Abstract
Genes homologous to the herpes simplex virus UL49.5 open reading frame are conserved throughout the Herpesviridae. In the alphaherpesvirus pseudorabies virus (PrV), the UL49.5 product is an O-glycosylated structural protein of the viral envelope, glycoprotein N (gN) (A. Jöns, H. Granzow, R. Kuchling, and T. C. Mettenleiter, J. Virol. 70:1237–1241, 1996). For functional characterization of gN, a gN-negative PrV mutant, PrV-gNβ, and the corresponding rescuant, PrV-gNβR, were constructed, gN-negative PrV was able to productively replicate on noncomplementing cells, and one-step growth in cell culture was only slightly reduced compared to that of wild-type PrV. However, penetration was significantly delayed. In indirect immunofluorescence assays with rabbit serum directed against baculovirus-expressed gN, specific staining of wild-type PrV-infected cells occurred only after permeabilization of cells, whereas live cells failed to react with the antiserum. This indicates the lack of surface accessibility of gN in the plasma membrane of a PrV-infected cell. Western blot analyses and radioimmunoprecipitation experiments under reducing and nonreducing conditions led to the discovery of a heteromeric complex composed of gM and gN. The complex was stable in the absence of 2-mercaptoethanol but dissociated after the addition of the reducing agent, indicating that the partners are linked by disulfide bonds. Finally, gN was absent from gM-negative PrV virions, whereas gM was readily detected in virions in the absence of gN. Thus, gM appears to be required for virion localization of gN.
Herpesviruses express a large number of glycoproteins, most of which are present in the mature virion. Inserted into the viral envelope, they play an important role in virus entry into and egress from cells, and they represent prime targets for the immune response of the infected host. In the alphaherpesvirus pseudorabies virus (PrV), 11 glycoproteins have been described so far; they are glycoprotein B (gB), gC, gD, gE, gG, gH, gI, gK, gL, gM, and gN. Homologous proteins are also present in other members of the Alphaherpesvirinae. With the exception of gG, the PrV glycoproteins are virion constituents (8, 19, 24, 36).
Several of the glycoproteins form complexes. The gB complex consists of two gB monomers linked via disulfide bonds, which are posttranslationally proteolytically cleaved into two subunits each (44). The heterodimeric complex consisting of gE and gI is noncovalently linked (48) and functions in direct cell-to-cell spread in culture and in the nervous system (11, 26, 46). The gE-gI complex of herpes simplex virus type 1 (HSV-1) binds the Fc part of immunoglobulin G (17, 18). Another heteromeric complex consists of the essential gH and gL. HSV-1 gL is required for proper folding and surface expression of gH (16). Consequently, an HSV-1 mutant unable to express gL is noninfectious and fails to incorporate gH into virions (40). Similarly, gL expressed in the absence of gH was not detected at the cell surface or in the virion (16) but was released into the medium (12). Virions of PrV mutants defective in gH expression do not contain gL (25); however, in contrast to HSV-1, PrV gL is not required for virion localization of gH (23). Nonetheless, gL-negative PrV mutants are noninfectious, indicating an active role for gL in the function of the gH-gL complex. Whereas alphaherpesvirus gH-gL complexes are noncovalently linked, the homologous glycoproteins of the betaherpesviruses human cytomegalovirus and human herpesvirus 6 form disulfide-linked complexes (21, 32). A third protein present in the gH-gL complex was identified in human cytomegalovirus (15, 28) and Epstein-Barr virus (29).
Genes homologous to HSV-1 UL49.5 are conserved within the Herpesviridae (2, 4, 6, 7, 30, 34, 39, 41, 47). gN, the product of the PrV UL49.5 gene, was characterized as an O-glycosylated structural protein of the virion envelope (19). In contrast, in bovine herpesvirus 1 (BHV-1) (31) and HSV-1 (1), the gene product does not appear to be glycosylated. However, the BHV-1 homolog is also a membrane protein and a constituent of mature BHV-1 virions. In addition, it was shown to form a complex with an unidentified 39-kDa virion protein (31).
The only nonessential glycoprotein conserved throughout the Herpesviridae is gM, the product of the HSV-1 UL10 gene. PrV gM is a 45-kDa structural component of virions (8). PrV mutants unable to express gM are impaired in replication in cell culture, exhibit a delay in penetration, and show significantly decreased virulence in pigs, PrV’s natural host (9). PrV gM is found in oligomeric forms, presumably primarily dimers (10).
To analyze gN in more detail, particularly to assay for a phenotype associated with the absence of gN, we isolated a gN− PrV mutant. Here we report the characterization of the mutant virus and demonstrate the existence of a gM-gN complex in mature PrV virions.
MATERIALS AND METHODS
Viruses and cells.
Virus mutants were derived from wild-type PrV strain Ka (20). They were propagated on porcine (PK15), bovine (MDBK), or rabbit kidney (RK13) cells. Cells were grown in minimum essential medium with Earle’s salts supplemented with nonessential amino acids and 10% fetal calf serum. Transfections by calcium phosphate precipitation (13) were performed with African green monkey kidney (Vero) or RK13 cells. Penetration kinetics were assayed on ith MDBK and Vero cells, and immunofluorescence studies were performed with RK13 cells.
For baculovirus infection, Estigmea agrae (EaA) cells grown in Grace’s insect medium with 10% fetal calf serum were used. Baculovirus DNA was transfected into Spodoptera frugiperda High Five (H5) cells grown in SF-900 medium (Gibco-BRL, Eggenstein, Germany).
Construction of gN− PrV mutant.
Two plasmids derived from a 2-kb BamHI/SalI fragment of genomic BamHI fragment 1 by nested deletion (19) were used (Fig. 1). A 1,214-bp insert of pBK41 which comprises 66 bp of the 3′ end of the UL49.5 gene and adjacent downstream sequences was joined to a 717-bp fragment of pUSS83 containing 204 bp of the 5′ end of the UL49.5 gene and adjacent upstream sequences, leading to the elimination of 24 bp from the UL49.5 gene. The cloning strategy resulted in the introduction of a unique SstI cleavage site between the two fragments, which was used for the insertion of a gG–β-galactosidase expression cassette (37), yielding pU8341β. To construct a gN-expressing cell line, a 405-bp fragment containing the entire UL49.5 gene was subcloned into expression vector pRc/CMV (Invitrogen, Leek, The Netherlands) and used for transfections of Vero cells. Stable clones were selected in medium containing 800 μg of geneticin (Sigma, Deisenhofen, Germany) per ml (38). The expression of gN was verified by Western blotting, and one clone, C4/2, was further used.
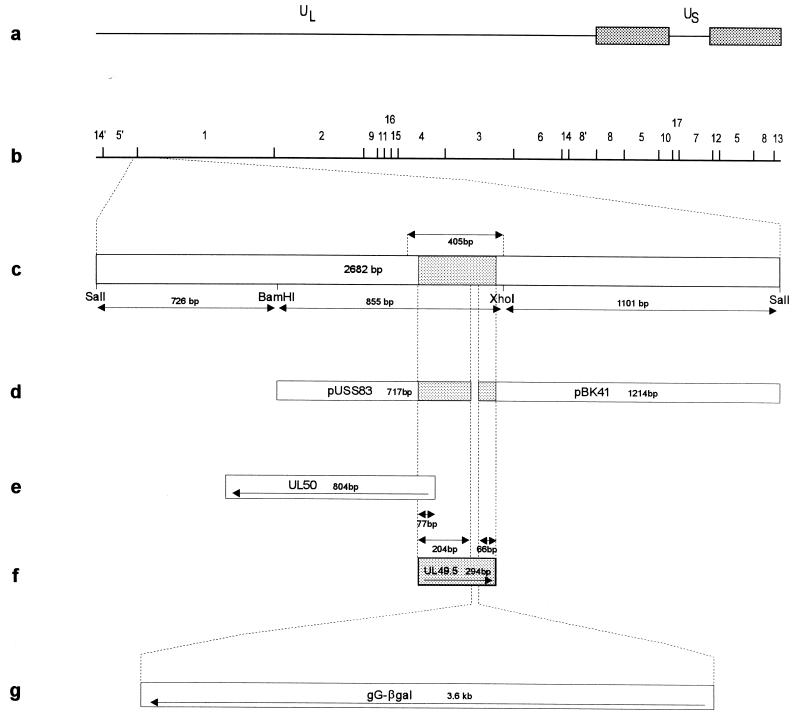
FIG. 1.
Schematic diagrams of the PrV genome and plasmids. (a) Diagram of the PrV genome, consisting of long (UL) and short unique (US) regions, with the latter bracketed by inverted repeats (stippled boxes). (b) BamHI restriction fragment map. (c) Relevant region of the genome is enlarged. The indicated 405-bp fragment was used for the establishment of the gN-expressing cell line. Inserts of plasmids pUSS83 and pBK41 (d) were joined to the gG–β-galactosidase (gG-βgal) cassette (g) for the construction of mutant PrV-gNβ. (e and f) Locations of UL50 and UL49.5 genes, respectively, are shown. The relevant restriction sites and sizes of fragments are indicated. The location of the UL49.5 open reading frame is shaded.
Wild-type PrV (Ka) DNA was cotransfected with pU8341β into C4/2 cells. Virus progeny were screened for the appearance of a blue-plaque phenotype under a Bluo-Gal (Bethesda Research Laboratories) agarose overlay. Blue plaques were picked by aspiration and purified five times. The genotype of mutant virus was verified by restriction analysis and Southern blotting. The lack of gN expression was monitored by Western blotting. One randomly selected isolate, PrV-gNβ, was further analyzed. To construct a corresponding rescuant, PrV-gNβ DNA was cotransfected with a 2.7-kbp fragment containing the UL49.5 gene. Virus progeny were assayed for a white-plaque phenotype under Bluo-Gal overlay. One isolate, PrV-gNβR, was randomly selected for further characterization.
Preparation of antiserum against baculovirus-expressed gN.
The UL49.5 gene was amplified by PCR with Deep Vent polymerase (New England Biolabs, Schwalbach, Germany) and the following primers: gN-1, 5′-TGGATCCAGGATGGTCTCCTCCG-3′ (positions 365 to 377 of GenBank accession no. U38547), and gN-2, 5′-CGAATTCGATCTTTATACGCGCCGCC-3′ (positions 82 to 62). By using the BamHI and EcoRI restriction sites contained in the primers, the PCR product was inserted into pVL1393 (PharMingen, San Diego, Calif.) under the control of the polyhedrin promoter, resulting in plasmid pVL1393gN. Ten days after cotransfection of BaculoGold DNA (PharMingen) and pVL1393gN into H5 cells, cells were freeze-thawed and supernatants were used for the infection of EaA cells. The expression of gN was monitored by Western blotting. Baculovirus-expressed gN appeared as a 12-kDa protein.
To prepare anti-gN serum, EaA cells were infected with recombinant gN-expressing baculovirus and lysed in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer 5 to 7 days after infection when the cytopathic effect was clearly visible. Proteins were separated by preparative SDS-PAGE on gels containing 14% polyacrylamide. The appropriate portion of the gel was excised, and gN was eluted with Biotrap (Schleicher & Schuell, Dassel, Germany). Eluted protein was dialyzed extensively against phosphate-buffered saline (PBS), emulsified with mineral oil, and used for immunization of a rabbit.
Metabolic labelling of proteins.
Confluent PK15 or RK13 cells were infected at a multiplicity of infection of 5. Six hours postinfection, the inoculum was replaced by minimal essential medium without either methionine, cysteine, or both, and cells were further incubated for 1 h at 37°C. Thereafter, the medium was replaced with fresh medium containing 50 μCi of Tran35S-label (3,000 Ci/mmol; ICN, Eschwege, Germany) per ml. Infected cells were labelled for 60 to 90 min. Thereafter, the medium was removed and cells were lysed in radioimmunoprecipitation assay buffer (10 mM sodium phosphate [pH 7.2], 10 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 40 mM sodium fluoride, 0.1% SDS, 1 μg of aprotinin per ml).
For the labelling of virion proteins, incubation with labelling medium was continued for 14 h. The supernatant was harvested, and virions were purified by centrifugation through a 30% sucrose cushion (5) and lysed by the addition of radioimmunoprecipitation assay buffer.
Radioimmunoprecipitation.
Radioimmunoprecipitation was performed essentially as previously described (33), except that cleared lysates were preadsorbed by the addition of 5 μl of rabbit preimmune serum. Proteins were separated by SDS-PAGE (27). For SDS-PAGE under nonreducing conditions, sample buffer without 2-mercaptoethanol was used. For two-dimensional SDS-PAGE, precipitates were separated under nonreducing conditions in the first dimension. After electrophoresis, one lane was cut off and incubated for 15 min in 0.5 M Tris-HCl (pH 6.8) and for 15 min in SDS-PAGE sample buffer with 10% 2-mercaptoethanol. The gel slice was placed horizontally on a second gel, and electrophoresis was performed. Proteins were visualized by fluorography.
Immunofluorescence.
RK13 cells grown on glass coverslips were infected at a multiplicity of infection of 0.01 and incubated for 16 h at 37°C. Thereafter, cells were fixed in acetone for 30 s, rehydrated with PBS for 10 min, and incubated with specific antisera for 2 h at room temperature in a humid chamber. After incubation with secondary antibody (goat anti-rabbit fluorescein isothiocyanate; Dianova, Hamburg, Germany) in a diluted (1:200) solution of 0.1% Evans’ blue for 1 h at room temperature, cells were washed twice with PBS and once with distilled water, air dried, and overlaid with mounting buffer containing 90% glycerol and 2.5% 1,4-diazabicyclo(2,2,2)-octane. For surface fluorescence assays, infected cells were washed with PBS and incubated with primary antibodies on ice for 1 h. Subsequent incubations were carried out as described above.
Penetration kinetics.
Entry kinetics were established as described before (35). Briefly, cells on six-well tissue culture dishes were infected with approximately 500 PFU of PrV(Ka), PrV-gNβ, or PrV-gNβR per well for 1 h at 4°C. The inoculum was removed, and cells were overlaid with prewarmed medium to initiate penetration. Immediately and at different times thereafter, extracellular virus was inactivated by treatment of the monolayer for 2 min with 40 mM citric acid–10 mM KCl–135 mM NaCl (pH 3.0). Cells were washed with PBS and overlaid with methylcellulose medium. After 2 days, plaques were counted and the percent penetration was established by comparison to PBS-treated cells.
RESULTS
Absence of gN impairs penetration.
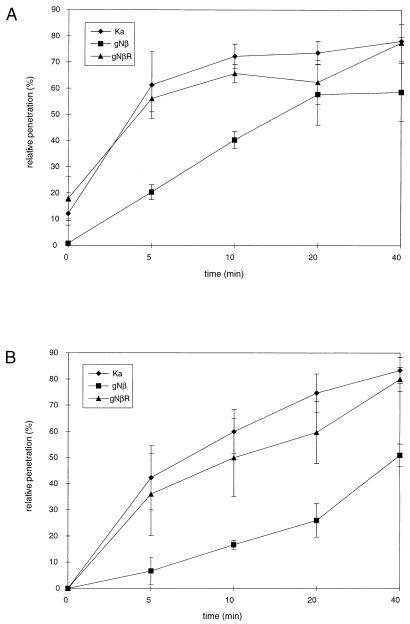
Mutant PrV-gNβ was originally isolated on complementing, gN-expressing cells. However, this virus mutant also replicated on noncomplementing cells, and one-step growth kinetics revealed only a slight reduction in the in vitro replication efficiency of PrV-gNβ compared to those of PrV(Ka) and PrV-gNβR. The final titers of PrV-gNβ consistently were approximately two- to fivefold lower (data not shown). To further search for phenotypic alterations associated with the absence of gN, the penetration kinetics of PrV-gNβ into MDBK and Vero cells were assayed in comparison with PrV(Ka) and PrV-gNβR. As shown in Fig. 2, the entry of PrV-gNβ was delayed in both cell lines, whereas PrV(Ka) and PrV-gNβR entered cells at similar rates. Thus, although gN is apparently not required for replication, its absence results in slower entry of PrV into target cells.
FIG. 2.
Penetration kinetics of gN− PrV. The entry kinetics of PrV(Ka), PrV-gNβ, and PrV-gNβR into MDBK (A) and Vero (B) cells were assayed by citrate inactivation of extracellular virus at different times after temperature shift. The percentage of plaques at a given time point calculated in comparison with PBS-treated control plates is shown. Data are averages of five independent experiments. Vertical lines indicate standard deviations.
Immunofluorescence in infected cells.
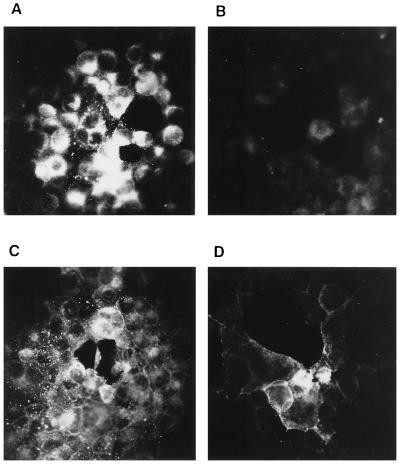
To test for intracellular presence and membrane localization of gN in infected cells, immunofluorescence assays were performed with anti-gN serum and permeabilized and live cells. As shown in Fig. 3A, permeabilized cells exhibited bright fluorescence, whereas gN was not stained by the antibodies in live nonpermeabilized cells (Fig. 3B). In contrast, gD was detected by a monospecific serum (22) in both permeabilized (Fig. 3C) and live (Fig. 3D) cells, demonstrating the localization of gD to the plasma membrane. This finding is surprising since gN appears to be accessible to antibodies in the virion envelope, as shown by immunoelectron microscopy (19). It is unclear whether gN is not translocated to the plasma membrane or whether it is present but not detectable by the antiserum used.
FIG. 3.
Immunofluorescence. Acetone-fixed (A and C) and live (B and D) RK13 cells infected with PrV(Ka) were incubated with anti-gN serum (A and B) or anti-gD serum (C and D). Bound primary antibodies were detected with a fluorescein isothiocyanate-conjugated secondary antibody, followed by fluorescence microscopy. Magnification, ×140.
gN forms a heteromeric disulfide-linked complex.
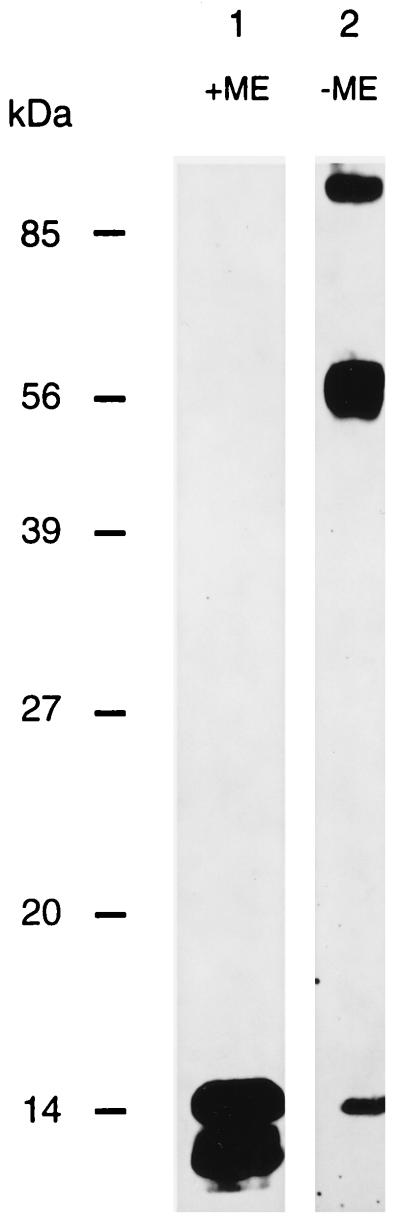
After electrophoresis under reducing conditions and Western blotting, anti-gN serum detected two proteins of 14 and 12 kDa in lysates of PrV(Ka) virions (Fig. 4, lane 1). The higher-molecular-weight protein represents mature O-glycosylated gN, whereas the lower-molecular-weight polypeptide presumably constitutes an under- or nonglycosylated form. Interestingly, under nonreducing conditions, only a small amount of 14-kDa gN was observed, whereas two other signals (at 56 and >85 kDa) appeared (Fig. 4, lane 2). This indicates that the major portion of intracellular gN is contained within disulfide-linked complexes. Since under reducing conditions only gN was detected, either the complexes consist solely of gN or they contain heterologous proteins which are not recognized by anti-gN serum after reduction.
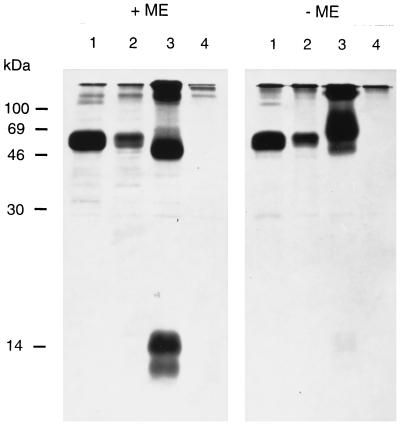
FIG. 4.
Western blot of purified virions. Gradient-purified PrV(Ka) virions were separated by SDS–13% PAGE under reducing (+ME) or nonreducing (−ME) conditions, transferred to nitrocellulose, and incubated with anti-gN serum. After incubation with peroxidase-conjugated secondary antibody, bound antibodies were detected by chemiluminescence. The positions of molecular mass markers are shown on the left. ME, 2-mercaptoethanol.
To identify the complex constituents, virion proteins were labelled with Tran35S-label (ICN), immunoprecipitated, and separated under reducing and nonreducing conditions (Fig. 5). Proteins of 14 and 12 kDa were detected after the precipitation of PrV(Ka) virions with anti-gN serum and electrophoresis under reducing conditions (Fig. 5, panel +ME, lane 3). In addition, proteins of 45 and >100 kDa were also precipitated. No gN protein was detected in precipitates from PrV-gNβ virions; the 45- and >100-kDa polypeptides were not detected (Fig. 5, panel +ME, lane 4). In contrast, gD was present in both virion preparations (Fig. 5, lanes 1 and 2). Under nonreducing conditions, prominent signals at 55 and >100 kDa were detected after the precipitation of PrV(Ka) virions with anti-gN serum, whereas minute amounts of 14-kDa gN and a 45-kDa protein were also observed. None of these proteins was precipitated from PrV-gNβ virions. Together, these data indicate that gN forms a disulfide-linked heteromeric 55-kDa complex with a 45-kDa protein. The >100-kDa signal may be derived from an oligomer of this complex.
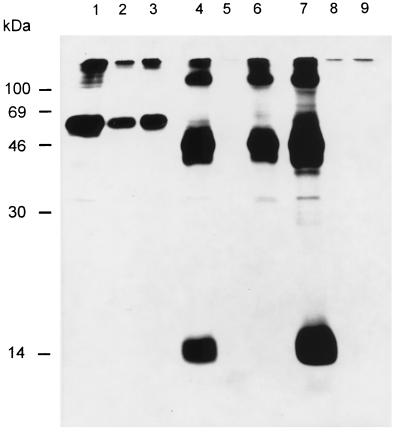
FIG. 5.
Radioimmunoprecipitation of virion proteins. PrV(Ka) (lanes 1 and 3) or PrV-gNβ (lanes 2 and 4) virions were labelled with Tran35S-label (ICN), purified, lysed, and precipitated with anti-gD serum (lanes 1 and 2) or anti-gN serum (lanes 3 and 4). Precipitates were separated by SDS–13% PAGE under reducing (+ME) or nonreducing (−ME) conditions and visualized by fluorography. The positions of molecular mass markers are shown on the left. ME, 2-mercaptoethanol.
To directly demonstrate that gN is contained in the 55-kDa complex, a two-dimensional SDS-PAGE of the virion proteins metabolically labelled with Tran35S-label (ICN) and precipitated with anti-gN serum was performed. The first dimension was run under nonreducing conditions in SDS–10% PAGE. As shown in Fig. 6A, the 55-kDa complex was precipitated from PrV(Ka) virions. Under these conditions, monomeric gN migrated with the gel front. A lane similar to the one shown in Fig. 6A was excised, placed horizontally on an SDS–13% acrylamide gel, and run under reducing conditions. As shown in Fig. 6B, 14-kDa gN was released from the 55-kDa complex. The broad smear ranging from 45 to 69 kDa presumably comprised the 45-kDa protein of the complex and possibly residual nonreduced 55-kDa complex. Monomeric gN was found at the expected position. Thus, gN is present in the 55-kDa complex, which appears to consist of a 45-kDa protein disulfide linked to gN.
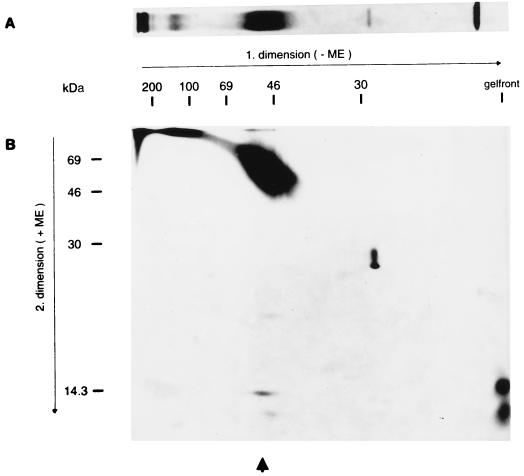
FIG. 6.
Radioimmunoprecipitation and two-dimensional electrophoresis. (A) The lysate of purified PrV(Ka) virions labelled with Tran35S-label (ICN) was precipitated with anti-gN serum. The precipitate was separated by SDS–10% PAGE under nonreducing (−ME) conditions and visualized by fluorography. (B) An identical lane was cut off the gel and placed horizontally on an SDS–13% acrylamide gel after incubation in reducing buffer (+ME). After electrophoresis, proteins were visualized by fluorography. The positions of molecular mass markers are indicated. The arrow denotes the position of the dissociated complex. ME, 2-mercaptoethanol.
The 45-kDa complex partner is gM.
The presence of a 45-kDa protein and a polypeptide with approximately twice that molecular mass is reminiscent of the appearance of gM on Western blots (8). To analyze whether gM and the 45-kDa complex partner are identical, radioimmunoprecipitations of cell lysates infected with PrV(Ka) (Fig. 7, lanes 1, 4, and 8), PrV-ΔgMβ (lanes 2, 5, and 9) (9), and PrV-gNβ (lanes 3, 6, and 10) were performed. As controls, noninfected-cell lysates were included (Fig. 7, lanes 7 and 11). Lysates were precipitated with anti-gD serum (Fig. 7, lanes 1 through 3), anti-gN serum (lanes 4 through 7), or anti-gM serum (lanes 8 through 11). Anti-gD serum precipitated similar amounts of gD from all lysates (Fig. 7, lanes 1 through 3). In addition, as expected, the 45-kDa mature gM and the 35-kDa precursor (10) were precipitated by anti-gM serum from PrV(Ka)- and PrV-gNβ-infected cells (Fig. 7, lanes 8 and 10) but were absent in PrV-ΔgMβ-infected cells (lane 9). Interestingly, proteins of 35 and 45 kDa were coprecipitated with gN from PrV(Ka)-infected cells (Fig. 7, lane 4) but were absent in PrV-ΔgMβ-infected cells (lane 5). From PrV-ΔgMβ-infected cells, only gN was precipitated by anti-gN serum. These results demonstrate that the 45- and 35-kDa proteins which were coprecipitated with gN by anti-gN serum from lysates of PrV(Ka)-infected cells indeed represent different forms of gM. Although in this exposure only the intracellular 12-kDa form of gN was visible, a longer exposure resulted in the detection of the 14-kDa mature gN and also showed coprecipitation of minor amounts of gN with anti-gM serum (data not shown).
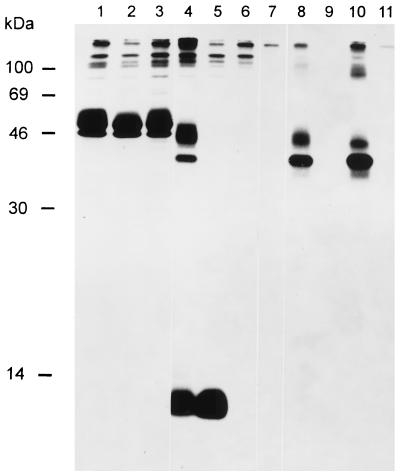
FIG. 7.
Demonstration of a gM-gN complex. Lysates of cells infected with PrV(Ka) (lanes 1, 4, and 8), PrV-ΔgMβ (lanes 2, 5, and 9), or PrV-gNβ (lanes 3, 6, and 10) or of noninfected cells (lanes 7 and 11) labelled with Tran35S-label (ICN) were precipitated with anti-gD serum (lanes 1 through 3), anti-gN serum (lanes 4 through 7), and anti-gM serum (lanes 8 through 11). Precipitates were separated by SDS–13% PAGE and visualized by fluorography. The positions of molecular mass markers are indicated on the left.
PrV-ΔgMβ virions lack gN.
To test for the presence of gN in PrV virions in the absence of gM, labelled PrV(Ka) (Fig. 8, lanes 1, 4, and 7), PrV-ΔgMβ (lanes 2, 5, and 8), and PrV-gNβ (lanes 3, 6, and 9) virions were precipitated with antiserum directed against gD (lanes 1 through 3), gM (lanes 4 through 6), or gN (lanes 7 through 9) and precipitates were separated under reducing conditions. Anti-gM serum coprecipitated 45-kDa mature gM and 14-kDa mature gN from PrV(Ka) virions (Fig. 8, lane 4). From PrV-gNβ virions, only gM was precipitated (Fig. 8, lane 6); from PrV-ΔgMβ virions, no protein was specifically precipitated (lane 5), as expected. Anti-gN serum also coprecipitated gM and gN from PrV(Ka) virions (Fig. 8, lane 7) and failed to precipitate a protein from PrV-gNβ virions (lane 9). However, anti-gN serum also failed to precipitate a protein from PrV-ΔgMβ virions (Fig. 8, lane 8). This indicates that the presence of gM is required for virion localization of gN, whereas gM is found in the virion in the presence and absence of gN.
FIG. 8.
Absence of gN in gM− virions. Purified virions of PrV(Ka) (lanes 1, 4, and 7), PrV-ΔgMβ (lanes 2, 5, and 8), and PrV-gNβ (lanes 3, 6, and 9) labelled with Tran35S-label were lysed and precipitated with anti-gD serum (lanes 1 through 3), anti-gM serum (lanes 4 through 6), and anti-gN serum (lanes 7 through 9). Precipitates were separated by SDS–13% PAGE and visualized by fluorography. The positions of molecular mass markers are indicated on the left.
DISCUSSION
We previously reported that the product of the PrV gene homologous to HSV-1 UL49.5 is represented by an O-glycosylated structural component of the virion, gN (19). The salient findings of the present study are as follows. (i) gN is nonessential for PrV replication in cell culture, but its absence leads to a delay in penetration. (ii) gN is not accessible to antibodies on the surfaces of infected cells, as analyzed by immunofluorescence. (iii) gN forms a stable, disulfide-linked complex with gM. (iv) gN requires gM for virion localization.
Although our initial attempts to isolate a gN-negative PrV mutant on normal Vero cells were not successful and the isolation of PrV-gNβ was achieved only on gN-expressing, complementing cells, gN proved to be nonessential for viral replication in tissue culture. One-step growth kinetics revealed only marginal growth impairment. The failure to isolate this mutant virus on noncomplementing cells may have been associated with its delayed penetration, which would lead to a selective advantage for the wild-type phenotype. In line with this finding, earlier attempts to isolate a UL49.5-negative HSV-1 mutant were also unsuccessful (3). Whether HSV-1 UL49.5 is indeed essential remains to be seen. It is interesting, however, that the BHV-1 homolog is dispensable for replication (30), as is the PrV protein (this report).
Previously, the only serological reagent available to detect PrV gN was a rabbit serum directed against a bacterially expressed gN fusion protein lacking the first 18 amino acids of the deduced gN sequence (19). Although that serum was valuable in the first detection of gN on Western blots, it was unsatisfactory in other serological assays. Therefore, a serum against baculovirus-expressed gN was generated; it detected gN in Western blot, immunoprecipitation, and immunofluorescence assays (this report) but did not neutralize PrV infectivity in either the absence or presence of complement (data not shown). The failure to neutralize virus infectivity parallels the lack of reactivity of this antiserum with the surfaces of virus-infected cells. In contrast, immunoelectron microscopy showed labelling by anti-gN serum of seemingly intact virion envelopes in negatively-stained preparations, which correlates with the hydrophobicity profile prediction of a type 1 membrane protein (19, 43). One possible explanation for this discrepancy is that the external domain of gN is masked by interactions with other virion structures, e.g., gM, and that this masking is eliminated during the preparation of samples for electron microscopy. In addition, in infected cells, gN may not be transported efficiently to the surface or may be retrieved from there. Since PrV derives its envelope from trans-Golgi network vesicles (14, 45), virion envelope proteins need not be present in the plasma membrane for virion localization. Therefore, the exact orientation of gN in the virion envelope still has to be established unequivocally.
The most important result of these studies is the demonstration of a complex between gN and gM. Both proteins are apparently conserved throughout the Herpesviridae, which is remarkable since both have been found to be nonessential for viral replication, at least in PrV. The existence of a complex was demonstrated by coprecipitation of a 45-kDa protein with anti-gN antibodies, a molecular mass which corresponds to that of mature gM, and by the absence of this protein in a gM deletion mutant. Disulfide linkage of the complex constituents was indicated by the sensitivity of the complex to reducing conditions and by the release of gN after reduction of the complex in situ, as demonstrated by two-dimensional gel electrophoresis.
For BHV-1, it was reported that the 8-kDa UL49.5 protein forms a disulfide-linked complex with another virion component of 39 kDa which may be located in the tegument, based on nonionic-detergent partition of purified virions (31). gM is predicted to be a multiply-membrane-spanning glycoprotein with eight predicted transmembrane domains (8). Thus, gM may interact tightly with tegument structures; this could influence its behavior in partition experiments. However, no characterization of the 39-kDa BHV-1 protein has so far been published. Given our data for the formation of a disulfide-linked complex between PrV gN and gM, it seems reasonable to speculate that BHV-1 gM is also part of a complex which involves the UL49.5 protein.
The deduced amino acid sequences of UL49.5 homologs (2–4, 6, 7, 19, 30, 39, 41, 42, 47) show poor overall conservation. The only strictly conserved amino acid is a cysteine residue in the N-terminal part of each deduced protein adjacent to the putative membrane anchor region, which corresponds to residue 49 in PrV gN. In the complex partner, gM, there is also one cysteine that is conserved in all gM homologs; it is located between the first and second hydrophobic domains (8). Since the single N-glycosylation site of PrV gM, which is the only N-glycosylation site conserved in all gM proteins, is also located in this region, this hydrophilic stretch must be exposed to the endoplasmic reticulum lumen and consequently to the exterior of infected cells and virions. Thus, the conserved cysteine must also be positioned on the outside of the virion. As predicted from the deduced amino acid sequence, gN is a type 1 membrane protein (4, 19), and the conserved cysteine is expected to be exposed on the virion surface. Thus, an interaction between the two conserved cysteine residues appears to be possible. Site-directed mutagenesis of respective codons should finally clarify this issue.
Coimmunoprecipitates of gN and gM from infected-cell lysates contained immature and mature forms of both proteins, indicating that complex formation occurs at an early stage in biosynthesis. Anti-gN serum precipitated the 12-kDa immature and 14-kDa mature forms of gN and the 35-kDa immature and 45-kDa mature forms of gM. Anti-gM serum also precipitated these forms. From cells infected with PrV-ΔgMβ, mainly the 12-kDa gN precursor was precipitated by anti-gN serum. This could be indicative of a delay in or inhibition of the maturation of gN in the absence of gM. A pulse-chase analysis of this phenomenon is under way. The results of our coprecipitation experiments (Fig. 7 and 8) also indicate that intracellularly (and possibly in virions) there is a significant amount of noncomplexed gM, whereas most of gN appeared to be complexed to gM.
Lastly, gN was not detected in PrV virions in the absence of gM. A similar situation is found for another glycoprotein complex, gH-gL. gL requires the presence of gH for virion localization. Most likely, gL is anchored in the virion envelope via gH since it lacks a predicted transmembrane anchor (25). In contrast, PrV gH reaches the envelope in the absence of gL. For the gM-gN complex, gN requires gM for virion localization, whereas gM is present in the virion in the absence of gN. So far, it is unclear why gN is excluded from virions in the absence of gM. As predicted from the secondary structure, gN specifies a carboxy-terminal transmembrane anchor. Thus, the dependence of gN on gM for anchoring in the virion envelope appears to be unlikely. However, formation of the complex early after synthesis may be required for proper intracellular sorting of gN, which eventually leads to integration into virions. In the absence of gM, this sorting may be impaired or abolished. In agreement with this assumption, mainly the immature 12-kDa precursor form of gN was detected in lysates of cells infected with gM-negative PrV.
The fact that gN requires gM for virion localization may have implications for the functional defects previously ascribed to the absence of gM (8, 9). gM-negative PrV mutants produce 10- to 50-fold-reduced titers in cell culture and exhibit a delay in penetration into MDBK and Vero cells. Whereas the gN− mutant did not appear to be significantly impaired in replication, it exhibited a similar delay in penetration. Thus, the replication defect of gM− PrV can be ascribed to the absence of gM, whereas the defect in penetration may be due to the lack of gM, gN, or both. Since virions containing gM but missing gN exhibit an impairment of penetration similar to that of virions lacking gM (and consequently gN), it seems reasonable to assume that gN alone or the gM-gN complex is involved in penetration.
gM-negative PrV also exhibited a strongly attenuated phenotype in its natural host, pigs (9). As discussed above, it is unclear whether this phenotype is attributable to gM or gN solely or to the gM-gN complex. Presumably, both the less efficient replication observed in the absence of gM and the penetration defect associated with the lack of gN contribute to the attenuated character of PrV-ΔgMβ.
Experiments to isolate a PrV mutant unable to express both gM and gN are under way to help in further elucidating the function of the gM-gN complex in PrV replication.
ACKNOWLEDGMENTS
We thank E. Mundt for help with the preparation of anti-gN serum.
This work was supported by grant Me 854/3-3 from the DFG.
REFERENCES
- 1.Adams, R., C. Cunningham, M. D. Davison, C. A. MacLean, and A. J. Davison. Personal communication.
- 2.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull T, Hudson G S, Satchwell S C, Seguin C, Tuffnell P S, Barrell B G. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature (London) 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 3.Barker D E, Roizman B. The unique sequence of the herpes simplex virus type 1 L component contains an additional translated open reading frame designated UL49.5. J Virol. 1992;66:562–566. doi: 10.1128/jvi.66.1.562-566.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnett B C, Dolan A, Telford E A R, Davison A J, McGeoch D. A novel herpes simplex virus gene (UL49A) encodes a putative membrane protein with counterparts in other herpesviruses. J Gen Virol. 1992;73:2167–2171. doi: 10.1099/0022-1317-73-8-2167. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Porat T, DeMarchi J, Kaplan A S. Characterization of defective interfering viral particles present in a population of pseudorabies virions. Virology. 1974;60:29–37. doi: 10.1016/0042-6822(74)90239-6. [DOI] [PubMed] [Google Scholar]
- 6.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C A, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein coding content of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 7.Davison A J, Scott J E. The complete DNA sequence of varicella zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 8.Dijkstra J M, Visser N, Mettenleiter T C, Klupp B G. Identification and characterization of pseudorabies virus glycoprotein gM as a nonessential virion component. J Virol. 1996;70:5684–5688. doi: 10.1128/jvi.70.8.5684-5688.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dijkstra J M, Gerdts V, Klupp B G, Mettenleiter T C. Deletion of glycoprotein gM of pseudorabies virus results in attenuation for the natural host. J Gen Virol. 1997;78:2147–2151. doi: 10.1099/0022-1317-78-9-2147. [DOI] [PubMed] [Google Scholar]
- 10.Dijkstra J M, Mettenleiter T C, Klupp B G. Intracellular processing of pseudorabies virus glycoprotein M (gM): gM of strain Bartha lacks N-glycosylation. Virology. 1997;237:113–122. doi: 10.1006/viro.1997.8766. [DOI] [PubMed] [Google Scholar]
- 11.Dingwell K S, Brunetti C R, Hendricks R L, Tang Q, Tang M, Rainbow A J, Johnson D C. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J Virol. 1994;68:834–845. doi: 10.1128/jvi.68.2.834-845.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubin G, Jiang H. Expression of herpes simplex virus type 1 glycoprotein L (gL) in transfected mammalian cells: evidence that gL is not independently anchored to cell membranes. J Virol. 1995;69:4564–4568. doi: 10.1128/jvi.69.7.4564-4568.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 14.Granzow H, Weiland F, Jöns A, Klupp B G, Karger A, Mettenleiter T C. Ultrastructural analysis of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huber M T, Compton T. Characterization of a novel third member of the human cytomegalovirus glycoprotein H-glycoprotein L complex. J Virol. 1997;71:5391–5398. doi: 10.1128/jvi.71.7.5391-5398.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutchinson L, Browne H, Wargent V, Davis-Poynter N, Primorac S, Goldsmith K, Minson A C, Johnson D C. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein gH (gH) and affects normal folding and surface expression of gH. J Virol. 1992;66:2240–2250. doi: 10.1128/jvi.66.4.2240-2250.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson D C, Feenstra V. Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J Virol. 1987;61:2208–2216. doi: 10.1128/jvi.61.7.2208-2216.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson D C, Frame M C, Ligas M W, Cross A M, Stow N D. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J Virol. 1988;62:1347–1354. doi: 10.1128/jvi.62.4.1347-1354.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jöns A, Granzow H, Kuchling R, Mettenleiter T C. The UL49.5 gene of pseudorabies virus codes for an O-glycosylated structural protein of the viral envelope. J Virol. 1996;70:1237–1241. doi: 10.1128/jvi.70.2.1237-1241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan A S, Vatter A. A comparison of herpes simplex and pseudorabies viruses. Virology. 1959;7:394–407. doi: 10.1016/0042-6822(59)90068-6. [DOI] [PubMed] [Google Scholar]
- 21.Kaye J F, Gompels U A, Minson A C. Glycoprotein H of human cytomegalovirus (HCMV) forms a stable complex with HCMV UL115 gene product. J Gen Virol. 1992;73:2693–2698. doi: 10.1099/0022-1317-73-10-2693. [DOI] [PubMed] [Google Scholar]
- 22.Klupp B, Visser N, Mettenleiter T C. Identification and characterization of pseudorabies virus glycoprotein H. J Virol. 1992;66:3048–3055. doi: 10.1128/jvi.66.5.3048-3055.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klupp B, Fuchs W, Weiland E, Mettenleiter T C. Pseudorabies virus glycoprotein L is necessary for virus infectivity but dispensable for virion localization of glycoprotein H. J Virol. 1997;71:7687–7695. doi: 10.1128/jvi.71.10.7687-7695.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klupp, B. G., J. Baumeister, P. Dietz, H. G. Granzow, and T. C. Mettenleiter. Submitted for publication.
- 25.Klupp B G, Baumeister J, Karger A, Visser N, Mettenleiter T C. Identification and characterization of a novel structural glycoprotein in pseudorabies virus, gL. J Virol. 1994;68:3868–3878. doi: 10.1128/jvi.68.6.3868-3878.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kritas S, Pensaert M, Mettenleiter T C. Role of gI, gp63 and gIII in the invasion and spread of Aujeszky’s disease virus (ADV) in the olfactory nervous pathway of pigs. J Gen Virol. 1994;75:2319–2327. doi: 10.1099/0022-1317-75-9-2319. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Li L, Nelson J A, Britt W J. Glycoprotein H-related complexes of human cytomegalovirus: identification of a third protein in the gCIII complex. J Virol. 1997;71:3090–3097. doi: 10.1128/jvi.71.4.3090-3097.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q, Turk S M, Hutt-Fletcher L M. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J Virol. 1995;69:3987–3994. doi: 10.1128/jvi.69.7.3987-3994.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang X, Tang M, Manns B, Babiuk L A, Zamb T J. Identification and deletion mutagenesis of the bovine herpesvirus 1 dUTPase gene and a gene homologous to herpes simplex virus UL49.5. Virology. 1993;195:42–50. doi: 10.1006/viro.1993.1344. [DOI] [PubMed] [Google Scholar]
- 31.Liang X, Chow B, Raggo C, Babiuk L A. Bovine herpesvirus 1 UL49.5 homolog gene encodes a novel viral envelope protein that forms a disulfide-linked complex with a second virion structural protein. J Virol. 1996;70:1448–1454. doi: 10.1128/jvi.70.3.1448-1454.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu D X, Gompels U A, Nicholas J, Lelliott C. Identification and expression of the human herpesvirus 6 glycoprotein H and interaction with an accessory 40K glycoprotein. J Gen Virol. 1993;74:1847–1857. doi: 10.1099/0022-1317-74-9-1847. [DOI] [PubMed] [Google Scholar]
- 33.Lukács N, Thiel H-J, Mettenleiter T C, Rziha H-J. Demonstration of three major species of pseudorabies virus glycoproteins and identification of a disulfide-linked glycoprotein complex. J Virol. 1985;53:166–173. doi: 10.1128/jvi.53.1.166-173.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 35.Mettenleiter T C. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology. 1989;171:623–625. doi: 10.1016/0042-6822(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 36.Mettenleiter T C. Pseudorabies (Aujeszky’s disease) virus: state of the art. Acta Vet Hung. 1994;42:153–177. [PubMed] [Google Scholar]
- 37.Mettenleiter T C, Rauh I. A glycoprotein gX-β-galactosidase fusion gene as insertional marker for rapid identification of pseudorabies virus mutants. J Virol Methods. 1990;30:55–66. doi: 10.1016/0166-0934(90)90043-f. [DOI] [PubMed] [Google Scholar]
- 38.Rauh I, Weiland F, Fehler F, Keil G M, Mettenleiter T C. Pseudorabies virus mutants lacking the essential glycoprotein gII can be complemented by glycoprotein gI of bovine herpesvirus 1. J Virol. 1991;65:621–631. doi: 10.1128/jvi.65.2.621-631.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riggio M P, Onions D E. DNA sequence of a gene cluster in the equine herpesvirus-4 genome which contains a newly identified herpesvirus gene encoding a membrane protein. Arch Virol. 1993;133:171–178. doi: 10.1007/BF01309752. [DOI] [PubMed] [Google Scholar]
- 40.Roop C, Hutchinson L, Johnson D C. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J Virol. 1993;67:2285–2297. doi: 10.1128/jvi.67.4.2285-2297.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Telford E A R, Watson M S, McBride K, Davison A J. The DNA sequence of equine herpesvirus-1. Virology. 1992;189:304–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- 42.Virgin H W, Latreille P, Wamsley P, Hallsworth K, Weck K, Dal Canto A, Speck S. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whealy M E, Robbins A K, Enquist L W. The export pathway of the pseudorabies virus gB homolog gII involves oligomer formation in the endoplasmic reticulum and protease processing in the Golgi apparatus. J Virol. 1990;64:1946–1955. doi: 10.1128/jvi.64.5.1946-1955.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whealy M E, Card J P, Meade R P, Robbins A K, Enquist L W. Effect of brefeldin A on alphaherpesvirus membrane protein glycosylation and virus egress. J Virol. 1991;65:1066–1081. doi: 10.1128/jvi.65.3.1066-1081.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whealy M E, Card J P, Robbins A K, Dubin J R, Rziha H-J, Enquist L W. Specific pseudorabies virus infection of the rat visual system requires both gI and gp63 glycoproteins. J Virol. 1993;67:3786–3797. doi: 10.1128/jvi.67.7.3786-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yanagida N, Yoshida S, Nazerian K, Lee L F. Nucleotide and predicted amino acid sequences of Marek’s disease virus homologues of herpes simplex virus major tegument proteins. J Gen Virol. 1993;74:1837–1845. doi: 10.1099/0022-1317-74-9-1837. [DOI] [PubMed] [Google Scholar]
- 48.Zuckermann F, Mettenleiter T C, Schreurs C, Sugg N, Ben-Porat T. Complex between glycoproteins gI and gp63 of pseudorabies virus: its effect on virus replication. J Virol. 1988;62:4622–4626. doi: 10.1128/jvi.62.12.4622-4626.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]