Abstract
Oncogenic mutations accumulating in many chromatin-associated proteins have been identified in different tumor types. With a mutation rate from 10 to 57%, ARID1A has been widely considered a tumor suppressor gene. However, whether this role is mainly due to its transcriptional-related activities or its ability to preserve genome integrity is still a matter of intense debate. Here, we show that ARID1A is largely dispensable for preserving enhancer-dependent transcriptional regulation, being ARID1B sufficient and required to compensate for ARID1A loss. We provide in vivo evidence that ARID1A is mainly required to preserve genomic integrity in adult tissues. ARID1A loss primarily results in DNA damage accumulation, interferon type I response activation, and chronic inflammation leading to tumor formation. Our data suggest that in healthy tissues, the increased genomic instability that follows ARID1A mutations and the selective pressure imposed by the microenvironment might result in the emergence of aggressive, possibly immune-resistant, tumors.
ARID1A loss in adult tissues affects genome integrity without altering enhancer-dependent transcriptional regulation.
INTRODUCTION
In the past 10 years, a considerable amount of data has been generated by cancer genomic studies showing that genes encoding for chromatin-modifying enzymes are among the most frequently mutated. The activity of SWItch/Sucrose Non-Fermentable (SWI/SNF) chromatin remodeling complexes is often affected by loss-of-function mutations involving several subunits, the most frequently mutated being ARID1A subunit (1). Mammalian SWI/SNF is a class of evolutionary-conserved, chromatin-associated, multiprotein complexes playing a crucial role in modulating transcription, DNA replication, and repair by promoting DNA accessibility. Encoded by 29 genes, SWI/SNF subunits assemble in three different complexes, the canonical BRG1/BRM associated factor (cBAF), noncanonical BAF (ncBAF), and polybromo-associated BAF (PBAF) complex. The three complexes share the mutually exclusive, adenosine triphosphate–dependent, catalytic subunits SMARCA4 and SMARCA2, through which they catalyze sliding and eviction of nucleosomes. Each complex is characterized by the presence of distinct subunits and distinct genomic targeting. ARID1A/ARID1B and DPF2 exclusively assemble into cBAF and mainly localize at enhancers. ARID2, PBRM1 and BRD7 can be exclusively found in PBAF and GLTSCR1/GLTSCR2 and BRD9 in ncBAF and are mainly recruited at promoters (2, 3).
The chromatin remodeling activity of these complexes is of fundamental importance to establishing and preserving cell identity. More than 20% of tumors harbor mutations in genes encoding for SWI/SNF subunits. ARID1A (also known as BAF250A) is the subunit mostly mutated with rates of 10 to 57%. Loss-of-function mutations in this subunit can be found, among others, in gastric, ovarian, uterine, bladder, colon, and liver cancer (1, 3). In vivo and in vitro experiments suggest that ARID1A loss promotes tumor formation by affecting enhancer-promoter regulation, accordingly to the specific genomic distribution of cBAF (4–7). However, crucial structural roles have also been described in vitro for ARID1A-containing cBAF, such as DNA damage repair, telomeres cohesion, chromosome decatenation, and R-loop resolution (8–12). Although altered enhancer-mediated gene expression being predominantly considered the oncogenic consequence of Arid1a mutations, the available data are mostly correlative, and in vivo causative evidence is still missing. The use of organoids or engineered mouse models suggests that ARID1A loss per se is not sufficient to drive intestinal, pancreatic, and gastric cancer (13–15). Notably, two important exceptions have been described. In colonic epithelial cells, ARID1A loss is sufficient to drive invasive carcinomas, although with long latency, while preventing adenomas formation in ApcMin mice (4). In hepatocytes, ARID1A loss favors liver regeneration and prevents tumor initiation (7, 16). The heterogeneity of these data has been largely ascribed to its role in preserving cell type–specific enhancers. Here, we describe the early oncogenic events following ARID1A loss, demonstrating that they are unrelated to enhancer-dependent transcriptional regulation and shared by all the tissues examined. ARID1A loss predominantly leads to DNA damage accumulation and genomic instability, interferon type I response activation, and chronic inflammation, ultimately favoring tumor formation.
RESULTS
ARID1A loss favors hepatocellular carcinoma formation
To thoroughly analyze the consequences of ARID1A loss in liver homeostasis, we generated tamoxifen-inducible, liver-specific, Arid1a knockout mice by crossing AlbCre-ERT2 and Arid1afl/fl mice. Tamoxifen injection efficiently abrogates Arid1a expression exclusively in hepatocytes leaving its expression in all the other cells unaltered (fig. S1, A and B). While ARID1A has been considered a tumor suppressor gene, recently published works suggest that it restricts liver regenerative capacities and promotes hepatocellular carcinoma (HCC) formation (7, 16). We, therefore, decided to evaluate whether liver-specific Arid1a-deficient animals were prone to develop tumors. Despite several established liver tumor models have been generated, few of them lead to the formation of tumors closely resembling human HCC (17–19). We decided to use a nongenotoxic stimulus that mimics a transient and stochastic activation of the detoxification pathway, a physiological biochemical cascade involved in the inactivation of potentially hazardous exogenous and endogenous molecules (20). The master regulator of this pathway, the nuclear receptor NR1I3, has been activated using the highly specific agonist 1,4-bis-[2-(3,5-dichloropyridyloxy)]benzene, administered 1 week and 2 months after tamoxifen injection (Fig. 1A). NR1I3 activation induces a transient hepatomegaly that in knockout animals persists for a longer time (fig. S1D). This effect was not due to a stronger activation of the xenobiotic response since the main NR1I3 target genes involved in this process were similarly activated (fig. S1E). After 5 months, the animals have been sacrificed, liver explanted, and used for histological analysis. In these mice, multiple macroscopic nodules were identified just by a gross examination in 8 of 10 animals (Fig. 1, B and C). Histological analysis revealed that dysplastic nodules were characterized by a compact or trabecular pattern of proliferating HNF4α-positive cells, with no sign of intranodular collagen deposition (Picrosirius red) and a peculiar CK19 positivity, all features reminiscent of a particular class of aggressive HCC (Fig. 1B) (21). Arid1a knockout was preserved in both normal and dysplastic cells (Fig. 1B), except for sporadic patches of Arid1a wild-type normal cells spreading from the periportal area (fig. S1C), suggesting the activation of the cholangiocytes-driven regenerative programs frequently observed upon chronic liver damage as a consequence of impaired hepatocyte-driven regeneration (22). Hyperplastic ductules can also be observed, while no intrahepatic cholangiocarcinomas have been identified. To identify the transcriptional programs possibly involved in the phenotype observed, we performed RNA sequencing (RNA-seq) analysis after 30 days from NR1I3 activation in Arid1a-deficient and Arid1a-proficient livers. We identified 876 differentially expressed genes (610 up- and 266 down-regulated) (Fig. 1D). Gene ontology (GO) term analysis revealed that several genes involved in negative regulation of mitotic cell cycle phase transition and chromosome segregation were up-regulated, suggesting the activation of G2/M checkpoint (Fig. 1E) as also supported by increased p53 levels (fig. S1F). To investigate whether liver-specific Arid1a−/− mice were prone to develop liver tumors in the absence of exogenous stimuli, we monitored knockout mice over a longer period of time. After 15 months from tamoxifen injection, mice start to show signs of distress and must be suppressed for ethical reasons. Arid1a-deficient mice (five of five) showed multiple liver nodules, suggesting that Arid1a-deficient mice are prone to spontaneously develop liver tumors also in the absence of NR1I3 activation (Fig. 1F). These data indicate that, by affecting G2/M transition, ARID1A loss is sufficient to sensitize hepatocytes to endogenous and exogenous insults, favoring liver tumor formation.
Fig. 1. ARID1A loss sensitizes to HCC formation.
(A) Experimental scheme. (B) Gross examination and histological and immunohistochemical analysis of wild-type and Arid1a−/− liver 5 months from NR1L3 agonist injection showing liver tumor formation (arrowheads). (C) Number of animals that developed tumors upon sporadic activation of NR1L3. (D) Volcano plot showing transcriptional alterations observed by RNA-seq analysis in Arid1a−/− compared to wild-type animals 30 days from NR1L3 agonist administration. (E) Bar plot showing overrepresented biological process (GO terms) of up-regulated genes in Arid1a−/− compared to wild-type animals 30 days from NR1L3 agonist administration. (F) Gross examination of wild-type and Arid1a−/− liver 15 months from tamoxifen injection showing liver tumor formation (arrowheads). N, nontumoral; T, tumor; H&E, hematoxylin and eosin.
ARID1A is largely dispensable for preserving tissue-specific enhancers
Given the extensive literature indicating that ARID1A-containing cBAF prevents tumor formation by preserving enhancer-dependent transcriptional regulation and cell identity, we analyzed the epigenetic and transcriptional alterations imposed by ARID1A loss in adult hepatocytes. ARID1A predominantly localizes at distal regulatory elements (DREs) accordingly with its role in preserving enhancer activity (fig. S2A) and poorly colocalizes with CTCF-occupied regions (fig. S2B) as previously described in human cancer cell lines (23). We, therefore, focused our attention on the consequence of ARID1A loss primarily in these regions. To monitor proximal and DRE activity, we mapped H3K27Ac levels, a typical histone mark reported to directly correlate with transcriptional activity and outperform chromatin accessibility in predicting and monitoring active regulatory regions (24). We then divided ARID1A-occupied distal elements into four quantiles based on the H3K27Ac intensity ratio. Although to a different extent, they are all characterized by high levels of H3K27Ac, indicating high levels of activity of these regulatory elements. Unexpectedly, ARID1A loss does not result in a global inactivation of these regulatory elements (Fig. 2A), despite an efficient deletion of exon 9 has been achieved (fig. S2C). While on half of them a considerable H3K27Ac reduction can be observed (−52.3% and −19.7% in the first and second quantiles, respectively), a modest increase (+6.5%) and a gain (+48.9%) of acetylation can be observed for the third and fourth quantiles, respectively (fig. S2D). Similarly, H3K4Me1 levels are reduced only on regulatory elements belonging to the first quantile (Fig. 2A and fig. S2D). We also noted that the lower the basal acetylation levels, the greater the regions were affected by ARID1A loss suggesting a stronger ARID1A dependency over these regions. These data show that ARID1A-containing cBAF complex is required to preserve only one-fourth of its targeted distal elements. To investigate whether ARID1B can compensate for ARID1A loss, we generated AlbCre-ERT2Arid1Afl/flArid1Bfl/fl mice. The analysis of ARID1A-occupied DREs reveals that concomitant loss of ARID1B significantly affects H3K27Ac and H3K4Me1 (Fig. 2A and fig. S2D), confirming the redundant activity of ARID1B-containing cBAF complex on most of these regions. No further reduction of H3K27Ac and H3K4Me1 is observed on regions belonging to the first quantile, suggesting that ARID1A-containing cBAF complex is required to preserve chromatin status over these regions. To further corroborate the limited effect of ARID1A loss on chromatin accessibility at DREs, we analyzed the occupancy of C/EBPα, a nonpioneer transcription factor required for liver development and homeostasis (25, 26). In the absence of ARID1A, reduced C/EBPα occupancy can be observed only on the first quantile regions, thus supporting a role of ARID1A mainly at these regions (Fig. 2, B and C). Accordingly, RNA-seq analysis performed to evaluate the functional impact of ARID1A loss identified a relatively small number of differentially expressed genes, while concomitant loss of both ARID1A and ARID1B markedly alters hepatocyte transcriptome (Fig. 2, D and E, and fig. S2E). GO analysis revealed that ARID1A/B loss abrogates the expression of liver-specific genes, mainly involved in metabolic functions. Increased transcription of genes involved in wound healing and matrix deposition (fibrosis) was observed, suggesting strong activation of regenerative processes and an extensive remodeling of liver architecture (fig. S3A and table S1). Notably, no differences have been observed in HNF4α expression, suggesting that cell identity is preserved in cBAF-defective cells (fig. S3B). Such extensive transcriptional alterations severely compromise liver functions. After around 20 days, Arid1a/b null mice became jaundiced and moribund and must be suppressed for ethical reasons. Histological analysis of liver samples revealed the presence of aggressive hepatitis characterized by extensive fibrosis, infiltration of IBA1-positive macrophages, increased CDKN1A levels, and cell death (Fig. 2F). Notably, while concomitant ARID1A/B loss is efficiently achieved using these mice, knockout cells are rapidly counter-selected, and only sporadic Arid1a or Arid1b single knockout patches of cells can be identified after 20 days from Cre activation (fig. S3C). To further investigate the transcriptional consequences of ARID1A loss, we integrated RNA-seq and chromatin immunoprecipitation sequencing (ChIP-seq) data. The number of differentially expressed genes showing ARID1A occupancy at promoters in wild-type cells (direct ARID1A targets) is extremely reduced and equally distributed between up- and down-regulated genes. Transcriptional alterations can be observed in around 1.6% of ARID1A target genes (fig. S4A). To analyze ARID1A-specific, enhancer-dependent transcriptional regulation, we applied an activity-by-contact (ABC) model (27) to predict active enhancers regulating differentially expressed genes. A total of 144 and 244 DREs have been predicted regulating down- and up-regulated genes, respectively. The overlap of these regions with ARID1A-targeted DREs belonging to the different quantiles was limited (fig. S4B), suggesting that other sets of DREs might compensate for ARID1A loss. Similar results have been obtained by predicting the genes regulated by all the ARID1A-occupied DREs belonging to the different quantiles. Few of the differentially expressed genes have been found to overlap with ARID1A-regulated genes (fig. S4C). Moreover, the intersection between the differentially expressed genes under the control of the different enhancers of each quantile revealed a high degree of redundancy, with enhancers belonging to different quantiles coregulating same set of genes (Fig. 2G). Except for a set of genes whose expression is preserved exclusively by DREs of the first quantile, no correlations have been observed between transcriptional changes and the DREs in all the other quantiles (Fig. 2G). Together, these data demonstrate that ARID1A loss has limited direct consequences on enhancer-dependent transcriptional regulation due to a high degree of redundancy between DREs and the compensatory effect of ARID1B that fully preserves hepatocyte-specific transcriptional programs.
Fig. 2. ARID1A is largely dispensable for preserving enhancer-dependent transcriptional regulation.
(A) Heatmap of H3K27Ac and H3K4Me1 levels on ARID1A-bound distal regions in wild-type, Arid1a−/− and Arid1a/b−/− hepatocytes. The four sets of regions (quantiles) were generated on the basis of the H3K27Ac intensity ratio (Arid1a−/− versus wild type; n = 2, see Materials and Methods). (B) Heatmap and (C) density plot showing C/EBPα occupancy on ARID1A-bound distal regions in wild-type, Arid1a−/− hepatocytes. (D) Volcano plots showing transcriptional alterations observed by RNA-seq analysis in Arid1a−/− compared to wild type at 9 and 30 days from tamoxifen injection. (E) Volcano plot showing transcriptional alterations observed by RNA-seq analysis in Arid1a/b−/− liver compared to wild type at 9 days from tamoxifen injection. (F) Histological and immunohistochemical analysis of wild-type and Arid1a/b−/− liver samples. (G) UpSet plots representing the number of differentially expressed genes (Arid1a−/− versus wild type) regulated by the ARID1A-bound enhancers in the four different quantiles. Only intersections ≥ 3 are shown. Boxplots represent transcriptional changes of each specific set of genes in Arid1a−/− liver compared to wild type.
Type I interferon response is activated upon ARID1A loss
To gain more insights into the early events that follow ARID1A loss, we thoroughly analyzed the transcriptome of proficient and deficient cells. The oncogenic activity of Arid1a mutations has been recently ascribed to a subtle up-regulation of cytochrome genes (7, 16). We did not observe substantial differences in the expression of cytochrome genes between wild-type and knockout cells (fig. S4D), suggesting that a transcriptional modulation of this set of genes may not be an early event. Notably, GO term analysis performed using differentially expressed genes revealed that up-regulated genes were enriched for genes involved in the interferon response, while no defined pathway was significantly enriched among down-regulated genes (Fig. 3A and table S1). In particular, among the activated genes, several interferon-stimulated genes (ISGs), proinflammatory chemokines such as Cxcl10, and master transcription factors such as Irf7 have been observed (table S1). The up-regulation of these genes was further confirmed using purified hepatocytes from wild-type and Arid1a knockout livers (Fig. 3B). Interferon target gene activation was not observed in ARID1B-defective cells (fig. S5A) and is preserved in double-knockout hepatocytes further supporting the specificity of this response (fig. S5B). To evaluate whether transcriptional activation of interferon target genes was a tissue-specific effect, we analyzed datasets obtained from Arid1a-proficient and Arid1a-deficient small intestinal crypts (15), HCT116 (4), 2′,3′-dideoxycytidine (DDC)–treated Arid1a−/− hepatocytes (28), and MCF7 (5) cell lines. GO term analysis of up-regulated genes revealed that, besides specific signatures (fig. S5C), genes involved in the interferon response were shared between all these different models (Fig. 3C and fig. S5D). These data suggest that basal common mechanisms became activated in the absence of Arid1a. To investigate the biological relevance of the up-regulated proinflammatory cytokines, we performed immunohistochemistry analysis using specific immune cells markers. Increased recruitment of CD8+ T cells and IBA1-positive macrophages as well as a widespread accumulation of ISG15 in ARID1A-defective livers was observed after 30 days from tamoxifen administration (Fig. 3D), suggesting that ARID1A loss promotes liver inflammation. This observation suggests that ARID1A-mutant tumors might evolve under the selective pressure of chronically active interferon signaling in a locally inflamed microenvironment. This implies that ARID1A mutations might be already acquired before tumor onset. Accordingly, the analysis we performed using chronically injured nontumoral liver tissues from 44 patients revealed that ARID1A-positive hepatocytes are completely lost in 7 of 44 (15.9%) and severely reduced in 6 of 44 (13.6%) samples (fig. S6).
Fig. 3. ARID1A loss induces interferon response activation.
(A) GO terms enriched between up-regulated genes in Arid1a−/− livers 9 days and 30 days from tamoxifen injection. (B) qPCR analysis of interferon target genes in purified hepatocytes 9 days from tamoxifen injection. (C) Common GO terms enriched in up-regulated genes in Arid1a−/− liver (this work), HCT116 (4), intestinal crypts (15), MCF7 (5), and liver from DDC-treated mice (28). (D) Immunohistochemistry analysis of wild-type and Arid1a−/− liver sections using anti ISG15, anti-CD8, and anti-IBA1 and quantification of CD8+ T cells and IBA1+ macrophages (unpaired t test, two-tailed).
ARID1A loss promotes DNA damage accumulation and cirrhosis
Chronic liver inflammation is a well-known risk factor promoting fibrosis and cirrhosis, ultimately favoring tumor formation (29). To evaluate the long-term consequence of ARID1A loss, we analyzed AlbCre-ERT2Arid1Afl/fl 5 months after tamoxifen injection. Gross examination of ARID1A-defective livers revealed the typical nodular aspect resulting from increased fibrosis (Fig. 4A). Histological examination using hematoxylin and eosin and Picrosirius red staining confirms loss of lobule structure, the presence of thin fibrotic septa, strong inflammation, and cell death (Fig. 4B). Immunohistochemical analysis confirmed that cell identity is preserved in the absence of ARID1A even after 5 months since no HNF4α-negative hepatocytes have been observed (Fig. 4C). Accordingly, with the cirrhotic phenotype observed, IBA1+ macrophages, CD8+ T cells, and B220+ B cells were strongly increased in defective livers, indicating that ARID1A loss promotes a chronic inflammatory state that gradually degenerates in cirrhosis. Notably, a substantial increase of γH2AX-positive cells was also observed together with widespread up-regulation of CDKN1A (Fig. 4C), both markers commonly observed in chronically injured liver (22). To evaluate whether they might be a direct consequence of ARID1A loss, we analyzed liver sections from Arid1a conditional knockout mice 9 days from tamoxifen injection. A significant increase of both γH2AX and CDKN1A levels can be observed throughout the parenchyma, together with scattered KI67-positive proliferating cells (Fig. 5A), suggesting the activation of compensatory proliferation. Increased DNA damage in Arid1a−/− hepatocytes was also confirmed by comet assay (Fig. 5B) and p53BP1 staining (fig. S7A). It has been shown that ARID1A loss promotes tumor formation in different tissues (3, 4, 14). These tumors have a long latency, suggesting that other mutations need to be gradually acquired to fully transform mutant cells. This let us hypothesize that common, slow-acting mechanisms, other than transcriptional alterations, might underlie tumor formation upon ARID1A loss. To investigate whether DNA damage accumulation was a common consequence shared by different tissues, we focused our attention on gastric and small intestinal epithelium by generating Prom1CreERT2Arid1Afl/fl animals and by analyzing VillinCRE/Arid1Afl/fl mice (15), respectively. In Prom1CreERT2Arid1Afl/fl mice, Cre expression can be observed in the stem/progenitor compartment of several different tissues, including gastric epithelium, prostate epithelium, lung, and kidney, among others (30, 31). Using this model, we extended our observation to Arid1A−/− gastric epithelial cells, confirming the increase of CDKN1A and γH2AX levels also in this compartment (Fig. 5C). We further extended these observations to intestinal epithelium confirming the accumulation of CDKN1A- and γH2AX-positive cells also in this tissue (Fig. 5D). Considering these data, it is reasonable to consider DNA damage accumulation a general consequence of ARID1A loss.
Fig. 4. ARID1A loss induces chronic inflammatory disease and DNA damage.
(A) Gross examination of wild-type and Arid1a−/− livers, 5 months from tamoxifen injection. (B) Histological and (C) immunohistochemical analysis of wild-type and Arid1a−/− liver 5 months from tamoxifen injection.
Fig. 5. ARID1A loss promotes DNA damage accumulation.
(A) Immunohistochemical analysis of wild-type and Arid1a−/− liver 9 days from tamoxifen injection using CDKN1A, γH2AX, and KI67 antibodies. Quantifications are shown for three antibodies (unpaired t test, two-tailed). (B) Comet assay performed using wild-type and Arid1a−/− hepatocytes. The quantification of the tail moment is shown (n > 50 per condition. Unpaired t test, two-tailed). AU, arbitrary units. (C) Immunohistochemical analysis of gastric sections from Prom1CreERT2 wild-type and Arid1a−/− mice, 7 days after tamoxifen injection using CDKN1A and γH2AX antibodies. Quantification is shown for the two antibodies (unpaired t test, two-tailed). (D) Immunohistochemical staining of small intestine sections from Arid1Afl/fl (15).
ARID1A-defective cells show nuclear abnormalities and micronuclei accumulation
Loss of ARID1A in established cancer cell lines increases DNA damage by altering multiple cellular processes (8–12, 32). This increases genomic instability, as shown by micronuclei accumulation. To identify these small DNA-containing cytoplasmic structures in vivo, we used known micronuclei markers such as DAPI/LAMIN A/C staining, γH2AX, and H3K27Me3 (33, 34). Arid1a null hepatocytes show profound nuclear envelope malformations, characterized by the presence of deep invaginations, nuclear inclusions, and membrane bending and blebbing (fig. S7, B and C). A significant increase in micronuclei-containing cells can be easily observed both in vivo and in vitro (Fig. 6A and fig. S7, B and D). Accordingly, mitotic defects, such as chromosomal bridges, can be easily scored in ARID1A-defective livers 30 days after a single dose of NR1L3 agonist (Fig. 6B). To verify the integrity of micronuclei membrane, we performed serial block face electron microscopy. Ultrastructural three-dimensional (3D) analysis confirmed the presence of micronuclei-containing cells and revealed the presence of membrane lesions in around 15% of them (Fig. 6C and fig. S7E). Genomic instability can be scored by analyzing the expression of a specific set of genes constituting the so-called CIN70 signature (35). To investigate whether increased tumor predisposition observed in Arid1a−/− mice can be ascribed to increased genomic instability, we interrogated the RNA-seq data (Fig. 1D) by performing gene set enrichment analysis. The CIN70 signature was highly enriched in samples from Arid1a−/− mice (Fig. 6D), further corroborating the role of ARID1A in preserving genome integrity. It has been shown that ARID1A loss increases genomic instability and sensitizes HCT116 to ATR inhibitors (32). Given the increased DNA damage observed in Prom1CreERT2Arid1Afl/fl mice (Fig. 5C), we decided to test the efficacy of 10 μM berzosertib or ceralasertib on patient-derived ARID1A wild-type and mutant human gastric organoids. Forty-eight hours of treatment selectively reduced the viability of ARID1A mutant organoids, confirming the results obtained using cancer cell lines (Fig. 6E). These data strongly suggest that, by increasing DNA damage and genomic instability, ARID1A loss favors the acquisition of additional cooperating mutations leading to tumor formation.
Fig. 6. ARID1A loss promotes micronuclei formation and sensitizes to ATR inhibitors.
(A) Immunohistochemical staining of wild-type and Arid1a−/− liver 9 days from tamoxifen injection using H3K27Me3 and γH2AX antibodies (arrowheads show micronuclei-containing cells) and quantification of micronuclei-containing cells. (B) Representative images of Arid1a-proficient and Arid1a-deficient hepatocytes during anaphase after 30 days from a single NR1L3 agonist injection. (C) 3D reconstruction obtained by 185 serial sections of nuclei (green) and micronuclei (purple) of hepatocyte in 16,650 μm3 of Arid1a−/− liver 9 days from tamoxifen injection. N, nucleus; MN, micronucleus. Two representative images in which the micronuclear envelope lesion (arrowheads) of the micronuclei present in the 3D reconstruction are visible. Scale bars, 1 μm. (D) Gene set enrichment analysis of differentially expressed genes in Fig. 1D performed using the CIN70 signature. (E) Representative images and cell viability of ARID1A wild-type and mutant human gastric cancer organoids treated with 10 μM berzosertib or ceralasertib for 48 hours. DMSO, dimethyl sulfoxide; NES, normalized enrichment score.
Whole-exome sequencing reveals oncogenic mutation cooperating with ARID1A loss
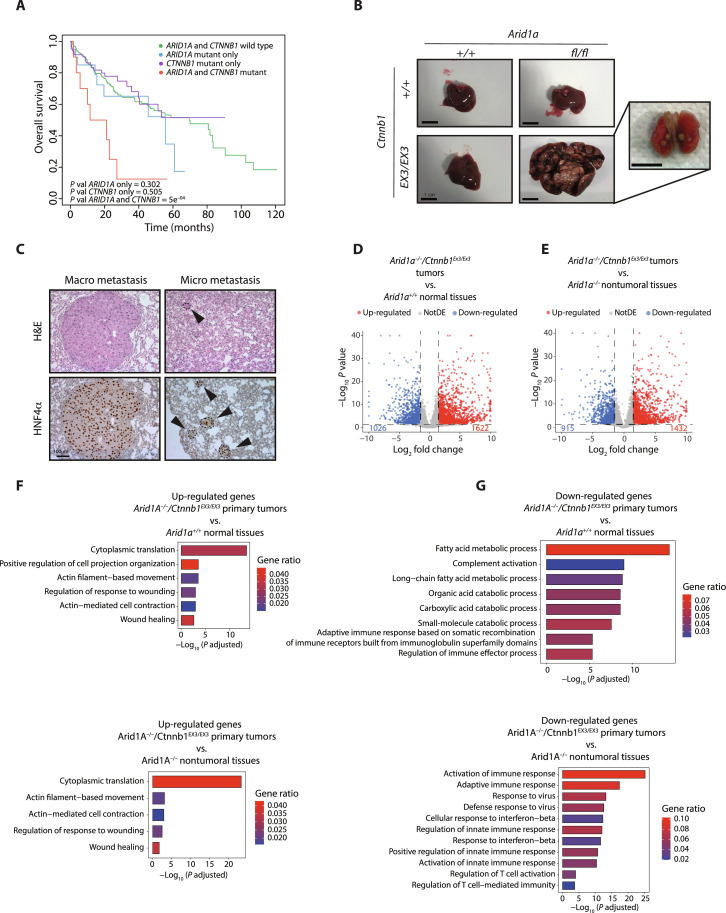
To identify these mutations, we performed whole-exome sequencing analysis of four tumor nodules, each derived from a different Arid1a-deficient animal treated with NR1L3 agonist (Fig. 1B). The nodules have been visually identified, carefully dissected, and used to extract genomic DNA and perform histological analysis. Only one of five nodules isolated has been classified as nontumoral and excluded from the analysis. Copy number variation study identified focally deleted or amplified genomic regions (Fig. 7A and fig. S8A). Single-nucleotide variant (SNV) analysis revealed a number of SNVs per megabase and a classification highly similar to human HCC (Fig. 7B and fig. S8B); a high degree of similarity between murine tumors and human HCCs was also observed by performing a comparative analysis using The Cancer Genome Atlas (TCGA) datasets (Fig. 7C). Notably, the mutational signature we identified in ARID1A-defective murine tumors showed a high degree of similarity with signatures frequently found in human HCCs (Fig. 7D). We also identified several genes recurrently mutated in human HCC (36). In particular, we observed Tert, Myc, and Yap1 amplification, Pten deletion, and Ctnnb1 oncogenic mutation, among others (Fig. 7E). We functionally validated Ctnnb1 mutation (leading to S33A substitution) by performing immunohistochemical analysis. Compared to normal tissues and to a Ctnnb1 wild-type tumor in which CTNNB1 is mainly localized at the cell membrane, cells accumulating nuclear CTNNB1 were clearly visible in tumors harboring S33A mutation (Fig. 7F). Co-occurrence of ARID1A and CTNNB1 mutations was also confirmed in human HCC by TCGA analysis. Concomitant ARID1A mutation confers a worse prognosis in CTNNB1-mutated HCC patients, markedly decreasing the overall survival (Fig. 8A). TCGA analysis also revealed a 2.6-fold increase in the proportion of HCV-derived liver tumors in the ARID1A mutant cohort (fig. S8C) possibly suggesting a selective advantage of mutant clones during HCV infection. To investigate whether Arid1a and Ctnnb1 mutations synergize in vivo to promote aggressive HCC, we decided to cross AlbCre-ERT2Arid1Afl/fl mice with Ctnnb1EX3/EX3 mice, a widely used mouse model allowing the conversion of the endogenous, wild-type, β-catenin into a constitutively stable, oncogenic form (37). Liver-specific overactivation of β-catenin severely affects periportal-specific metabolism, blunting the expression of enzymes involved in the urea cycle and leading to lethal hyperammonemia (38, 39). To overcome this problem and to mimic the stochastic acquisition of Arid1a and Ctnnb1 mutations, we took advantage of the very low leakiness of the AlbCre-ERT2 mice, an approach we recently used successfully (40). At 10 months of age, stochastic (spontaneous) activation of Cre recombinase leads to the formation of multiple liver tumors in AlbCre-ERT2 Arid1Afl/fl Ctnnb1EX3/EX3 mice, while all single-mutant mice appear completely normal and indistinguishable from wild-type animals (Fig. 8B). The tumors showed a trabecular pattern characterized by neoplastic, HNF4α-positive, hepatocytes arranged in two to four thick trabeculae with necrotic areas that can be easily identified. They are characterized by the absence of fibrotic tissue, a strong β-catenin immunoreactivity, the presence of proliferating cells, and the absence of Arid1a expression (fig. S9A). Notably, 20% of these mice showed poorly proliferative micro-and macrometastatic lesions in the lungs (Fig. 8C and fig. S9B), as commonly observed in patients with HCC. To characterize tumor-specific transcriptional programs, we performed RNA-seq from primary tumor samples. The data obtained have been compared with normal wild-type tissues (Fig. 8D) and normal Arid1a−/− tissues (Fig. 8E). In both cases, around 1500 up-regulated and 1000 down-regulated genes have been identified. GO term analysis revealed that genes involved in protein synthesis and cytoskeletal remodeling were enriched between up-regulated genes in Arid1a−/− Ctnnb1ΔEx3 tumors compared with normal wild-type and Arid1a−/− tissues (Fig. 8F and table S1). Genes involved in fatty acid metabolism, complement activation, and catabolic processes were specifically down-regulated in tumors compared to wild-type tissues. Of note, the interferon type I response characterizing ARID1A-defective hepatocytes was blunted in Arid1a−/-Ctnnb1ΔEx3 tumors (Fig. 8G). Together, these data show that ARID1A loss favors the gradual acquisition of additional oncogenic mutations. Among these, β-catenin–activating mutations cooperate with ARID1A loss to promote highly aggressive, metastatic HCC formation.
Fig. 7. ARID1A-defective murine tumors recapitulate aggressive human HCC.
(A) Copy number alterations in an Arid1a−/− defective tumor. (B) SNV classification in Arid1a−/− murine tumors and human HCC from TCGA datasets. (C) Cosine similarity between human HCCs and Arid1a−/− murine tumors. (D) Mutational signature of Arid1a−/− tumors and relative contribution of known single base substitution (SBS) signatures (from COSMIC). (E) Table of representative genomic alterations of Arid1a−/− murine tumors. (F) β-Catenin staining of normal tissue and Ctnnb1 wild-type and Arid1a−/− tumors. mad, median absolute deviation.
Fig. 8. Concomitant Arid1a and Ctnnb1 mutations promote metastatic liver tumors.
(A) Kaplan-Meier curves showing overall survival of patients with HCC harboring ARID1A and CTNNB1 mutations compared with single-mutant tumors. (B) Gross appearance of spontaneously develop tumors and lung metastasis in AlbCre-ERT2Arid1afl/flCtnnb1EX3/EX3 mice at 10 months of age. (C) Histological and immunohistochemical analysis of metastatic lesions in AlbCre-ERT2Arid1afl/flCtnnb1EX3/EX3 mice. (D) Volcano plot showing transcriptional alterations observed by RNA-seq analysis in AlbCre-ERT2Arid1afl/flCtnnb1EX3/EX3 primary tumors compared to wild-type normal tissues. (E) Volcano plot showing transcriptional alterations observed by RNA-seq analysis in AlbCre-ERT2Arid1afl/flCtnnb1EX3/EX3 primary tumor compared to Arid1a−/− nontumoral tissues. (F) Bar plot showing overrepresented biological process (GO terms) between up-regulated and (G) down-regulated genes in the AlbCre-ERT2Arid1afl/flCtnnb1EX3/EX3 primary tumors compared to wild-type normal tissues or Arid1a−/− nontumoral tissues.
DISCUSSION
ARID1A loss-of-function mutations have been identified in a significant number of tumors making it the most commonly mutated subunit within the SWI/SNF complex (1). The role of ARID1A has been predominantly investigated using already established cancer cell lines and only recently by using in vivo mouse models. While in vitro models provided a substantial amount of data, they failed to provide reliable evidence regarding the role of ARID1A mutations in tumor onset. Despite clinical data strongly indicating a tumor suppressor role of ARID1A, in vivo mouse models and 3D organoids provided very heterogeneous results suggesting a high context and tissue-dependent role. Loss of ARID1A promotes liver regeneration (16) but alters small intestine architecture (15). It prevents gastric and intestinal organoid formation (15, 41) and promotes spontaneous formation of invasive colon cancer but prevents oncogene-driven tumor initiation in the liver and colon (4, 7). ARID1A deletion per se is not sufficient to drive gastric cancer formation but cooperates to promote tumor progression in an overactive-Notch gastric adenoma model (6). Similar results have also been obtained in the pancreas, where the loss of ARID1A is able to promote the formation of neoplastic lesions but require concomitant KRAS mutations to evolve in ductal adenocarcinoma (13, 14). Such highly heterogeneous results have been ascribed to the ability of ARID1A-containing BAF complex to regulate transcription by promoting accessibility at enhancers, whose activity is highly context and tissue specific. SWI/SNF chromatin remodeling complexes exert fundamental roles in different cell processes regulating transcription, DNA repair, and cell cycle, among others (3). Such a pleiotropic activity and the possibility of acting redundantly impose multiple layers of complexity. Transcription-related and nonrelated activities of ARD1A-containing cBAF complex have been characterized in the past 10 years. While ARID1A loss has been shown to alter DNA accessibility at enhancers, it also severely compromises the ability of the cells to preserve genome integrity (9–12). While causative in vivo evidence of a tumor-promoting role of deregulated ARID1A-targeted enhancers is still insufficient, the role of ARID1A in preserving genome integrity in vivo has been largely underinvestigated. Therefore, the relative contribution of these two main activities in preserving cell homeostasis remains elusive and is still a matter of intense debate. We found that ARID1A is largely dispensable to preserve hepatocyte-specific chromatin landscape in vivo, mainly due to the redundant activity of the ARID1B-containing cBAF complex. Loss of cBAF activity, resulting from the concomitant loss of both ARID1A/B, completely blunted enhancer activity severely affecting liver-specific gene transcription leading to acute liver failure, similar to what has been observed by abrogating the expression of FOXA pioneer transcription factors (42). Considering also the limited transcriptional alterations observed in knockout cells, we disfavored the hypothesis that the reduced activity of a limited fraction of ARID1A target enhancers could fully account for the protumorigenic effect observed upon its loss. Oncogenic ARID1A mutations have been identified in nontumoral tissues (43–46). This corroborates the idea that ARID1A mutations play a major role in tumor onset. The presence of these clones is usually associated with chronic inflammation, suggesting that ARID1A mutations confer selective advantages under these conditions. Despite their oncogenic potential, they expand without necessarily giving rise to tumors, implying that additional mutations are required to fully transform these cells. We observed that ARID1A loss can already be found in nontumoral tissues from patients with chronic inflammatory liver disease. By using complementary approaches, we also showed that the early transcriptional events following ARID1A loss are mainly related to the activation of a cell-autonomous interferon type I response. We confirmed these data by analyzing published datasets generated from isogenic ARID1A-defective cells in vivo and in vitro (4, 5, 15, 28), demonstrating that interferon response activation is the only transcriptional consequence shared by all the different cell/tissue types analyzed. The activation of this transcriptional program might also justify the identification of ARID1A in several genetic screenings aimed at identifying genes involved in promoting microbial infection or resistance to T cell-mediated cancer cell death (47, 48). Interferon response activation is typically triggered by the activation of pattern recognition receptors (PRRs) a class of sensors able to recognize two main classes of molecules, pathogen-associated molecular patterns and damage-associated molecular patterns. Activation of PRRs is commonly observed upon extensive DNA damage or increased genomic instability, representing one of the underlying mechanisms of antitumor therapies (49). We found that hepatocytes, gastric epithelial cells, and intestinal epithelial cells accumulate DNA damage and up-regulate CDKN1A upon ARID1A loss in line with the transcriptional response we observed. Several works performed in cultured cells provided evidence of a pleiotropic role of ARID1A-containing cBAF complexes in preserving genomic integrity (8–12). Accordingly, together with increased γH2AX-positive cells, we also observed a significant increase of defective mitotic figures and micronucleated hepatocytes, suggesting that its ability to preserve genome integrity could be essential to preserve cell homeostasis in vivo. The formation of micronuclei is a typical sign of increased genomic instability that further sensitize cells to the accumulation of potentially oncogenic mutations upon exogenous or endogenous stimuli. We show that liver-specific Arid1a-knockout mice are prone to developing liver tumors highly resembling human HCCs. Whether these tumors are derived from micronucleated cells or from other ARID1A-defective cells remains to be addressed; however, these data clearly demonstrate the oncosuppressive role of the ARID1A-containing cBAF complex. By performing whole-exome sequencing and by generating a specific genetically modified mouse model, we also characterize the synergistic effect of Arid1a and Ctnnb1 mutations. Constitutively, stable β-catenin and ARID1A loss synergize to form aggressive, metastatic HCCs in mice, in line with a strong reduction in the overall survival of patients with HCC where the two mutations co-occur. These data strongly suggest that ARID1A loss sets the stage for the development of aggressive, possibly metastatic, disease. While we do not formally exclude an oncogenic role of deregulated enhancers, our data strongly indicate that the tumor-promoting activities of Arid1a mutations are mainly related to its role in preserving genome integrity, at least in the tissues we analyzed. These data are also consistent with p53 loss being able to rescue growth defects of ARID1A-deficient organoids (41) and the subtle accumulation of preneoplastic defects observed in gastric mucosa and pancreatic tissues (6, 13, 14). Our data suggest that Arid1a mutations play a dual role in promoting tumor formation, by directly affecting genomic integrity and by indirectly favoring local inflammation. Considering our data, it is reasonable to speculate that, well before tumor onset, the expansion of ARID1A mutant clones might be restrained by the activity of the immune system and/or by a reduced fitness compared with neighboring normal cells. Cancer driver mutations in nontumoral tissues are often associated with inflammatory disorders (50, 51). The selective pressure imposed by the environment might gradually favor the acquisition of mutations able to bypass immunosurveillance and provide growth advantages, especially in the context of chronic inflammation. Despite the molecular mechanism remains to be characterized, it is noteworthy that in Arid1a−/− Ctnnb1ΔEx3 liver tumors, the interferon type I transcriptional program activated in Arid1a−/− nontumoral tissues was completely blunted. The functional contribution of the interferon response to the selection of ARID1A mutant clones is far from being understood and deserves further investigation especially considering the role played in cancer cell dissemination and immune therapy response (52–54). From a therapeutic point of view, our data strongly support the use of ATR inhibitors for the treatment of ARID1A mutant tumors and the possibility of targeting other SWI/SNF subunits (e.g., ARID1B) to promote tumor regression (11, 32, 55). They also suggest that ARID1A mutations might have a prognostic value in the context of liver tumors.
MATERIALS AND METHODS
Mice
Animals have been housed under specific pathogen–free conditions at 22°C with free access to food and water and adequate environmental enrichment. Animals have been checked daily. To induce Cre-dependent recombination, 10- to 12-week-old male animals have been treated with a single intraperitoneal injection tamoxifen (80 mg/kg; Caiman, 13258) dissolved in corn oil (Merck Life Sci., C8267). AlbuminCreERT2 mice (56) (provided by P. Chambon) or Prom1CreERT2 (The Jackson Laboratory, stock no. 017743) have been crossed with Arid1Afl/fl (The Jackson Laboratory, stock no. 027717) and Arid1Bfl/fl (The Jackson Laboratory, stock no. 032061). Ctnnb1ex3/ex3 have been provided by M. Taketo (37). Mice have been identified using ear punch combination and the tissue derived used for polymerase chain reaction (PCR) genotyping. 1,4-Bis-[2-(3,5-dichloropyridyloxy)]benzene (Caiman,14140) has been dissolved in corn oil and administered intraperitoneally at 3 mg/kg. At least three mice per biological replicate have been used. Intestinal samples from Arid1afl/fl and VillinCREArid1Afl/fl have been previously described (15). Experiments involving animals were approved by the University of Trento and the European Institute of Oncology ethical committee, performed in accordance with the Italian Laws (D. lgs. 26/2014), which enforces Dir. 2010/63/EU (Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes), and authorized by the Italian Minister of Health.
Histology, immunohistochemistry, and immunofluorescence
For histological analysis, tissues have been explanted and immediately fixed overnight in 4% formaldehyde at 4°C. They have been dehydrated and paraffin-embedded. Five-micrometer-thick sections have been generated using standard rotary microtome (Leica) and stained using hematoxylin and eosin or Picrosirius red. For immunohistochemistry, we used proteinase K–mediated antigen unmasking or heat-induced antigen unmasking using tris-EDTA (pH 8). Mouse-on-mouse blocking reagent (Abcam, ab269452) or 5% normal goat serum has been used before incubation with primary antibodies. The list of primary and secondary antibodies used has been provided (table S2). Images have been collected using light microscopy (Nikon). Immunofluorescence images were analyzed with a custom Fiji Macro through a nuclei segmentation and a dot analysis. Mean gray value (the sum of the gray values of all the pixels divided by the number of the pixels) is used to graph the results obtained.
Serial block face scanning electron microscopy
Tissues were dissected and fixed using 2.5% glutaraldehyde [Electron Microscopy Sciences (EMS)] and 2% paraformaldehyde (EMS) in sodium cacodylate buffer 0.15 M (pH7.4) (EMS). Liver pieces (1 mm3) have been postfixed for an additional 24 hours at 4°C. The samples were prepared as previously described (57). The blocks were imaged and cut by a Thermo Fisher Scientific–FEI Teneo Volumescope scanning electron microscopy (SEM), operating at an accelerating voltage of 2 kV, a beam current of 50 pA, upon low-vacuum conditions (30 Pa), and with a cutting step of 100 nm. Serial SEM images were acquired by collecting the backscattered electron signal by a dedicated detector, with a magnification of ×3.500, a dwell time of 10 μs per pixel, and an image resolution of 6144 × 4096 pixels, corresponding to a final voxel size of 18.7 nm × 18.7 nm × 100 nm. For 3D reconstruction, rendering, and analysis, serial SEM images were manually segmented using AMIRA software package (FEI Company).
Cell culture
ARID1A-proficient and ARID1A-deficient (Q456*/Q456*) HCT116 cell lines (HD 104-049, parental cell line CCL-247) were purchased from Horizon Discovery Ltd. and maintained according to the manufacturer’s instructions. Human gastric cancer organoids have been previously described (58). They have been treated 48 hours with 10 μM berzosertib (RayBiotech Inc.) or Ceralasertib (ApexBio Tech.). Cell viability has been measured by the trypan blue exclusion assay.
RNA extraction and qPCR analysis
To obtain single primary hepatocytes, we performed in situ collagenase digestion by perfusing liver immediately after mouse suppression. Purified single hepatocytes have been separated by low-speed centrifugation and percoll gradient sedimentation. Total RNA has been extracted using TRIzol (Life Tech., 15596018) coupled with an RNAeasy extraction kit (Qiagen, 74106). One microgram of extracted RNA has been retrotranscribed using random hexamers and the ImProm-II Reverse Transcription System (Promega, A3800). cDNA has been used for quantitative PCR (qPCR) analysis using GoTaq qPCR master mix (Promega, A6002) and specific primer pairs (table S2).
Comet assay
A Comet Assay kit (Abcam, ab238544) was used to quantify DNA damage. Purified hepatocytes were resuspended with ice-cold phosphate-buffered saline at 1 × 105 cells/ml, combined with Comet Agarose at 1:10 ratio, and transferred on the top of Comet Agarose Base Layer onto comet slides. After solidification, comet slides were immersed in the lysis buffer for 45 min at 4°C in the dark. Next, buffer was replaced with cold alkaline solution for 30 min at 4°C in the dark. Slides were transferred in an electrophoresis chamber with freshly prepared alkaline electrophoresis solution, and 19 V was applied. After 15 min, slides were cleaned twice with cold water and fixed with 70% ethanol for 5 min. Following air drying, cells were stained with Vista Green DNA Dye, and images were collected at ×10 magnification. At least 50 Comets for conditions were analyzed by Comet Score 2.0 software (TriTek Corp., Sumerduck, VA). Data were reported as tail moment combining tail length and %DNA in the tail.
ChIP-seq, RNA-seq, and whole-exome sequencing experiments
ChIP-seq analysis has been performed as previously described (59) with the exception that chromatin extracted from Drosophila melanogaster S2 cells (5% of the total) has been spiked in for a more accurate quantification of histone modifications. Briefly, purified hepatocytes were immediately fixed in 1% formaldehyde for 10 min. Cells were washed and resuspended in SDS buffer [50 mM tris (pH 8.1), 0.5% SDS, 100 mM NaCl, 5 mM EDTA, and protease inhibitors]. The pellet was resuspended in immunoprecipitation buffer [100 mM tris (pH 8.6), 0.3% SDS, 1.7% Triton X-100, and 5 mM EDTA] and sonicated to an average length of 500 to 1000 bp. Sonicated chromatin was incubated with a 10-μg primary antibody (see table S2) overnight, and the immunocomplexes recovered using protein A–conjugated Sepharose beads (Life Tech., 101090). After extensive washes, chromatin was de-crosslinked overnight in 0.1 M NaHCO3 and 1% SDS. DNA was purified using a PCR purification kit (Qiagen, 28004) and used for library preparation as previously described (60). For RNA-seq, total RNA was extracted from whole liver tissue and isolated tumors as already described, and libraries were prepared using the TruSeq RNA Sample Preparation Kit (Illumina, 20020595) following the manufacturer’s instructions. At least three mice have been used for each ChIP-seq or RNA-seq analysis. For whole-exome sequencing analysis, tumors have been dissected from freshly collected organs and pulverized in liquid nitrogen, and DNA has been extracted and used to generate library using SureSelect XT–Low Input (Agilent, G9703-90050) and SureSelectXT Mouse All Exon library for postcapture processing (Agilent, #5190-4641). Libraries have been sequenced using Illumina NovaSeq 6000.
RNA-seq data analysis
Bulk paired-end RNA-seq has been performed to profile the transcriptome of the different models. First, the pair-end raw reads were trimmed using fastp v0.20.0 (61) with parameters “-t 1 -A -Q -L.” Afterward, the trimmed reads were aligned with STAR v2.7.3 (62) against the mouse genome (mm10 - GRCm38.p6). Duplicated reads were removed with SAMBLASTER (63). At this point, we quantified the gene expression using featureCounts v1.6.4 (64) with parameters “-s 0 -t exon -p -g gene_name.” The annotation file (gtf) for the counting was downloaded from Gencode (Release M21 - GRCm38.p6). Once obtained the gene expression tables, differential expression analysis was performed with the R package DESeq2 v1.32 (65). Moreover, shrinkage of fold changes for lowly expressed genes was achieved with the apeglm package (66), and P values were false discovery rate–corrected and weighted on the basis of the base mean through the IHW package (67).
The thresholds for significance were set to 0.05 for adjusted P values and 1.5 for the fold change. Last, gene set enrichment analysis was carried out with the R package cluster Profiler (68).
Chip-seq data analysis
For the ChIP-Rx-seq data analysis, we first trimmed the pair-end reads with fastp v0.20.0 (61), setting as parameters “-t 1 -A -Q -L,” Next, the trimmed reads were aligned with bowtie (69) against the mouse genome (mm10 - GRCm38.p6) and against the D. melanogaster genome (dm6 - Release 6 plus ISO1 MT). The parameters used for bowtie are “--chunkmbs 1024 -m 1 --best -S --no-unal -q -I 10 -X 1000.” From the two generated bam files, we removed the ambiguous reads, i.e., those aligning on both the genomes. The bam files from the replicates (N = 2) were merged with SAMtools. At this point, peak calling was performed with MACS2 v2.2.7.1 in narrow mode (70), setting the parameters to “--keep-dup all -m 3 30 --format BAMPE --pvalue 1e-05.” The called peaks were further filtered with a P value threshold of 1 × 10−10 and annotated using the package ChIPseeker v1.28.3 (71), with a range to define a promoter peak of ±−2.5 kb. Peaks found on blacklisted regions (mm10-blacklist.v2) and over noncanonical chromosomes were excluded. To generate the BigWig files, bamCoverage from deepTools v3.5.1 (72) was used with parameters “--binSize 50 --extendReads.” To calculate scaling factors, Trimmed Mean of M values (TMM) normalization (73) was applied on the count matrix of called peaks (shared between conditions). Next, the bamCoverage parameter “--scaleFactor” was set equal to 106/(TMM_NormFactor × LibSize) for the BigWig files creation. Heatmaps were generated with “computeMatrix reference-point -a 5000 -b 5000 -binSize 20” and plotHeatmap with custom settings (“ --sortUsing max”). Last, regions’ intensity values for the boxplots were retrieved from the BigWigs through the deepTools function multiBigWigSummary in BED file mode. To split the enhancers in quantiles, we computed the log2 ratio of H3K27Ac levels between Arid1a−/− and Arid1a+/+ samples. The Mann-Whitney U nonparametric test was used for the boxplot comparisons. Effect sizes were also calculated (wilcox_effsize from rstatix).
Enhancer-target gene prediction
We sought to predict and annotate the enhancers’ target genes. To do so, we took advantage of mouse liver deoxyribonuclease sequencing (DNase-seq) data from the ENCODE consortium (code ENCSR000CNI). Specifically, we downloaded the bam files for all the replicates and called the peaks using MACS2 with default parameters. Sample ENCFF455NQG was excluded due to its extremely low read depth. Next, we applied the ABC model v0.2.2 from the Broad Institute (27), providing as input the DNase-seq peak files, the H3K27Ac WT bam file (this work), and the gene expression matrix (this work). For the calculation of the ABC score, we set the parameter “--score_column powerlaw.Score.” Given the enhancer-target gene output table, we filtered out enhancers with low activity score (<2.5) and those within ±−2.5 kb from the transcription start site of an annotated gene (TxDb.Mmusculus.UCSC.mm10.knownGene v3.10.0). Last, the UpSet plot was generated with the R package ComplexHeatmap v2.14.0 (74).
TCGA analyses
For the survival analysis, we downloaded the data from cBioPortal (https://cbioportal.org) (69, 70). Specifically, we selected the Liver Hepatocellular Carcinoma dataset (TCGA, Firehose Legacy) and divided the samples into those with mutations in both ARID1A and CTNNB1 and those without. The R package “survival” was used for the analysis. We introduced two covariates in the Cox proportional hazard regression model, namely, age and sex. cBioPortal was also used for the gene’s mutually exclusivity analysis of ARID1A at the pan-cancer level (TCGA, Firehose Legacy).
Whole-exome sequencing data analysis
Whole-exome sequencing data were profiled for somatic point mutations and copy number status. First, the raw reads were trimmed with Trimmomatic v0.32 (75). Second, bwa v0.7.12 was used for read alignment (76). Third, SAM files were merged with Picard v2.17.4 (Broad Institute) and converted to sorted BAM files with SAMtools (77). Fourth, duplicated reads were removed with the Picard function MarkDuplicates, followed by realignment of indels and base quality score recalibration with GATK tools v3.1 (78). MuTect v1.1.7 (79) was used to detect SNVs, which were annotated with SnpEff 5.0 (80). DNA copy number analysis was performed with the R package CopywriteR (81) version 2.10.0 and CNVKit (82) 0.9.10 with the default parameters.
Mutational pattern analysis
To study the prevalence of annotated signatures in our whole-exome sequencing mouse data, we used the R package MutationalPattern (83). As reference signatures, we selected the single base substitution signatures from COSMIC (v3.2, March 2021). The mouse mutational profiles were compared to the Liver Hepatocellular Carcinoma dataset (TCGA, Firehose Legacy) by using the cosine similarity.
Acknowledgments
We would like to acknowledge IEO Genomic unit and animal facility and CIBIO animal facility for providing invaluable support; M. Roccuzzo and CIBIO imaging facility for technical assistance and support; S. Rodighiero for technical assistance in preparing and acquiring SBFSEM images; G. Natoli, A. Lunardi, F. Demichelis, L. Fava, S. Campaner, and members of Pasini and Demichelis labs for helpful discussion; and A. Lunardi and M. Simonatto for critically reading the manuscript. We thank P. Chambon for providing AlbCreERT2 mice. We dedicate this work to the memory of V. Graziani and to her family.
Funding: The work of the Chiacchiera Laboratory is supported by the Italian Association for Cancer Research, AIRC (MFAG-20344), by the Worldwide Cancer Research, WWCR, (grant no. 23-0321) and by the European Union under NextGenerationEU. PRIN 2022 (prot. no. 2022PWKZXE). A.D.A. and E.F. were supported by the Veronica Graziani AIRC fellowship for Italy. The work in Pasini Lab was supported by AIRC (IG-2017-20290) and by European Research Council (EC-H2020-ERC-CoG-DissectPcG:725268). Department CIBIO Core Facilities are supported by the European Regional Development Fund (ERDF) 2014–2020.
Author contributions: A.D.A., E.F., E.S., P.M., S.C., and G.F. performed experiments; D.B., F.R., C.B., and A.R. performed bioinformatic analysis; E.V. and A.F. performed SBF SEM experiments; S.B. provided support to the experimental work; F.Ca., C.S., T.D., G.B., and M.B. assembled tissue microarrays from human samples and performed histopathology and immunohistochemical analysis on human samples. A.D.A., E.F., P.M., S.C., D.B., F.R., C.B., A.F., E.V., A.R., and F.Ch. analyzed the data. S.C. provided gastric organoids. A.F. provided Arid1afl/fl and VillinCre Arid1afl/fl intestinal samples. D.P. supervised the work of P.M., F.R., and C.B. and provide invaluable scientific support; F.Ch. conceived the project and all the experiments; F.Ch. and D.B. conceived all the analysis; F.Ch., D.B., and E.F. wrote the manuscript and edited the figures.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq and ChIP-seq data of this work have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO series accession number GSE212970. Whole-exome sequencing data have been deposited in NCBI’s BioProject under accession number PRJNA879319. Published raw expression data were retrieved and reanalyzed from GSE71511 (HCT116), GSE124227 (MCF7), GSE111501 (liver DDC), and GSE110181 (intestinal crypts). Raw ChIP-seq data were reanalyzed from GSE65167 (ARID1A, C/EBPα) and ENCSR000CBU (CTCF). Further inquiries and requests for reagents should be directed to the corresponding author: Fulvio Chiacchiera (Fulvio.chiacchiera@unitn.it).
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Legends for tables S1 and S2
Other Supplementary Material for this manuscript includes the following:
Tables S1 and S2
REFERENCES AND NOTES
- 1.Centore R. C., Sandoval G. J., Soares L. M. M., Kadoch C., Chan H. M., Mammalian SWI/SNF chromatin remodeling complexes: Emerging mechanisms and therapeutic strategies. Trends Genet. 36, 936–950 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Cenik B. K., Shilatifard A., COMPASS and SWI/SNF complexes in development and disease. Nat. Rev. Genet. 22, 38–58 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Mittal P., Roberts C. W. M., The SWI/SNF complex in cancer - biology, biomarkers and therapy. Nat. Rev. Clin. Oncol. 17, 435–448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathur R., Alver B. H., San Roman A. K., Wilson B. G., Wang X., Agoston A. T., Park P. J., Shivdasani R. A., Roberts C. W. M., ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat. Genet. 49, 296–302 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu G., Chhangawala S., Cocco E., Razavi P., Cai Y., Otto J. E., Ferrando L., Selenica P., Ladewig E., Chan C., da Cruz Paula A., Witkin M., Cheng Y., Park J., Serna-Tamayo C., Zhao H. Y., Wu F., Sallaku M., Qu X., Zhao A., Collings C. K., D’Avino A. R., Jhaveri K., Koche R., Levine R. L., Reis-Filho J. S., Kadoch C., Scaltriti M., Leslie C. S., Baselga J., Toska E., ARID1A determines luminal identity and therapeutic response in estrogen-receptor-positive breast cancer. Nat. Genet. 52, 198–207 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loe A. K. H., Francis R., Seo J., du L., Wang Y., Kim J. E., Hakim S. W., Kim J. E., He H. H., Guo H., Kim T. H., Uncovering the dosage-dependent roles of Arid1a in gastric tumorigenesis for combinatorial drug therapy. J. Exp. Med. 218, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun X., Wang S. C., Wei Y., Luo X., Jia Y., Li L., Gopal P., Zhu M., Nassour I., Chuang J.-C., Maples T., Celen C., Nguyen L. H., Wu L., Fu S., Li W., Hui L., Tian F., Ji Y., Zhang S., Sorouri M., Hwang T. H., Letzig L., James L., Wang Z., Yopp A. C., Singal A. G., Zhu H., Arid1a has context-dependent oncogenic and tumor suppressor functions in liver cancer. Cancer Cell 32, 574–589 e576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayona-Feliu A., Barroso S., Munoz S., Aguilera A., The SWI/SNF chromatin remodeling complex helps resolve R-loop-mediated transcription-replication conflicts. Nat. Genet. 53, 1050–1063 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Dykhuizen E. C., Hargreaves D. C., Miller E. L., Cui K., Korshunov A., Kool M., Pfister S., Cho Y. J., Zhao K., Crabtree G. R., BAF complexes facilitate decatenation of DNA by topoisomerase IIα. Nature 497, 624–627 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen J., Ju Z., Zhao W., Wang L., Peng Y., Ge Z., Nagel Z. D., Zou J., Wang C., Kapoor P., Ma X., Ma D., Liang J., Song S., Liu J., Samson L. D., Ajani J. A., Li G. M., Liang H., Shen X., Mills G. B., Peng G., ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat. Med. 24, 556–562 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen J., Peng Y., Wei L., Zhang W., Yang L., Lan L., Kapoor P., Ju Z., Mo Q., Shih I. M., Uray I. P., Wu X., Brown P. H., Shen X., Mills G. B., Peng G., ARID1A deficiency impairs the DNA damage checkpoint and sensitizes cells to PARP inhibitors. Cancer Discov. 5, 752–767 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao B., Lin J., Rong L., Wu S., Deng Z., Fatkhutdinov N., Zundell J., Fukumoto T., Liu Q., Kossenkov A., Jean S., Cadungog M. G., Borowsky M. E., Drapkin R., Lieberman P. M., Abate-Shen C. T., Zhang R., ARID1A promotes genomic stability through protecting telomere cohesion. Nat. Commun. 10, 4067 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura Y., Fukuda A., Ogawa S., Maruno T., Takada Y., Tsuda M., Hiramatsu Y., Araki O., Nagao M., Yoshikawa T., Ikuta K., Yoshioka T., Wang Z., Akiyama H., Wright C. V., Takaori K., Uemoto S., Chiba T., Seno H., ARID1A maintains differentiation of pancreatic ductal cells and inhibits development of pancreatic ductal adenocarcinoma in mice. Gastroenterology 155, 194–209 e192 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Wang S. C., Nassour I., Xiao S., Zhang S., Luo X., Lee J., Li L., Sun X., Nguyen L. H., Chuang J. C., Peng L., Daigle S., Shen J., Zhu H., SWI/SNF component ARID1A restrains pancreatic neoplasia formation. Gut 68, 1259–1270 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiramatsu Y., Fukuda A., Ogawa S., Goto N., Ikuta K., Tsuda M., Matsumoto Y., Kimura Y., Yoshioka T., Takada Y., Maruno T., Hanyu Y., Tsuruyama T., Wang Z., Akiyama H., Takaishi S., Miyoshi H., Taketo M. M., Chiba T., Seno H., Arid1a is essential for intestinal stem cells through Sox9 regulation. Proc. Natl. Acad. Sci. U.S.A. 116, 1704–1713 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun X., Chuang J. C., Kanchwala M., Wu L., Celen C., Li L., Liang H., Zhang S., Maples T., Nguyen L. H., Wang S. C., Signer R. A. J., Sorouri M., Nassour I., Liu X., Xu J., Wu M., Zhao Y., Kuo Y. C., Wang Z., Xing C., Zhu H., Suppression of the SWI/SNF component arid1a promotes mammalian regeneration. Cell Stem Cell 18, 456–466 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friemel J., Frick L., Unger K., Egger M., Parrotta R., Böge Y. T., Adili A., Karin M., Luedde T., Heikenwalder M., Weber A., Characterization of HCC mouse models: Towards an etiology-oriented subtyping approach. Mol. Cancer Res. 17, 1493–1502 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Brown Z. J., Heinrich B., Greten T. F., Mouse models of hepatocellular carcinoma: An overview and highlights for immunotherapy research. Nat. Rev. Gastroenterol. Hepatol. 15, 536–554 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Dow M., Pyke R. M., Tsui B. Y., Alexandrov L. B., Nakagawa H., Taniguchi K., Seki E., Harismendy O., Shalapour S., Karin M., Carter H., Font-Burgada J., Integrative genomic analysis of mouse and human hepatocellular carcinoma. Proc. Natl. Acad. Sci. U.S.A. 115, E9879–E9888 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong B., Lee J. S., Park Y. Y., Yang F., Xu G., Huang W., Finegold M. J., Moore D. D., Activating CAR and β-catenin induces uncontrolled liver growth and tumorigenesis. Nat. Commun. 6, 5944 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Govaere O., Komuta M., Berkers J., Spee B., Janssen C., de Luca F., Katoonizadeh A., Wouters J., van Kempen L. C., Durnez A., Verslype C., de Kock J., Rogiers V., van Grunsven L. A., Topal B., Pirenne J., Vankelecom H., Nevens F., van den Oord J., Pinzani M., Roskams T., Keratin 19: A key role player in the invasion of human hepatocellular carcinomas. Gut 63, 674–685 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raven A., Lu W. Y., Man T. Y., Ferreira-Gonzalez S., O’Duibhir E., Dwyer B. J., Thomson J. P., Meehan R. R., Bogorad R., Koteliansky V., Kotelevtsev Y., ffrench-Constant C., Boulter L., Forbes S. J., Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature 547, 350–354 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michel B. C., D’Avino A. R., Cassel S. H., Mashtalir N., McKenzie Z. M., McBride M. J., Valencia A. M., Zhou Q., Bocker M., Soares L. M. M., Pan J., Remillard D. I., Lareau C. A., Zullow H. J., Fortoul N., Gray N. S., Bradner J. E., Chan H. M., Kadoch C., A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat. Cell Biol. 20, 1410–1420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z., Chivu A. G., Choate L. A., Rice E. J., Miller D. C., Chu T., Chou S. P., Kingsley N. B., Petersen J. L., Finno C. J., Bellone R. R., Antczak D. F., Lis J. T., Danko C. G., Prediction of histone post-translational modification patterns based on nascent transcription data. Nat. Genet. 54, 295–305 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karagianni P., Moulos P., Schmidt D., Odom D. T., Talianidis I., Bookmarking by Non-pioneer transcription factors during liver development establishes competence for future gene activation. Cell Rep 30, 1319–1328.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berasain C., Arechederra M., Argemi J., Fernandez-Barrena M. G., Avila M. A., Loss of liver function in chronic liver disease: An identity crisis. J. Hepatol. 78, 401–414 (2023). [DOI] [PubMed] [Google Scholar]
- 27.Fulco C. P., Nasser J., Jones T. R., Munson G., Bergman D. T., Subramanian V., Grossman S. R., Anyoha R., Doughty B. R., Patwardhan T. A., Nguyen T. H., Kane M., Perez E. M., Durand N. C., Lareau C. A., Stamenova E. K., Aiden E. L., Lander E. S., Engreitz J. M., Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 51, 1664–1669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W., Yang L., He Q., Hu C., Zhu L., Ma X., Ma X., Bao S., Li L., Chen Y., Deng X., Zhang X., Cen J., Zhang L., Wang Z., Xie W.-F., Li H., Li Y., Hui L., A homeostatic Arid1a-dependent permissive chromatin state licenses hepatocyte responsiveness to liver-injury-associated YAP signaling. Cell Stem Cell 25, 54–68.e55 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Ramadori P., Kam S., Heikenwalder M., T cells: Friends and foes in NASH pathogenesis and hepatocarcinogenesis. Hepatology 75, 1038–1049 (2022). [DOI] [PubMed] [Google Scholar]
- 30.Zhu L., Gibson P., Currle D. S., Tong Y., Richardson R. J., Bayazitov I. T., Poppleton H., Zakharenko S., Ellison D. W., Gilbertson R. J., Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature 457, 603–607 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu L., Finkelstein D., Gao C., Shi L., Wang Y., López-Terrada D., Wang K., Utley S., Pounds S., Neale G., Ellison D., Onar-Thomas A., Gilbertson R. J., Multi-organ mapping of cancer risk. Cell 166, 1132–1146 e1137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williamson C. T., Miller R., Pemberton H. N., Jones S. E., Campbell J., Konde A., Badham N., Rafiq R., Brough R., Gulati A., Ryan C. J., Francis J., Vermulen P. B., Reynolds A. R., Reaper P. M., Pollard J. R., Ashworth A., Lord C. J., ATR inhibitors as a synthetic lethal therapy for tumours deficient in ARID1A. Nat. Commun. 7, 13837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crasta K., Ganem N. J., Dagher R., Lantermann A. B., Ivanova E. V., Pan Y., Nezi L., Protopopov A., Chowdhury D., Pellman D., DNA breaks and chromosome pulverization from errors in mitosis. Nature 482, 53–58 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agustinus A. S., al-Rawi D., Dameracharla B., Raviram R., Jones B. S. C. L., Stransky S., Scipioni L., Luebeck J., di Bona M., Norkunaite D., Myers R. M., Duran M., Choi S., Weigelt B., Yomtoubian S., McPherson A., Toufektchan E., Keuper K., Mischel P. S., Mittal V., Shah S. P., Maciejowski J., Storchova Z., Gratton E., Ly P., Landau D., Bakhoum M. F., Koche R. P., Sidoli S., Bafna V., David Y., Bakhoum S. F., Epigenetic dysregulation from chromosomal transit in micronuclei. Nature 619, 176–183 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carter S. L., Eklund A. C., Kohane I. S., Harris L. N., Szallasi Z., A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 38, 1043–1048 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Research Network. Electronic address: wheeler@bcm.edu, Cancer Genome Atlas Research Network , Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 169, 1327–1341 e1323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harada N., Tamai Y., Ishikawa T., Sauer B., Takaku K., Oshima M., Taketo M. M., Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J. 18, 5931–5942 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benhamouche S., Decaens T., Godard C., Chambrey R., Rickman D. S., Moinard C., Vasseur-Cognet M., Kuo C. J., Kahn A., Perret C., Colnot S., Apc tumor suppressor gene is the "zonation-keeper" of mouse liver. Dev. Cell 10, 759–770 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Harada N., Miyoshi H., Murai N., Oshima H., Tamai Y., Oshima M., Taketo M. M., Lack of tumorigenesis in the mouse liver after adenovirus-mediated expression of a dominant stable mutant of beta-catenin. Cancer Res. 62, 1971–1977 (2002). [PubMed] [Google Scholar]
- 40.Bisso A., Filipuzzi M., Gamarra Figueroa G. P., Brumana G., Biagioni F., Doni M., Ceccotti G., Tanaskovic N., Morelli M. J., Pendino V., Chiacchiera F., Pasini D., Olivero D., Campaner S., Sabò A., Amati B., Cooperation between MYC and β-Catenin in liver tumorigenesis requires Yap/Taz. Hepatology 72, 1430–1443 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Lo Y. H., Kolahi K. S., du Y., Chang C. Y., Krokhotin A., Nair A., Sobba W. D., Karlsson K., Jones S. J., Longacre T. A., Mah A. T., Tercan B., Sockell A., Xu H., Seoane J. A., Chen J., Shmulevich I., Weissman J. S., Curtis C., Califano A., Fu H., Crabtree G. R., Kuo C. J., A CRISPR/Cas9-engineered ARID1A-deficient human gastric cancer organoid model reveals essential and nonessential modes of oncogenic transformation. Cancer Discov. 11, 1562–1581 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reizel Y., Morgan A., Gao L., Lan Y., Manduchi E., Waite E. L., Wang A. W., Wells A., Kaestner K. H., Collapse of the hepatic gene regulatory network in the absence of FoxA factors. Genes Dev. 34, 1039–1050 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim S. K., Takeda H., Takai A., Matsumoto T., Kakiuchi N., Yokoyama A., Yoshida K., Kaido T., Uemoto S., Minamiguchi S., Haga H., Shiraishi Y., Miyano S., Seno H., Ogawa S., Marusawa H., Comprehensive analysis of genetic aberrations linked to tumorigenesis in regenerative nodules of liver cirrhosis. J. Gastroenterol. 54, 628–640 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Ross-Innes C. S., Becq J., Warren A., Cheetham R. K., Northen H., O'Donovan M., Malhotra S., di Pietro M., Ivakhno S., He M., Weaver J. M. J., Lynch A. G., Kingsbury Z., Ross M., Humphray S., Bentley D., Fitzgerald R. C., for the Oesophageal Cancer Clinical and Molecular Stratification (OCCAMS) Study Group, Hayes S. J., Ang Y., Welch I., Preston S., Oakes S., Save V., Skipworth R., Tucker O., Davies J., Crichton C., Schusterreiter C., Underwood T., Noble F., Stacey B., Kelly J., Byrne J., Haydon A., Sharland D., Owsley J., Barr H., Lagergren J., Gossage J., Davies A., Mason R., Chang F., Zylstra J., Sanders G., Wheatley T., Berrisford R., Bracey T., Harden C., Bunting D., Roques T., Nobes J., Loo S., Lewis M., Cheong E., Priest O., Parsons S. L., Soomro I., Kaye P., Saunders J., Pang V., Welch N. T., Catton J. A., Duffy J. P., Ragunath K., Lovat L., Haidry R., Miah H., Kerr S., Eneh V., Butawan R., Roques T., Lewis M., Cheong E., Kumar B., Igali L., Walton S., Dann A., Safranek P., Hindmarsh A., Sudjendran V., Scott M., Cluroe A., Miremadi A., Mahler-Araujo B., Nutzinger B., Peters C., Abdullahi Z., Crawte J., MacRae S., Noorani A., Elliott R. F., Bower L., Edwards P., Tavare S., Eldridge M., Bornschein J., Secrier M., Yang T. P., O'Neill J. R., Adamczuk K., Lao-Sirieix P., Grehan N., Smith L., Lishman S., Beardsmore D., Dawson S., Whole-genome sequencing provides new insights into the clonal architecture of Barrett's esophagus and esophageal adenocarcinoma. Nat. Genet. 47, 1038–1046 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimizu T., Marusawa H., Matsumoto Y., Inuzuka T., Ikeda A., Fujii Y., Minamiguchi S., Miyamoto S.’i., Kou T., Sakai Y., Crabtree J. E., Chiba T., Accumulation of somatic mutations in TP53 in gastric epithelium with Helicobacter pylori infection. Gastroenterology 147, 407–417.e3 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Olafsson S., Intyre R. E. M., Coorens T., Butler T., Jung H., Robinson P. S., Lee-Six H., Sanders M. A., Arestang K., Dawson C., Tripathi M., Strongili K., Hooks Y., Stratton M. R., Parkes M., Martincorena I., Raine T., Campbell P. J., Anderson C. A., Somatic evolution in Non-neoplastic IBD-affected colon. Cell 182, 672–684.e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan D., Kobayashi A., Jiang P., Ferrari de Andrade L., Tay R. E., Luoma A. M., Tsoucas D., Qiu X., Lim K., Rao P., Long H. W., Yuan G. C., Doench J., Brown M., Liu X. S., Wucherpfennig K. W., A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 359, 770–775 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ding S., Diep J., Feng N., Ren L., Li B., Ooi Y. S., Wang X., Brulois K. F., Yasukawa L. L., Li X., Kuo C. J., Solomon D. A., Carette J. E., Greenberg H. B., STAG2 deficiency induces interferon responses via cGAS-STING pathway and restricts virus infection. Nat. Commun. 9, 1485 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Motwani M., Pesiridis S., Fitzgerald K. A., DNA sensing by the cGAS-STING pathway in health and disease. Nat. Rev. Genet. 20, 657–674 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Acha-Sagredo A., Ganguli P., Ciccarelli F. D., Somatic variation in normal tissues: Friend or foe of cancer early detection? Ann. Oncol. 33, 1239–1249 (2022). [DOI] [PubMed] [Google Scholar]
- 51.Herms A., Jones P. H., Somatic mutations in normal tissues: New perspectives on early carcinogenesis. Annu. Rev. Cancer Biol. 7, 189–205 (2023). [Google Scholar]
- 52.Larson R. C., Kann M. C., Bailey S. R., Haradhvala N. J., Llopis P. M., Bouffard A. A., Scarfó I., Leick M. B., Grauwet K., Berger T. R., Stewart K., Anekal P. V., Jan M., Joung J., Schmidts A., Ouspenskaia T., Law T., Regev A., Getz G., Maus M. V., CAR T cell killing requires the IFNγR pathway in solid but not liquid tumours. Nature 604, 563–570 (2022). [DOI] [PubMed] [Google Scholar]
- 53.Bakhoum S. F., Ngo B., Laughney A. M., Cavallo J. A., Murphy C. J., Ly P., Shah P., Sriram R. K., Watkins T. B. K., Taunk N. K., Duran M., Pauli C., Shaw C., Chadalavada K., Rajasekhar V. K., Genovese G., Venkatesan S., Birkbak N. J., McGranahan N., Lundquist M., LaPlant Q., Healey J. H., Elemento O., Chung C. H., Lee N. Y., Imielenski M., Nanjangud G., Pe’er D., Cleveland D. W., Powell S. N., Lammerding J., Swanton C., Cantley L. C., Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma P., Hu-Lieskovan S., Wargo J. A., Ribas A., Primary, Adaptive, and Acquired resistance to cancer immunotherapy. Cell 168, 707–723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Helming K. C., Wang X., Wilson B. G., Vazquez F., Haswell J. R., Manchester H. E., Kim Y., Kryukov G. V., Ghandi M., Aguirre A. J., Jagani Z., Wang Z., Garraway L. A., Hahn W. C., Roberts C. W. M., ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat. Med. 20, 251–254 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schuler M., Dierich A., Chambon P., Metzger D., Efficient temporally controlled targeted somatic mutagenesis in hepatocytes of the mouse. Genesis 39, 167–172 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Birolini G., Valenza M., di Paolo E., Vezzoli E., Talpo F., Maniezzi C., Caccia C., Leoni V., Taroni F., Bocchi V. D., Conforti P., Sogne E., Petricca L., Cariulo C., Verani M., Caricasole A., Falqui A., Biella G., Cattaneo E., Striatal infusion of cholesterol promotes dose-dependent behavioral benefits and exerts disease-modifying effects in Huntington's disease mice. EMBO Mol. Med. 12, e12519 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Corso S., Isella C., Bellomo S. E., Apicella M., Durando S., Migliore C., Ughetto S., D’Errico L., Menegon S., Moya-Rull D., Cargnelutti M., Capelôa T., Conticelli D., Giordano J., Venesio T., Balsamo A., Marchiò C., Degiuli M., Reddavid R., Fumagalli U., de Pascale S., Sgroi G., Rausa E., Baiocchi G. L., Molfino S., Pietrantonio F., Morano F., Siena S., Sartore-Bianchi A., Bencivenga M., Mengardo V., Rosati R., Marrelli D., Morgagni P., Rausei S., Pallabazzer G., de Simone M., Ribero D., Marsoni S., Sottile A., Medico E., Cassoni P., Sapino A., Pectasides E., Thorner A. R., Nag A., Drinan S. D., Wollison B. M., Bass A. J., Giordano S., A comprehensive PDX gastric cancer collection captures cancer cell-intrinsic transcriptional MSI traits. Cancer Res. 79, 5884–5896 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Pivetti S., Fernandez-Perez D., D’Ambrosio A., Barbieri C. M., Manganaro D., Rossi A., Barnabei L., Zanotti M., Scelfo A., Chiacchiera F., Pasini D., Loss of PRC1 activity in different stem cell compartments activates a common transcriptional program with cell type-dependent outcomes. Sci Adv 5, eaav1594 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blecher-Gonen R., Barnett-Itzhaki Z., Jaitin D., Amann-Zalcenstein D., Lara-Astiaso D., Amit I., High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat. Protoc. 8, 539–554 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Chen S., Zhou Y., Chen Y., Gu J., fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Faust G. G., Hall I. M., SAMBLASTER: Fast duplicate marking and structural variant read extraction. Bioinformatics 30, 2503–2505 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu A., Ibrahim J. G., Love M. I., Heavy-tailed prior distributions for sequence count data: Removing the noise and preserving large differences. Bioinformatics 35, 2084–2092 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ignatiadis N., Klaus B., Zaugg J. B., Huber W., Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat. Methods 13, 577–580 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., Fu X., Liu S., Bo X., Yu G., clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu G., Wang L. G., He Q. Y., ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
- 72.Ramirez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dündar F., Manke T., deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gu Z., Eils R., Schlesner M., Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 75.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup , The sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M. A., The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cibulskis K., Lawrence M. S., Carter S. L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E. S., Getz G., Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., Wang L., Land S. J., Lu X., Ruden D. M., A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kuilman T., Velds A., Kemper K., Ranzani M., Bombardelli L., Hoogstraat M., Nevedomskaya E., Xu G., de Ruiter J., Lolkema M. P., Ylstra B., Jonkers J., Rottenberg S., Wessels L. F., Adams D. J., Peeper D. S., Krijgsman O., CopywriteR: DNA copy number detection from off-target sequence data. Genome Biol. 16, 49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Talevich E., Shain A. H., Botton T., Bastian B. C., CNVkit: Genome-wide copy number detection and visualization from targeted DNA sequencing. PLOS Comput. Biol. 12, e1004873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Blokzijl F., Janssen R., van Boxtel R., Cuppen E., MutationalPatterns: Comprehensive genome-wide analysis of mutational processes. Genome Med. 10, 33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Legends for tables S1 and S2
Tables S1 and S2