Abstract
A seroprevalence survey was conducted for simian immunodeficiency virus (SIV) antibody in household pet monkeys in Gabon. Twenty-nine monkeys representing seven species were analyzed. By using human immunodeficiency virus type 2 (HIV-2)/SIVsm, SIVmnd, and SIVagm antigens, one red-capped mangabey (RCM) (Cercocebus torquatus torquatus) was identified as harboring SIV-cross-reactive antibodies. A virus isolate, termed SIVrcm, was subsequently established from this seropositive RCM by cocultivation of its peripheral blood mononuclear cells (PBMC) with PBMC from seronegative humans or RCMs. SIVrcm was also isolated by cocultivation of CD8-depleted RCM PBMC with Molt 4 clone 8 cells but not with CEMx174 cells. The lack of growth in CEMx174 cells distinguished this new SIV from all previously reported sooty mangabey-derived viruses (SIVsm), which grow well in this cell line. SIVrcm was also successfully transmitted (cell free) to human and rhesus PBMC as well as to Molt 4 clone 8 cells. To determine the evolutionary origins of this newly identified virus, subgenomic pol (475 bp) and gag (954 bp) gene fragments were amplified from infected cell culture DNA and sequenced. The position of SIVrcm relative to those of members of the other primate lentivirus lineages was then examined in evolutionary trees constructed from deduced protein sequences. This analysis revealed significantly discordant phylogenetic positions of SIVrcm in the two genomic regions. In trees derived from partial gag sequences, SIVrcm clustered independently from all other HIV and SIV strains, consistent with a new primate lentivirus lineage. However, in trees derived from pol sequences, SIVrcm grouped with the HIV-1/SIVcpz lineage. These findings suggest that the SIVrcm genome is mosaic and possibly is the result of a recombination event involving divergent lentiviruses in the distant past. Further analysis of this and other SIVrcm isolates may shed new light on the origin of HIV-1.
Phylogenetic analyses of simian immunodeficiency virus (SIV) isolates reveal that they belong to five distinct lineages of the lentivirus family of retroviruses (46). These five SIV lentivirus lineages form a distinct subgroup, because primate viruses are more closely related to each other than to lentiviruses from nonprimate hosts (46). Importantly, only simian species indigenous to the African continent are naturally infected (4, 13, 28, 35). Thus far, natural SIV infections in Africa have been documented in the sooty mangabey (SM), (Cercocebus torquatus atys) (SIVsm strains) in Liberia (30), in Sierra Leone (4, 5), and the Ivory Coast (43); in all four subspecies of African green monkeys (Cercopithecus aethiops) (1, 21, 22, 25, 33, 34, 39) (SIVagm strains) in eastern, central, and western Africa; in the Sykes monkey (Cercopithecus mitis) (SIVsyk strains) (9) in Kenya; in the mandrill (Mandrillus sphinx) (SIVmnd strains) (38, 50) in Gabon; and in chimpanzees (Pan troglodytes) (SIVcpz strains) (19, 20, 41, 42) in Gabon. Because these SIVs and their simian hosts are highly divergent from each other and are widely distributed across Africa, it is believed that the SIV family evolved and established itself in African simians long before AIDS appeared in humans (4, 15, 18, 19, 21, 30, 37, 47).
One common characteristic among all SIVs is that none are associated with immunodeficiency or any other disease in their natural hosts (9, 13, 22, 28, 30, 35, 38). This finding is in marked contrast to AIDS, which occurs in humans and macaques infected with primate lentiviruses (2, 7, 8, 27, 35). This lack of disease in the natural SIV hosts may be an example of long-term evolution toward avirulence (16), which supports the hypothesis that SIV has infected African simians for a relatively long time.
Human AIDS is caused by two distinct primate lentiviruses, human immunodeficiency virus type 1 (HIV-1) and HIV-2 (2, 7). Interestingly, the phylogeny of HIV is markedly different from that of SIV, because genetic analyses have shown that the human viruses do not represent separate sixth and seventh lineages of primate lentiviruses but instead are members of two of the five existing SIV lineages (37, 46). HIV-1 is in the HIV-1/SIVcpz group (19, 51), and HIV-2 belongs to the HIV-2/SIVsm family (18, 23). These phylogenetic data on the HIV-1 and HIV-2 lineages have long suggested separate simian origins for HIV-1 and HIV-2 (37, 46).
Molecular studies of naturally occurring SIVsm and HIV-2 strains from rural West Africa have provided convincing evidence for a simian origin of HIV-2. A close genetic relationship has been established between the HIV-2 D and E subtypes and SIVsm strains found in household pet SMs in West Africa (4, 14, 15). Moreover, all six known subtypes of HIV-2, including a new subtype, F (3), are found only within the natural range of SIV-infected SMs in West Africa. No other area of Africa or of the rest of the world has all six known HIV-2 subtypes. Together, these data provide strong support for independent transmissions of SIVsm from naturally infected SMs to humans.
In contrast, there is much less information to support a simian origin for HIV-1. Although SIVcpz is the closest relative to HIV-1, there are only a few isolates, thus raising questions as to the likely primate reservoir. Only three SIVcpz strains have thus far been identified (20, 41, 42, 51). The first one was isolated from a single, household pet chimpanzee in Gabon that was not part of a primate research colony (42). An additional SIVcpz strain was found in a captive chimpanzee which was wild-caught in Zaire and thus likely was infected in Africa (41, 51). Finally, PCR data suggested the existence of a third SIVcpz strain, again from a wild-caught chimpanzee from Gabon (20). Thus, although based on limited data, the hypothesis that HIV-1 is derived from members of a larger SIVcpz lineage remains plausible. However, additional SIVs within the HIV-1/SIVcpz lineage must be found to better understand the origin of the HIV-1 family.
Here, we report the isolation and genetic characterization of a new SIV from a red-capped mangabey (RCM), Cercocebus torquatus torquatus. This RCM was a household pet in Lambarene, Gabon, and was not part of a primate colony, a zoo, or a research facility. Phylogenetic analysis of SIVrcm pol gene sequences surprisingly showed a close relationship with the HIV-1/SIVcpz group of viruses. However, analysis of gag gene sequences indicated a new lineage, independent from previously characterized SIVs. Based on these phylogenetic data and the geographic location of the new mangabey host, SIVrcm may have been generated by an ancient recombination involving a member of an independent (sixth) SIV lineage and an ancestor of today’s HIV-1/SIVcpz group.
MATERIALS AND METHODS
Animals and specimens.
Ten-milliliter samples of heparinized whole blood were collected from household pet monkeys on site under ketamine anesthesia (10 mg/kg). Table 1 shows the number of each monkey species that were tested. Each pet monkey was tattooed with a unique number so that no animal was inadvertently sampled more than once and so that the monkeys could be identified for follow-up specimen collection. Peripheral blood mononuclear cells (PBMC) and plasma were separated in the field by centrifugation with Lymphocyte Separation Medium (Organon Teknika, Inc., Durham, N.C.), as previously described (4). Cynomolgus macaques, Macaca fascicularis, were housed at the Centre International de Recherches Medicales (CIRMF), Franceville, Gabon.
TABLE 1.
Seroprevalence survey for HIV-2-reactive antibodies in monkeys kept as household pets in Gabon
| Common namea | Scientific namea | No. positive/no. testedb |
|---|---|---|
| Gray-cheeked mangabey | Cercocebus albigena | 0/2 |
| RCM | Cercocebus torquatus torquatus | 1/9 |
| Moustached monkey | Cercopithecus cephus | 0/4 |
| Greater white-nosed monkey | Cercopithecus nictitans | 0/5 |
| Black-and-white colobus | Colobus guereza | 0/1 |
| Mandrill | Mandrillus sphinxc | 1/7 |
| Tallapoin | Miopithecus tallapoin | 0/1 |
EIA and Western blots.
An HIV-2 enzyme immunoassay (EIA) was used as described previously (3) for initial screening of plasma collected in the field. Samples that were positive by EIA were retested with Western blot strips containing HIV-2 (Sanofi Diagnostics, Pasteur, Inc., Paris, France), SIVsmLib1 (a West African-derived SIVsm) (30), SIVmnd (a gift from Jo Ann Yee, University of California, Davis), and SIVagm (a gift from Jonathan Allan, Southwest Foundation for Biomedical Research, San Antonio, Tex.) antibodies HIV-1 Western blot strips (Diagnostic Biotechnology Ltd., Singapore) contained HIV-1 viral proteins plus an antigenic peptide derived from the HIV-2 transmembrane (TM) protein. Positive control plasma and sera were from HIV-1 or HIV-2 antibody-positive humans, SIVmac251-infected macaques, SIVmnd-infected mandrills, and SIVagm-infected African green monkeys. Negative control sera and plasma were from humans, mangabeys, and macaques.
Treatment of RCM PBMC to remove CD8-bearing cells.
To increase isolation efficiency, RCM PBMC were treated with a monoclonal antibody and magnetic beads designed to remove CD8+ cells. PBMC were separated on Lymphocyte Separation Medium as described above and resuspended in phosphate-buffered saline with 5% fetal calf serum, penicillin, streptomycin, and 20 mM HEPES buffer. IOT8a antibody (Becton Dickinson and Co., Los Angeles, Calif.) was added at a 1:50 final dilution and incubated with 2 × 107 PBMC per ml for 30 min at 4°C with gentle agitation. After incubation with IOT8a antibody, the PBMC were washed in phosphate-buffered saline and resuspended with beads (two beads per cell) that attach to the cell-bound IOT8a antibody (Dynabeads M450; Dynal, Inc., Lake Success, N.Y.). After incubation for 30 min at 4°C, the beads were bound to a magnet and cells bound by the magnet were removed. The treated PBMC were used immediately for cocultivation with Molt 4 clone 8 or CEMx174 cells.
Tissue culture.
For propagation of virus, uninfected human and RCM PBMC (106/ml) were stimulated for 3 days with 10 μg of concanavalin A (ConA) per ml in RPMI 1640 medium plus antibiotics and 10% fetal calf serum. Rhesus PBMC were prepared as described above except that 10 μg of staphylococcal enterotoxin β per ml was used as the mitogen. The infection of cultures was monitored by SIV p27 antigen assay as described previously (31). Virus-containing tissue culture fluids were stored in liquid nitrogen and shipped in dry ice for virus isolation. Cell lines CEMx174 and Molt 4 clone 8 were maintained and cocultivated with monkey PBMC as described previously (29).
Extraction and PCR amplification of DNA.
DNA was isolated from infected cell cultures as reported previously (3). pol primers were designed from a highly conserved area of the pol gene; these were UNIPOL1 (5′-AGTGGATTCATAGAAGCAGAAGT-3′) and UNIPOL2 (5′-CCCCTATTCCTCCCCTTCTTTTAAAA-3′) (32). Additional pol primers were used to extend the pol sequence toward the 3′ end of the pol gene; these were SS1 (5′-CAAGGAGTAGTGGAAAGCATG-3′) and SS2 (5′-TACTGCCCCTTCACCTTTC-3′). PCR conditions were as reported previously (32). The leftward primer (SS1) was homologous with the newly sequenced SIVrcm pol fragment, and the rightward primer (SS2) was a conserved sequence from SIVcpzANT pol positions 4424 to 4406. gag was amplified with primers gag A (5′-AGGTTACGGCCCGGCGGAAAGAAAA-3′) and gag B (5′-CCTACTCCCTGACAGGCCGTCAGCATTTCTTC-3′) as described previously (14). These primers have previously been shown to be highly cross-reactive, amplifying both HIV-1 and HIV-2 strains (14).
Sequencing and phylogenetic analysis.
PCR products were cloned, sequenced, and analyzed as described previously (3, 11, 12, 14). Proviral DNA sequences were aligned by using Clustal W and adjusted by eye with the multiple-aligned-sequence editor (10). The genetic distances given in Table 2 were calculated by using DOTS (26). No primer sequences were included in the phylogenetic analyses. The reproducibility of the branching orders was determined by bootstrap analysis with 1,000 replicates. Trees were plotted by using Treetool.
TABLE 2.
gag and pol nucleotide distances between SIVrcm and primate lentivires
| Virus |
gag (top) and pol (bottom) nucleotide distances (%) froma:
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SIVrcm95Gab1 | HIV1U455 | HIV1ANT70 | HIV1MVP5180 | HIV1HXB2 | HIV1JRCSF | HIV1RF | HIV1NDK | SIVcpzANT | SIVcpzGAB | HIV2ROD | HIV2D205 | SIVagm677 | SIVagmTYO-1 | SIVmac251 | SIVsmH4 | SIVstm | SIVmndGB-1 | SIVsyk173 | |
| SIVrcm95Gab1 | 36.5 | 35.2 | 37.6 | 38.7 | 39.6 | 38.8 | 38.8 | 39.0 | 38.1 | 31.7 | 29.1 | 29.0 | 29.8 | 31.8 | 30.0 | 29.0 | 45.3 | 38.1 | |
| HIV-1U455 | 18.2 | 14.6 | 13.3 | 8.0 | 6.0 | 7.3 | 13.4 | 34.8 | 24.8 | 37.1 | 36.8 | 40.4 | 39.5 | 37.8 | 36.9 | 38.4 | 39.0 | 48.3 | |
| HIV-1ANT70 | 24.3 | 27.9 | 10.1 | 27.4 | 27.0 | 27.7 | 30.5 | 34.5 | 26.9 | 40.8 | 41.7 | 38.3 | 40.8 | 42.8 | 40.9 | 40.6 | 40.4 | 45.9 | |
| HIV-1MVP5180 | 24.3 | 25.8 | 3.3 | 26.0 | 25.6 | 25.6 | 29.1 | 37.0 | 26.3 | 42.9 | 43.2 | 39.3 | 41.2 | 45.3 | 43.4 | 43.1 | 39.7 | 46.7 | |
| HIV-1HXB2 | 21.6 | 12.2 | 16.7 | 16.7 | 3.7 | 4.7 | 8.1 | 34.5 | 24.0 | 39.0 | 38.4 | 42.2 | 40.5 | 39.6 | 38.7 | 40.6 | 39.0 | 49.5 | |
| HIV-1JRCSF | 21.0 | 12.3 | 13.3 | 14.7 | 4.7 | 6.4 | 8.1 | 35.1 | 23.6 | 39.2 | 38.7 | 43.1 | 41.4 | 40.1 | 39.7 | 40.4 | 39.0 | 49.3 | |
| HIV-1RF | 22.2 | 12.3 | 15.2 | 16.6 | 6.0 | 5.3 | 9.8 | 36.1 | 24.4 | 39.4 | 38.0 | 42.0 | 41.0 | 40.4 | 39.2 | 40.7 | 39.5 | 48.3 | |
| HIV-1NDK | 20.8 | 5.3 | 13.3 | 13.3 | 6.0 | 4.0 | 6.6 | 33.3 | 26.1 | 40.1 | 38.9 | 40.6 | 39.3 | 40.1 | 39.6 | 40.0 | 39.5 | 50.0 | |
| SIVcpzANT | 26.4 | 18.7 | 21.2 | 19.5 | 19.5 | 18.7 | 18.8 | 18.7 | 35.3 | 43.5 | 42.2 | 34.0 | 38.3 | 41.5 | 41.0 | 40.2 | 42.5 | 49.7 | |
| SIVcpzGAB | 22.8 | 8.0 | 13.2 | 12.6 | 9.3 | 7.3 | 11.2 | 7.2 | 17.2 | 44.3 | 43.0 | 34.7 | 39.1 | 41.5 | 41.0 | 40.2 | 38.8 | 46.5 | |
| HIV-2ROD | 37.2 | 42.2 | 45.3 | 43.5 | 45.0 | 42.2 | 44.6 | 43.2 | 39.8 | 40.2 | 11.9 | 31.8 | 33.2 | 12.8 | 10.7 | 12.7 | 35.4 | 41.4 | |
| HIV-2D205 | 38.4 | 43.9 | 44.6 | 43.2 | 43.2 | 41.9 | 43.0 | 43.0 | 40.0 | 39.7 | 12.8 | 30.3 | 29.8 | 12.0 | 8.9 | 11.9 | 37.2 | 39.9 | |
| SIVagm677 | 28.1 | 34.0 | 33.3 | 32.2 | 34.3 | 33.6 | 33.3 | 33.3 | 39.3 | 38.4 | 39.6 | 37.9 | 21.1 | 32.1 | 30.6 | 29.4 | 32.6 | 44.2 | |
| SIVagmTYO-1 | 33.8 | 40.0 | 38.7 | 38.0 | 41.3 | 38.0 | 40.4 | 39.1 | 41.3 | 42.4 | 38.5 | 39.6 | 19.7 | 33.8 | 32.0 | 31.4 | 36.7 | 41.6 | |
| SIVmac251 | 34.5 | 40.8 | 42.9 | 42.2 | 43.5 | 40.8 | 43.2 | 41.9 | 43.2 | 42.6 | 10.1 | 11.5 | 38.2 | 37.2 | 5.4 | 9.4 | 34.0 | 39.9 | |
| SIVsmH4 | 35.6 | 40.5 | 43.9 | 43.2 | 41.9 | 40.5 | 41.6 | 41.6 | 42.2 | 42.2 | 8.8 | 8.8 | 37.9 | 38.3 | 2.7 | 6.7 | 33.8 | 41.2 | |
| SIVstm | 37.0 | 42.6 | 43.9 | 43.21 | 42.6 | 41.2 | 42.3 | 42.3 | 41.5 | 43.0 | 11.5 | 10.1 | 37.9 | 38.9 | 6.8 | 5.4 | 34.5 | 39.0 | |
| SIVmndGB-1 | 33.1 | 44.6 | 43.6 | 45.1 | 44.8 | 45.3 | 45.9 | 45.9 | 45.0 | 45.2 | 46.7 | 45.4 | 45.7 | 46.6 | 44.2 | 44.7 | 45.9 | 45.3 | |
| SIVsyk173 | 43.8 | 41.8 | 40.4 | 39.7 | 41.5 | 39.7 | 41.5 | 40.1 | 41.4 | 40.1 | 46.5 | 45.5 | 32.9 | 39.5 | 43.8 | 45.5 | 46.2 | 39.2 | |
Genetic distances for gag and pol were calculated for the PCR-derived SIVrcm gag and pol sequences and compared with the values from the corresponding sequences from the database (36). Sequences were aligned by using Clustal W and adjusted with the multiple-aligned-sequence editor (10), and distances were calculated by using DOTS (26).
Nucleotide sequence accession numbers.
Nucleotide sequences were submitted to GenBank and are available under accession numbers AF028607 and AF028608.
RESULTS
Identification of seropositive pet monkeys.
Table 1 shows data for 29 nonhuman primates that were being kept as household pets in Gabonese villages near the town of Lambarene. Seven different species were identified, all of which were indigenous to Gabon (52). With the permission of the owners, 10 ml of heparinized blood was collected from each animal. Monkeys were tattooed with a unique number (001 through 029) so that they could be relocated and identified for follow-up specimen collections.
Plasma samples from 2 of the 29 monkeys contained antibody that reacted in the HIV-2 antibody detection assay. One animal was a mandrill, and the other was an RCM, Cercocebus torquatus torquatus. Attempts to isolate virus from the mandrill were unsuccessful (data not shown). Moreover, this mandrill could not be relocated for a second, follow-up blood sample. The focus of the study thus was on the seropositive mangabey, RCM 9, which was the first SIV-seropositive individual of its species to be identified. RCM 9 was relocated for follow-up studies.
RCM 9 was a young female which had been captured from a free-living troop of RCMs in the coastal region near Lambarene, Gabon. She weighed 2.7 kg. According to her owner, RCM 9 had been kept alone for about 1 year. Based on her weight, apparent sexual immaturity, and information from the owner, her age was estimated at 2 to 3 years at the time of the first sampling in 1995. Physical examination confirmed that the animal was healthy, showing no evidence of lymphadenopathy, wasting, or other observable signs of illness.
Analysis of RCM antibodies by Western blotting.
To test for SIV-cross-reacting antibodies, RCM 9 plasma was retested by Western blotting with HIV-2, SIVsm, SIVmnd, and SIVagm antigens (Fig. 1). The results in Fig. 1 showed that RCM 9 plasma reacted with p26/p27 of all four viruses as well as with the TM antigens of HIV-2 and SIVagm. Binding to p27 on SIVsm and SIVmnd immunoblots (Fig. 1) was weaker than binding to the HIV-2 and SIVagm core antigens but was still detectable. The plasma specimens listed in Table 1 were also tested by Western blotting with HIV-1 antigens. The results were similar to those in Fig. 1 (data not shown). These data thus provide evidence for a present or past infection of RCM 9 with a virus related to previously characterized primate lentiviruses. However, Western blot results are not reliably predictive of phylogenetic relationships between viruses, as shown in our previous work (4). The identification of the lentivirus lineage to which this RCM virus belonged thus required sequencing of part of the viral genome and phylogenetic analysis.
FIG. 1.
Western blots showing SIVrcm antibody reactivity with various primate lentiviruses, i.e., HIV-2, SIVsm, SIVmnd, and SIVagm. Plasma (1:100 dilution) from an HIV/SIV-seropositive RCM (RCM 9), (Cercocebus torquatus torquatus) (Table 1) was used. For the SIVagm immunoblot, two dilutions of RCM plasma, 1:50 and 1:100, were used. Both of these blots are shown as lanes RCM. For all other blots, 1:100 dilutions of plasma were used. HIV-2-positive (+) and -negative (−) control sera were supplied by the manufacturer of the HIV-2 blots. SIVsm-, SIVagm-, and SIVmnd-positive (+) and -negative (−) control sera were from positive or negative macaques, African green monkeys, and mandrills, respectively. Molecular weight markers for each gel were used to calculate the molecular weights of viral proteins for SIVsm, SIVmnd, and SIVagm. Viral proteins for HIV-2 were identified by the manufacturer of the immunoblots. gp105 is a trimer of the HIV-2 transmembrane protein. The viral polypeptides do not comigrate in the different gels because they were run under different conditions.
Transmission of the SIV-related virus to a seronegative RCM and to cynomolgus macaques.
The SIV-seropositive RCM 9 was relocated in a second visit to Lambarene, and its identity was confirmed from the tattoo. The pet was obtained from the owner and transferred to the primate facility at CIRMF. To serve as uninfected control mangabeys and sources of uninfected RCM PBMC, two seronegative RCMs identified in Table 1, RCM 6 and RCM 15, were also obtained from their owners in Lambarene.
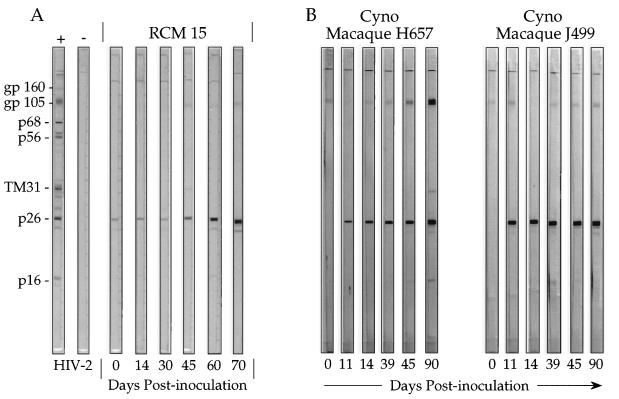
To determine if the naturally seropositive RCM 9 was harboring an active virus infection that was transmissible to uninfected monkeys, 10 ml of whole heparinized blood from RCM 9 was inoculated intravenously into RCM 15 and also into two cynomolgus macaques, all of which were housed at CIRMF. Figure 2A depicts the HIV-2-reactive antibody pattern from day 0 through day 70 after blood transfusion of RCM 15. On day 0, plasma of RCM 15 had a nonspecific reaction with the core antigen of HIV-2 (Fig. 2). We have previously reported similar nonspecific activity for mangabey plasma collected in West Africa (4), which also occurs in human plasma and is unrelated to lentivirus infections (4). By day 70, RCM 15 had developed a strong reaction to HIV-2 p26 and also a reaction to the HIV-2 gp105 TM trimer. This serological result at day 70 postinoculation of RCM 15 was the same as that seen in the naturally infected RCM 9 (Fig. 1, HIV-2 lanes). The two inoculated cynomolgus macaques also seroconverted by 11 days postinoculation (Fig. 2B). By 90 days after inoculation, macaque H657 had developed a strong response to p26 and to the gp105 trimer of HIV-2. The second macaque, J499, developed a strong p26 response, but its response to gp105 was only slightly more than the background level. Nevertheless, both macaques showed evidence of infection after transfusion with blood from RCM 9. The results showed that RCM 9 had an active infection with an SIV-HIV-related virus that was transmissible to another RCM and to two cynomolgus macaques. Because the blood from RCM 9 was inoculated directly into both the macaques and RCM 15 without laboratory manipulation, inadvertent laboratory contamination could be excluded. Clinical observation for 1 year has shown that both infected mangabeys (RCM 9 and RCM 15) remained clinically normal, with white cell counts and hematocrits in the expected range for healthy monkeys (data not shown). Moreover, no signs of disease have been observed in the macaques after 1 year of observation.
FIG. 2.
Inoculation of an RCM and two cynomolgus macaques with blood from RCM 9, an SIV-seropositive RCM. (A) Seroconversion of an RCM (RCM 15) transfused with blood from RCM 9. HIV-2 Western blots were used to test for seroconversion as described previously (3). RCM 15 was a household pet tested and found to be seronegative (Table 1). Tests were on days 0, 14, 30, 45, 60, and 70 after inoculation of RCM 15 with blood from seropositive RCM 9. Lanes + and −, positive and negative HIV-2 control sera, respectively. (B) Seroconversion of cynomolgus macaques H657 and J499 after transfusion with blood from RCM 9. Immunoblots at days 0, 11, 14, 39, 45, and 90 postinoculation with RCM 9 blood are shown. Lanes + and −, positive and negative HIV-2 control sera, respectively.
Isolation of SIVrcm.
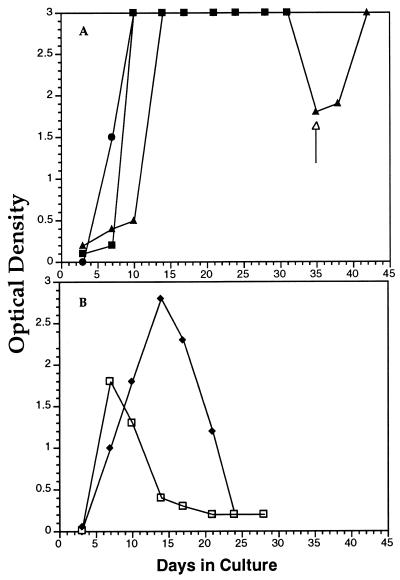
RCM 9 PBMC were cultured in vitro for virus isolation. RCM 9 and RCM 15 PBMC (106/ml) were cocultivated with mitogen-stimulated human PBMC (106/ml) (Fig. 3A). The infection was monitored by testing for SIVmac p27 antigen in the culture fluids at regular intervals. The virus grew readily and yielded maximal levels of SIVmac p27 reactivity by 8 days postinfection (Fig. 3A). The PBMC culture media continued to be antigen positive for 30 days, when the cultures were terminated. The p27 antigen was not quantified in nanograms per milliliter, because the assay is specific for SIVmac and quantification of the p27 of an unknown SIV would require homologous standards, which are not yet available. Nevertheless, the SIVmac p27 assay was a reliable qualitative assay for infection and was used to show that the virus readily replicated in vitro in human PBMC.
FIG. 3.
Isolation of SIVrcm from RCM PBMC. (A) PBMC from RCM 9 (▪) and from RCM 15 on day 11 postinoculation (•) were cocultivated with human PBMC. Uninfected PBMC from RCM 6 (▴) were cultured with RCM 15 PBMC collected on day 45 postinoculation. Cell culture fluids were monitored for SIVmac p27 at regular intervals. p27 antigen is shown in optical density units. The optical density readings were over 3.0 from day 8 through 31. The arrow marks the addition of fresh uninfected RCM 6 PBMC. RCM 9 (□) and RCM 15 (day 11 postinoculation) (⧫) PBMC were stimulated with ConA and cultured alone.
SIVrcm was also isolated from RCM PBMC without addition of PBMC from seronegative animals. ConA-stimulated PBMC from RCM 9 and from the newly infected RCM 15 (Fig. 3B) both produced virus. PBMC from RCM 15 were taken 11 days after inoculation with blood from RCM 9. Day 11 was chosen because this is the time of peak viremia observed in macaques when inoculated intravenously with SIVmac (44). The kinetics of SIV p27 accumulation in the supernatant fluids (Fig. 3B) were similar to those obtained when RCM 9 PBMC were cocultivated with human PBMC (Fig. 3A). However, the p27 levels were not sustained, as would be expected for cell cultures not supplemented with additional uninfected PBMC.
SIVrcm was also grown by cultivation of seronegative RCM PBMC with seropositive RCM 15 cells. RCM 6 was confirmed as seronegative by reaction of its plasma with an HIV-2 immunoblot (data not shown). PBMC were collected from RCM 15 on day 45 postinoculation and cocultured with mitogen-treated PBMC from RCM 6. Figure 3A shows that large amounts of p27 antigen were detected in the second week after cocultivation with RCM 6 cells. By day 35, when SIV antigen amounts had declined, fresh RCM 6 PBMC were added, and p27 antigen levels rapidly increased to maximum optical density readings. In contrast, cocultivation of PBMC from the two inoculated macaques did not yield SIV p27 activity when they were cocultured with either human or RCM PBMC. This may have been due to a low virus load in the inoculated macaques or to a cleared infection. Nevertheless, the data in Fig. 3 show that an SIV, whose core antigen serologically cross-reacted with that of SIVmac, was readily isolated from the PBMC of the infected mangabey RCM 9 and that this virus was also recovered from RCM 15 after inoculation with blood from RCM 9.
Adaptation of SIVrcm to human T-cell lines.
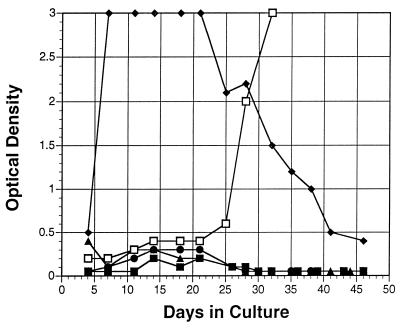
To obtain sufficient amounts of virus for genetic characterization, attempts were made to grow the virus in CEMx174 and Molt 4 clone 8 cells. These two human T-cell lines were chosen because they readily support growth of various divergent SIVs from African primates (4, 25, 39, 50). PBMC (106/ml) from RCM 9, the chronically infected household pet, were cocultured with cells (106/ml) from each of the two T-cells lines shown in Fig. 4. After 14 days of cocultivation, small amounts of p27 antigen were detected in both cell lines; however, viral replication was minimal and transient. No cytopathic effect was observed, and both cell lines grew exponentially until they were terminated 46 days later.
FIG. 4.
Cocultivation of RCM PBMC with human T-cell lines before and after treatment with CD8-binding beads. ▪ and •, cocultivation of RCM 9 PBMC with CEMx174 (▪) and Molt 4 clone 8 (•) cells. ▴ and ⧫, cocultivation of CEMx174 (▴) and Molt 4 clone 8 (⧫) cells with RCM 9 PBMC that were treated with CD8-binding beads. □, cell-free virus was passaged to uninfected Molt 4 clone 8 cells by inoculation with cell-free supernatant fluids from SIV-positive Molt 4 cell cultures.
Further attempts to adapt the virus to T cells were made by cocultivating the T-cell lines with RCM 9 PBMC that had been treated to remove CD8-bearing cells. CEMx174 cell cultures still did not produce p27 antigen when maintained for 46 days (Fig. 4). However, Molt 4 clone 8 cells supported growth of the virus and had sustained p27 optical density values for 3 weeks before a decline was observed. Cell-free fluids from these Molt 4 clone 8 cells were successfully transmitted to uninfected Molt 4 cells and human PBMC, as well as rhesus PBMC (data not shown). Rhesus PBMC cultures yielded the highest p27 level recorded in our laboratory. The 50% tissue culture infective dose of the RCM virus stock was 3.2 × 104/ml when tested with human PBMC.
Amplification of pol and gag fragments from cell cultures infected with SIVrcm.
To characterize the newly found SIV at the molecular level, DNA was extracted from the infected Molt 4 clone 8 cell culture shown in Fig. 4. Attempts to amplify SIVrcm DNA with primers from conserved SIVsm/HIV-2 env regions (3) were negative. However, conserved pol primers (32) yielded a 288-bp fragment of DNA. Pairwise sequence comparison to corresponding pol sequences of other primate lentiviruses revealed an unexpected close relationship to HIV-1 and SIVcpz (data not shown). To confirm this finding and to obtain additional pol sequences for phylogenetic analyses, we extended the pol sequence to 475 bp by using additional pol primers, SS1 and SS2. The leftward primer was taken from positions 179 to 199 of the newly sequenced 288-bp pol fragment, while the rightward primer was taken from a highly conserved sequence of the SIVcpzANT pol gene (positions 4424 to 4406). The SS1 and SS2 primers amplified a 314-bp fragment, which yielded 187 bp of new SIVrcm sequence (the new 314-bp fragment showed the expected sequence when it was aligned with the homologous part of the 288-bp fragment).
To amplify a second genomic region, DNA was extracted from SIVrcm-infected human PBMC and used for PCR amplification with conserved gag primers A and B (14). A 954-bp fragment was obtained and cloned (14). Both pol and gag fragments were sequenced and subjected to phylogenetic analyses.
Phylogenetic analysis of SIVrcm.
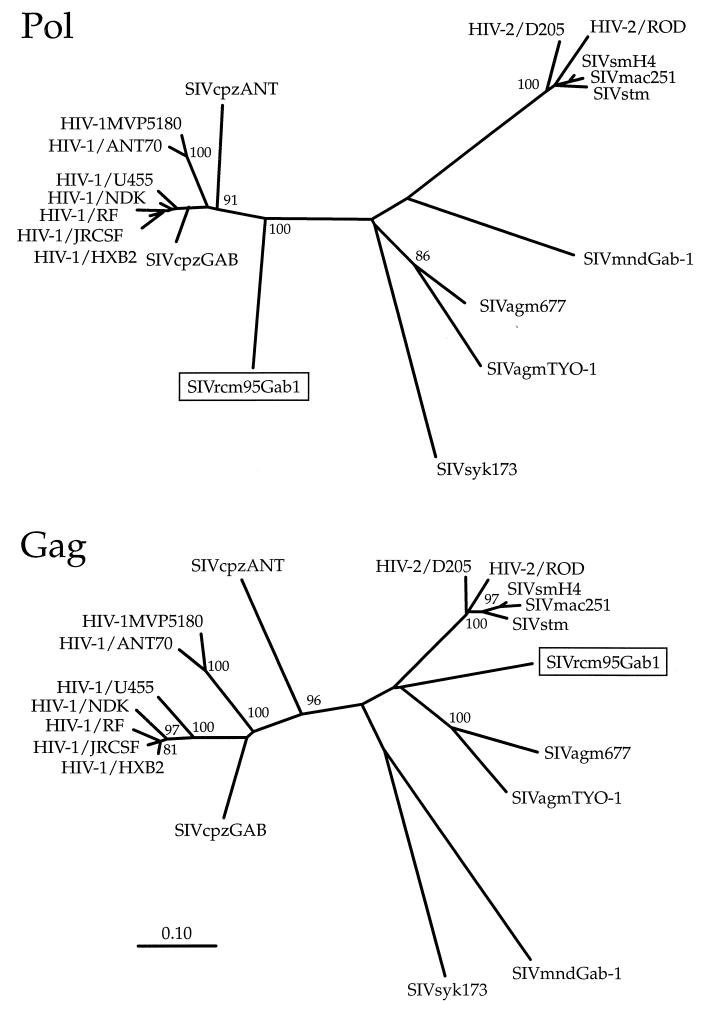
Nucleotide distances were calculated for the two genome fragments as described in Materials and Methods. Table 2 shows that the SIVrcm pol fragment was 28.1 to 38.4% distant from the SM, African green monkey, and mandrill lineages. The greatest distance, 43.8%, was from SIVsyk. However, the nucleotide distances from human and chimpanzee immunodeficiency viruses were less, ranging from 18.2 to 26.4%. Results from amino acid sequence comparisons were similar (data not shown). In marked contrast, the gag fragment was 35.2 to 39.0% distant from HIV-1 and SIVcpz. This gag fragment was most distant (45.3%) from SIVmnd and was about equidistant from the SIVagm and SIVsm lineages, (29.81 and 31.8%, respectively) (Table 2).
To define the phylogenetic relationships of SIVrcm to members of the other primate lentivirus lineages, trees were constructed from partial gag and pol protein sequences. This analysis revealed significantly discordant phylogenetic positions for SIVrcm in the two genomic regions (Fig. 5). In the pol tree, SIVrcm grouped with the HIV-1/SIVcpz lineage, and this grouping was supported by 100% of bootstrap values (Fig. 5). However, in the gag tree, SIVrcm clustered separately from all other HIV and SIV strains and formed a new independent lineage. These findings suggest that the SIVrcm genome is mosaic and possibly is the result of an ancient recombination event involving a member of an independent (sixth) SIV lineage and an ancestor of today’s HIV-1/SIVcpz strains. However, the phylogenies shown in Fig. 5 are based on only small sequence fragments, and definitive conclusions concerning the evolutionary history of SIVrcm must await sequence analysis of the entire SIVrcm genome. The new virus was designated SIVrcm95Gab1, according to reference 3, to indicate the host species, the year (1995) and country (Gabon) of origin, and the isolate number (no. 1).
FIG. 5.
Phylogenetic relationships of SIVrcm to members of other primate lentivirus lineages. Trees were constructed from partial pol (155 amino acids) and gag (318 amino acids) protein sequences derived by PCR from infected-cell DNA. Sequences were aligned by using CLUSTAL, with minor manual adjustments. Pairwise distances were estimated by using Kimura’s two-parameter method (equation 4.14 in reference 24) to correct for superimposed hits. Gaps were excluded from the alignments. Phylogenetic trees were constructed by the neighbor-joining method as before (4, 14), and their reliabilities were estimated from 1,000 bootstrap samples. These methods were implemented by CLUSTAL W and programs from the PHYLIP package (12). Branch lengths are drawn to scale. Values at nodes indicate the percentages of bootstraps in which the cluster is supported. Clusters found in >80% of 1,000 bootstraps are indicated. The position of the newly identified SIVrcm strain is boxed.
DISCUSSION
In this study we describe a new primate lentivirus which we discovered during a seroprevalence survey of household pet monkeys in Gabon. Detailed characterization revealed that this virus has a number of unique features that distinguish it from other primate lentiviruses. First, SIVrcm was identified in an RCM, a species not previously known to harbor primate lentiviruses. Second, in vitro culture studies indicated that this virus can readily infect human PBMC. This is not the case for most of the other primate lentiviruses (e.g., SIVagm), presumably because of incompatibility of host proteins which need to interact with viral proteins (47, 48). Third, evolutionary tree analysis revealed that SIVrcm clusters discordantly in different regions of its genome. Although mosaic genomes have been described for various HIV and SIV strains in the past (4, 14, 21), SIVrcm is the only monkey virus (other than SIVcpz), that has significant sequence homology with HIV-1.
The relationship of SIVrcm to SIVcpz in the pol region raises the question whether SIVrcm may be the result of a recombination event including a member of an independent (sixth) SIV lineage and a divergent SIVcpz strain. In Lambarene, Gabon, where the SIV-positive RCM was found, we did not observe contact between RCM 9 and chimpanzees. Moreover, the owners denied a history of chimpanzee contact with RCM 9. In addition, there are no SIVcpz-seropositive chimpanzees at CIRMF, and RCM 9 was tested before its transport to CIRMF. Therefore, a recent cross-species infection of RCM 9 with a chimpanzee-derived SIV is very unlikely. However, we obviously cannot exclude past cross-species transmission of viruses between chimpanzees and RCMs or between RCMs and other primate species. Thus, additional isolates from RCMs and chimpanzees are needed to understand the increasingly complex HIV-1–SIVcpz–SIVrcm lineage, the extent of sequenced divergence among its members, and potential recombinational events. Moreover, it will be important to determine whether these SIVs occur in free-ranging members of these species.
In vitro studies showed that SIVrcm readily infected human and macaque PBMC. The in vitro yields as measured by 50% tissue culture infective dose were similar to those for infections with SIVmac251 (31). Moreover, in vitro studies showed that SIVrcm replicated in Molt 4 clone 8 cells but not in CEMx174 cells. This finding suggests that SIVrcm and SIVmac251 may not share the same T-cell coreceptors. Indeed, preliminary experiments showed that SIVrcm used the SIV coreceptor “Bonzo” as well as CCR2b, but not rhesus or human CCR5 (26a). This is in contrast to SIVcpz, which uses CCR5 for entry (6).
In vivo studies showed that inoculation of blood from the SIV-positive RCM into two cynomolgus macaques and one seronegative RCM resulted in seroconversion of all three monkeys. The infection of the macaques and in vitro susceptibility of macaque PBMC suggest that SIVrcm could become pathogenic in this species. Thus far, no disease has been observed in the two cynomolgus macaques, and the virus was not recovered from their PBMC. In contrast, rhesus macaques can be infected, and SIVrcm was recovered from them (47a). Additional in vivo passage experiments will be necessary to address this question.
With the addition of SIVrcm to the family of primate lentiviruses, two distinct SIVs have now been reported to infect household mangabeys in West Africa, i.e., viruses infecting SMs and RCMs. SMs and RCMs are closely related monkeys and represent two of three members of the torquatus group, which lives along the West African coast. The third member is the white-collared mangabey (WCM), Cercocebus torquatus lunulatus (52). Most primatologists consider these to represent three races of a single species (52). However, despite the close phylogenetic relationship of their host, SIVrcm95Gab1 is highly divergent from SIVsm (Fig. 5). SIVsm and SIVrcm are about 30 to 35% distant from each other in the pol and gag fragments tested (Table 2 and Fig. 5). Therefore, SIVrcm is no closer to SIVsm than it is to SIVs of monkeys from entirely different genera. This finding is consistent with the gaps in their natural ranges. The SM range is about 1,500 km to the northwest of the RCM range. Moreover, the RCM range is discontiguous from the SM and WCM ranges, due to an 800-km coastal gap separating the RCMs from both SMs and WCMs (4, 45).
This 800-km gap is known as the Dahomey gap and is a savannah barrier between the Upper and Lower Guinea Forest (45) that separates the ranges of the torquatus group of mangabeys. The gap most likely formed about 5,000 years ago when the climate became arid. Therefore, although RCMs and SMs are members of the same species, they have been separated for thousands of years. The phylogenetic data on this group of monkeys suggest that the distinct races emerged 100,000 years ago (17), but data are limited. The amount of divergence found between SIVsm and SIVrcm is consistent with their hosts being isolated geographically for an extended time. However, there is not enough information to determine if an ancestor of SIVrcm and SIVsm entered their respective hosts before the monkeys diverged.
The WCM, the third member of the torquatus group, has been reported to be infected with an SIV that is distinct from SIVsm (49). However, this SIVwcm is closely related to SIVagmTYO (39). It is unlikely that SIVwcm represents a natural infection of WCMs in their home range in West Africa, because the SIV-positive WCMs had been housed for many years in East Africa at a primate facility where SIVagmTYO occurs in the African green monkey colony (40). No blood specimens were available before transport to East Africa. Thus, there is no independent confirmation that the SIVagm-like virus was a preexisting infection of the WCMs. Therefore, it is not known whether the WCMs naturally harbor SIV. The discovery of SIVrcm thus emphasizes the need to study WCMs in their natural range, which is adjacent to the SM range. Identification and characterization of a naturally occurring SIV in the WCM would be important for understanding the SIVs that infect the torquatus group of mangabeys.
Cross-species virus transmissions continue to be a major concern as a potential source of new human viruses, including new HIV variants as well as nonretroviruses such as Ebola virus. Further research should include human seroprevalence and virological studies in the home range of RCMs to determine if SIVrcm has infected humans.
ACKNOWLEDGMENTS
We thank Jonathan Allan and Jo Ann Yee for providing materials, Agegnehu Gettie, Christine Russo, and Amara Luckay for technical assistance, Theresa Secrist for secretarial and administrative assistance, and Gordon Cook for graphics.
This work was funded by grants from the National Institutes of Health to P.A.M. (AI38573-02 and AI27698-05) and B.H.H. (R01 AI25291 and NO1 AI35170).
REFERENCES
- 1.Allan J S, Short M, Taylor M E, Su S, Hirsch V M, Johnson P R, Shaw G M, Hahn B H. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J Virol. 1991;65:2816–2828. doi: 10.1128/jvi.65.6.2816-2828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barre-Sinoussi F, Chermann J C, Rey F, Nugeyre M T, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 3.Chen Z, Luckay A, Sodora D L, Telfer P, Reed P, Gettie A, Kanu J M, Yee J A, Ho D D, Zhang L, Marx P A. Human immunodeficiency virus type 2 (HIV-2) seroprevalence and characterization of a new HIV-2 genetic subtype (F) within the natural range of simian immunodeficiency virus-infected sooty mangabeys. J Virol. 1997;71:3953–3960. doi: 10.1128/jvi.71.5.3953-3960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Z, Telfer P, Reed P, Gettie A, Zhang L, Ho D D, Marx P A. Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J Virol. 1996;70:3617–3627. doi: 10.1128/jvi.70.6.3617-3627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Z, Telfer P, Reed P, Zhang L, Gettie A, Ho D D, Marx P A. Isolation and characterization of the first simian immunodeficiency virus from a feral sooty mangabey (Cercocebus atys) in West Africa. J Med Primatol. 1995;24:108–115. doi: 10.1111/j.1600-0684.1995.tb00155.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z, Zhou P, Ho D D, Landau N R, Marx P A. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clavel F, Guetard D, Brun-Vezinet F, Chamaret S, Rey M A, Santos-Ferreira M O, Laurent A G, Dauguet C, Katlama C, Rouzioux C, Klatzmann D, Champalimaud J L, Montagnier L. Isolation of a new human retrovirus from West African patients with AIDS. Science. 1986;233:343–346. doi: 10.1126/science.2425430. [DOI] [PubMed] [Google Scholar]
- 8.Daniel M D, Letvin N L, King N W, Kannagi M, Sehgal P K, Hunt R D, Kanki P J, Essex M, Desrosiers R C. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science. 1985;228:1201–1204. doi: 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- 9.Emau P, McClure H M, Isahakia M, Else J G, Fultz P N. Isolation from African Sykes’ monkeys (Cercopithecus mitis) of a lentivirus related to human and simian immunodeficiency viruses. J Virol. 1991;65:2135–2140. doi: 10.1128/jvi.65.4.2135-2140.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faulkner D M, Jurka J. Multiple aligned sequence editor (MASE) Trends Biochem Sci. 1988;13:321–322. doi: 10.1016/0968-0004(88)90129-6. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 13.Fultz P N, McClure H M, Anderson D C, Swenson R B, Anand R, Srinivasan A. Isolation of a T-lymphotropic retrovirus from naturally infected sooty mangabey monkeys (Cercocebus atys) Proc Natl Acad Sci USA. 1986;83:5286–5290. doi: 10.1073/pnas.83.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao F, Yue L, Robertson D L, Hill S C, Hui H, Biggar R J, Neequaye A E, Whelan T M, Ho D D, Shaw G M, Sharp P M, Hahn B H. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J Virol. 1994;68:7433–7447. doi: 10.1128/jvi.68.11.7433-7447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao F, Yue L, White A T, Pappas P G, Barchue J, Hanson A P, Greene B M, Sharp P M, Shaw G M, Hahn B H. Human infection by genetically diverse SIVsm-related HIV-2 in West Africa. Nature (London) 1992;358:495–499. doi: 10.1038/358495a0. [DOI] [PubMed] [Google Scholar]
- 16.Garnett G P, Antia R. Population biology of virus-host interactions. In: Morse S, editor. The evolutionary biology of viruses. New York, N.Y: Raven Press; 1994. pp. 51–73. [Google Scholar]
- 17.Grubb L. Refuges and dispersal in the speciation of African forest mamamals. In: Prance G T, editor. Biological diversification in the tropics. New York, N.Y: Columbia University Press; 1982. pp. 537–553. [Google Scholar]
- 18.Hirsch V M, Olmsted R A, Murphey-Corb M, Purcell R H, Johnson P R. An African primate lentiviruses (SIVsm) closely related to HIV-2. Nature (London) 1989;339:389–392. doi: 10.1038/339389a0. [DOI] [PubMed] [Google Scholar]
- 19.Huet T, Cheynier R, Meyerhans A, Roelants G, Wain-Hobson S. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature (London) 1990;345:356–359. doi: 10.1038/345356a0. [DOI] [PubMed] [Google Scholar]
- 20.Janssens W, Fransen K, Peeters M, Heyndrickx L, Motte J, Bedjabaga L, Delaporte E, Piot P, Van Der Groen G. Phylogenetic analysis of a new chimpanzee lentivirus SIVcpz-gab2 from a wild-captured chimpanzee from Gabon. AIDS Res Hum Retroviruses. 1994;10:1191–1192. doi: 10.1089/aid.1994.10.1191. [DOI] [PubMed] [Google Scholar]
- 21.Jin M J, Hui H, Robertson D L, Muller M C, Barre-Sinoussi F, Hirsch V M, Allan J S, Shaw G M, Sharp P M, Hahn B H. Mosaic genome structure of simian immunodeficiency virus from West African green monkeys. EMBO J. 1994;13:2935–2947. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson P R, Fomsgaard A, Allan J, Gravell M, London W T, Olmsted R A, Hirsch V M. Simian immunodeficiency viruses from African green monkeys display unusual genetic diversity. J Virol. 1990;64:1086–1092. doi: 10.1128/jvi.64.3.1086-1092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kestler H W, Li Y, Naidu Y M, Butler C V, Ochs M F, Jaenel G, King N W, Daniel M D, Desrosiers R C. Comparison of simian immunodeficiency virus isolates. Nature (London) 1988;331:619–622. doi: 10.1038/331619a0. [DOI] [PubMed] [Google Scholar]
- 24.Kimura M. The neutral theory of molecular evolution. Cambridge, United Kingdom: Cambridge University Press; 1983. [Google Scholar]
- 25.Kraus G, Werner A, Baier M, Binniger D, Ferdinand F J, Norley S, Kurth R. Isolation of human immunodeficiency virus-related simian immunodeficiency viruses from African green monkeys. Proc Natl Acad Sci USA. 1989;86:2892–2896. doi: 10.1073/pnas.86.8.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kusumi K, Conway B, Cunningham S, Berson A, Evans C, Iversen A K N, Colvin D, Gallo M V, Coutre S, Shpaer E G, Faulkner D V, deRonde A, Volkman S, Williams C, Hirsch M S, Mullins J I. Human immunodeficiency virus type 1 envelope gene structure and diversity in vivo and after cocultivation in vitro. J Virol. 1992;66:875–885. doi: 10.1128/jvi.66.2.875-885.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Kwon, D., et al. Unpublished data.
- 27.Letvin N L, Daniel M D, Sehgal P K, Desrosiers R C, Hunt R D, Waldron L M, MacKey J J, Schmidt D K, Chalifoux L V, King N W. Induction of AIDS-like disease in macaque monkeys with T-cell tropic retrovirus STLV-III. Science. 1985;230:71–73. doi: 10.1126/science.2412295. [DOI] [PubMed] [Google Scholar]
- 28.Lowenstine L J, Pedersen N C, Higgins J, Pallis K C, Uyeda A, Marx P A, Lerche N W, Munn R J, Gardner M B. Seroepidemiologic survey of captive Old World primates for antibodies to human and simian retroviruses and isolation of a lentivirus from sooty mangabeys (Cercocebus atys) Int J Cancer. 1986;38:563–574. doi: 10.1002/ijc.2910380417. [DOI] [PubMed] [Google Scholar]
- 29.Marx P A, Compans R W, Gettie A, Staas J K, Gilley R M, Mulligan M J, Dexiang C, Eldridge J H. Protection against vaginal SIV transmission with microencapsulated vaccine. Science. 1993;260:1323–1327. doi: 10.1126/science.8493576. [DOI] [PubMed] [Google Scholar]
- 30.Marx P A, Li Y, Lerche N W, Sutjipto S, Gettie A, Yee J A, Brotman B, Prince A M, Hanson A, Webster R G, Desrosiers R C. Isolation of simian immunodeficiency virus related to human immunodeficiency virus type 2 from a West African pet sooty mangabey. J Virol. 1991;65:4480–4485. doi: 10.1128/jvi.65.8.4480-4485.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marx P A, Spira A I, Gettie A, Dailey P J, Veazey R S, Lackner A A, Mahoney C J, Miller C J, Claypool L E, Ho D D, Alexander N J. Progesterone implants enhance SIV vaginal transmission and early virus load. Nat Med. 1996;2:1084–1089. doi: 10.1038/nm1096-1084. [DOI] [PubMed] [Google Scholar]
- 32.Miura T, Sakuragi J, Kawamura M, Fukasawa M, Moriyama E N, Gojobori T, Ishikawa K, Mingle J A A, Nettey V B A, Akari H, Enami M, Tujimoto H, Hayami M. Establishment of a phylogenetic survey system for AIDS-related lentiviruses and demonstration of a new HIV-2 subgroup. AIDS. 1990;4:1257–1261. doi: 10.1097/00002030-199012000-00012. [DOI] [PubMed] [Google Scholar]
- 33.Mojun J J, Huxiong H, Robertson D L, Muller M C, Barre-Sinoussi F, Hirsch V M, Allan J S, Shaw G M, Sharp P M, Hahn B H. Mosaic genome structure of simian immunodeficiency virus from West African Green monkeys. EMBO J. 1994;13:2935–2947. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller M C, Saksena N K, Nerrienet E, Chappey C, Herve V M, Durand J P, Legal P, Lang-Campodonico M C, Digoutte J P, Georges A J, Georges M, Sonigo P, Barre-Sinoussi F. Simian immunodeficiency viruses from central and western Africa: evidence for a new species-specific lentivirus in tantalus monkeys. J Virol. 1993;67:1227–1235. doi: 10.1128/jvi.67.3.1227-1235.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphey-Corb M, Martin L N, Rangan S R, Baskin G B, Gormus B J, Wolf R H, Andes W A, West M, Montelaro R C. Isolation of an HTLV-III related retrovirus from macaques with simian AIDS and its possible origin in asymptomatic mangabeys. Nature (London) 1986;321:435–437. doi: 10.1038/321435a0. [DOI] [PubMed] [Google Scholar]
- 36.Myers G, Hahn B H, Mellors J W, Henderson L E, Korber B, Jeang K-T, McCutchan F E, Pavlakis G N. Human retorviruses and AIDS. A compilation and analysis of nucleaic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1995. [Google Scholar]
- 37.Myers G, MacInnes K, Korber B. The emergence of simian/human immunodeficiency viruses. AIDS Res Hum Retroviruses. 1992;8:373–386. doi: 10.1089/aid.1992.8.373. [DOI] [PubMed] [Google Scholar]
- 38.Nerrienet, E., X. Amouretti, V. Poaty-Mavoungou, M. C. Muller-Truwin, I. Bedjebaga, H. Thi Nguyen, G. Dubreuil, S. Corbet, E. J. Wickings, F. Barre-Sinoussi, A. J. Georges, and M.-C. Georges-Courbot. Phylogenetic analysis of SIV and STLV-I in a mandrill (Mandrillus sphinx) colony indicates a low rate of sexual transmission. AIDS Res. Human Retroviruses, in press. [DOI] [PubMed]
- 39.Ohta Y, Masuda T, Tsujimoto H, Ishikawa K, Kodama T, Morikawa S, Nakai M, Honjo S, Hayami M. Isolation of simian immunodeficiency virus from African green monkeys and seroepidemiologic survey of the virus in various non-human primates. Int J Cancer. 1988;41:115–122. doi: 10.1002/ijc.2910410121. [DOI] [PubMed] [Google Scholar]
- 40.Otsyula M, Yee J, Jennings M, Suleman M, Gettie A, Tarara R, Isahakia M, Marx P, Lerche N. Prevalence of antibodies against simian immunodeficiency virus (SIV) and simian T-lymphotropic virus (STLV) in a colony of non-human primates in Kenya, East Africa. Ann Trop Med Parisitol. 1996;90:65–70. doi: 10.1080/00034983.1996.11813027. [DOI] [PubMed] [Google Scholar]
- 41.Peeters M, Fransen K, Delaporte E, Van den Haesevelde M, Gershy-Damet G-M, Kestens L, Van der Groen G, Piot P. Isolation and characterization of a new chimpanzee lentivirus (simian immunodeficiency virus isolate cpz-ant) from a wild-captured chimpanzee. AIDS. 1992;6:447–451. doi: 10.1097/00002030-199205000-00002. [DOI] [PubMed] [Google Scholar]
- 42.Peeters M, Honore C, Huet T, Bedjabaga L, Ossari S, Bussi P, Cooper R W, Delaporte E. Isolation and partial characterization of an HIV-related virus occurring naturally in chimpanzees in Gabon. AIDS. 1989;3:625–630. doi: 10.1097/00002030-198910000-00001. [DOI] [PubMed] [Google Scholar]
- 43.Peeters M, Janssens W, Fransen K, Brandful J, Hendrickx L, Koffi K, Delaporte E, Piot P, Gershy-Damet G M, Van der Groen G. Isolation of simian immunodeficiency viruses from two sooty mangabeys in Cote d’Ivoire: virological and genetic characterization and relationship to other HIV type 2 and SIVsm/mac strains. AIDS Res Hum Retroviruses. 1994;10:1289–1294. doi: 10.1089/aid.1994.10.1289. [DOI] [PubMed] [Google Scholar]
- 44.Reimann K A, Tenner-Racz K, Racz P, Montefiori D C, Yasutomi Y, Lin W, Ransil B J, Letvin N L. Immunopathogenic events in acute infection of rhesus monkeys with simian immunodeficiency virus of macaques. J Virol. 1994;68:2362–2370. doi: 10.1128/jvi.68.4.2362-2370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robbins C B. The Dahomey Gap—a reevaluation of its significance as a faunal barrier to West African high forest mammals. Bull Carnegie Mus Nat Hist. 1978;6:168–174. [Google Scholar]
- 46.Sharp, P. M., D. L. Robertson, F. Gao, and B. H. Hahn. 1994. Origins and diversity of human immunodeficiency viruses. AIDS 8(Suppl.):S27–S42.
- 47.Simon, J. H. M., D. L. Miller, R. A. M. Fouchier, M. A. Soares, K. Peden, and M. H. Malim. Unpublished data.
- 47a.Smith, S., and P. Marx. Unpublished data.
- 48.Stivahtis G L, Soares M A, Vodicka M A, Hahn B H, Emerman M. Conservation and host specificity of Vpr-mediated cell cycle arrest suggest a fundamental role in primate lentivirus evoluation and biology. J Virol. 1997;71:4331–4338. doi: 10.1128/jvi.71.6.4331-4338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomonaga K, Katahira J, Fukasawa M, Hassan M A, Kawamura M, Akari H, Miura T, Goto T, Nakai M, Suleman M, Isahakia M, Hayami M. Isolation and characterization of simian immunodeficiency virus from African white-crowned mangabey monkeys (Cercocebus torquatus lunulatus) Arch Virol. 1993;129:77–92. doi: 10.1007/BF01316886. [DOI] [PubMed] [Google Scholar]
- 50.Tsujimoto H, Cooper R W, Kodama T, Fukasawa M, Miura T, Ohta Y, Ishikawa K I, Nakai M, Frost E, Roelants G E, Roffi J, Hayami M. Isolation and characterization of simian immunodeficiency virus from mandrills in Africa and its relationship to other human and simian immunodeficiency viruses. J Virol. 1988;62:4044–4050. doi: 10.1128/jvi.62.11.4044-4050.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vanden Haesevelde M M, Peeters M, Jannes G, Janssens W, Van Der Groen G, Sharp P M, Saman E. Sequence analysis of a highly divergent HIV-1-related lentivirus isolated from a wild captured chimpanzee. Virology. 1996;221:346–350. doi: 10.1006/viro.1996.0384. [DOI] [PubMed] [Google Scholar]
- 52.Wolfheim J H. Primates of the world. Seattle: University of Washington; 1983. [Google Scholar]