Dear Editor,
In 2020, kidney cancer is the 14th most common malignancy with an estimated 431 288 new cases globally, according to the Global Cancer Observatory1. Currently, prognostic studies on renal cell carcinoma are conducted extensively. Newly developed prognostic models by Wang et al.2 at the International Journal of Surgery significantly increase our confidence in clinical decisions and offer practical assistance in the management of patients with renal cell carcinoma. However, the etiologic research on renal neoplasms including benign tumors is limited. It is reported that gut microbiota could be a preventive or risk factor for cancers, especially for gastrointestinal malignancies3. Thus, whether intestinal microbiota as a potentially modifiable factor is causally associated with renal neoplasms still needs further investigation.
Theoretically, randomized controlled trial (RCT) is regarded as the greatest approach to establishing a causal link between intestinal microbiota and renal neoplasms. Nevertheless, it is not pragmatic to determine the causal effect of gut microbiota on renal neoplasms using an RCT. Mendelian randomization (MR) is an epidemiological approach in which the exposures and outcomes are replaced by single nucleotide polymorphisms (SNPs) and corresponding instrumental variables (IVs). This method has been widely applied for causal inferences to avoid confounding and reverse causal biases originating from small sample sizes and cross-sectional design4. Here, we performed this two-sample MR analysis to determine the causal links between gut microbiota and renal neoplasms based on the summary statistics of a genome-wide association study (GWAS).
Summary statistics of gut microbiota in humans were retrieved from the MiBioGen consortium, in which 18 340 individuals from 24 cohorts were included. Meanwhile, summary statistics of cases with renal neoplasms (malignant and benign) were retrieved from FinnGen (https://r7.finngen.fi/). For benign neoplasm of the kidney, 581 cases and 308 573 controls were included. For malignant neoplasm of the kidney, 1631 cases and 307 523 controls were enrolled. All participants were of European descent. Of note, cancer cases (in other organs of any type) in the controls were all removed in the sensitivity analyses for further verification. Finally, 248 356 controls for benign renal tumor and 238 678 controls for renal malignancy were enrolled in the sensitivity analyses. Six methods were applied to investigate the effects of gut microbiota on kidney neoplasms, including inverse variance weighted (IVW), maximum likelihood (ML), weighted median, MR-Egger, pleiotropy residual sum and outlier (MR-PRESSO), and robust adjusted profile score (MR.RAPS). All IVs were assumed to be valid IVs in IVW models and then were merged using meta-technique. Cochran Q was selected to explore the heterogeneity of adopted IVs, and P-value <0.05 represented the existence of heterogeneity, and the random-effects IVW method was applied. MR-Egger and MR-PRESSO global tests were utilized to detect the horizontal pleiotropy. The SNPs strength was quantified by F-statistics of bacterial taxon5. The F-statistic greater than 10 indicated the existence of bias. All statistical analyses in this study were conducted using R 4.0.3 (R Foundation for Statistical Computing, Vienna, Austria).
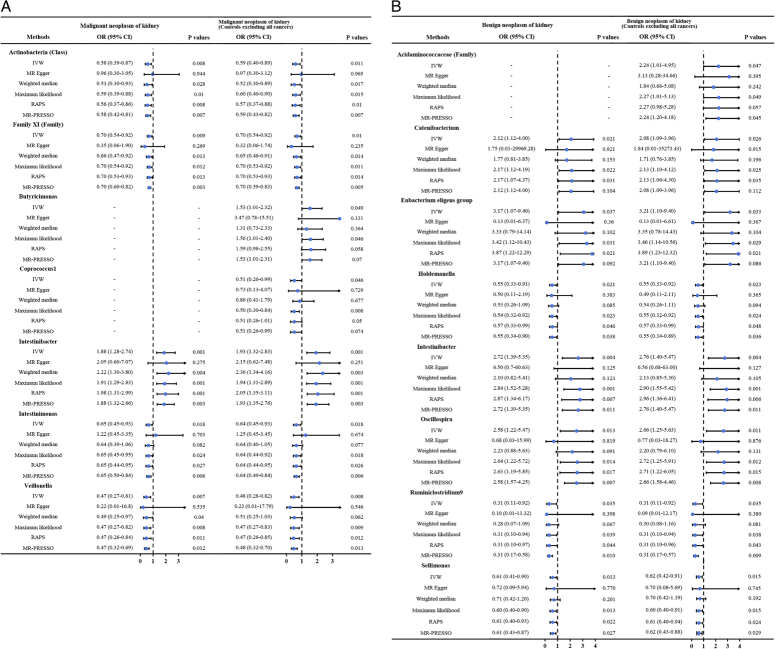
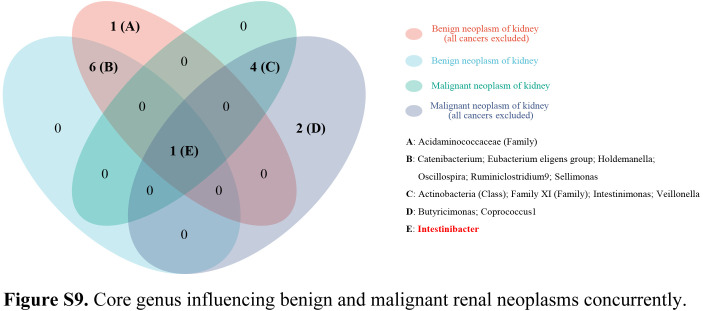
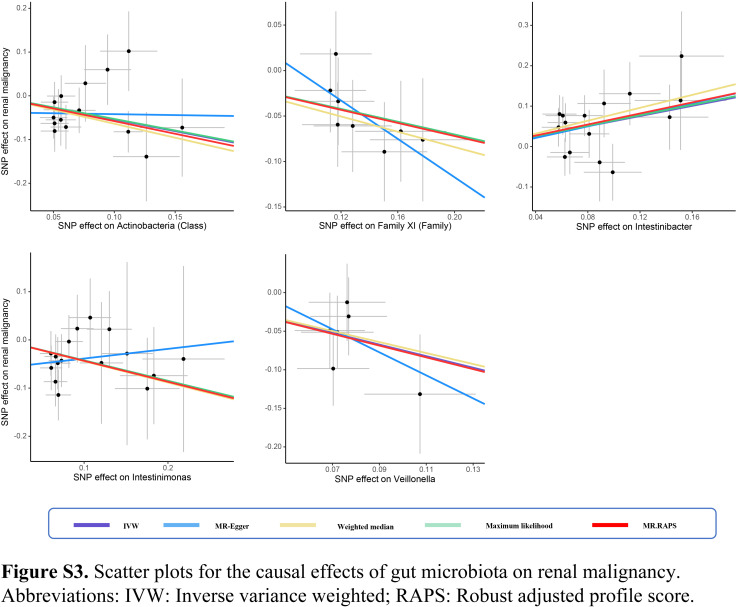
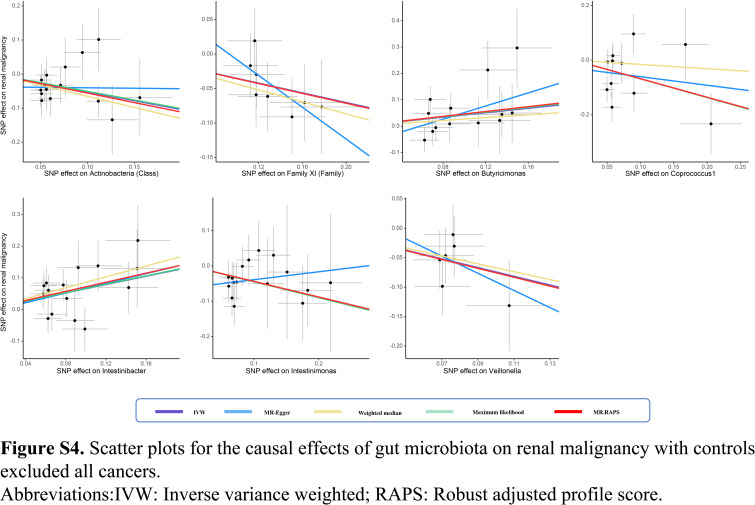
In this two-sample MR study, we found that gut microbiota might have causal effects on kidney neoplasms. Significantly protective effects of Class Actinobacteria (OR: 0.58), Family XI (OR: 0.70), Intestinimonas (OR: 0.65), and Veillonella (OR: 0.47) against renal malignancy were detected (all P<0.05). Moreover, Intestinibacter showed causality on the risk of renal malignancy (OR: 1.88, P<0.001). The protective effects against benign renal tumor were identified in the Holdemanella (OR: 0.55), Ruminiclostridium9 (OR: 0.31), and Sellimonas (OR: 0.61) (all P<0.05). Additionally, Catenibacterium (OR: 2.12), Eubacterium eligens group (OR: 3.17), Intestinibacter (OR: 2.72), and Oscillospira (OR: 2.58) revealed a causal association with increased risk of benign renal tumor (all P<0.05) (Fig. 1). The effect sizes and direction remained consistent in the other five methods. No heterogeneity and pleiotropy were identified by Cochran’s Q test and global test (all P>0.05) (Supplementary Figs S1–S8, Supplemental Digital Content 1, http://links.lww.com/JS9/B686, Supplemental Digital Content 2, http://links.lww.com/JS9/B687, Supplemental Digital Content 3, http://links.lww.com/JS9/B688, Supplemental Digital Content 4, http://links.lww.com/JS9/B689, Supplemental Digital Content 5, http://links.lww.com/JS9/B690, Supplemental Digital Content 6, http://links.lww.com/JS9/B691, Supplemental Digital Content 7, http://links.lww.com/JS9/B692, Supplemental Digital Content 8, http://links.lww.com/JS9/B693) (Supplementary Tables S1–S4, Supplemental Digital Content 9, http://links.lww.com/JS9/B694, Supplemental Digital Content 10, http://links.lww.com/JS9/B695, Supplemental Digital Content 11, http://links.lww.com/JS9/B696, Supplemental Digital Content 12, http://links.lww.com/JS9/B697). We further evaluated whether there were bacteria that can simultaneously affect benign and malignant kidney neoplasms. In all, only one genus, Intestinibacter, could increase the risks of developing both benign (OR: 2.72) and malignant renal neoplasm (OR: 1.93), indicating the core role of Intestinibacter in the onset of kidney neoplasms (Supplementary Figure S9, Supplemental Digital Content 13, http://links.lww.com/JS9/B698). More efforts are necessary to clarify the molecular mechanisms underlying these links.
Figure 1.
(A) The results of MR estimating the causal association between intestinal microbiota and malignant neoplasm of the kidney. (B) The results of MR estimating the causal association between intestinal microbiota and benign neoplasm of the kidney. CI, confidence interval; IVW, inverse variance weighted; MR, Mendelian randomization; OR, odds ratio; PRESSO, pleiotropy residual sum and outlier; RAPS, robust adjusted profile score.
Ethics statement
Ethical approval is not needed in this study.
Consent
Consent is not applicable in this study.
Sources of funding
The source of funding has been declared in the text. Our work was supported by the National Natural Science Foundation of China (No. 81872077).
Author contribution
F.Z., Y.X., and B.Z.: conception, design, and manuscript writing; B.Z.: administrative support; F.Z. and Y.X.: provision of study materials or patients, collection and assembly of data, and data analysis and interpretation. All authors were involved in the final approval of the manuscript.
Conflicts of interest disclosure
All authors report no conflicts of interest in this work.
Research registration unique identifying number (UIN)
Not applicable.
Guarantor
Bo Zhang.
Data availability statement
The datasets are available from the corresponding author on reasonable request.
Provenance and peer review
Our paper was not invited.
Supplementary Material
Supplementary Material
Acknowledgements
We appreciate the participants and investigators of the FinnGen study and the MiBioGen consortium for releasing the GWAS summary statistics.
Footnotes
Fuxun Zhang and Yang Xiong contributed equally to this work.
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal’s website, www.lww.com/international-journal-of-surgery.
Published online 15 January 2024
Contributor Information
Fuxun Zhang, Email: 45312425@qq.com.
Yang Xiong, Email: cqmu_xy@163.com.
Bo Zhang, Email: zhangbo_tduro@163.com.
References
- 1. Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209–249. [DOI] [PubMed] [Google Scholar]
- 2. Wang Y, Xuan Y, Su B, et al. Predicting recurrence and survival in patients with non-metastatic renal-cell carcinoma after nephrectomy: a prospective population-based study with multicenter validation. Int J Surg 2023. doi: 10.1097/JS9.0000000000000935. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tilg H, Adolph TE, Gerner RR, et al. The intestinal microbiota in colorectal cancer. Cancer Cell 2018;33:954–964. [DOI] [PubMed] [Google Scholar]
- 4. Greenland S. An introduction to instrumental variables for epidemiologists. Int J Epidemiol 2000;29:722–729. [DOI] [PubMed] [Google Scholar]
- 5. Xiong Y, Zhang F, Zhang Y, et al. Insights into modifiable risk factors of erectile dysfunction, a wide-angled Mendelian Randomization study. J Adv Res 2023. doi: 10.1016/j.jare.2023.05.008. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets are available from the corresponding author on reasonable request.