Abstract
Stressed cells secret misfolded proteins lacking signaling sequence via an unconventional protein secretion (UcPS) pathway, but how misfolded proteins are targeted selectively in UcPS is unclear. Here, we report that misfolded UcPS clients are subject to modification by a ubiquitin-like protein named ubiquitin-fold modifier 1 (UFM1). Using α-synuclein (α-Syn) as a UcPS model, we show that mutating the UFMylation sites in α-Syn or genetic inhibition of the UFMylation system mitigates α-Syn secretion, whereas overexpression of UFBP1, a component of the endoplasmic reticulum–associated UFMylation ligase complex, augments α-Syn secretion in mammalian cells and in model organisms. UFM1 itself is cosecreted with α-Syn, and the serum UFM1 level correlates with that of α-Syn. Because UFM1 can be directly recognized by ubiquitin specific peptidase 19 (USP19), a previously established UcPS stimulator known to associate with several chaperoning activities, UFMylation might facilitate substrate engagement by USP19, allowing stringent and regulated selection of misfolded proteins for secretion and proteotoxic stress alleviation.
UFMylation facilitates the secretion of α-synuclein, which may influence its seeding and propagation in Parkinson’s disease.
INTRODUCTION
Protein secretion is critical for animal development and cell homeostasis. In eukaryotic cells, most secretory proteins carry an N-terminal signal sequence (SS), which cotranslationally targets nascent polypeptides to the lumen of the endoplasmic reticulum (ER). Within the ER, proteins undergo folding and maturation and then move through the Golgi before reaching their final destinations (1). However, it has been noticed that many proteins lacking SS can also be exported from cells via a collection of unconventional protein secretion (UcPS) mechanisms (2). Among them, the type I and type III UcPS pathways are best characterized (3). Type I UcPS translocates cargos directly across the plasma membrane (4), while for type III UcPS, cargos first enter an intermediate membrane compartment such as autophagosomes, lysosomes, or a Golgi-associated compartment termed Compartment for UcPS (CUPS) before entering the cell exterior (5–7).
The sorting signals that target UcPS cargos for secretion are not fully defined. For type I UcPS, cargos appear to contain specific protein sequences that facilitate membrane binding and translocation (8), whereas two consensus targeting sequences were identified for some type III UcPS cargos (9). By contrast, for lysosome-mediated type III UcPS, cargos first enter the lumen of lysosomes via the endosomal sorting complexes required for transport–dependent multivesicular body formation, which uses ubiquitin as a sorting signal (10).
UcPS mechanisms are now widely observed in various cell types and organisms, and their roles in diverse physiological and pathological settings are emerging (11). We previously reported a type III UcPS mechanism termed misfolding-associated protein secretion (MAPS), which exports misfolded cytosolic proteins to alleviate proteotoxic stress (12). This process is initiated when misfolded proteins are recognized by the ER-associated ubiquitin specific peptidase 19 (USP19), which transfers cargos to DNAJC5, a J-domain–containing cochaperone for heat shock cognate 71-kDa protein (HSC70). DNAJC5 is associated with a Golgi-associated membrane compartment and endolysosomes (13–15). USP19 can form stable interactions with HSC70 and heat shock protein 90 (HSP90) (16). In addition, it itself also contains a holdase activity capable of distinguishing misfolded proteins (12). We postulated that USP19 may use these chaperoning activities to select misfolded proteins for secretion. Since α-synuclein (α-Syn) and Tau are MAPS cargos (12, 13, 17), deregulation of MAPS may contribute to the seeding and propagation of these proteins in neurodegenerative diseases.
In this study, we show that MAPS substrates are modified with a single moiety of the small ubiquitin-like protein ubiquitin-fold modifier 1 (UFM1), which functions as a triaging signal to promote cargo secretion. UFM1 itself is cosecreted with misfolded proteins. In humans, the serum UFM1 level is reduced in senior patients, correlating with reduced α-Syn secretion. These observations raise the possibility of using UFM1 as a biomarker for disease diagnosis or therapeutic evaluation. Protein UFMylation has recently been established as a key protein homeostasis regulatory hub at the ER that controls translocation-associated quality control (18–20) and ERphagy (21–24). Our results suggest that similar to ubiquitin, UFM1 can also function as a sorting signal in membrane trafficking.
RESULTS
MAPS substrates are mono-UFMylated in mammalian cells
To identify MAPS regulators, we used cross-linking–based mass spectrometry (MS) to search for proteins that bind GFP1-10, a truncated green fluorescence protein (GFP) variant undergoing USP19-dependent UcPS (12). We focused on membrane proteins because previous studies suggested that GFP1-10 is translocated into a membrane compartment in an SS-independent manner before secretion (12, 14). The total membrane fraction from cells overexpressing FLAG-tagged GFP1-10 and mCitrine-USP19 were treated with a low concentration of the reversible cross-linker formaldehyde and then fractionated into a NP40 soluble and insoluble fraction. Both fractions were subject to affinity purification with FLAG beads. As a negative control, cells without GFP1-10 expression were processed in parallel. MS identified multiple peptides matching UFM1-specific ligase 1 (UFL1), UFM1-binding protein 1 (UFBP1), and cyclin-dependent kinase 5 (CDK5) regulatory subunit–associated protein 3 (CDK5RAP3) in both fractions from GFP1-10– and USP19-expressing cells, whereas no or fewer peptides were found in the corresponding control samples (fig. S1A). Since UFL1, UFBP1, and CDK5RAP3 form a protein UFMylation ligase complex (25, 26), the coidentification of these proteins strongly indicated them as a GFP1-10 interactor. Additional immunoprecipitation and immunoblotting confirmed the interaction of GFP1-10 with UFL1 but not an abundant ER membrane protein calnexin (fig. S1B). When USP19 was present, we detected reproducibly an increase in the relative amount of GFP1-10–bound UFL1 (fig. S1B), consistent with the observation that USP19 recruits GFP1-10 to the ER surface during MAPS (12).
Our findings raised the possibility that MAPS cargos might be UFMylated. To test this possibility, we expressed FLAG–GFP1-10 together with hemagglutinin (HA)–tagged UFM1 in human embryonic kidney (HEK) 293T cells. UFM1 overexpression facilitates the detection of UFMylated products by overcoming the robust deUFMylating activities in cells (22). In accordance with our hypothesis, FLAG immunoprecipitation under denaturing conditions revealed a protein whose molecular weight and immunoreactivity are consistent with a modified GFP1-10 species carrying a single UFM1 moiety (Fig. 1A, lane 2).
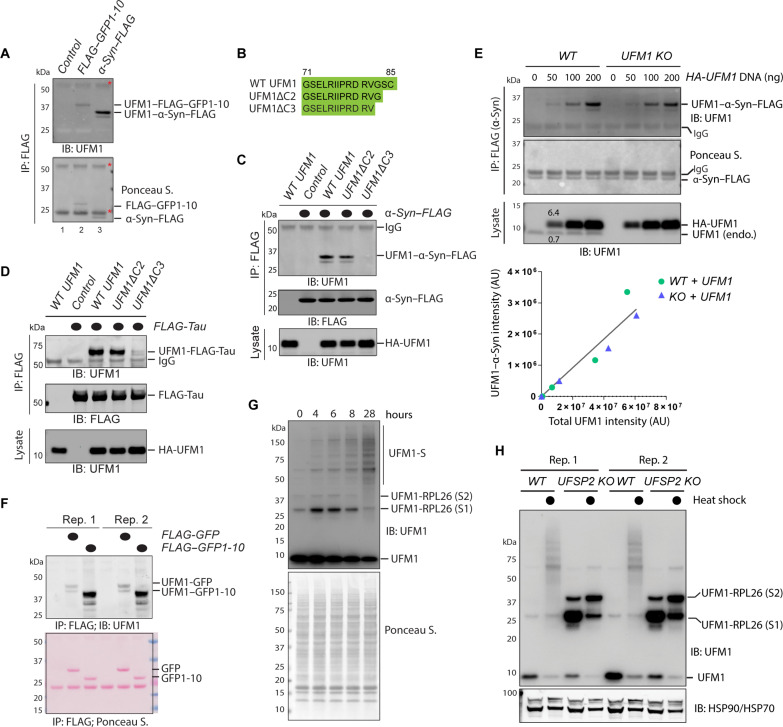
Fig. 1. Misfolded MAPS substrates are mono-UFMylated.
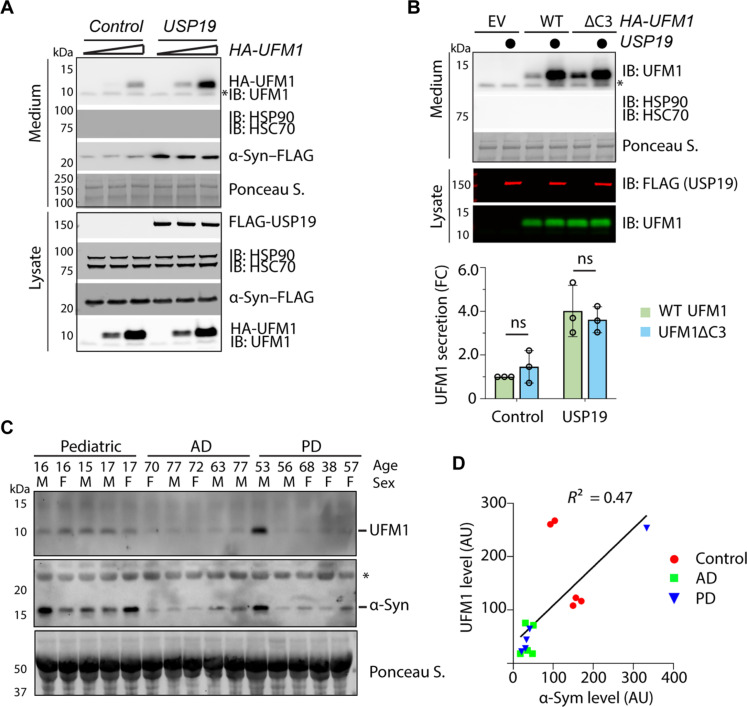
(A) UFMylation of GFP1-10 and α-Syn in HEK293T cells. Cells transfected with HA-tagged UFM1 together with either an empty vector control or FLAG–GFP1-10 or α-Syn–FLAG were subject to immunoprecipitation (IP) by FLAG beads under denaturing conditions. Precipitated proteins were analyzed by immunoblotting (IB). Asterisks indicate immunoglobulin G (IgG). (B) The amino acid sequence of the C terminus of WT UFM1 and the mutants used in the study. (C) UFMylation of α-Syn requires G83 of UFM1. As in (A), except that cells were transfected with the indicated plasmids. Where indicated, 15% of the total lysates were analyzed together with the immunoprecipitated samples. (D) UFMylation of Tau in HEK293T cells. As in (C), except that FLAG-tagged Tau was used. (E) The level of UFMylated α-Syn correlates with UFM1 concentrations in cells. α-Syn–FLAG immunoprecipitated from HEK293T cells transfected with the indicated plasmids were analyzed by immunoblotting. The graph shows a linear correlation between the level of UFMylated α-Syn and UFM1. AU, arbitrary units. (F) UFMylation of GFP1-10 is more efficient than full-length GFP. Cells transfected with HA-UFM1 together with the indicated GFP variants were analyzed as in (A). Shown is a blot with two biological repeats (Rep.). (G) Heat shock induces protein UFMylation. HEK293T cells treated at 43°C for the indicated time points were lysed in sample buffer and analyzed by immunoblotting with UFM1 antibodies. UFM1-S, heat-induced UFMylated substrates. (H) Protein UFMylation under the heat shock condition is not regulated by UFSP2. Whole-cell extracts from untreated or heat-treated (16 hours, indicated by filled circles) WT or UFSP2 CRISPR KO cells were analyzed by immunoblotting. Note that UFSP2 KO increases ribosome UFMylation but inhibits heat-induced UFMylation.
To see whether other MAPS substrates are also UFMylated, we tested α-Syn and Tau, two misfolding-prone proteins known to be secreted at least in part by a type III UcPS mechanism (12, 13, 15, 17). To demonstrate the specificity of the assay, we expressed these substrates together with either wild-type (WT) UFM1 or UFM1 mutants lacking two (ΔC2) or three (ΔC3) amino acids from the C terminus (Fig. 1B). WT UFM1 and UFM1ΔC2 but not UFM1ΔC3 are expected to be fully active because they contain G83 required for UFMylation (27). We found that both α-Syn and Tau were readily mono-UFMylated in cells expressing WT or UFM1ΔC2, but not in UFM1ΔC3-expressing cells (Fig. 1, C and D).
To exclude the possibility of UFM1 overexpression artifact, we reconstituted UFM1 expression in WT and UFM1 knockout (KO) cells by titrating the amount of transfected DNA. We detected α-Syn UFMylation with HA-UFM1 expressed at just a few folds higher than endogenous UFM1 (Fig. 1E). A linear correlation between UFMylated α-Syn and free UFM1 was observed over a large signal dynamic range, suggesting that the lack of α-Syn modification by endogenous UFM1 was likely caused by a detection sensitivity issue (Fig. 1, C and D). After optimizing the coimmunoprecipitation and immunoblotting conditions, we could detect a small amount of UFM1-modified α-Syn without exogenous UFM1 (fig. S2A). Thus, a fraction of MAPS substrates undergoes mono-UFMylation in cells.
We next compared the UFMylation efficiency of GFP1-10 to that of GFP because the latter is a folded counterpart of GFP1-10 and is secreted at a lower level compared to GFP1-10 (12). Immunoprecipitation consistently detected more UFMylated GFP1-10 than UFMylated GFP (Fig. 1F), raising the possibility that misfolded proteins might be a preferred UFMylation target. To further test this model, we examined the global UFMylation profile in heat-treated WT and UFM1-specific protease 2 (UFSP2) KO cells by immunoblotting. In nonstressed cells, the UFMylation system is highly specific, primarily modifying ribosome protein L26 (RPL26) in translation-stalled ribosomes at the ER (28, 29). Heat treatment initially increased UFMylated RPL26 in WT cells, probably due to heat-induced ribosome stalling. However, prolonged treatment reduced both mono- and di-UFMylated RPL26; meanwhile, a time-dependent accumulation of high–molecular weight UFMylated species was observed (Fig. 1G and fig. S2B). In UFSP2 KO cells, UFMylated RPL26 was substantially up-regulated under both normal and heat-stressed conditions. By contrast, heat-induced UFMylation was not observed (Fig. 1H). These findings suggest that heat-induced UFMylation and RPL26 UFMylation are competing pathways regulated by distinct deUFMylases. Since immunoblotting detected no increase in the UFMylating enzymes [e.g., ubiquitin-fold modifier-conjugating enzyme 1 (UFC1) and UFL1] after heat treatment (fig. S2B), the accumulation of UFMylated proteins after prolonged heat treatment is probably caused by heat-induced protein denaturation. Together, our results suggest that mono-UFMylation occurs on a fraction of misfolded proteins in mammalian cells, a notion consistent with the curated Biological General Repository for Interaction Datasets, which reveals several heat shock proteins as potential UFL1 interactors (30).
UFMylation facilitates unconventional secretion of α-Syn
To test the role of protein mono-UFMylation in UcPS, we first used site-directed mutagenesis to determine the UFMylation site(s) in α-Syn. α-Syn is a small protein containing 15 lysine residues. Most lysine residues are in the N-terminal amphipathic repeat region, but two lysine residues are found in the middle non–amyloid-β component (NAC) domain and two in the C-terminal acidic region (Fig. 2A). As anticipated, when all lysine residues were mutated to arginine, α-Syn UFMylation was abolished (fig. S3A), suggesting that UFMylation requires lysine residues. However, when we substituted individual lysine residues to arginine, none of the K-to-R mutations affected α-Syn UFMylation (fig. S3B), suggesting that similar to ubiquitination, multiple lysine residues can serve as conjugation sites (31, 32). When we analyzed two additional α-Syn mutants: one with the nine lysine residues on the N-terminal half mutated to arginine (N_KR) and the other with the six lysine residues on the C-terminal half mutated (C_KR) (Fig. 2A), we noticed that compared to WT α-Syn, UFMylation of N_KR and C_KR was reduced by ~75 and ~60%, respectively (Fig. 2B). Although UFMylation can take place at multiple sites, no multi-mono-UFMylated α-Syn was detected. How cells control the number of UFM1 moieties attached to a substrate remains to be elucidated.
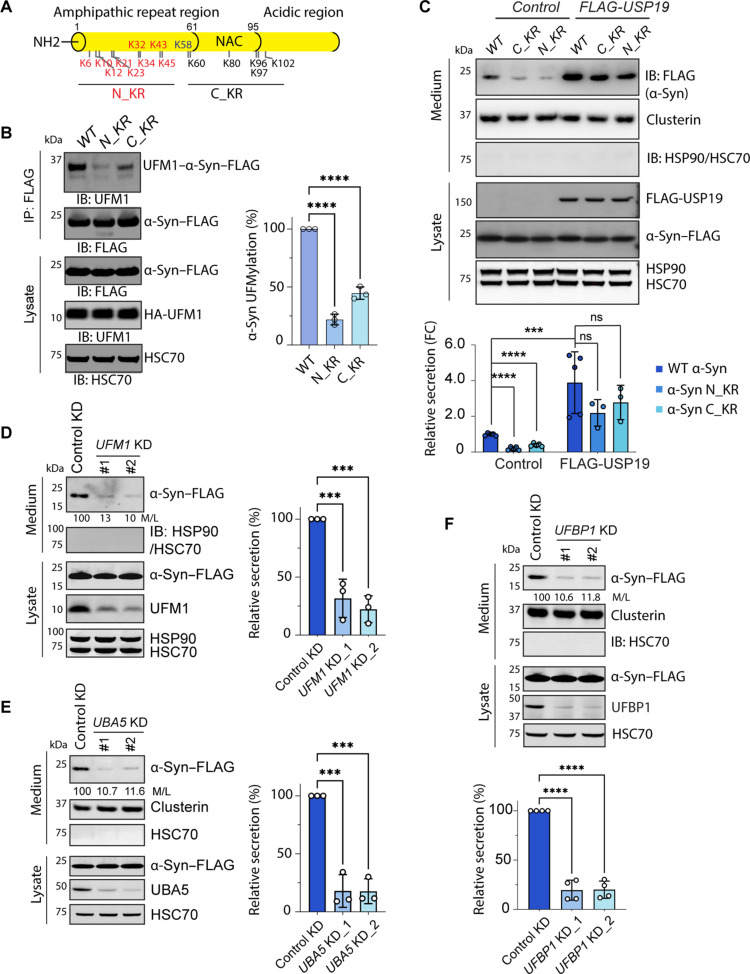
Fig. 2. UFMylation of α-Syn promotes its secretion.
(A) A schematic diagram showing the lysine positions in α-Syn and the mutants tested. (B) Mono-UFMylation of α-Syn can occur on multiple lysine residues. HEK293T cells were transfected with HA-UFM1 together with the indicated α-Syn lysine mutants and then subject to immunoprecipitation and immunoblotting. The graph shows the quantification of three biological repeats. Error bars indicate means ± SD. ****P < 0.0001 by one-way analysis of variance (ANOVA) with Dunnett’s multiple comparison test. (C) Reduced secretion of α-Syn lysine mutants. Immunoblotting analysis of condition medium and lysates from HEK293T cells transfected with the indicated plasmids. The graph shows the quantification of the relative ratio of α-Syn in medium normalized to that in the corresponding lysate (M/L). The dots indicate the number of biological repeats. FC, fold change. Error bars indicate means ± SD. ****P < 0.0001; ***P < 0.001; ns, not significant by one-way ANOVA. (D) Knockdown (KD) of UFM1 reduces α-Syn secretion. Conditioned medium and lysates from cells transfected with α-Syn–FLAG together with the indicated siRNAs were analyzed by immunoblotting. The numbers and the graph show the relative ratio of medium α-Syn versus that in lysates (M/L). Error bars indicate means ± SD. ***P < 0.001 by one-way ANOVA, n = 3 biological repeats. (E) Knockdown of UBA5 reduces α-Syn secretion. Same as (D), except that two UBA5-specific siRNAs were used. Error bars indicate means ± SD. ***P < 0.001 by one-way ANOVA, n = 3 biological repeats. (F) Knockdown of UFBP1 reduces α-Syn secretion. Same as (D), except that two UFBP1-targeting siRNAs were used and that n = 4 biological repeats. Error bars indicate means ± SD. ****P < 0.001 by one-way ANOVA.
We next compared the secretion efficiency of α-Syn N_KR and C_KR mutants to that of WT α-Syn. To this end, we transfected HEK293T cells with WT α-Syn or α-Syn mutants either with or without USP19 coexpression. We determined the secretion level by normalizing α-Syn in conditioned medium to that in cell lysates. We also immunoblotted HSP90 and HSC70, two abundant cytosolic chaperones whose absence from the medium indicated plasma membrane intactness. We found that both the N_KR and C_KR α-Syn mutants were secreted at much reduced levels compared to WT α-Syn (Fig. 2C). Overall, the level of α-Syn secretion mirrors its UFMylation efficiency. When USP19 was coexpressed to enhance α-Syn secretion, the secretion defect of these lysine mutants became less notable (Fig. 2C) (see Discussion).
Because lysine can also serve as ubiquitin acceptor, lysine mutations may affect α-Syn secretion by blocking ubiquitination. To rule out this possibility, we tested the effect of a potent ubiquitin activating enzyme (E1) inhibitor (TAK-243) on α-Syn secretion (33). As expected, cells treated with TAK-243 had notably reduced ubiquitin chains. However, TAK-243 treatment stimulated α-Syn secretion, similarly as USP19 overexpression (fig. S3C). These findings suggest that ubiquitination is dispensable for α-Syn secretion. Therefore, the secretion defect of the α-Syn lysine mutants cannot be attributed to lack of ubiquitination.
To further explore the role of UFMylation in α-Syn secretion, we used small interfering RNAs (siRNAs) to specifically knock down UFMylation pathway genes. In addition to UFM1, UFBP1, and UFL1, we also knocked down ubiquitin-like modifier activating enzyme 5 (UBA5), which encodes the sole UFM1-activating enzyme. To control off-target effects, we chose two siRNAs for each gene. Immunoblotting analyses showed that depletion of any of these UFMylation pathway genes reduced α-Syn secretion substantially (Fig. 2, D to F, and fig. S3D). This phenotype could not be explained by the role of UFM1 in ER protein biogenesis because α-Syn secretion from CRISPR-engineered HEK293T cells lacking the C terminus of RPL26, therefore, ribosome UFMylation (28), was similar to that of WT cells (fig. S3E). Collectively, these results suggest that UFMylation positively regulates UcPS.
Overexpression of UFBP1 stimulates α-Syn secretion
Next, we tested whether overexpression of the UFM1 ligase components, either individually or in combination, affects α-Syn secretion. Immunoblotting analyses showed that overexpression of UFBP1 alone was sufficient to induce the secretion of WT α-Syn (Fig. 3A) or a Parkinson’s disease–associated α-Syn mutant (fig. S4A). As expected, the UcPS-stimulating activity of UFBP1 required its N-terminal membrane localization sequence (MLS) but was independent of K267, a known UFMylation site in UFBP1 (Fig. 3B). WT UFBP1 but not the MLS-deleted UFBP1 mutant also promoted Tau secretion (Fig. 3C). By contrast, expression of UFL1 or CDK53RAP1 did not affect either the basal or UFBP1-stimulated secretion (Fig. 3A). Thus, UFBP1’s UcPS-stimulating activity does not require the whole UFL1-UFBP1-CDK53RAP1 complex.
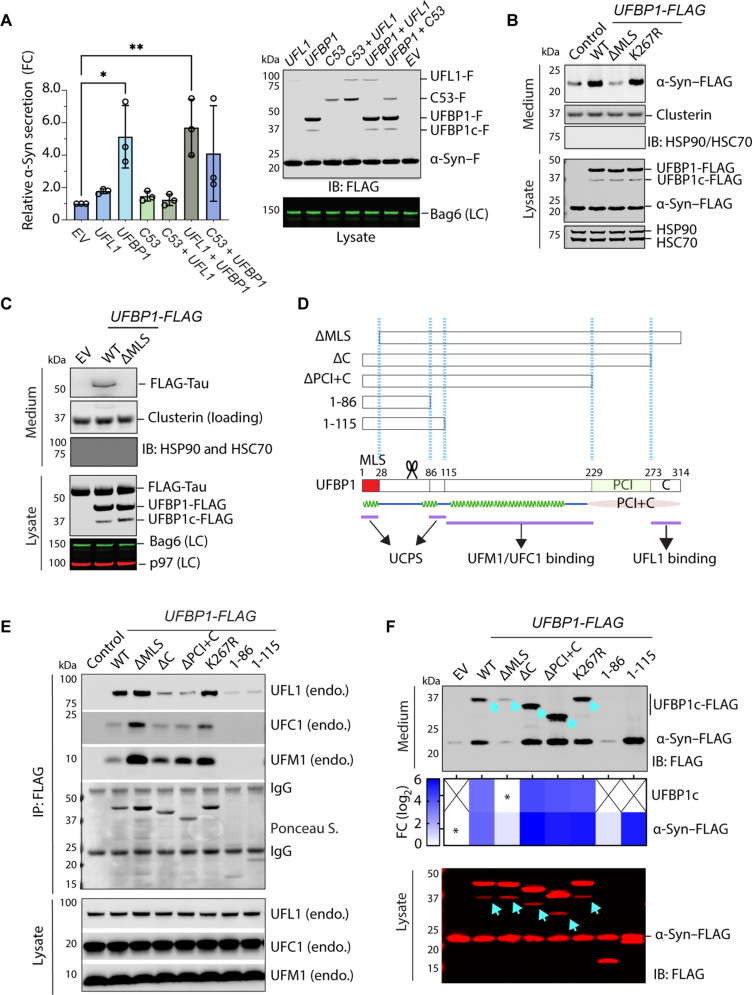
Fig. 3. Overexpression of UFBP1 induces α-Syn secretion in mammalian cells.
(A) Overexpression of UFBP1 stimulates α-Syn secretion. Conditioned medium and lysates from HEK293T cells transfected with α-Syn–FLAG together with the indicated plasmids were analyzed by immunoblotting. F, FLAG tag; C53, CDK53RAP1; EV, empty vector control. The graph shows secreted α-Syn normalized by α-Syn in cell lysates (right). Error bars indicate means ± SD. *P < 0.05; **P < 0.01 by unpaired two-tailed t test. n = 3 biological repeats. (B) As in (A), except that α-Syn–FLAG was cotransfected with the indicated plasmids. (C) As in (A), except that FLAG-tagged Tau was expressed together with the indicated UFBP1 variants. LC, loading control. (D) A schematic diagram shows the UFBP1 variants tested in (E) and (F). The bottom graph shows the predicted secondary and domain structure of UFBP1. Domains involved in known protein-protein interactions are marked. (E) The interactions of UFBP1 variants with the endogenous UFM1, UFC1, and UFL1 were analyzed by coimmunoprecipitation, followed by immunoblotting. (F) UFBP1 is cosecreted with α-Syn. Shown is a representative immunoblotting analysis of conditioned medium and lysates from cells transfected with α-Syn–FLAG together with the indicated UFBP1 variants. Note that a cleaved UFBP1 fragment (UFBP1c-FLAG) was cosecreted with α-Syn with the exception for UFBP1 1-86 and 1-115. The heatmap shows the relative secretion of UFBP1c and α-Syn. Asterisks indicate the samples used for fold change normalization. n = 2 biological repeats. X = not detected. Arrows denote UFBP1c.
To further define the cis-element involved in UFBP1-stimulated UcPS, we generated a series of UFBP1 truncation mutants (Fig. 3D). We first determined the impact of these deletions on UFBP1’s interactions with various UFM1 pathway components by coimmunoprecipitation, which confirmed the middle helical domain as the major UFM1- and UFC1-binding region (Fig. 3E). As reported previously, the C-terminal 40 amino acids harbor a UFL1 binding site (34), but its deletion did not completely abolish UFL1 binding, consistent with another study showing an interaction between UFBP1 and UFL1 independent of this sequence (21). Cell-based UFMylation assay showed that overexpression of UFBP1 or the UFBP1ΔC mutant did not reduce α-Syn UFMylation (fig. S4B), suggesting that these proteins do not interfere with the interaction between α-Syn and the endogenous UFM1 ligase. α-Syn secretion experiments showed that deleting up to two-third of UFBP1 from the C terminus did not abolish its UcPS-stimulating activity. To the contrary, the mutant proteins (ΔC, ΔPCI+C and 1-115) were more active (Fig. 3F). Further deleting residues 86 to 115 abolished the UcPS-stimulating activity. Since neither UFL1 nor UFC1 binding is required for UFBP1’s UcPS-stimulating activity, UFBP1 may also have a UFMylation-independent function in UcPS.
Overexpressed UFBP1 was cleaved by an unknown protease, generating a truncated 37-kDa product (UFBP1c) that was also secreted (Fig. 3, B, C, and F). The secretion of cleaved UFBP1 was not caused by overexpression because we observed a similar UFBP1c fragment in cell lysate and condition medium from HEK293T cells expressing endogenously tagged UFBP1-GFP (fig. S4C). The size and immunoreactivity of this truncated UFBP1c species suggested that the cleavage site is close to the N terminus (Fig. 3D). The secretion of UFBP1c was not affected by brefeldin A, a Golgi-disrupting drug (fig. S4D), suggesting that it itself is a UcPS substrate. Notably, although this fragment was present at a level substantially lower than α-Syn in the cell, conditioned medium had similar amount of UFBP1c and α-Syn (Fig. 3F), suggesting that UFBP1c is secreted more efficiently than α-Syn. We did not observe any cleavage for the 1-86 and 1-115 UFBP1 mutants, neither did we detect any secretion of these mutants (Fig. 3F). By contrast, the UFBP1 ΔMLS mutant was cleaved as efficiently as WT UFBP1, but UFBP1c cleaved from the ΔMLS mutant was not secreted efficiently. These results suggest that UFBP1 cleavage is necessary but not sufficient for UFBP1c secretion.
UFBP1 and UFM1 regulate α-Syn secretion in model organisms
We next tested whether UFBP1 could regulate α-Syn secretion in vivo. To this end, we took advantage of the fact that the UFMylation system is conserved in Caenorhabditis elegans (35) and that proteins secreted from C. elegans body wall muscle cells are usually engulfed by and accumulated in the macrophage-like coelomocytes (Fig. 4A). We analyzed a WT strain expressing yellow fluorescence protein (YFP)–tagged α-Syn in body wall muscle and detected coelomocyte-localized YFP signal in ~24% of the worms (Fig. 4, B and C). In worms coexpressing C. elegans UFBP1 (encoded by ZK1236.7) and YFP–α-Syn in body wall muscle cells, more than 70% of the worms had YFP-positive coelomocytes. By contrast, overexpressing the UFBP1 ΔMLS mutant did not increase the number of worms with YFP-positive coelomocytes (Fig. 4C). As expected, in worms overexpressing WT UFBP1-mCherry, we also detected UFBP1-mCherry in coelomocytes, colocalizing with α-Syn (Fig. 4C). Thus, UFBP1 overexpression also stimulates α-Syn secretion in C. elegans.
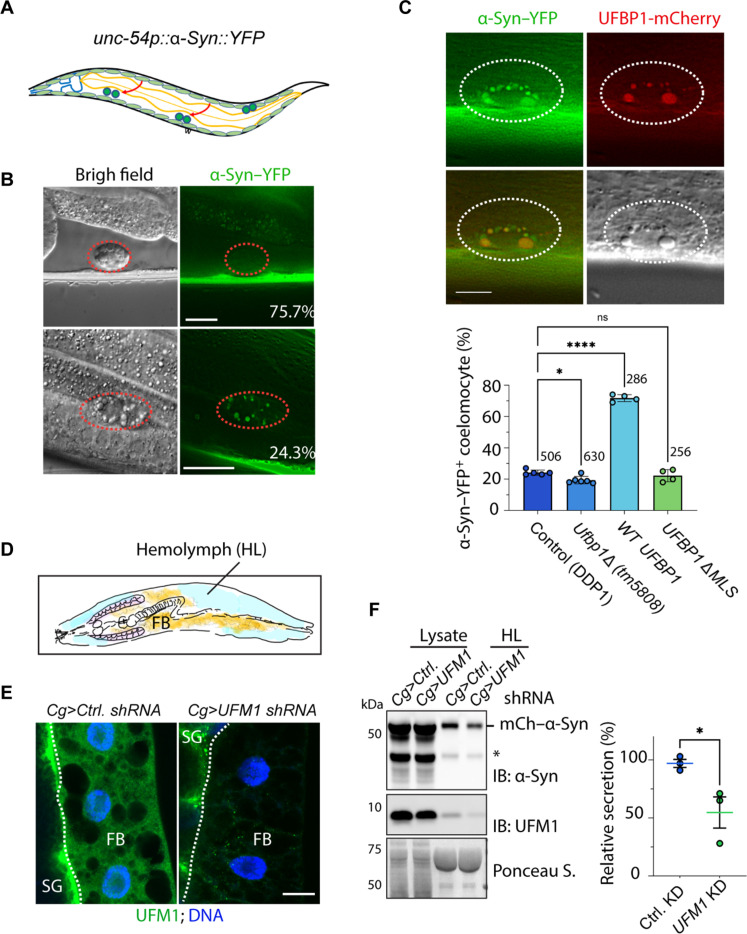
Fig. 4. The UFMylation system facilitates α-Syn secretion in model organisms.
(A) A schematic diagram of the worm-based secretion assay. (B) α-Syn is secreted from body wall muscle cells and internalized by coelomocytes in C. elegans. Worms bearing a Venus (YFP)–tagged α-Syn were imaged by a fluorescence microscope. The images represent WT worms with two different phenotypes. Scale bars, 10 μm. (C) UFBP1 promotes α-Syn secretion in C. elegans. Top: Representative images of coelomocyte-accumulated α-Syn–YFP and UFBP1-mCherry. The graph shows the quantification of worms of the indicated genotypes with α-Syn–YFP–positive (+) coelomocyte. Dots indicate the number of repeats, whereas the numbers indicate total worms counted. Error bars indicate means ± SD. *P < 0.05; ****P < 0.0001, by one-way ANOVA. Scale bar, 5 μm. (D) A schematic illustration of the fly UcPS model. FB, fat body; HL, hemolymph. (E) Immunostaining confirms fat body–specific knockdown of UFM1. Fat bodies isolated from third instar larvae of the indicated genotypes were stained with a UFM1 antibody (green) and 4′,6-diamidino-2-phenylindole (blue). Ctrl. shRNA, control shRNA. Note that the UFM1 signal is only specifically reduced in fat body. SG, salivary gland. Scale bar, 20 μm. (F) Fat body–specific knockdown of UFM1 reduces α-Syn secretion. Hemolymph and hemolymph-depleted larva lysates were analyzed by immunoblotting. The asterisk indicates a cleaved α-Syn species. The graph shows the quantification of three biological repeats. Error bars indicate means ± SD. *P < 0.05 by unpaired two-tailed t test. n = 3 biological repeats.
We next analyzed α-Syn secretion in a homozygous mutant strain bearing a 352–base pair deletion in the C. elegans ufbp1 locus (ufbp1Δ and tm5808). Since the deletion takes out part of the ufbp1 promoter and the MLS-coding sequence, homozygous mutant worms are expected to have no functional UFBP1. We detected coelomocyte-localized YFP–α-Syn in ~19% mutant adult worms. Although the reduction in worms with coelomocyte-localized YFP–α-Syn was small, it was reproducible and statistically significant (Fig. 4C), suggesting that UFBP1 has a function in UcPS in worm muscle cells but is not essential.
Because the worm-based assay does not report the precise amount of secreted α-Syn, we developed a quantitative α-Syn secretion assay to further explore the in vivo function of the UFMylation system in UcPS. To this end, we used the upstream activating sequence (UAS)–GAL4 system to express mCherry-tagged α-Syn in fat body, a major secretory tissue in fruit flies (see Materials and Methods). We then analyzed the amount of α-Syn in third instar larval hemolymph and that in larvae after hemolymph removal by immunoblotting (Fig. 4D and fig. S5A), which readily detected mCherry–α-Syn in hemolymph of flies expressing a control small hairpin RNA (shRNA) in fat body. Noticeably, like in Ufbp1-deficient worms, fat body–specific depletion of UFM1 reduced but did not abolish α-Syn secretion (Fig. 4, E and F), further supporting the existence of both UFM1-dependent and UFM1-independent mechanisms for α-Syn secretion in animals (see Discussion).
UFM1 is cosecreted with α-Syn
While UcPS cargos are UFMylated in cells, the secreted proteins were detected in non-UFMylated state, suggesting that UFM1 conjugates might be removed by a protease, while substrates are being exported. If this cleavage event occurs late in this process, then free UFM1 might be cosecreted with MAPS cargos. While analyzing α-Syn secretion in flies, we detected a fraction of unconjugated UFM1 in larval hemolymph (Fig. 4F). When we knocked down UFM1 in fat body, the UFM1 level in hemolymph was modestly reduced (fig. S5A), suggesting that tissues other than fat body also contribute to UFM1 secretion.
To test whether UFM1 is secreted by mammalian cells, we expressed HA-tagged UFM1 in HEK293T cells either alone or together with USP19. We then analyzed the cell lysates and conditioned medium by immunoblotting, which detected a fraction of HA-UFM1 in the medium (Fig. 5A). As expected, HA-UFM1 secretion is enhanced by WT USP19 but not by a USP19 mutant lacking the C-terminal transmembrane domain (ΔTM) (Fig. 5A and fig. S5B). A USP19 catalytically inactive mutant only partially increased UFM1 secretion (fig. S5B). Notably, the secretion of UFM1 is not dependent on G83, suggesting that UFM1 can be secreted without picky backing modified substrates (Fig. 5B). This observation suggests that UFM1 might serve as a triaging signal to promote UcPS.
Fig. 5. Endogenous UFM1 is cosecreted with α-Syn.
(A) USP19 promotes UFM1 secretion. Conditioned medium and lysates from HEK293T cells transfected with different amounts of HA-UFM1 together with an empty vector control or FLAG-USP19 were analyzed by immunoblotting. (B) The secretion of UFM1 does not require G83. The indicated UFM1 variants or an empty vector control were transfected together with (indicated by filled circles) or without USP19. The graph shows the quantification of three biological repeats. Error bars indicate means ± SD. (C) Endogenous UFM1 and α-Syn are detected in human serum. Serum from patients with Alzheimer’s disease (AD) and Parkinson’s disease (PD) were analyzed together with a group of pediatric serum. (D) Quantification of the UFM1 and α-Syn levels in human serum samples.
Several lines of evidence indicate that endogenous UFM1 secretion also occurs in mammals under physiological conditions. First, immunoblotting using UFM1 antibodies detected untagged UFM1 in conditioned medium, which originated from fetal bovine serum (FBS) (Fig. 5, A and B) and therefore not subject to regulation by overexpressed USP19. The amount of UFM1 in FBS exceeded cell-secreted UFM1 substantially (fig. S5C), and, thus, masked UFM1 secreted endogenously from these cells. Immunoblotting a panel of human serum samples also detected UFM1 (Fig. 5C). The serum UFM1 level partially correlated with that of α-Syn (Fig. 5C), further supporting the notion of cosecretion. We also noticed that the level of UFM1 was higher in pediatric samples than geriatric samples, with the highest level detected in FBS (Fig. 5C and fig. S5, C and D), suggesting that the secretion of UFM1 and MAPS cargoes may be regulated by aging. Whether the secreted UFM1 can have a physiological function in animal development or aging is unclear.
UFM1 interacts with and regulates USP19
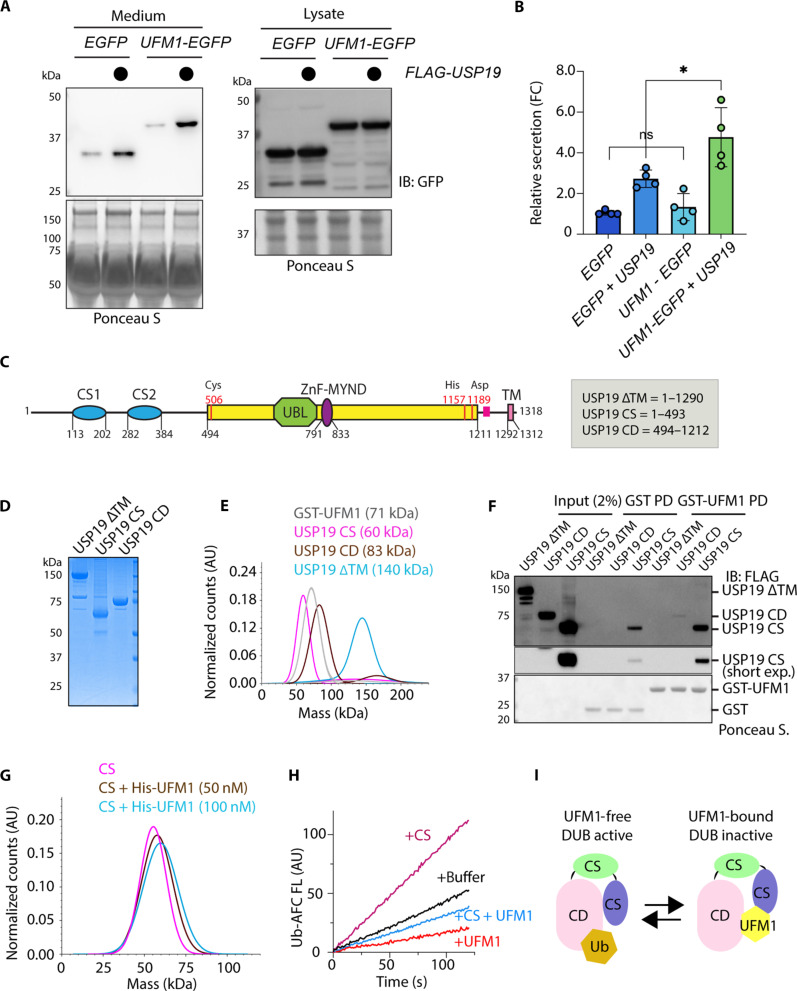
To demonstrate a direct involvement of UFM1 in triaging misfolded proteins via UcPS, we asked whether appending UFM1 to enhanced GFP (EGFP) as a chimeric protein would enhance the secretion of EGFP, which, otherwise, is secreted inefficiently compared to misfolded proteins (12). UFM1-EGFP secretion was detected at a similar level as that of EGFP in WT cells. However, when USP19 was coexpressed, the secretion of UFM1-EGFP was more efficient than EGFP (Fig. 6, A and B). The result suggests USP19 as a rating limiting factor in UFMylation-dependent UcPS: Only in USP19 high cells, increased UFMylation can lead to more secretion, demonstrating a functional interplay between USP19 and UFM1 in UcPS.
Fig. 6. UFM1 binds and regulates USP19.
(A) USP19 promotes the secretion of UFM1-EGFP. Conditioned medium and lysates from cells transfected as indicated were analyzed by immunoblotting. (B) Quantification of the experiments shown in (A). Error bars indicate means ± SD. *P < 0.05 by unpaired two-tailed t test. n = 4 biological repeats. (C) A diagram showing the USP19 domain structure and the truncation variants tested. (D) A Coomassie blue–stained gel showing the purified proteins. (E) Mass photometry confirms the molecular mass of the purified proteins. (F) A GST pull-down (PD) assay confirms the interaction of USP19 CS with UFM1. (G) Mass photometry demonstrates the interaction of His-UFM1 with USP19 CS (50 nM). (H) USP19 CS stimulates the deubiquitinating activity of USP19 CD, which is antagonized by UFM1. The deubiquitinating activity of USP19 CD (20 nM) was measured together with either a buffer control, or USP19 CS (200 nM), or UFM1 (5 μM) or the combination of USP19 CS (200 nM) and UFM1 (5 μM). (I) A toggle switch model showing USP19 in two functional states, a UFM1-free DUB active, and a UFM1-bound DUB-inactive form.
We next explored the possibility that USP19 might directly bind UFM1. To this end, we expressed and purified a USP19 variant lacking the ΔTM, USP19 CS (1 to 493), and the USP19 catalytic domain (CD; 494 to 1212) to >90% homogeneity (Fig. 6, C and D). Single-molecule mass photometry showed that these proteins all form monomer with predicted molecular mass (Fig. 6E). Pull-down using glutathione S-transferase (GST)–UFM1 fusion protein as a bait and GST as a negative control showed that USP19 CS readily coprecipitated with UFM1, whereas USP19 CD only weakly bound to UFM1 (Fig. 6F). The interaction of USP19 CS with UFM1 was further confirmed by mass photometry (Fig. 6G). Although these methods failed to detect an interaction between USP19 ΔTM and UFM1 (Fig. 6F and fig. S6A), there appears to be a transient interaction between these molecules because at a high concentration, recombinant UFM1 could inhibit the deubiquitinating activity of USP19 ΔTM (fig. S6B). Furthermore, cross-linking–based immunoprecipitation using HEK293T cells expressing FLAG-tagged USP19 revealed an interaction between full-length USP19 and endogenous UFM1 (fig. S6C). This interaction, albeit weak, was specific because we did not detect the abundant ER chaperone calnexin by USP19 pull-down (fig. S6D, E). In addition, when cells were serum-starved to inhibit MAPS, more UFM1 could be cross-linked to USP19 (fig. S6E), suggesting that MAPS inhibition traps more substrates in proximity to USP19.
Our results raised the possibility that USP19 might function as a “toggle switch,” regulated by an intramolecular interaction between the CS and CD. The notion is supported by an α-fold predicted USP19 structure, which shows that the CS domains are in proximity to the CD (fig. S6F). Furthermore, when we incubated USP19 CS with purified USP19 CD, USP19 CS could dose-dependently stimulate the deubiquitinating activity of USP19 CD, which was antagonized by recombinant UFM1 (Fig. 6H and fig. S6G).
DISCUSSION
In this study, we present several lines of evidence that establish UFM1 as an MAPS regulator, including mono-UFMylation of misfolded proteins, the requirement of lysine residues in α-Syn for efficient secretion, the modulation of α-Syn secretion by knockdown of UFM1 pathway genes or overexpression of UFBP1, and, last, the secretion of endogenous UFM1 and a cleaved UFBP1 fragment by UcPS. Given the observed functional interplays between USP19 and UFM1 and the established chaperoning activities associated with USP19 (12, 16), we propose a dual recognition mechanism that ensures stringent selection of abnormal substrates for UcPS. In this model, USP19 uses an associated chaperone activity to bind a misfolded protein bearing exposed hydrophobic motifs. Meanwhile, USP19 also queries the UFMylation status of the bound substrate, which may serve as a “checkpoint” to license the translocation of cargos into the Golgi-associated CUPS for secretion. Since the UFM1 ligase complex and USP19 are both localized to the ER surface and because ER makes extensive membrane contacts with other organelles including the Golgi, USP19 may cooperate with the UFMylation machinery, coupling substrate UFMylation to membrane translocation at the CUPS. This model would explain the multiple roles of UFBP1 in MAPS: In addition to serving as a UFL1 adaptor, it might also function as a key that links UFMylation to a downstream factor that channels substrates to the CUPS, a mechanism akin to the recently proposed ER retrieval mechanism in mitochondrial protein targeting (36).
Conceivably, the proposed substrate recognition model would work the best when USP19 concentrations are low because under these conditions, productive interaction only occurs when USP19 contacts both substrate and UFM1. The Human Protein Atlas database suggests that USP19 expression is generally low in various tissues except for the skeletal muscle (37). USP19 high cells appear to depend less on endogenous UFMylation for MAPS, likely because its chaperone activity would be dominant in substrate selection under this condition.
Dual substrate recognition may also render more regulation on USP19 in MAPS. We previously identified an autoinhibitory domain in USP19, consisting of a ubiquitin-like (UBL) domain and the zinc finger myeloid, Nervy, and DEAF-1 (Zn-MYDN) domain that breaks the USP19 CD into two parts (13). The α-fold predicted USP19 structure and our biochemical results suggest a transient intramolecular interaction between the CD and the USP19 CS domains, which masks the UFM1 binding site in the CS domain–containing region. Conversely, UFM1 binding would disrupt this interaction to convert USP19 into a different functional state (Fig. 6I).
In model organisms with the UFMylation system severely ablated, we could still detect α-Syn secretion from muscle cells in worms or from fat body in fly larvae. These observations suggest the existence of a UFM1-independent mechanism(s) that governs α-Syn secretion in vivo. Secretion of UcPS cargos by parallel pathways has been reported for several cargos including Tau and interleukin-1β (9, 38). Another limitation of the study is that the low level of mono-UFMylated model substrates in cells without exogenous UFM1 often escape detection. This is likely due to the presence of an unknown deUFMylating activity, which requires future validation.
UFM1 was discovered about two decades ago (39). Mice defective in UFMylation are embryonic lethal (40, 41), whereas humans bearing a defective allele of UFMylation genes often suffer from severe neurological disorders (42). However, the function of protein UFMylation has been elusive until recent studies that linked UFMylation to translocation-associated protein quality control at the ER (19, 20, 28), ERphagy (21–24), and DNA repair (43, 44). Our study has revealed a protein triaging function for UFMylation, and defects in this function may contribute to human diseases genetically associated with the UFMylation system. Several recent studies have linked UFM1 and USP19 to the intercellular transfer of neurodegenerative disease–associated neurotoxic proteins. Specifically, in UFM1-deficient induced pluripotent stem cell–derived neurons and in USP19 KO mice, the seeding and propagation of misfolded Tau and α-Syn are inhibited (45–47). Although the underlying mechanisms are unclear, these studies and our work consolidate an emerging theme that the dynamics of protein aggregation may be influenced by a functional interplay between USP19 and UFM1 in either MAPS or a related UcPS process.
The secretion of misfolded proteins by MAPS is enhanced under proteasome stress conditions (12, 48), which is essential for cell adaptation to declined proteasome function (12). As recent studies revealed a reverse correlation between aging and the fitness of the proteasome system (49), cells, particularly neurons in aged individuals, may depend more on UFM1-mediated MAPS for protein homeostasis maintenance. Given that the level of extracellular UFM1 mirrors the levels of MAPS, we envision the possibility of using serum UFM1 as a measure to gauge the proteostasis fitness of a multicellular organism during aging. In this view, since the serum UFM1 and α-Syn levels are similarly reduced in geriatric serum samples, it is possible that the MAPS capacity may diminish during aging or under disease conditions, which may contribute to aging- or disease-associated decline in proteostasis maintenance.
MATERIALS AND METHODS
Chemicals, antibodies, plasmids, and cells
Chemicals, antibodies, and plasmids used in this study are listed in table S1. Patient serum was purchased from Medix Biochemica USA.
Pre-validated siRNAs were purchased from Thermo Fisher Scientific. The sequences are UFM1_1 s28344, GUGCAAUUAUUACCAAUGAtt; UFM1_2 s28345, CAGACUGCUGGAAAUGUUUtt; UBA5_1 s36548, CCGUAGCAAUAGUAGGUGUtt; UBA5_2 s36550, GCAUUGAAACGAAUGGGAAtt; UFL1_1 s23682, GGAACUUGUUAAUAGCGGAtt; UFL1_2 s23683, GAGGAGUAAUUUUUACGGAtt; UFBP1_1 s35321, GAAAAUUGGAGCUAAGAAAtt; UFBP1_2 s35322, CCAUAAAUCGCAUCCAGGAtt.
Polymerase chain reaction (PCR)–based site-directed mutagenesis was used to make plasmids expressing different α-Syn lysine mutants. To make plasmids expressing various UFBP1 truncation mutants, we amplified the DNA segments encoding the sequence as shown in Fig. 3 by PCR. Amplified DNAs were digested with Asi SI and Mlu I and then cloned into the pCMV6-entry vector. pRK-USP19 ΔTM, pRK-USP19 CS, and pRK-USP19 CD were reported previously (12). The UFM1 and UFSP2 CRISPR KO cells and RPL26ΔC cells were also reported previously (12).
GFP tagging of endogenous UFBP1 was carried out using a CRISPR-Cas12–assisted PCR-based tagging system as described (www.pcr-tagging.com) (50). Briefly, the PCR cassette was amplified from pMaCTag-05 plasmid by the AccuPrime Pfx DNA Polymerase with the primers, M1_UFBP1 and M2_UFBP1, as listed below. The PCR product was gel-purified with the QIAGEN Gel Extraction Kit. HEK293T cells were transiently transfected with 1 μg of the PCR cassette and 1 μg of pCAG-enAsCas12a-HF1(E174R/N282A/S542R/K548R)-NLS(nuc)-3×HA (AAS1815) (a gift from Keith Joung & Benjamin Kleinstiver, Addgene, plasmid #107942) using TransIT293 (Mirus) according to the manufacturer’s protocol. The GFP-positive cells were sorted 2 weeks later by flow cytometry. M1_UFBP1, 5′-CAGCGGGGCCGGGTGTCCATCGCCGAGCTTGCCCAAGCCAGCAACTCCCTCATCGCCTGGGGCCGGGAGTCCCCTGCCCAAGCCCCAGCCTCAGGTGGAGGAGGTAGTG-3′; M2_UFBP1, 5′-ATCACTTCCCCAGGATGGTGGGGAGGGATGAAGATGTATAGCCAGGTAGGCCACAAAAAAACCAACTCTGAGTCCAAGAGGATCTACAAGAGTAGAAATTAGCTAGCTGCATCGGTACC-3′.
Identification of the UFL1 complex as a GFP1-10 interactor
To identify proteins interacting with the MAPS substrate GFP1-10, we transfected HEK293T cells in eight 10-cm dishes with 5 μg of FLAG–GFP1-10– and mCi-USP19–expressing plasmids. As negative controls, cells were transfected with an empty vector. Seventy-two hours after transfection, cells were harvested and washed with ice-cold phosphate-buffered saline (PBS). Cells were permeabilized in 6 ml of 1× PB buffer containing 25 mM Hepes (pH 7.3), 115 mM KOAc, 2.5 mM MgCl2, 5 mM NaOAc, 0.5 mM EGTA, 1 mM dithiothreitol (DTT), and 0.055% digitonin on ice for 5 min. Cytosol was removed by centrifugation at 1000g for 5 min at 4°C. The membrane-containing pellet fraction was washed with 4 ml of PB buffer without DTT. The membrane pellet was then treated with 0.33% formaldehyde at 37°C for 25 min. Membranes were recentrifuged and washed two times by the PB buffer without digitonin. The membrane pellet was resuspended in 8 ml of NP40 lysis buffer and incubated at 4°C for 1 hour. Cleared cell extracts were subject to immunoprecipitation with FLAG beads. The NP40-insoluble fraction was resuspended in 1.6 ml of buffer containing 25 mM tris-HCl (pH 7.4), 150 mM NaCl, 1% SDS, and 5 mM DTT and incubated at 37°C for 30 min. Extracted proteins were diluted fivefold with the NP40 lysis buffer and then subject to immunoprecipitation by FLAG beads. After immunoprecipitation, beads were extensively washed with the NP40 lysis buffer. The bound proteins were eluted with 150 μl of PB buffer containing FLAG peptide (0.2 mg/ml; Sigma-Aldrich). Purified samples were reduced and alkylated in 8 M urea buffer, then digested by Lys-C for 4 hours at 37°C and followed by trypsin overnight digestion with urea concentration reduced to 1.5 M. Peptide digests were desalted using Vivapure spin columns (Sartorius) before MS analysis.
Liquid chromatography–MS/MS analysis
Liquid chromatography (LC)–MS/MS was carried out by nanoflow reverse-phase LC (Eksigent, CA) coupled on-line to a Linear Ion Trap (LTQ)–Orbitrap XL mass spectrometer (Thermo Fisher Scientific). The LC analysis was performed using a capillary column (100 μm in inner diameter × 150 mm in length) packed with Inertsil ODS-3 resin (GL Sciences), and the peptides were eluted using a linear gradient of 2 to 35% B in 85 min at a flow of 400 nl/min (solvent A: 100% H2O/0.1% formic acid; solvent B: 100% acetonitrile/0.1% formic acid). A cycle of one full fourier transform scan mass spectrum [350 to 1800 mass/charge ratio (m/z) and resolution of 60,000 at m/z 400] followed by 10 data-dependent MS/MS scans was acquired in the linear ion trap with normalized collision energy (setting of 35%), with a dynamic exclusion for 30 s. LC MS/MS data were extracted and submitted to protein prospector (UCSF) for database searching against a concatenated database consisting of the normal and random form of the human protein database. Trypsin was set as the enzyme with a maximum of two missed cleavage sites. The mass tolerance for parent ion was set as ±20 parts per million, whereas ±0.6-Da tolerance was chosen for the fragment ions. Chemical modifications such as protein N-terminal acetylation, methionine oxidation, N-terminal pyroglutamine, and deamidation of asparagine were selected as variable modifications during database search. The search compare program in protein prospector was used for summarization, validation, and comparison of results.
Protein purification and in vitro assays
To purify USP19 variants, we transfected HEK293T cells in nine 10-cm dishes with 6 μg of DNA using TransIT293. Twenty-four hours after transfection, we moved cells to three large multilayer tissue culture flasks to amplify cells. We harvested cells in PBS 72 hours after transfection. Cells were incubated in a hypotonic buffer [20 mM tris (pH 7.4) and 10 mM KCl] with a protease inhibitor cocktail and 1 mM tris(2-carboxyethyl)phosphine (TCEP) on ice for 10 min. We then used a Dounce homogenizer to break the cells. After centrifugation at 20,000g for 15 min to remove the nuclei and membrane fractions, we added NaCl to cleared cytosolic fractions to 120 mM and then incubated the cytosolic extracts with FLAG beads to purify USP19 proteins. After extensive washing with a buffer containing 20 mM tris (pH 7.4), 120 mM NaCl, 2 mM MgCl2, 1 mM EDTA, and 1 mM TCEP, we eluted the proteins in a buffer containing 20 mM tris (pH 7.4), 120 mM NaCl, 1 mM TCEP, and 3×FLAG peptide (0.2 μg/ml). Elute proteins were pooled, passed through a PD-10 buffer exchange column (Bio-Rad) to remove the FLAG peptide. Proteins were concentrated to ~0.5 mg/ml and stored in −80°C.
Mass photometry was performed on an OneMP instrument (Refeyn, UK) at room temperature following the published protocol (51). Briefly, microscope coverslips (24 × 50 mm; Thermo Fisher Scientific) were rinsed consecutively in isopropanol and H2O and blow-dried in a stream of clean nitrogen. Proteins were mixed with the indicated concentrations in PBS. After applying 10 μl of solution on the slide, the mass distribution plot was obtained with the software provided by the instrument manufacturer.
GST pull-down experiments were performed by immobilizing 10 μg of GST or GST-UFM1 on glutathione beads. After washing the beads with PBS, we incubated the beads with 2 μg of purified USP19 variant proteins in 400 μg of PBS at 4°C for 60 min. Beads were washed two times with PBS. Bound proteins were eluted with 40 μl of Laemmli buffer.
We measured USP19 deubiquitinase activity using a previously published protocol (16). Briefly, we incubated 30 nM USP19 in a buffer (200 μl) containing 50 mM tris-HCl (pH 7.4), 20 mM KCl, 5 mM MgCl2, 1 mM DTT, and 0.5 μM ubiquitin-AFC (7-Amino-4-trifluoromethylcoumarin) at room temperature. Fluorescence intensity at excitation/emission = 400/505 nm was recorded in real time by Fluoromax-4 spectrofluorometer.
Immunoprecipitation and immunoblotting
To detect UFMylation of misfolded α-Syn and other substrates, we transfected cells with a FLAG-tagged plasmid expressing the reported substrate together with HA-tagged UFM1. Forty-eight hours after transfection, cells were harvested and washed with ice-cold PBS. Cells were then lysed in 100 μl of denaturing buffer containing 2% SDS and 5 mM DTT. The samples were heated at 95°C for 5 min to disrupt all protein-protein interactions, followed by a 10-fold dilution with an NP40 lysis buffer containing 50 mM tris-HCl (pH 7.4), 150 mM NaCl, 2 mM MgCl2, 0.5% Igepal, 2 mM EDTA, and 1 mM DTT. After removal of insoluble materials by centrifugation at 20,000g for 10 min, FLAG-tagged substrates were immunoprecipitated by FLAG beads and analyzed by immunoblotting.
To detect total UFMylated proteins in heat-treated cells, cells (4 million) harvested after PBS rinse were immediately lysed in 200 μl of 1× Laemmli buffer at 95°C for 30 min. Cell lysates were fractionated by 4 to 12% bis-tris gradient gel and analyzed by immunoblotting following the standard protocol. For serum samples, 100 μl of serum was centrifugated at 20,000g for 10 min. The supernatant fractions were mixed with 4x Laemmli buffer and diluted appropriately before gel analysis. For immunoblotting, horseradish peroxidase–conjugated secondary antibodies were used for detection.
To detect the interaction of USP19 with UFM1, cells expressing FLAG-tagged USP19 were washed with ice-cold PBS and then treated with 0.05% formaldehyde in 800 μl of PBS at 37°C for 15 min. Cells were washed three times with PBS, then resuspended in 200 μl of PBS containing 1% SDS and 5 mM DTT plus a protease inhibitor cocktail. Cells were treated at 37°C for 30 min to disrupt protein-protein interactions. NP40 lysis buffer (800 μl) with a protease inhibitor cocktail was then added to dilute the sample. The samples were then centrifuged at 20,000g for 10 min. The supernatant fraction was subject to immunoprecipitation with FLAG beads. The precipitated materials were analyzed by immunoblotting.
C. elegans and fly experiments
The PVZ-1 vector was used to make the constructs to express WT UFBP1 and the ΔMLS mutant in C. elegans body wall muscle cells. The body wall muscle–specific expression of mCherry-tagged UFBP1 (encoded by ZK1236.7) and UFBP1ΔMLS (PVZ-1-myo3p-ufbp1-mCherry-unc-54 3′UTR and pVZ-1-myo3p-ufbp1ΔMLS-mcherry-unc-54 3′UTR) were made by inserting PCR-amplified C. elegans UFBP1 cDNA fragments into the Xba I and Not I sites of the vector.
The following strains were used to assay α-Syn secretion in C. elegans. WT (N2), ufbp1(tm5808), and DDP1 that carries a body wall muscle–specific expressed α-Syn tagged with YFP (Venus) (52). KM581 and KM582 strains are DDP1 carrying the PVZ-1-myo3-UFBP1-mcherry-unc45 3′UTR construct. KM583 and KM584 strains are DDP1 carrying the PVZ-1-myo3-UFBP1ΔMLS-mcherry-unc45 3′UTR construct. The ufbp1 (tm5808) strain was backcrossed eight times with WT and then crossed into DDP1 to generate two independent strains for both ufbp1(tm5808) alone and DDP1;ufbp-1 (tm5808) strains.
Transgenic animals harboring body wall muscle–specific expressed constructs were generated by standard DNA injection techniques (53). The plasmid was injected at the concentration of 20 ng/μl with a transgenic marker (100 ng/μl; pRF4) into WT (N2) animals. As the DDP1 strain alone has an unhealthy phenotype after 3 days at the adult stage, DDP1 was crossed into two independent ufbp1-expressing transgenic worms, and two independent DDP1 strains carrying a body wall muscle–specific expressed ufbp-1 construct as described above were obtained to assay α-Syn secretion. The accumulation of fluorescence protein (Venus and mCherry) in coelomocytes from expressed ufbp-1 reporter constructs was checked in 2-day-old adult animals. Microscopic images for a coelomocyte were deconvoluted by Nikon (NIS-elements) software and were adjusted by Adobe Photoshop.
Transgenic flies harboring UAS–mCherry–α-Syn were generated by Rainbow Transgenic. The p{Cg-GAL4.A}2 (7011), UAS-control shRNA p{TRiP.JF01355}attP2 (31603), and UAS-UFM1 shRNA (39054) p{TRiP.HMS01974}attP40 lines were purchased from Bloomington Drosophila Stock Center. The p{Cg-GAL4.A}2 line, which expresses the GAL4 transcription factor under the control of a fat body–specific collagen promoter, was used to drive the expression of mCherry–α-Syn and shRNA in the same tissue. Fly larva dissection and tissue staining were done as previously described (54). To detect mCherry–α-Syn or UFM1 in third instar larvae hemolymph, six third instar larvae (three male and three female) were thoroughly rinsed first in ultrapure water and then by PBS. We then bleed the larvae by poking at the posterior end with a 25-gauge needle in a drop of PBS (45 μl). Hemolymph was collected and centrifuged at 10,000g for 10 min. The supernatant fraction (40 μl) was mixed with 2× Laemmli buffer and heated at 95°C for 5 min before SDS–polyacrylamide gel electrophoresis (PAGE) and immunoblotting analysis. The hemolymph-depleted larvae were placed in 40 μl PBS with addition of 60 μl of 2× Laemmli buffer. The samples were heated at 95°C for 25 min before SDS-PAGE and immunoblotting analyses.
Protein secretion experiments
Protein secretion experiments were performed following a well-established protocol (55). Briefly, we seeded 0.2 × 106 HEK293T cells per well to a poly-d-lysine–coated 12-well plate on day 1. Cells were transfected with a plasmid (300 ng) expressing either α-Syn or other MAPS substrates on day 2. Twenty-four hours later, the culture medium was carefully removed. Cells were replenished with 1 ml of complete Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS. Cells were cultured for another 15 hours before conditioned medium was harvested. Cells were washed with PBS and lysed in 300 μl of NP40 lysis buffer containing 50 mM tris-HCl (pH 7.4), 150 mM NaCl, 2 mM MgCl2, 0.5% Igepal, 2 mM EDTA, and 1 mM DTT plus a protease inhibitor cocktail. The conditioned medium was subject to two rounds of centrifugation, first at 1000g for 5 min and then at 10,000g for 40 min. The clear supernatant fractions were collected and mixed with 4× Laemmli buffer.
To control for cell lysis, we analyzed the medium fractions by immunoblotting with antibodies against abundant cytosolic chaperones such as HSC70 and HSP90. In most experiments, we do not detect these proteins in the medium, suggesting that the cells were intact.
Software and data analyses
Immunoblotting enhanced chemiluminescence signals were detected by a ChemiDoc Imager (Bio-Rad). Protein bands were quantified by ImageLab (Bio-Rad). Shown immunoblotting gels are representative of at least two independent biological repeats. Graphpad Prism 9.0 was used to generate graphs. Statistical analyses were conducted with either Excel or Graphpad Prism 9.0.
Acknowledgments
We thank the CGC (funded by NIH Office of Research Infrastructure Programs P40 OD010440) and the Japanese National Bioresource Project for C. elegans strains.
Funding: This research is supported by an intramural research program of the NIDDK (DK075143) at the National Institutes of Health to Y.Y. and by a grant NIHR35GM145249 to L.H.
Author contributions: Conceptualization: J.-G.L., X.W., L.W., and Y.Y. Methodology: L.W., Y.X., T.F., L.S., X.W., M.K., C.Y., and Y.Y. Investigation: J.-G.L., L.W., Y.X., C.Y., L.S., T.F., X.W., and Y.Y. Visualization: L.W. and Y.Y. Supervision: M.K., L.H., and Y.Y. Writing—original draft: L.W., C.Y., and Y.Y. Writing—review and editing: L.W., C.Y., M.K., and Y.Y.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Requests for cells or plasmids should be sent to yihongy@mail.nih.gov.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Table S1
REFERENCES AND NOTES
- 1.Benham A. M., Protein secretion and the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 4, a012872 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rabouille C., Pathways of unconventional protein secretion. Trends Cell Biol. 27, 230–240 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Nickel W., Rabouille C., Unconventional protein secretion: Diversity and consensus. Semin. Cell Dev. Biol. 83, 1–2 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Steringer J. P., Lange S., Čujova S., Šachl R., Poojari C., Lolicato F., Beutel O., Müller H. M., Unger S., Coskun Ü., Honigmann A., Vattulainen I., Hof M., Freund C., Nickel W., Key steps in unconventional secretion of fibroblast growth factor 2 reconstituted with purified components. eLife 6, e28985 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.New J., Thomas S. M., Autophagy-dependent secretion: Mechanism, factors secreted, and disease implications. Autophagy 15, 1682–1693 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee J., Ye Y., The roles of endo-lysosomes in unconventional protein secretion. Cell 7, 198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malhotra V., Unconventional protein secretion: An evolving mechanism. EMBO J. 32, 1660–1664 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Legrand C., Saleppico R., Sticht J., Lolicato F., Muller H. M., Wegehingel S., Dimou E., Steringer J. P., Ewers H., Vattulainen I., Freund C., Nickel W., The Na,K-ATPase acts upstream of phosphoinositide PI(4,5)P2 facilitating unconventional secretion of Fibroblast Growth Factor 2. Commun. Biol. 3, 141 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang M., Liu L., Lin X., Wang Y., Li Y., Guo Q., Li S., Sun Y., Tao X., Zhang D., Lv X., Zheng L., Ge L., A translocation pathway for vesicle-mediated unconventional protein secretion. Cell 181, 637–652.e15 (2020). [DOI] [PubMed] [Google Scholar]
- 10.MacDonald C., Buchkovich N. J., Stringer D. K., Emr S. D., Piper R. C., Cargo ubiquitination is essential for multivesicular body intralumenal vesicle formation. EMBO Rep. 13, 331–338 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J., Gee H. Y., Lee M. G., Unconventional protein secretion - New insights into the pathogenesis and therapeutic targets of human diseases. J. Cell Sci. 131, jcs213686 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Lee J. G., Takahama S., Zhang G., Tomarev S. I., Ye Y., Unconventional secretion of misfolded proteins promotes adaptation to proteasome dysfunction in mammalian cells. Nat. Cell Biol. 18, 765–776 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y., Cui L., Dibello A., Wang L., Lee J., Saidi L., Lee J. G., Ye Y., DNAJC5 facilitates USP19-dependent unconventional secretion of misfolded cytosolic proteins. Cell Discov. 4, 11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J., Xu Y., Saidi L., Xu M., Zinsmaier K., Ye Y., Abnormal triaging of misfolded proteins by adult neuronal ceroid lipofuscinosis-associated DNAJC5/CSPα mutants causes lipofuscin accumulation. Autophagy 19, 204–223 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu S., Hernandez Villegas N. C., Sirkis D. W., Thomas-Wright I., Wade-Martins R., Schekman R., Unconventional secretion of α-synuclein mediated by palmitoylated DNAJC5 oligomers. eLife 12, e85837 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J. G., Kim W., Gygi S., Ye Y., Characterization of the deubiquitinating activity of USP19 and its role in endoplasmic reticulum-associated degradation. J. Biol. Chem. 289, 3510–3517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fontaine S. N., Zheng D., Sabbagh J. J., Martin M. D., Chaput D., Darling A., Trotter J. H., Stothert A. R., Nordhues B. A., Lussier A., Baker J., Shelton L., Kahn M., Blair L. J., Stevens S. M. Jr., Dickey C. A., DnaJ/Hsc70 chaperone complexes control the extracellular release of neurodegenerative-associated proteins. EMBO J. 35, 1537–1549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L., Ye Y., Clearing traffic jams during protein translocation across membranes. Front. Cell Dev. Biol. 8, 610689 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L., Xu Y., Yun S., Yuan Q., Satpute-Krishnan P., Ye Y., SAYSD1 senses UFMylated ribosome to safeguard co-translational protein translocation at the endoplasmic reticulum. Cell Rep. 42, 112028 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scavone F., Gumbin S. C., Da Rosa P. A., Kopito R. R., RPL26/uL24 UFMylation is essential for ribosome-associated quality control at the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 120, e2220340120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang J. R., Lingeman E., Luong T., Ahmed S., Muhar M., Nguyen T., Olzmann J. A., Corn J. E., A Genome-wide ER-phagy screen highlights key roles of mitochondrial metabolism and ER-resident UFMylation. Cell 180, 1160–1177.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishimura R., El-Gowily A. H., Noshiro D., Komatsu-Hirota S., Ono Y., Shindo M., Hatta T., Abe M., Uemura T., Lee-Okada H. C., Mohamed T. M., Yokomizo T., Ueno T., Sakimura K., Natsume T., Sorimachi H., Inada T., Waguri S., Noda N. N., Komatsu M., The UFM1 system regulates ER-phagy through the ufmylation of CYB5R3. Nat. Commun. 13, 7857 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephani M., Picchianti L., Dagdas Y., C53 is a cross-kingdom conserved reticulophagy receptor that bridges the gap betweenselective autophagy and ribosome stalling at the endoplasmic reticulum. Autophagy 17, 586–587 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B., Niu F., Zeng Y., Tse M. K., Deng C., Hong L., Gao S., Lo S. W., Cao W., Huang S., Dagdas Y., Jiang L., Ufmylation reconciles salt stress-induced unfolded protein responses via ER-phagy in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 120, e2208351120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tatsumi K., Sou Y. S., Tada N., Nakamura E., Iemura S., Natsume T., Kang S. H., Chung C. H., Kasahara M., Kominami E., Yamamoto M., Tanaka K., Komatsu M., A novel type of E3 ligase for the Ufm1 conjugation system. J. Biol. Chem. 285, 5417–5427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peter J. J., Magnussen H. M., DaRosa P. A., Millrine D., Matthews S. P., Lamoliatte F., Sundaramoorthy R., Kopito R. R., Kulathu Y., A non-canonical scaffold-type E3 ligase complex mediates protein UFMylation. EMBO J. 41, e111015 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millrine D., Cummings T., Matthews S. P., Peter J. J., Magnussen H. M., Lange S. M., Macartney T., Lamoliatte F., Knebel A., Kulathu Y., Human UFSP1 is an active protease that regulates UFM1 maturation and UFMylation. Cell Rep. 40, 111168 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L., Xu Y., Rogers H., Saidi L., Noguchi C. T., Li H., Yewdell J. W., Guydosh N. R., Ye Y., UFMylation of RPL26 links translocation-associated quality control to endoplasmic reticulum protein homeostasis. Cell Res. 30, 5–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walczak C. P., Leto D. E., Zhang L., Riepe C., Muller R. Y., DaRosa P. A., Ingolia N. T., Elias J. E., Kopito R. R., Ribosomal protein RPL26 is the principal target of UFMylation. Proc. Natl. Acad. Sci. U.S.A. 116, 1299–1308 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oughtred R., Rust J., Chang C., Breitkreutz B. J., Stark C., Willems A., Boucher L., Leung G., Kolas N., Zhang F., Dolma S., Coulombe-Huntington J., Chatr-Aryamontri A., Dolinski K., Tyers M., The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 30, 187–200 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petroski M. D., Deshaies R. J., Context of multiubiquitin chain attachment influences the rate of Sic1 degradation. Mol. Cell 11, 1435–1444 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Zenko D., Marsh J., Castle A. R., Lewin R., Fischer R., Tofaris G. K., Monitoring α-synuclein ubiquitination dynamics reveals key endosomal effectors mediating its trafficking and degradation. Sci. Adv. 9, eadd8910 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyer M. L., Milhollen M. A., Ciavarri J., Fleming P., Traore T., Sappal D., Huck J., Shi J., Gavin J., Brownell J., Yang Y., Stringer B., Griffin R., Bruzzese F., Soucy T., Duffy J., Rabino C., Riceberg J., Hoar K., Lublinsky A., Menon S., Sintchak M., Bump N., Pulukuri S. M., Langston S., Tirrell S., Kuranda M., Veiby P., Newcomb J., Li P., Wu J. T., Powe J., Dick L. R., Greenspan P., Galvin K., Manfredi M., Claiborne C., Amidon B. S., Bence N. F., A small-molecule inhibitor of the ubiquitin activating enzyme for cancer treatment. Nat. Med. 24, 186–193 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Ishimura R., Ito S., Mao G., Komatsu-Hirota S., Inada T., Noda N. N., Komatsu M., Mechanistic insights into the roles of the UFM1 E3 ligase complex in ufmylation and ribosome-associated protein quality control. Sci. Adv. 9, eadh3635 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hertel P., Daniel J., Stegehake D., Vaupel H., Kailayangiri S., Gruel C., Woltersdorf C., Liebau E., The ubiquitin-fold modifier 1 (Ufm1) cascade of Caenorhabditis elegans. J. Biol. Chem. 288, 10661–10671 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen K. G., Aviram N., Laborenz J., Bibi C., Meyer M., Spang A., Schuldiner M., Herrmann J. M., An ER surface retrieval pathway safeguards the import of mitochondrial membrane proteins in yeast. Science 361, 1118–1122 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Uhlén M., Fagerberg L., Hallstrom B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjöstedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C. A.-K., Odeberg J., Djureinovic D., Takanen J. O., Hober S., Alm T., Edqvist P.-H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J. M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., Ponteén F., Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Merezhko M., Uronen R.-L., Huttunen H. J., The cell biology of tau secretion. Front. Mol. Neurosci. 13, 569818 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Komatsu M., Chiba T., Tatsumi K., Iemura S., Tanida I., Okazaki N., Ueno T., Kominami E., Natsume T., Tanaka K., A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J. 23, 1977–1986 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tatsumi K., Yamamoto-Mukai H., Shimizu R., Waguri S., Sou Y. S., Sakamoto A., Taya C., Shitara H., Hara T., Chung C. H., Tanaka K., Yamamoto M., Komatsu M., The Ufm1-activating enzyme Uba5 is indispensable for erythroid differentiation in mice. Nat. Commun. 2, 181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai Y., Pi W., Sivaprakasam S., Zhu X., Zhang M., Chen J., Makala L., Lu C., Wu J., Teng Y., Pace B., Tuan D., Singh N., Li H., UFBP1, a key component of the Ufm1 conjugation, is essential for ufmylation-mediated regulation of erythroid development. PLOS Genet. 11, e1005643 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerakis Y., Quintero M., Li H., Hetz C., The ufmylation system in proteostasis and beyond. Trends Cell Biol. 29, 974–986 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z., Gong Y., Peng B., Shi R., Fan D., Zhao H., Zhu M., Zhang H., Lou Z., Zhou J., Zhu W. G., Cong Y. S., Xu X., MRE11 UFMylation promotes ATM activation. Nucleic Acids Res. 47, 4124–4135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin B., Yu J., Nowsheen S., Wang M., Tu X., Liu T., Li H., Wang L., Lou Z., UFL1 promotes histone H4 ufmylation and ATM activation. Nat. Commun. 10, 1242 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schorova L., Bedard N., Khayachi A., Ho H.-H., Bolivar-Pedroso J., Huynh J., Piccirelli M., Wang Y., Plourde M., Luo W., Del Cid-Pellitero E., Shlaifer I., Castellanos-Montiel M. J., Yu Z., Valenzuela D. V. C., Lacalle-Aurioles M., Kriz A., Ye Y., Durcan T. M., Wing S. S., USP19 deubiquitinase inactivation regulates α-synuclein ubiquitination and inhibits accumulation of Lewy body-like aggregates in mice. NPJ Parkinsons Dis. 9, 157 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.C. Parra Bravo, A. Maria Giani, J. Madero Perez, Z. Zhao, A. Samelson, M. Y. Wong, A. Evangelisti, L. Fan, T. Pozner, M. Mercedes, P. Ye, T. Patel, A. Yarahmady, G. Carling, V. M. Y. Lee, M. Sharma, S.-A. Mok, W. Luo, M. Zhao, M. Kampmann, S. Gong, L. Gan, Human iPSC 4R tauopathy model uncovers modifiers of tau propagation. bioRvix 2023.06.19.544278 [Preprint] (22 June 2023). 10.1101/2023.06.19.544278. [DOI]
- 47.A. J. Samelson, N. Arigat, J. McKetney, G. Rohanitazangi, C. Parra Bravo, D. Goodness, R. Tian, P. Grosjean, R. Abskharon, D. S. Eisenberg, N. M. Kanaan, L. Gan, C. Condello, D. L. Swaney, M. Kampmann, CRISPR screens in iPSC-derived neurons reveal principles of tau proteostasis. bioRxiv 2023.06.16.545386 [Preprint] (26 June 2023). 10.1101/2023.06.16.545386. [DOI]
- 48.Lee H. J., Patel S., Lee S. J., Intravesicular localization and exocytosis of α-synuclein and its aggregates. J. Neurosci. 25, 6016–6024 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saez I., Vilchez D., The mechanistic links between proteasome activity, aging and age-related diseases. Curr. Genomics 15, 38–51 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kleinstiver B. P., Sousa A. A., Walton R. T., Tak Y. E., Hsu J. Y., Clement K., Welch M. M., Horng J. E., Malagon-Lopez J., Scarfo I., Maus M. V., Pinello L., Aryee M. J., Joung J. K., Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 37, 276–282 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu D., Piszczek G., Measuring the affinity of protein-protein interactions on a single-molecule level by mass photometry. Anal. Biochem. 592, 113575 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bodhicharla R., Nagarajan A., Winter J., Adenle A., Nazir A., Brady D., Vere K., Richens J., O'Shea P., Bell D. R., de Pomerai D., Effects of α-synuclein overexpression in transgenic Caenorhabditis elegans strains. CNS Neurol. Disord. Drug Targets 11, 965–975 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., Efficient gene transfer in C. elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye Y., Fortini M. E., Characterization of Drosophila Presenilin and its colocalization with Notch during development. Mech. Dev. 79, 199–211 (1998). [DOI] [PubMed] [Google Scholar]
- 55.Lee J., Ye Y., Studying unconventional secretion of misfolded proteins in cultured cells and primary neurons. Methods Mol. Biol. 2473, 349–366 (2022). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Table S1