Abstract
An antibody phage display library was constructed from RNA extracted from lymph node cells of a simian immunodeficiency virus (SIV)-infected long-term-nonprogressor macaque. Seven gp120-reactive Fabs were obtained by selection of the library against SIV monomeric gp120. Although each of the Fabs was unique in sequence, there were two distinct groups based on epitope recognition, neutralizing activity in vitro, and molecular analysis. Group 1 Fabs did not neutralize SIV and bound to a linear epitope in the V3 loop of the SIV envelope. In contrast, two of the group 2 Fabs neutralized homologous, neutralization-sensitive SIVsm isolates with high efficiency but failed to neutralize heterologous SIVmac isolates. Based on competition enzyme-linked immunosorbent assays with mouse monoclonal antibodies of known specificity, these Fabs reacted with a conformational epitope that includes domains V3 and V4 of the SIV envelope. These neutralizing and nonneutralizing Fabs provide valuable standardized and renewable reagents for studying the role of antibody in preventing or modifying SIV infection in vivo.
Simian immunodeficiency virus (SIV) infection of macaques is a relevant and widely used animal model for human immunodeficiency virus type 1 (HIV-1) infection. SIV-infected macaques develop an AIDS-like syndrome characterized by declining peripheral CD4+ cell counts, opportunistic infections, and wasting (11). As also observed in HIV-1-infected individuals, the duration of clinical latency in SIV-infected macaques varies widely, dependent upon the virulence of the infecting virus, as well as undefined host factors, which include the effectiveness of the immune response. The importance of host factors is underscored by the observation that animals experimentally infected with molecularly cloned SIV also exhibit considerable variation in disease outcome (15). While the majority of animals infected with pathogenic SIV isolates develop AIDS by 1 to 2 years postinfection, a small fraction of infected animals remain clinically healthy and appear to be the SIV macaque equivalent of human HIV-1 infected long-term nonprogressors (LTNP) (6, 35, 43). Although a vigorous humoral and cellular immune response accompanies both progressive and nonprogressive SIV infections, the mechanisms by which host immunity actually controls or protects against infection is not known. A role for neutralizing antibodies is suggested by the rapid death of animals that do not mount a SIV-specific antibody response (15, 53). However, the temporal association of measurable effector cytolytic CD8 suppression with down modulation of viral replication in the postacute phase of infection is also consistent with a role of cell-mediated immunity in controlling infection (27, 52).
The role of neutralizing antibody can be directly assessed through passive immunoprophylaxis experiments utilizing the SIV-infected macaque model. However, in considering SIV-infected macaques as a model to study the role of neutralizing antibody, one must take into consideration the fact that the immunodominant epitopes of the SIV and HIV envelope glycoproteins may differ. For example, although the V3 loop is an immunodominant region of both the HIV-1 and SIV envelope glycoproteins (31, 44), there are major differences in the characters of the V3 immune responses to these two viruses. This region of the HIV-1 envelope is antigenically variable and appears to represent a linear, type-specific, principal neutralizing domain of T-cell line-adapted isolates (18, 36) but not of primary isolates (48). Like primary HIV-1 isolates, the V3 analog of the SIV envelope is generally highly conserved and does not appear to constitute a neutralization epitope for SIV. Conformational epitopes may be the target of more broadly neutralizing antibody responses. Neutralizing antibodies to conformational epitopes are prevalent in sera from HIV-1-infected individuals (34, 45) and probably also in sera of SIV-infected macaques (19). Thus, 90% of the neutralizing activity in the serum of SIV-infected macaques is absorbed by a 45-kDa fragment of gp120 that encompasses domains V3 to V5 (20). In terms of the specific conformational epitopes of the two viruses, the majority of neutralizing monoclonal antibodies generated to HIV-1 appear to bind a conformational epitope involving the CD4 binding domain, whereas, this region has not been identified as a neutralizing domain of SIV. However, the repertoire of monoclonal antibodies generated to the SIV envelope is considerably less extensive than that of monoclonal antibodies generated to the HIV-1 envelope. Two neutralizing epitopes on SIV gp120 have been defined by mouse monoclonal antibodies, a linear V2 epitope and a conformational epitope that includes residues in the V3 and V4 domains (3, 22, 23, 25) but appears to be distinct from the CD4 binding site (8, 19, 20). The apparent importance of conformational epitopes as critical targets for neutralization of both HIV-1 and SIV suggests that the SIV-infected macaque model would make a relevant system for dissection of the role of antibody in protection.
A number of immunoprophylaxis trials have been conducted with serum or plasma collected from SIV-infected animals but have yielded conflicting results. In two studies, infusion of a plasma pool from SIVmac-infected animals or a cocktail of four neutralizing mouse monoclonal antibodies (13, 24) failed to protect any of the recipient macaques from a homologous SIV challenge. However, two other trials reported partial protection against a homologous intravenous virus challenge following administration of a plasma pool from SIVsm- or SIVmne-infected macaques (29, 38) and one reported partial protection against a heterologous SIVsm/B670 challenge (9). Protection of two of three neonatal macaques from oral challenge as a result of transplacentally acquired SIV antibodies also suggests a beneficial role for antibodies (49). Finally, in a postexposure immunotherapy trial, administration of purified immunoglobulin G (IgG) (SIVIG) from an SIV-infected LTNP macaque had a beneficial effect in modifying subsequent disease progression (14). Unfortunately, the neutralizing antibodies in these various pools differed significantly in character and breadth and therefore the conflicting results of many of these trials cannot be resolved.
The SIV-infected macaque model offers a unique system for dissection of the antiviral antibody response at the molecular level and assessment of the role of protective epitopes by in vivo experimentation. For this reason, we initiated a study to produce SIV envelope monoclonal antibodies with high-level neutralizing activity that could serve as standardized, renewable reagents. Since the humoral immune response to SIV infection mounted by LTNP might be predictive of an effective immune response and based upon the beneficial clinical effect of purified SIVIG from such a macaque (14), this nonprogressor macaque was chosen as a source of immune cells to extract RNA for the purpose of generating a combinatorial phage display library. The present report describes the recovery by phage display technology of envelope-specific Fabs with high-level homologous neutralizing activity.
MATERIALS AND METHODS
Animal.
An inguinal lymph node was biopsied from a rhesus macaque, E544, at 6 years post-SIVsmF236 challenge and was used as a source of mRNA for construction of a combinatorial phage display Fab library. This healthy monkey was also the source of a plasma pool used in a postexposure passive immunotherapeutic trial that demonstrated a beneficial effect of simian antibodies (14). The lymph node biopsy was gently disrupted and stored as a viable single-cell suspension in 10% dimethyl sulfoxide and 10% fetal calf serum in liquid nitrogen.
Construction of a lymph node γ1/κ antibody phage display library.
Total RNA isolated from 1.3 × 107 lymph node cells (RNA Isolation Kit; Stratagene, La Jolla, Calif.) was reverse transcribed into cDNA by using an oligo(dT) primer. Thirty cycles of 94°C for 15 s, 52°C for 50 s, and 72°C for 90 s were performed. Macaque κ-chain genes were amplified with primers specific for human κ light-chain genes. The Fd segment (variable and first constant domains) of macaque heavy-chain genes was amplified with a combination of family-specific human VH primers based at the 5′ end and a macaque isotype γ1-specific primer based at the 3′ end (16, 40), as listed in Table 1. A total of 27 heavy-chain and 21 κ-chain amplification reactions, each with a single pair of primers, were conducted.
TABLE 1.
Summary of oligonucleotide primers used to amplify the Fab library
| Primer(s) | Sequence |
|---|---|
| 5′ primers with SacI site-kappa light chain | 5′-GACATCGAGCTCACCCAGTCTCCA-3′ |
| 5′-GACATCGAGCTCACCCAGTCTCC-3′ | |
| 5′-GATATTGAGCTCACTCAGTCTCCA-3′ | |
| 5′-GAAATTGAGCTCACGCAGTCTCCA-3′ | |
| 5′-GAAATTGAGCTCAC(G/A)CAGTCTCCA-3′ | |
| 5′-GAGCCGCACGAGCCCGAGCTCCAGATGACCCAGTCTCC-3′ | |
| 5′-GAGCCGCACGAGCCCGAGCTCGTG(A/T)TGAC(A/G)CAGTCTCC-3′ | |
| 3′ primer with XbaI site-kappa light chain | 5′-GCGCCGTCTAGAATTAACACTCTCCCCTGTTGAAGCTCTTTGTGACGGGCGAACTCAG-3′ |
| 5′ primers with XhoI site-heavy chain | 5′-CAGGTGCAGCTCGAGCAGTCTGGG-3′ |
| 5′-CAGGTGCAGCTGCTCGAGTCTGGG-3′ | |
| 5′-CAGGTGCAGCTACTCGAGTCGGG-3′ | |
| 5′-GAGGTGCAGCTCGAGGAGTCTGGG-3′ | |
| 5′-GAGGTGCAGCTGCTCGAGTCTGGG-3′ | |
| 5′-CAGGTACAGCTCGAGCAGTCAGG-3′ | |
| 5′-AGGTGCAGCTGCTCGAGTCTGG-3′ | |
| 5′-CAGGTGCAGCTGCTCGAGTCGGG-3′ | |
| 5′-CAGGTGCAGCTACTCGAGTGGGG-3′ | |
| 3′ primer with SpeI site-heavy chain | 5′-AGGTTTACTAGTACCACCACATGTTTTGATCTC′-3′ |
Purification, restriction digestion, and sequential ligation of light- and heavy-chain DNAs into the pCOMB3H phage display vector and transformation of electrocompetent Escherichia coli XL-1 Blue (Stratagene) were performed essentially as previously described (2, 7). Briefly, 450 ng of κ-chain DNA digested with SacI and XbaI was ligated into 1,500 ng of pCOMB3H which had been digested with SacI and XbaI, and the ligated product was transformed into E. coli XL-1 Blue. Transformants were propagated overnight at 37°C by solid-phase amplification (28) in 3 liters of 0.3% SeaPrep agarose (FMC BioProducts, Rockland, Maine) in TerrificBroth (Gibco/BRL, Gaithersburg, Md.) supplemented with 1% glucose, 50 μg of carbenicillin per ml, and 10 μg of tetracycline per ml. Phagemid DNA (κ-chain pCOMB3H) was isolated from this overnight culture. Nine hundred nanograms of κ1 chain digested with XhoI and SpeI was ligated into 3,000 ng of similarly digested κ-chain pCOMB3H, and the ligated product was transformed into E. coli XL-1 Blue. Transformants were expanded in a volume of 5 liters by solid-phase amplification as described above. The final library of 3 × 107 clones was stored in 12.5% glycerol–Luria-Bertani (LB) broth at −80°C until use.
Panning and ELISA reagents.
SIVsmH4 recombinant glycoprotein 130 (rgp130) expressed in CHO cells was a gift from Nancy Haigwood, Bristol-Myers Squibb Pharmaceutical Research Institute (37, 42). SIVmac251 rgp120 expressed in baculovirus was purchased from Intracel Inc. (Cambridge, Mass.), and the following reagents were obtained through the AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases: SIVmac239 rgp140 produced by vaccinia virus provided by the Vaccine Research and Development Branch, Division of AIDS, National Institute of Allergy and Infectious Diseases; HIV-2 ST rgp120 expressed in S2 drosophila cells from Margery Chaikin, SmithKline Beecham Pharmaceuticals (4, 17, 26); and mouse monoclonal antibodies KK45 and KK46, specific for the V3 loop of SIV gp120, from Karen Kent and Caroline Arnold (25). In all panning and enzyme-linked immunosorbent assay (ELISA) experiments, recombinant envelope antigens were diluted to 2 to 8 μg/ml in phosphate-buffered saline (PBS) and adsorbed to EIA/RIA A/2 (ELISA) plates (Costar, Cambridge, Mass.) overnight at 4°C.
Library screening.
A total of 109 bacteria of the library stock were thawed and grown for 90 min at 37°C in 100 ml of 2×YT broth (Gibco/BRL) supplemented with 1% glucose, 100 μg of carbenicillin per ml, and 10 μg of tetracycline per ml. Bacteria were pelleted and resuspended in 100 ml of 2×YT broth with 100 μg of carbenicillin per ml and 10 μg of tetracycline per ml, and helper phage VCS M13 (Stratagene) was added at a multiplicity of infection of 50. The culture was expanded to 1,000 ml and shaken for 5 to 6 h at 37°C (30). Phage particles were precipitated overnight at 4°C from the clarified supernatant with 4% polyethylene glycol and 0.5 M NaCl, recovered by centrifugation, and resuspended in 2% skim milk powder in PBS with 0.05% sodium azide. ELISA wells were coated with the selection antigen, incubated overnight, and blocked with 2% skim milk powder in PBS and 0.05% sodium azide for 1 h at room temperature prior to addition of phage particles. Unbound phage particles were removed after 1 h by washing each well 10 times with PBS containing 0.05% Tween 20. Bound phage particles were eluted with 100 mM triethylamine (Sigma, St. Louis, Mo.) and immediately neutralized with 2 M Tris (pH 7.2). Exponentially growing E. coli XL-1 Blue was infected with the eluted phage and expanded by solid-phase amplification as described above. The eluted phage particles (clones) were enumerated after each round of panning and when a 5- to 10-fold increase was encountered (typically after the third round), phagemid DNA was extracted. The gene III-encoding segment of the phagemid was removed by restriction digestion with SpeI and NheI, and the phagemid DNA was self-ligated and transformed into E. coli XL-1 Blue. A total of 30 to 60 single colonies were picked, and each was inoculated into 200 μl of LB broth supplemented with 1% glucose and 100 μg of carbenicillin per ml in 96-well microtiter plates and grown overnight at 30°C. Fab production was induced by growing the colonies in 200 μl of LB broth with 100 μg of carbenicillin per ml and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Gold Biotechnology, St. Louis, Mo.) for 5 h. Crude bacterial supernatants were assayed directly by ELISA for reactivity with the same antigen used for selection.
Large-scale Fab production and purification.
To facilitate purification of Fabs, a six-residue histidine tail (6×HIS) was added to the C terminus of the heavy chain by overlapping PCR. Primers 5′-CAG GTG CAG CTG CTC GAG TCG GG-3′ and 5′-TCC CAG ATC TGC GGC CGC TTA AAT TAA TTA ATG GTG ATG G-3′ were used to amplify the heavy-chain gene from Fab clone 201. The primary PCR product was reamplified with primers 5′-CAG GTG CAG CTG CTC GAG TCG GG-3′ and 5′-TAA TTA ATG GTG ATG GTG ATG GTG GCT AGC ACC ACC ACA TGT TTT G-3′. The resulting fragment was cut with restriction enzymes XhoI and NotI and ligated back into Fab clone 201 which had been linearized with XhoI and NotI to create Fab 201-6×HIS. Fab clone 201-6×HIS was subsequently used as a cloning vector to introduce 6×HIS tails into Fab clones 101, 102, 202, 203, and 204 by subcloning the SacI-to-SauI fragment.
Bacterial cultures of 1 to 2 liters were grown in LB broth (Gibco/BRL) with 1% glucose and 100 μg of carbenicillin per ml at 30°C. At an optical density at 600 nm of approximately 0.2, the bacteria were pelleted and resuspended in LB broth supplemented with 100 μg of carbenicillin per ml and 0.1 mM IPTG. Cultures were induced for 5 h at room temperature. For Fabs with a 6×HIS tail, the bacterial pellet was resuspended in 20 to 40 ml of PBS–0.1 mM phenylmethylsulfonyl fluoride and subjected to three cycles of freezing (−80°C) and thawing (37°C). The extract was clarified by centrifugation and purified by chelate chromatography on a nitrilotriacetic acid column (Novagen, Madison, Wis.) by following the supplier’s instructions. For Fabs without a 6×HIS-tail, the bacterial pellet was resuspended in 20 to 40 ml of 20 mM Tris (pH 8.6) and subjected to three cycles of freezing (−80°C) and thawing (37°C). The clarified extract was applied to a 10-ml open DEAE–Affi-Gel Blue column (Bio-Rad, Hercules, Calif.) and washed with 20 column volumes of 20 mM Tris (pH 8.6). The column was eluted stepwise in 20-ml fractions containing 100, 250, and 500 mM NaCl. All fractions were tested in an ELISA for reactivity with antigen. Most macaque Fabs eluted in the 100 mM NaCl fraction when tested by ELISA for gp120 reactivity. Purified Fab was concentrated and dialyzed against PBS. Fab purity was judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and concentration was determined with a BCA Protein Assay Reagent kit (Pierce, Rockford, Ill.) with bovine serum albumin as the standard. The concentration of DEAE–Affi-Gel Blue-purified Fabs was estimated with an antigen capture ELISA using unconjugated and alkaline phosphatase-conjugated goat anti-human Fab antibodies (Pierce). Nitrilotriacetic acid column-purified Fab served as a standard.
ELISA analysis of Fab reactivity and cross-reactivity.
The following protein antigens were diluted and adsorbed to polystyrene plates as described above: SIV or HIV-2 gp120 at 2 μg/ml, carbonic anhydrase at 10 μg/ml, ovalbumin at 10 μg/ml, and thyroglobulin (Sigma) at 100 μg/ml. Plates were blocked with 3% bovine serum albumin–PBS for 1 h, and 50 μl of crude or purified Fab was subsequently added to the wells. Following 1 h of incubation, Fab binding was detected with a 1:1,000 goat anti-human F(ab′)2-alkaline phosphatase conjugate (Pierce). The assay was developed with 1 mg of p-nitrophenyl phosphate (Sigma) per ml.
Epitope mapping.
In initial experiments, Fabs 102 and 202 were tested in a competition ELISA as described previously (23). A panel of mouse monoclonal antibodies that define four epitope clusters on the SIV envelope glycoprotein were used: a conformation-dependent neutralization epitope in the V3-V4 region (KK5, KK9, KK17, KK44, KK56, and KK57), a linear epitope in the V3 loop (KK45 and KK46), a linear epitope in the V1-V2 region (KK13), and an epitope within gp41 (KK41). In a second series of experiments, the epitope specificities of Fabs 101, 102, 103, 201, 202, 203, and 204 were determined by using KK45 and 201-IgG1. Antibody 201-IgG1 is Fab 201 converted into a whole macaque IgG1 and expressed in COS cells (unpublished data). Subsaturating concentrations of KK45 and 201-IgG1 were mixed with serial dilutions of Fabs. The mixture was transferred to plates coated with SIVmac251 gp120 at 2 μg/ml and incubated for 2 h. Binding of mouse monoclonal antibody and 201-IgG1 was detected with a goat anti-mouse Ig-alkaline phosphatase conjugate (Jackson ImmunoResearch, West Grove, Pa.) and a goat anti-human IgG (heavy and light chains)-alkaline phosphatase conjugate, respectively. The latter conjugate does not cross-react with macaque Fabs expressed in bacteria; thus, it only detects whole macaque IgG. Fab binding was measured in parallel by setting up duplicate dilution series.
Peptide mapping.
Twenty-mer peptides, with an overlap of 10 amino acids, spanning the whole of SIVmac251 gp120 (EVA774) were obtained from the Reagent Program of the European Vaccine against AIDS. The reactivity of Fab 102 against these peptides was tested in an ELISA.
Assay of SIV-neutralizing antibodies.
SIV neutralization was measured by using a cell-killing assay as previously described (15, 32). All neutralization experiments were performed with purified Fabs or heat-inactivated plasma samples. SIVsmH4, SIVsmE543-3, and SIVmac239 are molecularly cloned viruses, whereas SIVsmE660, SIVsmB670, and SIVmac251 are uncloned virus stocks. Virus stocks were produced in H9 cells (SIVsmH4, SIVsmB670, and SIVmac251) or CEM×174 cells (SIVsmE543-3 and SIVmac239).
Nucleic acid sequencing and analysis.
Nucleic acid sequencing was carried out on a 373A automated DNA sequencer (Applied Biosystems) using a Taq fluorescent dideoxynucleotide terminator cycle sequencing kit. The following sequencing primers were used: heavy chain, 5′-ATTGCCTACGGCAGCCGCTGG-3′ (HC1) and 5′-GGAAGTAGTCCTTGACCAGGC-3′ (HC4); κ chain, 5′-ACAGCTATCGCGATTGCAGTG-3′ (LC1) and 5′-CACCTGATCCTCAGATGGCGG-3′ (LC4). Resulting sequences were analyzed by using MacVector (International Biotechnologies, New Haven, Conn.) and GeneWorks (IntelliGenetics, Campbell, Calif.) software. Sequence similarity searches were performed with V-base, a compilation of all available human variable-segment Ig germ line sequences obtained from I. Tomlinson (10).
RESULTS
Macaque SIV gp120-specific Fabs.
RNA extracted from an inguinal lymph node of SIV-infected LTNP rhesus macaque E544 was used as the template for first-strand cDNA synthesis. With the primers shown in Table 1, the κ light-chain gene and the Fd segment of the γ1 heavy-chain gene were amplified and cloned into a phagemid vector. The resulting Fab phage library was then selected against monomeric gp120 preparations of SIVsmH4, SIVmac251, or HIV-2/ST, and seven SIV gp120-specific Fabs were isolated. Every clone, except Fab 204, was recovered at least twice in independent panning experiments, showing the reproducibility of the propagation procedure employed in this study. To increase the diversity of the Fabs recovered or enrich for rarer Fabs, two epitope-masking experiments were conducted. Masking of the epitope recognized by Fab 201 resulted in the reisolation of group 1 clones solely. The masking of two epitopes (Fabs 101 and 201) resulted in the isolation of a variety of clones of which only one was new, Fab 204. In addition, two Fabs (1LNRhE544 and 2LNRhE544) were selected from the unscreened library and used as negative controls in subsequent experiments.
SIV gp120-specific Fabs form two distinct groups.
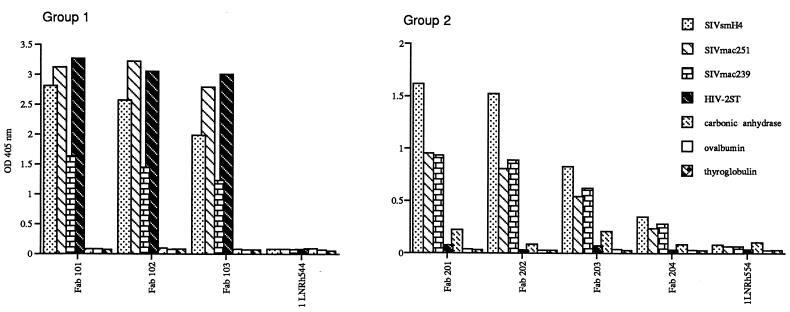
The specificity of the selected Fabs was assessed by ELISA using a panel of SIV and HIV-2 recombinant envelope proteins. As shown in Fig. 1, all seven Fabs were envelope specific, with no evidence of cross-reactivity to a panel of randomly chosen, unrelated protein antigens. Three of the Fabs were broadly reactive, binding to recombinant gp120 from SIVsmH4, SIVmac251, SIVmac239, and HIV-2/ST, whereas the reactivity of the other four Fabs was restricted to the SIV envelope preparations and did not include HIV-2 rgp120, suggesting that two distinct groups of Fabs had been recovered. Significantly, one of the Fabs with broad reactivity had been obtained by panning with HIV-2/ST gp120. Based upon this binding specificity, the Fab clones were numbered according to these two groups as 101, 102, and 103 for the broadly reactive Fabs and 201 to 204 for those specific for SIV envelopes alone.
FIG. 1.
Graphical representation of ELISA results. Reactivity of Fabs (10 μg/ml) to monomeric rgp120 from various SIV and HIV-2 isolates and a panel of irrelevant protein antigens. Fabs 101, 102, 103, 201, 202, 203, and 204 were selected by panning of the library against monomeric SIV rgp120, whereas 1LNRh544 is a randomly picked clone from the unscreened library.
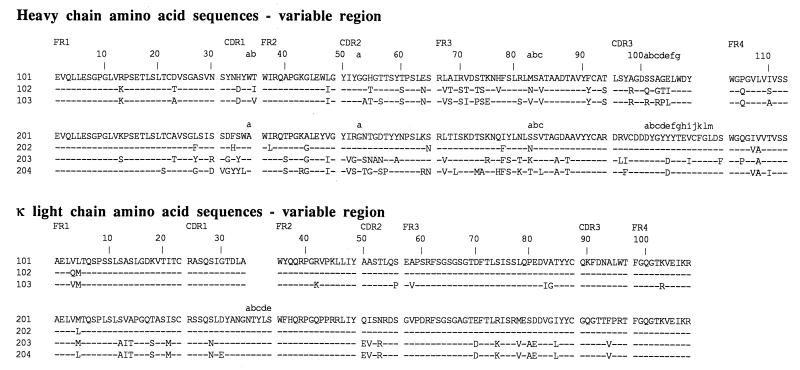
Sequence analysis confirmed the grouping based upon antigen binding. As shown in Fig. 2, the deduced amino acid sequence of the VH-D-JH and Vκ-Jκ segments illustrated relatedness among the Fabs within each group. Most notably, the heavy-chain CDR3 (HCDR3) regions within a group were conserved in terms of amino acid sequence and length. Within group 1, the heavy-chain sequences of Fabs 101, 102, and 103 shared 77 to 87% identity overall, with the greatest divergence observed in CDR2, FR3, and CDR3. The κ chains of clones 101 and 102 were identical, with the exception of two amino acid differences at the amino terminus due to the use of different primers for amplification. The κ chain of clone 103 was only slightly more divergent, differing from the latter two clones by six amino acid substitutions.
FIG. 2.
Alignment of the deduced amino acid sequences of the variable domains of the heavy and κ chains. Substitutions relative to either Fab 101 or 201 are shown in single amino acid code. Identical residues are indicated by dashes. Amino acids are numbered in accordance with Kabat et al. (21), and complementary determining regions (CDR1, CDR2, and CDR3) and framework regions (FR1, FR2, FR3, and FR4) are shown above the sequence alignments.
Within group 2, Fabs 201 and 202 were highly related, differing in only nine residues in the heavy chain and one residue in the kappa chain (due to the use of a different PCR primer). These clones had identical HCDR3 sequences. Fab 204 was the most divergent clone within this group, differing in 36 residues from either Fab 201 or 202. Despite the overall divergence, each of the Fabs within this group showed remarkably little variation in the HCDR3; most of the amino acid differences were seen within HCDR1, HCDR2, and FR3. An additional unique feature of each of the group 2 Fabs was the presence of two cysteine residues in HCDR3 that probably are bridged by a disulfide bond, thus forming an extra loop.
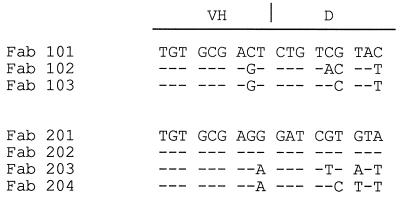
The striking length and sequence uniformity of the CDR3 regions within each group of Fabs suggested that each group might represent somatic variants of a common precursor. However, there were a number of differences at the nucleotide level at the VH-D junctional regions of the group 1 Fabs (Fig. 3), despite conserved amino acid sequences, indicating that the clones probably arose from independent recombinational events. In group 2, Fabs 201 and 202 had identical VH-D junctional regions and were probably somatic variants, but Fabs 203 and 204 appeared distinct and also may have arisen from independent precursors.
FIG. 3.
Alignment of the nucleotide sequences of the VH-D junctions from various SIV rgp120-specific Fab clones. Three codons on either side of the VH-D junction of Fab 101 or 201 are shown. Substitutions in clones 102, 103, 202, 203, and 204 are indicated by the appropriate nucleotides, and identity is indicated by dashes.
The occurrence of specific Fabs with relatively conserved HCDR3 regions but considerable variability throughout the rest of the VH gene has been described previously for anti HIV-1 gp41 Fabs (5). This could arise from multiple crossover events occurring during PCR amplification of antibody genes in library construction. A more likely explanation is that it represents convergent evolution driven by an HCDR3 motif giving high-affinity binding to a molecular feature on gp120.
To tentatively establish the germ line origin of the Fab clones, we relied upon the extensive homology between macaque and human Igs to conduct similarity searches of all known human Ig genes. As shown in Table 2, all of the heavy-chain sequences exhibited homology to the human VH4 family of germ line segments. All of the group 1 Fabs were most related to VH4.39. The heavy chains of Fabs 201 and 202 showed the best match to human VH segment 4.42 (50), whereas Fabs 203 and 204 showed the best alignment to human VH segments DP-67 (51) and DP-71 (47), respectively. However, in all four homology searches, a high score was obtained for human VH segment DP-67. Significant similarity to any of the human D segments was not found by searching V-base or all of GenBank.
TABLE 2.
Homology of macaque heavy and kappa light chains of various SIV-specific Fabs with human Ig germ line genesa
| Fab clone | VH | D | JH | VK | JK |
|---|---|---|---|---|---|
| 101 | 4.39 | ND | J4b | DPK4 | JK1 |
| 102 | 4.39 | ND | J4b | DPK4 | JK1 |
| 103 | 4.39 | ND | J4b | DPK4 | JK1 |
| 201 | 4.42 | ND | J6b | DPK18 | JK1 |
| 202 | 4.42 | ND | J6b | DPK18 | JK1 |
| 203 | DP-67 | ND | J6b | DPK18 | JK1 |
| 204 | DP-71 | ND | J6b | DPK18 | JK1 |
The human germ line segment with the highest score by DNA matrix analysis in MacVector is identified. ND, not determined due to lack of any identifiable homolog.
Macaque Fabs react with two distinct regions of SIV gp120.
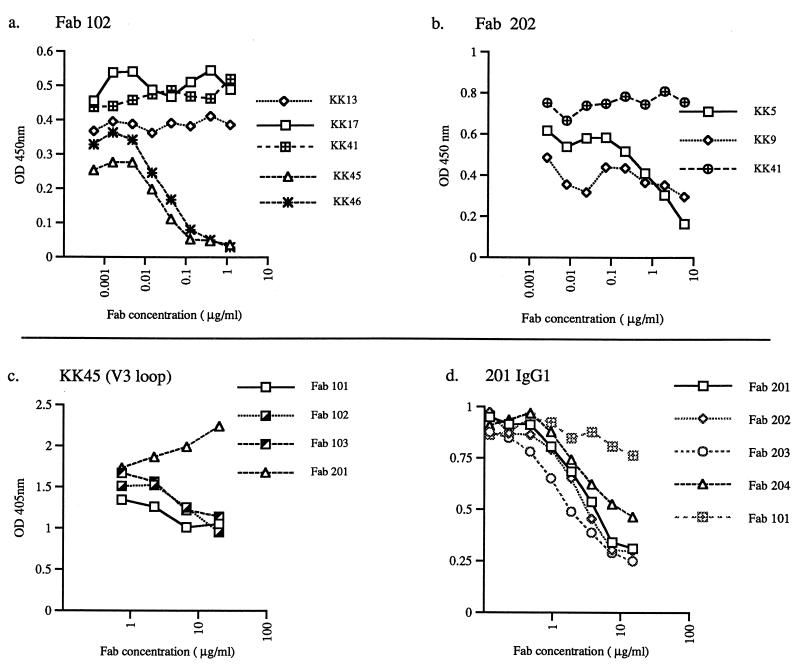
To identify the epitopes recognized by these macaque Fabs, we tested their ability to compete for binding in an ELISA with mouse monoclonal antibodies of known specificity. We utilized a series of mouse monoclonal antibodies that were recovered by Kent et al. following immunization with gp120 of SIVmac (23, 25), which shares 82% identity with SIVsm, the strain that induced the Fabs. As seen in Fig. 4a, Fab clone 102 competed with the binding of MAbs KK45 and KK46, which both recognize a linear epitope in the V3 loop. Peptide mapping indicated that Fab 102 reacted with two overlapping peptides (EVA774.30 and EVA774.31 (1) in the V3 loop (data not shown). Thus, it appeared that the epitope recognized by Fab 102 was contained within 10-mer peptide VTIMSGLVFH (amino acids 322 to 331), of which the last seven residues are perfectly conserved in SIVmac, SIVsm, and HIV-2, thereby accounting for the cross-reactivity of this Fab with these three envelope preparations. As expected based on sequence similarity, each of the group 1 Fabs also competed with V3 loop-specific monoclonal antibody KK45 (Fig. 4c). In contrast, Fab 202 competed with two neutralizing monoclonal antibodies, KK5 and KK9 (Fig. 4b), that target another epitope cluster (KK5, KK9, KK17, KK44, KK56, and KK57) (22, 23, 25) and recognize a conformation-dependent epitope in the V3-to-V4 region of the SIV envelope (8). Whole antibody 201-IgG1 inhibited the binding of all group 2 Fabs, consistent with recognition of a common region by these Fabs (Fig. 4d).
FIG. 4.
Identification of epitope specificity of macaque Fabs by competition ELISA with mouse monoclonal antibodies of defined specificity. Competition for binding of mouse monoclonal antibodies with a dilution series of Fab 102 (a) and Fab 202 (b) as the competitor. (c) Competition of Fabs 101, 102, 103, and 104 with V3 loop-specific monoclonal antibody KK45 as measured by the binding of the mouse monoclonal antibody. (d) Competition of Fabs 201, 202, 203, and 204 with antibody 201-IgG1 as measured by binding of the whole macaque antibody. OD, optical density.
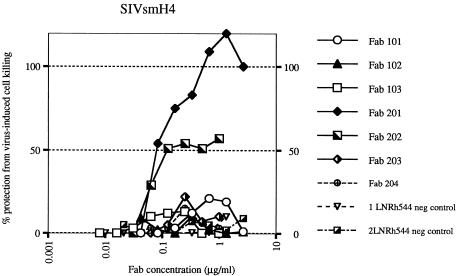
Neutralizing activity of macaque Fabs.
All seven SIV-specific Fabs, including the two randomly chosen Fabs from the unscreened library, were purified and tested for neutralizing activity in vitro. Only Fabs 201 and 202 demonstrated neutralizing activity against the homologous molecularly cloned virus SIVsmH4, as illustrated in Fig. 5. Although related to these clones, Fab clones 203 and 204 did not exhibit neutralizing activity. Subsequently, Fabs 201 and 202 were evaluated against a broader panel of viruses belonging to the SIVsm and SIVmac lineage. As shown in Table 3, the homologous virus SIVsmB670 was readily neutralized by Fabs 201 and 202 at a concentration of about 40 ng/ml, whereas heterologous viruses such as SIVmac251 and SIVmac239 were refractory to neutralization in vitro. A recently characterized neutralization-resistant infectious clone, SIVsmE543-3 (15), that also belongs to the SIVsm lineage, and shares 92% envelope identity with SIVsmH4, was resistant to neutralization by Fab 201 or 202 (data not shown). The type specificity of neutralization by these macaque Fabs was similar to the neutralizing profile observed for SIVIG purified from this donor macaque; the polyclonal SIVIG neutralized homologous SIVsmE660 and SIVsmB670 at a concentration of approximately 1 μg/ml but required a 40-fold higher concentration to neutralize the heterologous virus SIVmac251 (Table 3).
FIG. 5.
Neutralization titration curve of macaque Fabs against SIVsmH4. The Fab concentration is plotted versus the resulting protection from virus-induced killing in CEM×174 cells. Two Fabs (201 and 202) neutralized SIV in a dose-dependent fashion, whereas all other Fabs, including two irrelevant Fabs, 1LNRh544 and 2LNRh544 (negative [neg] controls); from the unscreened library, failed to neutralize SIV, even at high concentrations.
TABLE 3.
Profile of neutralizing activities of Fabs 201 and 202 against various SIV isolatesa
| Prepn | Concn (μg/ml) of Fab or IgG needed to neutralize indicated SIV strain
|
||||
|---|---|---|---|---|---|
| Homologous strains
|
Heterologous strains
|
||||
| smH4 | smB670 | smE660 | SIVmac251 | SIVmac239 | |
| Fab 201 | 0.078 | <0.042 | ND | >5.53 | >5.53 |
| Fab 202 | 0.117 | 0.038 | ND | >2.00 | >2.00 |
| SIVIG | ND | 1.35 | 0.78 | 40.4 | >50.0 |
The concentrations of Fab or SIVIG that resulted in 50% protection of CEM×174 cells from virus-induced cell killing are shown. ND, not determined.
DISCUSSION
This study demonstrates the usefulness of the antibody phage display library approach for recovering antigen-specific macaque antibodies from a SIV-infected animal and describes the first fully homologous macaque Fabs to be generated by this technique. In the present study, seven unique SIV gp120-specific macaque Fabs were obtained by screening a lymph node cDNA library from a healthy long-term survivor of SIV infection (rhesus macaque E544). Based on nucleotide sequence similarity, pattern of gp120 reactivity, and epitope-mapping analysis, as defined by competition with previously characterized mouse monoclonal antibodies, the Fab clones formed two distinct groups. One group of Fabs recognized a nonneutralization, linear epitope in the V3 loop, whereas the other recognized a neutralization, conformation-dependent epitope in the V3-to-V4 region of gp120. Neutralizing Fabs 201 and 202 were highly effective in neutralizing virus isolates very closely related to the virus that had infected this macaque (SIVsmH4 and SIVsmB670) but failed to neutralize the heterologous virus SIVmac251. Not unexpectedly, the Fabs also failed to neutralize a homologous, molecularly cloned, neutralization-resistant SIVsm (SIVsmE543-3 [15]). These findings underscore the observation that a biologically important neutralization epitope recognized by SIV-infected animals is within the V3-to-V4 region (8, 20, 22, 23, 25).
The specificity of binding of the Fabs to various gp120 preparations was not predictive of the ability to neutralize the isolates from which these envelopes were cloned. For example, group 2 Fabs bound to SIVmac and SIVsm gp120 preparations but neutralized only SIVsm isolates. This disparity between the ability to bind monomeric gp120 and neutralization might be due to differences in affinity for the oligomeric envelope as expressed on virions. This disparity has precedents in other viral systems, including HIV-1. For example, affinity of monoclonal antibodies for the mature oligomeric form of the envelope glycoprotein is more predictive of neutralization of HIV-1 primary isolates than is binding to monomeric forms of this envelope antigen (12, 33, 39, 41, 46). Interestingly, the preferential ability of the Fabs to neutralize homologous isolates was similar to the profile of a contemporaneous IgG preparation purified from this macaque, suggesting that the neutralizing Fabs recovered from this combinatorial library were representative of the antibody response of this animal. Although this library is unlikely to represent the full antibody repertoire, this technique shows promise as a method for the recovery of macaque SIV-specific neutralizing Fabs. Potential factors that could limit the repertoire are the restricted size of the library (3 × 107 clones) and the exclusive use of kappa light chains and γ1 heavy chains. Finally, since this is a systemic infection, the actual choice of tissue for generation of the library could affect the overall representation of clones. However, in an independently screened bone marrow library prepared from the same animal, all but one of the Fab clones grouped both biologically and genetically with those obtained from the lymph node library (data not shown), suggesting that the tissue source was not a major determinant of the antibody repertoire representation.
In summary, the SIV envelope Fabs generated from a SIV-infected LTNP provide further insight into the epitopes recognized by such animals. Only two epitopes were recognized by this particular animal, one of which constituted a neutralization epitope. A third Fab generated from the bone marrow library recognized a third epitope in the V1 region of gp120 which did not induce neutralizing antibodies (data not shown). This animal exhibited tightly restricted viral replication and, perhaps as a consequence, had a low-to-moderate serum neutralizing antibody titer which was fairly type specific. The spectrum of epitopes recognized by sera of SIV-infected progressor macaques may be more extensive. We are currently generating IgG1 macaque monoclonal antibodies which express the representative Fabs of each group. Such macaque monoclonal antibodies should prove valuable in defining the role of neutralizing antibody in preventing or ameliorating SIV infection in vivo. Additionally, the availability of molecularly cloned SIV stocks that are sensitive or resistant to neutralization by this antibody will facilitate studies intended to define the in vivo role of neutralizing antibodies that are identified by in vitro assays.
ACKNOWLEDGMENTS
We thank Robert Chanock for his support and for critical reading of the manuscript, Russ Byrum and Anna Hahn for technical support, and Mats Persson for sharing macaque Ig sequence information data prior to publication.
REFERENCES
- 1.Almond N, Jenkins A, Slade A, Cranage H A M, Kitchin P. Population sequence analysis of simian immunodeficiency (32H reisolate of SIVmac251): a virus stock used for international vaccine studies. AIDS Res Hum Retroviruses. 1992;8:77–88. doi: 10.1089/aid.1992.8.77. [DOI] [PubMed] [Google Scholar]
- 2.Barbas C F, III, Kang A S, Lerner R A, Benkovic S J. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc Natl Acad Sci USA. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benichou S, Legrand R, Nakagawa N, Taure T, Traincard F, Vogt G, Dormont D, Tiollais P, Kieny M-P, Madaule P. Identification of a neutralizing domain in the external envelope glycoprotein of simian immunodeficiency virus. AIDS Res Hum Retroviruses. 1992;8:1165–1170. doi: 10.1089/aid.1992.8.1165. [DOI] [PubMed] [Google Scholar]
- 4.Benos D J, Hahn B H, Bubien J K, Ghosh S K, Mashburn N A, Chaikin M A, Shaw G M, Benveniste E N. Envelope glycoprotein gp120 of human immunodeficiency virus type 1 alters ion transport in astrocytes: implications for AIDS dementia complex. Proc Natl Acad Sci USA. 1994;91:494–498. doi: 10.1073/pnas.91.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binley J M, Ditzel H J, Barbas III C F, Sullivan N, Sodroski J, Parren P W, Burton D R. Human antibody responses to HIV type 1 glycoprotein 41 cloned in phage display libraries suggest three major epitopes are recognized and give evidence for conserved antibody motifs in antigen binding. AIDS Res Hum Retroviruses. 1996;12:911–924. doi: 10.1089/aid.1996.12.911. [DOI] [PubMed] [Google Scholar]
- 6.Buchbinder S P, Katz M H, Hessol N A, O’Malley P M, Holmberg S D. Long-term HIV-1 infection without immunological progression. AIDS. 1994;8:1123–1128. doi: 10.1097/00002030-199408000-00014. [DOI] [PubMed] [Google Scholar]
- 7.Burton D R, Barbas III C F, Persson M A A, Koening S, Chanock R M, Lerner R A. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci USA. 1991;88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi W S, Collignon C, Thiriart C, Burns D P W, Stott E J, Kent K A, Desrosiers R C. Effects of natural sequence variation on recognition by monoclonal antibodies that neutralize simian immunodeficiency virus infectivity. J Virol. 1994;68:5395–5402. doi: 10.1128/jvi.68.9.5395-5402.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clement J E, Montelaro R C, Zink M C, Amedee A M, Miller S, Trichel A M, Jagerski B, Hauer D, Martin L N, Bohm R P, Murphey-Corb M. Cross-protective immune responses induced in rhesus macaques by immunization with attenuated macrophage-tropic simian immunodeficiency virus. J Virol. 1995;69:2737–2744. doi: 10.1128/jvi.69.5.2737-2744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook G P, Tomlinson I M. The human immunoglobulin VH repertoire. Immunol Today. 1995;16:237–242. doi: 10.1016/0167-5699(95)80166-9. [DOI] [PubMed] [Google Scholar]
- 11.Desrosiers R C. The simian immunodeficiency viruses. Annu Rev Immunol. 1990;8:557–578. doi: 10.1146/annurev.iy.08.040190.003013. [DOI] [PubMed] [Google Scholar]
- 12.Fouts T R, Binley J M, Trkola A, Robinson J E, Moore J P. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J Virol. 1997;71:2779–2785. doi: 10.1128/jvi.71.4.2779-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner M, Rosenthal A, Jennings M, Yee J, Antipa L, Robinson E. Passive immunization of rhesus macaques against SIV infection and disease. AIDS Res Hum Retroviruses. 1995;11:843–854. doi: 10.1089/aid.1995.11.843. [DOI] [PubMed] [Google Scholar]
- 14.Haigwood N L, Watson A, Sutton W F, McClure J, Lewis A, Ranchis J, Travis B, Voss G, Letvin N, Hu S-L, Hirsch V M, Johnson P R. Passive immune globulin therapy in the SIV/macaque model: early intervention can alter disease profile. Immunol Lett. 1996;51:107–114. doi: 10.1016/0165-2478(96)02563-1. [DOI] [PubMed] [Google Scholar]
- 15.Hirsch V, Adger-Johnson D, Campbell B, Goldstein S, Brown C, Elkins W R, Montefiori D C. A molecularly cloned, pathogenic, neutralization-resistant simian immunodeficiency virus, SIVsmE543-3. J Virol. 1997;71:1608–1620. doi: 10.1128/jvi.71.2.1608-1620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huse W D, Sastry L, Iverson S A, Kang A S, Alting-Mees M, Burton D R, Benkovic S J, Lerner R A. Generation of a large combinatorial library of the immunoglobulin repertoire in phage lambda. Science. 1989;246:1275–1281. doi: 10.1126/science.2531466. [DOI] [PubMed] [Google Scholar]
- 17.Ivey-Hoyle M, Culp J S, Chaikin M A, Hellmig B D, Matthews T J, Sweet R W, Rosenberg M. Envelope glycoproteins from biologically diverse isolates of human immunodeficiency viruses have widely different affinities for CD4. Proc Natl Acad Sci USA. 1991;88:512–516. doi: 10.1073/pnas.88.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Javaherian K, Langlois A J, McDanal C, Ross K L, Eckler L I, Hellis C L, Profy A T, Rusche J R, Bolognesi D P, Putney S D, Matthews T J. Principal neutralizing domain of the human immunodeficiency virus type 1 envelope protein. Proc Natl Acad Sci USA. 1989;86:6768–6772. doi: 10.1073/pnas.86.17.6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Javaherian K, Langlois A J, Montefiori D C, Kent K A, Ryan K A, Wyman P D, Stott J, Bolognesi D P, Murphey-Corb M, Larosa G J. Studies of the conformation-dependent neutralizing epitopes of simian immunodeficiency virus envelope protein. J Virol. 1994;68:2624–2631. doi: 10.1128/jvi.68.4.2624-2631.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Javaherian K, Langlois A J, Schmidt S, Kaufmann M, Cates N, Langedijk J P M, Meloen R H, Desrosiers R C, Burns D P W, Bolognesi D P, LaRosa G J, Putney S D. The principal neutralization determinant of simian immunodeficiency virus differs from that of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:1418–1422. doi: 10.1073/pnas.89.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabat E A, Wu T T, Perry H M, Gottesman K S, Foeller C. Sequences of proteins of immunological interest. 5th ed. Bethesda, Md: U.S. Department of Health and Human Services; 1991. [Google Scholar]
- 22.Kent K, Powell C, Corcoran T, Jenkins A, Almond N, Jones I, Robinson J, Stott E J. Huitieme colloque des cent gardes. Lyon, France: Fondation Marcel Merieux; 1993. Characterisation of neutralising epitopes on simian immunodeficiency virus envelope using monoclonal antibodies; pp. 167–172. [Google Scholar]
- 23.Kent K A, Gritz L, Stallard G, Crangage M P, Collignon C, Thiriart C, Corcoran T, Silvera P, Stott E J. Production and characterisation of monoclonal antibodies to simian immunodeficiency virus envelope glycoproteins. AIDS. 1991;5:829–836. doi: 10.1097/00002030-199107000-00006. [DOI] [PubMed] [Google Scholar]
- 24.Kent K A, Kitchen P, Mills K H G, Page M, Taffs F, Corcoran T, Silvera P, Flanagan B, Powell C, Rose J, Ling C, Aubertin A M, Stott E J. Passive immunization of cynomolgus macaques with immune sera or a pool of neutralizing monoclonal antibodies failed to protect against challenge with SIVmac251. AIDS Res Hum Retroviruses. 1994;10:189–194. doi: 10.1089/aid.1994.10.189. [DOI] [PubMed] [Google Scholar]
- 25.Kent K A, Rud E, Corcoran T, Powell C, Thiriart C, Collignon C, Stott E J. Identification of two neutralizing and 8 non-neutralizing epitopes on simian immunodeficiency virus envelope using monoclonal antibodies. AIDS Res Hum Retroviruses. 1992;8:1147–1151. doi: 10.1089/aid.1992.8.1147. [DOI] [PubMed] [Google Scholar]
- 26.Kong L I, Lee S, Kappes J C, Parkin J S, Decker D, Hoxie J A, Hahn B H, Shaw G M. West African HIV-2-related human retrovirus with attenuated cytopathicity. Science. 1988;240:1525–1529. doi: 10.1126/science.3375832. [DOI] [PubMed] [Google Scholar]
- 27.Koup R A, Safrit J T, Cao Y, Andrews C A, McLeod G, Borkowsky W, Farthing C, Ho D D. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kriegler M. Gene transfer and expression: a laboratory manual. W. H. New York, N.Y: Freeman & Company; 1990. [Google Scholar]
- 29.Lewis M G, Elkins W R, McCutchan F, Benveniste R E, Lai C Y, Montefiori D C, Burke D S, Eddy G A, Shafferman A. Passively transferred antibodies directed against conserved regions of SIV envelope protect macaques from SIV infection. Vaccine. 1993;11:1347–1355. doi: 10.1016/0264-410x(93)90106-8. [DOI] [PubMed] [Google Scholar]
- 30.Marks J D, Hoogenboom H R, Bonnert T P, McCafferty J, Griffiths A D, Winter G. Human antibodies from V-gene libraries displayed on phage. J Mol Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 31.Miller M A, Murphey-Corb M, Montelaro R C. Identification of broadly reactive continuous antigenic determinants of simian immunodeficiency virus glycoproteins. AIDS Res Hum Retroviruses. 1992;8:1153–1164. doi: 10.1089/aid.1992.8.1153. [DOI] [PubMed] [Google Scholar]
- 32.Montefiori D C, Robinson W E, Jr, Schuffman S S, Mitchell W M. Evaluation of antiviral drugs and neutralizing antibodies to human immunodeficiency virus by rapid and sensitive microtiter infection assay. J Clin Microbiol. 1988;26:231–235. doi: 10.1128/jcm.26.2.231-235.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore J P, Cao Y, Qing L, Sattentau Q J, Pyati J, Koduri R, Robinson J, Barbas III C F, Burton D R, Ho D D. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J Virol. 1995;69:101–109. doi: 10.1128/jvi.69.1.101-109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore J P, Ho D D. Antibodies to discontinuous or conformationally sensitive epitopes on the gp120 glycoprotein of human immunodeficiency virus type 1 are highly prevalent in sera of infected humans. J Virol. 1993;67:863–875. doi: 10.1128/jvi.67.2.863-875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munoz A, Kirkby A J, He Y D, Margolick J B, Visscher B R, Rinaldo C R, Kaslow R A, Phair J P. Long-term survivors with HIV-1 infection: incubation period and longitudinal patterns of CD4+ lymphocytes. J Acquired Immune Defic Syndr. 1995;8:496–505. doi: 10.1097/00042560-199504120-00010. [DOI] [PubMed] [Google Scholar]
- 36.Palker T J, Clark M E, Langlois A J, Matthews T J, Winhold K J, Randall R R, Bolognesi D P, Haynes B F. Type-specific neutralization of the human immunodeficiency virus with antibodies to env-encoded synthetic peptides. Proc Natl Acad Sci USA. 1988;85:1932–1936. doi: 10.1073/pnas.85.6.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Planelles V, Haigwood N L, Marthas M L, Mann K A, Scandella C, Lidster W D, Schuster J R, van Kuyk R, Marx P A, Gardner M B, Luciw P A. Functional and immunological characterization of SIV envelope glycoprotein produced in genetically engineered mammalian cells. AIDS Res Hum Retroviruses. 1991;7:889–896. doi: 10.1089/aid.1991.7.889. [DOI] [PubMed] [Google Scholar]
- 38.Putkonen P, Thorstensson R, Ghavamzadeh L, Albert J, Hild K, Biberfeld G, Norrby E. Prevention of HIV-2 and SIVsm infection by passive immunization in cynomolgus monkeys. Nature. 1991;352:436–438. doi: 10.1038/352436a0. [DOI] [PubMed] [Google Scholar]
- 39.Roben P, Moore J P, Thali M, Sodroski J, Barbas III C F, Burton D R. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding sites of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol. 1994;68:4821–4828. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samuelsson A, Chiodi F, Ohman P, Putkonen P, Norrby E, Persson M A. Chimeric macaque/human Fab molecules neutralize simian immunodeficiency virus. Virology. 1995;207:495–502. doi: 10.1006/viro.1995.1109. [DOI] [PubMed] [Google Scholar]
- 41.Sattentau Q J, Moore J P. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J Exp Med. 1995;182:185–196. doi: 10.1084/jem.182.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scandella V, Kilpatrick J, Lidster W, Parker C, Moore G K, Mann K A, Brown P, Coates S, Chapman B A, Maziarz F, Steimer K S, Haigwood N L. Non-affinity purification of recombinant gp120 for use in AIDS vaccine development. AIDS Res Hum Retroviruses. 1993;9:1227–1238. doi: 10.1089/aid.1993.9.1233. [DOI] [PubMed] [Google Scholar]
- 43.Sheppard H W, Lang W, Ascher M S, Vittinghoff E, Winkelstein W. The characterization of non-progressors: long-term HIV-1 infection with stable CD4 22 T-cell levels. AIDS. 1993;7:1159–1166. [PubMed] [Google Scholar]
- 44.Silvera P, Flanagan B, Kent K, Rud E, Powell C, Corcoran T, Bruck C, Thiriart C, Haigwood N L, Stott E J. Fine analysis of humoral antibody response to envelope glycoprotein of SIV in infected and vaccinated macaques. AIDS Res Hum Retroviruses. 1994;10:1295–1304. doi: 10.1089/aid.1994.10.1295. [DOI] [PubMed] [Google Scholar]
- 45.Steimer K S, Scandella C J, Skiles P V, Haigwood N L. Neutralization of divergent HIV-1 isolates by conformation-dependent human antibodies to gp120. Science. 1991;254:105–108. doi: 10.1126/science.1718036. [DOI] [PubMed] [Google Scholar]
- 46.Sullivan N, Sun Y, Li J, Hofman W, Sodroski J. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J Virol. 1995;69:4413–4422. doi: 10.1128/jvi.69.7.4413-4422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomlinson I M, Walter G, Marks J D, Llewelyn M B, Winter G. The repertoire of human germline VH segment reveals about fifty groups of VH segments with different hypervariable loops. J Mol Biol. 1992;227:776–798. doi: 10.1016/0022-2836(92)90223-7. [DOI] [PubMed] [Google Scholar]
- 48.Vancott T C, Polonis V R, Loomis L D, Michael N L, Nara P L, Birx D L. Differential role of V3-specific antibodies in neutralization assays involving primary and laboratory-adapted isolates of HIV type 1. AIDS Res Hum Retroviruses. 1995;11:1379–1390. doi: 10.1089/aid.1995.11.1379. [DOI] [PubMed] [Google Scholar]
- 49.Van Rompay K K A, Otsyula M G, Tarara R P, Canfield D R, Berardi C, McChesney M B, Marthas M L. Vaccination of pregnant macaques protects newborns against mucosal simian immunodeficiency virus infection. J Infect Dis. 1996;173:1327–1335. doi: 10.1093/infdis/173.6.1327. [DOI] [PubMed] [Google Scholar]
- 50.Weng N, Snyder J G, Yu-Lee L, Marcus D M. Polymorphism of human immunoglobulin VH4 germ-line genes. Eur J Immunol. 1992;22:1075–1082. doi: 10.1002/eji.1830220430. [DOI] [PubMed] [Google Scholar]
- 51.Winkler T H, Fehr H, Kalden J R. Analysis of immunoglobulin variable region genes from human IgG anti-DNA hybridomas. Eur J Immunol. 1992;22:1719–1728. doi: 10.1002/eji.1830220709. [DOI] [PubMed] [Google Scholar]
- 52.Yasutomi Y, Reiman K, Lord C, Miller M, Letvin N. Simian immunodeficiency virus-specific CD8+ lymphocyte response in acutely infected rhesus monkeys. J Virol. 1993;67:1707–1711. doi: 10.1128/jvi.67.3.1707-1711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J, Martin L N, Watson E A, Montelaro R C, West M, Epstein L, Murphey-Corb M. Simian immunodeficiency virus/Delta-induced immunodeficiency disease in rhesus monkeys: relation of antibody response and antigenemia. J Infect Dis. 1988;158:1277–1286. doi: 10.1093/infdis/158.6.1277. [DOI] [PubMed] [Google Scholar]