Summary
We use the implementation science framework RE-AIM (reach, effectiveness, adoption, implementation, and maintenance) to describe outcomes of In Our DNA SC, a population-wide genomic screening (PWGS) program. In Our DNA SC involves participation through clinical appointments, community events, or at home collection. Participants provide a saliva sample that is sequenced by Helix, and those with a pathogenic variant or likely pathogenic variant for CDC Tier 1 conditions are offered free genetic counseling. We assessed key outcomes among the first cohort of individuals recruited. Over 14 months, 20,478 participants enrolled, and 14,053 samples were collected. The majority selected at-home sample collection followed by clinical sample collection and collection at community events. Participants were predominately female, White (self-identified), non-Hispanic, and between the ages of 40–49. Participants enrolled through community events were the most racially diverse and the youngest. Half of those enrolled completed the program. We identified 137 individuals with pathogenic or likely pathogenic variants for CDC Tier 1 conditions. The majority (77.4%) agreed to genetic counseling, and of those that agreed, 80.2% completed counseling. Twelve clinics participated, and we conducted 108 collection events. Participants enrolled at home were most likely to return their sample for sequencing. Through this evaluation, we identified facilitators and barriers to implementation of our state-wide PWGS program. Standardized reporting using implementation science frameworks can help generalize strategies and improve the impact of PWGS.
Keywords: implementation science, population screening, CDC Tier 1 conditions, inherited cancer, familial hypercholesterolemia
We evaluated In Our DNA SC, a population-wide genomic screening program, through the RE-AIM framework. In 14 months, over 20,000 individuals enrolled, providing 14,053 samples. Pathogenic or likely pathogenic variants for a CDC Tier 1 condition were found in 137 individuals, with the majority of individuals agreeing to genetic counseling.
Introduction
Hereditary breast and ovarian cancer syndrome (HBOC), Lynch syndrome (LS), and familial hypercholesterolemia (FH) are recognized by the Centers for Disease Control and Prevention (CDC) as Tier 1 applications for genomic screening, with a significant potential for positive impact on public health.1,2,3,4 Individuals who are identified with a pathogenic or likely pathogenic variant in genes associated with each of these conditions are at a substantially elevated risk of serious, yet avoidable disease. Well-established preventive services are available to reduce associated morbidity and mortality for the 1%–2% of the US population affected by a genetic predisposition to CDC Tier 1 conditions (https://www.cdc.gov/genomics/implementation/toolkit/tier1.htm).1,2,3 Unfortunately, these hereditary conditions are poorly identified in the general population, with most individuals being unaware or discovering their condition only after a related disease diagnosis, representing a missed opportunity to prevent unnecessary suffering and potentially death (https://www.nationalacademies.org/our-work/genomics-and-population-health-action-collaborative).
Population-wide genomic screening (PWGS) for Tier 1 conditions can transform population health by identifying those who are genetically predisposed to cancer and cardiovascular disease before symptoms arise. Given the high penetrance and clinical actionability of variants in genes associated with Tier 1 conditions, in 2018, the National Academies of Science, Engineering, and Medicine’s Genomics Public Health Action Collaborative shared recommendations for implementing PWGS for genes associated with Tier 1 conditions (https://www.nationalacademies.org/our-work/genomics-and-population-health-action-collaborative). Further, the inclusion of diverse PWGS participants is essential for promoting health equity in genomic-informed preventive services and clinical care.5
To date, at least 14 PWGS programs screening for CDC Tier 1 conditions have been identified in the US.6 Established PWGS programs offer a prime opportunity to study the implementation of PWGS and inform a deeper understanding of the factors to enhance successful implementation and ensure scalable programs to maximize population impact.7 Incorporation of implementation science methods offers a pragmatic approach to evaluate the impact of population-based screening in specific settings and the impact of this approach on population health broadly.8,9,10,11,12 Our study team proactively developed a strategic implementation agenda that uses implementation science methods to evaluate the In Our DNA SC program at the Medical University of South Carolina (MUSC).13 In the current report, we focus on reporting RE-AIM (reach, effectiveness, adoption, implementation, and maintenance) outcomes of the first cohort of participants of the In Our DNA SC program.
Subjects and methods
Setting and sample collection
We describe findings from initial implementation of the In Our DNA SC program (November 8, 2021) through reaching our program milestone of enrolling at least 20,000 individuals (January 6, 2023). Eligibility to participate in the study included being over 18, having the ability to speak English, and not having primary residency in New York State. Participants could enroll and provide their saliva samples in several ways: (1) clinical appointments at 12 MUSC-affiliated outpatient clinics, (2) community events, or (3) at-home sample collection. The program is considered a research study, and the protocol was approved by Ethical & Independent Review Services (Salus IRB #21143-03).
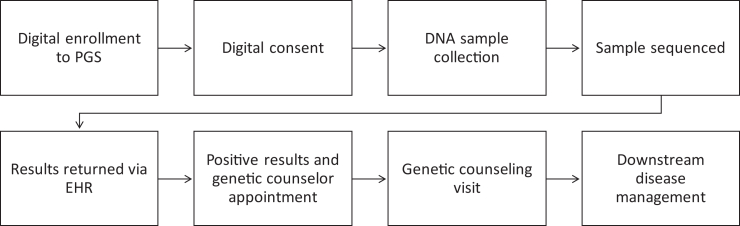
Clinical sites were selected to support enrollment based on the proportion of the patient population with active Epic MyChart accounts, geographic distribution, and patient volume. Individuals with a clinical visit at a participating clinic within the subsequent 7 days, received a message through MyChart alerting them of their eligibility to participate in the study. If individuals did not respond to the initial message, a follow-up message was sent through MyChart three days before their visit. If an individual expressed interest through their MyChart account, a study team member then sent a follow-up message through the patient portal with detailed instructions about enrollment and initiated a phone call. Once the self-consent process was completed via Research Eletronic Data Capture (REDCap), a standing order was automatically generated for sample collection during the upcoming clinical appointment. Participants were provided with instructions about the process for completing sample collection at their appointment. Trained clinical staff provided the specimen collection kit at the participant’s appointment and returned the completed kit to the Helix laboratory for processing (Figure 1).
Figure 1.
In Our DNA SC workflow
Community events included MUSC-sponsored events that took place on a reoccurring basis in four different regions of South Carolina (Charleston, Columbia, Florence, and Lancaster), as well as community-hosted events in partnership with local colleges and businesses (e.g., health fairs, senior expos) or community groups (e.g., Healthy Me Healthy SC, church groups). Individuals would consent into the study through MyChart; if they did not have an existing MyChart they were able to create one. Participants were provided with instructions about the process for completing sample collection at the event. A study-team member provided the specimen collection kit to the participant and returned the completed kit to the Helix laboratory for processing.
Participants who elected for an at-home sample collection during the consenting process were mailed a test kit to the address provided in their MyChart within one month of signing the consent form. At-home participants were provided instructions for completing sample collection within their kit. The participant then returned the completed kit to the Helix laboratory for processing with the provided shipping label.
Participants received notification of their results for the CDC Tier 1 conditions via MyChart approximately 8–12 weeks after a successful collection. All results were also recorded in the participant’s medical record. Results without a pathogenic or likely pathogenic variant are released automatically into the participant’s MyChart account as soon as they are available. For individuals with a pathogenic or likely pathogenic variant for one of the hereditary conditions, positive results were released to the participant’s medical records after two weeks, during which time a study team member attempted to contact the participant at least three times by phone. If the study team member was unable to reach the individual, they sent a certified letter via mail. Those who were identified with a pathogenic or likely pathogenic variant were offered free genetic counseling through MUSC where they can receive detailed information about their results, recommendations for follow-up care, and resources specific to their condition. Individuals who elected not to receive genetic counseling received additional resources, including a gene guide describing lifestyle recommendations, health risks, medical management, and resources about relevant support groups.
Sample sequencing
The three conditions included in our study are the CDC Tier 1 conditions. These are genomic applications that are considered to have the most evidence to support their early detection and intervention: HBOC, LS, and FH.14 The participant’s sample is sequenced for the CDC Tier 1 conditions by Helix. The sequencing includes the evaluation of BRCA1, BRCA2, MLH1, MSH2, MSH6, PMS2, EPCAM, APOB, LDLR, LDLRAP1, and PCSK9.
The participants’ raw genomic data are generated by Helix’s next-generation sequencing laboratory, CLIA-certified (CLIA #05D2117342) and CAP accredited (CAP #9382893), meaning that results can be returned to the participants’ electronic health record once sequencing is complete. Helix currently runs the Exome+ assay, a panel-grade clinical exome with a microarray backbone. The assay has been validated to support comprehensive and highly uniform coverage (>99.5% call rate at ≥ 20× for clinically relevant regions), clinically validated intragenic and multigenic copy number variants (CNVs) (100% sensitivity for ≥2 exons), clinically validated star allele calls for pharmacogenetic regions (accurate detection of >100 CYP2D6 star alleles), array-equivalent genome-wide imputation of tens of millions of high-confidence single nucleotide polymorphisms (SNPs) for discovery and polygenic risk scores, and inclusion of the full mitochondrial genome.
Variant calling is completed using a customized version of Sentieon’s DNAseq software, requiring 20× coverage for validated variant calls. Designation of pathogenicity for variants is performed using ACMG/AMP guidelines.15 CNVs are called using a proprietary bioinfomatics pipeline that compared the coverage profile of the sample with the coverage profiles of other reference set samples. Mayo Clinic GeneGuide then analyzed the generated variant data for the exons and 10 bp of flanking intronic sequence (and select tagged intronic variants) of the 11 genes included from the Helix Secure Database. The sample is reviewed for single-nucleotide variants (SNVs), indels up to 20 bp in length, and CNVs that are known or predicted to be pathogenic (https://cdn.shopify.com/s/files/1/2718/3202/files/Helix_Exome_Performance_White_Paper.pdf?v=1585153941).
Design and data collection
We proactively developed an evaluation plan to assess RE-AIM outcomes (Table 1).13,16 RE-AIM is an implementation science framework that focuses on outcomes to identify factors that facilitate or inhibit the translation of innovations. Through the assessment of factors at the individual and organizational level, RE-AIM provides a pragmatic approach to evaluate implementation of interventions.17,18 We previously described ten data collection methods used to regularly assess the In Our DNA SC program’s impact; for the current report, we use data from the data dashboard, which is a structured query language (SQL) database that extracts data from the electronic health record, which included demographic, lab, enrollment, and visit information for all individuals associated with In Our DNA SC as outlined by RE-AIM.13 The database is refreshed three times per week to support reporting needs for both the internal MUSC research team and third-party colleagues at our lab partner, Helix. This database enables secure access to clinical data related to the study population, as well as efficient and targeted analysis of the data to identify patterns and trends.
Table 1.
RE-AIM outcomes, definitions, and measures
| RE-AIMaOutcome | Definition | Measure |
|---|---|---|
| Reach | number and representativeness of participants compared to the intended audience | # eligible individuals reached |
| # declined | ||
| # non-responsive | ||
| # withdraw | ||
| # excluded | ||
| # enrolled | ||
| Effectiveness | degree to which an intervention changes a health outcome | # who complete in our DNA SC |
| # positives identified | ||
| # positive individuals who were referred to genetic counselor | ||
| Adoption | number of opportunities for individuals to participate | Total number of MUSC and community sites enrolling |
| Implementation | How well the intervention or program was delivered at the setting level (operationalized at the setting and individual level) | setting level: number of adaptations |
| individual level: among those who enrolled in In Our DNA SC: # samples collected, # recollected, # results sent, timelines of return of results | ||
| Maintenance | Continual impact of program (operationalized at the site and individual level) | setting level: sites where In Our DNA SC is continued |
| individual level: positive individuals who complete counseling; high-risk management of positive individuals |
RE-AIM: reach, effectiveness, adoption, implementation, maintenance.
Reach was defined as the number and representativeness of participants compared to the intended audience (number of eligible individuals reached, number who withdrew, number excluded by investigator, number who declined, number who were non-responsive, number enrolled). Effectiveness, or the degree to which the intervention changes a health outcome, was based on the number of individuals who completed the program (i.e., results were returned), the proportion of participants who were identified with a pathogenic or likely pathogenic variant for CDC Tier 1 conditions, and those that agreed to a genetic counseling referral. Adoption was defined as the number of opportunities for individuals to participate. We measured this as the number of opportunities for individuals to enroll at clinics, events, or through at-home options. Implementation focused on how well the intervention or program was delivered, which was operationalized at the setting and individual level, with the primary analysis at the individual level. At the setting level, we tracked adaptations made during implementation. At the individual level, we assessed the number of samples collected, number needing recollection, number of results sent to participants, and timeliness of return of results. Finally, maintenance was operationalized at the setting and individual level. At the site level, we considered maintenance to be the number of enrollment opportunities for In Our DNA SC participants and high-risk management of individuals identified with a pathogenic or likely pathogenic variant. For those who were identified with HBOC or LS variants, we considered high-risk management to be completion of a genetic counseling visit. We also tracked attendance at the Hereditary Cancer Clinic, which is designed to offer centralized services and referrals for individuals with pathogenic variants. For those who were identified with an FH variant, we considered high-risk management to be completion of genetic counseling (after referral from research coordinator).
Data analysis
Data were downloaded from the In Our DNA SC SQL database and REDCap databases. All analyses were completed in SAS v. 9.4. Data were merged based on common identifiers and cleaned prior to analysis. Descriptive analysis included mean and standard deviation, frequency, and percentage. We also assessed differences by enrollment type and sociodemographics using chi-square tests.
Results
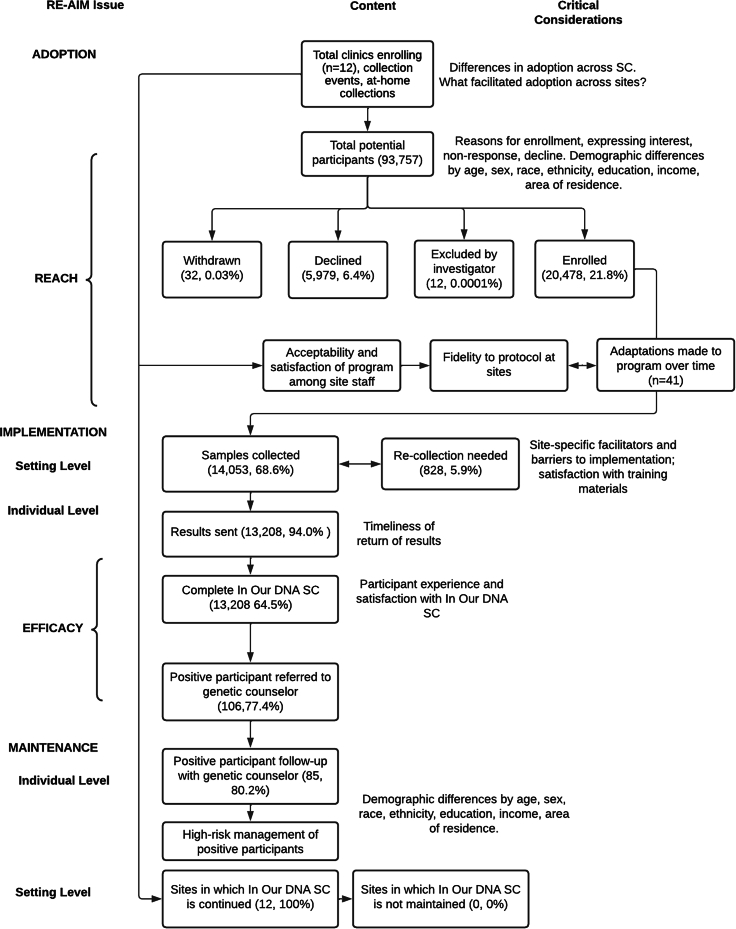
An adapted consolidated standards of reporting trials (CONSORT) diagram is provided (Figure 2) to outline site participation, study participation, and key outcomes. This CONSORT diagram was used throughout the study to track changes to the program and opportunities for enhancements.19
Figure 2.
CONSORT diagram of In Our DNA SC participants
Reach
All individuals in South Carolina who meet the eligibility criteria as mentioned above are eligible to participate in the In Our DNA SC program. In total, we met our milestone of enrolling 20,000 individuals of the 100,000 enrollment goal for the program in 14 months. We enrolled 20,478 participants between November 8, 2021 and January 6, 2023. Of those enrolled, the majority enrolled through the at-home option (n = 11,602, 56.7%), followed by clinical settings (n = 4,618, 22.6%), and community events (n = 2,037, 9.9%) (Table 2). We were missing enrollment type data from 10.8% of our participants. Other indicators of reach include the number of participants who withdrew from the study (n = 32, 0.03%), number of individuals who declined (n = 5,979, 6.4%), and number of individuals who were excluded by the investigator (n = 12, 0.0001%) (Figure 2). We found differences in enrollment by sociodemographic characteristics (Table 3). Participants who enrolled were more likely to be female (n = 15,011, 73.3%, p < 0.0001), White (n = 15,644, 76.4%, p < 0.0001), non-Hispanic (n = 17,348, 84.7%, p < 0.0001), and between the ages of 40 and 49 (n = 4,193, 20.5%, p < 0.0001). There were also sociodemographic differences across types of enrollment (clinic, community event, at home). Across enrollment types, participants were most likely to be female (clinical setting, n = 3,428, 74.2%, p < 0.0001; community events, n = 1,474, 72.4%, p < 0.0001; at home, n = 8,496, 73.2%, p < 0.0001), White (clinical setting, n = 3,895, 84.3%, p < 0.0001; community events, n = 1,333, 65.4%, p < 0.0001; at home, n = 8,566, 73.8%, p < 0.0001), and non-Hispanic/Latino (clinical settings, n = 4,445, 96.3%, p < 0.0001; community events, n = 1,650, 81.0%, p < 0.0001; at home, n = 9,259, 79.8%, p < 0.0001). There were different ages across enrollment type, with the clinical settings most frequent age being 30–39 years old (n = 882, 19.1%, p < 0.0001), community events being 18–29 years old (n = 454, 22.3%, p < 0.0001), and at home being 40–49 (n = 2,632, 22.7%, p < 0.0001) (Table 3).
Table 2.
Overview of participants by collection type
| Total | Clinic | Community Event | At home | Missing collection type | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | n | % | |
| Enrolled | 20,478 | 100.0 | 4,618 | 22.6 | 2,037 | 9.9 | 11,602 | 56.7 | 2,221 | 10.8 |
| Samples collected | 14,053 | 68.6 | 2,696 | 58.4 | 1,397 | 68.6 | 8,627 | 74.4 | 1,333 | 60.0 |
| Results returned | 13,208 | 94.0 | 2,547 | 94.5 | 1,320 | 94.5 | 8,051 | 93.3 | 1,290 | 96.8 |
| Positive results | 137 | 1.0 | 23 | 0.9 | 20 | 1.5 | 89 | 1.1 | 5 | 0.4 |
| Referral made to genetic counselor | 106 | 77.4 | 19 | 82.6 | 16 | 80.0 | 69 | 77.5 | 2 | 40.0 |
Enrolled: total number of individuals who consented; total number consented by type (in clinic, at community event, at home, or missing collection type).
Samples collected: number and proportion of those who enrolled that provided a saliva sample, and the sample was received at the Helix lab (accessioned) (sample collection/enrolled).
Results returned: number and proportion of those who provided a saliva sample (were accessioned) that have had their results returned to their electronic health record at time of data pull (results returned/samples collected).
Positive results: number and proportion of those who provided a saliva sample that received positive results for CDC Tier 1 conditions of hereditary breast and ovarian cancer, Lynch syndrome, and familial hypercholesterolemia (positive results/results returned).
Referral made to genetic counselor: number and proportion of those who received positive results for CDC Tier 1 conditions that agreed to a referral to genetic counseling (referral made/positive results).
Table 3.
Sociodemographic differences across collection type
|
Overall participants |
Clinic |
Community event |
At home |
Missing collection information |
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Enrolled |
Samples collecteda |
Proportion collected |
Enrolled |
Samples collected |
Proportion collected |
Enrolled |
Samples collected |
Proportion collected |
Enrolled |
Samples collected |
Proportion collected |
Enrolled |
Samples collected |
Proportion collected |
||||||||||||
|
N = 20,478 |
N = 14,053 |
N = 4,618 |
N = 2,696 |
N = 2,037 |
N = 1,397 |
N = 11,602 |
N = 8,627 |
N = 2,221 |
N = 1,333 |
|||||||||||||||||
| N | % | N | % | % | N | % | N | % | % | N | % | N | % | % | N | % | N | % | % | N | % | n | % | % | ||
| Sex | female | 15,011 | 73.3 | 10,156 | 72.3 | 67.7 | 3,428 | 74.2 | 1,973 | 73.2 | 57.6 | 1,474 | 72.4 | 988 | 70.7 | 67.0 | 8,496 | 73.2 | 6,246 | 72.4 | 73.5 | 1,613 | 72.6 | 949 | 71.2 | 58.8 |
| male | 5,464 | 26.7 | 3,894 | 27.7 | 71.3 | 1,190 | 25.8 | 723 | 26.8 | 60.8 | 561 | 27.5 | 407 | 29.1 | 72.6 | 3,105 | 26.8 | 2,380 | 27.6 | 76.7 | 608 | 27.4 | 384 | 28.8 | 63.2 | |
| Race | Black | 2,188 | 10.7 | 1,311 | 9.3 | 59.9 | 520 | 11.3 | 260 | 9.6 | 50.0 | 349 | 17.1 | 212 | 15.2 | 60.7 | 1,151 | 9.9 | 754 | 8.7 | 65.5 | 168 | 7.6 | 85 | 6.4 | 50.6 |
| White | 15,644 | 76.4 | 10,863 | 77.3 | 69.4 | 3,895 | 84.3 | 2,312 | 85.8 | 59.4 | 1,333 | 65.4 | 918 | 65.7 | 68.9 | 8,566 | 73.8 | 6,474 | 75.0 | 75.6 | 1,850 | 83.3 | 1,159 | 87.0 | 62.7 | |
| Asian | 272 | 1.3 | 188 | 1.3 | 69.1 | 56 | 1.2 | 33 | 1.2 | 58.9 | 54 | 2.7 | 39 | 2.8 | 72.2 | 124 | 1.1 | 95 | 1.1 | 76.6 | 38 | 1.7 | 21 | 1.6 | 55.3 | |
| other | 587 | 2.9 | 382 | 2.7 | 65.1 | 119 | 2.6 | 68 | 2.5 | 57.1 | 73 | 3.6 | 48 | 3.4 | 65.8 | 331 | 2.9 | 231 | 2.7 | 69.8 | 64 | 2.9 | 35 | 2.6 | 54.7 | |
| missing | 1,787 | 8.7 | 1,309 | 9.3 | 73.3 | 28 | 0.6 | 23 | 0.9 | 82.1 | 228 | 11.2 | 180 | 12.9 | 79.0 | 1,430 | 12.3 | 1,073 | 12.4 | 75.0 | 101 | 4.5 | 33 | 2.5 | 32.7 | |
| Ethnicity | Hispanic/Latino | 479 | 2.3 | 326 | 2.3 | 65.6 | 126 | 2.7 | 66 | 2.5 | 52.4 | 65 | 3.2 | 39 | 2.8 | 60.0 | 251 | 2.2 | 187 | 2.2 | 74.5 | 55 | 2.5 | 34 | 2.6 | 61.8 |
| non-Hispanic/Latino | 17,348 | 84.7 | 11,816 | 84.1 | 68.1 | 4,445 | 96.3 | 2,600 | 96.4 | 58.5 | 1,650 | 81.0 | 1,108 | 79.3 | 67.2 | 9,259 | 79.8 | 6,875 | 79.7 | 74.3 | 1,994 | 89.8 | 1,233 | 92.5 | 61.8 | |
| missing | 2,633 | 12.9 | 1,911 | 13.6 | 72.6 | 47 | 1.0 | 30 | 1.1 | 63.8 | 322 | 15.8 | 250 | 17.9 | 77.6 | 2,092 | 18.0 | 1,565 | 18.1 | 74.8 | 172 | 7.7 | 66 | 5.0 | 38.4 | |
| Age | 18–29 years | 2,795 | 13.6 | 1,714 | 12.2 | 61.3 | 558 | 12.1 | 263 | 9.8 | 47.1 | 454 | 22.3 | 317 | 22.7 | 69.8 | 1,532 | 13.2 | 1,025 | 11.9 | 66.9 | 251 | 11.3 | 109 | 8.2 | 43.4 |
| 30–39 years | 4,039 | 19.7 | 2,601 | 18.5 | 64.4 | 882 | 19.1 | 469 | 17.4 | 53.2 | 335 | 16.4 | 203 | 14.5 | 60.6 | 2,405 | 20.7 | 1,703 | 19.7 | 70.8 | 417 | 18.8 | 226 | 17.0 | 54.2 | |
| 40–49 years | 4,193 | 20.5 | 2,786 | 19.8 | 66.4 | 769 | 16.7 | 419 | 15.5 | 54.5 | 345 | 16.9 | 222 | 15.9 | 64.4 | 2,632 | 22.7 | 1,889 | 21.9 | 71.8 | 447 | 20.1 | 256 | 19.2 | 57.3 | |
| 50–59 years | 3,612 | 17.6 | 2,536 | 18.1 | 70.2 | 771 | 16.7 | 475 | 17.6 | 61.6 | 336 | 16.5 | 243 | 17.4 | 72.3 | 2,114 | 18.2 | 1,570 | 18.2 | 74.3 | 391 | 17.6 | 248 | 18.6 | 63.4 | |
| 60–69 years | 3,293 | 16.1 | 2,447 | 17.4 | 74.3 | 835 | 18.1 | 526 | 17.1 | 63.0 | 322 | 15.8 | 217 | 15.5 | 67.4 | 1,711 | 14.7 | 1,404 | 16.3 | 82.1 | 425 | 19.1 | 300 | 22.5 | 70.6 | |
| 70–79 years | 2,209 | 10.8 | 1,709 | 12.2 | 77.4 | 681 | 14.7 | 461 | 17.1 | 67.7 | 211 | 10.4 | 164 | 11.7 | 77.7 | 1,060 | 9.1 | 908 | 10.5 | 85.7 | 257 | 11.6 | 176 | 13.2 | 68.5 | |
| 80–89 years | 321 | 1.6 | 246 | 1.8 | 76.6 | 118 | 2.6 | 80 | 3.0 | 67.8 | 32 | 1.6 | 29 | 2.1 | 90.6 | 140 | 1.2 | 120 | 1.4 | 85.7 | 31 | 1.4 | 17 | 1.3 | 54.8 | |
| 90+ years | 16 | 0.1 | 14 | 0.1 | 87.5 | 4 | 0.1 | 3 | 0.1 | 75.0 | 2 | 0.1 | 2 | 0.1 | 100.0 | 8 | 0.1 | 8 | 0.1 | 100.0 | 2 | 0.1 | 1 | 0.1 | 50.0 | |
Sociodemographics are based on information provided in the electronic health record.
Samples collected: number and proportion of those who enrolled that provided a saliva sample and the sample was received at the Helix lab (accessioned).
Effectiveness
To assess effectiveness or whether the program achieved its intended public health goal of identifying individuals with CDC Tier 1 conditions, we assessed the number of individuals who completed the program (n = 13,208, 64.5% of those who enrolled). We also considered the total number of individuals who were identified with a pathogenic or likely pathogenic variant for a CDC Tier 1 condition (n = 137, 1.0% of results returned). We identified 56 individuals (40.9%) with pathogenic or likely pathogenic variants related to HBOC, 33 individuals with pathogenic or likely pathogenic variants related to LS (24.1%), and 48 individuals (35%) with pathogenic or likely pathogenic variants related to FH. Of the individuals who were identified with a pathogenic or likely pathogenic variant, 106 agreed to genetic counseling (77.4%). We found non-significant differences (p = 0.3211) across type of condition with those identified with LS variants being most likely to agree to genetic counseling (n = 28, 84.8%), followed by HBOC (n = 44, 78.6%), and FH (n = 34, 70.8%) (Table 4).
Table 4.
Individuals with a pathogenic or likely pathogenic variant for CDC tier 1 conditions
|
Total |
Hereditary breast and ovarian cancer |
Lynch syndrome |
Familial hypercholesterolemia |
|||||
|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | |
| Identified with pathogenic or likely pathogenic variant for Tier 1 condition | 137 | 100 | 56 | 40.9 | 33 | 24.1 | 48 | 35.0 |
| Agreed to genetic counseling | 106 | 77.4 | 44 | 78.6 | 28 | 84.8 | 34 | 70.8 |
| Completed genetic counseling | 85 | 80.2 | 40 | 90.9 | 27 | 96.4 | 18 | 52.9 |
Adoption
Adoption was considered at the organizational level, with the total number of clinics participating (n = 12). These clinics were selected to participate by the ambulatory leadership team based on clinic volume, high buy-in and engagement from clinical leadership, rate of MyChart use, clinic sociodemographic diversity, and clinical representation from across MUSC divisions. Initially, 10 clinics were included in the program as part of the pilot phase (results previously described).16,20 Staff and providers at the pilot phase clinics went live with the In Our DNA SC workflow at the same time (November 8, 2021). After the initial pilot phase, which ended in March 2022, we expanded to include community collection events (n = 108). Community collection events included standing events at key MUSC community clinics and community-focused partnership events. The study team worked closely with the project’s community advisory board to identify community organizations and events.
Implementation
Implementation was assessed at the setting level through tracking the number of adaptations made to the program over time (n = 41). These adaptations were coded according to the Framework for Reporting Adaptations and Modifications-Enhanced (FRAME).21 Of the 41 adaptations identified, the majority were changes to the content of the program (20%) that impacted the individual level (70%). The nature of the changes was primarily adding a component to the program (55%). We report detailed information about the types of adaptations made during the pilot phase of the program elsewhere.22
We also assessed implementation at the individual level. This was tracked through the number of samples collected, differences in sample collection across type of enrollment (clinic, community event, at home), and difference in samples collected based on sociodemographics (Table 3). In total, 14,053 samples were collected (68.6% of enrolled participants). There were differences in sample collection rates across the type of enrollment. At-home collection resulted in the highest proportion of samples collected among those enrolled (n = 8,627, 74.4%), followed by community events (n = 1,397, 68.6%), and clinical settings (n = 2,696, 58.4%). The majority of samples collected were from female participants (n = 10,156, 72.3%), White (n = 10,863, 77.3%), non-Hispanic (n = 11,816, 84.1%) and ages 40–49 years (n = 2,786, 19.8%). Regardless of sample collection type, the majority of participants with a sample collected were female (clinic, n = 1,973, 73.2%; community n = 988, 70.7%; at home n = 6,246, 72.4%), White (clinic, n = 2,312, 85.8%; community, n = 918, 65.7%, and at home 6,474, 75.0%), and non-Hispanic (clinic, n = 2,600, 96.4%, community event n = 1,108, 79.3%, at home n = 6,875, 79.7%). The age of participants varied across the type of sample collection. In clinical settings, the majority of participants with samples collected were between 50 and 59 years old (n = 475, 17.6%), community settings were more likely to collect samples from individuals between 18 and 29 years old (n = 317, 22.7%), and individuals collecting samples at home were more likely to be between 40 and 49 years old (n = 1,889, 21.9%).
Additional measures of implementation included number of samples needing to be recollected, number of results returned from the laboratory to participants, and timeliness of return of results. A total of 828 samples required recollection (5.9% of those received by the laboratory). Overall, 94.0% of results have been returned to participants (n = 13,208). The rates of results being returned were consistent across type of sample collection (clinic, n = 2,547 [94.5%], community, n = 1,320 [94.5%], at home 8,051 [93.3%]).
Maintenance
At the site level, clinical sites that enroll In Our DNA SC participants are continuing to enroll. We are actively expanding our community partnerships and providing additional opportunities to provide samples through laboratory settings. At the individual level, we are tracking high-risk management of individuals identified with a pathogenic or likely pathogenic variant (Table 4). The majority (78.6%) of individuals identified with an HBOC variant agreed to genetic counseling (n = 44). Of those who attended genetic counseling (n = 40), the majority also completed a follow-up visit with the Hereditary Cancer Clinic (n = 26, 65%) to discuss additional options for prevention. Of those who received referrals to genetic counseling for an LS variant (n = 28), the majority (n = 27, 96.4%) attended genetic counseling. Only one-third of individuals with an LS variant attended the Hereditary Cancer Clinic after genetic counseling (n = 9, 33.3%). Among those identified with an FH variant (n = 48), 34 (70.8%) agreed to genetic counseling and 18 (52.9%) completed genetic counseling after being referred.
Discussion
We completed interim analysis of a large-scale PWGS program based on pre-identified aims using the RE-AIM implementation science framework. Assessment of implementation outcomes from the first approximately 20,000 individuals recruited provides the opportunity to evaluate the impact of the program to date and plan for additional future modifications. This initiative demonstrates the value of the intersection of implementation science and genomics to understand the utility of the program and translation of genomic screening into practice.
A key goal for In Our DNA SC is to ensure that all individuals in South Carolina have access to PWGS. As part of this effort, we sought to ensure those who participate in the program are representative of the demographics of the state. Our efforts to enroll the first 20,000 individuals of our total 100,000 enrollment goal were greatly expanded from the initial pilot phase enrollment period, which involved only enrollment through messages in the electronic health record.16 In terms of racial diversity, during the pilot phase, 85.2% (n = 1,572 of 1,845) of those who enrolled were White (self-identified) and the majority of samples collected were also from White participants (86.7%, 851 of 982 samples collected). Upon expanding the type of collection options to include at home and community events, we saw an increase in racial diversity among those who enrolled. Overall, our enrollees were more racially diverse (76.4% White) compared to the pilot phase (85.2% White). We found similar patterns of enrollment among gender (74.6% female in pilot, 73.3% in current), ethnicity (96.2% non-Hispanic in pilot, 84.7% in current), and age (20.7% between 30 and 39 years in pilot, 20.5% between 40 and 49 years in current) between the pilot phase and current enrollment.
We found the least racial diversity among individuals who enrolled in clinical settings (84.8% White). Although we selected clinical sites at MUSC that are representative of the diverse South Carolina population, we continued to see poor uptake among American Indian or Alaska Natives, Asian, Black or African American, and Native Hawaiian or other Pacific Islander individuals. Clinical site recruitment strategies relied on deployment of Epic’s patient portal, which has been shown to result in bias toward younger, White populations.23,24,25,26 On the other hand, we found the most racial diversity among individuals who participated in community events (17.4% Black, 2.7% Asian, 3.6% other, and 65.4% White). Community events were successful in recruiting a younger group of enrollees (22.3% age 18–29) compared to all other enrollment types. We enrolled 9.9% of our participants (n = 2,037) in community settings compared to at home settings (n = 11,602, 56.7%) and clinical settings (n = 4,618, 22.6%). Offering enrollment in community settings appears to be effective at increasing racial diversity among those who enroll in the project; however, this approach is time and personnel intensive. Maximizing recruitment in community settings could help improve diversity and representativeness of participants in PWGS, but only if backed with appropriate resources to ensure scalability. Increasing the number of community events the In Our DNA SC project participates in would require additional staff time to identify partners and attend events. Currently, the program is supported by four research coordinators and one research manager. It is important to consider approaches for recruitment through community events while also balancing practical constraints (e.g., budget and staff). High-touch outreach and equipping established, trusted groups to partner in research recruitment have been recommended and successful in other smaller-scale genomic research studies; however, there are many opportunities to consider the feasibility of being able to scale these approaches as we continue to implement large-scale PWGS.27,28,29,30,31
Another key indicator of success of our program is implementation measured as the rate of DNA samples collected among those who enroll in the project. Overall, 68.6% of individuals who enrolled in In Our DNA SC provided saliva samples, which is an improvement from the pilot phase of our program, where only 53.2% of samples were returned.16 We found clinical settings to have the lowest rate of samples collected (58.4%), and at-home sample collection had the greatest rate of sample collection (74.4%). These low rates in clinical settings may have been due to several factors. Our process of outreach through clinical sites involved contacting individuals via the patient portal prior to an upcoming appointment to provide them with information about In Our DNA SC. After consent, a standing order was automatically generated prior to the clinical visit so that the sample could be collected during the upcoming appointment by a clinical staff person. It is possible individuals may have re-scheduled appointments or that the sample collection was not completed by staff during their planned visit. Additionally, while staff at clinical sites were provided with training and resources about the program, our prior analysis of sites included in the pilot phase of the program demonstrated low uptake of training among provider champions and modest knowledge change about In Our DNA SC pre- and post-training. This prior work also found implementation readiness as the most common barrier throughout implementation of the program (mentioned during 90% of technical assistance calls).20 Enhancing clinical site readiness to implementation of the program by enhancing the training for provider champions and clinical site leads, development of a training toolkit, and improving education about the importance of the program are key initiatives to enhancing clinical site engagement and ultimately clinical site sample collection.32
We also found differences in likelihood to provide samples across sociodemographic characteristics. Black individuals were most likely to return their samples through at-home collection (65.5%) and least likely to return their samples in clinical settings (50.0%). Younger individuals were most likely to return their samples in community settings (69.8% of 18–29 year olds returned) compared to all other recruitment strategies. Older adults and men were also more likely to return samples through at-home collection.
Our findings contribute to the literature that demonstrates at-home specimen collection is an effective approach to expand research participation and accessibility and contribute to the evidence regarding decentralized trial designs that became commonplace during the COVID-19 pandemic.33,34,35 Earlier studies of at-home saliva collection have reported a 41%–67% return rate among participants, with variability in likelihood to return samples across age and race.36,37,38,39,40 Comfort with at-home sample collection has grown since the COVID-19 pandemic, with recent surveys finding that individuals were more willing to collect saliva samples or throat swab samples at home than in clinical settings, citing ease and comfort with self-collection.41,42 The at-home approach for In Our DNA SC was highly successful in returning samples compared to all other approaches. Notably, we did not provide additional follow-up or outreach (e.g., phone calls) to individuals who elected to participate via at-home sample collection. Once an individual selects to participate in the study and receive an at-home kit, they receive instructions to verify their mailing address within MyChart and then receive their saliva sample kit with instructions on how to provide their sample within 7–10 business days. To improve return rate and accessibility of at-home kits, we plan to further engage participants through reminder messages, direct outreach, and providing instructions in multiple languages.
Our current sample collection approach includes only saliva sample, which was found to be a more feasible approach to sample collection at our site than blood samples.33 The recollection rate, another measure of implementation, was low, with only 828 (5.9%) of samples requiring recollection. Ultimately, of those who consented to participate, 13,208 (64.5%) of individuals received their results and completed In Our DNA SC at the time of analysis. Another recent statewide population-based screening program, the Healthy Oregon Project, found lower levels of sample failure (104/13,774 sequenced); however, the project’s methods for sample collection involved mouthwash sample collection methods.43 Additionally, our studies were similar in the overall completion rates, with 13,670 variants interpreted of 21,300 orders placed (64.6%).
Early identification of individuals with a pathogenic or likely pathogenic variant for Tier 1 conditions is a key outcome of In Our DNA SC and major public health goal of PWGS programs. Overall, we found 1.3% (n = 137) of participants who completed the program to have a predisposition to a Tier 1 condition. This rate of detection is aligned with other population screening programs, which have reported between 1.0% and 5.0% of individuals identified with pathogenic variants.7,44,45,46,47,48 The program that identified 5.0% of individuals with inherited cancer syndromes was potentially enriched beyond the carrier frequency to enroll individuals with prior cancer diagnosis and individuals with family history of disease.
Of those newly identified, the majority agreed to participate in genetic counseling (77.4%) with those identified with a predisposition to LS being the most likely to agree to being referred to genetic counseling (84.8%). This genotype-first service delivery has unique considerations and potential implications for downstream care. Although there were high rates of agreement to participate in genetic counseling following a pathogenic or likely pathogenic variant result, the likelihood of completing genetic counseling varied drastically across conditions. Nearly all individuals identified with a predisposition to LS (96.4%) completed genetic counseling compared to only 52.9% of those with a predisposition to FH completing genetic counseling. Better understanding of why individuals did not participate could be important to consider to ensure all individuals receive appropriate follow-up information. For example, individuals may elect to not participate in genetic counseling if they were already aware of their condition prior to participating in In Our DNA SC, or they may elect not to participate in genetic counseling because they do not perceive the results to be actionable to them. These reasons for non-participation in genetic counseling are important to ensure appropriate education and linkages to services.
Additional follow-up high-risk management beyond genetic counseling is essential to ensuring the benefit of population-based screening.49 While we found uptake of some additional services among those identified with HBOC and LS variants through participation in MUSC's Hereditary Cancer Clinic, further follow up is needed to identify downstream impact. The majority (70%) of individuals identified with clinically actionable Tier 1 conditions through Geisinger’s MyCode program completed a risk-management procedure after results disclosure, and 13% received a relevant clinical diagnosis following disclosure.50 Given that the PWGS approach shifts focus from risk assessment and pre-test counseling toward only post-test counseling, it is especially important to ensure pathways for those newly identified. Our approach removes the requirement for pre-test counseling, emphasizes shared decision making for the individual and connection to appropriate services for follow-up care, and ideally reduces overall cost and streamlines the workflow.51,52,53 However, further careful assessment of family history, whether individuals met existing genetic testing criteria, clinical impact, cost, and cascade testing are important considerations for the future. These considerations are important as genotype-first approaches such as PWGS continue to become more common.
Our study is not without limitations. We used RE-AIM, which is a comprehensive framework for evaluating our program; however, ensuring full assessment of each domain was challenging. For example, tracking adoption (the absolute number, proportion, and representativeness of intervention agents who are willing to initiate a program, and why) outcomes focused primarily on quantitative assessment of participating clinical and community sites. Additional qualitative assessment of acceptability and satisfaction among program and site staff could help better understand the adoption of the program. Maintenance is considered the number of sites who continue the workflow and high-risk management among individuals who are identified with a pathogenic or likely pathogenic variant. We will measure the longer-term management in future assessment of the program (e.g., individuals enrolled for one year); however, clinical management may be challenging to assess because individuals are not required to receive care at MUSC and information would likely be self-reported. In addition, we had high rates of missing information. This included missing information about the way participants enrolled (clinical settings, community events, at home) as well as race and ethnicity information. All individuals are required to have a MyChart account when they enroll in the In Our DNA SC program so their results can be returned; however, race and ethnicity are not mandatory fields when creating a new MyChart. This likely resulted in high levels of missing information, particularly among those who enrolled with at-home sample collection (i.e., individuals who did not have an existing MUSC MyChart account and would not have information on race and ethnicity from previous encounters with MUSC available). Further, the required use of MyChart may have impacted the likelihood individuals who are not familiar with MUSC to enroll. Although enrollment is open to individuals not affiliated with MUSC, data have shown that recruitment via patient portals may limit enrollment, especially among diverse populations.54 Our current evaluation efforts do not focus on the clinical utility of PWGS, and more research is needed to assess the impact of screening on clinical outcomes.50 Finally, In Our DNA SC is an ongoing program. Additional efforts will be needed to evaluate the utility of other components of the program that are underway. For example, we will assess the impact of the In Our DNA SC research database being developed for approached researchers at our institution.
PWGS for CDC Tier 1 conditions is considered a key example of a precision public health intervention that could offer substantial benefit by identifying individuals at higher risk for hereditary conditions and connecting them to services. By using an implementation science framework, we were able to prioritize and assess outcomes for this program and identify facilitators and barriers to implementation. We will continue using the RE-AIM approach to evaluate the impact of In Our DNA SC and identify opportunities to enhance the program. As PWGS programs expand nationally, use of implementation science frameworks will be essential to improving program roll out. Use of standardized reporting of findings using implementation science frameworks could help generalize the impact of PWGS across multiple study sites and provide information about the clinical- and population-level impact of these programs over the long term. Moving toward reporting across programs will help simultaneously generate and synthesize evidence to help reduce the translational gap in implementation of PWGS programs.
Data and code availability
De-identified dataset and code supporting the current study have not been deposited into a public repository due to ongoing study recruitment but are available upon request from the corresponding author. Genomic data for the In Our DNA SC program are available to investigators at the Medical University of South Carolina through a formal data request and approval by the Research Governance Committee. Table S1 includes a list of pathogenic and likely pathogenic variants identified in our cohort.
Acknowledgments
C.G.A. was supported by the National Cancer Institute (K00CA253576).
Author contributions
Conceptualization, C.G.A.; methodology, C.G.A., K.J.H., L.L.M., L.L., and D.P.J.; writing – original draft, C.G.A., K.J.H., and S.N.; writing – review & editing, C.G.A., K.J.H., S.N., A.J., L.L.M., D.P.J., K.L.G., K.F., L.M., P.M., K.P., C.B., and L.L.; data curation, C.G.A., K.J.H., C.T., J.T.C., K.K., K.F., S.N., L.L., and L.M.; formal analysis, C.G.A., S.N., and K.J.H.; funding acquisition, C.G.A., L.L.M., and C.B.
Declaration of interests
The authors declare no competing interests.
Published: February 1, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2024.01.004.
Supplemental information
Each variant is listed once for each time it was observed in the cohort in seemingly unrelated individuals. No further information is available regarding the breakpoints for deletions or duplications affecting one or more exons.
References
- 1.Manickam K., Buchanan A.H., Schwartz M.L.B., Hallquist M.L.G., Williams J.L., Rahm A.K., Rocha H., Savatt J.M., Evans A.E., Butry L.M., et al. Exome Sequencing-Based Screening for BRCA1/2 Expected Pathogenic Variants Among Adult Biobank Participants. JAMA Netw. Open. 2018;1 doi: 10.1001/jamanetworkopen.2018.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saadatagah S., Jose M., Dikilitas O., Alhalabi L., Miller A.A., Fan X., Olson J.E., Kochan D.C., Safarova M., Kullo I.J. Genetic basis of hypercholesterolemia in adults. NPJ Genom. Med. 2021;6:28. doi: 10.1038/s41525-021-00190-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Win A.K., Jenkins M.A., Dowty J.G., Antoniou A.C., Lee A., Giles G.G., Buchanan D.D., Clendenning M., Rosty C., Ahnen D.J., et al. Vol. 26. 2017. Prevalence and Penetrance of Major Genes and Polygenes for Colorectal Cancer; pp. 404–412. (Cancer Epidemiology, Biomarkers & Prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Austin M.A., Hutter C.M., Zimmern R.L., Humphries S.E. Genetic causes of monogenic heterozygous familial hypercholesterolemia: a HuGE prevalence review. Am. J. Epidemiol. 2004;160:407–420. doi: 10.1093/aje/kwh236. [DOI] [PubMed] [Google Scholar]
- 5.Sirugo G., Williams S.M., Tishkoff S.A. The Missing Diversity in Human Genetic Studies. Cell. 2019;177:1080–1131. doi: 10.1016/j.cell.2019.04.032. [DOI] [PubMed] [Google Scholar]
- 6.Williams M.S. Population Screening in Health Systems. Annu. Rev. Genom. Hum. Genet. 2022;23:549–567. doi: 10.1146/annurev-genom-111221-115239. [DOI] [PubMed] [Google Scholar]
- 7.Jones L.K., Strande N.T., Calvo E.M., Chen J., Rodriguez G., McCormick C.Z., Hallquist M.L.G., Savatt J.M., Rocha H., Williams M.S., et al. A RE-AIM Framework Analysis of DNA-Based Population Screening: Using Implementation Science to Translate Research Into Practice in a Healthcare System. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.883073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambers D.A. Commentary: Increasing the Connectivity Between Implementation Science and Public Health: Advancing Methodology, Evidence Integration, and Sustainability. Annu. Rev. Publ. Health. 2018;39:1–4. doi: 10.1146/annurev-publhealth-110717-045850. [DOI] [PubMed] [Google Scholar]
- 9.Chambers D.A., Feero W.G., Khoury M.J. Convergence of Implementation Science, Precision Medicine, and the Learning Health Care System: A New Model for Biomedical Research. JAMA. 2016;315:1941–1942. doi: 10.1001/jama.2016.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ginsburg G.S., Cavallari L.H., Chakraborty H., Cooper-DeHoff R.M., Dexter P.R., Eadon M.T., Ferket B.S., Horowitz C.R., Johnson J.A., Kannry J., et al. Establishing the value of genomics in medicine: the IGNITE Pragmatic Trials Network. Genet. Med. 2021;23:1185–1191. doi: 10.1038/s41436-021-01118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ginsburg G.S., Horowitz C.R., Orlando L.A. What will it take to implement genomics in practice? Lessons from the IGNITE Network. Per. Med. 2019;16:259–261. doi: 10.2217/pme-2019-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sperber N.R., Dong O.M., Roberts M.C., Dexter P., Elsey A.R., Ginsburg G.S., Horowitz C.R., Johnson J.A., Levy K.D., Ong H., et al. Strategies to Integrate Genomic Medicine into Clinical Care: Evidence from the IGNITE Network. J. Personalized Med. 2021;11 doi: 10.3390/jpm11070647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen C.G., Judge D.P., Levin E., Sterba K., Hunt K., Ramos P.S., Melvin C., Wager K., Catchpole K., Clinton C., et al. A pragmatic implementation research study for In Our DNA SC: a protocol to identify multi-level factors that support the implementation of a population-wide genomic screening initiative in diverse populations. Implement. Sci. Commun. 2022;3:48. doi: 10.1186/s43058-022-00286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray M.F., Evans J.P., Angrist M., Chan K., Uhlmann W.R., Doyle D.L., Fullerton S.M., Ganiats T.G., Hagenkord J., Imhof S., et al. National Academy of Medicine; Washington, DC: 2018. A Proposed Approach for Implementing Genomics-Based Screening Programs for Healthy Adults: Discussion Paper. NAM Perspectives. [Google Scholar]
- 15.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen C.G., Lenert L., Hunt K., Jackson A., Levin E., Clinton C., Clark J.T., Garrison K., Gallegos S., Wager K., et al. Lessons Learned from the Pilot Phase of a Population-Wide Genomic Screening Program: Building the Base to Reach a Diverse Cohort of 100,000 Participants. J. Personalized Med. 2022;12 doi: 10.3390/jpm12081228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glasgow R.E., Huebschmann A.G., Brownson R.C. Expanding the CONSORT Figure: Increasing Transparency in Reporting on External Validity. Am. J. Prev. Med. 2018;55:422–430. doi: 10.1016/j.amepre.2018.04.044. [DOI] [PubMed] [Google Scholar]
- 18.Glasgow R.E., Klesges L.M., Dzewaltowski D.A., Estabrooks P.A., Vogt T.M. Evaluating the impact of health promotion programs: using the RE-AIM framework to form summary measures for decision making involving complex issues. Health Educ. Res. 2006;21:688–694. doi: 10.1093/her/cyl081. [DOI] [PubMed] [Google Scholar]
- 19.Glasgow R.E., Chambers D. Developing robust, sustainable, implementation systems using rigorous, rapid and relevant science. Clin. Transl. Sci. 2012;5:48–55. doi: 10.1111/j.1752-8062.2011.00383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen C.G., Hunt K.J., Jackson A., Baierl J., McMahon L.L., Judge D.P. Applying the R = MC(2) implementation science heuristic to assess the impact of readiness on reach and implementation of a population-wide genomic screening program. J. Genet. Counsel. 2023 doi: 10.1002/jgc4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stirman S.W., Miller C.J., Toder K., Calloway A. Development of a framework and coding system for modifications and adaptations of evidence-based interventions. Implement. Sci. 2013;8:65. doi: 10.1186/1748-5908-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen C.G., Judge D.P., Nietert P.J., Hunt K.J., Jackson A., Gallegos S., Sterba K.R., Ramos P.S., Melvin C.L., Wager K., et al. Anticipating adaptation: tracking the impact of planned and unplanned adaptations during the implementation of a complex population-based genomic screening program. Transl. Behav. Med. 2023;13:381–387. doi: 10.1093/tbm/ibad006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obeid J.S., Shoaibi A., Oates J.C., Habrat M.L., Hughes-Halbert C., Lenert L.A. Research participation preferences as expressed through a patient portal: implications of demographic characteristics. JAMIA Open. 2018;1:202–209. doi: 10.1093/jamiaopen/ooy034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otte-Trojel T., de Bont A., Rundall T.G., van de Klundert J. What do we know about developing patient portals? a systematic literature review. J. Am. Med. Inf. Assoc. 2016;23:e162–e168. doi: 10.1093/jamia/ocv114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graetz I., Gordon N., Fung V., Hamity C., Reed M.E. The Digital Divide and Patient Portals: Internet Access Explained Differences in Patient Portal Use for Secure Messaging by Age, Race, and Income. Med. Care. 2016;54:772–779. doi: 10.1097/MLR.0000000000000560. [DOI] [PubMed] [Google Scholar]
- 26.Irizarry T., Shoemake J., Nilsen M.L., Czaja S., Beach S., DeVito Dabbs A. Patient Portals as a Tool for Health Care Engagement: A Mixed-Method Study of Older Adults With Varying Levels of Health Literacy and Prior Patient Portal Use. J. Med. Internet Res. 2017;19:e99. doi: 10.2196/jmir.7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kikut A., Sanyal M., Vaughn M., Ridley-Merriweather K.E., Head K., Salowe R., Lomax-Reese S., Lewis M., Ross A.G., Cui Q.N., et al. Learning from Black/African American Participants: Applying the Integrated Behavioral Model to Assess Recruitment Strategies for a Glaucoma Genetic Study. Health Commun. 2020;37:515–524. doi: 10.1080/10410236.2020.1853897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kikut A.I., O'Brien J.M. A Collaborative Community Model for Including Minorities in Genetic Research. JAMA Ophthalmol. 2018;136:313–314. doi: 10.1001/jamaophthalmol.2018.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scherr C.L., Ramesh S., Marshall-Fricker C., Perera M.A. A Review of African Americans' Beliefs and Attitudes About Genomic Studies: Opportunities for Message Design. Front. Genet. 2019;10:548. doi: 10.3389/fgene.2019.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisher E.R., Pratt R., Esch R., Kocher M., Wilson K., Lee W., Zierhut H.A. The role of race and ethnicity in views toward and participation in genetic studies and precision medicine research in the United States: A systematic review of qualitative and quantitative studies. Mol. Genet. Genomic Med. 2020;8:e1099. doi: 10.1002/mgg3.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allen C.G., Bethea B.J., McKinney L.P., Escoffery C., Akintobi T.H., McCray G.G., McBride C.M. Health promotion practice; 2021. Exploring the Role of Community Health Workers in Improving the Collection of Family Health History: A Pilot Study. [DOI] [PubMed] [Google Scholar]
- 32.Watson A.K., Hernandez B.F., Kolodny-Goetz J., Walker T.J., Lamont A., Imm P., Wandersman A., Fernandez M.E. Using Implementation Mapping to Build Organizational Readiness. Front. Public Health. 2022;10 doi: 10.3389/fpubh.2022.904652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansen T.V., Simonsen M.K., Nielsen F.C., Hundrup Y.A. Vol. 16. 2007. Collection of Blood, Saliva, and Buccal Cell Samples in a Pilot Study on the Danish Nurse Cohort: Comparison of the Response Rate and Quality of Genomic DNA; pp. 2072–2076. (Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology). [DOI] [PubMed] [Google Scholar]
- 34.McDermott M.M., Newman A.B. Remote Research and Clinical Trial Integrity During and After the Coronavirus Pandemic. JAMA. 2021;325:1935–1936. doi: 10.1001/jama.2021.4609. [DOI] [PubMed] [Google Scholar]
- 35.Dahne J., Hawk L.W., Jr. Health Equity and Decentralized Trials. JAMA. 2023;329:2013–2014. doi: 10.1001/jama.2023.6982. [DOI] [PubMed] [Google Scholar]
- 36.Dykema J., DiLoreto K., Croes K.D., Garbarski D., Beach J. Factors Associated with Participation in the Collection of Saliva Samples by Mail in a Survey of Older Adults Get access Arrow. Publ. Opin. Q. 2016;81:57–85. [Google Scholar]
- 37.Bhutta M.F., Hobson L., Lambie J., Scaman E.S.H., Burton M.J., Giele H., Jamieson S.E., Furniss D. Alternative recruitment strategies influence saliva sample return rates in community-based genetic association studies. Ann. Hum. Genet. 2013;77:244–250. doi: 10.1111/ahg.12009. [DOI] [PubMed] [Google Scholar]
- 38.Gatny H.H., Couper M.P., Axinn W.G. New strategies for biosample collection in population-based social research. Soc. Sci. Res. 2013;42:1402–1409. doi: 10.1016/j.ssresearch.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chartier K.G., Martinez P., Cummings C., Riley B.P., Karriker-Jaffe K.J. Recruiting for diversity: a pilot test of recruitment strategies for a national alcohol survey with mail-in genetic data collection. J. Community Genet. 2021;12:459–468. doi: 10.1007/s12687-020-00502-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osmond D.H., Catania J., Pollack L., Canchola J., Jaffe D., MacKellar D., Valleroy L. Obtaining HIV test results with a home collection test kit in a community telephone sample. J. Acquir. Immune Defic. Syndr. 2000;24:363–368. doi: 10.1097/00126334-200008010-00011. [DOI] [PubMed] [Google Scholar]
- 41.O'Laughlin K., Espinosa C.C., Smith-Jeffcoat S.E., Koh M., Khalil G.M., Hoffman A., Rebolledo P.A., Schechter M.C., Stewart R.J., da Silva J., et al. Specimen self-collection for SARS-CoV-2 testing: Patient performance and preferences-Atlanta, Georgia, August-October 2020. PLoS One. 2022;17 doi: 10.1371/journal.pone.0264085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hall E.W., Luisi N., Zlotorzynska M., Wilde G., Sullivan P., Sanchez T., Bradley H., Siegler A.J. Willingness to Use Home Collection Methods to Provide Specimens for SARS-CoV-2/COVID-19 Research: Survey Study. J. Med. Internet Res. 2020;22 doi: 10.2196/19471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Brien T.D., Potter A.B., Driscoll C.C., Goh G., Letaw J.H., McCabe S., Thanner J., Kulkarni A., Wong R., Medica S., et al. Population screening shows risk of inherited cancer and familial hypercholesterolemia in Oregon. Am. J. Hum. Genet. 2023;110:1249–1265. doi: 10.1016/j.ajhg.2023.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu C., Hart S.N., Gnanaolivu R., Huang H., Lee K.Y., Na J., Gao C., Lilyquist J., Yadav S., Boddicker N.J., et al. A Population-Based Study of Genes Previously Implicated in Breast Cancer. N. Engl. J. Med. 2021;384:440–451. doi: 10.1056/NEJMoa2005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Natarajan P., Gold N.B., Bick A.G., McLaughlin H., Kraft P., Rehm H.L., Peloso G.M., Wilson J.G., Correa A., Seidman J.G., et al. Aggregate penetrance of genomic variants for actionable disorders in European and African Americans. Sci. Transl. Med. 2016;8:364ra151. doi: 10.1126/scitranslmed.aag2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mazor K.M., Street R.L., Jr., Sue V.M., Williams A.E., Rabin B.A., Arora N.K. Assessing patients' experiences with communication across the cancer care continuum. Patient Educ. Couns. 2016;99:1343–1348. doi: 10.1016/j.pec.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.East K.M., Kelley W.V., Cannon A., Cochran M.E., Moss I.P., May T., Nakano-Okuno M., Sodeke S.O., Edberg J.C., Cimino J.J., et al. A state-based approach to genomics for rare disease and population screening. Genet. Med. 2021;23:777–781. doi: 10.1038/s41436-020-01034-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grzymski J.J., Elhanan G., Morales Rosado J.A., Smith E., Schlauch K.A., Read R., Rowan C., Slotnick N., Dabe S., Metcalf W.J., et al. Population genetic screening efficiently identifies carriers of autosomal dominant diseases. Nat. Med. 2020;26:1235–1239. doi: 10.1038/s41591-020-0982-5. [DOI] [PubMed] [Google Scholar]
- 49.Murray M.F., Evans J.P., Angrist M., Chan K., Uhlmann W., Doyle D.L., Fullerton S.M., Ganiats T., Hagenkord J., Imhof S., et al. NAM Perspectives DIscussion; 2018. A Proposed Approach for Implementing Genomics-Based Screening Programs for Healthy Adults. [Google Scholar]
- 50.Buchanan A.H., Lester Kirchner H., Schwartz M.L., Kelly M.A., Schmidlen T., Jones L.K., Hallquist M.L., Rocha H., Betts M., Schwiter R., et al. Clinical outcomes of a genomic screening program for actionable genetic conditions. Genet. Med. 2020;22:1874–1882. doi: 10.1038/s41436-020-0876-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwartz M.L.B., Buchanan A.H., Hallquist M.L.G., Haggerty C.M., Sturm A.C. Genetic counseling for patients with positive genomic screening results: Considerations for when the genetic test comes first. J. Genet. Counsel. 2021;30:634–644. doi: 10.1002/jgc4.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santos Gonzalez F., Mordaunt D., Stark Z., Dalziel K., Christodoulou J., Goranitis I. Microcosting diagnostic genomic sequencing: A systematic review. Genet. Med. 2023;25 doi: 10.1016/j.gim.2023.100829. [DOI] [PubMed] [Google Scholar]
- 53.Guzauskas G.F., Garbett S., Zhou Z., Spencer S.J., Smith H.S., Hao J., Hassen D., Snyder S.R., Graves J.A., Peterson J.F., et al. Cost-effectiveness of Population-Wide Genomic Screening for Hereditary Breast and Ovarian Cancer in the United States. JAMA Netw. Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.22874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller H.N., Lindo S., Fish L.J., Roberts J., Stover J., Schwark E.H., Eberlein N., Mack D., Falkovic M., Makarushka C., Chatterjee R. Describing current use, barriers, and facilitators of patient portal messaging for research recruitment: Perspectives from study teams and patients at one institution. J. Clin. Transl. Sci. 2023;7:e96. doi: 10.1017/cts.2023.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Each variant is listed once for each time it was observed in the cohort in seemingly unrelated individuals. No further information is available regarding the breakpoints for deletions or duplications affecting one or more exons.
Data Availability Statement
De-identified dataset and code supporting the current study have not been deposited into a public repository due to ongoing study recruitment but are available upon request from the corresponding author. Genomic data for the In Our DNA SC program are available to investigators at the Medical University of South Carolina through a formal data request and approval by the Research Governance Committee. Table S1 includes a list of pathogenic and likely pathogenic variants identified in our cohort.