Abstract
The mRNA encoding the M2 protein of respiratory syncytial (RS) virus contains two open reading frames (ORFs). ORF1 encodes the 22-kDa structural protein, M2, and ORF2 has the potential to encode a 10-kDa protein (90 amino acids). Using a vaccinia virus T7 expression system, we examined the RNA synthetic activities of mono- and dicistronic subgenomic replicons of RS virus by direct metabolic labeling of RNA in the presence and absence of the products of ORF1 and ORF2. In the absence of ORF1 and ORF2, the negative- and positive-sense products of genomic RNA replication and positive-sense polyadenylated mRNA(s) were synthesized. Expression of the whole M2 transcription unit (containing ORF1 and ORF2) or ORF1 alone caused an increase in the synthesis of polyadenylated mRNA, the majority of which was due to a substantial increase in the quantity of polycistronic mRNAs generated by the polymerase failing to terminate at gene end signals. In agreement with previous reports, the ORF2 product was found to inhibit viral RNA replication and mRNA transcription. These data show that the M2 protein functions as a transcriptional antiterminator that enhances the ability of the viral RNA polymerase to read through intergenic junctions. The role of such a function during the viral life cycle is discussed.
Human respiratory syncytial (RS) virus is a member of the pneumovirus genus of the Paramyxoviridae and is the leading viral cause of pediatric pneumonia and bronchiolitis. During transcription of the negative-sense RNA genome (15,222 nucleotides), 10 mRNAs that encode at least 11 proteins are made (10, 13). The genomic RNA is present in infected cells and virions as a ribonucleocapsid complex, in which the RNA is tightly bound by the nucleocapsid (N) protein (21). Also associated with the ribonucleocapsid are the phosphoprotein (P) and large polymerase protein (L) (21). These two proteins are the viral components required for transcription of the RS virus mRNAs and, along with the N protein, for replication of the genome (20, 34). Recent work indicates that the RS virus M2 protein is also involved in transcription (9).
The genome of RS virus possesses a single polymerase entry site at or near the 3′ end, and UV mapping has shown transcription of the genes occurs in a sequential polar fashion from 3′ to 5′ (15). Each gene is flanked by conserved gene start and gene end signals, and the genes are separated by intergenic regions of varying size and sequence which are not represented in the mRNAs (7, 24). The exact mechanisms of initiation and termination of transcription for nonsegmented negative-strand RNA viruses are not understood. However, the gene start is believed to signal transcription initiation and mRNA capping, and the gene end signals transcription termination and polyadenylation (23, 25). The level of each RS virus mRNA correlates with its distance from the 3′ promoter in the genome; i.e., the 3′-most gene gives rise to the greatest amount of mRNA (2, 15). By analogy with other nonsegmented negative-strand RNA viruses, this regulation is thought to be achieved by transcriptional attenuation at each gene junction. It is postulated that some of the polymerase molecules which have terminated transcription of the upstream gene do not reinitiate transcription of the downstream gene (22). In vesicular stomatitis virus, where this has been examined directly (3, 22), transcriptional attenuation has been measured at approximately 30% at each junction.
The M2 gene, found only in the pneumoviruses, encodes an mRNA containing two open reading frames (ORFs). ORF1 encodes the 22-kDa structural protein designated M2, whereas ORF2 has the capacity to produce a 90-amino-acid polypeptide (5, 14), detection of which has not been reported. The M2 protein contains a Cys3-His1 motif near the amino terminus which is highly conserved in the M2 proteins of the members of the pneumovirus genus (1, 5, 14, 27, 35). This motif in other proteins has been shown to bind zinc ions (33). Also, interaction between the M2 and N proteins has been observed upon coexpression and in infected cells (17). During dissociation of virions with detergent and high salt concentrations, M2 was removed from the nucleocapsid cores under similar conditions to the matrix (M) protein and was therefore termed matrix-like (21). Recent reports have shown that the M2 protein is required for recovery of infectious RS virus from a cDNA clone (8), and it has been proposed that the M2 protein enhances the processivity of the polymerase and thus acts as a transcription elongation factor (9, 20).
Here we report an analysis of the roles played by the products of ORF1 and ORF2 of the M2 mRNA in RS virus RNA transcription and replication by direct observation of the RNA, using metabolic labeling. Results presented here show that expression of the product of ORF1, the M2 protein, led to an enhancement in transcriptional readthrough at gene end signals. This finding demonstrates yet another role of the M2 protein in addition to that previously reported (9). The implications of a transcriptional antitermination activity for the replication cycle of the virus are discussed.
MATERIALS AND METHODS
cDNA constructs.
A cDNA encoding a subgenomic replicon of RS virus A-2 strain was generated as previously described by Yu et al. (34). The subgenomic replicon WT5 shown in Fig. 1 was derived from the previously described WT (wild-type) analog (34). The cDNA encoding the subgenomic WT replicon was generated by fusing the 5′ 1,182 nucleotides of the RS virus genome (nucleotides 14040 to 15222 in the positive sense; a single nucleotide deletion is present in the WT replicon at a position corresponding to 15211 in the RS virus genome) to the 3′ 413 nucleotides of the genome (nucleotides 1 to 413), using MunI sites located in the cDNA clones of the NS1 and L genes. The cDNA encoding WT5 (pWT5 [Fig. 1]) was generated by digestion of the WT cDNA with BclI and BglII. This removed a 860-bp fragment of L sequence (nucleotides 14146 to 15005 of the RS virus genome). Upon religation, this construct contained 745 nucleotides of RS virus sequence. It was predicted that the subgenomic replicon RNA synthesized from this cDNA by T7 RNA polymerase followed by autocatalytic cleavage by the hepatitis delta ribozyme would contain two non-RS virus G residues at the 5′ end preceding the trailer sequence and would therefore be 747 nucleotides in length. This subgenomic replicon contained a G-to-C change at position 4 in the leader sequence which causes enhanced RNA synthesis from this template (11). A cDNA encoding a subgenomic replicon containing two transcription units, pM/SH, was made by the insertion of a BsmI fragment containing the M/SH gene junction (nucleotides 3895 to 4497 of the RS virus genome) into a unique BsmI site (nucleotide 319 of the WT5 sequence) in pWT5 (see Fig. 4A).
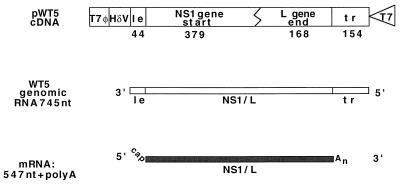
FIG. 1.
Diagram of plasmid pWT5 and the RNAs predicted to be synthesized from the monocistronic subgenomic replicon that it encodes. The subgenomic replicon transcribed from pWT5 includes the RS virus 3′ and 5′ termini flanking a fused partial NS1 and L gene. The 3′ leader (le) sequence includes the G-to-C mutation previously described (11). These sequences were inserted between a T7 RNA polymerase promoter (T7) and a copy of the self-cleaving hepatitis delta ribozyme (HδV). Thus, transcription by T7 polymerase followed by self-cleavage gave rise to a negative-sense copy of the subgenomic replicon RNA. This RNA can act as a template from which the RS virus RNA polymerase can synthesize a 547-nucleotide (nt) mRNA [not including the poly(A) tail] or a positive-sense product of replication. tr, trailer.
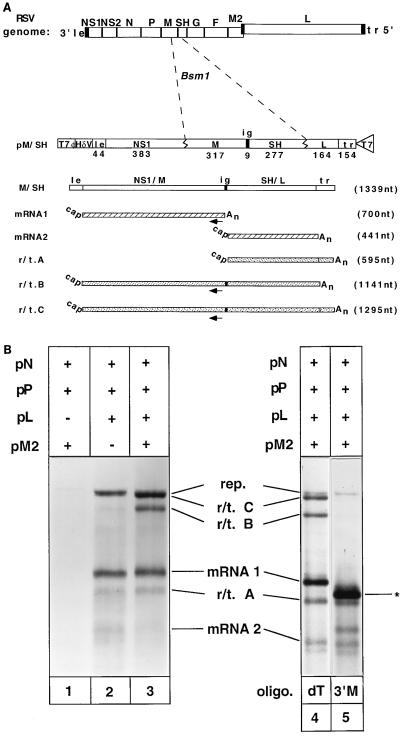
FIG. 4.
(A) Diagram of pM/SH and the RNAs synthesized from the dicistronic subgenomic replicon that it encodes. pM/SH was generated by inserting a DNA fragment encompassing the M/SH intergenic junction into the BsmI site of pWT5. The location of the oligonucleotide 3′M, used for RNase H analysis, is shown by the arrow. le, leader; tr, trailer; nt, nucleotides. (B) Products of RNA synthesis from M/SH genome analog in the presence of pN, pP, and pM2 but absence of pL (lane 1), presence of pN, pP, and pL but absence of pM2 (lane 2), or presence of pN, pP, pL, and pM2 (lane 3). Lanes 4 and 5 show the products of RNase H digestion of RNAs synthesized in the presence of pM2. RNAs in lane 4 were digested in the presence of oligo(dT); RNAs in lane 5 were digested in the presence of 3′M and oligo(dT). The asterisk indicates the 5′ products of RNase H digestion of mRNA1, r/t.B and r/t.C in the presence of 3′M, which comigrate.
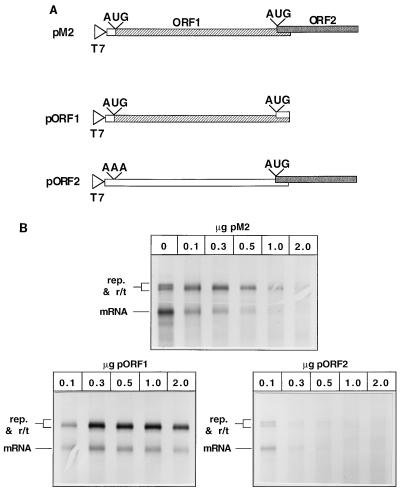
The cDNAs encoding the RS virus N, P, and L proteins were described by Yu et al. (34). A cDNA, pM2, encoding the complete M2 mRNA (both ORF1 and ORF2) of the A-2 strain of human RS virus was generated by reverse transcription of a single strand of cDNA from genomic RNA isolated from virus-infected cells which was amplified by PCR. The resulting DNA was cloned between the BamHI site of pGEM3 in the positive orientation with respect to the T7 promoter, as confirmed by sequence analysis. Individual cDNAs encoding either ORF1 or ORF2 of the M2 gene were generated. The M2 ORF1 cDNA (pORF1) was made by PCR amplification of the first 595 bp of the M2 mRNA sequence and was cloned between the BamHI and HindIII sites of pGEM3. The M2 ORF2 cDNA (pORF2) was generated by PCR from the pM2 cDNA. The entire M2 mRNA sequence was amplified, but the 5′ primer changed the first AUG of ORF1 to AAA. This prevented expression of the ORF1 product yet maintained ORF2 in its authentic context for translation (Fig. 5).
FIG. 5.
Effects of ORF1 or ORF2, expressed individually, on RNA synthesis from the WT5 subgenomic replicon. (A) Diagram of pM2, pORF1, and pORF2 cDNAs described in Materials and Methods. (B) Increasing concentrations of plasmids pM2, pORF1, and pORF2 were included in transfections. Labeled, actinomycin D-resistant RNAs were analyzed by agarose-urea gel electrophoresis and visualized by fluorography. rep., replication product.
DNA transfections, radioactive labeling, and electrophoretic analysis of RNA.
HEp-2 cells were grown in minimum essential medium (GIBCO Laboratories) supplemented with 5% heat-inactivated fetal bovine serum in 60-mm-diameter dishes. HEp-2 cells, infected with recombinant vaccinia virus expressing T7 RNA polymerase (vTF7-3), were transfected with plasmids expressing the subgenomic replicons and plasmids that expressed the trans-acting proteins necessary for RNA encapsidation, replication, and transcription (6, 34). For the RNA synthesis assays, cells were transfected with 5 μg of pWT5 or pM/SH, 5 μg of pN, 2 μg of pP, 2 μg of pL, and 0 or 0.1 to 2 μg of pM2, pORF1, or pORF2. Sixteen hours posttransfection, RS virus-specific RNAs were metabolically labeled with [3H]uridine (33 μCi/ml) for 5 h in the presence of actinomycin D (10 μg/ml).
Cells were harvested and cytoplasmic extracts were prepared as previously described (29). RNA was purified either directly or after immunoprecipitation of ribonucleocapsids, using RS virus-specific antiserum (34). Purified RNAs were analyzed by electrophoresis in 1.75% agarose-urea gels (31) and detected by fluorography (26).
Immunoprecipitation of encapsidated RNA.
Ribonucleocapsid complexes were immunoprecipitated from cytoplasmic extracts with goat anti-RS virus antiserum (Chemicon) and protein G-Sepharose (Pharmacia). RNA was purified by phenol extraction of precipitated complexes and analyzed as described above.
Oligo(dT) chromatography.
RNA purified by phenol extraction was subjected to oligo(dT) chromatography [oligo(dT)-cellulose; New England Biolabs]. Polyadenylated RNA was bound to the oligo(dT)-cellulose in a high-salt buffer (10 mM Tris-HCl [pH 7.5], 0.4 M NaCl, 1 mM EDTA, 0.02% sodium dodecyl sulfate) and eluted in low salt (100 mM Tris-HCl [pH 7.5], 1 mM EDTA). The bound and unbound fractions were recovered by ethanol precipitation, analyzed by 1.75% agarose-urea gel electrophoresis, and detected by fluorography.
In vitro transcription.
RNA was generated in vitro by using T7 RNA polymerase (GIBCO-BRL) according to the manufacturer’s instructions, with minor modifications (32). The ribonucleoside triphosphate concentrations were increased to 2.5 mM and supplemented with [3H]UTP (80 μCi/ml). RNA products of in vitro synthesis were resolved in 1.75% agarose-urea gels.
RNase H analysis of RNA products.
Phenol-extracted RNAs were incubated at 25°C for 20 min with an equal volume (20 μl) of 2× RNase H buffer (3, 4) and 10 μl of 1× RNase H buffer with or without 1 μg of an appropriate oligonucleotide. RNase H digestions were performed by the addition of 2 U of RNase H (GIBCO-BRL) and incubation for 30 min at 37°C. Digested RNAs were recovered by ethanol precipitation. RNAs were identified by electrophoretic mobility shifts in 1.75% agarose-urea gels following RNase H digestion. The oligonucleotides used are as follows (all designations are given from 5′ to 3′ of the positive or negative replication product of the subgenomic replicon WT5): oligo(dT), T12–15 (anneals to polyadenylated RNA); 3′L, 5′-GTTAGTGTATAGCTATGGG-3′ (anneals to positive-sense WT5 RNA from nucleotides 546 to 564); −tr, 5′-AACCAATTAGATTAGGG-3′ (anneals to positive-sense WT5 RNA from nucleotides 675 to 691); 5′NS1, 5′-GAATTGCTGCCCATCTC-3′ (anneals to positive-sense WT5 RNA from nucleotides 96 to 112); and +tr, 5′-CATTTTAATCTTGGAGG-3′ (anneals to negative-sense WT5 RNA from nucleotides 84 to 100).
RESULTS
Effect of M2 mRNA products on RNA synthesis from the WT5 template.
The effect of the M2 protein(s) on RNA synthesis was analyzed initially by using a subgenomic replicon, WT5, containing a single transcription unit (Fig. 1). This was generated by fusing the 3′ end of the RS virus genome, containing the leader and NS1 gene start signal, to the 5′ end of the genome, including the L gene end signal and the trailer sequence (745 nucleotides in total [Fig. 1]). Thus, the entire central portion of the RS virus genome was deleted, and a single transcription unit designed to express an mRNA of 547 nucleotides not including poly(A) was left (Fig. 1).
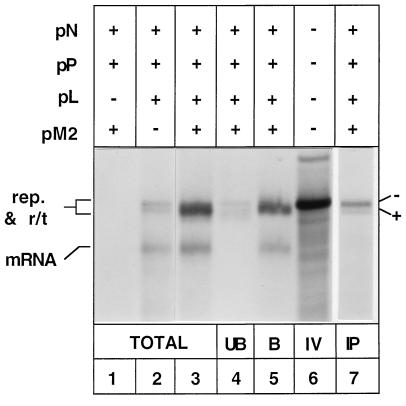
Expression of this genomic analog in vTF7-3-infected HEp-2 cells in the presence of N, P, and L proteins resulted in the actinomycin D-resistant synthesis of three RNA species (Fig. 2, lane 2). Upon addition of pM2 (encoding ORF1 and ORF2 of the M2 mRNA) to transfections, a significant increase in the quantity of slower-migrating RNA was observed (Fig. 2, lane 3). The RNA species were preliminarily identified by (i) immunoprecipitation of N encapsidated genomic RNA replication products, (ii) comigration of RNA with a negative-sense in vitro transcript of WT5, and (iii) oligo(dT) chromatography. The two slower-migrating RNAs (seen readily in lane 2 but masked in lane 3 by another RNA species [see below]) were identified as products of replication (negative and positive sense), as they were immunoprecipitated with anti-RS virus serum in the presence (Fig. 2, lane 7) and absence (data not shown) of pM2. This result indicated that they were encapsidated to form a ribonucleoprotein complex (28, 34). The upper band of the doublet was provisionally identified as the negative-sense product of replication, as it comigrated with an in vitro transcript of the negative-sense subgenomic replicon WT5 (Fig. 2, lane 6). While the negative- and positive-sense replication products are the same size, they can be separated by agarose-urea gel electrophoresis due to the difference in base composition (28).
FIG. 2.
Products of RNA synthesis from WT5 subgenomic replicon. Cells were infected with vTF7-3, transfected with cDNAs pWT5, pN, pP, pL, and pM2 (as indicated), and exposed to [3H]uridine in the presence of actinomycin D. Total RNA (lanes 1 to 3) was analyzed by agarose-urea gel electrophoresis. RNA extracted from cells transfected with pWT5, pN, pP, pL, and pM2 was subjected to oligo(dT) chromatography (lanes 4 and 5) or immunoprecipitation with anti-RS virus serum (lane 7) before agarose-urea gel electrophoresis; 2.5 times more cellular material was subjected to immunoprecipitation than was used for the total samples. A 3H-labeled T7 in vitro transcription product from pWT5 was run as a marker for the position of WT5 genomic RNA (lane 6). The − and + beside lane 7 signify the polarity of the replication products (rep.) in lanes 4 and 7 identified by the comigration of the negative-sense product with the in vitro transcript of WT5 in lane 6. UB, unbound fraction of oligo(dT) chromatography; B, bound fraction; IV, WT5 in vitro transcript; IP, RNA immunoprecipitated by anti-RSV serum.
Oligo(dT) chromatography was performed on RNA extracted from cells transfected with pWT5, pN, pP, pL, and pM2. The presence of a doublet of RNA in the unbound fraction (Fig. 2, lane 4) further confirmed the identity of the replication products and showed that the increase in RNA migrating in this position was not due to an increase in replication. Two major RNA species were retained by oligo(dT)-cellulose (Fig. 2, lane 5). Neither of these RNAs was immunoprecipitated by anti-RS virus serum (Fig. 2, lane 7). The faster-migrating RNA was produced in the presence and absence of pM2 (Fig. 2, lanes 2 and 3) and was tentatively identified as the predicted mRNA [547 nucleotides not including poly(A)]. The slower-migrating species was present in increased quantities in the presence of pM2 (Fig. 2, lane 3) and comigrated with the products of replication. However, as this RNA was not immunoprecipitated and was retained by oligo(dT)-cellulose, we postulated that it was a product of readthrough mRNA transcription into trailer. It was therefore provisionally designated r/t.
Primer extension analysis on positive-sense RNA confirmed the presence of RNAs with 5′ termini corresponding to those of the mRNA initiating at the NS1 start site and the positive-sense product of replication (data not shown). In the absence of the L protein (Fig. 2, lane 1), no [3H]uridine-labeled RNA was detected, demonstrating the requirement for the viral RNA-dependent RNA polymerase in order to generate RS virus-specific RNAs in the presence of actinomycin D.
Identification of RNA products by RNase H analysis.
The similar electrophoretic mobilities of the genomic RNAs and the polyadenylated r/t RNA synthesized in greater abundance upon addition of pM2 prevented the use of Northern blot analysis to definitively identify these RNAs. Therefore, digestion with RNase H following annealing of oligonucleotides specific for regions of the genome, antigenome, or mRNA was used to identify the products of RNA synthesis (3, 4). These analyses were performed on RNAs harvested from transfected cells in which neither M2 ORF1 nor ORF2 was present, or in which both ORFs of the M2 mRNA (pM2) were present, in order to determine their effect on viral RNA synthesis.
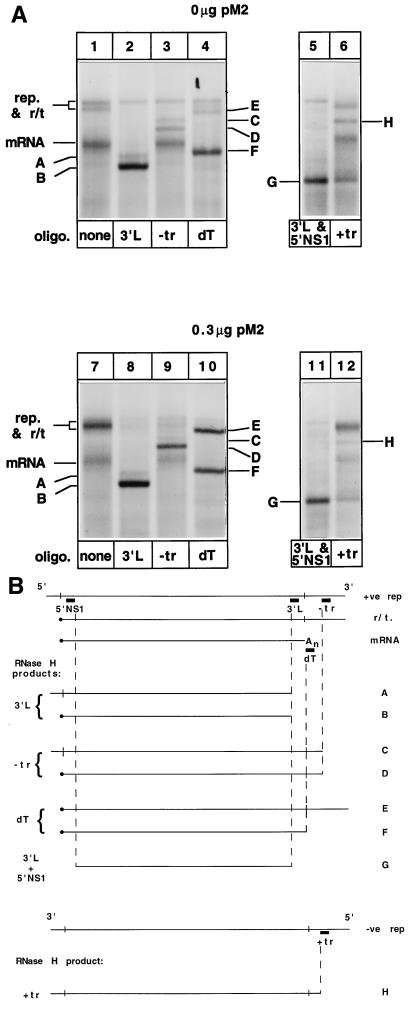
Figure 3 shows the identification of the products of RNA synthesis from the WT5 subgenomic replicon in the presence and absence of pM2 (encoding ORF1 and ORF2 of the M2 mRNA) and confirms the effect of pM2 on the pattern of RNA synthesis (Fig. 3A, lanes 1 and 7). These data also show that the products of RNA synthesis are not digested by RNase H in the absence of an oligonucleotide (Fig. 3A, lanes 1 and 7). However, in the presence of specific oligonucleotides, certain RNAs became susceptible to RNase H digestion, which allowed them to be identified.
FIG. 3.
Identification of RNAs produced from the WT5 template. (A) Labeled RNAs from transfections excluding (upper panel) or including (lower panel) 0.3 μg of pM2 (encoding ORF1 and ORF2 of M2 mRNA) were incubated with specific oligonucleotides (oligo.) as indicated, digested with RNase H, and separated by agarose-urea gel electrophoresis. Labels below the lanes designate which oligonucleotide was present during RNase H digestion. rep., replication products. (B) Diagrammatic representation of oligonucleotide binding sites and major products of RNase H digestion corresponding to RNAs A to H in panel A. +ve, positive; −ve, negative.
The RNA species identified as the mRNA was predicted to be polyadenylated and contain L sequence. This was confirmed by its sensitivity to RNase H digestion following annealing with oligo(dT) or an oligonucleotide which annealed to positive-sense L mRNA sequence (3′L). Digestion in the presence of oligo(dT) to remove the poly(A) tail caused a slight shift in migration and sharpening of the mRNA band, giving rise to band F (Fig. 3A, lanes 4 and 10). In the presence of the 3′L oligonucleotide, the mRNA band was digested by RNase H, giving rise to band B (Fig. 3A, lanes 2 and 8). The predicted 547-nucleotide product of transcription terminates at the L gene end signal and hence should be insensitive to RNase H digestion following incubation with an oligonucleotide which annealed to positive-sense trailer sequence (−tr). Digestion in the presence of −tr caused no shift in migration of the mRNA band (Fig. 3A, lanes 3 and 9), indicating that the positive-sense trailer sequence was not present in this RNA. Thus, we concluded that this RNA is the expected 547-nucleotide, polyadenylated mRNA product of transcription.
The positive-sense replication product is predicted to contain positive-sense leader, NS1 gene, L gene, and trailer sequences. Sensitivity to RNase H digestion following annealing of the 3′L and −tr oligonucleotides was used initially to identify this RNA. Following annealing with 3′L, all products of RNA synthesis except the negative-sense replication product (provisionally identified above) were sensitive to RNase H digestion, which produced bands A and B (Fig. 3A, lanes 2 and 8). Digestion in the presence of −tr did not alter the migration of the negative-sense replication product or the mRNA but did give rise to bands C and D (Fig. 3A, lanes 3 and 9). These products of RNase H digestion were of sizes consistent with the predicted products outlined in Fig. 3B. It was postulated that bands A and C migrated more slowly than B and D due to the presence of leader sequence (Fig. 3B) and, hence, represented the RNase H digestion products of the positive-sense replication product. To determine if this supposition was correct, a digestion was performed in the presence of oligonucleotides 3′L and 5′NS1 in order to remove both the 3′ and 5′ ends of all positive-sense RNAs. This digestion gave rise to a single band, G (Fig. 3A, lanes 5 and 11), indicating that the slower migration of bands A and C was due to the presence of leader sequence at their 5′ ends.
The r/t RNA, synthesized in greater abundance in the presence of pM2, and which migrated near the genomic RNAs but was retained by oligo(dT)-cellulose, was identified by RNase H digestion following annealing with oligonucleotide −tr. This digestion caused the loss of the positive replication product and r/t (Fig. 3A, lanes 3 and 9) and gave rise to bands C and D, indicating the presence of trailer sequence in these RNAs. The fact that band D migrated faster than band C indicated that it did not contain leader sequence; hence, it was concluded that band D was a digestion product of a readthrough transcript (band D migrated more slowly than the mRNA, as 83 nucleotides of trailer sequence still remain at its 3′ end following RNase H digestion). The level of band D, but not C, was increased when the cDNA encoding the M2 mRNA was included in transfections, thus indicating that a product encoded by the M2 mRNA was enhancing transcriptional readthrough at the L/trailer junction.
The final RNA species identified was the negative-sense product of replication. Following incubation with the oligonucleotide +tr, which anneals to negative-sense trailer sequence (Fig. 3B), RNase H digestion led to the disappearance of the slower-migrating replication product and the appearance of band H (Fig. 3A, lanes 6 and 12). No change in the migration of the positive-sense product of replication, r/t RNA, or the mRNA was seen. This result confirmed the slower-migrating product of replication was negative sense.
These analyses allowed us to identify the negative- and positive-sense products of replication, the predicted mRNA, and a readthrough product of transcription which failed to terminate at the L gene end and read through into trailer. This readthrough product was synthesized primarily in the presence of the M2 mRNA ORF1 and ORF2 (pM2). Although the trailer sequence does not contain an RS virus polyadenylation signal, the data show that the readthrough transcript was polyadenylated by its ability to bind to oligo(dT)-cellulose and its sensitivity to RNase H digestion in the presence of oligo(dT) (band E [lanes 4 and 10]). We postulate that the vaccinia virus poly(A) polymerase adds poly(A) to the 3′-terminal hydroxyl group of this readthrough transcript (18, 19). Encapsidation of the replication products probably prevents polyadenylation of these RNAs by this enzyme. Polyadenylation at the L/trailer junction by the RS virus polymerase followed by readthrough rather than termination can be eliminated as a possibility because if this were the case, exposure to RNase H in the presence of oligo(dT) would cause the digestion of the poly(A) tract and removal of the 3′ trailer sequence; therefore, product E (Fig. 3A, lanes 4 and 10) would not be observed.
Readthrough at an internal gene junction.
These results with a subgenomic replicon encoding one transcription unit demonstrated that the presence of M2 mRNA (ORF1 and ORF2) caused significant readthrough of the L gene transcription termination signal to give rise to an mRNA containing the trailer sequence. However, the L/trailer junction is not representative of the other RS virus gene junctions, as the L gene end signal is not followed by a downstream gene start signal. This may affect the efficiency of transcriptional termination and the behavior of the viral transcriptase. Therefore, the effect of the products of the M2 mRNA on transcription was analyzed by using a subgenomic replicon encoding two transcription units, separated by the authentic M/SH gene junction (Fig. 4A).
The predicted products of replication and transcription from the dicistronic subgenomic replicon, M/SH, as well as the possible readthrough products that could be generated are shown in Fig. 4A. The data in Fig. 4B show that in the absence of pM2 (lane 2), the major products of RNA synthesis were the positive and negative replication products (which comigrate), mRNA1 and mRNA2. In the presence of pM2 (Fig. 4B, lane 3), there was a significant change in the products of transcription. An RNA identified (see below) as the product of readthrough at the end of mRNA1 into mRNA2 (r/t.B) was significantly more abundant. A product of readthrough of mRNA1 into mRNA2 and then into trailer (r/t.C) was also more abundant. Readthrough of mRNA2 into trailer (r/t.A) was increased slightly.
The products of RNA synthesis were identified by RNase H analysis. Lanes 4 and 5 in Fig. 4B show the results of RNase H digestion of RNAs synthesized in the presence of pM2. The RNAs in lane 4 were digested in the presence of oligo(dT). This discriminated r/t.C from the products of replication, as it migrated faster when the poly(A) tail (probably added by a vaccinia enzyme) was removed. The RNAs in lane 5 were digested in the presence of oligo(dT) and an oligonucleotide (3′M) which annealed to sequence near the 3′ end of mRNA1 (the position at which this oligonucleotide anneals is indicated in Fig. 4A by arrows). Following annealing of 3′M, both r/t.B and r/t.C, in addition to mRNA1, were digested by RNase H, indicating they contained mRNA1 sequence as predicted. The 5′ products of digestion of mRNA1, r/t.B and r/t.C, comigrate and give rise to the major band in lane 5 (indicated by the asterisk). The RNA migrating between r/t.A and mRNA2 in lane 5 corresponds to the 3′ RNase H digestion product of r/t.B. Similar analysis using an oligonucleotide specific to mRNA2 resulted in RNase H digestion of r/t.A, r/t.B, and r/t.C in addition to mRNA2 (data not shown). This result confirmed the presence of mRNA2 sequence in each of the readthrough RNAs. Further RNase H analysis was performed to determine the identity of each RNA species; however, for the sake of brevity, those data are not shown.
The large increase in the quantity of r/t.B showed that a product of the M2 mRNA caused readthrough at an internal gene junction. Also, the presence of r/t.C at such high levels upon expression of the M2 gene demonstrated that the polymerase can read through more than one termination signal when the M2 mRNA products are present.
Assignment of readthrough function to ORF1 product.
Increased transcriptional readthrough of gene end signals was observed when the products of the M2 mRNA (ORF1 and ORF2) were present in transfections. To determine which of the two ORF products was responsible for causing this readthrough, cDNAs that expressed either ORF1 or ORF2 were generated (Fig. 5A). The cDNAs pM2 (encoding ORF1 and ORF2), pORF1, and pORF2 were added incrementally to transfections in order to determine the effects of increasing levels of ORF1 and ORF2 products. Figure 5B shows that the presence of pM2 caused an increase in readthrough when it was present at low levels in transfections. However, as the concentration of cDNA in transfections was increased, a corresponding decrease in viral RNA synthesis resulted. In contrast, expression of ORF1 alone resulted in an increase in readthrough transcription, but increasing the amount of cDNA, which led to a corresponding increase in protein expression (9, 20a, 29), did not have an inhibitory effect on RNA synthesis (Fig. 5B). ORF2 alone, however, significantly inhibited viral RNA synthesis even when expressed at low levels, as previously observed by Collins et al. (9). These data show that the product of ORF1, the M2 protein, caused the RS virus polymerase to read through transcription termination signals.
DISCUSSION
We have used an approach whereby the effects of trans-acting factors on replication and transcription of RS virus can be analyzed by direct metabolic labeling in the absence of helper virus (34). The products of transcription and replication of RS virus subgenomic replicons containing one or two transcription units were compared in the absence and presence of the cDNA pM2, encoding both ORF1 and ORF2 of the M2 mRNA. It was found that the expression of ORF1 and ORF2 caused a change in the pattern of RNA synthesis. Analysis of the RNA products by RNase H digestion in the presence of specific oligonucleotides demonstrated that the observed change in RNA synthesis was due to an increase in the production of readthrough transcripts (Fig. 3 and 4). Further, it was shown, using the M/SH dicistronic subgenomic replicon, that a product of either ORF1 or ORF2 allowed the viral polymerase to read through intergenic junctions as well as the L/trailer junction. Also, readthrough could occur at more than one termination signal to produce a transcription product containing mRNA1, mRNA2, and trailer sequence. This effect has not been reported in previous work using similar systems (9, 24).
Using cDNAs encoding only ORF1 or ORF2 of the M2 mRNA, we assigned the readthrough function to the product of ORF1, the 22-kDa M2 protein which is also a structural component of the RS virus virion. In contrast, the product of ORF2 dramatically downregulated both replication and transcription as previously reported (9).
Our conclusions concerning the function of the M2 protein differ from previous reports that M2 functions to enhance processive transcription but does not affect readthrough of transcription termination signals (9). We postulate that our different results may, in part, be due to the differences in the systems used: (i) these studies used direct RNA analysis rather than Northern blotting, (ii) the time of analysis posttransfection (a direct 5-h label at 16 h posttransfection in this report, compared to Northern blotting of total RNA harvested 42 h posttransfection in previous reports); and (iii) the agarose-urea gel system used here gave a higher resolution. Our finding that the M2 protein acts primarily as a transcription antiterminator does not conflict with but rather extends the basic finding by Collins et al. (9) that M2 enhances polymerase processivity. It has been demonstrated in other biological systems that factors which give rise to antitermination also increase polymerase processivity as part of the antitermination process (16).
During a natural RS virus infection, readthrough transcription occurs approximately 10% of the time (13, 24). This is significantly higher than observed with the prototypic nonsegmented negative-sense RNA virus, vesicular stomatitis virus, where readthrough occurs approximately 3% of the time (22). In fact, the readthrough transcripts of RS virus are so abundant that they were used to develop the first map of the gene order (13). Indirect evidence has also suggested that the efficiency of transcription termination at the end of the NS1 and NS2 genes is lower than for other RS virus genes (2, 23). These observations lend circumstantial support to the concept of RS virus encoding a protein which enhances transcriptional readthrough.
Examining the role played by transcription antitermination factors in other systems may help us understand why RS virus requires such a factor. Proteins such as N and Q of phage lambda allow the Escherichia coli RNA polymerase to access genes downstream of a termination site where the polymerase normally dissociates from the template (16). These factors play an essential role in balancing gene expression during the life cycle of the phage. The RS virus genome contains 10 transcriptional units (excluding the leader gene), and so the polymerase encounters nine transcription termination signals before it reaches the polymerase gene start site. Assuming that transcriptional attenuation occurs at each gene junction (15, 22, 24), this will lead to a diminishingly small number of polymerase molecules which reach the polymerase (L) gene start site. An antitermination factor that allows readthrough of transcription termination signals would counteract the severity of attenuation. This would allow more polymerase molecules to access promoter-distal genes as transcriptional attenuation is reduced, which may explain the requirement of the M2 protein in order to obtain recovery of virus from a full-length cDNA clone (8). We postulate that the M2 protein may function to allow more polymerase molecules deeper access into the genome, thus maintaining an optimal ratio of proteins by regulating transcription. A protein equivalent to M2 has not been identified in members of the Mononegavirales, which possess only five or six transcriptional units, and if our hypothesis is correct, one would not be required since transcriptional attenuation would not as severely restrict polymerase access to promoter distal genes.
The gene start signal for the L gene occurs prior to the gene end signal for the M2 gene, which means the two transcription units overlap. Previous reports have suggested that this overlap downregulates transcription of the L mRNA (12) in order to prevent overexpression of the L protein, which is believed to be detrimental to the virus (30). This would appear to be in direct contradiction to the theory we propose in which the M2 protein increases polymerase access to the L gene start. It is possible that the two regulatory mechanisms described above work as a check and balance to temporally control the level of L protein expression.
The finding that M2 enhances transcriptional readthrough suggests that RS virus possesses a second level of transcription regulation in addition to attenuation at the gene junctions. There are a number of possible mechanisms by which the M2 protein could prevent the polymerase from terminating transcription. We are currently investigating the specific manner by which the M2 protein causes transcriptional antitermination.
ACKNOWLEDGMENTS
We thank members of our laboratory and the L. A. Ball laboratory for advice and constructive criticism.
This work was supported by Public Health Service grants AI12464 and AI20181 from the NIH and NIAID to G.W.W.
REFERENCES
- 1.Alansari H, Potgieter L N D. Nucleotide sequence analysis of the ovine respiratory syncytial virus G glycoprotein. Virology. 1993;196:873–877. doi: 10.1006/viro.1993.1549. [DOI] [PubMed] [Google Scholar]
- 2.Barik S. Transcription of human respiratory syncytial virus genome RNA in vitro: requirement of cellular factor(s) J Virol. 1992;66:6813–6818. doi: 10.1128/jvi.66.11.6813-6818.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr J N, Whelan S P J, Wertz G W. Role of the intergenic dinucleotide in vesicular stomatitis virus RNA transcription. J Virol. 1997;71:1794–1801. doi: 10.1128/jvi.71.3.1794-1801.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavanagh D, Barrett T. Pneumovirus like characteristics of the mRNA and protein of turkey rhinotracheitis virus. Virus Res. 1988;11:241–256. doi: 10.1016/0168-1702(88)90086-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins P, Hill M, Johnson P. The two open reading frames of the 22K mRNA of human respiratory syncytial virus: sequence comparison of antigenic subgroups A and B and expression in vitro. J Gen Virol. 1990;71:3015–3020. doi: 10.1099/0022-1317-71-12-3015. [DOI] [PubMed] [Google Scholar]
- 6.Collins P L. The molecular biology of human respiratory syncytial virus (RSV) of the genus Pneumovirus. In: Kingsbury D, editor. The paramyxoviruses. New York, N.Y: Plenum Press; 1991. pp. 103–162. [Google Scholar]
- 7.Collins P L, Dickens L E, Buckler-White A, Olmstead R A, Spriggs M K, Camargo E, Coelingh K V W. Nucleotide sequences for the gene junctions of human respiratory syncytial virus reveal distinctive features of intergenic structure and gene order. Proc Natl Acad Sci USA. 1986;83:4594–4598. doi: 10.1073/pnas.83.13.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins P L, Hill M G, Camargo E, Grosfeld H, Chanock R M, Murphy B R. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci USA. 1995;92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins P L, Hill M G, Cristina J, Grosfeld H. Transcription elongation factor of respiratory syncytial virus, a non-segmented negative-strand RNA virus. Proc Natl Acad Sci USA. 1996;93:81–85. doi: 10.1073/pnas.93.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins P L, Huang Y T, Wertz G W. Identification of a tenth mRNA of respiratory syncytial virus and assignment of polypeptides to the 10 viral genes. J Virol. 1984;49:572–578. doi: 10.1128/jvi.49.2.572-578.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins P L, Mink M A, Stec D S. Rescue of synthetic analogs of respiratory syncytial virus genomic RNA and effect of truncations and mutations on the expression of a foreign reporter gene. Proc Natl Acad Sci USA. 1991;88:9663–9667. doi: 10.1073/pnas.88.21.9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins P L, Olmstead R A, Spriggs M K, Johnson P R, Buckler-White A J. Gene overlap and attenuation of transcription of the viral polymerase L gene of human respiratory syncytial virus. Proc Natl Acad Sci USA. 1987;84:5134–5138. doi: 10.1073/pnas.84.15.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins P L, Wertz G W. cDNA cloning and transcriptional mapping of nine polyadenylated RNAs encoded by the genome of human respiratory syncytial virus. Proc Natl Acad Sci USA. 1983;80:3208–3212. doi: 10.1073/pnas.80.11.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins P L, Wertz G W. The envelope-associated 22K protein of human respiratory syncytial virus: nucleotide sequence of the mRNA and a related polytranscript. J Virol. 1985;54:65–71. doi: 10.1128/jvi.54.1.65-71.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickens L E, Collins P L, Wertz G W. Transcriptional mapping of human respiratory syncytial virus. J Virol. 1984;52:364–369. doi: 10.1128/jvi.52.2.364-369.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freidman D I, Court D L. Transcription antitermination: the λ paradigm updated. Mol Microbiol. 1995;18:191–200. doi: 10.1111/j.1365-2958.1995.mmi_18020191.x. [DOI] [PubMed] [Google Scholar]
- 17.Garcia J, Garcia-Barreno B, Vivo A, Melero J. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that co-express the nucleoprotein, the phosphoprotein, and the 22K protein. Virology. 1993;195:243–247. doi: 10.1006/viro.1993.1366. [DOI] [PubMed] [Google Scholar]
- 18.Gershon P D, Moss B. Stimulation of poly (A) tail elongation by the VP39 subunit of the vaccinia virus-encoded poly (A) polymerase. J Biol Chem. 1993;268:2203–2210. [PubMed] [Google Scholar]
- 19.Gershon P D, Moss B. Uridylate-containing RNA sequences determine specificity of binding and polyadenylation by the catalytic subunit of vaccinia virus poly (A) polymerase. EMBO J. 1994;12:4705–4714. doi: 10.1002/j.1460-2075.1993.tb06159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grosfeld H, Hill M, Collins P L. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J Virol. 1995;69:5677–5686. doi: 10.1128/jvi.69.9.5677-5686.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Hardy, R. W. Unpublished data.
- 21.Huang Y T, Collins P L, Wertz G W. Characterization of the 10 proteins of human respiratory syncytial virus: identification of a fourth envelope associated protein. Virus Res. 1985;2:157–173. doi: 10.1016/0168-1702(85)90246-1. [DOI] [PubMed] [Google Scholar]
- 22.Iverson L E, Rose J K. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell. 1981;23:477–484. doi: 10.1016/0092-8674(81)90143-4. [DOI] [PubMed] [Google Scholar]
- 23.Kuo L, Fearns R, Collins P L. Analysis of the gene start and gene end signals of human respiratory syncytial virus: quasi-templated initiation at position 1 of the encoded mRNA. J Virol. 1997;71:4944–4953. doi: 10.1128/jvi.71.7.4944-4953.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo L, Fearns R, Collins P L. The structurally diverse intergenic regions of respiratory syncytial virus do not modulate sequential transcription by a dicistronic minigenome. J Virol. 1996;70:6143–6150. doi: 10.1128/jvi.70.9.6143-6150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo L, Grosfeld H, Cristina J, Hill M G, Collins P L. Effect of mutations in the gene-start and gene-end sequence motifs on transcription of monocistronic and dicistronic minigenomes of respiratory syncytial virus. J Virol. 1996;70:6892–6901. doi: 10.1128/jvi.70.10.6892-6901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laskey R. The use of intensifying screens or organic scintillators for visualizing radioactive molecules resolved by gel electrophoresis. Methods Enzymol. 1980;65:363–371. doi: 10.1016/s0076-6879(80)65047-2. [DOI] [PubMed] [Google Scholar]
- 27.Ling R, Easton A J, Pringle C R. Sequence analysis of the 22K, SH and G genes of the turkey rhinotracheitis virus and their intergenic regions reveals a gene order different from that of other pneumoviruses. J Gen Virol. 1992;73:1709–1715. doi: 10.1099/0022-1317-73-7-1709. [DOI] [PubMed] [Google Scholar]
- 28.Pattnaik A K, Ball L A, LeGrone A W, Wertz G W. Infectious defective interfering particles of VSV from transcipts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 29.Pattnaik A K, Wertz G W. Replication and amplification of defective interfering particle RNAs of vesicular stomatitis virus in cells expressing viral proteins from vectors containing cloned cDNAs. J Virol. 1990;64:2948–2957. doi: 10.1128/jvi.64.6.2948-2957.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schubert M, Harmison G G, Richardson C D, Meier E. Expression of a cDNA encoding a functional 241-kilodalton vesicular stomatitis virus RNA polymerase. Proc Natl Acad Sci USA. 1985;82:7984–7988. doi: 10.1073/pnas.82.23.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wertz G W, Davis N L. Charecterization and mapping of RNaseIII cleavage sites in vesicular stomatitis virus genome RNA. Nucleic Acids Res. 1981;9:6487–6503. doi: 10.1093/nar/9.23.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whelan S P J, Ball L A, Barr J N, Wertz G T W. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci USA. 1995;92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Worthington M T, Amann B T, Nathans D, Berg J M. Metal binding properties and secondary structure of the zinc binding domain of Nup475. Proc Natl Acad Sci USA. 1996;93:13754–13759. doi: 10.1073/pnas.93.24.13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu Q, Hardy R W, Wertz G W. Functional cDNA clones of the human respiratory syncytial (RS) virus N, P, and L proteins support replication of RS virus genomic RNA analogs and define the minimal trans-acting requirements for RNA replication. J Virol. 1995;69:2412–2419. doi: 10.1128/jvi.69.4.2412-2419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zamora M, Samal S K. Sequence analysis of M2 mRNA of bovine respiratory syncytial virus obtained from an F-M2 dicistronic mRNA suggests structural homology with that of human respiratory syncytial virus. J Gen Virol. 1992;73:737–741. doi: 10.1099/0022-1317-73-3-737. [DOI] [PubMed] [Google Scholar]