Abstract
Pancreatic cancer will soon become the second cause of death by cancer in Western countries. The main barrier to increase the survival of patients with this disease requires the development of novel and efficient therapeutic strategies that better consider tumor biology. In this context, oncolytic viruses emerge as promising therapeutics. Among them, the fibrotropic minute virus of mice prototype (MVMp) preferentially infects migrating and undifferentiated cells that highly resemble poorly differentiated, basal-like pancreatic tumors showing the worst clinical outcome. We report here that MVMp specifically infects, replicates in, and kills pancreatic cancer cells from murine and human origin with a mesenchymal, basal-like profile, while sparing cancer cells with an epithelial phenotype. Remarkably, MVMp infection, at a dose that does not provoke tumor growth inhibition in athymic mice, shows significant antitumoral effect in immune-competent models; extended mouse survival; and promoted the massive infiltration of tumors by innate, myeloid, and cytotoxic T cells that exhibit a less terminally exhausted phenotype. Collectively, we demonstrate herein for the first time that MVMp is specific and oncolytic for pancreatic tumors with mesenchymal, basal-like profile, paving the way for precision-medicine opportunities for the management of the most aggressive and lethal form of this disease.
Keywords: MT: Regular Issue, oncolytic virus, pancreatic cancer, MVMp, mesenchymal phenotype, targeted virotherapy, immune mobilization
Graphical abstract

Cordelier and colleagues describe the selectivity of the minute virus of mice oncolytic virus for pancreatic cancer cells with a mesenchymal phenotype, which leads to tumor growth inhibition and innate and adaptive immune engagement in mice models of cancer.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the most common form of pancreatic cancer and has often been viewed as a disease with a poor prognosis.1 Over the last decade, multiple groups have characterized the complex molecular landscape of PDAC to reveal several distinct classes of disease.2 Two main molecular classifications emerged, namely a classical subtype and a basal-like gene expression program that resemble the basal subtype of breast and bladder cancers, with the basal-like subtype showing the poorer clinical outcome.3,4,5 However, the question remains as to how the recent progress in molecular stratification may translate into tangible benefit for patients with PDAC. With this in mind, we asked whether clinically relevant PDAC molecular subtypes may respond differently to innovative therapies such as oncolytic virotherapy.
Oncolytic virus (OV) exploits the frequent defects in antiviral signaling in cancer cells to prosper and kill cancer cells, leaving normal cells unharmed.6 Virotherapy is now recognized as a promising multi-mechanistic therapy for the treatment of cancer, with the first OV product, talimogene laherparepvec (T-Vec, Imlygic) approved by the US FDA in 2015 for the treatment of advanced melanoma.7,8 OVs come in different sizes and flavors; among them, the minute virus of mice prototype (MVMp) is a non-enveloped, small, single-stranded DNA virus with an icosahedral capsid of 25 nm.9 MVMp is member of the Protoparvovirus genus, is nonpathogenic in humans, and has potential utility as cancer therapeutic in experimental models of glioma.10
MVMp tropism for cancer cells is still not well understood. The role of type-I interferons in limiting MVMp infection is highly debated11,12; interestingly, as antiviral signaling is commonly silenced in PDAC cells,13 this may provide favorable ground for MVMp replication. Recent works demonstrate the role of sialic acid structures14 and galectin-315 in MVMp cell surface binding and entry. Remarkably, MVMp infection is promoted by epithelial-mesenchymal transition (EMT),16 a situation that closely resemble basal-like phenotype of PDAC tumors, as MVMp preferably enters the leading edge of migrating cells by endocytosis close to filopodial and focal adhesions areas.16 During this work, we asked whether PDAC cells with basal-like, mesenchymal phenotype might be target for MVMp infection. We identified a five-gene classifier (FAK, N-cadherin, E-cadherin, and β-catenin) that predicts MVMp cytotoxic activity in PDAC cells from murine and human origin with mesenchymal profile, when cells with a more epithelial phenotype were spared by the virus. MVMp tropism was maintained in vivo, as MVMp replicated in, inhibited the progression, and induced cell death by apoptosis of PDAC experimental tumors with mesenchymal phenotype only. Last, we found that MVMp partners with the immune system to inhibit PDAC growth, with evidence of adaptive immunity engagement. Collectively, we demonstrate here for the first time that MVMp is oncolytic for the most aggressive form of PDAC, paving the way for new precision-medicine opportunities for the therapy of this disease.
Results
Isolation of mesenchymal and epithelial subtypes of pancreatic cancer cells
Recent studies demonstrate that basal-like and classical cell types coexist within PDAC.17 To interrogate how these subtypes respond to virotherapy, we generated long-term cultures of murine R211 and R259 PDAC cells that derive from a KRAS P53 Cre (KPC) spontaneous mouse model of cancer (Pdx1- Cre, lox-stop-lox-KrasG12D/+, lox-stop-lox-tp53R172H/+).18 We identified clusters of differentiated cells with an epithelial phenotype coexisting with fibroblast-like, less-differentiated cells, highly resembling human tumors (Figures 1A and S1A). Cells from each phenotype were separated by differential adhesion to plastic and thereafter were named R211-M and R259-M for mesenchymal and R211-E and R259-E for epithelial (Figures 1A and S1A). We then performed western blot analysis for markers of EMT (FAK, N-cadherin, E-cadherin, β-catenin, Slug, Snail) and used β-actin and GAPDH as loading controls. Molecular exploration demonstrates that mesenchymal markers, such as FAK, N-cadherin, and Snail, are expressed at high levels in R211-M as compared to R211-E cells (Figure 1B). Conversely, epithelial markers such as E-cadherin and β-catenin are absent in cells with the mesenchymal phenotype but expressed at high levels in cells with the epithelial phenotype (Figure 1B). We next addressed the ability of these cultures to form tumors in immune-deficient mice. As shown in Figures 1C and 1D, engraftment in the pancreas of both R211-M and R211-E led to robust tumor formation, confirming the neoplastic nature of these cultures, with R211-M tumors growing faster than R211-E tumors. Macroscopic analysis of tumors at endpoint reveals that both phenotypes are conserved within experimental tumors, with R211-M and R259-M cells giving rise to poorly differentiated PDAC with extensive desmoplastic stroma, and R211-E and R259-E cells at the origin of moderately differentiated PDAC embedded in desmoplastic stroma (Figures 1E and S1B).
Figure 1.
Isolation of mesenchymal and epithelial subtypes of murine pancreatic cancer cells
R211-E and R211-M cells were isolated by differential adherence to plastic as mentioned in the “materials and methods” section. (A) Representative images of long-term culture of R211 cells (top) and of isolated R211-E (middle) and R211-M (bottom) cells. (B) Western blot analysis for the indicated proteins in LA9, parental R211, R211-E, and R211-M cells. Representative of two independent experiments. Representative images of tumor echography (C) and tumor growth kinetics (D) analyzed by echography of R211-M and R211-E tumors engrafted in NSG mice. N = 3–8 mice per group. (E) Hematoxylin and eosin (H&E) staining of orthotopic R211-E and R211-M tumors engrafted in nude mice at endpoint. Representative of n = 3 mice per group.
Murine pancreatic cancer cells with mesenchymal phenotype are preferential targets for MVMp infection
We next addressed the permissiveness of R211-M and R211-E cells to MVMp infection. For comparison, LA9 cells, the prototypical cellular model for the study for MVMp infection, were used as control. Oncolytic cell death was monitored in real time using the Cytotox Green dye and the Incucyte Zoom. As expected, LA9 cells were highly sensitive to MVMp oncolytic effect (Figures 2A and S2A), with more than 20-fold increase in cell death as compared to control, 72 h following infection (Figure 2B). Remarkably, MVMp infection induced strong cell killing of R211-M cells at MOI = 100 (8-fold increase, p < 0.001; Figure 2B), while R211-E cells largely resisted viral-induced oncolysis when challenged with a similar dose of virus. Western blot analysis for poly-(ADP-ribose) polymerase (PARP) cleavage indicate that MVMp infection induces cell death by apoptosis in R211-M cells (Figure 2C) but not in R211-E cells (Figure S2B). Plaque assay confirmed that R211-M cells are more sensitive to MVMp infection, albeit at lower level as compared to LA9 cells (Figure S2C). We also found that R259 with mesenchymal phenotype were killed by the virus when cells with epithelial phenotype resisted MVMp infection (Figures S2D and S2E). Thus, we demonstrate that infection by MVMp specifically kills murine PDAC cells with a mesenchymal phenotype in vitro.
Figure 2.
MVM is oncolytic in murine and human PDAC cells with mesenchymal phenotype
R211-M and R211-E cells were infected with MVMp at MOI = 100. Control cells were incubated with infection medium. LA9 wells were used as positive control. Representative images (A), real-time cell death analysis (B), and western blot for PARP cleavage of cells infected by MVMp or mock-treated cells, up to 72 h (A and B) or 48 h (C) following infection. Results are mean ± SEM or representative of at least three independent experiments performed in duplicate. ∗∗∗∗p < 0.001, multiple unpaired Student’s t test. R211-M and R211-E cells were infected with MVMp at MOI = 100, when LA9 were treated with 0.1 M MVMp. (D) Cellular content in viral genomes was analyzed by qPCR for NS1 viral gene at the time indicated. Results are mean ± SEM of at three independent experiments performed in duplicate. ∗p < 0.05, ∗∗∗:p < 0.005, multiple unpaired Student’s t test. (E) Cell supernatant was collected at the time indicated and analyzed in real time for oncolytic activity on LA9 reporter cells. Results are mean ± SEM of three independent experiments performed in duplicate. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001, multiple unpaired Student’s t test. (F) Western blot analysis for the indicated proteins in human primary PDAC cells PDAC087T, PDAC015T, PDAC051T, and PDACT1. Representative of two independent experiments. PDAC087T, PDAC015T, PDAC051T, and PDACT1 were infected with MVMp at MOI = 100 when control cultures were incubated with infection medium. E, epithelial; M, mesenchymal. Representative images (G) and live cell death analysis (H) of human primary PDAC cells 72 h following infection by MVMp. Results are mean ± SEM of three independent experiments performed in duplicate. ∗∗p < 0.01, unpaired Student’s t test.
MVMp preferentially replicates in murine pancreatic cancer cells with mesenchymal phenotype
We next questioned whether murine PDAC cells may support viral replication. Thus, R211-M and R211-E cells were infected with MVMp at an MOI of 10. As control, LA9 cells were infected with MOI = 0.1 of MVMp. Cells were collected at different times following infection and intracellular viral load was analyzed by qPCR. Figure 2D shows that LA9 cells replicate MVMp genome at very high level (more than 30,000-fold increase as compared to viral input, 72 h following infection, p < 0.005; Figure 2D). On the contrary, R211-E cells failed to replicate MVMp genome (Figure 2D). Remarkably, R211-M cells significantly supported MVMp genome replication following infection, yet at lower level as compared to LA9 cells (130-fold increase as compared to viral input, 72h following infection, p < 0.001; Figure 2D). We then asked whether the mesenchymal phenotype is associated with enhanced viral entry. We found a trend in the acceleration of MVM entry in cells with mesenchymal phenotype that did not reach statistical significance (Figures S2F and S2G). These results demonstrate that murine PDAC cells with mesenchymal phenotype are permissive to MVMp infection.
Viral progeny from MVMp-infected murine pancreatic cancer cells with mesenchymal phenotype is infectious
One key advantage of OVs such as MVMp is that infected cells become viral factories allowing for several consecutive rounds of infection of neighboring cancer cells. Thus, we analyzed whether R211-M cells may produce and secrete infectious MVMp progeny following initial infection. R211-M and R211-E cells were infected with MVMp at an MOI of 10. As control, LA9 cells were infected with MOI = 0.1 of MVMp. Following infection, cells were carefully washed with PBS and treated by proteinase K to remove viral input. Culture supernatants were collected at 0, 24, 48, and 72 h following infection, and LA9 cells were used as reporter cells for oncolytic activity. Results shown in Figure 2E indicate that cell supernatants from LA9 cells show lytic activity on reporter cultures, when supernatants collected from R211-E cells were ineffective. On the contrary, supernatant collected from R211-M cells infected by MVMp demonstrate a time-dependent increase lytic activity on reporter cells, indicating the presence of a fully oncolytic viral progeny (Figure 2E). Compared with cells with an epithelial phenotype, PDAC cells with a mesenchymal phenotype that corresponds to the most aggressive PDAC phenotype can be infected by MVMp to produce infectious viral progeny.
MVMp preferentially targets human primary pancreatic cancer cells with mesenchymal phenotype
To validate and extend these findings in patient-derived resources, we selected human PDAC primary cell cultures that presented with high expression of FAK, N-cadherin, and Snail1 that are indicative of the mesenchymal phenotype (PDAC015T and PDACT1; Figure 2F) or high expression of E-cadherin and β-catenin, which indicate the epithelial phenotype (PDAC087T, PDAC051T; Figure 2F). Primary cultures were infected with MVMp at MOI = 100. PDAC087T and PDAC051T largely resisted infection, while PDACT1, and to a lesser extent PDAC051T, died from MVMp infection (Figures 2G and 2H). These further argue for a stronger MVMp tropism for mesenchymal-like cultures in human PDAC.
MVMp shows antitumoral potential only in murine pancreatic cancer tumors with mesenchymal phenotype
To determine the antitumoral potential of MVMp in vivo, immune-deficient mice with orthotopic R211-E or R211-M tumors were injected intravenously (i.v.) with 106 or 107 particle-forming units (p.f.u.) of MVMp in two separate, injections 4 days apart (Figure S3A). Control mice received PBS as placebo. Tumor growth monitoring by echography showed no impact of virotherapy on R211-E tumor growth (Figure S3B). On the other hand, MVMp injection significantly reduced R211-M tumor growth in a dose-dependent manner (−35% ± 3%, p < 0.001; Figure 3A). qPCR for MVMp genome shows increased level of viral load in both tumor models (Figure 3B) and in the spleen of mice with tumors (Figure S3C). We then analyzed R211-M tumors 9 days after infection and found that they expressed detectable levels of NS1 and viral capsid mRNAs (Figure 3C). This latter result strongly suggests that MVMp replicates in experimental PDAC tumors in vivo. To identify key tumor-cell-intrinsic factors mediating MVMp antitumoral effect, we examined differentially expressed bulk transcriptomes from R211-M tumors receiving placebo or treated with 10e7 p.f.u. of the virus. Gene set enrichment analysis (GSEA) highlighted over-expression of pathways related to cell senescence and tumor necrosis factor (TNF) signaling (Figure 3D; Table S1) and inhibition of mitochondrial activity and DNA replication (Figure 3D; Table S2). In addition, these transcriptomes were enriched in genes and pathways related to apoptosis (Table S3). Consistent with this, western blot analysis revealed PARP cleavage in R211-M tumors treated by MVMp only, as compared to R211-E tumors (Figure 3E). Collectively, these data further argue for the selectivity of MVMp for mesenchymal-like PDAC tumors in vivo.
Figure 3.
MVM is oncolytic in murine models of PDAC with mesenchymal phenotype
R211-M or R211-E tumors were induced in the pancreas of NSG mice as described in the “materials and methods” section. Eleven days later, when tumors reached 100 mm3 in size, mice were randomized and received two sequential i.v. injections of MVMp (10e6 or 10e7 p.f.u.) in two separate injections 4 days apart (D0 = day of the first injection). N = 3 mice were used per group. (A) R211-M tumor volume by echography, 9 days after starting treatment. Results are mean ± SEM or representative of three mice per group. ∗∗∗∗p < 0.001, unpaired Student’s t test. At endpoint, tumors were sampled and tumor viral genome content was quantified by qPCR (B, results are shown as mean of three independent experiments. ∗p < 0.05, ∗∗∗p > 0.005). (C) Total RNA from R211-M tumors was analyzed for the presence of NS1 and viral capsid mRNA by qPCR. RNA-seq followed by gene set enrichment analysis (GSEA, D) and western blot for PARP cleavage (E) were performed. NES, normalized enrichment score. Results are mean ± SEM or representative of three mice per group. ∗p < 0.05, ∗∗∗p < 0.005, unpaired Student’s t test.
MVMp shows increased antitumoral efficacy in immune-competent models of pancreatic cancer
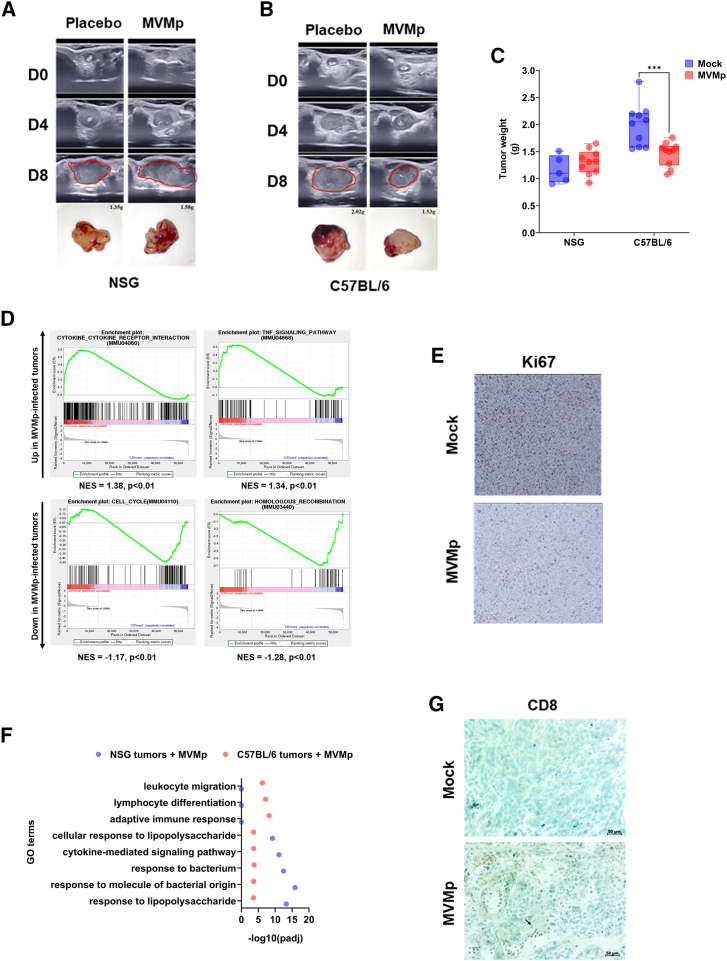
OV replication within tumors can cause direct lysis of cancer cells, leading to decrease of the bulk of the tumor. Along this line, preclinical and clinical data consistently suggest that OVs also utilize other anticancer mechanisms to combat tumors.19 Previous work demonstrated that MVMp-infected glioma cells activate immune cells in vitro and protect from tumor outgrowth in vivo in immunocompetent animals.10 These data suggest that MVMp could partner with the immune system to control tumor growth. However, the ability of MVMp to break immune tolerance in curative strategies has not been tested to date, especially in experimental models of PDAC. Accordingly, we generated orthotopic R211-M tumors in immune-deficient (NOD scid gamma [NSG]) and immune-competent (C56BL/6) mice models (Figure S3D). Mice received two sequential i.v. injections of a dose of MVMp (10e4 or 10e6 p.f.u.) that has no effect on tumor growth in immunodeficient models. Control mice received PBS as placebo. At the lowest dose tested (10e4 p.f.u.) MVMp showed no effect on the growth of R211-M tumors engrafted in immune-deficient or immune-competent mouse recipients (data not shown). At the highest dose tested (10e6 p.f.u), MVMp increased tumor cell death by apoptosis (Figure S3E), was detected in tumors (Figure S3F), but failed to inhibit R211-M experimental tumors growth when engrafted in immune-deficient mice (Figures 4A and 4C), consistent with our previous findings. Remarkably, 10e6 p.f.u of MVMp significantly impaired the growth of very aggressive R211-M tumors in mice with intact immune system (−32% ± 5%, p < 0.005; Figures 4B and 4C). Mice survival was significantly extended when R211-M tumors were treated by the virus, as compared to control mice (15 days vs. 11 days following initial virus injection, log rank Mantel-Cox test, p < 0.01). We failed to detect tumor cell death induced by virotherapy (Figure S3E) or to retrieve viral genomes from infected R211-M tumors in C57BL/6 mice (Figure S3F). On the other hand, antibodies against MVMp were readily detected by dot blot in the serum of infected mice (Figure S3G). These findings strongly suggest that infected cells are rapidly sensed and cleared in immune-competent animals.
Figure 4.
MVMp shows increased antitumoral efficacy in immune-competent models of pancreatic cancer
R211-M tumors were induced in the pancreas of NSG mice or of C57BL/6 mice as described in the “materials and methods” section. When tumors reached 100 mm3 in size, mice were randomized and received two sequential i.v. injections of MVMp (10e4 or 10e6 p.f.u.) in two separate injections 4 days apart (D0 = day of the first injection). Eight to 10 mice were used per group. Representative echography images and photographs with weight of R211-M tumors grown in NSG (A) or C57Bl6 (B) mice, up to 8 days following initial administration of MVMp. Tumors are circled in red. (C) Tumor weight at endpoint. Results are mean ± SEM or representative of 8–10 mice per group. ∗∗∗p < 0.005, unpaired Student’s t test. At endpoint, tumors were sampled and RNA-seq followed by GSEA (D), immunochemistry for Ki67 (E, R211-M tumors in C57Bl/6 mice only), GO terms analysis (F), and immunochemistry for Ki67 (G, R211-M tumors in C57Bl/6 mice only) were performed in mice receiving 10e6 p.f.u. of the virus. Results are mean ± SEM or representative of 8–10 mice per group.
Next, we performed RNA sequencing (RNA-seq) on R211-M tumors engrafted in immune-competent mice receiving placebo or 10e6 p.f.u. i.v. of the virus. Tumor-treated transcriptomes were enriched in genes and pathways related to immune recruitment/function, while pathways indicative of cell proliferation were repressed (Figure 4D; Table S4). Tumor staining for Ki67 confirmed that tumor cell proliferation was significantly decreased following MVMp injection (−47.5% ± 5%, p < 0.01; Figures 4E and S3H). To better understand the molecular basis for reduced tumor growth in the syngenic model of PDAC, we compared Gene Ontology (GO) terms between MVMp-infected R211-M tumors engrafted in NSG mice or in C57BL/6 mice. We found that pathways related to innate immune cell recruitment and function were similarly enriched in both models, while several pathways related to adaptive immunity were only enriched in immune-competent mice with tumors following MVMp administration (Figure 4F; Table S5). Immunochemistry analysis confirmed a dramatic increase in intratumoral CD8+ T cells in R211-M tumors engrafted in C57BL/6 mice and treated by MVMp, as compared to control tumors (Figure 4G).
We additionally analyzed the immune infiltrate in tumors treated with the virus using cell cytometry. We found that tumors receiving virotherapy demonstrated significantly higher numbers of viable cells (+80% ± 6%, p < 0.05; Figure 5A), which correlated with higher numbers of CD45-positive cells (+65% ± 11%, p < 0.05; Figure 5B). Apart neutrophils (Figure 5C), we identified that MVMp treatment induced a remarkable increase in tumoral natural killer (NK) cells (3-fold ±0.1, p < 0.01; Figure 5D), conventional dendritic cells (cDCs, 3.1-fold ± 0.1, p < 0.05; Figure 5E), CCR2+ monocytes (1.8-fold ± 0.2, p < 0.05; Figure 5F), and CCR2− macrophages (1.25-fold ± 0.2, p < 0.05; Figure 5G). Importantly, we also identified a significant increase in tumoral CD4+ (3.3-fold ± 0.1, p < 0.05; Figure 5H), and CD8+ T cells (2.7-fold ± 0.1, p < 0.05; Figure 5I) following virotherapy. We further analyzed the expression of immune checkpoints in intratumoral cytotoxic T cells. We found that MVMp treatment did not change the percentage of CD8+ T cells expressing either programmed cell death protein 1 (PD-1) or T cell immunoreceptor with immunoglobulin and ITIM domain (TIGIT) (Figures 6A and 6B). On the contrary, MVMp infection resulted in significantly lower numbers of intratumoral CD8+ T cells expressing LAG-3 (−66% ± 25%, p < 0.05; Figure 6C) or TIM-3 (−52% ± 14%, p < 0.05; Figure 6D). Collectively, these data demonstrate that MVMp virotherapy is more effective to inhibit the growth of autologous PDAC models and that MVMp infection significantly increases immune cell infiltration in immune-competent models of PDAC.
Figure 5.
MVMp infection increases the infiltration of immune cells into experimental pancreatic tumors
R211-M tumors were induced in the pancreas of C57BL/6 mice as described in the “materials and methods” section. When tumors reached 100 mm3 in size, mice were randomized and received two sequential i.v. injections of MVMp (10e6 p.f.u.) in two separate injections 4 days apart (D0 = day of the first injection). At endpoint, tumors were resected, minced, and single-cell suspensions were analyzed by cytometry for live cells (A), CD45+ cells (B), neutrophils (C), NKp46+ cells (D), conventional dendritic cells (cDC, E), CCR2+ monocytes (F), CCR2-macrophages (G), CD4+ (H), and CD8+ T cells (I). Results are mean ± SEM of three to six mice per group ∗p < 0.005, unpaired Student’s t test.
Figure 6.
MVMp infection reduces the expression of selected immune checkpoint by intratumoral CD8+ T cells
R211-M tumors were induced in the pancreas of C57BL/6 mice as described in the “materials and methods” section. When tumors reached 100 mm3 in size, mice were randomized and received two sequential i.v. injections of MVMp (10e6 p.f.u.) in two separate injections 4 days apart (D0 = day of the first injection). Five to eight mice were used per group. At endpoint, tumors were resected, minced, and single-cell suspensions were analyzed by cytometry for CD8+ T cells that were co-stained with PD-1 (A), TIGIT (B), LAG-3 (C), and TIM-3 (D) immune checkpoints. Results are mean ± SEM of three to six mice per group. ∗p < 0.005, Student’s t test. ∗p < 0.005, unpaired Student’s t test.
Discussion
The present study illustrates for the first time the high selectivity of MVMp OV for a particular subtype of PDAC, a disease that is highly resist to conventional therapies. MVMp was previously found to enter the leading edge of migrating cells by endocytosis close to filopodial and focal adhesions areas, notably following induction of EMT.16 Interestingly, two main molecular subtypes have recently been described in PDAC, namely the classical/pancreatic progenitor subtype associated with a better prognosis and the basal-like/squamous subtype associated with resistance to treatment and poorer clinical prognosis, including immune escape.20 During this study, we built on the characterization of murine PDAC cells with either mesenchymal or epithelial phenotypes that highly resemble the basal-like and classical phenotypes found in patients with PDAC.2 We demonstrate that infection by MVMp was cytotoxic and productive only in murine PDAC cells with mesenchymal phenotype, both in vitro and in vivo, as compared to cells with an epithelial phenotype that largely resisted virotherapy. To our knowledge, these data establish only the second example of a direct relationship between the mesenchymal phenotype of cancer cells with the increase in susceptibility to oncolytic infection. Indeed, Chen et al.. previously found that EMT enhances response to herpes simplex virus 1 (HSV-1) OV therapy, probably due to enhanced cell surface expression of the viral receptor nectin-1.21 Here, we went one step further as we identified a five-protein classifier (FAK, N-cadherin, E-cadherin, β-catenin, Snail) that predicts response to MVMp virotherapy in murine PDAC models. We speculate that MVMp hijacks the uptake and lysosomal/proteasomal degradation of expired focal-adhesion molecules that are generated following physical tension of stress fibers during the very dynamic process of mesenchymal cell migration, as previously described.16 Considering non-migrating (epithelial) cells, where focal-adhesion molecules remain very steadily attached and scarcely degraded, our data on viral replication indicate that the virus is either blocked outside the cell, trapped in endosomes, or brought back to the cell surface, unable to reach the nucleus and establish an infection. We observed a tendency of faster MVM entry in cells with a mesenchymal phenotype that did not achieve statistical significance. Electron microscopy experiments may help to better understand the mode of entry of MVMp in PDAC cells.
We also established an instructive role of this classifier in predicting MVMp lytic efficacy in patient-derived primary pancreatic tumor cells, albeit the “mesenchymal status” does not perfectly match the classical/basal-like molecular subtyping found in the literature.22 Here, Slug expression seems not to be as important as in murine PDAC cells to predict MVMp efficacy. Further investigations are warranted to confirm the predictive value of the MVMp signature in more PDAC samples but also in other tumor types with similar mesenchymal phenotype. As we and others documented that fine-needle material can be sampled and analyzed following endoscopic exploration,23 this finding opens the way for personalized, precision virotherapy for patients with PDAC.
Moreover, while R211-M cells successfully infected with the virus died by apoptosis, the non-infected cell population proliferated aggressively so that inhibition of cell proliferation was at best partial. The latter observation could be due to suboptimal viral replication and spread. Indeed, while detectable and infectious, viral progeny production by R211-M would be limiting to generate a second round of infection, as it may get diluted in the fast-growing cell population and eventually degraded. Importantly, MVMp establishes replication at cellular DNA damage sites that can be provoked by viral NS1 protein expression,9 which provides replication and expression machinery.24 Unfortunately, we failed to identify MVMp-induced DNA damage signaling in both R211-M and R211-E infected cells (western blot for phosphorylated ATM and CHECK1, personal observation). One avenue to increase MVMp therapeutic efficacy will be to combine virotherapy with clinically relevant drugs such as gemcitabine or 5-fluorouacil (5-FU) that induce DNA damage and that block cancer cells in G2/M or in S-phase to create a situation that is more favorable to viral replication.
The present data not only build on a growing body of evidence implicating direct oncolysis of cancer cells but also reveal novel insights into the immune potential of MVMp in experimental models of PDAC. This is of prime importance as pancreatic tumors are considered immunologically dead and have not yet succumbed to immunotherapies.1 To get one step closer to the clinical situation, we used unique experimental models where tumors were engrafted in the pancreas of recipient mice and the virus administrated systemically. Despite this very challenging setting, MVMp significantly inhibited the progression of experimental R211-M tumors when R211-E tumor growth was not affected by virotherapy, further demonstrating the more pronounced specificity of MVMp for the most aggressive subtype of PDAC. In immune-compromised mice, MVMp genomes were detected within tumors but also in the spleen of animals. Noteworthy, we detected viral transcripts within tumors, strongly suggesting active replication of the virus in tumor cells. MVMp infection was associated with tumor cell death by apoptosis, with evidence of expression of TNFα that is both involved in viral sensing and type-1 antitumoral responses, inhibition of cancer cell proliferation, and engagement of innate immunity. Remarkably, MVMp infection, at a dose that does not provoke tumor growth inhibition in athymic mice, shows significant antitumoral effect in immune-competent models, extended mice survival and promoted the massive infiltration of tumors by innate, myeloid, and cytotoxic T cells, which probably accounted for the increase in live intratumoral cells following virotherapy. Furthermore, our findings demonstrate that MVMp infection has no impact on the percentage of CD8+ T cells expressing PD-1 or TIGIT. However, it significantly reduces the percentage of cytotoxic T cells expressing TIM-3 or LAG-3. These data are important on several counts: first and foremost, they strongly suggest that cytotoxic cells exhibit a less terminally exhausted phenotype in the presence of the virus, so they might be more active against tumor cells. Second, they indicate that it would be more advisable to prioritize combinations of viruses and immunotherapies targeting PD-1 or TIGIT rather than those targeting TIM-3 or LAG-3. Our data confirm the general view that MVMp efficacy relies on the immune system, as previously found in experimental models of glioblastoma.10 Nevertheless, MVMp virotherapy comes with a cost. First, and as expected, mice produced antibodies against the virus following infection, a situation that may also occur in patients as MVMp is not a human pathogen. As active immune sensing of MVMp may blunt virotherapy efficacy, this finding argues for a safe use of this biotherapy in immune-competent patients. Second, our data suggest that infected cells could be rapidly cleared by the adaptive immune system following infection, as no viral genome or apoptotic cells were detected in tumors established in immune-competent animals. This could result from the relatively modest oncolysis of tumor cells and of the probable expression of viral epitopes at the surface of infected cells. One could speculate that the presence of newly activated antiviral T cells may invigorate the pool of newly primed, cancer-specific cytotoxic effector T cells within the tumor bed. Indeed, recent studies showed that pathogen-related CD4+ T cell memory populations can be re-engaged to support cytotoxic CD8+ T cells, converting a weak primary anti-tumor immune response into a stronger one. Besides the depletion of the different immune cell types that might be involved, it would be useful to proceed with T cell receptor sequencing to determine the target of such potential cytotoxic activity. Along this line, immunization with the Newcastle disease virus (NDV) strongly increases both antiviral and antitumoral CD8+ T cells numbers and tumor clearance before local treatment with NDV.25 Consequently, vaccination strategies may improve the antitumoral activity of MVMp in syngenic models of PDAC.
In summary, the present study describes for the first time the oncolytic potential of MVMp in experimental models of PDAC. We show here that MVMp’s stronger selectivity for cancer cells with a mesenchymal phenotype may reveal a novel molecular vulnerability for patients with the most aggressive form of PDAC. Hence, we have identified a five-gene classifier that may help select patients that would benefit the most from virotherapy. Last, we describe the immune potential of MVMp in syngenic tumor models, which can call for combination with immunotherapies. Utilizing this innovative strategy could be crucial for overcoming therapeutic resistance in patients with PDAC.
Materials and methods
Cell culture
LA9 cells were a kind gift of Dr Nelly Panté (The University of British Columbia, Vancouver, BC, Canada). R211 and R259 cells, which derive from spontaneous tumors generated in the KPC mice model (LSL-KrasG12D/+; LSL-Trp53R172H/+; Pdx-1-Cre), were obtained from D. Sauer (TUM, Munich, Germany). LA9, R211, and R259 cells were grown in DMEM supplemented with 4.5% glucose, 10% FBS, and 1% penicillin-streptomycin (Pen/Strep). Murine PDAC cells with mesenchymal phenotype (hereafter called R211-M or R259-M) were loosely attached to the culture dish and isolated following repeated pipetting in warm PBS. Murine cancer cells with epithelial phenotype (hereafter called R211-E or R259-E) strongly adhere to the plastic culture and were isolated following trypsinization. Pancreatic cancer patient-derived primary cells were obtained and cultured as previously described.26 Cells were incubated at 37°C with 5% CO2.
Virus production and purification
The initial MVMp stock was a kind gift of Nelly Panté (The University of British Columbia, Vancouver, BC, Canada). All agreements for using MVMp were obtained from the French Ministry of Research, with authorization number DUO2333. Adherent LA9 cells were infected in high-containment biological facility (Bl3, CRCT technological cluster) with MVMp at an MOI of 10−2, grown for 4 days at 37°C and 5% CO2, and harvested by scraping in PBS. After a centrifugation at 2,000 × g for 5 min, the cells were resuspended in TE (pH 8,7) and lysed through three cycles of freeze and thaw in liquid nitrogen. Further cell lysis was obtained by direct sonication at 10 W (4 times for 3 min) on ice, and cell debris was pelleted by centrifugation at 15,000 × g for 1 h. The virus was then precipitated by addition of CaCl2 (25 mM final), incubated on ice for 30 min, then spun at 10,000 × g for 15 min at 4°C and resuspended in DNase I (50 U/mL) to eliminate free viral DNA. After a 20-min incubation at 37°C, the precipitation step was repeated and the pellet of virus was resuspended in TE. The process of sonication was then repeated as described above, and cell debris was eliminated by centrifugation at 15,000 × g for 1 h at 4°C. The virus was finally loaded on sucrose/CsCl gradients for a 24-h centrifugation at 100,000 × g and 4°C. The virus was extracted and dialyzed three times overnight against TE, and then filtered through a 0.2-μm PES syringe filter.
Plaque assay
Cells were seeded in 6-cm dishes at 6e3 (R211) or 2e5 (LA9) cells/mL (5 mL/dish) in complete DMEM (4.5% glucose, 10% FBS, and 1% Pen/Strep) and grown at 37°C and 5% CO2 for 24 h. After a brief wash in PBS, the cells were incubated for 1 h at 37°C with 10× dilutions (in infection medium: DMEM 1% FBS and 1 mM HEPES) of purified MVMp, starting at an MOI of 100. As a control (mock), cells were incubated in infection medium alone. The supernatant was then removed and 7 mL of overlay medium (MEM containing 0.5% glucose, 5% FBS, 2% gentamycin, 1% tryptose phosphate, and 0.75% low-melting-point agarose) were added to each dish and allowed to solidify for 30 min at room temperature (RT). The cells were then grown at 37°C and 5% CO2 for 4 (R211) or 5 (LA9) days, then fixed in 4% formaldehyde for 30 min and stained with 0.3% methylene blue for 30 min.
Virus infections and analysis
Cells were seeded in six-well plates at 5.103 (R211) or 2.104 (LA9) cells/mL (2 mL/well) in complete DMEM and grown at 37°C and 5% CO2 for 24 h. After a brief wash in PBS, the cells were incubated for 1 h at 37°C with purified MVMp diluted in infection medium at the indicated MOIs. As a control (mock), cells were incubated in infection medium alone. The supernatant was then removed, 2 mL of complete DMEM containing 10 nM Cytotox Green (Sartorius) were added to each well, and the cells were grown at 37°C and 5% CO2 in an Incucyte Zoom (Sartorius). Cell death was monitored hourly.
MVMp secretion analysis
Cells were seeded in 10-cm dishes at 3.104 (R211) or 7.104 (LA9) cells/mL in complete DMEM and grown at 37°C and 5% CO2 for 24 h. After a brief wash in PBS, the cells were incubated for 1 h at 37°C with purified MVMp diluted in infectious medium, at an MOI of 10 (R211) or 0.1 (LA9). As a control (mock), cells were incubated in infectious medium only. The supernatant was then removed and the cells were incubated for 30 min at RT in PBS containing 0.1 mg/mL proteinase K (1 mL/plate) for 30 min at RT. The cells were subsequently collected, spun at 2,000 × g for 5 min, and washed in 1 mL of PBS. After another centrifugation at 2,000 × g for 5 min, the cells were resuspended in complete DMEM and finally reseeded in 10-cm dishes and grown at 37°C and 5% CO2. Culture supernatants were collected at 0, 24, 48, and 72 h post reseeding for Incucyte-based analysis of MVMp secretion using LA9 as a reporter cell. For this latter experiment, LA9 cells were seeded in six-well plates at 2.104 cells/mL in complete DMEM (2 mL/well) and grown at 37°C and 5% CO2 for 24 h. After a brief wash in PBS, the cells were incubated for 1 h at 37°C with 1 μL of each collected supernatant diluted 1:100 in infection medium. The supernatants were then aspirated and 2 mL of complete DMEM containing 10 nM Cytotox Green (Sartorius) were added to each well. The cells were grown at 37°C and 5% CO2 in an Incucyte Zoom (Sartorius) where cell death was monitored hourly.
Viral entry analysis
Here, 1.5e5 R211-E or M cells were seeded in 24-well plates with 500 μL of complete medium and incubated for 24 h at 37°C to let the well surfaces become fully covered by a monolayer of cells. Then the medium was aspirated and cells were infected with MVMp at MOI of 0.1, 1, or 5 by addition of 250 μL of infection medium or supplemented with Opti-MEM only and incubated at 37°C, 5% CO2. For each time point, cells were washed four times with PBS, detached with 250 μL of trypsin for 5 min at 37°C, then the trypsin activity was stopped by adding 250 μL of complete medium/PBS (1:1). Cells were lysed by three cycles of freeze and thaw and viral genomes were quantified by qPCR for NS1 genes.
Western and dot blots
For western blot analysis, cells were scraped in PBS, spun at 2,000 × g for 5 min at 4°C, resuspended in radioimmunoprecipitation assay (RIPA) lysis buffer (Bio Basic, CA) containing protease inhibitors (Sigma), and incubated on ice for 30 min. After a 10-min centrifugation at 15,000× g and 4°C, supernatants were collected and protein concentration determined by Bradford assay (Bio-Rad). For the preparation of tumor samples, 2- to 3-mm cubes of tumors were mixed with 1-mm glass beads and 300 μL of ice-cold RIPA lysis buffer containing protease inhibitors, then lysed using Precellys tissue homogenizer (Bertin) through four cycles of 10 s at 800 rpm. After a 10-min centrifugation at 15,000 × g and 4°C, supernatants were collected and protein concentration determined by Bradford assay (Bio-Rad). Depending on the proteins of interest, 8% or 14% SDS-PAGE gels were loaded with 40 or 80 μg of total proteins (diluted in 5× sample buffer and boiled for 5 min at 96°C) and run for 2 h at 140 V. After semi-dry transfer of the proteins onto nitrocellulose membranes (Bio-Rad), blocking was performed in PBS 5% skim milk for 1 h at RT. Primary antibodies (Cell Signaling Technology) were incubated overnight at 4°C as recommended by the manufacturer, and horseradish peroxidase (HRP)-coupled secondary antibodies (Bio-Rad) for 2 h at RT. For dot-blot studies, 1-μL drops containing 104 p.f.u. of purified MVMp were dried onto a nitrocellulose membrane. After blocking in PBS 5% skim milk for 1 h at RT, membranes were incubated overnight at 4°C with serums from MVMp or mock-treated mice diluted 1:1,000 in PBS. After three washes in PBS, membranes were incubated with HRP-coupled anti-mouse secondary antibodies (1/104) for 2 h at RT and then washed again three times before revelation by electrochemoluminescence (Bio-Rad) using the Chemi-doc XRS+ (Bio-Rad).
Viral genome quantification
Cells were seeded in 10-cm dishes at 3.104 (R211) or 7.104 (LA9) cells/mL (10 mL/dish) in complete DMEM and grown at 37°C and 5% CO2 for 24 h. After a brief wash in PBS, the cells were incubated for 1 h at 37°C with purified MVMp diluted in infectious medium at MOIs of 10 (R211) or 0.1 (LA9). The cells were subsequently grown at 37°C and 5% CO2 and harvested by scraping in 1 mL of PBS at 0, 24, 48, or 72 h post infection, then spun at 2,000 × g for 5 min. Cell pellets were resuspended in ice-cold TE (pH 8.7) and lysed through three cycles of freeze and thaw in liquid nitrogen. For the preparation of tumor samples, 2–3 mm3 of tumors were mixed with 1-mm glass beads and 300 μL of ice-cold TE, then lysed using a Precellys tissue homogenizer (Bertin) through four cycles of 10 s at 800 rpm. After a 30-min centrifugation at 15,000 × g and 4°C, 10 μL of supernatant was mixed with 30 μL of 1 M NaOH (diluted in TE) and immediately incubated at 56°C for 30 min to release viral genomes. The reaction was finally stopped by the addition of 960 μL of 40 mM HCl. For qPCR analysis, 1 μL of sample was mixed in a final 20 μL of SoFast-Green reaction mix containing 10 nM forward (GCGCGGCAGAATTCAAACT) and reverse (CCACCTGGTTGAGCCATCA) primers for MVMp NS1 gene amplification using the StepOnePlus real-time PCR system (Thermo Fisher Scientific). Primers targeting GAPDH were used for normalization (forward, GGCCACGCTAATCTCATTTT; reverse, AAGGCGGAGTTACCAGAGGT). Relative quantity of viral DNA was calculated by the comparative threshold cycle (CT) method as 2–ΔCT.
Viral mRNA quantification
RNA from tumor single-cell suspensions were extracted using TRIzol reagent (Invitrogen, Thermo Fisher Scientific) according to the manufacturer’s instructions. RNA was quantified using the NanoDrop device. Then 500 ng of RNA was treated with 0.5 units of DNase I, RNase free (Thermo Fisher Scientific) at 37°C for 30 min. DNase reaction was stopped by the addition of EDTA for 10 min at 65°C. Reverse transcription of the DNase-treated RNA was performed using random hexamer primers and RevertAid first-strand cDNA synthesis kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. For qPCR analysis, reverse transcription product was diluted at 1:20 and 2 μL of sample was mixed in a final 20 μL of SoFast-Green reaction mix containing the following forward and reverse primers at 10 mM. For NS1 mRNA quantification: forward, GCGCGGCAGAATTCAAACT; reverse, CCACCTGGTTGAGCCATCA. For viral capsid VP mRNA quantification: forward, ACAGGCCTGAAATCACTTGGT; reverse, AAAGCGTTGATCAGCAGCAG. Primers targeting the hypoxanthine phosphoribosyltransferase 1 (hprt) mRNA were used for normalization (forward, GGCCAGACTTTGTTGCATTTG; reverse, TGCGCTATCTTAGGCTTTGT). Relative quantity of viral RNA was calculated by the CT method as 2−ΔCT.
Animal experimentation
Experimental procedures performed on mice were approved by the ethical committee of INSERM CREFRE US006 animal facility and authorized by the French Ministry of Research: APAFIS#3600-2015121608386111v3. Six- to 8-week-old NSG or C57BL/6 mice were surgically grafted orthotopically with 5.104 R211-M or R211-E cells diluted in 50 μL of total PBS per animal. One week later, tumor formation was measured non-invasively by echography using the Aixplorer imaging device (Supersonic Imagine). Four days later, echography was performed and groups of mice with comparable average tumor sizes were made. Randomized mice were injected intravenously with 50 μL of purified MVMp (diluted 1:10 in PBS) in a high-containment animal facility (A3 level). As placebo, mice received a similar volume of TE diluted 1:10 in PBS. Tumor volume was measured at the indicated days using the Aixplorer imaging device. For dot-blot experiments, blood was sampled and spun at 2, 000 × g for 5 min, and serum was frozen immediately.
Immunohistochemistry
Samples were incubated at 60°C for 1 h, then incubated 3× in xylene for 5 min. Samples were then rehydrated by incubation in 100% ethanol (3 × 5 min), 90% ethanol (2 × 5 min), 70% ethanol for 2 min, 50% ethanol for 5 min, then 2× in dH2O for 5 min each. After a 10-min permeabilization step in 0.1% Triton X-100 in PBS, samples were washed 3× in milliQ distilled water (dH2O). For antigen retrieval, samples were autoclaved at 121°C for 12 min in citrate buffer (20 mM citric acid, 1 mM sodium citrate), allowed to cool for 1 h, and incubated 3 × 5 min in PBST. For sample staining, slides were placed in humid chambers, blocked in protein block (Dako) for 10 min at RT, then incubated overnight at 4°C with KI67 primary antibody (Cell Signaling Technology) diluted 1:400 or CD8a primary antibody (BioLegend) diluted 1:200 in antibody diluent signal stain (Dako). After three wash of 5 min in PBS-Tween, peroxidase activity was inhibited by incubation of the samples in PBST-H2O2 (3%) for 15 min at RT. The samples were then washed twice in PBS-Tween, once in PBS-Tween 1% BSA (5 min each), and subsequently incubated with SignalStain Boost IHC detection reagent (Cell Signaling Technology) for 45 min at RT in humid chambers. The samples were then washed 3× in PBS 1% BSA, once in PBS-Tween, incubated in AEC (Dako) for 5 min, and washed twice for 5 min in PBST. For counterstaining, samples were incubated for 2 min in hematoxylin (Merck) and washed for 5 min in dH2O, then mounted in Glycergel (Dako) and analyzed using an Axiolab A1 inverted microscope (Zeiss).
Analysis of intratumoral immune cells by cytometry
Tumors were resected, minced, and single-cell suspensions were obtained using tumor dissociation kit and gentleMACS octo dissociator (Miltenyi Biotec) according to the manufacturer’s instructions. Cells were stained for 30 min at 4°C in PBS, 5 mM EDTA, 10 μg/mL mouse FcBlock (rat anti-mouse CD16/CD32, BD Biosciences) with fluorochrome-conjugated monoclonal antibodies (Table 1). Dead cells were excluded from analyses by electronic gating of fixable viability dye (Live/Dead fixable blue from Invitrogen, Thermo Fisher Scientific). Data were collected on an LSR Fortessa X-20 (BD Biosciences) and analyzed with FlowJo Software.
Table 1.
Antibodies for flow cytometry used during this study
| Target | Fluorochrome | Clone | Company | Reference |
|---|---|---|---|---|
| CCR2 | APC-Vio770 | REA538 | Miltenyi | 130-117-501 |
| CD4 | APC | RM4-5 | BD | 553051 |
| CD8a | APC-Vio770 | REA601 | Miltenyi | 130-120-737 |
| CD11b | BV605 | M1/70 | BD | 563015 |
| CD11c | PE-Cyanine7 | N418 | BioLegend | 117318 |
| CD25 | PE-Dazzle594 | PC61 | BioLegend | 102048 |
| CD45.2 | BUV737 | 104 | BD | 612778 |
| F4/80 | BV421 | T45-2342 | BD | 565411 |
| I-A/I-E | BV510 | 2G9 | BD | 743871 |
| LAG-3 | VioBright FITC | C9B7W | Miltenyi | 130-120-170 |
| Ly-6G | Alexa Fluor 700 | 1A8 | BD | 561236 |
| NK1.1 | BV510 | PK136 | BD | 563096 |
| NKp46 | PE-Cyanine7 | 29A1.4 | BioLegend | 137618 |
| PD-1 | BV421 | J43 | BD | 562584 |
| TCRb | PE-Cyanine5 | H57-597 | BioLegend | 109210 |
| TIGIT | PE | 1G9 | BioLegend | 142104 |
| TIM-3 | BV786 | 5D12/TIM-3 | BD | 747621 |
RNA isolation and gene expression analysis
Tumors were sampled and minced and total RNA was extracted from tumors with RNeasy kit (Qiagen) according to the manufacturer’s instructions; quality and quantity were measured on a NanoDrop system (Thermo Fisher Scientific). RNA-seq analysis (poly(A) capture) was performed using 1 μg of total RNA and subcontracted to Novogene. Briefly, RNA degradation and contamination was monitored on 1% agarose gels. RNA purity was checked using the NanoPhotometer spectrophotometer (IMPLEN, CA, USA). RNA integrity and quantitation were assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). A total amount of 1 μg of RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext Ultra RNA. Library Prep Kit for Illumina (NEB, USA) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5×). First-strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-). Second-strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3′ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure were ligated to prepare for hybridization. In order to select cDNA fragments of preferentially 150–200 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). Then 3 μL of USER Enzyme (NEB, USA) was used with size-selected, adaptor-ligated cDNA at 37°C for 15 min followed by 5 min at 95°C before PCR. Then PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer. At last, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using PE Cluster Kit cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina platform and paired-end reads were generated.
Raw data (raw reads) of FASTQ format were first processed through fastp. In this step, clean data (clean reads) were obtained by removing reads containing adapter and poly-N sequences and reads with low quality from raw data. At the same time, Q20, Q30, and guanosine cytosine (GC) content of the clean data were calculated. All the downstream analyses were based on the clean data with high quality. Reference genome and gene model annotation files were downloaded from genome website browser (NCBI/UCSC/Ensembl) directly. Paired-end clean reads were mapped to the reference genome using HISAT2 software. HISAT2 uses a large set of small graph FM index (GFM) indexes that collectively cover the whole genome. These small indexes (called local indexes), combined with several alignment strategies, enable rapid and accurate alignment of sequencing reads. Differential expression analysis between two conditions/groups (three biological replicates per condition) was performed using DESeq2 R package. DESeq2 provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting p values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate (FDR). Genes with an adjusted p value <0.05 found by DESeq2 were assigned as differentially expressed. GO enrichment analysis of differentially expressed genes was implemented by the clusterProfiler R package, in which gene length bias was corrected. GO terms with corrected p value less than 0.05 were considered significantly enriched by differential expressed genes. We used clusterProfiler R package to test the statistical enrichment of differential expression genes in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
Ethics reporting
Experimental procedures performed on mice were approved by the ethics committee of INSERM CREFRE US006 animal facility and authorized by the French Ministry of Research: APAFIS#3600-2015121608386111v3.
Statistical analysis
Grouped analyses were performed using Student’s t tests, including multiple comparisons using GraphPad Prism 9 software. Survival analyses were performed using a log rank (Mantel-Cox) test. Values are presented as ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. Error bars are SEM unless otherwise stated. The experiments were performed with a sample size (n) greater than or equal to three replicates, with the exception of Figure S2E, and the results from representative experiments were confirmed in at least two independent experiment repeats. When monitoring tumor growth, the investigators were blinded to the group allocation but were aware of group allocation when assessing the outcome. No data were excluded from the analyses.
Data and code availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
The authors are grateful to Dr Nelly Panté (The University of British Columbia, Vancouver, BC, Canada) for the kind gift of LA9 cells and MVMp virus, and to Dr. Pierre Garcin for his scientific input. The authors are grateful for the financial support of Fondation ARC (PG fellowship, 2016), Programme Equipes labellisées Fondation ARC (2023–2026), and of Inserm Transfert. The authors thank Ms. Emilie Martin and Catherine Zanibellato for technical assistance. We are grateful to the Non-Invasive Exploration service (UMS006/CREFRE, Anexplo Platform, Toulouse) for giving us the access to the Aixplorer, which was acquired with financial support from ITMO Cancer AVIESAN (Alliance Nationale pour les Sciences de la Vie et de la Santé, National Alliance for Life Sciences & Health) within the framework of the Cancer Plan. The authors are grateful to Loïc Van Den Berghe and Tiphaine Fraineau (Vectorology Facility, Technology Cluster of the Cancer Research Center of Toulouse, INSERM-UMR1037) for their technical assistance.
Author contributions
Conceptualization, P.C., M.V., N.D., H.L., and L.B.; investigations, M.V., A.N., G.L., C.L., H.L., and P.C.; data analysis, M.V., N.D., L.B., and P.C.; supervision, P.C.; writing – original draft, P.C.; writing – review & editing, P.C. and M.V.; funding acquisition, P.C.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omton.2024.200780.
Supplemental information
References
- 1.Park W., Chawla A., O’Reilly E.M. Pancreatic Cancer: A Review. JAMA. 2021;326:851–862. doi: 10.1001/jama.2021.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martens S., Lefesvre P., Nicolle R., Biankin A.V., Puleo F., Van Laethem J.L., Rooman I. Different shades of pancreatic ductal adenocarcinoma, different paths towards precision therapeutic applications. Ann. Oncol. 2019;30:1428–1436. doi: 10.1093/annonc/mdz181. [DOI] [PubMed] [Google Scholar]
- 3.Moffitt R.A., Marayati R., Flate E.L., Volmar K.E., Loeza S.G.H., Hoadley K.A., Rashid N.U., Williams L.A., Eaton S.C., Chung A.H., et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015;47:1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puleo F., Nicolle R., Blum Y., Cros J., Marisa L., Demetter P., Quertinmont E., Svrcek M., Elarouci N., Iovanna J., et al. Stratification of Pancreatic Ductal Adenocarcinomas Based on Tumor and Microenvironment Features. Gastroenterology. 2018;155:1999–2013.e3. doi: 10.1053/j.gastro.2018.08.033. [DOI] [PubMed] [Google Scholar]
- 5.Nicolle R., Blum Y., Marisa L., Loncle C., Gayet O., Moutardier V., Turrini O., Giovannini M., Bian B., Bigonnet M., et al. Pancreatic Adenocarcinoma Therapeutic Targets Revealed by Tumor-Stroma Cross-Talk Analyses in Patient-Derived Xenografts. Cell Rep. 2017;21:2458–2470. doi: 10.1016/j.celrep.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quillien L., Buscail L., Cordelier P. Pancreatic cancer cell and gene biotherapies; past, present and future. Hum. Gene Ther. 2023;34:150–161. doi: 10.1089/hum.2022.210. [DOI] [PubMed] [Google Scholar]
- 7.Andtbacka R.H.I., Kaufman H.L., Collichio F., Amatruda T., Senzer N., Chesney J., Delman K.A., Spitler L.E., Puzanov I., Agarwala S.S., et al. Talimogene Laherparepvec Improves Durable Response Rate in Patients With Advanced Melanoma. J. Clin. Oncol. 2015;33:2780–2788. doi: 10.1200/JCO.2014.58.3377. [DOI] [PubMed] [Google Scholar]
- 8.Chesney J., Puzanov I., Collichio F., Singh P., Milhem M.M., Glaspy J., Hamid O., Ross M., Friedlander P., Garbe C., et al. Randomized, Open-Label Phase II Study Evaluating the Efficacy and Safety of Talimogene Laherparepvec in Combination With Ipilimumab Versus Ipilimumab Alone in Patients With Advanced, Unresectable Melanoma. J. Clin. Oncol. 2018;36:1658–1667. doi: 10.1200/JCO.2017.73.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartley A., Kavishwar G., Salvato I., Marchini A. A Roadmap for the Success of Oncolytic Parvovirus-Based Anticancer Therapies. Annu. Rev. Virol. 2020;7:537–557. doi: 10.1146/annurev-virology-012220-023606. [DOI] [PubMed] [Google Scholar]
- 10.Grekova S.P., Raykov Z., Zawatzky R., Rommelaere J., Koch U. Activation of a glioma-specific immune response by oncolytic parvovirus Minute Virus of Mice infection. Cancer Gene Ther. 2012;19:468–475. doi: 10.1038/cgt.2012.20. [DOI] [PubMed] [Google Scholar]
- 11.Paglino J.C., Andres W., van den Pol A.N. Autonomous parvoviruses neither stimulate nor are inhibited by the type I interferon response in human normal or cancer cells. J. Virol. 2014;88:4932–4942. doi: 10.1128/JVI.03508-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ventoso I., Berlanga J.J., Almendral J.M. Translation control by protein kinase R restricts minute virus of mice infection: role in parvovirus oncolysis. J. Virol. 2010;84:5043–5051. doi: 10.1128/JVI.02188-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muthalagu N., Monteverde T., Raffo-Iraolagoitia X., Wiesheu R., Whyte D., Hedley A., Laing S., Kruspig B., Upstill-Goddard R., Shaw R., et al. Repression of the Type I Interferon Pathway Underlies MYC- and KRAS-Dependent Evasion of NK and B Cells in Pancreatic Ductal Adenocarcinoma. Cancer Discov. 2020;10:872–887. doi: 10.1158/2159-8290.CD-19-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nam H.-J., Gurda-Whitaker B., Gan W.Y., Ilaria S., McKenna R., Mehta P., Alvarez R.A., Agbandje-McKenna M. Identification of the sialic acid structures recognized by minute virus of mice and the role of binding affinity in virulence adaptation. J. Biol. Chem. 2006;281:25670–25677. doi: 10.1074/jbc.M604421200. [DOI] [PubMed] [Google Scholar]
- 15.Garcin P.O., Nabi I.R., Panté N. Galectin-3 plays a role in minute virus of mice infection. Virology. 2015;481:63–72. doi: 10.1016/j.virol.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 16.Garcin P.O., Panté N. Cell migration is another player of the minute virus of mice infection. Virology. 2014;468–470:150–159. doi: 10.1016/j.virol.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Juiz N., Elkaoutari A., Bigonnet M., Gayet O., Roques J., Nicolle R., Iovanna J., Dusetti N. Basal-like and classical cells coexist in pancreatic cancer revealed by single-cell analysis on biopsy-derived pancreatic cancer organoids from the classical subtype. Faseb. J. 2020;34:12214–12228. doi: 10.1096/fj.202000363RR. [DOI] [PubMed] [Google Scholar]
- 18.Hingorani S.R., Wang L., Multani A.S., Combs C., Deramaudt T.B., Hruban R.H., Rustgi A.K., Chang S., Tuveson D.A. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 19.Achard C., Surendran A., Wedge M.-E., Ungerechts G., Bell J., Ilkow C.S. Lighting a Fire in the Tumor Microenvironment Using Oncolytic Immunotherapy. EBioMedicine. 2018;31:17–24. doi: 10.1016/j.ebiom.2018.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou X., Hu K., Bailey P., Springfeld C., Roth S., Kurilov R., Brors B., Gress T., Buchholz M., An J., et al. Clinical Impact of Molecular Subtyping of Pancreatic Cancer. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.743908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C.-H., Chen W.-Y., Lin S.-F., Wong R.J. Epithelial–Mesenchymal Transition Enhances Response to Oncolytic Herpesviral Therapy Through Nectin-1. Hum. Gene Ther. 2014;25:539–551. doi: 10.1089/hum.2013.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rashid N.U., Peng X.L., Jin C., Moffitt R.A., Volmar K.E., Belt B.A., Panni R.Z., Nywening T.M., Herrera S.G., Moore K.J., et al. Purity Independent Subtyping of Tumors (PurIST), A Clinically Robust, Single-sample Classifier for Tumor Subtyping in Pancreatic Cancer. Clin. Cancer Res. 2020;26:82–92. doi: 10.1158/1078-0432.CCR-19-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bournet B., Gayral M., Torrisani J., Selves J., Cordelier P., Buscail L. Role of endoscopic ultrasound in the molecular diagnosis of pancreatic cancer. World J. Gastroenterol. 2014;20:10758–10768. doi: 10.3748/wjg.v20.i31.10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Majumder K., Wang J., Boftsi M., Fuller M.S., Rede J.E., Joshi T., Pintel D.J. Parvovirus minute virus of mice interacts with sites of cellular DNA damage to establish and amplify its lytic infection. Elife. 2018;7 doi: 10.7554/eLife.37750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ricca J.M., Oseledchyk A., Walther T., Liu C., Mangarin L., Merghoub T., Wolchok J.D., Zamarin D. Pre-existing Immunity to Oncolytic Virus Potentiates Its Immunotherapeutic Efficacy. Mol. Ther. 2018;26:1008–1019. doi: 10.1016/j.ymthe.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraunhoffer N.A., Abuelafia A.M., Bigonnet M., Gayet O., Roques J., Telle E., Santofimia-Castaño P., Borrello M.T., Chuluyan E., Dusetti N., Iovanna J. Evidencing a Pancreatic Ductal Adenocarcinoma Subpopulation Sensitive to the Proteasome Inhibitor Carfilzomib. Clin. Cancer Res. 2020;26:5506–5519. doi: 10.1158/1078-0432.CCR-20-1232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.