Abstract
Background.
Few-shot learning (FSL) is a class of machine learning methods that require small numbers of labeled instances for training. With many medical topics having limited annotated text-based data in practical settings, FSL-based natural language processing (NLP) holds substantial promise. We aimed to conduct a review to explore the current state of FSL methods for medical NLP.
Methods.
We searched for articles published between January 2016 and October 2022 using PubMed/Medline, Embase, ACL Anthology, and IEEE Xplore Digital Library. We also searched the preprint servers (e.g., arXiv, medRxiv, and bioRxiv) via Google Scholar to identify the latest relevant methods. We included all articles that involved FSL and any form of medical text. We abstracted articles based on the data source, target task, training set size, primary method(s)/approach(es), and evaluation metric(s).
Results.
Fifty-one articles met our inclusion criteria—all published after 2018, and most since 2020 (42/51; 82%). Concept extraction/named entity recognition was the most frequently addressed task (21/51; 41%), followed by text classification (16/51; 31%). Thirty-two (61%) articles reconstructed existing datasets to fit few-shot scenarios, and MIMIC-III was the most frequently used dataset (10/51; 20%). 77% of the articles attempted to incorporate prior knowledge to augment the small datasets available for training. Common methods included FSL with attention mechanisms (20/51; 39%), prototypical networks (11/51; 22%), meta-learning (7/51; 14%), and prompt-based learning methods, the latter being particularly popular since 2021. Benchmarking experiments demonstrated relative underperformance of FSL methods on biomedical NLP tasks.
Conclusion.
Despite the potential for FSL in biomedical NLP, progress has been limited. This may be attributed to the rarity of specialized data, lack of standardized evaluation criteria and the underperformance of FSL methods on biomedical topics. The creation of publicly-available specialized datasets for biomedical FSL may aid method development by facilitating comparative analyses.
Keywords: few-shot learning, natural language processing, machine learning, biomedical informatics
Graphical Abstract
1. Introduction
Few-shot learning (FSL), also referred to as low-shot learning, is a class of machine learning methods that attempt to learn to execute tasks using small numbers (i.e., few) of labeled training examples [1–3]. In supervised learning (i.e., learning from labeled data) settings with limited training instances, the application of traditional machine learning methods typically leads to overfitting (i.e., the learner is incapable of generalizing the characteristics of the training data) [4, 5]. Learning from small numbers of training instances is challenging for machine learning models, although it is conceptually possible since humans are often capable of generalizing learned concepts with limited exposure or using only partial information [6] (e.g., recognizing numbers or pictures [3]). Thus, true replication of human behavior by artificial intelligence (AI) requires the development of models that can learn to generalize from small numbers of training instances—an objective that FSL aims to achieve.
For many natural language processing (NLP) tasks, particularly within the medical domain (e.g., for rare or novel diseases), the availability of labeled data is often limited. Even when large labeled datasets are created for targeted tasks, due to restrictions associated with data privacy and patient security, it can be difficult or impossible to release or share them if they originate from medical sources, such as electronic health records (EHRs). Oftentimes, the data available for manual annotation is insufficient. Limited data is often associated with specific subpopulations (e.g., racial minorities), and machine learning models often underperform for such subpopulations [7]. Even when sufficient data is available, manually annotating them can be time-consuming, error-prone, and costly. This is particularly true for medical free text as manual annotation requires the annotators to read and interpret texts prior to assigning labels, and reliable annotations can only be obtained from high-skilled annotators (e.g., doctors). Sometimes, multiple rounds of annotations are required on the same data, further increasing the costs of such annotation. With the application domain being healthcare, it is critical to develop machine learning strategies that can address these practical limitations while ensuring high performance.
Over recent years, deep neural network-based approaches (a.k.a., deep learning) have seen high adoption and have achieved state-of-the-art results in many supervised learning tasks, sometimes achieving human-level performances [8]. Such methods require large amounts of labeled training data, which restricts their utility to only tasks for which such large labeled datasets are available. In the absence of large, manually annotated datasets, dictionary or lexicon-based approaches are commonly used in biomedical NLP tasks, such as named entity recognition (NER). These lexicon-based approaches utilize lists of biomedical terms to identify relevant expressions in texts, usually via string matching techniques. Lexicon- and rule-based methods typically work well compared to deep learning methods for NER tasks when the number of annotated expressions is small, and the texts do not contain too many lexical variants (e.g., misspellings). However, these approaches are not very scalable. In cases where concepts are expressed using a variety of expressions (e.g., generic vs. brand names for medications) or concept expressions are ambiguous, these methods are less reliable. Rule-based systems can also become very complex and challenging to maintain as the number of rules increases, rendering them difficult to adapt to new domains or situations.
The limitations of lexicon-/rule-based and deep learning-based approaches necessitate the development of alternative methods, such as FSL, which can effectively learn from small amounts of labeled data. FSL methods can potentially adapt to new situations by fine-tuning models on few examples without having to modify existing rules. FSL has numerous potential applications within the biomedical NLP space. For example, FSL techniques can be used to develop personalized medicine models that provide tailored treatment recommendations based on individuals’ medical history, genetic information, and limited available data for specific conditions. FS-based NLP methods may also be leveraged to reduce inequities in the application of artificial intelligence by enabling the optimization of models on data from minority groups (e.g. American Indian and Alaskan Native women) who are underrepresented in health systems. The problems that FSL methods attempt to solve are closely aligned with the practical challenges many medical NLP tasks face. While several FSL strategies have been explored for medical texts by distinct research communities (e.g., health informatics, computational linguistics), there is currently no review that summarizes the current state of the art. Also, no existing article has compiled the reported performances of FSL methods on distinct medical NLP data/tasks. We attempt to address these gaps in this review. Our specific contributions are highlighted below:
We present the first comprehensive review of FSL for medical text, comprising 51 articles.
We characterize each reviewed article in terms of the type of task (e.g., text classification, NER), primary aim(s), dataset(s), evaluation metrics, and other relevant aspects to provide a systematic resource for the research community.
We outline the commonly-used methods and current trends, and present suggestions for conducting evaluations.
We illustrate the current limitations of FSL by benchmarking several prominent methods on medical NER tasks.
We discuss primary challenges, current limitations, essential future research, and opportunities for progressing research in this space.
2. Background
2.1. Few-shot Learning in NLP
FSL research progress in NLP has been notably slower compared to other fields such as image processing, primarily due to more significant difficulties posed by natural language data and the lack of unified benchmarks in few-shot NLP [9]. Unlike images, text-based data may contain ambiguities and connotations that make generalization complicated. The presence of domain-specific terminologies, expressions, and associations in medical texts further exacerbates the difficulties of FSL [10]. As only small numbers of labeled examples are available in the training data, a typical FSL approach, including for NLP, is to develop innovative mechanisms of incorporating prior knowledge—knowledge that can be provided to the learner before training [11].
Using prior knowledge, FSL models can potentially generalize to new tasks effectively, and a small number of training instances may be sufficient for fine-tuning them for a given task [12]. Wang et al. [12] divide FSL methods into three categories based on how prior knowledge is incorporated: (i) data—approaches that attempt to incorporate prior knowledge by augmenting the training data; (ii) model—those that incorporate prior knowledge to constrain hypothesis space; and (iii) algorithm—those that use prior knowledge to guide how parameters are obtained. The relative effectiveness of these categories for FSL-based NLP is not yet conclusively determined, but all these mechanisms are topics of ongoing research attention. Recent advances have seen the applications of FSL for parsing text [13], machine translation [14], and classification [15, 16], among others. Several application domains have also been explored for FSL in NLP, such as legal [17] and biomedical, the latter being the focus of this review. Before diving into our review of FSL-based biomedical NLP, below we outline, with visual examples, some key developments in FSL, and their application in NLP. We encourage the reader read the cited articles for in-depth explanations of the methods.
2.2. Few-shot Learning Approaches
2.2.1. Metric Learning
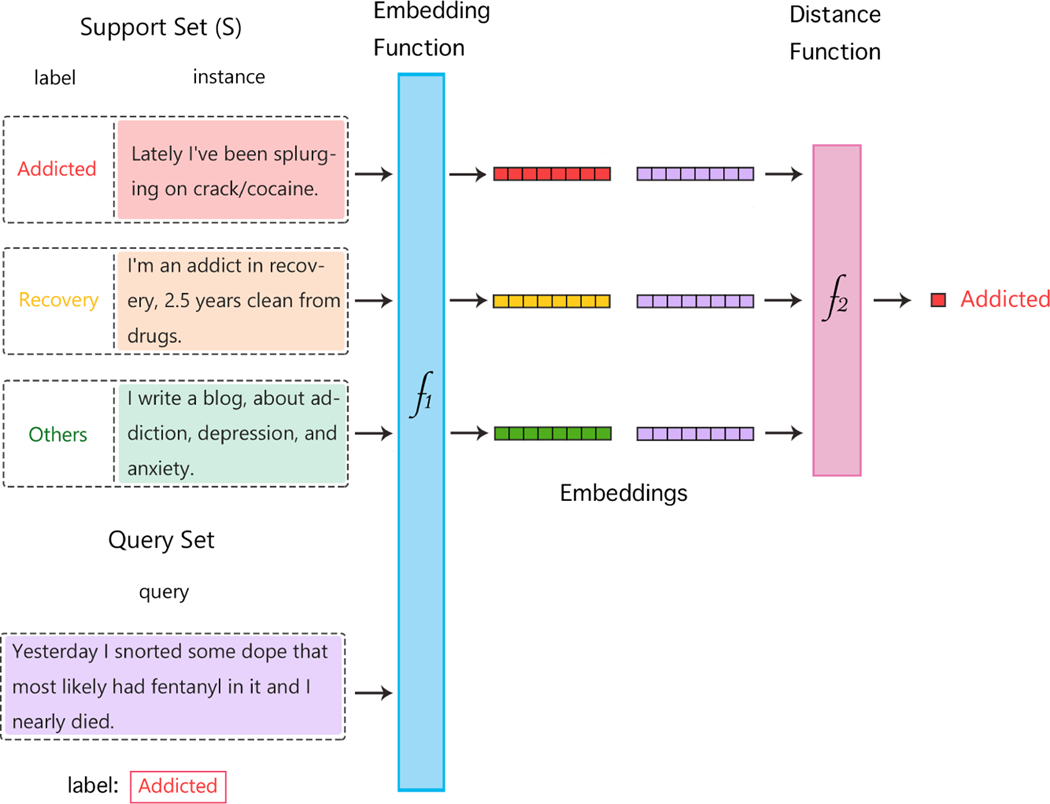
Metric learning is a class of FSL methods that employs distance-based metrics (e.g., nearest neighbor) to compute similarity or dissimilarity between data points. Given a support set (i.e., set of labelled examples for each of the classes, a.k.a. the training set), metric learning methods typically produce weighted nearest neighbor classifiers, such as via non-linear transformations in an embedding space. Features are extracted from the support set and the query set (i.e., set of samples on which the model attempts to generalize, a.k.a. the test set), to compute the distance between the instances in the embedding space. This distance function can be any distance metric such as Euclidean distance or cosine similarity. The labels of the examples in the support set that are closest to the query example based on the metric used are applied to the latter, imitating how humans use similar examples or analogies to learn. Figure 1 illustrate the architecture of metric learning method.
Figure 1:
Architecture for metric learning: the support set is used to generate embeddings using the embedding function f1. The embeddings of the query set, also generated using f1, are compared with the support set embeddings using a suitable distance function f2. Depending upon the task, the label of the most similar (or dissimilar) support set samples is assigned to the query set samples.
2.2.2. Matching Networks
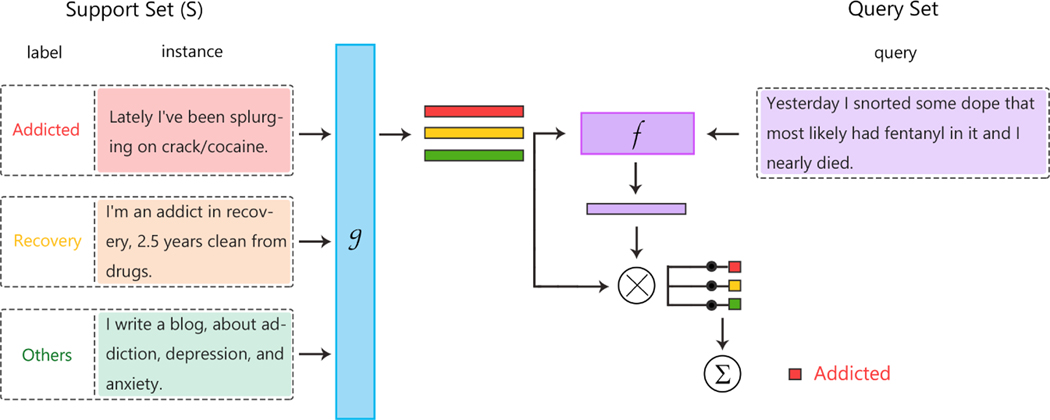
Matching networks, another class of FSL methods, attempt to use two embedding functions—one for the support set and one for the query set—to imitate how humans generalize the knowledge learned from examples. The matching network architecture uses memory-augmented neural networks [19, 20]) comprising an external memory and an attention mechanism for accessing the memory. The framework attempts to optimize the two embedding functions from the support set and the query set, and attempts to measure how well the trained model can generalize [21, 22]. Figure 2 illustrates the functionality of matching networks in a simplified manner. A variant of matching networks utilizes active learning by adding a sample selection step that augments the training data by labeling the most beneficial unlabeled sample to incorporate model-level prior knowledge. Matching networks [21] are unique in FSL in that they were the first to train and test with K-Shot-N-Way settings, which is a popular way to represent data in FSL, where “-shot” applies to the number of examples per category, and the suffix “-way” refers to the number of possible categories.
Figure 2:
Architecture for matching networks: a small support set contains some instances with their labels (one instance per label in the figure). Given a query, the goal is to calculate a value that indicates if the instance is an example of a given class. For a similarity metric, two embedding functions, f() and g(), need to take similarity based on the feature space. The function f(), which is a neural network, is applied first, and then the embedding function g() is applied to each instance to process the kernel for each support set. (Note: example uses the DASH 2020 Drug Data [18]).
2.2.3. Prototypical Networks
Prototypical networks [2], another class of FSL approaches, particularly attempt to address the issue of overfitting due to small training samples by generating prototype representations of classes from the training samples, similar to how humans summarize knowledge from examples. Prototypical networks are based on the concept that there exists an embedding in which several points cluster around a single prototype representation for each class. The aim is to learn per-class prototypes based on sample averaging in the feature space. Prediction of unknown data samples can be performed by computing distances to the class prototypes (e.g., support set means) and choosing the nearest one as the predicted label. Figure 3 illustrates the functionality of a prototypical network.
Figure 3:
Architecture for prototypical networks: a class’s prototype is the mean of its support set in the embedding space. Given a query, its distance to each class’s prototype is computed to decide its label. (Note: example uses the DASH 2020 Drug Data [18]).
2.2.4. Transfer Learning
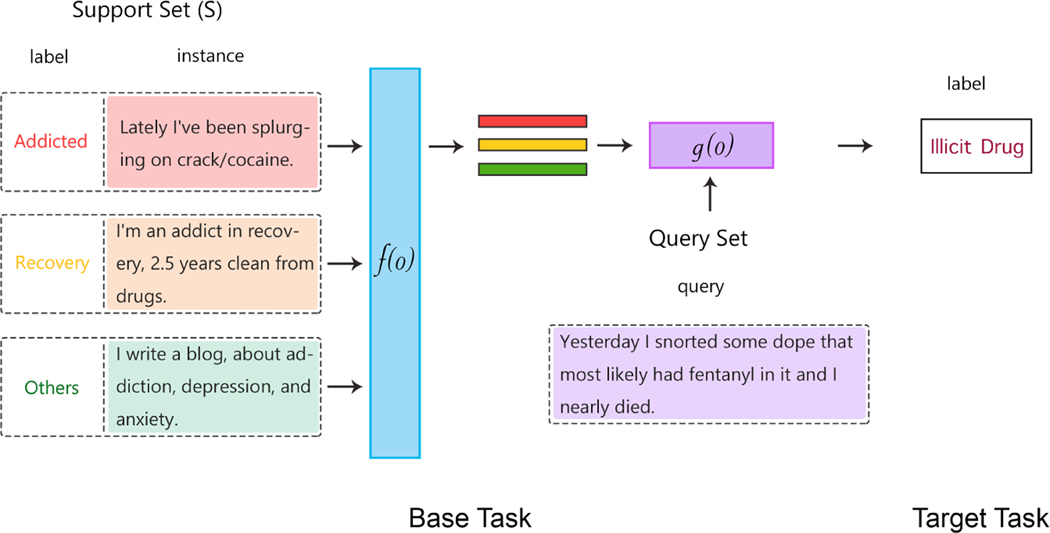
Transfer learning is a commonly used approach in FSL that incorporates prior knowledge at the data level, as knowledge learned from data in prior tasks are transferred to new few-shot tasks [23]. At first, a base network is trained on the base dataset and task. Then, it is reused to transfer the learned features to a second target network for training or fine-tuning on the target dataset and task. Transfer learning is seen to work better when the features are general (i.e., applicable to both the base task and the target task [24]). Figure 4 shows how transfer learning works.
Figure 4:
Architecture for transfer learning: in the context of few-shot learning, transfer learning involves using a base task to train the base classifier (f()). In this example, the base classifier is trained on the task of addiction/recovery detection (text classification). The learned embeddings from the base classifier are used to produce embeddings with data-level prior knowledge. The embeddings are used to train the target classifier (g()) on a different, but related text classification task: illicit drug detection.
2.2.5. Meta-Learning
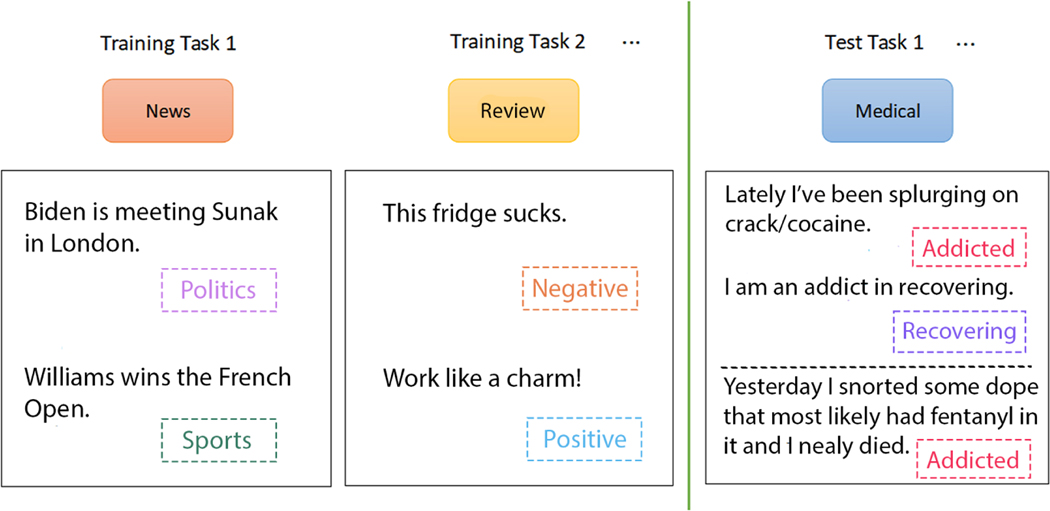
A more challenging subset of promising FSL approaches involves meta-learning (a.k.a., “learning to learn” [25]). It is a branch of metacognition, which is concerned with learning about one’s own learning and learning processes [26]. In contrast to classical learning frameworks, in the meta-learning framework, a model is trained using a set of training tasks, not data, and model performance is evaluated on a set of test tasks. Thus, the learner attempts to obtain prior knowledge by incorporating generic knowledge across different tasks (i.e., algorithm-level prior knowledge). A small number of labeled instances for the target task is then used to fine-tune the model. Figure 5 illustrates the meta-learning framework using a simple example—an entity recognition model is trained on tasks involving news and music data, and is evaluated on a medical task.
Figure 5:
Architecture for meta-learning: each task mimics the few-shot scenario and can be completely non-overlapping. Support sets are used to train; query sets are used to evaluate the model. In this example, several text classification tasks on different datasets (and label sets) are used to train the meta-learner. Finally, the test task (medical domain) is used for generalizing the meta-learner to the test task.
3. Methods
3.1. Experimental Design
We followed the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) protocol to conduct this review [27]. FSL for NLP is a relatively recent research topic, so we concentrated on a short time range for our literature search—January 2016 to October 2022. While there have been past research focusing on learning from small numbers of examples, in a preliminary search, we did not find any notable article specific to this topic before 2018. Consequently, we chose 2016 as the beginning of our date range—a two-year window to find any notable article missed during our initial search. We searched the following bibliographic databases to identify relevant articles: (1) PubMed/Medline, (2) Embase, (3) IEEE Xplore Digital Library, (4) ACL Anthology, and (5) Google Scholar, the latter being a meta-search engine, not a database. We included ACL Anthology (the primary source for the latest NLP research) and IEEE Xplore, in addition to EMBASE and PubMed/Medline, because much of the methodological progress in FSL has been published in non-medical journals and conference proceedings. At the time of searching (October 2022), ACL Anthology hosted over 82,000, and IEEE Xplore hosted over 5.8 million articles, although most articles in the latter did not focus on NLP or medicine. Over recent years, preprint servers (e.g., arXiv, bioRxiv, and medRxiv) have emerged as major sources of the latest information regarding research progress in computer science and NLP. For example, description of the widely popular pretrained model RoBERTa is available via arXiv [28]. We used Google Scholar primarily to search these preprint servers or published articles from other sources. Note that we also searched the ACM Digital Library1, but discovered no additional articles. Hence, we do not report it as a data source for our review.
3.2. Search Strategy
We applied marginally different search strategies depending on the database to account for the differences in their contents. We used three types of queries:
Queries focusing on the technical field of research (phrases included: ‘natural language processing’, ‘text mining’, ‘text classification’, ‘named entity recognition’, and ‘concept extraction’);
Queries focusing on the learning strategy (phrases included: ‘few-shot’, ‘low-shot’, ‘one-shot’, and ‘zero-shot’); and
Queries focusing on the domain of interest (phrases included: ‘medical’, ‘clinical’, ‘biomedical’, ‘health’, and ‘health-related’).
All articles on PubMed and Embase fall within the broader biomedical domain, so we used combinations of the phrases in 1 and 2 above for searching these two databases, leaving out the phrases in 3. All articles in the ACL Anthology involve NLP, so we used phrases from 2 and 3 for this source. For IEEE Xplore and Google Scholar, the articles can be from any domain and on any topic, so we used combinations of all three sets of phrases for searching. PubMed, Embase, and IEEE only returned articles that entirely matched the queries; ACL Anthology and Google Scholar retrieved larger sets of articles and ranked them by relevance. For ACL Anthology, the articles retrieved were reviewed sequentially in decreasing order of relevance. For each query combination, we continued reviewing candidate articles until we came across at least two pages (about 20 articles) of no relevant articles, at which point we decided that no relevant articles would be found on the following pages. We used Google Scholar as an auxiliary search engine to identify potentially relevant articles indexed in such preprint servers and similar public sources (e.g., Open Review2).
3.3. Study Selection and Exclusion Criteria
All articles shortlisted from initial searches were screened for eligibility by three authors (YGe, YGuo, and AS). While it was always possible to identify the technical field/topic (NLP or not) from the titles and abstracts, to determine the domain, we had to review full articles because some articles included multiple datasets, and only a subset of these datasets were from the medical domain. We excluded articles if none of the datasets were related to medicine/health, or they did not explicitly focus on NLP in few/low-shot settings, and reviewed the remaining articles. Because of the relatively small number of articles that were eventually included in the review, we did not attempt to compare inter-reviewer agreements regarding article relevance. Instead, the authors discussed each included/excluded article to reach consensus.
3.4. Data Abstraction and Synthesis
We abstracted the following details from each article, if available: publication year, data source, primary research aim(s), training set size(s), number of entities/classes, entity type for training, entity type for evaluation/testing, primary method(s), and evaluation metric. For articles including data from multiple sources, we only abstracted those related to health/medicine. In terms of primary aim(s), some articles reported multiple objectives, and we abstracted all the NLP-oriented ones (e.g., text classification, concept extraction). For training set sizes, we abstracted information about the number of instances used for training and, if applicable, how larger datasets were reconstructed to create few-shot samples. We also extracted the number of labels for each study/task; for NER/concept extraction methods, we identified the number of entities/concepts, and for classification, we identified the type of classification (multi-label or multi-class) along with the number of classes. We also noted the training domain(s) and test/evaluation domain(s) for each few-shot method, when applicable. Abstracting the primary approach(es) and evaluation methodology was more challenging due to the complexities of some of the model implementations, and we reviewed and summarized the descriptions provided in each article, including the strategies and performances reported for evaluations.
4. Results
4.1. Data Collection Results
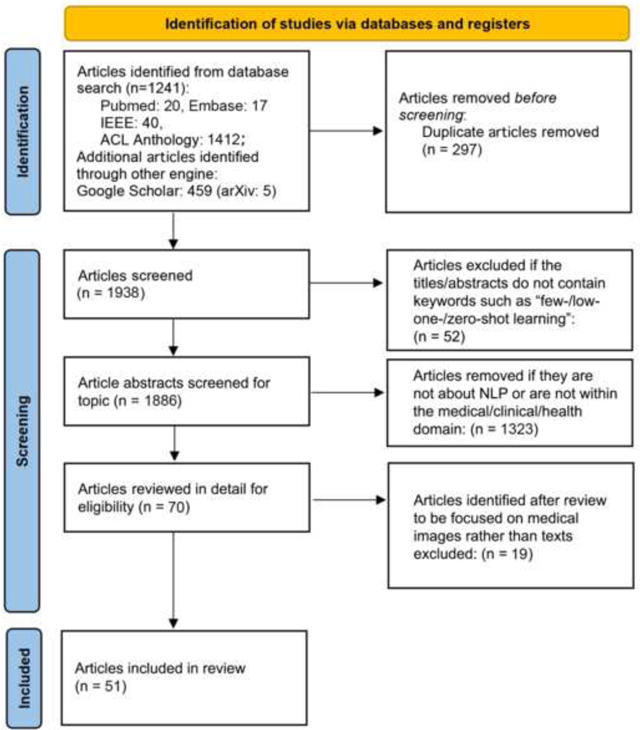
Fifty-one articles met our inclusion criteria. Initial searches retrieved 1241 articles from PubMed, Embase, IEEE Xplore, and ACL Anthology, and an additional 459 from Google Scholar. Figure 6 presents the screening procedures and numbers at each stage. After initial filtering, we reviewed 70 full-text articles for eligibility, excluding 19 from the final review. The first included article was from 2018, and most articles (43/51; 84%) were from 2020 to 2022, although only articles published prior to October 31 were included for 2022.
Figure 6:
PRISMA flow diagram depicting the number of articles at each stage of collection and the filtering process.
4.2. Study Characteristics
Table 1 summarizes some fundamental characteristics of each article (authors, year of publication, data source, research aims, training set sizes, number of entities/classes, and training and evaluation domains). In terms of training data sizes, 14/51 (27%) articles included zero-shot scenarios (i.e., prediction without any supervision) into their research scope, including two on zero-shot learning only. 1-shot, 5-shot, and 10-shot were the most common ‘shot’ settings, representing 17/51 (33%) of the reviewed articles. 9/51 (18%) reviewed articles used samples of larger datasets for training, often specified in percentages (e.g., 25%, 50%). 6/51 (12%) articles did not explicitly specify shot values. Two articles did not perform experiments in accordance with traditional few-shot scenarios, and divided all labels into three categories according to the frequency of occurrences (frequent group contained all labels occurring more than 5 times; few-shot group contained labels occurring 1–5 times; zero-shot group included labels that never occurred in the training data), causing some labels to have large numbers of annotated samples. 10/51 (20%) articles involved cross-domain transfer, with different domains of training and test/evaluation data.
Table 1:
Articles published for few-shot learning on medical data, their publication years, data sources, search engine, which downstream tasks the literature focus, size of the training set (number of the shot), number of the entities (for few-shot NER tasks), number of the classes (for few-shot classification tasks), type of data in training and test domain.
| Study | Data source | Research aim | Size of training set | Number of entities / classes | Dataset domain |
|---|---|---|---|---|---|
| Rios et al., 2018 [16] | MIMIC II [29] MIMIC III [30] | Multi-label Text Classification | Frequent group (all labels that occur >5 times), the few-shot group (labels that occur between 1 and 5 times), and the zero-shot group (labels that never occur in the training dataset), reconstructed | Not mentioned 1 | Medical, discharge summaries annotated with a set of ICD-9 diagnosis and procedure labels |
| Rios et al., 2018 [31] | MIMIC II [29] MIMIC III [30] | Multi-label Text Classification | Original dataset, with no reconstruction | Not mentioned 1 | Medical |
| Hofer et al., 2018 [10] | i2b2 2009 32 i2b2 2010 33 i2b2 2012 34 CoNLL-2003 [35] BioNLP-2016 [36] MIMIC-III [30] UK CRIS [37, 38] | NER | 10-shot | Not mentioned 1 | Medical and nonmedical (e.g, news) |
| Pham et al., 2018 [39] | The Europarl datasets [40] IWSLT17[41] The UFAL Medical Corpus HIML2017 dataset 3 | Neural Machine Translation (NMT) | One-Shot | N/A 2 | German→English: medical; English→Spanish, the proceedings of the European Parliament and data from TED |
| Yan et al., 2018 [42] | Multigames dataset [43] HCR dataset [44] SS-Tweet dataset [45] SemEval-2013 Dataset (SemEval b) [46] | Text Classification | Few-shot, but reconstructed | Multi games: 3 HCR: 5 SS-Tweet: 3 SemEval-2013 Dataset: 3 | Tweets about sentiment and games, and health Care Reform (HCR) data |
| Manousogiannis et al., 2019 [47] | Tweets (provided by SMM4H 2019) [48] | NER | Original dataset, with no reconstruction | 1, ADR with 319 MEDDRA codes | Medical (ADR) |
| Gao et al., 2019 [49] | FewRel dataset [50] | Relation Classification | 5-Way 1-Shot / 5-Way 5-Shot / 10-Way 1-Shot / 10-Way 5-Shot | 25 | Wikipedia corpus 10 and Wikidata knowledge bases |
| Lara-Clares et al., 2019 [51] | MEDDOCAN shared task dataset [52] | NER | 500 clinical cases, with no reconstruction | 29 | Clinical |
| Ferré et al., 2019 [53] | BB-norm dataset [54] | Entity Nor mal izat io n | Original dataset with no reconstruction and zero-shot | Not mentioned 1 | Biological |
| Hou et al., 2020 [55] | Snips dataset [56] | Slot Tagging (NER) | 1-shot and 5-shot | 7 | Six of Weather, Music, PlayList, Book (including biomedical), Search Screen (including biomedical), Restaurant and Creative Work. 11 |
| Sharaf et al., 2020 [57] | Ten different datasets collected from the Open Parallel Corpus (OPUS) [58] | Neural Machine Translation (NMT) | Sizes ranging from 4k to 64k training words (200 to 3200 sentences), but reconstructed | N/A 2 | Bible, European Central Bank, KDE, Quran, WMT news test sets, Books, European Medicines Agency (EMEA), Global Voices, Medical (ufal-Med), TED talks |
| Lu et al., 2020 [59] | MIMIC II [29] MIMIC III [30] EU legislation dataset [60] | Multi-label Text Classification | 5-shot for MIMIC II and III, 50-shot for EU legislation | MIMIC II: 9 MIMIC III: 15 EU legislation: 5 | Medical |
| Jia et al., 2020 [61] | BioNLP13PC BioNLP-13CG [62] CoNLL-2003 dataset [35] Broad Twitter dataset [63] Twitter dataset [64] CBS SciTech News dataset [65] | NER | Four few-shot (reconstructed) and zero-shot | CoNLL: 4 Broad Twitter: 3 Twitter: 4 BioNLP13PC: >=3 BioNLP13CG: >=3 CBS News: 4 | For the BioNLP dataset, BioNLP13PC as the source domain dataset; In the Broad Twitter dataset, the CoNLL-2003 as the source domain dataset; In the Twitter dataset,the CoNLL- 2003 as the source domain dataset |
| Chalkidis et al., 2020 [66] | EURLEX5TK [60] MIMIC III [30] AMAZON13K [67] | Multi-label Text Classification | The labels are divided into frequent (≥ 50), few-shot (≤ 50), and zero-shot | Not mentioned 1 | English legislative documents, English discharge summaries from US hospitals, English product descriptions from Amazon |
| Lwowski et al., 2020 [68] | Tweets about COVID-19 [69] | Text Classification | 100 tweets, with no reconstructed | 4 | Tweets about COVID-19 |
| Hou et al., 2020 [9] | Dialogue utterances from the AIUI open dialogue platform of iFlytek4 | Dialogue Language Understanding: includes two sub-tasks: Intent Detection (classification) and Slot Tagging (sequence labeling) | 1-shot, 3-shot, 5-shot and 10-shot | Train Domains: 45 Dev Domains: 5 Test Domains: 9 | General dialogue (including health domain) |
| Chen et al., 2020 [70] | WIKIBIO dataset [71] | Natural Language Generation (NLG) | Dataset sizes: 50, 100, 200 and 500, with no reconstruction | N/A 2 | Books, Songs and Human domain (including biomedical) |
| Vaci et al., 2020 [72] | UK-CRIS system that provides a means of searching and analysing deidentified clinical case records from 12 National Health Service Mental Health Trusts [37, 38] | NER | Original dataset, with no reconstruction | 7 | Clinical |
| Huang et al., 2020 [73] | 10 public datasets | NER | 5-shot, 10%, 100% | CoNLL: 4 Onto: 18 WikiGold: 4 WNUT: 6 Movie: 12 Restaurant: 8 SNIPS: 53 AT IS: 79 Multiwoz: 14 I2B2: 23 | 10 public datasets, different domains |
| Chen et al., 2020 [74] | MRI image dataset MRI text reports5 | Text Classifie ait ion | Original dataset, with no reconstruction | Not mentioned 1 | MRI data |
| Yin et al., 2020 [75] | MLEE [76] BioNLP’13-GE [62] | Sequence Tagging (NER) | 5-way-10-shot, 5-way-15-shot, and 5-way-20-shot | 5 | Biological event |
| Goodwin et al., 2020 [77] | Tensor Flow DataSets catalogue6 | Abstractive Summarization | Zero-shot and 10-shot | N/A 2 | 3 general domain & 1 consumer health |
| Yang et al., 2020 [78] | OntoNotes 5.0 [79] CoNLL-2003 [35] I2B2 2014 [80] WNUT 2017 [81]11 | NER | 1-shot and 5-shot | Onto: 18 CoNLL: 4 I2B2–14: 23 WNUT: 6 | Three of general, news, medical and social media |
| Hartmann et al., 2021 [82] | The IULA dataset [83] The NUBES dataset [84] The FRENCH dataset [85] Negation Scope Resolution datasets | NER | Zero-shot, with no reconstruction | 1, Negation | No training data for the clinical datasets |
| Fi vez et al., 2021 [86] | SNOMED-CT17 ICD-10 | Name Normalization | Zero-shot, with no reconstructed | N/A 2 | Biomedical |
| Lu et al., 2021 [87] | Constructed a dataset 8 based on Wei bo for the research of few-shot rumor detection, and use PHEME dataset [88] | Rumor Detection (NER) | For the Weibo dataset: 2-way 3-event 5-shot 9-query; for PHEME dataset: 2-way 2-event 5-shot 9-query | Weibo: 14 PHEME: 5 | Source posts and comments from Sina Weibo related to COVID-19 |
| Ma et al., 2021 [89] | CCLE CERES-corrected CRISPR gene disruption scores G DSC1000 dataset PDTC dataset PDX dataset9 | Drug-response Predictions | 1-shot, 2-shot, 5-shot, and 10-shot | N/A 2 | Biomedical |
| Kormilitzin et al., 2021 [90] | MIMIC-III [30] UK-CRIS datasets [37, 38] | NER | 25%, 50%, 75% and 100% of the training set, with no reconstruction | 7 | Electronic health record |
| Guo et al., 2021 [91] | BioNLP Shared Task 2011 and 2019 [54] structured biological datasets | NER | 100%, 75%, 50%, 25%, 0% of training set, with no reconstructed | Not mentioned 1 | Biomedical entities |
| Lee et al., 2021 [92] | COVID19-Scientific [93] COVID19-Social [94] FEVER [95] | Fact- Checki ng (close to Text Classification) | 2-shot, 10-shot, and 50-shot | Not mentioned 1 | Facts about COVID-19 |
| Fi vez et al., 2021 [96] | ICD-10 SNOMED-CT7 | Name Nor mal izat io n | 15-shot | N/A 2 | Biomedical |
| Xiao et al., 2021 [97] | FewRel dataset | Relation Classification | 5-Way-1-Shot 5-Way-5-Shot 10-Way-l-Shot 10-Way-5-Shot | Not mentioned 1 | Wikipedia and Wikidata |
| Ziletti et al., 2021 [98] | MedDRA ontology | Medical Coding / classification | Zero-shot Few-shot | 26,000 distinct classes | Synonyms and biomedical text |
| Ye et al., 2021 [99] | Huggingface Datasets | Cross-task Generalization | Few-shot More-shot | N/A 2 | 160 datasets |
| Aly et al., 2021 [100] | OntoNotes-ZS MedMentions-ZS | NER and classification | zero-shot | Train: 19 classes Dev: 12 classes Test: 12 classes | General, Biomedical |
| Wright et al., 2021 [101] | Curated a dataset of paired sentences from abstracts and associated press releases, labeled by experts for exaggeration based on their claim strength, and ScienceDaily | Information Extraction | 100-shot | N/A 2 | A science reporting website which aggregates and re-releases press releases from a variety of sources |
| Lee et al., 2021 [102] | C0NLLO3 Ontonotes 5.0 BC5CDR | NER | 2 5-shot and 50-shot | Not mentioned 1 | General 12 |
| Wang et al., 2022 [103] | i2b2 2010 dataset i2b2 2012 dataset MIMIC-III dataset BioScope NegEx Chia | Classification | Whole datasets but few-shot classes | Not mentioned 1 | Annotates a corpus 13 of assertions |
| Yan et al., 2022[104] | 677 full-text articles were obtained as neuroimaging corpora | NER | Whole datasets | 10 categories of neuroimaging entities and 55 categories of neuroimaging interactions | Neuroimaging entities and their interactions |
| Lin et al., 2022 [105] | Neuroimaging event mention set | Information Extraction | Whole datasets but few-shot classes | 788 “Activate” event mentions, 128 “Deactivate” event mentions, 1169 “Effect” event mentions, 665 “Perform Experiment” event mentions, 266 “Acquisition” event mentions, and 315 “Perform Analysis” event mentions. | Ne uro imaging event |
| Riveland et al., 2022 [106] | Psychophysical tasks | Classfication | Zero-shot | 4 categories | Psychophysical tasks |
| Navarro et al., 2022 [107] | 27 recorded conversations between general practitioners and patients at Primary Care facilities | Abstractive summarization | Zero-shot 10-shot 20-shot 50-shot | N/A 2 | Medical dialogues from various online chats |
| Das et al., 2022 [108] | OntoNotes C0NLLO3 WNUTir GUM Few-NERD | NER | 1–2 shot-5-way 5–10 shot-5-way 1–2 shot-10-way 5–10 shot-10-way | Not Mentioned 1 | General (OntoNotes 5.0), Medical (I2B2), News (C0NLLO3), Social (WNUT17) 14 |
| Ma et al., 2022 [109] | C0NLLO3 WNUT1T JNLPBA NCBI-disease 12B2–14 datasets | NER | 1-shot, 5-shot, 20-shot, 50-shot | Not mentioned 1 | General, Social, Biomedical |
| Par mar et al., 2022 [110] | 32 datasets | Multi-Task Learning | 32-shot, 100-shot, 1000-shot, 2000-shot | Not mentioned 1 | Biomedical and health data |
| Boulanger et al., 2022 [111] | I2B2 CoNLL | NER | 50-shot, 100-shot, 250-shot, 500-shot, 1000-shot | Not mentioned 1 | General and Biomedical data |
| Yeh et al., 2022 [112] | ChemProt dataset | Relation Extraction | Zero-shot | Not mentioned 1 | Scientific paper abstracts annotated with 6 relation types between the chemicals and genes in sentences |
| Pan et al., 2022 [113] | MoviesQA NewsQA BioQA CovidQA | Question Answering | Zero-shot | Not mentioned 1 | Movies, News, Biomedical, and COVID-19 domains |
| Wadden et al., 2022 [114] | Scientific claim verification datasets | Scientific claim verification | Zero-shot few-shot | N/A 2 | SCIFACT, Health Ver, COVIDFact, FEVER, EVIDENCE- INFERENCE, PUBMEDQA |
| Li et al., 2022[115] | FewRel 1.0 FewRel 2.0 | Relation classification | 5-way-1-shot, 5-way-5-shot, 10-way-1-shot, 10-way-5-shot | 100 relations split into training, validation and test sets with respectively 64, 16 and 20 relations without overlapping | Relations from PubMed articles |
| Zhang et al., 2022 [116] | 7 STS tasks STS 2012–2016 STS Benchmark S ICK-Relatedness | Natural Language Inference (NLI) | 16 labeled instances per class | Not mentioned 1 | News, Biomedical, Search snippets and Social media data |
The research aim of this paper is text classification or NER, but the size of training set is Not mentioned in the paper.
The research aim of this paper is neither text classification nor NER.
UFAL Medical Corpus v.1.0 and HIML2017 dataset: http://aiui.xfyun.cn/index-aiui. Last accessed November 22, 2021.
iFlytek: http://aiui.xfyun.cn/index-aiui. Last accessed November 22, 2021.
Those datasets are not released.
TensorFlow DataSets: https://www.tensorflow.org/datasets. Last accessed November 22, 2021.
SNOMED-CT1: https://www.snomed.org. Last accessed November 22, 2021.
A novel dataset proposed by this paper: https://github.com/jncsnlp/Sina-Weibo-Rumors-for-few-shot-learning-research. Last accessed November 22, 2021.
Links are provided in the original paper.
Test data is biomedical literature with UMLS, a large-scale biomedical knowledge base.
The remaining one class is used at test time.
PubMed articles and chemical-disease texts are used as additional test data.
Patient eligibility data and 3 assertion types are used for test data: Present, Absent & Possible
GUM, Few-NERD used as test data.
Table 2 summarizes the proposed methods and their evaluations. Variants of neural network-based (deep learning) algorithms, such as Siamese Convolutional Neural Networks) [42], were the most common. Only 4/51 (8%) articles proposed new datasets, and 3/51 (6%) presented benchmarks for comparing multiple few-shot methods. Evaluation strategies had considerably less diversity. Almost all evaluation methodologies for classification and NER tasks involved standard metrics such as accuracy, precision, recall, and F1-scores.
Table 2:
A summary table showing primary few-shot approaches and evaluation methodologies.
| Study | Task | Primary approach(es) | Evaluation metric(s) |
|---|---|---|---|
| Rios et al. [16] | Multi-label Text Classification | Neural architecture suitable for handling few- and zero-shot labels in the multi-label setting where the output label space satisfies two constraints: (1) the labels are connected forming a DAG; (2) each label has a brief natural language descriptor. | R@k (Recall@k), P@k (Precision@k), Macro-F1 scores |
| Rios et al. [31] | Multi-label Text Classification | Semi-parametric neural matching network for diagnosis/procedure code prediction from EMR narratives. | Precision, Recall, F1-scores, AUC (PR), AUC (ROC), P@k, R@k |
| Hofer et al. [10] | NER | Five improvements on NER tasks when only 10 annotated examples are available: 1. Layer-wise initialization with pre-trained weights (single pre-training); 2. Hyperparameter tuning; Combining pre-training data; Custom word embeddings; Optimizing out-of-vocabulary (OOV) words. | F1-score |
| Pham et al. [39] | Neural Machine Translation (NMT) | A generic approach to address the challenge of rare word translation in NMT by using external phrase-based models to annotate the training data as experts. A pointer network is used to control the model-expert interaction. The trained model is able to copy the annotations into the output consistently. | BLEU score, SUGGESTION (SUG), SUGGESTION ACCURACY (SAC) |
| Yan et al. [42] | Text Classification | Short text classification framework based on Siamese CNNs and few-shot learning, to learn the discriminative text encoding for helping classifiers distinguish obscure or informal sentences. The different sentence structures and different descriptions of a topic are learned by few-shot learning strategy to improve the classifier’s generalization. | Accuracy |
| Manousogiannis et al. [47] | Concept Extraction | A simple few-shot learning approach, based on pre-trained word embeddings and data from the UMLS, combined with the provided training data. | Relaxed and strict Precision/Recall/F1-scores |
| Gao et al. [49] | Relation Classification | Propose FewRel 2.0, a new task containing two real-world issues that FewRel ignores: few-shot domain adaptation, and few-shot none-of-the-above detection. | Accuracy |
| Lara-Clares et al. [51] | NER | Hybrid Bi-LSTM and CNN model to recognize multi-word entities. Learns high level features from datasets using a few-shot learning model. Wikipedia2vec is used for automatic extraction and classification of keywords. | F1-score |
| Ferré et al. [53] | Entity Normalization | A new neural approach (C-Norm) which synergistically combines standard and weak supervision, ontological knowledge integration and distributional semantics. | The offcial evaluation tool of the BB-norm task: a similarity score and a strict exact match score. |
| Hou et al. [55] | Slot Tagging (NER) | Introduction of a collapsed dependency transfer mechanism into CRF to transfer abstract label dependency patterns in the form of transition scores. The emission score of CRF is computed as the word similarity with respect to each label representation. A Label-enhanced Task-Adaptive Projection Network (L-TapNet) based on TapNet is used to compute the similarity by leveraging label name semantics in representing labels. | F1-score |
| Sharaf et al. [57] | Neural Machine Translation (NMT) | Framing the adaptation of NMT systems as a meta-learning problem. The model can learn to adapt to new unseen domains based on simulated offline meta-training domain adaptation tasks. | BLEU, SacreBLEU (to measure case-sensitive de-tokenized BLEU) |
| Lu et al. [59] | Multi-label Text Classification | A simple multi-graph aggregation model that fuses knowledge from multiple label graphs encoding different semantic label relationships to incorporate aggregated knowledge in multi-label zero/few-shot document classification. Three kinds of semantic information are used: pre-trained word embeddings; label description; pre-defined label relations. | Recall@K, nDCG@K |
| Jia et al. [61] | NER | Creation of distinct feature distributions for each entity type across domains, which improves transfer learning power, as compared to representation networks that do not explicitly differentiate between entity types. | F1-score |
| Chalkidis et al. [66] | Multi-label Text Classification | Hierarchical methods based on Probabilistic Label Trees (PLTs); Combines BERT with LWAN; Use of structural information from thelabel hierarchy in LWAN. Leverages label hierarchy to improve few and zero-shot learning. | R-Precision@K (a top-K version of R-Precision of each document), nDCG@K |
| Lwowski et al. [68] | Text Classification | A self-supervised learning algorithm to monitor COVID-19 Twitter using an autoencoder to learn the latent representations. Knowledge transfer to COVID-19 infection classifier by fine-tuning the Multi-Layer Perceptron (MLP) using fewshot learning. | Accuracy, Precision, Recall, F1-score |
| Hou et al. [9] | Dialogue Language Understanding with two sub-tasks: Intent Detection (classification) and Slot Tagging (sequence labeling) | A novel few-shot learning benchmark for NLP (FewJoint). Introduces few-shot joint dialogue language understanding, which additionally covers the problems of structure prediction and multi-task reliance. | Intent Accuracy, Slot F1-score, Sentence Accuracy |
| Chen et al. [70] | Natural Language Generation (NLG) | The design of the model architecture is based on two aspects: content selection from input data and language modeling to compose coherent sentences, which can be acquired from prior knowledge. | BLEU-4, ROUGE-4 (F-measure) |
| Vaci et al. [72] | Concept Extraction | Used a combination of methods to extract salient information from electronic health records. First, clinical experts define the information of interest and subsequently build the training and testing corpora for statistical models. Second, built and finetuned the statistical models using active learning procedures. | Precision, Recall, F1-score |
| Huang et al. [73] | NER | The first systematic study for few-shot NER. Three distinctive schemes (and their combinations) are investigated: (1) meta-learning to construct entity prototypes; (2) supervised pre-training to obtain generic entity representations; (3) self-supervised training to utilize unlabeled in-domain data. | F1-score |
| Chen et al. [74] | Classification | A classification and diagnosis method for Alzheimer’s patients based on multi-modal feature fusion and small sample learning. The compressed interactive network is then used to explicitly fuse the extracted features at the vector level. Finally, the KNN attention pooling layer and the convolutional network are used to construct a small sample learning network to classify the patient diagnosis data. | Accuracy, F1-score |
| Yin et al. [75] | Sequence Tagging (Event trigger identification) | Combination of a prototypical network and a relation network module to model the task of biomedical event trigger identification. In addition, to make full use of the external knowledge base to learn the complex biological context, a self-attention mechanism is introduced. | F1-score |
| Goodwin et al. [77] | Abstractive Summarization | Highly-abstractive multi-document summarization conditioned on user-defined query using BART, T5, and PEGASUS. | ROUGE-1, ROUGE-2, ROUGE-L F1-scores, BLEU-4, Repetition Rate |
| Yang et al. [78] | NER | Uses an NER model trained under supervision on source domain for feature extraction. Structured decoding is used with nearest neighbor learning instead of expensive CRF training. | F1-score |
| Hartmann et al. [82] | Concept Extraction | A universal approach to multilingual negation scope resolution: zero-shot cross-lingual transfer for negation scope resolution in clinical text. Exploits data from disparate sources by data concatenation, or in an MTL setup. | Percentage of correct spans (PCS), F1-score over scope tokens |
| Fivez et al. [86] | Name Normalization | Propose truly robust representations, which capture more domain-specific semantics while remaining universally applicable across different biomedical corpora and domains. Use conceptual grounding constraints which more effectively align encoded names to pretrained embeddings of their concept identifiers. | For synonym retrieval: Mean average precision (mAP) over all synonyms. For concept mapping: Accuracy (Acc) and Mean reciprocal rank (MRR) of the highest ranked correct synonym. |
| Lu et al. [87] | Rumor Detection1 | A few-shot learning-based multi-modality fusion model named for COVID-19 rumor detection. Includes text embedding modules with pre-trained BERT model, a feature extraction module with multilayer Bi-GRUs, and a multi-modality feature fusion module with a fusion layer. Uses a metalearning based few-shot learning paradigm. | Accuracy |
| Ma et al. [89] | Drug-response Predictions | Applied the few-shot learning paradigm to three context-transfer challenges: transfer of a predictive model learned in one tissue type to the distinct contexts of other tissues; transfer of a predictive model learned in tumor cell lines to patient-derived tumor cell (PDTC) cultures in vitro; transfer of a predictive model learned in tumor cell lines to the context of patient-derived tumor xenografts (PDXs) in mice in vivo. | Accuracy, Pearson’s correlation, AUC |
| Kormilitzin et al. [90] | NER | Self-supervised training of deep neural network language model using the cloze-style approach. Synthetic training data with noisy labels is created using weak supervision. All constituent components are combined into an active learning approach. | Accuracy, Precision, Recall, F1-score |
| Guo et al. [91] | Extract Entity Relations | A Siamese graph neural network (BioGraphSAGE) with structured databases as domain knowledge to extract biological entity relations from literature. | Precision (P-value), Recall (R-value), F1-score |
| Lee et al. [92] | Fact-Checking (Text Classification) | Propose evidence-conditioned perplexity, a novel way of leveraging the perplexity score from LMs for the few-shot fact-checking task. | Accuracy, Macro-F1-score |
| Fivez et al. [96] | Name Normalization | A scalable few-shot learning approach for robust biomedical name representations. Training a simple encoder architecture in a few-shot setting using small subsamples of general higher-level concepts which span a large range of fine-grained concepts. | Spearman’s rank correlation coefficient |
| Xiao et al. [97] | Relation Classification | Adaptive prototypical networks with label words and joint representation learning based on metric learning for FSRC, which performs classification by calculating the distances in the learned metric space. | Accuracy |
| Ziletti et al. [98] | Medical Coding (classification) | Combines traditional BERT-based classification with task-aware representation of sentences, a zero/few-shot learning approach that leverages label semantics. | Accuracy |
| Ye et al. [99] | Cross-task Generalization | Present CROSSFIT, a few-shot learning challenge to acquire, evaluate and analyze cross-task generalization in a realistic setting. Additionally, introduce the NLP Few-shot Gym, a repository of 160 few-shot NLP tasks gathered from open-access resources. | Average Relative Gain (ARG) |
| Aly et al. [100] | NER and classification (NERC) | Present the first approach for zero-shot NERC by using transformers with cross-attention to leverage naturally occurring entity type descriptions. The negative class is modeled by: (1) description-based encoding, and (2) independent (direct) encoding (3) class-aware encoding. | F1-score |
| Wright et al. [101] | Exaggeration Detection1 (Information Extraction) | Propose multi-task Pattern Exploiting Training (MT-PET) to leverage knowledge from auxiliary cloze-style QA tasks for few-shot learning. Present a set of labeled press release/abstract pairs from existing expert-annotated studies on exaggeration in the press releases of scientific papers suitable for benchmarking the performance of machine learning models. | Precision, Recall, F1-score |
| Lee et al. [102] | NER | Present a simple demonstration-based learning method for NER, which lets the input be prefaced by task demonstrations for in-context learning, and perform a systematic study on demonstration strategy regarding what to include, how to select the examples, and what templates to use. | F1-score |
| Wang et al. [103] | Classification | Propose a prompt-based learning approach, which treats the assertion classification task as a masked language auto-completion problem. | Comprehensiveness, Sufficiency (for measuring to what extent the model adheres to human rationales.) |
| Yan et al. [104] | NER | Proposes a text mining pipeline for enabling the FAIR neuroimaging study. In order to avoid fragmented information, the Brain Informatics provenance model is redesigned based on NIDM (Neuroimaging Data Model) and FAIR facets. | Precision, Recall, F1-score |
| Lin et al. [105] | Information Extraction | Proposes a literature mining-based approach for research sharing-oriented neuroimaging provenance construction. A joint extraction model based on deep adversarial learning, called AT-NeuroEAE, is proposed to realize the event extraction in a few-shot learning scenario. | Precision, Recall, F1-score |
| Riveland et al. [106] | Classfication | Present neural models of one of humans’ most astonishing cognitive feats: the ability to interpret linguistic instructions in order to perform novel tasks with just a few practice trials. Models are trained on a set of commonly studied psychophysical tasks, and receive linguistic instructions embedded by transformer architectures pretrained on natural language processing. | Accuracy |
| Navarro et al. [107] | Abstractive summarization2 | Fine-tuned several state-of-the-art (SOTA) models in a newly created medical dialogue dataset of 143 snippets, based on 27 general practice conversations paired with their respective summaries. | ROUGE scores |
| Das et al. [108] | NER | Present CONTAINER, a novel contrastive learning technique that optimizes the intertoken distribution distance for Few-Shot NER. Instead of optimizing class-specific attributes, CONTAINER optimizes a generalized objective of differentiating between token categories based on their Gaussian-distributed embeddings. | F1-score |
| Ma et al. [109] | NER | Leveraging the semantic information in the names of the labels as a way of giving the model additional signal and enriched priors. Propose a neural architecture consisting of two BERT encoders, one for document encoding and another for label encoding. | F1-score |
| Parmar et al. [110] | Multi-Task Learning2 | Explores the impact of instructional prompts for biomedical MTL. Introduce BoX, a collection of 32 instruction tasks for Biomedical NLP across various categories. Propose a unified model (In-BoXBART) using this meta-dataset, that can jointly learn all BoX tasks without any task-specific modules. | ROUGE-L, F1-score |
| Boulanger et al. [111] | NER | Use the generative capacity of LLMs to create unlabelled synthetic data. Semi-supervised learning is used for NER in a low resource setup. | F1-score |
| Yeh et al. [112] | Relation Extraction | Present a simple yet effective method to systematically generate comprehensive prompts that reformulate the relation extraction task as a cloze-test task under a simple prompt formulation. In particular, experiment with different ranking scores for prompt selection. | F1-score |
| Pan et al. [113] | Question Answering | Supervised pretraining on source-domain data to reduce sample complexity on domain-specific downstream tasks. Zero-shot performance on domain-specific reading comprehension tasks is evaluated by combining task transfer with domain adaptation to fine-tune a pre-trained model with no labelled data from the target task. | F1-score |
| Wadden et al. [114] | Scientific Claim Verification | Present MULTIVERS, which predicts a fact-checking label and identifies rationales in a multitask fashion based on a shared encoding of the claim and full document context using weakly-supervised domain adaptation. | Precision, Recall, F1-score |
| Li et al. [115] | Relation Classification | Learn a prototype encoder from relation definition text in a way that is useful for relation instance classification. Use a joint training approach to train both a prototype encoder from definition and an instance encoder. | Accuracy |
| Zhang et al. [116] | Natural Language Inference (NLI) | An instance discrimination based approach to bridge semantic entailment and contradiction understanding with high-level categorical concept encoding (PairSupCon). | Clustering Accuracy |
Denotes papers where a new non-biomedical FSL dataset is introduced.
Denotes papers where a new FSL dataset specific to the biomedical domain is introduced.
4.3. Data Characteristics
We grouped the datasets described into three categories: (i) publicly downloadable data; (ii) datasets from shared tasks; and (iii) new datasets specifically created for the target tasks. We found that datasets belonging to (ii) and (iii) were often difficult to obtain—shared task data unavailable after their completion, and specialized datasets often not made public (e.g., if they contained protected health information). Articles involving datasets from category (i) often reported performances on multiple datasets, consequently making the evaluations more comparable. However, overall, the overlap of datasets among distinct articles was relatively low, making comparative analyses difficult. The MIMIC-III (Medical Information Mart for Intensive Care) dataset [30] was the most frequently used across articles (10/51; 20%), particularly for few-shot classification and NER tasks. This was likely due to the public availability of the dataset and the presence of many labels in it (7000) [31]. Six articles used datasets from shared tasks, of which 4 were from BioNLP [54, 62], one from the Social Media Mining for Health Applications (SMM4H) [48], and one from the Medical Document Anonymization (MEDDOCAN) shared task [52]. Only 4 articles created new datasets, reflecting the paucity of corpora built to support FSL for medical NLP.
4.3.1. Reconstruction of Datasets
32/51 (63%) reviewed articles reconstructed existing datasets for conducting experiments in fewshot settings (i.e., subsets of labeled instances were extracted from larger datasets). For multi-label text classification tasks, especially when the number of labels is large, and for few-shot NER tasks, reconstructing datasets can be complex. A popular way to represent data in FSL is K-Shot-N-Way, which means that each of N classes or entities contains K labeled samples. For multi-label classification tasks, each instance may have more than one label, often making it difficult to ensure that the reconstructed datasets included only K labeled samples for each class. The same challenge exists for NERt tasks as each text segment may have overlapping entities. 12/51 (24%) articles did not construct special datasets to represent few-shot settings. 16% (8/51) used existing datasets with high class imbalances, and the few-shot algorithms were focused on sparsely-occurring labels.
The differing training data sizes across articles demonstrate that there are currently no unified standards for FSL datasets. However, for articles published between late 2021 to 2022, we found that 80% (16/20) made explicit the specific number of shots, or used zero-shot instead of using the term ”few-shot” vaguely. This shift possibly demonstrates that the topic of FSL is gradually becoming standardized within the broader biomedical domain.
4.4. Summary of Methodologies and Applications
Text classification and NER/concept extraction were the most common FSL applications (37/51 articles; 73%), only 14 (27%) focused exclusively on other tasks such as summarization or machine translation. Incorporating prior knowledge being a hallmark of FSL, we found that the reviewed articles employed a wide variety of strategies. Biomedical resources such as SNOMED-CT, Med-Mentions, EHRs, and UMLS were reported to be used to incorporate domain knowledge [47, 91]. 39/51 (77%) articles attempted to incorporate data-level prior knowledge to augment the small datasets available for training. 19 of these chose to augment the training data with other available annotated datasets; or through transfer learning, aggregating and adjusting input-output pairs from larger datasets. For example, due to the scarcity of samples, Manousogiannis et al. [47] attempted to incorporate prior or domain knowledge into their approach by adding concept codes from MEDDRA (Medical Dictionary for Regulatory Activities). Five articles used pre-trained models learned from other tasks and then refined parameters on the given training data, and 6 articles learned a meta-learner as optimizer or refined meta-learned parameters (algorithm-level prior knowledge). Some articles incorporated prior knowledge from more than one source to increase within-domain generalizability.
4.4.1. Few-shot Text Classification
16/51 articles (31%) focused on few-shot classification; 56% (9/16) specified the approximate number of classes and half involved multi-label classification. Multi-label classification is a popular task because the associated datasets generally contain some very low-frequency classes. 11/16 (70%) articles incorporated data level prior knowledge. 11/16 (70%) classification articles proposed deep learning algorithms, and 5/16 (30%) were inspired by label-wise attention mechanisms. 3/16 (20%) combined few-shot tasks with graphs, such as similarity or co-occurrence graphs, or hierarchical structures that encode relationships between labels for knowledge aggregation. While convolutional neural networks have been popular for FSL, transformer-based models such as BERT [117] and RoBERTa [28] rarely appeared in these articles. Only one article [66] mentioned applying BERT to generate instance embeddings and then passing top-level output representations into a label-wise attention mechanism.
4.4.2. Few-shot NER or Concept Extraction
14 reviewed articles were described as NER; 7 as concept extraction. Generally, articles that described themselves as concept extraction applied distinct methodologies compared to each other and involved task-specific configurations based on the characteristics of the data and extraction objectives. Five articles incorporated data level, two incorporated model level, and two incorporated algorithm level prior knowledge. 50% (7/14) of the articles described as NER employed transfer learning, with training and testing data from different domains. Articles commonly used the BIO (beginning, inside, outside) or IO tagging schemes. Two articles investigated both BIO and IO tagging schemes, concluding that systems trained using IO schemes outperform those trained using BIO schemes. Articles reported that the O (outside) tag was often ill-defined, as specific entities (e.g., time entities such as ‘today’, ‘tomorrow’) would be tagged as O if they were not the primary focus of the dataset, while the same entities would be tagged as B or I for other datasets. Five articles used BIO schemes, while one considered only the entity names without any tagging schemes. The NLP/machine learning strategies employed varied substantially and included, for example, the application of fusion layers for combining features [87], biological semantic and positional features [91], prototypical representations and nearest neighbor classifiers [78], transition scorers for modeling transition probabilities between abstract labels [55, 73, 78], self-supervised methods [68, 73, 90], noise networks for auxiliary training [61, 90], and LSTM cells for encoding multiple entity type sequences [61].
4.4.3. Overview of Other Methods
7/51 (14%) articles applied meta-learning strategies, and 20/51 (39%) articles demonstrated the advantages of attention mechanisms in few-shot scenarios, such as handling the difficulty of recognizing multiple unseen labels. Among the latter, 5/20 used self-attention-related methods, and 4/20 used label-wise attention mechanisms. 11/51 (22%) articles reproduced prototypical networks or added enhancements to them. Only 1 article used matching networks, and 2 articles included them as baselines. Since its proposal by Liu et al. [118] in 2021, prompting has gained popularity in the field of few-shot learning. Among the 20 articles we reviewed from the second half of 2021 to the present, 4 articles proposed prompt-based learning methods with promising results. Based on the trends we observed from our review, it is likely that such methods will receive increasing attention in the near future.
4.5. Performance Metrics
14/51 (27%) articles used accuracy, and the reported values on medical datasets or datasets that included medical texts varied between 67.4% and 96%. Two-thirds (10/14) reported accuracies higher than 70%. For the 29/51 (57%) articles that reported F1-score, performance variations were even larger—from 31.8% to 95.7% (median: 68.6%). We were unable to determine in most cases if the performance differences were due to the effectiveness of the FSL methods, or if the dataset characteristics were primarily responsible.
For the vast majority of articles, reported performances on medical datasets were relatively low compared to nonmedical datasets. For articles that reported good performances, we investigated their methods as described and found that, in most cases, they did not mention how many training examples they used or had large (e.g., in the hundreds) training sizes. While these approaches may still be considered few-shot learning, comparing these reported performances with those obtained in truly low-shot settings (e.g., 5-shot) does not constitute a fair comparison. We also observed that some articles reporting high F1-scores included datasets from different domains and only reported aggregated performances rather than dataset-specific ones. In the next section, we present head-to-head comparisons of several FSL systems on the same datasets as part of the Discussion.
5. Discussion
5.1. Summary of Findings
In this review, we systematically compared 51 articles focusing on FSL for biomedical NLP. Similar to its progress in the general domain, FSL research in the medical domain has primarily been in computer vision [12]. Over two-thirds of the articles included in our review were published in the 24 months preceding the review, which demonstrates the rapidly growing interest on the topic. Despite the relatively small number of articles that met our inclusion criteria, several observations were fairly consistent across articles: (i) under the same experimental parameters, the performances reported on medical data were worse than those reported on data from other domains [42, 73, 78]; (ii) incorporating prior knowledge via transfer learning or using specialized training datasets typically produced better results; and (iii) systems generally reported better performances on datasets with formal texts compared to those with noisy texts (e.g., from social media) [55, 68, 78].
Using just the information in the publications, we found it difficult to perform head-to-head comparisons of the proposed methods due to the use of distinct or non-standardized evaluation strategies, training/test data, and experimental settings. For example, Chalkidis et al. [66] used 50 or fewer instances in their few-shot setting, while Rios and Kavuluru [16] used 5 or fewer, making it impossible to perform meaningful comparisons of their proposed methods. In the absence of specialized datasets for FSL, K-Shot-N-Way settings were commonly reported for simulating few-shot scenarios. In such synthetically created datasets, the number of instances for training is predetermined. Such consistency in characteristics is seldom the case with real-world text-based medical data. Though this design attempts to make a direct comparison between different methods or tasks easier, only speculative estimates can be made about how the proposed methods may perform if deployed in real-world settings. It was also typically impossible to compare the performances of FSL methods with the state-of-the-art systems reported in prior literature, as FSL methods were expected to underperform compared to methods trained using larger training sets.
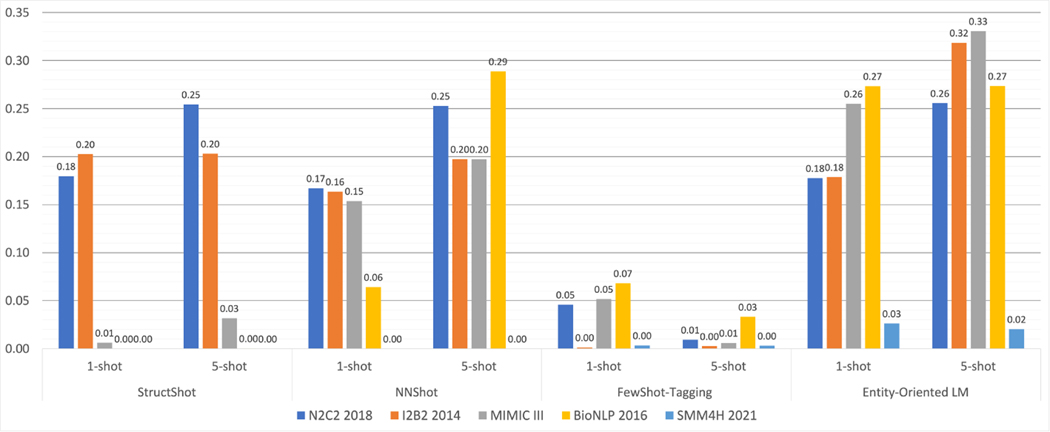
Due to the absence of standardized datasets that enable head-to-head comparisons of all systems, we benchmarked several FSL methods with publicly available implementations on multiple datasets. We focused on the task of NER for this since that was the most commonly addressed task in our review. Figure 7 presents the performance comparisons between 4 FSL NER models on 5 datasets. The results of 3 of the models (StructShot, NNShot and Few-shot Tagging) come from our previous article [119], while the Entity-Oriented LM [120] is a new prompt-based few-shot learning method and comes from our recent ongoing experiments. The benchmarking results demonstrated that all models achieve significantly lower performances compared to the state-of-the-art. More research is clearly required to develop FSL methods that are applicable in practical settings. The results also show how these models underperform for medical texts and specifically for noisy medical texts such as those from social media.
Figure 7:
F1-scores for four FSL NER models on five different medical texts datasets. Further details are reported in a recent publication [119].
Few articles reported the creation of new datasets specialized for FSL or provided benchmarks that future research could use for comparison. The paucity of standardized datasets and the consequent need to reconstruct datasets for simulating few-shot scenarios is a notable obstacle to progress. Since FSL for biomedical NLP is an under-explored field, such datasets and benchmarks are essential for promoting future development. Goodwin et al. [77] echo this need for FSL datasets to advance biomedical NLP. FSL datasets specialized for biomedical NLP need to contain entities/classes that are naturally sparsely occurring, and the distribution of classes/entities need to reflect real-life data. These conditions are necessary for ensuring that developed systems can be compared directly, and that the system performances reflect what is expected in practical settings. Reconstructed datasets often use randomly sampled subsets for evaluation, making direct comparisons between systems difficult (since the specific training and test instances may not be known), and increasing the potential for biased performance estimates.
5.2. Recommendations and Best Practices for Evaluation
In light of the weaknesses and inconsistencies in FSL system evaluations discussed in the review, below we provide three key recommendations.
When reconstructing existing datasets to simulate few-shot settings, the specific training and testing instances used should be made explicit. Ideally, the average and standard deviation over multiple runs should be reported along with the instances involved in each run.
Whenever possible, the natural distribution of the data should be used in the experiments. This means that if the proportion of the positive classes/entities is extremely low compared to the negative classes, the experimental setting must incorporate that difficulty. Performances reported over artificially balanced datasets, particularly in few-shot settings, are not achievable in real-life settings.
Learning curves of performances should be presented, particularly when simulating few-shot settings. This means that performances (using standard metrics such as F1-score and accuracy) should be reported for 1-shot, 5-shot, 10-shot, … , 10%, 20%, … 100% of the training data. It is expected that FSL systems will perform better in low-shot settings while traditional machine learning systems will outperform them when large training datasets are available. Knowledge of the data size at which traditional systems overtake FSL systems is crucial for potential future users of the system.
5.3. Future Directions
Our review showed that FSL for biomedical NLP is still very much in its infancy, and reported performances are typically low with high variance. Importantly, the review enabled us to identify future research activities that will be most impactful in moving this sub-field of research forward. We outline these in the following subsections.
5.3.1. Specialized Datasets for Few-shot Learning
To improve the state of the art in FSL for medical text, the most crucial activity currently is perhaps the creation of specialized, standardized, publicly available datasets. Ideally, such datasets should replicate real-world scenarios and pose practical challenges for FSL. The creation of such datasets will enable the direct comparison of distinct FSL strategies, and of FSL methods with traditional methods (e.g., deep neural networks). Public datasets have helped progress NLP and machine learning research over the years, such as through shared tasks [48]. Our review, however, did not find any current shared task that provides specialized datasets for FSL-based biomedical NLP.
5.3.2. System Comparisons and Benchmarking
FSL methods for NLP comprise a wide variety of approaches [12]. Facilitated by standardized datasets, articles need to focus on comparing distinct categories of FSL for biomedical NLP tasks and identify promising methods that need exploration. Benchmarking articles can customize existing datasets and compare distinct FSL methods on identical evaluation sets, similar to the experiments reported in this paper. Researchers proposing new FSL methods for biomedical NLP should also take the steps necessary to enable head-to-head comparisons and reproducible research, following the guidelines presented in the earlier subsection.
5.3.3. Opportunities
The paucity of research in this space means there are many potential opportunities. Domain-independent FSL methods have benefited by incorporating prior knowledge to compensate for the low numbers of training instances [12]. FSL methods for biomedical NLP can follow the same path. Over the years, medical NLP researchers have created many resources to support NLP methods, such as the Unified Medical Language System (UMLS) [121, 122]. However, limited efforts have been made to innovatively incorporate such knowledge in FSL methods. Effectively incorporating prior knowledge by utilizing such domain-specific knowledge sources is a particularly attractive opportunity.
In the recent review by Wang et al. (2020) [12], the authors specified multi-modal data augmentation as a potential opportunity for improving the state-of-the-art in FSL. The same opportunity also exists in the medical domain. To enable FSL systems to achieve performance levels suitable for deployment, future research may focus on augmenting information derived from medical texts with other information, such as images and ontologies. Existing FSL techniques for medical free-text data usually incorporate prior knowledge from one single modality (text), and it is generally not possible to incorporate information from other types of data, such as images. Multi-modal strategies that combine knowledge from several sources (e.g., texts, images, knowledge bases, ontologies) may enable FSL methods to achieve the performance levels needed to be applicable in real-world medical settings. Intuitively, multi-modal learning models are more akin to human learning. Unsurprisingly data augmentation methods in NLP have recently seen growing interest [123]. Notwithstanding this recent rise, this space is still comparatively under-explored, possibly due to the difficulties in augmentation of natural language data in general, and medical free text in particular.
The widespread popularity and usage of large language models (LLMs) such as GPT and biomedical domain-specific BERT models presents the unique opportunity to evaluate the capability of LLMs in few-shot settings. Nori et al [124] take a step in this direction and find that GPT-4 outperforms GPT-3.5 for biomedical NLP tasks. Most LLM-based approaches, however, fall outside the inclusion timeframe of this review, and future reviews should investigate this emerging space.
5.4. Responsible AI, Ethics, and Privacy in the Context of FSL
We end this review by providing a brief discussion of how the concepts of responsible AI, ethics, and privacy apply to the emerging field of FSL. Responsible AI calls for the development of AI systems that promote common good, and take great care to identify and evaluate any potential harms [125, 126]. Specifically within the field of medicine, responsible AI demands, among other things, that machine learning models are not biased against any subpopulation even if data for such populations are limited. Minority subpopulations (e.g., racial/ethnic minorities) are often underrepresented in clinical data, leading to the development of systems that are biased and/or suboptimal for these subpopulations. FSL methods have the potential of alleviating these problems by being able to effectively learn from small samples. Recent years have seen the growing usage of pretrained models, including LLMs, which add another layer of complexity from the perspective of bias and equity. The underlying data used to train such pretrained models are primarily from the majority population, and minority populations are underrepresented. FSL systems relying on such pretrained models are likely to be impacted by the biases encoded in such models. Ideally, as FSL research matures, such methods will produce performances comparable to state-of-the-art machine learning approaches that learn from big data (e.g., deep neural networks), without suffering from the problems of bias. Our review and benchmarking experiments, however, demonstrate that substantial research advances are required in order to move the state of the art in FSL to that level.
FSL approaches also present risks and opportunities from the perspective of privacy and security. On one hand, due to small data sizes, FSL models may be relatively more vulnerable to inference attacks, leaking personal information in training data and harming user privacy [126]. On the other hand, with the promised flexibility of FSL, it is possible to build light-weight FSL systems that only require the user’s data to train and can operate locally on the user’s device accurately [127, 128]. Such personalized deployment alleviates the issue of user information leakage because the data and model would never need to leave the devices. The principles of responsible AI apply across the spectrum of machine learning research, and the specific considerations are unique to the research being conducted. Consequently, as FSL research in this space evolves, researchers must ensure that ethical implications are carefully considered—particularly from the perspective of privacy, security and equity.
6. Conclusion
FSL approaches have substantial promise for NLP in the medical domain, as many medical datasets naturally have low numbers of annotated instances. Some promising approaches have been proposed in the recent past, most of which focused on classification or NER. Meta-learning and transfer learning were commonly used strategies, and a number of articles reported on the benefits of incorporating attention mechanisms. Typical performances of FSL-based medical NLP systems are not yet good enough to be suitable for real-world application, and further research on improving performance is required. The lack of public datasets specialized for FSL, and the absence of standard evaluation frameworks present obstacles to progressing research on the topic, and future research should consider creating such datasets and benchmarks for comparative analyses.
Highlights.
Comprehensive review of few-shot learning for medical text, comprising 51 artcles
Systematc resource of research aims, datasets, evaluaton metrics, and methodology
Best practce recommendatons for evaluaton of few-shot methods for medical text
Benchmarking of medical named entty recogniton using several few-shot methods
Current state of the feld, open research opportunites, and challenges associated
Acknowledgments
We thank the generous support of the National Institute on Drug Abuse of the National Institutes of Health.
Funding
Research reported in this publication was supported by the National Institute on Drug Abuse (NIDA) of the National Institutes of Health (NIH) under award number R01DA057599. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
https://dl.acm.org/. Last accessed September 9, 2022.
https://openreview.net/. Last accessed March 9, 2023.
Declaration of interests
The authors declare that they have no known competng fnancial interests or personal relatonships that could have appeared to infuence the work reported in this paper.
Ethical Approval
This work does not involve human subjects and does not require approval from the IRB.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability
All the research articles reviewed have been listed in Table 1. There is no other data related to this work.
References
- [1].Sung F, Yang Y, Zhang L, Xiang T, Torr PH, and Hospedales TM, “Learning to compare: Relation network for few-shot learning,” Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 1199–1208, 2018. eprint: https://openaccess.thecvf.com/content_cvpr_2018/papers/Sung_Learning_to_Compare_CVPR_2018_paper.pdf. [Google Scholar]
- [2].Snell J, Swersky K, and Zemel RS, “Prototypical networks for few-shot learning,” Advances in Neural Information Processing Systems, vol. 30, 2017. [Online]. Available: https://proceedings.neurips.cc/paper/2017/file/cb8da6767461f2812ae4290eac7cbc42-Paper.pdf. [Google Scholar]
- [3].Lake BM, Salakhutdinov R, and Tenenbaum JB, “One-shot learning by inverting a compositional causal process,” Advances in Neural Information Processing Systems, vol. 26, 2013. eprint: https://papers.nips.cc/paper/2013/file/52292e0c763fd027c6eba6b8f494d2eb-Paper.pdf. [Google Scholar]
- [4].Dong N. and Xing EP, “Few-Shot Semantic Segmentation with Prototype Learning,” in British Machine Vision Conference (BMVC), vol. 3, 2018. eprint: http://bmvc2018.org/contents/papers/0255.pdf. [Google Scholar]
- [5].Li W, Wang L, Xu J, Huo J, Gao Y, and Luo J, “Revisiting Local Descriptor based Image-to-Class Measure for Few-shot Learning,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2019, pp. 7260–7268. eprint: https://openaccess.thecvf.com/content_CVPR_2019/papers/Li_Revisiting_Local_Descriptor_Based_Image-To-Class_Measure_for_Few-Shot_Learning_CVPR_2019_paper.pdf. [Google Scholar]
- [6].Lake BM, Salakhutdinov R, Gross J, and Tenenbaum JB, “One shot learning of simple visual concepts,” in Proceedings of the Annual Meeting of the Cognitive Science Society, vol. 33, 2011. eprint: https://escholarship.org/content/qt4ht821jx/qt4ht821jx.pdf. [Google Scholar]
- [7].Thompson HM et al. , “Bias and fairness assessment of a natural language processing opioid misuse classifier: detection and mitigation of electronic health record data disadvantages across racial subgroups,” Journal of the American Medical Informatics Association, vol. 28, no. 11, pp. 2393–2403, Aug. 2021, issn: 1527–974X. doi: 10.1093/jamia/ocab148. eprint: https://academic.oup.com/jamia/article-pdf/28/11/2393/40576428/ocab148.pdf. [Online]. Available: 10.1093/jamia/ocab148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Young T, Hazarika D, Poria S, and Cambria E, “Recent trends in deep learning based natural language processing,” IEEE Computational Intelligence Magazine, vol. 13, no. 3, pp. 55–75, 2018. [Online]. Available: https://ieeexplore.ieee.org/abstract/document/8416973. [Google Scholar]
- [9].Hou Y. et al. , “FewJoint: A Few-shot Learning Benchmark for Joint Language Understanding,” arXiv preprint arXiv:2009.08138, 2020. [Online]. Available: https://arxiv.org/abs/2009.08138. [Google Scholar]
- [10].Hofer M, Kormilitzin A, Goldberg P, and Nevado-Holgado A, “Few-shot Learning for Named Entity Recognition in Medical Text,” arXiv preprint arXiv:1811.05468, 2018. [Online]. Available: https://arxiv.org/abs/1811.05468. [Google Scholar]
- [11].Schmidt HK, Rothgangel M, and Grube D, “Prior knowledge in recalling arguments in bioethical dilemmas,” Frontiers in psychology, vol. 6, p. 1292, 2015. doi: 10.3389/fpsyg.2015.01292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang Y, Yao Q, Kwok JT, and Ni LM, “Generalizing from a few examples: A survey on few-shot learning,” ACM Computing Surveys (CSUR), vol. 53, no. 3, pp. 1–34, 2020. doi: 10.1145/3386252. [DOI] [Google Scholar]
- [13].Joshi V, Peters ME, and Hopkins M, “Extending a Parser to Distant Domains Using a Few Dozen Partially Annotated Examples,” in Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), 2018, pp. 1190–1199. [Online]. Available: https://aclanthology.org/P18-1110. [Google Scholar]
- [14].Kaiser L, Nachum O, Roy A, and Bengio S, “Learning to Remember Rare Events,” in International Conference on Learning Representations, 2017. [Online]. Available: https://arxiv.org/abs/1703.03129. [Google Scholar]
- [15].Yu M. et al. , “Diverse Few-Shot Text Classification with Multiple Metrics,” in Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long Papers), New Orleans, Louisiana: Association for Computational Linguistics, Jun. 2018, pp. 1206–1215. doi: 10. 18653/v1/N18–1109. [Online]. Available: https://aclanthology.org/N18-1109. [Google Scholar]
- [16].Rios A. and Kavuluru R, “Few-shot and zero-shot multi-label learning for structured label spaces,” in Proceedings of the Conference on Empirical Methods in Natural Language Processing. Conference on Empirical Methods in Natural Language Processing, NIH Public Access, vol. 2018, 2018, p. 3132. [Online]. Available: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6375489/. [PMC free article] [PubMed] [Google Scholar]
- [17].Hu Z, Li X, Tu C, Liu Z, and Sun M, “Few-shot charge prediction with discriminative legal attributes,” in Proceedings of the 27th International Conference on Computational Linguistics, 2018, pp. 487–498. [Online]. Available: https://aclanthology.org/C18-1041. [Google Scholar]
- [18].Ghosh S, Misra J, Ghosh S, and Podder S, “Utilizing Social Media for Identifying Drug Addiction and Recovery Intervention,” in 2020 IEEE International Conference on Big Data (Big Data), 2020, pp. 3413–3422. doi: 10.1109/BigData50022.2020.9378092. [Online]. Available: https://ieeexplore.ieee.org/abstract/document/9378092. [DOI] [Google Scholar]
- [19].Weston J, Chopra S, and Bordes A, “Memory networks,” in 3rd International Conference on Learning Representations, ICLR 2015, 2015. [Google Scholar]
- [20].Graves A, Wayne G, and Danihelka I, “Neural turing machines,” arXiv preprint arXiv:1410.5401, 2014. [Google Scholar]
- [21].Vinyals O, Blundell C, Lillicrap T, and Wierstra D, “Matching networks for one shot learning,” Advances in Neural Information Processing Systems, vol. 29, pp. 3630–3638, 2016. doi: 10.5555/3157382.3157504. [DOI] [Google Scholar]
- [22].Bachman P, Sordoni A, and Trischler A, “Learning algorithms for active learning,” in International Conference on Machine Learning (ICML), PMLR, 2017, pp. 301–310. eprint: http://proceedings.mlr.press/v70/bachman17a/bachman17a.pdf. [Google Scholar]
- [23].Pan SJ and Yang Q, “A survey on transfer learning,” IEEE Transactions on Knowledge and Data Engineering, vol. 22, no. 10, pp. 1345–1359, 2009. doi: 10.1109/TKDE.2009.191. [DOI] [Google Scholar]
- [24].Yosinski J, Clune J, Bengio Y, and Lipson H, “How transferable are features in deep neural networks?” Advances in neural information processing systems, vol. 27, 2014. [Google Scholar]
- [25].Hospedales TM, Antoniou A, Micaelli P, and Storkey AJ, “Meta-Learning in Neural Networks: A Survey,” IEEE Transactions on Pattern Analysis and Machine Intelligence, pp. 1–1, 2021. doi: 10.1109/TPAMI.2021.3079209. [DOI] [PubMed] [Google Scholar]
- [26].Schmidhuber J, “Evolutionary Principles in Self-Referential Learning. On Learning now to Learn: The Meta-Meta-Meta…-Hook,” Diploma Thesis, Technische Universitat Munchen, Germany, May 1987. [Online]. Available: http://www.idsia.ch/~juergen/diploma.html. [Google Scholar]
- [27].Moher D, Liberati A, Tetzlaff J, Altman DG, and Group TP, “Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement,” PLoS medicine, vol. 6, no. 7, pp. 1549–1676, 2009. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liu Y. et al. , “RoBERTa: A Robustly Optimized BERT Pretraining Approach,” arXiv preprint arXiv:1907.11692, 2019. [Online]. Available: https://arxiv.org/abs/1907.11692. [Google Scholar]
- [29].Jouhet V. et al. , “Automated classification of free-text pathology reports for registration of incident cases of cancer,” Methods of Information in Medicine, vol. 51, no. 03, pp. 242–251, 2012. doi: 10.3414/ME11-01-0005. [DOI] [PubMed] [Google Scholar]
- [30].Johnson AE et al. , “MIMIC-III, a freely accessible critical care database,” Scientific data, vol. 3, no. 1, pp. 1–9, 2016. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Rios A. and Kavuluru R, “EMR coding with semi–parametric multi–head matching networks,” in Proceedings of the conference. Association for Computational Linguistics. North American Chapter. Meeting, NIH Public Access, vol. 2018, 2018, p. 2081. [Online]. Available: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7720861/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Uzuner Ö, Solti I, and Cadag E, “Extracting medication information from clinical text,” Journal of the American Medical Informatics Association, vol. 17, no. 5, pp. 514–518, 2010. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Uzuner Ö, South BR, Shen S, and DuVall SL, “2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text,” Journal of the American Medical Informatics Association, vol. 18, no. 5, pp. 552–556, 2011. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sun W, Rumshisky A, and Uzuner O, “Evaluating temporal relations in clinical text: 2012 i2b2 Challenge,” Journal of the American Medical Informatics Association, vol. 20, no. 5, pp. 806–813, 2013. doi: 10.1136/amiajnl-2013-001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tjong Kim Sang EF and De Meulder F, “Introduction to the CoNLL-2003 Shared Task: Language-Independent Named Entity Recognition,” in Proceedings of the Seventh Conference on Natural Language Learning at HLT-NAACL 2003, 2003, pp. 142–147. [Online]. Available: https://aclanthology.org/W03-0419. [Google Scholar]
- [36].Chiu B, Crichton G, Korhonen A, and Pyysalo S, “How to train good word embeddings for biomedical NLP,” in Proceedings of the 15th Workshop on Biomedical Natural Language Processing, 2016, pp. 166–174. doi: 10.18653/v1/W16–2922. eprint: https://aclanthology.org/W16-2922.pdf. [Google Scholar]
- [37].Callard F. et al. , “Developing a new model for patient recruitment in mental health services: a cohort study using Electronic Health Records,” BMJ open, vol. 4, no. 12, e005654, 2014. doi: 10.1136/bmjopen-2014-005654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Stewart R. et al. , “The South London and Maudsley NHS foundation trust biomedical research centre (SLAM BRC) case register: development and descriptive data,” BMC psychiatry, vol. 9, no. 1, pp. 1–12, 2009. doi: 10.1186/1471-244X-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pham N-Q, Niehues J, and Waibel A, “Towards one-shot learning for rare-word translation with external experts,” in Proceedings of the 2nd Workshop on Neural Machine Translation and Generation, Melbourne, Australia: Association for Computational Linguistics, Jul. 2018, pp. 100–109. doi: 10.18653/v1/W18–2712. [Online]. Available: https://aclanthology.org/W18-2712. [Google Scholar]
- [40].Koehn P, “Europarl: A parallel corpus for statistical machine translation,” in Proceedings of machine translation summit x: papers, 2005, pp. 79–86. eprint: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.459.5497&rep=rep1&type=pdf. [Google Scholar]
- [41].Cettolo M, Girardi C, and Federico M, “WIT3: Web Inventory of Transcribed and Translated Talks,” in Conference of European Association for Machine Translation, 2012, pp. 261–268. [Online]. Available: http://hdl.handle.net/11582/104409. [Google Scholar]
- [42].Yan L, Zheng Y, and Cao J, “Few-shot learning for short text classification,” Multimedia Tools and Applications, vol. 77, no. 22, pp. 29799–29810, 2018. doi: 10.1007/s11042-018-5772-4. [DOI] [Google Scholar]
- [43].Yan L, Zheng W, Zhang H, Tao H, and He M, “Learning Discriminative Sentiment Chunk Vectors for Twitter Sentiment Analysis,” Journal of Internet Technology, vol. 77, no. 22, pp. 29799–29810, 2018. doi: 10.6138/JIT.2017.18.7.20170410. [DOI] [Google Scholar]
- [44].Speriosu M, Sudan N, Upadhyay S, and Baldridge J, “Twitter Polarity Classification with Label Propagation over Lexical Links and the Follower Graph,” in Proceedings of the First Workshop on Unsupervised Learning in NLP, 2011, pp. 53–63. eprint: https://aclanthology.org/W11-2207.pdf. [Google Scholar]
- [45].Thelwall M, Buckley K, and Paltoglou G, “Sentiment strength detection for the social web,” Journal of the American Society for Information Science and Technology, vol. 63, no. 1, pp. 163–173, 2012. doi: 10.1002/asi.21662. [DOI] [Google Scholar]
- [46].Nakov P. et al. , “Developing a successful SemEval task in sentiment analysis of Twitter and other social media texts,” Language Resources and Evaluation, vol. 50, no. 1, pp. 35–65, 2016. doi: 10.1007/s10579-015-9328-1. [DOI] [Google Scholar]
- [47].Manousogiannis M, Mesbah S, Santamaria SB, Bozzon A, and Sips R-J, “Give it a shot: Few-shot learning to normalize ADR mentions in Social Media posts,” in Proceedings of the Fourth Social Media Mining for Health Applications (# SMM4H) Workshop & Shared Task, 2019, pp. 114–116. eprint: https://aclanthology.org/W19-3219.pdf. [Google Scholar]
- [48].Weissenbacher D. et al. , “Overview of the fourth social media mining for health (SMM4H) shared tasks at ACL 2019,” in Proceedings of the Fourth Social Media Mining for Health Applications (#SMM4H) Workshop & Shared Task, 2019, pp. 21–30. eprint: https://aclanthology.org/W19-3203.pdf. [Google Scholar]
- [49].Gao T. et al. , “FewRel 2.0: Towards More Challenging Few-Shot Relation Classification,” in Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), Hong Kong, China: Association for Computational Linguistics, Nov. 2019, pp. 6250–6255. doi: 10.18653/v1/D19–1649. [Online]. Available: https://aclanthology.org/D19-1649. [Google Scholar]
- [50].Han X. et al. , “FewRel: A Large-Scale Supervised Few-Shot Relation Classification Dataset with State-of-the-Art Evaluation,” in Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, Brussels, Belgium: Association for Computational Linguistics, 2018, pp. 4803–4809. doi: 10.18653/v1/D18–1514. [Online]. Available: https://aclanthology.org/D18-1514. [Google Scholar]
- [51].Lara-Clares A. and Garcia-Serrano A, “Key Phrases Annotation in Medical Documents: MEDDOCAN 2019 Anonymization Task.,” in IberLEF@ SEPLN, 2019, pp. 755–760. eprint: http://ceur-ws.org/Vol-2421/MEDDOCAN_paper_15.pdf. [Google Scholar]
- [52].Marimon M. et al. , “Automatic De-identification of Medical Texts in Spanish: the MEDDOCAN Track, Corpus, Guidelines, Methods and Evaluation of Results.,” in IberLEF@ SEPLN, 2019, pp. 618–638. eprint: http://ceur-ws.org/Vol-2421/MEDDOCAN_overview.pdf. [Google Scholar]
- [53].Ferré A, Deléger L, Bossy R, Zweigenbaum P, and Nédellec C, “C-Norm: a neural approach to few-shot entity normalization,” BMC Bioinformatics, vol. 21, pp. 1–9, 2020. doi: 10.1186/s12859-020-03886-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Bossy R, Deléger L, Chaix E, Ba M, and Nédellec C, “Bacteria biotope at BioNLP open shared tasks 2019,” in Proceedings of the 5th Workshop on BioNLP Open Shared Tasks, 2019, pp. 121–131. doi: 10.18653/v1/D19–5719. [Google Scholar]
- [55].Hou Y. et al. , “Few-shot Slot Tagging with Collapsed Dependency Transfer and Label-enhanced Task-adaptive Projection Network,” in Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Online: Association for Computational Linguistics, Jul. 2020, pp. 1381–1393. doi: 10.18653/v1/2020.acl-main.128. [Online]. Available: https://aclanthology.org/2020.acl-main.128. [Google Scholar]
- [56].Coucke A. et al. , “Snips voice platform: an embedded spoken language understanding system for private-by-design voice interfaces,” arXiv preprint arXiv:1805.10190, 2018. [Online]. Available: https://arxiv.org/abs/1805.10190. [Google Scholar]
- [57].Sharaf A, Hassan H, and Daumé H, “Meta-learning for few-shot NMT adaptation,” in Proceedings of the Fourth Workshop on Neural Generation and Translation, Online: Association for Computational Linguistics, Jul. 2020, pp. 43–53. doi: 10.18653/v1/2020.ngt-1.5. [Online]. Available: https://aclanthology.org/2020.ngt-1.5. [Google Scholar]
- [58].Tiedemann J, “Parallel Data, Tools and Interfaces in OPUS,” in Louisiana Real Estate Commission (LREC), Citeseer, vol. 2012, 2012, pp. 2214–2218. eprint: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.673.2874&rep=rep1&type=pdf. [Google Scholar]
- [59].Lu J, Du L, Liu M, and Dipnall J, “Multi-label Few/Zero-shot Learning with Knowledge Aggregated from Multiple Label Graphs,” in Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP), Online: Association for Computational Linguistics, Nov. 2020, pp. 2935–2943. doi: 10.18653/v1/2020.emnlp-main.235. [Online]. Available: https://aclanthology.org/2020.emnlp-main.235. [Google Scholar]
- [60].Chalkidis I, Fergadiotis E, Malakasiotis P, and Androutsopoulos I, “Large-scale multilabel text classification on EU legislation,” in Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, Florence, Italy: Association for Computational Linguistics, Jul. 2019, pp. 6314–6322. doi: 10.18653/v1/P19–1636. [Online]. Available: https://aclanthology.org/P19-1636. [Google Scholar]
- [61].Jia C. and Zhang Y, “Multi-cell compositional LSTM for NER domain adaptation,” in Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Online: Association for Computational Linguistics, Jul. 2020, pp. 5906–5917. doi: 10.18653/v1/2020.acl-main.524. [Online]. Available: https://aclanthology.org/2020.aclmain.524. [Google Scholar]
- [62].Nédellec C. et al. , “Overview of BioNLP Shared Task 2013,” in Proceedings of the BioNLP Shared Task 2013 Workshop, 2013, pp. 1–7. eprint: https://aclanthology.org/W13-2001.pdf. [Google Scholar]
- [63].Derczynski L, Bontcheva K, and Roberts I, “Broad twitter corpus: A diverse named entity recognition resource,” in Proceedings of COLING 2016, the 26th International Conference on Computational Linguistics: Technical Papers, 2016, pp. 1169–1179. eprint: https://aclanthology.org/C16-1111.pdf. [Google Scholar]
- [64].Lu D, Neves L, Carvalho V, Zhang N, and Ji H, “Visual Attention Model for Name Tagging in Multimodal Social Media,” in Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Melbourne, Australia: Association for Computational Linguistics, Jul. 2018, pp. 1990–1999. doi: 10.18653/v1/P18–1185. [Online]. Available: https://aclanthology.org/P18-1185. [Google Scholar]
- [65].Jia C, Liang X, and Zhang Y, “Cross-domain NER using cross-domain language modeling,” in Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, Florence, Italy: Association for Computational Linguistics, Jul. 2019, pp. 2464–2474. doi: 10.18653/v1/P19–1236. [Online]. Available: https://aclanthology.org/P19-1236. [Google Scholar]
- [66].Chalkidis I, Fergadiotis M, Kotitsas S, Malakasiotis P, Aletras N, and Androutsopoulos I, “An Empirical Study on Large-Scale Multi-Label Text Classification Including Few and Zero-Shot Labels,” in Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP), 2020, pp. 7503–7515. eprint: https://aclanthology.org/2020.emnlp-main.607.pdf. [Google Scholar]
- [67].Lewis DD, Yang Y, Rose TG, and Li F, “Rcv1: A new benchmark collection for text categorization research,” Journal of Machine Learning Research, vol. 5, no. Apr, pp. 361–397, 2004. eprint: https://www.jmlr.org/papers/volume5/lewis04a/lewis04a.pdf. [Google Scholar]
- [68].Lwowski B. and Najafirad P, “COVID-19 Surveillance through Twitter using Self-Supervised and Few Shot Learning,” in Proceedings of the 1st Workshop on NLP for COVID-19 (Part 2) at EMNLP 2020, Online: Association for Computational Linguistics, Dec. 2020. doi: 10. 18653/v1/2020.nlpcovid19–2.9. [Online]. Available: https://aclanthology.org/2020.nlpcovid19-2.9. [Google Scholar]
- [69].Lamsal R, “Coronavirus (COVID-19) Tweets Dataset,” 2020. doi: 10.21227/781w-ef42. [Online]. Available: 10.21227/781w-ef42. [DOI] [Google Scholar]
- [70].Chen Z, Eavani H, Chen W, Liu Y, and Wang WY, “Few-Shot NLG with Pre-Trained Language Model,” in Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Online: Association for Computational Linguistics, Jul. 2020, pp. 183–190. doi: 10.18653/v1/2020.acl-main.18. [Online]. Available: https://aclanthology.org/2020.acl-main.18. [Google Scholar]
- [71].Lebret R, Grangier D, and Auli M, “Neural Text Generation from Structured Data with Application to the Biography Domain,” in Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, Austin, Texas: Association for Computational Linguistics, Nov. 2016, pp. 1203–1213. doi: 10.18653/v1/D16–1128. [Online]. Available: https://aclanthology.org/D16-1128. [Google Scholar]
- [72].Vaci N. et al. , “Natural language processing for structuring clinical text data on depression using UK-CRIS,” Evidence-based mental health, vol. 23, no. 1, pp. 21–26, 2020. doi: 10.1136/ebmental-2019-300134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Huang J. et al. , “Few-Shot Named Entity Recognition: An Empirical Baseline Study,” in Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing, Online and Punta Cana, Dominican Republic: Association for Computational Linguistics, Nov. 2021, pp. 10408–10423. doi: 10.18653/v1/2021.emnlp-main.813. [Online]. Available: https://aclanthology.org/2021.emnlp-main.813. [Google Scholar]
- [74].Chen D, Zhang L, and Ma C, “A Multimodal Diagnosis Predictive Model of Alzheimer’s Disease with Few-shot Learning,” in 2020 International Conference on Public Health and Data Science (ICPHDS), IEEE, 2020, pp. 273–277. doi: 10.1109/ICPHDS51617.2020.00060. [DOI] [Google Scholar]
- [75].Yin S, Zhao W, Jiang X, and He T, “Knowledge-aware Few-shot Learning Framework for Biomedical Event Trigger Identification,” in 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), IEEE, 2020, pp. 375–380. doi: 10.1109/BIBM49941. 2020.9313195. [Google Scholar]
- [76].Pyysalo S, Ohta T, Miwa M, Cho H-C, Tsujii J, and Ananiadou S, “Event extraction across multiple levels of biological organization,” Bioinformatics, vol. 28, no. 18, pp. i575–i581, 2012. doi: 10.1093/bioinformatics/bts407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Goodwin TR, Savery ME, and Demner-Fushman D, “Flight of the PEGASUS? Comparing Transformers on Few-Shot and Zero-Shot Multi-document Abstractive Summarization,” in Proceedings of the 28th International Conference on Computational Linguistics, NIH Public Access, vol. 2020, 2020, pp. 5640–5646. eprint: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7720861/pdf/nihms-1650639.pdf. [PMC free article] [PubMed] [Google Scholar]
- [78].Yang Y. and Katiyar A, “Simple and Effective Few-Shot Named Entity Recognition with Structured Nearest Neighbor Learning,” in Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP), Online: Association for Computational Linguistics, Nov. 2020, pp. 6365–6375. doi: 10.18653/v1/2020.emnlp-main.516. [Online]. Available: https://aclanthology.org/2020.emnlp-main.516. [Google Scholar]
- [79].Weischedel R. et al. , “Ontonotes release 5.0,” Linguistic Data Consortium, Philadelphia, PA., 2013. doi: 10.35111/xmhb-2b84. [DOI] [Google Scholar]
- [80].Stubbs A. and Uzuner Ö, “Annotating longitudinal clinical narratives for de-identification:¨ The 2014 i2b2/UTHealth corpus,” Journal of biomedical informatics, vol. 58, S20–S29, 2015. doi: 10.1016/j.jbi.2015.07.020. eprint: https://aclanthology.org/W17-4418.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Derczynski L, Nichols E, van Erp M, and Limsopatham N, “Results of the WNUT2017 shared task on novel and emerging entity recognition,” in Proceedings of the 3rd Workshop on Noisy User-generated Text, 2017, pp. 140–147. doi: 10.18653/v1/W17–4418. eprint: https://aclanthology.org/W17-4418.pdf. [Google Scholar]
- [82].Hartmann M. and Søgaard A, “Multilingual Negation Scope Resolution for Clinical Text,” in Proceedings of the 12th International Workshop on Health Text Mining and Information Analysis, 2021, pp. 7–18. eprint: https://aclanthology.org/2021.louhi-1.2.pdf. [Google Scholar]
- [83].Marimon M, Vivaldi J, and Bel N, “Annotation of negation in the IULA Spanish Clinical Record Corpus,” Blanco E, Morante R, Saurıé R, editors. SemBEaR 2017. Computational Semantics Beyond Events and Roles; 2017 Apr 4; Valencia, Spain. Stroudsburg (PA): ACL; 2017. p. 43–52, [Online]. Available: http://hdl.handle.net/10230/33296. [Google Scholar]
- [84].Lima S, Perez N, Cuadros M, and Rigau G, “NUBes: A corpus of negation and uncertainty in Spanish clinical texts,” in Proceedings of The 12th Language Resources and Evaluation Conference, 2020, pp. 5772–5781. [Online]. Available: https://arxiv.org/abs/2004.01092. [Google Scholar]
- [85].Dalloux C, Grabar N, and Claveau V, “Détection de la négation : corpus français et apprentissage supervisé,” Revue des Sciences et Technologies de l’Information-Série TSI: Technique et Science Informatiques, pp. 1–21, 2019. eprint: https://hal.archives-ouvertes.fr/hal02402913/document. [Google Scholar]
- [86].Fivez P, Suster S, and Daelemans W, “Conceptual Grounding Constraints for Truly Robust Biomedical Name Representations,” in Proceedings of the 16th Conference of the European Chapter of the Association for Computational Linguistics: Main Volume, 2021, pp. 2440–2450. doi: 10.18653/v1/2021.eacl-main.208. [Google Scholar]
- [87].Lu H.-y., Fan C, Song xiaoning, and Fang W, “A novel few-shot learning based multimodality fusion model for COVID-19 rumor detection from online social media,” PeerJ Computer Science, vol. 7, e688, 2021. doi: 10.7717/peerj-cs.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Zubiaga A, Liakata M, and Procter R, “Learning Reporting Dynamics during Breaking News for Rumour Detection in Social Media,” arXiv preprint arXiv:1610.07363, 2016. [Online]. Available: https://arxiv.org/abs/1610.07363. [Google Scholar]
- [89].Ma J. et al. , “Few-shot learning creates predictive models of drug response that translate from high-throughput screens to individual patients,” Nature Cancer, vol. 2, no. 2, pp. 233–244, 2021. doi: 10.1038/s43018-020-00169-2. [Online]. Available: https://www.nature.com/articles/s43018-020-00169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Kormilitzin A, Vaci N, Liu Qiang, and Nevado-Holgado A, “Med7: A transferable clinical natural language processing model for electronic health records,” Artificial Intelligence in Medicine, vol. 118, p. 102086, 2021. doi: 10.1016/j.artmed.2021.102086. [DOI] [PubMed] [Google Scholar]
- [91].Guo S, Huang L, Yao G, Wang Y, Guan H, and Bai T, “Extracting Biomedical Entity Relations using Biological Interaction Knowledge,” Interdisciplinary Sciences: Computational Life Sciences, vol. 13, no. 2, pp. 312–320, 2021. doi: 10.1007/s12539-021-00425-8. [Online]. Available: https://link.springer.com/article/10.1007/s12539-021-00425-8. [DOI] [PubMed] [Google Scholar]
- [92].Lee N, Bang Y, Madotto A, and Fung P, “Towards Few-shot Fact-Checking via Perplexity,” in Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Online: Association for Computational Linguistics, Jun. 2021, pp. 1971–1981. doi: 10.18653/v1/2021.naacl-main.158. [Online]. Available: https://aclanthology.org/2021.naacl-main.158. [Google Scholar]
- [93].Su D, Xu Y, Yu T, Siddique FB, Barezi E, and Fung P, “CAiRE-COVID: A Question Answering and Query-focused Multi-Document Summarization System for COVID-19 Scholarly Information Management,” in Proceedings of the 1st Workshop on NLP for COVID-19 (Part 2) at EMNLP 2020, Online: Association for Computational Linguistics, Dec. 2020. doi:10.18653/v1/2020.nlpcovid19–2.14. [Online]. Available: https://aclanthology.org/2020.nlpcovid19-2.14. [Google Scholar]
- [94].Alhindi T, Petridis S, and Muresan S, “Where is your Evidence: Improving Fact-checking by Justification Modeling,” in Proceedings of the First Workshop on Fact Extraction and VERification (FEVER), 2018, pp. 85–90. doi: 10.18653/v1/W18–5513. eprint: https://aclanthology.org/W18-5513.pdf. [Google Scholar]
- [95].Thorne J, Vlachos A, Christodoulopoulos C, and Mittal A, “FEVER: a large-scale dataset for Fact Extraction and VERification,” in Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long Papers), 2018, pp. 809–819. [Online]. Available: https://arxiv.org/abs/1803.05355. [Google Scholar]
- [96].Fivez P, Suster S, and Daelemans W, “Scalable Few-Shot Learning of Robust Biomedical Name Representations,” in Proceedings of the 20th Workshop on Biomedical Language Processing, 2021, pp. 23–29. doi: 10.18653/v1/2021.bionlp-1.3. [Google Scholar]
- [97].Xiao Y, Jin Y, and Hao K, “Adaptive Prototypical Networks With Label Words and Joint Representation Learning for Few-Shot Relation Classification,” IEEE Transactions on Neural Networks and Learning Systems, vol. 34, no. 3, pp. 1406–1417, 2023. doi: 10.1109/TNNLS.2021.3105377. [DOI] [PubMed] [Google Scholar]
- [98].Ziletti A, Akbik A, Berns C, Herold T, Legler M, and Viell M, “Medical Coding with Biomedical Transformer Ensembles and Zero/Few-shot Learning,” in Proceedings of the 2022 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies: Industry Track, Hybrid: Seattle, Washington: + Online: Association for Computational Linguistics, Jul. 2022, pp. 176–187. doi: 10.18653/v1/2022.naacl-industry.21. [Online]. Available: https://aclanthology.org/2022.naaclindustry.21. [Google Scholar]
- [99].Ye Q, Lin BY, and Ren X, “CrossFit: A Few-shot Learning Challenge for Cross-task Generalization in NLP,” in Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing, Online and Punta Cana, Dominican Republic: Association for Computational Linguistics, Nov. 2021, pp. 7163–7189. doi: 10.18653/v1/2021.emnlpmain.572. [Online]. Available: https://aclanthology.org/2021.emnlp-main.572. [Google Scholar]
- [100].Aly R, Vlachos A, and McDonald R, “Leveraging type descriptions for zero-shot named entity recognition and classification,” in Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (Volume 1: Long Papers), 2021, pp. 1516–1528. doi: 10.18653/v1/ 2021.acl-long.120. eprint: https://aclanthology.org/2021.acl-long.120.pdf. [Google Scholar]
- [101].Wright D. and Augenstein I, “Semi-Supervised Exaggeration Detection of Health Science Press Releases,” in Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing, Online and Punta Cana, Dominican Republic: Association for Computational Linguistics, Nov. 2021, pp. 10824–10836. doi: 10.18653/v1/2021.emnlp-main.845. [Online]. Available: https://aclanthology.org/2021.emnlp-main.845. [Google Scholar]
- [102].Lee D-H et al. , “Good Examples Make A Faster Learner: Simple Demonstration-based Learning for Low-resource NER,” in Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Dublin, Ireland: Association for Computational Linguistics, May 2022, pp. 2687–2700. doi: 10.18653/v1/2022.acllong.192. [Online]. Available: https://aclanthology.org/2022.acl-long.192. [Google Scholar]
- [103].Wang S. et al. , “Trustworthy assertion classification through prompting,” Journal of biomedical informatics, vol. 132, p. 104139, 2022. doi: 10.1016/j.jbi.2022.104139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Yan J. et al. , “Neuroimaging-ITM: A Text Mining Pipeline Combining Deep Adversarial Learning with Interaction Based Topic Modeling for Enabling the FAIR Neuroimaging Study,” Neuroinformatics, pp. 1–26, 2022. doi: 10.1007/s12021-022-09571-w. [DOI] [PubMed] [Google Scholar]
- [105].Lin S, Xu Z, Sheng Y, Chen L, and Chen J, “AT-NeuroEAE: A Joint Extraction Model of Events With Attributes for Research Sharing-Oriented Neuroimaging Provenance Construction,” Frontiers in neuroscience, vol. 15, 2021. doi: 10.3389/fnins.2021.739535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Riveland R. and Pouget A, “A neural model of task compositionality with natural language instructions,” bioRxiv, 2022. [Online]. Available: https://web.archive.org/web/20220424102915id_/https://www.biorxiv.org/content/biorxiv/early/2022/02/24/2022.02.22.481293.full.pdf. [Google Scholar]
- [107].Navarro DF, Dras M, and Berkovsky S, “Few-shot fine-tuning SOTA summarization models for medical dialogues,” in Proceedings of the 2022 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies: Student Research Workshop, 2022, pp. 254–266. doi: 10.18653/v1/2022.naacl-srw.32. eprint: https://aclanthology.org/2022.naacl-srw.32.pdf. [Google Scholar]
- [108].Das SSS, Katiyar A, Passonneau R, and Zhang R, “CONTaiNER: Few-Shot Named Entity Recognition via Contrastive Learning,” in Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Dublin, Ireland: Association for Computational Linguistics, May 2022, pp. 6338–6353. doi: 10.18653/v1/ 2022.acl-long.439. [Online]. Available: https://aclanthology.org/2022.acl-long.439. [Google Scholar]
- [109].Ma J. et al. , “Label Semantics for Few Shot Named Entity Recognition,” in Findings of the Association for Computational Linguistics: ACL 2022, Dublin, Ireland: Association for Computational Linguistics, May 2022, pp. 1956–1971. doi: 10.18653/v1/2022.findings-acl.155. [Online]. Available: https://aclanthology.org/2022.findings-acl.155. [Google Scholar]
- [110].Parmar M, Mishra S, Purohit M, Luo M, Mohammad M, and Baral C, “In-BoXBART: Get Instructions into Biomedical Multi-Task Learning,” in Findings of the Association for Computational Linguistics: NAACL 2022, Seattle, United States: Association for Computational Linguistics, Jul. 2022, pp. 112–128. doi: 10.18653/v1/2022.findings-naacl.10. [Online]. Available: https://aclanthology.org/2022.findings-naacl.10. [Google Scholar]
- [111].Boulanger H, Lavergne T, and Rosset S, “Generating unlabelled data for a tri-training approach in a low resourced NER task,” in Third Workshop on Deep Learning for Low-Resource Natural Language Processing, Association for Computational Linguistics, 2022, pp. 30–37. doi: 10.18653/v1/2022.deeplo-1.4. [Google Scholar]
- [112].Yeh H-S, Lavergne T, and Zweigenbaum P, “Decorate the Examples: A Simple Method of Prompt Design for Biomedical Relation Extraction,” in Proceedings of the Thirteenth Language Resources and Evaluation Conference, Marseille, France: European Language Resources Association, Jun. 2022, pp. 3780–3787. [Online]. Available: https://aclanthology.org/2022.lrec-1.403. [Google Scholar]
- [113].Pan X, Sheng A, Shimshoni D, Singhal A, Rosenthal S, and Sil A, “Task Transfer and Domain Adaptation for Zero-Shot Question Answering,” in Proceedings of the Third Workshop on Deep Learning for Low-Resource Natural Language Processing, Hybrid: Association for Computational Linguistics, Jul. 2022, pp. 110–116. doi: 10.18653/v1/2022.deeplo-1.12. [Online]. Available: https://aclanthology.org/2022.deeplo-1.12. [Google Scholar]
- [114].Wadden D, Lo K, Wang L, Cohan A, Beltagy I, and Hajishirzi H, “Multivers: Improving scientific claim verification with weak supervision and full-document context,” in Findings of the Association for Computational Linguistics: NAACL 2022, 2022, pp. 61–76. doi: 10.18653/v1/2022.findings-naacl.6. eprint: https://aclanthology.org/2022.findingsnaacl.6.pdf. [Google Scholar]
- [115].Zhenzhen L, Zhang Y, Nie J-Y, and Li D, “Improving Few-Shot Relation Classification by Prototypical Representation Learning with Definition Text,” in Findings of the Association for Computational Linguistics: NAACL 2022, 2022, pp. 454–464. doi: 10.18653/v1/2022. findings-naacl.34. eprint: https://aclanthology.org/2022.findings-naacl.34.pdf. [Google Scholar]
- [116].Zhang D. et al. , “Pairwise Supervised Contrastive Learning of Sentence Representations,” in Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing, Online and Punta Cana, Dominican Republic: Association for Computational Linguistics, Nov. 2021, pp. 5786–5798. doi: 10.18653/v1/2021.emnlp-main.467. [Online]. Available: https://aclanthology.org/2021.emnlp-main.467. [Google Scholar]
- [117].Devlin J, Chang M-W, Lee K, and Toutanova K, “BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding,” in Proceedings of the 2019 North American Chapter of the Association for Computational Linguistics: Human Language Technologies, 2019. eprint: https://www.aclweb.org/anthology/N19-1423.pdf. [Google Scholar]
- [118].Liu P, Yuan W, Fu J, Jiang Z, Hayashi H, and Neubig G, “Pre-train, prompt, and predict: A systematic survey of prompting methods in natural language processing,” ACM Computing Surveys, vol. 55, no. 9, pp. 1–35, 2023. doi: 10.1145/3560815. eprint: https://dl.acm.org/doi/pdf/10.1145/3560815. [DOI] [Google Scholar]
- [119].Ge Y, Guo Y, Yang Y−-C, Al–Garadi MA, and Sarker A, “A comparison of few-shot and traditional named entity recognition models for medical text,” in 2022 IEEE 10th International Conference on Healthcare Informatics (ICHI), IEEE, 2022, pp. 84–89. doi: 10. 1109/ICHI54592.2022.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Ma R. et al. , “Template-free prompt tuning for few-shot NER,” in Proceedings of the 2022 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Seattle, United States: Association for Computational Linguistics, Jul. 2022, pp. 5721–5732. doi: 10.18653/v1/2022.naacl-main.420. [Online]. Available: https://aclanthology.org/2022.naacl-main.420. [Google Scholar]
- [121].Bodenreider O, “The Unified Medical Language System (UMLS): integrating biomedical terminology,” Nucleic Acids Research, vol. 32, no. suppl-1, pp. D267–D270, Jan. 2004, issn: 0305–1048. doi: 10.1093/nar/gkh061.eprint: https://academic.oup.com/nar/article-pdf/32/suppl\_1/D267/7621558/gkh061.pdf. [Online]. Available: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Lu CJ, Payne A, and Mork JG, “The Unified Medical Language System SPECIALIST Lexicon and Lexical Tools: Development and applications,” Journal of the American Medical Informatics Association, May 2020, issn: 1067–5027. doi: 10.1093/jamia/ocaa056. [Online]. Available: https://academic.oup.com/jamia/advance-article/doi/10.1093/jamia/ocaa056/5848743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Feng SY et al. , “A Survey of Data Augmentation Approaches for NLP,” in Findings of the Association for Computational Linguistics: ACL-IJCNLP 2021, Online: Association for Computational Linguistics, Aug. 2021, pp. 968–988. doi: 10.18653/v1/2021.findings-acl.84. [Online]. Available: https://aclanthology.org/2021.findings-acl.84. [Google Scholar]
- [124].Nori H, King N, McKinney SM, Carignan D, and Horvitz E, “Capabilities of gpt-4 on medical challenge problems,” arXiv preprint arXiv:2303.13375, 2023. [Google Scholar]
- [125].Mikalef P, Conboy K, Lundstr JEöm, and A. Popovič, “Thinking responsibly about responsible AI and ‘the dark side’ of AI,” European Journal of Information Systems, vol. 31, no. 3, pp. 257–268, 2022. doi: 10.1080/0960085X.2022.2026621. eprint: https://www.tandfonline.com/doi/epdf/10.1080/0960085X.2022.2026621?needAccess=true&role=button. [DOI] [Google Scholar]
- [126].Theodorou A. and Dignum V, “Towards ethical and socio-legal governance in AI,” Nature Machine Intelligence, vol. 2, no. 1, pp. 10–12, 2020. doi: 10.1038/s42256-019-0136-y. [DOI] [Google Scholar]
- [127].Shokri R, Stronati M, Song C, and Shmatikov V, “Membership inference attacks against machine learning models,” in 2017 IEEE Symposium on Security and Privacy (SP), IEEE, 2017, pp. 3–18. doi: 10.1109/SP.2017.41. eprint: https://ieeexplore.ieee.org/stamp/stamp.jsp?tp=&arnumber=7958568. [DOI] [Google Scholar]
- [128].Research M. “Bucket of Me: Using Few-Shot Learning to Realize Teachable AI Systems.,” Youtube. (2022), [Online]. Available: www.youtube.com/watch?v=WpvOkIJjg-A. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the research articles reviewed have been listed in Table 1. There is no other data related to this work.