Summary
Variants of uncertain significance (VUSs) in BRCA2 are a common result of hereditary cancer genetic testing. While more than 4,000 unique VUSs, comprised of missense or intronic variants, have been identified in BRCA2, the few missense variants now classified clinically as pathogenic or likely pathogenic are predominantly located in the region encoding the C-terminal DNA binding domain (DBD). We report on functional evaluation of the influence of 462 BRCA2 missense variants affecting the DBD on DNA repair activity of BRCA2 using a homology-directed DNA double-strand break repair assay. Of these, 137 were functionally abnormal, 313 were functionally normal, and 12 demonstrated intermediate function. Comparisons with other functional studies of BRCA2 missense variants yielded strong correlations. Sequence-based in silico prediction models had high sensitivity, but limited specificity, relative to the homology-directed repair assay. Combining the functional results with clinical and genetic data in an American College of Medical Genetics (ACMG)/Association for Molecular Pathology (AMP)-like variant classification framework from a clinical testing laboratory, after excluding known splicing variants and functionally intermediate variants, classified 431 of 442 (97.5%) missense variants (129 as pathogenic/likely pathogenic and 302 as benign/likely benign). Functionally abnormal variants classified as pathogenic by ACMG/AMP rules were associated with a slightly lower risk of breast cancer (odds ratio [OR] 5.15, 95% confidence interval [CI] 3.43–7.83) than BRCA2 DBD protein truncating variants (OR 8.56, 95% CI 6.03–12.36). Overall, functional studies of BRCA2 variants using validated assays substantially improved the variant classification yield from ACMG/AMP models and are expected to improve clinical management of many individuals found to harbor germline BRCA2 missense VUS.
Keywords: cancer, BRCA2, VUS, HDR, variant classification, predisposition genes
Combining results from a homology-directed repair functional assay with clinical and genetic data in an American College of Medical Genetics (ACMG)/AMP-like variant classification framework classified 97.5% of the evaluated BRCA2 non-splicing missense variants as pathogenic/likely pathogenic or benign/likely benign. The results should prove useful for clinical management of individuals with germline BRCA2 missense variants.
Introduction
Germline pathogenic variants (PVs) in several cancer predisposition genes are associated with increased breast cancer risk.1 The rapid application of genetic testing has provided benefits to individuals found to harbor PVs in predisposition genes, such as enhanced screening, improved clinical management, and cancer surveillance for relatives. However, the identification of many variants of uncertain significance (VUSs) has led to difficulties in breast cancer risk evaluation and clinical management of individuals with these variants. The majority of VUSs are missense variants that are individually rare in the general population but collectively common, with over 4,000 such variants identified in BRCA2 alone. In the past, a limited number of VUSs in BRCA1 and BRCA2 were classified as pathogenic or benign based on multifactorial prediction models using a prior probability of pathogenicity and information on personal and family history of cancer and cosegregation of variants with cancer.2,3,4 More recently, American College of Medical Genetics (ACMG)/Association for Molecular Pathology (AMP) guidelines that incorporate variant frequency, family history, in silico prediction models, and other evidence have been used by clinical testing centers to classify several additional variants as pathogenic or benign. However, the great majority of missense variants remain classified as VUSs due to limited phenotype and genotype information.
Information from functional assays validated using known pathogenic and benign variants can also be incorporated into the ACMG/AMP variant classification framework. Assays evaluating the impact of variants on homologous recombination DNA repair activity, cell survival, and response to DNA-damaging agents have been reported for BRCA2.5,6,7,8,9,10,11 Results from a homology-directed repair (HDR) cell-based assay that can provide strong evidence of pathogenicity (PS3/BS3) for missense variants have recently been combined with other ACMG/AMP rules-based data to classify approximately 90% of targeted VUSs from the BRCA2 DNA binding domain (DBD) in studies from clinical testing laboratories.12,13 Thus, functional assay data, when added to the ACMG/AMP rules-based classification framework, can potentially reclassify large numbers of VUSs in BRCA2.
Here, we report on functional evaluation of 462 BRCA2 missense variants affecting the DBD using a BRCA2 HDR assay that measures the influence of the variants on the homologous recombination DNA repair activity of BRCA28 and has been associated with strong odds of pathogenicity for BRCA2 non-functional variants under the PS3/BS3 rule from the ACMG/AMP variant classification framework.12,13 We provide comparisons with results from other functional assays and in silico sequence-based prediction models for subsets of these variants. The HDR assay results were combined with other clinical and genetic data for variant classification using an ACMG/AMP classification framework.13 Additionally, we estimated the risk of breast and ovarian cancer associated with variants classified as pathogenic or benign. The clinical classification of 431 BRCA2 missense variants and the estimation of risks of breast and ovarian cancer associated with these variants may advance the utility of genetic testing and improve clinical management for individuals harboring these variants and their families.
Material and methods
Variant selection and annotation
The 462 BRCA2 missense variants affecting the DBD represent a summary of all variants evaluated by HDR functional analysis in the Couch laboratory, including 174 variants not reported in prior studies (Table S1).7,12,13,14,15,16 Briefly, missense variants affecting the BRCA2 DBD (amino acids 2481–3186) were selected for HDR evaluation based on ClinVar/gnomAD database observations, predicted functional effects by the BayesDel and/or BRCA_ML sequence-based in silico prediction models, prior evaluation in other functional studies, and direct requests from physicians and clinical testing labs. All variants were labeled by the Human Genome Variation Society (HGVS) nomenclature (RefSeq transcript ID: NM_000059.3) and annotated with functional scores from the BayesDel17 and BRCA_ML (https://github.com/Steven-N-Hart/BRCA-ML) in silico prediction models, as well as SpliceAI (https://github.com/Illumina/SpliceAI) scores predicting the effects of nucleotide changes on splice donor and acceptor gain or loss.
HDR assay
The BRCA2 missense HDR assay has been described previously.13 This functional assay provides strong evidence of pathogenicity for BRCA2 missense variants affecting the DBD (odds of pathogenicity > 18.7) and has been incorporated into the ACMG/AMP guidelines for BRCA2 VUS classification.12,13 Importantly, this odds of pathogenicity for the assay was estimated using pathogenic and benign missense standards that were classified independently of functional evidence. Briefly, full-length BRCA2 cDNA in a mammalian expression vector was subjected to site-directed mutagenesis to introduce each of the 462 BRCA2 missense variants. Variants were verified by Sanger sequencing. Production of full-length BRCA2 was confirmed by western blot using anti-BRCA2 antibody (OP95, Calbiochem). BRCA2 variant constructs were co-transfected with iSce1 expression vector into 1 × 106 brca2-deficient V-C8 cells containing the DR-GFP reporter.3,7,8,13,14,15,16 The proportion of viable cells displaying green fluorescent protein (GFP+) was quantified by flow cytometry after 72 h. Fold changes in GFP+ cells were normalized and rescaled to a 1–5 range based on the c.8167G>C (p.Asp2723His) standard PV scored as 1 and the wild-type BRCA2 scored as 5. All experiments were conducted in duplicate. Stability of HDR scores across time is shown for six representative missense variants (Figure S1). A Bayesian regression model for the log HDR scores was used to estimate the distribution of HDR scores8,13 and 95% confidence intervals for all variants and previously defined 99.9% probability thresholds for both pathogenic (<1.49) and benign (>2.50) variants were applied.12 Variants scoring between 1.49 and 2.50 were categorized as intermediate.
Comparisons of the HDR assay with other functional assays and in silico prediction models
Results for BRCA2 missense variants from the HDR assay were compared with results from other functional studies, including mouse embryonic stem cell (mESC)-based functional analysis of homologous recombination activity and sensitivity to therapeutic agents,5,11,18,19,20 BRCA2-deficient cell-line-based drug response assays using four DNA damaging agents (MANO-B),10 and a high-throughput prime-editing-based saturation genome editing analysis of exons 15 and 17.6 In addition, results from the HDR assay were compared with several in silico prediction scores including BayesDel,17 which has been selected by the ClinGen Variant Curation Expert Panel (VCEP) as the model for assessment of BRCA1/2 variants (BayesDel Ambry Genetics BRCA2-specific threshold of >0.431 for functionally abnormal variants and <0.056 for functionally normal variants); the BRCA_ML ensemble model that was trained on functional data18; EVE, an unsupervised deep-learning method implementing a generative variational autoencoder21; ESM1b, a deep protein language model shown to be effective in predicting variant effects22; and AlphaMissense, which is based on unsupervised protein language modeling and incorporates structural context from an AlphaFold-derived system.23
Clinical classification of variants using an ACMG/AMP framework
Data for an Ambry Genetics ACMG/AMP-like variant classification framework were assembled and used for variant classification by both a qualitative categorization13 and a points-based classification system.24 Briefly, for pathogenicity scoring, the following ACMG/AMP codes were evaluated: PS3 (functional study), PS4 (case-control analysis), PM1 (mutational hotpot), PM2 (rarity), PM3 (in trans with pathogenic BRCA2 variants in subjects affected by Fanconi anemia), PM5 (Grantham-informed hotspot), PP1 (cosegregation), PP3 (in silico BayesDel Ambry Genetics BRCA2-specific threshold: >0.431), and PP3 (in silico BayesDel-ClinGen general thresholds: strong >0.5, moderate 0.27 to 0.5, supporting 0.13 to 0.27) were applied.25 For benign impact, the following ACMG/AMP codes were evaluated: BA1 (population frequency >0.1%), BS1 (population frequency >0.01%), BS3 (functional study), BP2 (in trans with pathogenic BRCA2 variants in subjects unaffected by Fanconi anemia), BP4 (in silico BayesDel Ambry Genetics BRCA2-specific threshold: <0.056), and BP4 (in silico ClinGen general thresholds: supporting −0.36 to −0.18, moderate <−0.36) were applied. The strength ascribed to each code is designated within the second letter as supporting evidence (P), moderate (M), and strong (S) evidence.
Statistical analysis
The sensitivities and specificities of different assays for variants classified by the HDR assay were determined. Associations (odds ratios [ORs], 95% confidence intervals, and p values) between functionally evaluated variants and breast and ovarian cancer were estimated by logistic regression of the frequencies of selected variants in breast and ovarian cases subjected to clinical germline testing by Ambry Genetics between 2012 and 2017 and in female, non-cancer reference controls from the gnomAD and female, non-cancer controls from the CARRIERS population-based study,1 weighted by race and ethnicity. Absolute risks of breast cancer and ovarian cancer to age 80 were calculated for non-Hispanic white individuals harboring these variants by combining gene-specific ORs with age-specific breast cancer incidence rates for non-Hispanic white women from the NCI SEER (SEER21) Program.1 All-cause mortality was included as a competing event based on the US Centers for Disease Control and Prevention mortality rates among women (https://wonder.cdc.gov/). All statistical analyses were performed using R (version 3.5.2), and all tests were two sided.
Informed consent and ethics approval
The study was approved by the Mayo Clinic institutional review board, and the analysis of the clinical-testing cohort was deemed exempt from review by the Western Institutional Review Board.
Results
HDR analysis of BRCA2 missense variants affecting the DBD
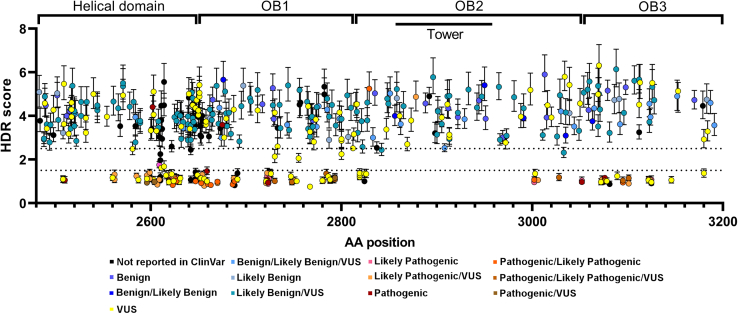
The HDR functional assay was previously shown to have 100% sensitivity (95% CI 79%–100%) and 100% specificity (95% CI 93%–100%) for pathogenic missense variants in the BRCA2 DBD based on 21 known non-splicing pathogenic/likely pathogenic and 35 known benign/likely benign missense variants.12 Thresholds for 99.9% probability for pathogenic and benign effects were estimated based on these standards, and an HDR score <1.49 was considered functionally abnormal (nonfunctional) with probability of pathogenicity >0.99, whereas variants with HDR score >2.50 were considered functionally normal (functional) with probabilities of neutrality >0.99. Here, we report on HDR assay analysis of 462 missense variants, affecting the DBD between amino acid (aa) residues 2508 and 3180, that includes 403 variants previously reported in ClinVar (Figure 1). Among these variants, 137 (30.4%) had HDR scores <1.49 (ranging from 0.76 to 1.43), similar to pathogenic missense variants, and were designated as functionally abnormal. In contrast, 313 had HDR scores >2.5 (ranging from 2.52 to 6.14), similar to benign missense variants, and were designated as functionally normal. Another 12 variants had HDR scores between 1.49 and 2.5 and were designated as functionally intermediate (Figure 1; Table S1). When excluding 8 observed splice variants, 135 were designated functionally abnormal, 12 functionally intermediate, and 307 functionally normal.
Figure 1.
HDR score of 462 BRCA2 missense variants
Plot of 462 BRCA2 missense variants affecting the DBD ordered by amino acid position (x axis) against HDR score with 95% CIs. The thresholds for functionally abnormal (HDR < 1.49) and functionally normal (HDR > 2.5) were plotted as horizontal dotted lines. Variants reported in ClinVar were categorized by color.
The BRCA2 DBD helical subdomain (HD; residues 2482–2668) contained 55 of the 137 (40%) of the functionally abnormal variants along with 125 functionally normal and 7 functionally intermediate variants; the oligonucleotide/oligosaccharide binding domain 1 (OB1 aa2670–aa2803) contained 46 (34%) of the functionally abnormal variants along with 67 functionally normal and 3 functionally intermediate variants; OB3 (aa3055–aa3184) had 25 (18%) of the functionally abnormal variants along with 40 functionally normal variants. In contrast, only 11 of the functionally abnormal variants (8%) were located in the OB2 domain (aa2808–aa3049) (excluding the aa2838–aa2961 tower domain) along with 42 functionally normal variants and 1 functionally intermediate variant. Furthermore, no functionally abnormal variants, 1 functionally intermediate variant, and 39 functionally normal variants were found in the tower domain, a 130 aa region that adopts a tower-like structure protruding from the OB2 (Figure 1). Additionally, 50 variants in the DBD were in residues that interact with DSS1, which is thought to stabilize the BRCA2 DBD, and 9 variants located between aa2833 to aa3126 were in residues that interact with single-strand DNA during homologous recombination repair of DNA damage (Figure 2; Table S1).
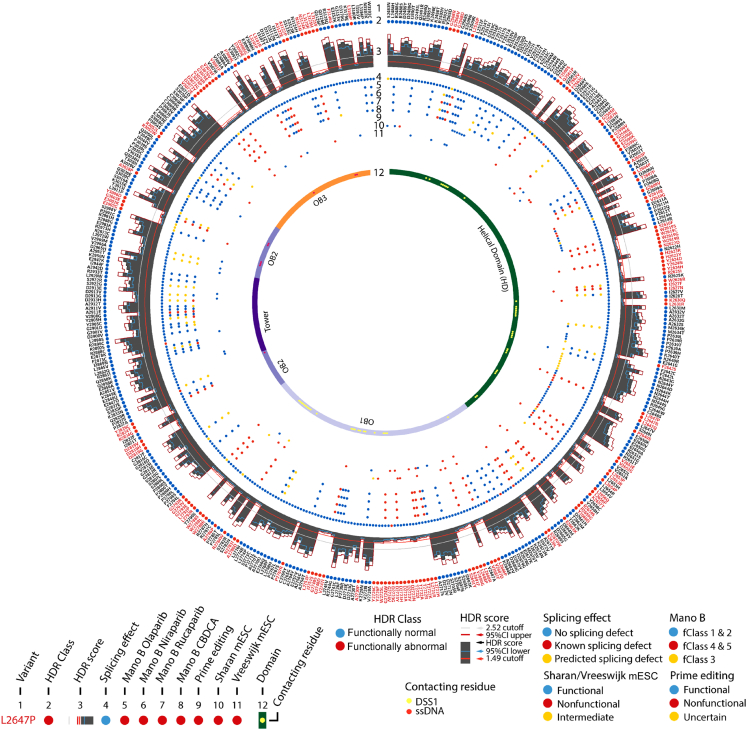
Figure 2.
Characteristics of 462 BRCA2 missense variants
CIRCOS plot of BRCA2 missense variants. (1) Variant; (2) HDR class (blue: functionally normal, red: functionally abnormal); (3) histogram of HDR score with individual HDR 95% CI upper (red) and 95% CI lower (blue) (orange line indicates 1.49 cutoff point, gray line indicates 2.52 cutoff point); (4) splicing effect (blue: no splicing effect; red: known splicing effect; yellow: predicted splicing effect by SpliceAI); (5–8) MANO-B four drugs classification (olaparib, niraparib, rucaparib, and carboplatin [CBDCA]); (9) prime editing classification (blue: functional, red: nonfunctional, yellow: uncertain); (10–11) mESC functional assay result (Sharan5,18,19 and Vreeswijk11; blue: functional, red: nonfunctional, yellow: intermediate); (12) protein structure domain (HD, OB1, tower, OB2, OB3) with yellow dot indicating DSS1 contacting residue and red dot indicating single-stranded DNA (ssDNA) contacting residue.
Because ACMG/AMP guidelines for classification of variants include a PM5 variant hotspot rule, where a variant located at the same aa position as a known functionally abnormal variant may be used as evidence of pathogenicity, residues containing multiple different functionally evaluated variants were evaluated. In the current study, 106 of the 137 HDR functionally abnormal variants (77.4%), 8 of the 12 functionally intermediate variants (66.7%), and 169 of the 313 HDR functionally normal variants (54%) were in the same aa as at least one other HDR evaluated variant (Table S1). Six variants located in residue 2723 all demonstrated non-functional effects in the HDR assay. Similarly, 30 other hotspot residues harbored only functionally abnormal or intermediate variants, and 56 hotspot residues harbored only functionally normal variants (Table S1). Furthermore, 23 hotspot residues contained both functionally normal and abnormal variants (Table S1). These data suggest that the PM5 variant hotspot rule for predicting PVs should be re-evaluated.
Comparisons between HDR results and in silico prediction models
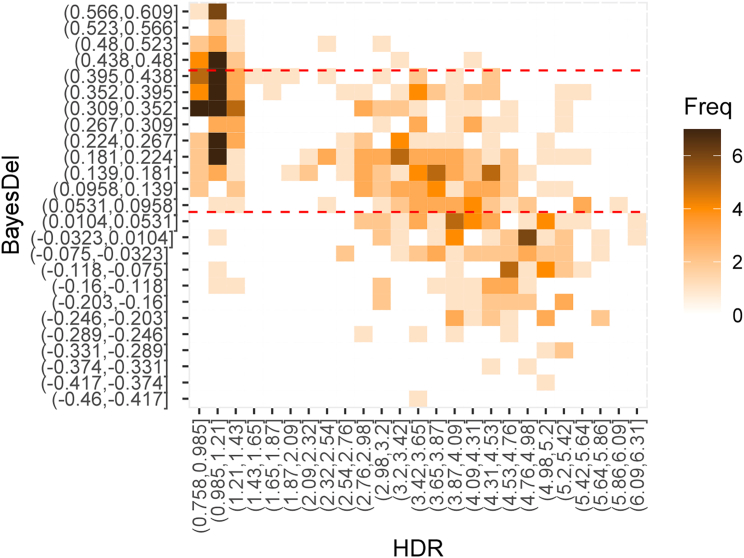
The HDR assay results were also compared with functional effects predicted by the in silico BayesDel and BRCA_ML models (Figures 3 and S2; Table S1). BayesDel displayed 42.9% (21.8%–66.0%) sensitivity and 100% (92.1%–100.0%) specificity for 21 known BRCA2 pathogenic and 45 known benign non-splicing missense standard variants when applying the Ambry Genetics BRCA2-specific BayesDel threshold of >0.431 for functionally abnormal variants and ≤0.431 for functionally normal and intermediate variants. Interestingly, when the intermediate BayesDel category (0.431–0.056) was combined with the functionally abnormal category (>0.431), the sensitivity improved to 95.2% (76.2%–99.9%), whereas specificity fell to 80% (65.4%–90.4%). When applied to the full 454 variants scored by HDR assays, the combined functionally abnormal and intermediate categories of BayesDel yielded 97.3% (93.1%–99.3%) sensitivity and 45.0% (39.3%–50.7%) specificity. In contrast, when applying ClinGen genome-wide thresholds to BayesDel, a sensitivity of 92.59% (86.8%–96.39%) and specificity of 56.74% (51.1%–62.25%) was achieved. Application of the BRCA_ML in silico model to the 454 variants after exclusion of training data yielded 78.2% (68.0%–86.3%) sensitivity and 75.7% (69.4%–81.2%) specificity (Figure S2). Based on general thresholds for functionally abnormal variants (<−7.5 damaging; >−7.5 neutral), ESM1b yielded sensitivity of 89.6% (83.2%–94.2%) and specificity of 53.3% (47.7%–58.9%) for the 454 variants with HDR scores. EVE yielded 98.5% (94.8%–99.8%) sensitivity and 21.6% (17.2%–26.7%) specificity when compared to the 454 HDR results. Finally, AlphaMissense yielded sensitivity of 95.56% (90.58%–98.35%) and specificity of 71.47% (66.18%–76.37%) when compared to the 454 HDR results. Thus, when considering that high specificity is most important for clinical testing in order to avoid misclassification of a variant as functionally abnormal and pathogenic, the BRCA_ML and AlphaMissense models perform the best.
Figure 3.
Correlation of in silico prediction model scores with HDR
Heatmaps indicating the correlation between HDR scores and the BayesDel in silico prediction model for BRCA2 effect. The heatmap contains 25 equal categories of BayesDel scores (y axis) and 25 categories for HDR score (x axis). Red dashed lines reflect thresholds for Ambry BRCA2-specific BayesDel functionally normal (0.056) and functionally abnormal (0.431). HDR score thresholds are <1.50 for functionally abnormal and >2.50 for functionally normal.
Comparisons between functional assays
A total of 216 variants evaluated by the HDR assay were previously evaluated by other functional assays including an mESC based assay (70 of 462), MANO-B drug assay (120 of 462), prime-editing-based cell survival assay (59 of 462), and DLD BRCA2 (−/−) homologous recombination assay (10 of 462) (Figure 2; Table S2; Figure S3). The sensitivity of these assays for the functionally abnormal variants in the HDR assay, based on functional/nonfunctional/intermediate or uncertain categories for each assay (consistent with terminology used in respective studies), or functional classes for the Mano B assay ranged from 93% to 100%, whereas the specificity ranged from 38% to 100% (Figure 2; Table S3). The prime-editing-based high-throughput cell survival assay displayed sensitivity of 100% but had the lowest specificity of 38% due to a large number of uncertain variants that were functionally normal in the HDR assay. The mESC-based studies all showed sensitivity of 100% and specificity of 95%–100% except one study with specificity of 40%5 (Table S3). The BRCA2/DSS1 binding assay study as well as the DLD BRCA2 (−/−) HR assay both showed 100% sensitivity and 100% specificity when compared to HDR assay, although these comparisons were based on small numbers of variants (Table S1). The Mano B drug assay, which evaluated response to four different drugs (olaparib, niraparib, rucaparib, and CBDCA) had high sensitivities ranging from 93% to 98% but relatively lower specificities ranging from 69% to 81% caused by variants with intermediate effects in Mano B that were predominantly functionally normal in the HDR assay (Table S3). Thus, the selected functional assays appear to be highly effective in identifying variants with functionally abnormal effects but are less effective in distinguishing intermediate variants from functionally normal variants, in part because of challenges in calibrating assays.
Clinical classification of BRCA2 missense variants
An ACMG/AMP classification framework developed by Ambry Genetics for BRCA2 variant classification that incorporated functional HDR data and used point scoring rather than qualitative estimates for each classification rule was applied to 442 variants (excluding the known splicing variants and functionally intermediate variants) (Figures S4 and S5; Table S4).3,4,8,9 In parallel, an ACMG/AMP classification model incorporating a PP3/BP4 rule based on different ClinGen-approved thresholds for the BayesDel in silico prediction model was also used for variant classification (Table S4). Out of the 442 variants, 300 were classified as benign or likely benign variants, 126 were classified as pathogenic or likely pathogenic variants, and 16 remained as VUSs (−1 to +5 points) by either classification model (Table S4). Importantly, there was no discordance in point classifications when using either the Ambry Genetics BRCA2-specific or the ClinGen general in silico BayesDel thresholds. Of the 386 variants reported in ClinVar, 289 were reported as VUSs by at least one ClinGen-approved testing laboratories (Figure 1), and another five were reported by other laboratories. In the current study, 281 of these variants were successfully classified as either benign/likely benign or pathogenic/likely pathogenic when incorporating the HDR functional data into the ACMG/AMP classification models (Figures S4 and S5; Table S4). When combining all classifications from ClinVar, multifactorial/VarCall models, previous ACMG classification, and ACMG points classification from the current study, a total of 431 BRCA2 missense variants were classified (78 benign, 224 likely benign, 58 pathogenic, and 71 likely pathogenic), while 11 remained as VUSs (Figure 4; Table S4).
Figure 4.
Reclassification of missense variants in ClinVar
Changes in classification of 442 missense variants including 386 from ClinVar (left side) to the updated classification from all sources (updated; right side). Categories of variants are labeled by color.
Case-control association analysis
To better understand the contributions to cancer risk of the 135 HDR functionally abnormal non-splicing missense variants and the 129 of these variants that were classified as pathogenic/likely pathogenic, associations between pooled functionally abnormal variants and breast cancer were evaluated. Specifically, the frequency of the pooled 135 functionally abnormal variants in female, primary breast cancer cases subjected to hereditary cancer genetic testing by Ambry Genetics between 2012 and 2017 was compared to the frequency in female, non-cancer reference controls from gnomAD and female, non-cancer controls from the CARRIERS population-based study.1 The 135 functionally abnormal variants with HDR < 1.49 yielded an OR for breast cancer of 5.20 (95% CI 3.47–7.9) and the 129 missense variants classified as pathogenic/likely pathogenic using the Ambry Genetics ACMG classification model yielded similar results (OR 5.15, 95% CI 3.43–7.83) (Table 1). These ORs were attenuated compared to results for established standard pathogenic missense variants (OR 8.83, 95% CI 4.34–18.37), and DBD protein truncating mutations (OR 8.56, 95% CI 6.03–12.36), although the differences were not significantly different (Table 1). In contrast, exon 11 protein-truncating variants (PTVs), which are located in the ovarian cancer cluster region of BRCA2 and are associated with lower risks of breast cancer than other PTVs, were similar to the missense variants that were functionally impaired in the HDR assay and that were classified as likely pathogenic or pathogenic by the ACMG/AMP points-classification system (OR 5.53, 95% CI 4.72–6.48) (Table 1). Importantly, the missense variants scored as functionally normal by the HDR assay and classified as benign using the ACMG/AMP framework were not associated with increased breast cancer risk (OR 1.04, 95% CI 0.98–1.11). To further investigate the apparently attenuated effects, the functionally abnormal missense variants were evaluated in three categories based on strength of effect in the HDR assay: HDR < 1.2, HDR between 1.2 and 1.49, and HDR < 1.49. The results showed that the functionally abnormal missense variants with HDR < 1.2 (OR 5.25, 95% CI 3.34–8.39) had similar associations with breast cancer as those with HDR between 1.2 and 1.49 (OR 5.00, 95% CI 2.05–12.70) and all functionally abnormal variants with HDR < 1.49 (OR 5.20, 95% CI 3.47–7.9).
Table 1.
Case-control association analysis for BRCA2 missense variants affecting the DBD and PTVs in breast cancer and ovarian cancer
|
Case |
Controls |
Cancer risk |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| # mut | # tested | Freq (%) | # mut | # tested | Freq (%) | ORa | 95% CI | p value | ||
| Breast cancer | ||||||||||
| Functionally abnormal missense (HDR <1.2) | 84 | 82,372 | 0.11 | 24 | 123,464 | 0.02 | 5.25 | 3.34–8.39 | 1.63 × 10−15 | |
| Functionally abnormal missense (HDR 1.2–1.49) | 20 | 82,372 | 0.02 | 6 | 123,464 | 0.00 | 5.00 | 2.05–12.70 | 1.76 × 10−4 | |
| Functionally abnormal missense (HDR <1.49) | 104 | 82,372 | 0.13 | 30 | 123,464 | 0.02 | 5.2 | 3.47–7.9 | 9.99 × 10−19 | |
| Classified B or LB missense variants | 1,645 | 82,372 | 2.00 | 2,363 | 123,464 | 1.91 | 1.04 | 0.98–1.11 | 0.18 | |
| Classified P or LP missense variants | 103 | 82,372 | 0.13 | 30 | 123,464 | 0.02 | 5.15 | 3.43–7.83 | 1.88 × 10−18 | |
| Pathogenic missense standards | 53 | 82,372 | 0.06 | 9 | 123,464 | 0.01 | 8.83 | 4.34–18.37 | 2.07 × 10−13 | |
| DBD PTV | 211 | 82,372 | 0.26 | 37 | 123,464 | 0.03 | 8.56 | 6.03–12.36 | 1.37 × 10−48 | |
| Exon 11 PTV | 732 | 82,372 | 0.89 | 199 | 123,464 | 0.16 | 5.53 | 4.72–6.48 | 6.94 × 10−128 | |
| Ovarian cancer | ||||||||||
| Functionally abnormal missense (HDR < 1.2) | 22 | 10,960 | 0.2 | 24 | 123,464 | 0.02 | 10.34 | 5.69–18.43 | 1.25 × 10−12 | |
| Functionally abnormal missense (HDR 1.2–1.49) | 2 | 10,960 | 0.02 | 6 | 123,464 | 0.00 | 3.76 | 0.55–19.63 | 0.13 | |
| Functionally abnormal missense (HDR < 1.49) | 24 | 10,960 | 0.22 | 30 | 123,464 | 0.02 | 9.02 | 5.15–15.84 | 9.02 × 10−13 | |
| Classified B or LB missense variants | 232 | 10,960 | 2.13 | 2,363 | 123,464 | 1.91 | 1.11 | 0.97–1.27 | 0.14 | |
| Classified P or LP missense variants | 23 | 10,960 | 0.21 | 30 | 123,464 | 0.02 | 8.64 | 4.85–15.33 | 4.95 × 10−12 | |
| Pathogenic missense standards | 12 | 10,960 | 0.1 | 9 | 123,464 | 0.01 | 15.03 | 6.19–37.18 | 1.25 × 10−8 | |
| DBD PTV | 41 | 10,960 | 0.37 | 37 | 123,464 | 0.03 | 12.5 | 7.92–19.66 | 2.58 × 10−24 | |
| Exon 11 PTV | 221 | 10,960 | 2.02 | 199 | 123,464 | 0.16 | 12.63 | 10.38–15.33 | 3.07 × 10−124 | |
Cases: Ambry Genetics breast cancer or ovarian cancers; controls: gnomAD 2.1 and 3.1 non-cancer females and CARRIERS population-based controls.
OR, odds ratio calculated by Fisher’s exact test; HDR, homology directed repair; CI, confidence interval; Mut, number with variants of interest; Freq, frequency; DBD, DNA binding domain; PTV, protein truncating variant; P, pathogenic; LP, likely pathogenic; B, benign; LB, likely benign.
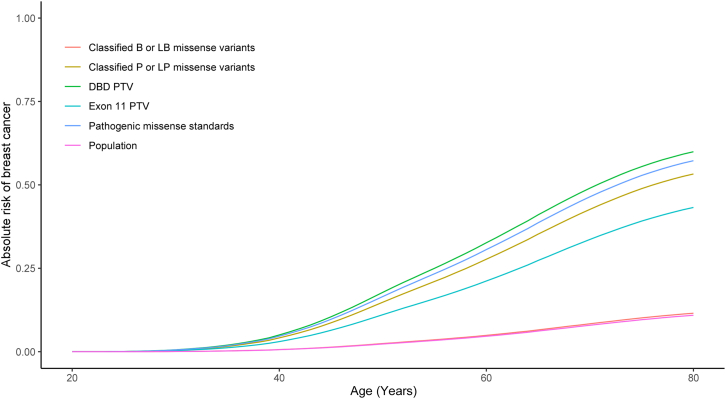
Comparisons of lifetime risk estimates for breast cancer were estimated using ORs from the current study and SEER rates of disease (1975–2020) (https://seer.cancer.gov/data/access.html). The classified pathogenic missense variants had lifetime risks of 53% to age 80, consistent with 57% risks for established pathogenic missense variant standards, 60% risks for DBD protein-truncating mutations, and 12% risks for classified benign missense variants (Figure 5; Table S5). A similar analysis of associations with ovarian cancer risk found that the pooled functionally abnormal missense variants with HDR < 1.49 were associated with substantially increased risks of ovarian cancer (OR 9.02, 95% CI 5.15–15.84) (Table 1). These results were similar to those for the classified pathogenic missense variants (OR 8.64, 95% CI 4.85–15.33) but lower than the established pathogenic missense variant standards (OR 15.03, 95% CI 6.19–37.18), results for DBD PTVs (OR 12.5, 95% CI 7.92–19.66), and exon 11 PTVs (OR 12.63, 95% CI 10.38–15.33). In contrast with ORs for breast cancer, when stratified by HDR score, variants scoring between 1.2 and 1.49 had relatively lower risk (OR 3.76, 95% CI 0.55–19.63) than the group of HDR < 1.2 (OR 10.34, 95% CI 5.69–18.43) and the overall group with HDR < 1.49 (OR 9.02, 95% CI 5.15–15.84). Ovarian cancer lifetime risk by age 80 was also estimated at 13% for the classified pathogenic missense variants from the current study and 17% for the DBD PTVs (Table S5; Figure S6).
Figure 5.
Lifetime breast cancer absolute risks associated with BRCA2 variants
Lifetime risks were estimated by combining case-control OR estimates for non-Hispanic White breast cancer subjects (breast cancer subjects subjected to cancer testing by Ambry Genetics versus gnomAD reference controls) with prevalence in the SEER registry. Lifetime risk curves include BRCA2 missense variants classified as benign (B) or likely benign (LB) (classified B or LB missense variants); BRCA2 missense variants classified as pathogenic (P) or likely pathogenic (LP) (classified as P or LP missense variants); pathogenic missense standards; BRCA2 protein truncating variants (PTVs) including frameshift and stop-gain variants affecting the DNA binding domain (DBD PTV); in exon 11 (exon 11 PTV); and general population risk of breast cancer (general population).
Discussion
There are over 20,000 possible missense variants in the full-length BRCA2 gene, and over 7,000 different individual missense variants have been observed and reported to ClinVar. This study represents a comprehensive functional analysis of 462 BRCA2 missense variants affecting the DBD domain, which is the only part of BRCA2 containing pathogenic missense variants in ClinVar. Of these, 137 variants exhibited loss of HDR function and were designated as functionally abnormal while 313 were found to be functionally normal, and 12 were functionally intermediate. When combining the functional data from 442 variants (after removing the splicing and functionally intermediate variants) with other genetic and clinical data in an Ambry Genetics ACMG/AMP classification framework, a total of 129 variants were classified as pathogenic/likely pathogenic, 302 were classified as benign or likely benign variants, and 11 remained as VUSs. The classification of 97.5% of the functionally evaluated variants in this study compares well with earlier studies of the HDR assay where 132 of 154 (85.7%) observed VUSs were classified using an earlier Ambry Genetics ACMG model13 and 62 of 67 (92.5%) observed VUSs were classified using a GeneDx classification model.12 Importantly, there are subtle differences in various ACMG/AMP classification frameworks. For instance, among the 442 evaluated BRCA2 missense variants, 289 variants have at least one uncertain significance interpretation in ClinVar reported from ClinGen-approved laboratories. Incorporation of these HDR data into the BRCA1/2 variant classification rules developed by the Evidence-based Network for the Interpretation of Germline Mutation Alleles (ENIGMA) consortium (http://enigmaconsortium.org) and ClinGen BRCA1/2 VCEP and BRCA1/2 classification frameworks developed by other entities will further inform on the status of the many variants reported in this study and is expected to improve risk assessment and management for many individuals and their family members who are known or will be found to harbor these variants.
The HDR assay results were very consistent with results from other functional studies, including mESC survival assays, Mano B drug response assays, prime-editing analysis, and DLD−/− cells. Each of these functional assays have been approved by the BRCA1/2 VCEP (https://cspec.genome.network/cspec/ui/svi/doc/GN092) as high-quality assays that can be utilized for BRCA2 variant curation. Relative to the HDR assay, each of the other assays yielded sensitivity for HDR functionally abnormal variants of >90%. While most of these assays have not yet been validated relative to established BRCA2 pathogenic and benign missense standards, the strong correlation with the HDR assay suggests that each method can be used to identify functionally abnormal variants for incorporation in ACMG/AMP/ClinGen classification frameworks. However, several of these assays show poor specificity relative to the HDR assay.
Comparisons between the HDR assay and in silico models were also performed. BayesDel is currently used as the in silico model of preference by the ClinGen BRCA1/2 VCEP (https://cspec.genome.network/cspec/ui/svi/doc/GN092). While BayesDel had high sensitivity (97.3%) using either Ambry Genetics or ClinGen thresholds this model yielded poor specificity (43.6% or 56.7%, respectively) for HDR scored variants. Given that high specificity is needed to avoid misclassification of variants as functionally abnormal and pathogenic, where errors may lead to unnecessary prophylactic surgery for individuals harboring improperly classified variants, reconsideration of BayesDel as the in silico model in the ClinGen ACMG/AMP BRCA1/2 classification model may be needed. In contrast, BRCA_ML, which was trained on HDR functional data, yielded 78.2% sensitivity and 75.7% specificity, and AlphaMissense yielded 95.6% sensitivity and 71.1% specificity for the 454 variants (excluding known splicing variants). Both methods may be of value for selection of certain variants for further evaluation. However, high-throughput multiplex assays of variant effect for BRCA2 are likely to nullify the utility of these prediction methods in the near future. The HDR functional assay results can serve as a validation dataset for these studies and for any individual variant that may have a discrepancy or uncertain result from the high-throughput analysis, providing a valuable cross-validation method.
The case-control association studies of pooled variants revealed that established pathogenic missense standards, likely pathogenic/pathogenic variants (as classified by an ACMG/AMP points system), and HDR functionally abnormal variants all showed high risks for breast and ovarian cancer and high lifetime risks of these cancers, similar to DBD PTVs. Furthermore, variants designated as functionally abnormal fClass 4/5 in the separate Mano B assay were also associated with a high risk of breast cancer (OR 5.32, 95% CI 3.36–8.81) (Table S6). These findings suggest that BRCA2 missense variants designated as functionally abnormal by HDR functional analysis are associated with high risks of breast cancer. However, estimates of breast cancer risk for ACMG/AMP-classified pathogenic missense variants were lower than risks for known pathogenic missense standards and also for PTVs located in the BRCA2 DBD region. These lower risks are consistent with a recent family-based study suggesting that BRCA2 missense variants affecting the DBD had approximately 70% of the risk of PTVs.26 The reduced risk relative to known pathogenic missense standards may be a reflection of the very strong effects of the standards, which were classified because they were detected in multiple large families and segregated well with cancer. Other functionally abnormal missense variants may not have equivalently strong risk effects. Similarly, missense variants had attenuated ovarian cancer risks relative to PTVs. Additional studies are needed to confirm these differences in risk by variant type. As testing and variant observations increase, it may become possible to estimate risks for individual variants.
One variant, c.7466A>G (p.Asp2489Gly), showed functionally normal activity in the HDR assay but was reported as nonfunctional in an mESC-based functional assay.5 The SpliceAI prediction for this variant was not significant, and cBROCA analysis reported no splicing effect.27 Furthermore, a multifactorial model for variant classification, based on co-segregation, family history of cancer, and in silico prediction models that has been used to establish pathogenic missense variants3 gave an intermediate posterior probability for this variant. Further evaluation is needed to resolve the discrepancies between these results.
In summary, HDR functional analysis and other validated functional assays provide a systematic and standardized approach for evaluating BRCA2 missense variants. The results will be incorporated into ACMG/AMP/ClinGen classification models to successfully classify BRCA2 VUSs. These results are expected to improve risk assessment and risk management for a large number of individuals found to harbor these VUSs with those found to have PVs potentially benefitting from enhanced cancer screening modalities, risk prediction for a variety of cancer, survival and chemo prevention measures, and access to the poly (ADP-ribose) polymerase (PARP) inhibitors for treatment in the setting of a cancer diagnosis. Furthermore, those found to have benign variants can benefit by knowing that the identified VUS is not driving risk of cancer for the tested individual or their family members.
Data and code availability
The published article includes all HDR scores and data for case-control associations and ACMG-like variant classifications. CARRIERS data is available at dbGAP (phs002820). Code for the analysis is available through Github at https://github.com/nboddicker/BRCA2_462_variant.git. Registered users can access the SEER data at https://seer.cancer.gov/data/access.html.
Acknowledgments
This work was supported by the Breast Cancer Research Foundation (F.J.C. and S.M.D.); National Institutes of Health (R35CA253187, R01CA225662, and P50 CA116201 Specialized Program of Research Excellence [SPORE] for breast cancer to F.J.C.).
Declaration of interests
R.K., T.P., J.D.W., C.M.C., C.A.S., C.C.Y., K.F., Z.H., and M.E.R. are employees of Ambry Genetics Inc. All other authors declare no competing interests.
Published: February 27, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2024.02.002.
Supplemental information
References
- 1.Hu C., Hart S.N., Gnanaolivu R., Huang H., Lee K.Y., Na J., Gao C., Lilyquist J., Yadav S., Boddicker N.J., et al. A Population-Based Study of Genes Previously Implicated in Breast Cancer. N. Engl. J. Med. 2021;384:440–451. doi: 10.1056/NEJMoa2005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldgar D.E., Easton D.F., Deffenbaugh A.M., Monteiro A.N.A., Tavtigian S.V., Couch F.J., Breast Cancer Information Core BIC Steering Committee Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am. J. Hum. Genet. 2004;75:535–544. doi: 10.1086/424388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindor N.M., Guidugli L., Wang X., Vallée M.P., Monteiro A.N.A., Tavtigian S., Goldgar D.E., Couch F.J. A review of a multifactorial probability-based model for classification of BRCA1 and BRCA2 variants of uncertain significance (VUS) Hum. Mutat. 2012;33:8–21. doi: 10.1002/humu.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsons M.T., Tudini E., Li H., Hahnen E., Wappenschmidt B., Feliubadaló L., Aalfs C.M., Agata S., Aittomäki K., Alducci E., et al. Large scale multifactorial likelihood quantitative analysis of BRCA1 and BRCA2 variants: An ENIGMA resource to support clinical variant classification. Hum. Mutat. 2019;40:1557–1578. doi: 10.1002/humu.23818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas K., Lipton G.B., Stauffer S., Sullivan T., Cleveland L., Southon E., Reid S., Magidson V., Iversen E.S., Jr., Sharan S.K. A computational model for classification of BRCA2 variants using mouse embryonic stem cell-based functional assays. NPJ Genom. Med. 2020;5:52. doi: 10.1038/s41525-020-00158-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erwood S., Bily T.M.I., Lequyer J., Yan J., Gulati N., Brewer R.A., Zhou L., Pelletier L., Ivakine E.A., Cohn R.D. Saturation variant interpretation using CRISPR prime editing. Nat. Biotechnol. 2022;40:885–895. doi: 10.1038/s41587-021-01201-1. [DOI] [PubMed] [Google Scholar]
- 7.Guidugli L., Pankratz V.S., Singh N., Thompson J., Erding C.A., Engel C., Schmutzler R., Domchek S., Nathanson K., Radice P., et al. A classification model for BRCA2 DNA binding domain missense variants based on homology-directed repair activity. Cancer Res. 2013;73:265–275. doi: 10.1158/0008-5472.CAN-12-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guidugli L., Shimelis H., Masica D.L., Pankratz V.S., Lipton G.B., Singh N., Hu C., Monteiro A.N.A., Lindor N.M., Goldgar D.E., et al. Assessment of the Clinical Relevance of BRCA2 Missense Variants by Functional and Computational Approaches. Am. J. Hum. Genet. 2018;102:233–248. doi: 10.1016/j.ajhg.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hart S.N., Polley E.C., Shimelis H., Yadav S., Couch F.J. Prediction of the functional impact of missense variants in BRCA1 and BRCA2 with BRCA-ML. NPJ Breast Cancer. 2020;6:13. doi: 10.1038/s41523-020-0159-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikegami M., Kohsaka S., Ueno T., Momozawa Y., Inoue S., Tamura K., Shimomura A., Hosoya N., Kobayashi H., Tanaka S., Mano H. High-throughput functional evaluation of BRCA2 variants of unknown significance. Nat. Commun. 2020;11:2573. doi: 10.1038/s41467-020-16141-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mesman R.L.S., Calléja F.M.G.R., Hendriks G., Morolli B., Misovic B., Devilee P., van Asperen C.J., Vrieling H., Vreeswijk M.P.G. The functional impact of variants of uncertain significance in BRCA2. Genet. Med. 2019;21:293–302. doi: 10.1038/s41436-018-0052-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu C., Susswein L.R., Roberts M.E., Yang H., Marshall M.L., Hiraki S., Berkofsky-Fessler W., Gupta S., Shen W., Dunn C.A., et al. Classification of BRCA2 Variants of Uncertain Significance (VUS) Using an ACMG/AMP Model Incorporating a Homology-Directed Repair (HDR) Functional Assay. Clin. Cancer Res. 2022;28:3742–3751. doi: 10.1158/1078-0432.CCR-22-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richardson M.E., Hu C., Lee K.Y., LaDuca H., Fulk K., Durda K.M., Deckman A.M., Goldgar D.E., Monteiro A.N.A., Gnanaolivu R., et al. Strong functional data for pathogenicity or neutrality classify BRCA2 DNA-binding-domain variants of uncertain significance. Am. J. Hum. Genet. 2021;108:458–468. doi: 10.1016/j.ajhg.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu K., Hinson S.R., Ohashi A., Farrugia D., Wendt P., Tavtigian S.V., Deffenbaugh A., Goldgar D., Couch F.J. Functional evaluation and cancer risk assessment of BRCA2 unclassified variants. Cancer Res. 2005;65:417–426. [PubMed] [Google Scholar]
- 15.Farrugia D.J., Agarwal M.K., Pankratz V.S., Deffenbaugh A.M., Pruss D., Frye C., Wadum L., Johnson K., Mentlick J., Tavtigian S.V., et al. Functional assays for classification of BRCA2 variants of uncertain significance. Cancer Res. 2008;68:3523–3531. doi: 10.1158/0008-5472.CAN-07-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hart S.N., Hoskin T., Shimelis H., Moore R.M., Feng B., Thomas A., Lindor N.M., Polley E.C., Goldgar D.E., Iversen E., et al. Comprehensive annotation of BRCA1 and BRCA2 missense variants by functionally validated sequence-based computational prediction models. Genet. Med. 2019;21:71–80. doi: 10.1038/s41436-018-0018-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng B.J. PERCH: A Unified Framework for Disease Gene Prioritization. Hum. Mutat. 2017;38:243–251. doi: 10.1002/humu.23158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biswas K., Das R., Alter B.P., Kuznetsov S.G., Stauffer S., North S.L., Burkett S., Brody L.C., Meyer S., Byrd R.A., Sharan S.K. A comprehensive functional characterization of BRCA2 variants associated with Fanconi anemia using mouse ES cell-based assay. Blood. 2011;118:2430–2442. doi: 10.1182/blood-2010-12-324541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biswas K., Das R., Eggington J.M., Qiao H., North S.L., Stauffer S., Burkett S.S., Martin B.K., Southon E., Sizemore S.C., et al. Functional evaluation of BRCA2 variants mapping to the PALB2-binding and C-terminal DNA-binding domains using a mouse ES cell-based assay. Hum. Mol. Genet. 2012;21:3993–4006. doi: 10.1093/hmg/dds222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuznetsov S.G., Liu P., Sharan S.K. Mouse embryonic stem cell-based functional assay to evaluate mutations in BRCA2. Nat. Med. 2008;14:875–881. doi: 10.1038/nm.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frazer J., Notin P., Dias M., Gomez A., Min J.K., Brock K., Gal Y., Marks D.S. Disease variant prediction with deep generative models of evolutionary data. Nature. 2021;599:91–95. doi: 10.1038/s41586-021-04043-8. [DOI] [PubMed] [Google Scholar]
- 22.Brandes N., Goldman G., Wang C.H., Ye C.J., Ntranos V. Genome-wide prediction of disease variant effects with a deep protein language model. Nat. Genet. 2023;55:1512–1522. doi: 10.1038/s41588-023-01465-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng J., Novati G., Pan J., Bycroft C., Žemgulytė A., Applebaum T., Pritzel A., Wong L.H., Zielinski M., Sargeant T., et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science. 2023;381 doi: 10.1126/science.adg7492. [DOI] [PubMed] [Google Scholar]
- 24.Tavtigian S.V., Greenblatt M.S., Harrison S.M., Nussbaum R.L., Prabhu S.A., Boucher K.M., Biesecker L.G., ClinGen Sequence Variant Interpretation Working Group ClinGen SVI Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genet. Med. 2018;20:1054–1060. doi: 10.1038/gim.2017.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pejaver V., Byrne A.B., Feng B.-J., Pagel K.A., Mooney S.D., Karchin R., O’Donnell-Luria A., Harrison S.M., Tavtigian S.V., Greenblatt M.S., et al. Calibration of computational tools for missense variant pathogenicity classification and ClinGen recommendations for PP3/BP4 criteria. Am. J. Hum. Genet. 2022;109:2163–2177. doi: 10.1016/j.ajhg.2022.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., Engel C., Hoya M.d.l., Peterlongo P., Yannoukakos D., Livraghi L., Radice P., Thomassen M., Hansen T.V.O., Gerdes A.M., et al. Risks of breast and ovarian cancer for women harboring pathogenic missense variants in BRCA1 and BRCA2 compared with those harboring protein truncating variants. Genet. Med. 2022;24:119–129. doi: 10.1016/j.gim.2021.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casadei S., Gulsuner S., Shirts B.H., Mandell J.B., Kortbawi H.M., Norquist B.S., Swisher E.M., Lee M.K., Goldberg Y., O'Connor R., et al. Characterization of splice-altering mutations in inherited predisposition to cancer. Proc. Natl. Acad. Sci. USA. 2019;116:26798–26807. doi: 10.1073/pnas.1915608116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all HDR scores and data for case-control associations and ACMG-like variant classifications. CARRIERS data is available at dbGAP (phs002820). Code for the analysis is available through Github at https://github.com/nboddicker/BRCA2_462_variant.git. Registered users can access the SEER data at https://seer.cancer.gov/data/access.html.