Abstract
Polyomavirus middle T antigen (MT) is phosphorylated on serine residues. Partial proteolytic mapping and Edman degradation identified serine 257 as a major site of phosphorylation. This was confirmed by site-directed mutagenesis. Isoelectric focusing of immunoprecipitated MT from transfected 293T cells showed that phosphorylation on wild-type MT occurred at near molar stoichiometry at S257. MT was previously shown to be associated with 14-3-3 proteins, which have been connected to cell cycle regulation and signaling. The association of 14-3-3 proteins with MT depended on the serine 257 phosphorylation site. This has been demonstrated by comparing wild-type and S257A mutant MTs expressed with transfected 293T cells or with Sf9 cells infected with recombinant baculoviruses. The 257 site is not critical for transformation of fibroblasts in vitro, since S257A and S257C mutant MTs retained the ability to form foci or colonies in agar. The tumor profile of a virus expressing S257C MT showed a striking deficiency in the induction of salivary gland tumors. The basis for this defect is uncertain. However, differences in activity for the wild type and mutant MT lacking the 14-3-3 binding site have been observed in transient reporter assays.
Polyomavirus middle T antigen (MT) is necessary (11, 39, 61) and often sufficient (63) for transformation. It is associated with membranes (35, 54), and its transforming ability depends on that association (11). MT has no known enzymatic activity but instead appears to function by association with cellular proteins involved in signal transduction. It has been a particularly useful model because mutations that affect particular associations have generally had clearly identifiable phenotypes. In contrast, single knockouts in receptors have often not been sufficient to generate clear-cut answers. Among the MT-associated proteins are protein tyrosine kinases of the Src family (Src, Yes, and Fyn) (14, 18, 33, 37). As a consequence of association with activated tyrosine kinase, MT is phosphorylated on tyrosine residues 250, 315, and 322 (10, 32, 34, 51). Mediated by these tyrosine phosphorylations, MT interacts with SHC (9, 19), PI-3 kinase (16, 36, 59), and phospholipase Cγ1 (57), respectively.
Not all MT associations are dependent on tyrosine phosphorylation. The binding to protein phosphatase 2A (PP2A) is one example of this (48, 64). This association appears to be important for the ability of MT to associate with protein tyrosine kinases (8). Not surprisingly, mutants defective in PP2A binding are defective in transformation. All three polyomavirus T antigens contain sequences resembling a DnaJ domain (12). As expected, MT also associates with hs70 (47). This interaction apparently occurs when MT either cannot bind PP2A or when the level of expression exceeds that of the cellular PP2A. The significance of the heat shock interaction is not known.
MT also binds proteins of the 14-3-3 family (45, 46). These abundant cell proteins clearly function in protein-protein associations. Among important proteins involved in cellular growth regulation and signal transduction, Raf (23, 25, 28), PI-3 kinase (4), and cdc25 phosphatase (17) are all known to associate with the 14-3-3 proteins. There is also evidence that 14-3-3 association can modulate apoptosis (67). While the evidence for association is strong, the role of these proteins has not been completely clarified (1, 7, 42). Most thinking has focused on the possibility that these proteins, through their ability to form dimers (40, 65), could function as adaptors.
In mammalian systems, 14-3-3 associates with Raf in a manner that depends on serine phosphorylation (44). A similar conclusion has been reached for the association of 14-3-3 with BAD (67). The work presented here shows that the association of MT with 14-3-3 is mediated by serine phosphorylation at residue 257. Further, while mutation of the 14-3-3 binding site has no striking effect on MT transforming ability in vitro, it leads to a specific alteration in the tumor profile and also alters the action of MT in assays of promoter activity.
MATERIALS AND METHODS
Cells, plasmids, and viruses.
NIH 3T3 and derived cell lines were grown in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% calf serum (HyClone). 10.1 are p53-negative murine 3T3 cells (31) and were maintained in 10% fetal calf serum. 293T cells were grown in Dulbecco’s medium supplemented with 10% calf serum. Sf9 (Spodoptera frugiperda) insect cells were maintained in Grace’s complete medium at 27°C as described by Summers and Smith (58a).
MT cDNA was reconstructed into an expression vector bearing the human cytomegalovirus (CMV) immediate-early promoter. To do this, 5′-GCGCGGATCCATCATGGATAGAGTTCTGAGC-3′ and 5′-GCGCGGATCCCTAGAAATGCCGGGAACGTT-3′ were used as outside primers for PCR. PCR was carried out by using Vent polymerase (New England Biolabs) in a Perkin-Elmer thermal cycler for 25 cycles of 1 min at 96°C, 2 min at 55°C, and 3 min at 72°C. After BamHI digestion, the fragment was ligated into the BamHI site of the pCMV NeoBam parent vector (49). Serine 257 was converted to alanine by using overlap PCR with wild-type CMV MT as a template. The 5′ primer (5′-GCGCGGATCCATCATGGATAGAGTTCTGAGC-3′) and a 257 noncoding primer (5′-TCGGGTTGGGGGATAGGCGTGGCTCCTCATAAC-3′) were used as one pair and a 257 coding primer (5′-GTTATGAGGAGCCACGCCTATCCCCCAACCCGA-3′) and the 3′ primer (5′-GCGCGGATCCCTAGAAATGCCGGGAACGTT-3′) were used as the other pair for the initial PCR. To regenerate virus bearing a mutation at 257, residue 257 was also converted to a cysteine so that the large T antigen reading frame could be conserved. The primer 5′-ATGAGGAGCCACTGCCTATCCCCCAACCCGA-3′ was used for this construction as described previously (26).
Myc-tagged 14-3-3 was made in a PRK5 (21, 41) background by amplification of ovine zeta cDNA from plasmid pHAF603 by PCR. Glutathione S-transferase (GST)-14-3-3 was made in Escherichia coli DH5α. Bovine zeta cDNA from plasmid pHAF603 was amplified by PCR and ligated into pGEX-2T cleaved with BamHI and EcoRI. The GST-14-3-3 protein was purified on glutathione agarose by standard methods.
Rous sarcoma virus β-galactosidase (RSV β-gal) (2, 22) was kindly provided by Amy Yee.
Metabolic labeling and MT Immunoprecipitation.
In vivo labeling was carried out by published procedures (51). Briefly, cells were rinsed with phosphate-free medium for [32P]orthophosphate labeling or with methionine-free medium or Hanks’ salts for [35S]methionine labeling, and then medium containing the label was added for 2 h. Typically, 25 to 100 μCi of [35S]methionine (Express label) (Dupont-NEN) or 200 to 1,000 μCi of [32P]orthophosphate (Dupont-NEN) was used in a volume of 1.5 to 2 ml on a 100-mm-diameter dish of cells.
Immunoprecipitations were done as previously described (50). T antigens were extracted with TEB (T-Ag extraction buffer) consisting of 0.137 M NaCl, 0.020 M Tris (pH 9.0), 0.00092 M CaCl2, 0.00049 M MgCl2, 1% (vol/vol) Nonidet P-40 (NP-40), and 10% (vol/vol) glycerol. After extraction for 20 min at 4°C, the lysate was spun for 15 min at 10,000 × g. After incubation with antibody and protein A-Sepharose for 60 min at 4°C with mixing, the beads were washed twice with phosphate-buffered saline, twice with 0.5 M LiCl–0.1 M Tris (pH 8.0), and once with distilled water, with 5 ml for each wash.
Electrophoresis.
Samples were routinely analyzed on discontinuous buffer-sodium dodecyl sulfate (SDS) gels (38). Western blot analysis (62) was carried out after transfer to nitrocellulose membranes. The blot was then blocked in Tris-buffered saline–Tween (500 mM Tris-Cl [pH 7.5], 1.5 M NaCl, 0.5% [vol/vol] Tween-20) containing 5% (wt/vol) dried milk (Carnation) for 1 h at room temperature. The blot was incubated with 1:50 PN-116 monoclonal anti-T antibody followed by 1:5,000 anti-mouse horseradish peroxidase-conjugated antibody (Amersham). Protein was detected by enhanced chemiluminescence (Amersham).
Isoelectric focusing (IEF) samples were prepared by treatment with Garrels’ sample buffer (9.5 M urea, 2% ampholytes [Pharmacia-LKB] [pH 3 to 10: pH 5 to 7: pH 6 to 8 in a 1:2:2 ratio], 100 mM dithiothreitol, 4% NP-40) (29). After incubation on ice for 30 min the samples were loaded onto IEF cylinders. First-dimension IEF gels composed of 4% bis/acrylamide (1.62/28.3 [wt/wt]), 8.0 M urea, 4% NP-40, and 2% ampholytes in the ratios described above were poured to a height of 15 cm. Samples were run at 400 to 500 V per h for 9,000 V · h. At that point, the gels were turned up to 800 V for 1 h. Second-dimension electrophoresis of the focused proteins was done on 5 or 7.5% acrylamide discontinuous buffer SDS slab gels.
Protein analysis.
To examine phosphorylations of various wild-type and mutant MTs, partial V8 protease digestion (51, 53) was performed. Immunoprecipitated proteins separated on a cylindrical gel (15 cm by 2 mm) were digested with Staphylococcus aureus V8 protease (U.S. Biochemicals, Cleveland, Ohio) prior to a second dimension of SDS electrophoresis. To prepare samples for Edman degradation, cyanogen bromide digestion was carried out as described previously (10).
Manual Edman degradation was performed according to the method of Sullivan and Wong (58). The peptide to be sequenced was dissolved in 30 μl of 50% acetonitrile and spotted onto a 1,4-phenylene diisothiocyanate-Sequelon (Millipore) disc. The disc was placed on a Mylar sheet on top of a heating block set at 50°C for 5 min. Covalent linkage was accomplished by adding a solution of N-methylmorpholine to the diisothiocyanate disc. After 30 min at room temperature the disc was washed extensively with water, extracted five times with trifluoroacetic acid (TFA) to remove unbound peptide, and then extracted with methanol. Edman degradation of the immobilized peptide was carried out in 1.5-ml centrifuge tubes with washing done by gentle vortexing. The following cycles were performed: (i) incubation with 0.5 ml of coupling reagent (methanol-water-triethylamine-phenylisothiocyanate; 7:1:1:1, vol/vol/vol/vol) for 10 min at 50°C, (ii) five washes with 1 ml of methanol, (iii) drying under vacuum for 5 min, (iv) incubation with 0.5 ml of TFA for 6 min at 50°C, (v) extraction with 1 ml of TFA–42.5% phosphoric acid (9:1, vol/vol), and (vi) Cerenkov counting of disc and of washes from steps 4 and 5. The disc was washed six times with 1 ml of methanol before the next cycle was started.
Reporter assays.
10.1 cells were transfected (13) at 15 to 25% confluence with pCMV-based MT plasmids and either RSV β-gal or a chloramphenicol acetyltransferase (CAT) reporter plasmid by N,N-bis[2-hydroxyethyl]-2-aminoethanesulfonic acid-buffered saline-mediated-CaPO4 precipitation in a total volume of 1.5 ml. A total of 1 ml of each precipitate was added to a 100-mm-diameter dish for the CAT assay. For transient expression, BES-buffered saline CaPO4 transfection of NIH 3T3 cells was employed. Cells were harvested 48 h posttransfection, and β-gal was measured by standard techniques. CAT activity was measured chromatographically (30). Thin-layer plates were quantitated with ImageQuant software (Molecular Dynamics) to determine the percentage of acetylated [14C]chloramphenicol versus all forms.
RESULTS
MT is phosphorylated on serine 257.
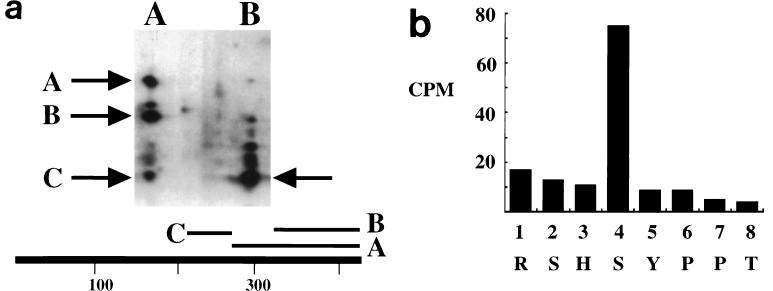
It has long been known that MT is phosphorylated on serine residues as well as tyrosine residues (43, 50, 51, 53, 55). In fact, the principal phosphoamino acid recovered after in vivo labeling is phosphoserine. Figure 1a shows that the major site(s) of serine phosphorylation in vivo resides in V8 fragment C; this fragment has been previously mapped as arising from residues 225 to 275 (53). To localize the site of phosphorylation further, a cyanogen bromide 32P-labeled fragment starting at residue 253 (10) was isolated from acrylamide gels. This peptide was then subjected to manual Edman degradation (Fig. 1b). There was specific release of label at turn 4 indicating the phosphorylation of MT at serine residue 257.
FIG. 1.
(a) Phosphorylation of MT. MT was labeled in vivo with 32PO4 (right) or in vitro with [32P]ATP (left). After resolution on cylindrical gels, the MT was digested with S. aureus V8 protease in the second dimension. The major peptides are indicated by the arrows, and their positions in the MT sequence are indicated below the gel. (b) Edman degradation of phosophrylated MT produced in Sf9 cells. The 32P released by each turn of Edman degradation of the cyanogen bromide fragment was measured by Cerenkov counting. Each cycle of Edman degradation is shown with the corresponding Py MT sequence indicated underneath.
To confirm that serine 257 was a phophosphorylation site, the residue was converted to an alanine by site-directed mutagenesis using overlap PCR. Figure 2 shows a mapping of wild-type and 257 mutant MT labeled in vivo with 32P. While the metabolic labeling of peptides A and B, which contain the tyrosine phosphorylation sites at residues 315 and 322, respectively, was similar for the two proteins, there was a dramatic decrease in phosphorylation of peptide C. This means that mutation of serine 257 gave the expected decrease of phosphorylation. The remaining phosphorylation of peptide C could well arise from tyrosine 250, a known phosphorylation site (32).
FIG. 2.
In vivo phosphorylation of wild-type and 257 mutant MT. 3T3 cells were transfected with CMV vectors expressing wild-type or 257 mutant MT. Cells were labeled with 32PO4 for 2 h. MT immunoprecipitates were resolved on cylindrical gels and then digested with V8 protease in the second dimension. The arrows indicate the positions of the V8 peptides shown in Fig. 1. WT, wild type.
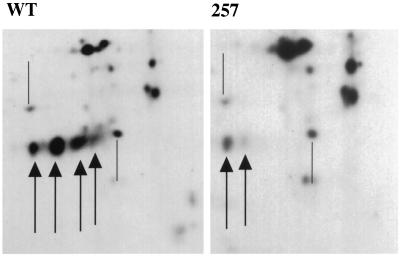
IEF suggested that the phosphorylation of MT at residue 257 was almost stoichiometric in 293T cells. MT does not focus on IEF gels, perhaps because of the membrane anchor. However, there is a fragment somewhat greater than 40 kDa known to lack the C terminus (52) that can be focused. In Fig. 3, the focusing patterns for this fragment are compared for 257 mutant and wild-type labeling. The multiple forms came from phosphorylation. When labeled with 32PO4, increasingly acidic forms had a higher specific activity (not shown). For wild-type MT, the most abundant fragment corresponded to a singly phosphorylated species. By contrast, when the 257 mutant was analyzed, it was clear that the pattern shifted one step to the basic direction compared to wild type. In other words the 257 mutant MT had on average one less mole of phosphate than the wild type per mole of protein. The predominant 257 mutant species was an unphosphorylated form.
FIG. 3.
IEF of a proteolytic fragment of MT. 293T cells were transfected with CMV vectors expressing wild-type or 257 mutant MT. Cells were labeled with 35S-Express label for 2 h. MT immunoprecipitates were resolved on IEF gels as described previously (15, 16). Portions of the IEF gels showing the resolved forms of the MT fragments for wild-type (left) and S257A (right) MTs are shown. Arrows indicate the major species; from left to right are increasingly acidic forms. For comparison of the two gels, the positions of two nonspecific cellular bands are shown by the lines.
The 257 mutant is defective in association with 14-3-3 proteins but retains associated tyrosine kinase activity.
MT is known to bind to 14-3-3 proteins (45, 46). Analysis of 14-3-3 binding suggested a consensus RSXSXP binding sequence (44), consistent with the MT sequence around residue 257. Both in vivo and in vitro binding assays show that the 257 phosphorylation site is important for binding of 14-3-3.
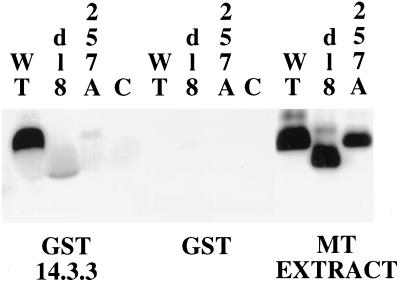
Recombinant baculoviruses were used to express MTs for the in vitro binding assay (Fig. 4). GST-14-3-3 zeta fusions bound to agarose beads were incubated with extracts from wild type, dl8 (carrying a deletion of residues 253 to 282), or 257A baculovirus-infected cells. Wild type bound to the 14-3-3 fusion but not GST; neither the point mutant 257A or the deletion mutant dl8 showed significant binding.
FIG. 4.
Binding of MT to 14.3.3 in vitro. Extracts from Sf9 cells infected with baculoviruses expressing control, wild-type, dl8, or 257 mutant MT were incubated with GST or a GST fusion with 14-3-3 bound to GST beads. After incubation for 90 min the beads were washed and the proteins were resolved on an SDS gel and blotted with antibody (F4) recognizing MT. WT, wild type.
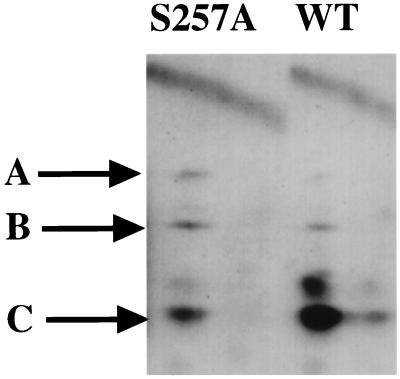
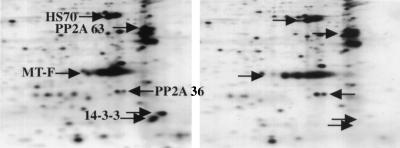
The same results were obtained from in vivo experiments. IEF represents a convenient method for analyzing MT-associated proteins. IEF of the 257A mutant showed that the 14-3-3 polypeptides were absent, while the spots for heat shock and the PP2A 63- and 36-kDa subunits were present (Fig. 5). In confirmation of this result, wild-type, but not mutant, MT was precipitated with 9E10 antibody recognizing a Myc epitope tag when a Myc-14-3-3 fusion was cotransfected (not shown).
FIG. 5.
Binding of MT to 14.3.3 in vivo. 293T cells were transfected with CMV vectors expressing wild-type (left) or 257 mutant (right) MT. Cells were labeled with 35S-Express label for 2 h. MT immunoprecipitates were resolved on IEF gels as described previously (15, 16). Proteins specifically associated with MT are indicated by the arrows. These include hsc70 (HS70), the 36 and 63-kDa subunits of PP 2A, as well as the 14-3-3 proteins.
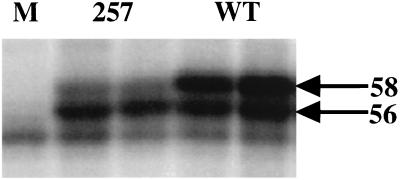
The association of MT with members of the Src tyrosine kinase family is restricted and occurs in a relatively slow fashion (3, 15). Given the high apparent level of serine 257 phosphorylation and the known abundance of 14-3-3 proteins, it was important to check MT-associated tyrosine kinase activity. Figure 6 shows that there was little difference in the amount of tyrosine phosphorylation of wild-type and mutant MTs. Peptide mapping confirmed that wild type and mutant were phosphorylated at similar sites (not shown). One difference that was noted was a reduction in the amount of the 58-kDa form of MT (Fig. 6). This was not surprising, since the 58-kDA form has previously been shown to depend on serine phosphorylation in this part of the MT molecule (51). Support of this interpretation can also be seen in the in vivo labeling shown in Fig. 2, where there is a reduction in the amount of the peptide derived from the 58-kDa form migrating more slowly than peptide C.
FIG. 6.
Comparison of in vitro kinase activity for wild-type and 257 mutant MTs. MT immunoprecipitates from control (lane (M) 1) and cells expressing 257 mutant (lanes 2 and 3) or wild type (lane 4 and 5) were labeled in vitro with [32P]ATP. Proteins were then resolved on a 7.5% acrylamide-SDS gel. The 56 and 58-kDa forms of MT are indicated by the arrows.
MT mutant in 14-3-3 binding retains the ability to transform cells and to induce most tumors.
The most direct functional test of the associated tyrosine kinase activity is transformation. To test transforming activity, NIH 3T3 cells were transfected with empty vector DNA, wild-type MT, or mutant DNAs (S257A or NG59, a classic nontransforming mutant), and dense foci were scored 9 days later. The S257A mutant was rather similar to wild type in the number of foci induced (43 versus 77, respectively, per 100-mm-diameter dish). Cells transfected with empty vector DNA or NG59 did not induce foci. It did appear that the foci were perhaps slightly smaller with the mutant. Whether this very modest difference resulted from slightly lower levels of MT is uncertain.
To provide a more complete test of 14-3-3 association, polyomavirus was reconstructed by using the PTA strain bearing a mutation changing serine 257 to a cysteine. This alteration was used instead of alanine so that the large T antigen reading frame remained intact. The virus obtained grew similarly to wild type as judged by the virus titers obtained after infection of baby mouse kidney cells. Table 1 shows the results of transformation assays on F111 rat embryo fibroblast cells. Assayed either by focus formation or growth in soft agar, the S257C virus behaved like wild type.
TABLE 1.
Viral transformation of F111 rat embryo cellsa
| Polyomavirus | PFU | TFU (foci) | TFU (agar) | PFU/TFU |
|---|---|---|---|---|
| Wild type | 2.7 × 106 | 1.2 × 103 | 1.5 × 103 | ∼2.0 × 103 |
| S257C mutant | 7.8 × 106 | 2.9 × 103 | 4.5 × 103 | ∼2.1 × 103 |
Virus stocks were grown on primary baby mouse kidney cells. Plaque titers (PFU) were determined on NIH 3T3 cells and transforming titers (TFU) were determined on F111 cells (24).
The mutant virus was then examined for its ability to induce tumors in mice. Newborn animals were infected with mutant virus as described previously (27). As shown in Table 2, all infected animals developed tumors. Virus recovered from tumor-bearing animals contained the expected mutation. The data for S257C can be compared to that previously obtained with wild type. Overall, the profiles are similar, with tumors of both epithelial and mesenchymal origin arising at approximately the same time and at similar frequencies at most sites. One notable exception was in the salivary glands, where the mutant failed to induce any tumors despite the fact that such tumors occurred in over half of the wild-type-infected animals. Fibrosarcomas were induced by the mutant but at a lower frequency than wild type. These findings are reminiscent of results with other MT mutants showing differential effects on tumors at different sites (6, 27, 66).
TABLE 2.
Tumor induction by virus with S257C mutationa
| Characteristic | PTA with S257C mutation | PTA |
|---|---|---|
| No. of mice with tumors/total no. of mice | 48/48 | 256/259 |
| Mean age (days) at necropsy (range) | 83 (53–227) | 108 (43–381) |
| No. of epithelial tumors (%) | ||
| Hair follicle | 40 (83) | 229 (88) |
| Thymus | 33 (69) | 227 (88) |
| Mammary gland | 23 (48) | 126 (49) |
| Salivary gland | 0 (0) | 148 (57) |
| No. of mesenchymal tumors (%) | ||
| Kidney | 17 (35) | 123 (47) |
| Bone | 12 (25) | 58 (22) |
| Subcutaneous connective tissue | 2 (4) | 56 (22) |
MT mutant in 14-3-3 binding showed altered behavior in promoter assays.
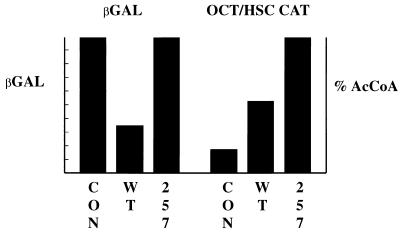
In searching for a possible role for 14-3-3 interaction with MT, the possibility that it modulates transcription was tested with 10.1 3T3 cells. Figure 7 shows that cotransfected wild-type MT can reduce expression from an RSV β-gal reporter. Wild-type MT increases expression from some synthetic promoters developed by Taylor and colleagues (60); Fig. 7 shows the results for one containing an octamer binding site, but similar results have been obtained for AP1 or ATF (not shown). The S257A mutant MT gave higher promoter activity than wild type, reflected as either loss of repression of the RSV construct or greater activation of the octamer construct. It could be imagined that wild-type MT simply titrates cellular 14-3-3. However, the difference between wild type and mutant was not overcome by the additional coexpression of 14-3-3 (not shown).
FIG. 7.
Reporter assays. 3T3 10.1 cells were transfected with an RSV β-gal (2, 22) or an octamer/heat shock CAT (OCT/HSC CAT) (60) reporter and CMV empty vector (CON) CMV wild type (WT), or S257A mutant MT. Activity was measured 48 h after transfection. Values are averages from 10 experiments (β-gal) or 5 experiments (CAT) done in duplicate or triplicate and are normalized to 100%.
DISCUSSION
This work establishes that serine residue 257 is a major site of phosphorylation of MT. Further, phosphorylation on serine residue 257 is the primary basis for association of MT with the 14-3-3 family of proteins. Based on the IEF experiments, phosphorylation of serine 257 appears to be stoichiometric. The kinase responsible for this phosphorylation is unknown. Given the extreme overexpression in 293T cells, it must have a very high capacity. In the same cells, it appears that the binding of 14-3-3 to MT appears to be less than stoichiometric. This conclusion is based on the observation that cotransfection of 14-3-3 increases the amount of protein associated with MT immunoprecipitates. Since 14-3-3 proteins are reported to be very abundant, this result might seem surprising. However, it can be imagined that much of the pool of cellular 14-3-3 is already associated with cellular phosphoproteins.
While the biochemistry of 14-3-3 association is clear, the consequences of the binding to MT complexes are less clear. There is a restriction in the amount of wild-type MT associated with Src family members and therefore downstream targets such as SHC and PI 3′-kinase (3, 9, 15). Evidence suggests that the NPTY motif starting at 248 is the SHC binding site (9). A little further upstream (residues 185 to 210) are elements required for Src binding (5, 20). It is therefore natural to ask whether 14-3-3 is part of the restriction. However, there is no indication that MT of mutant 257 has associated kinase activity significantly greater than wild type (Fig. 6). Similarly, 14-3-3 binding does not prevent association with SHC, since SHC can be found in 14-3-3 immunoprecipitates from MT transformed cells (not shown). Transformation assays in fibroblasts also support the conclusion that association with tyrosine kinases or their targets such as SHC does not depend substantially on the 14-3-3 interaction. 257 mutants were neither defective nor hyperactive in transformation. A very recent report raises the possibility that this region of MT is involved in multimerization of MT complexes (56).
The ability of 257 mutant MT to transform in vitro was broadly reflected in the ability of virus to induce tumors in animals. All infected animals had tumors with a broad range of tumor types represented. However, the absence of salivary gland tumors and the reduction in the subcutaneous connective tissues was dramatic. This underlines the fact that viral tumorigenesis is a complex process, reflecting contributions of the viral control regions, viral capsid proteins, and the transforming proteins. This differential effect on certain tumor types by mutation in MT in particular repeats a theme that has been seen before in examination of MTs with mutations at residues 315 (27) or 250 (6, 66). In those cases, the specific molecular defect can be traced to loss of binding of a specific signalling pathway. For 257, we can show that the 14-3-3 association alters the ability of MT to affect the activity of at least some promoters. Sorting out the precise basis for the effect represents an interesting problem for the future.
ACKNOWLEDGMENTS
This work was supported by NIH grants to B.S.S. (PO-CA50661 and R37-CA34722), T.R. (PO1-CA50661), T.B. (PO1-CA50661 and R35-CA44343) and D.P. (CA57327) as well as by the Markey Foundation (B.S.S.).
We thank Ken Dower and John Carroll for their assistance.
REFERENCES
- 1.Aitken A. 14-3-3 proteins on the MAP. Trends Biochem Sci. 1995;20:95–97. doi: 10.1016/s0968-0004(00)88971-9. [DOI] [PubMed] [Google Scholar]
- 2.Antonucci T K, Wen P, Rutter W J. Eukaryotic promoters drive gene expression in Escherichia coli. J Biol Chem. 1989;264:17656–17659. [PubMed] [Google Scholar]
- 3.Bolen J B, Thiele C J, Israel M A, Yonemoto W, Lipsich L A, Brugge J S. Enhancement of cellular src gene product associated tyrosyl kinase activity following polyoma virus infection and transformation. Cell. 1984;38:767–777. doi: 10.1016/0092-8674(84)90272-1. [DOI] [PubMed] [Google Scholar]
- 4.Bonnefoy-Berard N, Liu Y C, von Willebrand M, Sung A, Elly C, Mustelin T, Yoshida H, Ishizaka K, Altman A. Inhibition of phosphatidylinositol 3-kinase activity by association with 14-3-3 proteins in T cells. Proc Natl Acad Sci USA. 1995;92:10142–10146. doi: 10.1073/pnas.92.22.10142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brewster C, Glover H, Dilworth S M. pp60c-src binding to polyomavirus middle T antigen (MT) requires residues 185 to 210 of the MT sequence. J Virol. 1997;71:5512–5520. doi: 10.1128/jvi.71.7.5512-5520.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bronson R, Dawe C, Carroll J, Benjamin T. Tumor induction by a transformation-defective polyoma virus mutant blocked in signaling through shc. Proc Natl Acad Sci USA. 1997;94:7954–7958. doi: 10.1073/pnas.94.15.7954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burbelo P D, Hall A. 14-3-3 proteins. Hot numbers in signal transduction. Curr Biol. 1995;5:95–96. doi: 10.1016/s0960-9822(95)00022-4. [DOI] [PubMed] [Google Scholar]
- 8.Campbell K, Auger K, Hemmings B, Roberts T, Pallas D. Identification of regions in polyomavirus middle T and small t antigens important for association with protein phosphatase 2A. J Virol. 1995;69:3721–3728. doi: 10.1128/jvi.69.6.3721-3728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell K S, Ogris E, Burke B, Su W, Auger K R, Druker B J, Schaffhausen B S, Roberts T M, Pallas D C. Polyoma middle T antigen interacts with SHC via the NPTY motif in middle T. Proc Natl Acad Sci USA. 1994;91:6344. doi: 10.1073/pnas.91.14.6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carmichael G, Schaffhausen B S, Mandel G, Liang T J, Benjamin T L. Transformation by polyoma virus is drastically reduced by substitution of phenylalanine for tyrosine at residue 315 of middle-sized tumor antigen. Proc Natl Acad Sci USA. 1984;81:679–683. doi: 10.1073/pnas.81.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carmichael G C, Schaffhausen B S, Dorsky D I, Oliver D B, Benjamin T L. Carboxy terminus of polyoma middle-sized tumor antigen is required for attachment to membranes, associated protein kinase activities, and cell transformation. Proc Natl Acad Sci USA. 1982;79:3579–3583. doi: 10.1073/pnas.79.11.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheetham M E, Brion J P. Human homologues of the bacterial heat-shock protein DnaJ are preferentially expressed in neurons. Biochem J. 1992;284:469–476. doi: 10.1042/bj2840469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C A, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng S H, Harvey R, Espino P C, Semba K, Yamamoto, T. K. T, Smith A E. Peptide antibodies to the human c-fyn gene product demonstrate pp59c-fyn is capable of complex formation with the middle-T antigen of polyomavirus. EMBO J. 1988;7:3845–3855. doi: 10.1002/j.1460-2075.1988.tb03270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen B, Liu Y, Druker B, Roberts T M, Schaffhausen B S. Characterization of pp85, a target of oncogenes and growth factor receptors. Mol Cell Biol. 1990;10:2909–2915. doi: 10.1128/mcb.10.6.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen B, Yoakim M, Piwnica-Worms H, Roberts T, Schaffhausen B. Tyrosine phosphorylation is a signal for the trafficking of pp85, a polypeptide associated with phosphatidylinositol kinase activity. Proc Natl Acad Sci USA. 1990;87:4458–4462. doi: 10.1073/pnas.87.12.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conklin D S, Galaktionov K, Beach D. 14-3-3 proteins associate with cdc25 phosphatases. Proc Natl Acad Sci USA. 1995;92:7892–7896. doi: 10.1073/pnas.92.17.7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Courtneidge S A, Smith A E. Polyoma virus transforming protein associates with the product of the c-src cellular gene. Nature. 1983;303:435–439. doi: 10.1038/303435a0. [DOI] [PubMed] [Google Scholar]
- 19.Dilworth S M, Brewster C E, Jones M D, Lanfrancone L, Pelicci G, Pelicci P G. Transformation by polyoma virus middle T-antigen involves the binding and tyrosine phosphorylation of Shc. Nature. 1994;367:87–90. doi: 10.1038/367087a0. [DOI] [PubMed] [Google Scholar]
- 20.Dilworth S M, Horner V P. Novel monoclonal antibodies that differentiate between the binding of pp60c-src or protein phosphatase 2A by polyomavirus middle T antigen. J Virol. 1993;67:2235–2244. doi: 10.1128/jvi.67.4.2235-2244.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eaton D, Wood W, Eaton D, Hass P, Hollingshead P, Wion K, Mather J, Lawn R, Vehar G, Gorman C. Construction and characterization of an active factor VIII variant lacking the central one-third of the molecule. Biochemistry. 1986;25:8343–8347. doi: 10.1021/bi00374a001. [DOI] [PubMed] [Google Scholar]
- 22.Edlund T, Walker M D, Barr P J, Rutter W J. Cell-specific expression of the rat insulin gene: evidence for role of two distinct 5′ flanking elements. Science. 1985;230:912–916. doi: 10.1126/science.3904002. [DOI] [PubMed] [Google Scholar]
- 23.Fantl W J, Muslin A J, Kikuchi A, Martin J A, MacNicol A M, Gross R W, Williams L T. Activation of Raf-1 by 14-3-3 proteins. Nature. 1994;371:612–614. doi: 10.1038/371612a0. [DOI] [PubMed] [Google Scholar]
- 24.Fluck M M, Benjamin T L. Comparison of two early gene functions essential for transformation in polyoma virus and SV40. Virology. 1979;96:205–225. doi: 10.1016/0042-6822(79)90185-5. [DOI] [PubMed] [Google Scholar]
- 25.Freed E, Symons M, Macdonald S G, McCormick F, Ruggieri R. Binding of 14-3-3 proteins to the protein kinase Raf and effects on its activation. Science. 1994;265:1713–1716. doi: 10.1126/science.8085158. [DOI] [PubMed] [Google Scholar]
- 26.Freund R, Bronson R T, Benjamin T L. Separation of immortalization from tumor induction with polyoma large T mutants that fail to bind the retinoblastoma gene product. Oncogene. 1992;7:1979–1987. [PubMed] [Google Scholar]
- 27.Freund R, Dawe C J, Carroll J P, Benjamin T L. Changes in frequency, morphology, and behavior of tumors induced in mice by a polyoma virus mutant with a specifically altered oncogene. Am J Pathol. 1992;141:1409–1425. [PMC free article] [PubMed] [Google Scholar]
- 28.Fu H, Xia K, Pallas D C, Cui C, Conroy K, Narsimhan R P, Mamon H, Collier R J, Roberts T M. Interaction of the protein kinase Raf-1 with 14-3-3 proteins. Science. 1994;266:126–129. doi: 10.1126/science.7939632. [DOI] [PubMed] [Google Scholar]
- 29.Garrels J I. Two dimensional gel electrophoresis and computer analysis of proteins synthesized by clonal cell lines. J Biol Chem. 1979;254:7961–7977. [PubMed] [Google Scholar]
- 30.Gorman C, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harvey D M, Levine A J. p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine embryo fibroblasts. Genes Dev. 1991;5:2375–2385. doi: 10.1101/gad.5.12b.2375. [DOI] [PubMed] [Google Scholar]
- 32.Harvey R, Oostra G L, Belsham G J, Gillett P, Smith A E. An antibody to a synthetic peptide recognizes polyomavirus middle-T antigen and reveals multiple in vitro tyrosine phosphorylation sites. Mol Cell Biol. 1984;4:1334–1342. doi: 10.1128/mcb.4.7.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horak I D, Kawakami T, Gregory F, Robbins K C, Bolen J B. Association of p60fyn with middle tumor antigen in murine polyomavirus-transformed rat cells. J Virol. 1989;63:2343–2347. doi: 10.1128/jvi.63.5.2343-2347.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunter T, Hutchinson M A, Eckhart W. Polyoma middle-sized T antigen can be phosphorylated on tyrosine at multiple sites in vitro. EMBO J. 1984;3:73–80. doi: 10.1002/j.1460-2075.1984.tb01763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ito Y. Polyoma virus-specific 55K protein isolated from plasma membrane of productively infected cells is virus-coded and important for cell transformation. Virology. 1979;98:295–298. doi: 10.1016/0042-6822(79)90545-2. [DOI] [PubMed] [Google Scholar]
- 36.Kaplan D R, Whitman M, Schaffhausen B, Raptis L, Garcea R L, Pallas D, Roberts T M, Cantley L. Phosphatidylinositol metabolism and polyoma-mediated transformation. Proc Natl Acad Sci USA. 1986;83:3624–3628. doi: 10.1073/pnas.83.11.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kornbluth S, Sudol M, Hanafusa H. Association of the polyomavirus middle T antigen with c-yes protein. Nature. 1987;325:171–173. doi: 10.1038/325171a0. [DOI] [PubMed] [Google Scholar]
- 38.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Liang T J, Carmichael G G, Benjamin T L. A polyoma mutant that encodes small T antigen but not middle T antigen demonstrates uncoupling of cell surface and cytoskeletal changes associated with cell transformation. Mol Cell Biol. 1984;4:2774–2783. doi: 10.1128/mcb.4.12.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu D, Bienkowska J, Petosa C, Collier R J, Fu H, Liddington R. Crystal structure of the zeta isoform of the 14-3-3 protein. Nature. 1995;376:191–194. doi: 10.1038/376191a0. [DOI] [PubMed] [Google Scholar]
- 41.Luttrell L, Ostrowski J, Cotecchia S, Kendall H, Lefkowitz R. Antagonism of catecholamine receptor signaling by expression of cytoplasmic domains of the receptors. Science. 1993;259:1453–1457. doi: 10.1126/science.8383880. [DOI] [PubMed] [Google Scholar]
- 42.Marais R, Marshall C. 14-3-3 proteins: structure resolved, functions less clear. Structure. 1995;3:751–753. doi: 10.1016/s0969-2126(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 43.Matthews J, Benjamin T. 12-O-Tetradecanoylphorbol-13-acetate stimulates phosphorylation of the 58,000-Mr form of polyomavirus middle T antigen in vivo: implications for a possible role of protein kinase C in middle T function. J Virol. 1986;58:239–246. doi: 10.1128/jvi.58.2.239-246.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muslin A J, Tanner J W, Allen P M, Shaw A S. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 45.Pallas D C, Cherington V, Morgan W, DeAnda J, Kaplan D, Schaffhausen B, T. M R. Cellular proteins that associate with the middle and small T antigens of polyomavirus. J Virol. 1988;62:3934–3940. doi: 10.1128/jvi.62.11.3934-3940.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pallas D C, Fu H, Haehnel L C, Weller W, Collier R J, Roberts T M. Association of polyomavirus middle tumor antigen with 14-3-3 proteins. Science. 1994;265:535–537. doi: 10.1126/science.8036498. [DOI] [PubMed] [Google Scholar]
- 47.Pallas D C, Morgan W, Roberts T M. The cellular proteins which can associate specifically with polyomavirus middle T antigen in human 293 cells include the major human 70-kilodalton heat shock proteins. J Virol. 1989;63:4533–4539. doi: 10.1128/jvi.63.11.4533-4539.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pallas D C, Shahrik L K, Martin B L, Jaspers S, Miller T B, Brautigan D L, Roberts T M. Polyoma small and middle T antigens and SV40 small t antigen form stable complexes with protein phosphatase 2A. Cell. 1990;60:167–176. doi: 10.1016/0092-8674(90)90726-u. [DOI] [PubMed] [Google Scholar]
- 49.Pasleau F, Tocci M J, Leung F, Kopchick J J. Growth hormone gene expression in eukaryotic cells directed by the Rous sarcoma virus long terminal repeat or cytomegalovirus immediate-early promoter. Gene. 1985;38:227–232. doi: 10.1016/0378-1119(85)90221-5. [DOI] [PubMed] [Google Scholar]
- 50.Schaffhausen B, Silver J, Benjamin T. Tumor antigen(s) in cells productively infected by wild-type polyoma virus and mutant NG18. Proc Natl Acad Sci USA. 1978;75:79–83. doi: 10.1073/pnas.75.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schaffhausen B, Benjamin T L. Comparison of phosphorylation of two polyomavirus middle T antigens in vivo in vitro. J Virol. 1981;40:184–196. doi: 10.1128/jvi.40.1.184-196.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schaffhausen B S, Benjamin T L. Polyoma virus middle T antigen associated protein kinase. Cold Spring Harbor Conf Cell Proliferation. 1981;8:1281–1300. [Google Scholar]
- 53.Schaffhausen B S, Bockus B J, Berkner K L, Kaplan D, Roberts T M. Characterization of middle T antigen expressed by using an adenovirus expression system. J Virol. 1987;61:1221–1225. doi: 10.1128/jvi.61.4.1221-1225.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schaffhausen B S, Dorai H, Arakere G, Benjamin T L. Polyoma virus middle T antigen: relationship to cell membranes and apparent lack of ATP-binding activity. Mol Cell Biol. 1982;2:1187–1198. doi: 10.1128/mcb.2.10.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Segawa K, Ito Y. Differential subcellular localization of in vivo phosphorylation and nonphosphorylation middle-sized tumor antigen of polyoma virus and its relationship to middle-sized tumor antigen phosphorylation activity in vitro. Proc Natl Acad Sci USA. 1982;79:6812–6816. doi: 10.1073/pnas.79.22.6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Senften M, Dilworth S, Ballmer-Hofer K. Multimerization of polyomavirus middle-T antigen. J Virol. 1997;71:6990–6995. doi: 10.1128/jvi.71.9.6990-6995.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Su W, Liu W, Schaffhausen B, Roberts T. Association of polyomavirus middle tumor antigen with phospholipase C-gamma 1. J Biol Chem. 1995;270:12331–12335. doi: 10.1074/jbc.270.21.12331. [DOI] [PubMed] [Google Scholar]
- 58.Sullivan S, Wong T W. A manual sequencing method for identification of phosphorylated amino acids in phosphopeptides. Anal Biochem. 1991;197:65–68. doi: 10.1016/0003-2697(91)90356-x. [DOI] [PubMed] [Google Scholar]
- 58a.Summers M D, Smith G. A manual of methods for baculovirus vector and insect cell culture procedures. College Station, Tex: Texas Agricultural Experiment Station; 1987. [Google Scholar]
- 59.Talmage D A, Freund R, Young A T, Dahl J, Dawe C J, Benjamin T L. Phosphorylation of middle T by pp60c-src: a switch for binding of phosphatidylinositol 3-kinase and optimal tumorigenesis. Cell. 1989;59:55–65. doi: 10.1016/0092-8674(89)90869-6. [DOI] [PubMed] [Google Scholar]
- 60.Taylor I C, Kingston R E. Factor substitution in a human HSP70 gene promoter: TATA-dependent and TATA-independent interactions. Mol Cell Biol. 1990;10:165–175. doi: 10.1128/mcb.10.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Templeton D, Eckhart W. Mutation causing premature termination of the polyoma virus medium T antigen blocks cell transformation. J Virol. 1982;41:1014–1024. doi: 10.1128/jvi.41.3.1014-1024.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Treisman R, Novak U, Favaloro J, Kamen R. Transformation of rat cells by an altered polyoma virus genome expressing only the middle T protein. Nature. 1981;292:595–600. doi: 10.1038/292595a0. [DOI] [PubMed] [Google Scholar]
- 64.Walter G, Ruediger R, Slaughter C, Mumby M. Association of protein phosphatase 2A with polyoma virus medium tumor antigen. Proc Natl Acad Sci USA. 1990;87:2521–2525. doi: 10.1073/pnas.87.7.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiao B, Smerdon S J, Jones D H, Dodson G G, Soneji Y, Aitken A, Gamblin S J. Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature. 1995;376:188–191. doi: 10.1038/376188a0. [DOI] [PubMed] [Google Scholar]
- 66.Yi X, Peterson J, Freund R. Transformation and tumorigenic properties of a mutant polyomavirus containing a middle T antigen defective in Shc binding. J Virol. 1997;71:6279–6286. doi: 10.1128/jvi.71.9.6279-6286.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zha J, Harada H, Yang E, Jockel J, Korsmeyer S J. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]