Abstract
Herein, we focus on native mass spectrometry (nMS) combined with a fast, tunable gas-phase charge reduction, electron capture charge reduction (ECCR), and illustrate its utility in the characterization of protein complex topology and glycoprotein heterogeneity. ECCR is illustrated to effectively spread the charge states of tetradecameric GroEL, illustrating Orbitrap m/z measurement out to greater than 100,000 m/z. For both the pentameric C-reactive protein and tetradecameric GroEL, our novel device combining ECCR with surface induced dissociation (SID) lowers the charge states and produces more topologically informative fragmentation. While more native-like fragmentation has previously been illustrated for complexes charge reduced by proton abstraction in solution, this is the first illustration that ECCR can lead to more native-like SID fragmentation of protein complexes. Application to protein glycosylation, one of the most common and diverse protein posttranslational modifications, is also illustrated because glycosylation is important for structural and functional properties and plays essential roles in many key biological processes. The immense heterogeneity resulting from variability in glycosylation sites and glycan composition and structure poses significant analytical challenges that hinder a mechanistic understanding of the biological role of glycosylation. Data for stabilized heavily glycosylated SARS-CoV-2 spike protein trimer and thyroglobulin dimer illustrate that ECCR enables significantly improved resolution of glycan heterogeneity. Without ECCR, the charge states of a glycoprotein complex are not resolved and average mass determination is available only through the use of charge detection mass spectrometry or mass photometry. With ECCR after narrow m/z selection, multiple glycoform m/z values are apparent, providing quick global, glycoform profiling and providing a future path for glycan localization on individual intact glycoforms (e.g., though top-down dissociation).
Graphical Abstract
INTRODUCTION
Proteins and other biological macromolecules rarely work alone. The assembly and regulation of active cellular machinery is largely dependent on structure, which is modulated by subunit interactions, protein posttranslational modifications (PTMs), and ligand binding among many other structure modifying mechanisms. PTMs are covalent modifications of proteins and play a key role in numerous biological processes, affecting both structure and function. Full characterization of protein and nucleoprotein complexes with and without PTMs, especially large or complicated systems with many small and large partners, remains an analytical challenge. Of the potential modifications a protein can undergo, glycosylation is one of the most diverse and common PTMs (with up to one-fifth of proteins potentially being glycosylated)1, both with respect to the amino acids that can be modified and the modification structures. Monosaccharides can combine in a variety of ways with varying sequences, lengths, anomeric natures, and positions and branching points of linkages. In addition, the same site can be occupied by different glycosylation events. Characterization of glycosylation is essential as many therapeutic proteins are derived from endogenous glycoproteins, and a substantial number of currently approved protein therapeutics need to be properly glycosylated to exhibit optimal therapeutic efficacy.2
Native mass spectrometry (nMS) has emerged as a powerful tool for structural biology and enables characterization of soluble and membrane protein complexes as well as heterogeneous assemblies, such as glycoprotein complexes, not readily suitable for complete high resolution structural characterization.3–6 Although native MS does not typically provide atomic level structure information, it does have advantages in speed, sensitivity, selectivity, and the ability to simultaneously measure many components of a heterogeneous assembly or mixture.7 However, extreme heterogeneity in PTMs, assembly composition, and adduction of non-volatile salts can severely hinder mass determination and structure characterization by nMS. Several approaches have been presented in recent years to overcome this issue. These include charge manipulation approaches and charge detection mass spectrometry (CDMS), in which the charge and mass-to-charge ratio are simultaneously measured. CDMS overcomes the need for resolved charge states and provides insights into heterogeneous glycoproteins and large heterogeneous systems.8–10 Charge manipulation approaches typically involve reducing the charge either in solution or the gas-phase. This moves the charge state distribution to lower charges where the spacing between charge states is larger and enables resolution of heterogenous systems.11,12
Ion-neutral reactions and ion-ion reactions have been exploited to reduce the charge acquired by denatured proteins, native proteins, and protein complexes. Smith and co-workers demonstrated that gas-phase collisions between desolvated protein ions and ammonia, methylamines, and ethylamines resulted in effective charge reduction under denaturing and native conditions.13–15 Additionally, the pioneering work by McLuckey and Stephenson demonstrated the utility of ion-ion proton transfer reactions (PTR) for the study of ion chemistry as well as enhanced analytical capabilities for polypeptide characterization.16–21 In these experiments, a multiply charged polypeptide cation was reacted with a singly-charged reagent anion in an ion trapping device to generate a cation with reduced charge and a neutral reagent molecule. PTR enabled analysis of mixtures of low, medium, and high molecular weight peptides and proteins by reducing spectral complexity introduced by multiple charging from electrospray ionization.20 Additionally, the development of “ion parking” techniques made it possible to inhibit ion-ion reaction rates via manipulation of ion velocities within ion trapping devices.22 This capability was demonstrated by concentration of ions originally present in a range of charge states into a selected charge state using PTR22 as well as inhibiting sequential electron transfer dissociation (ETD) reactions to enrich first-generation ETD product ions23. Commercially, PTR and the related proton transfer charge reduction (PTCR) has been offered on various instrument platforms.24–29 Similarly, Bush and coworkers have used a glow-discharge source to generate anions for PTR with m/z-selected ions of native proteins and complexes to enable charge assignment and mass determination.30 Sandoval and coworkers have recently demonstrated the utility of PTCR and gas-phase fractionation for the analysis of intact glycoproteins.31
Ion-ion and ion-electron reactions have predominantly been used to enable and enhance the characterization of peptides and denatured proteins by inducing covalent fragmentation for sequencing and PTM localization. However, several groups have utilized electron-based fragmentation to characterize higher order protein structure.32–34 Electron-based fragmentation methods don’t generally impart sufficient energy to disrupt noncovalent interactions in addition to protein backbone fragmentation. Thus, ETD and its ion-electron reaction counterpart, electron capture dissociation (ECD), enable mapping of surface exposed and flexible regions of protein structure.35–37 Supplemental activation via collisions with, e.g., inert gas or infrared photons are needed to release additional fragments and obtain greater sequence coverage.38–41 Without supplemental activation to disrupt noncovalent interactions in larger proteins and protein complexes, ECD and ETD can yield abundant charge reduction without dissociation of the complementary N- and C- terminal peptide backbone cleavage product ions. To date, non-dissociative electron capture or electron transfer events have generally been regarded as reaction by-products to be minimized to enhance yield of peptide backbone cleavage products. However, some work on intact proteins and complexes has sought to utilize this by-product for the characterization of heterogeneous samples. Sobott and coworkers have demonstrated the utility of electron-based charge reduction to resolve heterogeneous oligomers of αβ-crystallin,42 while Ujma and co-workers have shown its utility to resolve AAV capsids.43 The Smith group utilized corona discharge or alpha-particle sources to generate anions used for charge manipulation via proton abstraction of electrosprayed proteins.44,45
Treated as an analytical tool, tunable electron transfer or electron capture charge reduction could be used to improve the apparent resolving power for large heterogeneous proteins and protein complexes by shifting ions to lower charge and higher m/z, as previously demonstrated using PTR. However, one challenge encountered with charge-reduced proteins and protein complexes is that they do not fragment as effectively as their high-charge counterparts. It is difficult to perform effective tandem mass spectrometry (MS/MS) via collisions with an inert gas, i.e., collision induced dissociation (CID)46 at energies accessible within practical limitations of the mass spectrometer hardware and electronics. Native protein complexes activated by CID dissociate via a monomer unfolding/complex restructuring mechanism. Typical products of CID are unfolded or elongated monomers with a disproportionate amount of charge and the corresponding (n-1) multimer; while these products confirm stoichiometry the provide limited information on overall complex topology/connectivity.47 Surface collisions, i.e., surface induced dissociation (SID), on the other hand proceed via fast, high-energy deposition to yield more structurally informative subcomplexes, with the weakest protein complex interfaces cleaving first, producing products with compact structures and more proportionate charge partitioning.48–54 The Wysocki group has recently developed a simple yet novel split lens geometry SID device that was readily adapted and integrated into Q-IM-TOF, FTICR, and Orbitrap mass spectrometry platforms.55
Here, we combine electron capture charge reduction with SID, via a novel ExD cell40,41 designed for improved transmission of high m/z ions and facile integration of split lens SID electrodes at the exit of the ExD cell. The cell provides effective SID, extensive and tunable electron capture charge reduction (ECCR), or a combination of the two (ECCR-SID), with each of these modes illustrated for large protein complexes. Tetradecameric GroEL (800 kDa) captured more than 50 electrons in a single pass through the cell and enabled observation of charge reduced peaks at m/z greater than 100,000. Charge-reduced GroEL was also subjected to SID to determine whether the gas-phase charge reduced precursors gave more native fragmentation. ECCR was also applied to heterogeneous glycoproteins, including a construct of the SARS-CoV-2 spike protein and thyroglobulin enabling resolution of overlapping glycoform charge state distributions and hence mass determination.
EXPERIMENTAL SECTION
Materials and Sample Preparation.
C-reactive protein, GroEL, β-Amylase (BAM), and thyroglobulin were purchased from Sigma Aldrich. VFLIP (a stable, covalently linked SARS-coV-2 spike trimer with the D614G mutation of the Wuhan variant) was expressed and purified as previously described.56 GroEL was refolded based on a previously described protocol.52 CRP, thyroglobulin, GroEL and VFLIP were buffer exchanged into 200 mM ammonium acetate using BioRad micro P6 columns. VFLIP for narrow window selection was buffer exchanged into 200 mM ammonium acetate using 50 kDa molecular weight cut-off (MWCO) Amicon® Ultra Centrifugal Filter (Sigma Aldrich). Samples were run at approximately 1 μM complex concentration for native mass spectrometry, 500 nM for CDMS, and 10 nM for mass photometry.
Mass Spectrometry.
All experiments were performed using a Q Exactive UHMR Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) fitted with a hybrid device enabling electron-based fragmentation and surface induced dissociation, here referred to as the ExD-SID cell (cell generated by modifying ExD from e-MSion Inc., Corvallis, OR with SID, see results). The ExD-SID cell replaced the transfer multipole between the quadrupole mass filter and C-trap as previously described for the standard ExD cell37,40,41 and standard 4cm SID cell.57 Static nanoelectrospray ionization was performed using uncoated glass capillary emitters pulled in-house from borosilicate glass with a filament (O.D 1mm I.D 0.78 mm) using a Sutter P-97 micropipette puller. A platinum wire was inserted into the emitter, and the electrospray voltage was directly applied to the analyte solution. Typical electrospray voltage was 900 V. In-source trapping collision energy for desolvation was optimized for each sample and was generally between −30 and −100 V. An inlet capillary temperature of 250 °C, S-lens RF of 200, and a fixed ion injection time of 50−1000 ms were used for all experiments. Approximately 2–5 minutes of averaging was used to produce ECCR, SID, ECCR-SID spectra. Deconvolution and average charge state analysis was performed using UniDec.58 The UniDec parameters for Figure 5C are Charge Range: 1–60, Mass range 400–600 kDa, Sample Mass Every (Da) 1, Peak FWHM(Th) 200.0, Beta: 50.0, Charge Smooth Width: 1.0, Point Smooth Width: 10.0, Mass Smooth Width: 0.0, Maximum # of Iterations: 100, Peak Detection 5000 Da, Peak Detection Threshold 0.5. For the rest narrow quadrupole selections, the parameters were Charge Range: 1–50, Mass range 400–600 kDa, Sample Mass Every (Da) 1, Peak FWHM(Th) 5.0, Beta: 0, Charge Smooth Width: 1.0, Point Smooth Width: 1, Mass Smooth Width: 0, Maximum # of Iterations: 100, Peak Detection 500 Da, Peak Detection Threshold 0.1. The CDMS data were acquired at 200,000 resolving power atm/z 400 and trapping gas 2 using Direct Mass Technology mode. The CDMS datasets were processed and deconvolved by STORIboard software (Thermo Fisher Scientific).59 We note here that with extensive use of the ExD-SID cell, some surface contamination is detected; work is in progress to define the problem and resolve it with a cell redesign, although we continue using the device to solve real-world problems.
Figure 5.
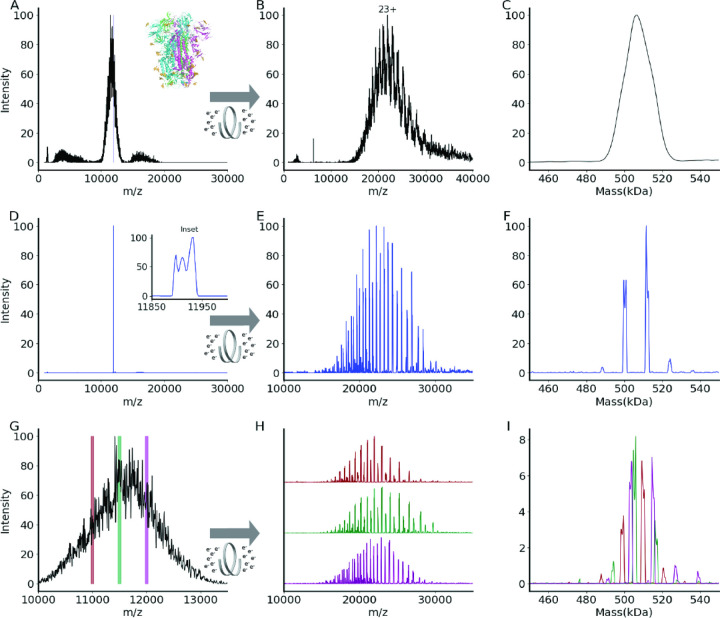
(A) An unresolved native mass spectrum of the heterogeneously glycosylated spike trimer VFLIP.56 Inset shows the ribbon structure of SARS-CoV-2 spike trimer (PDB: 6X79). Three protomers are shown in pink, green, or blue, and N-glycans are shown in gold. (B) A charge state-resolved native mass spectrum of the spike trimer after electron capture charge reduction (ECCR, voltage 7V). (C) The average mass is 506 kDa deconvolved using UniDec.58 (D) Selection of a narrow m/z window (11900–11950) of the spike trimer without charge reduction. (E) A charge state-resolved native mass spectrum after the m/z selection of D and electron capture charge reduction (ECCR, voltage 7 V). (F) The detected masses are 499,799 Da, 500,950 Da, 511,614 Da, 512,771 Da. This was deconvolved using UniDec. 58 (G) Zoom in for spike trimer VFLIP from A with the positions of three narrow quadrupole selections shown. H) Plots showing ECCR corresponding to the three narrow window isolations (10975–11025 (maroon); 11475–11525 (green); 11975–12025 (magenta), respectively. (I) Overlayed deconvolved mass spectra for data shown in panel G. The intensity was not normalized to better reflect the original intensity in panel G. The detected masses for first narrow window selection (maroon) are 499,645 Da, 509,355 Da, 520,689 Da; those for the second (green) are 494,331 Da, 504,835 Da, 506,070 Da, 516,276 Da, and 517,408 Da; those for the third selection (magenta) are 503,916 Da, 514,746, and 526,832 Da.
RESULTS AND DISCUSSION
In previous work in our lab and others, when it has been desirable to obtain topological/connectivity information on protein complexes low charge states have been used or the charge has been intentionally reduced. Charge reduction has been accomplished with solution additives (e.g., addition of the more basic triethylammonium acetate to the typical ammonium acetate electrolyte used for nanoelectrospray) or an alternative electrolyte (e.g., ethylene diammonium diacetate).60–62 While this accomplishes the needed charge reduction, solution additives particularly triethylammonium acetate often cause peak broadening, influencing mass accuracy. While adducts can be removed by collisional activation, this activation can restructure protein complexes, leading to non-native fragmentation of the complex, obscuring desirable topology information. There has thus been a strong need to accomplish charge reduction without adducting and peak broadening.
Hybrid ExD-SID Cell.
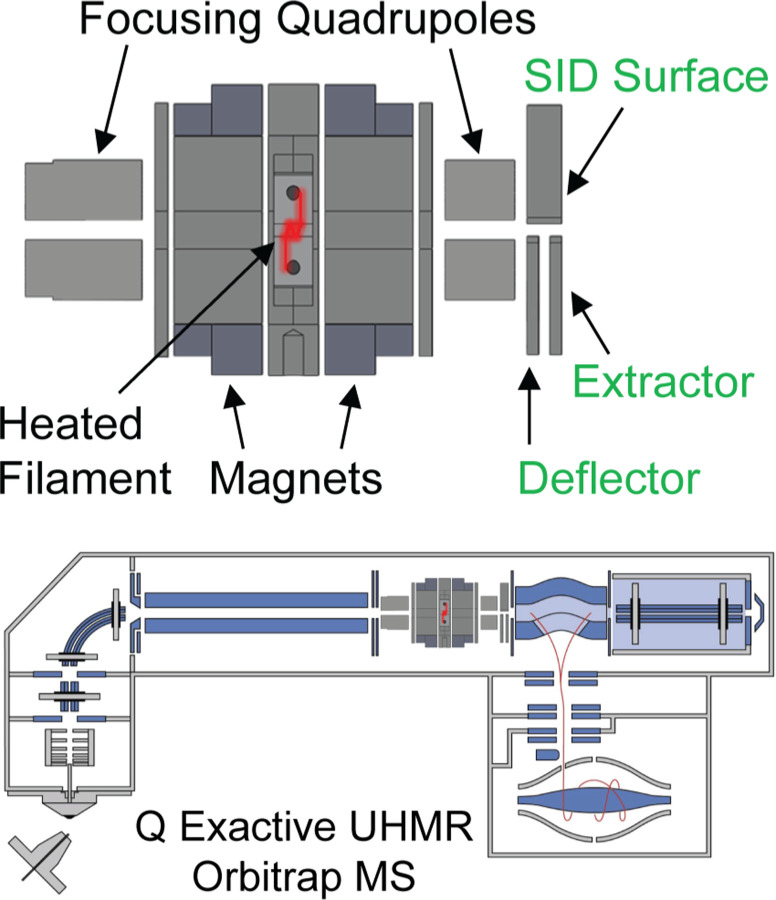
An ExD cell designed and sold by eMSion for the Thermo Scientific Q Exactive Orbitrap platform was modified, through the addition of short focusing quadrupoles before and after the electron producing cell and through the addition of a split lens SID device with the goal of improving transmission of a broad and high m/z range and incorporation of electrodes to perform SID. Figure 1 shows schematics of the hybrid ExD-SID cell and integration of the cell into the Q Exactive UHMR Orbitrap mass spectrometer. Thick electrostatic lenses at each end of the standard ExD cell were replaced with quadrupole ion guides. The quadrupoles were constructed using 3/16-inch diameter precision ground stainless steel round rods with a rod radius to inscribed radius ratio of 1.14. DC voltages were supplied to the ExD portion of the cell using a commercial power supply and software from eMSion. The SID electrode design and integration was similar to that previously described by Snyder and coworkers55 and illustrated recently in combination with CDMS e.g., for GroEL, proteasome, and AAV863. Briefly, the three-electrode split-lens design was integrated at the exit of the cell. Half of the split-lens is the SID surface electrode with a thickness of 3 mm. The other half is composed of the deflector and extractor electrodes with 1 mm thickness and separated by a 1 mm insulating spacer. The split lens aperture is 2 mm in diameter. The C-trap DC offset during injection steps was supplied by an external power supply to allow a larger range of voltages to be applied and therefore increasing the SID voltage range (defined as the difference in voltage between the bent flatapole and surface voltage), as previously described.57 C-Trap DC offset and SID device voltages were supplied via external power supplies (Ardara Technologies, Ardara, PA) and controlled by Tempus software (Ardara Technologies, Ardara, PA). The device works well as an SID device, as illustrated by fragmentation of the 50S ribosome subunit spectrum shown in Figure S1.
Figure 1.
Schematics of the ExD-SID cell and integration of the cell into the Q Exactive UHMR Orbitrap mass spectrometer.
Tunable Electron Capture Charge Reduction (ECCR).
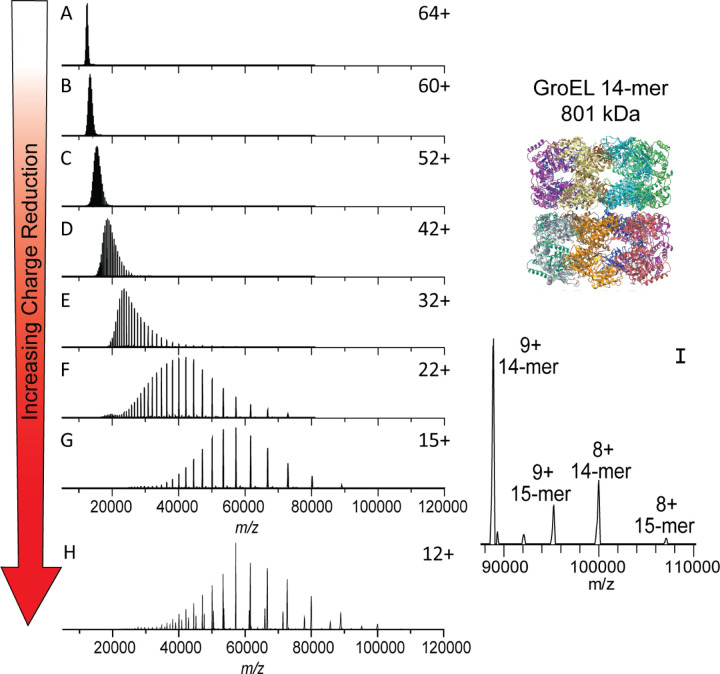
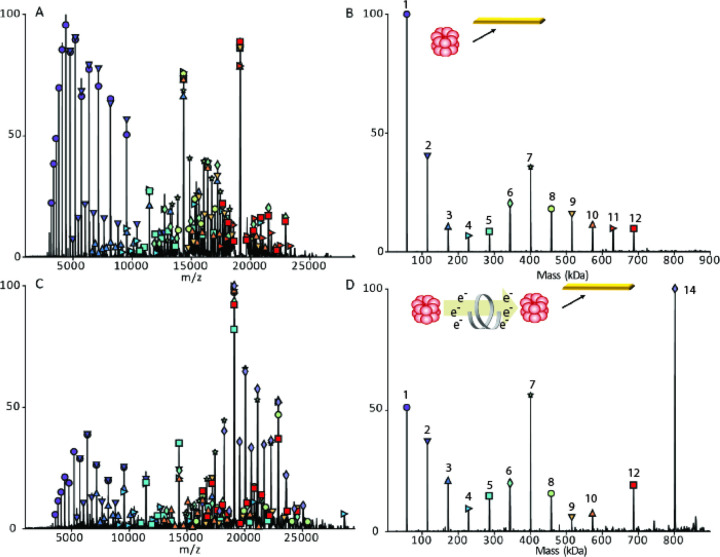
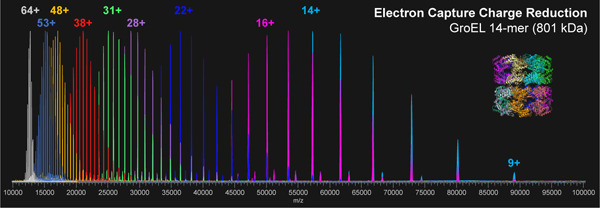
The first goal of this work was to develop an approach for tunable gas phase charge reduction to improve the effective resolving power of heterogeneous protein complexes. By reducing the charge of the heterogeneous complex ions, the species detected are shifted to a greater m/z range thus increasing the Δ m/z for overlapping peaks, an approach that has been used by others to better resolve large or heterogeneous systems.12,31,64–66 For initial method development and proof of principle for ECCR, we first used GroEL.67–69 GroEL is an ~801 kDa homo-tetradecameric protein composed of two stacked seven-member rings and has been thoroughly studied by native mass spectrometry.47,61,70,71 GroEL prepared in 200 mM ammonium acetate and ionized by nano-electrospray ionization produced the “native charge” mass spectrum shown at the top of Figure 2. The intensity weighted average charge was 64+ with a charge state distribution, for peaks greater than 5% relative abundance, ranging from 67+ to 62+. Charge reduction was achieved via ion-electron reactions in which the multiply charged cations of GroEL captured multiple low energy electrons while flying through the ExD-SID cell. For peptides and some proteins, ECD products (covalent bond cleavages) are the major expected products, but for large complexes, the major product is the intact non-covalent complex with multiple lower charge states. Detection of the intact mass of the charge-reduced species relies on preservation of GroEL subunit intramolecular and intermolecular non-covalent interactions. This is achieved using minimal collisional activation throughout the experiment. The extent of ECCR was modulated by adjusting the DC voltages applied to the center of the ExD device namely the two magnetic lenses and filament holder (representative tuning conditions are given in Table S1 along with a labeled schematic of the ExD-SID cell). The voltages were incrementally increased to reduce the velocity of the ions in transit. The lower panels of Figure 2 show incremental increases in ECCR with maximum ECCR yielding reduction in the average charge of GroEL from 64 to 12+, with an average of 52 electrons captured at maximum ECCR. The width of the charge state distribution increased from 7 to 11 charge states with minimal ECCR and plateaued at approximately 22 for higher levels of ECCR before returning to 12 charge states above 5% intensity at maximum ECCR. This reflects the distribution of ion kinetic energies in the ion beam and the strong dependence of electron capture kinetics on the ion kinetic energy and charge state. All experiments were performed using in-source trapping (−75 to −100 V) for desolvation.72 This process involves collisional activation follow by storage of the ions in the source region of the instrument for a few milliseconds. The momentum dampening and desolvation capabilities enabled by in-source trapping greatly increase transmission and sensitivity for high m/z ions, but some distribution in ion kinetic energy is unavoidable due to gas dynamics in the atmospheric pressure interface and spatial distribution of ions trapped in the injection flatapole. The extent of charge reduction exhibited a linear relationship when voltages are incrementally linearly increased (data shown in SI Figure S2).
Figure 2.
Electron capture charge reduction of GroEL. The top panel shows the mass spectrum of GroEL with the typical charge distribution generated by spraying from ammonium acetate (i.e., no gas phase charge reduction). Panels B-G show results from increases in electron capture charge reduction, with the device voltage that initiates ECCR increasing in two-volt steps (in source trapping of −75V and trap gas setting of 7). Panel H is from a different sample from A-G and shows more extensive charge reduction with peaks up to 107K m/z; more 15-mer of GroEL (additional, lower abundance peaks) was present in this sample, an older sample that had aged after refolding (in source trapping of −100 V and trap gas setting of 8). Panels I and J are zoomed in on the high m/z range of panels G and H, respectively.
Coupling ECCR and Surface Induced Dissociation
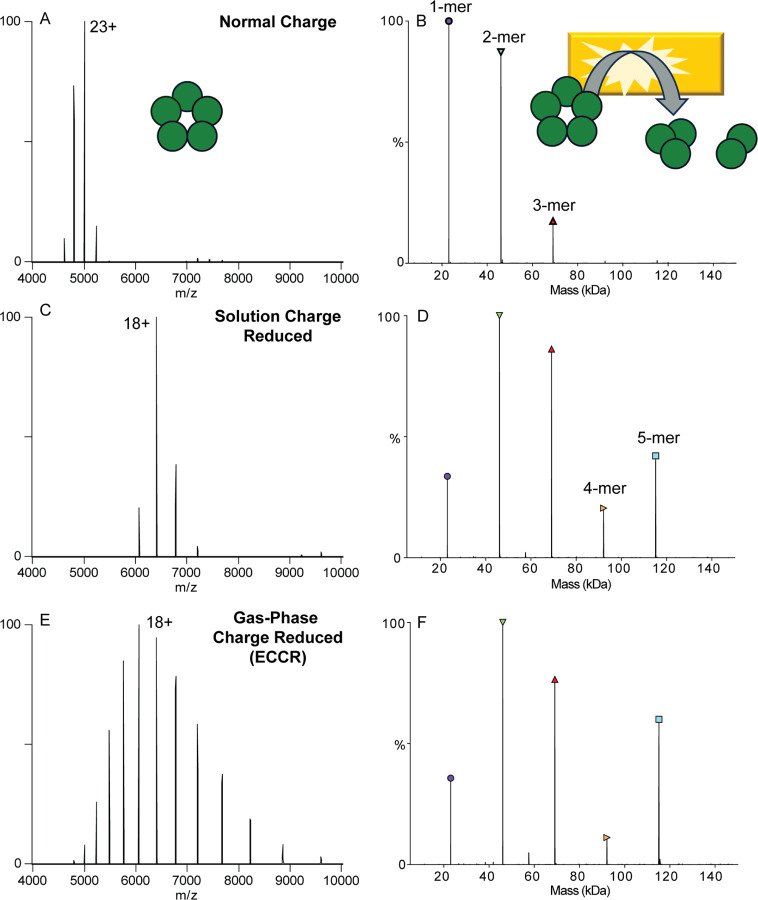
Initial experiments with GroEL highlighted that ECCR is tunable, which could be advantageous for heterogeneous samples (as demonstrated with glycoproteins below) but we also wanted to determine whether ECCR affects the protein quaternary structure and if structurally relevant information could still be obtained from the protein complex after ECCR. This is especially important because many structural studies using nMS already employ charge reducing conditions (typically with solution phase additives), as lower charge states can give more stable, native-like structures and fragmentation products.60,73 To probe the structure of ions after ECCR we used SID. It has been shown that protein complex dissociation via a surface collision can occur, depending on the complex, with greatly reduced subunit or subcomplex unfolding compared to traditional inert gas collision induced dissociation, providing information consistent with the native structure.51,74 To investigate the effect of gas phase charge reduction on SID, we compared dissociation of the pentameric C-reactive protein complex (CRP) under normal charge (ammonium acetate), solution charge reduction, and gas phase charge reduction conditions (Figure 3). Analysis of CRP from 200 mM ammonium acetate yields a weighted average charge state of 23+ which will be referred to as native charge for CRP (Figure 3A). SID of CRP at 40 V (average energy of 920 eV) produced primarily monomer and dimer, with lower levels of trimer and tetramer in agreement with previous studies (Figure 3B).73,75 For a cyclic complex like CRP, we expect all oligomeric states between monomer and tetramer at low SID energies, with relatively high abundance due to the equal interfaces between all subunits. Analysis in 160 mM AmAc and 40 mM TEAA yielded solution charge reduction of CRP and produced a charge state distribution with a weighted average charge of 16+ (Figure 3C), SID of this distribution at 60 V (average energy of 960 eV) produced monomer and tetramer, and dimer and trimer at high intensity (Figure 3D) consistent with the native cyclic structure and with previous SID studies of CRP.51,75,76 Finally, gas-phase charge reduction via ECCR of the native charge distribution yielded a weighted average charge state of 18+ (Figure 3E), which when subjected to SID at 40V (average energy of 720 eV) (Figure 3F) produced a similar SID spectra to that obtained from solution-phase charge reduction (Figure 3D). Previous studies have shown solution charge reducing agents, such as TEAA, do not markedly change the gas-phase structure of precursor ions,51 but help stabilize noncovalently associated complexes and direct the course of gas-phase dissociation.62 Therefore, the high degree of similarity between solution charge reduced and gas-phase charge reduced CRP indicate modulation of charge driven processes during dissociation rather than gas-phase structural changes resulting from charge neutralization via ECCR. These results demonstrate that ECCR-SID can be used to probe the topology of protein complexes with dissociation occurring in a manner consistent with the solved structure.
Figure 3.
Comparison of C-reactive protein MS and SID patterns for A&B) normal charge (acquired in ammonium acetate), C&D) solution charge reduced (acquired with an 80:20 ratio of ammonium acetate to triethylammonium acetate) E&F) gas-phase charge reduced (ECCR). Acquired with in-source trapping −30 V, and SID voltage of 40 V (normal charge) or 60 V (solution phase and gas-phase charge reduced).
We further investigated gas phase charge reduction (ECCR) to produce more native-like topology/connectivity information by comparing SID and ECCR followed by SID for the tetradecamer of GroEL. Figures 4A and B show the SID mass spectrum and charge deconvolved spectrum for native charge (68+ weighted average charge) GroEL. The monomer of GroEL is by far the most abundant product ion with lower levels of dimer through 12mer. However, gas-phase charge reduction of native charge GroEL (46+ weighted average charge) followed by SID (Figure 4C and D) yielded a significantly lower relative abundance of monomer and very abundant 7mer corresponding to dissociation at the interface of the two stacked 7mer rings. It should be noted that SID was performed at the same voltage in each case, corresponding to different SID energies because charge is reduced in the ExD-SID device and charge and electric field determine kinetic energy. To confirm that the kinetic energy difference did not cause the spectral differences, we lowered the kinetic energy for the normal charge GroEL and that did not yield increased 7mer.
Figure 4.
GroEL SID at 230 V under (A) raw data for native charge (68+ weighted average charge) and (B) deconvolved data for native charge (C) raw data for gas-phase charge reduction (46+ weighted average charge) and (D) deconvolved data for gas-phase charge reduction.
Application of ECCR to Glycoproteins
Posttranslational modifications pose significant challenges to mass spectrometric analysis at the intact protein and protein complex levels. For glycosylation the challenge results from the immense complexity arising from glycosylation site heterogeneity and glycan heterogeneity at each site. Electrospray ionization mass spectra of heterogeneously glycosylated proteins and protein complexes contain complex overlapping distributions of peaks resulting from the observation of multiple charge states and varying states of glycosylation.31 Heterogeneity is often sufficient to preclude mass determination due to the inability to resolve distinct isotopic and or charge state distributions. An example of this is shown in Figure 5 for VFLIP, a stable construct of the SARS-CoV-2 spike protein trimer with disulfide bonding between monomers and native-like glycosylation which is of interest as an improved tool for diagnostics and vaccine design.56 Figure 5A shows a broad and unresolved m/z distribution corresponding to the intact complex for which mass determination is not possible by traditional mass spectrometry methods. To resolve charge states of VFLIP, ECCR was applied to the entire m/z distribution of the intact complex, moderately reducing the charge in the gas-phase. The resulting mass spectrum in Figure 5B shows resolved charge states of the VFLIP complex which enable mass determination. An average mass of 506 kDa was determined from UniDec consistent with the expected mass based on sequence and known glycosylation profile (expected mass 510 kDa). The application of ECCR to the heterogeneous glycoprotein complex VFLIP illustrates the capability to determine mass and resolve sources of heterogeneity when traditional native MS approaches don’t provide any information. There are, however, alternative approaches that can be used for such heterogeneous samples. Mass photometry utilizes the linear relationship between light scattered from a single particle and this particle’s mass to measure masses of molecules.77–79 This instrument provides single molecule level studies in solution for proteins without labels.80 The mass was measured using MP and the mass distribution centered at 510 kDa (see Figure S3A). Charge detection mass spectrometry (CDMS)81 and the recently commercialized version of CDMS, Direct Mass Technology (DMT)82 for Orbitrap mass analyzers, enables simultaneous detection of m/z and charge (z) of analytes. This approach is particularly useful for mass determination of very large biomolecules, e.g., megadalton size viral capsids, where heterogeneity in composition and incomplete desolvation yield very broad and unresolved spectra and for heterogeneous spike proteins.8,63,81 CDMS was applied to a VFLIP spike protein and mass distributions centered at 513 kDa (HCD 0 V) and 509 kDa (HCD 200 V) were determined (see Figure S3B and C).
While we can determine the average mass with CDMS, ECCR, or mass photometry of the full charge state distribution, more detailed information on the different glycoforms present can be obtained using narrow window isolations and ECCR across the unresolved region, enabling better resolution of the different glycoforms (Figure 5). Three narrow window isolations across the unresolved charge state distribution, followed by ECCR, give results illustrated in Figures 5G and H. ECCR of the narrow window isolation shows well resolved peaks that can easily be deconvolved to gain more accurate mass measurements. Within each m/z selection window, 3–6 different mass species are observed (Figure 5F, I), reflecting the heterogeneity of the VFLIP glycosylation. For a protein complex with a lower total glycosylation mass, e.g., thyroglobulin, each m/z selection window results in a less complex distribution of glycoforms (see Figure S4) and narrow m/z selection windows reveal glycoforms. Overlaying the deconvolved mass peaks from multiple narrow window isolations for the spike protein (e.g., three shown in Fig. 5I) reconstructs the original, unresolved VFLIP mass distribution, providing a global glycan profile. This method of narrow-window m/z selection coupled to ECCR could be utilized as a quick screen for glycan complexity under different expression conditions for therapeutic proteins or various variants of concern in infectious diseases and could be coupled with in-depth glycoproteomics studies (top down or bottom up) when the variable identities at each glycosylation site are required.
CONCLUSIONS
In this study, a prototype device combining ion-electron reactions and surface induced dissociation was developed and incorporated into a commercial ultra-high mass range Q-Orbitrap mass spectrometer for the characterization of native protein complexes. The new device enabled efficient and tunable gas-phase electron capture charge reduction (ECCR) as demonstrated for GroEL by enabling detection of 8+ tetradecamer and pentadecamer at greater than 100,000 m/z. Furthermore, we demonstrated that gas-phase charge reduction produces charge states that give native-like surface induced dissociation fragments for C-reactive protein and GroEL. Additionally, ECCR yields mass spectra that enable mass determination and better resolved heterogeneity in the glycoproteins stabilized VFLIP spike and thyroglobulin. This rapid and tunable gas-phase charge reduction technique is complementary to charge detection mass spectrometry techniques for very large and heterogeneous biomolecular assemblies and is expected to enhance future MS-based structural biology approaches. Work is in progress that applies this approach to SARS-CoV-2 spike proteins from multiple variants of concern and to other structural biology problems.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Yury Vasil’ev and Joe Beckman for helpful conversations on the ExD device, Dalton Snyder (OSU) for helpful conversations on SID device geometry and tuning, Andrew Arslanian for helping to collect the CDMS spectra in the supporting information, and Jacelyn Greenwald (OSU) for refolding the GroEL samples. The authors gratefully acknowledge funding from the National Institutes of Health awarded to J.B.S (R43GM140749), E.O.S. (P01AI165072), and V.H.W (P41GM128577 and RM1GM149374).
REFERENCES
- (1).Khoury G. A.; Baliban R. C.; Floudas C. A. Proteome-Wide Post-Translational Modification Statistics: Frequency Analysis and Curation of the Swiss-Prot Database. Sci Rep 2011, 1 (1), 90. 10.1038/srep00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Solá R. J.; Griebenow K. Glycosylation of Therapeutic Proteins: An Effective Strategy to Optimize Efficacy. BioDrugs 2010, 24 (1), 9–21. 10.2165/11530550-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Heck A. J. R. Native Mass Spectrometry: A Bridge between Interactomics and Structural Biology. Nat. Meth. 2008, 5 (11), 927–933. 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- (4).Barrera N. P.; Robinson C. V. Advances in the Mass Spectrometry of Membrane Proteins: From Individual Proteins to Intact Complexes. Annu. Rev. Biochem. 2011, 80 (1), 247–271. 10.1146/annurev-biochem-062309-093307. [DOI] [PubMed] [Google Scholar]
- (5).Allison T. M.; Reading E.; Liko I.; Baldwin A. J.; Laganowsky A.; Robinson C. V. Quantifying the Stabilizing Effects of Protein–Ligand Interactions in the Gas Phase. Nat Commun 2015, 6 (1), 8551. 10.1038/ncomms9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Uetrecht C.; Barbu I. M.; Shoemaker G. K.; van Duijn E.; Heck A. J. R. Interrogating Viral Capsid Assembly with Ion Mobility–Mass Spectrometry. Nature Chem 2011, 3 (2), 126–132. 10.1038/nchem.947. [DOI] [PubMed] [Google Scholar]
- (7).Lössl P.; Snijder J.; Heck A. J. R. Boundaries of Mass Resolution in Native Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2014. 10.1007/s13361-014-0874-3. [DOI] [PubMed] [Google Scholar]
- (8).Stiving A. Q.; Foreman D. J.; VanAernum Z. L.; Durr E.; Wang S.; Vlasak J.; Galli J.; Kafader J. O.; Li X.; Schuessler H.; Richardson D. Dissecting the Heterogeneous Glycan Profiles of Recombinant Coronavirus Spike Proteins with Individual Ion Mass Spectrometry. 2022. 10.26434/chemrxiv-2022-9vnch. [DOI] [PubMed] [Google Scholar]
- (9).Jarrold M. F. Applications of Charge Detection Mass Spectrometry in Molecular Biology and Biotechnology. Chem. Rev. 2022, 122 (8), 7415–7441. 10.1021/acs.chemrev.1c00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Miller L. M.; Barnes L. F.; Raab S. A.; Draper B. E.; El-Baba T. J.; Lutomski C. A.; Robinson C. V.; Clemmer D. E.; Jarrold M. F. Heterogeneity of Glycan Processing on Trimeric SARS-CoV-2 Spike Protein Revealed by Charge Detection Mass Spectrometry. J Am Chem Soc 2021, 143 (10), 3959–3966. 10.1021/jacs.1c00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Pacholarz K. J.; Barran P. E. Use of a Charge Reducing Agent to Enable Intact Mass Analysis of Cysteine-Linked Antibody-Drug-Conjugates by Native Mass Spectrometry. EuPA Open Proteomics 2016, 11, 23–27. 10.1016/j.euprot.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Bobst C. E.; Sperry J.; Friese O. V.; Kaltashov I. A. Simultaneous Evaluation of a Vaccine Component Microheterogeneity and Conformational Integrity Using Native Mass Spectrometry and Limited Charge Reduction. J. Am. Soc. Mass Spectrom. 2021, 32 (7), 1631–1637. 10.1021/jasms.1c00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Loo R. R. O.; Loo J. A.; Udseth H. R.; Fulton J. L.; Smith R. D. Protein Structural Effects in Gas Phase Ion/Molecule Reactions with Diethylamine. Rapid Commun. Mass Spectrom. 1992, 6 (3), 159–165. 10.1002/rcm.1290060302. [DOI] [PubMed] [Google Scholar]
- (14).Ogorzalek Loo R. R.; Winger B. E.; Smith R. D. Proton Transfer Reaction Studies of Multiply Charged Proteins in a High Mass-to-Charge Ratio Quadrupole Mass Spectrometer. J. Am. Soc. Mass Spectrom. 1994, 5 (12), 1064–1071. 10.1016/1044-0305(94)85067-4. [DOI] [PubMed] [Google Scholar]
- (15).Ogorzalek Loo R. R.; Smith R. D. Investigation of the Gas-Phase Structure of Electrosprayed Proteins Using Ion-Molecule Reactions. J. Am. Soc. Mass Spectrom. 1994, 5 (4), 207–220. 10.1016/1044-0305(94)85011-9. [DOI] [PubMed] [Google Scholar]
- (16).McLuckey S. A.; Stephenson J. L.; Asano K. G. Ion/Ion Proton-Transfer Kinetics: Implications for Analysis of Ions Derived from Electrospray of Protein Mixtures. Anal. Chem. 1998, 70 (6), 1198–1202. 10.1021/ac9710137. [DOI] [PubMed] [Google Scholar]
- (17).Stephenson J. L.; McLuckey S. A. Ion/Ion Reactions in the Gas Phase: Proton Transfer Reactions Involving Multiply-Charged Proteins. J. Am. Chem. Soc. 1996, 118 (31), 7390–7397. 10.1021/ja9611755. [DOI] [Google Scholar]
- (18).Stephenson J. L.; McLuckey S. A. Anion Effects on Storage and Resonance Ejection of High Mass-to-Charge Cations in Quadrupole Ion Trap Mass Spectrometry. Anal. Chem. 1997, 69 (18), 3760–3766. 10.1021/ac970399i. [DOI] [Google Scholar]
- (19).Stephenson J. L.; McLuckey S. A. Ion/Ion Proton Transfer Reactions for Protein Mixture Analysis. Anal. Chem. 1996, 68 (22), 4026–4032. 10.1021/ac9605657. [DOI] [PubMed] [Google Scholar]
- (20).Stephenson J. L.; McLuckey S. A. Ion/Ion Reactions for Oligopeptide Mixture Analysis: Application to Mixtures Comprised of 0.5–100 kDa Components. J. Am. Soc. Mass Spectrom. 1998, 9 (6), 585–596. 10.1016/S1044-0305(98)00025-7. [DOI] [PubMed] [Google Scholar]
- (21).Stephenson J. L.; McLuckey S. A. Simplification of Product Ion Spectra Derived from Multiply Charged Parent Ions via Ion/Ion Chemistry. Anal. Chem. 1998, 70 (17), 3533–3544. 10.1021/ac9802832. [DOI] [PubMed] [Google Scholar]
- (22).McLuckey S. A.; Reid G. E.; Wells J. M. Ion Parking during Ion/Ion Reactions in Electrodynamic Ion Traps. Anal. Chem. 2002, 74 (2), 336–346. 10.1021/ac0109671. [DOI] [PubMed] [Google Scholar]
- (23).Chrisman P. A.; Pitteri S. J.; McLuckey S. A. Parallel Ion Parking: Improving Conversion of Parents to First-Generation Products in Electron Transfer Dissociation. Anal. Chem. 2005, 77 (10), 3411–3414. 10.1021/ac0503613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Hartmer R.; Kaplan D. A.; Gebhardt C. R.; Ledertheil T.; Brekenfeld A. Multiple Ion/Ion Reactions in the 3D Ion Trap: Selective Reagent Anion Production for ETD and PTR from a Single Compound. International Journal of Mass Spectrometry 2008, 276 (2), 82–90. 10.1016/j.ijms.2008.05.002. [DOI] [Google Scholar]
- (25).Rožman M.; Schneider A.; Gaskell S. J. Proton Transfer Reactions for Improved Peptide Characterisation. Journal of Mass Spectrometry 2011, 46 (6), 529–534. 10.1002/jms.1920. [DOI] [PubMed] [Google Scholar]
- (26).Huguet R.; Mullen C.; Srzentić K.; Greer J. B.; Fellers R. T.; Zabrouskov V.; Syka J. E. P.; Kelleher N. L.; Fornelli L. Proton Transfer Charge Reduction Enables High-Throughput Top-Down Analysis of Large Proteoforms. Anal. Chem. 2019, 91 (24), 15732–15739. 10.1021/acs.analchem.9b03925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Kline J. T.; Mullen C.; Durbin K. R.; Oates R. N.; Huguet R.; Syka J. E. P.; Fornelli L. Sequential Ion–Ion Reactions for Enhanced Gas-Phase Sequencing of Large Intact Proteins in a Tribrid Orbitrap Mass Spectrometer. J. Am. Soc. Mass Spectrom. 2021, 32 (9), 2334–2345. 10.1021/jasms.1c00062. [DOI] [PubMed] [Google Scholar]
- (28).Bailey A. O.; Huguet R.; Mullen C.; Syka J. E. P.; Russell W. K. Ion–Ion Charge Reduction Addresses Multiple Challenges Common to Denaturing Intact Mass Analysis. Anal. Chem. 2022, 94 (9), 3930–3938. 10.1021/acs.analchem.1c04973. [DOI] [PubMed] [Google Scholar]
- (29).Sanders J. D.; Mullen C.; Watts E.; Holden D. D.; Syka J. E. P.; Schwartz J. C.; Brodbelt J. S. Enhanced Sequence Coverage of Large Proteins by Combining Ultraviolet Photodissociation with Proton Transfer Reactions. Anal. Chem. 2020, 92 (1), 1041–1049. 10.1021/acs.analchem.9b04026. [DOI] [PubMed] [Google Scholar]
- (30).Laszlo K. J.; Bush M. F. Analysis of Native-Like Proteins and Protein Complexes Using Cation to Anion Proton Transfer Reactions (CAPTR). J. Am. Soc. Mass Spectrom. 2015, 26 (12), 2152–2161. 10.1007/s13361-015-1245-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Schachner L. F.; Mullen C.; Phung W.; Hinkle J. D.; Beardsley M. I.; Bentley T.; Day P.; Tsai C.; Sukumaran S.; Baginski T.; DiCara D.; Agard N.; Masureel M.; Gober J.; ElSohly A.; Syka J. E. P.; Huguet R.; Marty M. T.; Sandoval W. Exposing the Molecular Heterogeneity of Glycosylated Biotherapeutics. bioRxiv May 11, 2023, p 2023.05.10.540271. 10.1101/2023.05.10.540271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Breuker K.; Oh H.; Horn D. M.; Cerda B. A.; McLafferty F. W. Detailed Unfolding and Folding of Gaseous Ubiquitin Ions Characterized by Electron Capture Dissociation. J. Am. Chem. Soc. 2002, 124 (22), 6407–6420. 10.1021/ja012267j. [DOI] [PubMed] [Google Scholar]
- (33).Skinner O. S.; McLafferty F. W.; Breuker K. How Ubiquitin Unfolds after Transfer into the Gas Phase. J. Am. Soc. Mass Spectrom. 2012, 23 (6), 1011–1014. 10.1007/s13361-012-0370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Harvey S. Dissecting the Dynamic Conformations of the Metamorphic Protein Lymphotactin. The journal of physical chemistry. B 2014. 10.1021/jp504997k. [DOI] [PubMed] [Google Scholar]
- (35).Lermyte F.; Sobott F. Electron Transfer Dissociation Provides Higher-Order Structural Information of Native and Partially Unfolded Protein Complexes. PROTEOMICS 2015, 15 (16), 2813–2822. 10.1002/pmic.201400516. [DOI] [PubMed] [Google Scholar]
- (36).Li H.; Nguyen H. H.; Ogorzalek Loo R. R.; Campuzano I. D. G.; Loo J. A. An Integrated Native Mass Spectrometry and Top-down Proteomics Method That Connects Sequence to Structure and Function of Macromolecular Complexes. Nat. Chem. 2018, 10 (2), 139–148. 10.1038/nchem.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Zhou M.; Liu W.; Shaw J. B. Charge Movement and Structural Changes in the Gas-Phase Unfolding of Multimeric Protein Complexes Captured by Native Top-Down Mass Spectrometry. Anal. Chem. 2020, 92 (2), 1788–1795. 10.1021/acs.analchem.9b03469. [DOI] [PubMed] [Google Scholar]
- (38).Horn D. M.; Ge Y.; McLafferty F. W. Activated Ion Electron Capture Dissociation for Mass Spectral Sequencing of Larger (42 kDa) Proteins. Anal. Chem. 2000, 72 (20), 4778–4784. https://doi.org/doi: 10.1021/ac000494i. [DOI] [PubMed] [Google Scholar]
- (39).Riley N. M.; Westphall M. S.; Coon J. J. Sequencing Larger Intact Proteins (30–70 kDa) with Activated Ion Electron Transfer Dissociation. J. Am. Soc. Mass Spectrom. 2018, 29, 140–149. 10.1007/s13361-017-1808-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Shaw J. B.; Malhan N.; Vasil’ev Y. V.; Lopez N. I.; Makarov A.; Beckman J. S.; Voinov V. G. Sequencing Grade Tandem Mass Spectrometry for Top–Down Proteomics Using Hybrid Electron Capture Dissociation Methods in a Benchtop Orbitrap Mass Spectrometer. Anal. Chem. 2018, 90 (18), 10819–10827. 10.1021/acs.analchem.8b01901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Shaw J. B.; Liu W.; Vasil′ev Y. V.; Bracken C. C.; Malhan N.; Guthals A.; Beckman J. S.; Voinov V. G. Direct Determination of Antibody Chain Pairing by Top-down and Middle-down Mass Spectrometry Using Electron Capture Dissociation and Ultraviolet Photodissociation. Anal. Chem. 2020, 92 (1), 766–773. 10.1021/acs.analchem.9b03129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Lermyte F.; Williams J. P.; Brown J. M.; Martin E. M.; Sobott F. Extensive Charge Reduction and Dissociation of Intact Protein Complexes Following Electron Transfer on a Quadrupole-Ion Mobility-Time-of-Flight MS. J. Am. Soc. Mass Spectrom. 2015, 26 (7), 1068–1076. 10.1007/s13361-015-1124-z. [DOI] [PubMed] [Google Scholar]
- (43).Ujma J.; Giles K.; Anderson M.; Richardson K. ENHANCED DECLUSTERING AND CHARGE-STRIPPING ENABLES MASS DETERMINATION OF AAVS IN TOF MS. 310509 (MP 481), Proceedings of the 70th ASMS Conference on Mass Spectrometry and Allied Topics, Minneapolis, Minnesota. June5-9, 2022. [Google Scholar]
- (44).Scalf M.; Westphall M. S.; Krause J.; Kaufman S. L.; Smith L. M. Controlling Charge States of Large Ions. Science 1999, 283 (5399), 194–197. 10.1126/science.283.5399.194. [DOI] [PubMed] [Google Scholar]
- (45).Ebeling D. D.; Westphall M. S.; Scalf M.; Smith L. M. Corona Discharge in Charge Reduction Electrospray Mass Spectrometry. Anal. Chem. 2000, 72 (21), 5158–5161. 10.1021/ac000559h. [DOI] [PubMed] [Google Scholar]
- (46).Lorenzen K.; Versluis C.; van Duijn E.; van den Heuvel R. H. H.; Heck A. J. R. Optimizing Macromolecular Tandem Mass Spectrometry of Large Non-Covalent Complexes Using Heavy Collision Gases. International Journal of Mass Spectrometry 2007, 268 (2), 198–206. 10.1016/j.ijms.2007.06.012. [DOI] [Google Scholar]
- (47).Sobott F.; Robinson C. V. Characterising Electrosprayed Biomolecules Using Tandem-MS—the Noncovalent GroEL Chaperonin Assembly. International Journal of Mass Spectrometry 2004, 236 (1), 25–32. 10.1016/j.ijms.2004.05.010. [DOI] [Google Scholar]
- (48).Jones C. M.; Beardsley R. L.; Galhena A. S.; Dagan S.; Cheng G.; Wysocki V. H. Symmetrical Gas-Phase Dissociation of Noncovalent Protein Complexes via Surface Collisions. J. Am. Chem. Soc. 2006, 128 (47), 15044–15045. 10.1021/ja064586m. [DOI] [PubMed] [Google Scholar]
- (49).Blackwell A. E.; Dodds E. D.; Bandarian V.; Wysocki V. H. Revealing the Quaternary Structure of a Heterogeneous Noncovalent Protein Complex through Surface-Induced Dissociation. Anal. Chem. 2011, 83 (8), 2862–2865. 10.1021/ac200452b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Zhou M.; Huang C.; Wysocki V. H. Surface-Induced Dissociation of Ion Mobility-Separated Noncovalent Complexes in a Quadrupole/Time-of-Flight Mass Spectrometer. Anal. Chem. 2012, 84 (14), 6016–6023. 10.1021/ac300810u. [DOI] [PubMed] [Google Scholar]
- (51).Zhou M.; Dagan S.; Wysocki V. H. Protein Subunits Released by Surface Collisions of Noncovalent Complexes: Nativelike Compact Structures Revealed by Ion Mobility Mass Spectrometry. Angew. Chem. Int. Ed. 2012, 51 (18), 4336–4339. 10.1002/anie.201108700. [DOI] [PubMed] [Google Scholar]
- (52).Zhou M.; Jones C. M.; Wysocki V. H. Dissecting the Large Noncovalent Protein Complex GroEL with Surface-Induced Dissociation and Ion Mobility–Mass Spectrometry. Anal. Chem. 2013, 85 (17), 8262–8267. 10.1021/ac401497c. [DOI] [PubMed] [Google Scholar]
- (53).Harvey S. Relative Interfacial Cleavage Energetics of Protein Complexes Revealed by Surface Collisions. Proceedings of the National Academy of Sciences of the United States of America 2019. 10.1073/pnas.1817632116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Snyder D. T.; Harvey S. R.; Wysocki V. H. Surface-Induced Dissociation Mass Spectrometry as a Structural Biology Tool. Chemical Reviews 2021. 10.1021/acs.chemrev.1c00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Snyder D. T.; Panczyk E. M.; Somogyi A.; Kaplan D. A.; Wysocki V. Simple and Minimally Invasive SID Devices for Native Mass Spectrometry. Anal. Chem. 2020, 92 (16), 11195–11203. 10.1021/acs.analchem.0c01657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Olmedillas E.; Mann C. J.; Peng W.; Wang Y.-T.; Avalos R. D.; Bedinger D.; Valentine K.; Shafee N.; Schendel S. L.; Yuan M.; Lang G.; Rouet R.; Christ D.; Jiang W.; Wilson I. A.; Germann T.; Shresta S.; Snijder J.; Saphire E. O. Structure-Based Design of a Highly Stable, Covalently-Linked SARS-CoV-2 Spike Trimer with Improved Structural Properties and Immunogenicity; preprint; Microbiology, 2021. 10.1101/2021.05.06.441046. [DOI] [Google Scholar]
- (57).VanAernum Z. L.; Gilbert J. D.; Belov M. E.; Makarov A. A.; Horning S. R.; Wysocki V. H. Surface-Induced Dissociation of Noncovalent Protein Complexes in an Extended Mass Range Orbitrap Mass Spectrometer. Anal. Chem. 2019, 91 (5), 3611–3618. 10.1021/acs.analchem.8b05605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Marty M. T.; Baldwin A. J.; Marklund E. G.; Hochberg G. K. A.; Benesch J. L. P.; Robinson C. V. Bayesian Deconvolution of Mass and Ion Mobility Spectra: From Binary Interactions to Polydisperse Ensembles. Anal. Chem. 2015, 87 (8), 4370–4376. 10.1021/acs.analchem.5b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Kostelic M. M.; Zak C. K.; Liu Y.; Chen V. S.; Wu Z.; Sivinski J.; Chapman E.; Marty M. T. UniDecCD: Deconvolution of Charge Detection-Mass Spectrometry Data. Anal. Chem. 2021, 93 (44), 14722–14729. 10.1021/acs.analchem.1c03181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Hall Z.; Politis A.; Bush M. F.; Smith L. J.; Robinson C. V. Charge-State Dependent Compaction and Dissociation of Protein Complexes: Insights from Ion Mobility and Molecular Dynamics. J. Am. Chem. Soc. 2012, 134 (7), 3429–3438. 10.1021/ja2096859. [DOI] [PubMed] [Google Scholar]
- (61).Dyachenko A.; Gruber R.; Shimon L.; Horovitz A.; Sharon M. Allosteric Mechanisms Can Be Distinguished Using Structural Mass Spectrometry. PNAS 2013, 110 (18), 7235–7239. 10.1073/pnas.1302395110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Pagel K.; Hyung S.-J.; Ruotolo B. T.; Robinson C. V. Alternate Dissociation Pathways Identified in Charge-Reduced Protein Complex Ions. Anal. Chem. 2010, 82 (12), 5363–5372. 10.1021/ac101121r. [DOI] [PubMed] [Google Scholar]
- (63).Du C.; Cleary S. P.; Kostelic M. M.; Jones B. J.; Kafader J. O.; Wysocki V. H. Combining Surface-Induced Dissociation and Charge Detection Mass Spectrometry to Reveal the Native Topology of Heterogeneous Protein Complexes. Anal. Chem. 2023, 95 (37), 13889–13896. 10.1021/acs.analchem.3c02185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Lermyte F.; Williams J. P.; Brown J. M.; Martin E. M.; Sobott F. Extensive Charge Reduction and Dissociation of Intact Protein Complexes Following Electron Transfer on a Quadrupole-Ion Mobility-Time-of-Flight MS. J. Am. Soc. Mass Spectrom. 2015, 26 (7), 1068–1076. 10.1007/s13361-015-1124-z. [DOI] [PubMed] [Google Scholar]
- (65).Yang Y.; Niu C.; Bobst C. E.; Kaltashov I. A. Charge Manipulation Using Solution and Gas-Phase Chemistry to Facilitate Analysis of Highly Heterogeneous Protein Complexes in Native Mass Spectrometry. Anal. Chem. 2021, 93 (7), 3337–3342. 10.1021/acs.analchem.0c05249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Pacholarz K. J.; Barran P. E. Use of a Charge Reducing Agent to Enable Intact Mass Analysis of Cysteine-Linked Antibody-Drug-Conjugates by Native Mass Spectrometry. EuPA Open Proteom 2016, 11, 23–27. 10.1016/j.euprot.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Vasil’ev Y. V.; Du C.; Harvey S. R.; Snyder D. T.; Wysocki V. H.; Shaw J. B. Native Top-Down Characterization of Protein Complexes with a Hybrid ECD-SID Device. 311519 (WP176), Proceedings of the 70th ASMS Conference on Mass Spectrometry and Allied Topics, Minneapolis, Minnesota. June5-9, 2022. [Google Scholar]
- (68).Shaw J. B.; Harvey S. R.; Du C.; Wysocki V. H. Protein Complex Heterogeneity and Structure Revealed by Native MS with ECCR and SID. 316163 (WOA pm 03:50), Proceedings of the 71st ASMS Conference on Mass Spectrometry and Allied Topics, Houston, Texas. June 4-8, 2023. [Google Scholar]
- (69).Le Huray K. I. P.; Woerner T. P.; Reinhardt-Szyba M.; Fort K. L.; Sobott F.; Makarov A. A. To 200k m/z and beyond: Native Electron-Capture Charge Reduction Resolves Heterogeneous Signals in Large Biopharmaceutical Analytes, a New Orbitrap Record. 313520 (WOA pm 03:10), Proceedings of the 71st ASMS Conference on Mass Spectrometry and Allied Topics, Houston, Texas. June 4-8, 2023. [Google Scholar]
- (70).Zhou M.; Jones C. M.; Wysocki V. H. Dissecting the Large Noncovalent Protein Complex GroEL with Surface-Induced Dissociation and Ion Mobility–Mass Spectrometry. Anal. Chem. 2013, 85 (17), 8262–8267. 10.1021/ac401497c. [DOI] [PubMed] [Google Scholar]
- (71).Walker T. E.; Shirzadeh M.; Sun H. M.; McCabe J. W.; Roth A.; Moghadamchargari Z.; Clemmer D. E.; Laganowsky A.; Rye H.; Russell D. H. Temperature Regulates Stability, Ligand Binding (Mg2+ and ATP), and Stoichiometry of GroEL-GroES Complexes. J Am Chem Soc 2022, 144 (6), 2667–2678. 10.1021/jacs.1c11341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Fort K. L.; van de Waterbeemd M.; Boll D.; Reinhardt-Szyba M.; Belov M. E.; Sasaki E.; Zschoche R.; Hilvert D.; Makarov A. A.; Heck A. J. R. Expanding the Structural Analysis Capabilities on an Orbitrap-Based Mass Spectrometer for Large Macromolecular Complexes. Analyst 2018, 143 (1), 100–105. 10.1039/C7AN01629H. [DOI] [PubMed] [Google Scholar]
- (73).Zhou M.; Dagan S.; Wysocki V. H. Impact of Charge State on Gas-Phase Behaviors of Noncovalent Protein Complexes in Collision Induced Dissociation and Surface Induced Dissociation. Analyst 2013, 138 (5), 1353–1362. 10.1039/C2AN36525A. [DOI] [PubMed] [Google Scholar]
- (74).Harvey S. R.; Seffernick J. T.; Quintyn R. S.; Song Y.; Ju Y.; Yan J.; Sahasrabuddhe A. N.; Norris A.; Zhou M.; Behrman E. J. Relative Interfacial Cleavage Energetics of Protein Complexes Revealed by Surface Collisions. Proceedings of the National Academy of Sciences 2019, 116 (17), 8143–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Harvey S.; VanAernum Z.; Wysocki V. Surface-Induced Dissociation of Anionic vs Cationic Native-like Protein Complexes. 2021. 10.26434/chemrxiv.13547837.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Busch F.; VanAernum Z. L.; Ju Y.; Yan J.; Gilbert J. D.; Quintyn R. S.; Bern M.; Wysocki V. H. Localization of Protein Complex Bound Ligands by Surface-Induced Dissociation High-Resolution Mass Spectrometry. Anal. Chem. 2018, 90 (21), 12796–12801. 10.1021/acs.analchem.8b03263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Cole D.; Young G.; Weigel A.; Sebesta A.; Kukura P. Label-Free Single-Molecule Imaging with Numerical-Aperture-Shaped Interferometric Scattering Microscopy. ACS Photonics 2017, 4 (2), 211–216. 10.1021/acsphotonics.6b00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Young G.; Hundt N.; Cole D.; Fineberg A.; Andrecka J.; Tyler A.; Olerinyova A.; Ansari A.; Marklund E. G.; Collier M. P.; Chandler S. A.; Tkachenko O.; Allen J.; Crispin M.; Billington N.; Takagi Y.; Sellers J. R.; Eichmann C.; Selenko P.; Frey L.; Riek R.; Galpin M. R.; Struwe W. B.; Benesch J. L. P.; Kukura P. Quantitative Mass Imaging of Single Molecules. Science 2018, 360 (6387), 423–427. 10.1126/science.aar5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Wu D.; Piszczek G. Standard Protocol for Mass Photometry Experiments. Eur Biophys J 2021, 50 (3–4), 403–409. 10.1007/s00249-021-01513-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Sonn-Segev A.; Belacic K.; Bodrug T.; Young G.; VanderLinden R. T.; Schulman B. A.; Schimpf J.; Friedrich T.; Dip P. V.; Schwartz T. U.; Bauer B.; Peters J.-M.; Struwe W. B.; Benesch J. L. P.; Brown N. G.; Haselbach D.; Kukura P. Quantifying the Heterogeneity of Macromolecular Machines by Mass Photometry. Nat Commun 2020, 11 (1), 1772. 10.1038/s41467-020-15642-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Keifer D. Z.; Pierson E. E.; Jarrold M. F. Charge Detection Mass Spectrometry: Weighing Heavier Things. Analyst 2017, 142 (10), 1654–1671. 10.1039/C7AN00277G. [DOI] [PubMed] [Google Scholar]
- (82).Kafader J. O.; Beu S. C.; Early B. P.; Melani R. D.; Durbin K. R.; Zabrouskov V.; Makarov A. A.; Maze J. T.; Shinholt D. L.; Yip P. F.; Kelleher N. L.; Compton P. D.; Senko M. W. STORI Plots Enable Accurate Tracking of Individual Ion Signals. J Am Soc Mass Spectrom 2019, 30 (11), 2200–2203. 10.1007/s13361-019-02309-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.