Keywords: Ascaridida, Baylisascaris, genetic data, morphology, phylogeny, species delimitation, zoonotic nematode
Abstract
Species of Baylisascaris (Nematoda: Ascarididae) are of great veterinary and zoonotic significance, owing to cause Baylisascariosis or Baylisascariasis in wildlife, captive animals and humans. However, the phylogenetic relationships of the current 10 Baylisascaris species remain unclear. Moreover, our current knowledge of the detailed morphology and morphometrics of the important zoonotic species B. procyonis is still insufficient. The taxonomical status of B. procyonis and B. columnaris remains under debate. In the present study, the detailed morphology of B. procyonis was studied using light and scanning electron microscopy based on newly collected specimens from the raccoon Procyon lotor (Linnaeus) in China. The results of the ASAP analysis and Bayesian inference (BI) using the 28S, ITS, cox1 and cox2 genetic markers did not support that B. procyonis and B. columnaris represent two distinct species. Integrative morphological and molecular assessment challenged the validity of B. procyonis, and suggested that B. procyonis seems to represent a synonym of B. columnaris. Molecular phylogenetic results indicated that the species of Baylisascaris were grouped into 4 clades according to their host specificity. The present study provided new insights into the taxonomic status of B. procyonis and preliminarily clarified the phylogenetic relationships of Baylisascaris species.
Introduction
Species of Baylisascaris (Nematoda: Ascarididae) mainly occur in the digestive tract of carnivores and omnivores worldwide (Sprent, 1968, 1970; Bauer, 2013), which are of great veterinary and zoonotic significance, owing to cause Baylisascariosis or Baylisascariasis in wildlife, captive animals and humans (Sato et al., 2004; Bauer, 2013; Graeff-Teixeira et al., 2016). The life cycle of Baylisascaris spp. seems to be heteroxenous, and the second-stage larvae in eggs are considered to be the infective stage (Bauer, 2013). When infective eggs are ingested by small mammals including humans, the second-stage larvae hatch from eggs, then penetrate the intestine wall and migrate into various organs and tissues causing ocular larva migrans (OLM), visceral larva migrans (VLM) and neural larva migrans (NLM) (Gavin et al., 2005; Bauer, 2013; Graeff-Teixeira et al., 2016).
According to Sprent (1968) and Bauer (2013), 9 species were assigned to Baylisascaris, namely B. transfuga (Rudolphi, 1819), B. laevis (Leidy, 1856), B. columnaris (Leidy, 1856), B. melis (Gedoelst, 1920), B. schroederi (McIntosh, 1939), B. procyonis (Stefanski and Zarnowski, 1951), B. devosi (Sprent, 1952), B. tasmaniensis Sprent, 1970 and B. ailuri (Wu et al., 1987). Later, B. potosis Tokiwa et al., 2014 and B. venezuelensis Mata et al., 2016 were described from Japan and Venezuela, respectively (Tokiwa et al., 2014; Mata et al., 2016). In addition, Li et al. (2016) treated B. ailuri as a synonym of B. transfuga. Consequently, the genus Baylisascaris currently includes 10 recognized species. Although the phylogenetic analyses of Baylisascaris species were investigated using different genetic data in some previous studies (Nadler, 1992; Nadler and Hudspeth, 1998, 2000; Xie et al., 2011a, 2011b; Tokiwa et al., 2014; Mata et al., 2016; Camp et al., 2018; Li et al., 2018; Sharifdini et al., 2021), the phylogenetic relationships of these 10 Baylisascaris species remain unclear, because all of the previous phylogenetic studies only included a limited number of species.
The raccoon roundworm B. procyonis, is widely distributed in North America, and can cause severe clinical disease in humans and animals, due to extensive larval migration through host tissues (Kazacos and Boyce, 1989; Kazacos, 2001; Gavin et al., 2002, 2005; Graeff-Teixeira et al., 2016). Although the morphological characters of B. procyonis have been reported by some previous studies (Stefanski and Zarnowski, 1951; Hartwich, 1962; Sprent, 1968; Overstreet, 1970; Kikuchi and Oshima, 1977; Kazacos and Turek, 1982; Snyder, 1989), our current knowledge of the detailed morphology and morphometrics of B. procyonis is still insufficient. Furthermore, in the genus Baylisascaris, B. procyonis is highly similar to B. columnaris morphologically and genetically. Some recent studies based on different genetic data considered that B. procyonis and B. columnaris are closely related, but distinct species (Franssen et al., 2013; Choi et al., 2017); however, the results of other molecular studies did not support the current species partition of the 2 species (Camp et al., 2018). The hypothesis that B. procyonis and B. columnaris represent 2 separate species still needs to be further tested using different methods or based on different genetic data and broader samples collected from different localities.
In the present study, several adults of B. procyonis were collected from the raccoon Procyon lotor (Linnaeus) (Mammalia: Carnivora) in the Zoo of Kunming, Yunnan Province, China. The detailed morphology of B. procyonis was further studied using light and scanning electron microscopy. The ASAP (Assemble Species by Automatic Partitioning) analysis and Bayesian inference (BI) were employed for delimitation of B. procyonis and B. columnaris based on different nuclear [large ribosomal DNA (28S) and internal transcribed spacer (ITS)] and mitochondrial [cytochrome c oxidase subunit 1 (cox1) and 2 (cox2)] genetic markers. Moreover, to evaluate the evolutionary relationships of Baylisascaris species, phylogenetic analyses including the most comprehensive taxa sampling of Baylisascaris to date, were performed based on the ITS and 28S + ITS + cox1 + cox2 sequence data using maximum likelihood (ML) and Bayesian inference (BI) methods, respectively.
Materials and methods
Specimen collection and morphological study
Single Procyon lotor died naturally in the Zoo of Kunming, Yunnan Province, China, which was opportunistically dissected for parasites. Some nematode specimens were isolated from the small intestine of this raccoon. Nematodes were fixed and stored in 70% ethanol until the study. For light microscopy studies, nematodes were cleared in glycerine for examination using a Nikon® optical microscope. For scanning electron microscopy (s.e.m.), specimens were re-fixed in a 4% formaldehyde solution, post-fixed in 1% OsO4, dehydrated via an ethanol series (50, 70, 80, 90, 100, 100%) and acetone (100%), and then critical point dried. Samples were coated with gold at about 20 nm and examined using a Hitachi S-4800 scanning electron microscope at an accelerating voltage of 20 kV. Measurements (range, followed by mean in parentheses) are given in millimetres (mm) unless otherwise stated. Voucher specimens were deposited in College of Life Sciences, Hebei Normal University, Hebei Province, China.
Molecular procedures
The mid-body parts of 2 randomly selected nematode specimens (1 male, 1 female) were used for molecular analysis. Genomic DNA from each sample was extracted using a Column Genomic DNA Isolation Kit (Shanghai Sangon, China) according to the manufacturer's instructions. The partial 28S region was amplified by PCR using the forward primer 28S-F (5′-AGC GGA GGA AAA GAA ACT AA −3′) and the reverse primer 28S-R (5′-ATC CGT GTT TCA AGA CGG G −3′) (Nadler and Hudspeth, 1998). The partial cox1 region was amplified by PCR using the forward primer COI-F (5′-TTT TTT GGT CAT CCT GAG GTT TAT −3′) and the reverse primer COI-R (5′-ACA TAA TGA AAA TGA CTA ACA AC −3′) (Lazarova et al., 2006). The partial cox2 region was amplified by PCR using the forward primer COII-F (5′-AAT TTT AAT TGT AGT CTT TTG TTT GG −3′) and the reverse primer COII-R (5′-CTA TGA TTA GCA CCA CAA ATC −3′) (Nadler and Hudspeth, 1998). The partial ITS-1 region was amplified by PCR using the forward primer SS1 (5′- GTT TCC GTA GGT GAA CCT GCG −3′) and the reverse primer SS2R (5′- AGT GCT CAA TGT GTC TGC AA −3′). The ITS-2 region of nuclear rDNA was amplified by PCR using the forward primer NC13 (5′-ATC GAT GAA GAA CGC AGC-3′) and the reverse primer NC2 (reverse: 5′-TTA GTT TCT TTT CCT CCG CT-3′) (Zhu et al., 2000).
All PCRs of nematodes were performed in 50 μL of PCR reaction buffer with 10 mm Tris–HCl at pH 8.4, 50 mm KCl, 3.0 mm MgCl2, 250 μm of each dNTP, 50 pmol of each primer and 1.5 U of Taq polymerase [Takara Biotechnology (Dalian) Co. Ltd., Japan] in a thermocycler (2720, Applied Biosystems) under the following conditions: the partial 28S region: 94°C, 5 min (initial denaturation), followed by 30 cycles of 94°C, 30 s (denaturation), 55°C, 30 s (annealing), 72°C, 70 s (extension), and a final extension of 72°C for 7 min; the partial ITS region: 94°C, 5 min (initial denaturation), followed by 30 cycles of 94°C, 30 s (denaturation), 55°C, 30 s (annealing), 72°C, 30 s (extension), and a final extension of 72°C for 7 min; the partial cox1 region: 94°C, 5 min (initial denaturation), followed by 30 cycles of 94°C, 30 s (denaturation), 55°C, 30 s (annealing), 72°C, 45 s (extension), and a final extension of 72°C for 7 min; the partial cox2 region: 95°C, 15 min (initial denaturation), followed by 35 cycles of 95°C, 60 s (denaturation), 45°C, 60 s (annealing), 72°C, 75 s (extension), and a final extension of 72°C for 7 min.
PCR products were checked on GoldView-stained 1.5% agarose gels and purified with Column PCR Product Purification Kit (Shanghai Sangon, China). Sequencing for each sample was carried out on both strands. Sequences were aligned using ClustalW2. The DNA sequences obtained herein were compared (using the algorithm BLASTn) with those available in the National Centre for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov). The 28S, ITS, cox1 and cox2 sequences data of Baylisascaris procyonis were deposited in the GenBank (http://www.ncbi.nlm.nih.gov).
Species delimitation
The ASAP method (Puillandre et al., 2021) and Bayesian inference were used for species delimitation of Baylisascaris procyonis and B. columnaris based on the 28S, ITS, cox1, and cox2 sequences, respectively. The BI trees were inferred using MrBayes 3.2.7 (Ronquist et al., 2012) under the K81UF model for cox2, MTART + I + F for ITS, HKY + F + I for 28S and TRN for cox1 (two parallel runs, 1 000 000 generations). Baylisascaris laevis was chosen as the out-group. The ASAP analyses was conducted using the ASAP online server (https://bioinfo.mnhn.fr/abi/public/asap) under the Kimura (K80) ts/tv model. The results of ASAP with lowest scores were considered as the optimal group number, except the optimal results recommended by ASAP.
Phylogenetic analyses
Phylogenetic analyses were performed based on the ITS and ITS + 28S + cox1 + cox2 sequence data using maximum likelihood (ML) inference with IQTREE v2.1.2 (Minh et al., 2020) and Bayesian inference (BI) with MrBayes 3.2.7 (Ronquist et al., 2012), respectively. Toxascaris leonina (Ascaridoidea: Ascarididae) was chosen as the out-group. The in-group included 9 Baylisascaris species. The species B. melis has no published sequence data; consequently, it was not included in the present phylogenetic analyses. The detailed information on Baylisascaris nematodes included in the present phylogenetic analyses is provided in Table 1.
Table 1.
Species of Baylisascaris with detailed genetic information included in the phylogenetic analyses
| Species | Host | Locality | GenBank ID | References | |||
|---|---|---|---|---|---|---|---|
| ITS | 28S | cox1 | cox2 | ||||
| B. procyonis (Stefanski and Zarnowski, 1951) | Procyon lotor | China | OR453333–OR453334 | OR457646–OR457647 | OR453235–OR453236 | OR463271–OR463272 | Present study |
| B. transfuga (Rudolphi, 1819) | Ursus arctos; Melursus ursinus | Netherlands | KC543489 | KC543471 | KC543477 | KC543483 | Franssen et al. (2013) |
| B. laevis (Leidy, 1856) | Marmota vancouverensis; Urocitellus columbianus | Canada; USA | ON982744 | ON994376 | ON982731 | ON988170 | Barrera et al. (2022) |
| B. columnaris (Leidy, 1856) | Mephitis mephitis | USA; Netherlands | KC543484 | KC543466 | KY580736 | KC543478 | Choi et al. (2017), Franssen et al. (2013) |
| B. schroederi (McIntosh, 1939) | Ailuropoda melanoleuca | China | MH030599 | MG937777 | MH795152 | MH469667 | Camp et al. (2018), Hoberg et al. (2018) |
| B. procyonis (Stefanski and Zarnowski, 1951) | Procyon lotor | Norway | KC543488 | KC543470 | KC543476 | KC543482 | Franssen et al. (2013) |
| B. devosi (Sprent, 1952) | Pekania pennanti | Canada | MH030598 | MG937776 | MH795151 | MH469666 | Camp et al. (2018), Hoberg et al. (2018) |
| B. tasmaniensis Sprent, 1970 | Sarcophilus harrisii | Australia | MH030603 | MG937781 | MH795156 | MH469671 | Camp et al. (2018), Hoberg et al. (2018) |
| B. potosis Tokiwa et al. 2014 | Potos flavus | Guyana | AB901104 | AB893608 | AB893609 | Tokiwa et al. (2014) | |
| B. venezuelensis Mata et al. 2016 | Tremarctos ornatus | Venezuela | KX151726 | Mata et al. (2016) | |||
The nucleotide sequences were aligned in batches using MAFFT v7.313 under iterative refinement method of E-INS-I (Katoh and Standley, 2013), poorly aligned regions were excluded using BMGE v1.12 (h = 0.4) (Criscuolo and Gribaldo, 2010). Furthermore, partially ambiguous bases were manually inspected and removed. Substitution models were compared and selected according to the Bayesian Information Criterion by using ModelFinder (Kalyaanamoorthy et al., 2017). The HKY + F + I and HKY + I model were identified as the optimal nucleotide substitution model for the ML and BI inference of ITS sequences. The partitioning schemes and the optimal nucleotide substitution model selected for each combination of partition for the ML and BI inference of ITS + 28S + cox1 + cox2 sequences are shown in Table 2. Reliabilities for maximum likelihood inference were tested using 1000 bootstrap replications and Bayesian Information Criterion analysis was run for 5 × 106 MCMC generations.
Table 2.
The partitioning schemes and the optimal model selected for each combination of partition for the ML and BI inference based on the ITS + 28S + cox1 + cox2 sequences
| Subset | Best Model-BI | Best Model-ML | Number of sites | Partitioning schemes |
|---|---|---|---|---|
| 1 | K80 + I | K2P + FQ + I | 1067 | 28S |
| 2 | HKY + I | K2Pu + F + I | 1685 | cox1; cox2 |
| 3 | HKY + I | GTR + F + I + G4 | 927 | ITS |
In the ML tree, the bootstrap (BS) values ≥90 were considered to constitute strong nodal support, whereas BS values ≥70 and <90 were considered to constitute moderate nodal support. In the BI tree, the Bayesian posterior probabilities (BPP) values ≥0.90 were considered to constitute strong nodal support, whereas BPP values ≥0.70 and <0.90 were considered to constitute moderate nodal support. The BS values ≥70 and BPP values ≥0.70 were shown in the phylogenetic trees.
Results
Morphology of Baylisascaris procyonis
General
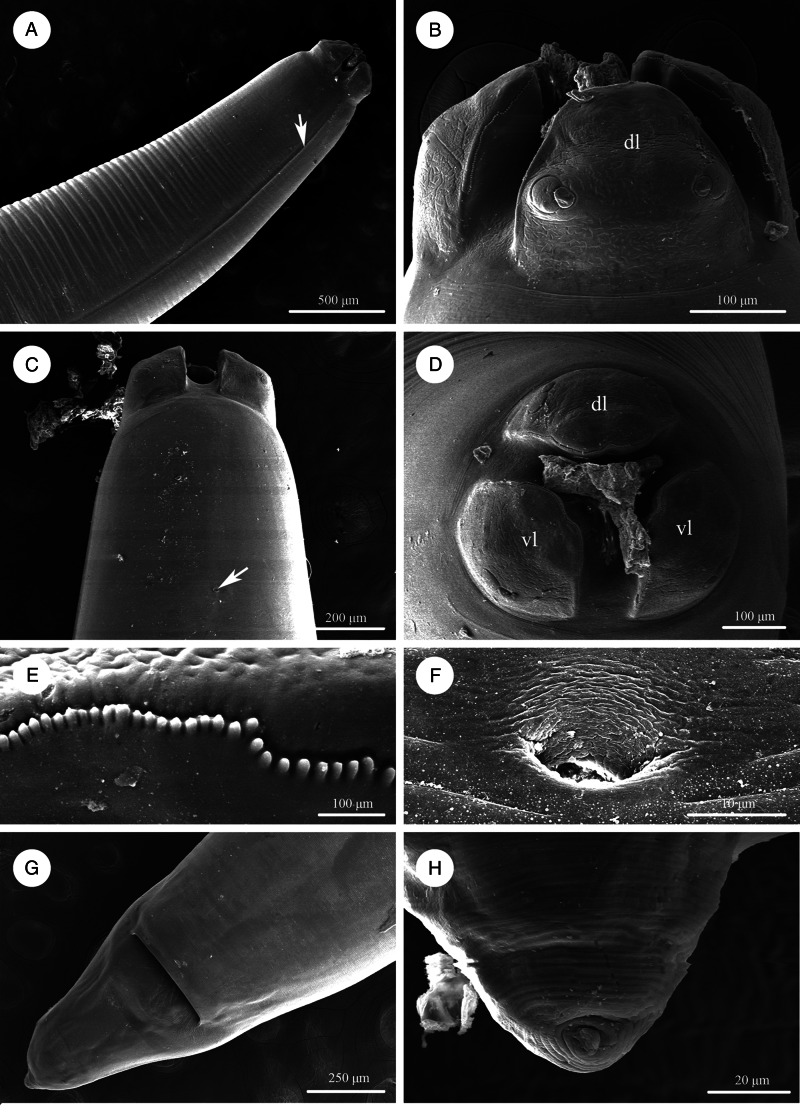
Large, whitish nematodes with finely transversely striated cuticle (Stefanski and Zarnowski, 1951) (Figures 1–3, Table 3). Maximum width of body at about mid-body. Cervical alae very narrow, starting from some distance from the base of ventrolateral lips and extending to anterior 1/4–1/2 of oesophageal length (Figs 1A, 2A); caudal alae absent. Cephalic extremity with 3 roughly trapezoid lips, postlabial grooves inconspicuous (Figs 1A, B; 2A–D, 3A, B). Dorsal lip with 1 pair of large double papillae; ventrolateral lips each with single double papilla, small papilla and amphid (Figs 2B, D, 3A, B). Each lip with distal ridge is armed with 140–150 small denticles (Figs 1B, D, 2B, D, E). Anterior margin of each lip with a small obtusely triangular medio-apical notch (Figs 1A, B, 2D, 3B). Interlabia absent. Oesophagus muscular, nearly cylindrical, distinctly broader posteriorly than anteriorly. Nerve-ring at about 20% of oesophageal length. Excretory pore just posterior to nerve-ring. Ventriculus, intestinal caecum and ventricular appendix absent. Tail of both sexes conical (Figs 1C, H; 2G, H, 3C, G, H).
Figure 1.
Scanning electron micrographs of Baylisascaris procyonis collected from Procyon lotor (Mammalia: Carnivora) in China, male. (A) Anterior part of body (lateral ala arrowed), lateral view. (B) Cephalic end, apical view. (C) Tail, ventrolateral view. (D) Magnified image of labial denticles. (E) Magnified image of cloacal area (medio-ventral precloacal papilla arrowed), ventral view. (F) Posterior end of body, ventral view. (G) Magnified image of postcloacal papillae. (H) Tail (medio-ventral precloacal papilla arrowed), ventral view. Abbreviations: pp, paracloacal double papillae; ps1, first pair of postcloacal double papillae; ps2-4, second to fourth pairs of postcloacal single papillae.
Figure 2.
Scanning electron micrographs of Baylisascaris procyonis collected from Procyon lotor (Mammalia: Carnivora) in China, female. (A) Anterior part of body (lateral ala arrowed), lateral view. (B) Cephalic end, dorsal view. (C) Anterior part of body (excretory pore arrowed), ventral view. (D) Cephalic end, apical view. (E) Magnified image of labial denticles. (F) Magnified image of excretory pore. (G) Posterior end of body, ventral view. (H) Magnified image of tail tip. Abbreviations: dl: dorsal lip; vl: ventrolateral lip.
Figure 3.
Baylisascaris procyonis collected from Procyon lotor (Mammalia: Carnivora) in China. (A) Anterior part of male, dorsal view. (B) Cephalic end of male, apical view. (C) Posterior end of male, lateral view. (D) Spicules. (E) Egg. (F) Magnified image of cloacal area, ventral view. (G) Posterior end of male, ventral view. (H) Posterior end of female, ventral view.
Table 3.
Morphometric comparisons of Baylisascaris procyonis (measurements in millimetres)
| Characteristics | Present study | Stefanski and Zarnowski (1951) | Hartwich (1962) | Sprent (1968) | Overstreet (1970) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | Male | Female | Male | Female | Male | Female | Male | Female | Male | Female | |
| BL | 66–72 | 99–152 | 62–76 | 90–122 | 62–77 | 90–200 | 90 | 200 | 57–99 | 87–160 | 57–119 | 49–189 |
| OL | 3.24–3.44 | 3.78–4.27 | 3.90 | 4.50 | 3.30–4.60 | – | – | – | 2.23–4.15 | 2.96–4.54 | – | – |
| LSL | 0.41–0.49 | N/A | 0.51–0.55 | N/A | 0.51–0.55 | N/A | 0.61 | N/A | 0.38–0.57 | N/A | 0.38–0.62 | N/A |
| RSL | 0.44–0.53 | N/A | 0.53–0.59 | N/A | 0.53–0.61 | N/A | 0.61 | N/A | 0.39–0.52 | N/A | 0.38–0.62 | N/A |
| PRP | 33–42 pairs | N/A | 40–44 pairs | N/A | 27–43 pair | N/A | 43 pair | N/A | 36–50 pairs | N/A | 28–49 pairs | N/A |
| PDP | 1 pair | N/A | 1 pair | N/A | 1 pair | N/A | – | N/A | 1 pair | N/A | 1 pair | N/A |
| PSP | 1 pair double + 3 pairs single | N/A | 1 pair double + 3 pairs single | N/A | 1 pair double + 3 pairs single | N/A | – | N/A | 1 pair double + 3 pairs single | N/A | 1 pair double + 3 pairs single | N/A |
| VA | N/A | 31–42 | N/A | 21–28 | N/A | 21–28 | N/A | – | N/A | 31.4–49.5 | N/A | – |
| VA/BL (%) | N/A | 27.6–31.3 | N/A | – | N/A | – | N/A | – | N/A | 30–36 | N/A | 28.0–36.0 |
| TL | 0.42–0.45 | 0.59–1.02 | 0.35–0.40 | 0.74–0.94 | 0.35–0.52 | 0.74–0.94 | 0.52 | 0.94 | 0.35–0.52 | 0.77–1.24 | 0.36–0.59 | 0.54–1.22 |
| SE | N/A | 0.07–0.08 × 0.05–0.07 | N/A | 0.08 × 0.05–0.06 | N/A | 0.07–0.08 × 0.05–0.07 | N/A | 0.08 × 0.07 | N/A | 0.07–0.08 × 0.05–0.07 | N/A | – |
| OL/BL (%) | 4.78–4.91 | 2.49–3.87 | – | – | – | – | – | – | 3.50–4.70 | 2.70–3.50 | 3.09–5.20 | 2.90–5.60 |
| Host | Procyon lotor | Procyon lotor | Procyon lotor | Procyon lotor | Potos flavus | Procyon lotor | ||||||
| Locality | China | Poland | ? | USA, Canada | USA | USA | ||||||
BL, length of body; OL, length of oesophagus; RSL, length of right spicule; LSL, length of left spicule; PRP, number of precloacal papillae; PDP, number of paracloacal double papillae; PSP, number of postcloacal papillae; SE, size of egg; VA, distance from vulva to anterior extremity; TL, length of tail; SE, size of eggs. N/A represents ‘Not applicable’.
Male (based on 2 mature specimens)
Body 66.0–72.0 (69.0) long, maximum width 1.70–1.76 (1.73). Dorsal and ventrolateral lips approximately equal in size, 0.23–0.28 (0.26) long, 0.30–0.32 (0.31) wide. Oesophagus 3.24–3.44 (3.34) long, 0.49–0.59 (0.54) in maximum width, representing 4.78–4.91 (4.85) % of body length. Nerve-ring and excretory pore 0.69–0.79 (0.74) and 0.93–1.15 (1.04) from anterior extremity, respectively. Cervical alae about 1.60 long. Posterior end of body curves ventrally. Spicules robust, ornamented with remarkable sculptures on surface, without alae, slightly unequal in length, distal end rounded, left spicule 0.44–0.53 (0.49) long, representing 0.61–0.80 (0.71) % body length, right spicule 0.41–0.49 (0.45) long and representing 0.57–0.74 (0.66) % body length, (Figs 1C, E, H, 3C, D, F, G). Gubernaculum absent. Caudal papillae 45–65 pairs in total, arranged as follows: 33–42 pairs precloacal, 1 pair paracloacal double papillae (slightly posterior to cloaca) and 4 pairs postcloacal (distal) (1st pair being double papillae, 3rd pair distinctly smaller than the others and 4th pair located sub-laterally) (the nomenclature of caudal papillae according to Fagerholm, 1991) (Figs 1C, E–H, 3C, G). Single medio-ventral precloacal papilla present (Figs 1E, H, 3F, G). Precloacal and postcloacal regions ornamented with rows of cuticular protuberance (commonly called ‘rugose area’ in previous studies). Precloacal ornamentation nearly meniscus-like, with 4–5 rows of cuticular protuberance; postcloacal ornamentation more or less semicircular, with 9–11 rows of cuticular protuberance (Figs 1C, E, H, 3F). Tail 0.42–0.45 (0.44) long, with rounded tip (Figs 1C, H, 3C, G).
Female (based on 3 gravid specimens)
Body 99.0–152.0 (127.0) long; maximum width 1.76–2.44 (1.80). Dorsal and ventro-lateral lips approximately equal in size, 0.23–0.29 (0.26) long, 0.32–0.35 (0.33) wide. Oesophagus 3.78–4.27 (3.96) long, 0.40–0.98 (0.74) in maximum width, representing 2.49–3.87 (3.21) % of body length. Nerve-ring and excretory pore 0.60–0.94 (0.73) and 0.96–1.41 (1.11) from anterior extremity, respectively. Cervical alae about 0.98 long. Vulva slit-like, pre-equatorial, 31.0–42.0 (36.5) mm from anterior extremity, at 27.6–31.3 (29.5) % of body length. Vagina muscular, directed posteriorly from vulva. Eggs oval, with rough shell, 0.07–0.13 × 0.05–0.08 (n = 20) (Fig. 3E). Tail 0.59–1.02 (0.80) long, with small button-shaped tip (Figs 2G, H, 3H). Phasmids 0.17–0.26 (0.23) from tail tip.
Species delimitation of B. procyonis and B. columnaris
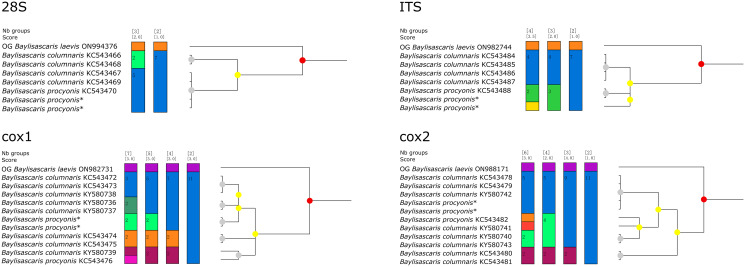
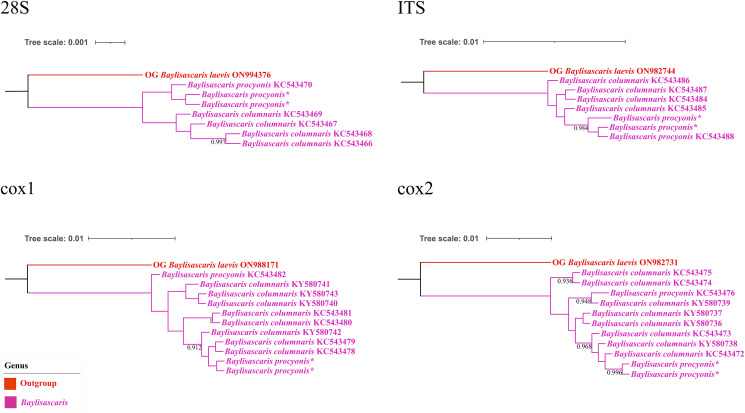
Molecular analysis of B. procyonis and B. columnaris revealed the presence of a very low level of nucleotide divergence between the 2 species in the 28S, ITS, cox1 and cox2 regions (Figures 4, 5; please see Tables 4–7 for the details). ASAP analyses of B. procyonis and B. columnaris using the 28S, ITS, cox1, and cox2 sequence data all did not support the current species partition of these 2 species (Fig. 4). Bayesian inference analyses based on the ITS, cox1, and cox2 sequence data also showed samples of B. procyonis mixed with B. columnaris (Fig. 5), which are accordant with the ASAP results. Although the result of BI analysis based on the 28S sequence data displayed B. procyonis and B. columnaris formed 2 distinct clades with weak support, the present molecular analysis revealed the presence only 1 polymorphic loci between the partial 28S region of these 2 species (Table 4).
Figure 4.
Assemble species by automatic partitioning (ASAP) analyses of Baylisascaris procyonis and B. columnaris based on 4 different nuclear and mitochondrial genetic markers. Abbreviations: cox1, cytochrome c oxidase subunit I; cox2, cytochrome c oxidase subunit II; ITS, internal transcribed spacer; 28S, large ribosomal subunit; OG, out-group;. Asterisk indicated the genetic data of samples obtained in the present study.
Figure 5.
Bayesian inference analyses of Baylisascaris procyonis and B. columnaris based on 4 different nuclear and mitochondrial genetic markers, respectively. Bayesian posterior probabilities values ≥0.70 were shown on nodes. Asterisk indicated the genetic data of samples obtained in the present study.
Table 4.
Base difference in the partial 28S region between B. columnaris and B. procyonis
| Species | 28S fragment (717 bp) | |
|---|---|---|
| 49 | 487 | |
| B. procyonis*_OR457646 | T | C |
| B. procyonis*_OR457647 | T | C |
| B. procyonis_KC543470 | T | C |
| B. columnaris_KC543466 | C | G |
| B. columnaris_KC543467 | C | C |
| B. columnaris_KC543468 | C | G |
| B. columnaris_KC5434869 | C | C |
Asterisks indicate the sequences obtained herein.
Table 5.
Base difference in the partial ITS region between B. columnaris and B. procyonis
| Species | ITS fragment (651 bp) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 158 | 201 | 271 | 540 | 541 | 542 | 543 | 544 | 545 | |
| B. procyonis*_OR453333 | – | C | A | A | G | A | G | A | G |
| B. procyonis*_OR453334 | – | C | G | A | G | A | G | A | G |
| B. procyonis_KC543488 | – | C | G | A | G | A | G | A | G |
| B. columnaris_KC543484 | – | T | G | – | – | – | – | – | – |
| B. columnaris_KC543485 | A | T | G | A | G | – | – | – | – |
| B. columnaris_KC543486 | – | T | G | – | – | – | – | – | – |
| B. columnaris_KC5434847 | A | T | G | A | G | – | – | – | – |
Asterisks indicate the sequences obtained herein.
Table 6.
Base difference in the partial cox1 region between B. columnaris and B. procyonis
| Species | cox1 fragment (413 bp) | ||||||
|---|---|---|---|---|---|---|---|
| 59 | 61 | 79 | 91 | 229 | 259 | 353 | |
| B. procyonis*_OR453235 | C | G | T | A | A | G | T |
| B. procyonis*_OR453236 | C | G | T | A | A | G | T |
| B. procyonis_KC543476 | T | A | T | G | A | A | G |
| B. columnaris_KC543472 | C | G | T | A | A | G | G |
| B. columnaris_KC543473 | C | G | T | A | A | G | G |
| B. columnaris_KC543474 | T | G | C | A | G | G | G |
| B. columnaris_KC543475 | T | G | C | A | G | G | G |
| B. columnaris_KY580736 | T | G | T | A | A | G | G |
| B. columnaris_KY580737 | T | G | T | A | A | G | G |
| B. columnaris_KY580738 | C | G | T | A | A | G | G |
| B. columnaris_KY580739 | T | A | T | G | A | G | G |
Asterisks indicate the sequences obtained herein.
Table 7.
Base difference in the partial cox2 region between B. columnaris and B. procyonis
| Species | cox2 fragment (483 bp) | ||||
|---|---|---|---|---|---|
| 66 | 117 | 246 | 280 | 480 | |
| B. procyonis*_OR463271 | G | A | T | C | G |
| B. procyonis*_OR463272 | G | A | T | C | G |
| B. procyonis_KC543482 | A | A | T | T | G |
| B. columnaris_KC543478 | G | A | T | C | G |
| B. columnaris_KC543479 | G | A | T | C | G |
| B. columnaris_KC543480 | G | G | T | T | T |
| B. columnaris_KC543481 | G | G | T | T | T |
| B. columnaris_KY580740 | G | A | C | T | G |
| B. columnaris_KY580741 | A | A | C | T | G |
| B. columnaris_KY580742 | G | A | T | C | G |
| B. columnaris_KY580743 | G | A | C | T | G |
Asterisks indicate the sequences obtained herein.
Phylogenetic analyses of Baylisascaris spp.
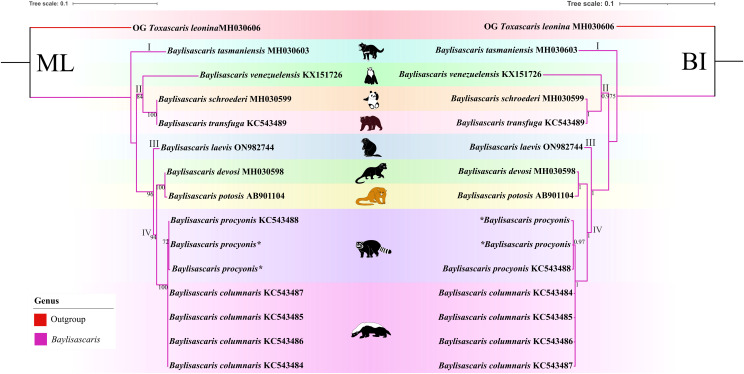
(Figures 6, 7). The results of phylogenetic analyses based on the ITS sequence data using ML and BI methods were more or less identical in topology, which displayed species of Baylisascaris divided into 4 clades (clade I, II, III and IV) (Fig. 6). Baylisascaris tasmaniensis located at the base of the phylogenetic trees representing clade I, which formed a sister relationship with the other species of Baylisascaris. Clade II included B. transfuga, B. schroederi and B. venezuelensis, all parasitic in the ursid hosts. Among them, B. transfuga and B. schroederi showed closer relationship than B. venezuelensis. Clade III contained only B. laevis reported from the rodent definitive hosts. Clade IV comprising B. devosi, B. potosis, B. columnaris and B. procyonis, parasitising the mustelid and procyonid hosts. Among them, B. procyonis clustered together with B. columnaris, and B. devosi is a sister to B. potosis.
Figure 6.
Phylogenetic relationships of representatives of the Baylisascaris using maximum likelihood and Bayesian inference analyses based on the ITS sequences. Toxascaris leonina (Ascaridomorpha: Ascarididae) was chosen as the out-group. Bootstrap values ≥70 and Bayesian posterior probabilities values ≥0.70 were shown on nodes in the phylogenetic trees. Asterisk indicated the genetic data of samples obtained in the present study.
Figure 7.
Phylogenetic relationships of representatives of the Baylisascaris using maximum likelihood and Bayesian inference analyses based on the ITS + 28S + cox1 + cox2 sequences. Toxascaris leonina (Ascaridomorpha: Ascarididae) was chosen as the out-group. Bootstrap values ≥70 and Bayesian posterior probabilities values ≥0.70 were shown on nodes in the phylogenetic trees. Asterisk indicated the genetic data of samples obtained in the present study.
Phylogenetic results based on the 28S + ITS + cox1 + cox2 sequence data using ML and BI methods were slightly different from the phylogenetic results based on the ITS data, which showed B. tasmaniensis is a sister to B. transfuga + B. schroederi + B. venezuelensis (Fig. 7). Phylogenetic relationships of these species B. laevis, B. devosi, B. potosis, B. procyonis and B. columnaris agreed well with that of the phylogenetic trees based on the ITS data (Fig. 7).
Discussion
Stefanski and Zarnowski (1951) described Ascaris procyonis from the intestine of P. lotor from a zoo in Poland. Sprent (1968) transferred this species to the genus Baylisascaris. Although B. procyonis has been studied extensively with respect to development and life cycle (Kazacos, 1983; Donnelly et al., 1989; Sakla et al., 1989), population genetic structure (Blizzard et al., 2010; Al-Warid et al., 2018), mitochondrial genome (Xie et al., 2011a, 2011b), prevalence and epidemiology of host (Snyder and Fitzgerald, 1987; Matoba et al., 2006; Kresta et al., 2010; Hernandez et al., 2013; Jardine et al., 2014; Page et al., 2016; Al-Warid et al., 2017; Thornton et al., 2020), human infection and pathologies (Fox et al., 1985; Murray, 2002; Gavin et al., 2005; Hung et al., 2012; Bauer, 2013); the morphology of this species has received little attention since its original description, and only the compilations of Hartwich (1962), Sprent (1968), Overstreet (1970), Kazacos and Turek (1982) and Snyder (1989) included it.
The morphology and morphometric data of the present specimens more or less agreed with the previous descriptions of B. procyonis by Stefanski and Zarnowski (1951), Hartwich (1962), Sprent (1968) and Overstreet (1970), including the lengths of body and oesophagus, the morphology of lips and lateral alae, the morphology and length of spicules, the number and arrangement of precloacal papillae, the morphology of cloacal ornamentations, the relative position of vulva, the size of eggs, and the length of tail (See Table 1 for details). In addition,
our specimens are also collected from the type of host P. lotor. Consequently, we considered our specimens to be B. procyonis. However, the length of spicules in our specimens is slightly shorter than that in some previous descriptions (Stefanski and Zarnowski, 1951; Hartwich, 1962; Sprent, 1968), but it is accordant with Overstreet's (1970) description. Moreover, Stefanski and Zarnowski (1951) and Overstreet (1970) both considered the tail of male with small spike-like tip. However, we did not observe that in our specimens using s.e.m.
The morphology of B. procyonis is very similar to B. columnaris, and some previous phylogenetic studies also supported B. procyonis and B. columnaris have a close affinity (Franssen et al., 2013; Tokiwa et al., 2014; Mata et al., 2016; Camp et al., 2018; Sharifdini et al., 2021). Overstreet (1970) considered the morphology of lips and first pair of postcloacal papillae to be important characters for distinguishing B. procyonis from B. columnaris (lips possessing remarkable medio-apical notch and the first pair of postcloacal papillae being double in B. procyonis vs the medio-apical notch of lips unconspicuous and the first pair of postcloacal papillae not being double in B. columnaris). The present s.e.m. observations confirmed the presence of small obtusely triangular medio-apical notch on each lip and first pair of postcloacal papillae being double in B. procyonis. However, the s.e.m. observations of B. procyonis by Kazacos and Turek (1982) found the absence of medio-apical notch on the lips of their specimens from P. lotor. Kikuchi and Oshima (1977) observed the detailed morphology of B. columnaris based on specimens collected from a skunk using s.e.m., and revealed the presence of small obtusely triangular medio-apical notch on the lips. Additionally, the number and morphology of postcloacal papillae of ascaridoid nematodes often vary between different individuals collected from a same or different hosts, even between the rows of postcloacal papillae in single individual (Uni and Takada, 1981; Li et al., 2016; Zhao et al., 2017). For example, some previous studies reported the presence of 2 closely associated single papillae instead of the postcloacal double papillae in their material of B. transfuga (Baylis and Daubney, 1922; Okoshi et al., 1961; Tenora et al., 1989). Snyder (1989) also pointed out that the first pair of postcloacal papillae of some of his specimens of B. procyonis was not double papillae and appeared as 2 single closely associated papillae based on s.e.m. observations. Moreover, Kikuchi and Oshima (1977) confirmed the first pair of postcloacal papillae of B. columnaris was double papillae in their material. Consequently, it is not reasonable and reliable to use the absence or presence of medio-apical notch on lip and the morphology of first pair of postcloacal papillae (single or double) as criteria for delimitation of B. procyonis and B. columnaris.
Franssen et al. (2013) and Choi et al. (2017) reported some loci of nucleotide polymorphisms in both mitochondrial (i.e. cox1, cox2, ND2, and several tRNA genes) and nuclear markers (ITS) and considered that these loci of nucleotide polymorphisms could be used as a tool to differentiate B. procyonis from B. columnaris. However, the present results of ASAP and BI analyses do not support that B. procyonis and B. columnaris represent 2 distinct species, and indicated that these loci of nucleotide polymorphisms in the 28S, ITS, cox1, and cox2 regions do not represent fixed differences between B. procyonis and B. columnaris. The validity of the important zoonotic species B. procyonis previously supported by morphological features (Hartwich, 1962; Sprent, 1968; Overstreet, 1970) and molecular data (Franssen et al., 2013; Choi et al., 2017) was challenged by the present study. Our results are consistent with the previous study (Camp et al., 2018). Due to the current morphological studies and genetic analyses of B. procyonis and B. columnaris, it is possibly premature to treat B. procyonis as a synonym of B. columnaris. A more rigorous study integrating compared morphological study and molecular analyses with broader samples of B. procyonis and B. columnaris collected from different localities and hosts worldwide is required to solve the taxonomical status of these 2 species.
Among the 9 species of Baylisascaris, B. tasmaniensis showed sister relationship to the remaining Baylisascaris in the phylogenetic trees based on the ITS data, which are accordant with the previous phylogenetic studies (Sharifdini et al., 2021; Barrera et al., 2022), but are in conflict with other phylogeny (Camp et al., 2018). The phylogenetic relationships of B. tasmaniensis and the others Baylisascaris spp. can be easily understood, because B. tasmaniensis only parasitises the marsupial carnivores in Australia (i.e. Sarcophilus harrisii, Dasyurus viverrinus and Dasyurops maculatus) and possesses some particular morphological features (i.e. the presence of fan-shaped lips, 3 pairs of postcloacal double papillae and slender spicules) (Sprent, 1970). Phylogenetic construction based on the ITS and 28S + ITS + cox1 + cox2 sequence data all supported these 3 species B. transfuga, B. schroederi and B. venezuelensis all parasitic in the ursid hosts, have a close affinity, that is identical to the previous studies (Mata et al., 2016; Sharifdini et al., 2021). Barrera et al. (2022) showed the systematic status of B. laevis based on molecular phylogeny for the first time. Baylisascaris laevis is the only known species in this genus to parasitise the rodent definitive hosts, e.g. marmot and ground squirrel (Rodentia: Sciuridae) (Sprent, 1968; Barrera et al., 2022), which showed sister relationship with the 4 Baylisascaris species (B. devosi + B. potosis + B. procyonis + B. columnaris) parasitic in the carnivorous definitive hosts including mustelids and procyonids. Our phylogenetic results also revealed B. devosi is a sister to B. potosis for the first time. However, B. procyonis and B. potosis both hosted the procyonids, did not display a close relationship (the similar situation also occurring in B. devosi and B. columnaris), that possibly indicated that it is not reasonable to use the specific groups of definitive hosts (Procyonidae or Mustelidae) as a criterion for distinguishing procyonis from B. columnaris.
The present study revealed some previously unreported morphological features of B. procyonis. The results of morphological study and ASAP and BI analyses all did not support that B. procyonis and B. columnaris represent 2 distinct species and the validity of B. procyonis was challenged. Molecular phylogeny provided new insights into the evolutionary relationships of Baylisascaris spp.
Acknowledgements
The authors are grateful to Dr Frantisek Moravec (Institute of Parasitology, Biology Centre of the Czech Academy of Sciences, Czech Republic), Professor Hideo Hasegawa (Oita University, Japan) and Dr Vitaliy Kharchenko (I. I. Schmalhausen Institute of Zoology, National Academy of Sciences of Ukraine, Ukraine) for providing important literatures of Baylisascaris.
Data availability statement
The nuclear and mitochondrial DNA sequences of Baylisascaris procyonis obtained in the present study were deposited in GenBank database [sequences of B. procyonis under the accession numbers: OR457646, OR457647 (28S), OR453333, OR453334 (ITS), OR453235, OR453236 (cox1), OR463271, OR463272 (cox2)]. Voucher specimens of B. procyonis were deposited in the College of Life Sciences, Hebei Normal University, Hebei Province, China (HBNU-N-2023M006G-L).
Author contributions
XHG and LL contributed to the study design and identification of the nematode specimens. XHG, HXC and LL sequenced and analyzed genetic data. XHG, HXC and LL conducted the ASAP and phylogenetic analyses. JJH provided nematode specimens. XHG and LL wrote the manuscript. All authors read and approved the final manuscript.
Financial support
This study was supported by the National Natural Science Foundation of China (Grant No. 32170442), National Key R&D Programme of China (Grant No. 2022YFC2601200) and the National Parasitic Resources Centre, and the Ministry of Science and Technology fund (Grant No. NPRC-2019-194-30).
Competing interest
The authors declare that they have no competing interests.
Ethical standards
This study was conducted under the protocol of Hebei Normal University. All applicable national and international guidelines for the protection and use of animals were followed.
References
- Al-Warid HS, Belsare A, Straka K and Gompper ME (2017) Baylisascaris procyonis roundworm infection patterns in raccoons (Procyon lotor) from Missouri and Arkansas, USA. Helminthologia 54, 113–118. [Google Scholar]
- Al-Warid HS, Belsare A, Straka K, Gompper ME and Eggert LS (2018) Genetic polymorphism of Baylisascaris procyonis in host infrapopulations and component populations in the Central USA. Parasitology International 67, 392–396. [DOI] [PubMed] [Google Scholar]
- Barrera MA, Janes JK and Gorrell JC (2022) Molecular phylogenetics and systematics of two enteric helminth parasites (Baylisascaris laevis and Diandrya vancouverensis) in the Vancouver Island marmot (Marmota vancouverensis). International Journal for Parasitology-Parasites and Wildlife 19, 301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C (2013) Baylisascariosis—infections of animals and humans with ‘unusual’ roundworms. Veterinary Parasitology 193, 404–412. [DOI] [PubMed] [Google Scholar]
- Baylis HA and Daubney R (1922) Report on the parasitic nematodes in the collection of the zoological survey of India. Memoirs of the Indian Museum 7, 263–347. [Google Scholar]
- Blizzard EL, Davis CD, Henke S, Long DB, Hall CA and Yabsley MJ (2010) Distribution, prevalence, and genetic characterization of Baylisascaris procyonis in selected areas of Georgia. Journal of Parasitology 96, 1128–1133. [DOI] [PubMed] [Google Scholar]
- Camp LE, Radke MR, Shihabi DM, Pagan C, Yang G and Nadler SA (2018) Molecular phylogenetics and species-level systematics of Baylisascaris. International Journal for Parasitology-Parasites and Wildlife 7, 450–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Mason S, Ahlborn M, Zscheile B and Wilson E (2017) Partial molecular characterization of the mitochondrial genome of Baylisascaris columnaris and prevalence of infection in a wild population of striped skunks. International Journal for Parasitology-Parasites and Wildlife 6, 70–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo A and Gribaldo S (2010) BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evolutionary Biology 10, e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly JJ, Sakla AA, Khatami M and Rockey JH (1989) Baylisascaris procyonis (Stefanski and Zarnowski, 1951) Ascarididae: Nematoda. II. Third stage larvae, morphogenesis and migratory behaviour. Assiut Veterinary Medical Journal 21, 77–91. [Google Scholar]
- Fagerholm HP (1991) Systematic implications of male caudal morphology in ascaridoid nematode parasites. Systematic Parasitology 19, 215–228. [Google Scholar]
- Fox AS, Kazacos KR, Gould NS, Heydemann PT, Thomas C and Boyer KM (1985) Fatal eosinophilic meningoencephalitis and visceral larva migrans caused by the raccoon ascarid Baylisascaris procyonis. New England Journal of Medicine 312, 1619–1623. [DOI] [PubMed] [Google Scholar]
- Franssen F, Xie K, Sprong H and van der Giessen J (2013) Molecular analysis of Baylisascaris columnaris revealed mitochondrial and nuclear polymorphisms. Parasites Vectors 6, e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin PJ, Kazacos KR, Tan TQ, Brinkman WB, Byrd SE, Davis AT, Mets MB and Shulman ST (2002) Neural larva migrans caused by the raccoon roundworm Baylisascaris procyonis. Pediatric Infectious Disease Journal 21, 971–975. [DOI] [PubMed] [Google Scholar]
- Gavin PJ, Kazacos KR and Shulman ST (2005) Baylisascariasis. Clinical Microbiology Reviews 18, 703–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeff-Teixeira C, Morassutti AL and Kazacos KR (2016) Update on Baylisascariasis, a highly pathogenic zoonotic infection. Clinical Microbiology Reviews 29, 375–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwich G (1962) On the raccoon roundworm Ascaris procyonis Stefanski et Zarnowski 1951, and its position in the system of the Ascaroidea (Nematoda). Ceskoslovenska Parasitologie 9, 239–256. [Google Scholar]
- Hernandez SM, Galbreath B, Riddle DF, Moore AP, Palamar MB, Levy MG, DePerno CS, Correa MT and Yabsley MJ (2013) Baylisascaris procyonis in raccoons (Procyon lotor) from North Carolina and current status of the parasite in the USA. Parasitology Research 112, 693–698. [DOI] [PubMed] [Google Scholar]
- Hoberg EP, Burek-Huntington K, Beckmen K, Camp LE and Nadler SA (2018) Transuterine infection by Baylisascaris transfuga: neurological migration and fatal debilitation in sibling moose calves (Alces alces gigas) from Alaska. International Journal for Parasitology-Parasites and Wildlife 7, 280–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Neafie RC and Mackenzie IR (2012) Baylisascaris procyonis infection in elderly person, British Columbia, Canada. Emerging Infectious Diseases 18, 341–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine CM, Pearl DL, Puskas K, Campbell DG, Shirose L and Peregrine AS (2014) The impact of land use, season, age, and sex on the prevalence and intensity of Baylisascaris procyonis infections in raccoons (Procyon lotor) from Ontario, Canada. Journal of Wildlife Diseases 50, 784–791. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Haeseler A and Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14, 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K and Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazacos KR (1983) Life cycle studies on Baylisascaris procyonis in raccoons. Abstract Proceeding 64th Conform Research Work Animal Diseases, abstract 130.
- Kazacos KR (2001) Baylisascaris procyonis and related species. In Samuel WM, Pybus MJ and Kocan AA (eds), Parasitic Diseases of Wild Mammals, 2nd Edn. Ames (IA): Iowa State University Press, pp. 301–341. [Google Scholar]
- Kazacos KR and Boyce WM (1989) Baylisascaris larva migrans. Journal of the American Veterinary Medical Asso ciation 195, 894–903. [PubMed] [Google Scholar]
- Kazacos KR and Turek JJ (1982) Scanning electron microscopy of the labia of Baylisascaris procyonis (Nematoda). Journal of Parasitology 68, 634–641. [PubMed] [Google Scholar]
- Kikuchi S and Oshima T (1977) Scanning electron microscopy of an ascarid Baylisascaris columnaris (Leidy, 1856) from a skunk. Japanese Journal of Parasitology 26, 376–383. [Google Scholar]
- Kresta AE, Henke SE and Pence DB (2010) Baylisascaris procyonis in raccoons in Texas and its relationship to habitat characteristics. Journal of Wildlife Diseases 46, 843–853. [DOI] [PubMed] [Google Scholar]
- Lazarova SS, Malloch G, Oliveira CMG, Hübschen J and Neilson R (2006) Ribosomal and mitochondrial DNA analyses of Xiphinema americanum-group populations. Journal of Nematology 38, 404–410. [PMC free article] [PubMed] [Google Scholar]
- Li L, Gibson DI and Zhang LP (2016) An annotated catalogue of the ascaridoid nematode parasites of Chinese vertebrates. Systematic Parasitology 93, 1–35. [DOI] [PubMed] [Google Scholar]
- Li L, Lü L, Nadler SA, Gibson DI, Zhang LP, Chen HX, Zhao WT and Guo YN (2018) Molecular phylogeny and dating reveal a terrestrial origin in the early carboniferous for ascaridoid nematodes. Systematic Biology 67, 888–900. [DOI] [PubMed] [Google Scholar]
- Mata PA, Pérez GH and Parra GJ (2016) Morphological and molecular description of Baylisascaris venezuelensis, n. sp. from a natural infection in the South American spectacled bear Tremarctos ornatus Cuvier, 1825 in Venezuela. Neotropical Helminthology 10, 85–103. [Google Scholar]
- Matoba Y, Yamada D, Asano M, Oku Y, Kitaura K, Yagi K, Tenora F and Asakawa M (2006) Parasitic helminths from feral raccoons (Procyon lotor) in Japan. Helminthologia 43, 139–146. [Google Scholar]
- Minh BQ, Hahn MW and Lanfear R (2020) New methods to calculate concordance factors for phylogenomic datasets. Molecular Biology and Evolution 37, 2727–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray WJ (2002) Human infections caused by the raccoon roundworm, Baylisascaris procyonis. Clinical Microbiology Newsletter 24, 1–7. [Google Scholar]
- Nadler SA (1992) Phylogeny of some ascaridoid nematodes, inferred from comparison of 18S and 28S rRNA sequences. Molecular Biology and Evolution 9, 932–944. [DOI] [PubMed] [Google Scholar]
- Nadler SA and Hudspeth DS (1998) Ribosomal DNA and phylogeny of the Ascaridoidea (Nemata: Secernentea): implications for morphological evolution and classification. Molecular Phylogenetics and Evolution 10, 221–236. [DOI] [PubMed] [Google Scholar]
- Nadler SA and Hudspeth DS (2000) Phylogeny of the Ascaridoidea (Nematoda: Ascaridida) based on three genes and morphology: hypotheses of structural and sequence evolution. Journal of Parasitology 86, 380–393. [DOI] [PubMed] [Google Scholar]
- Okoshi S, Takashio M and Nakagawa S (1961) Toxascaris transfuga (Rudolphi, 1819) Baylis & Daubney 1922, found in captive bears in Japan. The Japanese Journal of Veterinary Science 24, 151–155. [Google Scholar]
- Overstreet RM (1970) Baylisascaris procyonis (Stefanski and Zarnowski, 1951) from the kinkajou, Potos flavus, in Colombia. Journal of Parasitology 37, 192–195. [Google Scholar]
- Page LK, Delzell DA, Gehrt SD, Harrell ED, Hiben M, Walter E, Anchor C and Kazacos KR (2016) The structure and seasonality of Baylisascaris procyonis populations in raccoons (Procyon lotor). Journal of Wildlife Diseases 52, 286–292. [DOI] [PubMed] [Google Scholar]
- Puillandre N, Brouillet S and Achaz G (2021) ASAP: assemble species by automatic partitioning. Molecular Ecology Resources 21, 609–620. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA and Huelsenbeck JP (2012) MrBayes 32: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61, 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakla AA, Donnelly JJ, Khatami M and Rockey JH (1989) Baylisascaris procyonis (Stefanski and Zarnowski, 1951) Ascarididae: Nematoda. I. Embryonic development and morphogenesis of second stage larvae. Assiut Veterinary Medical Journal 21, 68–76. [Google Scholar]
- Sato H, Matsuo K, Osanai A, Kamiya H, Akao N, Owaki S and Furuoka H (2004) Larva migrans by Baylisascaris transfuga: fatal neurological diseases in Mongolian jirds, but not in mice. Journal of Parasitology 90, 774–781. [DOI] [PubMed] [Google Scholar]
- Sharifdini M, Heckmann RA and Mikaeili F (2021) The morphological and molecular characterization of Baylisascaris devosi Sprent, 1952 (Ascaridoidea, Nematoda), collected from Pine marten (Martes martes) in Iran. Parasites Vectors 14, e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder DE (1989) Scanning electron microscopic observations of adult Baylisascaris procyonis (Nematoda). International Journal for Parasitology 19, 571–574. [DOI] [PubMed] [Google Scholar]
- Snyder DE and Fitzgerald PR (1987) Contaminative potential, egg prevalence, and intensity of Baylisascaris procyonis-infected raccoon (Procyon lotor) from Illinois, with a comparison to worm intensity. Proceedings of the Helminthological Society of Washington 54, 141–145. [Google Scholar]
- Sprent JFA (1968) Notes on Ascaris and Toxascaris, with a definition of Baylisascaris gen. nov. Parasitology 58, 185–198. [DOI] [PubMed] [Google Scholar]
- Sprent JFA (1970) Baylisascaris tasmaniensis sp. nov. in marsupial carnivores – heirloom or souvenir. Parasitology 61, 75–86. [Google Scholar]
- Stefanski W and Zarnowski E (1951) Ascaris procyonis n. sp. from the intestine of Procyon lotor L. Annales Musei Zoologica Polonici 14, 199–203. [Google Scholar]
- Tenora F, Mituch J and Hovorka I (1989) Scanning electron microscopic study on Toxascaris transfuga (Rudolphi, 1819) (Nematoda). Folia Parasitol (Praha) 36, 371–374. [PubMed] [Google Scholar]
- Thornton GL, French SK, Peregrine AS and Jardine CM (2020) Prevalence of Baylisascaris procyonis in raccoon latrines in southern Ontario, Canada. Veterinary Parasitology Regional Studies and Reports 20, e100392. [DOI] [PubMed] [Google Scholar]
- Tokiwa T, Nakamura S, Taira K and Une Y (2014) Baylisascaris potosis n. sp., a new ascarid nematode isolated from captive kinkajou, Potos flavus, from the cooperative Republic of Guyana. Parasitology International 63, 591–596. [DOI] [PubMed] [Google Scholar]
- Uni S and Takada S (1981) Comparison of scanning electron microscopy on Baylisascaris transfuga, Toxascaris leonina and Ascaris lumbricoides (Nematoda: Ascaridida). Japanese Journal of Parasitology 30, 187–195. [Google Scholar]
- Xie Y, Zhang Z, Niu L, Wang Q, Wang C, Lan J, Deng J, Fu Y, Nie H, Yan N, Yang D, Hao G, Gu X, Wang S, Peng X and Yang G (2011a) The mitochondrial genome of Baylisascaris procyonis. PLoS ONE 6, e27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Zhang Z, Wang C, Lan J, Li Y, Chen Z, Fu Y, Nie H, Yan N, Gu X, Wang S, Peng X and Yang G (2011b) Complete mitochondrial genomes of Baylisascaris schroederi, Baylisascaris ailuri and Baylisascaris transfuga from giant panda, red panda and polar bear. Gene 482, 59–67. [DOI] [PubMed] [Google Scholar]
- Zhao JY, Zhao WT, Ali AH, Chen HX and Li L (2017) Morphological variability, ultrastructure and molecular characterisation of Hysterothylacium reliquens (Norris & Overstreet, 1975) (Nematoda: Raphidascarididae) from the oriental sole Brachirus orientalis (Bloch & Schneider) (Pleuronectiformes: Soleidae). Parasitology International 66, 831–838. [DOI] [PubMed] [Google Scholar]
- Zhu XQ, D'Amelio S, Paggi L and Gasser RB (2000) Assessing sequence variation in the internal transcribed spacers of ribosomal DNA within and among members of the Contracaecum osculatum complex (Nematoda: Ascaridoidea: Anisakidae). Parasitology Research 86, 677–683. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The nuclear and mitochondrial DNA sequences of Baylisascaris procyonis obtained in the present study were deposited in GenBank database [sequences of B. procyonis under the accession numbers: OR457646, OR457647 (28S), OR453333, OR453334 (ITS), OR453235, OR453236 (cox1), OR463271, OR463272 (cox2)]. Voucher specimens of B. procyonis were deposited in the College of Life Sciences, Hebei Normal University, Hebei Province, China (HBNU-N-2023M006G-L).