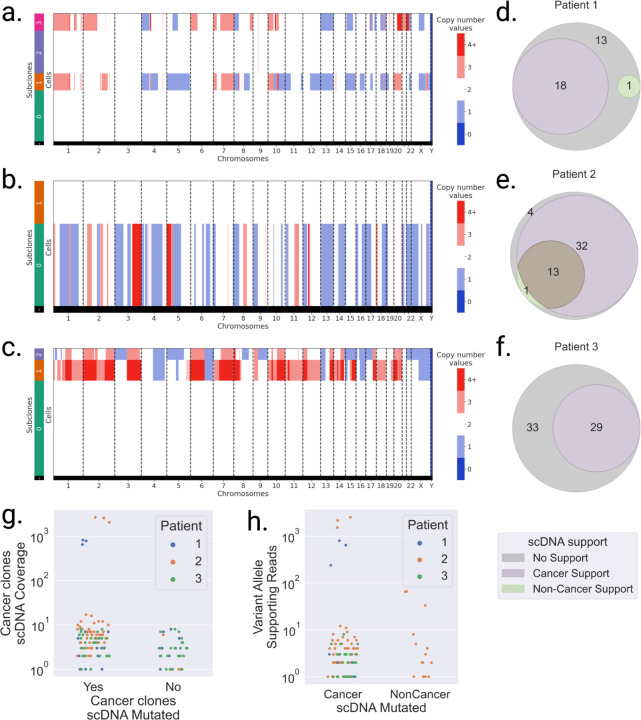
Figure 3: Somatic SNVs detected in scRNA are validated as somatic in scDNA.
a,b,c. scDNA-seq copy number values per subclone in a. patient 1, b. patient 2, and c. patient 3 data. Subclones with multiple copy number alterations are aneuploid (likely cancer), while copy number neutral subclones are diploid (likely non-cancer). d,e,f Venn diagrams of somatic SNVs supported (VAF>10%) in scDNA cancer subclones (purple), scDNA non-cancer subclones (green), and both (brown). g. scDNA cancer subclones coverage per somatic locus identified in scRNA, categorized by whether they are found mutated in cancer subclones (Yes) or not (No). h. Number of mutated reads in scDNA subclones per somatic loci identified in LR scRNA, categorized by cancer and non-cancer scDNA subclones.