Figure 3.
Serology suggests that sACE2 impairs generation of antibodies preventing RBD-ACE2 interaction
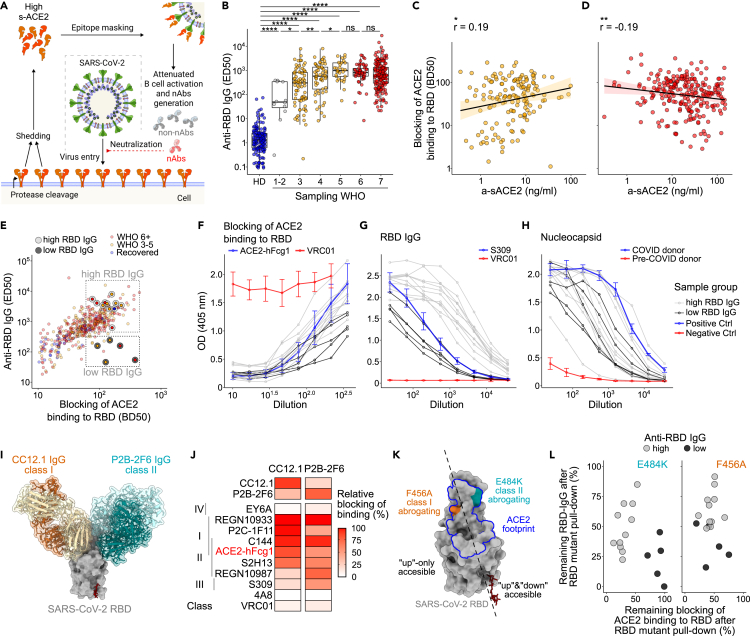
(A) Scheme depicting SARS-CoV-2 spike epitope masking by sACE2.
(B) Serum IgG binding to RBD (ED50, serum dilution corresponding to 50% of maximum binding activity) of HDs (blue, N = 151), moderate (yellow, N = 120) and severe (red, N = 198) patients collected >20 PSO. Patients are stratified into the respective WHO class at the time of sampling. Statistical analyses by two-sided Mann–Whitney test (∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05; ns, not significant). Boxplots depict median +/− interquartile range. Whisker length is 1.5 interquartile ranges.
(C) Blocking of ACE2-RBD interaction (BD50, serum dilution corresponding to 50% of maximum blocking activity) plotted against a-sACE2 for moderate and (D) severe COVID-19. Data for individual samples are shown; samples drawn after 15 days PSO are selected. Spearman correlation performed with a 95% confidence interval.
(E) ED50 of anti-RBD IgG plotted against blocking of ACE2-binding (BD50).
(F) ACE2 competition, (G) anti-RBD IgG, and (H) anti-nucleocapsid IgG confirmed for two sample groups with high (gray, N = 12) versus low (black line, N = 5) RBD titers. Positive and negative controls in blue and red, respectively.
(I) Structural depiction of the RBD bound to CC12.1 and P2B-2F6. PDB: 6XDG, 7BWJ, and 6XC2.
(J) RBD binding of recombinant antibodies in competition with class 1 nAb CC12.1 and class 2 nAb P2B-2F6.
(K) Localization of class 1 and 2 abrogating RBD mutations F456A and E484K. ACE2 footprint shown in blue.
(L) RBD high versus low binding sera cleared by a pull-down with RBD WT, RBD-F456A, RBD-E484K, and MERS-CoV RBD as control. Remaining IgG binding to RBD and ACE2 competition is depicted in %.