Abstract
We studied the temporal relationship between human immunodeficiency type 1 (HIV-1) quasispecies in tissues and in peripheral blood mononuclear cells (PBMC) of infected individuals. Sequential PBMC and tissue samples from various organs obtained at autopsy from three patients who died of AIDS-related complications were available for analysis. Biological HIV-1 clones were isolated from PBMC samples, and cellular tropism and syncytium-inducing (SI) capacity were determined. Genomic DNA was isolated from 1 cm3 of organ tissue, and proviral DNA was amplified by means of PCR and cloned with the PGEM-T vector system. A 185-bp region encompassing the third variable domain of the virus envelope, known to influence HIV-1 biological properties, was sequenced. HIV-1 could be amplified from all PBMC and organ samples, except from liver tissue for two patients. Both SI and non-syncytium-inducing (NSI) genotypes could be detected in the different tissues. Tissue-specific quasispecies were observed in brain, lung, and testis. Lymphoid tissues, such as bone marrow, lymph node, and spleen, harbored several different variants similar to those detected in blood in the last PBMC samples. In general, only tissues in which macrophages are likely to be the main target cell for HIV-1 harbored NSI HIV-1 sequences that clustered separately. Both SI and NSI sequences that clustered with sequences from late-stage PBMC were present in other tissues, which may indicate that the presence of HIV-1 in those tissues is secondary to lymphocyte infiltration rather than to tissue tropism of HIV-1 itself. These data suggest that the viral reservoir may be limited, which will have important implications for the success of HIV-1 eradication.
The human immunodeficiency virus type 1 (HIV-1) variants can differ with respect to syncytium-inducing (SI) capacity, replication rate, and cellular tropism (2, 7, 51, 52). In asymptomatic individuals, slowly replicating, preferentially macrophage-tropic nonsyncytium-inducing (NSI) HIV-1 variants predominant (4, 10, 42, 45, 54, 58). With progression of disease, more rapidly replicating, preferentially T-cell-tropic viruses emerge, in 50% of individuals coinciding with the emergence of SI HIV-1 variants (28, 45, 52). The presence of preferentially macrophage-tropic HIV-1 in the asymptomatic phase, when host immunity is relatively intact, has led to the suggestion that macrophage tropism is a mechanism for HIV-1 variants to escape immune surveillance (45). Alternatively or in addition, the persistence of macrophage-tropic NSI variants during all stages of disease may point to a reservoir function for HIV-1-infected macrophages (46) from which new variants are generated. The presence of SI HIV-1 in lymph nodes in the absence of this phenotype in peripheral blood indeed indicates that new virus variants may be generated in lymphoid tissues (34, 50). The lymphoid tissues have been shown to be active sites of replication throughout HIV infection also during the asymptomatic phase (14, 39). In situ hybridization, however, showed that only a very few macrophages in lymph nodes were infected (23). This can mean either that macrophages constitute a reservoir at another body site or that this cell type does not constitute a major reservoir in HIV-1 infection.
With the availability of potent treatment regimens, the eradication of HIV-1 seems feasible. To achieve this, however, all compartments relevant for virus replication should be accessible to the antiretroviral compounds. The exact identification of the viral reservoir in the body is therefore of the utmost importance. The presence of HIV-1 in nonlymphoid tissues may be attributed to infected macrophages. These cells could thus constitute a separate compartment of viral replication, with subsequent separate evolution of HIV-1 quasispecies. Alternatively, the presence of HIV-1 in these nonlymphoid tissues may result only from lymphocyte infiltration secondary to infections with opportunistic pathogens coinciding with the onset of AIDS (13). If the first scenario were true, HIV-1 quasispecies in nonlymphoid tissues would probably be more closely related to the quasispecies that were present in peripheral blood mononuclear cells (PBMC) early in infection than to those late in infection, as the two compartments would have evolved separately from a common precursor. Conversely, close resemblance of the HIV-1 quasispecies in the nonlymphoid tissues to the peripheral quasispecies after the onset of AIDS would indicate similar virus turnover with continuous exchange in tissues and periphery or very late spread of the virus to the nonlymphoid tissues. The latter case, although mediated through a different mechanism, would not influence the outcome of antiretroviral intervention, as the spread to the nonlymphoid tissues would be limited.
To investigate the relation between virus variants in periphery and in lymphoid and nonlymphoid tissues and to identify the reservoir function of macrophages for HIV-1 in tissues, we analyzed HIV-1 populations in sequential blood samples and several organ tissues obtained at autopsy. Phylogenetic analyses were performed to study the relation between infectious cellular quasispecies present in PBMC over time and virus variants present in tissue compartments at the time of death.
MATERIALS AND METHODS
Patient samples.
Tissues from various lymphoid and nonlymphoid organs were obtained at autopsy, carried out within 2 days of death, from three individuals, one female and two males, who died of AIDS-related complications. Clinical, immunological, and viral load information for the years prior to death are summarized in Table 1 and Fig. 1. Samples of brain, lung, liver, kidney, spleen, lymph node, bone marrow, and/or testis were washed with phosphate-buffered saline–10% trinatrium citrate, dissected into 1-cm3 pieces, and stored at −70°C (Table 2). Because of the absence of HIV-related neurological symptoms in patient ACH0208, no brain autopsy was performed. From the two other patients, samples of cerebrospinal fluid (CSF) were also obtained. These were cultured in a limiting dilution fashion on phytohemagglutinin (PHA)-stimulated PBMC for 4 weeks (see below). PBMC samples were available from all three individuals spanning at least the period from AIDS onset to death (in subjects ACH6052 and HIVAms198) or the time from seroconversion to death (in subject ACH0208).
TABLE 1.
Characteristics of the three subjects under study
| Patient | Sex | Duration of phase
(mo)
|
Pathology (affected tissue) | |
|---|---|---|---|---|
| Asymp- tomatic | AIDS | |||
| p198 | Female | ≥10a | 42 | B-cell non-Hodgkin’s lymphoma (brain), endocarditis, lung embolism, pneumonia, carcinoma (vulva) |
| 6052 | Male | ≥7a | 40 | B-cell non-Hodgkin’s lymphoma (brain), Kaposi’s sarcoma (skin), Mycobacterium avium (lymph nodes), cytomegalovirus (adrenal gland), Pseudomonas aeruginosa (lung) |
| 0208 | Male | 54 | 39 | Generalized Kaposi’s sarcoma (lung, skin, liver, intestines) causing lung bleeding and respiratory insufficiency |
No known seroconversion date.
FIG. 1.
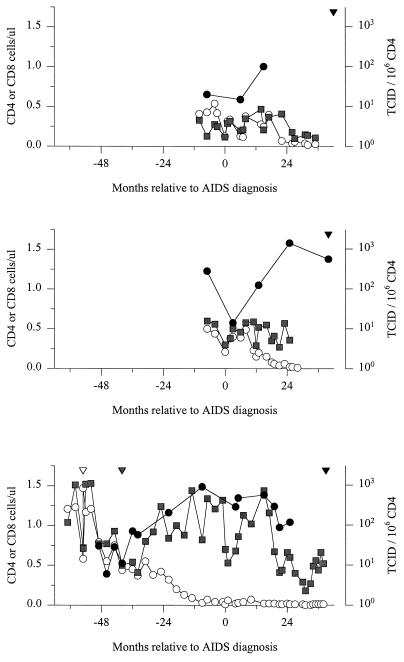
Changes in the numbers of CD4 (○) and CD8 ( ) T cells and cellular infectious load (•) relative to the moment of AIDS diagnosis for the three subjects under study (HIVAms198 [top], ACH6052 [middle], and ACH0208 [bottom]). White, grey, and black inverted triangles denote the moment of seroconversion, emergence of SI variants, and death, respectively.
TABLE 2.
List of samples analyzed
| Patient | PBMC | Cardiac blood | Lymph nodes | Spleen | Bone marrow | Liver | Kidney | Testis | Lung | Brain | CSF |
|---|---|---|---|---|---|---|---|---|---|---|---|
| p198 | + | − | + | + | + | + | + | NAa | + | + | + |
| 6052 | + | + | + | + | − | + | + | + | + | + | + |
| 0208 | + | − | + | + | − | + | + | + | + | −b | −b |
NA, not applicable.
No brain autopsy was performed on patient ACH0208.
Analysis of CD4+ T-cell counts.
T-lymphocyte immunophenotyping for CD4+ and CD8+ T cells was carried out at 3-month intervals by flow cytofluorometry. PBMC were stained with CD4 or CD8 monoclonal antibody according to standard procedures for fluorescence-activated cell sorting analysis.
Preparation of DNA from patient tissues.
Extraction of DNA from the tissues was carried out by resuspending small pieces of tissue in lysis buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 50 mM EDTA, 1% sodium-n-lauroylsarcosine, 100 μg of proteinase K per ml) as described elsewhere (13). The digestion process was allowed to continue for 2 to 3 h at 65°C. This was followed by phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation overnight. DNA pellets were dried and resuspended in 200 μl of distilled water.
Cloning and sequencing of PCR products.
Envelope sequences were amplified by PCR with primers V1V2-1 (5′-GCCTGTGTACCCCACAGACCCCAA-3′, nucleotide position 6463, sense) (22) and PS-D (5′-ATTACAGTAGAAAAATTCCCC-3′, position 7381, antisense) (49) in the presence of 3 mM MgCl2 in the first reaction and primers V1V2-2 (5′-GAGGATATAATCAGTTTATGGGAT-3′, position 6562, sense) (22) and PS-C (5′-CTGGGTCCCCTCCTGAGG-3′, position 7331, antisense) (49) in the presence of 3 mM MgCl2 in the nested reaction. For amplification, the following PCR amplification cycles were used: 5 min at 95°C once and 1.5 min at 95°C, 1 min at 55°C, and 1 min at 72°C, repeated 25 times, followed by an extra 5 min of extension at 72°C and subsequent cooling to 4°C. The resulting ±800-bp products from three to six nested PCRs were pooled and purified with a Geneclean kit (Bio 101, Inc., Vista, Calif.). These purified products were cloned into PGEM-T vector (Promega) according to the manufacturer’s protocol. Fifty microliters of competent Escherichia coli cells (strain JM109) was then transformed with 4 to 7 μl of ligation mix. The recombinants were plated on Luria broth agar with ampicillin (75 mg/ml), incorporating 50 μl of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 40 mg/ml) and 20 μl of isopropyl-3-d-thiogalactopyranoside (IPTG; 100 ng/ml), and incubated overnight at 37°C. A total of 20 to 55 individual clones were picked and expanded overnight in Luria broth at 37°C. Bacterial cells were pelleted, DNA was extracted with the Vacman system (Qiagen), and samples were digested with restriction enzymes SphI and PstI (GIBCO-BRL) for 1 h at 37°C and checked for the presence of insert on a 1% agarose gel. Clones containing insert were sequenced directly with sense primer PS-B (49) by the dideoxy chain termination method with Sequenase (U.S. Biochemical, Cleveland, Ohio) according to the instructions from the manufacturer.
Isolation of biological virus clones from patient PBMC.
Limiting dilution cultures were performed as described elsewhere (45). Briefly, participant PBMC (0.5 × 104 to 4 × 104 cells per well, 24 to 96 replicates per concentration) or CSF samples (1-in-5 dilution series) were cocultivated with PHA-stimulated healthy donor PBMC (105 per well) in 96-well microtiter plates. Every week for 5 weeks, one-third of each culture supernatant was collected for detection of p24 antigen by an in-house p24 antigen capture enzyme-linked immunosorbent assay. At the same time, one-third of the culture volume, containing half of the cells, was transferred to new 96-well plates and 105 fresh PHA-stimulated healthy donor PBMC were added to propagate the culture. The proportion of productively infected CD4+ T cells was estimated by the formula for Poisson distribution, F = −ln (F0), in which F0 is the fraction of negative cultures. PBMC from wells testing positive were transferred to 25-ml culture flasks containing 5 × 106 fresh PHA-stimulated PBMC in 5 ml of culture medium to grow virus stocks. Virus-containing cell-free culture supernatant was stored at −70°C until further use, cells were frozen, and approximately 106 cells were used for isolation of DNA. SI capacity of virus clones was determined by cocultivation with MT-2 cells (106). Biological virus clones from patient HIVAms198 and patient ACH6052 were all isolated on target cells from the same healthy blood donor. For patient ACH0208, additional isolations were performed on target cells derived from three additional healthy blood donors. In our experience, target cells derived from different healthy blood donors do not influence the clonal composition of the viruses isolated from one sample, except when the healthy blood donor is heterozygous or homozygous for the 32-bp deletion in CCR5 (data not shown).
Infection of monocyte-derived macrophages.
Virus-containing culture supernatants derived from the CSF-derived virus clones from patients HIVAms198 and ACH6052 and PBMC-derived virus clones from patient ACH0208 were tested for their capacity to infect monocyte-derived macrophages from different healthy blood donors as described elsewhere (46, 54).
DNA isolation, PCR, and sequencing of biological virus clones.
Total DNA from PBMC harboring the biological HIV-1 clones was isolated as described elsewhere (6). Envelope V3 sequences were amplified by PCR as described elsewhere (49), with primers PS-A and PS-H in the first reaction and PS-B and PS-G in the nested reaction. The resulting ±700-bp products were purified with a Geneclean kit (Bio 101, Inc.), and 185 bp were sequenced directly with sense primer PS-B (49) by the dideoxy chain termination method with Sequenase (U.S. Biochemical), both according to the instructions from the manufacturers.
Sequence data analysis.
Alignment of the sequences was straightforward, done with the PILEUP program, and checked manually, to ensure that codons remained intact. To check for contamination, a BLAST search of GenBank HIV-1 sequences was performed for sequences with the highest similarity. Phylogenetic analyses were done with the neighbor-joining program (43) as implemented in the PHYLIP package (16). For bootstrapping, the SEQBOOT, DNADIST, NEIGHBOR, and CONSENSE programs from this package were used. PHYLIP’s DRAWTREE program was used to produce the plots. For direct comparison of nucleotide sequences, Hamming distances were used. The distance matrix input for the neighbor-joining analysis was generated with Kimura’s two-parameter estimation for nucleotides (27). Estimation of the number of silent and nonsilent substitutions was done according to Nei and Gojobori’s method (37), as implemented in MEGA (32). To evaluate possible associations between certain amino acid changes and tissue tropisms, the following method was used. The tissues were divided into two groups (one dominated by T-cell-tropic HIV-1 [PBMC, lymph node, spleen, and bone marrow] and the other dominated by macrophage-tropic HIV-1 [brain, CSF, lung, and liver]). Kidney was regarded as a control for contamination with variants from the blood. For each position in the sequences, a pairwise comparison matrix consisting of zeros and ones was constructed, indicating for each pair of sequences whether they contained identical or different nucleotides at that position (position matrix). A second matrix contained a similar same-different index indicating whether each pair of sequences came from the same or a different tissue group (group matrix). A Pearson correlation coefficient was then calculated between corresponding cells of the position matrix and the tissue group matrix, yielding a position-wise indication of the association between sequence differences and tissue group (31).
Nucleotide sequence accession numbers.
All newly generated sequences have been deposited in GenBank (accession no. AF021367-AF021476 [HIVAms198], AF021477-AF021683 [ACH6052], and AF021684-AF021790 [ACH0208]).
RESULTS
Proviral HIV-1 sequences (185 bp encompassing the V3 loop) were obtained from all PBMC and tissue samples, except from the livers of patients ACH0208 and ACH6052. PCR amplification of the HIV-1 V3 region from DNA isolated from a liver tissue sample from patient HIVAms198 yielded only 1 positive reaction (of 14), which was used for ligation. In all samples tested, the sequences from clonal populations reflected bulk sequences from uncloned material (both PBMC and tissues), i.e., neither cloning procedure preferentially expanded rare variants (data not shown). SI capacity of biological clones isolated from peripheral blood was determined by cocultivation with the MT-2 cell line, in each case confirming the phenotype as predicted from the genotype (19). The 98% correlation between V3 genotype and HIV-1 biological phenotype with respect to SI capacity enabled us to predict the phenotype from the virus clones in different tissues based on their V3 sequence (18). In the phylogenetic tree for each patient, sequences from the two other patients were included, both as outgroups and to exclude contamination. In all instances, the patient sequences clustered separately from the outgroup sequences in 100 of 100 trees (data not shown). The patient sequences were also different from the laboratory strains available in our laboratory (HIV-1 BaL, HIV-1 ADA, HIV-1 IIIb, ACH0320.2A5/2A6, and ACH0172.Ba-L). Moreover, a BLAST similarity search of GenBank HIV-1 sequences did not provide evidence for contamination with virus strains obtained from other patients in the Amsterdam cohort (data not shown). Tissue sequences from all three patients were further analyzed for signature patterns pertaining to particular tissues, and for presumed T-cell-tropic HIV-1-dominated tissues (PBMC, lymph node, bone marrow, and spleen) versus compartments considered to be macrophage-tropic HIV-1 dominated (lung, brain, CSF, and liver). Although specific groupings were detected in each patient separately (see below), no specific signature pattern for either of these comparisons was detected (all Pearson correlations, ≤0.23). Moreover, no evidence for previously described signature patterns for brain and/or CSF sequences was observed (30, 35).
Results per patient.
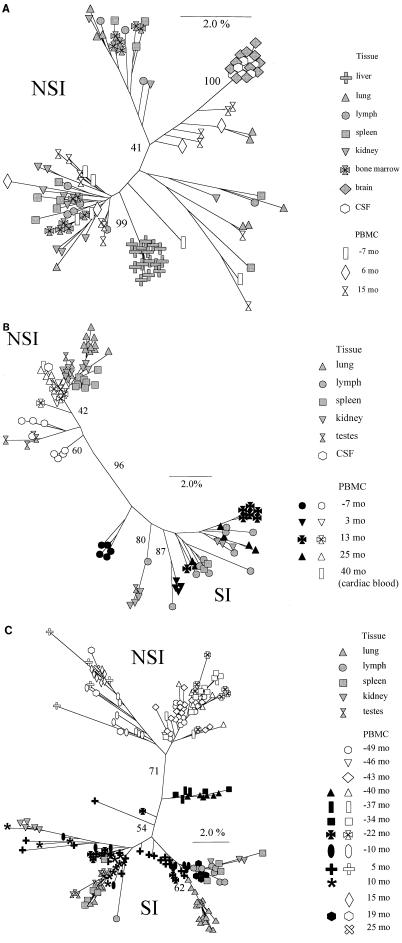
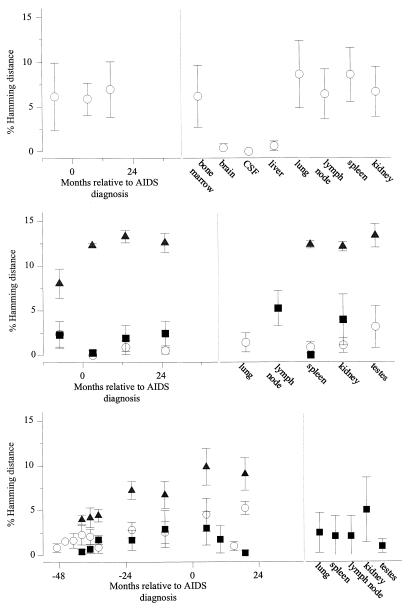
From patient HIVAms198, eight different tissues and three different PBMC samples (collected 7 months before and 6 and 15 months after AIDS diagnosis) were available for analysis (Table 2). From all body sites, only NSI variants were obtained, as judged from the MT-2 assay and/or predicted from V3 sequences. Alignment of all sequences relative to the consensus NSI sequences from the first PBMC sample is shown in Fig. 2A. With these sequences, neighbor-joining trees with bootstrap resampling were constructed (Fig. 3A). Brain and CSF quasispecies were highly homogeneous, clustered closely together, and were most closely related to some of the latest available sequences obtained from PBMC (15 months after AIDS diagnosis). In cell culture, these CSF variants were shown to be highly macrophage tropic, able to infect monocyte-derived macrophages from several different donors (data not shown). The liver quasispecies also clustered closely together; however, the liver sequences were all generated from only one positive PCR, and the homogeneity is possibly a reflection of low proviral copy numbers. The quasispecies from the other tissues and PBMC samples were heterogeneous and did not show a particular tissue- or time-specific clustering. Heterogeneity of the samples was further analyzed by calculating pairwise Hamming distances (Fig. 4). Hamming distances were less than 2% for brain, CSF, and liver but ranged up to 14% for sequences derived from the other tissues and from PBMC.
FIG. 2.
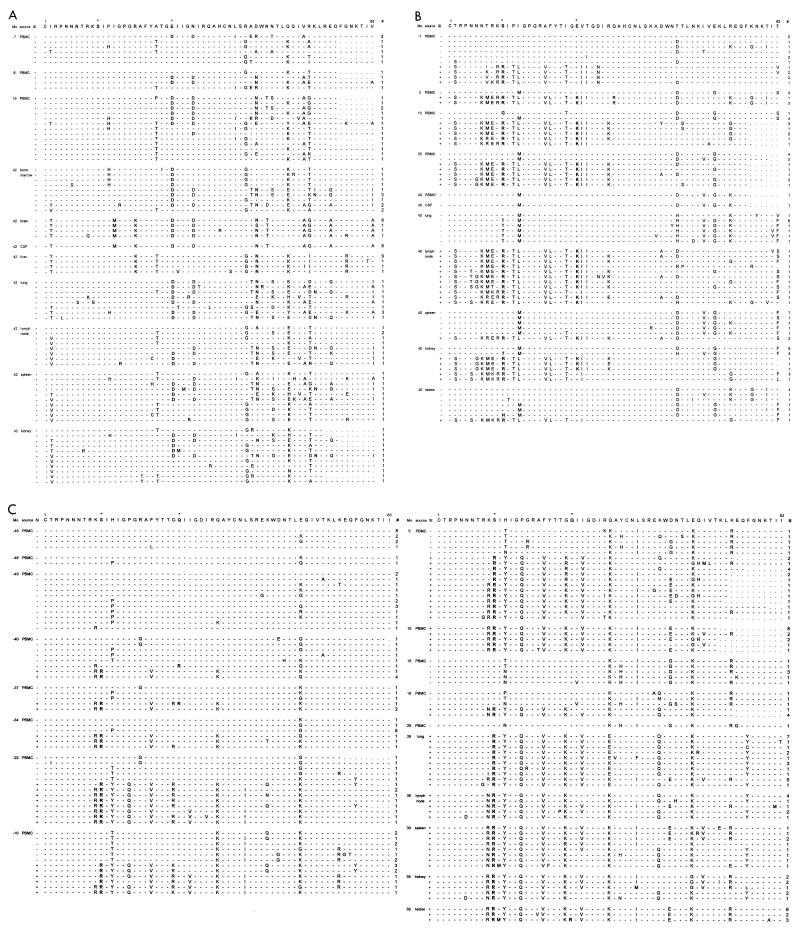
Deduced amino acid sequences of the V3 region. The sequences are aligned with the consensus sequence of the NSI variants present in the first sample for each patient. Amino acid positions involved in SI capacity are marked by asterisks. Dashes indicate identity with the reference sequence. Position 1 corresponds to amino acid 296 of the HXB2 envelope protein. Mo., number of months relative to AIDS diagnosis; #, number of virus clones with a specific sequence. Virus clones were obtained from PBMC and tissue samples of patient HIVAms198 (A), participant ACH6052 (B), and participant ACH0208 (C).
FIG. 3.
Results of phylogenetic analysis of the V3 region (neighbor-joining method, unrooted tree) from virus clones obtained during the course of infection. Bootstrap values indicate the percentages of trees showing the observed specific groupings. White and black symbols depict NSI and SI PBMC-derived sequences, respectively, and grey symbols depict tissue-derived sequences. Branch lengths are drawn to scale. (A) Subject HIVAms198; (B) subject ACH6052; (C) subject ACH0208.
FIG. 4.
Plots of Hamming distances between the V3 regions of the virus clones obtained from PBMC in the course of HIV-1 infection and those from several tissues obtained at autopsy from three subjects, HIVAms198 (top), ACH6052 (middle), and ACH0208 (bottom). Distances were calculated with DNADIST as implemented in the PHYLIP program. Means and standard deviations of comparisons between NSI variants (○), between SI variants (▪), and between SI and NSI variants (▴) from the same time point are shown.
From patient ACH6052, six different tissues and five PBMC samples (obtained 7 months before and 3, 13, and 25 months after AIDS diagnosis and postmortem from the heart [cardiac blood]) were available for analysis (Table 2). This patient had been infected for several years before entry into the study, which is reflected by the presence of SI variants already in the first PBMC sample, obtained 7 months before AIDS diagnosis (see alignments in Fig. 2B). At this time, the two populations (SI versus NSI) had already diverged (see neighbor-joining tree in Fig. 3B). The SI and NSI quasispecies seem to have continued to evolve as separate groups from the first time point onwards, reflected by the separate grouping of the early sequences and late sequences within their respective phenotypic groups. Interestingly, no new SI variants generated from late NSI variants were detected. Tissue-specific distribution was observed for lung and lymph node. The lymph node variants clustered with the late PBMC SI clones, whereas the lung variants clustered with the last available PBMC NSI clones. Spleen and testis harbored mostly NSI variants, clustering with late or early PBMC samples, respectively. Kidney, which was regarded as a control for contamination with variants from the blood, showed no such restrictions. Only one biological virus clone was obtained from CSF, reflecting a very low infectious load, and the sequence clustered with the latest blood sequence obtained at the time of death. Pairwise Hamming distances between SI and NSI sequences obtained from the same sample (either tissue or PBMC) ranged from 10 to 15%, while within the respective phenotypic groups Hamming distance was generally less than 5% (Fig. 4). In 12 clones, suggestive evidence for recombination of 3′ and 5′ ends between SI and NSI virus clones was found, involving viruses from the −7- and 3-month PBMC, lymph node, and testes (Fig. 2B).
Thirteen PBMC samples spanning the 74-month period between 6 months after seroconversion and up to 14 months before death were available from patient ACH0208, in addition to the five tissue samples obtained after death (Table 2). None of the biological virus clones obtained from PBMC samples were able to infect monocyte-derived macrophages obtained from two healthy blood donors (data not shown). Of note, this patient was shown to be heterozygous for a previously described 32-bp deletion (Δ32) in the CCR5 coreceptor gene. After seroconversion, this patient harbored only NSI variants for 15 months (=40 months before AIDS diagnosis) before developing SI variants (see alignments in Fig. 2C). Biological virus clones from the PBMC sample obtained 10 months after AIDS diagnosis were isolated on PHA-stimulated target cells from a healthy blood donor, who was retrospectively identified to be homozygous for the 32-bp deletion in CCR5, reflected by the isolation of only SI variants from this sample. Two possible intermediate NSI variants (so-called switch NSI) were detected 43 and 40 months before AIDS diagnosis with arginines at positions 10 and/or 25. Thereafter, the SI and NSI quasispecies seem to have continued to evolve as separate groups (see neighbor-joining tree in Fig. 3C). Again, no new SI variants generated from late NSI variants were detected. Between 34 and 22 months before AIDS diagnosis, there was a complete replacement of the SI virus population by SI variants with glutamine at amino acid position 16, and between 22 and 10 months before AIDS diagnosis, there was a replacement of the NSI virus population by NSI variants with threonine at position 13. Pairwise Hamming distances between SI and NSI sequences obtained from the same sample (either tissue or PBMC) increased over time up to 10%, while within the respective phenotypic groups Hamming distance was generally less than 5% (Fig. 4). NSI clones isolated from 10 months before AIDS diagnosis onwards may have resulted from a recombination of 5′ NSI and 3′ SI ends between SI and NSI virus clones found at the previous time point (Fig. 2C).
All tissues harbored only SI variants that were also present in PBMC between 10 and 19 months after AIDS diagnosis. The lymph node quasispecies clustered with the last available SI PBMC sequences obtained 19 months after AIDS diagnosis. While the testis quasispecies were rather homogeneous (three main variants, Hamming distance within 2%), the spleen, lung, and kidney quasispecies were more diverse (Hamming distances ranging from 0 to 8%).
DISCUSSION
In this study, we analyzed the temporal relationship between the clonal composition of HIV-1 populations in sequential PBMC samples and various tissue samples obtained at autopsy. Using phylogenetic analysis, we tried to identify the PBMC sample containing virus variants most closely similar to the viral quasispecies in lymphoid and nonlymphoid tissues, since this could shed light on the moment in infection at which HIV-1 was disseminated into the nonlymphoid tissue compartment or, alternatively, at which moment HIV-1 was shed from tissue into the circulation. Viral quasispecies from most lymphoid and nonlymphoid tissues clustered with the viral sequences obtained from late PBMC samples. This can indicate either that migration of HIV-1 into the nonlymphoid tissues is a late event in the clinical course of infection (13) or that there is a relatively high turnover of viral quasispecies with an active interplay between replication in all tissues and peripheral blood. The lymph node sequences clustered most closely with very late PBMC sequences. This is in agreement not only with the assumptions that plasma virions are produced in lymph nodes and are responsible for PBMC infection (24, 55) but also with the close correlation between plasma virus load and cellular infectious load (5). The most likely explanation for the presence of HIV-1 in nonlymphoid tissues is migration of HIV-1-infected lymphocytes, secondary to late-stage pathology in these tissues. Alternatively, the migration could be a consequence of disturbed lymphocyte homing due to the HIV-1-related destruction of lymph node architecture (36, 40, 41).
The viral quasispecies from brain tissue were very homogeneous and clustered separately from all other sequences, but most closely with some of the late PBMC sequences. However, the brain-CSF populations were distinctly different from the PBMC variants despite their clustering. These results are more suggestive of compartmentalization and early spread of HIV-1 to the brain, which is in agreement with previous findings (1, 3, 8, 15, 21, 26, 30, 38, 48, 56). The fact that some of the late PBMC sequences are more similar to this compartment may actually reflect leakage of virus produced in the brain into the circulation.
One individual in our present study harbored only NSI HIV-1 variants, both in PBMC and in all tissues analyzed. In the other two individuals, SI variants were present at least at later stages of infection. The emergence of SI variants seemed to be a unique event, as no new SI variants that could have evolved from later-stage NSI variants were detected (53), nor did the SI populations in tissues cluster separately from the PBMC SI variants. Rather, the SI and NSI quasispecies in all tissue compartments seemed to evolve separately from a common precursor NSI sequence, which was indeed reflected by the increasing Hamming distance between the coexisting NSI and SI HIV-1 populations, up to 14%.
Within the quasispecies of either NSI or SI variants, Hamming distances did not exceed the 5% level. This may indicate fierce competition between coexisting NSI and SI virus populations, in which only the fittest variant in each phenotypic group evolves further, generating the new homogeneous virus population that is subsequently present. The detection of possible recombinants between SI and NSI variants in the two patients who harbored both variants may indicate that SI and NSI variants have some overlapping target cell populations. Although the recombinants detected in the tissues could be an artifact due to PCR recombination, the fact that the same recombinants were detected in tissue and biological virus clones (with a clonal input for the PCRs) suggests that they are not solely the result of a PCR artifact.
In individual HIVAms198 harboring only NSI variants, brain and CSF harbored very homogeneous virus populations, whereas other compartments contained highly heterogeneous HIV-1 quasispecies with Hamming distances ranging up to 14%. Apparently, in some compartments, more variation may occur if these NSI HIV-1 quasispecies are able to occupy the niches from which they otherwise would have been excluded by competing coexisting SI variants. The homogeneity in virus populations in brain and CSF might then be explained by the fact that SI HIV-1 lacks the capacity anyway to replicate in these compartments and that only limited variation in NSI is allowed in replication at these sites. Alternatively, the limited variation may reflect slower turnover of the virus populations and infected cells in this compartment compared to that in the lymphoid tissues, compatible with the slow resistance development detected in the brain (48, 56).
Although we know that SI HIV-1 evolves from NSI HIV-1, it is still unclear in which compartment the generation of SI variants occurs. The presence of SI sequences in lymph nodes of individuals who showed future SI conversion has been reported elsewhere (34, 50). This indeed points to the lymph nodes as the most active site for viral replication. Here, we observed the presence of both NSI and SI HIV-1 in all tissues, although a tissue-specific abundance of specific phenotypes could not be excluded. The clustering of tissue SI HIV-1 sequences with SI sequences from late-stage-obtained PBMC and not with the first SI sequences obtained from PBMC recently after phenotype conversion exclude, in our opinion, the possibility that new virus variants are generated in tissues other than lymph nodes and then shed into the circulation.
Patient ACH6052 showed an SI prevalence in lymph node and a predominance of NSI HIV-1 in lung and CSF and to a lesser extent in spleen and testis. The preferential presence of NSI variants in testis is in accordance not only with a previous study that compared virus variants in blood and semen (59) but also with the hypothesis that HIV-1 infection is preferentially established by macrophage-tropic NSI variants (20, 58), which might be due to compartmentalization of macrophage-tropic HIV-1 in the donor. Kidney tissue from patient ACH6052, which was used as a control for blood contamination, indeed harbored an NSI-SI distribution that reflected the clonal composition in peripheral blood.
Despite the presence of NSI HIV-1 in peripheral blood at least up to 14 months prior to death, patient ACH0208 harbored only SI HIV-1 variants in all tissues analyzed. Although it may be that by the time of death only SI variants were present in PBMC as well, other explanations are also feasible. Interestingly, this individual was heterozygous for the previously described 32-bp deletion in the CCR5 gene which encodes the second receptor for primary HIV-1 (11, 44). PBMC from CCR5 Δ32 heterozygotes show impaired support of NSI HIV-1 replication (33), associated in vivo with lower levels of viral RNA load in serum (12, 25). SI HIV-1 is able to use CXCR4 as an alternative coreceptor and may therefore have a growth advantage over NSI HIV-1 in CCR5 Δ32 heterozygotes (17). However, if SI and NSI variants indeed remained present up to the time of death, tissue migration could be specific for CXCR4-expressing potential SI target lymphocytes. The absence of NSI HIV-1, which in general is more macrophage tropic (46), in tissues in which macrophages are considered the important target cell, might directly be associated with the impaired coreceptor expression in CCR5 Δ32 heterozygotes. This reduced expression together with the already low expression of CD4 on primary macrophages (9) may be insufficient to support replication even of macrophage-tropic NSI HIV-1. This latter observation also leads to the intriguing possibility that Δ32 heterozygous individuals do not have an important macrophage reservoir for HIV-1 replication. Indeed, none of the biological virus clones obtained from ACH0208 either early or late in infection was able to productively infect monocyte-derived macrophages.
Assuming that all subjects initially had a relatively homogeneous virus population (54, 57, 58), considerable diversification has occurred up to the moment of AIDS diagnosis. Thereafter, there seems to be no significant further increase in variability in any of the subjects. It has been postulated that on this V3 region of the viral envelope a considerable pressure is exerted, which is reflected in a higher rate of nonsynonymous than synonymous mutations (29, 47, 49). Assuming that at the moment of AIDS diagnosis the host immune system has collapsed, our finding may suggest that the selection pressure on V3 is predominantly exerted by immune control. However, the strong segregation of central nervous system-lymphoid quasispecies in both subjects where central nervous system virus could be evaluated and the emergence and dominance (in tissues) of the SI type sequences when Δ32 CCR5 was present show the strong influence of host cell availability on the V3 region.
In conclusion, apart from the brain, our study finds no evidence for the existence of true tissue reservoirs that are installed early in infection and from which new variants are continuously generated. Rather, the presence of HIV-1 in most nonlymphoid tissues seems to be a late-stage event and secondary to lymphocyte infiltration due to pathology. The absence of a large tissue reservoir for HIV-1 favors the possibility of completely eradicating HIV-1. The presence of a separate virus reservoir in the brain underscores, however, the importance of including in the combination of drugs used antiretroviral drugs which have good bioavailability in the brain.
ACKNOWLEDGMENTS
These studies were performed as part of the Amsterdam Cohort Studies of AIDS, a collaboration among the Municipal Health Service, The Academic Medical Centre, and the Central Laboratory of the Netherlands Red Cross Blood Transfusion Service, Amsterdam, The Netherlands. Proleukin (recombinant interleukin-2) was kindly provided by R. Rombouts, Chiron Benelux B.V., Amsterdam, The Netherlands. We are greatly indebted to Jan Weening, head of the Department of Pathology at the Academic Medical Centre, and his colleagues and especially to Iwan Nektar for outstanding assistance during the autopsy procedures; to Marijke Roos and colleagues for excellent technical assistance; and to Hetty Blaak, Frank van Engelenburg, Maarten Koot, and Frank Miedema for critical reading of the manuscript.
REFERENCES
- 1.Ait-Khaled M, McLaughlin J E, Johnson M A, Emery V C. Distinct HIV-1 long terminal repeat quasispecies present in nervous tissues compared to that in lung, blood and lymphoid tissues of an AIDS patient. AIDS. 1995;9:675–683. doi: 10.1097/00002030-199507000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Asjo B, Albert J, Karlsson A, Morfeldt-Månson L, Biberfeld G, Lidman K, Fenyö E M. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet. 1986;ii:660–662. [PubMed] [Google Scholar]
- 3.Ball J K, Holmes E C, Whitwell H, Desselberger U. Genomic variation of human immunodeficiency virus type 1 (HIV-1): molecular analyses of HIV-1 in sequential blood samples and various organs obtained at autopsy. J Gen Virol. 1994;75:867–879. doi: 10.1099/0022-1317-75-4-867. [DOI] [PubMed] [Google Scholar]
- 4.Blaak, H., M. Brouwer, L. J. Ran, F. De Wolf, and H. Schuitemaker. In vitro replication kinetics of HIV-1 variants in relation to viral load in long-term survivors of HIV-1 infection. [DOI] [PubMed]
- 5.Blaak, H., F. De Wolf, A. B. Van’t Wout, N. G. Pakker, M. Bakker, J. Goudsmit, and H. Schuitemaker. Temporal relationship between human immunodeficiency virus type 1 RNA levels in serum and cellular infectious load in peripheral blood. J. Infect. Dis., in press. [DOI] [PubMed]
- 6.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, Van der Noordaa J. A rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1991;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng-Mayer C, Seto D, Tateno M, Levy J A. Biologic features of HIV-1 that correlate with virulence in the host. Science. 1988;240:80–82. doi: 10.1126/science.2832945. [DOI] [PubMed] [Google Scholar]
- 8.Chiodi F, Valentin A, Keys B, Schwartz S, Åsjö B, Gartner S, Popovic M, Albert J, Sundqvist V-A, Fenyö E M. Biological characterization of paired human immunodeficiency virus type 1 isolates from blood and cerebrospinal fluid. Virology. 1989;173:178–187. doi: 10.1016/0042-6822(89)90233-x. [DOI] [PubMed] [Google Scholar]
- 9.Collman R, Godfrey B, Cutilli J, Rhodes A, Hassan N F, Sweet R, Douglas S D, Friedman H, Nathanson N, Gonzalez-Scarano F. Macrophage-tropic strains of human immunodeficiency virus type 1 utilize the CD4 receptor. J Virol. 1990;64:4468–4476. doi: 10.1128/jvi.64.9.4468-4476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connor R I, Mohri H, Cao Y, Ho D D. Increased viral burden and cytopathicity correlate temporally with CD4+T-lymphocyte decline and clinical progression in human immunodeficiency virus type 1-infected individuals. J Virol. 1993;67:1772–1777. doi: 10.1128/jvi.67.4.1772-1777.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O’Brien S J Hemophilia Growth and Development Study; Multicenter AIDS Cohort Study; Multicenter Hemophilia Cohort Study; San Francisco City Cohort; ALIVE study. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 12.De Roda Husman, A. M., M. Koot, M. Cornelissen, M. Brouwer, S. M. Broersen, M. Bakker, M. T. L. Roos, M. Prins, F. De Wolf, R. A. Coutinho, F. Miedema, J. Goudsmit, and H. Schuitemaker. Association between CCR5 genotype and the clinical course of HIV-1 infection. Ann. Intern. Med., in press. [DOI] [PubMed]
- 13.Donaldson Y K, Bell J E, Ironside J W, Brette R P, Robertson J R, Busuttil A, Simmonds P. Redistribution of HIV outside the lymphoid system with the onset of AIDS. Lancet. 1994;343:382–385. doi: 10.1016/s0140-6736(94)91222-x. [DOI] [PubMed] [Google Scholar]
- 14.Embretson J, Zupancic M, Ribas J L, Burke A, Racz P, Tenner-Racz K, Haase A T. Massive covert infection of helper T lymphocytes and macrophages by HIV during the incubation period of AIDS. Nature. 1993;362:359–361. doi: 10.1038/362359a0. [DOI] [PubMed] [Google Scholar]
- 15.Epstein L G, Kuiken C, Blumberg B M, Hartman S, Sharer L R, Clement M, Goudsmit J. HIV-1 V3 domain variation in brain and spleen of children with AIDS: tissue-specific evolution within host-determined quasispecies. Virology. 1991;180:583–590. doi: 10.1016/0042-6822(91)90072-j. [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein J. PHYLIP manual version 3.5. Berkeley, Calif: University Herbarium of the University of California at Berkeley; 1993. [Google Scholar]
- 17.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 18.Fouchier R A M, Brouwer M, Broersen S M, Schuitemaker H. Simple determination of human immunodeficiency virus type 1 syncytium-inducing genotype by PCR. J Clin Microbiol. 1995;33:906–911. doi: 10.1128/jcm.33.4.906-911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fouchier R A M, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillis S, Ferm M M, Ou W, Smith K A. T cell growth factor: parameters of production and a quantitative microassay for activity. J Immunol. 1978;120:2027–2032. [PubMed] [Google Scholar]
- 21.Goudsmit J, De Wolf F, Paul D A, Epstein L G, Lange J M A, Krone W J A, Speelman H, Wolters E C, Van der Noordaa J, Oleske J M, van der Helm H J, Coutinho R A. Expression of human immunodeficiency virus antigen (HIV-Ag) in serum and cerebrospinal fluid during acute and chronic infection. Lancet. 1986;ii:177–180. doi: 10.1016/s0140-6736(86)92485-2. [DOI] [PubMed] [Google Scholar]
- 22.Groenink M, Fouchier R A M, Broersen S, Baker C H, Koot M, Van’t Wout A B, Huisman H G, Miedema F, Tersmette M, Schuitemaker H. Relation of phenotype evolution of HIV-1 to envelope V2 configuration. Science. 1993;260:1513–1516. doi: 10.1126/science.8502996. [DOI] [PubMed] [Google Scholar]
- 23.Haase T A, Henry K, Zupanc M, Sedgewick G, Faust R A, Melroe H, Cavert W, Gebhard K, Staskus K, Zhang Z, Dailey P J, Balfour H H, Erice A, Perelson A S. Quantitative image analysis of HIV-1 infection in lymphoid tissue. Science. 1996;274:985–989. doi: 10.1126/science.274.5289.985. [DOI] [PubMed] [Google Scholar]
- 24.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 25.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau N R, Phair J, Ho D D, Koup R A. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 26.Hughes E S, Bell J E, Simmonds P. Investigation of the dynamics of the spread of human immunodeficiency virus to brain and other tissues by evolutionary analysis of sequences from the p17gag and envgenes. J Virol. 1997;71:1272–1280. doi: 10.1128/jvi.71.2.1272-1280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura M. A simple method for estimating evolutionary rates of base substitution through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 28.Koot M, Keet I P M, Vos A H V, De Goede R E Y, Roos M T L, Coutinho R A, Miedema F, Schellekens P T A, Tersmette M. Prognostic value of human immunodeficiency virus type 1 biological phenotype for rate of CD4+cell depletion and progression to AIDS. Ann Intern Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 29.Korber, B. T. M., E. E. Allen, A. D. Farmer, and G. L. Myers. 1995. Heterogeneity of HIV-1 and HIV-2. AIDS 9(Suppl. A):S5–S18. [PubMed]
- 30.Korber B T M, Kunstman K J, Patterson B K, Furtado M, McEvilly M M, Levy R, Wolinsky S M. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of barin-derived sequences. J Virol. 1994;68:7467–7481. doi: 10.1128/jvi.68.11.7467-7481.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuiken C L, Cornelissen M T E, Zorgdrager F, Hartman S, Gibbs A J, Goudsmit J. Consistent risk group-associated differences in human immunodeficiency virus type 1 vpr, vpu and V3 sequences despite independent evolution. J Gen Virol. 1996;77:783–792. doi: 10.1099/0022-1317-77-4-783. [DOI] [PubMed] [Google Scholar]
- 32.Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. CABIOS. 1994;10:189–192. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- 33.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:1–20. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 34.McGavin C H, Land S A, Sebire K L, Hooker D J, Gurusinghe A D, Birch C J. Syncytium-inducing phenotype and zidovudine susceptibility of HIV-1 isolates from post-mortem tissue. AIDS. 1996;10:47–53. doi: 10.1097/00002030-199601000-00007. [DOI] [PubMed] [Google Scholar]
- 35.Moses A V, Stenglein S G, Strussenberg J G, Wehrly K, Chesebro B, Nelson J A. Sequences regulating tropism of human immunodeficiency virus type 1 for brain capillary endothelial cells map to a unique region on the viral genome. J Virol. 1996;70:3401–3406. doi: 10.1128/jvi.70.6.3401-3406.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mueller B U, Sei S, Anderson B, Luzuriaga K, Farley M, Venzon D J, Tudor-Williams G, Schwartzentruber D J, Fox C, Sullivan J L, Pizzo P A. Comparison of virus burden in blood and sequential lymph node biopsy specimens from children infected with human immunodeficiency virus. J Pediatr. 1996;129:410–418. doi: 10.1016/s0022-3476(96)70074-4. [DOI] [PubMed] [Google Scholar]
- 37.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 38.Pang S, Vinters H V, Akashi T, O’Brien W A, Chan I S Y. HIV-1 env sequence variation in brain tissue of patients with AIDS-related neurologic disease. J Acquired Immune Defic Syndr. 1991;4:1082–1092. [PubMed] [Google Scholar]
- 39.Pantaleo G, Graziosi C, Demarest J F, Butini L, Montroni M, Fox C H, Orenstein J M, Kotler D P, Fauci A S. HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease. Nature. 1993;362:355–358. doi: 10.1038/362355a0. [DOI] [PubMed] [Google Scholar]
- 40.Pantaleo G, Graziosi C, Demarest J F, Cohen O J, Vaccarezza M, Gantt K, Muro-Cacho C, Fauci A S. Role of lymphoid organs in the pathogenesis of human immunodeficiency virus (HIV) infection. Immunol Rev. 1994;140:105–130. doi: 10.1111/j.1600-065x.1994.tb00867.x. [DOI] [PubMed] [Google Scholar]
- 41.Pantaleo G, Graziosi C, Fauci A S. Mechanisms of disease: the immunopathogenesis of human immunodeficiency virus infection. N Engl J Med. 1993;328:327–335. doi: 10.1056/NEJM199302043280508. [DOI] [PubMed] [Google Scholar]
- 42.Roos M T L, Lange J M A, De Goede R E Y, Coutinho R A, Schellekens P T A, Miedema F, Tersmette M. Viral phenotype and immune response in primary human immunodeficiency virus type 1 (HIV-1) infection. J Infect Dis. 1992;165:427–432. doi: 10.1093/infdis/165.3.427. [DOI] [PubMed] [Google Scholar]
- 43.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 44.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C-M, Saragosti S, Lapouméroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 45.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, De Goede R E Y, Van Steenwijk R P, Lange J M A, Eeftink Schattenkerk J K M, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuitemaker H, Kootstra N A, De Goede R E Y, De Wolf F, Miedema F, Tersmette M. Monocytotropic human immunodeficiency virus type 1 (HIV-1) variants detectable in all stages of HIV infection lack T-cell line tropism and syncytium-inducing ability in primary T-cell culture. J Virol. 1991;65:356–363. doi: 10.1128/jvi.65.1.356-363.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seibert S A, Howell C Y, Hughes M K, Hughes A L. Natural selection on the gag, pol, and env genes of human immunodeficiency virus 1 (HIV-1) Mol Biol Evol. 1995;12:803–813. doi: 10.1093/oxfordjournals.molbev.a040257. [DOI] [PubMed] [Google Scholar]
- 48.Sheehy N, Desselberger U, Whitwell H, Ball J K. Concurrent evolution of regions of the envelope and polymerase genes of human immunodeficiency virus type 1 observed during zidovudine (AZT) therapy. J Gen Virol. 1996;76:1071–1081. doi: 10.1099/0022-1317-77-5-1071. [DOI] [PubMed] [Google Scholar]
- 49.Simmonds P, Balfe P, Ludlam C A, Bishop J O, Brown A J L. Analysis of sequence diversity in hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J Virol. 1990;64:5840–5850. doi: 10.1128/jvi.64.12.5840-5850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamalet C, Lafeuillade A, Yahi N, Vignoli C, Tourres C, Pellegrino P, de Micco P. Comparison of viral burden and phenotype of HIV-1 isolates from lymph nodes and blood. AIDS. 1994;8:1083–1088. doi: 10.1097/00002030-199408000-00007. [DOI] [PubMed] [Google Scholar]
- 51.Tersmette M, De Goede R E Y, Al B J M, Winkel I N, Gruters R A, Cuypers H T M, Huisman H G, Miedema F. Differential syncytium-inducing capacity of human immunodeficiency virus isolates: frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J Virol. 1988;62:2026–2032. doi: 10.1128/jvi.62.6.2026-2032.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tersmette M, Gruters R A, De Wolf F, De Goede R E Y, Lange J M A, Schellekens P T A, Goudsmit J, Huisman J G, Miedema F. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J Virol. 1989;63:2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van’t Wout, A. B., H. Blaak, L. J. Ran, M. Brouwer, C. Kuiken, and H. Schuitemaker. Evolution of syncytium inducing and non-syncytium inducing biological virus clones in relation to replication kinetics during the course of HIV-1 infection. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 54.Van’t Wout A B, Kootstra N A, Mulder-Kampinga G A, Albrecht-van Lent N, Scherpbier H J, Veenstra J, Boer K, Coutinho R A, Miedema F, Schuitemaker H. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral and vertical transmission. J Clin Invest. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Saag M S, Shaw G M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 56.Wong J K, Ignacio C C, Torriani F, Havlir D, Fitch N J S, Richman D D. In vivo compartmentalization of human immunodeficiency virus: evidence from the examination of polsequences from autopsy tissues. J Virol. 1997;71:2059–2071. doi: 10.1128/jvi.71.3.2059-2071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang L Q, MacKenzie P, Cleland A, Holmes E C, Leigh-Brown A J, Simmonds P. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J Virol. 1993;67:3345–3356. doi: 10.1128/jvi.67.6.3345-3356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 59.Zhu T, Wang N, Carr A, Nam D S, Moor-Jankowski R, Cooper D A, Ho D D. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J Virol. 1996;70:3098–3107. doi: 10.1128/jvi.70.5.3098-3107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]