Abstract
The Epstein-Barr virus (EBV) EBNA2 protein is a transcriptional activator that regulates viral and cellular gene expression and is also essential for EBV-driven immortalization of B lymphocytes. The EBNA2-responsive enhancer in the viral latency C promoter (Cp) binds two cellular factors, CBF1 and CBF2. The precise role of the CBF2 protein for Cp enhancer function is presently unclear. CBF2 does not appear to interact with EBNA2 and binds just downstream of CBF1 between positions −339 and −368 in the Cp EBNA2 enhancer. Within this region an 8-bp sequence, CAGTGCGT, can be found, and a similar sequence is also located downstream of CBF1 binding sites in other EBNA2-responsive promoters. Previous studies have indicated that mutations and methylation in this sequence affect EBNA2 responsiveness. To investigate the requirements for CBF2 binding, we synthesized a series of oligonucleotides carrying double transversion mutations spanning both the conserved core sequence and outside flanking sequences. Surprisingly, mutations outside of the conserved core sequence in 4 bases immediately flanking the 5′ end, GGTT, had the most deleterious effect on CBF2 binding. Mutations in the conserved core had a gradient effect, with those near the 5′ end having the most deleterious effects on CBF2 binding. In addition, the affinities of CBF2 for binding to the LMP-1, LMP-2, and CD23 promoters were also measured. These promoters contain the conserved core but lack the 5′ flanking GGTT motif and bound CBF2 weakly or not at all. Using Cp reporter plasmids containing CBF2 mutant binding sites, we were also able to show that at lower doses of EBNA2, Cp transactivation required a functional CBF2 binding site but that higher doses of EBNA2 transactivated CBF2 mutant promoters to 40% of wild-type levels. These assays indicate that CBF2 is important for EBNA2-mediated transactivation of the viral latency Cp. In addition, CBF2 activity was found to be associated with two polypeptides of 27 and 33 kDa.
Epstein-Barr virus (EBV) has been consistently associated with a variety of B-cell malignancies, including endemic Burkitt’s lymphomas, immunoblastic lymphomas of the immunosuppressed, and a proportion of Hodgkin’s disease (33). EBV infection of primary B cells results in the establishment of a latent infection, in which only 11 of potentially more than 80 viral proteins are expressed (17). These gene products include EBNAs 1, 2, 3A, 3B, 3C, and 5 (leader protein); latent membrane proteins (LMP) 1, 2A, and 2B; and the small noncoding EBERs 1 and 2. EBV infection of primary B cells also leads to immortalization and outgrowth of continuously proliferating lymphoblastoid cell lines (LCL). Following EBV infection, the latency W promoter (Wp) is active initially and the first viral genes expressed are EBNA-LP and EBNA2 (1, 2, 39, 52). After EBNA2 expression, transcription initiation switches from the Wp to the upstream latency C promoter (Cp), which in turn leads to expression of the other members of the EBNA family: EBNA1, -3A, -3B, and -3C (1, 2, 31, 39, 52, 53). Cp activation is dependent on a functional EBNA2 protein (41, 52). In addition to activating expression of the Cp, EBNA2 activates expression of the viral LMP-1 and LMP-2 promoters, as well as the cellular CD23 and the c-fgr proto-oncogene promoters (11, 18, 27, 49, 51, 57).
EBNA2 is a transcriptional activator that does not directly bind DNA and interacts with target promoters through contact with the ubiquitous cellular Cp binding factor 1 (CBF1), also called RBPJκ (13, 14, 23, 55). CBF1 binding sites have been observed in all of the EBNA2-responsive enhancers (13, 14, 22, 23, 27, 55). Except for the viral LMP-1 promoter, all known EBNA2-responsive promoters require interaction between EBNA2 and CBF1 for activation in transient-transfection assays (14, 16, 22, 23, 56). EBNA2 possesses an acidic activation domain located at the carboxy terminus of the protein (7, 24) and functions at least in part through interactions with the basal transcription factors TFIIB, TAF40, TFIIH, and a novel protein, p100 (45–47). Recombinant viruses expressing a gene encoding a mutant EBNA2 protein unable to interact with CBF1 or containing a deletion of the transactivation domain fail to immortalize B cells, thus linking EBNA2 transactivation and immortalization functions (6, 54).
Regulation of Cp activity may be important for governing latency gene expression patterns established in the infected cell (26, 43). This idea is supported by the fact that in infectious mononucleosis, during primary infection, and in most EBV-positive cell lines (lymphoblastoid cell lines), Cp is active (44). Under these conditions EBV infection of B cells is characterized by latency group III gene expression. Latency group III is normally characterized by the activity of the Cp and expression of the complete set of viral latent proteins, including those of the EBNA family. In contrast, tumor cells associated with Burkitt’s lymphoma, nasopharyngeal carcinoma, and Hodgkin’s disease usually contain an inactive Cp. Cell lines or tissues from these EBV-associated malignancies display group I or II patterns of latent gene expression. Latency group I is characterized by expression of only EBNA1, with transcripts initiated primarily from the Q promoter, while latency group II is characterized by expression of EBNA1, LMP-1, and LMP-2, with transcripts initiating from the Q promoter and LMP promoters. Recently, a number of studies have demonstrated that the majority of EBV-infected cells in the peripheral blood of healthy, persistently infected individuals are resting in G0 (CD23− B7−) and express LMP-2A mRNA, but not other latency mRNAs, including those derived from Cp (9, 29, 30). Repression of Cp activity may be important to reduce EBNA expression and avoid immune surveillance mechanisms of the host. In contrast, the ability to reactivate to latency group III, inducing lymphocyte proliferation, for example, may also be important for transient expansion of the pool of infected lymphocytes as an important mechanism for EBV persistence. Consistent with this notion, several mechanisms that both positively and negatively regulate Cp activity have been described. In addition to EBNA2, glucocorticoids positively modulate Cp activity through glucocorticoid response elements located upstream of the EBNA2-responsive enhancer (19). In contrast, sequence-specific methylation may contribute to silencing of Cp activity (28, 37). EBNA3 also downregulates EBNA2-mediated transactivation of the Cp (and also the LMP-1 and LMP-2 promoters) through competition for CBF1 (25, 34, 50).
The EBNA2-responsive enhancer sequences from the LMP-1 and LMP-2 promoters have also been characterized. These enhancer sequences bind many cellular factors in addition to CBF1 (16, 40, 55). Deletion and point mutation analysis of the LMP-1 promoter have suggested that other cellular factors in addition to CBF1 may mediate or enhance EBNA2 transactivation. One of these factors, PU.1, has been proposed to mediate EBNA2 interaction with the LMP-1 promoter (16, 21, 40, 55). A proposed role for a POU domain protein required for EBNA2-mediated transactivation has also been suggested (40).
The Cp EBNA2 enhancer is structurally simple, interacting with only two cellular activities (23). A 100-bp sequence from −330 to −430 bases from the Cp transcription initiation site has been mapped as the EBNA2-responsive element (15). The two cellular activities associated with this fragment have been called CBF1 and CBF2 (23). Whereas CBF1 has been extensively investigated, the identity of CBF2 and its role in EBNA2-mediated transactivation have not been determined. Several lines of evidence suggest an important role for CBF2 in EBNA2 transactivation. First, clustered mutations between −340 to −360 in the Cp, where CBF2 binds, greatly reduces EBNA2 transactivation (15). Second, synthetic promoters containing both CBF1 and CBF2 binding sites respond to EBNA2 transactivation significantly more strongly than promoters containing CBF1 binding sites alone (23). Third, examination of the EBNA2 enhancer regions in the Cp and the LMP-1 and -2 promoters reveals that sequences contributing to EBNA2 transactivation have a common 5-bp sequence, CAGTG, or a similar expanded 8-bp sequence, CAGTG(C/T)G(T/G/C). Finally, some studies have suggested that the inactivity of the Cp in some EBV cell lines correlates with CpG methylation of the CBF2 binding site and that cell lines in which Cp was active lacked CpG methylation of this site (23, 35, 37, 38). DNase protection assays also showed that CBF2 binding is sensitive to methylation and that the methylated residue maps to position 6 in the 8-bp conserved sequence CAGTGCGT (37). In addition, a synthetic reporter plasmid containing a C-to-T change in this residue results in a 50-fold reduction in the ability of this enhancer to respond to EBNA2 transactivation (37).
In this study, we further define the sequence required for CBF2 binding and we evaluate the contribution of CBF2 towards EBNA2-mediated transactivation. These experiments provide further support for an important role for CBF2 in EBNA2-mediated transactivation. The data also indicates that the most important sequences that determine CBF2 binding lie outside of a conserved 8-bp sequence found in several EBNA2-responsive enhancers. Dissecting CBF2 participation in this process will further enhance our understanding of the mechanism by which EBNA2 controls latent gene expression and B-cell immortalization.
MATERIALS AND METHODS
Cell culture.
DG75 and CA46, EBV-negative Burkitt’s lymphoma cell lines, were maintained in RPMI 1640 supplemented with 10% fetal bovine serum and incubated in 5% CO2 at 37°C.
Plasmids.
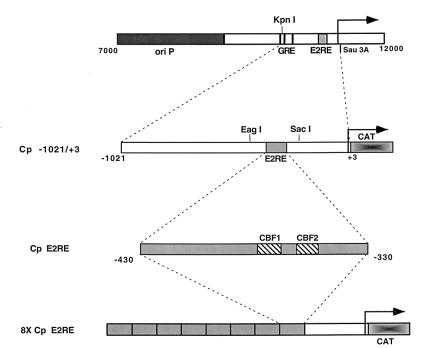
Reporter plasmids containing EBV sequences from nucleotides 10312 to 11336, which correspond to nucleotides −1021 to +3 relative to the start site of the BamHI Cp, were constructed (Fig. 1). A reporter vector, pPDL3, was constructed by subcloning a SalI fragment containing the chloramphenicol acetyltransferase (CAT) reporter gene (also contains a polyadenylation element) from the pCATB′ plasmid into the SalI site of pUC18. Plasmid pPDL5 carries the wild-type Cp sequence −1021 to +3 cloned as a KpnI/Sau3A fragment into the KpnI and BamHI sites in plasmid pPDL3. Mutations in the Cp (see Fig. 3B) were generated by site-directed mutagenesis as described by Chen and Przybyla (4). Briefly, PCR was performed with a 5′ primer that carries the desired mutation and a 3′ primer that binds downstream of the mutagenic site. The Cp amplified fragment was agarose gel purified and used as a primer together with another 5′ primer (upstream from the first 5′ primer) in a second PCR. This second product was agarose gel purified, digested with SacI and EagI, and cloned into the same sites in the wild-type Cp −1021-to-+3 fragment. Plasmid MA1 carries the wild-type Cp −1021-to-+3 fragment cloned as a KpnI/Sau3A fragment into the KpnI and BglII sites of the pGL2-Basic vector (Promega). The Sau3A/BglII junction results in retention of the BglII site. MA1 was used as the template for construction of most of the mutant fragments, except for pEFP 56 (mutant 12), in which pEFP 46 (mutant 3) was used as the template (see Fig. 1 and 3 for illustrations of constructions). Mutant promoters were digested with KpnI and BglII and inserted into the same sites of plasmid pPDL3. The following primers, with changes introduced into the wild-type sequence underlined, were used: OPL67 (mutant 3 primer), 5′-GGGAAAAAATTTATGGGGCAGTGCGTCGAGTGC-3′ (for construction of pEFP 46; Cp −1021-to-+3 mutant 3); OPL126 (mutant 11 primer), 5′-AATTTATGGTTCAGTGTGTCGAGTGCTATC-3′ (for construction of pEFP 52; Cp −1021-to-+3 mutant 11); OPL207 (mutant 12 primer), 5′-GTAAACACGCCGTGGGAAAAAATTTATTTGGCAGTGCGTCGAGTG-3′ (for construction of pEFP 56; Cp −1021-to-+3 mutant 12); and OPL213 (CBF1 mutant primer), 5′-GGTGTAAACACGCCGTTTGAAAAAATTTATGGTTC-3′ (for construction of pEFP 76; Cp −1021-to-+3 CBF1 mutant). The downstream 3′ primer used in the first round of PCR was OPL108, 5′-ACGTTGCTCCACCTCTAAGG-3′, and the upstream 5′ primer used in the second round of PCR was OPL109, 5′-CCTTGCGAACAATTATTAGT-3′.
FIG. 1.
Structure of Cp reporter constructions. A schematic illustration of the EBV genomic region containing the Cp and upstream sequences is shown at the top. Indicated are the episomal origin of replication (ori P), glucocorticoid response element (GRE), EBNA2 response element (E2RE), and the KpnI and Sau3A restriction sites. The arrow indicates the site of transcription initiation. Below are shown the Cp sequences cloned into reporter vectors (Cp −1021 to +3) and a multimerized version of the EBNA2 response element corresponding to positions −330 to −430 also cloned into a reporter vector (8X Cp E2RE).
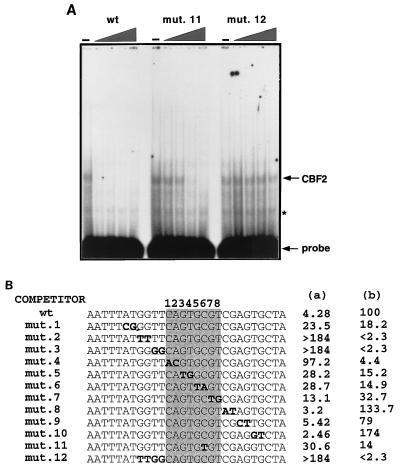
FIG. 3.
Mutagenesis analysis of the conserved 8-bp sequence in the Cp and identification of residues crucial for CBF2 binding. Nuclear extracts were mixed with 0-, 2.5-, 5-, 25-, and 125-fold molar excesses of the different unlabeled mutant oligonucleotides. A fixed amount of 32P-wild-type (wt) oligonucleotide was then added to the shift reaction mixture. The reaction mixtures were separated on 4.5% nondenaturing polyacrylamide gels, dried, and autoradiographed. Amounts of CBF2 complex were quantified with a Betascope 603 blot analyzer. (A) The EMSA gel shows binding of nuclear extract containing CBF2 to a 30-mer oligonucleotide probe from positions −339 to −368 alone or with increasing (triangle) amounts of cold competitor oligonucleotide of the same sequence (the wild type [wt]) or mutant 11 (mut. 11) or mutant 12 (mut. 12) oligonucleotide. The asterisk denotes a nonspecific band that does not compete with the CBF2-specific oligonucleotide probes and appears in lanes containing probe only. (B) Summary of competition results. The sequences shown represent the central 30 bp of the 36-mer oligonucleotide. The 8-nucleotide conserved sequence is shaded. The mutations consist of double transversion mutations, and they are shown in boldface type and underlined. The numbers in column a are concentrations of unlabeled mutant oligonucleotides required for 50% competition. The numbers in column b are percentages representing the ability of each oligonucleotide to compete for CBF2 relative to that of the wt element, which was set at 100%. Results are averages from three independent experiments.
The mutagenized Cp fragments were sequenced with the OPL109 and OPL108 primers to confirm the presence of the mutation and to confirm that no other changes were introduced.
Construction of plasmids carrying eight copies of the Cp EBNA2 enhancer region have been described previously (24). Plasmid pPDL84A contains eight copies of the wild-type Cp EBNA2-responsive element, which directs the expression of the CAT reporter (Fig. 1). DNA fragments containing mutants 3, 11, and 12 (plasmids pEFP 32, pEFP 63, and pEFP 68) were generated by PCR with primers that amplify the sequence from −330 to −430. The templates used were derived from the Cp −1021-to-+3 promoter constructions containing the appropriate mutation. Mutant CBF1 (GTCCGAA [with changes from the wild-type sequence underlined]) was generated by PCR, as previously described (4). The PCR product was digested with BglII and BamHI and cloned into the same sites of plasmid pGH56. Plasmid constructions with one copy of the Cp sequence −330 to −430 were sequenced with the forward universal primer. The Cp fragments containing sequence from −330 to −430 were then oligomerized as previously described (24), digested with BglII and BamHI, and cloned into the BglII site of the CAT expression vector pGH262. pPDL151 expresses EBNA2 and has been previously described (23).
EMSA.
Nuclear extracts were prepared from CA46 cells as previously described (23). Nuclear extracts from 30 liters of culture were fractionated by application to a heparin-Sepharose column and were eluted with a linear 0.1- to 1.0-M KCl gradient. The different fractions were tested for CBF2 activity by electrophoretic mobility shift assay (EMSA). For the EMSA, the protein-DNA complexes were formed as previously described (23). Two different DNA sequences from the Cp were used as probes: (i) the Cp EBNA2 enhancer, extending from −330 to −430 bases from the transcription initiation site, which was excised from pDL63 with BglII and BamHI, and (ii) the 30-bp oligonucleotide duplex corresponding to bases −339 to −368 of the Cp. The probe containing the sequence from −339 to −368 has been shown previously to bind CBF2 in gel mobility shift assays (23). Both fragments were end labeled with the Klenow fragment of DNA polymerase. The reaction mixtures were resolved under nondenaturing conditions on 4.5% polyacrylamide gels.
For competition assays, we synthesized a series of 30-mer oligonucleotides (and their complementary pairs) containing double transversion mutations in their sequences from −339 to −368. In addition to Cp sequences, these oligonucleotides contain BglII and BamHI overhangs that increase their overall length to 36 bp. These oligonucleotides carried mutations across the conserved 8-bp sequence and in the flanking regions (see Fig. 3B). Unlabeled mutant oligonucleotides were annealed and added to the binding reaction mixture at 2.5, 5.0, 25, and 125 molar excesses relative to the molarity of the labeled wild-type oligonucleotide probe. After EMSA, the amount of bound CBF2 activity was quantified with a Betascope 603 blot analyzer.
UV cross-linking.
An oligonucleotide containing the CBF2 binding site of the Cp (−339 to −368) and a 17-bp 3′ sequence homologous to the M13 forward sequencing primer was synthesized and annealed with the M13 forward universal primer. The sequence of the CBF2/M13 primer is 5′-AATTTATGGTTCAGTGCGTCGAGTGCTATGACTGGCCGTCGTTTTAC-3′. The oligonucleotide was converted to fully duplex form, radiolabeled, and purified as described previously (22). Binding reaction mixtures were prepared essentially as described above for EMSA, but 200,000 cpm of probe was used. After 30 min of incubation, samples were irradiated on a model TMW-20 transilluminator (UVP, Inc.) for 1 h. Digestion with 12 U of DNase I (Boehringer Mannheim) and 1 U of microccocal nuclease (Pharmacia) was then carried out for 15 min in the presence of a solution containing 7 mM Tris-HCl (pH 7.5), 7 mM MgCl2, and 7 mM CaCl2. Reaction products were electrophoresed on sodium dodecyl sulfate–12% polyacrylamide gels and visualized by autoradiography. Competition reactions were done with a 50-M excess of competitor.
Transfections and CAT assays.
DNA transfections were carried out by a DEAE-dextran method (24). Cells were transfected with the amounts of target and effector plasmids indicated in the figures. Transfections were harvested after 3 days of incubation, cells were lysed with reporter lysis buffer (Promega), and CAT assays were carried out as previously described (24). Acetylated forms of chloramphenicol in the chromatographic plates were detected by autoradiography and quantitated by excising the spot and counting in a liquid scintillation counter. A reporter vector constitutively expressing luciferase (GL3 control; Promega) was used as an internal control for transfections, and the values from CAT assays were normalized to the luciferase activity.
RESULTS
The CBF2 recognition element requires sequences that are unique to the Cp.
In gel mobility shift assays the Cp EBNA2 enhancer binds CBF1 and CBF2. By competition analysis the CBF2 binding element maps to sequences between −368 and −339 from the Cp transcription initiation site (Fig. 2A) (23). Comparison of these essential sequences with the EBNA2-responsive sequences of the viral LMP-1 and LMP-2A promoters and the cellular CD23 promoter reveals that all of these promoters have a common pentanucleotide, CAGTG, or a similar 8-bp sequence, CAGTGCGT (Fig. 2B). In addition, clustered mutations in this region of the Cp have been shown to decrease the responsiveness of this promoter to EBNA2-mediated transactivation (15). Therefore, this conserved 8-bp sequence represented a potential CBF2 recognition element. To map more precisely the sequences required for CBF2 binding in the Cp EBNA2 enhancer, we synthesized a series of oligonucleotides containing sequences from −368 to −339. Double transversion mutations were introduced into some of these oligonucleotides across the putative CBF2 binding site and in the 5′ and 3′ flanking regions (Fig. 3B). The affinity of each oligonucleotide for CBF2 was measured by its ability to compete for CBF2 binding with the wild-type sequence as evaluated by gel mobility shift assay. The wild-type oligonucleotide was radiolabeled, and its ability to bind CBF2 was measured in the presence of increasing amounts of the unlabeled wild type or mutant oligonucleotides. An example of the competitor assays is shown in Fig. 3A. A summary of data accumulated with all the mutant oligonucleotide competitors is shown in Fig. 3B. From this analysis we expected to find that regions flanking the conserved 8-bp sequence would have a moderate effect on CBF2 binding but that the most important sequences would be present in the conserved 8-bp conserved sequence. However, this analysis showed that although the conserved 8-bp sequence is important for binding, the changes that affected CBF2 binding the most lie in the 5′ flanking region, in sequences that are unique to the Cp (Fig. 2B). Paired mutations across the 4 bases flanking the 5′ side of the conserved core GGTT eliminated CBF2 binding (mutants 2 and 3), while mutant 1, which contains mutations even further upstream, reduced CBF2 binding by fivefold relative to that of the wild-type sequence. Each of the paired mutations introduced into the 3′ flanking region does not affect CBF2 binding, and some of the mutants in this region seem to be even better competitors than the wild type.
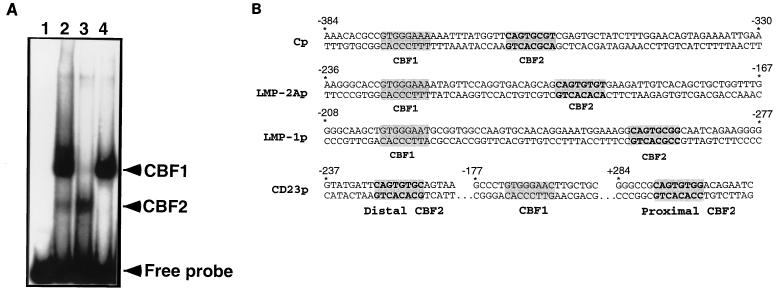
FIG. 2.
CBF1 and CBF2 bind distinct sequences in the Cp EBNA2 enhancer which are conserved in other EBNA2-responsive promoters. (A) Nuclear extracts from CA46 cells were incubated with a radiolabeled Cp sequence from positions −330 to −430 in the presence or absence of competitor oligonucleotides and analyzed by EMSA. Lanes: 1, probe only; 2, CA46 extract; 3, CA46 extract and cold 30-mer oligonucleotide competitor from positions −359 to −388 (CBF1 binding element); 4, CA46 extract and cold 30-mer oligonucleotide competitor from positions −339 to −368 (CBF2 binding element). (B) Comparison of the nucleotide sequences of EBNA2-responsive promoters and locations of their putative conserved CBF2 binding sequences. Shown also is the location of the CBF1 binding sites previously characterized in these promoters. The LMP-1 promoter (LMP-1p) contains in its natural context the consensus CBF1 binding site in the antisense orientation and is shown on top as the reverse complement strand to aid in the comparison of similarities. LMP-2Ap, LMP-2A promoter; CD23p, CD23 promoter.
Mutations inside the conserved 8-bp sequence do not completely abolish CBF2 binding, and the ability to compete for CBF2 is diluted as the paired changes are located farther from the 5′ flanking region. Mutant 4, the one with the greatest effect, lies in the 5′ border of the conserved 8-bp sequence. This mutant has 23-fold decreased affinity for CBF2. Mutations downstream of this site decreased activity by seven- to threefold.
We also tested other mutations that have been previously reported to affect the activity of the enhancer. The 4ch mutant (CGGCCGGT) contains four changes in the central part of the core and has a sixfold reduction in binding (data not shown), while mutant 11 has a sevenfold reduction. The result with mutant 11 is in agreement with that with mutant 6, whose affinity was also reduced sevenfold. Jin and Speck (15) reported that the 4ch mutant reduced the ability of the enhancer to respond to EBNA2 approximately eightfold in transient-transfection assays, but binding activities for this mutant were not reported. These results contrast with those of Robertson et al. (37), who reported that mutant 11 abolishes CBF2 binding. The differences for those results are not known, but CBF2 binding in the study by Robertson et al. was observed through direct binding to mutant DNA, a method which may not be as sensitive as our assays. Our results show that mutant 11 has an intermediate affinity for CBF2 (Fig. 3B; see below). On the other hand, mutations that lie in the 5′ flanking region were not able to compete with the wild-type oligonucleotide even at a concentration of 125-fold molar excess.
To confirm the competition results, mutant oligonucleotides 2, 3, 8, 10, and 11 were tested for their ability to directly bind CBF2 in a gel shift mobility assay. As expected, mutants 2 and 3 were unable to bind CBF2 while mutants 8 and 10 showed wild-type levels of binding. Mutant 11 retained some binding activity (data not shown). The full-length sequence of the EBNA2 enhancer was also tested for its ability to bind CBF2 in the presence of increasing amounts of some of the unlabeled mutant oligonucleotides, and results similar to those obtained with the 30-mer oligonucleotide probe were observed (data not shown).
CBF2 activity is associated only with the Cp and not with other EBNA2 enhancers.
In addition to that of the Cp, EBNA2 upregulates the expression of other viral and cellular promoters in EBV-immortalized B cells. The putative CBF2 binding sites present in the LMP-1 and LMP-2A viral promoters and the distal and proximal sites of the cellular CD23 promoter are shown in Fig. 2B. Although these promoters contain the conserved 8-bp sequence, their 5′ flanking regions are highly divergent from those of the Cp. We measured the ability of oligonucleotides containing the putative CBF2 recognition element from these other promoters for CBF2 binding by competition assays (Fig. 4). The results of these competitions show that the sequences from the other promoters fail to detectably bind CBF2. LMP-1 was able to compete at a very low level (37-fold-reduced affinity), while the other promoters could not compete even at the highest concentration of competitor tested. These sequences were also tested for their ability to directly bind CBF2 in a gel mobility shift assay, and CBF2 binding was undetectable (data not shown). This result is consistent with the result of our previous mutagenesis of the Cp, which indicated that the 5′ sequences flanking the conserved 8-bp sequence are crucial for CBF2 binding.
FIG. 4.
The LMP-1, LMP-2A, and CD23 promoters fail to bind CBF2. Competitions were carried out as described for Fig. 3. The sequences shown represent the central 30 bp of the 36-mer oligonucleotide. The 8-bp putative CBF2 binding sites are shaded. The natural changes between the sequences of the LMP-1, LMP-2A, and CD23 promoters and Cp are shown in boldface type and underlined. The numbers in column a are concentrations of unlabeled mutant oligonucleotides required for 50% competition. The numbers in column b are percentages representing the ability of each oligonucleotide to compete for CBF2 relative to that of the wild-type element, which was set at 100%. Results are averages from three independent experiments.
Activation of the Cp by EBNA2 is strongly dependent on CBF2.
To evaluate the importance of CBF2 for EBNA2-mediated transactivation of the Cp, we tested the ability of EBNA2 to transactivate wild-type and mutant Cp constructions in transient-transfection assays. Two different sequences from the Cp were used for this analysis (Fig. 1): (i) the 100-bp EBNA2 enhancer from −330 to −430 and (ii) a larger fragment spanning −1021 to +3 from the Cp. The 100-bp fragments containing −430 to −330 were multimerized to eight copies and cloned upstream of the adenovirus E1b TATA box in the reporter plasmid E1b CAT. The larger −1021-to-+3 fragment contains the natural TATA box and transcription initiation sites from the Cp, and they were cloned upstream of the CAT reporter gene.
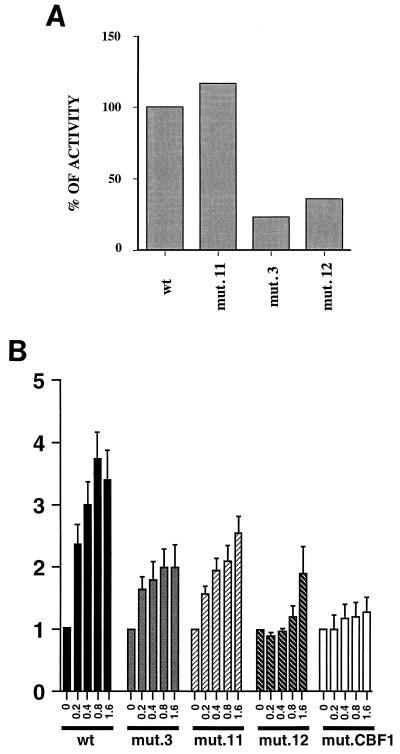
The results of these transfections are shown in Fig. 5 and summarized in Table 1. For transfection of plasmids with eight copies of the EBNA2 enhancer, 0.5 μg of EBNA2-expressing plasmid and 2 μg of each of the target plasmids were used. Fold inductions after addition of effector plasmid could not be calculated due to the low basal activity of these constructions, which was not higher than the background level given by the CAT expression vector alone, and transfections were performed with single doses of effector plasmid. Under these conditions, mutant 11 had no effect on responsiveness of the enhancer (Fig. 5A). Both mutant 3 and mutant 12, whose affinities for CBF2 are significantly weaker than that of the wild type, were transactivated by EBNA2 to 23 and 36% of wild-type levels, respectively.
FIG. 5.
Analysis of the EBNA2 responsiveness of wild-type (wt) and CBF2 Cp mutants. (A) Results of single-dose transfections to compare levels of EBNA2 transactivation of reporter plasmids containing eight tandem copies of the EBNA2 enhancer. DG75 cells were cotransfected with 2 μg of the target plasmid and 0.5 μg of the effector EBNA2-expressing plasmid. Results are averages from two independent experiments. mut., mutant. (B) Dose responses for comparison of levels of EBNA2 transactivation of reporter plasmids containing the Cp −1021-to-+3 sequence. Cells were transfected with 2 μg of target plasmid and 0.2, 0.4, 0.8, and 1.6 μg of effector EBNA2-expressing plasmid. Results are averages from five independent experiments. Standard errors of the means are indicated with T bars.
TABLE 1.
Summary of levels of transactivation of the wild-type and mutant Cps
| Plasmid | Fragment | Mutation | % of wild-type activity |
|---|---|---|---|
| pPDL84A | 8X E2REa | Wild type | 100 |
| pEFP 63 | Mutant 11 | 117 | |
| pEFP 32 | Mutant 3 | 23 | |
| pEFP 68 | Mutant 12 | 36 | |
| pPDL5 | Cp −1021 to +3 | Wild type | 100 |
| pEFP 52 | Mutant 11 | 40.3–63.3 | |
| pEFP 46 | Mutant 3 | 36.6–45.5 | |
| pEFP 56 | Mutant 12 | 0–36.2 | |
| pEFP 76 | Mutant CBF1 | 0–10.6 |
8X E2RE, a multimerized version of the EBNA2 response element corresponding to positions −330 to −430 and cloned into a reporter vector.
Figure 5B shows the results of transfections with the Cp −1021-to-+3 constructions. Table 1 shows the range of activities obtained with these plasmids in response to the different concentrations of EBNA2. In general, the activities of the mutants correlated well with the CBF2 binding affinities. Mutant 11 had an intermediate effect (up to 63% of the activity of the wild-type promoter), while mutant 3 was a slightly worse responder, with its maximal activity being 45% of wild-type levels. Interestingly, mutant 12 was not responsive at the lowest concentrations tested, but with increasing amounts of EBNA2, it was activated to 36% of wild-type levels. As expected, the CBF1 mutant was the one with the lowest activity and was not activated to a significant degree at any EBNA2 concentration tested. The results of these transfection experiments indicate that CBF2 binding contributes significantly to EBNA2 transactivation.
Two polypeptides of 33 and 25 kDa are associated with the Cp CBF2 recognition element.
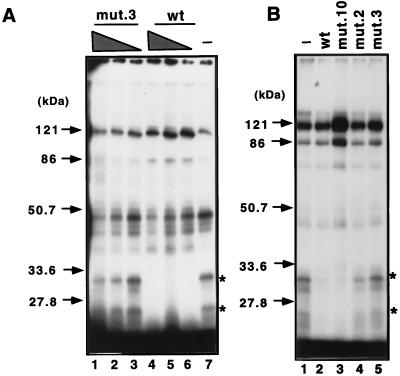
In order to identify proteins that specifically associate with the CBF2 site of the Cp, bromo-dUTP-containing oligonucleotide probes representing the sequences from −368 to −339 were incubated with the CA46 nuclear extracts under conditions similar to those employed in the shift assays. The samples were irradiated with UV light to cross-link the DNA-protein complexes formed during the binding reaction and digested with nucleases to remove unbound DNA. Figure 6A shows two polypeptides of 33 and 25 kDa specifically associated with the Cp DNA probe containing sequence −368 to −339. Competition with a 75-fold molar excess of unlabeled mutant 3 probe did not affect the binding of these peptides, while competition with only a 25-fold molar excess of the unlabeled wild-type probe was sufficient to abolish binding to the CBF2 response element. In an independent experiment, 50-fold molar excesses of mutants 2, 3, and 10 were also tested (Fig. 6B). As expected, while mutant 10 resembled the wild-type oligonucleotide probe in binding, mutant 2 and 3 oligonucleotides did not compete for binding. This result confirms the importance of the 5′ flanking sequences for CBF2 binding. These results suggest that the 33- and 25-kDa polypeptides are candidates for nuclear factors that may bind and mediate CBF2 activity.
FIG. 6.
UV-cross-linking analysis demonstrating that the CBF2 binding site specifically interacts with two polypeptides of 33 and 25 kDa. (A) Competition with different amounts of unlabeled competitors. Reaction mixtures containing UV-cross-linked proteins were resolved on sodium dodecyl sulfate–12.5% polyacrylamide gels, dried, and autoradiographed. To some cross-linking reaction mixtures cold competitor oligonucleotides were added as indicated. Lanes 1 to 3, competition with 75-, 50-, and 25-fold molar excesses, respectively, of mutant 3 (mut.3); lanes 4 to 6, competition with 75-, 50-, and 25-fold molar excesses, respectively, of the wild-type (wt) sequence; lane 7, no competition. (B) Competition with 50-fold molar excesses of wild-type, mutant 10, mutant 2, and mutant 3 oligonucleotides. Asterisks indicate specific bands of 33 and 25 kDa interacting with the CBF2 probe. Lane 1 contains no competitor.
DISCUSSION
Understanding the mechanisms of EBNA2-regulated gene expression during viral latency is essential to understanding EBV-driven B-cell immortalization. The viral latency Cp provides a simple model to study EBNA2-transactivating mechanisms. This enhancer binds two cellular proteins, CBF1 and CBF2. CBF1 is responsible for tethering EBNA2 to promoter DNA, and as such, CBF1 binding elements have been identified in all known EBNA2 enhancer elements.
In contrast, CBF2 has not been well-characterized. Sequence comparisons reveal similar 8-bp sequences, CAGT(C/T)G(T/G/C), in all known EBNA2-responsive promoters. The 8-bp conserved sequence in the Cp is protected in DNase I footprinting analysis with nuclear extracts from B lymphocytes (15, 23, 37). In addition, some mutations in the 8-bp conserved sequence affect transactivation by EBNA2 (15). Based on these observations we hypothesized (i) that several of the known EBNA2-responsive promoters bind CBF2, (ii) that the 8-bp conserved sequence is responsible for this binding, and (iii) that CBF2 plays a general role in EBNA2-transactivating function. Surprisingly, although we found that the conserved 8-bp core influenced binding, mutations adjacent to and upstream of this conserved region produced more-dramatic effects on CBF2 binding.
These results were obtained in both EMSA and UV-cross-linking analyses. These analyses identified the sequence GGTTCA as the basic core element required for CBF2 binding. Transversion changes in these sequences eliminated CBF2 binding. The sequences that flank GGTTCA were also found to decrease binding affinity up to sevenfold (e.g., mutations in the 5′ AT and those in the 3′ GTGC). Our data is also consistent with the results of the footprinting analysis by Jin and Speck (15), where the GGTTCA sequence was protected in the absence of CBF1 binding. Database searches with the basic hexanucleotide GGTTCA, the extended sequence ATGGTTCAGTGC, or the conserved core sequence have not been informative in determining whether other DNA binding proteins identified so far will recognize this sequence. The UV-cross-linking results also identified that CBF2 activity is associated with two polypeptides of 25 and 33 kDa. It is unclear whether CBF2 is a heterodimer of two proteins or if the smaller 25-kDa protein is a degradation product or some spliced variant of the larger 33-kDa protein. Alternatively, the two proteins may be differentially modified species of the same protein.
The putative binding sites present in other EBNA2 enhancers were not able to bind CBF2. Although these promoters contain the conserved 8-bp sequence, their 5′ flanking regions diverge from those of the Cp sequences. Because the conserved core is not sufficient to confer CBF2 binding activity and these enhancers do not contain crucial sequences required for CBF2 binding, we predict that CBF2 does not play a significant role in the EBNA2-mediated transcriptional activation of these promoters. Consistent with this hypothesis, Meitinger et al. introduced clustered mutations into the LMP-2A promoter, some of which overlap the conserved 8-bp core (27). These mutations had only small effects on EBNA2 transactivation of the LMP-2A promoter.
The level of EBNA2 transactivation of the Cp −1021-to-+3 reporter correlated with the relative affinity of the CBF2 binding site for CBF2. Mutant 11 reduced competitor efficiency to 86% of wild-type levels. Cp −1021-to-+3 promoter constructions containing this mutation were activated by EBNA2 to 40 to 60% of wild-type levels (Table 1). Mutants 3 and 12 reduced competitor efficiency by greater than 98%. Cp −1021-to-+3 promoter constructions containing these mutations were activated to 37 to 47% and 0 to 36%, respectively (Table 1). Interestingly, the requirement for a functional CBF2 element was most apparent at low concentrations of EBNA2 in the transient-transfection assays.
Optimal transactivation of the multimerized enhancer constructions also required a functional CBF2 binding site. Both mutants 3 and 12 were transactivated to 23 and 36% of wild-type levels, respectively. Mutant 11 activity, however, resembled that of the wild-type enhancer construction. This mutation only partially reduced the affinity for CBF2. The deleterious effect predicted for this mutant may be masked due to the presence of several copies of the enhancer and the number of EBNA2 molecules that can bind this artificial promoter, which results in high levels of activity. Our results differ from those of a previous publication that demonstrate virtual loss of activity from an identical construction (37). Recent studies, however, now confirm that a multimerized version of an EBNA2 response element containing mutant 11 and cloned into a reporter vector may have only small effects on EBNA2-mediated transactivation (3). Methylation of the natural cytosine in this position has been suggested to regulate Cp expression in different types of EBV-driven lymphomas (35, 36). In the more natural context of the Cp −1021-to-+3 construction, mutant 11 causes a reduction in EBNA2 transactivation of 47 to 60%. Methylation at this site together with methylation at other positions or the presence of additional inhibitory factors may be working together to inactivate Cp in the lymphoma-derived cell lines. Also, EBNA2 is expressed from the Cp and mechanisms that reduce Cp activity will reduce the concentration of EBNA2. Under these conditions, the Cp may be more sensitive to the presence of CBF2.
The regulatory mechanism of CBF2 action is at present unclear. Previous studies have been unable to demonstrate an interaction with EBNA2 (23). Multimerized CBF2 binding sites also had no effect on transcriptional activity of a basal promoter (23). Stabilizing interactions between CBF1 and CBF2 also remain a possibility. However, due to the low concentrations of CBF2 derived from nuclear extracts, it has thus far not been possible to obtain sufficient quantities of pure CBF2 to test this hypothesis (unpublished observations). Recent work has revealed a distinct class of proteins termed architectural transcription factors (20). Examples of such factors are the high-mobility-group proteins that include LEF-1 (48). Like CBF2, multimerized LEF-1 binding sites were unable to activate transcription of a basal promoter element. In the context of the T-cell receptor α enhancer, LEF-1 DNA bending appears to facilitate interaction of other promoter-bound factors and to further activate enhancer activity (48). Whether CBF2 is an architectural factor or targets an interaction of other important transcription factors awaits the identification of the gene product.
The presence of the 8-bp sequence conserved among the EBNA2 enhancers may be explained by several reasons. First, this core may represent the remains of a lost regulatory element. It is possible that CBF2 was required for the activation of these enhancers (e.g., LMP-1, LMP-2A, and CD23) but that binding to CBF2 was not necessary as the enhancers gained the ability to bind other cellular factors that could substitute for CBF2. As evidence for this, the LMP-2A promoter region of the closely related herpesvirus papio does not contain the conserved core (12). Alternatively, there may be cellular or viral activities, different from that of CBF2, that associate with the conserved 8-bp sequence. This activity may be present only under a given condition or stage of viral infection that has heretofore been undetected.
The role of the EBNA2-responsive enhancer for Cp expression has recently been challenged. Viral recombinants containing mutations in the Cp CBF1 binding site were viable, and some clones retained almost wild-type Cp activity (10). One possibility for this result is that the activity of the EBNA2-responsive enhancer was not sufficiently reduced by mutation of the CBF1 binding site alone and may require additional mutations, for example, in the CBF2 element. In vitro, viral recombinants containing large deletions in Cp are also immortalizing competent and use Wp (42). While Cp usage may not be required for immortalization in vitro, transcripts initiating from Cp in EBV-infected lymphocytes in vivo are detected in individuals with both primary and persistent infection (5, 32, 44). Several studies are also consistent with CBF2 having a significant role in Cp activation. Contreras-Brodin et al. (8) showed that Cp activity is restricted to B cells. Unlike CBF1, which is constitutively expressed in almost all cell types, CBF2 appears to be expressed only in lymphocytes (15, 52). It is intriguing to speculate that CBF2 may be an important factor required for Cp activity in lymphoid cells. In addition, site-specific methylation that prevents CBF2 binding but not CBF1 binding also correlates with Cp silencing in a Burkitt’s lymphoma cell line (28, 37). Treatment with 5-azacytidine results in demethylation, CBF2 binding, and activation of Cp. Future studies involving the introduction of mutations in the CBF2 binding element in the context of the viral genome will be informative in confirming the role of CBF2 for Cp activity.
Overall, mutagenesis of the CBF2 response element permitted us to determine that the sequence GGTTCA is essential for CBF2 binding, with the immediately flanking sequences influencing the affinity for this element. Furthermore, the transient-transfection analysis provided evidence for the importance of CBF2 function in the Cp EBNA2 enhancer. Further characterization and identification of the CBF2 gene will contribute to a better understanding of the mechanisms controlling Cp activity during latent infections.
ACKNOWLEDGMENTS
We thank R. T. Javier and A. P. Rice for their critical reading of the manuscript.
This work was supported by NIH grant R29 CA69437 to P.D.L. and a Leukemia Society Special Fellowship to P.D.L.
REFERENCES
- 1.Alfieri C, Birkenbach M, Kieff E. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology. 1991;181:595–608. doi: 10.1016/0042-6822(91)90893-g. [DOI] [PubMed] [Google Scholar]
- 2.Allday M J, Crawford D H, Griffin B E. Epstein-Barr virus latent gene expression during the initiation of B cell immortalization. J Gen Virol. 1989;70:1755–1764. doi: 10.1099/0022-1317-70-7-1755. [DOI] [PubMed] [Google Scholar]
- 3.Ambinder, R. F. Personal communication.
- 4.Chen B, Przybyla A E. An efficient site-directed mutagenesis method based on PCR. BioTechniques. 1994;17:657–659. [PubMed] [Google Scholar]
- 5.Chen F, Zou J Z, di Renzo L, Winberg G, Hu L F, Klein E, Klein G, Ernberg I. A subpopulation of normal B cells latently infected with Epstein-Barr virus resembles Burkitt lymphoma cells in expressing EBNA-1 but not EBNA-2 or LMP1. J Virol. 1995;69:3752–3758. doi: 10.1128/jvi.69.6.3752-3758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen J I. A region of herpes simplex virus VP16 can substitute for a transforming domain of Epstein-Barr virus nuclear protein 2. Proc Natl Acad Sci USA. 1992;89:8030–8034. doi: 10.1073/pnas.89.17.8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen J I, Kieff E. An Epstein-Barr virus nuclear protein 2 domain essential for transformation is a direct transcriptional activator. J Virol. 1991;65:5880–5885. doi: 10.1128/jvi.65.11.5880-5885.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Contreras-Brodin B, Karlsson A, Nilsson T, Rymo L, Klein G. B cell-specific activation of the Epstein-Barr virus-encoded C promoter compared with the wide-range activation of the W promoter. J Gen Virol. 1996;77:1159–1162. doi: 10.1099/0022-1317-77-6-1159. [DOI] [PubMed] [Google Scholar]
- 9.Decker L L, Klaman L D, Thorley-Lawson D A. Detection of the latent form of Epstein-Barr virus DNA in the peripheral blood of healthy individuals. J Virol. 1996;70:3286–3289. doi: 10.1128/jvi.70.5.3286-3289.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans T J, Farrell P J, Swaminathan S. Molecular genetic analysis of Epstein-Barr virus Cp promoter function. J Virol. 1996;70:1695–1705. doi: 10.1128/jvi.70.3.1695-1705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fahraeus R, Jansson A, Ricksten A, Sjoblom A, Rymo L. Epstein-Barr virus-encoded nuclear antigen 2 activates the viral latent membrane protein promoter by modulating the activity of a negative regulatory element. Proc Natl Acad Sci USA. 1990;87:7390–7394. doi: 10.1073/pnas.87.19.7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franken M, Annis B, Ali A N, Wang F. 5′ coding and regulatory region sequence divergence with conserved function of the Epstein-Barr virus LMP2A homolog in herpesvirus papio. J Virol. 1995;69:8011–8019. doi: 10.1128/jvi.69.12.8011-8019.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grossman S R, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 15.Jin X W, Speck S H. Identification of critical cis elements involved in mediating Epstein-Barr virus nuclear antigen 2-dependent activity of an enhancer located upstream of the viral BamHI C promoter. J Virol. 1992;66:2846–2852. doi: 10.1128/jvi.66.5.2846-2852.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johannsen E, Koh E, Mosislos G, Tong X, Kieff E, Grossman S R. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU.1. J Virol. 1995;69:253–262. doi: 10.1128/jvi.69.1.253-262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 107–172. [Google Scholar]
- 18.Knutson J C. The level of c-fgr RNA is increased by EBNA-2, an Epstein-Barr virus gene required for B-cell immortalization. J Virol. 1990;64:2530–2536. doi: 10.1128/jvi.64.6.2530-2536.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kupfer S R, Summers W C. Identification of a glucocorticoid-responsive element in Epstein-Barr virus. J Virol. 1990;64:1984–1990. doi: 10.1128/jvi.64.5.1984-1990.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- 21.Laux G, Dugrillon F, Eckert C, Adam B, Zimber S U, Bornkamm G W. Identification and characterization of an Epstein-Barr virus nuclear antigen 2-responsive cis element in the bidirectional promoter region of latent membrane protein and terminal protein 2 genes. J Virol. 1994;68:6947–6958. doi: 10.1128/jvi.68.11.6947-6958.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ling P D, Hsieh J J, Ruf I K, Rawlins D R, Hayward S D. EBNA-2 upregulation of Epstein-Barr virus latency promoters and the cellular CD23 promoter utilizes a common targeting intermediate, CBF1. J Virol. 1994;68:5375–5383. doi: 10.1128/jvi.68.9.5375-5383.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ling P D, Rawlins D R, Hayward S D. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc Natl Acad Sci USA. 1993;90:9237–9241. doi: 10.1073/pnas.90.20.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ling P D, Ryon J J, Hayward S D. EBNA-2 of herpesvirus papio diverges significantly from the type A and type B EBNA-2 proteins of Epstein-Barr virus but retains an efficient transactivation domain with a conserved hydrophobic motif. J Virol. 1993;67:2990–3003. doi: 10.1128/jvi.67.6.2990-3003.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall D, Sample C. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J Virol. 1995;69:3624–3630. doi: 10.1128/jvi.69.6.3624-3630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masucci M G, Ernberg I. Epstein-Barr virus: adaptation to a life within the immune system. Trends Microbiol. 1994;2:125–130. doi: 10.1016/0966-842x(94)90599-1. [DOI] [PubMed] [Google Scholar]
- 27.Meitinger C, Strobl L J, Marschall G, Bornkamm G W, Zimber-Strobl U. Crucial sequences within the Epstein-Barr virus TP1 promoter for EBNA2-mediated transactivation and interaction of EBNA2 with its responsive element. J Virol. 1994;68:7497–7506. doi: 10.1128/jvi.68.11.7497-7506.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minarovits J, Hu L F, Minarovits K S, Klein G, Ernberg I. Sequence-specific methylation inhibits the activity of the Epstein-Barr virus LMP1 and BCR2 enhancer-promoter regions. Virology. 1994;200:661–667. doi: 10.1006/viro.1994.1229. [DOI] [PubMed] [Google Scholar]
- 29.Miyashita E M, Yang B, Babcock G J, Thorley-Lawson D A. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J Virol. 1997;71:4882–4891. doi: 10.1128/jvi.71.7.4882-4891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyashita E M, Yang B, Lam K M, Crawford D H, Thorley-Lawson D A. A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell. 1995;80:593–601. doi: 10.1016/0092-8674(95)90513-8. [DOI] [PubMed] [Google Scholar]
- 31.Puglielli M T, Desai N, Speck S H. Regulation of EBNA gene transcription in lymphoblastoid cell lines: characterization of sequences downstream of BCR2 (Cp) J Virol. 1997;71:120–128. doi: 10.1128/jvi.71.1.120-128.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qu L, Rowe D T. Epstein-Barr virus latent gene expression in uncultured peripheral blood lymphocytes. J Virol. 1992;66:3715–3724. doi: 10.1128/jvi.66.6.3715-3724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2397–2446. [Google Scholar]
- 34.Robertson E S, Grossman S, Johannsen E, Miller C, Lin J, Tomkinson B, Kieff E. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein Jκ. J Virol. 1995;69:3108–3116. doi: 10.1128/jvi.69.5.3108-3116.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robertson K D, Ambinder R F. Regulation of the Epstein-Barr virus BamHI-C promoter by DNA methylation. Epstein-Barr Virus Rep. 1996;3:115–121. [Google Scholar]
- 36.Robertson K D, Ambinder R F. Mapping promoter regions that are hypersensitive to methylation-mediated inhibition of transcription: application of the methylation cassette assay to the Epstein-Barr virus major latency promoter. J Virol. 1997;71:6445–6454. doi: 10.1128/jvi.71.9.6445-6454.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robertson K D, Hayward S D, Ling P D, Samid D, Ambinder R F. Transcriptional activation of the Epstein-Barr virus latency C promoter after 5-azacytidine treatment: evidence that demethylation at a single CpG site is crucial. Mol Cell Biol. 1995;15:6150–6159. doi: 10.1128/mcb.15.11.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson K D, Swinnen L J, Zong J C, Gulley M L, Ambinder R F. CpG methylation of the major Epstein-Barr virus latency promoter in Burkitt’s lymphoma and Hodgkin’s disease. Blood. 1996;88:3129–3136. [PubMed] [Google Scholar]
- 39.Rooney C, Howe J G, Speck S H, Miller G. Influence of Burkitt’s lymphoma and primary B cells on latent gene expression by the nonimmortalizing P3J-HR-1 strain of Epstein-Barr virus. J Virol. 1989;63:1531–1539. doi: 10.1128/jvi.63.4.1531-1539.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sjoblom A, Jansson A, Yang S, Lain S, Nilsson T, Rymo L. PU box-binding transcription factors and a POU domain protein cooperate in the Epstein-Barr virus (EBV) nuclear antigen 2-induced transactivation of the EBV latent membrane protein 1 promoter. J Gen Virol. 1995;76:2679–2692. doi: 10.1099/0022-1317-76-11-2679. [DOI] [PubMed] [Google Scholar]
- 41.Sung N S, Kenney S, Gutsch D, Pagano J S. EBNA-2 transactivates a lymphoid-specific enhancer in the BamHI C promoter of Epstein-Barr virus. J Virol. 1991;65:2164–2169. doi: 10.1128/jvi.65.5.2164-2169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swaminathan S. Characterization of Epstein-Barr virus recombinants with deletions of the BamHI C promoter. Virology. 1996;217:532–541. doi: 10.1006/viro.1996.0148. [DOI] [PubMed] [Google Scholar]
- 43.Thorley-Lawson D A, Miyashita E M, Khan G. Epstein-Barr virus and the B cell: that’s all it takes. Trends Microbiol. 1996;4:204–208. doi: 10.1016/s0966-842x(96)90020-7. [DOI] [PubMed] [Google Scholar]
- 44.Tierney R J, Steven N, Young L S, Rickinson A B. Epstein-Barr virus latency in blood mononuclear cells: analysis of viral gene transcription during primary infection and in the carrier state. J Virol. 1994;68:7374–7385. doi: 10.1128/jvi.68.11.7374-7385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tong X, Drapkin R, Reinberg D, Kieff E. The 62- and 80-kDa subunits of transcription factor IIH mediate the interaction with Epstein-Barr virus nuclear protein 2. Proc Natl Acad Sci USA. 1995;92:3259–3263. doi: 10.1073/pnas.92.8.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tong X, Drapkin R, Yalamanchili R, Mosialos G, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol Cell Biol. 1995;15:4735–4744. doi: 10.1128/mcb.15.9.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tong X, Wang F, Thut C J, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain can interact with TFIIB, TAF40, and RPA70 but not with TATA-binding protein. J Virol. 1995;69:585–588. doi: 10.1128/jvi.69.1.585-588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function. Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 49.Tsang S F, Wang F, Izumi K M, Kieff E. Delineation of the cis-acting element mediating EBNA-2 transactivation of latent infection membrane protein expression. J Virol. 1991;65:6765–6771. doi: 10.1128/jvi.65.12.6765-6771.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waltzer L, Perricaudet M, Sergeant A, Manet E. Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-Jκ–EBNA2-activated transcription by inhibiting the binding of RBP-Jκ to DNA. J Virol. 1996;70:5909–5915. doi: 10.1128/jvi.70.9.5909-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang F, Kikutani H, Tsang S F, Kishimoto T, Kieff E. Epstein-Barr virus nuclear protein 2 transactivates a cis-acting CD23 DNA element. J Virol. 1991;65:4101–4106. doi: 10.1128/jvi.65.8.4101-4106.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woisetschlaeger M, Jin X W, Yandava C N, Furmanski L A, Strominger J L, Speck S H. Role for the Epstein-Barr virus nuclear antigen 2 in viral promoter switching during initial stages of infection. Proc Natl Acad Sci USA. 1991;88:3942–3946. doi: 10.1073/pnas.88.9.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woisetschlaeger M, Yandava C N, Furmanski L A, Strominger J L, Speck S H. Promoter switching in Epstein-Barr virus during the initial stages of infection of B lymphocytes. Proc Natl Acad Sci USA. 1990;87:1725–1729. doi: 10.1073/pnas.87.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yalamanchili R, Tong X, Grossman S, Johannsen E, Mosialos G, Kieff E. Genetic and biochemical evidence that EBNA 2 interaction with a 63-kDa cellular GTG-binding protein is essential for B lymphocyte growth transformation by EBV. Virology. 1994;204:634–641. doi: 10.1006/viro.1994.1578. [DOI] [PubMed] [Google Scholar]
- 55.Zimber-Strobl U, Kremmer E, Grasser F, Marschall G, Laux G, Bornkamm G W. The Epstein-Barr virus nuclear antigen 2 interacts with an EBNA2 responsive cis-element of the terminal protein 1 gene promoter. EMBO J. 1993;12:167–175. doi: 10.1002/j.1460-2075.1993.tb05642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zimber-Strobl U, Strobl L J, Meitinger C, Hinrichs R, Sakai T, Furukawa T, Honjo T, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-J kappa, the homologue of Drosophila Suppressor of Hairless. EMBO J. 1994;13:4973–4982. doi: 10.1002/j.1460-2075.1994.tb06824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zimber-Strobl U, Suentzenich K O, Laux G, Eick D, Cordier M, Calender A, Billaud M, Lenoir G M, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 activates transcription of the terminal protein gene. J Virol. 1991;65:415–423. doi: 10.1128/jvi.65.1.415-423.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]