mol Cell Biol 21, 7747–7760 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Fischer JW, Busa VF, Shao Y & Leung AKL Structure-Mediated RNA Decay by UPF1 and G3BP1. Mol Cell 78, 70–84.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Edupuganti RR et al. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol 24, 870–878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.He X, Yuan J & Wang Y G3BP1 binds to guanine quadruplexes in mRNAs to modulate their stabilities. Nucleic Acids Res 49, 11323–11336 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Oi N et al. Resveratrol induces apoptosis by directly targeting Ras-GTPase-activating protein SH3 domain-binding protein 1. Oncogene 34, 2660–2671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Soncini C, Berdo I & Draetta G Ras-GAP SH3 domain binding protein (G3BP) is a modulator of USP10, a novel human ubiquitin specific protease. Oncogene 20, 3869–3879 (2001). [DOI] [PubMed] [Google Scholar]
- 225.Cohen M, Stutz F, Belgareh N, Haguenauer-Tsapis R & Dargemont C Ubp3 requires a cofactor, Bre5, to specifically de-ubiquitinate the COPII protein, Sec23. Nat Cell Biol 5, 661–667 (2003). [DOI] [PubMed] [Google Scholar]
- 226.Nostramo R, Varia SN, Zhang B, Emerson MM & Herman PK The Catalytic Activity of the Ubp3 Deubiquitinating Protease Is Required for Efficient Stress Granule Assembly in Saccharomyces cerevisiae. Mol Cell Biol 36, 173–183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Liu J et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 147, 223–234 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Costa M, Ochem A, Staub A & Falaschi A Human DNA helicase VIII: a DNA and RNA helicase corresponding to the G3BP protein, an element of the ras transduction pathway. Nucleic Acids Res 27, 817–821 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Annibaldi A, Dousse A, Martin S, Tazi J & Widmann C Revisiting G3BP1 as a RasGAP binding protein: sensitization of tumor cells to chemotherapy by the RasGAP 317–326 sequence does not involve G3BP1. PLoS One 6, e29024 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Xu C et al. Integrative genomics in combination with RNA interference identifies prognostic and functionally relevant gene targets for oral squamous cell carcinoma. PLoS Genet 9, e1003169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Prentzell MT et al. G3BPs tether the TSC complex to lysosomes and suppress mTORC1 signaling. Cell 184, 655–674.e27 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Sahoo PK et al. Axonal G3BP1 stress granule protein limits axonal mRNA translation and nerve regeneration. Nature Communications 2018 9:1 9, 1–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Jin G et al. G3BP2: Structure and function. Pharmacol Res 186, 106548 (2022). [DOI] [PubMed] [Google Scholar]
- 234.Decker CJ, Burke JM, Mulvaney PK & Parker R RNA is required for the integrity of multiple nuclear and cytoplasmic membrane-less RNP granules. EMBO J 41, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Clemson CM et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33, 717–26 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Yamazaki T et al. Functional Domains of NEAT1 Architectural lncRNA Induce Paraspeckle Assembly through Phase Separation. Mol Cell 70, 1038–1053.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 237.Asano-Inami E et al. The association of UBAP2L and G3BP1 mediated by small nucleolar RNA is essential for stress granule formation. Commun Biol 6, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Fernandes Nikita, Buchan JR RPS28B mRNA Acts as a Scaffold Promoting Cis-Translational Interaction of Proteins Driving P-body Assembly. Nucleic Acids Res 48, 6265–6279 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Anders M et al. Dynamic m6A methylation facilitates mRNA triaging to stress granules. Life Sci Alliance 1, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Ries RJ et al. m6A enhances the phase separation potential of mRNA. Nature 571, 424–428 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Wang X et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Fu Y & Zhuang X m6A-binding YTHDF proteins promote stress granule formation. Nature Chemical Biology 2020 16:9 16, 955–963 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Jones JD, Monroe J & Koutmou KS A molecular-level perspective on the frequency, distribution, and consequences of messenger RNA modifications. Wiley Interdiscip Rev RNA 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zhao Z et al. QKI shuttles internal m7G-modified transcripts into stress granules and modulates mRNA metabolism. Cell 186, 3208–3226.e27 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Alriquet M et al. The protective role of m1A during stress-induced granulation. J Mol Cell Biol 12, 870–880 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Khong A, Matheny T, Huynh TN, Babl V & Parker R Limited effects of m6A modification on mRNA partitioning into stress granules. Nat Commun 13, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Zou Z, Sepich-Poore C, Zhou X, Wei J & He C The mechanism underlying redundant functions of the YTHDF proteins. Genome Biol 24, 1–13 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Arguello AE, Deliberto AN & Kleiner RE RNA Chemical Proteomics Reveals the N6-Methyladenosine (m6A)-Regulated Protein-RNA Interactome. J Am Chem Soc 139, 17249–17252 (2017). [DOI] [PubMed] [Google Scholar]
- 249.Van Treeck B et al. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proceedings of the National Academy of Sciences 115, 2734–2739 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Fay MM, Anderson PJ & Ivanov P ALS/FTD-Associated C9ORF72 Repeat RNA Promotes Phase Transitions In Vitro and in Cells. Cell Rep 21, 3573–3584 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Turner M et al. rG4detector, a novel RNA G-quadruplex predictor, uncovers their impact on stress granule formation. Nucleic Acids Res 50, 11426–11441 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Sheth U & Parker R Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300, 805–8 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Tibble RW, Depaix A, Kowalska J, Jemielity J & Gross JD Biomolecular condensates amplify mRNA decapping by biasing enzyme conformation. Nat Chem Biol 17, 615–623 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Decker CJ, Teixeira D & Parker R Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol 179, 437–49 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Eulalio A, Behm-Ansmant I, Schweizer D & Izaurralde E P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol 27, 3970–81 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Aizer A et al. Quantifying mRNA targeting to P-bodies in living human cells reveals their dual role in mRNA decay and storage. J Cell Sci 127, 4443–56 (2014). [DOI] [PubMed] [Google Scholar]
- 257.Pitchiaya S et al. Dynamic Recruitment of Single RNAs to Processing Bodies Depends on RNA Functionality. Mol Cell 74, 521–533.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Pelechano V, Wei W & Steinmetz LM Widespread Co-translational RNA Decay Reveals Ribosome Dynamics. Cell 161, 1400–1412 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Horvathova I et al. The Dynamics of mRNA Turnover Revealed by Single-Molecule Imaging in Single Cells. Mol Cell 68, 615–625.e9 (2017). [DOI] [PubMed] [Google Scholar]
- 260.Pizzo E et al. Ribonuclease/angiogenin inhibitor 1 regulates stress-induced subcellular localization of angiogenin to control growth and survival. J Cell Sci 126, 4308–4319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Wilbertz JH et al. Single-Molecule Imaging of mRNA Localization and Regulation during the Integrated Stress Response. Mol Cell 73, 946–958.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 262.Das S, Santos L, Failla AV & Ignatova Z mRNAs sequestered in stress granules recover nearly completely for translation. RNA Biol 19, 877 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Van Treeck B & Parker R Principles of Stress Granules Revealed by Imaging Approaches. Cold Spring Harb Perspect Biol 11, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Omer A et al. Autophagy and heat-shock response impair stress granule assembly during cellular senescence. Mech Ageing Dev 192, (2020). [DOI] [PubMed] [Google Scholar]
- 265.Hallacli E et al. The Parkinson’s disease protein alpha-synuclein is a modulator of processing bodies and mRNA stability. Cell 185, 2035–2056.e33 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Balak C et al. Rare De Novo Missense Variants in RNA Helicase DDX6 Cause Intellectual Disability and Dysmorphic Features and Lead to P-Body Defects and RNA Dysregulation. The American Journal of Human Genetics 105, 509–525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Hu S et al. Time-resolved proteomic profiling reveals compositional and functional transitions across the stress granule life cycle. Nat Commun 14, 7782 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Shabanov I & Buchan JR HARLEY mitigates user bias and facilitates efficient quantification and co-localization analyses of foci in yeast fluorescence images. Sci Rep 12, 12238 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Buchan JR, Yoon J-H & Parker R Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J Cell Sci 124, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Aulas A et al. Stress-specific differences in assembly and composition of stress granules and related foci. J Cell Sci 130, 927–937 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


