Abstract
Parvovirus B19 infections are associated with diverse clinical manifestations, ranging from no symptoms to severe symptoms. The virus shows an extreme tropism for replication in erythroid progenitor cells, possibly due to the activity of the only functional promoter (p6) of the B19 virus genome in combination with both cell- and cell cycle-specific factors and the trans-activator protein NS1. As presented here, p6 promoter sequences derived from several B19 virus isolates proved to be highly conserved. Furthermore, mutations did not affect any of the potential binding sites for transcription factors. One variation of the base at position 223 was identified only in B19 virus isolates derived from patients with persistent infection or chronic arthritis. To determine promoter activity and to characterize regulatory elements, sequences spanning the total p6 promoter and subfragments of them were introduced into a eukaryotic expression vector upstream of the luciferase gene (from Photinus pyralis). After transfection into HeLa, CEM, BJAB, and K562 cells, the p6 promoter was found to be highly active. When introduced into the erythroid cell line K562, p6-controlled transcription exceeded that of the simian virus 40 promoter-enhancer used as a control by more than 25-fold. Sequence elements relevant for promoter activity mapped to the regions from nucleotides (nt) 100 to 190 and 233 to 298. Also, the segment from nt 343 to 400 downstream of the TATA box was important for transcriptional activity in HeLa and K562 cells. By transfecting the promoter-luciferase constructs into a HeLa cell line stably carrying the viral NS1 gene under the control of an inducible promoter, transcriptional activity mediated by the p6 promoter rose significantly after induction of NS1 expression. The region from nt 100 to 160 proved to be essential for NS1-mediated transcriptional activation. Furthermore, NS1-mediated transactivation was dependent on the presence of two GC-rich elements arranged in tandem upstream of the TATA box. These data indicate that NS1-mediated p6 transactivation is dependent on a multicomponent complex combining NS1 with ATF, NF-κB/c-Rel, and GC-box binding cellular factors.
Parvovirus B19 is the only parvovirus known to be pathogenic in humans. Besides fifth disease (erythema infectiosum), common during childhood, it can cause acute and persistent arthritis in adults (1, 24), aplastic crisis in patients suffering from chronic hemolytic anemia (23), or hydrops fetalis after transplacental infection during pregnancy (6). It shows an extreme host and cell tropism and can induce productive infections only in human erythroid progenitor cells. Several reasons for this tropism have been discussed, yet its basis remains unclear. The viral receptor, the blood group antigen P, or globoside (5), is present on the surface of both erythroid cells and a variety of lymphoid and endothelial cells. In the latter, however, the virus is unable to replicate. The viral genome contains two promoter-like elements (3, 8, 9), of which only the one at map unit 6 proved to be functionally active in both permissive and nonpermissive cells (4, 15, 20). The p6 promoter was shown to confer autonomous replication competence and erythroid-cell specificity to adeno-associated virus 2 (32). It is transactivated by the nonstructural protein NS1 (9), which probably exerts its action in interplay with cellular factors, as has been shown for the NS1 protein of the murine parvovirus minute virus of mice (MVM) (13). Thus, cell specificity may at least in part be due to promoter strength influenced by cell-specific and cell cycle-specific factors in combination with the viral NS1 protein. Two polyadenylation sites located in the center and at the end of the genome, in combination with alternative splicing events, are responsible for the production of a total of nine viral transcripts, seven of which are used as mRNAs (21). Both processes may be regulated and influenced by cell-specific factors. In nonpermissive cells, a shift towards the production of functional transcripts from the 3′ half of the genome is obvious, resulting in NS1-specific mRNAs. This probably leads to the accumulation of the cytotoxic NS1 protein and to cell death (16, 22). Furthermore, virus replication was shown to be dependent on dividing cells and S-phase-specific host factors (33). However, it must be supposed that none of these reasons alone may account for the cell specificity of viral replication.
In the work presented here, the functionally cis-active elements of the p6 promoter were characterized in diverse cell types. p6 promoter regions derived from 11 different virus isolates were amplified by nested PCR, and the nucleotide sequences were determined. The promoter sequences were integrated into a vector system using firefly luciferase as a reporter. After transfection, transcriptional activity was analyzed by measuring the amount of reporter protein in four cell lines, namely, HeLa (epithelial cells), BJAB (Epstein-Barr virus [EBV]-negative B cells), CEM (CD4-positive T cells), and K562 (erythroleukemic cells). By using different sets of primers and PCR, promoter fragments truncated from both directions and mutations in individual potential binding sites were created and used to control the expression of the luciferase reporter in the four cell lines. To characterize the p6 promoter domains which are responsive to transactivation by the viral NS1 protein, the various promoter-luciferase constructs were introduced into HeLa cells stably carrying the NS1 gene under the control of an inducible promoter. These experiments allowed fine-mapping of the p6 promoter of parvovirus B19 and the characterization of cis-acting elements influenced by cell-specific factors in combination with the NS1 protein.
MATERIALS AND METHODS
Patients’ sera, nested PCR, and DNA sequencing.
Patients’ sera were supplied by the Institute for Medical Microbiology and Hygiene, University of Regensburg, Regensburg, Germany; the Institute for Virology, University of Vienna, Vienna, Austria; the Max-von-Pettenkofer Institute, Ludwig-Maximilians University, Munich, Germany; the Rheumaklinik at Olsberg, Olsberg, Germany; and the Institute for Clinical Virology and Immunology, St. Gallen, Switzerland. To amplify the B19 virus promoter regions even from sera with a low virus load, a sensitive nested-PCR technique was applied as described by Hemauer and coworkers (11). The two forward primers are homologous to positions 195 to 212 (outer forward primer) and 221 to 237 (inner forward primer) of the pVD6 sequence published by Deiss et al. (7). The backward primers are homologous to positions 494 to 475 (inner backward primer) and 532 to 509 (outer backward primer) of the pYT103 sequence published by Shade et al. (27).
The PCR products were sequenced by using a cycle-sequencing technique and a 373A Sequencer (Applied Biosystems, Weiterstadt, Germany). Only sequences which proved identical in two independent amplifications were considered to be valid.
Cell lines.
All cells were grown in media containing 10% fetal calf serum. HeLa cells were grown in Dulbecco’s modified Eagle’s medium with 0.11 g of sodium pyruvate per liter and 2 mM glutamine, with 100 U of penicillin per ml and 100 μg of streptomycin per ml. For growth of cell line HeLa/NS-21, which contained the episomal vector pGRE5-2/EBV-NS, and of cell line HeLa/pGRE, which carried the vector pGRE, 300 μg of hygromycin B per ml was added to select for cells containing the episomal vector (14). BJAB, CEM, and K562 cells were grown in Dulbecco’s RPMI 1640 with penicillin (100 U/ml) and streptomycin (100 μg/ml) or 100 μg of kanamycin per ml.
Construction of the B19 virus promoter-luciferase expression plasmids.
Promoter fragments and mutants (see Fig. 2 and 3) were created by PCR amplification of the respective segments, with plasmid pJB (kindly provided by J. P. Clewley, London, United Kingdom), which contains almost the complete parvovirus B19 genome, as template DNA. The primers were synthesized on an Expedite solid-phase synthesis system (Millipore, Eschborn, Germany). NheI and HindIII restriction sites were inserted at the ends of the PCR primers and used to clone the constructs into the respective sites of the luciferase vector pGL-3 Basic (Promega Corporation, Madison, Wis.). The smallest fragment, p4/2 (see Fig. 2), was represented by an oligonucleotide whose complementary strands were also synthesized by the solid-phase method, phosphorylated, annealed, and inserted into the vector via NheI and HindIII restriction sites. Mutations were inserted into the promoter sequences by an overlap extension technique described previously (12). As a control, the vector pGL-3 Control (also purchased from Promega), which carries the luciferase gene under control of the simian virus 40 (SV40) promoter-enhancer, was used.
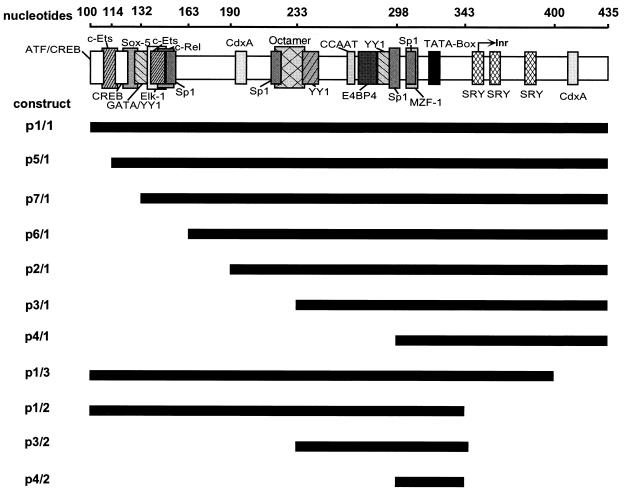
FIG. 2.
The p6 promoter fragments constructed and used for analysis of the promoter activity. Potential binding sites for transcription factors are indicated (shaded boxes). Inr, putative transcription start site. The lengths of the bars below the p6 promoter (top line) represent the lengths of the fragments.
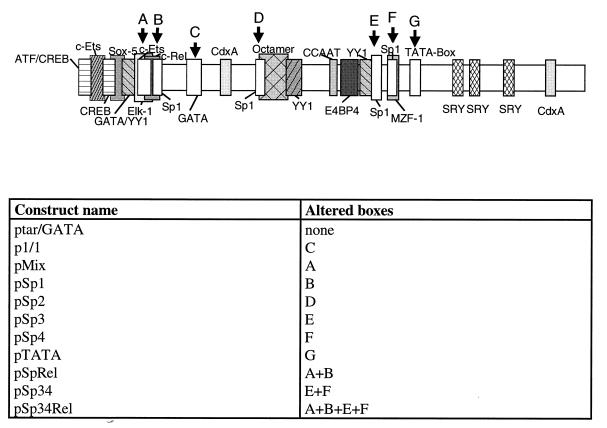
FIG. 3.
Mutant p6 promoter constructs tested. The binding sites for transcription factors which were altered (boxes A to G) are indicated, and the constructs and the boxes into which mutations were introduced are listed below the diagram.
DNA transfection and luciferase assay.
To analyze the luciferase activities with the Dual Luciferase System (Promega), production of a second type of luciferase derived from the marine coelenterate Renilla reniformis was achieved via the vector pRL-CMV (also purchased from Promega), which was cotransfected into the cells with the promoter constructs and used as an internal reference. HeLa/NS-21 cells were stably transfected with the expression vector pGRE5-2/EBV-NS, which carried the NS1 gene under the control of a dexamethasone-inducible glucocorticoid response element in five copies (14). HeLa/pGRE cells contained the same vector without the sequences encoding the NS1 gene (18). For determination of luciferase activity, all tests were performed in triplicate. Approximately 3 × 105 HeLa cells cultivated in six-well plates were transfected with the various vector constructs by the calcium phosphate method as described previously (2). HeLa/NS-21 and HeLa/pGRE cells were induced by addition of 10−6 M dexamethasone (Sigma-Aldrich, Steinheim, Germany) 1 day prior to transfection. HeLa cells were cotransfected with 0.5 μg of the respective promoter-luciferase constructs and 0.1 μg of pRL-CMV, each time in duplicate. When the various vector constructs were introduced into CEM, BJAB, or K562 cells, 107 cells were suspended in phosphate-buffered saline. The plasmids were introduced by electroporation, applying a current of 240 V and an electric field of 1,050 μF with an EasyJect Plus electroporator (Eurogentec, Seraing, Belgium). BJAB and K562 cells were transfected with 5 μg of promoter-luciferase constructs in combination with 1 μg of pRL-CMV, and CEM cells were transfected with 10 μg of the constructs and 2 μg of pRL-CMV. Two days after transfection, cells were harvested, washed in phosphate-buffered saline without Ca2+ and Mg2+ ions, and lysed in 100 μl of passive lysis buffer supplied by the manufacturer (Promega). Samples were rocked for 20 min at room temperature and freeze-thawed once in liquid nitrogen. A 20-μl sample of the lysate was used for analysis of the luciferase activity according to the instructions of the manufacturer (Promega) with a luminometer (Lumat LB9501; EG+G Berthold, Munich, Germany). This assay takes advantage of the successive determination of the amounts of two coexpressed luciferase enzymes: one is the luciferase of the firefly Photinus pyralis, used as the reporter protein, and the other (applied as an internal reference) is derived from the marine coelenterate R. reniformis and uses a different substrate.
Nucleotide sequence accession numbers.
The nucleotide sequence data from this study have been deposited with the EMBL database under accession no. Z95625 to Z95635.
RESULTS
Determination of promoter sequences of B19 virus isolates.
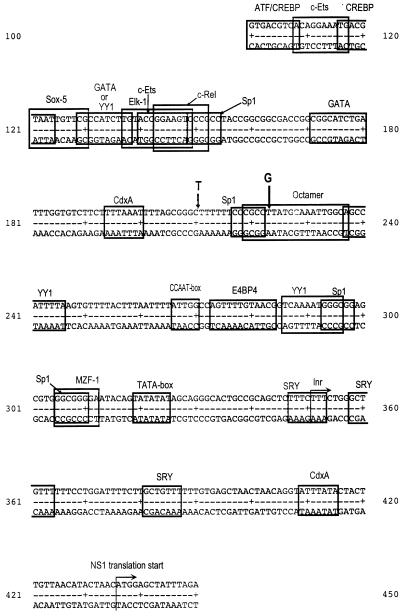
In order to test if variants of the p6 promoter sequences of virus isolated from patients with different B19 virus-associated diseases correlate with specific clinical manifestations, we determined the amount of sequence variability in the corresponding regions of the virus genome. Therefore, the p6 promoter domains of several B19 virus isolates were amplified from patients’ sera by nested PCR and subjected to DNA sequencing. Table 1 shows the clinical manifestations of the patients from whom the sera were obtained and the degree of variability observed. The promoter region of all sequenced isolates turned out to be highly conserved compared to the previously published B19 virus sequences (7, 27), with a degree of variability below 0.8%. Isolate S724 showed the insertion of an additional thymidine between nucleotides (nt) 210 and 211 (Fig. 1). In five of the sequenced isolates (5, A, V6, S758, and SP [Table 1]), the thymidine at position 223 was replaced by a guanosine. All of the patients from whom the virus isolates with differences in the p6 promoter sequence were obtained (S724, 5, A, V6, S758, and SP) suffered from long-lasting B19 virus-associated arthritis or chronic B19 virus infections of the hematopoietic system. Both clinical symptoms can be associated with persistent B19 virus infections (30, 31).
TABLE 1.
Variability of p6 promoter sequences from patients with different B19 virus-associated diseasesa
| Isolate | Disease and/or symptoms of patient | No. of mutations in p6 sequence/total no. of bases (%) |
|---|---|---|
| K | Acute infection, asymptomatic | 3/389 (0.77) |
| KS | Acute infection, asymptomatic | 3/388 (0.77) |
| 2/II | Acute infection | 0/388 (0) |
| A | Persistent infection | 1/389 (0.26) |
| B | Persistent infection | 0/389 (0) |
| 5 | Anemia after transplantation | 1/389 (0.26) |
| S471 | Arthritis | 0/389 (0) |
| V6 | Arthritis | 1/388 (0.26) |
| S758 | Arthritis | 2/388 (0.52) |
| SP | Arthritis | 2/388 (0.52) |
| S724 | Arthritis | 3/387 (0.78) |
Sera from patients with B19 virus infection were used for determination of the sequence of the p6 promoter to examine whether there was a correlation with the respective clinical manifestations and variability in the p6 promoter sequence. The sequences were compared to those published by Deiss et al. (7) and Shade et al. (27).
FIG. 1.
Map of the p6 promoter of parvovirus B19 with potential binding sites for transcription factors indicated (boxes). Inr, putative transcription start site. The position of the insertion of a T in one of the virus isolates and the locations of the exchange of a T for a G in five of the isolates sequenced are indicated (thick arrows).
The promoter sequences were analyzed for the presence of DNA elements known to interact with cellular transcription factors by using the program TFSEARCH 1.3 (Y. Akiyama, on-line at http//:www.genome.ad.jp/SIT/TFSEARCH.html), which recruits the TRANSFAC-MATRIX TABLE database 2.5 (32a). Various elements could be identified; however, none of the observed mutations affected any of them (Fig. 1). Therefore, it can be concluded that the activity of the p6 promoter is probably identical in all the B19 virus isolates, independent of the clinical course of infection.
Creation of promoter constructs.
To assay the activity of the complete p6 promoter, the region from nt 100 to 435 (referring to plasmid pYT103, construct p1/1 [Fig. 1 and 2]), derived from plasmid pJB, was used (27). For the p38 promoter of MVM, the respective GC-rich elements had been shown previously to be important for transactivation via the MVM-specific NS1 protein (13). To delete the GC boxes, the domain was shortened stepwise, beginning at the genomic 3′ end, resulting in constructs p2/1, p3/1, and p4/1 (Fig. 2). In order to analyze the influence of p6 promoter elements located downstream of the TATA box, constructs p1/3 and p1/2 were created (Fig. 2). In this case, potential transcription factor binding sites located between the TATA box and the first AUG used for translation of the NS1 protein were deleted. The importance of the sequences surrounding the TATA box was assayed by using constructs p3/2 and p4/2 (Fig. 2). p5/1 and p6/1 were created for fine-mapping of the NS1-responsive site.
By the overlap extension technique (12), mutations were introduced into the sequences encoding potential binding sites for transcription factors in order to determine their individual influence on the promoter activity. In construct ptar/GATA (Fig. 3), the trans-activating region (tar) and a potential GATA binding site were restored since these regions were deleted in plasmid pJB compared to the original B19 virus sequence (27) and to the virus isolates sequenced as described above. A similar tar identified in parvovirus H1 had been shown to serve as interaction site for the H1-specific NS1 protein (25). ptar/GATA served as a template for the construction of further mutant promoter sequences. In constructs pSp1, pSp2, pSp3, and pSp4, the four GC-rich boxes which had been identified were individually mutated (CCGCC to AAGAA [Fig. 3]). The mutation in pSp1 additionally affects one half-site of a c-Rel/NF-κB consensus site. Construct pSp34 represents a mutant destroying both GC boxes 3 and 4. pTATA lacks a functional TATA box (TATATA to GCGCGC). In construct pMix, a potential binding site for Ets and one half-site of a c-Rel/NF-κB consensus sequence were altered (GGAAGT to TCGCAG). The complete c-Rel/NF-κB site was altered, combining the mutations of pSp1 and pMix; the resulting construct was termed pRel. In pSp34Rel, the tandem GC boxes neighboring the TATA box were mutated in addition to the c-Rel/NF-κB site.
Activity of the p6 promoter in different cell lines in comparison to the SV40 promoter-enhancer.
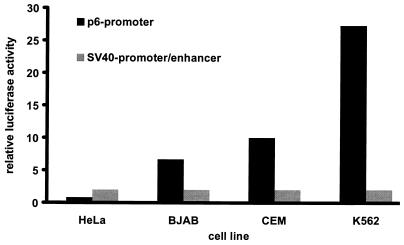
In order to analyze the activity of the complete p6 promoter of the B19 virus, construct p1/1, spanning nt 100 to 435 (Fig. 1 and 2), was introduced into four cell lines (HeLa, CEM, BJAB, and K562). In parallel, plasmid pGL-3 Control containing the luciferase gene under control of the SV40 promoter-enhancer region was used. The activities of both viral promoters were determined by assaying the amount of luciferase produced in the cells and compared. The results are shown in Fig. 4. In epithelial HeLa cells, the SV40 promoter turned out to be three to four times more active than the p6 promoter. However, the activity of the B19 virus promoter exceeded that of the SV40 promoter-enhancer by more than 25 times in K562 cells of hematopoietic origin. In the B-cell line BJAB and the T-cell line CEM, the B19 virus-derived promoter was about 6 and 10 times stronger than the SV40 promoter-enhancer, respectively. These data show that the p6 promoter of parvovirus B19 is also highly active in human cells which generally do not allow productive replication.
FIG. 4.
Comparison of the promoter strength of the p6 promoter from parvovirus B19 to that of the promoter-enhancer of SV40 in four cell lines. The values were calculated by dividing the amount of luciferase activity (normalized against the internal R. reniformis standard) of the p6 promoter by that of the SV40 promoter-enhancer in each cell line. The activity of the SV40 promoter-enhancer is therefore 1 in all cell lines tested.
Identification of p6 promoter sequence elements influenced by cell-specific factors.
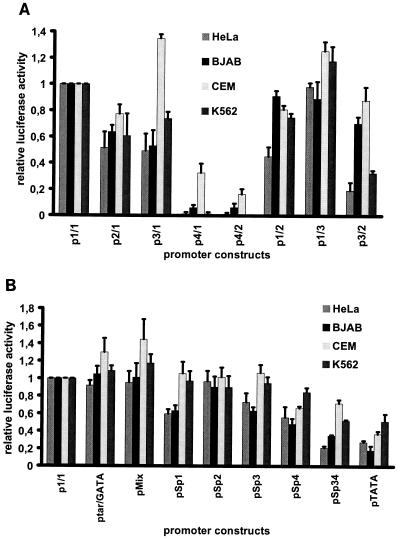
The various promoter constructs (Fig. 2 and 3) were introduced into HeLa, BJAB, CEM, and K562 cells. After 2 days, the respective transcriptional activities were determined in at least three independent tests using luciferase assays. The results are shown in Fig. 5 and represent the respective values in relation to the activity of the wild-type B19 virus promoter, determined by the use of construct p1/1, whose activity was taken as 100%. Truncation of the promoter by the first 90 nt from the upstream direction (construct p2/1 [Fig. 2]) displayed similar effects in all cell lines: the activity of the p6 promoter was reduced to values between 50 and 80% of the wild-type activity. Further truncation of the promoter by 42 nt (construct p3/1 [Fig. 2 and 5A]) had almost no additive effect when assayed in HeLa, BJAB, or K562 cells. However, a significant rise in promoter activity was observed in CEM cells when the same construct was used. When a further 59 nt were deleted (construct p4/1 [Fig. 2]), transcription was almost abolished in HeLa, BJAB, and K562 cells. In contrast, construct p4/1 retained more than 30% of promoter activity in CEM cells (Fig. 5A). Deletion of segments located close to the translation start site was possible without any loss in promoter activity in all cell lines tested, as shown by construct p1/3 (Fig. 2 and 5A). Fragment p1/2, however, which lacked further elements between the TATA box and the translation start site including the putative B19 virus transcription start site, reduced promoter activity in HeLa cells to 50% of the wild type. In the other cell lines, the effect was not as pronounced (reduction to about 80%). Construct p3/2, however, reduced activity to 50% compared to that of plasmid p3/1 (Fig. 5A) in both HeLa and K562 cells, while this central fragment (p3/2) retained almost the full p6 activity in CEM cells and about 70 to 80% activity in BJAB cells.
FIG. 5.
Comparison of the activities of the different wild-type (Fig. 2) (A) and mutant (Fig. 3) (B) p6 promoter constructs in cell lines HeLa, BJAB, CEM, and K562. Values are given in relation to that of construct p1/1, representing the unmodified p6 promoter, whose activity was arbitrarily set as 1 and whose standard deviation is therefore 0.
The promoter activity displayed by the construct ptar/GATA (Fig. 3 and 5B) was similar to that of the B19 wild-type virus in BJAB, HeLa, and K562 cells; a small increase could be measured only in CEM cells. Identical results were obtained with construct pMix (Fig. 3 and 5B). Mutation of the most upstream GC box (construct pSp1) revealed that the influence of this cis-acting element was significantly different in the four cell lines: promoter activity was unaffected in CEM and K562 cells but was reduced by 40% in both HeLa and BJAB cells. The mutation of the second GC box (construct pSp2 [Fig. 3]) had no effect in either cell line (Fig. 5B). In contrast, promoter activity was reduced in HeLa and BJAB cells, but not in CEM and K562 cells, when construct pSp3, in which the third of the GC-rich elements was altered, was used. The mutation of the fourth GC box (construct pSp4) had a negative effect on promoter strength in all cell lines tested (Fig. 3 and 5B). When the double mutation of the third and fourth GC boxes as represented by construct pSp34 (Fig. 3) was introduced into CEM cells, promoter activity was similar to that of pSp4, in contrast to the other cell lines, in which a further reduction could be observed. The mutation of the TATA box (construct pTATA) led to the loss of 50 to 80% of promoter activity in all cell lines (Fig. 5B).
All these data indicate that irrespective of the fact that the p6 promoter is highly active in a variety of human cells, transcriptional activity of this promoter is subject to cell-type-specific influences. Alterations in the DNA elements spanning nt 190 to 233 were shown to be highly sensitive for the regulation via cellular transactivators.
Identification of NS1 protein-responsive DNA elements in the p6 promoter.
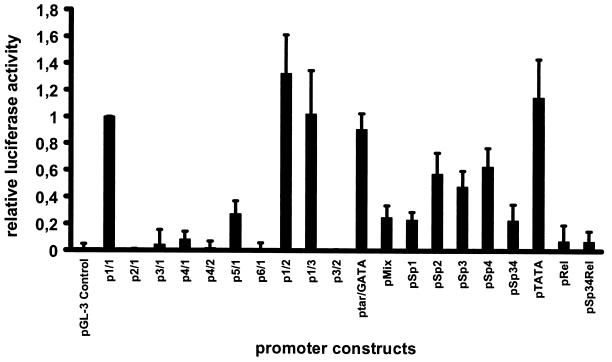
The various p6 promoter-luciferase constructs were introduced into cell lines HeLa/NS-21 and HeLa/pGRE, which had been treated with 10−6 M dexamethasone 1 day prior to transfection in order to induce expression of NS1 protein. Luciferase expression was measured 2 days after transfection. HeLa/pGRE cells were used as a reference. The synthesis of NS1 protein in HeLa/NS-21 cells was confirmed by Western blotting and indirect immunofluorescence using an NS1-specific rabbit serum (30). About 30% of the cells stained positive for NS1 production by indirect immunofluorescence (data not shown). When introduced into dexamethasone-induced and NS1 protein-producing HeLa/NS-21 cells, constructs which contained elements of the p6 promoter spanning nt 100 to 160 (p1/1, p1/2, p1/3, and p5/1 [Fig. 2]) showed a significant increase in luciferase expression (up to 50 times [Fig. 6]). In cases where this region had been deleted (constructs p6/1, p2/1, p3/1, p4/1, p3/2, and p4/2 [Fig. 2]), enhancement of promoter activity was completely abolished (Fig. 6). A reduction in transactivation of about 70% was detected when only the first 12 bases were deleted, as in construct p5/1 (Fig. 2), on which an ATF/CREB site is located (Fig. 6). When constructs pMix and pSp1 (Fig. 3) were introduced into HeLa/NS-21 cells, transactivation mediated by NS1 protein was reduced to about one-third of that of the wild-type p6 promoter (Fig. 6). The same amount of reduction was observed when construct pSp34, in which the third and fourth GC boxes had been altered, was used. In construct pRel, in which the complete NF-κB/c-Rel-like site was altered, combining the mutations of pSp1 and pMix, enhancement of transcription by NS1 could be abolished almost completely (Fig. 6). The same was true for construct pSp34Rel, in which both the third and fourth GC elements were mutated in addition to the NF-κB/c-Rel site. ptar/GATA and pTATA were fully transactivatable by NS1, indicating that in contrast to the case for MVM, the tar homology element of parvovirus B19 is not responsive to NS1-mediated transactivation. These results indicate that in addition to elements located between nt 100 and 160, the sequences spanning the promoter region of nt 233 to 298 are responsive to the transactivating effect of the NS1 protein.
FIG. 6.
Transactivation of the different p6 promoter constructs by the NS1 protein. Values were calculated as follows. The amount of transactivation by each construct was calculated by setting the luciferase activity of the respective construct in HeLa/NS-21 cells (normalized against the internal R. reniformis standard) relative to that in HeLa/pGRE cells. These values were then divided by the values obtained with construct p1/1, which represents the wild-type promoter. The value of transactivation of construct p1/1 is therefore 1 (with no standard deviation). pGL-3 Control contains the SV40 promoter-enhancer and was regarded as not transactivatable; therefore its value is the baseline.
DISCUSSION
The fact that infections with human parvovirus B19 are associated with a wide range of different clinical manifestations indicates that the virus can enter additional target cells besides the erythroid progenitor cells which are known to be productively infected and destroyed during erythema infectiosum. Globoside has been identified as the receptor for virus attachment to the cell surface (5). This molecule, also known as blood group antigen P, is, however, present on the surfaces of a variety of other cells which are not permissive to productive B19 virus infection. These observations allow the conclusion that in addition to other factors, cell-type-specific factors possibly influence the activity of the p6 promoter and thus contribute to the ability to replicate preferentially in erythroid progenitor cells.
To check if B19 virus isolate-specific sequence variations are associated with clinical symptoms, e.g., persistent B19 virus infection, the promoter regions of different virus isolates were amplified by PCR and sequenced. The data show a high degree of conservation of this region, indicating that parvovirus B19 exhibits only a very low degree of sequence variability (Fig. 1 and Table 1). This is in agreement with the results of other investigators who examined the coding sequences for the nonstructural and structural B19 virus proteins (10, 11). The exchange of a single base at position 223 in 5 of the 11 B19 virus isolates sequenced suggests an increased variability at this position. This change was found exclusively in patients with B19 virus-associated arthritis, chronic anemia, and persistent infection. Therefore, it may be proposed that this mutation, which lies in the vicinity of an octamer consensus site, may be associated with a tendency to enhanced viral persistence. Since, however, only 11 isolates were sequenced, it is not possible to draw statistical conclusions from these results.
In a computer analysis, the p6 promoter showed a high content of potential binding sites for erythroid-cell-specific trans-activating factors (GATA and MZF) as well as a variety of lymphoid-cell-specific factors such as Octamer, c-Ets, c-Rel, or NF-κB (Fig. 1 and 2). This may be the basis for the enhanced transcriptional activity of the p6 promoter which we found in hematopoietic K562 cells in contrast to epithelial HeLa cells (Fig. 4). The p6 promoter was also more active in erythroleukemic K562 cells, as in the lymphoid cell lines BJAB and CEM. In K562, BJAB, and CEM cells, the activity of the B19 virus promoter even exceeded that of the SV40 promoter-enhancer region, which is known to be a highly active control element for transcription in eukaryotic cells. It was shown earlier that the combination of several cis-acting DNA elements contributes to the cell type specificity of transactivating elements (26). Taking these results into account, the p6 promoter may indeed be important for viral cell tropism and specificity of replication in cell types in distinct stages of differentiation.
Four conclusions can be made from this study. (i) The promoter regions which were identified to be relevant for transcription in the different cell lines were nt 100 to 190. When deleted, a loss of activity was found in all cell lines (Fig. 5A). By use of footprint experiments, the promoter region spanning nt 137 to 156 had previously been shown to interact with proteins in HeLa and CEM cells (17). (ii) DNA elements located in the region between nt 190 and 233 could be shown to mediate transcriptional repression in CEM cells (Fig. 5A). However, in BJAB, HeLa, or K562 cells, the deletion of this region did not show any effect on the transcriptional activity of the p6 promoter. (iii) In contrast to those cell-specific regulation events, the region spanning nt 233 to 298 was essential for induction of transcription in all the cell lines tested. Liu and coworkers had identified nt 220 to 254 and 280 to 315 to be protected in both HeLa and CEM cells in DNA footprint assays (17). Two YY1 binding sites, which are functionally active as shown by electrophoretic mobility shift assay and transfection assay, are located in this region (20). (iv) We were able to show that the sequences between the TATA box and the translation start site are relevant for mRNA synthesis. nt 343 to 400 were important for effective transcription in HeLa and K562 cells. In contrast, sequences from nt 400 to the start site for NS1 translation at nt 435 appeared to be nonessential. The transcription rate was slightly enhanced when this 5′ untranslated domain was deleted (Fig. 5A).
The NS1 protein of parvovirus B19 is known to exert a variety of different functions during viral replication. Among other functions, the transactivation of the p6 promoter had been shown by Doerig and coworkers (9). The fact that the transactivation assays were conducted only with HeLa cells, which are not the natural host cells for B19 virus replication in vivo, probably limits our results to a certain extent. Nevertheless, some of the sequences responsible for transactivation by the viral NS1 protein in those cells could be clearly identified. It could be shown that neither the TATA box nor the putative NS1 transactivating region, which has similarity to that of parvovirus H1, is responsible for the transactivation of the p6 promoter by the NS1 protein. The sequence elements absolutely necessary for the transactivator function of the NS1 protein obviously lie within the region spanning nt 100 to 160 of the p6 promoter (Fig. 1). Loss of the first 12 bp, which contains an ATF/CREB consensus site, led to a reduction of transactivatability to only 30% of that of the wild-type promoter. Until now, direct interactions between the NS1 protein and DNA could not be shown for parvovirus B19. Therefore, it has to be assumed that the NS1 protein exerts its action not by binding to the DNA itself, but rather by interacting with transactivators, transcription or accessory factors bound to the nucleic acid. It has been shown that the NS1 protein of parvovirus B19 is able to transactivate the long terminal repeat promoter of human immunodeficiency virus type 1 only in combination with the Tat protein of human immunodeficiency virus (28). Recently, Moffatt and coworkers reported that the transactivation of the interleukin-6 promoter by the NS1 protein of parvovirus B19 is almost completely abolished by deletion of an NF-κB site (19). A sequence element similar to the NF-κB site is also present within the region shown to be important for NS1-specific transactivation of the p6 promoter: 5′-CCGGAAGTCCCGCC-3′ (nt 141 to 155). After mutation of either half of this element, transcriptional activation by the NS1 protein could be reduced to about 30% compared to that of the wild-type p6 promoter. Enhancement of transcription by NS1 transactivation was abolished almost completely by mutation of the complete NF-κB/Rel-like site. The 3′ half of this element resembles a consensus site for the transcription factor Sp1. According to Liu and coworkers, it is probably not Sp1 that binds to this site, since they found that the complete NF-κB consensus site was protected in footprint experiments and therefore complexed with protein (17). Transactivation by NS1 was also significantly reduced by the double mutation of both GC boxes arranged in tandem in proximity to the TATA box.
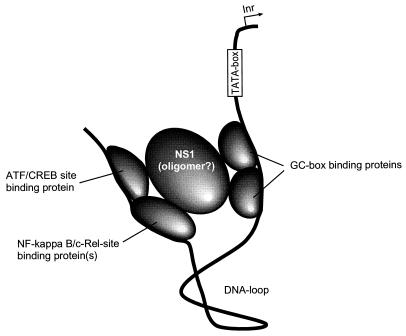
These results suggest that transcriptional enhancement mediated by the NS1 protein involves a complex of individually DNA-binding proteins, which are bridged by the nonstructural B19 virus protein. Thus, a model for the transactivating function of the NS1 protein which is similar to that of the Tax proteins of human T-cell leukemia virus may be proposed (Fig. 7). Tax protein exerts its transactivating action by binding to c-Rel and bridging between other factors, such as Sp1 and Ets (29). Likewise, NS1 activation seems to be dependent on proteins that bind at or near consensus sites for ATF/CREB, NF-κB/Rel, and distant GC boxes, which are arranged in tandem in the vicinity of the TATA box. The DNA segment in between could easily be looped out. In this way, the NS1 protein may bridge the distant promoter sites in the form of a monomer or an oligomer.
FIG. 7.
Model of the transactivating complex involving proteins bound to consensus sites for ATF/CREB, NF-κB/c-Rel, and Sp1 (GC boxes) which are bridged by NS1 monomer or oligomer. The DNA in between could easily be looped out. Inr, transcription start site.
ACKNOWLEDGMENTS
This work has been supported by the Deutsche Forschungsgemeinschaft DFG (grant Mo620/5-1). A.G. has been supported by a grant of the University of Regensburg (Förderung des wissenschaftlichen Nachwuchses), and A.H. has been supported by the Studienstiftung des Deutschen Volkes.
REFERENCES
- 1.Anderson M J, Jones S E, Fisher-Hoch S P, Lewis E, Hall S M, Bartlett C L R, Cohen B J, Mortimer P P, Pereira M S. Human parvovirus, the cause of erythema infectiosum (fifth disease)? Lancet. 1983;i:1378. doi: 10.1016/s0140-6736(83)92152-9. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel I, editor. Current protocols in molecular biology. 1 to 3. New York, N.Y: John Wiley and Sons; 1995. [Google Scholar]
- 3.Blundell M C, Astell C R. In vitro identification of a B19 parvovirus promoter. Virology. 1987;157:534–538. doi: 10.1016/0042-6822(87)90296-0. [DOI] [PubMed] [Google Scholar]
- 4.Blundell M C, Astell C R. A GC-box motif upstream of the B19 parvovirus unique promoter is important for in vitro transcription. J Virol. 1989;63:4814–4823. doi: 10.1128/jvi.63.11.4814-4823.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown K E, Anderson S M, Young N S. Erythrocyte P antigen: cellular receptor for the B19 virus. Science. 1993;262:1192–1196. doi: 10.1126/science.8211117. [DOI] [PubMed] [Google Scholar]
- 6.Brown T, Anand A, Ritchie L D, Clewley J P, Reid T M S. Intrauterine parvovirus infection associated with hydrops fetalis. Lancet. 1984;i:1033–1034. doi: 10.1016/s0140-6736(84)91126-7. [DOI] [PubMed] [Google Scholar]
- 7.Deiss V, Tratschin J-D, Weitz M, Siegl G. Cloning of the human parvovirus B19 genome and structural analysis of its palindromic termini. Virology. 1990;175:247–254. doi: 10.1016/0042-6822(90)90205-6. [DOI] [PubMed] [Google Scholar]
- 8.Doerig C, Beard P, Hirt B. A transcriptional promoter of the human parvovirus B19 active in vitro and in vivo. Virology. 1987;157:539–542. doi: 10.1016/0042-6822(87)90297-2. [DOI] [PubMed] [Google Scholar]
- 9.Doerig C, Hirt B, Antonietti J-P, Beard P. Nonstructural protein of parvoviruses B19 and minute virus of mice controls transcription. J Virol. 1990;15:387–396. doi: 10.1128/jvi.64.1.387-396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erdman D D, Durigon E L, Wang Q-Y, Anderson L. Genetic diversity of human parvovirus B19: sequence analysis of the VP1/VP2 gene from multiple isolates. J Gen Virol. 1996;77:2767–2774. doi: 10.1099/0022-1317-77-11-2767. [DOI] [PubMed] [Google Scholar]
- 11.Hemauer A, von Poblotzki A, Gigler A, Cassinotti P, Siegl G, Wolf H, Modrow S. Sequence variability among different parvovirus B19 isolates. J Gen Virol. 1996;77:1781–1785. doi: 10.1099/0022-1317-77-8-1781. [DOI] [PubMed] [Google Scholar]
- 12.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 13.Krady J K, Ward D C. Transcriptional activation by the parvoviral nonstructural protein NS1 is mediated via a direct interaction with Sp1. Mol Cell Biol. 1995;15:524–533. doi: 10.1128/mcb.15.1.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leruez-Ville M, Vassias I, Pallier C, Cecille A, Hazan U, Morinet F. Establishment of a cell line expressing human parvovirus B19 non-structural protein from an inducible promoter. J Gen Virol. 1997;78:215–219. doi: 10.1099/0022-1317-78-1-215. [DOI] [PubMed] [Google Scholar]
- 15.Liu J M, Fujii H, Green S W, Komatsu N, Young N S, Shimada T. Indiscriminate activity from the B19 parvovirus P6 promoter in nonpermissive cells. Virology. 1991;182:361–364. doi: 10.1016/0042-6822(91)90682-2. [DOI] [PubMed] [Google Scholar]
- 16.Liu J M, Green S, Shimada T, Young N S. A block in full length transcript maturation in cells nonpermissive for B19 parvovirus. J Gen Virol. 1992;66:4686–4692. doi: 10.1128/jvi.66.8.4686-4692.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J M, Green S W, Yu-Shu H, McDonagh K T, Young N, Shimada T. Upstream sequences within the terminal hairpin positively regulate the P6 promoter of B19 parvovirus. Virology. 1991;185:39–47. doi: 10.1016/0042-6822(91)90751-v. [DOI] [PubMed] [Google Scholar]
- 18.Mader S, White J H. A steroid-inducible promoter for the controlled overexpression of cloned genes in eukaryotic cells. Proc Natl Acad Sci USA. 1993;90:5603–5607. doi: 10.1073/pnas.90.12.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moffatt S, Tanaka N, Tada K, Nose M, Nakamura M, Muraoka O, Hirano T, Sugamura K. A cytotoxic nonstructural protein, NS1, of human parvovirus B19 induces activation of interleukin-6 gene expression. J Virol. 1996;70:8485–8491. doi: 10.1128/jvi.70.12.8485-8491.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Momoeda M, Kawase M, Jane S M, Miyamura K, Young N S, Kajigaya S. The transcriptional regulator YY1 binds to the 5′-terminal region of B19 parvovirus and regulates P6 promoter activity. J Virol. 1994;68:7159–7168. doi: 10.1128/jvi.68.11.7159-7168.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozawa K, Ayub J, Yu-Shu H, Kurtzman G, Shimada T, Young N. Novel transcription map for the B19 (human) pathogenic parvovirus. J Virol. 1987;61:2395–2406. doi: 10.1128/jvi.61.8.2395-2406.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozawa K, Ayub J, Kajigaya S, Shimada T, Young N. The gene encoding the nonstructural protein of B19 (human) parvovirus may be lethal in transfected cells. J Virol. 1988;62:2884–2889. doi: 10.1128/jvi.62.8.2884-2889.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pattison J R, Jones S E, Hodgson J, Davis L R, White J M, Stroud C E, Murtaza L M. Parvovirus infection and hypoplastic crises in sickle cell anemia. Lancet. 1981;i:664. doi: 10.1016/s0140-6736(81)91579-8. [DOI] [PubMed] [Google Scholar]
- 24.Reid D M, Brown T, Reid T M S, Rennie J A N, Eastmond C J. Human parvovirus-associated arthritis: a clinical and laboratory description. Lancet. 1985;i:422–425. doi: 10.1016/s0140-6736(85)91146-8. [DOI] [PubMed] [Google Scholar]
- 25.Rhode S L, III, Richard S. Characterization of the trans-activation-responsive element of the parvovirus H-1 P38 promoter. J Virol. 1987;61:2807–2815. doi: 10.1128/jvi.61.9.2807-2815.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schirm S, Jiricny J, Schaffner W. The SV40 enhancer can be dissected into multiple segments each with a different cell type specificity. Genes Dev. 1987;1:65–74. doi: 10.1101/gad.1.1.65. [DOI] [PubMed] [Google Scholar]
- 27.Shade O R, Blundell M C, Cotmore S F, Tattersall P, Astell C R. Nucleotide sequence and genome organization of human parvovirus B19 isolated from the serum of a child during aplastic crisis. J Virol. 1986;58:921–936. doi: 10.1128/jvi.58.3.921-936.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sol N, Morinet F, Alizon M, Hazan U. Trans-activation of the long terminal repeat of human immunodeficiency virus type 1 by the parvovirus B19 NS1 gene product. J Gen Virol. 1993;74:2011–2014. doi: 10.1099/0022-1317-74-9-2011. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki T, Hirai H, Yoshida M. Tax protein of HTLV-1 interacts with the Rel homology domain of NF-kappa B p65 and c-Rel protein bound to the NF-kappa B binding site and activates transcription. Oncogene. 1994;9:3099–3105. [PubMed] [Google Scholar]
- 30.von Poblotzki A, Gigler A, Lang B, Wolf H, Modrow S. Antibodies to parvovirus B19 NS-1 protein in infected individuals. J Gen Virol. 1995;76:519–527. doi: 10.1099/0022-1317-76-3-519. [DOI] [PubMed] [Google Scholar]
- 31.von Poblotzki A, Hemauer A, Gigler A, Puchhammer-Stöckl E, Heinz F-X, Pont J, Laczika K, Wolf H, Modrow S. Antibodies to the non-structural protein of parvovirus B19 in persistently infected patients: implications for pathogenesis. J Infect Dis. 1995;172:1356–1359. doi: 10.1093/infdis/172.5.1356. [DOI] [PubMed] [Google Scholar]
- 32.Wang X-S, Yoder M C, Zhou S Z, Srivastava A. Parvovirus B19 promoter at map unit 6 confers autonomous replication competence and erythroid specificity to adeno-associated virus 2 in primary human hematopoietic progenitor cells. Proc Natl Acad Sci USA. 1995;92:12416–12420. doi: 10.1073/pnas.92.26.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Wingender, E., R. Knueppel, P. Dietze, and H. Karas. TRANSFAC-MATRIX TABLE database 2.5.
- 33.Young N S, Mortimer P P, Moore J G, Humphries R K. Characterisation of a virus that causes transient aplastic crisis. J Clin Invest. 1984;73:224–230. doi: 10.1172/JCI111195. [DOI] [PMC free article] [PubMed] [Google Scholar]