Foretinib exhibits superior efficacy to approved drugs in AML with FLT3-ITD mutations and retains activity in AML with secondary FLT3 mutations that mediate resistance to clinical FLT3 inhibitors.
Abstract
FLT3 internal tandem duplication (FLT3-ITD) mutations are one of the most prevalent somatic alterations associated with poor prognosis in patients with acute myeloid leukemia (AML). The clinically approved FLT3 kinase inhibitors gilteritinib and quizartinib improve the survival of patients with AML with FLT3-ITD mutations, but their long-term efficacy is limited by acquisition of secondary drug-resistant mutations. In this study, we conducted virtual screening of a library of 60,411 small molecules and identified foretinib as a potent FLT3 inhibitor. An integrated analysis of the BeatAML database showed that foretinib had a lower IC50 value than other existing FLT3 inhibitors in patients with FLT3-ITD AML. Foretinib directly bound to FLT3 and effectively inhibited FLT3 signaling. Foretinib potently inhibited proliferation and promoted apoptosis in human AML cell lines and primary AML cells with FLT3-ITD mutations. Foretinib also significantly extended the survival of mice bearing cell-derived and patient-derived FLT3-ITD xenografts, exhibiting stronger efficacy than clinically approved FLT3 inhibitors in treating FLT3-ITD AML. Moreover, foretinib showed potent activity against secondary mutations of FLT3-ITD that confer resistance to quizartinib and gilteritinib. These findings support the potential of foretinib for treating patients with AML with FLT3-ITD mutations, especially for those carrying secondary mutations after treatment failure with other FLT3 inhibitors.
Significance:
Foretinib exhibits superior efficacy to approved drugs in AML with FLT3-ITD mutations and retains activity in AML with secondary FLT3 mutations that mediate resistance to clinical FLT3 inhibitors.
Introduction
Acute myeloid leukemia (AML) is an aggressive hematopoietic malignancy characterized by uncontrolled proliferation of hematopoietic stem and progenitor cells and failure to differentiate into mature blood cells (1, 2). Activating mutations in FMS-like tyrosine kinase 3 (FLT3) are the most prevalent genetic alterations in patients with AML and occur in approximately 30% of patients (3). In general, two types of FLT3 mutations can frequently be identified in AML. Approximately 23% of patients with AML acquire an internal tandem duplication mutation in or near the juxtamembrane domain of FLT3 (FLT3-ITD), which is associated with poor overall survival and relapse-free survival. In approximately 7% of patients with AML (4, 5), FLT3 point mutations have been detected within the activation loop of the tyrosine kinase domain (FLT3-TKD), with missense mutations at D835 being the most frequently occurring TKD mutation (6). FLT3-TKD mutations do not appear to affect the prognosis (7, 8). FLT3-ITD mutations promote ligand-independent receptor dimerization, resulting in autonomous phosphorylation and constitutive activation of the receptor with subsequent activation of downstream proliferative signaling cascades, including those of JAK/STAT5, MAPK, and PI3K/AKT (9–11), which ultimately block differentiation, inhibit apoptosis, and promote cytokine-independent proliferation of leukemia cells.
To date, multiple FLT3 kinase inhibitors have been developed, and some are approved for clinical use, such as midostaurin, quizartinib (AC220), and gilteritinib (12); however, most patients develop drug resistance just a few months after receiving treatment with FLT3 inhibitors. One particular mechanism of resistance involves the acquisition of additional mutations in the TKD (13–16). In a phase II trial of quizartinib monotherapy, 9 patients with AML with FLT3-ITD relapsed after remission due to secondary mutations at the D835 and Y842 residues, as well as at the “gatekeeper residue” F691 in the kinase domain (13, 17). During treatment with gilteritinib, a consistently active inhibitor against both FLT3-ITD-AML and FLT3-TKD-AML, secondary mutations at FLT3-F691 L have also been reported at relapse in patients after the initial response (18). In addition, it has been confirmed that subclones carrying both FLT3-ITD and D835 mutations already exist in leukemia initiating cells (19). Patients with FLT3-ITD mutations were prone to relapse due to the development of FLT3-TKD mutations during treatment with FLT3 kinase inhibitors (17). Therefore, it is highly desirable to develop new FLT3 inhibitors against both FLT3-ITD and FLT3-TKD mutations to treat refractory/relapsed FLT3-ITD–positive patients.
Foretinib is a novel tyrosine kinase inhibitor (TKI) designed to target c-MET and VEGFR2, while also has affinity for other tyrosine kinases, such as PDGFR (20). Foretinib showed good clinical activity in solid tumors with MET mutations, including gastric cancer, ovarian cancer, renal cell carcinoma, liver cancer, and non–small cell lung cancer (21–26). Currently, no studies have evaluated the activity of foretinib in AML, especially FLT3-ITD and TKD AML. Through virtually screening the ZINC in vivo library and analyzing the drug screening data of patients with AML in BeatAML database (27), we found that foretinib might be an effective FLT3 inhibitor.
Our results of the cellular thermal shift assay (CETSA) confirmed the direct binding of foretinib and FLT3. Foretinib exerted a potent inhibitory effect against leukemia cells expressing FLT3-ITD and overcame resistance to existing FLT3 inhibitors conferred by secondary mutations in FLT3 in vitro. Furthermore, we showed that foretinib treatment significantly prolonged the survival of mice in both the FLT3-ITD models and the drug-resistant models. Compared with gilteritinib and quizartinib, foretinib displayed more favorable therapeutic effects on primary AML cells with FLT3 mutations both in vitro and in mice xenografted with primary AML cells [patient-derived xenografts (PDX)].
Thus, we identify foretinib as a potent FLT3 inhibitor, and this finding provides a rationale for its application in patients with AML with FLT3-ITD mutations, as well as refractory/relapsed patients with secondary FLT3-TKD mutations.
Materials and Methods
Patient sample preparation
Bone marrow (BM) samples were collected from patients with AML (patient details are provided in Supplementary Table S1). Mononuclear cells were separated with Lymphoprep reagent (Stemcell Technologies) using density gradient centrifugation. They were subsequently cultured in StemSpan SFEM medium (Stemcell Technologies) supplemented with 10 ng/mL human stem cell factor, 10 ng/mL human IL3, 10 ng/mL human IL6 (all cytokines were purchased from R&D Systems), 100 U/mL penicillin, and 100 μg/mL streptomycin (both from BBI Life Sciences).
Written informed consents were obtained from all patients in accordance with the Declaration of Helsinki, and all manipulations were approved by the Institutional Review Board of Guangzhou First People's Hospital, School of Medicine, South China University of Technology (Guangzhou, P.R. China).
Compounds
Foretinib (Targetmol), quizartinib (Targetmol), and gilteritinib (Abmole) was dissolved in 100% DMSO to make a 10 μmol/L stock solution and stored at −20°C.
Virtual screening and docking
AutoDock Vina, AutoDock Tools, Schrödinger, Discovery Studio 2019, and PyMOL were used for the computational modeling experiment. The structure of FLT3-ITD (residues 587–947) was prepared on the basis of the sequence from the Protein Data Bank (accession number: 5×02). Schrödinger's Protein Preparation Wizard module was used to process the protein as follows: crystal water was removed, missing hydrogen atoms were added, missing bond information was repaired, missing peptides were repaired, and energy optimization was performed under the OPLS_2005 force field. Then, the original eutectic ligand was selected in the Grid Generation module to generate a grid file for the docking site. Small molecules from the in vivo library of the ZINC database, which consisted of 60,411 ligands by that time, were docked onto the ATP binding site of FLT3 using AutoDock Vina. For macromolecule preparation, all water molecules were deleted and all polar hydrogens were added to the structure. The docking of each ligand was performed five times, and each repeated simulation generated 20 poses, yielding a total of 100 poses for each ligand. The top 1% scoring pose (according to the binding free energy estimated by AutoDock Vina) was selected for Schrödinger standard precision docking. The lower the score was, the lower the binding free energy between the compound and the protein, and the higher the binding stability.
For the detailed docking of foretinib to FLT3, the OPLS_2005 force field and the Liand Docking module were used to perform XP precision docking. The LigPrep module was used to protonate small molecules at pH 7.0 ± 2.0. The results were visualized with Discovery Studio 2019 and PyMOL.
Public databases analysis
The gene mutation and drug susceptibility data of patients with AML were obtained from the BeatAML database including Vizome (http://vizome.org/aml2/) and BeatAML2 (https://biodev.github.io/BeatAML2/). In patients with AML, the high FLT3-ITD allelic ratio was defined as 0.5 or greater and the low ratio was less than 0.5.
Cell lines and cell culture
The construction of Ba/F3 cell lines stably expressing human FLT3 with different mutations (FLT3-ITD, FLT3-D835Y, FLT3-D835V, FLT3-N676K, FLT3-Y842C, FLT3-ITD-D835Y, FLT3-ITD-D835V, FLT3-ITD-Y842C, FLT3-ITD-F691L) has been described previously (17). Ba/F3 (DSMZ, RRID: CVCL_0161) cells were grown in RPMI1640 medium (Gibco) supplemented with 10% FBS (Gibco) and mouse interleukin IL3 (1 ng/mL; R&D Systems). By expressing the tyrosine kinase construct, the Ba/F3 cells were transformed into cells that could grow independent of IL3. Then, they were grown in the absence of IL3. MV4-11 (ATCC, RRID: CVCL_0064), MOLM13 (DSMZ, RRID: CVCL_2119), NB4 (DSMZ, RRID: CVCL_0005), THP1 (ATCC, RRID: CVCL_0006), HL60 (ATCC, RRID: CVCL_0002), OCI-AML2 (DSMZ, RRID: CVCL_1619) and OCI-AML3 (DSMZ, RRID: CVCL_1844), and K562 (ATCC, RRID: CVCL_0004) cell lines were cultured in RPMI1640 supplemented with 10% FBS. NRAS-WT containing MSCV-GFP-IRES (MGI) retroviral constructs were generated as described previously (28). NRAS-G12C mutants were generated from the NRAS-WT plasmid using Quickchange Site-Directed Mutagenesis Kit (Stratagene) with the forward primer 5′-TGGAGCATGTGGTGTTGGGAAAAGCGCACTGA-3′ and reverse primer 5′-CAACACCACATGCTCCAACCACCACCAGTTTG-3′. Then the lentivirus of NRAS-G12C mutants was produced and transduced to MOLM13 and MV4-11 cells as described previously (28). The MycoFluor mycoplasma detection kit (Invitrogen) was used for regular inspections, and the antibiotic Mycoplasma (Invivogen) was used to eliminate Mycoplasma contamination. Short tandem repeat profiling was performed on all cell lines. All the cell lines were used within 10 passages.
Cell viability assay
Cell viability assays were carried out using the CellTiter-Glo Luminescent cell viability assay as described previously (29). The cells were seeded into a 96-well cell culture plate at a density of 5,000 cells per well, and different concentrations of the designated drugs were added. After 48 hours of incubation, the cells were lysed by adding CellTiter Glo Reagent (Promega) for 30 minutes at room temperature. The luminescence signal produced by ATP molecules in living cells was measured by VICTOR Nivo (Revvity).
Cell apoptosis and cell cycle assay
MOLM13 and MV4-11 cells (2.0 × 105) were seeded in 6-well plates and incubated with foretinib or gilteritinib for 24 or 48 hours. For cell cycle analysis, cells were fixed with 70% ethanol at 4 °C overnight and then stained with propidium iodide (PI; Sigma-Aldrich). Cell cycle distribution was determined using flow cytometry (BD LSRFortessa). APC-Annexin V and PI (Invitrogen) were used for staining and detecting apoptotic cells. After washing off excess dye with PBS, flow cytometry was performed. Single dyes were used to adjust the compensation between instrument channels during the experiment. The results were analyzed using FlowJo software.
Western blotting analysis
Cell samples were counted and washed in PBS. They were then lysed in 2× SDS sample loading buffer (Sigma-Aldrich) supplemented with protease inhibitors (Targetmol). The protein samples were then subjected to sodium lauryl sulfate polyacrylamide electrophoresis and transferred to a nitrocellulose membrane. The following antibodies were used for this experiment: anti-phosphorylated (p)-FLT3 (catalog no. 3464, Cell Signaling Technology, RRID:AB_2107051), anti-FLT3 (catalog no. 3462, Cell Signaling Technology, RRID: AB_2107052), anti-pSTAT5 (catalog no. 9351, Cell Signaling Technology, RRID: AB_2315225), anti-STAT5 (catalog no. 9363, Cell Signaling Technology, RRID: AB_2196923), anti-ERK (catalog no. 4695, Cell Signaling Technology, RRID: AB_390779), anti-pERK (catalog no. 4370, Cell Signaling Technology, RRID: AB_2315112), anti-AKT (catalog no. 9272, Cell Signaling Technology, RRID: AB_329827) and anti-pAKT (catalog no. 4060, Cell Signaling Technology, RRID: AB_2315049), horseradish peroxidase–conjugated tubulin (catalog no. HRP-66031, Proteintech, RRID: AB_2687491), PARP1 (catalog no.13371-1-AP, Proteintech, RRID: AB_2160459) and caspase8 (catalog no.13423-1-AP, Proteintech, RRID: AB_2068463). Western blotting images were taken using the ChemiDoc MP (Bio-Rad).
CETSA
Twenty million Ba/F3 cells expressing FLT3-ITD were collected and incubated with foretinib (1 μmol/L) or DMSO for 90 minutes. Then, the cells were resuspended in PBS supplemented with protease inhibitor and evenly divided into seven parts to receive heat treatment at a temperature range of 43 °C to 55 °C for 3 minutes in a Veriti thermal cycler (Applied Biosystems). The heated cells were lysed using liquid nitrogen through freeze-thaw cycles. The mixtures were centrifuged, and the supernatants were subjected to SDS-PAGE for Western blotting analysis. The dose effect of foretinib on the thermal stability of FLT3 was evaluated as follows. Twenty million Ba/F3 FLT3-ITD cells were divided into eight portions and incubated with various concentrations of foretinib for 30 minutes. Subsequently, the cells were heated at 51.1 °C for 3 minutes and lysed in liquid nitrogen. The mixtures were then centrifuged, and the supernatants were subjected to immunoblotting analysis. ImageJ was used to quantify the density of the blot.
In vivo efficacy studies
Human AML cell lines (MOLM13, MV4-11 cells, 5 × 106) were injected into the tail vein of NOD/SCID gamma (NSG) mice (Shanghai Model Organisms Center) to construct mouse xenograft models. After 1 week, the mice were randomly administered with oral gilteritinib (30 mg/kg), quizartinib (10 mg/kg), foretinib (15 mg/kg), or control for 2 weeks. Three days after ending the treatment, cells from the BM and spleen (SP) were collected and stained with anti-human CD45 antibody (hCD45, HI30, BioLegend) to measure the percentage of leukemia cells using flow cytometry.
In the FLT3-ITD/-TKD Ba/F3 mouse model, 6-week-old female BALB/c mice (Charles River) were injected intravenously with Ba/F3 FLT3-ITD or FLT3-ITD-TKD cells (2 × 106), and the animals were randomly assigned to the control group or the treatment group. Foretinib (15 mg/kg), quizartinib (10 mg/kg), gilteritinib (30 mg/kg), or control were administered every day until the first mouse died in the vehicle group. To assess the leukemia burden, peripheral blood (PB) was collected, and 3 mice of each group were sacrificed to collect BM and SP cells. In the flow cytometry analysis, leukemia cells were defined as GFP-positive cells. Tissue morphology was visualized using hematoxylin and eosin (H&E) staining.
In the PDX model, 6-week-old female NOG mice (Charles River) were sublethally treated with busulfan (20 mg/kg) 24 hours before injection of 2 × 106 human AML cells harboring FLT3-ITD (PDX #1) or 2 × 106 human AML cells harboring FLT3-ITD, DNMT3A, and NPM1 (PDX #2). The percentages of primary human AML cells in PB of mice were followed monthly by flow cytometry analyses. When the percentage of human AML cells [human CD45-positive cells (hCD45+)] reached at least 90% in PB, BM, and SP cells were collected, pooled, and cryopreserved. The collected BM cells (2 × 106 PDX #1 cells/mouse and 5 × 105 PDX #2 cells/mouse) were injected by tail vein into 6-week-old female NOG mice pretreated with busulfan (20 mg/kg). Residual leukemia cells were defined as hCD45+ cells. PB was collected from the orbital vein at several timepoints, and 3 mice from each group were sacrificed to collect BM and SP cells. The percentage of AML cells was determined by flow cytometry. SP morphology was visualized using H&E staining and IHC with hCD45. The survival of mice was determined when the animals exhibited hindlimb paralysis and became moribund.
All animal studies were approved by the Animal Ethics Committee at South China University of Technology (Guangzhou, P.R. China). This study complied with all relevant ethical regulations regarding animal research.
Colony-forming assays
Human cord blood was obtained after full-term delivery with informed consent, under the approval of Institutional Review Board of Guangzhou First People's Hospital. Mononuclear cells were isolated with Lymphoprep reagent (Stemcell Technologies) through density gradient centrifugation and then seeded in MethoCult methylcellulose medium (H4435; Stemcell Technologies) supplemented with various concentrations of gilteritinib or foretinib at a density of 2 × 104 cells/plate. After 10 days, burst-forming unit-erythroid (BFU-E), colony-forming unit-granulocyte, macrophage (CFU-GM), and colony-forming unit–granulocyte, erythrocyte, monocyte/macrophage, megakaryocyte (CFU-GEMM) colonies were counted under the microscope.
Statistical analysis
GraphPad Prism 9 software was used for statistical analysis. A two-tailed unpaired Student t test was used for comparison between the two groups, whereas the Kaplan‒Meier survival curve and log-rank test were used to estimate survival. P values < 0.05 were considered statistically significant, and different levels were denoted as *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P <0.0001
Data availability
The gene mutation and drug susceptibility data of patients with AML were obtained from the BeatAML database including Vizome (http://vizome.org/aml2/) and BeatAML2 (https://biodev.github.io/BeatAML2/). All other raw data generated in this study are available upon request from the corresponding author.
Results
Identification of foretinib as a FLT3 inhibitor
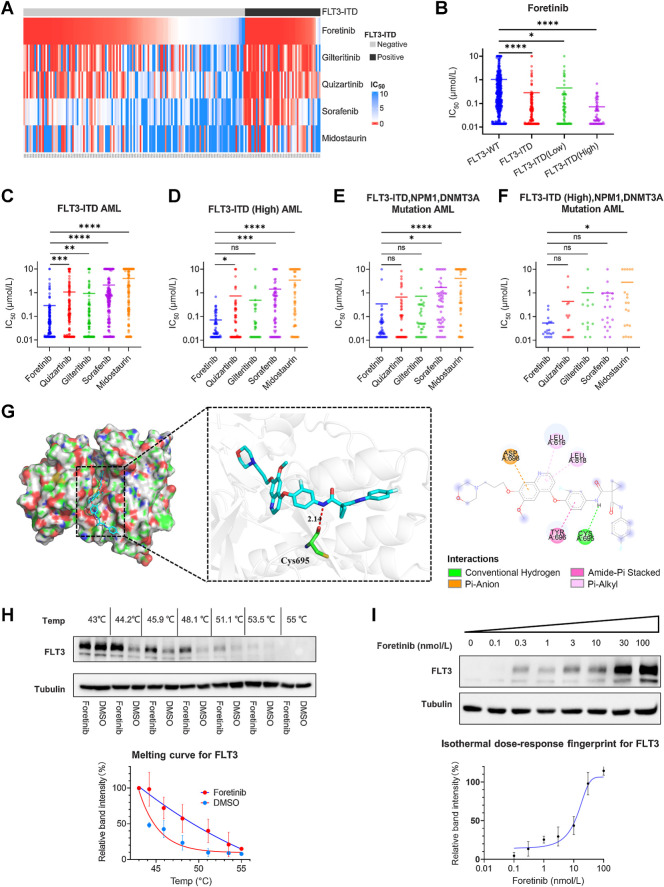
To find hit compounds targeting FLT3, we performed molecular docking simulations of the ZINC in vivo library (60,411 molecules) to the catalytic site of FLT3 (5×02). The docking results were ranked by predicted binding affinity, and the small molecules within the top 1% in terms of binding capacity (523 small molecules) were used for standard precision screening by Schrödinger software. After standard precision screening, there were 78 small molecules with higher affinity than quizartinib (Supplementary Fig. S1A; Supplementary Tables S2–S4). We next analyzed the 122 compounds in BeatAML and found that two of these compounds were included in the group of 78 small molecules. One of these compounds was ibrutinib, which has been identified as a FLT3 inhibitor (30); however, the effect of foretinib on FLT3 mutations has not been reported. Foretinib was developed as a c-MET/VGEFR2 inhibitor and has shown some therapeutic effects for the treatment of solid tumors in clinical trials (21, 23, 26). Through analyzing the drug screening results from the BeatAML database (27), we found that foretinib could specifically inhibit primary leukemia cells carrying the FLT3-ITD mutation, and the IC50 values were lower in patients with a high FLT3-ITD allele ratio (Fig. 1A and B; Table 1). Compared with FLT3 inhibitors used in clinical (gilteritinib, quizartinib, midostauin, and sorafenib), foretinib was the most potent in inhibiting the growth of FLT3-ITD primary cells (Fig. 1C and D; Supplementary Fig. S1B–S1E). Patients with FLT3-ITD, NPM1, and DNMT3A mutations have an extremely poor prognosis and are always resistant to chemotherapy (31). These patient samples were also more sensitive to foretinib (Fig. 1E and F).
Figure 1.
Foretinib is a potential FLT3 inhibitor. A, Heat map depicting the effect of FLT3-ITD mutations on the IC50 values of foretinib and other FLT3 inhibitors in the same 189 patient samples. Samples were grouped by FLT3 mutation status. B, Analysis of the effect of the FLT3-ITD mutation and allelic ratio on sensitivity to foretinib and its IC50. C–F, Comparison of the IC50 values of foretinib, gilteritinib, quizartinib, sorafenib, and midostaurin in patients with FLT3-ITD mutation (C), high allelic ratio of FLT3-ITD mutation (D), and FLT3-ITD (high allelic ratio), NPM1, DNMT3A mutation (E and F). IC50 values are the mean of all patient values. G, Computational modeling of foretinib binding to wild-type FLT3 (Protein Data Bank: 5×02). Foretinib formed a hydrogen bond with the amino acid residue Cys-695, with a distance of 2.14 Å. It also had hydrophobic interactions with Leu-616, Tyr-696, Asp-698, and Leu-818. H, Ba/F3 FLT3-ITD cells were treated with foretinib (1 μmol/L) or DMSO and subjected to heat treatment staring at 43.0°C and up to 55°C. Top, representative Western blotting of FLT3. Bottom, melting curve of FLT3. Quantitative analysis of FLT3 protein expression was measured using ImageJ. Data are presented as the mean ± SD of three independent experiments. I, Ba/F3 FLT3-ITD cells were treated with increasing concentrations of foretinib (0–100 nmol/L) and heated at 51.1°C. Top, representative blotting of FLT3. Bottom, isothermal dose‒response curve of FLT3. Quantitative analysis of FLT3 protein expression was measured using ImageJ. Data are presented as the mean ± SD of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.001.
Table 1.
IC50 values of various FLT3 inhibitors against patients with FLT3-ITD AML in BeatAML database.
| IC50 (μmol/L) | Foretinib | Quizartinib | Gilteritinib | Sorafenib | Midostaurin |
|---|---|---|---|---|---|
| FLT3-WT | 1.03 | 2.56 | 2.62 | 4.58 | 5.83 |
| FLT3-ITD | 0.28 | 1.06 | 0.92 | 2.09 | 3.97 |
| FLT3-ITD (Low) | 0.45 | 1.61 | 1.29 | 2.72 | 5.00 |
| FLT3-ITD (High) | 0.07 | 0.74 | 0.49 | 1.44 | 3.47 |
| FLT3-ITD, NPM1, DNMT3A | 0.34 | 0.66 | 0.72 | 1.71 | 4.10 |
| FLT3-ITD (High), NPM1, DNMT3A | 0.05 | 0.44 | 1.02 | 0.98 | 2.84 |
To better understand the inhibitory activity of foretinib against FLT3-ITD at the structural level, we carried out computational modeling of FLT3 (5×02) with foretinib (32). The docking study revealed nine putative binding positions of foretinib with the ATP binding sites of FLT3. We selected the position with the lowest binding energy (−13.01 kcal/mol) for analysis. The docking results indicated that foretinib bound to the FLT3 protein via amino acid residue Cys-695. The hydrogen bond between the nonpolar hydrogen atom of foretinib and the Cys-695 residue was 2.14 Å in length, which was far smaller than the 3.5 Å standard hydrogen bonds. The model also indicated that Tyr-696, Leu-616, Leu-818, and Asp-698 could form hydrophobic bonds with foretinib (Fig. 1G), which contributed to improving the stability of foretinib binding in the protein pocket.
To demonstrate whether the small molecule foretinib directly bound to the FLT3-ITD protein, we performed CESTA in Ba/F3 FLT3-ITD cells. Using DMSO treatment as a control, there was an obvious thermal shift of the melting curve following foretinib treatment. The temperature of 51.1°C was selected for isothermal dose‒response experiments, at which most FLT3 protein disappeared with DMSO treatment, but was detectable following foretinib treatment (Fig. 1H). Foretinib at 0.1 nmol/L to 100 nmol/L stabilized FLT3 in a dose-dependent manner, confirming the interaction between FLT3 and foretinib (Fig. 1I).
Foretinib is active against cell lines with FLT3-ITD mutations in vitro
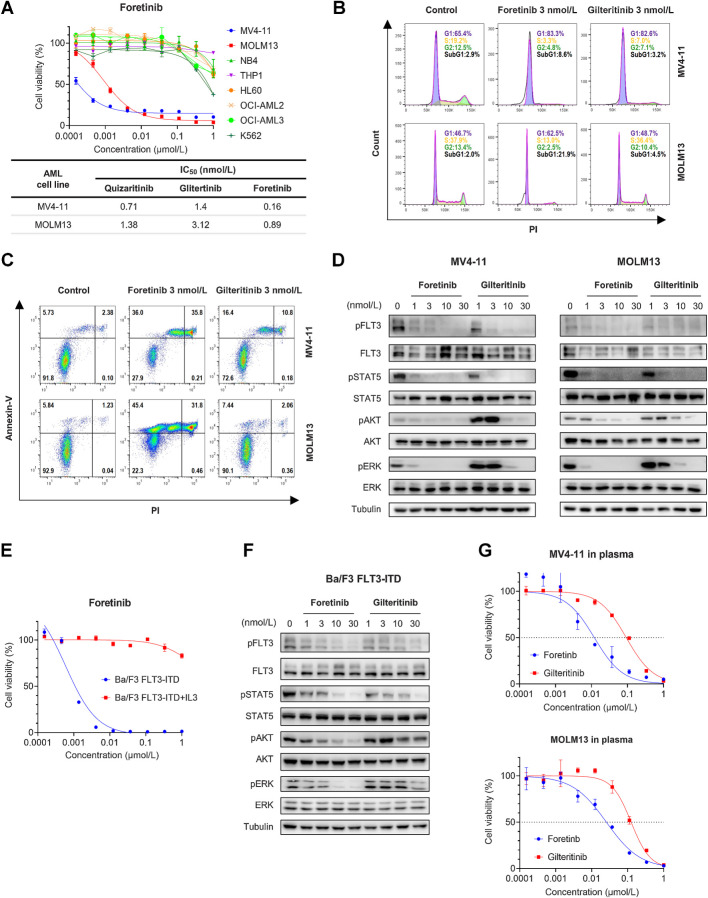
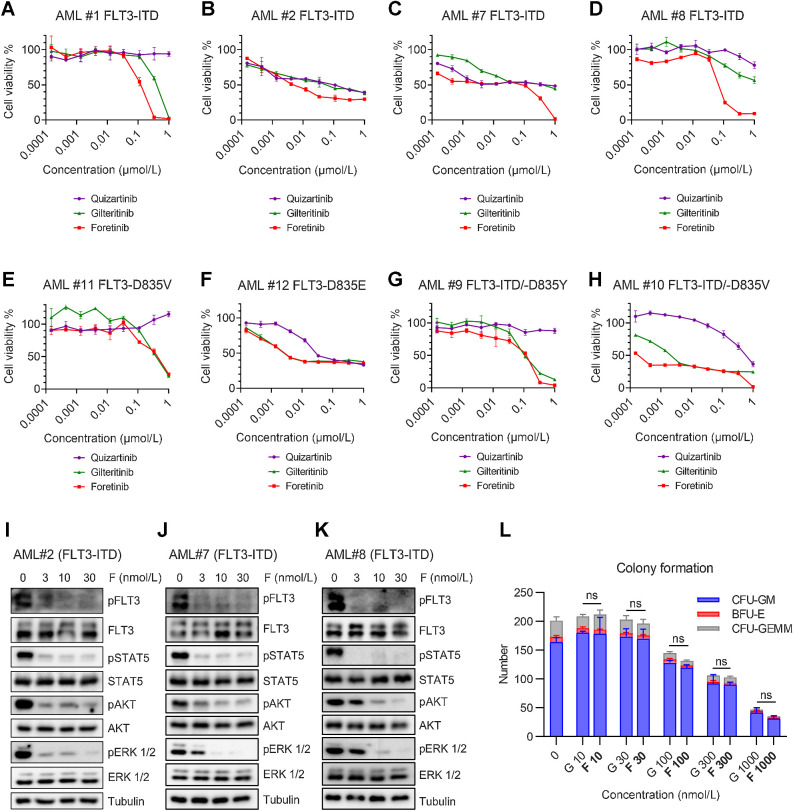
We tested the growth inhibitory effect of foretinib on human leukemia cell lines with FLT3-ITD mutation or wild-type FLT3. Foretinib effectively inhibited the growth of MV4-11 and MOLM13 cells carrying FLT3-ITD mutations with IC50 values of 0.16 and 0.89 nmol/L, respectively, whereas cell lines expressing wild-type FLT3 (NB4, THP1, HL60, OCI-AML2, OCI-AML3, and K562) were barely affected by foretinib (Fig. 2A). The cytotoxicity of foretinib on MV4-11 and MOLM13 cells was stronger than that of gilteritinib and quizartinib (Fig. 2A). Analysis of the cell cycle revealed that foretinib induced cell cycle arrest in MV4-11 and MOLM13 cells in a dose-dependent manner 24 hours after administration (Fig. 2B; Supplementary Fig. S2A). Significant apoptosis was observed in MV4-11 and MOLM13 cells after 48 hours of treatment with foretinib (Fig. 2C; Supplementary Fig. S2B), and simultaneous activation of caspase 8 and PARP1 cleavage was detected by Western blotting (Supplementary Fig. S2C). Notably, the cell cycle arrest and apoptosis-promoting effects of foretinib were superior to those of gliteritinib, which was consistent with the IC50 values. Immunoblotting showed that foretinib prominently inhibited the phosphorylation of FLT3 and its downstream targets STAT5, ERK, and AKT in MV4-11 and MOLM13 cells (Fig. 2D; Supplementary Fig. S2D). Foretinib had a stronger inhibitory effect than glitertinib on the activity of FLT3 signaling pathways. We also engineered Ba/F3 cells to express FLT3-ITD mutations, which rendered these Ba/F3 cells able to survive independent of IL3. Foretinib potently inhibited the growth of Ba/F3 FLT3-ITD cells. The selective and potent activity of foretinib was found to come from the inhibition of FLT3-ITD because antiproliferation was rescued in Ba/F3 FLT3-ITD cells in the presence of IL3 (Fig. 2E). In consistent with the results from the human cell lines, foretinib more significantly inhibited the activity of FLT3 pathways than gilteritinib in Ba/F3 FLT3-ITD cells (Fig. 2F; Supplementary Fig. S2E). It has been reported that plasma protein binding is a leading cause limiting the clinical efficacy of FLT3 inhibitors (33). We compared the IC50 values of foretinib and gilteritinib for MOLM13 and MV4-11 cells cultured in 100% AML patient plasma. The addition of human plasma increased the IC50 values of both foretinib and gilteritinib. However, the IC50 values of gilteritinib were apparently higher than those of foretinib for MV4-11 cells (92.39 vs. 11.98 nmol/L) and MOLM13 cells (124.3 vs. 25.57 nmol/L) in human plasma (Fig. 2G). As a control, the IC50 values of midostaurin in human plasma increased to more than 1 mmol/L, which accounted for the failure of midostaurin monotherapy in clinical (Supplementary Fig. S2F). These results indicated that foretinib specifically and potently inhibited the growth of cells with FLT3-ITD mutations in vitro. We also tested the inhibitory effect of foretinib on common clinically identified FLT3-TKD mutants. Ba/F3 cells expressing D835Y, D835V, Y842C, and N676K were all resistant to quizartinib. Foretinib inhibited all assessed TKD mutations with IC50 values comparable to gilteritinib, indicating that foretinib also has efficacy against cells carrying FLT3-TKD alone (Supplementary Fig. S3A–S3D).
Figure 2.
Foretinib shows potent antileukemia activity in AML cells harboring FLT3-ITD mutations. A, Dose‒response curves of eight AML cell lines treated with increasing concentrations of foretinib for 48 hours. Data are representative of three experiments. The IC50 values of quizartinib, gilteritinib, and foretinib in the AML cell lines harboring FLT3-ITD are shown. Data are presented as the mean of three experiments. B, Cell cycle analysis of MV4-11 and MOLM13 cells treated with foretinib (3 nmol/L) or gilteritinib (3 nmol/L) for 24 hours by flow cytometry. Data are representative of three experiments. C, Apoptosis analysis of MV4-11 and MOLM13 cells treated with foretinib (3 nmol/L) or gilteritinib (3 nmol/L) for 48 hours by flow cytometry. Data are representative of three experiments. D, The phosphorylation and total levels of FLT3, STAT5, ERK, and AKT protein in MV4-11 and MOLM13 cells detected by Western blotting after incubation with the indicated concentrations of foretinib and gilteritinib for 2 hours. Tubulin was used as a loading control. E, Dose‒response curve of Ba/F3 FLT3-ITD cells treated with foretinib for 48 hours in the presence or absence of IL3. Data are representative of three experiments. F, Western blotting analysis of pFLT3, FLT3, pSTAT5, STAT5, pAKT, AKT, pERK, and ERK in Ba/F3 FLT3-ITD cells treated with foretinib or gilteritinib for 2 hours. Tubulin was used as a loading control. G, Dose‒response curves of MV4-11 and MOLM13 cells in 100% AML patient plasma treated with increasing concentrations of foretinib or gilteritinib for 48 hours. Data are representative of three experiments.
Comparison of the antileukemic effects of foretinib, quizartinib, and gilteritinib in cell-derived leukemia mouse models
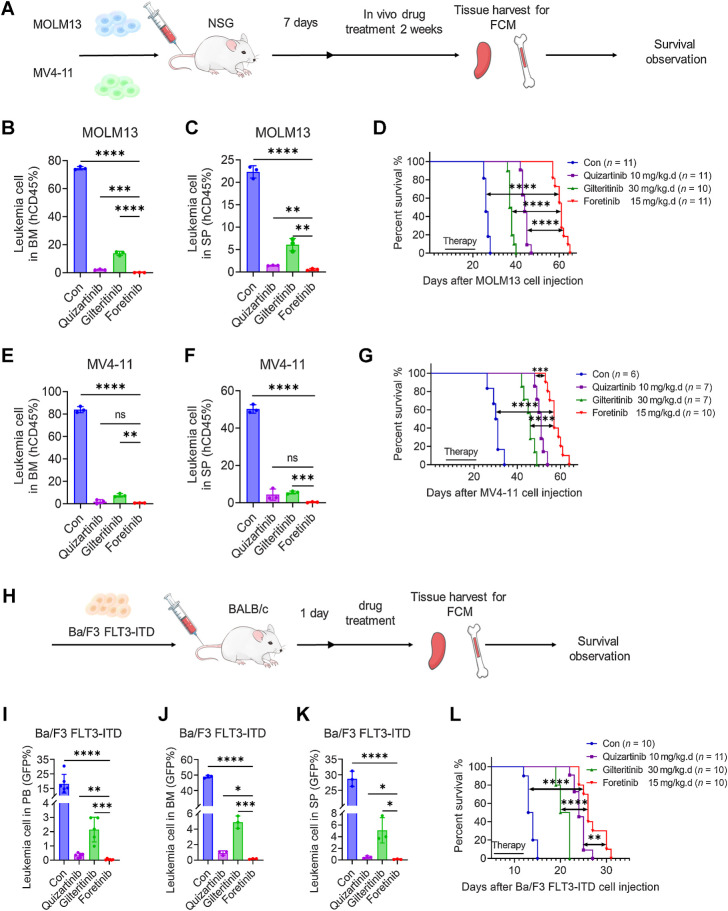
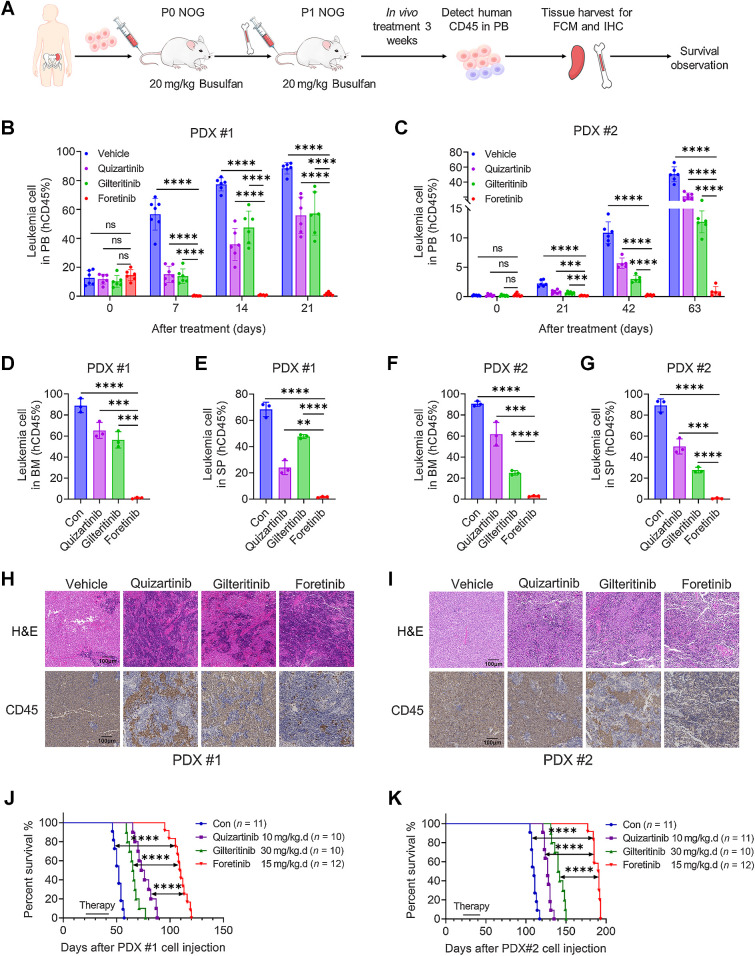
On the basis of the results of in vitro experiments, we further evaluated the efficacy of foretinib in vivo. NSG mice were injected with MOLM13 or MV4-11 cells and then subsequently received foretinib, gilteritinib, quizartinib, or control treatment randomly (Fig. 3A). The administration dose of each drug was determined on the previous studies. We detected the proportion of leukemia cells (hCD45) in the BM and SP of the mice by flow cytometry. In the MOLM13 animal model, leukemia cells in the BM were almost absent (0.07%) following treatment with foretinib; in comparison, leukemia cells made up 74.6% of cells in the control group, 2.13% of cells in the quizartinib group, and 13.8% of cells in the giltertinib group (Fig. 3B; Supplementary Fig. S4A). Consistent with the BM sample, leukemia cells in the SP were almost absent (0.56%) in the foretinb group, while they made up 22.3% of cells in the control group, 1.43% in the quizartinib group, and 6.08% in the giltertinib group (Fig. 3C; Supplementary Fig. S4A). The mice in the control group died at an average of 26 days after transplantation, while the mice treated with gilteritinib and quizartinib survived for 37.5 days and 44 days, respectively. The median survival time of mice in foretinib group was significantly extended to 61 days (P < 0.0001 compared with the control, gilteritinib and quizartinib groups; Fig. 3D). The same therapeutic advantages were observed in the MV4-11 animal model. All of the drugs reduced the leukemia burden in mice, while the foretinib group had a lower percentage of leukemia cells in the BM and SP than the quizatinib and gilteritinib groups (Fig. 3E and F; Supplementary Fig. S4B). Foretinib treatment prolonged the survival of MV4-11 diseased mice from 30.5 days in the control group, 46 days in the gilteritinib group and 51 days in the quizartinib group to 57 days (Fig. 3G). We further injected GFP-positive Ba/F3 FLT3-ITD cells into BALB/c mice, and drugs were administered randomly from the second day (Fig. 3H). The infiltration of leukemia cells in the PB, BM, and SP were reduced by gilteritinib while GFP-positive cells were almost undetectable in the quizartinib and foretinib groups (Fig. 3I–K; Supplementary Fig. S4C). The survival of mice in the foretinib group was significantly longer than that in the gilteritinib group and slightly longer than that in the quizartinib group (Fig. 3L). Collectively, these data support the superior efficacy of foretinib in the treatment of FLT3-ITD AML.
Figure 3.
In vivo antileukemia activity of foretinib in the FLT3-ITD-AML mouse model. A, Schematic illustration of the MV4-11 and MOLM13 xenograft AML mouse model. Female NSG mice engrafted with MV4-11 and MOLM13 cells were orally administered with vehicle, quizartinib (10 mg/kg), gilteritinib (30 mg/kg), or foretinib (15 mg/kg) daily for 14 days beginning on day 8. Three days after ending the treatment, three mice of each group were randomly sacrificed, and then the percentages of leukemia cells infiltrated in the BM (B and E) and SP (C and F) were determined by flow cytometry. Survival of mice engrafted with MOLM13 (D) and MV4-11 (G) cells is shown by Kaplan‒Meier analysis. H, Schematic illustration of the Ba/F3 FLT3-ITD mouse model. Female BALB/c mice engrafted with Ba/F3 FLT3-ITD cells were orally administered with vehicle, quizartinib (10 mg/kg), gilteritinib (30 mg/kg), or foretinib (15 mg/kg) daily from the second day until the first mouse in the vehicle group died. I, After 8 days of treatment, the percentages of leukemia cells that infiltrated the PB were determined by flow cytometry. J and K, After 9 days of treatment, three mice were randomly sacrificed, and the percentages of leukemia cells infiltrated in the BM (J) or SP (K) were determined by flow cytometry. L, Kaplan‒Meier analysis of the survival of mice engrafted with Ba/F3 FLT3-ITD cells. Data are presented as the mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.001; ns, nonsignificant.
Foretinib effectively inhibits clinical drug resistance mediated by secondary mutations both in vitro and in vivo
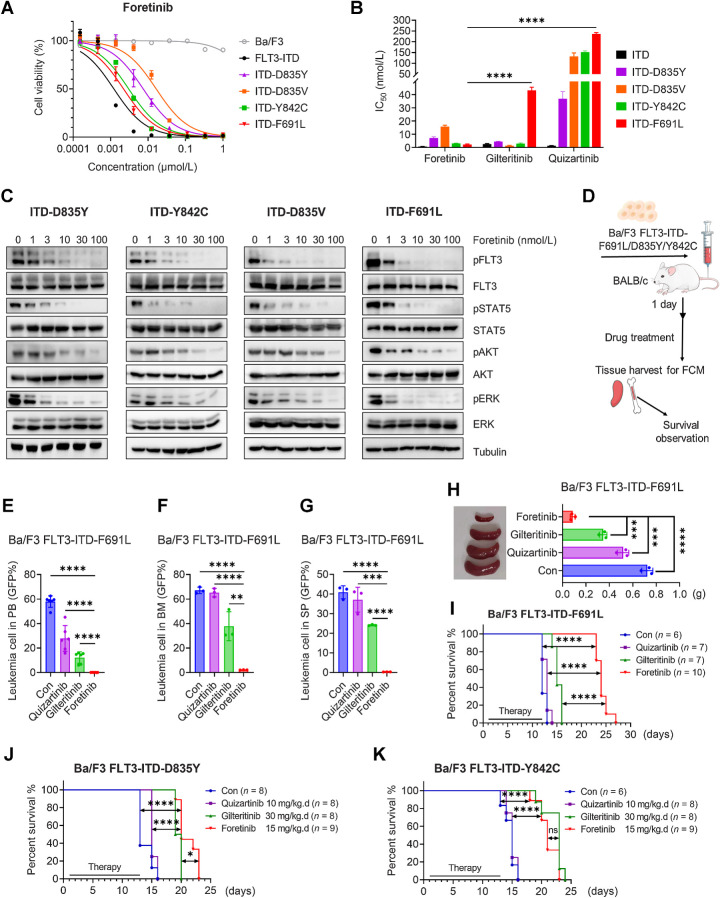
To evaluate whether foretinib can inhibit the growth of cells with various types of FLT3-ITD-TKD resistance mutations, especially the F691 L gatekeeper mutation, we engineered Ba/F3 cells stably expressing FLT3-ITD-D835Y/D835V/Y842C/F691L. As reported previously, gilteritinib was effective against cells with secondary mutations other than F691L, while cells with any secondary mutations were resistant to quizartinib. However, foretinib inhibited the growth of all tested cells, with IC50 values ranging from 0.65 to 16.4 nmol/L (Fig. 4A and B; Supplementary Fig. S5A and S5B). Consistently, foretinib effectively inhibited the phosphorylation of FLT3 and its downstream signaling pathways, including STAT5, ERK, and AKT, in Ba/F3 FLT3-ITD-D835Y/D835V/Y842C/F691 L cells (Fig. 4C). BALB/c mice were injected intravenously with Ba/F3 FLT3-ITD-F691L/D835Y/Y842C cells and were randomly divided into four groups to receive control, foretinib, quizartinib, or gilteritinib treatment (Fig. 4D). In the PB, BM, and SP of FLT3-ITD-F691 L mice, gilteritinib slightly inhibited the growth of GFP-positive cells, and quizartinib had no therapeutic effects; meanwhile, there were almost no leukemia cells following foretinib treatment (Fig. 4E–G; Supplementary Fig. S6A). The weight of SPs in foretinib-treated mice was lighter than those of mice in the quizartinib, gilteritinib, and control groups (Fig. 4H). H&E staining of the SPs and livers demonstrated that treatment with foretinib significantly reduced the infiltration of AML cells in these tissues, which is a characteristic of AML invasion and spreading (Supplementary Fig. S6B). Compared with control or quizartinib group, mice in the gilteritinib group had mildly increased survival rates. Importantly, foretinib showed significant therapeutic benefits on median survival than gilteritinib (24 vs. 15 days, P < 0.0001; Fig. 4I). During treatment, no significant weight loss (Supplementary Fig. S6C) or any other signs of toxicity were observed in any group. Foretinib was slightly better than gilteritinib at prolonging the survival time of mice in the D835Y model (Fig. 4J), and foretinib was comparable to gilteritinib in the Y842C animal model (Fig. 4K); meanwhile, quizartinib was ineffective in both animal models. Overall, our results showed that foretinib could overcome secondary mutations that occurred during the treatment of existing clinical drugs.
Figure 4.
Foretinib is effective in cells with FLT3-ITD-TKD mutations that confer resistance to approved FLT3 inhibitors. A, Dose‒response curves of Ba/F3 cells with or without the indicated FLT3 mutations and treated with increasing concentrations of foretinib for 48 hours. Data are representative of three experiments. B, The IC50 values of quizartinib, gilteritinib, and foretinib in Ba/F3 FLT3-ITD-TKD cells. Data are presented as the mean ± SD of three experiments. C, Western blotting analysis of pFLT3, FLT3, pSTAT5, STAT5, pAKT, AKT, pERK, and ERK in Ba/F3 FLT3-ITD-TKD cells treated with foretinib for 2 hours. Tubulin was used as a loading control. D, Schematic illustration of the Ba/F3 FLT3-ITD-TKD mouse model. BALB/c mice engrafted with Ba/F3 FLT3-ITD-F691 L cells were orally administered with vehicle, quizartinib (10 mg/kg), gilteritinib (30 mg/kg), or foretinib 15 (mg/kg) daily until the first mouse died in the vehicle group. E, After 7 days of treatment, the percentages of leukemia cells infiltrated in the PB were determined by flow cytometry. F and G, Ten days after cell injection, the percentages of leukemia cells that infiltrated the BM (F) and SP (G) of three mice in each group were determined by flow cytometry. H, The SP weight was measured. I, The survival of mice engrafted with Ba/F3 FLT3-ITD-F691 L cells was monitored by Kaplan‒Meier analysis. J and K, Kaplan‒Meier analysis of the survival of BALB/c mice engrafted with Ba/F3 FLT3-ITD-D835Y (J) or Ba/F3 FLT3-ITD-Y842C (K) cells with different treatments. Data are presented as the mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.001.
Activating NRAS mutations that cause resistance to gilteritinib and quizartinib also decrease the efficacy of foretinib
In addition to on-target resistance mutations, it has been reported that off-target mutations of other genes also account for resistance to FLT3 inhibitors, especially for type I FLT3 inhibitor (34). The genetic profile of patients following gilteritinib treatment showed that activating NRAS mutation is the most common mechanism of resistance (16). We tested the efficacy of foretinib against MV4-11 and MOLM13 cells expressing NRAS-G12C (Supplementary Fig. S7A). As expected, expression of NRAS-G12C significantly increased viabilities of MOLM13 and MV4-11 cells under gilteritinib and quizatinib treatment. Although foretinib retained a better and more stable inhibitory effect against cells harboring NRAS-G12C than gilteritinib and quizartinib, NRAS-G12C mutation also induced upward shift in the dose–response curves of foretinib, indicating that foretinib were also likely vulnerable to the emergence of off-target mutations (Supplementary Fig. S7B–S7G).
Foretinib shows potent antileukemia activity against primary AML blasts harboring FLT3 mutations
We evaluated the in vitro antileukemia effects of foretinib on primary AML cells from 10 patients with FLT3 mutations and 2 patients with FLT3-WT. In primary samples with FLT3-ITD mutations, foretinib effectively reduced cell viability in a dose-dependent manner, and its overall activity was more potent than that of gilteritinib and quizartinib (Fig. 5A–D; Supplementary Fig. S8A and S8B). In primary blast harboring FLT3-D835V/E point mutation or FLT3-ITD/-D835V/Y secondary mutation, foretinib and gilteritinib demonstrated comparable inhibitory effects while quizartinib showed much weaker efficacy (Fig. 5E–H). The two primary AML samples expressing FLT3-WT had no or little response to foretinib, gilteritinib, and quizartinib (Supplementary Fig. S8C and S8D). Western blotting showed that foretinib potently inhibited phosphorylation of FLT3 and its downstream signaling pathways in patient primary cells (Fig. 5I–K). These results indicated that foretinib exhibited significant therapeutic effect on AML blasts harboring FLT3 mutation in vitro. We also tested the effect of foretinib on PB mononuclear cells from healthy donors. The results showed that foretinib did not inhibit the cell viability of normal blood cells (Supplementary Fig. S8E). Moreover, the impact of foretinib on normal hematopoiesis was evaluated using human cord blood. Both foretinib and gilteritinib inhibited the colony formation in a dose-dependent manner and there was little difference in the number of colonies reduced by the two drugs, which suggests an acceptable toxicity of foretinib on normal hematopiesis (Fig. 5L; Supplementary Fig. S8F).
Figure 5.
Potent inhibition by foretinib in primary AML blasts harboring FLT3 mutations. A–D, Cell viabilities of four primary AML samples harboring FLT3-ITD after treatment with the indicated concentrations of quizartinib, gilteritinib, or foretinib for 48 hours. E and F, Cell viabilities of two primary AML samples harboring FLT3-D835 point mutation after treatment with the indicated concentrations of quizartinib, gilteritinib, or foretinib for 48 hours. G and H, Cell viabilities of two primary AML blasts with FLT3-ITD and FLT3-D835 point mutation after treatment with the indicated concentrations of quizartinib, gilteritinib, or foretinib for 48 hours. Cell viability data are representative of three experiments. I–K, Western blotting analysis of pFLT3, FLT3, pSTAT5, STAT5, pAKT, AKT, pERK, and ERK in three primary AML samples harboring FLT3-ITD after treatment with foretinib (F) for 2 hours. Tubulin was used as a loading control. L, Mononuclear cells isolated from cord blood were seeded in methylcellulose medium and treated with increasing concentrations of foretinib (F) or gilteritinib (G) for 10 days. CFU-GM, BFU-E, and CFU-GEMM colonies were counted after 10 days (n = 3). Data are presented as the mean ± SD. ns, nonsignificant, P > 0.05.
Antileukemia effects of foretinib in the FLT3-ITD-AML PDX model
To translate our experimental results into clinical applications, we tested the in vivo effects of foretinib on two AML PDX models. Mice in the first PDX (PDX #1) model were transplanted with cells from a patient with relapsed FLT3-ITD AML, and mice in the second PDX (PDX #2) model were injected with cells from a patient with refractory AML harboring FLT3-ITD, DNMT3A, and NPM1 mutations. Administration started 21 days after transplantation, and mice were treated with control, foretinib (15 mg/kg/day), quizartinib (10 mg/kg/day), or gilteritinib (30 mg/kg/day) for 3 weeks (Fig. 6A). The gilteritinib and quizartinib treatment groups had reduced hCD45 leukemia cells in the PB of mice, and there were almost no leukemia cells in the PB of the mice treated with foretinib in PDX #1 (Fig. 6B). The same results were observed in the refractory leukemia PDX #2 model (Fig. 6C). We detected hCD45 leukemia cells in the BM and SP by flow cytometry in PDX mice. Leukemia cells in the BM of PDX #1 were almost absent (1.1%) following treatment with foretinib; in comparison, leukemia cells made up 88.8% of cells in the control group, 65.3% of cells in the quizartinib group, and 56.3% of cells in the giltertinib group (Fig. 6D). Consistent with the BM samples, leukemia cells in the SP were almost absent (1.6%) in the foretinib group, while they made up 68.3% of cells were in the control group, 24% of cells in the quizartinib group, and 47.7% of cells in the giltertinib group (Fig. 6E). Similar results were observed in the PDX#2 animal model (Fig. 6F and G). Compared with the control, quizartinib and giltertinib, foretinib significantly reduced the SP size in both PDX models (Supplementary Fig. S9A and S9B). Results of H&E staining and hCD45 IHC of the SPs also demonstrated that there was almost no infiltration of leukemia cells in foretinib-treated mice (Fig. 6H and I). No differences in body weight were observed among groups during the dosing period (Supplementary Fig. S9C). Compared with control-treated mice, mice in the quizartinib or gilteritinb groups had prolonged survival and foretinib exhibits the most potent effect on prolonging the survival period in both PDX models (Fig. 6J and K). We treated BM cells from mice in the control groups of PDX models with foretinib or gilteritinib for 2 hours, and found that foretinib had stronger inhibition of pFLT3, pERK and pAKT than gilteritinib (Supplementary Fig. S9D and S9E). These results strongly suggest that foretinib had higher efficacy to eliminate primary FLT3-ITD AML cells in vivo.
Figure 6.
Foretinib has a potent therapeutic effect in two PDX models. A, Schematic illustration of the PDX model. Primary BM cells harboring FLT3-ITD were injected into busulfan (20 mg/kg)-pretreated NOG mice via the tail vein. After engraftment was confirmed, the BM cells were collected and then injected into busulfan-pretreated NOG mice for secondary transplantation. After secondary transplantation was confirmed, groups of mice were orally administered with vehicle, quizartinib (10 mg/kg), gilteritinib (30 mg/kg), or foretinib (15 mg/kg) daily for 3 weeks. B–G, Primary blasts expressing hCD45 infiltrated in PB (B and C), BM (D and F), or SP (E and G) were detected by flow cytometry. H and I, H&E staining and IHC results of hCD45 in the SPs of PDX mice are shown. J and K, The survival of mice in the two PDX models treated with vehicle, quizartinib, gilteritinib, or foretinib was analyzed. Data are presented as the mean ± SD. **, P < 0.01; ***, P < 0.001; ****, P < 0.001; ns, nonsignificant.
Discussion
FLT3 is one of the most promising targets for AML because FLT3 mutations are the most frequently identified genetic alterations in AML and are associated with poor prognosis (2, 35, 36). Among the multiple FLT3 inhibitors currently available, gilteritinib, quizartinib, and midostaurin have been approved for clinical application. Unfortunately, remission is short, and relapses usually occur quickly; for example, gilteritinib is the only monotherapy widely used in the clinic for refractory/relapsed patients, but approximately one-third of patients do not respond and the median survival is only 9.3 months (37). Resistance to FLT3 inhibitors as a result of acquired point mutations in the TKD or other mechanisms has limited the sustained efficacy of treatments, and a “gatekeeper” mutation (F691L) is known to cause resistance to all currently available FLT3 inhibitors (16, 17, 38, 39). Therefore, finding more effective FLT3 inhibitors that can overcome the drug resistance caused by F691 L and other mutations is still an urgent problem. Here, we demonstrated that foretinib is effective against cells with FLT3-ITD and drug-resistant FLT3-ITD-TKD mutations; in particular, it is effective against cells with FLT3-ITD-F691L, which is currently understood as the most difficult mutation to overcome.
First, we found that the multi-kinase inhibitor foretinib was a potential FLT3 inhibitor through virtual screening. CESTA confirmed the direct interaction between FLT3 and foretinib. Foretinib exerted effective inhibitory activities against FLT3-ITD AML cell lines and patient primary cells in vitro. In CDX and PDX leukemia animal models, foretinib displayed the most potent therapeutic effect among the tested FLT3 inhibitors. The BeatAML database also showed that foretinib was superior to the existing clinical drugs. The therapeutic advantages of foretinib may be due to its lower IC50 values and stronger inhibition of the downstream phosphorylation of ERK and AKT. Previous studies have shown that the activation of AKT and ERK impeded the efficacy of gilteritinib (16, 40). An important reason limiting the clinical efficacy of FLT3 inhibitors is plasma protein binding (33). In particular, previous studies and our results showed that the concentration at 1 mmol/L did not reach the IC50 values of midostaurin in plasma, which explains the little therapeutic effect of midostaurin monotherapy (41). In our tests, the IC50 values of foretinib were significantly lower than those of gilteritinib in 100% patient plasma, demonstrating the potential of foretinib to achieve clinical efficacy. In addition, foretinib was originally developed as an inhibitor of c-MET and VEGFR2 (20). It has been reported that MET signaling was aberrantly activated in AML and MET blockade inhibited the proliferation of leukemia cells (42, 43). Early studies have also shown that VEGFR2 can be used as a target for AML therapy (44, 45). These may also be part of the reasons why foretinib is superior to the existing clinical drugs in vivo. Interestingly, the FLT3-ITD, DNMT3A, and NPM1 AML PDX model was also more sensitive to foretinib. This represents a type of leukemia with a very poor prognosis that accounts for approximately 10% of all patients (31). This result is conducive to the screening of patients in subsequent clinical trials.
Although most of the clinically relevant on-target secondary mutations can be suppressed by gilteritinib, F691 L site mutations occur in relapsed patients following gilteritinib treatment and cannot be overcome by clinically used FLT3 inhibitors (16, 18). Through in vitro and in vivo experiments, we have shown that foretinib can overcome a variety of drug-resistant mutations in cells, including D835, F691, and Y842 site mutations, at concentrations far lower than the safe plasma concentration determined by clinical trials. These results suggest that foretinib may be an effective second-line TKI for patients with AML with acquired resistance to gilteritinib, quizartinib, or sorafenib due to secondary TKD mutations in F691, D835, and Y842. However, secondary off-target mutation of NRAS, which developed as a common resistance mechanism to gilteritinib, also decreased the efficacy of foretinib. Whether NRAS or other mutation arises to confer resistance after foretinib treatment deserves further clinical study.
Foretinib was originally developed for the treatment of various solid tumors and has been tested in clinical trials (22, 26, 46). On the basis of our preclinical data of foretinib, it would be reasonable to conduct clinical trials in patients with AML with FLT3 mutations, especially those who developed mutations for resistance to gilteritinib, quizartinib, or sorafenib. Although the current trials of foretinib in solid tumors have been terminated considering that its efficacy is no better than that of current clinical drugs, the drug dosage and pharmacokinetic data could provide some indications. In phase II clinical trials of patients with solid tumors, the two general dosing schedules were 240 mg once per day on days 1 through 5 every 14 days or 80 mg daily, both of which were well tolerated and showed similar safety profile (21, 23, 47). The pharmacokinetic data of phase I clinical trials in solid tumors showed that the maximum plasma drug concentrations (Cmax) of foretinib were 0.34 μmol/L/mL on day 8 after intermittent dosing with 240 mg and 0.072 μmol/L/mL on day22 after daily dosing with 80 mg (24, 48). As for gilteritinib, the Cmax is 0.51 μmol/L on day15 with the clinical dosage of 120 mg/day (49). Considering the IC50 values of foretinib and gilteritinib for patients with AML in BeatAML database are 0.28 μmol/L and 0.92 μmol/L, respectively, intermittent dosing with 240 mg may be a more suitable dosing regimen for further clinical trials of foretinib in patients with AML and is expected to show effective inhibition against AML blasts. We believe that foretinib may be a key advancement in the treatment of FLT3-mutated AML, and foretinib has promising potential to achieve durable responses in the clinic.
Supplementary Material
Supplementary Data
Table S2,S3,S4
Acknowledgments
This work was supported by National Natural Science Foundation of China (grant no. 82100178 to X. Xiao), Science and Technology Key Project of Guangzhou, China (grant no. 202102010037 to S. Wang), Guangzhou Health Technology Project (grant no. 20221A011037 to X. Xiao; 20231A011012 to P. Wang), Guangzhou Municipal Science and Technology Project (grant no. 202002030035 to S. Wang; 202201010999 to X. Xiao; 2023A04J0616 to P. Wang). The authors thank all the patients and their families for supporting this study and the health care providers who took care of these patients and contributed to the medical records.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
P. Wang: Conceptualization, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. Y. Zhang: Conceptualization, data curation, software, formal analysis, validation, investigation, visualization, writing–original draft, writing–review and editing. R. Xiang: Resources, validation, methodology. J. Yang: Data curation, validation, visualization. Y. Xu: Resources, software. T. Deng: Resources, investigation. W. Zhou: Resources, investigation. C. Wang: Investigation. X. Xiao: Conceptualization, supervision, funding acquisition, writing–original draft, writing–review and editing. S. Wang: Conceptualization, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lemonnier F, Inoue S, Mak TW. Genomic classification in acute myeloid leukemia. N Engl J Med 2016;375:900. [DOI] [PubMed] [Google Scholar]
- 3. Drexler HG. Expression of FLT3 receptor and response to FLT3 ligand by leukemic cells. Leukemia 1996;10:588–99. [PubMed] [Google Scholar]
- 4. Kihara R, Nagata Y, Kiyoi H, Kato T, Yamamoto E, Suzuki K, et al. Comprehensive analysis of genetic alterations and their prognostic impacts in adult acute myeloid leukemia patients. Leukemia 2014;28:1586–95. [DOI] [PubMed] [Google Scholar]
- 5. Kiyoi H, Naoe T, Yokota S, Nakao M, Minami S, Kuriyama K, et al. Internal tandem duplication of FLT3 associated with leukocytosis in acute promyelocytic leukemia. leukemia study group of the ministry of health and welfare (Kohseisho). Leukemia 1997;11:1447–52. [DOI] [PubMed] [Google Scholar]
- 6. Yamamoto Y, Kiyoi H, Nakano Y, Suzuki R, Kodera Y, Miyawaki S, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood 2001;97:2434–9. [DOI] [PubMed] [Google Scholar]
- 7. Sakaguchi M, Yamaguchi H, Kuboyama M, Najima Y, Usuki K, Ueki T, et al. Significance of FLT3-tyrosine kinase domain mutation as a prognostic factor for acute myeloid leukemia. Int J Hematol 2019;110:566–74. [DOI] [PubMed] [Google Scholar]
- 8. Bacher U, Haferlach C, Kern W, Haferlach T, Schnittger S. Prognostic relevance of FLT3-TKD mutations in AML: the combination matters–an analysis of 3082 patients. Blood 2008;111:2527–37. [DOI] [PubMed] [Google Scholar]
- 9. Brandts CH, Sargin B, Rode M, Biermann C, Lindtner B, Schwäble J, et al. Constitutive activation of Akt by Flt3 internal tandem duplications is necessary for increased survival, proliferation, and myeloid transformation. Cancer Res 2005;65:9643–50. [DOI] [PubMed] [Google Scholar]
- 10. Kornblau SM, Womble M, Qiu YH, Jackson CE, Chen W, Konopleva M, et al. Simultaneous activation of multiple signal transduction pathways confers poor prognosis in acute myelogenous leukemia. Blood 2006;108:2358–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rocnik JL, Okabe R, Yu JC, Lee BH, Giese N, Schenkein DP, et al. Roles of tyrosine 589 and 591 in STAT5 activation and transformation mediated by FLT3-ITD. Blood 2006;108:1339–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perrone S, Ottone T, Zhdanovskaya N, Molica M. How acute myeloid leukemia (AML) escapes from FMS-related tyrosine kinase 3 (FLT3) inhibitors? Still an overrated complication? Cancer Drug Resist 2023;6:223–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cortes J, Perl AE, Dh H, Kantarjian H, Martinelli G, Kovacsovics T, et al. Quizartinib, an FLT3 inhibitor, as monotherapy in patients with relapsed or refractory acute myeloid leukaemia: an open-label, multicentre, single-arm, phase 2 trial. Lancet Oncol 2018;19:889–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Joshi SK, Sharzehi S, Pittsenbarger J, Bottomly D, Tognon CE, McWeeney SK, et al. A noncanonical FLT3 gatekeeper mutation disrupts gilteritinib binding and confers resistance. Am J Hematol 2021;96:E226–E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smith CC, Paguirigan A, Jeschke GR, Lin KC, Massi E, Tarver T, et al. Heterogeneous resistance to quizartinib in acute myeloid leukemia revealed by single-cell analysis. Blood 2017;130:48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McMahon CM, Ferng T, Canaani J, Wang ES, Morrissette JJD, Eastburn DJ, et al. Clonal selection with RAS pathway activation mediates secondary clinical resistance to selective FLT3 inhibition in acute myeloid leukemia. Cancer Discov 2019;9:1050–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smith CC, Wang Q, Chin CS, Salerno S, Damon LE, Levis MJ, et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature 2012;485:260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tarver TC, Hill JE, Rahmat L, Perl AE, Bahceci E, Mori K, et al. Gilteritinib is a clinically active FLT3 inhibitor with broad activity against FLT3 kinase domain mutations. Blood Adv 2020;4:514–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Man CH, Fung TK, Ho C, Han HH, Chow HC, Ma AC, et al. Sorafenib treatment of FLT3-ITD(+) acute myeloid leukemia: favorable initial outcome and mechanisms of subsequent nonresponsiveness associated with the emergence of a D835 mutation. Blood 2012;119:5133–43. [DOI] [PubMed] [Google Scholar]
- 20. Qian F, Engst S, Yamaguchi K, Yu P, Won KA, Mock L, et al. Inhibition of tumor cell growth, invasion, and metastasis by EXEL-2880 (XL880, GSK1363089), a novel inhibitor of HGF and VEGF receptor tyrosine kinases. Cancer Res 2009;69:8009–16. [DOI] [PubMed] [Google Scholar]
- 21. Choueiri TK, Vaishampayan U, Rosenberg JE, Logan TF, Harzstark AL, Bukowski RM, et al. Phase II and biomarker study of the dual MET/VEGFR2 inhibitor foretinib in patients with papillary renal cell carcinoma. J Clin Oncol 2013;31:181–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fujino T, Suda K, Koga T, Hamada A, Ohara S, Chiba M, et al. Foretinib can overcome common on-target resistance mutations after capmatinib/tepotinib treatment in NSCLCs with MET exon 14 skipping mutation. J Hematol Oncol 2022;15:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shah MA, Wainberg ZA, Catenacci DV, Hochster HS, Ford J, Kunz P, et al. Phase II study evaluating 2 dosing schedules of oral foretinib (GSK1363089), cMET/VEGFR2 inhibitor, in patients with metastatic gastric cancer. PLoS One 2013;8:e54014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shapiro GI, McCallum S, Adams LM, Sherman L, Weller S, Swann S, et al. A phase 1 dose-escalation study of the safety and pharmacokinetics of once-daily oral foretinib, a multi-kinase inhibitor, in patients with solid tumors. Invest New Drugs 2013;31:742–50. [DOI] [PubMed] [Google Scholar]
- 25. Yau TCC, Lencioni R, Sukeepaisarnjaroen W, Chao Y, Yen CJ, Lausoontornsiri W, et al. A phase I/II multicenter study of single-agent foretinib as first-line therapy in patients with advanced hepatocellular carcinoma. Clin Cancer Res 2017;23:2405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zillhardt M, Park SM, Romero IL, Sawada K, Montag A, Krausz T, et al. Foretinib (GSK1363089), an orally available multikinase inhibitor of c-Met and VEGFR-2, blocks proliferation, induces anoikis, and impairs ovarian cancer metastasis. Clin Cancer Res 2011;17:4042–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018;562:526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao H, Liu P, Zhang R, Wu M, Li D, Zhao X, et al. Roles of palmitoylation and the KIKK membrane-targeting motif in leukemogenesis by oncogenic KRAS4A. J Hematol Oncol 2015;8:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ouchida AT, Li Y, Geng J, Najafov A, Ofengeim D, Sun X, et al. Synergistic effect of a novel autophagy inhibitor and quizartinib enhances cancer cell death. Cell Death Dis 2018;9:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu H, Hu C, Wang A, Weisberg EL, Wang W, Chen C, et al. Ibrutinib selectively targets FLT3-ITD in mutant FLT3-positive AML. Leukemia 2016;30:754–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Loghavi S, Zuo Z, Ravandi F, Kantarjian HM, Bueso-Ramos C, Zhang L, et al. Clinical features of de novo acute myeloid leukemia with concurrent DNMT3A, FLT3 and NPM1 mutations. J Hematol Oncol 2014;7:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yamaura T, Nakatani T, Uda K, Ogura H, Shin W, Kurokawa N, et al. A novel irreversible FLT3 inhibitor, FF-10101, shows excellent efficacy against AML cells with FLT3 mutations. Blood 2018;131:426–38. [DOI] [PubMed] [Google Scholar]
- 33. Levis M, Brown P, Smith BD, Stine A, Pham R, Stone R, et al. Plasma inhibitory activity (PIA): a pharmacodynamic assay reveals insights into the basis for cytotoxic response to FLT3 inhibitors. Blood 2006;108:3477–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lang TJL, Damm F, Bullinger L, Frick M. Mechanisms of resistance to small molecules in acute myeloid leukemia. Cancers 2023;15:4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu M, Li C, Zhu X. FLT3 inhibitors in acute myeloid leukemia. J Hematol Oncol 2018;11:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Larrosa-Garcia M, Baer MR. FLT3 inhibitors in acute myeloid leukemia: current status and future directions. Mol Cancer Ther 2017;16:991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Perl AE, Martinelli G, Cortes JE, Neubauer A, Berman E, Paolini S, et al. Gilteritinib or chemotherapy for relapsed or refractory FLT3-mutated AML. N Engl J Med 2019;381:1728–40. [DOI] [PubMed] [Google Scholar]
- 38. Eskazan AE. Midostaurin in FLT3-mutated acute myeloid leukemia. N Engl J Med 2017;377:1901. [DOI] [PubMed] [Google Scholar]
- 39. Kancha RK, Grundler R, Peschel C, Duyster J. Sensitivity toward sorafenib and sunitinib varies between different activating and drug-resistant FLT3-ITD mutations. Exp Hematol 2007;35:1522–6. [DOI] [PubMed] [Google Scholar]
- 40. Joshi SK, Nechiporuk T, Bottomly D, Piehowski PD, Reisz JA, Pittsenbarger J, et al. The AML microenvironment catalyzes a stepwise evolution to gilteritinib resistance. Cancer Cell 2021;39:999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Young DJ, Nguyen B, Li L, Higashimoto T, Levis MJ, Liu JO, et al. A method for overcoming plasma protein inhibition of tyrosine kinase inhibitors. Blood cancer discovery 2021;2:532–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kentsis A, Reed C, Rice KL, Sanda T, Rodig SJ, Tholouli E, et al. Autocrine activation of the MET receptor tyrosine kinase in acute myeloid leukemia. Nat Med 2012;18:1118–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen EC, Gandler H, Tosic I, Fell GG, Fiore A, Pozdnyakova O, et al. Targeting MET and FGFR in relapsed or refractory acute myeloid leukemia: preclinical and clinical findings, and signal transduction correlates. Clin Cancer Res 2023;29:878–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Santos SC, Dias S. Internal and external autocrine VEGF/KDR loops regulate survival of subsets of acute leukemia through distinct signaling pathways. Blood 2004;103:3883–9. [DOI] [PubMed] [Google Scholar]
- 45. Zhu Z, Hattori K, Zhang H, Jimenez X, Ludwig DL, Dias S, et al. Inhibition of human leukemia in an animal model with human antibodies directed against vascular endothelial growth factor receptor 2. Correlation between antibody affinity and biological activity. Leukemia 2003;17:604–11. [DOI] [PubMed] [Google Scholar]
- 46. Rayson D, Lupichuk S, Potvin K, Dent S, Shenkier T, Dhesy-Thind S, et al. Canadian Cancer Trials Group IND197: a phase II study of foretinib in patients with estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2-negative recurrent or metastatic breast cancer. Breast Cancer Res Treat 2016;157:109–16. [DOI] [PubMed] [Google Scholar]
- 47. Seiwert T, Sarantopoulos J, Kallender H, McCallum S, Keer HN, Blumenschein G Jr. Phase II trial of single-agent foretinib (GSK1363089) in patients with recurrent or metastatic squamous cell carcinoma of the head and neck. Invest New Drugs 2013;31:417–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Eder JP, Shapiro GI, Appleman LJ, Zhu AX, Miles D, Keer H, et al. A phase I study of foretinib, a multi-targeted inhibitor of c-Met and vascular endothelial growth factor receptor 2. Clin Cancer Res 2010;16:3507–16. [DOI] [PubMed] [Google Scholar]
- 49. Perl AE, Altman JK, Cortes J, Smith C, Litzow M, Baer MR, et al. Selective inhibition of FLT3 by gilteritinib in relapsed or refractory acute myeloid leukaemia: a multicentre, first-in-human, open-label, phase 1–2 study. Lancet Oncol 2017;18:1061–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Table S2,S3,S4
Data Availability Statement
The gene mutation and drug susceptibility data of patients with AML were obtained from the BeatAML database including Vizome (http://vizome.org/aml2/) and BeatAML2 (https://biodev.github.io/BeatAML2/). All other raw data generated in this study are available upon request from the corresponding author.