Abstract
Tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb), continues to be a major public health problem worldwide. The human immunodeficiency virus (HIV) is another equally important life-threatening pathogen. Further, co-infections with HIV and Mtb have severe effects in the host, with people infected with HIV being fifteen to twenty-one times more likely to develop active TB. The use of an appropriate animal model for HIV/Mtb co-infection that can recapitulate the diversity of the immune response in humans would be a useful tool for conducting basic and translational research in HIV/Mtb infections. The present study was focused on developing a humanized mouse model for investigations on HIV-Mtb co-infection. Using NSG-SGM3 mice that can engraft human stem cells, our studies showed that they were able to engraft human CD34+ stem cells which then differentiate into a full-lineage of human immune cell subsets. After co-infection with HIV and Mtb, these mice showed decrease in CD4+ T cell counts overtime and elevated HIV load in the sera, similar to the infection pattern of humans. Additionally, Mtb caused infections in both lungs and spleen, and induced the development of granulomatous lesions in the lungs, detected by CT scan and histopathology. Distinct metabolomic profiles were also observed in the tissues from different mouse groups after co-infections. Our results suggest that the humanized NSG-SGM3 mice are able to recapitulate the effects of HIV and Mtb infections and co-infection in the human host at pathological, immunological and metabolism levels, providing a dependable small animal model for studying HIV/Mtb co-infection.
Keywords: Humanized mouse model, HIV, Mycobacterium tuberculosis, NSG-SGM3 mice, HIV/Mtb-induced immunopathogenesis, HIV/Mtb-differentiated metabolites
INTRODUCTION
Tuberculosis (TB) remains one of the biggest public health problems worldwide, being the second cause of death in mankind in 2022, behind COVID-19 1. Over seven million people were newly diagnosed with TB in the past year and around 1.3 million people were killed by this deadly disease. There is a consensus that a quarter of the world population are infected with Mycobacterium tuberculosis (Mtb), the causative agent for TB1. The majority of Mtb-infected individuals remain latently infected without clinical signs (LTBI). However, around 10% of the infected patients will develop active TB, especially in conjunction with immunodeficiency caused by malnutrition, immunosuppressive therapy using steroids, or infection with immunosuppressive pathogens2. Among these infection with human immunodeficiency virus (HIV) plays a pivotal role, given that immunosuppression is the hallmark of HIV pathogenesis3. HIV is the etiological agent for acquired immunodeficiency syndrome (AIDS), another equally important public health concern responsible for the death of over 40 million people as of 20234. The synergy between HIV and Mtb in co-infection has been extensively examined, and compelling evidence showed that HIV exacerbates TB severity, and is the leading cause of death for people infected with Mtb4-6. This is likely because HIV-induced immunosuppression leads to a disruption of CD4 T cells, the main driver of Th-1 immunity in LTBI patients, resulting in active TB7.
Non-human primates (NHP) are routinely used as large animal models for HIV/Mtb research not only because the monkeys and humans have remarkably similar genomes, physiology, and immune systems, but also because the monkeys can be infected by both Mtb and Simian immunodeficiency virus (SIV)8. The latter is also a retrovirus and belongs to the same Lentivirus genus as HIV and causes HIV-like infection in NHPs. After co-infection, NHPs also display AIDS-like features as in humans, such as massive reduction of CD4+ T cells and a high viral load in the sera if without anti-retroviral treatment, as well as chronic immune activation in animals during extended observation 7, 9. Furthermore, the co-infected monkeys also recapitulate key aspects of human TB infection stages, including latent infection, chronic progressive infection, and acute TB, depending on the route and dose of infection10-12. Importantly, Mtb latently infected macaques co-infected with SIV results in reproducible LTBI reactivation13, providing a reliable model for HIV/Mtb research. However, NHPs require specialized infrastructure for experimentation and are cost-restrictive, and are not readily available in the majority of animal facilities14, 15,
The use of other small animal models, such as rodents poses different challenges. Although inbred and genetic knockout mice are easily available, and readily infective using Mtb, most strains of mice are not a natural host for HIV, which require human CD4+ T cells to establish infection. Whereas the use of mouse models for Mtb research has also been criticized due to their inability to form granulomas which are a hallmark of Mtb infection in humans16, certain mouse strains and infection protocols show the formation of granulomas. 17. Fortunately, humanized mouse models, the immunodeficient mice that have been reconstituted with a human immune system, appears to be a promising small animal model for HIV and Mtb reseach14, 15, 18, 19. They have been extensively used for evaluating HIV gene therapy and therapeutics20, 21, and recently, the NSG (NOD scid gamma)-based humanized BLT mice were developed for analyzing Mtb and HIV/Mtb co-infections15, 18, 22. However, humanized BLT mice need surgical transplantation (under the kidney capsule) of fetal liver, bone marrow and thymus tissues, and restriction of human fetal tissues used for research and the sophisticated surgery has markedly limited the use of this model. In addition, these mice have immature B cells with poor IgG class-switching and poor reconstitution of myeloid lineage of antigen-presenting cells (APCs)23, 24, posing a challenge for HIV/Mtb research because myeloid cells, especially macrophages, are important targets for both HIV and Mtb.
We demonstrate here that these deficiencies can be ameliorated in the newly developed NSG-SGM3 mice, which transgenically express three human cytokine/chemokine genes IL-3, GM-CSF, and KITLG. The expression of these genes improves the differentiation and maturation of the myeloid cells25-29. The present study is aimed at establishing a reliable new-generation, humanized mouse model for the HIV/Mtb co-infection research. We show that humanized NSG-SGM3 mice can differentiate CD34+ stem cells into a full-lineage of immune cell subsets, including both lymphoid and myeloid lineages. Importantly, we show that HIV/Mtb infections are reproducible in these mice with a spectrum of immunological, pathological, and metabolic changes when compared to uninfected mice.
MATERIALS AND METHODS
Bacterial and viral strains
Mtb H37Rv was obtained from BEI Resources (USA) and propagated in the biosafety level 3 (BSL-3) facilities at the University of Texas Health Science Center at Tyler (UTHSCT). It was cultured in 7H9 broth with 10% OADC supplement following standard Mtb culture procedures30. After 7 days of growth, the bacteria were collected and subjected to sonication three times, at an amplitude of 38%, for 10 seconds/each, with a 5-second interval, followed by low-speed centrifugation (1,100 RPM). Bacteria were diluted to an optical density (OD) value of ≈ 1 in sterile NaCl 0.9% and aliquots were made and frozen at −80 °C to be used as inoculum. Two weeks later, one aliquot was thawed, and the bacterial content was evaluated by plating ten-fold serial dilutions in 7H10 agar, supplemented with OADC. After 3 weeks of incubation, the colony forming units (CFU) per mL were calculated.
HIV-1 BaL strain was obtained from NIH AIDS Reagent Program, also prepared in the BSL-3 facilities at UTHSCT, following standard procedures 31. Briefly, frozen human PBMCs (STEMCELL Technologies, Vancouver, Canada) were thawed and seeded in a 75 cm2 flask at a concentration of 5 × 106 cells/mL in RPMI 1640 media (Corning Inc., Corning, NY) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, 1 μg/ml of PHA and 2 μg/ml polybrene (MilliporeSigma, Burlington, MA). After 3 days of stimulation, 4 × 107 cells were centrifuged and infected with HIV-1 BaL using an MOI (multiplicity of infection) of 0.1 (4 × 106 TCID50) in two adsorption cycles. Following the second adsorption cycle, the cells were seeded in two 75 cm2 flasks with 30 ml of media supplemented with FBS, antibiotics, and human IL-2 (20 Units/ml). Cell culture supernatant was collected every three days, with fresh media being added, until day 21 of culture and stored at −80 °C. A small aliquot from each collection will be used to titrate the virus using quantitative RT-PCR.
Animal experiment design
All animal procedures were approved by the UTHSCT Institutional Animal Care and Use Committee (IACUC) (Protocol #707). NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CMV-IL3,CSF2,KITLG)1Eav/MloySzJ (NSG-SGM3) mice were purchased from The Jackson laboratory (Bar Harbor, ME) and bred in the Vivarium facilities at UTHSCT. Pups were weaned at 21 days after birth and, 1-3 weeks after that, they were irradiated at a dose of 100 cgy/mouse, followed by intravenous injection with 2 × 105 CD34+ stem cells/mouse at 12 h post-irradiation. Humanization was monitored starting at 12 weeks after stem cell transplantation and again at 14 and 16 weeks. For this purpose, blood was drawn from the submandibular vein (100-150 μl, based on animal weight) and PBMCs were collected through density gradient centrifugation using Ficoll Paque (Cytiva, Marlborough, MA). After erythrocyte lysis, the PBMC from each animal were stained for human (hu) and mouse (mo) hematopoietic cell surface marker (CD45+), as well as lymphocytic and myeloid markers. Animals that showed a positive huCD45+/moCD45+ ratio, accompanied by differentiation of various immune cell populations, were selected for experimental infection.
Mice were randomly divided into four experimental groups: Uninfected (n=5), HIV-infected (n=8), Mtb-infected (n=8) and HIV/Mtb co-infected (n=7). Mtb infection was performed using aerosolized Mtb H37Rv through a Madison chamber, as previously described32, using an infection dose of 100 CFU/mouse. Three additional mice were included in the Madison chamber at the time of infection and were euthanized 24 hours after infection. The lungs were collected, macerated and plated on 7H10 agar, to confirm the initial bacterial implantation33.
One day after Mtb infection, the mice for the HIV alone and HIV/Mtb co-infection groups were subjected to intraperitoneal (IP) inoculation with 105 TCID50 of HIVBaL. Blood samples from all experimental groups were collected on the day of infection and at 15-, 28- and 35-days post infection (dpi). Serum samples from all the animals were separated and stored at −80 °C until further use. PBMCs were isolated and stained for flow cytometry analysis. At 35 dpi, the animals were terminally anesthetized, using a Ketamine/Xylazine mixture, in order to perform computed tomography (CT) scan and pulmonary function (PF) tests. Afterwards, the animals were euthanized and whole blood samples were collected through cardiac punction. During necropsy, lung and spleen samples were collected and macerated through a 70 μM cell strainer (Thermo Fisher scientific) in a final volume of 2 ml of PBS. Serial ten-fold dilutions of the organ macerates were plated in 7H10 agar, supplemented with OADC, to assess the bacterial load. The remaining volume of lung and spleen macerates were stored at −80 °C for further analysis.
For each experimental group, lung sample from one animal was selected for histopathological analysis and, therefore, not subjected to maceration and bacterial culture. Lungs were filled with 10% formalin, before being removed from the animal, and stored in the same media after the necropsy34, 35. Sample processing and Hematoxylin-Eosin (HE) staining was carried out at the histopathology core of UT southwestern.
CT scan and PF testing
Mice were intraperitoneally injected with ketamine/xylazine (100 mg/kg Ketamine, 20 mg/kg Xylazine). Once the correct anesthetic plane was achieved, the mice were intubated with a sterile, 20-gauge intravenous cannula through the vocal cords into the trachea. Following intubation, anesthesia was maintained using isoflurane.
Elastance (Ers), compliance (Crs), and total lung resistance (Rrs) was assessed for each mouse through the snapshot perturbation method, as previously described36. Measurements were performed in triplicates for each animal, using the FlexiVent system (SCIREQ, Tempe, AZ), with a tidal volume of 30 mL/kg at a frequency of 150 breaths/min against 2–3 cm H2O positive end-expiratory pressure.
After PF testing, the mice were subjected to CT scans for the measurements of lung volume, using the Explore Locus Micro-CT Scanner (General Electric, GE Healthcare, Wauwatosa, WI). CT scans were performed during full inspiration and at a resolution of 93 μm. Lung volumes were calculated from lung renditions collected at full inspiration. Microview software 2.2 (http://microview.sourceforge.net) was used to analyze lung volumes and render three-dimensional images.
RNA extraction and RT-qPCR
Serum samples from all experimental groups were extracted using the NucleoSpin RNA isolation kit (Macherey-Nagel, Allentown, PA). Following viral RNA extraction, samples were evaluated using RT-qPCR to determine the viral RNA load in each animal37. Control standards (obtained from NIH AIDS Reagent Program) with known quantities of HIV-1 genome copies were used as amplification controls, as well as to stablish a standard curve that was used to determine the viral RNA load, based on the cycle threshold (Ct) value.
Flow cytometry analysis
Flow cytometry was performed using the PBMCs from all experimental animals at the specified sampling timepoints. In all cases, the PBMCs isolated from each animal were divided into two wells of a 96-well U-shaped bottom plate (Corning Inc., Corning, NY), used for staining with two separate flow cytometry panels. Cells were washed and inoculated with Fc block (Biolegend, San Diego, CA) at 4 °C for 20 minutes, followed by another wash. Afterwards, cells were incubated with fluorescence-conjugated monoclonal antibodies. For the first flow cytometry panel, cells were incubated with antibodies against the following human surface markers: Alexa Fluor™ 405-CD45, FITC-CD3, APC-CD4, PE-CD8, PerCP-CD56, Alexa Fluor™ 510-CD19 (Biolegend, San Diego, CA). For the second flow cytometry panel, the antibodies against human cell surface markers were as follows: Alexa Fluor™ 405-CD45, Alexa Fluor™ 510-CD86, APC-CD11b, PE-CD11c, PerCP-HLA-DR, Alexa Fluor™ 700-CD14 (Biolegend, San Diego, CA). Additionally, for the second panel, the cells were also incubated with an FITC-labelled antibody against moCD45. After staining, the cells were washed and fixed for 1 hour, followed by another wash. Flow cytometry was performed using the Attune NxT flow cytometer (Invitrogen, Waltham, MA), including the corresponding isotype controls for each antibody. Analysis was carried out with the FlowJo software v10.6.1 (BD life sciences), using the isotype controls as guidelines for gating.
Immunofluorescence staining
Paraffin-embedded lung sections were used for immunofluorescent staining against human immune cell subsets38. Samples were deparaffined by submerging the slides in Xylene (Fisher bioreagents), followed by sequentially lower concentrations of ethanol. Afterwards, antigen retrieval and blocking of non-specific binding were performed, using 10mM sodium citrate buffer and PBS with 0.4% triton and 5% FBS, respectively. Primary antibody incubation was carried out overnight at 4 °C with human-CD68 monoclonal antibody (cat. No. 14-0688-82, Invitrogen) and CD19 Rabbit polyclonal antibody (cat. No. 27949-1-AP, Proteintech, Rosemont, USA), diluted in PBS + 0.4% triton + 1% FBS at the recommended dilutions. The following day, samples were incubated for 2 hours at room temperature with goat anti-mouse IgG1-Alexa Fluor™ 568 (cat. No. A21124, Invitrogen) and goat anti-rabbit IgG-Alexa Fluor™ 488 (cat. No. A11008, Invitrogen), at the recommended dilutions. The slides were mounted using DAPI-supplemented mounting medium (Abcam, Cambridge, UK) and images were captured with a LionheartLX automated microscope (Biotek, Winoovski, VT). Images were processed with the GEN5 software version 3.09 (Biotek) and the ImageJ software (NIH).
Multiplex assay for cytokine profiling
The cytokine profile in lung and spleen tissue macerate, as well as serum samples at 35 dpi, from all experimental groups were evaluated in duplicates using the Bio-Plex Pro™ Human Cytokine panel (Bio-Rad, Hercules, CA), according to the manufacturer’s instructions. Briefly, 50 μL of filtered tissue homogenate, or 1:4 diluted serum, were dispensed in a 96-well plate containing magnetic beads conjugated with antibodies for the detection of 27 different cytokines. Following incubation with detection antibodies and streptavidin-PE, the samples were analyzed in the Bio-Plex MAGPIX multiplex reader (Bio-Rad Laboratories Inc., CA). A regression curve, based on the values obtained from a set of standard dilutions, was used to convert the fluorescence values reported by the machine into cytokine concentrations (expressed as pg/mL).
The 27 cytokines and chemokines reported by the Bio-Plex Pro™ Human Cytokine panel were: Basic FGF, Eotaxin, G-CSF, GM-CSF, IFN-γ, IL-1β, IL-1Ra, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12, IL-13, IL-15, IL-17, IP-10, MCP-1, MIP-1α, MIP-1β, PDGF-BB, RANTES, TNF-α and VEGF.
Mouse blood sample handling for metabolomic analysis
Whole blood sample was collected from mice in all the experimental groups at the end of the study and plasma was separated through centrifugation. The samples were processed for collection of the metabolite pellet as follows: 50 μl of plasma were mixed with 950 μl of 80% ice-cold methanol, followed by centrifugation at >20.000 G for 15 minutes in a refrigerated centrifuge. Afterwards, the supernatant was transferred to a new tube and vacuum dried, using no heat. The metabolite pellet was analyzed at the metabolomic core facility at the Children’s Medical Center Research Institute at University of Texas Southwestern Medical Center (Dallas, TX, USA) using liquid chromatography-mass spectrometry (LC-MS), as previously described39.
Metabolome data analysis
Statistical analysis of metabolome profiles was performed in R environment (R version 4.1.0). Raw abundance values of metabolites were used as input for statistical analysis. The raw data was log2 transformed and normalized across the samples using ‘limma’ package40 by cyclically applying fast linear loess normalization with a 0.3 span of loess smoothing window and 10 iterations wherein each sample was normalized to pseudo-reference sample which was computed by averaging all samples. Principal components analysis was performed using ‘PCAtools’ package. Orthogonal partial least squares discriminant analysis (OPLS-DA) was performed and variable importance on projection (VIP) score were computed using ‘ropls’ package. VIP score of >1 is considered for feature selection. Hierarchical clustering was performed on normalized data after univariate scaling. Hierarchical clustering was performed using correlation to calculate clustering distance with averaging method for clustering. Differentially abundant metabolites (DAMs) were identified using student t test. The correlation between metabolite abundances and Mtb or HIV loads were analyzed using Pearson correlation method. For all hypothesis testing analyses, statistical significance was set 5% (p value = 0.05) to reject null hypothesis.
Statistical analysis
Statistical differences between groups were assessed using the Prism software version 8.3.0. for Windows (GraphPad Software, San Diego, California USA, www.graphpad.com). Unpaired, non-parametric, t-tests were employed for different comparisons between groups.
RESULTS
Human CD34+ HSCs-engrafted NSG-SGM3 mice can differentiate a full array of human immune cell phenotypes.
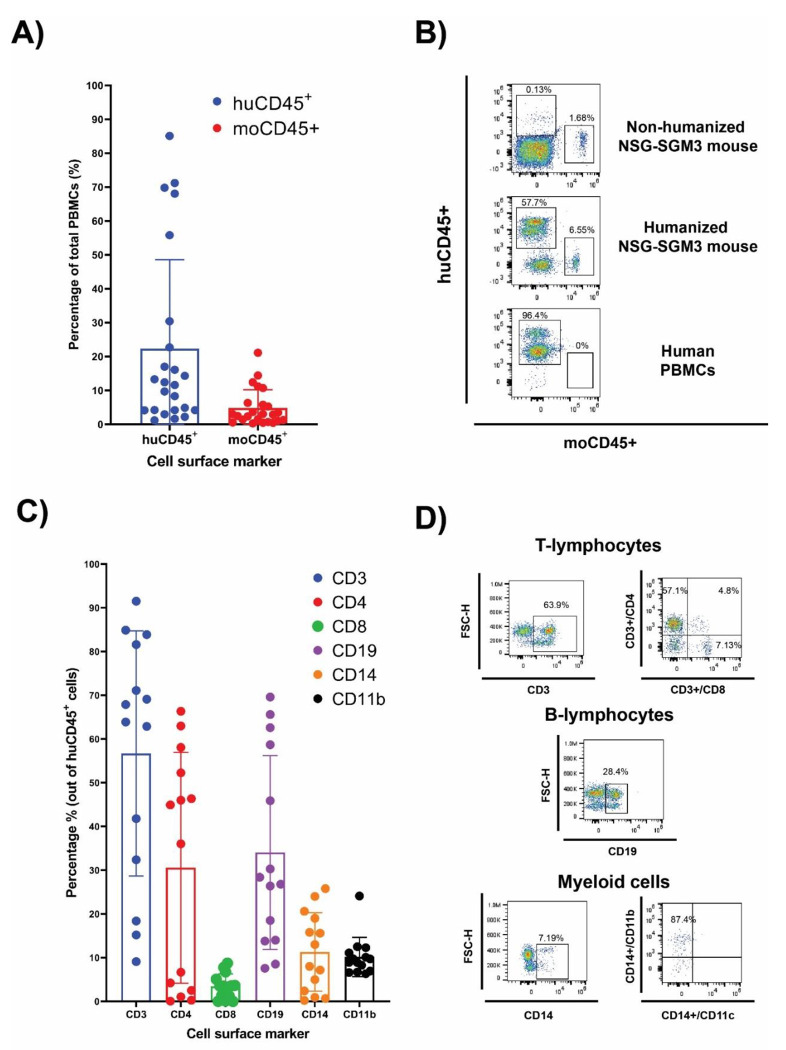
After 16 weeks of humanization, PBMCs from the hCD34+ HSCs-transplanted mice were evaluated by flow cytometry for human lymphoid and myeloid cell surface markers. The NSG-SGM3 mice allow stem cells to develop into human lymphoid lineages, such as T cells (CD3+, between 10-90%, including both CD4+ and CD8+ T cells) and B cells (CD19+, between 7-60%) (Fig. 1). Additionally, differentiation of human myeloid subsets (CD14+) was also observed, ranging between 1 and 25%. Within the myeloid lineage, we also detected CD11b+ macrophages (Fig. 1, Gating strategy is shown in Supplementary Fig. 1).
Figure 1. Human CD34+ hematopointci stem cells (HSC) engraftment and differentiation of human immune cells in the NSG-SGM3 mice.
(A and B): The differential expression of humanCD45 (huCD45) and mouseCD45 (moCD45) expressing cells in mice after 14 weeks of humanization. Percentages of human and mouse CD45+ cells are shown as histogram in A (n=27), and the representative flow cytometry dot plot of the comparative expression of human cell surface markers between the humanized NSG-SGM3 mice and human PBMCs are shown in B. (C) Percentages of human immune cell populations (n=27). (D) representative flow cytometry dot plot of T lymphocytes, B cells and myeloid cells.
Humanized NSG-SGM3 mice are susceptible to both HIV-1 and Mtb infections.
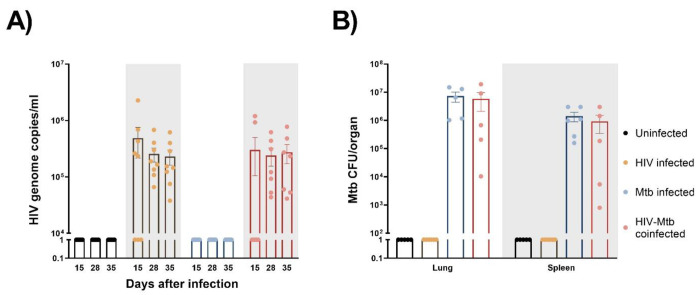
After HIV/Mtb infections, HIV viral RNA was detected in serum samples from the infected mice starting at 15 dpi, with most animals in the HIV single-infection group being positive at this time, while only two out of the seven mice in the HIV/Mtb co-infection group showed viral RNA (Fig. 2a). The viral RNA load detected in the positive animals at 15 dpi was between 2×105 and 2.2×106 copies/ml. However, all the HIV-infected animals were positive in subsequent samplings at 28 and 35 dpi. The HIV RNA load was between 3.7×104 and 6.8×105 copies/ml for animals with single HIV infection and between 4.1×104 and 7.7×105 copies/ml for the HIV/Mtb co-infected mice. No significant differences were detected in the viral RNA load between the two HIV-infected groups at these timepoints.
Figure 2. Establishment of HIV-1 and Mycobacterium tuberculosis (Mtb) infections in humanized mice.
(A) HIV-1 RNA load, expressed as genome copies/mL, was assessed in serum samples from all experimental groups at three different timepoints of the study (B) Mtb bacterial load in lungs and spleens, expressed as CFU/organ, was evaluated in all experimental groups at the end of the study.
The Mtb bacterial load was assessed in lung and spleen samples after euthanasia in the Mtb single infection group and the HIV/Mtb coinfected mice (Fig. 2b). In both groups, a higher bacterial load was found in lungs than in spleens. Moreover, the mean CFU count in the lungs and spleens from Mtb single infection group (7.3×106 and 1.4×106, respectively) was higher than the animals co-infected with HIV (5.8×106 for lung and 9.2×105 for spleen), even though their differences are not significant (Fig. 2b).
Immune phenotype changes in humanized mice after infection.
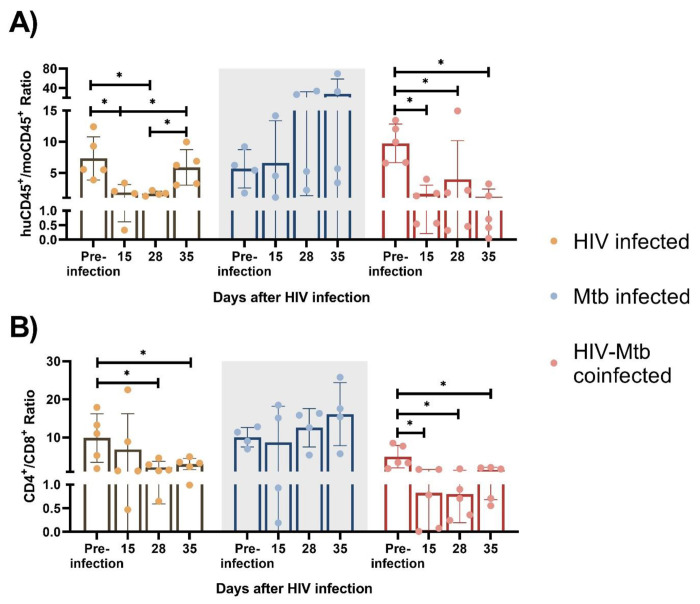
We also monitored the human immune cell population changes over time after HIV/Mtb infections. Starting from 15 dpi, huCD45+/moCD45+ ratio was significantly decreased (p<0.05) in the two HIV-infected groups (HIV single infection and HIV/Mtb co-infection), and the huCD45+/moCD45+ ratio decrease was sustained until the late stage of the experiment. Conversely, the Mtb single infection group showed similar or even increased huCD45+/moCD45+ ratio after infection (Fig. 3a).
Figure 3. Immune cell phenotype changes after HIV-1 and Mtb infections.
HuCD45+/moCD45+ ratio (A) and CD4+/CD8+ ratio (B) were calculated for each infected animal at different timepoints after infection. Asterisk indicates statistically significant differences (p<0.05, what test )
We observed significant CD4+ T cell depletion in the HIV-infected groups (HIV single infection and HIV/Mtb co-infection). We used CD4+/CD8+ ratio as an indicator for CD4+ T cell depletion, and we found a ~10-fold CD4+/CD8+ reduction (p<0.05) in the HIV/Mtb co-infected mice as early as 15 dpi, and this trend remained until the end of the experiment. In the single infection group, we also found a lower mean CD4+/CD8+ ratio since 15 dpi, while the subsequent samplings at 28 and 35 dpi showed significant decreases on CD4+/CD8+ ratio values. In contrast, there was no significant difference detected over time in the Mtb alone infection group (Fig. 3b).
Alterations in cytokines and chemokines production in humanized mice after infection
In serum sample, significant increases in G-CSF, MCP-1 and MIP-1α was detected in the Mtb single infection group, in comparison with both HIV-infected groups (Fig. 4a). Additionally, the serum concentration of IL-2 and IL-8 were also significantly increased in the Mtb single infection group, compared to the HIV/Mtb co-infection. The HIV/Mtb co-infected mice analyzed showed higher IP-10 than both the HIV and Mtb single infection mice (Fig. 4a).
Figure 4. Cytokine profiles (Heatmap) in serum, lung and spleen samples.
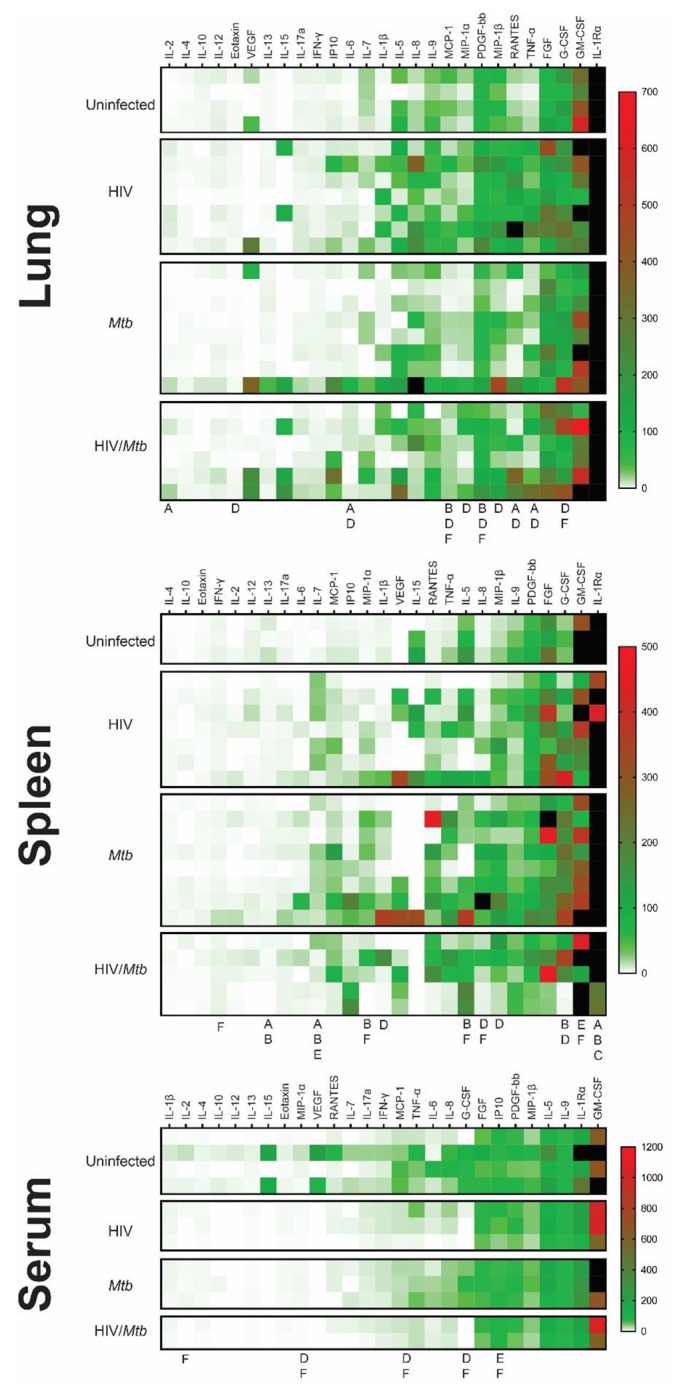
The Bio-Plex Pro™ Human Cytokine panel was used in the multiplex assay to evaluate the concentrations of 27 different human cytokines, which are expressed as pg/ml. (A) Cytokine profile of lung samples. (B) Cytokine profile of spleen samples. (C) Cytokine profile of serum samples. the letters under the columns show differences as follows: A: Difference between uninfected and HIV-infected, B: Difference between uninfected and Mtb-infected, C: difference between uninfected and HIV/Mtb-coinfected, D: Difference between HIV-infected and Mtb-infected, E: difference between HIV-infected and HIV/Mtb-coinfected, and F: Difference between Mtb-infected and HIV/Mtb-coinfected. (p<0.05; unpaired T test). Note: The black color on the right of heatmap shows the far high value that are out-of-range levels.
Lung macerate supernatants showed an increase in the concentration of IL-6, RANTES and TNF-α in the HIV single infection group compared to the uninfected control animals, as well as the Mtb single infection group (Fig. 4b). Additionally, IL-2 concentrations were also higher in the HIV-infected animals than in the uninfected mice. Moreover, HIV single infection also induced statistically higher levels of Eotaxin, MIP-1α and MIP-1β than single Mtb infection. Statistical analysis also revealed a decrease in MCP-1 and PDGF concentration in lung samples from Mtb infected mice, compared to the remaining three experimental groups (Fig. 4b).
In the case of spleen samples, macerates from the Mtb single-infection group were found to have significantly higher concentrations of IL-1β, G-CSF and MIP-1β than the HIV single-infection group (Fig. 4c). Similarly, the levels of IL-8 and MIP-1α were higher in the Mtb group than in both HIV-infected groups. In contrast, both the HIV and Mtb single infection groups showed lower concentrations of GM-CSF than the HIV/Mtb co-infected animals, while this group also had statistically higher amounts of IFN-γ than the Mtb group. All the infected groups showed a decrease in IL-1Rα and IL-13, compared to the uninfected control animals (Fig. 4c).
Mtb infection induced pathological changes in the lungs of humanized mice.
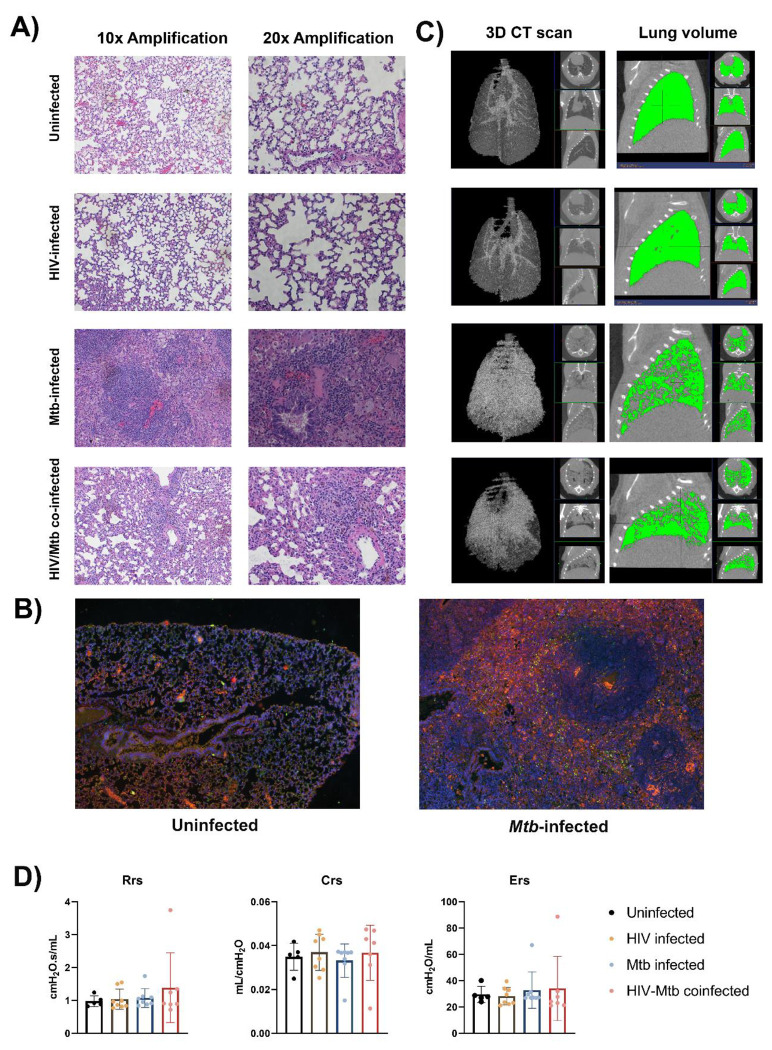
We stained the lung section with H&E staining, and we observed diffuse immune cell infiltration in lung sample from Mtb-infected mice. In some cases, immune cell infiltration was observed around a necrotic nucleus, in structures similar to TB granulomas. No such cellular aggregates were detected in either the uninfected or the HIV single infection groups (Fig. 5a). We stained lung sections from Mtb-infected humanized mice by immunofluorescent staining, and the result showed that the cell populations surrounding the necrotic area mostly corresponded with macrophages (CD68+), though other immune cell types, such as CD19+ B cells, were also found. However, no granuloma structure was observed in the lung section of the uninfected mice, even though a low proportion of cells expressing the human CD68+ and CD19+ surface markers was observed in the lung sections from uninfected mice (Fig. 5b).
Figure 5. Histopathological, radiological and functional changes in the lungs of NSG-SGM3 mice after HIV/Mtb infection and coinfection.
(A) Lung sections were obtained from formalin-fixed tissues of animals in all experimental groups (one animal for each group) and subjected to hematoxylin-eosin staining, two different amplifications are shown. (B) Immunofluorescence staining of surface markers for human macrophages (CD68-Alexafluor 568, in orange) and B-cells (CD19-Alexa 488, in green) in lung sections from uninfected and Mtb-infected mice. DAPI-supplemented mounting buffer (in blue) was used for nuclei staining. (C) Representative 3D renditions of CT scan and lung volume pictures obtained from animals in all experimental groups. (D) Pulmonary function test parameters: Resistance (Rrs), compliance (Crs) and elastance (Ers), were collected from animals in all experimental groups at the end of the trial (Uninfected: n=5; HIV-infected: n=8; Mtb-infected: n=8; HIV/Mtb-co-infected: n=7).
The CT scan showed an increase in high density areas in the Mtb-infected animals, regardless of their HIV-infection status, indicating the occurrence of inflammation and other pathological changes in the lungs (Fig. 5c). However, no significant differences were detected in the pulmonary function tests between the experimental groups (Fig. 5d).
Different plasma metabolome landscapes in healthy mice, HIV infection, Mtb infection and co-infection.
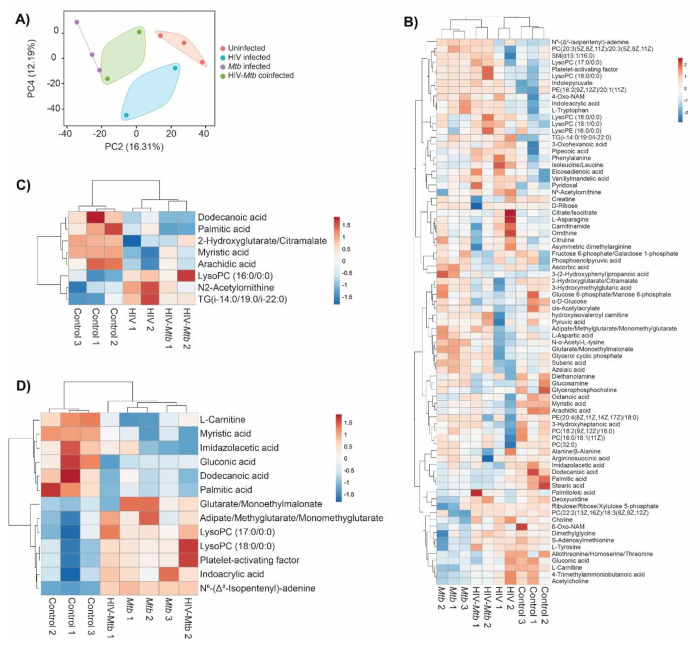
Plasma metabolome profiling was performed for a total of 10 samples including no infection (n=3), Mtb infection (n=3), HIV infection (n=2), and HIV/Mtb co-infection (n=2). Abundances of 175 metabolites were estimated. To enable comparison of metabolite abundances between different samples, data was normalized across the samples. To investigate differences in plasma metabolome landscape among the four categories of infection, principal components analysis (PCA) was performed. PCA is an unsupervised learning method suitable for dimensionality reduction of high dimensional metabolome data. Interestingly, the plasma metabolome profiles are stratified according to infection status in PCA (Fig. 6a). Mice with no infection appeared distinct from all infected mice. While the mice with infections were clustered separately from healthy mice, there was a clear distinction among HIV infection alone, Mtb infection alone, and HIV/Mtb co-infection. This suggests that the global plasma metabolome is distinctly altered based on infection status and type. Interestingly, the samples from HIV/Mtb co-infected mice clustered in between HIV infection alone and Mtb infection alone suggesting they show metabolic changes common for individual infections.
Figure 6. Metabolomics analysis of the plasma from healthy and HIV and/or Mtb-infected humanized mice.
(A) Principal components analysis of plasma metabolome profiles of mice from no infection (n=3), Mtb infection (n=3), HIV infection (n=2), and dual infection (n=2) categories. Two principal components were selected to plot a two-dimensional graph to depict variation across the sample categories. Variance explained by each of the two components was given in parenthesis. (B) Heatmap showing abundances of 75 metabolites with a VIP score > 1 computed in OPLS-DA on plasma metabolome profiles of mice from no infection (n=3), Mtb infection (n=3), HIV infection (n=2), and dual infection (n=2) categories. Normalized data was scaled using univariate scaling. Hierarchical clustering was performed using correlation to calculate clustering distance with averaging method for clustering. (C and D) Heatmap showing differentially abundant metabolites in (C) HIV infection and (D) Mtb infection compared with healthy mice. Normalized data was scaled using univariate scaling. Hierarchical clustering was performed using correlation to calculate clustering distance with averaging method for clustering.
To identify metabolites varying across the four categories, we performed OPLS-DA followed by computation of VIP scores on all 175 metabolites. OPLS-DA is a supervised analysis which helps in identifying variables that discriminate different categories of samples based on VIP score. There were 75 metabolites with a VIP score >1 (Supplementary Table 1). The abundances of these metabolites across all four categories were shown with hierarchical clustering (an unsupervised algorithm) in Fig. 6b. As expected, in concordance with PCA, dendrogram of hierarchical clustering showed that infection and no infection categories are distinct, while co-infection stratified between the two individual infections (Fig. 6b).
To identify metabolites that are differentially abundant in HIV infection, we compared healthy mice (n=3) to HIV infection mice (n=4; HIV infection alone and HIV/Mtb co-infection). We identified 8 DAMs in HIV infection with a p value <0.05 (Fig. 6c and Table 1). Similarly, we compared healthy mice (n=3) to Mtb infection mice (n=5; Mtb infection alone and HIV/Mtb co-infection) to identify metabolites differentially abundant in Mtb infection which yielded 13 DAMs (Fig. 6d and Table 2). Interestingly, three fatty acids, namely dodecanoic acid, palmitic acid and myristic acid exhibited reduced abundance in HIV infected mice as well as Mtb infection mice (Table 1 and 2).
Table 1:
Metabolites differentially abundant in HIV infection.
| Control_1 | Control_2 | Control_3 | HIV_1 | HIV_2 | HIV.Mtb_1 | HIV.Mtb_2 | Log2FC | P Values | |
|---|---|---|---|---|---|---|---|---|---|
| Dodecanoic acid | 30.84885 | 29.48828 | 29.18148 | 27.5704 | 28.27857 | 27.25359 | 27.25838 | −2.2493 | 0.030561 |
| Myristic acid | 27.62307 | 27.64284 | 27.60289 | 26.46469 | 26.14845 | 27.22307 | 26.72852 | −0.98175 | 0.022716 |
| Arachidic acid | 26.34177 | 26.28904 | 25.58546 | 25.05928 | 24.64442 | 25.58284 | 25.14256 | −0.96481 | 0.033856 |
| Palmitic acid | 32.91019 | 33.14279 | 32.61936 | 32.21672 | 32.58976 | 31.93621 | 31.97329 | −0.71178 | 0.022067 |
| 2-Hydroxyglutarate/Citramalate | 25.06593 | 25.04706 | 25.11372 | 24.06289 | 24.50181 | 24.48949 | 24.90492 | −0.58579 | 0.04128 |
| N2-Acetylornithine | 21.69086 | 21.75382 | 21.41989 | 22.18159 | 22.33382 | 22.04002 | 21.82825 | 0.474396 | 0.024962 |
| LysoPC(16:0/0:0) | 32.25447 | 32.3347 | 32.58121 | 32.82015 | 32.90289 | 32.72403 | 33.16134 | 0.511976 | 0.014624 |
| TG(i-14:0/19:0/i-22:0) | 26.03645 | 26.48443 | 26.03211 | 26.84162 | 27.13346 | 26.63223 | 26.67844 | 0.637108 | 0.02702 |
Table 2:
Metabolites differentially abundant in Mtb infection
| Control_1 | Control_2 | Control_3 | Mtb_1 | Mtb_2 | Mtb_3 | HIV.Mtb_1 | HIV.Mtb_2 | Log2FC | P Values | |
|---|---|---|---|---|---|---|---|---|---|---|
| Dodecanoic acid | 30.84885 | 29.48828 | 29.18148 | 28.63719 | 27.70525 | 28.08153 | 27.25359 | 27.25838 | −2.05235 | 0.036073 |
| L-Carnitine | 29.48852 | 29.10614 | 29.56318 | 27.76163 | 27.77312 | 28.5133 | 28.5302 | 28.77454 | −1.11539 | 0.004612 |
| Gluconic acid | 27.48128 | 26.69745 | 27.25191 | 26.19547 | 26.24986 | 26.29529 | 25.81699 | 26.40603 | −0.95082 | 0.038034 |
| Palmitic acid | 32.91019 | 33.14279 | 32.61936 | 32.25708 | 32.07672 | 32.22627 | 31.93621 | 31.97329 | −0.79686 | 0.020473 |
| Myristic acid | 27.62307 | 27.64284 | 27.60289 | 27.11416 | 26.69864 | 27.04336 | 27.22307 | 26.72852 | −0.66138 | 0.003101 |
| Imidazoleacetic acid | 22.10264 | 21.58731 | 21.79266 | 21.68153 | 21.05739 | 20.98498 | 21.58037 | 20.94255 | −0.57817 | 0.040855 |
| Glutarate/Monoethylmalonate | 23.40639 | 23.4395 | 23.50502 | 23.97748 | 23.97929 | 23.69304 | 23.38075 | 23.76985 | 0.309782 | 0.046714 |
| Adipate/Methyglutarate/Monomethylglutarate | 22.09507 | 22.23793 | 22.42262 | 22.54096 | 22.81072 | 22.38233 | 22.71625 | 22.58343 | 0.354862 | 0.037601 |
| Indoleacrylic acid | 25.11462 | 25.54944 | 25.52602 | 25.9564 | 25.74232 | 26.24451 | 26.02944 | 25.90877 | 0.579597 | 0.031357 |
| LysoPC(18:0/0:0) | 30.93056 | 31.40324 | 31.21889 | 31.77792 | 31.69742 | 31.71439 | 31.8485 | 32.31356 | 0.68613 | 0.014163 |
| Platelet-activating factor | 30.90966 | 31.38026 | 31.20963 | 31.7655 | 31.68164 | 31.70822 | 31.88227 | 32.29686 | 0.70038 | 0.013158 |
| LysoPC(17:0/0:0) | 26.47687 | 26.82401 | 27.24183 | 27.52935 | 27.56634 | 27.37764 | 27.9864 | 27.78227 | 0.80083 | 0.048066 |
| N6-(delta2-Isopentenyl)-adenine | −20.6891 | −18.9004 | −18.5067 | 10.33699 | 7.60551 | 8.233653 | 8.368181 | 8.958558 | 28.06597 | 5.36E-06 |
Metabolite abundances correlated with HIV and Mtb loads.
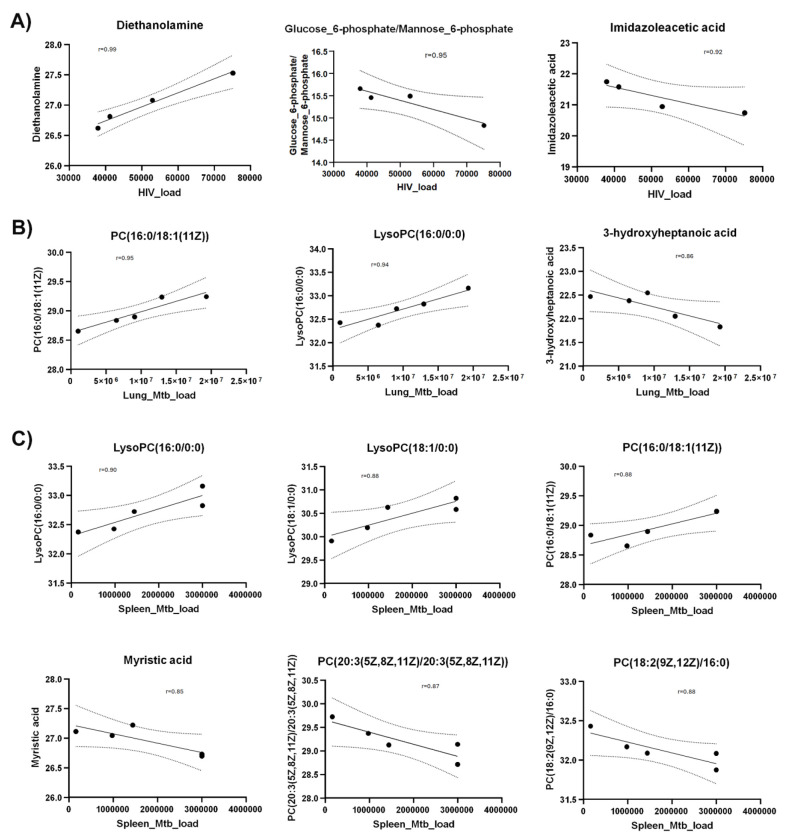
To identify metabolites correlating with HIV or Mtb load with metabolites, we used Pearson correlation analysis. HIV infection load (as detected by RNA copies/ml plasma) positively correlated with diethanolamine (r=0.99), and negatively correlated with glucose 6-phosphate/mannose 6-phosphate (r=−0.95) and imidazole acetic acid (r=−0.92) (Fig. 7A).
Figure 7. Scatter plots show Pearson correlation between metabolites and HIV/Mtb load in mice.
(A) Pearson correlation between metabolites and serum HIV load (viral copies/ml). (B) Pearson correlation between metabolites and Mtb load in lungs (CFU/lung). (C) Pearson correlation between metabolites and Mtb load in spleens (CFU/spleen). Y axis shows normalized metabolites abundance values. Dotted curves show 95% confidence interval of model fit. r denotes Pearson correlation coefficient.
Next, we observed that Mtb-infected mice did not show a strong correlation (r=0.68) between pathogen load (as measured by colony forming units per organ) in the lungs and spleens (Supplementary Fig. 2) underscoring the heterogeneity of Mtb distribution in these organs of the humanized mice. This is consistent with an earlier report41 showing that increase of Mtb load in the lungs and spleens follow different trajectories over the course of infection. Therefore, we analyzed the correlation between metabolites abundances and Mtb load in spleens and lungs separately.
Interestingly, none of the metabolites correlated with the HIV load (shown in Fig. 6c) exhibited correlation either positively or negatively with Mtb load in lung or spleen. However, PC(16:0/18:1(11Z)) and lysoPC(16:0/0:0) positively correlated with Mtb load in lung as well as spleen (Fig. 7b). In addition, 3-hydroxyheptanoic acid exhibited a strong negative correlation with Mtb load in lung (Fig. 7b). Similarly, LysoPC(18:1/0:0) showed strong positive correlation, and myristic acid, PC(20:3(5Z,8Z,11Z)/20:3(5Z,8Z,11Z)) and PC(18:2(9Z,12Z)/16:0) showed strong negative correlation with Mtb load of the spleens (Fig. 7c).
DISCUSSION
The development of animal models is a major requirement for developing drugs and vaccines for infectious diseases42-44. The lack of an ideal animal model can therefore delay the development of intervention strategies that can improve the outcome of disease in humans. The study of the interactions taking place during HIV/Mtb co-infection is particularly challenging due to a variety of factors, related to the nature of these pathogens, and the animal models. In this study, we demonstrated a reliable and reproducible small animal model for HIV/Mtb co-infection research using humanized NSG-SGM3 mice. We show that our model can recapitulate many aspects of HIV/Mtb co-infection in clinical settings, which will be helpful for characterizing the HIV/Mtb-induced immunopathogenesis, and to test therapeutics and vaccines.
A primary concern with using the mouse models for HIV/Mtb co-infection studies relates to the viral host range, which is naturally limited to humans and some NHPs45, 46. This poses restrictions on experimentation using NHPs, which require specialized infrastructure and personal training that is not widely available 8. However, this limitation has been circumvented to some extent by the use of immunocompromised mice strains that can engraft human stem cells and differentiate them into a variety of human immune cells, allowing for both HIV and Mtb infection and viral replication14, 15, 18, 19, 47. We show that the NSG-SGM3 mice allow stem cells to differentiate into a range of immune cells becoming susceptible to HIV infection and viral replication. This is due to the differentiation of human lymphoid lineage cell subsets, in particular generation of CD4+ T cells, which are the major target for HIV infection and replication. Moreover, the abundant differentiation of both lymphoid and myeloid lineage subsets allows for the assessment of immunological markers of disease relevance during HIV infection, and to measure vaccination-induced immune responses. A decreased CD4+/CD8+ ratio was observed in the humanized mice following HIV-1 infection, suggesting that our model reproduced similar immunological alterations observed during the natural infection of humans48, 49.
A comparative advantage that the NSG-SGM3 mice used in the present study over the previous generation of humanized mouse models is the transgenic expression of three human cytokine genes that enhance the differentiation and maturation of myeloid cell lineages and regulatory T cells14. This is particularly important, considering that these immune cells play important roles in controlling both HIV and Mtb growth and also serve as the target cells for these pathogens50-53, 54, 55. Moreover, the presence of granulomas, which are the hallmark of Mtb pathology, in the Mtb-infected humanized NSG-SGM3 mice is noteworthy, given that these structures are composed of multiple human immune cell populations from different lineages, that has not been seen in the C57BL/6 or BALB/c mice56. In addition, the previously reported humanized NSG-BLT mice required specialized surgical procedures in adult mice19, or the handling of newborns14. The humanization of NSG-SGM3 mice only requires a single intravenous injection of stem cells, which makes humanization much simpler to produce a viable small animal model for HIV/Mtb research.
We further note the differential expression of multiple human cytokines by the NSG-SGM3 humanized mice after HIV and Mtb single-infection or co-infection, which indicates that the reconstituted human immune cell subsets in these animals are functional and responsive during the infectious process. It should be noted that many of the cytokines that showed increased levels of expression in tissues after infection, were colony stimulating factors (G-CSF and GM-CSF) or chemoatractants (MCP-1, MIP-1α, MIP-1β), which have been implicated in human immune response against HIV and Mtb57-62. This indicates that immune cell recruitment and differentiation diverge according to the immune response induced by these pathogens in our model. Moreover, each tissue exhibited a different cytokine production profile. This could be due to the difference in cell types present in the tissues, as well as the viral/bacterial load and its effect on the immune response. In this regard, we noted that cytokine production did not increase in the lungs of the Mtb infection group, despite having a high bacterial load confirmed by culture. This is interesting and may suggest that Mtb suppresses lung immune responses to enhance its growth52, 63-65. Nevertheless, cytokine expression in spleens was increased in the Mtb-infected mice, indicating immune activation in this organ. Similarly, the results of the Pearson correlation in plasma metabolites from the HIV-infected mice likely reflect the immune modulation by the pathogen, considering the positive correlation of viral load with an immunostimulatory xenobiotic (diethanolamine)66, while an inverse correlation was found with a subproduct of histamine metabolism (Imidazoleacetic acid)67. Although additional investigations are required, these results suggests concurrent activation of immune response, and suppression of the inflammation pathway; this coincides with earlier reports which show that histamine release is inversely correlated to the number of HIV-infected CD4+ T cells in humans68. The differences in cytokine and metabolite production may also reflect different stages of disease, and further studies are needed to validate these hypotheses.
The metabolome data also provided insight into the disruptions of the immunometabolism after HIV/Mtb infections in the humanized mice. It is noteworthy that the majority of the DAMs detected in the present study for both HIV and Mtb infection are fatty acids or metabolites involved in their metabolism. In accordance with previous reports, triglycerides were found to be increased in the plasma of HIV-infected mice, regardless of Mtb infection status69. Thus, Lysophosphatidylcholines (LysoPC), such as LysoPC (16:0/0:0), have been found to be increased in HIV-infected individuals70. Paradoxically, the concentration of palmitic acid (16:0), the fatty acid attached to the C-1 position of LysoPC (16:0/0:0), was found to be decreased in HIV-infected mice compared to the uninfected controls, suggesting a disruption in fatty acid metabolism. Moreover, dodecanoic (12:0), myristic (14:0) and arachidic (20:0) acids were also decreased in the HIV-infected mice, in line with a previous study that reported a reduction in free fatty acid concentration in serum from people living with HIV, which increased after antiretroviral treatment71. On the other hand, Pearson correlation showed an inverse relation between HIV load and imidazoleacetic acid, an imidazole receptor stimulator. Given the anti-HIV potential of the imidazole derivatives72, 73, the higher concentration of imidazoleacetic acid may facilitate the imidazole receptor binding, thus activating the imidazole-mediated anti-HIV capacity, and a lower HIV load. In addition, glucose metabolic pathways in regulating HIV infection in CD4+ T cells have been extensively reported74, 75. HIV infection increases glucose uptake in CD4+ T cells, and consequently, a higher glucose uptake by the CD4+ T cells will result in a lower concentration of glucose left in the serum; therefore, it was not surprising to see a negative correlation between HIV load and the metabolite glucose/mannose 6-phosphate in the serum (Fig 7a).
In the case of Mtb infection, multiple DAMs related to TB pathogenesis were found in the plasma of infected mice (Table 2). Platelet-activating factor, increased in the Mtb-infected mice, has been previously shown to be an important part of TB immunopathology, and present in TB granulomas of humans and participating in the activation of other immune cell types during infection76. Meanwhile, N6-(Δ2-isopentenyl) adenine, a cytokinin previously thought to be produced only in plants, has been recently proven to be produced by Mtb (thus significantly increased in Mtb-infected mice), likely having a role in the protection of Mtb against nitric oxide77. Interestingly, three fatty acids (Dodecanoic acid, Myristic acid, and Palmitic acid) that were decreased in the HIV-infected mice were also decreased in plasma from Mtb-infected humanized mice, in addition to gluconic acid (6:0). The fatty acids alterations reflected the changes of mitochondrial function and β-oxidation, and this also is also evidenced by the reduction of L-carnitine, a metabolite necessary for the uptake of large chain fatty acids by the mitochondria78. We recall here that lipid-related metabolites have been reported to be decreased in humans co-infected with HIV and Mtb79. It has been reported that Mtb can alter lipid metabolism in macrophages, reducing the rate of ATP production, while at the same time, increasing their dependence on exogenous rather than endogenous fatty acids80. We therefore propose that the decrease of free fatty acids in the plasma of Mtb-infected animals might be related to sequestering of the pathogen in the macrophages81.
Collectively, our study shows that the NSG-SGM3 humanized mice can efficiently engraft human CD34+ stem cells which differentiate into a full lineage of functional immune cells. These mice are susceptible to both HIV-1 and Mtb infections, and the HIV/Mtb infections cause similar immunological, pathological, and metabolic changes in these mice as in humans. Therefore, the humanized NSG-SGM3 mice recapitulate the human-like immune responses to HIV/Mtb infections.
Supplementary Material
Acknowledgments
We thank Dr. Amy Tvinnereim for helping perform the following experiments: Irradiated the mice and performed Mtb infection of the humanized mice.
Funding:
This work was partially supported by the NIH Common funds and the National Institute of Allergy and Infectious Diseases grant UG3AI150550, and the National Heart, Lung, and Blood Institute grant R01HL125016 to G.Y., and National Institute of Allergy and Infectious Diseases grant 1RO1 AI161015 to C.J.
Footnotes
Competing interests
All the authors declare no competing interests.
Data availability
All data supporting the findings of this study are available in the manuscript. If there are any special requests or questions for the data, please contact the corresponding author (G.Y.).
References
- 1.WHO Global Tuberculosis Report; WHO: Geneva, Switzerland, 2023, 2023. [Google Scholar]
- 2.Palanivel J.; Sounderrajan V.; Thangam T.; Rao S. S.; Harshavardhan S.; Parthasarathy K., Latent Tuberculosis: Challenges in Diagnosis and Treatment, Perspectives, and the Crucial Role of Biomarkers. Curr Microbiol 2023, 80 (12), 392. [DOI] [PubMed] [Google Scholar]
- 3.Kaushal D.; Singh D. K.; Mehra S., Immune Responses in Lung Granulomas during Mtb/HIV Co-Infection: Implications for Pathogenesis and Therapy. Pathogens 2023, 12 (9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO, HIV and AIDS factsheet. WHO, Ed. Geneva, Switzerland, 2023. [Google Scholar]
- 5.Azevedo-Pereira J. M.; Pires D.; Calado M.; Mandal M.; Santos-Costa Q.; Anes E., HIV/Mtb Co-Infection: From the Amplification of Disease Pathogenesis to an “Emerging Syndemic”. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO Global Tuberculosis Report; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- 7.Sharan R.; Bucşan A. N.; Ganatra S.; Paiardini M.; Mohan M.; Mehra S.; Khader S. A.; Kaushal D., Chronic Immune Activation in TB/HIV Co-infection. Trends in Microbiology 2020, 28 (8), 619–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Estes J. D.; Wong S. W.; Brenchley J. M., Nonhuman primate models of human viral infections. Nature Reviews Immunology 2018, 18 (6), 390–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okoye A. A.; Picker L. J., CD4+ T-cell depletion in HIV infection: mechanisms of immunological failure. Immunological Reviews 2013, 254 (1), 54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter R. L.; Actor J. K.; Hwang S. A.; Khan A.; Urbanowski M. E.; Kaushal D.; Jagannath C., Pathogenesis and Animal Models of Post-Primary (Bronchogenic) Tuberculosis, A Review. Pathogens 2018, 7 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaushal D.; Mehra S.; Didier P. J.; Lackner A. A., The non-human primate model of tuberculosis. J Med Primatol 2012, 41 (3), 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cepeda M.; Salas M.; Folwarczny J.; Leandro A. C.; Hodara V. L.; de la Garza M. A.; Dick E. J. Jr.; Owston M.; Armitige L. Y.; Gauduin M. C., Establishment of a neonatal rhesus macaque model to study Mycobacterium tuberculosis infection. Tuberculosis (Edinb) 2013, 93 Suppl, S51–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganatra S. R.; Bucsan A. N.; Alvarez X.; Kumar S.; Chatterjee A.; Quezada M.; Fish A.; Singh D. K.; Singh B.; Sharan R.; Lee T. H.; Shanmugasundaram U.; Velu V.; Khader S. A.; Mehra S.; Rengarajan J.; Kaushal D., Antiretroviral therapy does not reduce tuberculosis reactivation in a tuberculosis-HIV coinfection model. J Clin Invest 2020, 130 (10), 5171–5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lepard M.; Yang J. X.; Afkhami S.; Nazli A.; Zganiacz A.; Tang S.; Choi M. W. Y.; Vahedi F.; Deshiere A.; Tremblay M. J.; Xing Z.; Kaushic C.; Gillgrass A., Comparing Current and Next-Generation Humanized Mouse Models for Advancing HIV and HIV/Mtb Co-Infection Studies. Viruses 2022, 14 (9), 1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nusbaum R. J.; Calderon V. E.; Huante M. B.; Sutjita P.; Vijayakumar S.; Lancaster K. L.; Hunter R. L.; Actor J. K.; Cirillo J. D.; Aronson J.; Gelman B. B.; Lisinicchia J. G.; Valbuena G.; Endsley J. J., Pulmonary Tuberculosis in Humanized Mice Infected with HIV-1. Scientific Reports 2016, 6 (1), 21522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter R.; Actor J.; Hwang S.-A.; Khan A.; Urbanowski M.; Kaushal D.; Jagannath C., Pathogenesis and Animal Models of Post-Primary (Bronchogenic) Tuberculosis, A Review. Pathogens 2018, 7 (1), 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunter R. L.; Olsen M.; Jagannath C.; Actor J. K., Trehalose 6,6’-Dimycolate and Lipid in the Pathogenesis of Caseating Granulomas of Tuberculosis in Mice. The American Journal of Pathology 2006, 168 (4), 1249–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calderon V. E.; Valbuena G.; Goez Y.; Judy B. M.; Huante M. B.; Sutjita P.; Johnston R. K.; Estes D. M.; Hunter R. L.; Actor J. K.; Cirillo J. D.; Endsley J. J., A Humanized Mouse Model of Tuberculosis. PLoS ONE 2013, 8 (5), e63331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biradar S.; Agarwal Y.; Lotze M. T.; Bility M. T.; Mailliard R. B., The BLT Humanized Mouse Model as a Tool for Studying Human Gamma Delta T Cell-HIV Interactions In Vivo. Frontiers in Immunology 2022, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denton P. W.; Garcia J. V., Humanized mouse models of HIV infection. AIDS Rev 2011, 13 (3), 135–48. [PMC free article] [PubMed] [Google Scholar]
- 21.Victor Garcia J., Humanized mice for HIV and AIDS research. Current Opinion in Virology 2016, 19, 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huante M. B.; Saito T. B.; Nusbaum R. J.; Naqvi K. F.; Chauhan S.; Hunter R. L.; Actor J. K.; Rudra J. S.; Endsley M. A.; Lisinicchia J. G.; Gelman B. B.; Endsley J. J., Small Animal Model of Post-chemotherapy Tuberculosis Relapse in the Setting of HIV Co-infection. Frontiers in Cellular and Infection Microbiology 2020, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lang J.; Kelly M.; Freed B. M.; McCarter M. D.; Kedl R. M.; Torres R. M.; Pelanda R., Studies of Lymphocyte Reconstitution in a Humanized Mouse Model Reveal a Requirement of T Cells for Human B Cell Maturation. The Journal of Immunology 2013, 190 (5), 2090–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Q.; He F.; Kwang J.; Chan J. K. Y.; Chen J., GM-CSF and IL-4 Stimulate Antibody Responses in Humanized Mice by Promoting T, B, and Dendritic Cell Maturation. The Journal of Immunology 2012, 189 (11), 5223–5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu C. I.; Martinek J.; Wu T. C.; Kim K. I.; George J.; Ahmadzadeh E.; Maser R.; Marches F.; Metang P.; Authie P.; Oliveira V. K. P.; Wang V. G.; Chuang J. H.; Robson P.; Banchereau J.; Palucka K., Human KIT+ myeloid cells facilitate visceral metastasis by melanoma. J Exp Med 2021, 218 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coughlan A. M.; Harmon C.; Whelan S.; O’Brien E. C.; O’Reilly V. P.; Crotty P.; Kelly P.; Ryan M.; Hickey F. B.; O’Farrelly C.; Little M. A., Myeloid Engraftment in Humanized Mice: Impact of Granulocyte-Colony Stimulating Factor Treatment and Transgenic Mouse Strain. Stem Cells Dev 2016, 25 (7), 530–41. [DOI] [PubMed] [Google Scholar]
- 27.Billerbeck E.; Barry W. T.; Mu K.; Dorner M.; Rice C. M.; Ploss A., Development of human CD4+FoxP3+ regulatory T cells in human stem cell factor-, granulocyte-macrophage colony-stimulating factor-, and interleukin-3-expressing NOD-SCID IL2Rgamma(null) humanized mice. Blood 2011, 117 (11), 3076–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janke L. J.; Imai D. M.; Tillman H.; Doty R.; Hoenerhoff M. J.; Xu J. J.; Freeman Z. T.; Allen P.; Fowlkes N. W.; Iacobucci I.; Dickerson K.; Mullighan C. G.; Vogel P.; Rehg J. E., Development of Mast Cell and Eosinophil Hyperplasia and HLH/MAS-Like Disease in NSG-SGM3 Mice Receiving Human CD34+ Hematopoietic Stem Cells or Patient-Derived Leukemia Xenografts. Vet Pathol 2021, 58 (1), 181–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terahara K.; Iwabuchi R.; Tsunetsugu-Yokota Y., Perspectives on Non-BLT Humanized Mouse Models for Studying HIV Pathogenesis and Therapy. Viruses 2021, 13 (5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X.; Barnes P. F.; Huang F.; Alvarez I. B.; Neuenschwander P. F.; Sherman D. R.; Samten B., Early Secreted Antigenic Target of 6-kDa Protein of <i>Mycobacterium tuberculosis</i> Primes Dendritic Cells To Stimulate Th17 and Inhibit Th1 Immune Responses. The Journal of Immunology 2012, 189 (6), 3092–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van ’t Wout A. B.; Schuitemaker H.; Kootstra N. A., Isolation and propagation of HIV-1 on peripheral blood mononuclear cells. Nat Protoc 2008, 3 (3), 363–70. [DOI] [PubMed] [Google Scholar]
- 32.Feng Y.; Kong Y.; Barnes P. F.; Huang F.-F.; Klucar P.; Wang X.; Samten B.; Sengupta M.; Machona B.; Donis R.; Tvinnereim A. R.; Shams H., Exposure to Cigarette Smoke Inhibits the Pulmonary T-Cell Response to Influenza Virus and<i>Mycobacterium tuberculosis</i>. Infection and Immunity 2011, 79 (1), 229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreira J. D.; Iakhiaev A.; Vankayalapati R.; Jung B.-G.; Samten B., Histone deacetylase-2 controls IL-1β production through the regulation of NLRP3 expression and activation in tuberculosis infection. iScience 2022, 25 (8), 104799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morton J.; Snider T. A., Guidelines for collection and processing of lungs from aged mice for histological studies. Pathobiology of Aging & Age-related Diseases 2017, 7 (1), 1313676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davenport M. L.; Sherrill T. P.; Blackwell T. S.; Edmonds M. D., Perfusion and Inflation of the Mouse Lung for Tumor Histology. J Vis Exp 2020, (162). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tucker T. A.; Jeffers A.; Alvarez A.; Owens S.; Koenig K.; Quaid B.; Komissarov A. A.; Florova G.; Kothari H.; Pendurthi U.; Mohan Rao L. V.; Idell S., Plasminogen Activator Inhibitor-1 Deficiency Augments Visceral Mesothelial Organization, Intrapleural Coagulation, and Lung Restriction in Mice with Carbon Black/Bleomycin–Induced Pleural Injury. American Journal of Respiratory Cell and Molecular Biology 2014, 50 (2), 316–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butler S. L.; Hansen M. S. T.; Bushman F. D., A quantitative assay for HIV DNA integration in vivo. Nature Medicine 2001, 7 (5), 631–634. [DOI] [PubMed] [Google Scholar]
- 38.Zaqout S.; Becker L.-L.; Kaindl A. M., Immunofluorescence Staining of Paraffin Sections Step by Step. Frontiers in Neuroanatomy 2020, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang B.; Mukherjee T.; Radhakrishnan R.; Paidipally P.; Ansari D.; John S.; Vankayalapati R.; Tripathi D.; Yi G., HIV-Differentiated Metabolite N-Acetyl-L-Alanine Dysregulates Human Natural Killer Cell Responses to Mycobacterium tuberculosis Infection. International Journal of Molecular Sciences 2023, 24 (8), 7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ritchie M. E.; Phipson B.; Wu D.; Hu Y.; Law C. W.; Shi W.; Smyth G. K., limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 2015, 43 (7), e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chackerian A. A.; Alt J. M.; Perera T. V.; Dascher C. C.; Behar S. M., Dissemination of<i>Mycobacterium tuberculosis</i>Is Influenced by Host Factors and Precedes the Initiation of T-Cell Immunity. Infection and Immunit 2002, 70 (8), 4501–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Domínguez-Oliva A.; Hernández-Ávalos I.; Martínez-Burnes J.; Olmos-Hernández A.; Verduzco-Mendoza A.; Mota-Rojas D., The Importance of Animal Models in Biomedical Research: Current Insights and Applications. Animals 2023, 13 (7), 1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukherjee P.; Roy S.; Ghosh D.; Nandi S. K., Role of animal models in biomedical research: a review. Laboratory Animal Research 2022, 38 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rong N.; Liu J., Development of animal models for emerging infectious diseases by breaking the barrier of species susceptibility to human pathogens. Emerging Microbes & Infections 2023, 12 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao F.; Bailes E.; Robertson D. L.; Chen Y.; Rodenburg C. M.; Michael S. F.; Cummins L. B.; Arthur L. O.; Peeters M.; Shaw G. M.; Sharp P. M.; Hahn B. H., Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 1999, 397 (6718), 436–441. [DOI] [PubMed] [Google Scholar]
- 46.Nakayama E. E.; Shioda T., TRIM5α and Species Tropism of HIV/SIV. Frontiers in Microbiology 2012, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brehm M. A.; Cuthbert A.; Yang C.; Miller D. M.; Diiorio P.; Laning J.; Burzenski L.; Gott B.; Foreman O.; Kavirayani A.; Herlihy M.; Rossini A. A.; Shultz L. D.; Greiner D. L., Parameters for establishing humanized mouse models to study human immunity: Analysis of human hematopoietic stem cell engraftment in three immunodeficient strains of mice bearing the IL2rγnull mutation. Clinical Immunology 2010, 135 (1), 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martínez-Sanz J.; Díaz-Álvarez J.; Rosas Cancio-Suarez M.; Ron R.; Iribarren J. A.; Bernal E.; Gutiérrez F.; Ruiz Sancho A.; Cabello N.; Olalla J.; Moreno S.; Serrano-Villar S.; CoRIS, Expanding HIV clinical monitoring: the role of CD4, CD8, and CD4/CD8 ratio in predicting non-AIDS events. eBioMedicine 2023, 95, 104773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Serrano-Villar S.; Sainz T.; Lee S. A.; Hunt P. W.; Sinclair E.; Shacklett B. L.; Ferre A. L.; Hayes T. L.; Somsouk M.; Hsue P. Y.; Van Natta M. L.; Meinert C. L.; Lederman M. M.; Hatano H.; Jain V.; Huang Y.; Hecht F. M.; Martin J. N.; Mccune J. M.; Moreno S.; Deeks S. G., HIV-Infected Individuals with Low CD4/CD8 Ratio despite Effective Antiretroviral Therapy Exhibit Altered T Cell Subsets, Heightened CD8+ T Cell Activation, and Increased Risk of Non-AIDS Morbidity and Mortality. PLoS Pathogens 2014, 10 (5), e1004078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mikulak J.; Di Vito C.; Zaghi E.; Mavilio D., Host Immune Responses in HIV-1 Infection: The Emerging Pathogenic Role of Siglecs and Their Clinical Correlates. Frontiers in Immunology 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thoulouze M. I.; Sol-Foulon N.; Blanchet F.; Dautry-Varsat A.; Schwartz O.; Alcover A., Human Immunodeficiency Virus Type-1 Infection Impairs the Formation of the Immunological Synapse. Immunity 2006, 24 (5), 547–561. [DOI] [PubMed] [Google Scholar]
- 52.Cao S.; Li J.; Lu J.; Zhong R.; Zhong H., Mycobacterium tuberculosis antigens repress Th1 immune response suppression and promotes lung cancer metastasis through PD-1/PDl-1 signaling pathway. Cell Death & Disease 2019, 10 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCaffrey E. F.; Donato M.; Keren L.; Chen Z.; Delmastro A.; Fitzpatrick M. B.; Gupta S.; Greenwald N. F.; Baranski A.; Graf W.; Kumar R.; Bosse M.; Fullaway C. C.; Ramdial P. K.; Forgó E.; Jojic V.; Van Valen D.; Mehra S.; Khader S. A.; Bendall S. C.; Van De Rijn M.; Kalman D.; Kaushal D.; Hunter R. L.; Banaei N.; Steyn A. J. C.; Khatri P.; Angelo M., The immunoregulatory landscape of human tuberculosis granulomas. Nature Immunology 2022, 23 (2), 318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kruize Z.; Kootstra N. A., The Role of Macrophages in HIV-1 Persistence and Pathogenesis. Frontiers in Microbiology 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lerner T. R.; Borel S.; Greenwood D. J.; Repnik U.; Russell M. R. G.; Herbst S.; Jones M. L.; Collinson L. M.; Griffiths G.; Gutierrez M. G., <i>Mycobacterium tuberculosis</i>replicates within necrotic human macrophages. Journal of Cell Biology 2017, 216 (3), 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cronan M. R., In the Thick of It: Formation of the Tuberculous Granuloma and Its Effects on Host and Therapeutic Responses. Frontiers in Immunology 2022, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin Y.; Gong J.; Zhang M.; Xue W.; Barnes P. F., Production of Monocyte Chemoattractant Protein 1 in Tuberculosis Patients. Infection and Immunity 1998, 66 (5), 2319–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mishra A.; Singh V. K.; Actor J. K.; Hunter R. L.; Jagannath C.; Subbian S.; Khan A., GM-CSF Dependent Differential Control of Mycobacterium tuberculosis Infection in Human and Mouse Macrophages: Is Macrophage Source of GM-CSF Critical to Tuberculosis Immunity? Frontiers in Immunology 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson R. T., T Cell Production of GM-CSF Protects the Host during Experimental Tuberculosis. mBio 2017, 8 (6), e02087–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo J.; Zhang M.; Yan B.; Li F.; Guan S.; Chang K.; Jiang W.; Xu H.; Yuan T.; Chen M.; Deng S., Diagnostic performance of plasma cytokine biosignature combination and MCP-1 as individual biomarkers for differentiating stages Mycobacterium tuberculosis infection. J Infect 2019, 78 (4), 281–291. [DOI] [PubMed] [Google Scholar]
- 61.Hilda J. N.; Narasimhan M.; Das S. D., Neutrophils from pulmonary tuberculosis patients show augmented levels of chemokines MIP-1α, IL-8 and MCP-1 which further increase upon in vitro infection with mycobacterial strains. Hum Immunol 2014, 75 (8), 914–22. [DOI] [PubMed] [Google Scholar]
- 62.Saukkonen J. J.; Bazydlo B.; Thomas M.; Strieter R. M.; Keane J.; Kornfeld H., β-Chemokines Are Induced by<i>Mycobacterium tuberculosis</i>and Inhibit Its Growth. Infection and Immunity 2002, 70 (4), 1684–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Almeida A. S.; Lago P. C. M.; Boechat N.; Huard R. C.; Lazzarini L. C. O.; Santos A. R.; Nociari M.; Zhu H.; Perez-Sweeney B. M.; Bang H.; Ni Q.; Huang J.; Gibson A. L.; Flores V. C.; Pecanha L. R.; Kritski A. N. L.; Lapa E Silva J. R.; Ho J. L., Tuberculosis Is Associated with a Down-Modulatory Lung Immune Response That Impairs Th1-Type Immunity. The Journal of Immunology 2009, 183 (1), 718–731. [DOI] [PubMed] [Google Scholar]
- 64.Gern B. H.; Adams K. N.; Plumlee C. R.; Stoltzfus C. R.; Shehata L.; Moguche A. O.; Busman-Sahay K.; Hansen S. G.; Axthelm M. K.; Picker L. J.; Estes J. D.; Urdahl K. B. ; Gerner M. Y., TGFβ restricts expansion, survival, and function of T cells within the tuberculous granuloma. Cell Host & Microbe 2021, 29 (4), 594–606.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Knaul J. K.; Jörg S.; Oberbeck-Mueller D.; Heinemann E.; Scheuermann L.; Brinkmann V.; Mollenkopf H.-J.; Yeremeev V.; Kaufmann S. H. E.; Dorhoi A., Lung-Residing Myeloid-derived Suppressors Display Dual Functionality in Murine Pulmonary Tuberculosis. American Journal of Respiratory and Critical Care Medicine 2014, 190 (9), 1053–1066. [DOI] [PubMed] [Google Scholar]
- 66.Gerson K. D.; Yang N.; Anton L.; Levy M.; Ravel J.; Elovitz M. A.; Burris H. H., Second trimester short cervix is associated with decreased abundance of cervicovaginal lipid metabolites. American Journal of Obstetrics and Gynecology 2022, 227 (2), 273.e1–273.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prell G. D.; Martinelli G. P.; Holstein G. R.; Matulić-Adamić J.; Watanabe K. A.; Chan S. L. F.; Morgan N. G.; Haxhiu M. A.; Ernsberger P., Imidazoleacetic acid-ribotide: An endogenous ligand that stimulates imidazol(in)e receptors. Proceedings of the National Academy of Sciences 2004, 101 (37), 13677–13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pedersen M.; Nielsen C. M.; Permin H., HIV antigen-induced release of histamine from basophils from HIV infected patients. Allergy 1991, 46 (3), 206–212. [DOI] [PubMed] [Google Scholar]
- 69.Grunfeld C.; Kotler D. P.; Hamadeh R.; Tierney A.; Wang J.; Pierson R. N., Hypertriglyceridemia in the acquired immunodeficiency syndrome. The American Journal of Medicine 1989, 86 (1), 27–31. [DOI] [PubMed] [Google Scholar]
- 70.Zhang J.; Jin H.-L.; Jian F.-B.; Feng S.-L.; Zhu W.-T.; Li L.-H.; Yuan Z.-W., Evaluation of lipid metabolism imbalance in HIV-infected patients with metabolic disorders using high-performance liquid chromatography-tandem mass spectrometry. Clinica Chimica Acta 2022, 526, 30–42. [DOI] [PubMed] [Google Scholar]
- 71.Bowman E. R.; Kulkarni M.; Gabriel J.; Mo X.; Klamer B.; Belury M.; Lake J. E.; Zidar D.; Sieg S. F.; Mehta N. N.; Playford M. P.; Kuritzkes D. R.; Andrade A.;Schmidt E. K.; Taylor C.; Overton E. T.; Willig A. L.; Lederman M. M.; Funderburg N. T., Plasma lipidome abnormalities in people with HIV initiating antiretroviral therapy. Translational Medicine Communications 2020, 5 (1). [Google Scholar]
- 72.Ganguly S.; Vithlani V. V.; Kesharwani A. K.; Kuhu R.; Baskar L.; Mitramazumder P.; Sharon A.; Dev A., Synthesis, antibacterial and potential anti-HIV activity of some novel imidazole analogs. Acta Pharm 2011, 61 (2), 187–201. [DOI] [PubMed] [Google Scholar]
- 73.Abdel-Meguid S. S.; Metcalf B. W.; Carr T. J.; Demarsh P.; DesJarlais R. L.; Fisher S.; Green D. W.; Ivanoff L.; Lambert D. M.; Murthy K. H.; et al. , An orally bioavailable HIV-1 protease inhibitor containing an imidazole-derived peptide bond replacement: crystallographic and pharmacokinetic analysis. Biochemistry 1994, 33 (39), 11671–7. [DOI] [PubMed] [Google Scholar]
- 74.Loisel-Meyer S.; Swainson L.; Craveiro M.; Oburoglu L.; Mongellaz C.; Costa C.; Martinez M.; Cosset F. L.; Battini J. L.; Herzenberg L. A.; Herzenberg L. A.; Atkuri K. R.; Sitbon M.; Kinet S.; Verhoeyen E.; Taylor N., Glut1-mediated glucose transport regulates HIV infection. Proc Natl Acad Sci U S A 2012, 109 (7), 2549–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hollenbaugh J. A.; Munger J.; Kim B., Metabolite profiles of human immunodeficiency virus infected CD4+ T cells and macrophages using LC-MS/MS analysis. Virology 2011, 415 (2), 153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kirwan D. E.; Chong D. L. W.; Friedland J. S., Platelet Activation and the Immune Response to Tuberculosis. Frontiers in Immunology 2021, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Samanovic M. I.; Tu S.; Novák O.; Iyer L. M.; McAllister F. E.; Aravind L.; Gygi S. P.; Hubbard S. R.; Miroslav S.; Darwin K. H., Proteasomal Control of Cytokinin Synthesis Protects Mycobacterium tuberculosis against Nitric Oxide. Molecular Cell 2015, 57 (6), 984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yano H.; Oyanagi E.; Kato Y.; Samejima Y.; Sasaki J.; Utsumi K., l-Carnitine is essential to β-oxidation of quarried fatty acid from mitochondrial membrane by PLA2. Molecular and Cellular Biochemistry 2010, 342 (1-2), 95–100. [DOI] [PubMed] [Google Scholar]
- 79.Herbert C.; Luies L.; Loots D. T.; Williams A. A., The metabolic consequences of HIV/TB co-infection. BMC Infectious Diseases 2023, 23 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cumming B. M.; Addicott K. W.; Adamson J. H.; Steyn A. J., Mycobacterium tuberculosis induces decelerated bioenergetic metabolism in human macrophages. eLife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Daniel J.; Maamar H.; Deb C.; Sirakova T. D.; Kolattukudy P. E., Mycobacterium tuberculosis Uses Host Triacylglycerol to Accumulate Lipid Droplets and Acquires a Dormancy-Like Phenotype in Lipid-Loaded Macrophages. PLoS Pathogens 2011, 7 (6), e1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available in the manuscript. If there are any special requests or questions for the data, please contact the corresponding author (G.Y.).