Take Home Message
This study highlights the significance of genetic ancestry in renal cell carcinoma (RCC) risk assessment, urges carful interpretation of CHEK2’s role in RCC predisposition, and uncovers understudied inherited mutations potentially increasing RCC risk.
Keywords: Renal cell carcinoma, Population stratification, Germline pathogenic variants, Cryptic splice variant, Copy number variant, CHEK2-associated cancer risk
Abstract
Background and objective
Previous germline studies on renal cell carcinoma (RCC) have usually pooled clear and non–clear cell RCCs and have not adequately accounted for population stratification, which might have led to an inaccurate estimation of genetic risk. Here, we aim to analyze the major germline drivers of RCC risk and clinically relevant but underexplored germline variant types.
Methods
We first characterized germline pathogenic variants (PVs), cryptic splice variants, and copy number variants (CNVs) in 1436 unselected RCC patients. To evaluate the enrichment of PVs in RCC, we conducted a case-control study of 1356 RCC patients ancestry matched with 16 512 cancer-free controls using approaches accounting for population stratification and histological subtypes, followed by characterization of secondary somatic events.
Key findings and limitations
Clear cell RCC patients (n = 976) exhibited a significant burden of PVs in VHL compared with controls (odds ratio [OR]: 39.1, p = 4.95e-05). Non–clear cell RCC patients (n = 380) carried enrichment of PVs in FH (OR: 77.9, p = 1.55e-08) and MET (OR: 1.98e11, p = 2.07e-05). In a CHEK2-focused analysis with European participants, clear cell RCC (n = 906) harbored nominal enrichment of low-penetrance CHEK2 variants—p.Ile157Thr (OR: 1.84, p = 0.049) and p.Ser428Phe (OR: 5.20, p = 0.045), while non–clear cell RCC (n = 295) exhibited nominal enrichment of CHEK2 loss of function PVs (OR: 3.51, p = 0.033). Patients with germline PVs in FH, MET, and VHL exhibited significantly earlier age of cancer onset than patients without germline PVs (mean: 46.0 vs 60.2 yr, p < 0.0001), and more than half had secondary somatic events affecting the same gene (n = 10/15, 66.7%). Conversely, CHEK2 PV carriers exhibited a similar age of onset to patients without germline PVs (mean: 60.1 vs 60.2 yr, p = 0.99), and only 30.4% carried somatic events in CHEK2 (n = 7/23). Finally, pathogenic germline cryptic splice variants were identified in SDHA and TSC1, and pathogenic germline CNVs were found in 18 patients, including CNVs in FH, SDHA, and VHL.
Conclusions and clinical implications
This analysis supports the existing link between several RCC risk genes and RCC risk manifesting in earlier age of onset. It calls for caution when assessing the role of CHEK2 due to the burden of founder variants with varying population frequency. It also broadens the definition of the RCC germline landscape of pathogenicity to incorporate previously understudied types of germline variants.
Patient summary
In this study, we carefully compared the frequency of rare inherited mutations with a focus on patients’ genetic ancestry. We discovered that subtle variations in genetic background may confound a case-control analysis, especially in evaluating the cancer risk associated with specific genes, such as CHEK2. We also identified previously less explored forms of rare inherited mutations, which could potentially increase the risk of kidney cancer.
1. Introduction
Renal cell carcinoma (RCC) is the ninth most common neoplasm in the USA, accounting for 2% of all cancers worldwide [1]. The Nordic Twin study has placed the genetic heritability of RCC as high as 38% [2]; however, only a fraction of the heritability is explained by the currently identified rare and common RCC risk loci. Moving beyond known RCC risk genes (Supplementary Table 1) [3], [4], several pan-RCC studies have reported rare germline pathogenic variants (PVs) in DNA damage repair (DDR) genes such as CHEK2, ATM, or BRCA1/2 [5], [6], [7], [8], [9], [10], suggesting that inherited defects in DDR may contribute to RCC risk. However, most studies lacked ancestry-matched cancer-free controls to formally test these hypotheses.
Two recent studies performed case-control gene-level burden analyses, in RCC alone [11] and across cancer types, finding a higher burden of germline PVs in CHEK2 in RCC patients than in matched controls [12]. However, these studies pooled all RCC subtypes together as one phenotype for association testing, although clear cell RCC (ccRCC) and non–clear cell RCC (nccRCC; eg, papillary and chromophobe) have distinct molecular and clinical features [13], [14]. Furthermore, additional analyses are necessary to account for fine-level population stratification within Europe to mitigate spurious association [15], especially when evaluating genes such as CHEK2, which is known to harbor many putative PVs that are founder variants from bottlenecked populations (eg, Ashkenazi Jewish [ASJ]) with highly variable allele frequencies between different European subpopulations.
Here, we first performed a germline variant discovery analysis of 1436 unselected RCC patients to characterize several types of genomic variation. Next, we performed a case-control association study of ccRCC and nccRCC in a subset (n = 1356) that was ancestry matched successfully with 16 512 cancer-free controls, and we utilized an ancestry-informed generalized linear model (GLM) to evaluate the major germline drivers of RCC risk. Furthermore, we performed a sub-European ancestry-focused meta-analysis of CHEK2 to address finer-level population stratification within European populations, and we evaluated associated tumor genomic data for concomitant somatic assessments of candidate PVs. Finally, we evaluated potential clinically relevant but underexplored germline variant types (cryptic splice and copy number variants [CNVs]) by using integrative genomic and transcriptomic analyses, all toward expanding and refining the landscape of germline pathogenic variation in RCC.
2. Patients and methods
2.1. RCC patient and cancer-free control cohorts
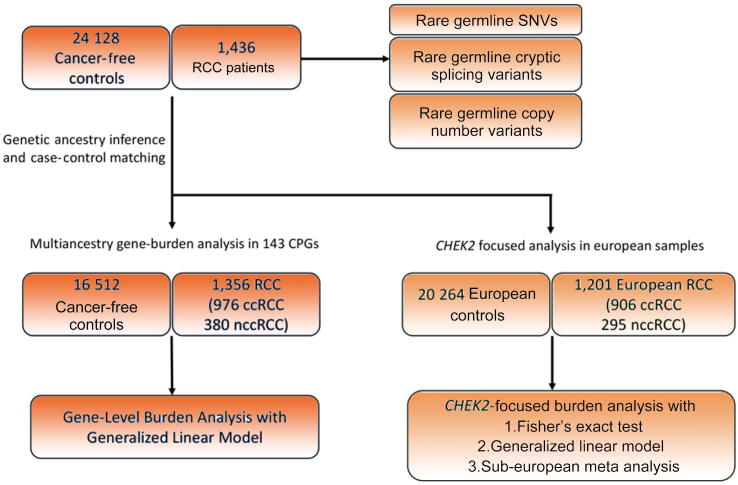
Whole-exome sequencing (WES) binary alignment maps (BAMs) aligned to Genome Research Consortium human build 37 (GRCh37) from 1436 RCC patients were collected from eight different RCC studies (Fig. 1, Table 1, Supplementary Table 2, and Supplementary material). WES BAMs from a total of 24 128 adult unrelated individuals without known cancer diagnosis were collected from five different studies available on the Database of Genotypes and Phenotypes (dbGAP; Supplementary material). All case and control samples underwent identical quality control procedures and were processed using the same analytical methods. This study was approved by the participating institutions where written consent from participants was collected. This study conforms to the Declaration of Helsinki.
Fig. 1.
Overview of the study. Rare germline PVs, cryptic splice variants, and copy number variants were characterized in 1436 RCC patients. The gene-level burden analysis was restricted to 1356 RCC patients ancestry matched with 16 512 cancer-free controls after genetic ancestry inference and case-control pair matching (Supplementary material). The CHEK2 focused analysis was restricted to 1201 European RCC patients and 20 264 European cancer-free controls. ccRCC = clear cell RCC; CPG = cancer predisposition gene; nccRCC = non–clear cell RCC; PV = pathogenic variant; RCC = renal cell carcinoma; SNV = single nucleotide variant.
Table 1.
Patient characteristics of all 1436 renal cell carcinoma patients
| Renal cell carcinoma case cohort (N = 1436) | |||
|---|---|---|---|
| n | % | ||
| Age at diagnosis | 10–19 | 1 | 0.1 |
| 20–29 | 10 | 0.7 | |
| 30–39 | 46 | 3.2 | |
| 40–49 | 191 | 13.3 | |
| 50–59 | 397 | 27.6 | |
| 60–69 | 469 | 32.7 | |
| 70–79 | 249 | 17.3 | |
| 80< | 55 | 3.8 | |
| Age unknown | 18 | 1.3 | |
| Self-identified gender | Male | 980 | 68.2 |
| Female | 456 | 31.8 | |
| Histology | Clear cell RCC | 1031 | 71.8 |
| Papillary RCC | 302 | 21.0 | |
| Chromophobe RCC | 103 | 7.2 | |
| Original study | CHECKMATE 025 | 446 | 31.1 |
| TCGA KIRC | 372 | 25.9 | |
| TCGA KIRP | 285 | 19.8 | |
| ICGC RECA EU | 95 | 6.6 | |
| TCGA KICH | 76 | 5.3 | |
| CHECKMATE 010 | 61 | 4.2 | |
| CHECKMATE 009 | 57 | 4.0 | |
| GENETECH Non ccRCC | 44 | 3.1 | |
| Inferred ancestry | European | 1201 | 83.6 |
| African | 131 | 9.1 | |
| Admixed American | 79 | 5.5 | |
| East Asian | 19 | 1.3 | |
| South Asian | 6 | 0.4 | |
ccRCC = clear cell RCC; ICGC = International Cancer Genome Consortium; RCC = renal cell carcinoma; TCGA = The Cancer Genome Atlas.
2.2. Evaluation of exome sequencing coverage
The average sample-level sequencing coverage was calculated using the genome analysis toolkit (GATK [16]; version 3.7) tool “DepthofCoverage” to ensure that all BAMs of cases and controls had sufficient read counts to confidently call germline variants. The exome-wide mean coverage of 10× was considered the minimum acceptable coverage to ensure confident germline variant detection. WES samples of 1436 RCC patients had a mean sample coverage of 116.82 (median = 114.96), and those of 16 512 ancestry-matched controls used for a burden analysis had a mean sample coverage of 96.69 (median = 90.48; Supplementary Fig. 1).
2.3. Germline variant detection from WES data
Germline variants were called from the BAM files using a deep learning–based variant discovery method, DeepVariant (version 0.8.0, docker: gcr.io/deepvariant-docker/deepvariant:0.8.0) [17], which had demonstrated superior sensitivity and specificity to GATK-based joint genotyping [18], [19]. Final sets of high-quality variants were merged into cohort-level VCF files using the GATK (version 3.7) tool “CombineVariants.” Subsequently, the “vt” tool (version 3.13) was used on the cohort VCF files to normalize and decompose multiallelic variants.
2.4. Genetic relatedness analysis
We performed a genetic relatedness analysis on the cohort VCF files in two steps. In the first step, we implemented the GENESIS (version 2.12.0) tool PC-AiR [20] to perform a principal component analysis (PCA) on the detected germline variants for the detection of population structure in the case and control cohorts, respectively. We then used the GENESIS tool “PC-Relate” [21] implemented in “Hail” (version 0.2.11; https://github.com/hail-is/hail) [22] to estimate kinship coefficients between every possible pair within a cohort. We removed one sample out of each pair that had a kinship coefficient above 0.125, which indicates genetic relatedness within second-degree relatives.
2.5. Genetic ancestry inference
First, cohort VCF files for cases and controls were combined with a cohort VCF file of 1000 Genomes Project [23] samples (n = 2504) with known continental ancestries. Next, the combined VCF file was loaded into a matrix table using Hail (version 0.2.11; https://github.com/hail-is/hail), and rare germline variants with a cohort allele frequency below 1% and deviating from Hardy-Weinberg equilibrium (chi-square p < 1 × 10–6) were excluded. We next performed linkage disequilibrium pruning using the Hail “ld_prune” method, and the resulting filtered germline variants were used for PCA using the Hail “hwe_normalized_pca” method. Finally, Sklearn (version 0.20.0) “RandomForestClassifier” function was applied to the top ten global principal components (PCs) of reference samples from the 1000 Genomes Project to train random forest classifiers for the five continental ancestries, which were used to uniformly assign continental ancestry to the cases and controls (Supplementary Fig. 2A).
2.6. Ancestry pair matching of cases and controls
Once continental ancestry was assigned, cases and controls were divided into each continental ancestry group, and the second round of PCA was performed to identify continental ancestry-specific PCs. We then used the “pairmatch” function of the R optmatch (version 0.9-14) package to identify control samples that were closest to each case based on the top ten PCs. To ensure an equivalent representation of each ancestry group, we applied a fixed 1:12 ratio between the number of cases and controls across all continental ancestry groups, and AMR (Admixed American) cases and controls were excluded in the gene-level burden analysis due to the limited number of AMR control samples failing to meet the case-control ratio (Supplementary Fig. 2B and 2C).
2.7. Sub-European ancestry inference and ASJ inference
For sub-European ancestry inference, the same inference approach was repeated, but only using samples identified as Europeans. The top ten PCs and sub-European ancestry labels from the 1000 Genomes European samples were used to train a random forest classifier. Since ASJ individuals were unable to be identified using the above approach, we used SNPweights [24] software with precalculated SNPweights from ASJ reference samples to identify ASJ individuals (Supplementary Fig. 3). Samples with ASJ proportion >0.5 were defined as ASJ. In the end, European cases and controls were divided into Northwestern Europeans (including Utah residents with Northern and Western Europeans [CEU] and British [GBR]), Southern Europeans (Iberian [IBS] and Toscani [TSI] excluding ASJ), Finnish, and ASJ.
2.8. Functional and clinical annotation and prioritization of germline variants
Germline variants in the cohort VCF files were annotated using Variant Effect Predictor (version 104.3) [25]. A curated list of 143 cancer predisposition genes (CPGs; Supplementary Table 3) was used to identify candidate rare (minor allele frequency [MAF] <1%) germline PVs. All identified variants were then classified using the American College of Medical Genetics classification [26] provided by VarSome [27] website (accessed between September and November 2022). Variants classified as likely pathogenic or pathogenic are collectively referred to as PVs.
2.9. Gene-level burden analysis with a GLM
To perform a gene-level burden analysis, a null model based on a GLM, as implemented in the Python “statsmodel” library (version 0.13.2) [28], was first constructed using the top ten global PCs from ancestry inference as covariates and RCC case status as the dependent variable. For each gene with at least one PV in RCC cases or controls, a corresponding extended model incorporating a burden indicator variable representing the presence of a PV in the gene for every sample was constructed. A likelihood ratio test was then performed between the null model and each extended model, and the resulting test statistics were adjusted for the false discovery rate (FDR) using the Benjamini-Hochberg procedure with FDR = 0.05. The burden of three low-penetrance CHEK2 variants defined by a recent study [12]—CHEK2 c.470T>C (p.Ile157Thr), c. 1283C>T (p.Ser428Phe), and c.1427C>T (p.Thr476Met)—were evaluated separately from the pathogenic loss of function (LOF) variants identified in CHEK2.
2.10. Statistical analysis and data visualization
Odds ratios (ORs), 95% confidence intervals (CIs), and p values for two-sided Fisher’s exact test were computed as implemented in the exact2x2 R package. Adjusted p values (q values) were computed based on the Benjamini-Hochberg procedure with FDR = 0.05. A one-way analysis of variance test was run using the “f_oneway” function from Python “scipy” library (version 1.5.2) [29], and post hoc pairwise comparisons were performed where applicable using the “pairwise_tukeyhsd” function from Python “statsmodels” library. Sample proportion CIs were calculated using the R “prop.test” function. For the meta-analysis, association statistics from sub-European groups were combined using a fixed-effect meta-analysis implemented using the “metafor“ R package [30]. All figures were generated using Python “Seaborn” (version 0.11.0) [31] and “Matplotlib” (version 3.3.2) packages, and were further refined using Adobe Photoshop 2021. The commutation plot summarizing the germline and somatic variants in RCC cases was generated using Python “CoMut” package (version 0.0.3; https://github.com/vanallenlab/comut) [32].
2.11. Identification of somatic variants and copy number events
For carriers of germline PVs in VHL, MET, FH, and CHEK2, somatic variant data provided from The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC) were downloaded from the Genomic Data Commons (GDC) data portal (https://portal.gdc.cancer.gov/) and ICGC data portal (https://dcc.icgc.org/), respectively (accessed: October 2022). For somatic variants, LOF truncating variants as well as missense variants with oncogenic annotation from OncoKB were included. For samples from CheckMate studies, somatic variants and copy number alterations were identified with the CGA WES characterization pipeline (https://github.com/broadinstitute/CGA_Production_AnalysisPipleline; Supplementary material).
2.12. Identification and validation of cryptic splice variants
SpliceAI (version 1.3.1; https://github.com/Illumina/SpliceAI) [33] was used to identify cryptic splice variants among the called germline variants detected by DeepVariant, and rare germline variants with SpliceAI score over 0.5 were defined as putative cryptic splice variants. For samples carrying a cryptic splice variant identified from SpliceAI, available tumor or germline mRNA BAM files were manually reviewed using Integrative Genomics Viewer (version 2.11.1) to visualize and evaluate their splicing patterns.
2.13. CNV detection from WES data
We applied GATK-gCNV to detect rare germline CNVs from exome sequencing data [34]. To minimize the discrepancy among the different exome sequencing baits used for different sources, the GATK (version 4.1.9.0) tool “CollectReadCounts” was used to gather read counts on the 8441 sequencing bait regions unique to seven major capture kits, and PCA was run to make batches of samples sequenced using the same sequencing bait (Supplementary Fig. 6A). From each identified batch, germline CNVs were detected using GATK-gCNV [34] following the best practices on the Terra platform (https://app.terra.bio/#workspaces/help-gatk/Germline-CNVs-GATK4). The detected CNVs were harmonized and filtered using the gCNV filtering R scripts downloaded from the gCNV repository (https://github.com/theisaacwong/talkowski/tree/master/gCNV; Supplementary material)
3. Results
3.1. Patient characteristics of RCC discovery case cohort
We collected WES data from 1436 RCC patients unselected for earlier age of disease onset or positive family history from eight independent RCC studies (Fig. 1 and Table 1). Of the patients, 71.8% had ccRCC (n = 1031), while the rest had nccRCC, including papillary (n = 302, 21.0%) and chromophobe (n = 103, 7.2%) RCC. Broad continental-level genetic ancestry inference (Supplementary Fig. 2) identified most of the cohort as being of predominantly European ancestry (83.6%, n = 1200), followed by African (9.1%, n = 131), Admixed American (5.6%, n = 80), East Asian (1.3%, n = 19), and South Asian (0.4%, n = 6) ancestry.
3.2. Prevalence of rare germline PVs in ccRCC and nccRCC
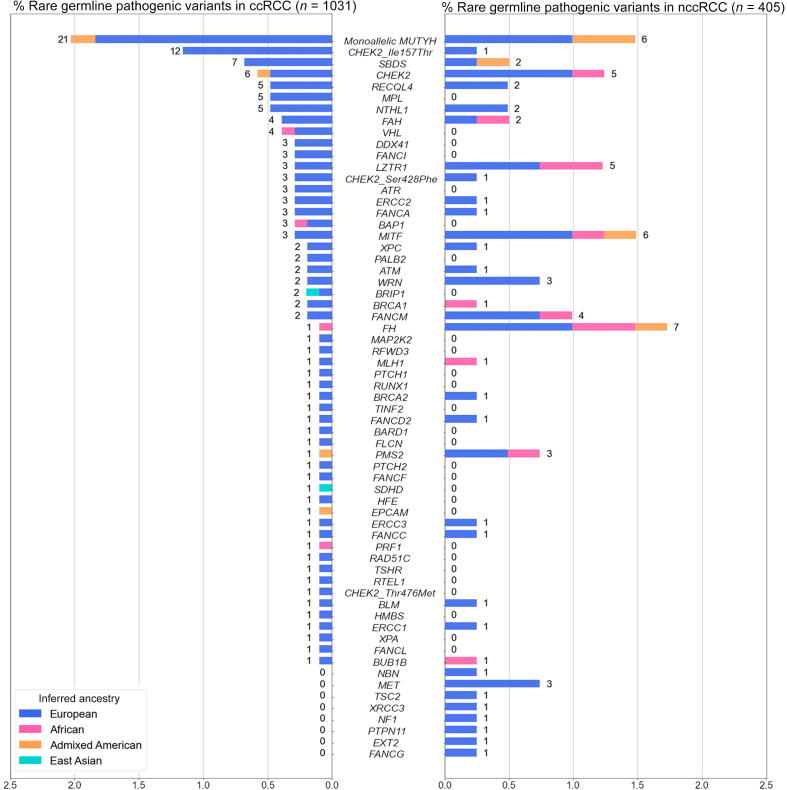
We first evaluated rare (MAF <1%) germline variants that met the existing clinical interpretation guidelines [26] as pathogenic or likely pathogenic in 1031 ccRCC patients (Fig. 2). In known RCC risk genes, we identified rare germline PVs in VHL (n = 4, 0.38%, 95% CI: 0.12–1.1%), BAP1 and MITF (n = 3 each, 0.29%, 95% CI: 0.075–0.92%), and FLCN, FH, and SDHD (n = 1 each, 0.097%, 95% CI: 0.0051–0.63%). When evaluating DDR genes (Supplementary Table 4), we identified 52 ccRCC patients who harbored rare germline PVs in homologous recombination or Fanconi Anemia genes such as CHEK2, RECQL4, FANCA, or BRCA1/2 (5.04%, 95% CI: 3.82–6.60%); 26 in base excision repair genes MUTYH and NTHL1 (2.52%, 95% CI: 1.69–3.73%); eight in nucleotide excision repair genes ERCC1, ERCC2, ERCC3, XPA, and XPC (0.77%, 95% CI: 0.36–1.59%); and two in mismatch repair genes MLH1 and PMS2 (0.19%, 95% CI: 0.033–0.79%). Overall, 131 ccRCC patients carried one or more heterozygous rare germline PVs (12.71%, 95% CI: 10.77–14.93%; Supplementary Table 5)—13 in previously established RCC risk genes (1.26%, 95% CI: 0.71–2.21%), 86 in DDR genes (8.34%, 95% CI:6.76-10.24%), and 37 in rest of the germline CPGs (3.59%, 95% CI: 2.57–4.96%). Among these, nine ccRCC patients carried rare germline PVs in two different CPGs (0.87%, 95% CI: 0.43–1.71%; Supplementary Table 7).
Fig. 2.
Rare germline pathogenic variants identified in the 1436 RCC patients. The proportion of ccRCC or nccRCC patients carrying rare (MAF <1%) germline pathogenic variants in one of the 143 germline cancer predisposition genes was tested. Three known CHEK2 low-penetrance variants were treated separately from the rest of the PVs in CHEK2. Numbers next to the bars indicate the counts of RCC patients carrying PVs in each gene. ccRCC = clear cell RCC; MAF = minor allele frequency; nccRCC = non–clear cell RCC; PV = pathogenic variant; RCC = renal cell carcinoma.
In parallel, we also characterized rare germline PVs in 405 nccRCC patients (Fig. 2). Rare germline PVs were found in the following known kidney cancer risk genes: seven in FH (1.72%, 95% CI: 0.76–3.69%), six in MITF (1.48%, 95% CI: 0.60–3.36%), three in MET (0.74%, 95% CI: 0.19–2.33%), and one in TSC2 (0.25%, 0.013–1.59%). Regarding DDR genes, 24 germline PVs were detected in homologous recombination or Fanconi Anemia genes (5.93%, 95% CI: 3.91–8.81%), eight in base excision repair genes with PVs in ccRCC—MUTYH and NTHL1 (1.98%, 95% CI: 0.92–4.01%), and four each in mismatch repair and nucleotide excision repair genes (0.99%, 95% CI: 0.32–2.69% each). Altogether, one or more rare pathogenic germline PVs were detected in 68 nccRCC patients (16.79%, 95% CI: 13.35–20.87%; Supplementary Table 6)—17 in known RCC risk genes (4.20%, 95% CI: 2.54–6.77%), 39 in DDR genes (9.63%, 95% CI: 7.02–13.03%), and 13 in other CPGs (3.21%, 95% CI: 1.79–5.57%). Four patients were identified with rare germline PVs in two different CPGs (0.99%, 95% CI: 0.32–2.69%; Supplementary Table 7). Thus, the relatively higher proportion of patients with identified germline PVs in DDR genes, which was mainly driven by rare germline PVs in CHEK2 and MUTYH (3.90%, n = 56/1436 across RCC subtypes, 95% CI: 2.98–5.07%), was consistent with the observations in the pan-RCC patients from previous RCC studies [6], [8], [9], [10].
3.3. Gene-level enrichment of rare germline PVs in ccRCC and nccRCC patients
To investigate whether the identified PVs predispose individuals to an increased risk of RCC, we performed genetic ancestry inference and case-control pair matching to link 1356 RCC patients (of the original 1436; 94.4% of the RCC cohort) with 16 512 cancer-free controls (Supplementary material and Supplementary Fig. 2), and compared the gene-level burden of rare germline PVs in ccRCC (n = 976) and nccRCC (n = 380) separately against the cancer-free controls. To further account for residual population stratification not captured in our ancestry matching procedure, we conducted a gene-level burden analysis for each gene using a GLM that accounts for continental ancestry (Supplementary material). As expected, ccRCC patients exhibited significantly higher enrichment of germline PVs in VHL than the ancestry-matched controls (OR: 39.1, 95% CI: 7.01–218.07, p = 4.95e-05, q value: 0.00584), and in the low-penetrance CHEK2 p.Ser428Phe variant (OR: 31.96, 95% CI: 6.23–163.89, p = 0.000385, q value: 0.0227) after multiple-hypothesis correction (Table 2). Patients with ccRCC also carried a nominally higher frequency of PVs in the two other known kidney cancer risk genes, BAP1 and SDHD, as well as in the common low-penetrance CHEK2 p.Ile157Thr variant (p < 0.05, q values >0.05; Supplementary Table 8).
Table 2.
Gene-level burden analysis of clear cell and non–clear cell RCC
| Gene/variant | FDR adjusted p value | p value | Odds ratio | OR 95% CI low | OR 95% CI high | Case PV carrier | Control PV carrier | |
|---|---|---|---|---|---|---|---|---|
| Clear cell RCC | VHL | 0.00584 | 4.95E-05 | 39.1 | 7.01 | 218.07 | 4 | 2 |
| CHEK2_S428F | 0.0227 | 0.000385 | 31.96 | 6.23 | 163.89 | 3 | 3 | |
| SDHD | 0.506 | 0.0141 | 1.73E + 10 | 0 | Inf | 1 | 0 | |
| BAP1 | 0.526 | 0.0314 | 5.57 | 1.44 | 21.52 | 3 | 9 | |
| CHEK2_I157T | 0.526 | 0.0356 | 2.04 | 1.11 | 3.73 | 12 | 122 | |
| Non–clear cell RCC | FH | 1.83E-06 | 1.55E-08 | 77.9 | 18.68 | 324.97 | 6 | 3 |
| MET | 2.07E-05 | 3.50E-07 | 1.98E + 11 | 0 | Inf | 3 | 0 | |
| LZTR1 | 0.244 | 0.00619 | 4.96 | 1.92 | 12.85 | 5 | 42 | |
| PMS2 | 0.653 | 0.0241 | 5.58 | 1.65 | 18.84 | 3 | 25 | |
| MITF | 0.653 | 0.0277 | 3.38 | 1.34 | 8.53 | 5 | 62 | |
| CHEK2_S428F | 0.707 | 0.036 | 27.73 | 2.78 | 276.8 | 1 | 3 | |
| EXT2 | 0.79 | 0.0483 | 22.36 | 2 | 250.27 | 1 | 2 |
CI = confidence interval; FDR = false discovery rate; Inf = infinity; OR = odds ratio; PV = pathogenic variant; RCC = Renal cell carcinoma.
Test statistics from the generalized linear model for genes with nominal enrichment. Genes with adjusted p < 0.05 were considered significant and highlighted. A table of results for all genes tested can be found in Supplementary Tables 8 and 9.
For nccRCC, patients had a significantly higher prevalence of germline PVs than the controls in FH (OR: 77.9, 95% CI: 18.68–324.97, p = 1.55e-08, q value: 1.83e-06) and MET (OR: 1.98e11, 95% CI: 0–inf, p = 2.07e-05, q value: 3.50e-07). LZTR1, PMS2, MITF, EXT2, and CHEK2 p.Ser428Phe also exhibited nominal enrichment of germline PVs but did not pass multiple hypothesis correction (p < 0.05, q values >0.05; Table 2). In contrast to prior studies [9], [11], [12], LOF variants in CHEK2 were not significantly enriched in ccRCC after excluding low-penetrance variants (OR: 1.01, CI: 0.41–2.52; p = 0.980, q value: 0.997) or nccRCC (OR: 2.82, CI: 1.13–7.05; p = 0.0536, q value: 0.79). No other DDR genes were enriched with PVs in ccRCC or nccRCC patients compared with the cancer-free controls (Supplementary Tables 8 and 9).
3.4. Evaluation of CHEK2 in RCC risk via accounting for fine-scale genetic differences in European subpopulations
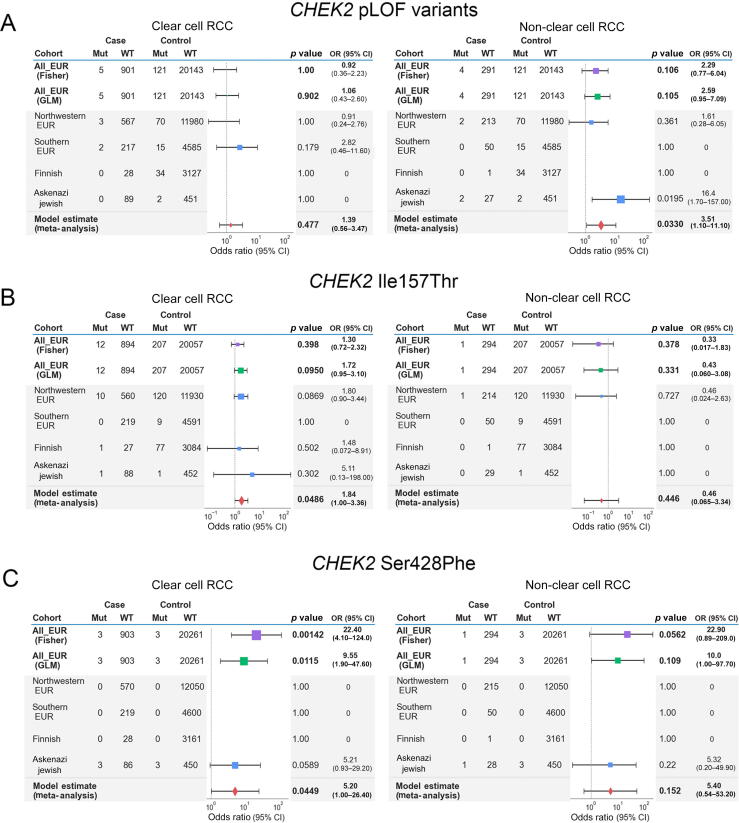
The three main germline PVs identified in CHEK2 in our study—c.1100del (p.Thr367Metfs), Ser428Phe, and Ile157Thr—are all founder variants from different European subpopulations, with substantial variation in population MAFs across different European populations (Supplementary Table 10) [35]. These variants were also identified at different frequencies in our cases and controls of different sub-European ancestries (Supplementary Fig. 4). Therefore, to explore whether subtle ancestry differences in European populations may have confounded CHEK2 rare variant analyses, we performed three additional CHEK2 burden analyses restricted to European samples to evaluate the impact of addressing fine-level population stratification on the role of CHEK2 as an RCC risk gene: (1) a Fisher’s exact-based association study on all Europeans pooled together, (2) a GLM-based burden analysis using the top ten genetic PCs from a European-only PCA, and (3) a meta-analysis combining test statistics from different sub-European populations (Fig. 3 and Supplementary material). CHEK2 germline LOF PVs did not demonstrate enrichment in ccRCC cases in all three tests, although they exhibited a nominal enrichment in nccRCC only in the meta-analysis (OR: 3.51, 95% CI: 1.10–11.10, combined p = 0.0330; Fig. 3A). In contrast, only ccRCC patients exhibited a nominally higher burden of the CHEK2 p.Ile157Thr variant in the meta-analysis (OR: 1.84, 95% CI: 1.00–3.36, combined p = 0.0486; Fig. 3B). Finally, this expanded statistical framework demonstrated that the CHEK2 p.Ser428Phe variant that was significantly enriched in the above multiancestry GLM-based burden analysis were only modestly enriched in these ccRCC patients (OR: 5.20, 95% CI: 1.00–26.40, combined p = 0.0449), reflecting the localized burden of the variant in the inferred ASJ RCC individuals in cases and cancer-free controls (Fig. 3C). Taken together, the results demonstrate that the risk assessment of CHEK2 germline variants requires careful consideration of population stratification due to the varying frequencies of founder variants in this gene.
Fig. 3.
Meta-analysis of CHEK2 risk in ccRCC and nccRCC in European samples. Tables and forest plots summarize the model estimate summary statistics from the sub-European meta-analysis for CHEK2 germline variants. The area of squares is proportional to the –log10 of the p values, and the horizontal bars indicate 95% confidence intervals for the estimated odds ratio. Test statistics from Fisher’s exact and GLM tests were plotted for comparison. (A) Summary tables for CHEK2 LOF excluding the low-penetrance variants. (B) Summary tables for the CHEK2 founder variant p.Ile157Thr. (C) Summary tables for the CHEK2 founder variant p.Ser428Phe. ccRCC = clear cell RCC; CI = confidence interval; EUR = European; GLM = generalized linear model; LOF = loss of function; Mut = mutations; nccRCC = non–clear cell RCC; OR = odds ratio; pLOF = putative loss of function; RCC = renal cell carcinoma; WT = wild type.
3.5. Prevalence of somatic second-hit variants in carriers of germline PVs
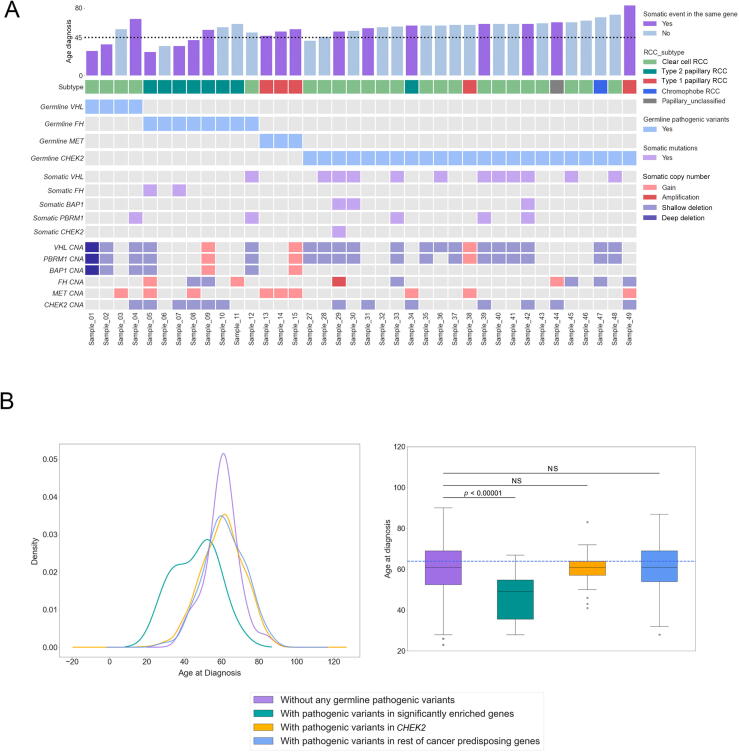
To further clarify the potential roles of the rare germline PVs identified in these analyses, we next investigated the available tumor samples from the RCC patients in our study to identify somatic events (truncating somatic variants or copy number alterations; Supplementary material) accompanying the germline PVs identified in our analyses (Fig. 4A and Supplementary Table 11). Tumors from ccRCC patients carrying germline PVs in tumor suppressor VHL had a somatic copy number deletion in chromosome 3 spanning VHL (n = 3/4, 75.0%, 95% CI: 21.94–98.68%) [36]. Regarding nccRCC, all three carriers of germline PVs in oncogene MET were patients with type 1 papillary RCC whose tumors had somatic copy number gains at chromosome 7 (spanning the MET locus), while 87.5% (n = 7/8, 95% CI: 46.68–99.34%) of patients with germline PVs in tumor suppressor FH were from type 2 papillary RCC whose tumors often had somatic variants or copy number deletions in FH (n = 4/7, 57.1%, 95% CI: 20.24–88.19%). Overall, ten patients (66.7%, 95% CI: 38.69–87.01%) carrying germline PVs in FH, MET, or VHL harbored identifiable secondary somatic events in the same genes, further indicating the importance of these genes in the RCC oncogenesis. In contrast, patients carrying germline variants in tumor suppressor CHEK2 were not limited to a specific RCC subtype, and only seven of 23 (30.4%, 95% CI: 14.06–53.01%) RCC patients with germline variants in CHEK2 had secondary somatic variants in CHEK2 (one patient with both somatic variant and copy number deletion in CHEK2; six patients with CHEK2 copy number deletions).
Fig. 4.
Somatic events in the carriers of pathogenic variants in genes with enrichment. Somatic alterations were characterized for the carriers of germline PVs in CHEK2, MET, FH, and VHL, and age of disease onsets were compared between groups of patients with different germline variant status. (A) Comutation plot summarizing germline and somatic variants in RCC patients with germline pathogenic variants in the three significantly enriched genes as well as CHEK2. For the somatic events, only variants in relevant genes are featured. The dotted horizontal line on the bar plot indicates the age of onset at 45 yr. (B) Density plot (left) and box plot (right) for the distribution of age of disease onset for the different pathogenic variant carrier groups. Adjusted p value was calculated after a one-way ANOVA with post hoc Tukey’s HDS test. ANOVA = analysis of variance; CNA = copy number alterations; HDS = honestly significant difference; NS = not significant; PV = pathogenic variant; RCC = renal cell carcinoma.
3.6. Age of RCC presentation for carriers of germline PVs in RCC risk genes
Rare germline PVs in genes known to cause hereditary cancer syndromes have been associated with an earlier onset of disease in different cancers including RCC [37], [38], [39], [40], [41], and detection of these variants with genetic testing can guide clinical management in at an elevated genetic risk for cancer [42]. To further characterize the clinical impact of the rare germline PVs identified in the genes with a significantly higher burden of PVs in RCC patients, we compared the age of disease onset between the groups defined by genetic status: (1) patients carrying germline PVs in the known RCC risk genes; FH, MET, and VHL; (2) patients carrying germline PVs in CHEK2; (3) patients carrying germline PVs in other CPGs without enrichment; and (4) patients carrying no germline PVs (Fig. 4B and Supplementary Table 12). The carriers of rare germline PVs in FH, MET, and VHL presented with disease at a significantly earlier age than the other three groups (mean: 46.0, median: 49.0 yr old, Tukey post hoc adjusted p < 0.01 for all three pairwise comparisons), and patients with germline and somatic biallelic events presented with disease at an earlier age (n = 10/15, mean: 44.6, median: 44.5 yr old; Fig. 4A). However, the age of disease onset for the patients carrying germline CHEK2 PVs showed no evidence of age difference from that of patients carrying no germline PVs (mean: 60.1, median: 61 yr old vs mean: 60.2, median: 61 yr old, Tukey post hoc adjusted p = 1.0) or patients carrying PVs in the rest of CPGs that did not exhibit enrichment in RCC (mean: 60.1, median: 61 yr old vs mean: 61.3, median: 61 yr old, Tukey post hoc adjusted p = 0.949). Similarly, RCC patients carrying both germline and somatic variants in CHEK2 did not present at an earlier age of onset compared with patients without any germline PVs (n = 7/23, mean: 62.1, median: 61.0 yr old). These results, taken together with the only modest enrichment of germline PVs in CHEK2, suggest caution for considering CHEK2 as a RCC predisposition gene.
3.7. Identifying additional forms of inherited genomic alterations in RCC risk genes
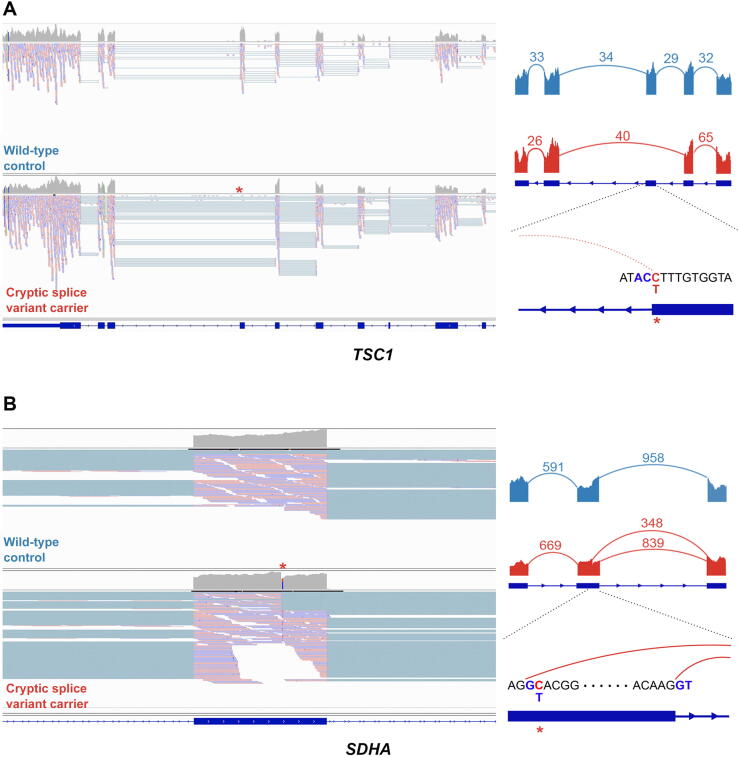
While 13.9% (n = 199/1436, 95% CI: 12.13–15.78%) of total RCC (ccRCC and nccRCC) patients carried rare germline PVs in CPGs, only 15 RCC patients (1.04%, 95% CI: 0.61–1.76%) harbored germline PVs in known RCC risk genes (FH, MET, and VHL), but the rest of the CPGs did not exhibit an increased burden of PVs in RCC patients in our analyses and thus are of uncertain biological significance in RCC pathogenesis. Thus, we hypothesized that RCC risk genes may also be disrupted through mechanisms that can escape the detection of conventional germline variant detection methods commonly used in clinical and research contexts, such as cryptic splice variants outside of the canonical splice sites and germline CNVs. Using existing computational methods to predict cryptic splice variants, we identified 109 candidate rare germline cryptic splice variants in CPGs in 102 RCC patients (Supplementary Fig. 5A, Supplementary Table 13, and Supplementary material). Of these, 86 patients had tumor and/or germline mRNA sequencing data available to validate these predicted splice variants. The available RNA sequencing data showed no evidence of aberrant splicing for 82 variants (95.35%, 95% CI: 87.87–98.50%). However, two cryptic splice variants in two RCC risk genes, TSC1 and SDHA, demonstrated a clear pattern of aberrant splicing (Fig. 5). The cryptic splice variant in TSC1 in a chromophobe RCC patient changed antisense cytosine upstream of a splice donor motif to thymine, leading to complete exon skipping of exon 21. In the papillary RCC patient with a variant in SDHA, the cryptic splice variant introduced a cryptic donor motif inside exon 13 and removed 15 amino acids at the end of the exon. Two other cryptic splice variants in TP53 and LZTR1 showed aberrant splicing, but the number of splice junction reads was too low to confidently conclude them as clear splice variants (Supplementary Fig. 5B and Supplementary material).
Fig. 5.
Examples of cryptic splice variants in established RCC risk genes. (A) Disruption of splice donor motif led to complete exon skipping in TSC1. (B) The cryptic splice variant in SDHA introduced a new splice donor motif GT inside an exon. Images at the left represent IGV screenshots of the tumor mRNA sequencing data of the wild-type control (Fig. 5A) and the carrier of cryptic splice variant (Fig. 5B). Images at the right represent Sashimi plots showing the pattern of splicing with the numbered split junction reads. IGV = Integrative Genomics Viewer; RCC = renal cell carcinoma.
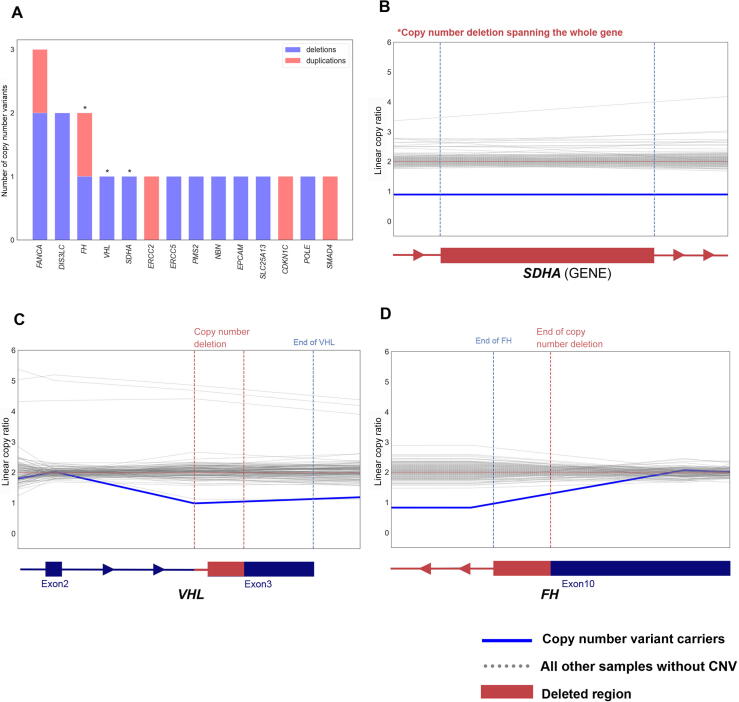
We next evaluated germline CNVs (Supplementary material). Collectively, we identified 2503 high-quality rare germline CNVs in 888 RCC samples (1211 deletions and 1292 duplications; Supplementary Fig. 6B and 6C, and Supplementary material). Of these, 18 heterozygous CNVs in 18 (1.25%, 95% CI: 0.77–2.02%) RCC patients affected 14 CPGs including RCC risk genes FH, VHL, and SDHA (Fig. 6A and Supplementary Table 14). For example, we found a ccRCC patient harboring a deletion spanning part of the last exon of VHL (Chr3:10191124-10192282, GRCh37; Fig. 6C), and another deletion was identified in a papillary RCC patient overlapping the last 761bp of FH (Chr1:240070386-241661618; Fig. 6D). We also identified a large 215 kbp deletion completely spanning SDHA in a different papillary RCC patient (Chr5:139251-354374; Fig. 6B), which disrupted a region similar to a variant described previously in the gnomAD structural variant database (gnomAD ID: DEL_5_54065, Chr5:135575-308762) [43]. Thus, by characterizing underappreciated variant types such as cryptic splice and CNVs, we identified six additional putative PVs in the established RCC risk genes, increasing the diagnostic yield of rare germline PVs in risk genes from 2.1% (n = 30/1436, 95% CI: 1.44–3.01%) to 2.5% (n = 36/1436, 95% CI: 1.79–3.49%).
Fig. 6.
Examples of rare germline copy number variants (CNVs) in established cancer-predisposing genes. (A) Bar-plot summarizing the counts of deletions and duplications including the germline cancer predisposing genes. (B–D) Denoised linear copy ratio (DCR) plots indicating heterozygous copy number deletion with a copy ratio of 1 for the carriers of CNVs. The blue line indicates DCR for the CNV carrier, and the dotted grey lines indicate DCR for the rest of wild-type samples.
4. Discussion
Thus far, 14 genes in the HIF, PI3K/mTOR, chromatin regulation, and cell cycle pathways have been identified as established RCC risk genes [44], but the estimated high genetic heritability of RCC is not fully explained by germline PVs detected in these genes. Multiple recent studies have built on these foundational discoveries to report frequencies of rare germline PVs in DDR genes in large RCC cohorts. However, analyses with proper cancer-free controls and statistical models accounting for population structure are necessary to determine whether these PVs are significantly enriched in RCC populations. Attempts to demonstrate association by comparing frequencies of PVs in RCC cases against public databases such as ExAC [45] or gnomAD [35] are fraught with major technical limitations, including that the samples are likely not sequenced using the same sequencing platform, variants were not called using the same variant discovery pipeline and were not processed identically, and cases and controls were not ancestry matched to ensure a robust statistical comparison. Recently, two studies leveraged case-control approaches to report an association of rare germline PVs in CHEK2 with an elevated risk of RCC [11], [12]. However, both studies treated different subtypes of RCC together as a single phenotype and included CHEK2 variants with distinct population properties. Although Sekine et al [46] conducted a comprehensive ancestry-matched burden analysis on ccRCC and nccRCC separately, their study was constrained to Japanese patients only. To address the gaps of knowledge in the field, we performed histology-specific and case-control analyses of rare germline PVs in RCC, finding that PVs in the three significantly enriched genes—VHL, MET, and FH—had no significant overlap in different RCC subtypes. This molecular difference was also clearly demonstrated when we investigated companion somatic variants and copy number events stratified by histological subtype.
Furthermore, while adjusting for finer-scale population stratification is the current standard in genome-wide association studies (GWAS), many case-control rare germline variant studies in cancer (including most RCC studies) adjusted only for population stratification by limiting their association analysis to cases and controls of European descent under the assumption that individuals of broad continental European ancestry would have a similar genetic background. Consistent with prior observations that population stratification can confound rare variant association studies in other disease contexts [47], [48], [49], [50], we found that a standard Fisher’s exact test–based association model without addressing finer-scale population stratification can lead to a false association in this context, especially for CHEK2 that has several founder variants with varying population frequency within Europe. For example, the most well-studied CHEK2 c.1100del variant’s population frequency varies substantially in different regions of Europe, ranging from ∼0% in Spain to 1.6% in the Netherlands, and has a relatively lower population frequency in North America than in Europe [51], [52], [53], [54], [55], [56], [57]. These factors may explain why in our study, the CHEK2 variant was detected in only 0.35% of RCC cases, where most patients were from the USA and patients were unselected for family history. Meanwhile, this variant was identified in >1% of RCC cases in a UK-based RCC association study [11], and significant enrichment of CHEK2 germline PVs was reported in studies with patients selected for a positive family history [9] or positive CHEK2 germline variant carrier status in panel sequencing [12]. The variation in population frequency of CHEK2 variants combined with prior studies lacking robust genetic ancestry inference and case-control matching may partially explain the disagreeing risk assessment within and across cancer types for this gene [58]. Our result warrants caution against the commonly used practice of treating all “White” or “Caucasian” individuals with predominantly European ancestry as one group in cancer genetic studies. This practice can substantially confound association studies, particularly for studies including participants from the US population, which consists of different European ancestry groups as well as non-European ancestry groups.
Furthermore, in this study, RCC patients with germline PVs in CHEK2 did not have frequent secondary somatic events, whereas tumors from carriers of PVs in bona fide RCC predisposition genes such as FH, MET, or VHL largely exhibited secondary somatic events in these genes. In addition, in this cohort of RCC patients unselected for the age of disease onset, RCC patients with germline PVs in CHEK2 did not exhibit an earlier age of RCC onset, contrasting the relatively early age of breast cancer onset for germline CHEK2 variant carriers reported in several studies [51], [53], [59], [60], [61], which suggests uncertainty regarding the role of CHEK2 germline PVs in RCC risk. Unlike in breast cancer, the biological role of DDR genes in Fanconi anemia or homologous recombination pathways such as CHEK2 has not definitively been demonstrated in RCC, and we did not identify any enrichment of germline PVs in other DDR genes besides CHEK2. Given these observations, we suggest caution in including CHEK2 or any other DDR genes as RCC risk genes. Critically, our analysis does not preclude the association of CHEK2 with RCC, but advises for addressing population stratification in larger cohorts that include different sub-European ancestry groups to clarify the role of this gene in RCC risk and heritability.
Moreover, the general focus on germline small nucleotide variants and small indels in coding sequences in prior studies may limit our understanding of potential PVs in established RCC risk genes or other candidate genes. Thus, we also investigated rare germline cryptic splice variants and germline CNVs that have not been well characterized in RCC or cancer germline studies. Cryptic splice variants can introduce or remove splicing donor or acceptor motifs inducing aberrant mRNA splicing and loss of protein function [62], [63]. However, these can be easily overlooked as nonpathogenic because these are usually annotated as nontruncating when using conventional annotation approaches. In this study, we identified two rare germline cryptic splice variants that induced aberrant splicing in RCC risk genes SDHA and TSC1, which appear to reduce wild-type transcript abundance based on our investigation of matched transcriptome sequencing data. To our knowledge, this is the first description of germline cryptic splice variants in RCC and may warrant incorporation of them into comprehensive clinical genetic testing strategies.
Lastly, to further augment the search space for germline inherited risk events, we systematically characterized rare germline CNVs in RCC patients. Rare germline CNVs are known to increase susceptibility for different cancers [64], [65], [66], [67], [68], [69] and a few RCC studies reported CNVs detected in FLCN and VHL in RCC patients [70], [71]. However, systematic characterization of germline CNVs using WES data has not been explored fully in RCC despite the wider availability of WES data and improved methods for germline CNV detection. Here, we successfully identified 18 rare germline copy number duplications and deletions in CPGs from the whole-exome sequenced samples, including four CNVs in RCC risk genes FH, SDHA, and VHL. With the widespread use of WES and improvement in CNV identification methods, investigation of germline single nucleotide variant and short insertions and deletions together with cryptic splice and CNVs should be considered a routine testing strategy for RCC inherited risk assessment and possibly across cancer types.
The current study has several limitations. First, the findings from our gene-burden analyses merit validation in additional independent case and control cohorts, particularly in larger and more diverse patient populations. Indeed, even for the sub-European ancestry identification, we did not have the means to distinguish Northwestern Europeans from Eastern or central Europeans who might have clustered together with the Northwestern Europeans in the 1000 Genomes Project–based inference, let alone for the myriad subpopulations in other non-European continents. In the future, more refined meta-analyses might take advantage of reference panels representing diverse ancestry groups to better address such subtle subcontinental differences. Further, we had to constrain the analysis to 143 CPGs, as opposed to a comprehensive whole-exome–wide analysis, due to the limited study power, further emphasizing the need for larger and more diverse patient cohorts. Lastly, the application of the new World Health Organization renal cancer classification, which categorizes type 2 papillary as a separate non-RCC entity [72], to pre-existing characterized samples poses substantial practical challenges. These are particularly pronounced when the required molecular and immunohistochemical correlations were either not performed or not readily available. Consequently, we opted to use the previous tumor classification system of type 1 and type 2 disease for papillary RCC. Further research should consider incorporating the updated classification, distinguishing chromophobe RCC from papillary RCC, and including other rare subtypes such as medullary or collecting duct RCC for a more thorough characterization of inherited risk events in heterogeneous nccRCC subtypes.
5. Conclusions
Taken together, this systematic population stratification–aware analysis supports the link between several RCC risk genes and elevated risk and describes distinct patterns of inherited germline and somatic variants in different RCC subtypes. Our results also call for caution when assessing the risk conferred by germline PVs in CHEK2. Finally, it broadens the definition of the RCC germline landscape of pathogenicity to incorporate previously underutilized germline variations.
Author contributions: Eliezer M. Van Allen had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Han, AlDubayan, Van Allen.
Acquisition of data: Han, Bakouny, Reardon, Moore, Labaki, Braun, Choueiri, Ricker, Park.
Analysis and interpretation of data: Han, AlDubayan.
Drafting of the manuscript: Han, AlDubayan, Van Allen.
Critical revision of the manuscript for important intellectual content: Gillani, Collins, Chu.
Statistical analysis: Han, Camp, AlDubayan, Chu, Moore, Kofman, Reardon.
Obtaining funding: AlDubayan, Van Allen.
Administrative, technical, or material support: Park.
Supervision: AlDubayan, Van Allen.
Other: None.
Financial disclosures: Eliezer M. Van Allen certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Eliezer M. Van Allen holds consulting roles with Tango Therapeutics, Genome Medical, Genomic Life, Enara Bio, Manifold Bio, Monte Rosa, Novartis Institute for Biomedical Research, Riva Therapeutics, and Serinus Bio; receives research support from Novartis, Bristol-Myers Squibb, and Sanofi; has equity in Tango Therapeutics, Genome Medical, Genomic Life, Syapse, Enara Bio, Manifold Bio, Microsoft, Monte Rosa, Riva Therapeutics, and Serinus Bio; and has filed institutional patents on chromatin mutations, immunotherapy response, and methods for clinical interpretation. Toni K. Choueiri reports institutional and personal, paid and/or unpaid support for research, advisory boards, consultancy, and honoraria from Alkermes, AstraZeneca, Aravive, Aveo, Bayer, Bristol Myers-Squibb, Calithera, Circle Pharma, Eisai, EMD Serono, Exelixis, GlaxoSmithKline, IQVA, Infinity, Ipsen, Jansen, Kanaph, Lilly, Merck, Nikang, Nuscan, Novartis, Pfizer, Roche, Sanofi/Aventis, Surface Oncology, Takeda, Tempest, Up-To-Date, and CME events (Peerview, OncLive, MJH, and others), outside the submitted work; institutional patents filed on molecular alterations and immunotherapy response/toxicity, and ctDNA; equity in Tempest, Pionyr, Osel, Precede Bio, and CureResponse; being in committees at NCCN, GU Steering Committee, ASCO/ESMO, ACCRU, and KidneyCan; medical writing and editorial assistance support may have been funded by communications companies in part; no speaker’s bureau; mentoring of several non-US citizens on research projects with potential funding (in part) from non-US sources/foreign components; the institution (Dana-Farber Cancer Institute) may have received additional independent funding of drug companies or/and royalties potentially involved in research around the subject matter; and also being supported in part by the Dana-Farber/Harvard Cancer Center Kidney SPORE (2P50CA101942-16) and Program 5P30CA006516-56, the Kohlberg Chair at Harvard Medical School and the Trust Family, Michael Brigham, Pan Mass Challenge, Hinda and Arthur Marcus Fund, and Loker Pinard Funds for Kidney Cancer Research at DFCI. David Braun reports nonfinancial support from Bristol Myers Squibb; honoraria from LM Education/Exchange Services; advisory board fees from Exelixis and AVEO; personal fees from Charles River Associates, Schlesinger Associates, Imprint Science, Insight Strategy, Trinity Group, Cancer Expert Now, Adnovate Strategies, MDedge, CancerNetwork, Catenion, OncLive, Cello Health BioConsulting, PWW Consulting, Haymarket Medical Network, Aptitude Health, ASCO Post/Harborside, Targeted Oncology, and AbbVie; and research support from Exelixis and AstraZeneca, outside of the submitted work. Riaz Gillani has equity in Google, Microsoft, Amazon, Apple, Moderna, Pfizer, and Vertex Pharmaceuticals. Brendan Reardon has filed institutional patents on methods for clinical interpretation. Ziad Bakouny receives research support from Bristol Myers Squibb and imCORE/Genentech, and also reports honoraria from UpToDate. Chris Labaki receives research funding from imCORE/Genentech outside the submitted work. The other authors declare no competing interests.
Funding/Support and role of the sponsor: This work was supported by The National Institutes of Health R37CA222574 (Eliezer M. Van Allen), R01CA227388 (Eliezer M. Van Allen), R50CA265182 (Jihye Park), Mark Foundation Emerging Leader Award, the Department of Defense Physician Research Award (W81XWH-21-1-0084, PC200150; Saud H. AlDubayan), the Department of Defense Idea Development Award—Early-Career Investigator (KC210042/W81XWH-22-1-0455; Saud H. AlDubayan), Alex’s Lemonade Stand Foundation Young Investigator Grant (Riaz Gillani), and the Wong Family Award in Translational Oncology (Riaz Gillani). The funding organizations were not responsible for the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Acknowledgments: We would like to thank all individuals who participated in the multiple studies from which we collected our raw sequencing data. We also would like to thank Isaac Wong and Jack Fu from Talkowski Lab for helping us trouble-shooting the GATK-gCNV. Finally, we would like to thank Dr. Alexander (Sasha) Gusev for his input on the Jewish inference analysis using SNPweights.
Ethics statement: All individuals in this study consented to institutional review board–approved protocol (IRB# 20-293) that allowed for a comprehensive genetic analysis of germline samples (Supplementary material). This study conforms to the Declaration of Helsinki.
Data sharing statement: All computation tools and packages in this study are publicly available. The docker image containing all GATK tools is available at https://hub.docker.com/r/broadinstitute/gatk/. The docker image containing the germline variant detection tool DeepVariant can be found at https://hub.docker.com/r/google/deepvariant. Tools and detailed usage for SpliceAI (https://github.com/Illumina/SpliceAI) and GATK-gCNV (https://github.com/theisaacwong/talkowski/tree/master/gCNV) can be found on the respective GitHub pages. All raw sequencing data for TCGA studies can be accessed with controlled access on the GDC data portal (https://portal.gdc.cancer.gov/) with approval. All raw sequencing data for the ICGC study can be accessed with controlled access on the ICGC data portal (https://dcc.icgc.org/) with approval. All raw sequencing data for CHECKMATE clinical studies and Genentech study can be downloaded from European Genome-Phenome Archive (dataset ID: EGAD00001001023) with approval. All raw sequencing data for cancer-free control samples can be accessed on dbGAP—Autism Sequencing Consortium (dbGAP:phs000298.v4.p3), Framingham Cohort (dbGAP:phs000007.v32.p1), MESA Cohort (dbGAP: phs000209.v13.p3), and NHLBI GO-ESP: Lung Cohorts Exome Sequencing Project (dbGAP: phs000291.v2.p1).
Associate Editor: M. Carmen Mir
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.euros.2024.02.006.
Contributor Information
Saud H. AlDubayan, Email: Saud_Aldubayan@dfci.harvard.edu.
Eliezer M. Van Allen, Email: EliezerM_VanAllen@dfci.harvard.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Padala S.A., Barsouk A., Thandra K.C., et al. Epidemiology of renal cell carcinoma. World J Oncol. 2020;11:79–87. doi: 10.14740/wjon1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mucci L.A., Hjelmborg J.B., Harris J.R., et al. Familial risk and heritability of cancer among twins in Nordic countries. JAMA. 2016;315:68–76. doi: 10.1001/jama.2015.17703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haas N.B., Nathanson K.L. Hereditary kidney cancer syndromes. Adv Chronic Kidney Dis. 2014;21:81–90. doi: 10.1053/j.ackd.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt L.S., Linehan W.M. Genetic predisposition to kidney cancer. Semin Oncol. 2016;43:566–574. doi: 10.1053/j.seminoncol.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen K.A., Syed J.S., Espenschied C.R., et al. Advances in the diagnosis of hereditary kidney cancer: Initial results of a multigene panel test. Cancer. 2017;123:4363–4371. doi: 10.1002/cncr.30893. [DOI] [PubMed] [Google Scholar]
- 6.Carlo M.I., Mukherjee S., Mandelker D., et al. Prevalence of germline mutations in cancer susceptibility genes in patients with advanced renal cell carcinoma. JAMA Oncol. 2018;4:1228–1235. doi: 10.1001/jamaoncol.2018.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J., Wang H., Ricketts C.J., et al. Germline mutations of renal cancer predisposition genes and clinical relevance in Chinese patients with sporadic, early-onset disease. Cancer. 2019;125:1060–1069. doi: 10.1002/cncr.31908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartman T.R., Demidova E.V., Lesh R.W., et al. Prevalence of pathogenic variants in DNA damage response and repair genes in patients undergoing cancer risk assessment and reporting a personal history of early-onset renal cancer. Sci Rep. 2020;10:13518. doi: 10.1038/s41598-020-70449-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abou Alaiwi S., Nassar A.H., Adib E., et al. Trans-ethnic variation in germline variants of patients with renal cell carcinoma. Cell Rep. 2021;34:108926. doi: 10.1016/j.celrep.2021.108926. [DOI] [PubMed] [Google Scholar]
- 10.Truong H., Sheikh R., Kotecha R., et al. Germline variants identified in patients with early-onset renal cell carcinoma referred for germline genetic testing. Eur Urol Oncol. 2021;4:993–1000. doi: 10.1016/j.euo.2021.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yngvadottir B., Andreou A., Bassaganyas L., et al. Frequency of pathogenic germline variants in cancer susceptibility genes in 1336 renal cell carcinoma cases. Hum Mol Genet. 2022;31:3001–3011. doi: 10.1093/hmg/ddac089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bychkovsky B.L., Agaoglu N.B., Horton C., et al. Differences in cancer phenotypes among frequent CHEK2 variants and implications for clinical care-checking CHEK2. JAMA Oncol. 2022;8:1598–1606. doi: 10.1001/jamaoncol.2022.4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato Y., Yoshizato T., Shiraishi Y., et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 14.Motzer R.J., Jonasch E., Boyle S., et al. NCCN guidelines insights: kidney cancer, version 1.2021. J Natl Compr Canc Netw. 2020;18:1160–1170. doi: 10.6004/jnccn.2020.0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Persyn E., Redon R., Bellanger L., Dina C. The impact of a fine-scale population stratification on rare variant association test results. PLoS One. 2018;13:e0207677. doi: 10.1371/journal.pone.0207677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKenna A., Hanna M., Banks E., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poplin R., Chang P.C., Alexander D., et al. A universal SNP and small-indel variant caller using deep neural networks. Nat Biotechnol. 2018;36:983–987. doi: 10.1038/nbt.4235. [DOI] [PubMed] [Google Scholar]
- 18.AlDubayan S.H., Conway J.R., Camp S.Y., et al. Detection of pathogenic variants with germline genetic testing using deep learning vs standard methods in patients with prostate cancer and melanoma. JAMA. 2020;324:1957–1969. doi: 10.1001/jama.2020.20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camp S.Y., Kofman E., Reardon B., et al. Evaluating the molecular diagnostic yield of joint genotyping-based approach for detecting rare germline pathogenic and putative loss-of-function variants. Genet Med. 2021;23:918–926. doi: 10.1038/s41436-020-01074-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conomos M.P., Miller M.B., Thornton T.A. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet Epidemiol. 2015;39:276–293. doi: 10.1002/gepi.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conomos M.P., Reiner A.P., Weir B.S., Thornton T.A. Model-free estimation of recent genetic relatedness. Am J Hum Genet. 2016;98:127–148. doi: 10.1016/j.ajhg.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hail Team. Hail 0.2. 2021. https://github.com/hail-is/hail.
- 23.1000 Genomes Project Consortium, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed]
- 24.Chen C.Y., Pollack S., Hunter D.J., Hirschhorn J.N., Kraft P., Price A.L. Improved ancestry inference using weights from external reference panels. Bioinformatics. 2013;29:1399–1406. doi: 10.1093/bioinformatics/btt144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLaren W., Gil L., Hunt S.E., et al. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopanos C., Tsiolkas V., Kouris A., et al. VarSome: the human genomic variant search engine. Bioinformatics. 2019;35:1978–1980. doi: 10.1093/bioinformatics/bty897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seabold S, Perktold J. Statsmodels: econometric and statistical modeling with python. Presented at the Proceedings of the 9th Python in Science Conference; 2010.
- 29.Virtanen P., Gommers R., Oliphant T.E., et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viechtbauer W. Conducting meta-analyses in R with the metafor Package. 2010.
- 31.Waskom M.L. Seaborn: statistical data visualization. J Open Source Software. 2021;6(60):3021. doi: 10.21105/joss.03021. [DOI] [Google Scholar]
- 32.Crowdis J., He M.X., Reardon B., Van Allen E.M. CoMut: visualizing integrated molecular information with comutation plots. Bioinformatics. 2020;36:4348–4349. doi: 10.1093/bioinformatics/btaa554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535–548. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 34.Babadi M, Fu JM, Lee SK, et al. GATK-gCNV: a rare copy number variant discovery algorithm and its application to exome sequencing in the UK Biobank. bioRxiv 2022.
- 35.Karczewski K.J., Francioli L.C., Tiao G., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jonasch E., Walker C.L., Rathmell W.K. Clear cell renal cell carcinoma ontogeny and mechanisms of lethality. Nat Rev Nephrol. 2021;17:245–261. doi: 10.1038/s41581-020-00359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melhem-Bertrandt A., Bojadzieva J., Ready K.J., et al. Early onset HER2-positive breast cancer is associated with germline TP53 mutations. Cancer. 2012;118:908–913. doi: 10.1002/cncr.26377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearlman R., Frankel W.L., Swanson B., et al. Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol. 2017;3:464–471. doi: 10.1001/jamaoncol.2016.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y.L., Cadoo K.A., Maio A., et al. Early age of onset and broad cancer spectrum persist in MSH6- and PMS2-associated Lynch syndrome. Genet Med. 2022;24:1187–1195. doi: 10.1016/j.gim.2022.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reckamp K.L., Behrendt C.E., Slavin T.P., et al. Germline mutations and age at onset of lung adenocarcinoma. Cancer. 2021;127:2801–2806. doi: 10.1002/cncr.33573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stadler ZK, Maio A, Padunan A, et al. Germline mutation prevalence in young adults with cancer. Presented at American Association for Cancer Research Virtual Annual Meeting II. 2020.
- 42.Weitzel J.N., Blazer K.R., MacDonald D.J., Culver J.O., Offit K. Genetics, genomics, and cancer risk assessment: state of the art and future directions in the era of personalized medicine. CA Cancer J Clin. 2011;61:327–359. doi: 10.3322/caac.20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collins R.L., Brand H., Karczewski K.J., et al. A structural variation reference for medical and population genetics. Nature. 2020;581:444–451. doi: 10.1038/s41586-020-2287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ricketts C.J., Crooks D.R., Sourbier C., Schmidt L.S., Srinivasan R., Linehan W.M. SnapShot: renal cell carcinoma. Cancer Cell. 2016;29:610–610.e1. doi: 10.1016/j.ccell.2016.03.021. [DOI] [PubMed] [Google Scholar]
- 45.Karczewski K.J., Weisburd B., Thomas B., et al. The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res. 2017;45:D840–D845. doi: 10.1093/nar/gkw971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sekine Y., Iwasaki Y., Aoi T., et al. Different risk genes contribute to clear cell and non-clear cell renal cell carcinoma in 1532 Japanese patients and 5996 controls. Hum Mol Genet. 2022;31:1962–1969. doi: 10.1093/hmg/ddab345. [DOI] [PubMed] [Google Scholar]
- 47.Tintle N., Aschard H., Hu I., Nock N., Wang H., Pugh E. Inflated type I error rates when using aggregation methods to analyze rare variants in the 1000 Genomes Project exon sequencing data in unrelated individuals: summary results from Group 7 at Genetic Analysis Workshop 17. Genet Epidemiol. 2011;35(Suppl 1):S56–S60. doi: 10.1002/gepi.20650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathieson I., McVean G. Differential confounding of rare and common variants in spatially structured populations. Nat Genet. 2012;44:243–246. doi: 10.1038/ng.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang Y., Epstein M.P., Conneely K.N. Assessing the impact of population stratification on association studies of rare variation. Hum Hered. 2013;76:28–35. doi: 10.1159/000353270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y., Guan W., Pan W. Adjustment for population stratification via principal components in association analysis of rare variants. Genet Epidemiol. 2013;37:99–109. doi: 10.1002/gepi.21691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.CHEK2 Breast Cancer Case-Control Consortium. CHEK2*1100delC and susceptibility to breast cancer: a collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies. Am J Hum Genet 2004;74:1175–82. [DOI] [PMC free article] [PubMed]
- 52.Offit K., Pierce H., Kirchhoff T., et al. Frequency of CHEK2*1100delC in New York breast cancer cases and controls. BMC Med Genet. 2003;4:1. doi: 10.1186/1471-2350-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mateus Pereira L.H., Sigurdson A.J., Doody M.M., et al. CHEK2:1100delC and female breast cancer in the United States. Int J Cancer. 2004;112:541–543. doi: 10.1002/ijc.20439. [DOI] [PubMed] [Google Scholar]
- 54.Neuhausen S., Dunning A., Steele L., et al. Role of CHEK2*1100delC in unselected series of non-BRCA1/2 male breast cancers. Int J Cancer. 2004;108:477–478. doi: 10.1002/ijc.11385. [DOI] [PubMed] [Google Scholar]
- 55.Osorio A., Rodriguez-Lopez R., Diez O., et al. The breast cancer low-penetrance allele 1100delC in the CHEK2 gene is not present in Spanish familial breast cancer population. Int J Cancer. 2004;108:54–56. doi: 10.1002/ijc.11414. [DOI] [PubMed] [Google Scholar]
- 56.Vahteristo P., Bartkova J., Eerola H., et al. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am J Hum Genet. 2002;71:432–438. doi: 10.1086/341943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laitman Y., Kaufman B., Lahad E.L., Papa M.Z., Friedman E. Germline CHEK2 mutations in Jewish Ashkenazi women at high risk for breast cancer. Isr Med Assoc J. 2007;9:791–796. [PubMed] [Google Scholar]
- 58.Stolarova L., Kleiblova P., Janatova M., et al. CHEK2 germline variants in cancer predisposition: stalemate rather than checkmate. Cells. 2020;9:2675. doi: 10.3390/cells9122675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Margolin S., Eiberg H., Lindblom A., Bisgaard M.L. CHEK2 1100delC is prevalent in Swedish early onset familial breast cancer. BMC Cancer. 2007;7:163. doi: 10.1186/1471-2407-7-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rashid M.U., Jakubowska A., Justenhoven C., et al. German populations with infrequent CHEK2*1100delC and minor associations with early-onset and familial breast cancer. Eur J Cancer. 2005;41:2896–2903. doi: 10.1016/j.ejca.2005.04.049. [DOI] [PubMed] [Google Scholar]
- 61.Oldenburg R.A., Kroeze-Jansema K., Kraan J., et al. The CHEK2*1100delC variant acts as a breast cancer risk modifier in non-BRCA1/BRCA2 multiple-case families. Cancer Res. 2003;63:8153–8157. [PubMed] [Google Scholar]
- 62.Lee M., Roos P., Sharma N., et al. Systematic computational identification of variants that activate exonic and intronic cryptic splice sites. Am J Hum Genet. 2017;100:751–765. doi: 10.1016/j.ajhg.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cooper T.A., Wan L., Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walker L.C., Pearson J.F., Wiggins G.A., Giles G.G., Hopper J.L., Southey M.C. Increased genomic burden of germline copy number variants is associated with early onset breast cancer: Australian breast cancer family registry. Breast Cancer Res. 2017;19:30. doi: 10.1186/s13058-017-0825-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laitinen V.H., Akinrinade O., Rantapero T., Tammela T.L., Wahlfors T., Schleutker J. Germline copy number variation analysis in Finnish families with hereditary prostate cancer. Prostate. 2016;76:316–324. doi: 10.1002/pros.23123. [DOI] [PubMed] [Google Scholar]
- 66.Yoshihara K., Tajima A., Adachi S., et al. Germline copy number variations in BRCA1-associated ovarian cancer patients. Genes Chromosomes Cancer. 2011;50:167–177. doi: 10.1002/gcc.20841. [DOI] [PubMed] [Google Scholar]
- 67.Brea-Fernandez A.J., Fernandez-Rozadilla C., Alvarez-Barona M., et al. Candidate predisposing germline copy number variants in early onset colorectal cancer patients. Clin Transl Oncol. 2017;19:625–632. doi: 10.1007/s12094-016-1576-z. [DOI] [PubMed] [Google Scholar]
- 68.Shi J., Zhou W., Zhu B., et al. Rare germline copy number variations and disease susceptibility in familial melanoma. J Invest Dermatol. 2016;136:2436–2443. doi: 10.1016/j.jid.2016.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park R.W., Kim T.M., Kasif S., Park P.J. Identification of rare germline copy number variations over-represented in five human cancer types. Mol Cancer. 2015;14:25. doi: 10.1186/s12943-015-0292-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schneider M., Dinkelborg K., Xiao X., et al. Early onset renal cell carcinoma in an adolescent girl with germline FLCN exon 5 deletion. Fam Cancer. 2018;17:135–139. doi: 10.1007/s10689-017-0008-8. [DOI] [PubMed] [Google Scholar]
- 71.Matsuda D., Khoo S.K., Massie A., et al. Identification of copy number alterations and its association with pathological features in clear cell and papillary RCC. Cancer Lett. 2008;272:260–267. doi: 10.1016/j.canlet.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 72.Moch H., Amin M.B., Berney D.M., et al. The 2022 World Health Organization classification of tumours of the urinary system and male genital organs—part A: renal, penile, and testicular tumours. Eur Urol. 2022;82:458–468. doi: 10.1016/j.eururo.2022.06.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.