Abstract
Defective interfering (DI) particles have been found in many RNA and DNA viruses of bacteria, plants, and animals since their first discovery in influenza virus. However, this fundamental phenomenon has not been demonstrated in human natural infections. Using a new approach, here we provide the first experimental evidence for the existence of DI-like viruses in human chronic carriers of hepatitis B virus (HBV). Functional characterization of naturally occurring core internal deletion (CID) variants of HBV revealed all of the features of DI particles. When equal amounts of wild-type and CID variant DNAs were cotransfected into a human hepatoma cell line, Huh7, a three- to fivefold enrichment of CID variants was most often observed. The fluctuations of the virus populations between CID variants and helper HBV in three chronic carriers are reminiscent of the cycling phenomenon in other DI viral systems. This finding has important implications for chronic viral hepatitis and other chronic progressive viral diseases.
“Incomplete particles” were discovered during successive undiluted passages of the influenza viruses (39). In general, these incomplete particles contain a less than full-length genome and are replication defective. They can be rescued by, and interfere with, the replication of homologous helper viruses. Another important characteristic of incomplete particles is their ability to enrich their proportion in the total viral yield in cells infected with wild-type and incomplete viruses (reviewed in references 7, 14, and 30). Based on these properties, Huang and Baltimore defined these biologically active incomplete particles as defective interfering (DI) particles and the replication-competent homologous virions as standard viruses (19). DI particles are widespread among many DNA and RNA viruses in bacteria, plants, and animals. In tissue culture, DI viruses are capable of establishing persistent viral infections (14). In animal models, some DI viruses have been shown to modulate the course of disease by attenuating the virulence of standard viruses (3, 8, 35). Coinfection with DI RNA of tomato bushy stunt virus can modify the course of disease induced by the wild-type tomato bushy stunt virus infection (13). In humans, it has been proposed that DI particles might be responsible for the chronic recurrence of viral diseases. Perturbation of the balance between DI and standard viruses could trigger a new episode of disease manifestation (19).
Despite the extensive research on DI viruses, the molecular basis leading to a DI phenotype is often unclear. Most DI studies have demonstrated a correlation between a genomic deletion and the DI phenotype. There is no formal proof, beyond correlation, that a deletion is indeed the cause, entirely or in part, of the DI phenotype (7). To date, most, if not all, DI particles have been discovered in laboratory settings. Although human DI viruses have been found to occur during serial high-multiplicity-of-infection passages in tissue culture, it is not known if DI particles also exist in natural infections (3, 7, 14, 30).
HBV is one of the most common infectious agents in humans. However, the molecular and cellular mechanisms of pathogenesis and chronicity of HBV infection remain to be elucidated (33, 37). Recently, naturally occurring HBV variants containing a core antigen internal deletion (CID) were found to be geographically ubiquitous and highly prevalent in chronic HBV carriers (1, 16, 26, 40). These deletions found in CID mutants are often in frame, variable in size, map to the central portion of core antigen (HBcAg), and coincide with a known T-cell epitope (20, 24, 25, 38). Interestingly, CID variants have never been found in acute hepatitis patients (2, 9).
Many DI viruses are known to contain deletions in their structural protein genes (3, 7, 14, 30). Recently, we have demonstrated that HBV CID variants are replication defective and can be rescued by a wild-type core antigen expression vector (42). Although these properties of CID variants are similar to that of DI particles, it has been unclear if CID variants also have the interference and enrichment properties a DI particle is supposed to have. In this report, our results demonstrate that CID variants behave like the DI particles described in other viral systems (7, 14, 30).
MATERIALS AND METHODS
Plasmid constructs.
The expression vector for wild-type HBV (pWT) has been described as pSV2NeoHBV2x elsewhere (32). To construct pDEL85 and pDEL109, DNA fragments from nucleotides (nt) 1636 to 2688 containing the HBV deleted core gene were PCR amplified from total DNA of hepatoma samples T85 and T109 (16). The two oligonucleotide primers used in PCR amplification for T85 and T109 are as follows. One primer is a 30-mer (5′-A AGG GCA AAT ATT TGG TAA GGT TAG GAT AG-3′) containing HBV minus-strand DNA sequences from nt 2659 to nt 2688. The other primer is a 27-mer (5′-AGA AAT ATT GCC CAA GGT CTT ACA TAA-3′) containing HBV plus-strand DNA sequences from nt 1636 to nt 1659. The underlined sequences represent an SspI cleavage site used for subcloning into the wild-type expression vector. One microgram of tumor DNA and 100 ng of each primer were used in a 10-μl PCR mixture consisting of a denaturation step at 94°C (20 s) followed by a 40-cycle amplification at 94°C (1 s), 47°C (1 s), and 72°C (40 s). The amplified target sequence (0.9 kb) was subcloned into the pGEM-T vector (Promega Co.). The DNA fragments containing CID mutations were gel purified after digestion with SspI and used to replace both copies of the normal counterpart in the wild-type HBV genome carried on a pUC12-HBV tandem dimer plasmid.
Mutant TGAGC.
The HBV TGAGC mutant contains a G-to-A change at nt 1897, creating a TGA stop codon at precore antigen codon 28, and a G-to-C change at nt 1899. Both mutations affect the RNA encapsidation signal (E), and this mutant is thus replication defective and nonrescuable by transcomplementation (41).
Calcium phosphate cotransfection.
The human hepatoma cell lines Huh7 and HepG2 were maintained in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum at 37°C in the presence of 5.5% CO2. Approximately 2 × 106 to 3 × 106 Huh7 or HepG2 cells were seeded in each 10-cm-diameter dish 12 to 16 h before transfection (32). In each transfection, the total amount of donor DNA was kept constant (35 μg of DNA/2 × 106 to 3 × 106 cells/10-cm dish/transfection). Carrier DNA of Huh7 origin was used to adjust the final amount of donor DNA in each transfection to a constant amount of 35 μg total. Donor DNA was removed at approximately 6 h posttransfection, and cells were fed with fresh Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum.
DNA probes for Southern analysis. (i) Full-length HBV probe.
The full-length 3.1-kb HBV DNA fragment was purified from pWT by EcoRI digestion. Approximately 25 ng of the 3.1-kb DNA fragment was radiolabelled with a random-primed DNA labelling kit (Boehringer Mannheim Co.).
(ii) Wild-type-specific DNA probe.
A wild-type-specific DNA fragment of 135 nt (from nt 2141 to nt 2275) was synthesized by PCR with pWT as the DNA template. The oligonucleotides used to amplify the wild-type-specific fragment are both 21-mers (5′-TCT AGA GAC CTA GTA GTC AGT-3′ and 5′-CCA CAC TCC GAA AGA CAC CAA-3′).
(iii) DEL85-specific DNA probe.
The DEL85-specific DNA probe is 181 nt in length (from nt 2041 to nt 2365, with a deletion of 144 nt) and was synthesized by PCR with pDEL85 as a DNA template. The oligonucleotides used to amplify the DEL85-specific DNA fragment were 5′-GGA CCT GCC TCG TCG TCT AAC AAC AGT AGT-3′ and 5′-CAT TGT TCA CCT CAC CAT ACA-3′.
(iv) DEL109-specific DNA probe.
The DEL109-specific probe is 208 nt in length from nt 2041 to nt 2365, with a deletion of 123 nt, and was made with the same primers used for the DEL85-specific probe. These DNA fragments were radiolabelled by PCR with a reaction mixture containing 1 ng of DNA template; 200 μM dATP, dTTP, and dGTP; 10 μM dCTP; 150 μCi of [32P]dCTP (3,000 Ci/mmol [2 μM]); 100 ng of each primer; 2.5 U of TaqI polymerase; 4 mM MgCl2; and 10× PCR buffer (200 mM Tris-HCl, 500 mM KCl). The PCR procedure consisted of a denaturation step at 94°C (20 s) followed by a 40-cycle amplification step at 94°C (1 s), 55°C (1 s), and 72°C (40 s). Core particles and core-associated DNA from transfected cultures for Southern analysis were prepared as described elsewhere (41).
PCR analysis of core-associated DNA.
Either 1 ng of plasmid DNA or 1/20 of the core-associated DNA from 106 cells 5 days after transfection was used for PCR amplification. DNA transfection and purification of core-associated DNA were performed as described elsewhere (41). The oligonucleotide primers containing HBV consensus sequences that were used to amplify the core DNA fragment were described previously (16). The purification and PCR amplification of HBV DNA from the sera of patients were performed as described previously (16).
Calculation of the interference effects.
The interference effect was calculated by comparing the relative intensities of the wild-type population with and without cotransfection with CID variants. For example, in Fig. 1C, when equal amounts of pWT and pDEL85 were cotransfected, the signal intensity with wild-type-specific probe was 14 relative to 100 for pWT alone. We define the apparent interference effect as 7.2-fold (100/14) at an equal DNA mass ratio. However, since the molecular weight of pWT in pSV2Neo vector is approximately 12.6 kb while the pDEL85 in pUC12 vector is approximately 8.8 kb, the molar ratio between pDEL85 and pWT is approximately 1.4. Thus, when the 7.2-fold apparent effect is corrected by 1.4, the actual interference effect at the equal mass ratio is about 5.1-fold.
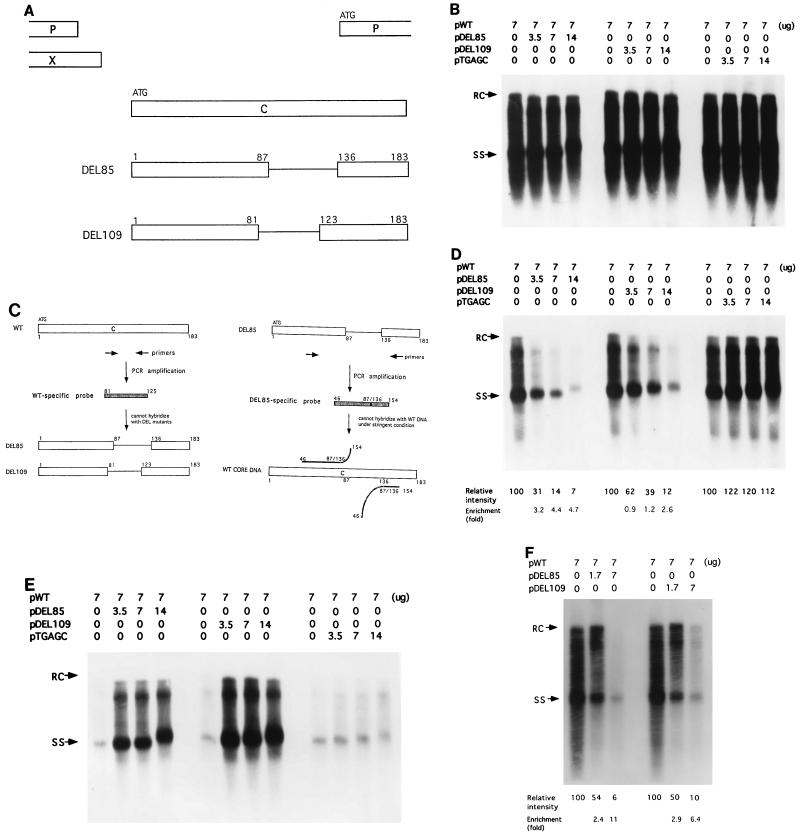
FIG. 1.
A DI-like phenomenon of HBV CID variants in human hepatoma cell lines Huh7 and HepG2. (A) In-frame deletions in the HBV core antigen of two different CID variants (Materials and Methods). DEL85 has a deletion of amino acids 88 to 135, while DEL109 has a deletion of amino acids 82 to 122. (B) Seven micrograms of wild-type HBV plasmid pWT was transfected into Huh7 cells or cotransfected with increasing amounts of pDEL85, pDEL109, or pTGAGC separately. Replicative viral DNAs from intracellular core particles were harvested 5 days after transfection and subjected to Southern blot analysis with the 3.1-kb full-length HBV DNA fragment as a probe. Relaxed-circular (RC) DNA at the 4.0-kb position and single-stranded (SS) DNA at the 1.5-kb position are indicated by arrows. The numbers at the top indicate the amount of plasmid DNA used for transfection. (C) The wild-type-specific and DEL mutant-specific probes used in panels D and E, respectively. (D) The same filter used in panel B was reprobed with a radiolabelled wild-type-specific DNA fragment (Materials and Methods), after removal of the 3.1-kb full-length HBV probe. The relative intensity of the replicative intermediates, as indicated at the bottom, was measured by scanning the entire lane below 4.0 kb by densitometer image analysis. Calculation of the interference and enrichment effects is described in Materials and Methods. (E) The nitrocellulose filter used in panel D was reprobed with a mixture of DEL85- and DEL109-specific DNA fragments (Materials and Methods). (F) The HepG2 human hepatoblastoma cell line was used in the same assay with a wild-type-specific probe as described for panel D.
Calculation of the enrichment effects and statistical analysis.
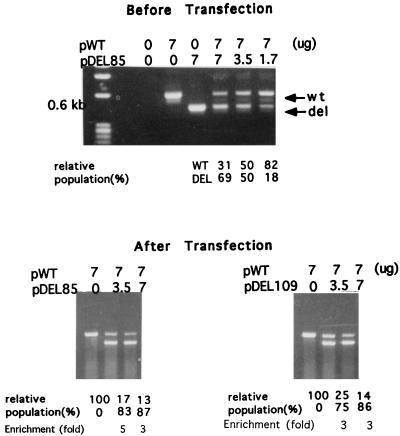
The calculation of the enrichment effect was done by comparing the signal ratios between the wild-type and CID variants before and after transfections (see Fig. 1 to 3). For example, in Fig. 1C, when equal amounts of pWT and pDEL85 were cotransfected, the signal intensity was 14 relative to 100 for pWT alone. Based on the results of Fig. 1B with the full-length HBV probe, the total signal intensity in the lane of pWT alone is comparable to that in the lane containing 7 μg each of pDEL85 and pWT. Therefore, it is reasonable to assume that the ratio between the de novo synthesized DEL85 and wild-type DNA in this cotransfection experiment on day 5 after transfection is approximately (100 − 14)/14 = 86/14 = 6. The molar ratio between the DEL85 (12.6 kb) and wild-type (8.8 kb) DNAs is about 1.4 as calculated above. The enrichment factor is calculated to be 6/1.4 = 4.4-fold. As another example, in Fig. 3, when 7 μg of pWT and 3.5 μg of pDEL85 were cotransfected, the apparent relative intensity of wild-type versus DEL85 donor DNA before transfection was equal to 1. However, 5 days after transfection, the relative intensity of core particle-associated DNAs between the deletion mutant and wild-type bands shifted to 83/17 = 5. The enrichment factor is calculated to be 5/1 = fivefold. (The amplified deletion mutant bands are present at a higher molarity than the donor DNA mass ratios would indicate, which is due in part to the preferential amplification over the wild-type band under these PCR conditions). Statistical analysis of the enrichment factors from four independent equal-dose (7 μg each) cotransfection experiments gives the following values: mean ± standard deviations of 7.90 ± 3.81 for DEL85 and 3.86 ± 2.60 for DEL109. The experiments included here for statistical analysis were arbitrarily chosen from a total of 12 independent repeat experiments.
FIG. 3.
Comparison of the relative abundance of wild-type (wt) and CID mutants (del) via PCR coamplification analysis with HBV core gene-specific primers (16). (Top) An aliquot of the premixed donor plasmid DNAs (pWT and pDEL85) was amplified by PCR before transfection. The results for pWT and pDEL109 (data not shown) are similar to those for pWT and pDEL85. (Bottom) Seven micrograms of pWT was cotransfected with increasing amounts of pDEL85 (left) or pDEL109 (right) into Huh7 cells, and core particle-associated DNAs were harvested 5 days after transfection. Identical PCR conditions were used for amplifications of both the plasmid and core particle-associated DNAs. The relative intensities of full-length nd deleted core gene fragments were measured by densitometric scanning.
HBV carriers.
The HBV serum sample we used was from a male hepatoma patient of Korean origin followed up at Fox Chase Cancer Center, Philadelphia, Pa., from 1989 to 1992. His serum alanine aminotransferase level during this period was only slightly above normal (50 to 80 IU/liter). Patient serum samples F090063 and F090245 were from HBV vaccine failure children identified in the Taiwanese HBV mass immunization program (17). To define a vaccine failure infection that results in HBsAg carriage, the patients should have received a hepatitis B immunization on schedule and subsequently should have become HBsAg positive during the follow-up period from 1986 to 1991.
RESULTS
One-filter, three-probe Southern blot analysis.
Two plasmids (pDEL85 and pDEL109) containing CID in their HBV genomes were constructed (Fig. 1A) (Materials and Methods). Both CID variants (pDEL85 and pDEL109) were replication defective when transfected into a human hepatoma cell line, Huh7. The defect in their DNA synthesis is correlated with the absence of a detectable HBcAg protein and can be rescued with a wild-type HBcAg expression vector (42). The rescued CID variant particles, which utilize the wild-type core protein, presumably are enveloped properly, since they can be secreted into the medium and band at a position similar to the mature wild-type Dane particles by gradient centrifugation analysis (42). To test the hypothesis that CID variants can function like DI particles, in addition to their replication defect and rescuability, one would need to demonstrate the phenomena of interference and enrichment that are characteristic of DI particles (7, 19, 30). When increasing amounts of CID variants were cotransfected with a constant amount of wild-type HBV, the replication activity of the total HBV population in cells did not change significantly, as measured by Southern blot analysis with the full-length HBV DNA as a probe (approximately 70 to 90% of the wild-type level) (Fig. 1B). To measure the proportional yields of wild-type and CID virus, we removed the full-length HBV probe from the filter of Fig. 1B and reprobed it with a wild-type-specific DNA fragment derived from part of the deleted region of the HBcAg gene in the CID mutant (Fig. 1C and D). Surprisingly, when CID variant and wild-type DNAs were cotransfected in equal amounts, wild-type HBV replication was reduced by seven- and threefold for DEL85 and DEL109, respectively (Fig. 1D). A packaging- and replication-defective HBV mutant TGAGC (41) was cotransfected with wild-type HBV DNA as a control for nonspecific effects that may arise from the squelching of limiting amounts of cellular transcription factors by excessive mutant HBV DNAs. Since mutant TGAGC contains a cis defect in the packaging signal, it cannot be rescued by the cotransfected wild-type virus. As shown in Fig. 1D, no apparent reduction of wild-type replication was observed when the wild type was cotransfected with the control mutant TGAGC. When the same filter was rehybridized with probes specific for DEL85 and DEL109, it became clear that HBV replication activity in the cotransfected culture was largely due to the replication of the CID variants (Fig. 1E). The CID-specific probes used here hybridized very weakly with the wild-type HBV DNA, since a very light signal was observed in the absence of CID variant DNA (Fig. 1E). The small amount of cross-hybridization under the stringent conditions used here is probably due to the presence of a 126-nt homology, from amino acids 46 to 87, between the CID-specific probe and the wild-type HBV DNA (Fig. 1C, right panel). In addition to Huh7 cells, the same result was observed with HepG2 cells in the cotransfection assay (Fig. 1F). A similar experiment was then performed to analyze the wild-type and CID variant virus populations in the media (Fig. 2A and B). The secreted CID variant viruses were found to be predominant over wild-type HBV, as was observed in the intracellular fraction (Fig. 1). Taken together, these results suggest that HBV CID variants behave like a DI virus which can replicate at the expense of helper virus and become the predominant viral population.
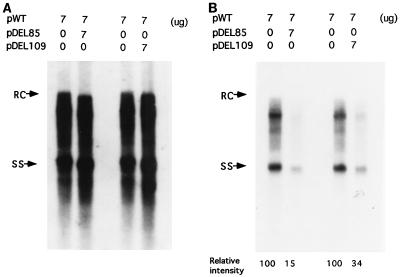
FIG. 2.
Secreted extracellular HBV particles showing the DI-like phenomenon analyzed by the replication assay. The conditioned media from the transfected culture were collected 5 days after transfection, and viral particles were pelleted through a 20% sucrose cushion. The DNA extracted from the extracelular viral particles was then subjected to Southern blot analysis. (A) The full-length 3.1-kb HBV DNA was used as a probe. (B) The same filter was reprobed with the wild-type-specific DNA fragment after removal of the probe used for panel A.
PCR coamplification analysis before and after cotransfection.
To confirm the results obtained by Southern blot analysis (Fig. 1 and 2), a PCR assay was used to measure directly the relative populations of wild-type helper virus and CID variants in the cotransfection experiment. When a mixture of the donor DNAs (pWT and pDEL85) in the 2:1 dose ratio was used in the PCR assay, the wild-type-specific DNA fragments exhibited an intensity similar to that of the DEL85-deleted fragment after amplification (Fig. 3, top). This is probably because the shorter (deleted) DNA fragments tend to be amplified favorably during PCR. However, the relative intensity between amplified wild-type and mutant DEL85 DNA fragments shifted from 1:1 (before cotransfection) to 1:5 (after cotransfection), suggesting the preferential de novo replication of CID variants over wild-type HBV in vivo (Fig. 3, bottom).
A cycling-like phenomenon of CID variants.
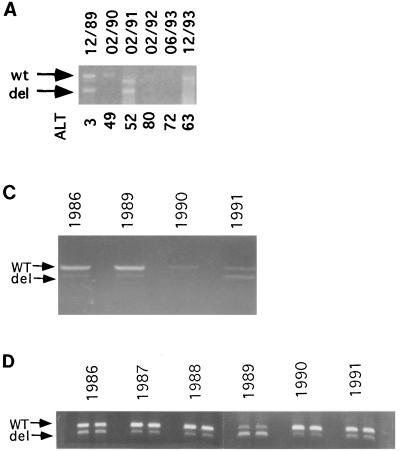
Previously, a cycling phenomenon between DI and helper virus populations has been reported in other viral systems in the laboratory setting (7, 19, 30). To look for a similar phenomenon in natural HBV infections, we collected serum samples from three chronic carriers during a longitudinal study and found the deleted core mutation in HBV by PCR (Fig. 4). The deletions in these DNA fragments were found to occur in the central part of HBcAg via sequencing. Furthermore, in the case of the Korean patient, the same CID variant population appeared to predominate during the 4-year follow-up period (Fig. 4B). The total HBV DNA titer was seen to fluctuate over time and sometimes even dropped to a very low or undetectable level (Fig. 4A and C). Interestingly, the relative abundance of CID variants and helper viruses also appeared to vary over time (Fig. 4). The relative intensity of helper virus-specific DNA was often greater than that of the CID variant DNA. However, at some time points, the reverse was observed (Fig. 4A, February 1991 [sample 02/1991]; C, 1991; D, 1989). It should be mentioned here that in Fig. 4A, C, and D, we are not measuring the absolute amount of the PCR product. Rather, we are comparing the ratios between the wild-type and CID variant populations at different time points. Therefore, the quantity of each band is internally controlled by the other band.
FIG. 4.
Fluctuation phenomenon of HBV wild-type (wt) and CID mutants (del) in serially collected serum samples from three chronic HBV carriers. These PCR data suggest a dynamic equilibrium between the CID mutants and helper HBV in vivo, where their relative abundance appears to vary over time. Purified DNAs from sera were used to amplify the core region by PCR (Materials and Methods) (16). Amplified DNA fragments, including the full-length (upper band) and deleted (lower band) core gene fragments, were separated by agarose gel electrophoresis. The dates of sampling are shown at the top. (A) Results for the Korean patient serum samples from December 1989 to December 1993. (B) Sequence analysis of the fluctuating CID variants in the Korean patient’s serum samples. Identical deletions are present in DEL85, which was derived from a Taiwanese patient, and the CID variants of the Korean patient. The letter “Z” represents a translational stop codon, and the letter “X” represents deletions. The slash symbol represents frameshift mutations. Subtype-specific sequence heterogeneity is depicted by an asterisk. The boxed regions of HBcAg mutational domains IV and V were described in reference 16. (C) Results for serum from patient F090245 from 1986 to 1991. Serum samples from 1987 and 1988 were not available. (D) Results for serum from patient F090063 from 1986 to 1991. Each sample was PCR amplified and then gel analyzed in duplicate.
These results indicate that a dynamic equilibrium could exist between the CID mutants and helper HBV in vivo. This observed waxing and waning of DI-like and helper viruses are reminiscent of the cycling phenomenon reported in other DI viral systems, such as rabies virus and vesicular stomatitis virus (21, 29).
DISCUSSION
HBV CID variants exhibit a strong DI phenotype.
We have characterized two different CID mutants (DEL85 and DEL109) isolated from two different patients with two different hepatoma cell lines (Huh7 and HepG2). Cytotoxicity caused by calcium phosphate during transfection cannot be an explanation for interference because of the simultaneous enrichment of the CID variants. In addition, when duck hepatitis B virus (DHBV) DNA was included in the transfection experiment as a control, no interference with DHBV replication by DEL85 was observed (data not shown). Thus, species-specific interference and enrichment cannot be explained by nonspecific cytotoxicity. Our results led to the conclusion that CID variants have all of the characteristics of DI particles: they have a deleted genome, are replication defective, are rescuable by standard helper virus, interfere with production of standard virus, and have a relative enrichment of DI particles compared with standard virus.
Quantitative aspects of interference and enrichment.
While the interference effect in a single cycle of transfection is apparent at a 1:1 donor DNA dose ratio between wild-type and CID variants, increasing the dose of the CID variant DNA results in a more obvious interference effect. It is therefore informative if we can directly measure the extent of increase of DI particles (i.e., enrichment), in addition to measuring the decrease of helper viruses (i.e., interference). In general, direct measurement of DI particles has been difficult because it is hard to separate DI particles from their helper viruses, and there have been no convenient or direct assays for DI particles. Here, we demonstrated the direct measurement of DI particles by comparing the ratios between helper virus and CID variants before and after transfection (Fig. 1, 2, and 3). According to our calculations (Materials and Methods), DEL85 has an average enrichment effect of 3- to 5-fold in Huh7 cells (Fig. 1D and 3) and up to 11-fold in HepG2 cells (Fig. 1F). Unlike DEL85, DEL109 has a less-pronounced enrichment effect (1.2- to 3.0-fold) in Huh7 cells (Fig. 1D and 3). However, the enrichment effect of DEL109 became more enhanced, up to 6.4-fold, when the HepG2 cells were used (Fig. 1F). It is also worth pointing out that the weaker enrichment effect of DEL109 in Huh7 cells in Fig. 1D was in part caused by the experimental variations. A mean of 3.9-fold of enrichment was obtained for DEL109 at an equal dose ratio in Huh7 cells (Materials and Methods). The approximately two- to threefold difference in the enrichment effect obtained from Huh7 and HepG2 cells is consistent with the previous reports that host cells can play an important role in the DI effect (14). We speculate that the difference in interference and enrichment between DEL85 and DEL109 could be related to the different sizes of their deletions. Finally, it is worth mentioning that enrichment and interference properties are not always coupled. Some DI particles of other viruses exhibited interference but no apparent enrichment effect (14, 29, 30). In this regard, we tentatively consider DEL109 to be DI-like as well, because it replicates at the expense of wild-type virus and displays a defective interfering phenotype. In summary, the interference and enrichment effects vary, depending on the specific DI variant, the host cell lines, the specificity of the probes, and the relative doses of DI and helper viruses used in the assay. The effects observed in a single-cycle experiment in vitro by transfection could, in theory, expand exponentially during sequential cycles of infection in vivo.
The long-sought DI virus in nature.
DI viruses in previous studies were always generated by either brief or continuous passages in tissue culture, chicken eggs, or animal models in a laboratory setting (3, 14, 30). Physically defective genomes of viruses have been found in clinical specimens. However, direct functional proof that the same defective viruses are indeed replication defective, interfering, or able to enrich themselves has not been provided (28, 34). According to the standard definition of DI particles (19), structurally defective viruses are not necessarily interfering functionally. The demonstration here of the CID mutants of HBV in human chronic carriers is the first example of DI-like particles in the family Hepadnaviridae, which replicates its DNA genome through an RNA intermediate (36). More importantly, this is, to the best of our knowledge, the first report of DI-like particles found in natural infections (3, 14, 30).
The one-filter, three-probe method: a new approach.
Previous approaches to identifying DI particles mainly relied on the plaque assay in tissue culture or passage in animal models. At present, there is neither a plaque assay nor a reliable in vitro infection system for HBV (11). Another major difficulty in conventional DI virus research is in the separation of DI particles from standard viruses. Although DI particles of vesicular stomatitis virus can be separated from their standard viruses (6, 18), this is more of an exception than a rule compared to other viruses. In addition, the DI population could be rather heterogeneous in an individual animal in some viral systems (3). This also contributes to the variability of DI particles from preparation to preparation. To circumvent these classic problems, we developed a new approach for the study of DI virus. Using a novel combination of methods, including gene cloning, cotransfection, and the one-filter, three-probe Southern assay (Fig. 1), we demonstrated here that mutations occurring in natural human HBV infections can result in a DI-like phenotype.
Mechanisms of enrichment and interference.
Previous studies of other DI viruses have established a correlation between the deletion genotype and the DI phenotype (7, 14). However, no cause-effect relationship has been demonstrated between the deletion and the DI phenomenon. In our study, we can attribute the entire DI phenomenon to a specific CID mutation alone. In the case of HBV CID mutants, it also remains unclear if the mechanisms of enrichment and interference in viral replication are related or independent. If they are independent, are they caused by two or more separate mutations or by a single pleiotropic mutation? When the nucleotide sequences of the CID mutation-containing DNA fragments of DEL85 and DEL109 were compared (data not shown), the only common mutations between these two different CID mutants were the core internal deletion as well as an A→T change and a G→A change at nt 1762 and 1764, respectively (referred to as TA mutations). These TA mutations occurred within the basal core promoter/X gene (27). A recent report suggested that TA mutants are replication competent and might replicate slightly better than the wild type (4). The modest effect of TA mutations on the replication of HBV cannot explain the dramatic interference, which can be up to a 16-fold effect (Fig. 1F). Whether these TA mutations in the context of CID mutants can contribute to the DI phenotype awaits further investigation.
The enrichment and interference phenomenon of CID mutants could result from a difference in response to possible DI-induced interferon-like soluble factors, to which the wild-type virus is perhaps more sensitive (10, 23). Inconsistent with this hypothesis, addition of the conditioned media from CID mutant-transfected culture to the wild-type HBV-transfected culture did not reduce wild-type HBV replication (data not shown). As mentioned above, when DHBV DNA was cotransfected with human CID mutants, no apparent decrease in DHBV DNA replication was observed (data not shown). Therefore, the interference effect from HBV CID mutants appears to be homotypic, species specific, and unlikely to be related to interferon-like factors.
Dominant negative mutants of hepadnaviruses have been artificially created previously (15, 31). Although these dominant negative mutants can interfere with the replication of wild-type virus, they are not able to enrich themselves at the expense of the wild type. Therefore, it is theoretically unlikely that the DI phenomenon is mediated through a simple dominant negative effect. Consistent with this hypothesis, cotransfection of wild-type HBV with an expression vector encoding the CID mutant core protein does not affect wild-type HBV replication (42). Taken together, this result and the unstable nature of the deleted core protein (42) suggest that the interference by CID variants cannot be explained by a dominant negative effect of the deleted core protein.
In summary, the DI phenotype of HBV CID mutants appears to be caused mainly by a single deletional mutation. It is tempting to speculate that interference by CID mutants is secondary to enrichment. In other studies of DI viruses, the advantage of DI over standard viruses appears to be related to more efficient replication and packaging of DI particles (22). The exact nature of the competitive edge of the HBV-CID variants over the wild type remains to be investigated.
Cycling-like phenomenon of CID variants in patients.
In the sample dated December 1989 (12/89) of Fig. 4A, a minor band appeared to migrate slightly faster than the wild-type band. We have not yet cloned and sequenced this minor band. However, it is common that the CID variants from the same individual can contain more than one predominant size of deletions. For example, in Fig. 1B of reference 42, CID variants of samples T85 and T109 also contain two predominant sizes of deletions. Since CID variants have to depend on the helper virus for replication, it is worth investigating why the wild-type band of helper virus in sample 02/91 of Fig. 4A was not apparent by ethidium bromide staining. When we cloned DNA from where the wild-type band would migrate, we obtained nondeleted core gene sequences (Fig. 4B). This result demonstrates that the wild-type DNA in sample 02/91 indeed is present, albeit at lower abundance. In other DI virus systems in vitro, DI particles can outnumber full-length-genome particles by large amounts. Our in vivo data from sample 02/91 would appear to be consistent with previous findings from other DI viruses in vitro.
As shown in Fig. 4B, the in vivo cycling of variants in the Korean patient happens to contain a predominant CID variant population which shares an identical deletion with DEL85 and two other CID variants independently identified by two other research groups (Fig. 5). Despite the identical CID deletion in the core region, DEL85 and the CID variants in the Korean patient are different from each other in the nondeleted portion by a total of six amino acids (data not shown). Thus, PCR contamination between DEL85 and the Korean sample is highly unlikely. In summary, the behavior of the cloned variant DEL85 probably can be directly related to the in vivo cycling data. Finally, according to the definition of cycling, the ratio between helper and DI particles does not remain constant. Depending on the timing of the sampling, the ratio between the helper and DI viruses is expected to vary.
FIG. 5.
The same CID deletion as DEL85 was also observed in serum samples from a British patient (23a), a patient from Hong Kong (1b), and a Korean patient (Fig. 4B) (also see Addendum in Proof).
It should also be noted that while both HepG2 and Huh7 cell lines are fully permissive for transfection and intracellular HBV replication, they are not permissive for extracellular viral infection. Thus, it would be interesting to see if CID variants also exhibit the DI phenotype in an in vitro infection system, when it is established. Furthermore, it remains to be demonstrated that HBV CID variants can contribute to the persistent infection of HBV in chronic carriers by attenuating the replication of wild-type helper viruses. Finally, it will be interesting in the future to see if there is a correlation between the clinical spectrum of chronic HBV infection (5) and the fluctuating patterns of HBV DI-like variants in patients. Perhaps a similar phenomenon can be found in other virus-associated chronic progressive diseases, such as subacute sclerosing panencephalitis and AIDS (12).
ACKNOWLEDGMENTS
We thank D. Walker, S. Baron, W. T. London, R. Goldblum, and colleagues in C. Shih’s laboratory for careful reading of the manuscript. We are grateful to A. O’Connell and W. T. London for providing the serum samples of the Korean patient. We acknowledge invaluable serum samples from the Hepatitis Control Committee, Department of Health, Executive Yuan, Taiwan (ROC).
This work was supported in part by Public Health Service grant RO1 CA70336 to C.S. from the National Institutes of Health. C.S. is a recipient of an NIH Research Career Development Award.
ADDENDUM IN PROOF
A CID deletion identical to our DEL85 and to several clones from the Korean patient (Fig. 4B) was also observed by W. Mason, T. London, and A. Evans at the Fox Chase Cancer Center, Philadelphia, Pa. (personal communication).
REFERENCES
- 1.Ackrill A M, Naoumov N V, Eddleston A L W F, Williams R. Specific deletions in the hepatitis B virus core open reading frame in patients with chronic active hepatitis B. J Med Virol. 1993;41:165–169. doi: 10.1002/jmv.1890410213. [DOI] [PubMed] [Google Scholar]
- 1a.Akarca U S, Lok A S F. Naturally occurring core-gene-defective hepatitis B viruses. J Gen Virol. 1995;76:1821–1826. doi: 10.1099/0022-1317-76-7-1821. [DOI] [PubMed] [Google Scholar]
- 2.Aye T T, Uchida T, Becker S O, Hirashima M, Shikata T, Komine F, Moriyama M, Arakawa Y, Mima S, Mizokami M, Lau J Y N. Variations of hepatitis B virus precore/core gene sequence in acute and fulminant hepatitis B. Dig Dis Sci. 1994;39:1281–1287. doi: 10.1007/BF02093794. [DOI] [PubMed] [Google Scholar]
- 3.Barret A D T, Dimmoch N J. Defective interfering virus and infections of animals. Curr Top Microbiol Immunol. 1986;128:55–84. doi: 10.1007/978-3-642-71272-2_2. [DOI] [PubMed] [Google Scholar]
- 4.Buckwold V E, Xu Z, Chen M, Yen T S B, Ou J-H. Effects of a naturally occurring mutation in the hepatitis B virus basal core promoter on precore gene expression and viral replication. J Virol. 1996;70:5845–5851. doi: 10.1128/jvi.70.9.5845-5851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen D S. From hepatitis to hepatoma: lessons from type B viral hepatitis. Science. 1993;262:369–370. doi: 10.1126/science.8211155. [DOI] [PubMed] [Google Scholar]
- 6.Crick J, Cartwright B, Brown F. Interfering components of vesicular stomatitis virus. Nature. 1966;211:1204–1205. doi: 10.1038/2111204a0. [DOI] [PubMed] [Google Scholar]
- 7.Dimmock N J. Antiviral activity of defective interfering influenza virus in vivo. In: Myint S, Taylor-Robinson D, editors. Viral and other infections of the human respiratory tract. London, England: Chapman 8 Hall, Inc.; 1996. pp. 421–445. [Google Scholar]
- 8.Doyle M, Holland J J. Prophylaxis and immunization in mice by use of virus-free defective T particles to protect against intracerebral infection by vesicular stomatitis virus. Proc Natl Acad Sci USA. 1973;70:2105–2108. doi: 10.1073/pnas.70.7.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehata T, Omata M, Chuang W L, Yokosuka O, Ito Y, Hosoda K, Ohto M. Mutations in core nucleotide sequence of hepatitis B virus correlate with fulminant and severe hepatitis. J Clin Invest. 1993;91:1206–1213. doi: 10.1172/JCI116281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fultz P N, Shadduck J A, Kang C-Y, Streilein J W. Mediators of protection against lethal systemic vesicular stomatitis virus infection in hamsters: defective interfering particles, polyinosinate-polycytidylate, and interferon. Infect Immun. 1982;37:679–686. doi: 10.1128/iai.37.2.679-686.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gripon P, Diot C, Guguen-Guillouzo C. Reproducible high level infection of cultured adult human hepatocytes by hepatitis B virus: effect of polyethylene glycol on adsorption and penetration. Virology. 1993;192:534–540. doi: 10.1006/viro.1993.1069. [DOI] [PubMed] [Google Scholar]
- 12.Haywood A M. Patterns of persistent viral infections. N Engl J Med. 1986;315:939–948. doi: 10.1056/NEJM198610093151506. [DOI] [PubMed] [Google Scholar]
- 13.Hillman B I, Carrington J C, Morris T J. A defective interfering RNA that contains a mosaic of plant virus genome. Cell. 1987;51:427–433. doi: 10.1016/0092-8674(87)90638-6. [DOI] [PubMed] [Google Scholar]
- 14.Holland J J. Defective interfering rhabdoviruses. In: Wagner R, editor. The rhabdoviruses. New York, N.Y: Plenum Publishing Co.; 1987. p. 297. [Google Scholar]
- 15.Horwich A L, Furtak K, Pugh J, Summers J. Synthesis of hepadnavirus particles that contain replication-defective duck hepatitis B virus genome in cultured Huh7 cells. J Virol. 1990;64:642–650. doi: 10.1128/jvi.64.2.642-650.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosono S, Tai P C, Wang W, Ambrose M, Hwang D, Yuan T T, Peng B H, Yang C S, Lee C S, Shih C. Core antigen mutations of human hepatitis B virus in hepatomas accumulate in MHC class II-restricted T cell epitopes. Virology. 1995;212:151–162. doi: 10.1006/viro.1995.1463. [DOI] [PubMed] [Google Scholar]
- 17.Hsu H M, Chen D S, Chuang C H, et al. Efficiency of a mass hepatitis B vaccination program in Taiwan. JAMA. 1988;260:2231–2235. [PubMed] [Google Scholar]
- 18.Huang A S, Greenwalt J W, Wagner R R. Defective T particles of vesicular stomatitis virus. I. Preparation, morphology, and some biologic properties. Virology. 1966;30:161–172. doi: 10.1016/0042-6822(66)90092-4. [DOI] [PubMed] [Google Scholar]
- 19.Huang A S, Baltimore D. Defective viral particles and viral disease processes. Nature. 1970;226:325–327. doi: 10.1038/226325a0. [DOI] [PubMed] [Google Scholar]
- 20.Jung M C, Diepolder H M, Spengler U, Wierenga E A, Zachoval R, Hoffmann R M, Eichenlaub D, Fro̊sner G, Will H, Pape G R. Activation of a heterogeneous hepatitis B (HB) core and e antigen-specific CD4+ T-cell population during seroconversion to anti-HBe and anti-HBs in hepatitis B virus infection. J Virol. 1995;69:3358–3368. doi: 10.1128/jvi.69.6.3358-3368.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawai A, Matsumoto S, Tanabe K. Characterization of rabies viruses recovered from persistently infected BHK cells. Virology. 1975;67:520–523. doi: 10.1016/0042-6822(75)90452-3. [DOI] [PubMed] [Google Scholar]
- 22.Levis R, Weiss B G, Tsiang M, Huang H, Schlesingers S. Deletion mapping of Sindbis virus DI RNAs derived from cDNAs defines the sequences essential for replication and packaging. Cell. 1986;44:137–145. doi: 10.1016/0092-8674(86)90492-7. [DOI] [PubMed] [Google Scholar]
- 23.Marcus P I, Gaccione C. Interferon induction by viruses. Vesicular stomatitis virus—New Jersey: high multiplicity passages generate interferon-inducing, defective-interfering particles. Virology. 1989;171:630–633. doi: 10.1016/0042-6822(89)90637-5. [DOI] [PubMed] [Google Scholar]
- 23a.Marinos G, Torre F, Gunther S, Thomas M G, Will H, Williams G, Naoumov N V. Hepatitis B virus variants with core gene deletions in the evolution of chronic hepatitis B infection. Gastroenterology. 1996;111:183–192. doi: 10.1053/gast.1996.v111.pm8698197. [DOI] [PubMed] [Google Scholar]
- 24.Menne S, Maschke J, Tolle T K, Lu M, Roggendorf M. Characterization of T-cell response to woodchuck hepatitis virus core protein and protection of woodchucks from infection by immunization with peptides containing a T-cell epitope. J Virol. 1997;71:65–74. doi: 10.1128/jvi.71.1.65-74.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milich D R, McLachlan A, Moriarty A, Thornton G B. Immune response to hepatitis B virus core antigen (HBcAg): localization of T cell recognition sites within HBcAg/HBeAg. J Immunol. 1987;139:1223–1231. [PubMed] [Google Scholar]
- 26.Okamoto H, Tsuda F, Mayumi M. Defective mutants of hepatitis B virus in the circulation of symptom-free carriers. Jpn J Exp Med. 1987;57:217–222. [PubMed] [Google Scholar]
- 27.Okamoto H, Tsuda F, Akahane Y, Sugai Y, Yoshiba M, Moriyama K, Tanaka T, Miyakawa Y, Mayumi M. Hepatitis B virus with mutations in the core promoter for an e antigen-negative phenotype in carriers with antibody to e antigen. J Virol. 1994;68:8102–8110. doi: 10.1128/jvi.68.12.8102-8110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pedley S, Hundley F, Chrystie I, McCrae M A, Desselberger U. The genomes of rotaviruses isolated from chronically infected immunodeficient children. J Gen Virol. 1984;65:1141–1150. doi: 10.1099/0022-1317-65-7-1141. [DOI] [PubMed] [Google Scholar]
- 29.Roux L, Holland J J. Viral genome synthesis in BHK 21 cells persistently infected with Sendai virus. Virology. 1980;100:53–64. doi: 10.1016/0042-6822(80)90551-6. [DOI] [PubMed] [Google Scholar]
- 30.Roux L, Simon A E, Holland J J. Effects of defective interfering viruses on virus replication and pathogenesis in vitro and in vivo. Adv Virus Res. 1991;40:181–211. doi: 10.1016/S0065-3527(08)60279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scaglioni P P, Melegari M, Wands J R. Characterization of hepatitis B virus core mutants that inhibit viral replication. Virology. 1994;205:112–120. doi: 10.1006/viro.1994.1625. [DOI] [PubMed] [Google Scholar]
- 32.Shih C, Li L S, Roychoudhury S, Ho M H. In vitro propagation of human hepatitis B virus in a rat hepatoma cell line. Proc Natl Acad Sci USA. 1989;86:6323–6327. doi: 10.1073/pnas.86.16.6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shih C, Tai P-C, Whitehead W, Hosono S, Lee C-S, Yang C-S. Hepatitis B and C viruses and liver cancer. In: Bertino J R, editor. Encyclopedia of cancer. II. New York, N.Y: Academic Press, Inc.; 1996. pp. 824–834. [Google Scholar]
- 34.Siegl G, Nüesch J P F, de Chastonay J. Defective interfering particles of hepatitis A virus in cell cultures and clinical specimens. In: Brinton M A, Heinz F X, editors. New aspects of positive-strand RNA viruses. Washington, D.C: American Society for Microbiology; 1990. pp. 102–107. [Google Scholar]
- 35.Spandidos D A, Graham A F. Generation of defective virus after infection of newborn rats with reovirus. J Virol. 1976;20:234–247. doi: 10.1128/jvi.20.1.234-247.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Summers J, Mason W S. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 37.Tiollais P, Pourcel C, Dejean A. The hepatitis B virus. Nature. 1985;317:489–495. doi: 10.1038/317489a0. [DOI] [PubMed] [Google Scholar]
- 38.Tsai S L, Chen M H, Yeh C T, Chu C M, Lin A N, Chiou F H, Chang T H, Liaw Y F. Purification and characterization of a naturally processed hepatitis B virus peptide recognized by CD 8 cytotoxic T lymphocytes. J Clin Invest. 1996;97:577–584. doi: 10.1172/JCI118450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Magnus P. Studies on interference in experimental influenza. I. Biological observations. Ark Kemi Mineral Geol. 1947;24:1. [Google Scholar]
- 40.Wakita T, Kakumu S, Shibata M, Yoshioka K, Ito Y, Shinagawa T, Ishikawa T, Takayanagi M, Morishima T. Detection of pre-c and core region mutants of hepatitis B virus in chronic hepatitis B virus carriers. J Clin Invest. 1991;88:1793–1801. doi: 10.1172/JCI115500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan T T, Faruqi A, Shih J W K, Shih C. The mechanism of natural occurrence of two closely-linked HBV precore predominant mutations. Virology. 1995;211:144–156. doi: 10.1006/viro.1995.1387. [DOI] [PubMed] [Google Scholar]
- 42.Yuan, T. T.-T., M.-H. Lin, S. M. Qiu, and C. Shih. Functional characterization of naturally-occurring variants of human hepatitis B virus containing the core internal deletion mutation. J. Virol., in press. [DOI] [PMC free article] [PubMed]