Summary
Identifying immune modulators that impact neutralizing antibody responses against severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) is of great relevance. We postulated that high serum concentrations of soluble angiotensin-converting enzyme 2 (sACE2) might mask the spike and interfere with antibody maturation toward the SARS-CoV-2-receptor-binding motif (RBM). We tested 717 longitudinal samples from 295 COVID-19 patients and showed a 2- to 10-fold increase of enzymatically active sACE2 (a-sACE2), with up to 1 μg/mL total sACE2 in moderate and severe patients. Fifty percent of COVID-19 sera inhibited ACE2 activity, in contrast to 1.3% of healthy donors and 4% of non-COVID-19 pneumonia patients. A mild inverse correlation of a-sACE2 with RBM-directed serum antibodies was observed. In silico, we show that sACE2 concentrations measured in COVID-19 sera can disrupt germinal center formation and inhibit timely production of high-affinity antibodies. We suggest that sACE2 is a biomarker for COVID-19 and that soluble receptors may contribute to immune suppression informing vaccine design.
Subject areas: Human specimen, Properties of biomolecules, Immune response, In silico biology
Graphical abstract
Highlights
-
•
Soluble ACE2 can reach up to 1 μg/mL in serum of severe COVID-19 patients
-
•
The enzymatic activity of sACE2 is inhibited in 50% of COVID-19 patients’ sera
-
•
The level of sACE2 correlates inversely with RBM-directed antibodies in severe COVID-19
-
•
In silico, high concentrations of sACE2 hamper germinal center performance
Human specimen; Properties of biomolecules; Immune response; In silico biology
Introduction
Neutralizing antibodies (nAbs) specific for the severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) crucially contribute to protective immunity against the virus.1,2,3,4 A prominent target of nAbs is the receptor-binding motif (RBM), which is part of the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein.5,6,7 The RBM represents the direct binding interface of the virus spike to its host cell receptor angiotensin-converting enzyme 2 (ACE2). Many potent nAbs bind the RBM, thereby interfering with virus entry into cells.4,8,9 Such ACE2-competing antibodies are therefore desired products after virus infection and vaccination.
ACE2 is an essential component of the renin-angiotensin system and contributes to the regulation of blood pressure.10 The disintegrin and metalloprotease 17 (ADAM17) cleaves the ACE2 ectodomain of the membrane-bound receptor, inducing its shedding into the extracellular space.11 ACE2 thus exists as a membrane-bound and soluble protein in the human body. For SARS-CoV-1, in vitro studies have shown that binding of the viral spike increases ADAM17-mediated release of soluble ACE2 (sACE2),12,13 which may consequently lead to an excess of sACE2 during SARS-CoV infection. Hypoxia and low oxygen conditions, which are common features of SARS, were shown to upregulate the expression and protease activity of ADAM17 in vitro.14 Furthermore, elevated blood concentrations of circulating sACE2 were associated with cardiovascular disease, high waist-to-hip ratio, diabetes, chronic kidney disease, and male sex.15,16,17,18 These preconditions are known risk factors for severe COVID-19.19,20 Interestingly, previous reports suggested elevated sACE2 in COVID-19.21,22,23,24,25
Circulating virus receptors could potentially mask crucial viral epitopes recognized by B cells and thus interfere with the generation of neutralizing antibodies, which can be weak in COVID-19.6,26,27 Antigen binding by ectopically applied or pre-existent antibodies can downregulate the maturation of B cells and immunoglobulins specific for the covered epitope,28,29,30 which was recently also shown for active and passive SARS-CoV-2 vaccinations.31,32 The fragment crystallizable (Fc) constant part of antibodies and Fc-receptors were suggested to be dispensable for epitope masking.30 Consequently, any high-affinity ligand may mask B cell recognition of a particular epitope, and ACE2 was shown to bind the SARS-CoV-2 spike with high affinity.33,34,35
In this study, we show that large amounts of sACE2 circulate in severe COVID-19 and demonstrate in an in silico model that masking by the soluble virus receptor can negatively impact the maturation of germinal center B cells.
Results
Soluble ACE2 concentration is increased in moderate and severe COVID-19 patients
To study sACE2 in COVID-19, we optimized the detection of enzymatically active sACE2 (a-sACE2). Cleavage of an ACE2-specific fluorescent substrate was previously measured in plasma15,16,18 and, less frequently, in serum samples.17 Chelating agents in plasma can decrease catalytic activity of the ACE2 metalloprotease18 and might explain some of the variability observed in previous studies. Therefore, we collected serum and plasma samples from three randomly selected, severe COVID-19 patients and three healthy controls. Substrate cleavage velocity in serum outperformed plasma due to a wider and sample-independent optimum of the enzyme cofactor zinc (Figure S1, Table S1). Assay specificity was confirmed by the ACE2 inhibitor DX600.36 Several factors were adjusted to improve assay sensitivity (STAR Methods section).
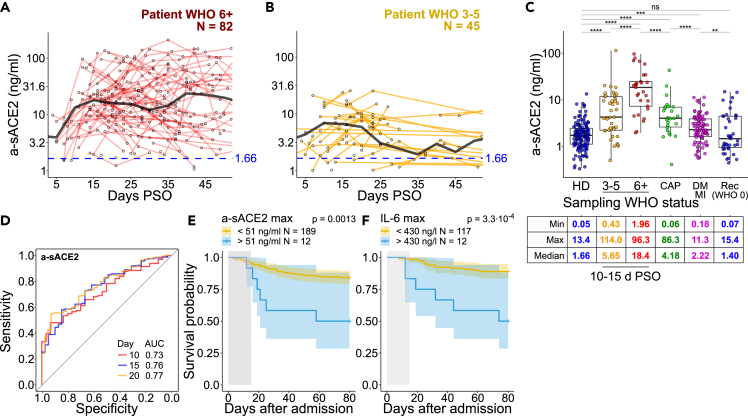
Next, we measured a-sACE2 in 717 serum samples from 295 PCR-confirmed COVID-19 patients collected between March 4th, 2020 until January 5th, 2021 over a time course of up to eight months post-symptom onset (PSO) plus 151 samples from donors tested negatively for SARS-CoV-2 (one sample each) (Table S2). COVID-19 severity was assessed at each sampling point and ranged from mild with ordinal scale WHO 1–2 (14.9%, outpatients), moderate WHO 3–5 (46.1%, hospitalized), and severe WHO 6+ cases requiring intubation and intensive care (39.0%). To address the dynamics of a-sACE2, we studied sera of 45 patients that reached moderate severity and 82 patients with a severe course of infection that provided multiple blood samples over time. COVID-19 patients with a severe disease course displayed highly elevated a-sACE2 concentrations during the first two months PSO, reaching 17.9 ng/mL at 15 days PSO (median, N = 314 samples; min 0.01, max 210.3 ng/mL; Figure 1A) when compared with 6.6 ng/mL for moderate cases (15 days PSO, N = 104; min 0.01, max 25.6 ng/mL, Figure 1B) and 1.7 ng/mL a-sACE2 in the healthy donor group (N = 151; min 0.1, max 13.4 ng/mL; Figures 1A and 1B).
Figure 1.
Circulating a-sACE2 correlates with mortality in COVID-19
(A and B) Concentration of a-sACE2 in (A) 82 severe and (B) 45 moderate patients up to 50 days post-symptom onset (PSO). Patients who donated blood more than once are selected here, WHO severity class corresponds to the highest severity level that a patient reached over the whole disease course. Black lines represent median values for the selected patients calculated as a trend over the whole course with five days binning; healthy donors (HD) median sACE2 level over the whole recruitment period is depicted in blue.
(C) Levels of a-sACE2 in healthy donors (151 samples, one sample per donor, no time-based selection performed), COVID-19 patients (717 samples from 295 patients), patients with commonly acquired pneumonia (CAP, 69 samples from 25 patients), and persons with self-reported history of diabetes mellitus and/or myocardial infarction (DM/MI, 91 samples, one sample per donor, NAKO study). WHO severity class is assessed at the time of sampling. Rec denotes recovered COVID-19 patients. For COVID-19 and CAP patients, a mean level within a time frame 10–20 days PSO is depicted. Time frame selection is based on the a-sACE2 level change over the disease course and samples availability, depicted in (A) and (B). Minimum (Min), maximum (Max), and median values by patient/donor group are shown.
(D) Receiver operating characteristic (ROC) curves for predicting death in all patients. Area under curve (AUC) calculated for maximum a-sACE2-concentrations within indicated days post-hospital admission.
(E and F) Cox regression models with two-sided 95% confidence interval for a-sACE2 and IL-6 showing overall survival probability based on maximum marker concentrations within 15 days post-admission (gray area). N = 12 patients in (E) and (F) are chosen independently based on a-sACE2 and IL-6 level and do not represent the same group. Statistical analyses by two-sided Mann–Whitney test (∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05; ns, not significant). In (A–C), sample mean is calculated from technical duplicates. Boxplots depict median +/− interquartile range. Whisker length is 1.5 interquartile ranges.
Healthy donors and patients suffering from moderate and severe COVID-19 displayed significant differences in sACE2 levels measured in a time frame between 10 and 20 days (mean value within the time frame per donor, Figure 1C). Healthy donors included volunteering healthcare workers and covered a wide range of demographic factors. No association of a-sACE2 with age, sex, or body mass index (BMI) was observed in severe disease, but a-sACE2 concentration was associated with sex in moderate disease (Figure S2). Male patients with moderate COVID-19 displayed on average a 2.1-fold increase in a-sACE2 compared with moderate female patients (median 8.4 vs. 3.9 ng/mL, p < 0.05). A sex association and significantly lower quantities of a-sACE2 were detected in healthy individuals (1.2-fold, median 1.9 for male vs. 1.6 ng/mL for female, p < 0.05).
Reported medical treatments, such as dexamethasone, did not significantly impact a-sACE2 level in severe COVID-19 (Figure S3). Likewise, diabetes, chronic kidney disease, or cancer did not significantly affect a-sACE2, although we detected a 2.4-fold elevation in WHO 3–5 patients with cardiovascular disease (Figure S4). Cardiovascular disease diagnosis in WHO 6+ patients was not associated with a-sACE2 level differences. In a separate cohort of 91 participants self-reported to suffer from myocardial infarction and/or diabetes mellitus37 with no COVID-19 diagnosis, a-sACE2 level was 1.3-fold higher compared with COVID-19-negative participants (Figure 1C).
Of note, a peak of a-sACE2 in the first 10–20 days after admission was associated with an increased risk of death, indicated by the receiver operating characteristic (ROC) curve with an area under the curve of 73%–77% (Figures 1D and S5). Thus, besides laboratory markers such as interleukin-6 (IL-6) and C-reactive protein (CRP),38 a-sACE2 is an independent predictor of death in COVID-19 (Figures 1E, 1F, and S5). Interestingly, non-COVID pneumonia patients (26 patients, 2–3 longitudinal samples per donor) with disease severities corresponding to COVID-19 WHO classes displayed significantly lower a-sACE2 levels (Figures 1C, S6A, and S6B).
sACE2 activity is inhibited in sera of moderate and severe COVID-19 patients
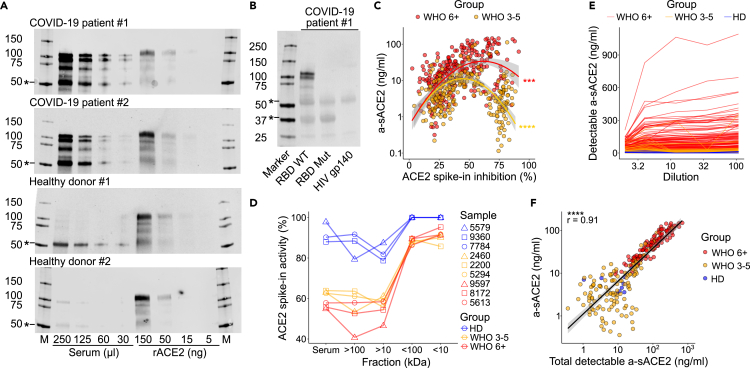
Determination of a-sACE2 in the blood might underestimate total sACE2 levels if enzymatically inactive sACE2 is present. Interferon (IFN)-inducible and enzymatically inactive ACE2 isoforms were recently described, which lack the SARS-CoV-2 binding site.39,40,41 To estimate total sACE2 capable of binding the spike, we conducted a serum pull-down using beads coated with the RBD, followed by western blotting. Surprisingly, sACE2 pull-down fractions largely exceeded the amounts of protein detected by enzymatic activity measurements (Figure 2A). The specificity of the pull-down and western blot was confirmed by an RBD mutant unable to bind sACE2 (Figure 2B). Moreover, the activity of recombinant ACE2 was inhibited after the addition to patient serum in 49.6% of COVID-19 samples (below median + 2-fold SD of healthy donors), further suggesting that enzyme activity measurements underestimate total sACE2 in blood samples (Figure S6C). When a-sACE2 was plotted against ACE2 inhibition, a likelihood-ratio test showed a significantly better U-shape fit compared with linear regression for both moderate and severe COVID-19, indicative of a negative feedback mechanism that may regulate ACE2 activity (Figure 2C). Inhibition was mediated by high-molecular-weight serum components >100 kDa (Figure 2D, Table S3). Compared with COVID-19, other pneumonia samples were lacking ACE2 inhibitory potential (Figure S6C). Importantly, upon serum dilution, an average 2-fold increase and amounts as high as 1,093 ng/mL a-sACE2 were observed (Figures 2E and 2F, Table S3). Together, our data indicate that ACE2 activity is dysregulated in COVID-19 and that large amounts of ACE2 virus receptor can circulate in patient blood.
Figure 2.
A large fraction of serum sACE2 is inactive and binds the SARS-CoV-2 spike
(A) ACE2 was pulled down from indicated volumes of serum from patients and HDs using RBD-coupled beads. Recombinant ACE2 (rACE2) was loaded as control. Markers (M) in kDa. ∗Unspecific bands.
(B) Specificity confirmed by a mutant RBD (A475R/G496R) lacking ACE2 binding and by an HIV gp140 control.
(C) Percent inhibition (%) of 50 ng/mL rACE2 after addition to serum plotted against a-sACE2. Significance indicates a likelihood ratio test to compare a two-degree polynomial model with linear regression for moderate or severe COVID-19.
(D) Activity of rACE2 spike-in (%) determined in serum fractions collected after ultrafiltration with indicated cut-off values.
(E) Determination of a-sACE2 in indicated reciprocal serum dilutions.
(F) Pearson correlation with a 95% confidence interval of a-sACE2 in undiluted samples and maximum a-sACE2 detected after serum dilution. (C), (E), and (F) include measurements of repeated longitudinal blood drawings. For gel source data, see Figure S11.
sACE2 level correlates inversely with ACE2-RBD interaction blocking potential in severe COVID-19 patients
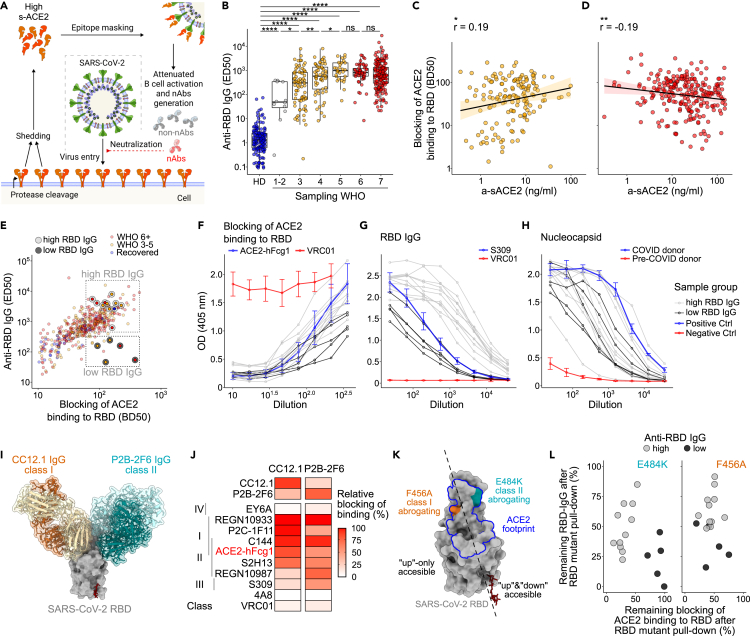
We next aimed to address if sACE2 in COVID-19 may mask the receptor-binding motif on the RBD and interfere with the antibody response (Figure 3A). We observed high levels of RBD-reactive immunoglobulin G (IgG) in both moderate and severe patients (Figures 3B and S7A). However, although the ACE2-RBD blocking potential of sera from moderate patients positively correlated with a-sACE2 (r = 0.25, p < 0.05, Figure 3C), severe patients showed a weak negative correlation (r = −0.19, p < 0.01, Figure 3D). The latter was not attributed to a weak humoral immune response as no correlation of a-sACE2 with anti-nucleocapsid IgG was observed (Figures S7B and S7C). It needs to be considered that blood levels of sACE2 may not represent the concentration of ligands available in ACE2-expressing tissues and may neither correspond to levels of the soluble receptor at the time when B cells encounter their antigen. Therefore, we aimed to determine if masking by sACE2 biases antibody recognition of RBD epitopes.
Figure 3.
Serology suggests that sACE2 impairs generation of antibodies preventing RBD-ACE2 interaction
(A) Scheme depicting SARS-CoV-2 spike epitope masking by sACE2.
(B) Serum IgG binding to RBD (ED50, serum dilution corresponding to 50% of maximum binding activity) of HDs (blue, N = 151), moderate (yellow, N = 120) and severe (red, N = 198) patients collected >20 PSO. Patients are stratified into the respective WHO class at the time of sampling. Statistical analyses by two-sided Mann–Whitney test (∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05; ns, not significant). Boxplots depict median +/− interquartile range. Whisker length is 1.5 interquartile ranges.
(C) Blocking of ACE2-RBD interaction (BD50, serum dilution corresponding to 50% of maximum blocking activity) plotted against a-sACE2 for moderate and (D) severe COVID-19. Data for individual samples are shown; samples drawn after 15 days PSO are selected. Spearman correlation performed with a 95% confidence interval.
(E) ED50 of anti-RBD IgG plotted against blocking of ACE2-binding (BD50).
(F) ACE2 competition, (G) anti-RBD IgG, and (H) anti-nucleocapsid IgG confirmed for two sample groups with high (gray, N = 12) versus low (black line, N = 5) RBD titers. Positive and negative controls in blue and red, respectively.
(I) Structural depiction of the RBD bound to CC12.1 and P2B-2F6. PDB: 6XDG, 7BWJ, and 6XC2.
(J) RBD binding of recombinant antibodies in competition with class 1 nAb CC12.1 and class 2 nAb P2B-2F6.
(K) Localization of class 1 and 2 abrogating RBD mutations F456A and E484K. ACE2 footprint shown in blue.
(L) RBD high versus low binding sera cleared by a pull-down with RBD WT, RBD-F456A, RBD-E484K, and MERS-CoV RBD as control. Remaining IgG binding to RBD and ACE2 competition is depicted in %.
ACE2-RBD binding can be prevented by so-called class 1 and class 2 nAbs, both targeting epitopes that overlap with the RBM.5,8 Although class 1 nAbs bind the RBD only in the “up” conformation of the spike, class 2 nAbs additionally recognize the RBD in the “down” state, which is inaccessible for ACE2 binding. Therefore, class 2 nAbs would be less affected by masking through sACE2. We selected 16 patients with similar ACE2 competition potential but low (N = 5) versus high (N = 11) RBD-specific IgG titers (Figure 3E). Patients with low and high RBD titers showed similar levels of nucleocapsid-specific IgG indicative of similar immune activation (Figures 3F–3H, Table S4). We addressed the contribution of class 1 versus 2 antibodies to RBD reactive serum antibodies in a competition ELISA, using the affinity matched class 1 nAb CC12.142 and class 2 nAb P2B-2F643 (Figure 3I, methods). We observed a similar blocking potential of selected sera for both antibodies (Figure S8A, Table S4). CC12.1 and P2B-2F6 recognize distinct RBD epitopes but moderately compete with each other, as also shown with a panel of class 1 and class 2 monoclonal antibodies (Figure 3J, Table S4). Therefore, it is difficult to decipher the contribution of individual nAb target sites to the ACE2 blocking potential by competition ELISA due to overlapping epitopes of class 1 and 2 nAbs. Hence, we made use of two RBD mutants, namely F456A and E484K, that lie within the class 1 or 2 nAb recognition sites, respectively (Figure 3K). Mutations at both residues are predominantly selected in RBD escape screenings in presence of convalescent plasma and have similar effects on loss of antibody binding.44 We aimed to address if RBD binding and competition for ACE2 binding to RBD correlate with titers of antibodies that target E484 or F456 by depleting sera with mutant RBDs. Interestingly, although antibodies remaining after E484K depletion separated the two patient groups with low and high RBD-specific antibody titers, no pattern could be observed for F456A depletions (Figures 3L and S8B–S8H, Tables S4 and S5). Although single mutant RBDs do not recapitulate the whole spectrum of class 1 and 2 nAbs, our data indicate that E484, which compared with F456 is also accessible when the RBD is in “down” state, may be a main determinant for blocking of ACE2 binding to RBD.
High sACE2 level inhibits germinal center evolution and neutralizing antibody production in silico
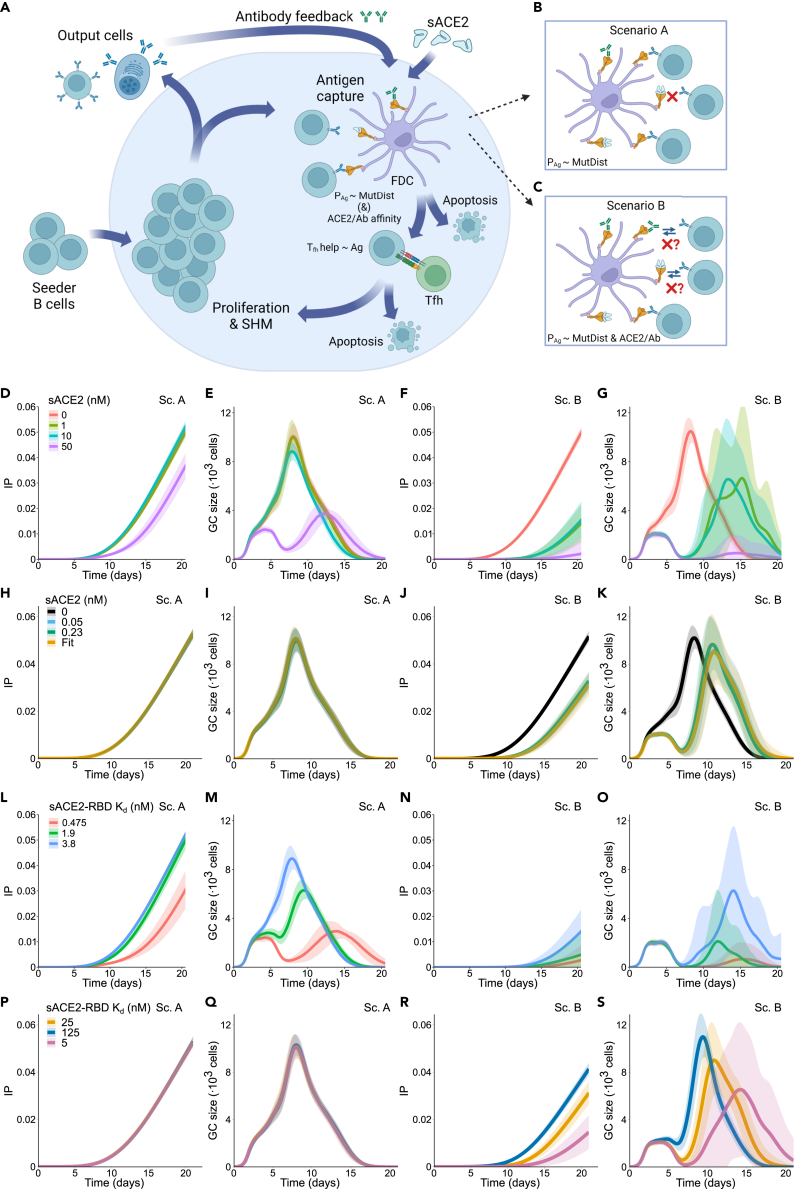
Given the limited availability of animal models that recapitulate severe COVID-19 with elevated levels of sACE2 over a long period of time, the effect of sACE2 on germinal center (GC) evolution was further investigated by adapting a previously published agent-based mathematical model.45,46 The model has been shown to quantitatively and qualitatively reproduce main GC characteristics by comparison with different sets of in vivo data.45,47,48,49 The broad schematic of the model is shown in Figure 4A. In the model, sACE2 as well as endogenous antibodies produced by the GC output cells inhibit B cell antigen (Ag) uptake. The effect of sACE2 on GC is modeled as an antibody feedback effect, which assumes that sACE2 acts like an antibody and alters antigen acquisition by B cells. The dissociation constant (KD) of sACE2 was set at 3.8 nM determined by bio-layer interferometry. As a fraction of sACE2 can be represented by monomeric enzyme, we also tested lower affinity of 25 nM as reported previously.50 We explored two scenarios: the epitope masking model (scenario A) and the epitope masking combined with a mean field increase of Ab-B cell competition (scenario B). Scenario A assumes that sACE2 masks antigen in an affinity-dependent manner. B cells only acquire unmasked antigen with a binding probability proportional to their B cell receptor (BCR) affinity (Figure 4B, see STAR Methods). In this scenario, masked antigen cannot be acquired by a B cell irrespective of its BCR affinity. Scenario B extends scenario A and entails that sACE2 reduces the binding probability of B cells for antigen as a function of sACE2 affinity (Figure 4C, see STAR Methods).
Figure 4.
Soluble ACE2 interferes with the germinal center reaction in silico
(A) Scheme depicting the germinal center (GC) response: GC reaction starts with an influx of low-affinity seeder cells. These cells are in a pro-apoptotic state and must acquire survival signals by antigen acquisition and T follicular helper cell (Tfh cell) signaling. Ag binding probability (PAg) is proportional to the mutational distance of the BCR to the Ag (MutDist, see STAR Methods and supplemental information). The Tfh cells preferentially provide signal to B cells that acquire larger amounts of antigen. Some of the selected B cells exit the GC and become output cells, whereas the rest recycle back and proliferate with mutations, resulting in daughter cells of higher affinity. These new cells again compete for survival signals, resulting in progressive B cell affinity increase. Serum sACE2 and the antibodies produced by output B cells can feedback to the GC by making antigen acquisition harder (mechanisms in B and C).
(B) Epitope masking mechanism (scenario A): sACE2 and antibodies can bind to the antigen and mask it, thus lowering the amount of antigen available for B cells.
(C) Epitope masking and mean field Ab-B cell competition mechanism (scenario B): BCR probability of binding the unmasked Ag is reduced depending on the average affinity of sACE2 and endogenous Abs (ACE2/Ab affinity).
(D–K) Simulation results for different sACE2 concentrations (0, 1, 10, and 50 nM constant concentration in D–G; 0, 0.05, 0.23 nM constant concentration and 0.05–0.23 nM linear increase in H–K): IP (D, H) and GC size (E, I) with scenario A; IP (F, J) and GC size (G, K) with mean field Ab-B cell competition. (L–S) Simulation results for different sACE2 affinities (0.475, 1.9, and 3.8 nM in L–O; 5, 25, 125 nM in P–S): IP (L, P) and GC size (M, Q) with epitope masking only; IP (N, R) and GC size (O, S) with scenario B. Mean (continuous lines) and standard deviation (shaded area) of simulations for a total of 20 simulated GCs are shown. Schemes (A–C) are created with BioRender.
First, we analyzed the impact of different concentrations of sACE2, including the value of 800 ng/mL (∼10 nM), observed in severe COVID-19 cases (Figure 2F). A fixed sACE2 concentration was injected at the start of the GC reaction and kept static over time: 1, 10, or 50 nM (Figures 4D–4G, 4L–4O, and S9). Additionally, to replicate the physiological range of sACE2 concentration and its temporal change, we injected 0.05 or 0.23 nM, the minimum and maximum value of the median sACE2 concentration observed in severe COVID-19 cases. In this case, the concentration was either kept static over time or linearly increased from 0.05 to 0.23 between day 5 and day 15 (Figures 4H–4K, 4P–4S, and S10). To measure the effect of sACE2 on antibody production, we quantified the immune power (IP) of the GC reaction (Figures 4D–4F, 4H, and 4J), which accounts for both affinity and quantity of endogenous antibodies51 (see STAR Methods). In scenario A, the sACE2 concentration affects the IP only at a concentration of 50 nM (∼4000 ng/mL) (Figure 4D). At the highest median concentration (0.23 nM, 18.4 ng/mL), the GC evolved as in the no-injection case (Figures 4H, 4I, and S10B) and could produce output cells of equal quantity and quality (Figures S10C and S10D).
On the contrary, in scenario B, the impact of sACE2 was evident even at the lowest concentration (0.05 nM, Figures 4J and 4K). Under every simulated sACE2 concentration, the IP was detectable five days later than in the no injection case and remained significantly lower (Figures 4F and 4J). Similarly, the GC expansion phase was delayed for all sACE2 concentrations (Figure 4K). Of note, despite a comparable GC peak size (Figure 4K) and increased affinity maturation of the GC and output cells (Figures S10E and S10G), the initial loss of output cells number could not be compensated before GC shutdown (Figure S10F).
Given that circulating sACE2 could include a mixture of various cleaved products, we tested a variation in sACE2 affinity (0.475–125 nM KD), whereas the sACE2 concentration either stayed constant (Figures 4L–4O) or followed the linear rise observed in the severe COVID-19 patients (Figures S10A, 4P–4S and S10H–S10M). Analogously to the variation of sACE2 concentration, under the assumption of scenario A, we did not detect a difference in the IP nor in the GC evolution (Figures 4H, 4I, 4P, 4Q, S9I, and S10H). Similarly, the quantity and quality of the output cells were altered only in the highest affinity of 0.475 nM (Figures S9G, S10I, and S10J). However, in scenario B, we found that the sACE2 affinity increase corresponded to a decrease in the IP (Figures 4J and 4R), due to the significant delay of the GC expansion phase (Figures 4K and 4S).
Our simulations show that concentrations and affinities of soluble receptors tested here are sufficient to produce significant suppression of the GC response. However, this was only observed in a scenario of intense feedback (scenario B). We conclude that soluble ACE2-mediated feedback is a possible mechanism for GC suppression in SARS-CoV-2 infections. However, a final proof requires further in vivo investigation.
Collectively, our data suggest that large amounts of sACE2 in COVID-19 may impair the antibody response to the RBM, which is a crucial target for neutralizing antibodies.
Discussion
Our work suggests that substantial shedding of the ACE2 ectodomain occurs during SARS-CoV-2 infection. Previous findings already reported elevated ACE2 in plasma of COVID-19 patients21,22,23 ranging from median values of 8.524 to 15.1 ng/mL25 in severe disease. Here, we observed a median a-sACE2 serum concentration of 17.9 ng/mL in patients with severe COVID-19 (15 days PSO), with the total ACE2 concentration reaching 1,093 ng/mL. We recently observed that COVID-19 sera inhibit ACE2 activity, which can explain the discrepancy between a-sACE2 and total sACE2 measurements.52 ACE2 reactive autoantibodies were described in multiple studies.53,54,55 However, our work suggests that anti-ACE2 IgG does not affect ACE2 enzymatic function, and its detection can be explained by polyreactivity.52
The dual role of soluble ACE2 in COVID-19 remains to be a subject of debate. Soluble ACE2 has been found to facilitate the SARS-CoV-2 entry into cells, and its increased level was associated with viremia.56,57,58 In a disease, a loss of ACE2 enzyme activity in tissues exacerbates acid aspiration or sepsis-induced acute lung injury.59 Massive shedding of ACE2 receptor into circulation suggests that tissue homeostasis of ACE2-dependent regulatory systems may be altered, including the renin-angiotensin, the kinin-kallikrein, and the coagulation system,60 thereby potentially contributing to inflammation, cardiovascular instability, or thrombosis in COVID-19. It was also assumed that endocytosis of virus-bound soluble ACE2 and membrane-bound ACE2 might exacerbate renin-angiotensin system dysregulation.23 In addition, it was suggested that circulating ACE2 contributes to hypotension and acute kidney disease.25 On the other hand, recombinant human ACE2 (rhACE2) was proposed as therapeutic agent for COVID-19 and decreased the viral load in a case study.61 A clinical trial, however, did not reveal a significant reduction of viremia (ClinicalTrials.gov: NCT04335136). Currently available data assume that although soluble ACE2 does not inhibit viral replication, rhACE2 therapy might be beneficial due to the restoration of the renin-angiotensin system function.62
SARS-CoV-2 neutralization by serum antibodies can be detected in most individuals after infection.6,26,27 Our in silico modeling suggests that levels of sACE2 detected in patients can interfere with the generation of nAbs that compete with ACE2 for binding to the RBM. A subset of sera exceeded 0.3 μg/mL sACE2. Assuming a dissociation constant of KD 3.8 nM, 1 μg/mL sACE2 (10 nM) may mask about 70% of RBM at viral loads of 1010 particles per milliliter considering up to 30 spikes per virion.63 Such masking resulted in a substantial reduction of germinal center size and immune power in simulations. Two simulation scenarios were investigated here, scenario A (epitope masking only) and scenario B (epitope masking with sACE2-affinity-dependent reduction in B cell binding for antigen). Although both are biologically plausible, scenario B was found to better agree with experiments.46 Of the two scenarios simulated, GCs in scenario B were much more susceptible to sACE2-mediated suppression. Therefore, our data might in part explain a disconnection of serum neutralization and RBD-specific antibodies64,65 and a delayed generation of neutralizing antibodies in severe COVID-19.27 Interestingly, individuals who received therapeutic monoclonal antibodies prior to SARS-CoV-2 vaccination displayed moderately reduced neutralizing antibody titers and an altered RBD target specificity of memory B cells.32 Likewise, high titers of high-affinity and mono-epitope-specific antibodies elicited during a primary response in mice attenuated cognate naive B cell recruitment into germinal centers.31
Immunodominance of class 2 over class 1 antibodies was suggested for convalescent plasma43 and was in part explained by accessibilities of the RBD in “up” and “down” conformations.66 As structural studies imply that around two-thirds of spikes display 1–2 RBDs in an ACE2-available “up”-state,55,59 we suggest that sACE2-mediated masking may further contribute to the dominance of class 2 antibodies.
Our data suggest that decoy receptors with high binding affinities can negatively impact humoral immunity by masking the most vulnerable sites of the virus. ACE2 can be released after spike binding,12 and the receptor is widely expressed in healthy tissue. We recently showed that an SARS-CoV-2 RBD designed to lack ACE2 receptor binding improves the neutralizing antibody response.67 As cognate receptors of other viruses are present at even higher concentrations and with broader expression profiles, our findings might be extrapolated beyond SARS-CoV-2 and inform vaccine design.
Limitations of the study
In this study, we analyzed samples from patients infected during early stages of the pandemic. All early variants of concern that circulated during the sampling period (March 2020–January 2021) featured a high-affinity Spike-ACE2 interaction.66,68,69,70,71,72,73,74 As Omicron variants were shown to bind to the host receptor with lower strength,75 a masking effect might be less pronounced. Furthermore, current study does not provide the information on viral load and ACE2 polymorphisms of the patients.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| EY6A | Zhou et al.76 | N/A |
| P2B-2F6 | Ge et al.43 | N/A |
| P2C-1F11 | Ge et al.43 | N/A |
| REGN10933 | Hansen et al.77 | N/A |
| REGN10987 | Hansen et al.77 | N/A |
| CC12.1 | Rogers et al.42 | N/A |
| C144 | Robbiani et al.1 | N/A |
| VRC01 | Wu et al.78 | RRID: AB_2491019 |
| S2H13 | Pinto et al.2 | N/A |
| S309 | Piccoli et al.5 | RRID: AB_2941328 |
| 4A8 | Chi et al.79 | N/A |
| Anti-human IgG-alkaline phosphatase (AP)-coupled antibody | SouthernBiotech | #2040-04 |
| Anti-human ACE2 antibody AF933 | R&D Systems | #AF933 |
| Anti-human ACE2 antibody F49433 | NSJ Bioreagents | #F49433 |
| Anti-human ACE2 antibody 3215 | ProSci Incorporated | #3215 |
| Anti-human ACE2 antibody AHP888 | Bio-Rad Laboratories | #AHP888 |
| IRDye® 800CW donkey anti-goat IgG | Li-Cor Biosciences | #925-32214 |
| IRDye® 800CW donkey anti-rabbit IgG | Li-Cor Biosciences | #925-32213 |
| IRDye® 800CW donkey anti-mouse IgG | Li-Cor Biosciences | #925-32212 |
| Biological samples | ||
| Pa-COVID-19 study human sera samples | Charité | DRKS00021688 |
| NAKO study human sera samples | NAKO, Peters et al.37 | N/A |
| CAPNETZ study human sera samples | CAPNETZ, capnetz.de | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant human ACE2 (N-terminus His-tagged) | In-house production | Uniprot: Q9BYF1 |
| Recombinant human ACE2 (hFcg1-tagged) | In-house production | Uniprot: Q9BYF1 |
| Recombinant SARS-CoV-2 Wuhan-Hu-1 RBD and Spike | Zhou et al.80 | GenBank: MN908947.3 |
| Recombinant MERS-CoV RBD | In-house production | Uniprot: R9UQ53 |
| Recombinant SARS-CoV-2 nucleocapsid protein | GeneTex | #GTX135357-PRO |
| (7-methoxycoumarin-4-yl)acetyl-Ala-Pro-Lys-2,4-dinitrophenyl (Mca-APK-Dnp) | Carbosynth Limited | #FM1-11000 |
| Captopril | Sigma-Aldrich | #PHR1307 |
| Bestatin hydrochloride | Sigma-Aldrich | #B8385 |
| Z-Pro-prolinal | Sigma-Aldrich | #SML0205 |
| DX600 | Enzo LifeSciences | #BV-9687-100 |
| Strep-Tactin®XT magnetic beads | IBA GmbH | #2-4090-002 or −010 |
| 4-Nitrophenyl phosphate disodium salt hexahydrate substrate | Sigma | #S0942-50TAB |
| Recombinant DNA | ||
| SARS-CoV-2 Wuhan-Hu-1 RBD and Spike | Zhou et al.80 | GenBank: MN908947.3 |
| human ACE2 | NCBI NIH NLM | Gene ID: 59272 |
| MERS-CoV Spike | NCBI NIH NLM | Gene ID: 14254594 |
| Software and algorithms | ||
| RStudio 2023.06.2 | posit.co | N/A |
| R 4.3.1 | r-project.org | N/A |
| In silico germinal center model | Meyer-Hermann et al.46,48,49 | Zenodo: https://doi.org/10.5281/zenodo.10605480 |
| GelAnalyzer 19.1 | gelanalyzer.com/?i=1 | N/A |
| UCSF ChimeraX v. 1.1 | Goddard et al.81 | N/A |
| Inkscape Graphics Editor | inkscape.org | N/A |
| Affinity Photo software | Serif (Europe) Ltd. | N/A |
| SWISS-MODEL protein modeling server | Waterhouse et al.82 | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kathrin de la Rosa (Kathrin.delarosa@mdc-berlin.de). Further information on the in silico GC modeling shall be addressed to Michael Meyer-Hermann (mmh@theoretical-biology.de).
Materials availability
The presented study did not generate new unique materials.
Data and code availability
-
•
Data: Data are provided in Tables S1, S2, S3, S4, and S5 including a-sACE2 measurements, patient age, gender, body mass index, sampling time point, and WHO score of patient samples, as well as raw data for the analyses performed.
-
•
Code: The mathematical model code is publicly available at Zenodo repository, https://doi.org/10.5281/zenodo.10605480. Any additional information is available upon request from the corresponding authors.
-
•
Other items: no other items were generated throughout the study.
Experimental model and study participants details
Sample donors and cohort characteristics
A total of 717 samples were obtained from COVID-19 patients, recruited as outpatients (37 donors, one sample each) or at hospitals (258 donors, 680 samples of the Pa-COVID-19 cohort study). Based on patient availability, single blood draws or longitudinal sampling up to 7 months post symptom onset were provided (160 patients of the Pa-COVID-19 cohort study). Patients included in this analysis were recruited from March 4th, 2020 until January 5th, 2021. Treatment followed the standard of care throughout the study period with the administration of dexamethasone for patients requiring respiratory support from mid-June, 2020 according to the preliminary results of the RECOVERY trial.83,84,85 Some patients received prednisolone or equivalents of other glucocorticoids before the introduction of dexamethasone or as a substitute for dexamethasone for individual reasons. Healthy donors group includes volunteering Charité healthcare workers tested negatively for SARS-CoV-2 (151 donors with single donations).
COVID-19 severity states were categorized at each sampling point according to WHO ordinal scale for clinical improvement: no clinical or virological evidence of infection (WHO0), ambulatory without limitation of activities (WHO1), ambulatory with limitations of activities (WHO2), hospitalized mild disease without oxygen therapy (WHO3), hospitalized mild disease with oxygen by mask or nasal prongs (WHO4), hospitalized severe disease with non-invasive ventilation or high-flow oxygen (WHO5), hospitalized severe disease with intubation and mechanical ventilation (WHO6), hospitalized severe disease with intubation, mechanical ventilation, and additional organ support - pressors, RRT, or ECMO (WHO7), death (WHO8). The Pa-COVID-19 study cohort was approved by the Charité Ethics Committee (EA2/066/20) and registered at the German Clinical Trials Register and WHO International Clinical Trials Registry Platform (DRKS00021688).86,87 The use of outpatient blood and further healthy donor samples was approved by the Charité Ethics Committee (EA2/092/20 and EA2/066/20). Outpatient subjects recovered from mild or moderate COVID-19 (WHO1-3) for at least 4 weeks before convalescent plasma donations.88 All samples were collected in compliance with the principles laid down in the 1964 Declaration of Helsinki and its later amendments. All patients gave written informed consent.
67% of hospitalized COVID-19 patients were male. Female patients had a median age of 59 years (min 18; max 99 years) and a median BMI of 27.8 kg/m2 (min 18; max 56 kg/m2). Male patients had a median age of 61 years (min 21; max 92 years) and a median BMI of 27.7 kg/m2 (min 16; max 54 kg/m2). COVID-19 outpatients were 43% male and had a median age of 45.4 years (min 20, max 64 years). The healthy donor group was 47% male. Female donors had a median age of 49 years (min 23; max 83 years) and a median BMI of 23.5 kg/m2 (min 17.4; max 37.9 kg/m2). Male donors had a median age of 49 years (min 20; max 86 years) and a median BMI of 25.4 kg/m2 (min 20.7; max 36.9 kg/m2). Information on ancestry, race, or ethnicity of study participants is not available.
69 serum samples were obtained from 25 pneumonia-affected individuals (SARS-CoV-2 PCR negative) recruited from February 2017 till June 2020 at hospitals within Community-Acquired Pneumonia Community Network (CAPNETZ). Based on patient availability, two (0 and 3 days after admission, 6 donors) or three blood draws were provided (0, 3, and 7 days after admission, 19 donors). Inclusion criteria were age ≥18 years, radiological proof of a new lung infiltrate, and either cough, fever, purulent sputum, or focal chest sign on auscultation. The median age of the patients was 67 years (range 26–85 years), 8 patients (32%) were female. Median BMI was 25.5 kg/m2 (range 17.3–38.1 kg/m2). Patients were classified into four groups according to the treatment administered: 1) invasive intubation and ICU, 2) non-invasive ventilation and ICU, 3) non-invasive ventilation only, 4) ICU only. The CAPNETZ study was approved by the Hannover Medical School committee, registration number 301–2008. All patients gave written informed consent. The diabetes mellitus and/or myocardial infarction samples are a part of the NAKO study and included single blood drawings from 33 volunteers with self-reported diabetes mellitus, 33 volunteers with self-reported myocardial infarction, and 25 with both myocardial infarction and diabetes mellitus. Diabetes mellitus group (DM) consisted of 23 men and 10 women with a mean age of 59.8 years. Myocardial infarction group (MI) comprised 25 men and 8 women with a mean age of 59.7 years. MI + DM group included 22 men and 3 women with a mean age of 62.8 years. The study was approved by the local ethics committee and is in accordance with national law and with the Declaration of Helsinki of 1975.37
Collection of sera and plasma
Sera were obtained from blood collected in tubes containing clot activator of silica particles, followed by centrifugation. Plasma was isolated from blood drawn using tubes or syringes pre-filled with EDTA (healthy donors) or heparin (patients), followed by 1:2 dilution with PBS/2mM EDTA and Ficoll density gradient centrifugation. Sera and plasma were stored at −80°C freezers and/or at 4°C. In NAKO, the sample collection and local processing was highly standardised by use of an automated liquid handling system for sample aliquoting.89 Samples used for this analysis had been stored in the vapor phase of liquid nitrogen at temperatures below −160°C in a fully automated sample handling robotic system.
Patient laboratory assessment
SARS-CoV-2 viral concentration was assessed by real-time RT-PCR in respiratory samples (nasopharyngeal swabs).90 Determination of C-reactive protein (CRP), ferritin, interleukin-6 (IL-6), and procalcitonin was carried out in accredited laboratories at Charité, Universitätsmedizin Berlin.
Ethical approval
The Pa-COVID-19 study and outpatient cohort were approved by the Charité Ethics Committee (EA2/066/20, EA2/092/20, EA2/066/20). The CAPNETZ study was approved by Hannover Medical School committee, registration number 301–2008. NAKO study was approved by the local ethics committee and is in accordance with national law and with the Declaration of Helsinki of 1975.
Method details
Cloning
Recombinant ACE2 with an N-terminal His-tag (rACE2-N’His) contained the Q18-V739 fragment of human ACE2 (Uniprot: Q9BYF1, GeneID: 59272, mRNA GenBank: AB046569.1) and was cloned into mammalian expression pcDNA3.1 vector containing a signal peptide (SP) (MGWSCIILFLVATATGVHS) and a molecular tag composed of eight histidine moieties (His-tag) connected to the N-terminus of ACE2 via a GSSGSSGSS-linker. We determined experimentally that C-end His-tag is not suitable for efficient protein purification likely due to the cleavage by proteases such as ADAM17 expressed by FreeStyle 293-F cells. Therefore, a His-tag was introduced between the SP and the N-terminus of the ACE2 fragment. The construct contained amino acids 615–732, which were previously shown to be crucial to form stable ACE2 dimers by Procko and colleagues.91 ACE2-hFcg1 was produced by fusing recombinant human ACE2 Q18-V739 fragment to human IgG1-Fc (E99-K330 portion, where 1st amino acid is G encoded by J-CH1 fusion). An RBD expression vector was kindly provided by Florian Krammer & colleagues.80 The mammalian expression vector pCAGGS containing the signal peptide spanning amino acids M1-Q14 and R319-F541 of the SARS-CoV-2 Wuhan-Hu-1 isolate (GenBank: MN908947.3) was complemented by a Twin-Strep-tag sequence (WSHPQFEKGGGSGGGSGGSAWSHPQFEK) upstream of the C-terminal hexahistidine tag. Two amino acid modifications, A475R and G496R, were introduced by PCR mutagenesis to create an RBD mutant that does not bind ACE2.80 Similarly, RBD with amino acid exchanges F456A or E484K were introduced to abrogate binding of class 1 or class 2 antibodies, respectively. Full spike comprising a 2-P-mutation (M1-Q1208 of the Wuhan SARS-CoV-2 variant GenBank: MN908947.3 with substitutions K986P and V987P) was cloned into the pcCDNA3.1 vector followed by a tobacco etch virus protease recognition site, GGGSG linker, fold-on trimerization domain (YIPEAPRDGQAYVRKDGEWVLLSTFL), Twin-Strep-tag and a hexahistidine encoding sequence. HIV gp140 BG505 NFL sequence92 was cloned into pcDNA3.1 with a 6xHis-tag. The signal peptide of SARS-CoV-2 (M1-Q14) was linked to an Avi-tag (GLNDIFEAQKIEWHE) followed by MERS-CoV RBD WT (G372-L588, GenBank: AFS88936.1) and C-terminal Twin-Strep-tag as well as hexahistidine tag. Heavy and light chain sequences of the following monoclonal antibodies were cloned into IgG1 heavy and kappa or lambda light chain expression vectors from Oxford Genetics (pSF-CMV-HuIgG1, pSF-CMV-HuLambda, and pSF-CMV-HuKappa): EY6A,93 P2B-2F6 and P2C-1F11,43 REGN10933 and REGN10987,77 CC12.1,42 C144,1 VRC01,78 S2H13,5 S309,2 4A8.79
Production of recombinant proteins
Cloning constructs were used to transfect FreeStyle 293-F cells that were grown in suspension using FreeStyle 293 expression medium (Life Technologies) at 37°C in a humidified 8% CO2 incubator rotating at 125 rpm. Cells were grown to a density of 2.5 million cells per ml, transfected using PEI (4 μg/ml in cell suspension) and DNA (1200 ng/ml in cell suspension), and cultivated for 3 days. The supernatants were harvested and proteins purified by His SpinTrap columns according to manufacturer’s instructions (Cytiva, 95056-290). The eluted protein was transferred to phosphate-buffered saline (PBS) via buffer exchange using Amicon Ultra-4 ultrafiltration column with 50 kDa cut-off (Millipore, UFC805008). Protein concentration was determined by His-tag specific ELISA using a mouse anti-His-tag antibody (Abcam, #ab18184) and a goat anti-mouse IgG Fc antibody conjugated to alkaline phosphatase (Southern Biotech, #SBA-1033-04) as detection reagent. Protein production was confirmed by SDS-PAGE and western blot using a mouse anti-His antibody (Abcam, #ab18184) and an IRDye 800CW donkey anti-mouse antibody (Li-Cor Biosciences, #925-32212).
Serum fractionation
Inhibitory capacity was measured in different serum size fractions by loading 100 μl on Amicon Ultra-0.5 ultrafiltration column with 10 or 100 kDa cut-off membrane (Merck, #UFC501024 or #UFC510024, respectively). The column was centrifuged at 20 000 g for 20 min (10 kDa cut-off) or at 10 000 g for 5 min (100 kDa cut-off). Flow-through (low molecular weight, MW) and supernatant (high MW) fractions were collected and tested along with the untreated serum samples for endogenous a-sACE2 enzymatic activity and inhibition of recombinant ACE2 spike-in as described below.
ACE2 enzymatic activity measurement
Enzymatic activity of ACE2 was determined by cleavage of the fluorescent substrate Mca-Ala-Pro-Lys(Dnp) (Mca = (7-methoxycoumarin-4-yl)acetyl; Dnp = (2,4-dinitrophenyl), Carbosynth Limited, #FM111000), which quenches the Mca fluorescence (Max Absorption/Emission upon cleavage = 325/393 nm) until the Pro-Lys linker is cleaved.94 The reaction was performed by mixing 7.5 μl serum in 1:2 - 1:54 dilutions with 7.5 μl PBS-1% BSA (or, when ACE2-inhibitory capacity was measured, 7.5 μl 100 ng/ml rACE2-N’His) or by using 15 μl plasma, followed by the addition of 85 μl of substrate solution. Substrate solution contained 8 μM Mca-Ala-Pro-Lys(Dnp) substrate, 75 mM trisaminomethane adjusted to pH 6.5 with hydrochloride, 1 M sodium chloride, 100 μM zinc chloride, and freshly added protease inhibitors: 10 μM Captopril (Sigma-Aldrich, #PHR1307), 10 μM bestatin hydrochloride (Sigma-Aldrich, #B8385), 10 μM Z-Pro-prolinal (Sigma-Aldrich, #SML0205). All fluorescence measurements were performed using 340 nm excitation and 420 nm emission light filters, (Cytation 5, BioTek). The sample baseline fluorescence (F340/4200) and absorbance at 450 nm were determined immediately after substrate addition, followed by incubation at 37°C in the dark. The second, third, and fourth fluorescence measurements were performed after 1h, 3-7h, and 20-24h (F340/420), 1-hour measurement was used for most of the samples, while subsequent time points were used only for samples in which the signal was below the detection limit. The number of fluorescence readings was minimized to avoid fluorescence bleaching. For data analysis, the amount of the substrate cleaved is calculated using the formula:
The absorbance at 450 nm was determined (A450), as samples displayed a wide variation in absorbance that affected the fluorescence measurement. The relation between 450 nm absorbance and 340/420 nm fluorescence was established by mixing different amounts of the low-density fraction of human serum (lipids fraction) with PBS-BSA buffer containing various amount of cleaved substrate. The RFU/[Mca] linear fit slope was determined to be equal to 202.94 – 123.3 ∗ log10(A450) by linear regression.
The concentration of a-sACE2 level was determined by interpolating the enzyme velocity (relative fluorescent units/time) of different amounts of self-made rACE2-N’His, calibrated with different commercial human recombinant ACE standards (Abcam, #ab151852; and Bon-Opus, #BP042) in the linear range of the calibration curve: initial enzyme velocity before fluorescence saturation. Recombinant ACE2 was diluted to various concentrations from 0.01 to 100 ng/ml to obtain a calibration curve via linear regression with a zero-point intercept. The slope coefficient of the fitted line was used to calculate the a-sACE2 activity in the samples using the following formula:
All samples were measured in duplicates or in serial dilutions. The specificity of ACE2 substrate cleavage in serum and plasma was confirmed using the ACE2 selective inhibitor DX600 (Enzo LifeSciences GmbH, BV-9687-100). Sample storage and freezing did not alter ACE2 activity.
ACE2 pull-downs
Recombinant RBD was expressed as described, containing the nucleotide sequence for the RBD of the Wuhan variant, codon-optimized for expression in human cells, and containing a Twin-Strep-tag (WSHPQFEKGGGSGGGSGGSAWSHPQFEK) - a peptide sequence allowing the recombinant protein to bind to Strep-Tactin. 10 μg SARS-CoV-2 WT RBD, a mutant RBD that abrogates ACE2 binding (A475R/G496R), or a HIV gp140 BG505 control antigen was mixed with 100 μl of 5% magnetic beads suspension covalently coated with Strep-Tactin (Strep-Tactin®XT #2-4090-002 or -010 from IBA GmbH) after equilibration according to manufacturer protocol. The mixture was incubated at 37°C for 1h on the orbital shaker set at 1400 rpm. The unbound RBD was removed by pelleting and resuspending the magnetic beads 5 times in 1 ml phosphate-buffered saline (PBS). The RBD-bound beads were resuspended in 500 μl freshly collected serum and incubated at +4°C overnight with constant mixing. The beads were pelleted and washed 3 times with 1 ml PBS with 0.05% Tween-20. Pelleted beads are resuspended in 20–40 μl buffer containing 94 mM trisaminomethane adjusted to pH 6.8 with hydrochloride, 12.5% glycerol, 2.3% SDS, and 0.005% bromphenol blue and incubated at 95°C for 30 minutes. Beads were pelleted, and supernatants were collected for western blot analysis.
RBD pull-downs
Antibody depletion from serum was performed with SARS-CoV-2 WT, MERS-CoV WT, or mutant RBDs (E484K, F456A) carrying amino acid exchanges to abrogate class 1 or class 2 antibody binding. After equilibration according to manufacturer protocol, 120μl of 5% Strep-Tactin®XT magnetic beads (IBA GmbH, #2-4090-002 or -010) were coupled with 24 μg of recombinant RBD via a C-terminal Twin-Strep-tag at 37°C for 1 hour. The unbound RBD was removed by pelleting the magnetic beads and washing the latter 3 times in 1 ml phosphate-buffered saline (PBS). Beads were blocked with PBS 1% BSA shaking at 37°C for 1 hour. After washing, serum was added to the beads to reach a final dilution of 1/10 in 200μl final volume and the mixture was incubated in a rotisserie shaker overnight at 4°C. The next day, tubes were placed on a magnetic holder and supernatants were collected. Depleted sera were analyzed by ELISA assays to determine RBD binding and ACE2 competition potential of the remaining antibodies. Percent of remaining antibodies after depletion with mutant RBDs were determined using MERS-CoV WT (corresponding to 100%) and SARS-CoV-2 WT (corresponding to 0%) control RBD pull downs.
ELISA
In-house produced recombinant SARS-CoV-2 RBD or commercial nucleocapsid protein (GeneTex, #GTX135357-PRO) was immobilized on a high-binding 96 well ELISA plate (Corning, #CLS3690) at 4 μg/ml in PBS (Sigma Aldrich, MO, USA) overnight at +4°C. Plates were blocked for 1h with 1% BSA (Thermo Fisher, Gibco, MA, USA) in PBS at room temperature. Sera were diluted in PBS 1% BSA to indicated serial dilutions, added to coated plates and incubated for 1h at room temperature. Plates were developed with an anti-human IgG-alkaline phosphatase (AP)-coupled antibody (SouthernBiotech #2040-04) diluted 1:500 in PBS 1% BSA. Bicarbonate buffer with 4-Nitrophenyl phosphate disodium salt hexahydrate substrate (Sigma, #S0942-50TAB) was added and absorbance was measured at 405 nm in a Cytation 5 device (BioTek). Between all indicated incubation steps, plates were washed 3 times with PBS 0.05% Tween-20. 50% of maximum IgG binding to RBD or nucleocapsid (ED50) was determined for each sample by sigmoid curve fitting with non-linear regression performed in R (stats package). For curve fitting, upper and lower plateaus of the S309 reference antibody (RBD) or of a COVID-19 WHO6 reference serum (Nucleocapsid) were applied to all samples. The positive control was used to normalize independent measurements.
Competition ELISA
Serial dilutions of sera or indicated concentrations of unlabeled monoclonal antibodies in PBS 1% BSA were added to RBD-coated 96-well plates. After 1h of incubation at room temperature, in-house produced biotinylated ACE2-hFcg1 was added to a final effective concentration 70 (EC70) in PBS 1% BSA. For class 1 and class 2 nAb competition assays, biotinylated CC12.1 or P2B-2F6 were added to a final EC30. After another hour at room temperature, plates were incubated with an AP-coupled streptavidin (Southern Biotech, #SBA-7100-04) at a 1:500 dilution in PBS 1% BSA to detect biotinylated ACE2-hFcg1, CC12.1 and P2B-2F6 antibodies that were not prevented by serum antibodies from binding to RBD. Similar to ED50, 50% of maximum ACE2-, CC12.1- and P2B-2F6-blocking potentials (BD50) were determined by sigmoid curve fitting with non-linear regression performed in R (stats package). The area under the curve (AUC) was determined by GraphPad Prism software. Upper and lower plateaus of the non-biotinylated ACE2-hFcg1, CC12.1 and P2B-2F6 controls served as a reference.
The equilibrium dissociation constant (KD) for binding SARS-CoV-2 spike or RBD ranged from 1-37 nM for ACE2 (KD = 1.2 nM, ka = 1.4×105 [1/Ms], kd = 1.6 × 10−4 [1/s]34; KD = 36.6 nM, ka = 1.36 × 105 [1/Ms], kd = 4.7 × 10−3 [1/s]32); 17 nM for CC12.1 (KD = 17 nM, ka = 8.62×104 [1/Ms], kd = 1.5 × 10−3 [1/s]95); and 5–16 nM for P2B-2F6 (KD = 5 nM4, and KD = 15.62 nM, ka = 6.42 × 106 [1/Ms], kd = 0.098 [1/s]43).
Western blotting
Different amounts of pull-down eluates corresponding to indicated volumes of the serum were loaded into the wells of a 4–15% gradient polyacrylamide gel (Bio-Rad, #4568083) together with the Precision Plus Protein Dual Color Standard (BioRad, #1610374). Proteins were transferred to a nitrocellulose membrane (BioRad, #1704159), blocked with 30 ml of 5% skimmed cow milk diluted in trisaminomethane-buffered saline with 0.1% Tween-20 (TBST) for an hour at room temperature on the orbital shaker. The membrane was stained in 5 ml anti-human ACE2 antibodies (R&D Systems, #AF933; #F49433, NSJ Bioreagents, #3215, ProSci Incorporated, #AHP888 Bio-Rad Laboratories) diluted to 0.2 μg/ml at +4°C overnight with constant mixing on the roller. The stained membrane was washed 5 times with 30 ml of TBST and stained with IRDye® 800CW donkey anti-goat IgG (Li-Cor Biosciences, #925-32214), donkey anti-rabbit IgG (Li-Cor Bioscience, #925-32213) or donkey anti-mouse IgG (Li-Cor Bioscience, #925-32212) antibodies at 0.2 μg/ml conjugated to the Infrared fluorescent dye 700DX for an hour at room temperature with constant mixing on the roller. The membrane was washed 3 times with 30 ml TBST and scanned with an infrared membrane scanner (Li-Cor Odyssey IR scanner). The ACE2 concentration was determined by densitometry analysis with GelAnalyzer 19.1 (http://www.gelanalyzer.com/?i=1) using 60-170 kDa portion of the sample lanes and comparing the intensity to loading controls: 150, 50, 15, and 5 ng of recombinant human rACE2-N’His produced in the laboratory.
Quantification and statistical analysis
Statistics
Tests were two-tailed, and a p value lower than 0.05 was considered statistically significant. A Shapiro-Wilk test was used for normality testing of continuous variables. An independent t-test was used when continuous data met the criteria of the normality test. Otherwise, the Mann-Whitney U test was used. Pearson and Spearman correlations were performed with a 95% confidence interval.
To model a-sACE2 and ACE2 spike-in activity relationship, we used the non-linear least squares method provided by stats (v4.0.3) R package. We tested linear and two-degree polynomial models for data stratified by severity state and used the likelihood ratio test to compare the models.
For the Cox regression model, we used the survival (v3.2.7) and survminer (v0.4.8) R packages. We set a “last follow-up date” to March 1st, 2021 for patients with missing outcome date and outcomes other than death and ignored events later than 80 days after admission. We further excluded undated samples, patients without outcome or undated deaths. We used patient gender, dexamethasone treatment, cardiovascular disease status, and lipid metabolism as covariates and tested against log2 measurement values. For the visualization in Kaplan-Meier plots, we used cut-offs in the 90-95th percentile range of mean measurement values per patient to define strata.
For the analysis of receiver operator characteristic (ROC) curves and calculation of the AUC with 95% confidence intervals, the pROC R package (v. 1.17) was used.96 Optimal cut-off thresholds were calculated using the OptimalCutpoints R package (v. 1.1) as follows: optimal cut-offs were calculated using a pre-set specificity value (0.9 or 0.95) as well as using the maximum Youden J-statistics.97
Calculation of a-sACE2 delta pre and post dexamethasone treatment
Donors were separated into two groups based on dexamethasone administration. 32 donors had at least one sample drawn during dexamethasone administration for which a-sACE2 was determined. On average, dexamethasone was administered between 8 (median, 5–11) days and 16 (median, 13–20) days post symptom onset (dexamethasone-time-window). The ACE2 delta was determined by subtracting the a-sACE2 value of samples directly prior to dexamethasone administration from those after dexamethasone treatment. Controls were determined by taking untreated patients (45 donors) of the same severity class (WHO3-5 or WHO6+) and calculating the a-sACE2 delta by subtracting a-sACE2 of the first sample within the dexamethasone time window from the consecutive data point of the respective patient.
Protein structure analysis
Protein structures were depicted with UCSF ChimeraX v. 1.181 using the following PDB: 6XDG for SARS-CoV-2 RBD, 7BWJ for P2B-2F6 IgG, 6XC2 for CC12.1 IgG. Mouse ACE2 protein was homology-modeled to human ACE2 (template: 6m18.1.B) via the SWISS-MODEL protein modeling server82 using the Uniprot sequence Q8R0I0 and further modified with UCSF Chimera X v. 1.1. Figures were generated using Inkscape and Affinity Photo software (Serif (Europe) Ltd., UK).
In silico model
A previously published agent-based model46 is used as the reference framework for the GC simulation (see supplemental information). To model sACE2 impact on GC evolution, the strategy developed for describing the Antibody feedback mechanism in Zhang et al. and Meyer-Hermann M45,46 is utilized.
In the model, endogenous antibodies (Abs) are produced by Output cells. Therefore, the antibody concentration depends on the Output cells’ differentiation rate and on the Ab production and degradation rates, and the Ab affinity is derived from the affinity of Output cells ([98] and supplemental information). A fixed sACE2 concentration is injected at the initial time point of the GC simulation, and it is assumed to be constant throughout the GC reaction. A range of concentrations were simulated: 0, 0.05, and 0.23 nM. The reference value of the dissociation constant KD of sACE2 is 25 nM as reported previously.50 To analyze the impact of possible increased affinity of sACE2 to the antigen, we simulated sACE2 with 5-fold change in affinities (KD = 5 nM and KD = 125 nM, respectively). The analysis was carried out with two scenarios. In the epitope masking model (scenario A), sACE2 and output cells-produced antibodies mask the antigen on follicular dendritic cells by forming immune complexes (ICs), according to a chemical kinetics equation (for the mathematical formulation, see [46] and supplemental information). The second model (scenario B) extends scenario A with a reduction of the Ag binding probability in dependence on the presence of sACE2.46 To model the latter mechanism, an additional feature from [46] is implemented here. B cells locally compete with the soluble antibodies for binding of free antigen. Without competition, the binding probability (or affinity) is assumed to depend on the Hamming distance d to the optimal clone in number of mutations in a four-dimensional shape space according to a Gaussian a = exp(−d2/w2) between 0 and 1 using a width of w = w0(1 − A), with w0 = 2.8 mutation.51 A is the average of affinity a of all produced antibodies and injected sACE2 calculated with the same Gaussian in the non-competition limit, i.e., with width w = w0. This model reflects antigen masking plus competitive binding of the remaining free antigen retained by higher-affinity antibodies. To measure the efficiency of the GC reaction, we used the immune power (IP) described in [51].
Acknowledgments
We thank the Pa-COVID-19 Study Group, especially Denise Treue, Tatjana Schwarz, Jan-Moritz Doehn, and Victor Corman for sample collection, testing, and biobanking. We thank the CAPNETZ Stiftung and the CAPNETZ study group with all investigators contributing to the study cohort. We thank Klaus Rajewsky for critically reading the manuscript and Florian Krammer for providing SARS-CoV-2 expression plasmids. We thank Anja Schütz and the MDC protein production facility for supporting large-scale purifications of recombinant proteins. This work was funded by the German Research Foundation (394523286; to K.D.L.R.); the European Research Council (948464; to K.D.L.R.); the Helmholtz Association's Initiative and Networking Fund (Project “Virological and immunological determinants of COVID-19 pathogenesis – lessons to get prepared for future pandemics; KA1-Co-02 “COVIPA”, to K.D.L.R.); the Berlin Institute of Health at Charité; and the German Federal Ministry of Education and Research (NaFoUniMedCovid19 – COVIM, NAPKON, FKZ: 01KX2021, PROVID - FKZ 01KI20160A, SYMPATH - (01ZX1906A), Pa-COVID-19 Study; to L.E.S., F.K., M.W.). The NAKO is funded by the Federal Ministry of Education and Research (BMBF) (project funding reference numbers: 01ER1301A/B/C and 01ER1511D), the federal states, and the Helmholtz Association, with additional financial support by the participating universities and the institutes of the Leibniz Association. A.G. has received funding from the Innovative Medicines Initiative 2 Joint Undertaking (JU) under grant agreement No 101007799. The JU receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA. M.S. was supported by the COSMIC Marie Sklodowska-Curie grant (765158).
Author contributions
C.R., M.L., and K.D.L.R. designed the experiments. A.G. and M.S. performed the in silico germinal center simulation. L.S., C.R., and M.L. expressed and purified the proteins. C.G., L.S., C.R., and M.L. cloned, sequenced, and expressed antibodies and performed ELISA binding assays. L.S. established the ACE2 ELISA competition assay. M.L. established and applied the a-sACE2 enzymatic activity assay, designed chimeric ACE2 as well as SARS-CoV-2 RBD variants, and visualized the data. C.R. designed mutant RBDs for pull-down experiments and established depletion and class-I/II nAb competition assays. C.V.G. performed ELISA binding assays and a-sACE2 enzymatic activity measurements. B.O. and J.W. performed survival analysis. L.H. collected and provided patient samples. P.S., C.T., M.W., L.E.S., F.K., I.M.V., and T.P. collected and provided samples and phenotypical data, performed clinical data curation, and discussed the data. C.R., M.L., and K.D.L.R. analyzed the data. M.M.H., F.K., L.E.S., and K.D.L.R. acquired funding. A.G., M.S., M.M.H., C.T., C.V.G., C.R., and M.L. reviewed and edited the draft. K.D.L.R. wrote the original draft, supervised, and conceptualized the work. All authors read and approved the manuscript.
Declaration of interests
The Max Delbrück Center for Molecular Medicine in the Helmholtz Association and the Charité - Universitätsmedizin Berlin have filed three patent applications in connection with this work on which M.L., F.K., L.E.S., and K.D.L.R. (EP21155584.2); C.V.G. and K.D.L.R. (EP22164013.9); and M.L., C.R., C.V.G., and K.D.L.R. (EP21194414.5) are inventors.
Published: February 24, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109330.
Contributor Information
Michael Meyer-Hermann, Email: mmh@theoretical-biology.de.
Kathrin de la Rosa, Email: kathrin.delarosa@mdc-berlin.de.
Supplemental information
References
- 1.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 3.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 4.Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 5.Piccoli L., Park Y.-J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.-W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 8.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.-M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S., et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R., et al. A Novel Angiotensin-Converting Enzyme–Related Carboxypeptidase (ACE2) Converts Angiotensin I to Angiotensin 1-9. Circ. Res. 2000;87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 11.Lambert D.W., Yarski M., Warner F.J., Thornhill P., Parkin E.T., Smith A.I., Hooper N.M., Turner A.J. Tumor necrosis factor-alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2) J. Biol. Chem. 2005;280:30113–30119. doi: 10.1074/jbc.m505111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haga S., Yamamoto N., Nakai-Murakami C., Osawa Y., Tokunaga K., Sata T., Yamamoto N., Sasazuki T., Ishizaka Y. Modulation of TNF-α-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-α production and facilitates viral entry. Proc. Natl. Acad. Sci. USA. 2008;105:7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glowacka I., Bertram S., Herzog P., Pfefferle S., Steffen I., Muench M.O., Simmons G., Hofmann H., Kuri T., Weber F., et al. Differential Downregulation of ACE2 by the Spike Proteins of Severe Acute Respiratory Syndrome Coronavirus and Human Coronavirus NL63. J. Virol. 2010;84:1198–1205. doi: 10.1128/jvi.01248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charbonneau M., Harper K., Grondin F., Pelmus M., McDonald P.P., Dubois C.M. Hypoxia-inducible Factor Mediates Hypoxic and Tumor Necrosis Factor α-induced Increases in Tumor Necrosis Factor-α Converting Enzyme/ADAM17 Expression by Synovial Cells. J. Biol. Chem. 2007;282:33714–33724. doi: 10.1074/jbc.m704041200. [DOI] [PubMed] [Google Scholar]
- 15.Epelman S., Tang W.H.W., Chen S.Y., Van Lente F., Francis G.S., Sen S. Detection of Soluble Angiotensin-Converting Enzyme 2 in Heart Failure Insights Into the Endogenous Counter-Regulatory Pathway of the Renin-Angiotensin-Aldosterone System. J. Am. Coll. Cardiol. 2008;52:750–754. doi: 10.1016/j.jacc.2008.02.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rice G.I., Jones A.L., Grant P.J., Carter A.M., Turner A.J., Hooper N.M. Circulating Activities of Angiotensin-Converting Enzyme, Its Homolog, Angiotensin-Converting Enzyme 2, and Neprilysin in a Family Study. Hypertension. 2006;48:914–920. doi: 10.1161/01.hyp.0000244543.91937.79. [DOI] [PubMed] [Google Scholar]
- 17.Úri K., Fagyas M., Kertész A., Borbély A., Jenei C., Bene O., Csanádi Z., Paulus W.J., Édes I., Papp Z., et al. Circulating ACE2 activity correlates with cardiovascular disease development. J. Renin-Angiotensin-Aldosterone Syst. JRAAS. 2016;17 doi: 10.1177/1470320316668435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anguiano L., Riera M., Pascual J., Valdivielso J.M., Barrios C., Betriu A., Mojal S., Fernández E., Soler M.J., NEFRONA study Circulating angiotensin-converting enzyme 2 activity in patients with chronic kidney disease without previous history of cardiovascular disease. Nephrol. Dial. Transplant. 2015;30:1176–1185. doi: 10.1093/ndt/gfv025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehra M.R., Desai S.S., Kuy S., Henry T.D., Patel A.N. Cardiovascular Disease, Drug Therapy, and Mortality in Covid-19. N. Engl. J. Med. 2020;382:e102. doi: 10.1056/nejmoa2007621. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Holman N., Knighton P., Kar P., O’Keefe J., Curley M., Weaver A., Barron E., Bakhai C., Khunti K., Wareham N.J., et al. Risk factors for COVID-19-related mortality in people with type 1 and type 2 diabetes in England: a population-based cohort study. Lancet Diabetes Endocrinol. 2020;8:823–833. doi: 10.1016/s2213-8587(20)30271-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel S.K., Juno J.A., Lee W.S., Wragg K.M., Hogarth P.M., Kent S.J., Burrell L.M. Plasma ACE2 activity is persistently elevated following SARS-CoV-2 infection: implications for COVID-19 pathogenesis and consequences. Eur. Respir. J. 2021;57 doi: 10.1183/13993003.03730-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kragstrup T.W., Singh H.S., Grundberg I., Nielsen A.L.-L., Rivellese F., Mehta A., Goldberg M.B., Filbin M.R., Qvist P., Bibby B.M. Plasma ACE2 predicts outcome of COVID-19 in hospitalized patients. PLoS One. 2021;16 doi: 10.1371/journal.pone.0252799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahman M.M., Hasan M., Ahmed A. Potential detrimental role of soluble ACE2 in severe COVID-19 comorbid patients. Rev. Med. Virol. 2021;31:1–12. doi: 10.1002/rmv.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Lier D., Kox M., Santos K., van der Hoeven H., Pillay J., Pickkers P. Increased blood angiotensin converting enzyme 2 activity in critically ill COVID-19 patients. ERJ Open Res. 2021;7 doi: 10.1183/23120541.00848-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reindl-Schwaighofer R., Hödlmoser S., Eskandary F., Poglitsch M., Bonderman D., Strassl R., Aberle J.H., Oberbauer R., Zoufaly A., Hecking M. ACE2 Elevation in Severe COVID-19. Am J Resp Crit Care. 2021;203:1191–1196. doi: 10.1164/rccm.202101-0142le. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aziz N.A., Corman V.M., Echterhoff A.K.C., Müller M.A., Richter A., Schmandke A., Schmidt M.L., Schmidt T.H., de Vries F.M., Drosten C., Breteler M.M.B. Seroprevalence and correlates of SARS-CoV-2 neutralizing antibodies from a population-based study in Bonn, Germany. Nat. Commun. 2021;12:2117. doi: 10.1038/s41467-021-22351-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lucas C., Klein J., Sundaram M.E., Liu F., Wong P., Silva J., Mao T., Oh J.E., Mohanty S., Huang J., et al. Delayed production of neutralizing antibodies correlates with fatal COVID-19. Nat. Med. 2021;27:1178–1186. doi: 10.1038/s41591-021-01355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henry C., Jerne N.K. Competition of 19S and 7S antigen receptors in the regulation of the primary immune response. J. Exp. Med. 1968;128:133–152. doi: 10.1084/jem.128.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reth M., Kelsoe G., Rajewsky K. Idiotypic regulation by isologous monoclonal anti-idiotope antibodies. Nature. 1981;290:257–259. doi: 10.1038/290257a0. [DOI] [PubMed] [Google Scholar]
- 30.Xu H., Heyman B. IgG-mediated suppression of antibody responses: Hiding or snatching epitopes? Scand. J. Immunol. 2020;92 doi: 10.1111/sji.12921. [DOI] [PubMed] [Google Scholar]
- 31.Tas J.M.J., Koo J.-H., Lin Y.-C., Xie Z., Steichen J.M., Jackson A.M., Hauser B.M., Wang X., Cottrell C.A., Torres J.L., et al. Antibodies from primary humoral responses modulate the recruitment of naive B cells during secondary responses. Immunity. 2022;55:1856–1871.e6. doi: 10.1016/j.immuni.2022.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaefer-Babajew D., Wang Z., Muecksch F., Cho A., Loewe M., Cipolla M., Raspe R., Johnson B., Canis M., DaSilva J., et al. Antibody feedback regulates immune memory after SARS-CoV-2 mRNA vaccination. Nature. 2023;613:735–742. doi: 10.1038/s41586-022-05609-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 35.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang L., Sexton D.J., Skogerson K., Devlin M., Smith R., Sanyal I., Parry T., Kent R., Enright J., Wu Q.l., et al. Novel Peptide Inhibitors of Angiotensin-converting Enzyme 2. J. Biol. Chem. 2003;278:15532–15540. doi: 10.1074/jbc.m212934200. [DOI] [PubMed] [Google Scholar]
- 37.Peters A., German National Cohort NAKO Consortium. Peters A., Greiser K.H., Göttlicher S., Ahrens W., Albrecht M., Bamberg F., Bärnighausen T., Becher H., et al. Framework and baseline examination of the German National Cohort (NAKO) Eur. J. Epidemiol. 2022;37:1107–1124. doi: 10.1007/s10654-022-00890-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leisman D.E., Ronner L., Pinotti R., Taylor M.D., Sinha P., Calfee C.S., Hirayama A.V., Mastroiani F., Turtle C.J., Harhay M.O., et al. Cytokine elevation in severe and critical COVID-19: a rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. Lancet Respir. Med. 2020;8:1233–1244. doi: 10.1016/s2213-2600(20)30404-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng K.W., Attig J., Bolland W., Young G.R., Major J., Wrobel A.G., Gamblin S., Wack A., Kassiotis G. Tissue-specific and interferon-inducible expression of nonfunctional ACE2 through endogenous retroelement co-option. Nat. Genet. 2020;52:1294–1302. doi: 10.1038/s41588-020-00732-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Onabajo O.O., Banday A.R., Stanifer M.L., Yan W., Obajemu A., Santer D.M., Florez-Vargas O., Piontkivska H., Vargas J.M., Ring T.J., et al. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat. Genet. 2020;52:1283–1293. doi: 10.1038/s41588-020-00731-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blume C., Jackson C.L., Spalluto C.M., Legebeke J., Nazlamova L., Conforti F., Perotin J.-M., Frank M., Butler J., Crispin M., et al. A novel ACE2 isoform is expressed in human respiratory epithelia and is upregulated in response to interferons and RNA respiratory virus infection. Nat. Genet. 2021;53:205–214. doi: 10.1038/s41588-020-00759-x. [DOI] [PubMed] [Google Scholar]
- 42.Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ge J., Wang R., Ju B., Zhang Q., Sun J., Chen P., Zhang S., Tian Y., Shan S., Cheng L., et al. Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry. Nat. Commun. 2021;12:250. doi: 10.1038/s41467-020-20501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greaney A.J., Starr T.N., Barnes C.O., Weisblum Y., Schmidt F., Caskey M., Gaebler C., Cho A., Agudelo M., Finkin S., et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat. Commun. 2021;12:4196. doi: 10.1038/s41467-021-24435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y., Meyer-Hermann M., George L.A., Figge M.T., Khan M., Goodall M., Young S.P., Reynolds A., Falciani F., Waisman A., et al. Germinal center B cells govern their own fate via antibody feedback. J. Exp. Med. 2013;210:457–464. doi: 10.1084/jem.20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyer-Hermann M. Injection of Antibodies against Immunodominant Epitopes Tunes Germinal Centers to Generate Broadly Neutralizing Antibodies. Cell Rep. 2019;29:1066–1073.e5. doi: 10.1016/j.celrep.2019.09.058. [DOI] [PubMed] [Google Scholar]
- 47.Garg A.K., Mitra T., Schips M., Bandyopadhyay A., Meyer-Hermann M. Amount of antigen, T follicular helper cells and affinity of founder cells shape the diversity of germinal center B cells: A computational study. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1080853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyer-Hermann M., Binder S.C., Mesin L., Victora G.D. Computer Simulation of Multi-Color Brainbow Staining and Clonal Evolution of B Cells in Germinal Centers. Front. Immunol. 2018;9:2020. doi: 10.3389/fimmu.2018.02020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyer-Hermann M., Mohr E., Pelletier N., Zhang Y., Victora G.D., Toellner K.-M. A Theory of Germinal Center B Cell Selection, Division, and Exit. Cell Rep. 2012;2:162–174. doi: 10.1016/j.celrep.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 50.Guo L., Bi W., Wang X., Xu W., Yan R., Zhang Y., Zhao K., Li Y., Zhang M., Cai X., et al. Engineered trimeric ACE2 binds viral spike protein and locks it in “Three-up” conformation to potently inhibit SARS-CoV-2 infection. Cell Res. 2021;31:98–100. doi: 10.1038/s41422-020-00438-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arulraj T., Binder S.C., Robert P.A., Meyer-Hermann M. Synchronous Germinal Center Onset Impacts the Efficiency of Antibody Responses. Front. Immunol. 2019;10:2116. doi: 10.3389/fimmu.2019.02116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lebedin M., García C.V., Spatt L., Ratswohl C., Thibeault C., Ostendorf L., Alexander T., Paul F., Sander L.E., Kurth F., de la Rosa K. Discriminating promiscuous from target-specific autoantibodies in COVID-19. Eur. J. Immunol. 2023;53 doi: 10.1002/eji.202250210. [DOI] [PubMed] [Google Scholar]
- 53.Hallmann E., Sikora D., Poniedziałek B., Szymański K., Kondratiuk K., Żurawski J., Brydak L., Rzymski P. IgG autoantibodies against ACE2 in SARS-CoV-2 infected patients. J. Med. Virol. 2023;95 doi: 10.1002/jmv.28273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arthur J.M., Forrest J.C., Boehme K.W., Kennedy J.L., Owens S., Herzog C., Liu J., Harville T.O. Development of ACE2 autoantibodies after SARS-CoV-2 infection. PLoS One. 2021;16 doi: 10.1371/journal.pone.0257016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Casciola-Rosen L., Thiemann D.R., Andrade F., Trejo-Zambrano M.I., Leonard E.K., Spangler J.B., Skinner N.E., Bailey J., Yegnasubramanian S., Wang R., et al. IgM anti-ACE2 autoantibodies in severe COVID-19 activate complement and perturb vascular endothelial function. JCI Insight. 2022;7 doi: 10.1172/jci.insight.158362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeung M.L., Teng J.L.L., Jia L., Zhang C., Huang C., Cai J.-P., Zhou R., Chan K.-H., Zhao H., Zhu L., et al. Soluble ACE2-mediated cell entry of SARS-CoV-2 via interaction with proteins related to the renin-angiotensin system. Cell. 2021;184:2212–2228.e12. doi: 10.1016/j.cell.2021.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo X., Cao J., Cai J.P., Wu J., Huang J., Asthana P., Wong S.K.K., Ye Z.W., Gurung S., Zhang Y., et al. Control of SARS-CoV-2 infection by MT1-MMP-mediated shedding of ACE2. Nat. Commun. 2022;13:7907. doi: 10.1038/s41467-022-35590-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y., Schneider A.M., Mehta A., Sade-Feldman M., Kays K.R., Gentili M., Charland N.C., Gonye A.L., Gushterova I., Khanna H.K., et al. SARS-CoV-2 viremia is associated with distinct proteomic pathways and predicts COVID-19 outcomes. J. Clin. Invest. 2021;131 doi: 10.1172/jci148635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H., et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sidarta-Oliveira D., Jara C.P., Ferruzzi A.J., Skaf M.S., Velander W.H., Araujo E.P., Velloso L.A. SARS-CoV-2 receptor is co-expressed with elements of the kinin–kallikrein, renin–angiotensin and coagulation systems in alveolar cells. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-76488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F., et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J., Zhao H., An Y. ACE2 Shedding and the Role in COVID-19. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.789180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Voss W.N., Hou Y.J., Johnson N.V., Delidakis G., Kim J.E., Javanmardi K., Horton A.P., Bartzoka F., Paresi C.J., Tanno Y., et al. Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes. Science. 2021;372:1108–1112. doi: 10.1126/science.abg5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Juno J.A., Tan H.-X., Lee W.S., Reynaldi A., Kelly H.G., Wragg K., Esterbauer R., Kent H.E., Batten C.J., Mordant F.L., et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020;26:1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- 66.Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ratswohl C., Vázquez García C., Ahmad A.U.W., Gonschior H., Lebedin M., Silvis C.E., Spatt L., Gerhard C., Lehmann M., Sander L.E., et al. A design strategy to generate a SARS-CoV-2 RBD vaccine that abrogates ACE2 binding and improves neutralizing antibody responses. Eur. J. Immunol. 2023;53 doi: 10.1002/eji.202350408. [DOI] [PubMed] [Google Scholar]
- 68.Yi H., Wang J., Wang J., Lu Y., Zhang Y., Peng R., Lu J., Chen Z. The Emergence and Spread of Novel SARS-CoV-2 Variants. Front. Public Health. 2021;9 doi: 10.3389/fpubh.2021.696664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perez-Gomez R. The Development of SARS-CoV-2 Variants: The Gene Makes the Disease. J. Dev. Biol. 2021;9:58. doi: 10.3390/jdb9040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang L., Jackson C.B., Mou H., Ojha A., Peng H., Quinlan B.D., Rangarajan E.S., Pan A., Vanderheiden A., Suthar M.S., et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020;11:6013. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tian F., Tong B., Sun L., Shi S., Zheng B., Wang Z., Dong X., Zheng P. N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. Elife. 2021;10 doi: 10.7554/elife.69091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gobeil S.M.-C., Janowska K., McDowell S., Mansouri K., Parks R., Stalls V., Kopp M.F., Manne K., Li D., Wiehe K., et al. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science. 2021;373 doi: 10.1126/science.abi6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen C., Boorla V.S., Banerjee D., Chowdhury R., Cavener V.S., Nissly R.H., Gontu A., Boyle N.R., Vandegrift K., Nair M.S., et al. Computational prediction of the effect of amino acid changes on the binding affinity between SARS-CoV-2 spike RBD and human ACE2. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2106480118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu L., Zhou L., Mo M., Liu T., Wu C., Gong C., Lu K., Gong L., Zhu W., Xu Z. SARS-CoV-2 Omicron RBD shows weaker binding affinity than the currently dominant Delta variant to human ACE2. Signal Transduct. Targeted Ther. 2022;7:8. doi: 10.1038/s41392-021-00863-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou T., Wang L., Misasi J., Pegu A., Zhang Y., Harris D.R., Olia A.S., Talana C.A., Yang E.S., Chen M., et al. Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529. Science. 2022;376 doi: 10.1126/science.abn8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu X., Yang Z.-Y., Li Y., Hogerkorp C.-M., Schief W.R., Seaman M.S., Zhou T., Schmidt S.D., Wu L., Xu L., et al. Rational Design of Envelope Identifies Broadly Neutralizing Human Monoclonal Antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou T., Teng I.-T., Olia A.S., Cerutti G., Gorman J., Nazzari A., Shi W., Tsybovsky Y., Wang L., Wang S., et al. Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., et al. Remdesivir for the Treatment of Covid-19 — Final Report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/nejmoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Group R.C., Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., Linsell L., Staplin N., Brightling C., Ustianowski A., et al. Dexamethasone in Hospitalized Patients with Covid-19. N. Engl. J. Med. 2020;384:693–704. doi: 10.1056/nejmoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kurth F., Roennefarth M., Thibeault C., Corman V.M., Müller-Redetzky H., Mittermaier M., Ruwwe-Glösenkamp C., Heim K.M., Krannich A., Zvorc S., et al. Studying the pathophysiology of coronavirus disease 2019: a protocol for the Berlin prospective COVID-19 patient cohort (Pa-COVID-19) Infection. 2020;48:619–626. doi: 10.1007/s15010-020-01464-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thibeault C., Mühlemann B., Helbig E.T., Mittermaier M., Lingscheid T., Tober-Lau P., Meyer-Arndt L.A., Meiners L., Stubbemann P., Haenel S.S., et al. Clinical and virological characteristics of hospitalised COVID-19 patients in a German tertiary care centre during the first wave of the SARS-CoV-2 pandemic: a prospective observational study. Infection. 2021;49:703–714. doi: 10.1007/s15010-021-01594-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schlickeiser S., Schwarz T., Steiner S., Wittke K., Al Besher N., Meyer O., Kalus U., Pruß A., Kurth F., Zoller T., et al. Disease Severity, Fever, Age, and Sex Correlate With SARS-CoV-2 Neutralizing Antibody Responses. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.628971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Petersmann A., Winter T., Lamp S., Nauck M. Relevant criteria for the selection of cryotubes. Experiences from the German National Cohort. J. Lab. Med. 2019;43:339–345. doi: 10.1515/labmed-2019-0172. [DOI] [Google Scholar]
- 90.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.es.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chan K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., Herbert A.S., Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;369:1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sharma S.K., de Val N., Bale S., Guenaga J., Tran K., Feng Y., Dubrovskaya V., Ward A.B., Wyatt R.T. Cleavage-Independent HIV-1 Env Trimers Engineered as Soluble Native Spike Mimetics for Vaccine Design. Cell Rep. 2015;11:539–550. doi: 10.1016/j.celrep.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou D., Duyvesteyn H.M.E., Chen C.-P., Huang C.-G., Chen T.-H., Shih S.-R., Lin Y.-C., Cheng C.-Y., Cheng S.-H., Huang Y.-C., et al. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat. Struct. Mol. Biol. 2020;27:950–958. doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- 94.Xiao F., Burns K.D. Measurement of Angiotensin Converting Enzyme 2 Activity in Biological Fluid (ACE2) Hypertension. 2017;1527:101–115. doi: 10.1007/978-1-4939-6625-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yuan M., Liu H., Wu N.C., Lee C.-C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.-C., Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.López-Ratón M., Rodríguez-Álvarez M.X., Suárez C.C., Sampedro F.G. OptimalCutpoints : An R Package for Selecting Optimal Cutpoints in Diagnostic Tests. J. Stat. Software. 2014;61 doi: 10.18637/jss.v061.i08. [DOI] [Google Scholar]
- 98.Figge M.T., Garin A., Gunzer M., Kosco-Vilbois M., Toellner K.-M., Meyer-Hermann M. Deriving a germinal center lymphocyte migration model from two-photon data. J. Exp. Med. 2008;205:3019–3029. doi: 10.1084/jem.20081160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data: Data are provided in Tables S1, S2, S3, S4, and S5 including a-sACE2 measurements, patient age, gender, body mass index, sampling time point, and WHO score of patient samples, as well as raw data for the analyses performed.
-
•
Code: The mathematical model code is publicly available at Zenodo repository, https://doi.org/10.5281/zenodo.10605480. Any additional information is available upon request from the corresponding authors.
-
•
Other items: no other items were generated throughout the study.