Summary
Disease-associated variants identified from genome-wide association studies (GWASs) frequently map to non-coding areas of the genome such as introns and intergenic regions. An exclusive reliance on gene-agnostic methods of genomic investigation could limit the identification of relevant genes associated with polygenic diseases such as Alzheimer disease (AD). To overcome such potential restriction, we developed a gene-constrained analytical method that considers only moderate- and high-risk variants that affect gene coding sequences. We report here the application of this approach to publicly available datasets containing 181,388 individuals without and with AD and the resulting identification of 660 genes potentially linked to the higher AD prevalence among Africans/African Americans. By integration with transcriptome analysis of 23 brain regions from 2,728 AD case-control samples, we concentrated on nine genes that potentially enhance the risk of AD: AACS, GNB5, GNS, HIPK3, MED13, SHC2, SLC22A5, VPS35, and ZNF398. GNB5, the fifth member of the heterotrimeric G protein beta family encoding Gβ5, is primarily expressed in neurons and is essential for normal neuronal development in mouse brain. Homozygous or compound heterozygous loss of function of GNB5 in humans has previously been associated with a syndrome of developmental delay, cognitive impairment, and cardiac arrhythmia. In validation experiments, we confirmed that Gnb5 heterozygosity enhanced the formation of both amyloid plaques and neurofibrillary tangles in the brains of AD model mice. These results suggest that gene-constrained analysis can complement the power of GWASs in the identification of AD-associated genes and may be more broadly applicable to other polygenic diseases.
Keywords: GWAS, amyloid plaque, neurofibrillary tangle, G protein, RGS protein, amyloid precursor protein, APP, G protein-coupled receptor, GPCR, Lodder-Merla syndrome
Nine genes that potentially enhance the risk of Alzheimer disease (AD) were identified using a novel gene-constrained analytical method. One gene encoded heterotrimeric G protein beta family member GNB5. In AD model mice, Gnb5 heterozygosity enhanced the development of AD neuropathology. This method may be applicable to other polygenic diseases.
Introduction
Disparities in the risk for polygenic diseases, such as type 2 diabetes mellitus (MIM: 125853), obesity (MIM: 601665), and cardiovascular disease (including MIM: 608446, 601367), result partly from differences in environmental stressors, socioeconomic determinants, and individual behaviors,1 but are also the result of genetic differences among populations with different ancestry.2 That being so, ancestry-specific pools of potentially impactful variants, including those affecting gene coding regions, constitute the genetic background for the vulnerability to polygenic diseases among populations in an ancestry-dependent manner. These considerations apply to Alzheimer disease (AD [MIM: 104300]), a progressive and debilitating polygenic neurodegenerative disease which is approximately twice as prevalent in individuals of African/African American ancestry (AFR) compared to White non-Hispanic/Europeans (EUR).3,4,5,6 The heritable component of AD risk due to cumulative genetic effects has been estimated to be ∼70% based on twin studies.7
The search for genetic variants associated with complex or polygenic traits and diseases has traditionally involved genome-wide association studies (GWASs). Until only recently GWASs have included mostly participants of EUR ancestry.8,9,10,11,12,13,14 Such bias in representation risks limiting our understanding of diseases such as AD shown to have an unequal impact on populations of different ancestry.15,16,17,18 Another feature of GWAS design that could potentially impair their ability to delineate the genetic basis for disease risk is the deliberately gene-agnostic approach for the detection of rare variants, such as single-nucleotide polymorphisms (SNPs) and short indels. It is not uncommon in GWASs that most SNPs detected end up mapping to noncoding regions of the genome.19,20
The potential functional impact of variants located in intergenic, regulatory, and intronic regions of the genome, though often representing the majority of signal in GWASs, may be challenging to predict or interpret in a mechanistic biological framework. Given this problem, methods of statistical fine mapping have been developed in recent years to refine GWAS signals, help select and prioritize genetic variants for further study, and better identify the variants that are truly causal to the phenotype.21 Although variants located in the gene coding regions themselves are not prioritized by GWASs and in fact represent only ∼10% of the entire GWAS signal,22 alternative methods of analysis that focus on this small set of variants may be desirable because such intragenic variants should be more easily interpretable in a functional context.
In the present study, we develop and apply a particular form of gene-constrained analysis for the identification of unknown AD-associated genes. Employing an analytical method that considers only moderate- and high-risk variants that affect gene coding sequences, denominated by the total number of variants affecting that gene, we were able to identify 660 genes potentially linked to the higher AD prevalence among AFR. In combination with transcriptome profiling of multiple brain regions, we ultimately concentrated on nine genes that potentially enhance the risk of AD and experimentally demonstrated that one of them, Gnb5 (GNB5 in humans [MIM: 604447]), exacerbated the development of Aβ plaque and neurofibrillary tangle (NFT) in a mouse model of AD. These results suggest that gene-constrained analysis can complement the power of GWASs in the identification of AD-associated genes and may be more broadly applicable to other complex polygenic human diseases.
Material and methods
Mouse husbandry and genotyping
The Gnb5 KO mice containing the germline deletion of exon 3 were a generous gift from Ching-Kang Jason Chen, PhD (Department of Molecular Medicine, The University of Texas Health Science Center at San Antonio).23 The amyloid precursor protein (APP [MIM: 104760])/presenilin 1 (PSEN1 [MIM: 104311]) transgenic mice (B6C3-Tg(APPswe, PSEN1dE9)85Dbo/Mmjax) as an Alzheimer disease model24 were purchased from Jackson laboratory (MMRRC Stock No: 34829-JAX). The APP/PSEN1 mice are doubly transgenic, expressing a chimeric mouse/human amyloid precursor protein (Mo/HuAPP695swe) and a mutant human presenilin 1 (PS1-dE9), both directed to CNS neurons.24 The breeding of C57BL/6 Gnb5+/− with the APP/PSEN1 mice generated the Gnb5+/−/APP/PSEN1, triple mutant mice. Mouse husbandry was according to the National Institutes of Health Guide for Care and Use of Laboratory Animals. Mouse studies were performed under the oversight of the NIDDK Animal Care and Use Committee, Animal Study Proposal K164-MDB. Mice were maintained in a pathogen-free facility in a temperature-controlled room with a 12-h light/dark cycle and free access to food and water. The mouse genotyping was conducted using the DirectPCR (Tail) (Viagen, Cat# 102-T) lysis buffer for crude DNA extraction and 2× PCR mix (Bioline, Cat# BIO25048) for PCR reactions. The primers and PCR conditions used for genotyping Gnb5+/− mice were described previously25 and those for APP/PSEN1 mice were as indicated at the vendor’s website (https://www.jax.org/strain/004462).
Aβ plaque and Tau-associated neurofibrillary tangle (NFT) staining
Mice were sacrificed and perfused as previously described.25 The mouse brains were excised and fixed in 4% paraformaldehyde in PBS for 2 h after perfusion and then stored in 5 mL 30% sucrose solution in a 15 mL centrifuge tube at 4°C for 2–3 days or until they settled to the bottom of the container. The brains were sectioned sagittally at 20 μm thickness using a Leica cryostat (Leica CM3050S, Leica Microsystems Inc.). For Aβ plaque staining, brain sections of desired genotypes from the same litter were processed in parallel and stained with Thioflavine S (Sigma, Cat# T1892) in accordance with the modified Guntern protocol.26 In brief, sections were stained for 8 min with 0.002% Thioflavine S in 1× TBS. Sections were then rinsed twice for 1 min each in 50% EtOH followed by 5 min in 1× TBS. For NFT staining, the brain sections were first permeabilized in 0.3% Triton X- for 30 min at room temperature, blocked in 4% BSA solution for 1 h at room temperature, and then incubated with anti-Tau antibody (EMD Millipore, now Millipore Sigma, Cat# MAB3420) in 4% BSA at 1:2,000 dilution overnight at 4°C. The sections were washed 3 × 10 min in 1% BSA/PBS and blocked again in 4% BSA for 30 min at room temperature prior to incubation with the secondary antibody incubation (Invitrogen, now Thermo Fisher Scientific, Cat# A11032) in 4% BSA at 1:450 dilution for 2 h at room temperature. Sections were finally washed 3 × 10 min in 1× PBS. All the sections were mounted with antifade mounting medium containing DAPI (Vectashield Cat# H-1200).
The stained sections were imaged on the Keyence fluorescent microscope (BZ-X810, Keyence). Quantifications of Aβ plaque and NFT numbers were performed by testers blind to the mouse genotypes. The Aβ plaque numbers for each section were manually quantified using the cell counter plugin on ImageJ while the NFT (>0.32258 μm) numbers were quantified automatically using the ImageJ cell counter by first converting the color images to black and white ones and setting the background threshold to 100/255. The data were analyzed using the Prism8 software (Graphpad Software).
Gene function impacting variant rate analysis
To perform a comprehensive analysis of whole-genome gene function impacting variant rate (GFIVR), we acquired counts variants associated with Alzheimer disease and the total variants located within gene and exon regions. These data were obtained through programmatic access to the NIAGADS Alzheimer’s GenomicsDB via REST services and by downloading the reference dataset from the gnomAD database (https://gnomad.broadinstitute.org/downloads). Both databases were generated based on the Genome Build, GRCh37/hg19, at the time of our analysis. After removing duplicate entries and applying quality control measures, a total of 20,324 genes common to both datasets were identified for analysis. The datasets were processed programmatically as follows: the total number of variants for each gene and their expected functional impact were extracted for two datasets. Functional annotations for the gene variants were generated using the Ensembl Variant Effect Predictor (VEP), an open-source toolkit.27 Variants predicted by VEP to have a moderate or high functional impact were categorized as gene function impacting variants (GFIVs) and counted for each gene in both datasets for subsequent analysis. GFIVRs were calculated for each gene by dividing the number of function-impacting variants (the sum of moderate and high-risk variants) by the total number of variants for that gene. The fold enrichment for each gene was obtained by comparing the GFIVR of a gene in the NIAGADS dataset to the GFIVR of the same gene in the gnomAD reference dataset.
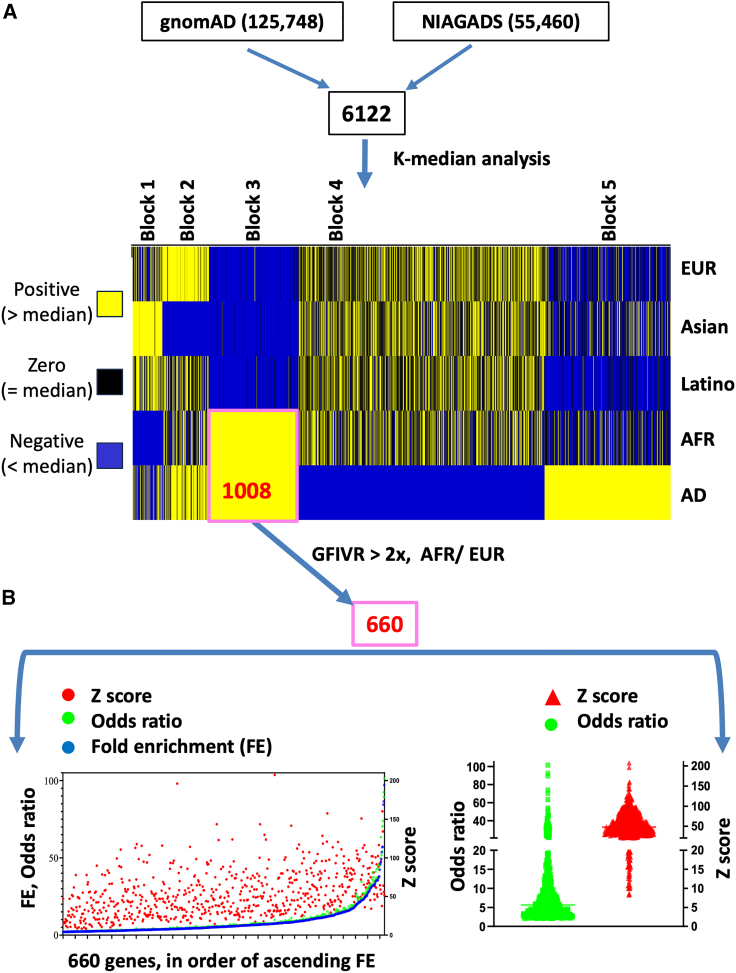
An initial quartile analysis was performed to identify candidate genes for further investigation. Specifically, genes that overlapped between the top 50% of genes based on the total number of variants and the top 75% of genes based on the GFIV were selected from both the NIAGADS and gnomAD reference datasets. This selection yielded 6,122 of 20,324 total genes. Subsequently, supervised K-median clustering analysis (open source Cluster 3.0 software)28 was conducted on these 6,122 selected genes across five distinct populations. These populations included the four ancestral subpopulations within gnomAD: AFR (African/African American), EUR (Ashkenazi Jewish, Finnish, and Non-Finnish European), Latino (Admixed Americans), and Asian (East Asian) and, as well as the NIAGADS dataset, treated as a single entity. Before conducting the analysis, the entire dataset consisting of 30,610 GFIVR values underwent normalization through global-median centering and log transformation. The resulting normalized dataset was visually represented using Java Treeview (https://jtreeview.sourceforge.net)29 utilizing a three-color scheme: yellow for positive values, black for zero values, and blue for negative values.
For Bonferroni correction of p values resulting from multiple comparisons, the chi-square p values were adjusted programmatically using the Bonferroni_p_value function available in the Python library and conducted on the Jupyter Notebook interface of Python 3.30
Transcriptome profiling of 23 human brain regions
The transcriptome profiles on Affymetrix microarray from 23 different regions of human brain tissues downloaded from NCBI/GEO database (www.ncbi.nlm.nih.gov/geo). The microarray raw data were imported into the software Partek Flow (http://www.partek.com/partek-flow/) and aligned on Genome Reference Consortium Human Build 37 (GRCh37) by modified STAR. The merged data table was quantified to an annotation model (Partek E/M) and normalized by its default method. One-way ANOVA analysis per brain region, chip type was considered as the random variable. The final list of the significantly differentially expressed genes was generated by the filters of the fold changes greater than 1.39 (AD vs. non-AD) with less than 0.05 FDR (false discovery rate). The bioinformatics analysis was conducted by the commercial Genomatix software suite (https://www.genomatix.de/solutions/genomatix-software-suite.html). The gene ontology analysis was performed by MetaCore in GeneGo (http://trials.genego.com/cgi/index.cgi). The 2D drawing of the relative locations of brain regions and the 3D diagram of the functional relationships between EC and HIP were created by Devon Art.
Gene network analysis
Network visualization and integrative analysis was performed using both the Ingenuity Pathway Analysis tool (IPA) software using the core analysis option (QIAGEN, Inc.; www.qiagenbioinformatics.com) and the web-based OmicsNet network visualization tool (www.omicsnet.ca) using the InnateDB option. The selected gene lists of interest were uploaded onto the corresponding bioinformatics system and subjected to unsupervised analysis by precisely following the corresponding instructions of each system.
Results
Comparative genomic analysis reveals 660 genes potentially associated with Alzheimer disease
Cognizant of the potential shortcomings of genome-wide SNP association studies described above, we developed a novel gene-constrained analysis, focusing on variants with the potential to impact gene function, for the identification of unknown AD-associated genes. Toward this end, we compared two large genomic datasets: (1) the summary statistics from the National Institute on Aging Genetics of Alzheimer Disease Data Storage Site (NIAGADS; Alzheimer’s Genomics Database) representing GWASs of both early- and late-onset AD31,32 and (2) the Genome Aggregation Database (gnomAD) which was utilized as a reference population.33,34 From each database, the total number of variants that mapped to a given gene was programmatically extracted, along with the expected functional impact for each genetic variant. The functional annotation for every variant was generated by the Ensembl Variant Effect Predictor (VEP),27 an open-source toolkit for annotating variants and predicting their effects on genes and regulatory elements. VEP uses a series of algorithms based on protein sequence conservation and other factors to predict pathogenicity and the functional impact of a variant.27,35 Variants predicted to have an effect impact categorized as either moderate or high by VEP were considered GFIVs, tallied for each gene in the two datasets, and used for subsequent analysis.
Preliminary filtering involved basic quartile analysis of the two genomic datasets. After removal of duplicates and performance of data quality control, 20,324 genes could be compared between the two datasets. Genes with the number of total variants in the top 50% and the number of GFIV in the top 75% in both the NIAGADS and gnomAD reference datasets were considered further. This yielded 6,122 of the 20,324 genes (Figure 1A).
Figure 1.
Comparative genomic analysis reveals 660 genes potentially associated with Alzheimer disease
(A) Quartile analysis of 20,324 unique genes derived from the National Institute on Aging Genetics of Alzheimer Disease Data Storage Site (NIAGADS; Alzheimer’s Genomics Database) and the Genome Aggregation Database (gnomAD) were conducted to obtain genes that are in top 50% total number of variants and the top 75% total number of gene function impacting variants (GFIVs) in both the NIAGADS and gnomAD reference datasets. This yielded 6,122 of the 20,324 genes. The GFIV rate (GFIVR) was then calculated, defined as the combined number of moderate and high-risk variants for each gene divided by the total number of variants in that gene for the 6,122 genes in the two large datasets. Heatmap showing results of K-median clustering analysis as described in results and performed as described in material and methods. For each of the 6,122 genes, the relationship to the median GFIVR in each of the five comparison groups is color coded: yellow, positive, above the median; black, zero, represents the median; blue, negative, below the median. A set of 1,008 genes was identified whose normalized GFIVRs were positive (yellow) in individuals of AFR ancestry within both gnomAD and the AD (NIAGADS) dataset as a whole (set boxed in pink). These 1,008 genes were compared to select 660 genes with a GFIVR fold enrichment of at least 2 in the AFR compared to EUR subpopulation (pink box).
(B) Left: dot plot of fold enrichment (in blue), odds ratio (in green), and Z score (in red) for each of the selected 660 genes. Right: distribution of the odds ratios (in green) and Z scores (in red) for the 660 genes.
Next, the GFIV rate (GFIVR), defined as the combined number of moderate- and high-risk variants for each gene divided by the total number of variants in that gene was calculated programmatically for the 6,122 genes in the two large datasets. Using the calculated GFIVR, we compared the summary statistics from NIAGADS with the gnomAD dataset on a gene-to-gene basis. Although longer genes might reasonably be expected to harbor more variants due solely to their larger genomic footprint, our method of gene-to-gene comparison between study populations controls for this. Furthermore, we found a high correlation in the average number of total variants per gene across subpopulations (see below).
We hypothesized that genes in the NIAGADS dataset with significantly higher GFIVR relative to the same gene in the gnomAD reference population might be associated with the AD phenotype. Note that the GFIVR analytical strategy presented here considers each gene, instead of each SNP or variant as in a GWAS, as the functional unit for comparison. This strategy also assumes that the overall rate of variation for a particular gene is relatively stable within a species. We tested this assumption by programmatically calculating the average number of total gene variants per individual across 20,324 genes among four subpopulations of different ancestry in gnomAD and found inter-ancestral correlation coefficients ≧ 0.93 (Figure S1). Because the GFIVR method aggregates all potentially impactful variants within a gene, instead of considering each SNP or variant in isolation as in a GWAS, GFIVR analysis may sensitize the detection of phenotypically relevant genes. This approach should also minimize analytical interference by variations of unrelated genes, and thus complement the power of GWAS analysis.
Because the prevalence of AD is some 2-fold higher in AFR than in EUR,3,4,5,6 it seemed likely that genes contributing to such divergence might have a higher GFIVR in the AFR subpopulation within the reference gnomAD population. To test this hypothesis, inter-ancestral K-median analysis was performed by comparing the GFIVR of the 6,122 potential AD-associated genes identified above among five groups: the four ancestral categories within the gnomAD reference dataset for which information was available (EUR, Asian, Latino, and AFR; see material and methods for further details of gnomAD ancestral classifications) and the NIAGADS dataset (as a whole). Consistent with our hypothesis, a subset of 1,008 genes from the 6,122 total could be identified in block 3 (Figure 1A, highlighted in pink box) whose normalized GFIVRs were highly correlated between individuals of AFR ancestry within gnomAD, and the NIAGADS dataset as a whole (Table S1). Among these 1,008 genes, 660 had a GFIVR in the AFR subpopulation of gnomAD more than 2-fold higher than in the EUR subpopulation (fold enrichment >2) and were considered further (Figure 1B) (Table S2).
We hypothesized that this subset of 660 genes, though representing only ∼3% of the starting set of 20,324 genes, would be enriched with AD-risk genes, especially those genes contributing to the higher prevalence of AD among individuals with AFR ancestry. As a check on this hypothesis, we considered a group of 76 loci and genes with genome-wide significant evidence of affecting AD risk compiled by the Alzheimer Disease Sequencing Project (ADSP) Gene Verification Committee (listed in Table S3; Figure S2A).36,37 We found that genes from among this set of 76 AD-risk genes were enriched throughout the steps of bioinformatic processing presented above; included among the final subset of 660 genes were 11 of the 76, supportive of the hypothesis (Figures S2B and S2C; chi-square two-tailed p < 0.0001).
This set of 660 genes was characterized further. By comparing the number of GFIV with the number of total variants on a gene-by-gene basis between individuals of AFR and EUR ancestry in gnomAD, fold enrichment (FE), odds ratios, and Z-scores were calculated (Figure 1B). The FE and odds ratios were highly correlated (R = 0.99) (Figure 1B, left). The median Z score was close to +50 for this set of genes when GFIVR were compared between AFR and EUR ancestral groups (Figure 1B, right).
Transcriptome profiling identifies 14 human brain regions with the most significantly altered gene expression in AD
We sought an orthogonal approach to define genes associated with AD pathogenesis that did not involve the analysis of germline genetic variants. An independent approach to uncovering genes relevant for AD pathogenesis could involve the identification of genes with significantly altered expression in the brains of individuals with AD relative to control subjects. This can be justified by imagining, for example, that elements of AD risk arising from non-genetic factors, such as environmental influences and individual behaviors, may converge in their impact on the same genes and pathways important for heritable AD risk. One limitation in the use of transcriptomic analysis for this purpose is the recognition that genes with significantly altered transcript levels may include not only primary molecular drivers of the earliest steps of AD, but also genes whose altered expression occurs only in the later stages of disease, representing secondary or tertiary adaptations or changes in molecular function and expression in response to disease progression.38 In comparison to the genomic analysis presented above, it could be argued that the functional impact of only transcriptionally down-regulated AD risk genes would parallel the effect of a GFIV of the same gene present in the germline (i.e., both alterations diminishing gene function). Whereas this might be true early in disease pathogenesis, it is conceivable that transcriptional up-regulation of critical AD risk genes might occur later in the disease process as compensatory secondary or tertiary responses. For this reason, and to reduce possible selection bias, we chose to consider genes with significantly altered expression, in either the upward or the downward direction, for the transcriptomic analysis of the brains of individuals with AD relative to control subjects.
To this end we performed transcriptome analysis of 23 brain regions, comparing data from 2,728 AD-affected and non-AD individuals using meta-data analysis of publicly available microarray datasets (Table S4). Using this approach, 12,776 transcripts were identified that had either significantly increased or decreased levels of expression in the brains of AD-affected individuals (Figure 2A). These transcripts implicate more than 50% of the protein-coding genes currently recognized in the human genome, a striking proportion that highlights the extensive impact of AD pathology on gene expression across multiple brain regions. As considered above, these transcripts presumably represent a combination of genes whose altered expression was of primary importance (occurring earlier and linked more closely to disease pathogenesis) and others with later, secondary or tertiary, association to AD brain pathology (Figure 2A).
Figure 2.
Transcriptome profiling identifies 14 human brain regions with the most significantly altered gene expression in AD
(A) Bar graph illustrating the number of genes with either 2-fold higher (depicted in green) or 2-fold lower (depicted in red) transcript expression in the brains of Alzheimer disease (AD)-affected individuals compared to non-AD control subjects. The x axis represents the abbreviated names of the 23 examined brain regions (for full names, refer to Table S6). The 14 brain regions to the left of the dashed blue line harbored the greatest number of significantly altered transcripts and were considered in subsequent clustering analysis.
(B) Uniform Manifold Approximation and Projection (UMAP) cluster analysis of the 3,419 genes with at least a 2-fold difference in expression between AD and non-AD brains in the entorhinal cortex (EC), hippocampus (HIP), and frontal pole (FP) regions, with individual genes indicated as dots. Color code for directionality of gene changes as in (A).
(D) Schematic representation of the human brain, highlighting the relative anatomical positions of the EC, HIP, and FP brain regions.
(E) Venn diagram illustrating a common set of 340 genes shared among HIP, EC, and FP brain regions, with at least a 2-fold difference in expression between AD and non-AD brains. The numbers for each brain region represent the genes with significant differences in at least two of the three brain regions: HIP (1,466 genes), EC (1,275 genes), and FP (1,759 genes).
The genes with altered expression varied among the 23 different brain regions analyzed. We arbitrarily chose to focus on the 14 brain regions that demonstrated the largest number of transcripts with altered expression in AD-affected individuals, hypothesizing that these 14 brain regions may play a more significant role in AD development (Figure 2A, regions to the left of the vertical dashed blue line; Table S5; see Table S6). Within these 14 brain regions were 3,419 genes whose expression levels were altered at least 2-fold in one or more regions in AD compared to control brains.
Analysis of the patterns of altered expression for the 3,419 genes in these 14 brain regions by unsupervised hierarchical clustering showed a pairing of the hippocampus (HIP) and entorhinal cortex (EC) (Figure 2B), suggesting that regulatory mechanisms may be shared by these two areas of the medial temporal lobe with well-established involvement in AD.39,40 In contrast, the genes in the frontal pole (FP), a brain region also implicated in AD39,41 exhibiting the largest number of altered gene transcripts in our analysis (Figure 2A), mapped to a cluster distinct from the HIP and EC regions (Figure 2B). Similar results were obtained through UMAP (Uniform Manifold Approximation and Projection for Dimension Reduction) analysis of the 3,419 genes as shown in Figure 2C. The EC and HIP regions, well established as centers of learning and memory, are in close anatomical proximity and participate in two-way communication through established neural circuits (Figure 2D).42,43,44,45,46,47,48 The EC, HIP, and FP are among the brain regions showing the earliest dysfunction in AD development, paralleling the early stages of AD during which affected individuals experience a diminished sense of space and time.47,49 Given the above, as well as a desire to further streamline the analysis, we arbitrarily decided to focus the remaining transcriptomic analysis on the HIP, EC, and FP brain regions and identified a set of 340 overlapping genes with significantly increased or decreased levels of expression that were shared among these three regions (Figures 2E and S3; Table S7).
Discovery of nine genes potentially linked to Alzheimer disease pathogenesis
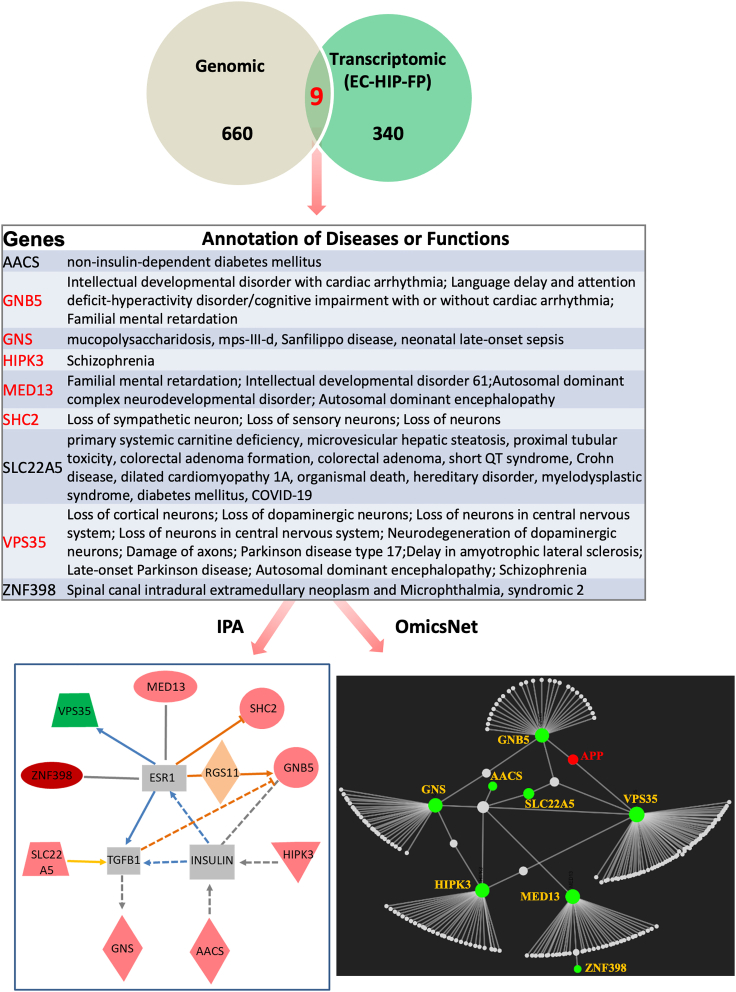
We juxtaposed the lists of candidate genes obtained by the two independent approaches. From among the 660 genes with germline GFIV enriched in subjects with AD from the NIAGADS dataset and those of AFR ancestry in the general gnomAD population (Figure 1B) and the 340 genes with significantly altered expression in the EC, HIP, and FP brain regions in AD (Figure 2E), there were nine shared genes (Figure 3, top and middle). Several of these nine, including GNB5, GNS (MIM: 607664), MED13 (MIM: 603808), and VPS35 (MIM: 601501), have established associations with various neurologic abnormalities (Figure 3, middle, in red font). Network visualization and integrative analyses identified direct or indirect interactions involving all nine genes using the Ingenuity Pathway Analysis (IPA) software (QIAGEN, Inc.) and interactions involving eight of the nine genes using the web-based OmicsNet network visualization tool (www.omicsnet.ca) (Figure 3 lower two panels). In the OmicsNet analysis using the innateDB option, both GNB5 and VPS35 showed direct interactions with the APP gene, a hallmark genetic risk factor for AD.
Figure 3.
Discovery of nine genes potentially linked to Alzheimer disease pathogenesis through combined genetic and transcriptomic analyses
Top: Venn diagram illustrating a set of nine shared genes from among the 660 genes with GFIVs enriched in both subjects with AD from the NIAGADS dataset and those of AFR ancestry in the general gnomAD population (refer to Figure 1), and the 340 genes with significantly altered expression in the EC, HIP, and FP brain regions in AD (refer to Figure 2E). Middle: a list detailing the reported associations and/or molecular functions of the nine identified genes. Genes in red font have established associations with neurologic abnormalities (see also main text). Lower: network visualization and integrative analyses of the nine genes using two different tools: the IPA package (displayed in the left panel) and the Omicsnet web-based bioinformatic tool (displayed in the right panel).
Heterozygosity of Gnb5 aggravates Aβ plaque and NFT development in an AD mouse model
Among the nine genes described above (AACS [MIM: 614364], GNB5, GNS, HIPK3 [MIM: 604424], MED13, SHC2 [MIM: 605217], SLC22A5 [MIM: 603377], VPS35, and ZNF398 [MIM: 618593]), one is GNB5, which encodes the highly evolutionarily conserved G protein β5 subunit (Gβ5), primarily expressed in neurons and essential for normal neuronal development in the mouse brain.25 In humans, homozygous or compound heterozygous loss of function of GNB5 has been associated with the Lodder-Merla syndrome (MIM: 617173) of neurodevelopmental and language delay with cardiac arrhythmia.50,51 Gβ5 forms heterodimers with one of four obligate R7 subfamily regulator of G protein signaling (RGS) binding partners (RGS6 [MIM: 603894], RGS7 [MIM: 602517], RGS9 [MIM: 604067], or RGS11 [MIM: 603895])23 to form a stable Gβ5/R7-RGS protein complex that acts as a GTPase-activating protein (GAP) targeting Gα proteins to restrain or dampen signaling downstream of G protein-coupled receptors (GPCRs).52 Using in vitro GAP assays employing purified recombinant complexes of Gβ5 bound to full-length RGS6 and RGS7, it has been shown that Gβ5/RGS7 acts exclusively on the Gαo subunit, and not on Gαq or even the closely related Gαi.53 In cell-based assays, however, the specificity of RGS7 may be less stringent. Both the RGS domain of RGS754,55 and full-length RGS7 complexed with Gβ556 inhibit Gαq-mediated calcium mobilization in transfected cells, suggesting that Gβ5/RGS7 may also regulate Gαq signaling. In Caenorhabditis elegans, the orthologs of mammalian Gβ5/R7-RGS dimers regulate both Gαi and Gαq signaling.57
The systemic examination of the transcript levels of Gnb5 and its partners in mouse brain using in situ hybridization with the RNAscope technology (ACD bio. Inc) demonstrated their wide expression consistent with a fundamental role in neuronal development and function25 (Figure S4). If such widespread expression of GNB5 and its partners is also present in human brain, it could well account for the deleterious neurodevelopmental impact of bi-allelic GNB5 loss of function described above. It is furthermore conceivable that heterozygous GFIV in GNB5 might have a phenotype, albeit more subtle, such as lowering the threshold for, or enhancing the risk of, polygenic neurologic disorders including AD.
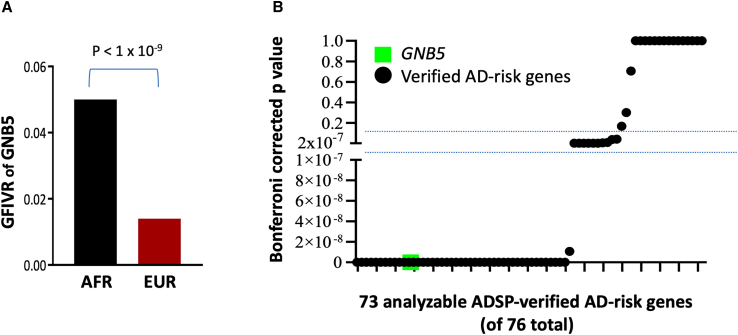
Since the prevalence of AD is 2-fold higher in individuals of AFR ancestry relative to EUR,3,4,5,6 we compared the prevalence of germline GFIV affecting GNB5 between these two ancestral groups in the general reference population. GFIVR analysis revealed that GNB5 had a 3.6-fold higher rate among individuals of AFR ancestry compared to EUR subjects within gnomAD (p < 1 × 10−9) (Figure 4A). The AFR and EUR subpopulations in gnomAD were further queried, computing the GFIVR and corresponding Bonferroni-corrected p values for 73 analyzable genes from among the 76 total genes and loci compiled by the ADSP Gene Verification Committee (Figure 4B; see also Table S3; Figure S2A).58,59,60 The majority of these 73 verified AD-risk genes had Bonferroni-corrected p values <1 × 10−9, like GNB5, demonstrating a disproportionate frequency of GFIV among subjects with AFR ancestry (Figure 4B). Taken together, these findings support the plausibility of GNB5 as a candidate genetic risk factor for AD.
Figure 4.
The GFIVR of GNB5 gene is higher in AFR than EUR populations in gnomAD
(A) Comparison of germline GFIVR of GNB5 between subjects of AFR and EUR ancestry within the reference (gnomAD) datasets (p < 1 × 10-9).
(B) Dot plot of the Bonferroni-corrected p values of the GFIVR differences for GNB5 (green square) and 73 analyzable genes (black dots) from among the 76 total loci and genes affecting AD risk compiled by the Alzheimer Disease Sequencing Project Gene Verification Committee (76 genes listed in Table S3; Figure S2A; the three non-analyzable genes omitted from the present analysis were SHARPIN/SIPL1, DOC2A, and SLC2A4RG).
According to the polygenic disease model, if GNB5 were an AD-risk gene then its haploinsufficiency would exacerbate or synergize with effects resulting from other pathogenic gene variants. To test this hypothesis, we mated Gnb5 mutant mice with double transgenic APP/PSEN1 AD model mice24 to create Gnb5+/−/APP/PSEN1 triple mutant mice and examined Aβ plaque and NFT formation in the brains of 8- to 11-month-old mice (Figure 5). Amyloid plaques and NFTs are hallmarks of AD progression and in humans the burden of Aβ plaques and NFTs largely correlates with the severity of AD symptoms.61,62,63 While no Aβ plaques or NFTs were found in wild-type or Gnb5+/− mice (Figures 5A–5E, 5I, 5M, 5B, 5F, 5J, and 5N), the APP/PSEN1/Gnb5+/− triple gene variant mice showed a significantly increased presence of both Aβ plaques (Figures 5D–5H, 5Q, and 5R) and NFTs (Figures 5L, 5P, 5S, and 5T) in both EC and HIP brain regions as compared to the APP/PSEN1 double transgenic mice (Figures 5C–5G, 5K, 5O, 5Q, 5R, 5S, and 5T). These results suggest that Gnb5 heterozygosity can synergize with other AD risk genes to aggravate AD-associated neuropathology, strengthening the candidacy of GNB5 as an AD-risk gene.
Figure 5.
Heterozygosity of GNB5 aggravates Aβ plaque and NFT development in AD mouse models
(A–H) Thioflavine S staining histological analyses of Aβ plaques in EC and HIP regions of 8- to 11-month-old mice with the indicated genotypes. All sections were counterstained with DAPI. Blue, DAPI; green, thioflavine S-positive Aβ plaques.
(I–P) Representative images of Tau-NFT antibody-stained EC and HIP sections of 8- to 11-month-old mice with the indicated genotypes. All sections were counterstained with DAPI. Blue, DAPI; red, Tau-NFT. Quantitative analyses of Aβ plaques observed in EC and HIP regions of mice with indicated genotypes are shown in (Q) and (R). ∗∗p = 0.005; ∗∗∗p = 0.0002. Quantitative analyses of Tau-NFT in EC and HIP regions of mouse brains with indicated genotypes are shown in (S) and (T). ∗p = 0.016; ∗∗∗∗p < 0.0001. All quantification of Aβ plaque and NFT staining was performed by observers blind to the mouse genotype.
Discussion
AD is a paradigmatic example of a polygenic neurodegenerative disorder with unequal impact on populations that differ in ancestry. Polygenic diseases such as AD reflect the functional status of multiple genes, each of which may contribute only minutely to disease development. The search for contributory genes has traditionally involved genome-wide SNP association studies. Recognizing that most disease-associated variants identified by GWASs map to intergenic, regulatory, and intronic regions without obvious biological relevance, we preferred to develop an alternative gene-constrained analysis that focused on variants with the potential to impact gene function, for the identification of unknown AD-associated genes. Although variants located in the genes themselves represent only a minor fraction of the entire GWAS signal, we focused on this small set of intragenic variants, hypothesizing that they should be more readily interpretable in a functional or mechanistic context. We further hypothesized that this approach might enhance the sensitivity of gene detection through cumulative “scoring” and aggregation of all potentially impactful disease-associated variants within a gene, instead of considering SNPs or variants in isolation from one another.
To test these hypotheses, we developed GFIVR analysis as a gene-constrained bioinformatic tool. From among 20,324 analyzable total genes, we identified a subset of 660 genes likely to be enriched with genes responsible for the higher AD prevalence among those of AFR relative to EUR ancestry through inter-ancestral GFIVR and bioinformatic clustering analysis. Comparison with an independently identified set of 340 genes highlighted by whole-brain transcriptomic analysis enabled the identification of nine candidate AD risk genes common to both sets with higher GFIVR in the germline and significantly altered expression levels in clinically relevant brain regions of AD-affected individuals: AACS, GNB5, GNS, HIPK3, MED13, SHC2, SLC22A5, VPS35, and ZNF398.
A potential weakness of the present approach to candidate gene identification is excessive stringency in the bioinformatic workflow and a related bias toward the detection of AD risk genes among subjects of AFR ancestry. In the K-median analysis, for example, we considered only the set of 1,008 genes in block 3 with normalized GFIVRs highly correlated between individuals of AFR ancestry within gnomAD, and the NIAGADS dataset considered as a whole (Figure 1A). We thus overlooked many potential AD-risk genes such as those in block 2 whose GFIVRs were elevated in both the EUR population in gnomAD and in the NIAGADS dataset. The elimination of two-thirds of the ADSP-verified AD-risk genes58,59,60 from the pool of genes retained for consideration at this stage of enrichment would be consistent with exclusionary excess at this step (Figure S2B). In the transcriptomic analysis also, by focusing on only 14 brain regions instead of considering significantly altered transcripts from all 23 areas, we may have been excessively stringent. Thus, the overall strategy presented here may have sacrificed sensitivity of gene detection in the quest for reliability in AD risk gene discovery.
Several of the nine candidate AD risk genes with higher GFIVR in the germline and significantly altered expression levels in the brains of AD-affected individuals have been previously associated with neurologic or neurodegenerative disease (Figure 3, middle). Variants in VPS35, for example, whose gene product functions in the endo-lysosomal pathway, have been linked to several neurodegenerative diseases, including Parkinson disease (MIM: 614203).64,65 Variants in MED13 have been associated with a severe neurodevelopmental disorder in infants (MIM: 618009).66,67,68 Bi-allelic loss of function of GNS has been associated with a lysosomal storage disease manifest by progressive neurodegeneration (MIM: 252940).69,70 As referenced above, bi-allelic loss of function of GNB5 causes the Lodder-Merla syndrome characterized by intellectual and neurodevelopmental delay with cardiac arrhythmia.50,51
As a proof of concept, we experimentally confirmed the relevance of one AD candidate risk gene, GNB5, by demonstrating a synergistic effect of Gnb5 heterozygosity on Aβ plaque and NFT formation in the APP/PSEN1 double transgenic AD mouse model. Though we did not try to correlate performance on behavioral tests of memory with the observed neuropathology in the present cohort of mice, it would be an important goal of future studies. One justification for the utilization of gene-constrained analysis like GFIVR presented here was the promise that the genes identified might be more easily interpretable in a functional context relative to the GWAS. We must therefore consider hypotheses regarding a possible role for GNB5 in the mechanism of AD pathogenesis.
A relevant clue may be our finding that GNB5 interacted with APP in the OmicsNet integrative analysis presented above (Figure 3, lower panel). Martemyanov and co-workers showed that the Gβ5/Rgs7 complex physically associates with metabotropic GABA-B receptors in hippocampal neurons71 and, via such interaction, regulates synaptic plasticity and memory.72,73 In separate studies, several labs have demonstrated the binding of APP to GABA-B receptors mediated by an extracellular sushi protein domain present on the receptor.74,75,76,77 Though it remains unclear whether APP binding modulates GABA-B receptor signaling,74,75,77 APP-GABA-B receptor complex formation may alter APP processing and affect amyloidogenesis.75 Thus it is conceivable that GNB5 might affect the amyloidogenic APP processing pathway by virtue of mutual interactions of Gβ5 and APP with the GABA-B GPCR in the hippocampus or elsewhere. Interestingly, potentially relevant non-canonical interactions of Gβ5 with GPCRs have also been demonstrated, including with the muscarinic M3 cholinergic receptor78,79,80,81 and the recently deorphanized receptor GPR158 (mGlyR).82,83
Another potential mechanism linking GNB5 with APP and amyloidogenesis centers on the Gαi/o-directed GAP activity of the Gβ5/R7-RGS protein complex by which it restrains or dampens Gi/o signaling.52 Nishimoto et al. reported some four decades ago that APP was a Go-coupled receptor.84 Subsequently it was shown that Aβ neurotoxicity may be mediated by the interaction of fibrillar Aβ (as an upstream activator) with APP, akin to the pathogenic mechanism of prions.85 Mutational or pharmacological blockade of Go/Gβγ signaling downstream of APP has been shown to block amyloidogenic processing of APP and interrupt a “feedforward” loop of amyloidogenesis driven by toxic forms of Aβ acting upstream of APP.86,87 There is little in the literature regarding possible effects of RGS proteins or GAP activity on Go signaling downstream of APP.88 Assuming however that the Gαi/o-directed GAP activity of the Gβ5/R7-RGS protein complex is agnostic with respect to the type of receptor driving Go activation (canonical seven transmembrane GPCR versus unconventional Go-coupled receptor like APP), then mutational inactivation of GNB5 would impair GAP activity, strengthen the Go signal, and promote Go/Gβγ-driven amyloidogenic activity downstream of APP.
Additional consideration regarding a possible role for GNB5 in AD pathogenesis must be given to indirect effects of GNB5 loss of function. Because a germline knockout mouse model of Gnb5 was employed, and because Gβ5 is also expressed outside the brain, including in sensory neurons89,90 and neuroendocrine tissue,91,92 the effect of Gnb5 heterozygosity on the neuropathological changes typical of AD may be indirect. Potential indirect mechanisms could include sensory deficits93 and/or metabolic or hormonal imbalance. In mice, for example, haploinsufficiency of Gnb5 results in adiposity, insulin resistance, and hepatic steatosis, hallmarks of the human metabolic syndrome (MIM: 605552).92 Metabolic syndrome is an established risk factor for AD.94,95 Indeed, indirect mechanisms of action for all nine candidate AD risk genes highlighted in this study need be entertained.
Because polygenic diseases such as AD reflect the influence of multiple genes, with each gene contributing only slightly to disease pathogenesis, the identification of relevant risk genes presents a major challenge. The challenge is magnified by the fact that disease onset and progression may be greatly influenced by environmental factors and individual behaviors, and these may reflect sociocultural and economic determinants. Our study reinforces the applicability of a gene-constrained analysis like the GFIVR method developed here for the discovery of candidate AD-associated genes possibly overlooked by GWAS approaches. The discovery here of nine candidate AD-risk genes including the preliminary validation of a role for GNB5 in AD development suggests that gene-constrained analysis like the GFIVR method can complement the power of GWASs and may be more broadly applicable to the study of other complex polygenic human traits.
Data and code availability
The datasets supporting the current study are available at the gnomAD database (https://gnomad.broadinstitute.org/downloads) and the NIAGADS Alzheimer’s Genomics Database (https://www.niagads.org/genomics/app). The program code generated and utilized for this study is available at Mendeley Data https://doi.org/10.17632/h9tgc9bw79.1.
Acknowledgments
The authors thank Harrison McNabb as well as other members of the Metabolic Diseases Branch, NIDDK for many helpful discussions and suggestions. We appreciate the generous gift of Gnb5 KO mice from Dr. Ching-Kang Jason Chen, PhD, Department of Molecular Medicine, The University of Texas Health Science Center at San Antonio, and the high-performance computing help from Drs. Susan Chacko and David Hoover at Center for Information Technology, NIH, Bethesda, Maryland. The Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases (ZIA DK043304-24) and the NIH National Institute on Aging Genetics of Alzheimer Disease Data Storage Site (U24AG041689) supported this research.
Declaration of interests
The authors declare no competing interests.
Published: February 13, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2024.01.005.
Contributor Information
Jianhua Zhang, Email: jianhuaz@mail.nih.gov.
William F. Simonds, Email: bills@niddk.nih.gov.
Web resources
OMIM, https://www.omim.org/
Supplemental information
References
- 1.Joseph J.J., Ortiz R., Acharya T., Golden S.H., López L., Deedwania P. Cardiovascular Impact of Race and Ethnicity in Patients With Diabetes and Obesity: JACC Focus Seminar 2/9. J. Am. Coll. Cardiol. 2021;78:2471–2482. doi: 10.1016/j.jacc.2021.06.020. [DOI] [PubMed] [Google Scholar]
- 2.Cromer S.J., Lakhani C.M., Mercader J.M., Majarian T.D., Schroeder P., Cole J.B., Florez J.C., Patel C.J., Manning A.K., Burnett-Bowie S.A.M., et al. Association and Interaction of Genetics and Area-Level Socioeconomic Factors on the Prevalence of Type 2 Diabetes and Obesity. Diabetes Care. 2023;46:944–952. doi: 10.2337/dc22-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chin A.L., Negash S., Hamilton R. Diversity and disparity in dementia: the impact of ethnoracial differences in Alzheimer disease. Alzheimer Dis. Assoc. Disord. 2011;25:187–195. doi: 10.1097/WAD.0b013e318211c6c9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cukier H.N., Kunkle B.W., Vardarajan B.N., Rolati S., Hamilton-Nelson K.L., Kohli M.A., Whitehead P.L., Dombroski B.A., Van Booven D., Lang R., et al. ABCA7 frameshift deletion associated with Alzheimer disease in African Americans. Neurol. Genet. 2016;2:e79. doi: 10.1212/NXG.0000000000000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayeda E.R., Glymour M.M., Quesenberry C.P., Whitmer R.A. Inequalities in dementia incidence between six racial and ethnic groups over 14 years. Alzheimers Dement. 2016;12:216–224. doi: 10.1016/j.jalz.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alzheimer’s_Association 2023 Alzheimer's disease facts and figures. Alzheimers Dement. 2023;19:1598–1695. doi: 10.1002/alz.13016. [DOI] [PubMed] [Google Scholar]
- 7.Karlsson I.K., Escott-Price V., Gatz M., Hardy J., Pedersen N.L., Shoai M., Reynolds C.A. Measuring heritable contributions to Alzheimer's disease: polygenic risk score analysis with twins. Brain Commun. 2022;4:fcab308. doi: 10.1093/braincomms/fcab308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sirugo G., Williams S.M., Tishkoff S.A. The Missing Diversity in Human Genetic Studies. Cell. 2019;177:26–31. doi: 10.1016/j.cell.2019.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bentley A.R., Callier S., Rotimi C.N. Diversity and inclusion in genomic research: why the uneven progress? J. Community Genet. 2017;8:255–266. doi: 10.1007/s12687-017-0316-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haga S.B. Impact of limited population diversity of genome-wide association studies. Genet. Med. 2010;12:81–84. doi: 10.1097/GIM.0b013e3181ca2bbf. [DOI] [PubMed] [Google Scholar]
- 11.Need A.C., Goldstein D.B. Next generation disparities in human genomics: concerns and remedies. Trends Genet. 2009;25:489–494. doi: 10.1016/j.tig.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 12.Bustamante C.D., Burchard E.G., De la Vega F.M. Genomics for the world. Nature. 2011;475:163–165. doi: 10.1038/475163a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei H., Wang Y., Zhou C., Lin D., Liu B., Liu K., Qiu S., Gong B., Li Y., Zhang G., et al. Distinct genetic alteration profiles of acute myeloid leukemia between Caucasian and Eastern Asian population. J. Hematol. Oncol. 2018;11:18. doi: 10.1186/s13045-018-0566-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng J., Chen H., Zhou D., Zhang J., Chen Y., Liu Q., Ai D., Zhu H., Chu L., Ren W., et al. Comparative genomic analysis of esophageal squamous cell carcinoma between Asian and Caucasian patient populations. Nat. Commun. 2017;8:1533. doi: 10.1038/s41467-017-01730-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popejoy A.B., Fullerton S.M. Genomics is failing on diversity. Nature. 2016;538:161–164. doi: 10.1038/538161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson G. Population genetics and GWAS: A primer. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2005485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hindorff L.A., Bonham V.L., Brody L.C., Ginoza M.E.C., Hutter C.M., Manolio T.A., Green E.D. Prioritizing diversity in human genomics research. Nat. Rev. Genet. 2018;19:175–185. doi: 10.1038/nrg.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spratt D.E., Chan T., Waldron L., Speers C., Feng F.Y., Ogunwobi O.O., Osborne J.R. Racial/Ethnic Disparities in Genomic Sequencing. JAMA Oncol. 2016;2:1070–1074. doi: 10.1001/jamaoncol.2016.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe K., Stringer S., Frei O., Umićević Mirkov M., de Leeuw C., Polderman T.J.C., van der Sluis S., Andreassen O.A., Neale B.M., Posthuma D. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 2019;51:1339–1348. doi: 10.1038/s41588-019-0481-0. [DOI] [PubMed] [Google Scholar]
- 20.Visscher P.M., Yengo L., Cox N.J., Wray N.R. Discovery and implications of polygenicity of common diseases. Science. 2021;373:1468–1473. doi: 10.1126/science.abi8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaid D.J., Chen W., Larson N.B. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet. 2018;19:491–504. doi: 10.1038/s41576-018-0016-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E., et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C.K., Eversole-Cire P., Zhang H., Mancino V., Chen Y.J., He W., Wensel T.G., Simon M.I. Instability of GGL domain-containing RGS proteins in mice lacking the G protein beta-subunit Gbeta5. Proc. Natl. Acad. Sci. USA. 2003;100:6604–6609. doi: 10.1073/pnas.0631825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Savonenko A., Xu G.M., Melnikova T., Morton J.L., Gonzales V., Wong M.P.F., Price D.L., Tang F., Markowska A.L., Borchelt D.R. Episodic-like memory deficits in the APPswe/PS1dE9 mouse model of Alzheimer's disease: relationships to beta-amyloid deposition and neurotransmitter abnormalities. Neurobiol. Dis. 2005;18:602–617. doi: 10.1016/j.nbd.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J.H., Pandey M., Seigneur E.M., Panicker L.M., Koo L., Schwartz O.M., Chen W., Chen C.K., Simonds W.F. Knockout of G protein beta5 impairs brain development and causes multiple neurologic abnormalities in mice. J. Neurochem. 2011;119:544–554. doi: 10.1111/j.1471-4159.2011.07457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guntern R., Bouras C., Hof P.R., Vallet P.G. An improved thioflavine S method for staining neurofibrillary tangles and senile plaques in Alzheimer's disease. Experientia. 1992;48:8–10. doi: 10.1007/BF01923594. [DOI] [PubMed] [Google Scholar]
- 27.Cunningham F., Allen J.E., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Austine-Orimoloye O., Azov A.G., Barnes I., Bennett R., et al. Ensembl 2022. Nucleic Acids Res. 2022;50:D988–D995. doi: 10.1093/nar/gkab1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Hoon M.J.L., Imoto S., Nolan J., Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 29.Saldanha A.J. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 30.Kluyver T., Ragan-Kelley B., Pérez F., Granger B., Bussonnier M., Frederic J., Kelley K., Hamrick J., Grout J., Corlay S., et al. In: Jupyter Notebooks – a publishing format for reproducible computational workflows, in Positioning and Power in Academic Publishing: Players, Agents and Agendas. Loizides F., Scmidt B., editors. IOS Press; 2016. Jupyter Notebooks – a publishing format for reproducible computational workflows; pp. 87–90. [Google Scholar]
- 31.Kuzma A., Valladares O., Cweibel R., Greenfest-Allen E., Childress D.M., Malamon J., Gangadharan P., Zhao Y., Qu L., Leung Y.Y., et al. NIAGADS: The NIA Genetics of Alzheimer's Disease Data Storage Site. Alzheimer's Dementia. 2016;12:1200–1203. [Google Scholar]
- 32.Greenfest-Allen E., Valladares O., Kuksa P.P., Gangadharan P., Lee W.P., Cifello J., Katanic Z., Kuzma A.B., Wheeler N., Bush W.S., et al. NIAGADS Alzheimer's GenomicsDB: A resource for exploring Alzheimer's disease genetic and genomic knowledge. Alzheimers Dement. 2023:1–14. doi: 10.1002/alz.13509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O'Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2020 doi: 10.1101/531210. Preprint at. [DOI] [Google Scholar]
- 35.Livesey B.J., Marsh J.A. Interpreting protein variant effects with computational predictors and deep mutational scanning. Dis. Model. Mech. 2022;15 doi: 10.1242/dmm.049510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee W.P., Choi S.H., Shea M.G., Cheng P.L., Dombroski B.A., Pitsillides A.N., Heard-Costa N.L., Wang H., Bulekova K., Kuzma A.B., et al. Association of Common and Rare Variants with Alzheimer's Disease in over 13,000 Diverse Individuals with Whole-Genome Sequencing from the Alzheimer's Disease Sequencing Project. medRxiv. 2023 doi: 10.1101/2023.09.01.23294953. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H., Dombroski B.A., Cheng P.L., Tucci A., Si Y.Q., Farrell J.J., Tzeng J.Y., Leung Y.Y., Malamon J.S., Alzheimer's Disease Sequencing P., et al. Structural Variation Detection and Association Analysis of Whole-Genome-Sequence Data from 16,905 Alzheimer's Diseases Sequencing Project Subjects. medRxiv. 2023 doi: 10.1101/2023.09.13.23295505. Preprint at. [DOI] [Google Scholar]
- 38.Sharma P., Srivastava P., Seth A., Tripathi P.N., Banerjee A.G., Shrivastava S.K. Comprehensive review of mechanisms of pathogenesis involved in Alzheimer's disease and potential therapeutic strategies. Prog. Neurobiol. 2019;174:53–89. doi: 10.1016/j.pneurobio.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 39.O'Brien J.T. Role of imaging techniques in the diagnosis of dementia. Br. J. Radiol. 2007;80 Spec No 2:S71–S77. doi: 10.1259/bjr/33117326. [DOI] [PubMed] [Google Scholar]
- 40.Duyckaerts C., Delatour B., Potier M.C. Classification and basic pathology of Alzheimer disease. Acta Neuropathol. 2009;118:5–36. doi: 10.1007/s00401-009-0532-1. [DOI] [PubMed] [Google Scholar]
- 41.Finger E., Zhang J., Dickerson B., Bureau Y., Masellis M., Alzheimer’s Disease Neuroimaging Initiative Disinhibition in Alzheimer's Disease is Associated with Reduced Right Frontal Pole Cortical Thickness. J. Alzheimers Dis. 2017;60:1161–1170. doi: 10.3233/JAD-170348. [DOI] [PubMed] [Google Scholar]
- 42.Ronaghi A., Zibaii M.I., Pandamooz S., Nourzei N., Motamedi F., Ahmadiani A., Dargahi L. Entorhinal cortex stimulation induces dentate gyrus neurogenesis through insulin receptor signaling. Brain Res. Bull. 2019;144:75–84. doi: 10.1016/j.brainresbull.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 43.Petrache A.L., Rajulawalla A., Shi A., Wetzel A., Saito T., Saido T.C., Harvey K., Ali A.B. Aberrant Excitatory-Inhibitory Synaptic Mechanisms in Entorhinal Cortex Microcircuits During the Pathogenesis of Alzheimer's Disease. Cereb. Cortex. 2019;29:1834–1850. doi: 10.1093/cercor/bhz016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Helton T.D., Zhao M., Farris S., Dudek S.M. Diversity of dendritic morphology and entorhinal cortex synaptic effectiveness in mouse CA2 pyramidal neurons. Hippocampus. 2019;29:78–92. doi: 10.1002/hipo.23012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chenani A., Sabariego M., Schlesiger M.I., Leutgeb J.K., Leutgeb S., Leibold C. Hippocampal CA1 replay becomes less prominent but more rigid without inputs from medial entorhinal cortex. Nat. Commun. 2019;10:1341. doi: 10.1038/s41467-019-09280-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwakowsky A., Calvo-Flores Guzmán B., Pandya M., Turner C., Waldvogel H.J., Faull R.L. GABA(A) receptor subunit expression changes in the human Alzheimer's disease hippocampus, subiculum, entorhinal cortex and superior temporal gyrus. J. Neurochem. 2018;145:374–392. doi: 10.1111/jnc.14325. [DOI] [PubMed] [Google Scholar]
- 47.Goyal A., Miller J., Watrous A.J., Lee S.A., Coffey T., Sperling M.R., Sharan A., Worrell G., Berry B., Lega B., et al. Electrical Stimulation in Hippocampus and Entorhinal Cortex Impairs Spatial and Temporal Memory. J. Neurosci. 2018;38:4471–4481. doi: 10.1523/JNEUROSCI.3049-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valero M., de la Prida L.M. The hippocampus in depth: a sublayer-specific perspective of entorhinal-hippocampal function. Curr. Opin. Neurobiol. 2018;52:107–114. doi: 10.1016/j.conb.2018.04.013. [DOI] [PubMed] [Google Scholar]
- 49.Parker A.F., Smart C.M., Scarapicchia V., Gawryluk J.R., the Alzheimer’s Disease Neuroimaging Initiative Identification of Earlier Biomarkers for Alzheimer's Disease: A Multimodal Neuroimaging Study of Individuals with Subjective Cognitive Decline. J. Alzheimers Dis. 2020;77:1067–1076. doi: 10.3233/JAD-200299. [DOI] [PubMed] [Google Scholar]
- 50.Lodder E.M., De Nittis P., Koopman C.D., Wiszniewski W., Moura de Souza C.F., Lahrouchi N., Guex N., Napolioni V., Tessadori F., Beekman L., et al. GNB5 Mutations Cause an Autosomal-Recessive Multisystem Syndrome with Sinus Bradycardia and Cognitive Disability. Am. J. Hum. Genet. 2016;99:704–710. doi: 10.1016/j.ajhg.2016.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shamseldin H.E., Masuho I., Alenizi A., Alyamani S., Patil D.N., Ibrahim N., Martemyanov K.A., Alkuraya F.S. GNB5 mutation causes a novel neuropsychiatric disorder featuring attention deficit hyperactivity disorder, severely impaired language development and normal cognition. Genome Biol. 2016;17:195. doi: 10.1186/s13059-016-1061-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson G.R., Posokhova E., Martemyanov K.A. The R7 RGS protein family: multi-subunit regulators of neuronal G protein signaling. Cell Biochem. Biophys. 2009;54:33–46. doi: 10.1007/s12013-009-9052-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Posner B.A., Gilman A.G., Harris B.A. Regulators of g protein signaling 6 and 7 - Purification of complexes with Gβ5 and assessment of their effects on G protein-mediated signaling pathways. J. Biol. Chem. 1999;274:31087–31093. doi: 10.1074/jbc.274.43.31087. [DOI] [PubMed] [Google Scholar]
- 54.DiBello P.R., Garrison T.R., Apanovitch D.M., Hoffman G., Shuey D.J., Mason K., Cockett M.I., Dohlman H.G. Selective uncoupling of RGS action by a single point mutation in the G protein alpha-subunit. J. Biol. Chem. 1998;273:5780–5784. doi: 10.1074/jbc.273.10.5780. [DOI] [PubMed] [Google Scholar]
- 55.Shuey D.J., Betty M., Jones P.G., Khawaja X.Z., Cockett M.I. RGS7 attenuates signal transduction through the G(alpha q) family of heterotrimeric G proteins in mammalian cells. J. Neurochem. 1998;70:1964–1972. doi: 10.1046/j.1471-4159.1998.70051964.x. [DOI] [PubMed] [Google Scholar]
- 56.Witherow D.S., Wang Q., Levay K., Cabrera J.L., Chen J., Willars G.B., Slepak V.Z. Complexes of the G protein subunit gbeta 5 with the regulators of G protein signaling RGS7 and RGS9. Characterization in native tissues and in transfected cells. J. Biol. Chem. 2000;275:24872–24880. doi: 10.1074/jbc.M001535200. [DOI] [PubMed] [Google Scholar]
- 57.Chase D.L., Patikoglou G.A., Koelle M.R. Two RGS proteins that inhibit Galpha(o) and Galpha(q) signaling in C. elegans neurons require a Gbeta(5)-like subunit for function. Curr. Biol. 2001;11:222–231. doi: 10.1016/s0960-9822(01)00071-9. [DOI] [PubMed] [Google Scholar]
- 58.Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B., et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Naj A.C., Jun G., Reitz C., Kunkle B.W., Perry W., Park Y.S., Beecham G.W., Rajbhandary R.A., Hamilton-Nelson K.L., Wang L.S., et al. Effects of multiple genetic loci on age at onset in late-onset Alzheimer disease: a genome-wide association study. JAMA Neurol. 2014;71:1394–1404. doi: 10.1001/jamaneurol.2014.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosenberg R.N., Lambracht-Washington D., Yu G., Xia W. Genomics of Alzheimer Disease: A Review. JAMA Neurol. 2016;73:867–874. doi: 10.1001/jamaneurol.2016.0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bensamoun D., Guignard R., Furst A.J., Derreumaux A., Manera V., Darcourt J., Benoit M., Robert P.H., David R. Associations between Neuropsychiatric Symptoms and Cerebral Amyloid Deposition in Cognitively Impaired Elderly People. J. Alzheimers Dis. 2016;49:387–398. doi: 10.3233/JAD-150181. [DOI] [PubMed] [Google Scholar]
- 62.Boublay N., Schott A.M., Krolak-Salmon P. Neuroimaging correlates of neuropsychiatric symptoms in Alzheimer's disease: a review of 20 years of research. Eur. J. Neurol. 2016;23:1500–1509. doi: 10.1111/ene.13076. [DOI] [PubMed] [Google Scholar]
- 63.Ehrenberg A.J., Suemoto C.K., França Resende E.d.P., Petersen C., Leite R.E.P., Rodriguez R.D., Ferretti-Rebustini R.E.d.L., You M., Oh J., Nitrini R., et al. Neuropathologic Correlates of Psychiatric Symptoms in Alzheimer's Disease. J. Alzheimers Dis. 2018;66:115–126. doi: 10.3233/JAD-180688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williams E.T., Chen X., Otero P.A., Moore D.J. Understanding the contributions of VPS35 and the retromer in neurodegenerative disease. Neurobiol. Dis. 2022;170 doi: 10.1016/j.nbd.2022.105768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muraleedharan A., Vanderperre B. The Endo-lysosomal System in Parkinson's Disease: Expanding the Horizon. J. Mol. Biol. 2023;435 doi: 10.1016/j.jmb.2023.168140. [DOI] [PubMed] [Google Scholar]
- 66.Snijders Blok L., Hiatt S.M., Bowling K.M., Prokop J.W., Engel K.L., Cochran J.N., Bebin E.M., Bijlsma E.K., Ruivenkamp C.A.L., Terhal P., et al. De novo mutations in MED13, a component of the Mediator complex, are associated with a novel neurodevelopmental disorder. Hum. Genet. 2018;137:375–388. doi: 10.1007/s00439-018-1887-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rogers A.P., Friend K., Rawlings L., Barnett C.P. A de novo missense variant in MED13 in a patient with global developmental delay, marked facial dysmorphism, macroglossia, short stature, and macrocephaly. Am. J. Med. Genet. 2021;185:2586–2592. doi: 10.1002/ajmg.a.62238. [DOI] [PubMed] [Google Scholar]
- 68.Trivisano M., De Dominicis A., Micalizzi A., Ferretti A., Dentici M.L., Terracciano A., Calabrese C., Vigevano F., Novelli G., Novelli A., Specchio N. MED13 mutation: A novel cause of developmental and epileptic encephalopathy with infantile spasms. Seizure. 2022;101:211–217. doi: 10.1016/j.seizure.2022.09.002. [DOI] [PubMed] [Google Scholar]
- 69.Mok A., Cao H., Hegele R.A. Genomic basis of mucopolysaccharidosis type IIID (MIM 252940) revealed by sequencing of GNS encoding N-acetylglucosamine-6-sulfatase. Genomics. 2003;81:1–5. doi: 10.1016/s0888-7543(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 70.Valstar M.J., Bertoli-Avella A.M., Wessels M.W., Ruijter G.J.G., de Graaf B., Olmer R., Elfferich P., Neijs S., Kariminejad R., Suheyl Ezgü F., et al. Mucopolysaccharidosis type IIID: 12 new patients and 15 novel mutations. Hum. Mutat. 2010;31:E1348–E1360. doi: 10.1002/humu.21234. [DOI] [PubMed] [Google Scholar]
- 71.Fajardo-Serrano A., Wydeven N., Young D., Watanabe M., Shigemoto R., Martemyanov K.A., Wickman K., Luján R. Association of Rgs7/Gbeta5 complexes with Girk channels and GABAB receptors in hippocampal CA1 pyramidal neurons. Hippocampus. 2013;23:1231–1245. doi: 10.1002/hipo.22161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ostrovskaya O., Xie K., Masuho I., Fajardo-Serrano A., Lujan R., Wickman K., Martemyanov K.A. RGS7/Gbeta5/R7BP complex regulates synaptic plasticity and memory by modulating hippocampal GABABR-GIRK signaling. Elife. 2014;3 doi: 10.7554/eLife.02053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ostrovskaya O.I., Orlandi C., Fajardo-Serrano A., Young S.M., Jr., Lujan R., Martemyanov K.A. Inhibitory Signaling to Ion Channels in Hippocampal Neurons Is Differentially Regulated by Alternative Macromolecular Complexes of RGS7. J. Neurosci. 2018;38:10002–10015. doi: 10.1523/JNEUROSCI.1378-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rice H.C., de Malmazet D., Schreurs A., Frere S., Van Molle I., Volkov A.N., Creemers E., Vertkin I., Nys J., Ranaivoson F.M., et al. Secreted amyloid-beta precursor protein functions as a GABA(B)R1a ligand to modulate synaptic transmission. Science. 2019;363 doi: 10.1126/science.aao4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dinamarca M.C., Raveh A., Schneider A., Fritzius T., Früh S., Rem P.D., Stawarski M., Lalanne T., Turecek R., Choo M., et al. Complex formation of APP with GABA(B) receptors links axonal trafficking to amyloidogenic processing. Nat. Commun. 2019;10:1331. doi: 10.1038/s41467-019-09164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tang B.L. Amyloid Precursor Protein (APP) and GABAergic Neurotransmission. Cells. 2019;8 doi: 10.3390/cells8060550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rem P.D., Sereikaite V., Fernández-Fernández D., Reinartz S., Ulrich D., Fritzius T., Trovo L., Roux S., Chen Z., Rondard P., et al. Soluble amyloid-beta precursor peptide does not regulate GABA(B) receptor activity. Elife. 2023;12 doi: 10.7554/eLife.82082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sandiford S.L., Slepak V.Z. The Gbeta5-RGS7 complex selectively inhibits muscarinic M3 receptor signaling via the interaction between the third intracellular loop of the receptor and the DEP domain of RGS7. Biochemistry. 2009;48:2282–2289. doi: 10.1021/bi801989c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sandiford S.L., Wang Q., Levay K., Buchwald P., Slepak V.Z. Molecular organization of the complex between the muscarinic M3 receptor and the regulator of G protein signaling, Gbeta(5)-RGS7. Biochemistry. 2010;49:4998–5006. doi: 10.1021/bi100080p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karpinsky-Semper D., Tayou J., Levay K., Schuchardt B.J., Bhat V., Volmar C.H., Farooq A., Slepak V.Z. Helix 8 and the i3 loop of the muscarinic M3 receptor are crucial sites for its regulation by the Gbeta5-RGS7 complex. Biochemistry. 2015;54:1077–1088. doi: 10.1021/bi500980d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karpinsky-Semper D., Volmar C.H., Brothers S.P., Slepak V.Z. Differential effects of the Gbeta5-RGS7 complex on muscarinic M3 receptor-induced Ca2+ influx and release. Mol. Pharmacol. 2014;85:758–768. doi: 10.1124/mol.114.091843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Patil D.N., Singh S., Laboute T., Strutzenberg T.S., Qiu X., Wu D., Novick S.J., Robinson C.V., Griffin P.R., Hunt J.F., et al. Cryo-EM structure of human GPR158 receptor coupled to the RGS7-Gbeta5 signaling complex. Science. 2022;375:86–91. doi: 10.1126/science.abl4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Laboute T., Zucca S., Holcomb M., Patil D.N., Garza C., Wheatley B.A., Roy R.N., Forli S., Martemyanov K.A. Orphan receptor GPR158 serves as a metabotropic glycine receptor: mGlyR. Science. 2023;379:1352–1358. doi: 10.1126/science.add7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nishimoto I., Okamoto T., Matsuura Y., Takahashi S., Okamoto T., Murayama Y., Ogata E. Alzheimer amyloid protein precursor complexes with brain GTP-binding protein G(o) Nature. 1993;362:75–79. doi: 10.1038/362075a0. [DOI] [PubMed] [Google Scholar]
- 85.Lorenzo A., Yuan M., Zhang Z., Paganetti P.A., Sturchler-Pierrat C., Staufenbiel M., Mautino J., Vigo F.S., Sommer B., Yankner B.A. Amyloid beta interacts with the amyloid precursor protein: a potential toxic mechanism in Alzheimer's disease. Nat. Neurosci. 2000;3:460–464. doi: 10.1038/74833. [DOI] [PubMed] [Google Scholar]
- 86.Bignante E.A., Ponce N.E., Heredia F., Musso J., Krawczyk M.C., Millán J., Pigino G.F., Inestrosa N.C., Boccia M.M., Lorenzo A. APP/Go protein Gbetagamma-complex signaling mediates Abeta degeneration and cognitive impairment in Alzheimer's disease models. Neurobiol. Aging. 2018;64:44–57. doi: 10.1016/j.neurobiolaging.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 87.Antonino M., Marmo P., Freites C.L., Quassollo G.E., Sánchez M.F., Lorenzo A., Bignante E.A. Abeta Assemblies Promote Amyloidogenic Processing of APP and Intracellular Accumulation of Abeta42 Through Go/Gbetagamma Signaling. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.852738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Copenhaver P.F., Kögel D. Role of APP Interactions with Heterotrimeric G Proteins: Physiological Functions and Pathological Consequences. Front. Mol. Neurosci. 2017;10:3. doi: 10.3389/fnmol.2017.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liapis E., Sandiford S., Wang Q., Gaidosh G., Motti D., Levay K., Slepak V.Z. Subcellular localization of regulator of G protein signaling RGS7 complex in neurons and transfected cells. J. Neurochem. 2012;122:568–581. doi: 10.1111/j.1471-4159.2012.07811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pandey M., Zhang J.H., Adikaram P.R., Kittock C., Lue N., Awe A., Degner K., Jacob N., Staples J., Thomas R., et al. Specific regulation of mechanical nociception by Gbeta5 involves GABA-B receptors. JCI Insight. 2023;8 doi: 10.1172/jci.insight.134685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nini L., Zhang J.H., Pandey M., Panicker L.M., Simonds W.F. Expression of the Gbeta5/R7-RGS protein complex in pituitary and pancreatic islet cells. Endocrine. 2012;42:214–217. doi: 10.1007/s12020-012-9611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Q., Levay K., Chanturiya T., Dvoriantchikova G., Anderson K.L., Bianco S.D.C., Ueta C.B., Molano R.D., Pileggi A., Gurevich E.V., et al. Targeted deletion of one or two copies of the G protein beta subunit Gbeta5 gene has distinct effects on body weight and behavior in mice. FASEB J. 2011;25:3949–3957. doi: 10.1096/fj.11-190157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang N.K., Zhang S.K., Zhang L.I., Tao H.W., Zhang G.W. Sensory processing deficits and related cortical pathological changes in Alzheimer's disease. Front. Aging Neurosci. 2023;15 doi: 10.3389/fnagi.2023.1213379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Atti A.R., Valente S., Iodice A., Caramella I., Ferrari B., Albert U., Mandelli L., De Ronchi D. Metabolic Syndrome, Mild Cognitive Impairment, and Dementia: A Meta-Analysis of Longitudinal Studies. Am. J. Geriatr. Psychiatry. 2019;27:625–637. doi: 10.1016/j.jagp.2019.01.214. [DOI] [PubMed] [Google Scholar]
- 95.Ezkurdia A., Ramírez M.J., Solas M. Metabolic Syndrome as a Risk Factor for Alzheimer's Disease: A Focus on Insulin Resistance. Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms24054354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the current study are available at the gnomAD database (https://gnomad.broadinstitute.org/downloads) and the NIAGADS Alzheimer’s Genomics Database (https://www.niagads.org/genomics/app). The program code generated and utilized for this study is available at Mendeley Data https://doi.org/10.17632/h9tgc9bw79.1.