Summary
Pathogenic variants in multiple genes on the X chromosome have been implicated in syndromic and non-syndromic intellectual disability disorders. ZFX on Xp22.11 encodes a transcription factor that has been linked to diverse processes including oncogenesis and development, but germline variants have not been characterized in association with disease. Here, we present clinical and molecular characterization of 18 individuals with germline ZFX variants. Exome or genome sequencing revealed 11 variants in 18 subjects (14 males and 4 females) from 16 unrelated families. Four missense variants were identified in 11 subjects, with seven truncation variants in the remaining individuals. Clinical findings included developmental delay/intellectual disability, behavioral abnormalities, hypotonia, and congenital anomalies. Overlapping and recurrent facial features were identified in all subjects, including thickening and medial broadening of eyebrows, variations in the shape of the face, external eye abnormalities, smooth and/or long philtrum, and ear abnormalities. Hyperparathyroidism was found in four families with missense variants, and enrichment of different tumor types was observed. In molecular studies, DNA-binding domain variants elicited differential expression of a small set of target genes relative to wild-type ZFX in cultured cells, suggesting a gain or loss of transcriptional activity. Additionally, a zebrafish model of ZFX loss displayed an altered behavioral phenotype, providing additional evidence for the functional significance of ZFX. Our clinical and experimental data support that variants in ZFX are associated with an X-linked intellectual disability syndrome characterized by a recurrent facial gestalt, neurocognitive and behavioral abnormalities, and an increased risk for congenital anomalies and hyperparathyroidism.
Keywords: behavioral problems, congenital anomalies, de novo mutation, developmental delay, hyperparathyroidism, hypotonia, intellectual disability, transcription factor, X-linked, ZFX, zinc finger
We describe a cohort of 18 individuals with germline variants in ZFX, which encodes a transcription factor not previously associated with a human disease. In addition to identifying recurrent clinical features, molecular characterization of variants in cultured cells, in silico modeling, and a zebrafish model suggest potential modes of pathogenicity.
Introduction
Syndromic and non-syndromic X-linked intellectual disability has been associated with more than 160 genes on the X chromosome.1,2,3,4 X-linked intellectual disability partially accounts for an excess of intellectual disability in males compared to females, and both dominant and recessive X-linked intellectual disability have the potential to cause disease in females as a result of skewed X-inactivation.4,5 The higher frequency of intellectual disability-associated genes on the X chromosome as compared to the autosomes is supported by an enrichment of brain-expressed transcripts on the X chromosome.4 Several of these genes are involved in transcriptional regulation, including the transcription factors (TFs) ZNF711 (MIM: 314990) and ARX (MIM: 300382) and the chromatin modifier MECP2 (MIM: 300005), demonstrating the potential for defects in transcriptional control to cause intellectual disability with concomitant pleomorphic phenotypes.6
ZFX (MIM: 314980), located in Xp22.11, encodes a conserved C2H2 zinc-finger TF that has not previously been reported in association with X-linked intellectual disability. ZFX is thought to primarily function by binding to CpG island promoter regions and activating the expression of many target genes.7 Structurally, ZFX is composed of an amino-terminal transactivation domain followed by 13 zinc-finger domains, of which the final three are necessary and sufficient for recruitment to promoter regions.8,9
Initially thought to play a role in sex determination,10,11,12 ZFX has more recently been investigated for its role in stem cell self-renewal and oncogenesis. ZFX is necessary for self-renewal of murine embryonic stem cells and the maintenance of murine adult hematopoietic stem cells.13 In mouse models, loss of Zfx was shown to impair the development of basal cell carcinoma, medulloblastoma, acute myeloid leukemia, and acute T-lymphoblastic leukemia, and it has been hypothesized that ZFX may promote metaplastic transformation.9,14,15 Male and female Zfx−/− mice displayed impaired embryonic growth and reduced adult body size and germ cell number.16 Partial neonatal lethality was observed, primarily in males, with only 10% of hemizygous knockout (KO) males surviving to weaning.16
ZFX is part of a gene family that includes ZFY (MIM: 490000), located in Yp11.2 and demonstrating 96% overall sequence similarity to ZFX, and ZNF711 (MIM: 314990), located in Xq21.1 and demonstrating 87% sequence similarity in the crucial DNA-contacting zinc-finger domains.9 Of note, variants in ZNF711 have previously been reported in individuals with X-linked intellectual disability (intellectual developmental disorder, X-linked 97; MIM: 300803).17,18 Genome-wide binding patterns of ZFX, measured by chromatin immunoprecipitation and sequencing (ChIP-seq), show considerable overlap with binding of ZFY and ZNF711.9 Cultured HEK293 cells lacking ZFX, ZFY, and ZNF711 demonstrated impaired proliferation and transcriptome alterations relative to wild-type (WT) cells.9
Recurrent somatic mutations in the final zinc finger of ZFX, resulting in substitution of arginine 786 with glutamine (p.Arg786Gln [c.2357G>A]) or leucine (p.Arg786Leu [c.2357G>T]), were identified in 6 of 130 samples in a sporadic parathyroid adenoma sequencing study.19 In the Catalog of Somatic Mutations in Cancer (COSMIC, https://cancer.sanger.ac.uk/cosmic), an additional 8 tumors with p.Arg786Gln missense variants are reported (COSMIC: COSV58447078): four endometrioid carcinomas, one melanoma, one acute lymphoblastic leukemia, one sarcoma, and one squamous cell carcinoma.
Germline variants in ZFX, however, remain largely uncharacterized. The NIH-sponsored ClinVar database of human genetic variation contains no nonsense or frameshift variants in the coding sequence of ZFX, and nine reported missense variants each with a single submitter (accessed October 6, 2023).20 The p.Arg786Gln variant, in particular, has not been reported in the human germline (dbSNP: rs748417793).
Variants in TFs like ZFX have the potential to cause broad and pleomorphic functional effects through diverse mechanisms. Truncated protein variants that lack a DNA-binding domain or trigger protein surveillance mechanisms such as nonsense-mediated mRNA decay (NMD) can be predicted to cause disease through protein insufficiency/loss-of-function mechanisms.21 Single-amino-acid variants in TF DNA binding domains have been shown to both abrogate and alter DNA binding specificity with the potential for both loss- and gain-of-function mechanisms, as well as cause dominant-negative interactions by interfering with DNA binding by other TFs.22,23
Here, we present detailed clinical and molecular data on 18 individuals with germline variants in ZFX, in two subgroups: 7 with truncating (frameshift or nonsense) variants predicted to result in loss of function via NMD of the resulting transcript or truncation of the DNA-binding zinc fingers, and 11 with missense variants in and around the two most carboxy-terminal zinc fingers responsible for DNA binding specificity, including two individuals with a germline p.Arg786Gln variant. The great majority of individuals exhibited developmental delay/intellectual disability, hypotonia, and overlapping facial features. We also observed increased incidence of congenital anomalies and hyperparathyroidism. We used in silico, in vitro, and in vivo approaches to functionally characterize the observed variants. For a subset of missense variants mapping within the DNA-contacting zinc fingers, we characterized the effects on genome-wide DNA binding by ChIP-seq and observed transcriptional perturbation in variants by RNA-seq. We also modeled the group of variants with truncations causing predicted loss of function in a zebrafish ZFX deletion model and characterized behavioral alterations consistent with an intellectual disability phenotype.
Material and methods
Probands
Through a collaborative effort involving clinicians and researchers from multiple institutions, we identified 18 individuals, including 3 individuals from one family, with missense and truncating predicted deleterious variants in ZFX. The connection between all collaborators was facilitated using GeneMatcher and virtual meetings.24 Several individuals were recruited through consortia and evaluated by their local medical geneticists, including the 100,000 Genomes Project, DECIPHER, and the Deciphering Developmental Disorders Study (details in Table S1).25 Exome sequencing (ES) or genome sequencing (GS) for other subjects was ordered as part of their clinical diagnostic workup for developmental delay/intellectual disability, congenital anomalies, and/or dysmorphic facial features by local specialists, typically clinical geneticists or neurologists. The probands or their guardians signed informed consent for publication, which was approved by the Institutional Review Board of their respective institutions.
Sequencing
All individuals had either ES or GS performed under a mixture of both clinical and research protocols. For specific sequencing details, see Table S1 (“Genetic Testing”) and the supplemental notes. Trio whole-genome sequencing was performed by Complete Genomics on a single individual from family 6 (proband 6B). Segregation of the coding c.2321A>G (p.Tyr774Cys) variant in ZFX was determined through gDNA extracted from blood of consented family members by Sanger sequencing (see supplemental notes; Figure 3). For proband 12, no causative variants were identified using panels, but subsequent panel-agnostic re-analysis using a previously described pipeline identified a ZFX variant (for details, see supplemental notes).26,27,28 The sequencing methodology and variant interpretation criteria were based on the local protocols of each laboratory. All variants were classified by the performing laboratories as variants of uncertain significance (VUSs) as is required for a candidate gene. In light of the evidence presented in this study (although ACMG criteria are not applicable for a novel disease classification), we putatively reclassified the variants according to the applicable ACMG standards and guidelines.29,30,31 Variant annotation was based on ZFX transcript GenBank: NM_003410.4. Sanger sequencing was used to confirm positive sequencing findings, except for probands 3 and 9. All ES and GS studies were performed as trios.
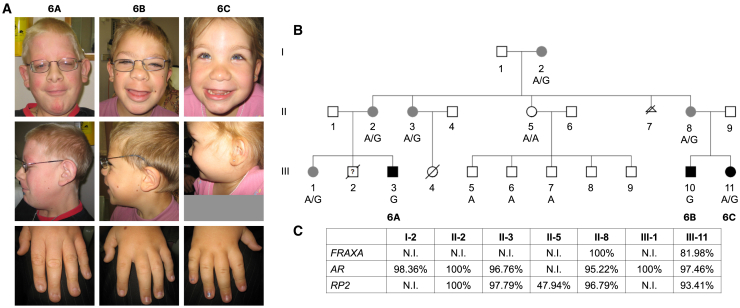
Figure 3.
Characterization of a family with an inherited ZFX variant
(A) Facial features and extremities of probands 6A–6C (see text and supplemental information for additional details).
(B) Three-generation pedigree of probands 6A–6C and family members. Dark black circles and squares indicate affected individuals; gray circles indicate carrier females diagnosed with hyperparathyroidism (except for III-1). A is the wild-type ZFX allele; G is the variant ZFX allele at same position (GRCh38 chrX: 24229396A>G, c.(2438A>G), p.Tyr774Cys).
(C) Results of X-inactivation studies showing skewing in all carrier females and random inactivation in a noncarrier female (II-5).
Evolutionary Action (EA)
We evaluated the impact of missense variants on protein function using the EA method.32 The EA scores come from solving a formal equation that states that the impact of each variant equals the importance of the variant residue times the magnitude of the change. The importance of each residue is calculated according to the Evolutionary Trace scores33,34 and the magnitude of change is calculated according to substitution odds. Objective assessments of the Critical Assessment of Genome Interpretation (CAGI) community have showed that EA performed consistently well among the state-of-the-art.35,36
For the evolutionary analysis of ZFX, we used 184 homologous sequences, obtained with BLASTp search37 using the NM_003410 sequence and the databases “non-redundant NCBI,” “UniRef100,” and “UniRef90.”38 The sequences included orthologs from distant species, such as zebra finch (Taeniopygia) and pufferfish, and we aligned them using MUSCLE.39 The ET scores were color-mapped on the 3D structure using PyMOL v.2.5.2 and the PyETV plugin.40 The 3D structure of ZFX was generated using AlphaFold41 and the sequence with Uniprot: P17010.
X-inactivation studies
For family 6, X-inactivation studies were performed following previously established protocols.42 Variable number tandem repeat loci adjacent to FMR1, AR, and RP2, which are subject to DNA methylation on the inactive X, were amplified by PCR from genomic DNA extracted from blood. The relative quantities of amplicons from gDNA digested with HpaII or untreated were compared by fragment analysis. Females with skewed X-inactivation will show greater than 90% loss of one allele where sizes are informative in the HpaII digested sample. Primers used in the assay were as follows: FMR1, forward 5′-GCTCAGCTCCGTTTCGGTTTCACTTCCGGT-3′ and reverse 5′-[HEX]AGCCCCGCACTTCCACCACCAGCTCCTCCA-3′; AR, forward 5′-TCCAGAATCTGTTCCAGAGCGTGC-3′ and reverse 5′-[FAM]GCTGTGAAGGTTGCTGTTCCTCAT-3′; RP2, forward 5′-[NED]TGACATAGCGAGACCCTGTG-3′ and reverse 5′-TGGTGGGTTCTCTAGCTGG-3′.
For proband 9, an X-inactivation study was performed by the Greenwood Diagnostic Laboratory (Greenwood, SC) using an androgen receptor X-inactivation assay.43
In vitro molecular characterization
Plasmid constructs
The plasmids described in this study were generated from the expression vector ZFX (GenBank: NM_003410) Human Tagged Open Reading Frame (ORF) Clone from Origene (Cat #RC214045). The parent vector for this construct is pCMV6-Entry (Origene #PS100001), in which expression is driven from the cytomegalovirus (CMV) promoter (this plasmid is also used as a negative control in relevant experiments). The same strategy, using two primers, was used to introduce each of the four single-nucleotide variants found in the cohort into the ZFX coding region. One primer contains the variant near the middle of the sequence, while the other primer amplifies in the other direction and abuts, but does not contain, the variant. For p.Arg786Gln and p.Arg764Trp, two primer pairs were used (denoted X01-3′ and X01-5′ or X02_3′ and X02_5′, respectively); see Table S2 for a list of primers used for cloning. For p.Tyr774Cys and p.Thr771Met, different primers were used to incorporate the variants (X04-5′ and X05_5′, respectively), but the same primer was used to amplify in the other direction (X05_04_3′). Complementary nucleotides were added to the ends of the primers so that plasmid assembly could occur via Gibson assembly and not rely on blunt-end ligation. Following amplification using the reverse-oriented primers to generate the entire vector sequence and verification by gel electrophoresis, fragments were purified by Ampure magnetic beads (Beckman Coulter #A63881). Assembly was carried out using Gibson Assembly mix (NEB #2611L), then a portion of each reaction was transformed into chemically competent CopyCutter E. coli (Lucigen #C400CH10). Plasmids were isolated from transformants, and the presence of the introduced variants was verified by Sanger sequencing (Azenta Life Sciences service facility). Large-scale plasmid preparations were subsequently isolated by different column-based protocols, but all utilized the induction of plasmid copy number in the Lucigen cells (Lucigen #C400CH10) that is required for these toxic plasmids.
Cell culture
The cell line used in this study, designated DKO, was derived from (female) HEK293T cells (which lack a Y chromosome and thus ZFY) in which the endogenous ZFX and ZNF711 genes were inactivated through CRISPR-mediated deletions.9 Cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (Thermo Fisher #10437036) plus 1% penicillin and 1% streptomycin at 37°C with 5% CO2. Cell lines were authenticated via the short tandem repeat (STR) method and validated to be mycoplasma free using a universal mycoplasma detection kit (ATCC #30-1012K).
Transactivation assays
To test the activity of the WT and variant ZFX proteins, a transient transfection assay using the ZFX expression vectors was performed using the DKO cell line. Transfections for RNA preparation were carried out using Lipofectamine 3000 (Thermo Fisher #L3000015), according to the manufacturer’s instructions. Much larger cell numbers were required for ChIP assays; the same transfection protocol was used but scaled appropriately. All transfections were performed in triplicate wells.
RNA preparation and RNA-seq
To recover RNA for expression analyses, cells were lysed 24 h after transfection in triplicate wells using TRI Reagent (Zymo #R2050-1-200), and RNA was recovered by isopropanol precipitation. RNA integrity was confirmed using a Bioanalyzer (Agilent) using an RNA integrity number (RIN) of 9.0 as a quality cutoff for library preparation. Samples were submitted to Novogene for their standard RNA-seq service (150 bp paired-end reads with a total minimum output of 6 Gb). Efficacy of transactivation prior to library preparation and sequencing was monitored by quantitative reverse-transcription PCR (RT-qPCR) of known responsive target genes, as described.9 All RNA-seq assays were performed in triplicate (see Table S3A).
Chromatin preparation, immunoprecipitation, and ChIP-seq
To obtain chromatin for ChIP-seq assays, cells were split 1:2 24 h after transfection and allowed an extra day of growth. After this additional 24 h, chromatin crosslinking and chromatin immunoprecipitation was carried out using established lab protocols.7,9 Briefly, cells were crosslinked in 1% formaldehyde for 10 min before quenching with 125 mM (final) glycine. Following washing in phosphate-buffered saline (PBS), crosslinked cells were swollen in cell lysis buffer (5 mM PIPES [pH 8.0], 85 mM KCl, 1% NP-40), then lysed by sonication in nuclei lysis buffer (0.1% sodium dodecyl sulfate [SDS], 50 mM Tris-HCl [pH 8], 10 mM ethylenediaminetetraacetic acid [EDTA]). All buffers included cOmplete Protease Inhibitor EDTA-free tablets (Roche #11836153001). Lysis volumes were based on cell pellet volumes. Sonication was carried out with a Diagenode Bioruptor Pico using conditions to generate fragments 400–800 bp in length. Chromatin and antibody amounts vary depending on the experiment; for the anti-FLAG ChIP-seq experiments, 100 μg soluble chromatin was diluted 1:5 in radioimmunoprecipitation assay buffer (RIPA, 150 mM NaCl, 50 mM Tris-HCl [pH 8.0], 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) and incubated overnight with 12.5 μL of anti-FLAG antibody (Cell Signaling Technology #5419S). Magnetic Protein A/G beads (Pierce #88803) were added for an additional 2 h of incubation. Bead-immunocomplexes were then washed twice with RIPA (150 mM NaCl, 50 mM Tris-HCl [pH 8.0], 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS), followed by washing three times with IP Wash Buffer II (100 mM Tris-Cl [pH 9.0], 500 mM LiCl, 1% NP-40, 1% sodium deoxycholate). Elution was performed in 150 μL of elution buffer (50 mM NaHCO3, 1% SDS), then ChIP samples and inputs (10 μL of precleared chromatin lysis plus 140 μL elution buffer) were reverse crosslinked overnight at 65°C. DNA was purified using Qiagen MinElute PCR clean up columns (cat #28006) and quantified spectrophotometrically. ChIP-seq libraries were prepared using the KAPA HyperPrep kit (Roche #KK8503) following the manufacturer’s protocol. Final cycle amplification numbers varied depending on input DNA and ChIP parameters. Libraries were quality checked by qPCR for target enrichment and visually by Bioanalyzer, then submitted to Novogene and sequenced on a Novaseq 6000, providing a minimum of 10 Gb of data in paired-end 150-bp format. All ChIP-seq assays were performed in duplicate (see Table S3B). Datasets from the two independent experiments were merged for downstream analyses.
Data analysis and visualization
RNA-seq
All RNA-seq analyses were carried out using Partek Flow modules (v.10.0.21.1103 at project initiation), as provided through the Norris Medical Library Bioinformatics Service. Following quality trimming, fastq files provided by Novogene were aligned using STAR v.2.7.8a with the default alignment parameters provided. Transcript and gene counts were generated using hg38 GENCODE Genes release 36. Gene counts were then analyzed for differential expression by DESeq2(R) 3.5. Default parameters were used except that fit type was set to “local” and not “parametric.”
ChIP-seq
All ChIP-seq data were aligned to hg38 using Bowtie2 software (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml). Only reads that aligned to a unique position in the genome with no more than two sequence mismatches were retained for further analysis. Duplicate reads that mapped to the same exact location in the genome were counted only once to reduce clonal amplification effects. Normalization was done across samples using an equal number of uniquely mapped reads. Biological replicates of ChIP-seq datasets for WT and each mutant were performed. ChIP-seq peaks were called using the narrowPeak setting in MACS2 (https://github.com/taoliu/MACS), using the ENCODE pipeline and Irreproducible Discovery Rate (IDR) method to determine reproducible peaks. Peaks were annotated using HOMER v.4.11 (http://homer.ucsd.edu/homer/), and average tag density plots were generated using CEAS software (https://liulab-dfci.github.io/software/).
Proportional Venn diagrams were generated using the web tool DeepVenn.44
In vivo characterization in zebrafish
Zebrafish husbandry
Animal experiments were conducted in accordance with protocols approved by the Animal Ethics Committee of Chungnam National University (202012A-CNU-170). Adult fish were reared under standard conditions with a 14 h/10 h light/dark cycle. We obtained embryos by natural mating of heterozygous adult zebrafish, and embryos were reared in egg water at 28.5°C. WT and KO zebrafish were obtained from the Zebrafish Center for Disease Modeling. All fish used in behavioral tests were fully grown, sexually mature, 3- to 12-month-old fish, ranging in size between 3.0 and 3.5 cm standard length.
Zebrafish generation
In zebrafish, zfx on chromosome 24 is the ortholog of human ZFX (Figure S2); we aligned the human ZFX (from transcript GenBank: NM_003410.4) and zebrafish zfx (Ensembl: ENSDART00000110652.4) protein sequences via the EMBOSS Needle tool and found 65.5% similarity and 53.6% identity. To understand the in vivo role of ZFX, we established a KO zebrafish model utilizing the CRISPR-Cas9 system.45 CRISPR single guide (sg) RNAs targeting zfx were identified using CRISPRScan (https://www.crisprscan.org/), and oligonucleotides were selected (Table S7). In vitro transcription was carried out using 150–200 ng of template and the MaxiScript T7 Kit (Ambion #AM1312). RNA was precipitated with isopropanol. Cas9 expression vector (Addgene #46757) was linearized with XbaI (NEB #R0145) and purified with an agarose gel DNA extraction kit (ELPIS #EBD-1009). Cas9 mRNA was transcribed with the mMESSAGE mMACHINE T3 Kit (Ambion #AM1348) and then purified by lithium chloride precipitation following the manufacturer’s protocol. One-cell-stage zebrafish embryos were injected with 300 ng/μL Cas9 mRNA and 150 ng/μL sgRNA. For genotyping F0, PCR products (20 μL) were re-annealed in a thermal cycler under the following conditions: 95°C for 2 min, 95°C–85°C at 2°C/s, 85°C–25°C at 0.1°C/s, then kept at 4°C. Part (16 μL) of the re-annealed mixture was incubated with 0.2 μL of T7 endonuclease I, 2 μL of NEB Buffer 2, and 1.8 μL of nuclease-free water at 37°C for 40 min. See Table S7 for genotyping primers.
Human ZFX variant over-expression in zebrafish
Human WT ZFX cDNA (GenBank: NM_003410.4) was subcloned into the pCS2+ expression vector. Missense variants were introduced by site-directed mutagenesis using WT ZFX pCS2+ vector as template. ZFX mRNA was transcribed with the mMESSAGE mMACHINE SP6 Transcription Kit (Ambion #AM1340) and then purified by lithium chloride following the manufacturer’s protocol. For over-expression, three different concentrations (100, 200, and 400 pg) of mRNAs were microinjected into 1- to 2-cell-stage zebrafish embryos.
Quantitative real-time PCR
Total RNA was isolated from brains of adult zfx KO and WT zebrafish via easy-Blue Total RNA isolation kit (iNtRON Biotechnology #17061). One microgram of RNA was converted to cDNA by SuperScript III First-Strand Synthesis System (Invitrogen #18080-51). RT-qPCR was conducted in triplicate using TB Green Premix Ex Taq II master mix (Takara #RR82LR) and a Thermal Cycler Dice Real Time System III (Takara, Japan). β-actin was used as a reference gene, and relative gene expression levels were calculated by the 2−Ct method.
Behavioral tests
Novel tank assay
A novel tank test was performed as previously described in Kim et al.46 to evaluate swimming activity and anxiety-like behavior. Each male WT (n = 8) or KO (n = 17) zebrafish was introduced to a tank (24 × 15 × 15 cm) filled with system water (up to 10 cm) and recorded with a video camera (Sony HDR-CX190) for 10 min. The videos were analyzed using EthoVision XT software. To examine exploratory activity, the tank was divided into three equally sized zones (top, middle, and bottom) as shown in Figure 6A. The percentage of average time spent in each zone and distance moved in first 5 min were analyzed.
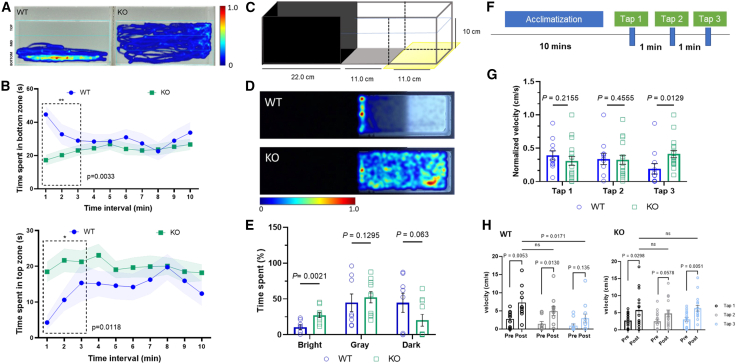
Figure 6.
Behavioral characterization of zfx knockout zebrafish
(A) Representative heatmap image for novel tank assay during the first 3 min. The lines indicate the boundaries of three vertically different zones (top, middle, and bottom).
(B) Quantification of average time spent in novel tank assay. Upper graph shows WT spent significantly increased time in bottom zone (t = 2.983, df = 23, p = 0.0033, n = 8); lower graph shows KO fish spent higher time in the top zone (t = 2.424, df = 23, p = 0.0118, n = 17).
(C) Test apparatus for the scototaxis assay modified with gray and bright zone. The external light source is placed underneath the tank to evaluate the specific preference of adult zebrafish.
(D) Representative heatmap image for scototaxis assay.
(E) Quantification of percentage of average time spent in bright, gray, and dark zones. KO zebrafish spent significantly increased time in the bright zone (t = 3.516, df = 16, p = 0.0021, n = 10) compared to WT siblings (n = 8).
(F) Behavioral paradigm showing the 10-min acclimatization in the round container followed by three tap stimuli with 1-min intervals.
(G) Quantification of normalized velocity (cm/s) post-tap. WT (n = 11) and KO (n = 17) indicated similar response for both the first and second tap stimuli. For the third tap, KO zebrafish displayed significantly increased response (t = 2.378, df = 26, p = 0.0129).
(H) Quantification of velocity change (pre- and post-tap). WT fish showed a trend of decreasing response to the tap stimuli, whereas KO fish showed significantly increased response to the third tap (t = 2.907, df = 16, p = 0.0051).
Scototaxis test
To assess anti-anxiety behavior, we evaluated scototaxis (dark/light preference) as previously described in Maximino et al.47 For this, we modified the experimental setup as shown in Figure 6C by dividing the light zone into gray and bright zones using an external light source underneath the tank. Each zebrafish was placed in the tank followed by a 5-min habituation period. Swimming activity was recorded from above for 10 min, and the percentage of average time spent in each region was analyzed for WT (n = 8) and KO (n = 10) zebrafish.
Adult startle tap test
To elicit an unconditioned startle response to a stimulus, a tap startle test was performed as described in Eddins et al.48 with adult zebrafish. We used only three tap stimuli, as the WT fish showed greater adaptation. Each fish was placed in a round container (95 mm in diameter and 40 mm in height) filled with system water to the height of 25 mm. The round arena was then placed in the observation chamber under bright light conditions (mimicking daylight). Fish were left for a 10-min acclimatization time, allowing for stable swimming bouts. After the acclimatization period, a sequence of three taps (stimulus intensity level at 5) were evoked at 1-min intervals (Figure 6F). The average velocity for 5 s before and after each tap was analyzed using DanioVision. The post-tap velocity data of each WT (n = 11) and KO (n = 17) zebrafish were normalized for the analysis.
Statistical analysis
All results were expressed as mean ± SEM. Statistical analysis was performed using GraphPad Prism software (v.8.01 for Windows, GraphPad Software Inc., USA). In all experiments, comparisons between WT and KO fish were done using a one-tailed Student’s t test. The effect size estimations were calculated using a web application (https://www.estimationstats.com/).49 The estimates were performed using 5,000 bootstraps, and the confidence interval was bias-corrected and accelerated. Group differences are normalized to Hedges’ g. Statistical significance is shown as follows: ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Results
We identified and recruited 18 individuals from 16 unrelated families for this study. Probands 6A, 6B, and 6C are relatives from the same family. Fourteen individuals were assigned male at birth (78%), while four were assigned female, with one self-identifying as male (referred to by their assigned sex throughout this manuscript). The mean age of subjects in our cohort was 16 years (range: 8–34 years). A full description of demographic, phenotypic, and genetic data is presented in Table S1, and a summary of the key findings in our cohort is found in Table 1 with additional details provided in the clinical findings section below.
Table 1.
Summary of demographic and clinical features in individuals with ZFX variants
| Demographic and clinical features | Frequencya (%) | ||
|---|---|---|---|
| Missense | Frameshift | Total | |
| Sex | males: 8/11 (73) | males: 6/7 (86) | males: 14/18 (78) |
| Mean Age (±SD) | 16.04 (±7.95) | 12.24 (±5.7) | 14.52 (±7.2) |
| De novo inheritance | 6/11 (55) | 4/7 (57) | 10/18 (56) |
| Development and neurobehavioral | |||
| Developmental delay/intellectual disability | 10/11 (91) | 7/7 (100) | 17/18 (94) |
| Gross motor delay | 9/11 (82) | 7/7 (100) | 16/18 (89) |
| Fine motor delay | 9/11 (82) | 7/7 (100) | 16/18 (89) |
| Speech delay | 10/11 (91) | 6/7 (86) | 16/18 (89) |
| ADHD | 5/11 (45) | 1/7 (14) | 6/18 (33) |
| Autism | 5/11 (45) | 1/7 (14) | 6/18 (33) |
| Autistic traitsb | 1/11 (9) | 2/7 (29) | 3/18 (17) |
| Other behavioral abnormalities | 9/11 (82) | 1/7 (14) | 10/18 (56) |
| Therapeutic services or special education | 10/11 (91) | 7/7 (100) | 17/18 (94) |
| Neurological | |||
| Hypotonia | 9/11 (82) | 4/7 (57) | 13/18 (72) |
| Abnormal brain MRI | 4/7 (57) | 6/6 (100) | 10/13 (77) |
| Sleep problems | 6/11 (55) | 2/7 (29) | 8/18 (44) |
| Epilepsy | 1/11 (9) | 1/7 (14) | 2/18 (11) |
| Growth parameters | |||
| Height 10%ile | 4/11 (36) | 4/7 (57) | 7/18 (39) |
| OFCc 90%ile | 4/10 (40) | 2/6 (33) | 6/16 (38) |
| Vision and hearing | |||
| Vision/eye abnormalities | 7/11 (64) | 6/7 (86) | 13/18 (72) |
| Hearing loss | 7/11 (64) | 2/7 (29) | 9/18 (50) |
| Dysmorphic features | |||
| Broad/thick eyebrows synophrys | 9/11 (82) | 6/7 (86) | 15/18 (83) |
| External eye findings | 9/11 (82) | 6/7 (86) | 15/18 (83) |
| Other face abnormalities | 10/11 (91) | 5/7 (71) | 15/18 (83) |
| Smooth long philtrum | 8/11 (72) | 6/7 (83) | 14/18 (78) |
| Ear abnormalities | 10/11 (91) | 4/7 (57) | 14/18 (78) |
| Thin upper lip | 10/11 (91) | 3/7 (43) | 13/18 (72) |
| Nose abnormalities | 6/11(55) | 6/7 (86) | 12/18 (67) |
| Forehead abnormalities | 6/11 (55) | 4/7 (57) | 10/18 (56) |
| Macroglossia | 7/11 (64) | 1/7 (14) | 8/18 (44) |
| Miscellaneous | |||
| Musculoskeletal findings | 10/11 (91) | 7/7 (100) | 17/18 (94) |
| Inguinal hernia | 9/11 (82) | 4/7 (57) | 13/18 (72) |
| Umbilical hernia | 8/11 (72) | 1/7 (14) | 9/18 (50) |
| Genitourinary congenital anomalies (males) | 7/8 (88) | 3/6 (50) | 10/14 (71) |
| Abnormal echocardiography | 4/9 (44) | 4/5 (80) | 8/14 (57) |
| Gastrointestinal abnormalities | 10/11 (91) | 2/7 (29) | 12/18 (67) |
| Abnormal renal ultrasound | 5/10 (50) | 4/7 (57) | 9/17 (53) |
| Hyperparathyroidism | 3/7 (43) | 0/5 (0) | 3/12 (25) |
Denominator can vary based on data availability
Autistic traits but no formal diagnosis with autism
OFC, occipital frontal circumference
Variant characterization
Four hemizygous or heterozygous ZFX variants were identified in 8 male and 3 female subjects: c.2290C>T (p.Arg764Trp), c.2312C>T (p.Thr771Met), c.2321A>G (p.Tyr774Cys), and c.2357G>A (p.Arg786Gln) (see Tables 2 and S1). In addition, 7 unrelated subjects (6 males and 1 female) were found to have truncating variants delineated in Table 2. None of these ZFX variants have been observed in the Genome Aggregation Database (gnomAD, last accessed on October 6, 2023).50
Table 2.
Table of ZFX variants
| Proband(s) | DNA (GenBank: NC_000023.11) | cDNA (GenBank: NM_003410.4) | Protein (GenBank: NP_003401.2) | CADD score | REVEL score | NMD predicted? | Putative applicable ACMG criteriaa | Consequence | Domain |
|---|---|---|---|---|---|---|---|---|---|
| 1, 2 | g.24211248C>T | c.(2290C>T) | p.Arg764Trp | 25 | 0.3 | – | PM2, PS2(2), PP3 | missense | ZF12 |
| 3, 4, 5 | g.24211270C>T | c.(2312C>T) | p.Thr771Met | 24.6 | 0.46 | – | PM2, PS2, PP3 | missense | ZF12-ZF13 linker |
| 6A–6C, 7 | g.24211279A>G | c.(2321A>G) | p.Tyr774Cys | 27.3 | 0.57 | – | PM2, PS2(7), PP3 | missense | ZF12-ZF13 linker |
| 8, 9 | g.24211315G>A | c.(2357G>A) | p.Arg786Gln | 24.6 | 0.34 | – | PM2, PS2(9), PP3 | missense | ZF13 |
| 10 | g.24207442A>AT | c.(768dup) | p.Lys257∗ | 29.3 | – | yes | PVS1, PM2 | truncation | acidic domain |
| 11 | g.24210271G>GT | c.(1319dup) | p.Leu440Phefs∗21 | 32 | – | no | PVS1_Strong, PS2, PM2 | truncation | ZF1 |
| 12 | g.24209008G>GGA | c.(1205_1206dup) | p.Arg403Glufs∗12 | 28.2 | – | no | PVS1_Strong, PS2, PM2 | truncation | acidic domain |
| 13 | g.24210953CAT>C | c.(1996_1997del) | p.Met666Valfs∗2 | 32 | – | no | PVS1_Strong, PS2, PM2 | truncation | ZF9 |
| 14 | g.24179650G>GT | c.(529dup) | p.Ser177Phefs∗12 | 27 | – | yes | PVS1, PS2, PM2 | truncation | acidic domain |
| 15 | g.24179543ATG>A | c.(423_424del) | p.Ser142∗ | 25 | – | yes | PVS1, PM2 | truncation | acidic domain |
| 16 | g.24179237CTG>C | c.(115_116del) | p.Val39Phefs∗14 | 26.2 | – | yes | PVS1, PM2 | truncation | N-terminal region |
CADD51 and REVEL52 scores were calculated for the missense variants, and putative applicable American College of Medical Genetics (ACMG) criteria are listed.29,30,31
For criteria that only apply to a subset of probands in each row, the applicable individuals(s) are indicated by superscript. PP3 was applied on the basis of CADD score. According to current ACMG criteria, PVS1 is applied for a gene where loss-of-function is a known mechanism of disease. However, PVS1 was used to upgrade the classification of the truncating variants based on evidence provided in this study.
Based on in silico analysis, the truncation variants lead to the creation of nonsense codons at or shortly downstream of the genetic change and are predicted to cause loss of function either through NMD or truncation of the essential DNA-binding zinc fingers (ZF11–ZF13). The results of in silico variant effect prediction tools for the missense variants are shown in Table 2. Three missense variants were maternally inherited in 5 subjects (including the 3 related subjects), and 6 subjects had de novo missense variants, while the truncating variants were de novo in 4 subjects and maternally inherited in 3 subjects. All missense variants were located within or between zinc fingers 12 and 13 in the ZFX protein (Figure 1).
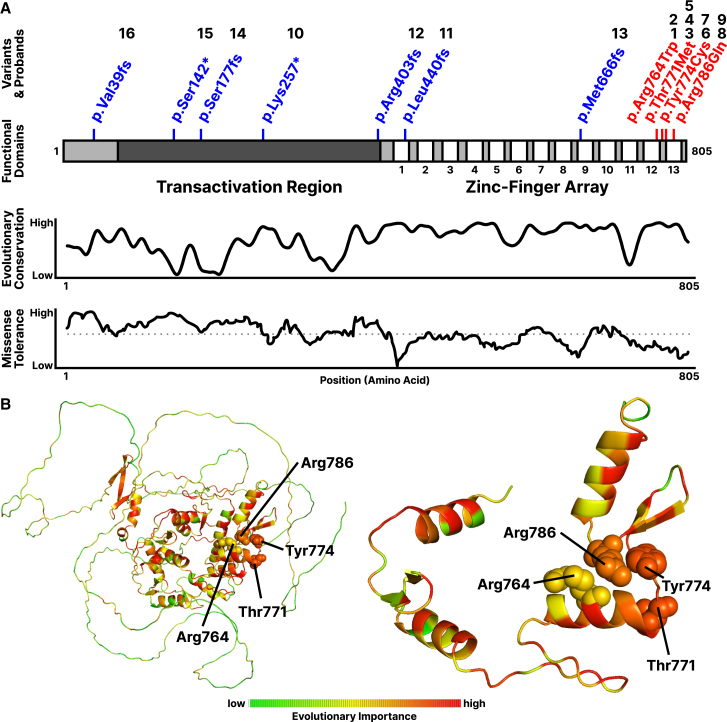
Figure 1.
Cohort variants and existing characterization of ZFX
(A) Distribution of cohort variants throughout the ZFX coding sequence, with missense variants in red and truncating variants in blue (numbers indicate the corresponding probands). Evolutionary sequence conservation shown for ZFX across 19 vertebrate species.53 Missense tolerance ratios (MTRs) are calculated from gnomAD v2.0 exomes; dotted line indicates 50th percentile for MTR.54.
(B) Evolutionary Action scores superimposed on a predicted ZFX structure, with residues colored proportional to evolutionary importance.
We analyzed the ZFX variants using the EA method (Figure 1B).32 Briefly, EA uses sequence homology and phylogenetic distances to prioritize the variants in each protein and score them on a scale from 100 (pathogenic) to 0 (benign). The missense sequence variants p.Arg764Trp, p.Thr771Met, p.Tyr774Cys, and p.Arg786Gln had EA scores of 56.77, 72.08, 72.47, and 54.03, respectively. These EA scores indicate intermediate to high impact on ZFX function, and they are larger than the average EA score that corresponds to missense variants due to random nucleotide changes (EA score of 42.27).
X-Inactivation
X-inactivation studies on gDNA extracted from blood were available for all informative females in family 6 (discussed below and in Figure 3C) and proband 9. Extreme skewing was detected in both carrier females in family 6 (p.Tyr774Cys) and in proband 9 (p.Arg786Gln), while random inactivation was observed in a non-carrier female in family 6. Early studies of ZFX reported it to escape X-inactivation, and recent single-cell RNA sequencing experiments have supported this claim.11,55
Clinical findings
Prenatal findings and growth
In this cohort, nine individuals (50%) were born via Cesarean section and two via induced vaginal delivery. The indications for these types of deliveries included fetal distress, pre-eclampsia, pregnancy-induced hypertension, fetal growth restriction, and oligohydramnios. Six of the participants (33%) were born prematurely (33–36 weeks of gestation). Furthermore, 4 subjects (22%) were born small for gestational age (defined as birth weight <10th percentile for gestational age) and 5 subjects (28%) had birth weight or length equal or above the 90th percentile, which along with additional clinical and physical findings prompted molecular testing for Beckwith-Wiedemann syndrome in 4 of these subjects.
Postnatal growth parameters showed a trend for shorter stature in 44% of subjects; 4/7 (57%) subjects with truncating variants as compared to 4 individuals (36%) with missense variants had height equal to or below the 10th percentile for age. Proband 14 receives growth hormone treatment, and his height on last assessment was within normal range, and proband 15 had borderline short stature with height at the 11th percentile. In addition, large head size (occipitofrontal circumference >90th percentile) was found in 38% of probands with available head-circumference measurements.
General findings
Nine subjects (50%) were found to have conductive or sensorineural hearing loss. Six subjects from the missense variant group required placement of ear tubes for recurrent ear infections and/or chronic serous otitis media, one subject from the truncation group was reported to have hyperacusis and chronic middle ear effusion, and another required ear tube placement.
Seven of the 11 subjects (64%) with missense variants and 6 probands with truncation variants (86%) had eye or vision abnormalities. These abnormalities included refractive errors, strabismus, astigmatism, nystagmus, optic nerve hypoplasia, and retinal detachment.
Neurodevelopmental and behavioral findings
All male probands in our cohort had developmental delay, intellectual disability, or learning disability; their intellectual disability was with variable severity ranging between borderline (low normal) to moderate. Two of the four females in this cohort were described as very intelligent, one had normal cognition (with relative weaknesses in visuo-motor coordination and executive functioning skills related to planning and decision-making), and one had borderline cognitive abilities. All subjects with truncating variants (including one female proband) and all male subjects as well as one female subject with missense variants had gross and fine motor delay. Of note, the other two female subjects with missense variants had normal developmental motor skills. In addition, with the exception of one female subject (proband 4), all probands with missense variants and six subjects with truncation variants had speech delay. All subjects, except one female proband (proband 4) with a missense variant, required therapeutic services, including physical, occupation, and speech therapies, with or without special education at school.
Neurobehavioral problems were also prevalent. A total of 50% of the probands were either diagnosed with autism spectrum disorder (ASD) (6/18) or reported to have autistic traits (3/18), and approximately 33% (6/18) of individuals had a diagnosis of attention-deficit/hyperactivity disorder (ADHD). Three individuals (17%) had ADHD as well as ASD. Other behavioral problems, such as anger and tantrums, were more common among individuals with missense variants than in those with truncating variants (82% vs. 14%; p = 0.006). Six participants (55%) with missense variants and two subjects (29%) with truncating variants had sleep problems or difficulty.
Hypotonia was found in 13/18 (72%) of the subjects, which may have played a role in the motor delay that was observed in these individuals. However, the hypotonia in two of the individuals was mild and resolved in late infancy/early childhood. Epilepsy was reported in two subjects only.
Abnormal but nonspecific brain magnetic resonance imaging findings, including cerebral atrophy, arachnoid cysts, delayed myelination, corpus callosum abnormalities, cerebellar atrophy, or pituitary abnormalities, were found in 10 out of 13 subjects (77%) who underwent brain imaging as part of their workup.
Facial features
Most cohort members exhibited recurrent facial features with an overlapping facial gestalt in a subset of individuals (Figures 2A, 2B, and 3A). Several individuals were described as having coarse facial features. The most prominent feature found in 15/18 individuals was broadening and/or thickening of eyebrows especially on the medial side with or without synophrys. Fifteen individuals (83%) had differences in the shape of the face: 7 individuals had pointed chins, 5 had long faces, and 2 individuals had midface hypoplasia. External eye findings were seen in 15/18 individuals, including 6 individuals with epicanthal folds and downslanting palpebral fissures in 8 individuals. Smooth and, in a subset of individuals, long philtrum was found in 14/18 individuals and thin upper lip in 13/18 individuals; the latter was mostly among individuals with a missense variant. Sixty-four percent (7/11) of individuals with missense variants had macroglossia, 3 of whom required tongue reduction surgery. Only one individual with a truncation variant had macroglossia (p = 0.04), and another had tongue protrusion. Forehead abnormalities were reported in 10/18 and included broad forehead, frontal bossing, and metopic ridging. Sixty-seven percent of individuals (12/18) had nasal anomalies: 5 had depressed nasal bridges, 8 had a bulbous/wide nasal tip, and 5 had hanging/prominent columella. Seventy-eight percent of individuals (14/18) had ear anomalies including abnormalities in size, helices, low set, posteriorly rotated, and prominent ears. A small subset of individuals had blond hair, but this trait was not well documented in other family members and we do not consider it at this time as one of the physical characteristics of this condition.
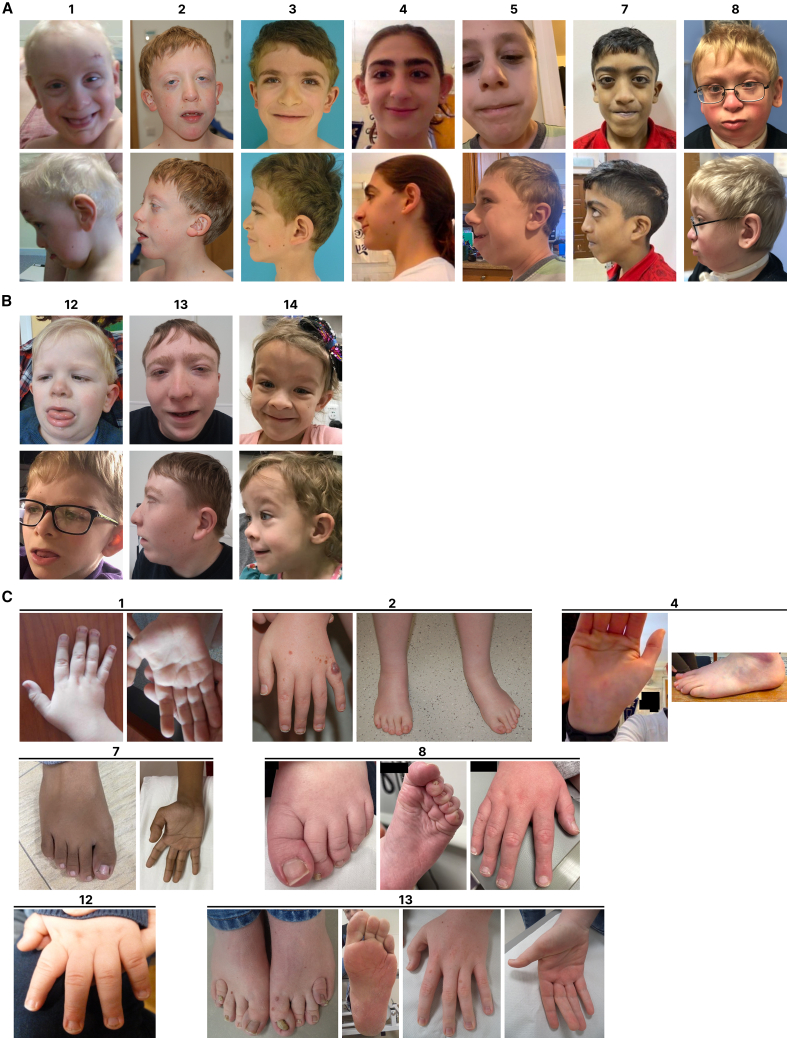
Figure 2.
Facial features of the presented individuals with ZFX variants
(A) Subjects with missense variants in the ZFX DNA binding domain.
(B) Subjects with truncating ZFX variants.
(C) Extremities of the indicated subjects. See supplemental information and Table S1 for additional details; images not available for all individuals.
Skeletal abnormalities were reported in all individuals except one. The most prominent skeletal findings were found in the hands (15 individuals) and feet (10 individuals) (Figure 2C). Hand findings included the shape of distal phalanges, abnormal shape and length of fingers, abnormal creases in hands, restriction or hypermobility in interphalangeal joints, and deep-seated fingernails. Feet findings included deep-seated toenails, abnormal or deep creases in soles, hallux valgus deformity, proximal insertion of a toe (1 individual) and polydactyly (1 individual). Joint hypermobility was found in 6 individuals with missense variants, one of whom met the diagnostic clinical criteria for Ehler Danlos syndrome-hypermobility type (proband 9). Three individuals with missense variants and 2 individuals with truncating variants had restriction in elbow extension. Pectus excavatum or carinatum was reported in 4 individuals. Spine anomalies were found in 6 individuals including 5 with scoliosis (all with missense variants) and sacral dysgenesis (1).
Other less common findings included skull abnormalities (5/18), high arched palate (4/18), micro/retrognathia (3/17), wide mouth (3/18), and short neck (2/18).
Congenital anomalies
Fifty-seven percent of individuals (8/14) who underwent echocardiography had abnormal findings. Four individuals with missense variants had variable structural cardiac abnormalities, while 4 individuals with truncation variants had atrial/ventricular septal defects and/or patent ductus arteriosus that spontaneously closed. Of note, probands 2 and 7 had a dilated aortic root, and proband 7 required a beta blocker to stop the progression.
A renal ultrasound was performed for all but one individual, and an abnormality was documented in 9 individuals (53%), but the majority of the findings were mild or transient, such as hydronephrosis or pelvicaliectasis. Interestingly, one individual had renal calculi and two had nephrocalcinosis.
Genitourinary anomalies were found in 69% of males, 7/8 males with missense variants (hypospadias in 6, cryptorchidism/retractable testes in 4) and in 3/6 males with truncating variants (hypospadias in 2, cryptorchidism in 1).
Seventy-eight percent of individuals had inguinal and/or umbilical hernia. Nine of 11 individuals with missense variants had inguinal hernias, seven of whom also had umbilical hernia, and one individual had epigastric, hiatal, and umbilical hernia. Four of 7 of the subjects with truncating variants had inguinal hernia, one of whom also had umbilical hernia.
Other rare congenital anomalies among individuals with ZFX variants included palate, dental, and upper airway anomalies. Cleft palate was found in two individuals with missense variants. Six subjects (33% of individuals) had dental anomalies: four had supernumerary teeth (one with delayed eruption), one had retained baby teeth, and one had crowded teeth. One individual was tracheostomy dependent, and another had mild tracheal stenosis and laryngomalacia requiring continuous positive airway pressure in infancy and arytenoid release surgery at 3 months of age.
Miscellaneous findings
Gastrointestinal abnormalities were observed in 12 individuals (67%), more common among individuals with missense variants (p = 0.008), and included feeding difficulties, gastroesophageal reflux, and constipation; two individuals required gastrostomy tube placement, and two additional individuals needed naso/orogastric tubes for a short period of time. Three individuals with missense variants and one individual with a truncation variant had pyloric stenosis.
Because of signs of overgrowth, umbilical hernia, macroglossia, and coarse facial features, four individuals underwent molecular testing for Beckwith-Wiedemann syndrome. Six individuals had molecular testing for other overgrowth syndromes (3/6 Simpson-Golabi-Behmel/GPC3, 2/6 Sotos/NSD1, 2/6 Costello), and three individuals had biochemical testing for mucopolysaccharidosis. All tests did not identify a genetic variant or biochemical evidence of the relevant disorder.
Hyperparathyroidism
Early in this study, an endocrine workup of proband 8 revealed inappropriately normal to high-normal levels of parathyroid hormone (PTH), which progressed to persistent PTH elevation, causing symptomatic hypercalcemia and eventually necessitating parathyroidectomy at 13 years of age. The mother of proband 8, who is a carrier for the same ZFX variant, also underwent parathyroidectomy for hyperparathyroidism. The histopathology in proband 8 and his mother was consistent with parathyroid hyperplasia.
Intriguingly, the same ZFX variant (c.2357G>A [p.Arg786Gln]) that was found in this family has been previously reported as a somatic variant in sporadic parathyroid adenoma.19 Based on this information, all collaborators were asked to share results of calcium and PTH levels if already drawn or order them in all probands. This approach led to the discovery of additional individuals with hyperparathyroidism. Proband 9, who carries the same variant (p.Arg786Gln) as proband 8, was diagnosed at 12 years of age with “familial hypocalciuric hypercalcemia” and at 15 years of age with parathyroid adenoma requiring parathyroidectomy. Proband 4, who carries the missense variant p.Thr771Met, exhibited biochemical abnormalities consistent with hyperparathyroidism (hypercalcemia and inappropriately normal PTH) and was evaluated for familial hypocalciuric hypercalcemia, but she is currently asymptomatic. Furthermore, four females with the p.Tyr774Cys variant from a three-generation family (including the mothers of probands 6A–6C; see Figure 3B and supplemental notes) were diagnosed with hyperparathyroidism and needed parathyroidectomy; the histology in one proband was consistent with parathyroid hyperplasia. None of the female relatives of probands 5A–5C and 7 with hyperparathyroidism exhibit the other clinical features seen in probands in this cohort. Notably, hypercalcemia secondary to hyperparathyroidism was observed only among individuals with missense variants. As previously discussed, X-inactivation studies were available for a subset of the individuals (for family 6, see Figure 3C); skewed X-inactivation may be a possible explanation for the observed phenotypes, but further sequencing studies are required.
Additional types of tumor and vascular anomalies were enriched in our cohort. Two individuals (probands 1 and 9) had multiple hemangiomas; proband 9 also had lymphatic malformations and was diagnosed at age 13 with thyroid papillary carcinoma. Proband 1 had hepatic angiomatosis, and proband 9 had hepatic adenoma. The mother of proband 8 was diagnosed with sarcoma. Proband 13 underwent a resection of benign suprarenal ganglioneuroma at 11 years of age. One of the female carriers in family 6 (III-1 in Figure 3C) had metastatic colorectal adenocarcinoma. Four individuals with missense variants had melanocytic nevi.
Missense ZFX variants demonstrate altered target gene expression
To explore the biochemical activities of several of the variant ZFX proteins present in the affected individuals, we individually introduced the p.Arg764Trp (probands 1 and 2), p.Thr771Met (probands 3, 4, and 5), p.Tyr774Cys (probands 6A–6C and 7), and p.Arg786Gln (probands 8 and 9) single-nucleotide changes into an expression vector containing a WT copy of a FLAG-tagged ZFX protein (Table S2). These four variants are located within the region of ZFX that we have previously shown to be required for DNA binding9; Arg786 and Arg764 are located within the finger loops of zinc finger 13 and zinc finger 12, respectively, whereas Thr771 and Tyr774 are located within the linker between zinc fingers 12 and 13 (Figure 1). The p.Arg786Gln, p.Arg764Trp, and p.Thr771Met variants have also been identified in The Cancer Genome Atlas database (TCGA, https://www.cancer.gov/ccg/research/genome-sequencing/tcga), indicating a possible role of these variants in carcinogenesis as well as in development. To evaluate the DNA binding and transactivation activities of the ZFX proteins harboring the individual amino acid changes, we used a cell line (DKO), derived from HEK293T cells, in which all endogenous ZFX family member protein expression has been eliminated through CRISPR-mediated deletion.9 The use of these cells allows a direct comparison of the binding pattern and activity of the transfected ZFX variant proteins to the activity of the transfected WT ZFX, without interference from endogenous proteins; see Table S3 for details concerning all genomic datasets used in this study.
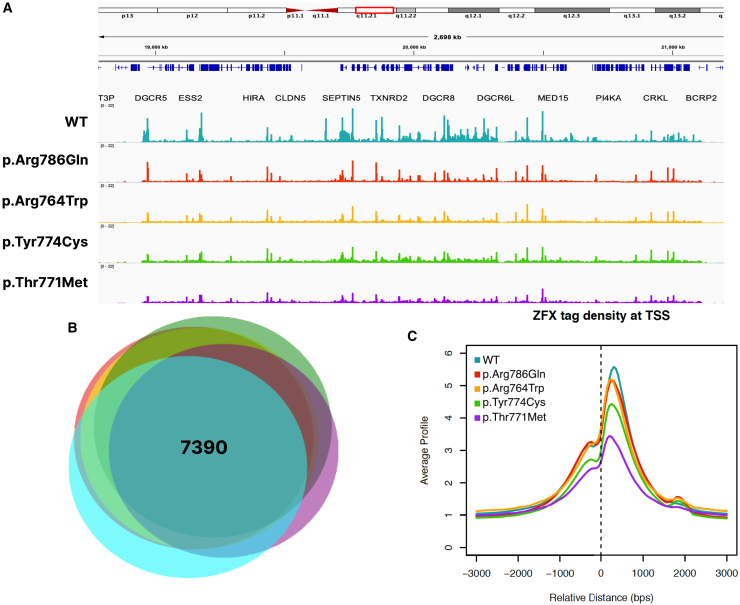
The DKO cell line was transfected with the different ZFX expression constructs or the parental vector as a control; 24 h after transfection, the cells were harvested, and the genomic DNA binding patterns for the variant ZFX proteins were analyzed and compared to the activity of WT ZFX. Genome-wide DNA binding profiles of the WT and variant ZFX proteins were determined by performing ChIP-seq assays using a FLAG antibody. Examination of the binding patterns revealed that, in general, the variant proteins had a very similar DNA binding pattern as did WT ZFX (Figure 4A); see also Figure S1B for heatmaps showing global comparisons of the binding patterns of the different ZFX proteins. However, we note that the variant ZFX proteins did appear to have slightly lower peaks than WT ZFX; this was especially noticeable for p.Thr771Met. These differences in peak height do not correlate with the expression levels of the variant proteins, as compared to the levels of WT ZFX; for example, p.Thr771Met has the most-reduced binding activity but is expressed at the same level as WT ZFX (Figure S1). As ZFX has previously been shown to be primarily localized to proximal promoter regions and the functional binding is confined to the sites located at +240 in the promoter region,7,9 the reproducible promoter peaks (ranked by peak score) located within ± 2 kb of known promoters were selected for WT ZFX and the mutant ZFX proteins for further analyses (peak files are provided in Table S4). Pairwise comparisons between the peak sets from the WT and variant ZFX proteins indicate a very high degree of overlap (Figure 4B), with all pairwise comparisons showing greater than 80% identity. Although, taken overall, the same promoter regions were bound by the WT and variant proteins, it was possible that the variants bound to different positions within the same promoters as did WT ZFX. To investigate this possibility, a tag density analysis of the promoter peaks for WT and variant ZFXs was performed (Figure 4C). We have previously shown that endogenous WT ZFX predominantly binds downstream of the transcription start site at position +240.9 Therefore, as expected, the transfected WT ZFX also displays this same pattern; we also found that each variant ZFX also has the same downstream binding pattern. Taken together, these data indicate that although the four missense variants are within the region of the ZFX protein required for DNA binding, these variants do not cause any major changes in genomic DNA binding patterns. However, we note that heatmaps of the ChIP-seq data suggest that the mutants have enhanced binding, as compared to WT ZFX, at a small set of promoters (Figure S1B).
Figure 4.
Characterization of wild-type and missense ZFX DNA binding by ChIP-seq
(A) Shown is a browser track displaying the genomic binding patterns of WT and variant ZFX proteins in a representative 2.4k-kb region of chromosome 22.
(B) Proportional Venn diagram of overlaps among the top 12,000 called peaks within 2 kb of the transcription start site (TSS) for WT and variant ZFX proteins. The number of peaks common to all ZFXs is indicated.
(C) Shown are the DNA binding profiles of WT and variant ZFX proteins from −2 kb to +2 kb from the TSS.
Although the missense variants had only subtle effects on DNA binding, it remained possible that they could affect the transcriptional activation properties of ZFX. Therefore, we next transfected DKO cells with the WT or variant ZFX expression plasmids or parental vector as a control and prepared RNA for RNA-seq. Genes with expression changes caused by transfection of the WT or variant ZFX proteins were identified using DEseq2 (R). Changes in the transcriptome caused by each ZFX construct, as compared to the parental vector, are visualized by volcano plots (Figure 5A), and lists of all genes used in these plots are provided in Table S5. These experiments clearly demonstrate that the four variant ZFX proteins are active in the transactivation assay and the majority of the differentially regulated genes are upregulated. These results are consistent with classification of ZFX as a transcriptional activator.7,9 Importantly, they also clearly demonstrate that the amino acid changes do not create a complete loss-of-function ZFX protein. However, there are fewer upregulated genes upon transfection of p.Tyr774Cys and p.Thr771Met, suggesting that the ZFX linker variants may have slightly lower transactivation ability than the WT ZFX or the ZFX finger variants; this may be due to the slightly lower occupancy of these TFs on the promoter regions, as shown by the tag density plots in Figure 4C.
Figure 5.
Characterization of differential expression in the context of missense ZFX variants
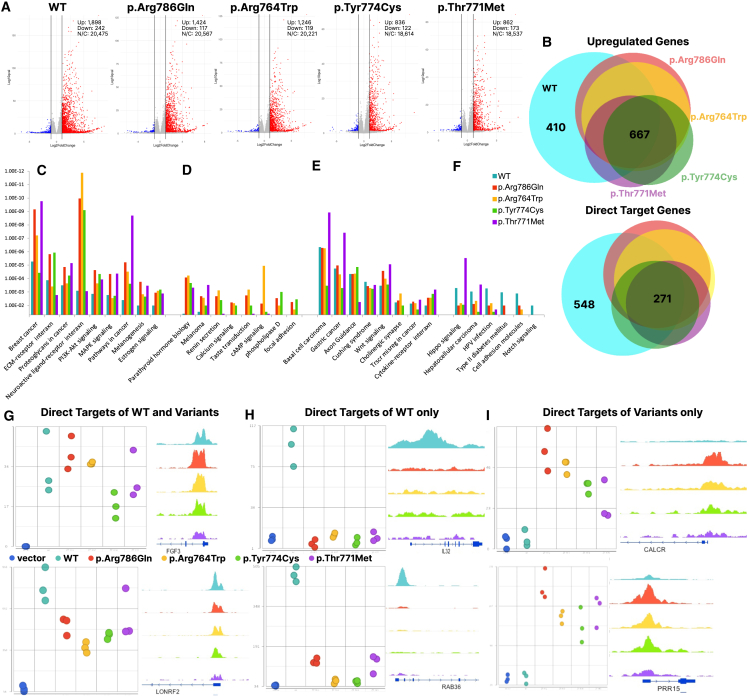
(A) Gene expression changes following transfection of WT or variant ZFX proteins into DKO cells. Volcano plots show gene expression changes in cells transfected with plasmids expressing the indicated ZFX compared to transfection with the vector alone. RNAs with increased expression are shown by red dots, RNAs with decreased expression are shown by blue dots; cut-offs used were a 2-fold change in expression and a q value < 0.05. The numbers of RNA with increased (Up), decreased (Down), and no change (N/C) in expression are displayed.
(B) Top: Overlap analysis of all genes activated by WT and variant ZFX proteins. The numbers of genes induced by all five proteins (667) and only by WT ZFX (410) are indicated. Bottom: Overlap analysis of direct targets activated by WT and variant ZFX proteins. The numbers of direct targets common to all five proteins (271) and direct targets unique to WT ZFX (548) are indicated.
(C–F) KEGG pathway analysis using the direct targets of WT and variant ZFX proteins. The top significant pathways are organized into four groups: (C) pathways more highly enriched in the set of genes regulated by the variant ZFXs than in the set of genes regulated by WT ZFX, (D) pathways only identified in sets of genes regulated by the variant ZFX proteins, (E) pathways enriched in both variant and WT gene sets, and (F) pathways enriched more in the sets of genes regulated by WT ZFX. Significance is plotted on the y axis.
(G–I) Examples of differences in expression and binding patterns of direct targets for WT ZFX vs. variant ZFX proteins; left panels show the expression level of the gene (values on the y axis represent normalized read counts mapping to all gene transcripts) in cells transfected with the WT and mutant ZFX proteins, whereas right panels show the ChIP-seq signals (browser shots) of the different ZFX WT and mutant constructs at the promoter of that gene. (G) Direct targets of both WT and variant ZFX, (H) direct targets of WT ZFX only, and (I) direct targets only of variant ZFX proteins.
A proportional Venn diagram of the overlaps of the sets of genes with increased expression (Figure 5B, top) shows that the linker variants (p.Thr771Met and p.Tyr774Cys) and the zinc finger variants (p.Arg786Gln and p.Arg764Trp) cause changes in expression of many of the same genes as regulated by WT ZFX. However, some genes whose expression is increased by WT ZFX are not regulated by the variant ZFX proteins, and the variant proteins also have their own distinct sets of regulated genes. We note that changes in the transcriptome mediated by the WT and variant ZFX proteins are a combination of altered expression of direct target genes (i.e., genes having a promoter bound by ZFX and that show upregulation of expression in the transactivation assay) and of genes in downstream signaling pathways controlled by the direct target genes. Although analysis of the entire set of altered genes can provide insight into the biological consequences of the variants, analysis of the set of altered direct target genes can provide insight into the mechanisms by which the pathogenic variants affect ZFX transcriptional activity. Therefore, we next examined the effects of the missense variants on the regulation of direct target genes. For these analyses, we combined the DNA binding profiles of the WT and variant ZFX proteins with the RNA expression profiles, selecting genes that show a greater than 2-fold increase in expression and that have a ZFX peak (taken from the top-ranked 12,000 promoter peaks) at their promoter region, to generate lists of direct target genes for each ZFX protein. From this analysis, we identified 1,300 direct targets for WT ZFX and 994, 879, 665, and 619 direct target genes for p.Arg786Gln, p.Arg764Trp, p.Tyr774Cys, and p.Thr771Met, respectively; lists of the direct target genes for WT ZFX and the variant ZFX proteins are provided in Table S6. To compare the sets of direct targets for the WT and ZFX variant proteins, a proportional Venn diagram was created (Figure 5B, bottom). Again, we found that the WT ZFX, the linker variant (p.Thr771Met and p.Tyr774Cys), and the finger variants (p.Arg786Gln and p.Arg764Trp) can regulate distinct sets of direct target genes, in addition to the 271 direct target genes activated by all ZFX proteins. Both sets of overlaps reveal that the transcriptional profiles of cells transfected with finger variants are more similar to each other than to the transcriptional profiles of cells transfected with the linker variants (and vice versa).
The fact that the finger variants p.Arg786Gln and p.Arg764Trp and the linker variants p.Thr771Met and p.Tyr774Cys increase the expression of some genes that are not affected by WT ZFX and also fail to increase the expression of some genes that are affected by WT ZFX suggests that the pathogenic variants may change the cellular phenotype. To assess the possible biological consequences of the variant-specific and WT-specific targets, we performed the Partek gene pathway-KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis. The pathways enriched in sets of genes regulated by WT and variant ZFX proteins were organized into four groups: pathways more highly enriched in the sets of genes regulated by the variant ZFXs than in the set of genes regulated by WT ZFX, pathways only identified in sets of genes regulated by the variant ZFX proteins, pathways identified in both variant and WT gene sets, and pathways enriched more in the sets of genes regulated by WT ZFX (Figures 5C–5F). These data, taken together with the gene expression and DNA binding data, suggest that altered activities of the variant ZFX proteins may not only cause a defect in certain phenotypes through an inability to activate certain genes but may also result in the acquisition of new cellular phenotypes in cells harboring the variants. The substantial enrichment of the neuroactive ligand physiology pathway in the sets of genes transfected with the variant ZFX proteins is of particular interest given the neurodevelopmental phenotypes observed in this cohort. Examples of genes that are regulated by both WT and variant ZFX, only WT ZFX, and only variant ZFX are shown in Figures 5G–5I; of interest is CALCR, which falls within the neuroactive ligand physiology pathway and is only activated by the variant ZFX proteins.
Loss-of-function ZFX zebrafish model supports neurocognitive phenotype
To model truncating loss-of-function ZFX variants in vivo, we generated a zfx KO zebrafish model and characterized neurocognitive behavioral changes in adult zebrafish. To generate zfx zebrafish, we injected Cas9 mRNA and CRISPR guide RNAs targeting zfx exon 2, screened for mosaic F0 founders, and established a mutant allele containing a 25-bp deletion in zfx, which is predicted to cause a frameshift with premature termination (Figure S2). Additionally, we sought to analyze the spatiotemporal mRNA expression of zfx in WT zebrafish larvae using a previously reported protocol.45 Whole-mount in situ hybridization indicated zfx expression in the brain regions of 24-hpf (hours postfertilization) and 48-hpf larvae (Figure S2C). In order to quantitate the mRNA expression level in KO organisms, real-time qPCR was conducted using RNA obtained from adult zebrafish brains. zfx mRNA expression was significantly reduced in zfx KO zebrafish compared to WT siblings (p = 0.008, Figure S2D).
KO zebrafish displayed no significant differences in terms of gross morphology compared to WT zebrafish during early developmental stages (Figure S3). Additionally, of note, over-expression of two ZFX missense variants (p.Arg764Trp and p.Arg786Gln) by microinjection of mRNA in WT zebrafish did not yield any developmental morphological phenotypes (Figure S4). However, the lack of gross morphological changes does not preclude the potential for more subtle alterations in adult zebrafish behavior.
To evaluate behavioral changes in adult zfx KO zebrafish relative to WT, we performed a battery of behavioral tests, including a novel tank test, social interaction assay, mirror biting test, scototaxis assay, and adult startle tap test. The novel tank diving test is used to assay swimming activity and evaluate anxiety (equivalent to the rodent open field test).56 When placed in a novel environment, zebrafish typically display a behavior called “novel tank diving” where they rapidly swim in the water column and explore their surroundings by diving and swimming along the bottom of the tank. This behavior is thought to be a response to an unfamiliar environment and can be used as a measure of anxiety-like behavior.57 As expected, sibling WT fish spent significantly more time in the bottom zone. The time spent in the top zone was significantly increased in zfx KO zebrafish compared to WT fish during the first 3 min (Figures 6A and 6B), suggesting reduced anxiety or an altered anxiolytic response. There was no significant difference in distance moved by WT and zfx KO, indicating normal motor functions of zfx KO zebrafish. Social interaction and mirror biting tests showed no significant difference in behavior between WT and zfx KO fish.
We next performed a scototaxis test to assay dark/gray/light preference. WT zebrafish prefer to spend more time in darker conditions.47 However, zfx KO zebrafish spent significantly increased time in the bright zone compared to WT fish (Figures 6C–6E). In the gray zone, both WT and zfx KO zebrafish spent equal time (t = 0.5461, df = 16, p = 0.1295). Although not statistically significant, WT fish spent more time in the dark zone compared to zfx KO zebrafish (t = 1.604, df = 16, p = 0.063). These results are consistent with the novel tank test and suggest decreased baseline anxiety or an altered anxiolytic response.
To examine the sensorimotor response and habituation of adult zebrafish, we also performed a startle tap test. In a startle tap test, the motor startle response is measured to determine an initial startle response and evaluate habituation with repeated testing.48 Typically, zebrafish can adapt to the startle response over time when exposed to repeated taps in the same environment.58,59 No significant differences were observed in responses for both the first and second tap stimuli between WT and zfx KO fish. However, for the third stimulus, zfx KO fish demonstrated a significantly increased response compared to WT fish (Figures 6F–6H). While WT fish showed a trend of significantly decreasing response by the third tap stimulus, zfx zebrafish showed the opposite response, indicating a lack of habituation to the external tap stimuli. Taken together, these experiments support that zfx KO zebrafish exhibit altered behavioral activity compared to WT fish, suggesting that loss of zfx function may result in neurocognitive abnormalities.
Discussion
Here, we present a large cohort with an overlapping syndromic intellectual disability phenotype linked by variants in the TF ZFX. These individuals share overlapping and recurrent facial features, neurocognitive abnormalities, behavioral problems, and congenital anomalies. A subset of individuals with missense variants in the DNA-binding domain demonstrate hyperparathyroidism. Molecular characterization of genome-wide binding profiles of the missense ZFX variants by ChIP-seq and transcriptional dysregulation by RNA-seq shows altered transcriptional activation of a small set of gene targets compared to WT ZFX. Furthermore, a zebrafish model of ZFX loss demonstrated abnormal anxiolytic and habituation behaviors in several tests, including a novel tank, scototaxis, and adult tap test—supporting altered neurocognitive behavior in association with truncating ZFX variants. Taken together, our data suggest that the individuals in this cohort represent a complex X-linked intellectual disability syndrome.
Individuals in this study cohort displayed considerable phenotypic variability, prompting clinicians to order non-targeted molecular studies, including ES and chromosomal microarray analysis, to evaluate the probands. The main indications for ordering such studies included developmental delay, intellectual disability, multiple congenital anomalies, and dysmorphic features, all of which are associated with remarkable locus heterogeneity (for example, there are numerous genes associated with syndromic neurodevelopmental disorders). Nevertheless, targeted testing for Fragile X syndrome, which is considered a first-tier testing candidate for individuals with developmental delay/intellectual disability, was ordered for several probands. In addition, four subjects underwent molecular testing for Beckwith-Wiedemann syndrome because of overgrowth, umbilical hernia, and macroglossia, and six additional individuals underwent molecular testing for other overgrowth syndromes including Simpson-Golabi-Behmel, Sotos, and Costello syndromes. Mucopolysaccharidosis has been also considered on the differential diagnosis of three probands based on coarse facial features, some overgrowth, and hernias. Hyperparathyroidism is an emerging phenotype associated with certain ZFX variants and, if present in individuals with syndromic neurodevelopmental disorder (NDD), should probably prompt consideration of this diagnostic entity.
Eight individuals had additional genetic findings of varying clinical significance (Table S1, supplemental notes), and a subset of them may have contributed to or modified the individuals’ phenotypes. Proband 9 was found to have a paternally inherited variant in KCNH2 (MIM: 152427), monoallelic variants in which are associated with long QT syndrome-2 (MIM: 613688). This finding is consistent with the proband’s personal and family history of prolonged QT syndrome, and it is not related to the phenotype associated with the ZFX variant. Proband 12 carries a pathogenic OCA2 (MIM: 611409) variant and a heterozygous missense variant in MC1R (MIM: 155555). The relevance of these variants to the proband’s phenotype is unclear. His mother is clinically affected with oculocutaneous albinism and carries 2 pathogenic variants in OCA2, but he does not display classical features of this condition, and his nystagmus is thought to be secondary to optic nerve hypoplasia. Proband 14 had a paternally inherited nonsense variant in ROBO1 (MIM: 602430), which can explain the proband’s pituitary hypoplasia and growth hormone deficiency (MIM: 620303; combined or isolated pituitary hormone deficiency-8). This is a typical example of a blended phenotype in the same individual. ES of proband 15 also revealed a VUS in TFAP2A (MIM: 107580), heterozygous pathogenic variants in which are associated with branchiooculofacial syndrome (MIM: 113620). The proband does have hearing loss, which has been described in this condition, but the absence of the other syndromic cardinal features made it seem unlikely that the TFAP2A variant is the cause of this proband’s phenotype. Four additional probands had small copy number variants of unclear clinical significance, but we cannot completely exclude a potential modifying effect.
Multiple lines of evidence support the pathogenicity of the observed ZFX variants, including shared and recurrent phenotypes in affected individuals, structural and in silico variant analysis, lack of these variants in the general population based on publicly available databases, and functional molecular data demonstrating altered transcriptional activity in missense variants and altered behavior in a zebrafish loss-of-function model. It is important to note that ZFX is a highly intolerant gene for loss-of-function variants, as demonstrated by high probability of loss-of-function intolerance score (pLI = 1.0), low loss-of-function observed/expected upper bound fraction (LOEUF = 0.16),60 and low haploinsufficiency score (HI index = 7.61%).61
Although we cannot yet estimate the frequency of ZFX-related disorder, ZFX variants appear to be very uncommon. GeneDx confirmed de novo missense or frameshift variants in ZFX for four affected individuals and maternally inherited missense variants in 12 affected hemizygotes out of 125,778 individuals undergoing clinical ES/GS for an indication of NDD. Of the de novo variants, there was one hemizygous missense variant, one heterozygous missense variant, and two heterozygous frameshift variants. Because of the design and scope of this study, we were unable to calculate the enrichment of ZFX variants in NDD. Nevertheless, none of the variants reported in this study were found in gnomAD (last accessed: October 6, 2023). ZFX is not only intolerant for loss-of-function variants (based on pLI, LOEUF, and HI index) but also highly constrained for missense variants. In fact, the missense constraint Z score for ZFX, which calculates the deviation of observed from predicted missense variants, is +3.15, with higher scores indicating that the transcript is more constrained and less tolerant of missense variation.62
Variants in TFs, by nature of their complex cellular functions, can result in pleiotropic phenotypes. Most fundamentally, TFs can be reduced to two functional domains: (1) a structured DNA-binding domain that interacts with a specific DNA motif and (2) a structured or unstructured transcriptional effector domain responsible for activation (the recruitment of transcriptional machinery to a target gene) and/or repression (the binding of corepressors and chromatin remodelers). Variants in TFs have the potential to impair or alter DNA binding specificity, inhibit transcriptional effector function, result in loss of TF expression entirely, or cause a gain of transcriptional function. In this ZFX cohort, we observe both predicted loss-of-function truncating variants and DNA-binding domain missense variants that exhibit the potential for gain-of-function behavior. This complexity is consistent with the observed pleiotropy of the individuals in this cohort, which may reflect variations in the particular gene targets of the different ZFX variants.
It is of particular interest that the missense variants cluster near the penultimate and ultimate zinc fingers of ZFX, which we have shown to be critical for genomic DNA binding activity.9 Although several variants fall within the zinc fingers, the variant proteins are not significantly impaired in their recruitment to promoter regions. Importantly, we show that the missense variants generally bind to the same promoters and activate the same genes as does WT ZFX, and thus they are not inactivating. However, it is also clear that some genes are (1) direct targets of WT ZFX but not the variant ZFX proteins and (2) direct targets of one or more of the variant ZFX proteins but not of the WT ZFX (Figures 5G–5I). Taken together, our studies suggest that the missense variants may result in both gain and loss of transcriptional function at a small set of genes.
In particular, the hyperparathyroidism observed across multiple individuals with three different nearby ZFX DNA-binding domain variants supports the possible role of altered binding affinity or activation in the pathogenesis of this phenotype. Hyperparathyroidism was also observed in carrier females of the p.Arg786Gln and p.Tyr774Cys variants who were otherwise phenotypically unaffected, suggesting that the parathyroid phenotype could result from an altered protein activity leading to mis-activation of downstream non-target genes. Our molecular studies of the missense variants demonstrating differential transcriptional activation (e.g., binding to and activation of CALCR, which encodes the calcitonin receptor, by missense ZFX proteins but not WT ZFX) support this hypothesis, but further molecular studies of these specific variants will be needed to investigate this link.
The prevalence of affected male individuals supports an X-linked recessive inheritance pattern, but the manifesting heterozygote females, including probands 4, 9, and 14 and members of the 6A–6C family, complicate this classification. Skewed mosaic X-inactivation in females, resulting from either random chance or selection during development, can vary the presence and severity of X-linked intellectual disability phenotypes.63,64 X-inactivation studies were only available for a subset of the cohort members, but for the 6A–6C family, several affected females displayed substantial X-inactivation skewing (see Figure 3C), and additional X-inactivation studies for proband 9 also demonstrated substantial skewing. It is important to note that only proband 6C from family 6 had clinical findings similar to the remainder of the cohort, whereas other females mainly had hyperparathyroidism. One possible explanation is variation of X-inactivation in different tissues, but the contribution of other genetic and nongenetic modifiers cannot be excluded. The consequence of this in their presented phenotype, and indeed the potential for disease in carrier females, will necessitate additional characterization.
The specific molecular pathomechanisms by which ZFX variants cause disease will require further studies, but this work reflects a comprehensive effort to collate a phenotypic profile of ZFX disease. The specific role of ZFX DNA-binding domain variants in the pathogenesis of neurocognitive phenotypes and hyperparathyroidism, in particular, is ripe for further characterization. Additionally, further investigation of the gene targets and transcriptional regulatory network of ZFX may yield substantial insights into molecular disease mechanisms.
A primary limitation of this study is the heterogeneity of clinical assessment of the participating individuals, with evaluations by different professionals resulting in the potential for interobserver bias. For instance, not all participants had a comprehensive and systematic neurocognitive evaluation, which restricts the potential for comparisons across some neurocognitive traits. Furthermore, due to lack of availability of biological specimens, molecular studies were not performed on all truncating variants, although their predicted loss-of-function phenotypes were modeled in zebrafish and assessed based on in silico models. Additionally, the small cohort size and the fact that the majority of the ZFX variants were private to each proband or family made the assessment of genotype-phenotype correlation difficult. Furthermore, because of the original design and scope of this study, we could not perform a formal statistical test of enrichment for ZFX variants in affected individuals as compared to controls. In the in vitro studies, co-deletion of partner TFs ZFY and/or ZNF711 was performed, although in this cohort these genes are unaffected. However, there is precedent for variants in only a single copy of a redundant X/Y gene pair to cause disease, such as the gene pair NLGN4X (MIM: 300427) and NLGN4Y (MIM: 400028), of which NLGN4X specifically is associated with X-linked intellectual disability (intellectual developmental disorder, X-linked, MIM: 300495). The need to delete homologous genes in models may just be a consequence of assay sensitivity.
In summary, both truncation and missense variants in the TF ZFX are associated with a complex X-linked intellectual disability syndrome with characteristic developmental and behavioral abnormalities. Further characterization of variants is needed to reveal the precise pathomechanism, particularly with regards to the recurrent hyperparathyroidism observed in a subset of affected individuals.
Data and code availability
All raw and some processed files used in this study are available at the Gene Expression Omnibus. The accession number for the data reported in this paper is GEO: GSE218691.
Acknowledgments
We thank the cohort members and their families for participating in this study. For additional acknowledgments, including detailed funding information, please see the supplemental acknowledgments.
Author contributions
Investigation: J.L.S., K.H., D.W.D., G.M., T.-I.C., C.A.A., D.J.A., S.B., D.G.B., L.D.B., D.A.C., R.C., J.C.-S., A.C., M.D., L.F., C.P.G., N.B.G., K.W.G., E.H., A.M.H., A.M.I., B.I., A.J., P.K., L.A.R.K., S.K., F.L., P.L., J. Maraval, N. Matsumoto, J. McCarrier, J. McCarthy, N. Miyake, L.H.M., A.H.N., E.Ø., R.P., K.P., J.E.P., R.E.S., M. Shaw, E.S., J.P.T., E. Wadman, E. Wakeling, S.M.W., L.C.W., J.R.L., O.L., M.A.C., J.G., C.M.N., P.J.F., C.-H.K., M. Shinawi; writing—original draft: J.L.S., D.W.D., P.K., C.M.N., M. Shinawi; writing—review & editing: J.L.S., K.H., D.W.D., G.M., C.A.A., D.J.A., S.B., D.G.B., L.D.B., D.A.C., J.C.-S., A.C., M.D., L.F., C.P.G., N.B.G., K.W.G., E.H., A.M.H., A.M.I., B.I., A.J., P.K., L.A.R.K., S.K., F.L., P.L., J. Maraval, N. Matsumoto, J. McCarrier, J. McCarthy, N. Miyake, L.H.M., A.H.N., E.Ø., R.P., K.P., J.E.P., R.E.S., E.S., J.P.T., E. Wadman, E. Wakeling, S.M.W., L.C.W., J.R.L., O.L., M.A.C., J.G., C.M.N., P.J.F., C.-H.K., M. Shinawi; resources: G.M., C.A.A., D.J.A., S.B., D.G.B., L.D.B., D.A.C., R.C., J.C.-S., M.D., L.F., C.P.G., N.B.G., K.W.G., E.H., A.M.H., A.M.I., B.I., A.J., L.A.R.K., G.E.R.C., S.K., F.L., P.L., J. Maraval, N. Matsumoto, J. McCarrier, J. McCarthy, N. Miyake, L.H.M., A.H.N., E.Ø., R.P., K.P., R.E.S., M. Shaw, E.S., E. Wadman, E. Wakeling, S.M.W., L.C.W., M.A.C., P.J.F., C.-H.K., M. Shinawi; supervision: O.L., J.G., P.J.F., C.-H.K., M. Shinawi; funding acquisition: J.G., P.J.F., C.-H.K., M. Shinawi; conceptualization: M. Shinawi; project administration: M. Shinawi.
Declaration of interests
D.A.C. and R.E.S. are employees of GeneDx, LLC. A.C. and J.P.T. are employees and shareholders of Illumina, Inc. L.D.B. performs advisory board, consulting, and speaking arrangement work unrelated to the present study for Sanofi S.A., Horizon Therapeutics, Amicus Therapeutics, and Chiesi Farmaceutici S.p.A. N.B.G. has received personal fees from Pfizer Inc. and RCG Consulting for work unrelated to the present study. J.R.L. has stock ownership in 23andMe and is a paid consultant to Genome International.
Published: February 6, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2024.01.007.
Contributor Information
Cheol-Hee Kim, Email: zebrakim@cnu.ac.kr.
Marwan Shinawi, Email: mshinawi@wustl.edu.
Supplemental information
References
- 1.Tejada M.I., Ibarluzea N. Non-syndromic X Linked Intellectual Disability: Current Knowledge in Light of the Recent Advances in Molecular and Functional Studies. Clin. Genet. 2020;97:677–687. doi: 10.1111/cge.13698. [DOI] [PubMed] [Google Scholar]
- 2.Stevenson R.E., Schwartz C.E., Rogers R.C. Malformations among the X-linked Intellectual Disability Syndromes. Am. J. Med. Genet. 2013;161:2741–2749. doi: 10.1002/ajmg.a.36179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz C.E., Louie R.J., Toutain A., Skinner C., Friez M.J., Stevenson R.E. X-Linked Intellectual Disability Update 2022. Am. J. Med. Genet. 2023;191:144–159. doi: 10.1002/ajmg.a.63008. [DOI] [PubMed] [Google Scholar]
- 4.Lubs H.A., Stevenson R.E., Schwartz C.E. Fragile X and X-Linked Intellectual Disability: Four Decades of Discovery. Am. J. Hum. Genet. 2012;90:579–590. doi: 10.1016/j.ajhg.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fieremans N., Van Esch H., Holvoet M., Van Goethem G., Devriendt K., Rosello M., Mayo S., Martinez F., Jhangiani S., Muzny D.M., et al. Identification of Intellectual Disability Genes in Female Patients with a Skewed X-Inactivation Pattern. Hum. Mutat. 2016;37:804–811. doi: 10.1002/humu.23012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaufman L., Ayub M., Vincent J.B. The Genetic Basis of Non-Syndromic Intellectual Disability: A Review. J. Neurodev. Disord. 2010;2:182–209. doi: 10.1007/s11689-010-9055-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rhie S.K., Yao L., Luo Z., Witt H., Schreiner S., Guo Y., Perez A.A., Farnham P.J. ZFX Acts as a Transcriptional Activator in Multiple Types of Human Tumors by Binding Downstream from Transcription Start Sites at the Majority of CpG Island Promoters. Genome Res. 2018;28:310–320. doi: 10.1101/gr.228809.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mardon G., Luoh S.W., Simpson E.M., Gill G., Brown L.G., Page D.C. Mouse Zfx Protein Is Similar to Zfy-2: Each Contains an Acidic Activating Domain and 13 Zinc Fingers. Mol. Cell Biol. 1990;10:681–688. doi: 10.1128/mcb.10.2.681-688.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ni W., Perez A.A., Schreiner S., Nicolet C.M., Farnham P. Characterization of the ZFX Family of Transcription Factors That Bind Downstream of the Start Site of CpG Island Promoters. Nucleic Acids Res. 2020;48:5986–6000. doi: 10.1093/nar/gkaa384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Page D.C., Mosher R., Simpson E.M., Fisher E.M., Mardon G., Pollack J., McGillivray B., de la Chapelle A., Brown L.G. The Sex-Determining Region of the Human Y Chromosome Encodes a Finger Protein. Cell. 1987;51:1091–1104. doi: 10.1016/0092-8674(87)90595-2. [DOI] [PubMed] [Google Scholar]
- 11.Schneider-Gädicke A., Beer-Romero P., Brown L.G., Nussbaum R., Page D.C. ZFX Has a Gene Structure Similar to ZFY, the Putative Human Sex Determinant, and Escapes X Inactivation. Cell. 1989;57:1247–1258. doi: 10.1016/0092-8674(89)90061-5. [DOI] [PubMed] [Google Scholar]
- 12.Schneider-Gädicke A., Beer-Romero P., Brown L.G., Mardon G., Luoh S.-W., Page D.C. Putative Transcription Activator with Alternative Isoforms Encoded by Human ZFX Gene. Nature. 1989;342:708–711. doi: 10.1038/342708a0. [DOI] [PubMed] [Google Scholar]
- 13.Galan-Caridad J.M., Harel S., Arenzana T.L., Hou Z.E., Doetsch F.K., Mirny L.A., Reizis B. Zfx Controls the Self-Renewal of Embryonic and Hematopoietic Stem Cells. Cell. 2007;129:345–357. doi: 10.1016/j.cell.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmer C.J., Galan-Caridad J.M., Weisberg S.P., Lei L., Esquilin J.M., Croft G.F., Wainwright B., Canoll P., Owens D.M., Reizis B. Zfx Facilitates Tumorigenesis Caused by Activation of the Hedgehog Pathway. Cancer Res. 2014;74:5914–5924. doi: 10.1158/0008-5472.can-14-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weisberg S.P., Smith-Raska M.R., Esquilin J.M., Zhang J., Arenzana T.L., Lau C.M., Churchill M., Pan H., Klinakis A., Dixon J.E., et al. ZFX Controls Propagation and Prevents Differentiation of Acute T-Lymphoblastic and Myeloid Leukemia. Cell Rep. 2014;6:528–540. doi: 10.1016/j.celrep.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luoh S.W., Bain P.A., Polakiewicz R.D., Goodheart M.L., Gardner H., Jaenisch R., Page D.C. Zfx Mutation Results in Small Animal Size and Reduced Germ Cell Number in Male and Female Mice. Development. 1997;124:2275–2284. doi: 10.1242/dev.124.11.2275. [DOI] [PubMed] [Google Scholar]
- 17.van der Werf I.M., Van Dijck A., Reyniers E., Helsmoortel C., Kumar A.A., Kalscheuer V.M., de Brouwer A.P., Kleefstra T., van Bokhoven H., Mortier G., et al. Mutations in Two Large Pedigrees Highlight the Role of ZNF711 in X-linked Intellectual Disability. Gene. 2017;605:92–98. doi: 10.1016/j.gene.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 18.Tarpey P.S., Smith R., Pleasance E., Whibley A., Edkins S., Hardy C., O’Meara S., Latimer C., Dicks E., Menzies A., et al. A Systematic, Large-Scale Resequencing Screen of X-chromosome Coding Exons in Mental Retardation. Nat. Genet. 2009;41:535–543. doi: 10.1038/ng.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soong C.-P., Arnold A. Recurrent ZFX Mutations in Human Sporadic Parathyroid Adenomas. Oncoscience. 2014;1:360–366. doi: 10.18632/oncoscience.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W., et al. ClinVar: Improving Access to Variant Interpretations and Supporting Evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Angelozzi M., Karvande A., Molin A.N., Ritter A.L., Leonard J.M.M., Savatt J.M., Douglass K., Myers S.M., Grippa M., Tolchin D., et al. Consolidation of the Clinical and Genetic Definition of a SOX4- Related Neurodevelopmental Syndrome. J. Med. Genet. 2022;59:1058–1068. doi: 10.1136/jmedgenet-2021-108375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma R., Sahoo S.S., Honda M., Granger S.L., Goodings C., Sanchez L., Künstner A., Busch H., Beier F., Pruett-Miller S.M., et al. Gain-of-Function Mutations in RPA1 Cause a Syndrome with Short Telomeres and Somatic Genetic Rescue. Blood. 2022;139:1039–1051. doi: 10.1182/blood.2021011980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoshino A., Boutboul D., Zhang Y., Kuehn H.S., Hadjadj J., Özdemir N., Celkan T., Walz C., Picard C., Lenoir C., et al. Gain-of-Function IKZF1 Variants in Humans Cause Immune Dysregulation Associated with Abnormal T/B Cell Late Differentiation. Sci. Immunol. 2022;7 doi: 10.1126/sciimmunol.abi7160. [DOI] [PubMed] [Google Scholar]
- 24.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: A Matching Tool for Connecting Investigators with an Interest in the Same Gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Vooren S.V., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faundes V., Newman W.G., Bernardini L., Canham N., Clayton-Smith J., Dallapiccola B., Davies S.J., Demos M.K., Goldman A., Gill H., et al. Clinical Assessment of the Utility of Sequencing and Evaluation as a Service (CAUSES) Study, Deciphering Developmental Disorders (DDD) Study, S. Banka, Histone Lysine Methylases and Demethylases in the Landscape of Human Developmental Disorders. Am. J. Hum. Genet. 2018;102:175–187. doi: 10.1016/j.ajhg.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson A., Banka S., Stewart H., Robinson H., Lovell S., Clayton-Smith J. Recurrent KCNT2 missense variants affecting p.Arg190 result in a recognizable phenotype. Am. J. Med. Genet. 2021;185:3083–3091. doi: 10.1002/ajmg.a.62370. [DOI] [PubMed] [Google Scholar]
- 28.Pagnamenta A.T., Jackson A., Perveen R., Beaman G., Petts G., Gupta A., Hyder Z., Chung B.H.-Y., Kan A.S.-Y., Cheung K.W., et al. Biallelic TMEM260 variants cause truncus arteriosus, with or without renal defects. Clin. Genet. 2022;101:127–133. doi: 10.1111/cge.14071. [DOI] [PubMed] [Google Scholar]
- 29.ACMG Laboratory Quality Assurance Committee, S. Richards. Aziz N., S. Bale D.B., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., E. Spector K.V., Rehm H.L. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abou Tayoun A.N., Pesaran T., DiStefano M.T., Oza A., Rehm H.L., Biesecker L.G., Harrison S.M. ClinGen Sequence Variant Interpretation Working Group (ClinGen SVI), Recommendations for Interpreting the Loss of Function PVS1 ACMG/AMP Variant Criterion. Hum. Mutat. 2018;39:1517–1524. doi: 10.1002/humu.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pejaver V., Byrne A.B., Feng B.-J., Pagel K.A., Mooney S.D., Karchin R., O’Donnell-Luria A., Harrison S.M., Tavtigian S.V., Greenblatt M.S., et al. ClinGen Sequence Variant Interpretation Working Group, Calibration of Computational Tools for Missense Variant Pathogenicity Classification and ClinGen Recommendations for PP3/BP4 Criteria. Am. J. Hum. Genet. 2022;109:2163–2177. doi: 10.1016/j.ajhg.2022.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katsonis P., Lichtarge O. A Formal Perturbation Equation between Genotype and Phenotype Determines the Evolutionary Action of Protein-Coding Variations on Fitness. Genome Res. 2014;24:2050–2058. doi: 10.1101/gr.176214.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lichtarge O., Bourne H.R., Cohen F.E. An Evolutionary Trace Method Defines Binding Surfaces Common to Protein Families. J. Mol. Biol. 1996;257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 34.Mihalek I., Res I., Lichtarge O. A Family of Evolution-Entropy Hybrid Methods for Ranking Protein Residues by Importance. J. Mol. Biol. 2004;336:1265–1282. doi: 10.1016/j.jmb.2003.12.078. [DOI] [PubMed] [Google Scholar]
- 35.Katsonis P., Lichtarge O. CAGI5: Objective Performance Assessments of Predictions Based on the Evolutionary Action Equation. Hum. Mutat. 2019;40:1436–1454. doi: 10.1002/humu.23873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katsonis P., Lichtarge O. Objective Assessment of the Evolutionary Action Equation for the Fitness Effect of Missense Mutations across CAGI-blinded Contests. Hum. Mutat. 2017;38:1072–1084. doi: 10.1002/humu.23266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 38.Suzek B.E., Huang H., McGarvey P., Mazumder R., Wu C.H. UniRef: Comprehensive and Non-Redundant UniProt Reference Clusters. Bioinformatics. 2007;23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 39.Edgar R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lua R.C., Lichtarge O. PyETV: A PyMOL Evolutionary Trace Viewer to Analyze Functional Site Predictions in Protein Complexes. Bioinformatics. 2010;26:2981–2982. doi: 10.1093/bioinformatics/btq566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thouin M.M., Giron J.M., Hoffman E.P. Detection of Nonrandom X Chromosome Inactivation. Curr. Protoc. Hum. Genet. 2002;35 doi: 10.1002/0471142905.hg0907s35. [DOI] [PubMed] [Google Scholar]
- 43.Allen R.C., Zoghbi H.Y., Moseley A.B., Rosenblatt H.M., Belmont J.W. Methylation of HpaII and HhaI Sites near the Polymorphic CAG Repeat in the Human Androgen-Receptor Gene Correlates with X Chromosome Inactivation. Am. J. Hum. Genet. 1992;51:1229–1239. [PMC free article] [PubMed] [Google Scholar]
- 44.T. Hulsen, DeepVenn – a Web Application for the Creation of Area-Proportional Venn Diagrams Using the Deep Learning Framework Tensorflow.Js,Preprint at arXiv. 10.48550/ARXIV.2210.04597. [DOI]
- 45.Lee Y.-R., Khan K., Armfield-Uhas K., Srikanth S., Thompson N.A., Pardo M., Yu L., Norris J.W., Peng Y., Gripp K.W., et al. Mutations in FAM50A Suggest That Armfield XLID Syndrome Is a Spliceosomopathy. Nat. Commun. 2020;11:3698. doi: 10.1038/s41467-020-17452-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim O.-H., Cho H.-J., Han E., Hong T.I., Ariyasiri K., Choi J.-H., Hwang K.-S., Jeong Y.-M., Yang S.-Y., Yu K., et al. Zebrafish Knockout of Down Syndrome Gene, DYRK1A, Shows Social Impairments Relevant to Autism. Mol. Autism. 2017;8:50. doi: 10.1186/s13229-017-0168-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maximino C., Marques de Brito T., Dias C.A. G.d.M., Gouveia A., Morato S. Scototaxis as Anxiety-like Behavior in Fish. Nat. Protoc. 2010;5:209–216. doi: 10.1038/nprot.2009.225. [DOI] [PubMed] [Google Scholar]
- 48.Eddins D., Cerutti D., Williams P., Linney E., Levin E.D. Zebrafish Provide a Sensitive Model of Persisting Neurobehavioral Effects of Developmental Chlorpyrifos Exposure: Comparison with Nicotine and Pilocarpine Effects and Relationship to Dopamine Deficits. Neurotoxicol. Teratol. 2010;32:99–108. doi: 10.1016/j.ntt.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ho J., Tumkaya T., Aryal S., Choi H., Claridge-Chang A. Moving beyond P Values: Data Analysis with Estimation Graphics. Nat. Methods. 2019;16:565–566. doi: 10.1038/s41592-019-0470-3. [DOI] [PubMed] [Google Scholar]
- 50.Chen S., Francioli L.C., Goodrich J.K., Collins R.L., Kanai M., Wang Q., Alföldi J., Watts N.A., Vittal C., Gauthier L.D., et al. A genomic mutational constraint map using variation in 76,156 human genomes. Nature. 2024;625:92–100. doi: 10.1038/s41586-023-06045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rentzsch P., Schubach M., Shendure J., Kircher M. CADD-Splice—Improving Genome-Wide Variant Effect Prediction Using Deep Learning-Derived Splice Scores. Genome Med. 2021;13:31. doi: 10.1186/s13073-021-00835-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D., et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang K.T., Guo J., di Ronza A., Sardiello M. Aminode: Identification of Evolutionary Constraints in the Human Proteome. Sci. Rep. 2018;8:1357. doi: 10.1038/s41598-018-19744-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silk M., Petrovski S., Ascher D.B., MTR-Viewer Identifying Regions within Genes under Purifying Selection. Nucleic Acids Res. 2019;47:W121–W126. doi: 10.1093/nar/gkz457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wainer Katsir K., Linial M. Human Genes Escaping X-inactivation Revealed by Single Cell Expression Data. BMC Genom. 2019;20:201. doi: 10.1186/s12864-019-5507-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kysil E.V., Meshalkina D.A., Frick E.E., Echevarria D.J., Rosemberg D.B., Maximino C., Lima M.G., Abreu M.S., Giacomini A.C., Barcellos L.J.G., et al. Comparative Analyses of Zebrafish Anxiety-Like Behavior Using Conflict-Based Novelty Tests. Zebrafish. 2017;14:197–208. doi: 10.1089/zeb.2016.1415. [DOI] [PubMed] [Google Scholar]
- 57.Blaser R., Gerlai R. Behavioral Phenotyping in Zebrafish: Comparison of Three Behavioral Quantification Methods. Behav. Res. Methods. 2006;38:456–469. doi: 10.3758/BF03192800. [DOI] [PubMed] [Google Scholar]
- 58.Wong K., Elegante M., Bartels B., Elkhayat S., Tien D., Roy S., Goodspeed J., Suciu C., Tan J., Grimes C., et al. Analyzing Habituation Responses to Novelty in Zebrafish (Danio Rerio) Behav. Brain Res. 2010;208:450–457. doi: 10.1016/j.bbr.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 59.Pittman J.T., Lott C.S. Startle Response Memory and Hippocampal Changes in Adult Zebrafish Pharmacologically-Induced to Exhibit Anxiety/Depression-like Behaviors. Physiol. Behav. 2014;123:174–179. doi: 10.1016/j.physbeh.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 60.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Exome Aggregation Consortium, Analysis of Protein-Coding Genetic Variation in 60,706 Humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang N., Lee I., Marcotte E.M., Hurles M.E. Characterising and Predicting Haploinsufficiency in the Human Genome. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The Mutational Constraint Spectrum Quantified from Variation in 141,456 Humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Plenge R.M., Stevenson R.A., Lubs H.A., Schwartz C.E., Willard H.F. Skewed X-Chromosome Inactivation Is a Common Feature of X-Linked Mental Retardation Disorders. Am. J. Hum. Genet. 2002;71:168–173. doi: 10.1086/341123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wongkittichote P., Wegner D.J., Shinawi M.S. Novel Exon-Skipping Variant Disrupting the Basic Domain of HCFC1 Causes Intellectual Disability without Metabolic Abnormalities in Both Male and Female Patients. J. Hum. Genet. 2021;66:717–724. doi: 10.1038/s10038-020-00892-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and some processed files used in this study are available at the Gene Expression Omnibus. The accession number for the data reported in this paper is GEO: GSE218691.