Abstract
Sex-biased genes offer insights into the evolution of sexual dimorphism. Sex-biased genes, especially those with male bias, show elevated evolutionary rates of protein sequences driven by positive selection and relaxed purifying selection in animals. Although rapid sequence evolution of sex-biased genes and evolutionary forces have been investigated in animals and brown algae, less is known about evolutionary forces in dioecious angiosperms. In this study, we separately compared the expression of sex-biased genes between female and male floral buds and between female and male flowers at anthesis in dioecious Trichosanthes pilosa (Cucurbitaceae). In floral buds, sex-biased gene expression was pervasive, and had significantly different roles in sexual dimorphism such as physiology. We observed higher rates of sequence evolution for male-biased genes in floral buds compared to female-biased and unbiased genes. Male-biased genes under positive selection were mainly associated with functions to abiotic stress and immune responses, suggesting that high evolutionary rates are driven by adaptive evolution. Additionally, relaxed purifying selection may contribute to accelerated evolution in male-biased genes generated by gene duplication. Our findings, for the first time in angiosperms, suggest evident rapid evolution of male-biased genes, advance our understanding of the patterns and forces driving the evolution of sexual dimorphism in dioecious plants.
Research organism: Other
Introduction
Sexual dimorphism is the condition where sexes of the same species exhibit different morphological, ecological, and physiological traits in gonochoristic animals and dioecious plants, despite male and female individuals sharing the same genome except for sex chromosomes or sex-determining loci (Mank, 2009; Barrett and Hough, 2013). Such sexual dimorphisms usually arise from differential expression of genes between the two sexes, i.e., sex-biased genes (including sex-specific genes expressed exclusively in one sex) that are located on autosomal chromosomes and sex chromosomes/or sex-determining regions (Ellegren and Parsch, 2007; Parsch and Ellegren, 2013; Grath and Parsch, 2016; Charlesworth, 2018; Tosto et al., 2023). Recently, some studies have begun to explore the strength and impact of evolutionary forces that shape different sexually dimorphic traits through sex-biased gene expression (Mank, 2009; Rowe et al., 2018; Naqvi et al., 2019; Cai et al., 2021; Mank, 2023; Murat et al., 2023; Singh et al., 2023). Previous studies revealed that sex-biased gene expressions were associated with the evolution of sexual dimorphisms in some animal species, although the extent of this bias exhibits great variation among taxa, tissues, and development stages (Mank, 2017; Harrison et al., 2015; Hsu et al., 2020; Khodursky et al., 2020; Lichilín et al., 2021; Toubiana et al., 2021; Djordjevic et al., 2022; Yue et al., 2023). Unlike most animals, the vast majority (~90%) of flowering plants (angiosperms) are hermaphroditic, while only a small fraction (~5%) are dioecious in which individuals have exclusively male or female reproductive organs (Renner, 2014). Most dioecious plants possess homomorphic sex-chromosomes that are roughly similar in size when viewed by light microscopy (Palmer et al., 2019). Furthermore, sexual dimorphism in dioecious plants is less common and less conspicuous than in most animals (Barrett and Hough, 2013). Hence, the study of sex-biased gene expression is of great interest to plant evolutionary biologists, as it is necessary to understand the evolution of sexual dimorphism in dioecious plants (Moore and Pannell, 2011).
A common pattern that has emerged from previous studies is that sex-biased genes, particularly male-biased genes, tend to evolve rapidly in protein sequence (the ratio of non-synonymous to synonymous substitutions, dN/dS) compared to unbiased genes (Ellegren and Parsch, 2007; Grath and Parsch, 2016). The rapid evolution of male-biased genes was first observed in Drosophila melanogaster (Zhang et al., 2004; Zhang and Parsch, 2005) and has been supported by recent investigations in a wider range of animals (Pröschel et al., 2006; Mank et al., 2007; Mank, 2017; Papa et al., 2017; Catalán et al., 2018; Toubiana et al., 2021). In recent years, there have been growing studies on the expression dynamics and molecular evolutionary rates of sex-biased genes in flowering plants, including hermaphroditic Arabidopsis thaliana (Gossmann et al., 2014; Gossmann et al., 2016), Solanum (Moyle et al., 2021), and dioecious Silene latifolia (Zemp et al., 2016), Salix viminalis (Darolti et al., 2018), Mercurialis annua (Cossard et al., 2019), Populus balsamifera (Sanderson et al., 2019), and Leucadendron (Scharmann et al., 2021). However, despite such advances, the molecular evolution pattern of sex-biased genes in plants remains inconsistent among the studied plant species (Muyle, 2019; Veltsos, 2019). In dioecious plants such as Mercurialis annua and Leucadendron, Cossard et al., 2019 and Scharmann et al., 2021 found no significant differences in evolutionary rates of proteins among female-biased, male-biased, and unbiased genes detected between male and female plants leaf tissues, although the expression of sex-biased genes was highly different from unbiased genes in leaves. Similar patterns have also been reported in dioecious Populus balsamifera, where evolutionary rates of male-biased, female-biased, and unbiased genes did not differ in reproductive tissues (Sanderson et al., 2019). However, in the dioecious Salix viminalis, male-biased genes have significantly lower evolutionary rates of proteins than female-biased and unbiased genes in catkin tissues (Darolti et al., 2018). To our knowledge, only the five above-mentioned studies have investigated expression differences and protein evolutionary rates of sex-biased genes in dioecious angiosperms. Moreover, these studies only compared gene expression in vegetative versus vegetative tissues and vegetative versus reproductive tissues, limiting our understanding of sexual selection at different floral development stages. Therefore, more studies and taxa are needed to explore the common patterns of sequence evolution in sex-biased genes, with more focus on comparing sex-biased gene expression in reproductive versus reproductive tissues, e.g., different floral development stages in dioecious angiosperms.
Evolutionary analyses indicate that different driving forces impact the rate of sequence evolution of sex-biased genes. These forces include positive selection, which promotes the spread and adaptive fixation of beneficial alleles; sexual selection, which results from male-male competition or female choice; and relaxed purifying selection, which reduces the removal of deleterious mutations (Grath and Parsch, 2016; Mank, 2017; Dapper and Wade, 2020). For example, in animal systems, particularly in Drosophila, the elevated sequence divergence rates of male-biased genes have often been interpreted as the signature of adaptive evolution, suggesting that sexual selection is the primary evolutionary force (Pröschel et al., 2006 ; Assis et al., 2012). In brown algae, female-biased and/or male-biased genes exhibited higher evolutionary rates than unbiased genes, suggesting that rapid evolution is partly driven by adaptive evolution or sexual selection (Lipinska et al., 2015; Cossard et al., 2022; Hatchett et al., 2023). However, studies in plants have never reported elevated rates of sex-biased genes.
An alternative explanation for the rapid evolution of sex-biased genes is a relaxation of purifying selection due to reduced constraints (Lahti et al., 2009; Dapper and Wade, 2020). In the model plant Arabidopsis thaliana, pollen genes were found to be evolving faster than sporophyte-specific genes due to relaxed purifying selection associated with the transition from outcrossing to selfing (Harrison et al., 2019). These trends were recently confirmed in Arabis alpina, which exhibits mating system variation across its distribution, suggesting that the efficacy of purifying selection on male gametophyte-expressed genes was significantly weaker in inbred populations (Gutiérrez-Valencia et al., 2022). Together, these findings in plants reinforce the idea that both adaptive (e.g. positive selection, sexual selection) and non-adaptive (e.g. relaxed selection) evolutionary processes differentially impact the sequence evolution of sex-biased genes. Hence, investigating the potential contribution of selection forces to the emergence of specific evolutionary patterns of sex-biased genes within a focal species is of great interest.
In the family Cucurbitaceae, there are about 96 genera and 1000 species, about 50% of species are dioecious, and the others are monoecious (Schaefer and Renner, 2011). Phylogenetic analyses of Cucurbitaceae suggest that dioecy is the ancestral state of the family, but transitions frequently to monoecy (Zhang et al., 2006). Trichosanthes pilosa (synonym: T. ovigera, 2n=22, Cucurbitaceae) is mainly distributed from Southwest and Southeast China to Japan, extending to Southeast Asia, New Guinea and Western Australia. It was suggested to have originated in the late Miocene (ca. 8–6 million-year ago) (de Boer et al., 2012; de Boer et al., 2015; Guo et al., 2020). Trichosanthes pilosa is a perennial, night-flowering, insect-pollinated dioecious vine that reproduces sexually and possesses a pair of heteromorphic sex chromosomes XX/XY (Ming et al., 2011). The male parts (e.g. anthers) of female flowers, and the female parts (e.g. pistil and ovaries) of male flowers are fully aborted. Its male and female flowers exhibit strong sexual dimorphism in floral morphological and phenological traits, such as racemose versus solitary (Figure 1), early-flowering versus late-flowering, and caducous versus long-lived (Wu et al., 2011).
Figure 1. Floral buds and flowers at anthesis of females (A, B) and males (C) in Trichosanthes pilosa.
To understand the evolution of sex-biased genes in dioecious T. pilosa, we collected floral buds and flowers at anthesis from male and female individuals and characterized their expression profiles using Illumina RNA sequencing. Our primary objectives are to (1) compare expression divergence between males and females at two floral development stages; (2) explore whether there are differences in the evolutionary rates of proteins among female-biased, male-biased, and unbiased genes; and if so, (3) determine the main selective forces that contribute to the differentiation of sequence evolution rates among gene categories.
Results
Transcriptome sequencing, de novo assembly, and annotation
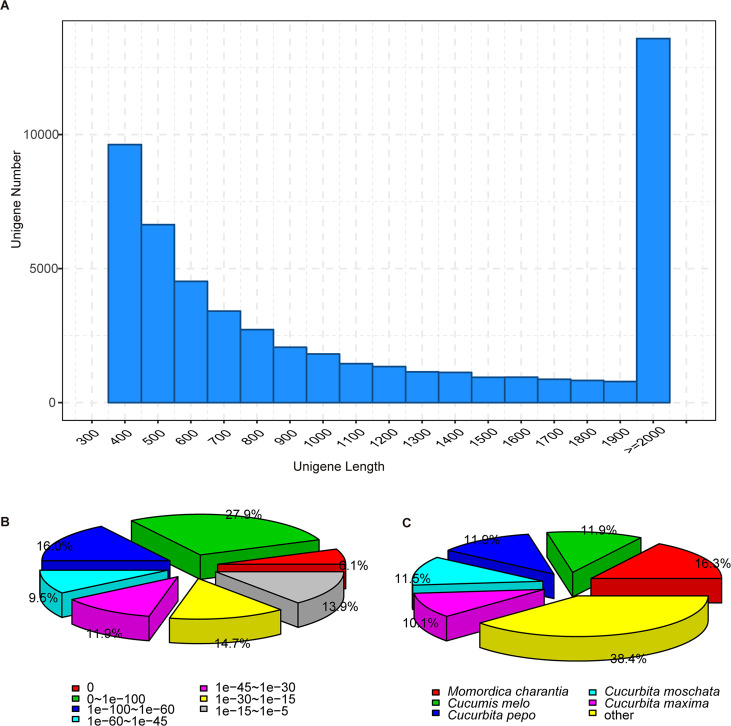
Using whole transcriptome shotgun sequencing, we sequenced floral buds and flowers at anthesis from females and males of dioecious T. pilosa. We set up three biological replicates from three female and three male plants, including 12 samples in total (six floral buds and six flowers at anthesis). We then generated a total of nearly 276 million clean reads (Supplementary file 1). Due to the absence of a reference genome, we performed de novo assembly of transcripts from all the clean reads, followed by clustering and filtering analysis, resulting in 59,051 unigenes (Figure 2—figure supplement 1). To evaluate the quality of the assembled unigenes, we used BUSCO assessments based on embryophyta_odb10 database, which showed the completeness of the reference transcriptome at 89.7% (Supplementary file 2). We then annotated them against protein databases including NR, KEGG, Swissport, PFAM, and GO using BLASTP and nucleotide database NT using BLASTN (Supplementary file 2). The e-value distribution of the best hits in the NR database suggested that 47,241 unigenes (80%) had strong homology, with an e-value smaller than 1.0e-15 (Figure 2—figure supplement 1). The majority of unigenes were annotated by homologs in species of Cucurbitaceae (61.6%, 36,375), such as Momordica charantia (16.3%, 9625), Cucumis melo (11.9%, 7027), Cucurbita pepo (11.9%, 7027), Cucurbita moschata (11.5%, 6791), Cucurbita maxima (10.1%, 5964), and other species (38.4%, 22,676) (Figure 2—figure supplement 1). Overall, our assessment suggested that we have generated high-quality reference transcriptomes.
Expression characteristics of sex-biased genes
We mapped the RNA-seq reads of floral buds and flowers at anthesis onto the reference transcriptome in dioecious T. pilosa, which resulted in approximately 75% read mappings per sample (Supplementary file 3). In floral buds, we identified 5096 (9.50%) female-biased genes and 4214 (7.86%) male-biased genes (Figure 2A). In contrast, only 380 (0.70%) female-biased genes and 233 (0.43%) male-biased genes were detected in flowers at anthesis (Figure 2B). Using hierarchical clustering analysis, we evaluated different levels of gene expression across sexes and tissues (Figure 2C). Gene expression for female floral buds clustered most distantly from expression in female flowers at anthesis. However, expression in male floral buds clustered with expression in female flowers at anthesis, suggesting that male floral buds maybe tend to feminization in the early stages of floral development. Furthermore, we observed that the number of sex-biased genes in floral buds was approximately 15 times higher than in flowers at anthesis, indicating that sex-biased genes associated with meiotic processes, sex differentiation, and sexually dimorphic traits are predominantly expressed in floral buds. We also analyzed sex-specific genes that were exclusively expressed in floral buds and flowers at anthesis of one sex. In floral buds, we found 253 out of 5096 (4.96%) female-specific genes and 465 out of 4214 (11.03%) male-specific genes. However, in flowers at anthesis, we only identified 26 out of 380 (6.84%) female-specific genes and 52 out of 233 (22.32%) male-specific genes. Taken together, sex bias is more prevalent in floral buds than in flowers at anthesis.
Figure 2. Sex-biased gene expression for floral buds and flowers at anthesis in males and females of Trichosanthes pilosa.
Volcano plots of average expression between female-biased, male-biased and unbiased genes in floral buds (A) and flowers at anthesis (B). M1 and F1 indicate male and female floral buds; M2 and F2 indicate male and female flowers at anthesis. The value of y coordinate represents -log10(FDR), and the value of x coordinate represents log2(Fold Change) identified by DESeq2. Heatmap of sex-biased gene expression (C) using hierarchical clustering analysis. Hierarchical gene clustering is based on Euclidean distance with an average of log2(FPKM) for differentially expressed genes. The color gradient represents from high to low (from red to green) gene expression.
Figure 2—figure supplement 1. The length distribution of unigenes (A) and the e-value distribution (B) and the species distribution (C) of BLAST hits for each unigene.
Tissue-biased/stage-biased gene expression
We compared the expression levels of transcripts in floral buds and flowers at anthesis within each sex to identify genes with tissue-biased expression. In male plants, the number (M2TGs: n=2795) of tissue-biased genes in male flowers at anthesis (M2TGs) was 1040 higher than that in male floral buds (M1TGs: n=1755, Figure 3A and B). However, in female plants, the number (F2TGs: n=660) of tissue-biased genes in female flowers at anthesis (F2TGs) was only 536 more than that in female floral buds (F1TGs: n=124, Figure 3C and D). Our results indicated that males had a higher tissue-bias relative to females. We also identified sex-biased genes that were expressed in both types of tissues by comparing tissue-biased genes with male-biased and female-biased genes, respectively. Few female-biased genes in floral buds (F1BGs: n=5096) overlapped with tissue-biased genes in female floral buds (F1TGs: n=124) and female flowers at anthesis (F2TGs: n=660), accounting for only 85 out of 5096 (1.67%) (Figure 3C). Similarly, few female-biased genes in flowers at anthesis (F2BGs: n=380) overlapped with tissue-biased genes in female floral buds (F1TGs: n=124) and female flowers at anthesis (F2TGs: n=660), occupying around 5 out of 380 (1.32%) (Figure 3D). However, a significant proportion of male-biased genes in floral buds (M1BGs: n=4214) overlapped with tissue-biased genes in male floral buds (M1TGs: n=1755) and male flowers at anthesis (M2TGs: n=2795), with 1010 out of 4214 (23.97%) (Figure 3A). A high proportion of male-biased genes in flowers at anthesis (M2BGs: n=233) overlapped with tissue-biased genes in male floral buds (M1TGs: n=1755) and male flowers at anthesis (M2TGs: n=2795), 145 out of 233 (62.23%) (Figure 3B).
Figure 3. The overlap between sex-biased and tissue-biased genes in two types of sexes and tissues.
Male-biased genes in floral buds (M1BGs) (A) or flowers at anthesis (M2BGs) (B) overlapped with tissue-biased genes in floral buds (M1TGs) and flowers at anthesis (M2TGs). Female-biased genes in floral buds (F1BGs) (C) or flowers at anthesis (F2BGs) (D) overlapped with tissue-biased genes in floral buds (F1TGs) and flowers at anthesis (F2TGs).
Figure 3—figure supplement 1. The overlap between male-biased genes with faster evolutionary rates and tissue-biased genes in floral buds.
Elevated protein evolutionary rates of male-biased genes in floral buds
We compared rates of protein evolution among male-biased, female-biased and unbiased genes in four species with phylogenetic relationships (((T. anguina, T. pilosa), T. kirilowii), Luffa cylindrica), including dioecious T. pilosa, dioecious T. kirilowii, monoecious T. anguina in Trichosanthes, together with monoecious Luffa cylindrica. To do this, we used the transcriptomes described above for T. pilosa. We also collected transcriptomes of T. kirilowii, as well as genomes of T. anguina and Luffa cylindrica (see Methods Section). We identified 1145 female-biased, 343 male-biased, and 2378 unbiased one-to-one orthologous groups (OGs) from floral buds. Additionally, we detected 45 female-biased, 13 male-biased, and 3782 unbiased one-to-one OGs from mature flowers in all four species. To quantify the rates of protein sequences, we separately calculated ω values for each sex-biased and unbiased orthologous gene using ‘two-ratio’ and ‘free-ratio’ branch models in juvenile and mature flowers (Figure 4 and Figure 4—figure supplement 1).
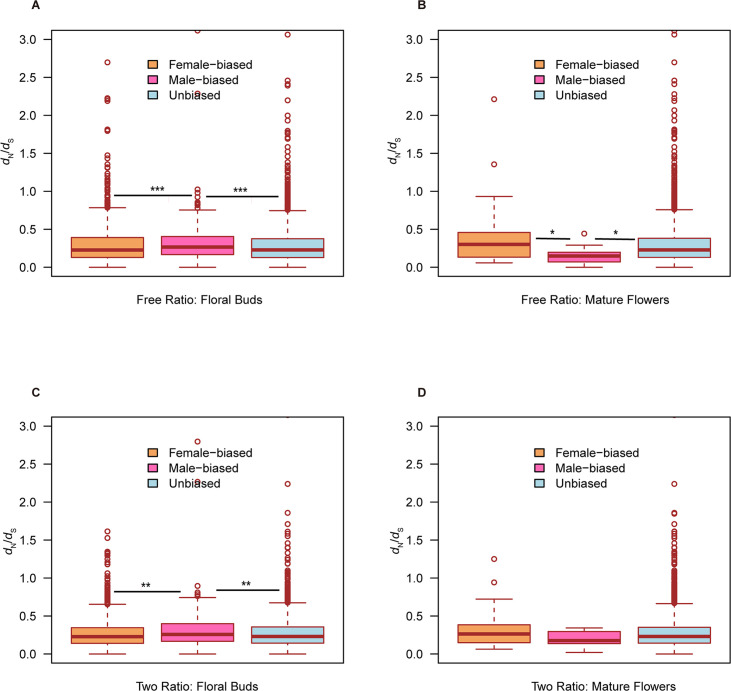
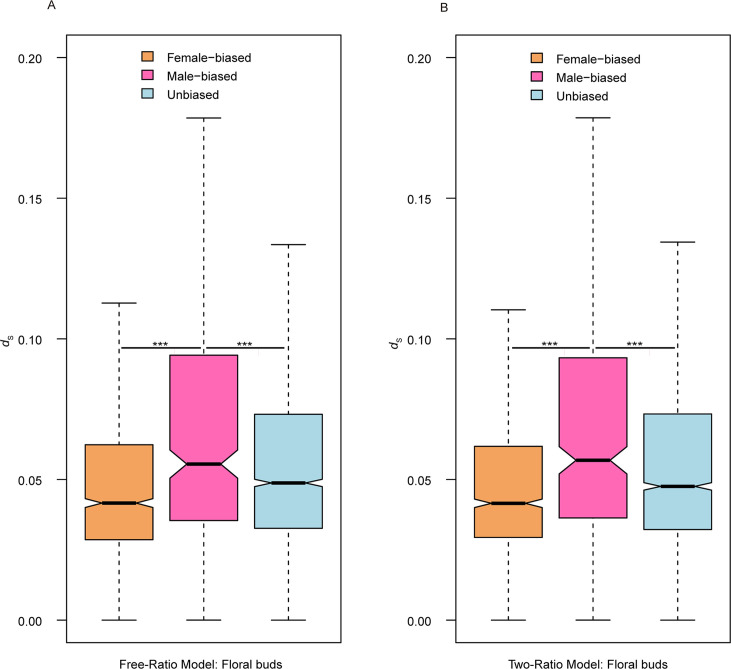
Figure 4. Violin plots of dN/dS values (0<ω<2) of female-biased, male-biased, and unbiased genes in floral buds and flowers at anthesis of Trichosanthes pilosa.
White dot indicates the median of dN/dS values for sex-biased and unbiased genes. Wilcoxon rank sum tests are used to test for significant differences (***p<0.0005, **p<0.005, and *p<0.05). The distributions of dN/dS values for female-biased, male-biased and unbiased genes in floral buds (A) and flowers at anthesis (B) using ‘two-ratio’ branch model. The distributions of dN/dS values for female-biased, male-biased and unbiased genes in floral buds (C) and flowers at anthesis (D) using ‘free-ratio’ branch model.
Figure 4—figure supplement 1. Boxplot of dN/dS values (including all ω values) of female-biased, male-biased, and unbiased genes in floral buds and flowers at anthesis of Trichosanthes pilosa.
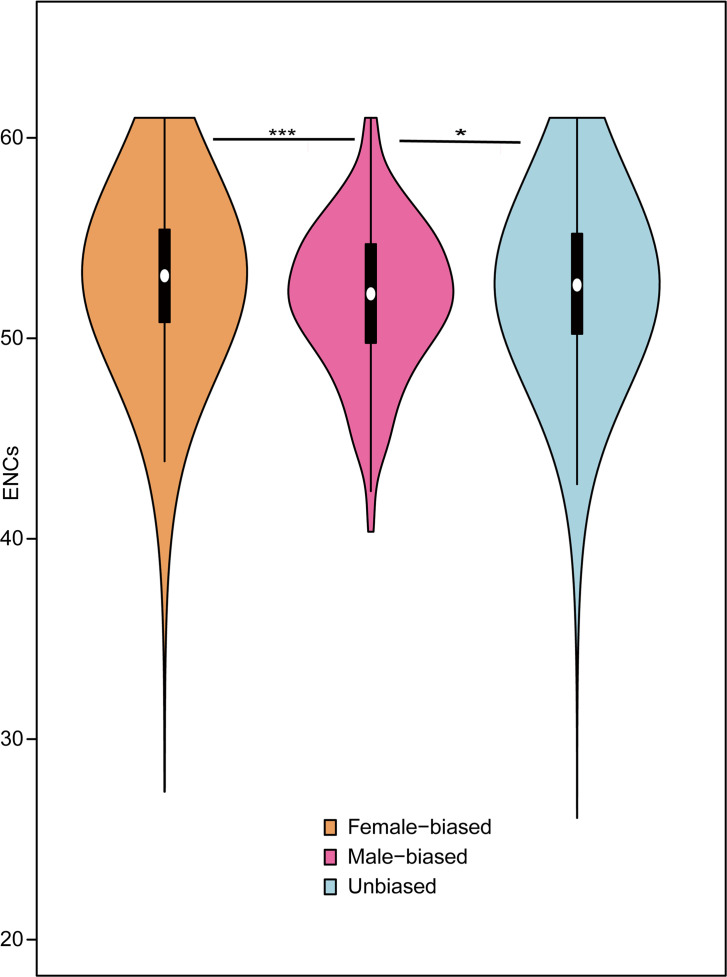
Figure 4—figure supplement 2. Violin plots of effective number of codons (ENCs) values of female-biased, male-biased and unbiased genes in floral buds.
Figure 4—figure supplement 3. Boxplot of dS values of female-biased, male-biased, and unbiased genes in floral buds of dioecious Trichosanthes pilosa using ‘free-ratio’ (A) and ‘two-ratio’ (B) branch model.
The two-ratio branch model, where the foreground (dioecious branches) has a different ω value relative to the background (all other branches), is better supported than the fixed-ratio branch model, where all branches are constrained to have the same ω value. In the results of the ‘two-ratio’ branch model, the median of ω values in female-biased, male-biased, and unbiased genes were 0.227, 0.257, and 0.230 in floral buds, respectively (Figure 4A and Supplementary file 4). We observed that male-biased genes had a 13.22% and 11.74% higher median than female-biased and unbiased genes in floral buds, respectively. The difference in the distribution of ω values between female-biased versus male-biased genes (p=0.0021) and male-biased versus unbiased genes (p=0.0051) was statistically significant in Wilcoxon rank sum tests. However, we did not find a significant difference in ω values between female-biased and unbiased genes in floral buds (Wilcoxon rank sum test, p=0.4618). In flowers at anthesis, the median of ω values for female-biased, male-biased, and unbiased genes were 0.269, 0.177, and 0.231, respectively (Figure 4B and Supplementary file 4). However, there was no statistically significant difference in the distribution of ω values using Wilcoxon rank sum tests for female-biased versus male-biased genes (p=0.0556), female-biased versus unbiased genes (p=0.0796), and male-biased versus unbiased genes (p=0.3296) possibly because of limited statistical power due to the low number of sex-biased genes in flowers at anthesis.
In free-ratios model, ω values are free to vary in each branch compared to fixed-ratio branch model and the two-ratio branch model. The ‘free-ratio’ branch model yielded interesting results. In floral buds, the median ω values for female-biased, male-biased, and unbiased genes were 0.222, 0.265, and 0.226, respectively (Figure 4C and Supplementary file 5). Male-biased genes had a significantly higher median relative to female-biased genes (19.37% higher, Wilcoxon rank sum test, p=0.0009) and unbiased genes (17.26% higher, Wilcoxon rank sum test, p=0.0004) in floral buds. However, there was no significant difference in ω values between female-biased and unbiased genes (Wilcoxon rank sum test, p=0.9862). In flowers at anthesis, the median ω values for female-biased, male-biased, and unbiased genes were 0.300, 0.148, and 0.227, respectively (Figure 4D and Supplementary file 5). Female-biased and unbiased genes had significantly higher ω values than male-biased (Wilcoxon rank sum test, p=0.0101, p=0.0146, respectively). However, there was no significant difference in ω values between female-biased and unbiased genes (Wilcoxon rank sum test, p=0.2887). Since the number of male-biased genes and evolutionary rates of male-biased genes in flowers at anthesis are lower than those in floral buds, we decided to focus on the latter in subsequent analyses. Additionally, we found that only in floral buds, there were significant differences in ω values in the results of ‘free-ratio’ model (female-biased versus male-biased genes, p=0.04282 and male-biased versus unbiased genes, p=0.01114) and ‘two-ratio’ model (female-biased versus male-biased genes, p=0.01992 and male-biased versus unbiased genes, p=0.02127, respectively) by permutation t-test, which is consistent with the results of Wilcoxon rank sum test.
Evidence of positive selection and relaxed selection for male-biased genes in floral buds
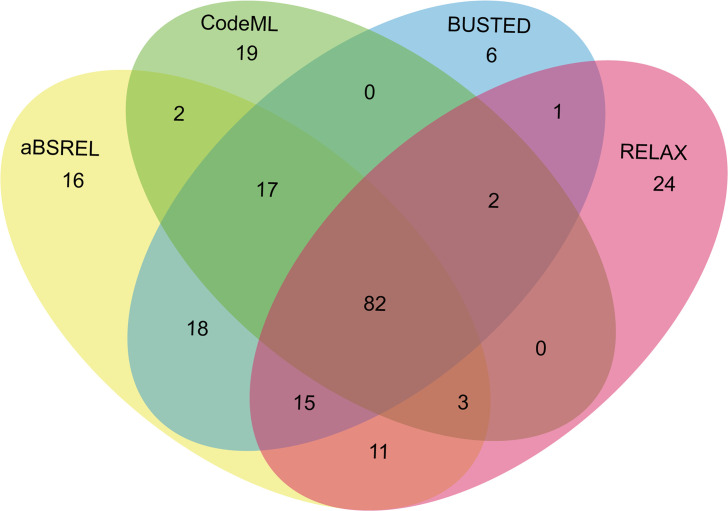
After comparing the alternative hypothesis (branch-site model A with estimated ω value) against the null model (branch-site model A with fixed ω=1) (see Methods section), we discovered that 39 out of 343 OGs (11.34%) in male-biased genes of floral buds exhibited strong evidence of having certain sites that evolved under positive selection based on foreground ω value, likelihood ratio tests (LRTs, p<0.05) and Bayes empirical Bayes (BEB) value (Figure 5 and Supplementary file 6). As a complementary approach, we utilized the aBSREL and BUSTED methods that are implemented in HyPhy v.2.5 software, which avoids false positive results by classical branch-site models due to the presence of rate variation in background branches, and detected significant evidence of positive selection. According to our findings, 84 out of 343 OGs (24.49%) were identified to be under episodic positive selection in male-biased genes of floral buds with a site proportion of 0.17–26.44% based on aBSREL (Supplementary file 7). In addition, 69 out of 343 OGs (20.01%) exhibited significant signs of positive selection with the site proportion of 0.28–32.65% in male-biased genes of floral buds according to BUSTED (Supplementary file 8). Among these, a total of 32 OGs (9.30%) were identified through our tests using CodeML, aBSREL, and BUSTED (Figure 5).
Figure 5. Venn diagrams of male-biased genes detected to be under positive selection using aBSREL, BUSTED, CodeML, and RELAX in floral buds.
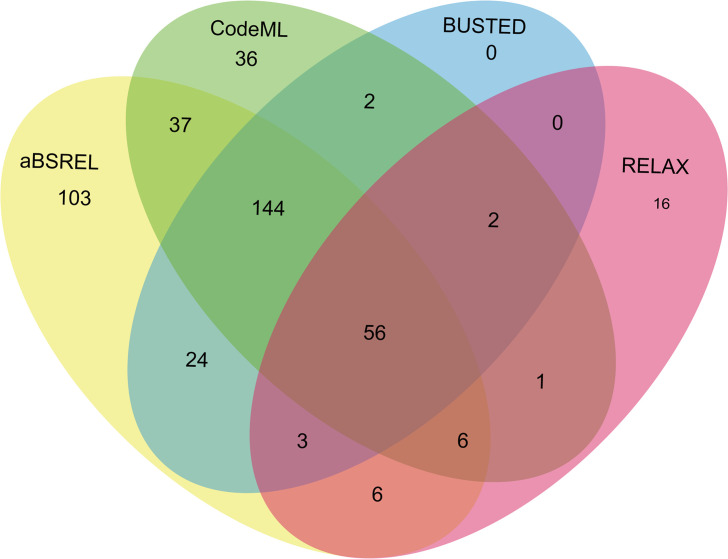
Figure 5—figure supplement 1. Venn diagrams of female-biased genes under positive selection in floral buds.
Figure 5—figure supplement 2. Venn diagrams of unbiased genes under positive selection in floral buds.
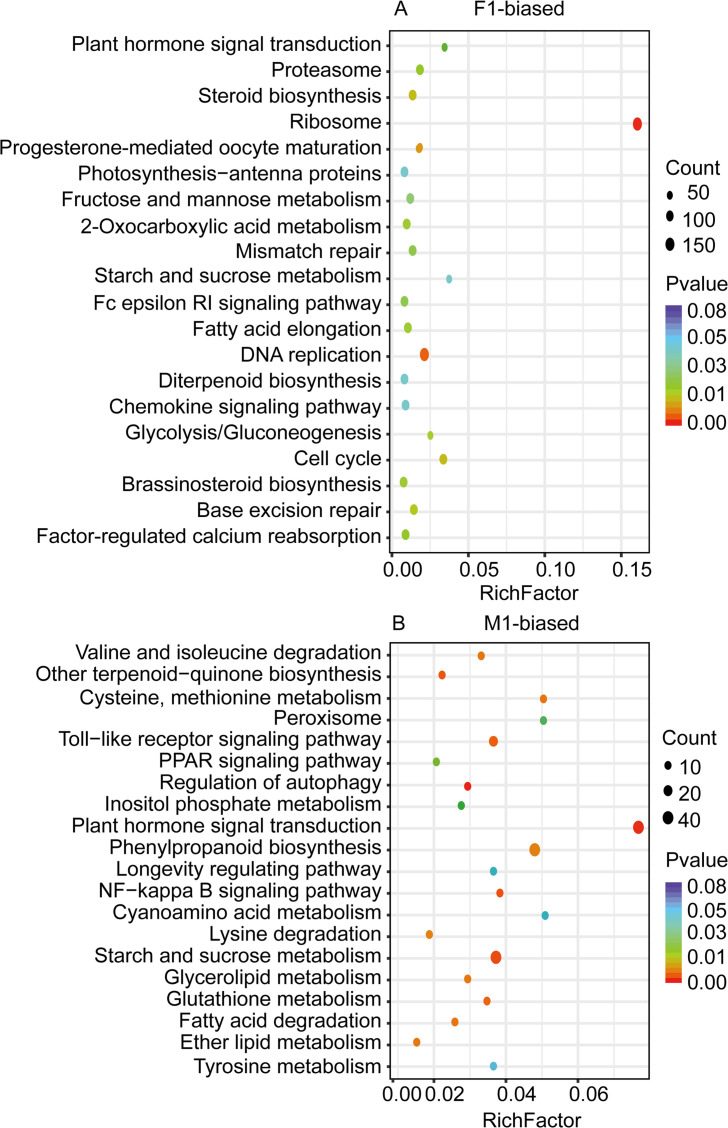
Figure 5—figure supplement 3. Scatterplots of KEGG pathway of sex-biased genes in female (A) and male (B) floral buds of the dioecious Trichosanthes pilosa.
Relaxed selection may occur when the efficiency of natural selection (e.g. the reduction of the strength of purifying selection) is reduced, leading to accumulations of deleterious mutations (Lahti et al., 2009; Dapper and Wade, 2020). This has been proposed as an explanation for the rapid evolution of sex-biased genes (Lahti et al., 2009; Mank, 2017). Using the RELAX model, we detected that 18 out of 343 OGs (5.23%) showed significant evidence of relaxed selection (K=0.0184–0.6497) (Supplementary file 9). Most of the 18 OGs were members of different gene families generated by gene duplication (Supplementary file 13). Additionally, we observed that 61 out of 343 OGs (17.73%) exhibited significant evidence of intensified positive selection (K=2.3363–50, ω2≥1) (Figure 5 and Supplementary file 10), which is consistent with the results obtained from CodeML, aBSREL, and BUSTED.
According to previous studies (Ellegren and Parsch, 2007; Catalán et al., 2018), genes that exhibit sex-biased expression with rapid evolutionary rates tend to display a lower codon bias compared to unbiased genes. In our results, we found that male-biased genes in floral buds had a significantly lower median effective number of codons (ENCs) than both female-biased and unbiased genes (Wilcoxon rank sum test, female-biased vs male-biased genes, p=0.0001 and male-biased vs unbiased genes, p=0.0123). This suggested that male-biased genes in floral buds exhibit stronger codon bias than both female-biased and unbiased genes (Figure 4—figure supplement 2). Similarly, given that dN/dS values of sex-biased genes were higher due to codon usage bias, lower dS rates would be expected in sex-biased genes relative to unbiased genes (Ellegren and Parsch, 2007; Parvathy et al., 2022). However, we exhibited that the median of dS values in male-biased genes was much higher than those in female-biased and unbiased genes in the results of ‘free-ratio’ (Figure 4—figure supplement 3, female-biased versus male-biased genes, p=6.444e-12 and male-biased versus unbiased genes, p=4.564e-13) and ‘two-ratio’ model (Figure 4—figure supplement 3, female-biased versus male-biased genes, p=2.2e-16 and male-biased versus unbiased genes, p=9.421e-08, respectively). In short, our analyses indicated that rapid evolutionary rates of male-biased genes in floral buds were not associated with a reduction in codon usage bias.
We also analyzed whether female-biased and unbiased genes underwent positive and relaxed selection in floral buds (Supplementary file 6–10). We identified 216 (18.86%) positively selected (Figure 5—figure supplement 1), and 69 (6.03%) relaxed selective female-biased genes from 1145 OGs, respectively. Similarly, we found 436 (18.33%) positively selected (Figure 5—figure supplement 2), and 43 (1.81%) unbiased genes under relaxed selection from 2378 OGs, respectively. Notably, male-biased genes have a higher proportion (10%) of positively selected genes compared to female-biased and unbiased genes. However, relaxed selective male-biased genes have a higher proportion (3.24%) than unbiased genes, but about 0.8% lower than that of female-biased genes. In summary, our analyses suggested that positive selection and relaxed selection likely drove the rapid evolutionary rates of male-biased genes compared to female-biased and unbiased genes in floral buds.
Functional analysis of sex-biased genes in floral buds
We conducted KEGG pathway enrichment analysis on sex-biased genes in floral buds. Our results showed that 699 genes were female-biased and 358 genes were male-biased, with significant enrichment (p<0.05) in 26 and 24 KEGG pathways, respectively (Supplementary file 11). In the floral bud stage, we observed that female-biased genes were mainly enriched in metabolic and signaling pathways, such as ribosome, Fatty acid elongation, photosynthesis, and plant hormone signal transduction (Figure 5—figure supplement 3 and Supplementary file 11). On the other hand, male-biased genes were significantly enriched in metabolic and signaling pathways, including inositol phosphate metabolism, starch, and sucrose metabolism, regulation of autophagy, plant hormone signal transduction, and Toll-like receptor signaling pathway (Figure 5—figure supplement 3 and Supplementary file 11).
We have also found that certain male-biased genes, which are evolving under positive selection and relaxed selection (Supplementary file 12 and 13), were related to abiotic stress and immune responses. For instance, mitogen-activated protein kinase kinase kinase 18 (MAPKKK18) (Zhang and Zhang, 2022), zinc finger CCCH domain-containing protein 20 (C3H20/TZF2) (Bogamuwa and Jang, 2014), and heat stress transcription factor B-3 (HSFB3) (Scharf et al., 2012) have been linked to stress. Additionally, ten male-biased genes with rapid evolutionary rates were associated with anther and pollen development. These genes include LRR receptor-like serine/threonine protein kinase (LRR-RLK) (Cui et al., 2022), pollen receptor-like kinase 3 (PRK3) (Muschietti and Wengier, 2018), autophagy-related protein 18 f (ATG18f) (Zhou et al., 2015; Li et al., 2020), and plant homeodomain (PHD) finger protein 3 (MALE STERILITY 3) (Hou et al., 2022) in floral buds of male plants.
Discussion
The Cucurbitaceae family, where half of the species are monoecious and half are dioecious, is an excellent model for studying the evolution of sexual systems of angiosperms, including sex-determination mechanism and sexual dimorphism (Schaefer and Renner, 2010; Boualem et al., 2015; Ma and Pannell, 2016). In this study, we compared the expression profiles of sex-biased genes between sexes and two tissue types, investigated whether sex-biased genes exhibited evidence of rapid evolutionary rates of protein sequences and identified the potential evolutionary forces responsible for the observed patterns in the dioecious Trichosanthes pilosa.
Sex-biased expression in floral buds
Several studies have shown that in dioecious plants, male-biased genes tend to outnumber female-biased genes, consistent with the patterns in most animals (Zhang et al., 2007; Djordjevic et al., 2022). For instance, insect-pollinated dioecious plants such as Asparagus officinalis (Harkess et al., 2015) and Silene latifolia (Zemp et al., 2016), exhibit a higher proportion of male-biased genes. In contrast, the wind-pollinated dioecious plant Populus balsamifera (Sanderson et al., 2019) has twice as many female-biased genes as male-biased genes. The differences in these studies could be partly attributed to the impact of sexual selection on secondary sexual traits in insect-pollinated dioecious plants, as opposed to wind-pollinated ones (Delph and Herlihy, 2012; Muyle, 2019; Sanderson et al., 2019). Similar to the above study of Populus balsamifera, our findings revealed that the number of female-biased genes in floral buds of the night-flowering, insect-pollinated dioecious plant Trichosanthes pilosa exceeded that of male-biased genes by 882 (~21%). This excess of female-biased expression could be due to lower energy consumption needs and reduced chemical defense capability against insect herbivores in short-lived male flowers (Sanderson et al., 2019). Indeed, functional enrichment analysis in chemical pathways such as terpenoid backbone and diterpenoid biosynthesis indicated that relative to male floral buds, female floral buds had more expressed genes that were equipped to defend against herbivorous insects and pathogens, except for growth and development (Vaughan et al., 2013; Ren et al., 2022; Figure 5—figure supplement 3 and Supplementary file 11 ). Additionally, our enrichment analysis showed that the photosynthesis, porphyrin, and chlorophyll metabolism pathways were more active in female floral buds compared to male floral buds (Figure 5—figure supplement 3 and Supplementary file 11), enabling them to acquire more resources such as carbon for fruit and seed production (Delph, 1999).
We identified functional enrichments in Toll-like receptor signaling, NF-kappa B signaling, and inositol phosphate metabolism pathways in male floral buds (Figure 5—figure supplement 3 and Supplementary file 11 ). We also found that male-biased genes with high evolutionary rates in male floral buds were associated with functions to abiotic stresses and immune responses (Supplementary file 12 and 13), which suggests that male floral buds through rapidly evolving genes are adapted to mountain climate and the environment in Southwest China relative to female floral buds through high gene expression. In addition, the enrichment in regulation of autophagy pathways could be associated with gamete development and the senescence of male floral buds (Supplementary file 14; Liu and Bassham, 2012; Li et al., 2020; Zhou et al., 2021). In fact, it was observed that male flowers senesced faster (Wu et al., 2011). We also found that homologous genes of two male-biased genes in floral buds (Supplementary file 14) that control the raceme inflorescence development (Teo et al., 2014) were highly expressed compared to female floral buds. Taken together, these results indicate that expression changes in sex-biased genes, rather than sex-specific genes play different roles in sexual dimorphic traits in physiology and morphology (Dawson and Geber, 1999).
Rapid evolution of male-biased genes in floral buds
It has been observed that, in most animals, sex-biased genes, particularly those biased towards males, often exhibit more rapid evolutionary rates than unbiased genes (Parsch and Ellegren, 2013; Grath and Parsch, 2016; Mank, 2017; Toubiana et al., 2021). However, in dioecious angiosperms, no evidence of rapid evolution in sex-biased genes relative to unbiased genes has been found (Zemp et al., 2016; Darolti et al., 2018; Cossard et al., 2019; Sanderson et al., 2019; Scharmann et al., 2021). In contrast, our findings indicated that male-biased genes experience higher evolutionary rates than both female-biased and unbiased genes in floral buds of dioecious T. pilosa. We proposed that positive selection and relaxed purifying selection may be responsible for the rapid sequence evolution of male-biased genes.
After analyzing the data, we found that around 28.57% (98 genes) of male-biased genes have undergone positive selection. Additionally, we observed that the proportion of male-biased genes under positive selection was about 10% higher than that of female-biased and unbiased genes. Furthermore, we discovered that some male-biased genes under positive selection were linked to abiotic stress and immune responses (Supplementary file 12). Our findings are consistent with studies on Drosophila and Ectocarpus (Zhang and Parsch, 2005; Lipinska et al., 2015), suggesting that adaptive evolution is one of the important driving forces for rapid evolutionary rates. Notably, we identified several male-biased genes under positive selection that are functionally related to early flowering (phyB) (Stephenson and Bertin, 1983; Forrest, 2014; Hajdu et al., 2015) and pollen development (Skogsmyr and Lankinen, 2002; Williams and Reese, 2019; Supplementary file 12–14). These findings indicate that a small fraction of male-biased genes may experience adaptive evolution due to sexual selection, driven by male-male competition.
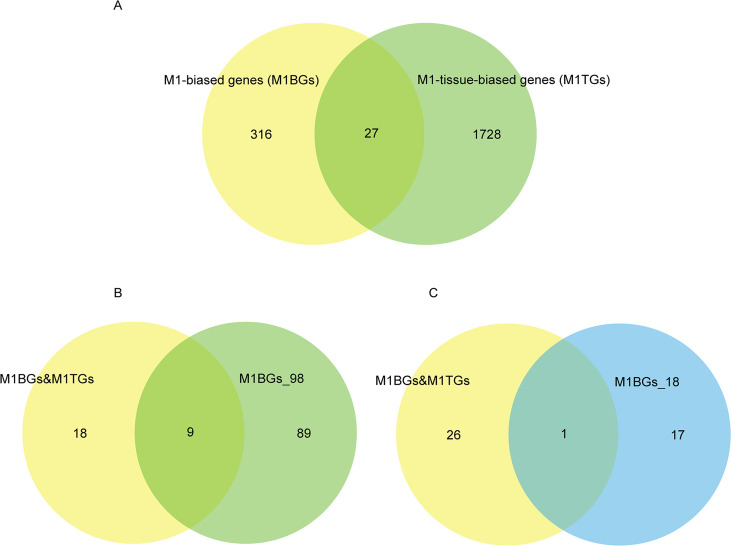
Alternatively, relaxed constraints could contribute to the rapid evolutionary rates of sex-biased genes through three key characteristics (Dapper and Wade, 2020; Tosto et al., 2023). First, sex-biased genes are often expressed solely in reproductive tissues of one sex (e.g. sex-specific genes), particularly in the haploid phase (Sandler et al., 2018; Immler, 2019; Beaudry et al., 2020). Sex-specific selection (e.g. relaxed purifying selection) acting on sex-specific genes could decrease the elimination of deleterious mutations (Mank, 2017), such as pollen-specific (Harrison et al., 2019; Arunkumar et al., 2013) or testes-specific genes (Gershoni and Pietrokovski, 2014). However, we observed male-biased genes but not male-specific genes undergoing relaxed purifying selection. Second, sex-biased genes are often expressed in few tissues (tissue-biased genes) (Meisel, 2011; Tosto et al., 2023), resulting in these genes rapidly evolving under positive selection or relaxed purifying selection due to low evolutionary constraints (Congrains et al., 2018; Whittle et al., 2021; Tosto et al., 2023). In our results, 343 male-biased genes (M1-biased genes, M1BGs) with faster evolutionary rates relative to female-biased and unbiased genes overlapped with 1755 tissue-biased genes in floral buds (M1-tissue-biased genes, M1TGs) (27 out of 343, 7.87%) (Figure 3—figure supplement 1). Furthermore, 27 out of 343 male-biased genes (that is, tissue-biased genes) in floral buds overlapped with nine out of 98 (9.18%) male-biased genes under positive selection (Figure 3—figure supplement 1), and one out of 18 (5.56%) male-biased genes under relaxed purifying selection (Figure 3—figure supplement 1). So, we obtained ten rapidly evolving tissue-biased genes which were also male-biased in male flower buds, suggesting that elevated evolutionary rates may partly be linked to low constraints, consistent with male-biased genes in Anastrepha and Fucus (Congrains et al., 2018; Hatchett et al., 2023). Finally, gene duplication has long been thought to promote functional divergences and phenotypic novelties by relaxing the constraints of purifying selection on the duplicated gene copy early in its history (Lynch and Conery, 2000; Lynch and Katju, 2004; Lahti et al., 2009). For instance, the progesterone receptor gene family in the human lineage Marinić and Lynch, 2020 and the CYP98A9 clade in Brassicales (Liu et al., 2016) have demonstrated rapid evolution and divergent function due to relaxed purifying selection. In our results, we identified only 18 out of 343 (5.25%) male-biased genes that underwent relaxed purifying selection using RELAX model (Supplementary file 13). Interestingly, the vast majority of genes under relaxed selection were members of different gene families generated by gene duplication (including whole-genome duplication), such as LOB domain-containing protein 18 (LBD18) (Zhang et al., 2020), WRKY transcription factor 72 (WRKY72) (Chen et al., 2017), and pollen receptor-like kinase 3 (PRK3) (Muschietti and Wengier, 2018).
Reducing codon usage bias could theoretically accelerate evolutionary rates of sex-biased genes by decreasing synonymous substitution rates. However, our results did not support this idea due to stronger codon usage bias in male-biased genes (Figure 4—figure supplement 2). Codon usage bias is influenced by many factors, such as levels of gene expression. Highly expressed genes have a stronger codon usage bias and could be encoded by optimal codons for more efficient translation (Frumkin et al., 2018; Parvathy et al., 2022), consistent with high levels of gene expression in males (that is, male-biased genes) in floral buds. Additionally, stronger codon usage bias may be related to higher synonymous substitution rates (Parvathy et al., 2022). Indeed, male-biased genes had significantly higher median dS values than female-biased and unbiased genes, both in the ‘free-ratio’ analysis (Figure 4—figure supplement 3) and ‘two-ratio’ branch model (Figure 4—figure supplement 3).
The presence of sex chromosomes may be a potential confounding factor for evolutionary rates of sex-biased genes which are X-linked, Y-linked, and autosomal genes (Hough et al., 2014; Sandler et al., 2018). We distinguished these sex-biased genes on sex chromosomes from autosomal chromosomes following the steps of Sandler et al., 2018, and computed the overall comparable proportions of sex-linked genes among male-biased (3/343=0.087%), female-biased (19/1145=1.66%) and unbiased genes (36/2378=1.51%). These analyses suggested that sex-linked genes may contribute relatively little to the rapid evolution of male-biased genes.
Several species have been observed to exhibit rapid evolutionary rates of sequences on sex chromosomes compared to autosomes, which has been related to the evolutionary theories of fast-X or fast-Z (Meisel and Connallon, 2013; Hough et al., 2014; Wright et al., 2015; Charlesworth et al., 2018; Darolti et al., 2023). Furthermore, the quantification of gene expression by bulk RNA-seq technology, relative to single-cell transcriptome analysis, has been shown to potentially obfuscate true signals in the evolution of sex-biased gene expression in complex aggregations of diverse cell types (Darolti and Mank, 2023; Tosto et al., 2023). Additionally, our samples were relatively small, and may provide low power to detect differential expression and evolutionary analysis. Therefore, investigation of these interesting issues related to sex-biased gene evolution in T. pilosa can only be conducted when whole genome sequences and population datasets become available in the near future.
Methods
Plant materials and RNA isolation
Floral buds (≤3 mm) and flowers at anthesis were sampled from three female and three male plants (Figure 1) from the mountainous regions of Anning (Qinglong Gorge), Yunnan Province in Southwest China. Floral buds from female and male plants were named F1 and M1, respectively. Similarly, flowers at anthesis from female and male plants were named F2 and M2, respectively (Supplementary file 1). To exclude possible bacterial contamination, all tissues were sterilized with 75% alcohol and immediately rinsed with purified water. All samples were then snap-frozen in liquid nitrogen, and stored at –80 °C. Total RNA was extracted from each sample using TRIzol reagent (Life Technologies, CA, USA) according to the manufacturer’s instructions. The quantification and qualification of RNA were assessed by the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
Illumina sequencing, de novo assembly, and annotation
To construct the library, approximately 2 μg of total RNA was used with the Illumina NEBNext UltraTM RNA Library Prep Kit. RNA sequencing was performed on the Illumina NovaSeq 6000, generating 150 bp paired-end reads. The resulting clean reads were obtained by removing adapters, reads containing N bases and low-quality reads using Trimmomatic v.0.39 (Bolger et al., 2014). These reads were deposited in the NCBI database (PRJNA899312).
De novo assembly for clean reads from all samples was performed using Trinity v.2.10.0 (Haas et al., 2013) with min_kmer_cov: 3 and all other default parameters. To eliminate contamination, all transcripts of de novo assembly were compared to bacterial genomes downloaded from NCBI databases using BLASTN with an e-value of 1.0e-05 in blast + 2.12.0 software. We used Corset v.4.6 (Davidson and Oshlack, 2014) to obtain high-quality, non-redundant consensus transcripts (unigenes). TransDecoder v.5.5.0 was run with -m 100 parameters, namely at least 100 amino acids, to predict the coding DNA and protein sequences (Haas et al., 2013).
To evaluate the accuracy and completeness of reference transcriptomes, we performed gene function annotations based on the following databases, using BLAST with a cutoff e-value of 1.0e-05: NR, NT, and Swissport (Shiryev et al., 2007). We mapped the unigenes to Pfam database using InterProScan v.5.41 (Jones et al., 2014), to the GO database using Blast2GO (Conesa et al., 2005), and to the KEGG database using KEGG automatic annotation server (Moriya et al., 2007). Additionally, we estimated the completeness of reference transcriptomes using BUSCO v.5.4.5 based on embryophyta_odb10 database (Seppey et al., 2019).
Detection of sex-biased genes
Clean reads were mapped onto all unigenes using Bowtie2 (Langmead and Salzberg, 2012). Read counts were normalized to FPKM (Fragments Per Kilobase Million) value for each unigene using RSEM (Li and Dewey, 2011) in different male and female samples. Genes with zero read counts (i.e. no expression) in both two sexes and tissues were excluded. Differential expression analysis between sexes and tissue types was performed using DESeq2 R package (Love et al., 2014). Unigenes with an FDR-adjusted p<0.05 and an absolute value of log2 ratio ≥1 identified by DESeq2 were considered as sex-biased genes. To perform KEGG functional enrichment, we used all KEGG annotation terms for all genes as the background and performed the analyses using KOBAS v.2.0.12 (Mao et al., 2005).
Evolutionary rate analyses
To quantify the evolutionary rates of sex-biased genes, we download published genome datasets for monoecious Trichosanthes anguina (Ma et al., 2020) and monoecious Luffa cylindrica which has a closer phylogenetic relationship with Trichosanthes (de Boer et al., 2012; Wu et al., 2020) from CuGenDB database (Zheng et al., 2019). Additionally, we also download published RNA sequencing reads of floral buds and flowers from CNCB (Accession CRA002313) and NCBI databases (Accession SRR5259239) for dioecious plant Trichosanthes kirilowii (Hu et al., 2020), and de novo assembled by previously described methods.
We identified one-to-one OGs using OrthoFinder v.2.3.3 with default parameters from T. anguina, T. pilosa, T. kirilowii, and Luffa cylindrica (Emms and Kelly, 2019). Then, we employed TranslatorX with -c 1 p M -g -b5 n parameters (i.e. the multiple alignment and the trimming using Muscle and GBlocks, respectively), translated nucleotide sequences and back-translated amino acid alignments into nucleotide alignments to ensure codon-to-codon alignment (Abascal et al., 2010). The remaining gapless alignments (≥100 bp in length) were retained.
To investigate the evolutionary rates of coding sequences, we estimated nonsynonymous substitution (dN), synonymous substitution (dS) rates, as well as protein substitution rates (dN/dS, ω), using two branch models from CodeML package in PAML v.4.9h with the F3 × 4 codon frequencies (CodonFreq = 2) (Yang, 2007). According to the phylogenetic relationships of Trichosanthes (de Boer et al., 2012; Guo et al., 2020), we set up tree structures ((T. anguina, T. pilosa), T. kirilowii, L. cylindrica) in the control file of CodeML. First, we employed a ‘two-ratio’ branch model (model = 2, Nssites = 0) that assumes the foreground (two dioecious species) has a different ω value from the background (two monoecious species) to estimate and compare the divergences of the foreground. Second, to reduce the potential bias of ω value due to the conflation of two dioecious species, we also implemented a ‘free-ratio’ branch model (model = 1, Nssites = 0), which assumes an independent ω ratio for each branch. Finally, to avoid the effects of saturation substitution, we used separately OGs with 0<ω<2 and all OGs with ω>0, plotted the distribution of ω values, and compared the median of ω values in female-biased, male-biased and unbiased orthologous genes of floral buds and flowers at anthesis. All comparisons between sex-biased and unbiased genes were tested using the Wilcoxon rank sum test in R software. Additionally, we also performed permutations t-tests with 100,000 permutations in the R package Deducer (Fellows, 2012).
Estimation of the strength of natural selection
The rapid evolutionary rates of sex-biased genes may be attributed to positive selection, relaxed selection, and lower codon usage bias (Catalán et al., 2018; Dapper and Wade, 2020). Therefore, we conducted separate analyses using classical branch-site models that assume different ω values both among branches and across sites (Álvarez-Carretero et al., 2023), the adaptive branch-site random effects likelihood (aBSREL) model (Smith et al., 2015), the branch-site unrestricted statistical test for episodic diversification (BUSTED) model (Murrell et al., 2015), the RELAX model (Wertheim et al., 2015), and the effective number of codons (ENC) in PAML v.4.9h (Yang, 2007), HyPhy v.2.5 (Kosakovsky Pond et al., 2020) and CodonW v.1.4.2 (http://codonw.sourceforge.net) to distinguish which evolutionary forces are driving the rapid evolutionary rates of sex-biased genes.
To determine if amino acid sites in the foreground, including the T. pilosa lineage have undergone positive selection (foreground ω>1) compared with the background for each OGs, we followed the steps of Zhang et al., 2005, and used branch-site model A (model = 2, Nssite = 2, fix_omega = 0, omega = 1.5) and branch-site model null (model = 2, Nssite = 2, fix_omega = 1, omega = 1). The classical branch-site model assumes four site classes (0, 1, 2 a, 2b), with different ω values for the foreground and background branches. In site classes 2 a and 2b, the foreground branch undergoes positive selection when there is ω>1. We examined the significance of likelihood ratio tests (LRTs, p<0.05) to identify positively selected sites between model A and model null by comparing LRTs to the Chi-square distribution with two degrees of freedom. We adjusted the LRTs p value for multiple comparisons using Benjamini and Hochberg’s (FDR) algorithm. When the p value was significant, we used Bayes Empirical Bayes (BEB) estimates to identify sites with a high posterior probability (pp ≥0.95) of being under positive selection (Yang et al., 2005).
To detect episodic positive selection at a proportion of sites on the foreground branch, we employed the aBSREL method (Smith et al., 2015) in the HyPhy v.2.5 packages to compare the fully adaptive model (ω>1) to the null model that allows no positive selection rate classes by LRTs, which is an improved algorithm of branch-site models in PAML. For relatively small datasets, such as those with fewer than 10 taxa, the aBSREL method may not have enough power to detect positive selection. Therefore, we also ran the BUSTED method to identify gene-wide evidence of episodic positive selection at least one site on at least one branch (Murrell et al., 2015). We set T. pilosa as the foreground and assessed the statistical significance (p<0.05) using LRTs with the Holm-Bonferroni correction.
To test the relaxation of selective strength, we utilized the RELAX model in the HyPhy v.2.5 software (Wertheim et al., 2015; Schrader et al., 2021). The RELAX model estimates three ω parameters (ω0≤ω1≤1 ≤ω2), and determines the proportion of sites in the test (foreground) and reference (background) branches using a branch-site model. The first two ω classifications indicate that sites have undergone purifying selection, and the third classification indicates that sites have been under positive selection. Additionally, the model introduces a selection intensity parameter (K value) to compare a null model (K=1) with an alternative model, thereby assessing the strength of natural selection. When K>1, it suggests intensified natural selection, when K<1, indicates relaxed natural selection in the test branch relative to the reference branch. We quantified the statistical confidence of K value (p<0.05) using LRTs and the Holm-Bonferroni correction.
To investigate codon usage bias, which refers to the differences in the frequency of occurrence of synonymous codons in coding DNA, we employed CodonW v.1.4.2. This program considers the ENC values from 20 to 61 as a measure of the departure of the genetic codes for a given gene (Wright, 1990), with lower ENC values represent stronger codon usage bias (Hambuch and Parsch, 2005). We performed a Wilcoxon rank sum test to determine if there were deviations in ENC values among female-biased, male-biased, and unbiased genes in floral buds.
Acknowledgements
We are very grateful to Spencer C H Barrett (University of Toronto, Canada) for his critical reading and suggestions on the manuscript. We also thank three anonymous reviewers for their comments on improving the quality of the manuscript. We are indebted to Ting Zhang, Zhi-Yun Yang, Jiang-Li Ma, Peng-Fei Ma, Xu-Kun Wu, Shi-Yu Lv, Zhen Peng, and other members of staff of Germplasm Bank of Wild Species for sampling. We also thank the iFlora HPC Center (iFlora High-Performance Computing Center) of Germplasm Bank of Wild Species for computational support on data analysis.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
De-Zhu Li, Email: dzl@mail.kib.ac.cn.
Hong-Tao Li, Email: lihongtao@mail.kib.ac.cn.
Vincent Castric, University of Lille, France.
Detlef Weigel, Max Planck Institute for Biology Tübingen, Germany.
Funding Information
This paper was supported by the following grants:
Chinese Academy of Sciences (CAS) Strategic Priority Research Program (XDB 31000000) to De-Zhu Li.
National Natural Science Foundation of China 32370233 to Hong-Tao Li.
Key R & D Program of Yunnan Province, China 202103AC100003 to De-Zhu Li.
Key Basic Research Program of Yunnan Province, China 202101BC070003 to Hong-Tao Li.
Science and Technology Basic Resources Investigation Program of China 2019FY100900 to Hong-Tao Li.
Open Research Project of “Cross-Cooperative Team” of the Germplasm Bank of Wild Species, CAS Kunming Institute of Botany to Hong-Tao Li.
National Wild Plant Germplasm Resource Center, China to Hong-Tao Li.
CAS Key Technology Talent Program to Hong-Tao Li.
National Natural Science Foundation of China 31570333 to Hong-Tao Li.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Resources, Data curation, Software, Formal analysis, Supervision, Investigation, Visualization, Methodology, Writing - original draft, Writing – review and editing.
Writing – review and editing.
Investigation.
Conceptualization, Supervision, Funding acquisition, Project administration, Writing – review and editing.
Conceptualization, Resources, Data curation, Supervision, Funding acquisition, Investigation, Project administration, Writing – review and editing.
Additional files
Two dioecious species Trichosanthes pilosa and T. kirilowii represent the foreground and two monoecious species T. anguina and Luffa cylindrica represent the background.
Two dioecious species Trichosanthes pilosa and T. kirilowii represent the foreground and two monoecious species, T. anguina and Luffa cylindrica represent the background.
The dioecious species Trichosanthes pilosa represents the foreground and other species represent the background.
The dioecious species Trichosanthes pilosa represents the foreground (unconstrained branch) and other species represent the background (constrained branch).
The dioecious species Trichosanthes pilosa represents the foreground (test) and other species represent the background (reference).
The dioecious species Trichosanthes pilosa represents the foreground (test) and other species represent the background (reference).
Data availability
All RNA-Sequencing clean reads have been deposited in the databases of the National Center for Biotechnology Information (NCBI) under BioProject ID PRJNA899312. The reference transcriptome, orthology data, and alignments are available as Source data 1.
The following dataset was generated:
Zhao L. 2022. Trichosanthes pilosa Transcriptome or Gene expression. NCBI BioProject. PRJNA899312
The following previously published datasets were used:
Xin J. 2020. Transcriptome sequencing and screening of genes related to sex determination of Trichosanthes kirilowii Maxim. Genome Sequence Archive. CRA002313
University of Nebraska-Lincoln 2017. RNA-Seq of Trichosanthes kirilowii for transcriptome assembly. NCBI Sequence Read Archive. SRR5259239
References
- Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Research. 2010;38:W7–W13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Carretero S, Kapli P, Yang Z. Beginner’s guide on the use of PAML to detect positive selection. Molecular Biology and Evolution. 2023;40:msad041. doi: 10.1093/molbev/msad041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunkumar R, Josephs EB, Williamson RJ, Wright SI. Pollen-Specific, but not sperm-specific, genes show stronger purifying selection and higher rates of positive selection than sporophytic genes in Capsella grandiflora. Molecular Biology and Evolution. 2013;30:2475–2486. doi: 10.1093/molbev/mst149. [DOI] [PubMed] [Google Scholar]
- Assis R, Zhou Q, Bachtrog D. Sex-biased transcriptome evolution in Drosophila. Genome Biology and Evolution. 2012;4:1189–1200. doi: 10.1093/gbe/evs093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SCH, Hough J. Sexual dimorphism in flowering plants. Journal of Experimental Botany. 2013;64:67–82. doi: 10.1093/jxb/ers308. [DOI] [PubMed] [Google Scholar]
- Beaudry FEG, Rifkin JL, Barrett SCH, Wright SI. Evolutionary genomics of plant gametophytic selection. Plant Communications. 2020;1:100115. doi: 10.1016/j.xplc.2020.100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogamuwa SP, Jang JC. Tandem CCCH zinc finger proteins in plant growth, development and stress response. Plant & Cell Physiology. 2014;55:1367–1375. doi: 10.1093/pcp/pcu074. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boualem A, Troadec C, Camps C, Lemhemdi A, Morin H, Sari MA, Fraenkel-Zagouri R, Kovalski I, Dogimont C, Perl-Treves R, Bendahmane A. A cucurbit androecy gene reveals how unisexual flowers develop and dioecy emerges. Science. 2015;350:688–691. doi: 10.1126/science.aac8370. [DOI] [PubMed] [Google Scholar]
- Cai ZY, Yang CC, Liao J, Song HF, Zhang S. Sex-biased genes and metabolites explain morphologically sexual dimorphism and reproductive costs in Salix paraplesia catkins. Horticulture Research. 2021;8:125. doi: 10.1038/s41438-021-00566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalán A, Macias-Muñoz A, Briscoe AD. Evolution of sex-biased gene expression and dosage compensation in the eye and brain of heliconius butterflies. Molecular Biology and Evolution. 2018;35:2120–2134. doi: 10.1093/molbev/msy111. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. Does sexual dimorphism in plants promote sex chromosome evolution? Environmental and Experimental Botany. 2018;146:5–12. doi: 10.1016/j.envexpbot.2017.11.005. [DOI] [Google Scholar]
- Charlesworth B, Campos JL, Jackson BC. Faster-X evolution: theory and evidence from Drosophila. Molecular Ecology. 2018;27:3753–3771. doi: 10.1111/mec.14534. [DOI] [PubMed] [Google Scholar]
- Chen F, Hu Y, Vannozzi A, Wu K, Cai H, Qin Y, Mullis A, Lin Z, Zhang L. The WRKY transcription factor family in model plants and crops. Critical Reviews in Plant Sciences. 2017;36:311–335. doi: 10.1080/07352689.2018.1441103. [DOI] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Congrains C, Campanini EB, Torres FR, Rezende VB, Nakamura AM, de Oliveira JL, Lima ALA, Chahad-Ehlers S, Sobrinho IS, Jr, de Brito RA. Evidence of adaptive evolution and relaxed constraints in sex-biased genes of South American and West Indies Fruit Flies (Diptera: Tephritidae) Genome Biology and Evolution. 2018;10:380–395. doi: 10.1093/gbe/evy009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossard GG, Toups MA, Pannell JR. Sexual dimorphism and rapid turnover in gene expression in pre-reproductive seedlings of a dioecious herb. Annals of Botany. 2019;123:1119–1131. doi: 10.1093/aob/mcy183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossard GG, Godfroy O, Nehr Z, Cruaud C, Cock JM, Lipinska AP, Coelho SM. Selection drives convergent gene expression changes during transitions to co-sexuality in haploid sexual systems. Nature Ecology & Evolution. 2022;6:579–589. doi: 10.1038/s41559-022-01692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui YW, Lu XT, Gou XP. Receptor-like protein kinases in plant reproduction: Current understanding and future perspectives. Plant Communications. 2022;3:100273. doi: 10.1016/j.xplc.2021.100273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapper AL, Wade MJ. Relaxed selection and the rapid evolution of reproductive genes. Trends in Genetics. 2020;36:640–649. doi: 10.1016/j.tig.2020.06.014. [DOI] [PubMed] [Google Scholar]
- Darolti I, Wright AE, Pucholt P, Berlin S, Mank JE. Slow evolution of sex‐biased genes in the reproductive tissue of the dioecious plant Salix viminalis. Molecular Ecology. 2018;27:694–708. doi: 10.1111/mec.14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darolti I, Fong LJM, Sandkam BA, Metzger DCH, Mank JE. Sex chromosome heteromorphism and the Fast‐X effect in poeciliids. Molecular Ecology. 2023;32:4599–4609. doi: 10.1111/mec.17048. [DOI] [PubMed] [Google Scholar]
- Darolti I, Mank JE. Sex-biased gene expression at single-cell resolution: cause and consequence of sexual dimorphism. Evolution Letters. 2023;7:148–156. doi: 10.1093/evlett/qrad013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson NM, Oshlack A. Corset: enabling differential gene expression analysis for de novoassembled transcriptomes. Genome Biology. 2014;15:410. doi: 10.1186/s13059-014-0410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson TE, Geber MA. In: Gender and Sexual Dimorphism in Flowering Plants. Geber MA, Dawson TE, Delph LF, editors. Berlin: Springer; 1999. Sexual Dimorphism in physiology and morphology; pp. 175–215. [DOI] [Google Scholar]
- de Boer HJ, Schaefer H, Thulin M, Renner SS. Evolution and loss of long-fringed petals: a case study using a dated phylogeny of the snake gourds, Trichosanthes (Cucurbitaceae) BMC Evolutionary Biology. 2012;12:108. doi: 10.1186/1471-2148-12-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer HJ, Steffen K, Cooper WE, Richardson J. Sunda to Sahul dispersals in Trichosanthes (Cucurbitaceae): a dated phylogeny reveals five independent dispersal events to Australasia. Journal of Biogeography. 2015;42:519–531. doi: 10.1111/jbi.12432. [DOI] [Google Scholar]
- Delph LF. In: Gender and Sexual Dimorphism in Flowering Plants. Geber MA, Dawson TE, Delph LF, editors. Berlin: Springer; 1999. Sexual Dimorphism in life history; pp. 149–173. [DOI] [Google Scholar]
- Delph LF, Herlihy CR. Sexual, fecundity, and viability selection on flower size and number in a sexually dimorphic plant. Evolution; International Journal of Organic Evolution. 2012;66:1154–1166. doi: 10.1111/j.1558-5646.2011.01510.x. [DOI] [PubMed] [Google Scholar]
- Djordjevic J, Dumas Z, Robinson-Rechavi M, Schwander T, Parker DJ. Dynamics of sex-biased gene expression during development in the stick insect Timema californicum. Heredity. 2022;129:113–122. doi: 10.1038/s41437-022-00536-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nature Reviews. Genetics. 2007;8:689–698. doi: 10.1038/nrg2167. [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biology. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellows I. Deducer: a data analysis GUI for R. Journal of Statistical Software. 2012;49:1–15. doi: 10.18637/jss.v049.i08. [DOI] [Google Scholar]
- Forrest JRK. Plant size, sexual selection, and the evolution of protandry in dioecious plants. The American Naturalist. 2014;184:338–351. doi: 10.1086/677295. [DOI] [PubMed] [Google Scholar]
- Frumkin I, Lajoie MJ, Gregg CJ, Hornung G, Church GM, Pilpel Y. Codon usage of highly expressed genes affects proteome-wide translation efficiency. PNAS. 2018;115:E4940–E4949. doi: 10.1073/pnas.1719375115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershoni M, Pietrokovski S. Reduced selection and accumulation of deleterious mutations in genes exclusively expressed in men. Nature Communications. 2014;5:4438. doi: 10.1038/ncomms5438. [DOI] [PubMed] [Google Scholar]
- Gossmann TI, Schmid MW, Grossniklaus U, Schmid KJ. Selection-driven evolution of sex-biased genes is consistent with sexual selection in Arabidopsis thaliana. Molecular Biology and Evolution. 2014;31:574–583. doi: 10.1093/molbev/mst226. [DOI] [PubMed] [Google Scholar]
- Gossmann TI, Saleh D, Schmid MW, Spence MA, Schmid KJ. Transcriptomes of Plant gametophytes have a higher proportion of rapidly evolving and young genes than sporophytes. Molecular Biology and Evolution. 2016;33:1669–1678. doi: 10.1093/molbev/msw044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grath S, Parsch J. Sex-biased gene expression. Annual Review of Genetics. 2016;50:29–44. doi: 10.1146/annurev-genet-120215-035429. [DOI] [PubMed] [Google Scholar]
- Guo J, Xu W, Hu Y, Huang J, Zhao Y, Zhang L, Huang CH, Ma H. Phylotranscriptomics in Cucurbitaceae reveal multiple whole-genome duplications and key morphological and molecular innovations. Molecular Plant. 2020;13:1117–1133. doi: 10.1016/j.molp.2020.05.011. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Valencia J, Fracassetti M, Horvath R, Laenen B, Désamore A, Drouzas AD, Friberg M, Kolář F, Slotte T, de Meaux J. Genomic signatures of sexual selection on pollen-expressed genes in Arabis alpina. Molecular Biology and Evolution. 2022;39:msab349. doi: 10.1093/molbev/msab349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature Protocols. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu A, Ádám É, Sheerin DJ, Dobos O, Bernula P, Hiltbrunner A, Kozma‐Bognár L, Nagy F. High‐level expression and phosphorylation of phytochrome B modulates flowering time in Arabidopsis. The Plant Journal. 2015;83:794–805. doi: 10.1111/tpj.12926. [DOI] [PubMed] [Google Scholar]
- Hambuch TM, Parsch J. Patterns of synonymous codon usage in Drosophila melanogaster genes with sex-biased expression. Genetics. 2005;170:1691–1700. doi: 10.1534/genetics.104.038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkess A, Mercati F, Shan HY, Sunseri F, Falavigna A, Leebens-Mack J. Sex-biased gene expression in dioecious garden asparagus (Asparagus officinalis) The New Phytologist. 2015;207:883–892. doi: 10.1111/nph.13389. [DOI] [PubMed] [Google Scholar]
- Harrison PW, Wright AE, Zimmer F, Dean R, Montgomery SH, Pointer MA, Mank JE. Sexual selection drives evolution and rapid turnover of male gene expression. PNAS. 2015;112:4393–4398. doi: 10.1073/pnas.1501339112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MC, Mallon EB, Twell D, Hammond RL. Deleterious mutation accumulation in Arabidopsis thaliana pollen genes: a role for A recent relaxation of selection. Genome Biology and Evolution. 2019;11:1939–1951. doi: 10.1093/gbe/evz127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatchett WJ, Jueterbock AO, Kopp M, Coyer JA, Coelho SM, Hoarau G, Lipinska AP. Evolutionary dynamics of sex-biased gene expression in a young XY system: insights from the brown alga genus fucus. The New Phytologist. 2023;238:422–437. doi: 10.1111/nph.18710. [DOI] [PubMed] [Google Scholar]
- Hou J, Fan W, Ma R, Li B, Yuan Z, Huang W, Wu Y, Hu Q, Lin C, Zhao X, Peng B, Zhao L, Zhang C, Sun L. MALE STERILITY 3 encodes a plant homeodomain‐finger protein for MALE fertility in soybean. Journal of Integrative Plant Biology. 2022;64:1076–1086. doi: 10.1111/jipb.13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough J, Hollister JD, Wang W, Barrett SCH, Wright SI. Genetic degeneration of old and young Y chromosomes in the flowering plant Rumex hastatulus. PNAS. 2014;111:7713–7718. doi: 10.1073/pnas.1319227111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SK, Jakšić AM, Nolte V, Lirakis M, Kofler R, Barghi N, Versace E, Schlötterer C. Rapid sex-specific adaptation to high temperature in Drosophila. eLife. 2020;9:e53237. doi: 10.7554/eLife.53237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu XQ, Liao ZY, Zhang B, Yue JJ, Wang Z, Jie X, Liu J. Transcriptome sequencing and screening of genes related to sex determination of Trichosanthes kirilowii Maxim. PLOS ONE. 2020;15:e0239230. doi: 10.1371/journal.pone.0239230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immler S. Haploid selection in “Diploid” organisms. Annual Review of Ecology, Evolution, and Systematics. 2019;50:219–236. doi: 10.1146/annurev-ecolsys-110218-024709. [DOI] [Google Scholar]
- Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong S-Y, Lopez R, Hunter S. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodursky S, Svetec N, Durkin SM, Zhao L. The evolution of sex-biased gene expression in the Drosophila brain. Genome Research. 2020;30:874–884. doi: 10.1101/gr.259069.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Poon AFY, Velazquez R, Weaver S, Hepler NL, Murrell B, Shank SD, Magalis BR, Bouvier D, Nekrutenko A, Wisotsky S, Spielman SJ, Frost SDW, Muse SV, Crandall K. HyPhy 2.5—a customizable platform for evolutionary hypothesis testing using phylogenies. Molecular Biology and Evolution. 2020;37:295–299. doi: 10.1093/molbev/msz197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahti DC, Johnson NA, Ajie BC, Otto SP, Hendry AP, Blumstein DT, Coss RG, Donohue K, Foster SA. Relaxed selection in the wild. Trends in Ecology & Evolution. 2009;24:487–496. doi: 10.1016/j.tree.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Yan H, Mei WM, Tse YC, Wang H. Boosting autophagy in sexual reproduction: a plant perspective. The New Phytologist. 2020;226:679–689. doi: 10.1111/nph.16414. [DOI] [PubMed] [Google Scholar]
- Lichilín N, El Taher A, Böhne A. Sex-biased gene expression and recent sex chromosome turnover. Philosophical Transactions of the Royal Society B. 2021;376:20200107. doi: 10.1098/rstb.2020.0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinska A, Cormier A, Luthringer R, Peters AF, Corre E, Gachon CMM, Cock JM, Coelho SM. Sexual dimorphism and the evolution of sex-biased gene expression in the brown alga ectocarpus. Molecular Biology and Evolution. 2015;32:1581–1597. doi: 10.1093/molbev/msv049. [DOI] [PubMed] [Google Scholar]
- Liu Y, Bassham DC. Autophagy: pathways for self-eating in plant cells. Annual Review of Plant Biology. 2012;63:215–237. doi: 10.1146/annurev-arplant-042811-105441. [DOI] [PubMed] [Google Scholar]
- Liu Z, Tavares R, Forsythe ES, André F, Lugan R, Jonasson G, Boutet-Mercey S, Tohge T, Beilstein MA, Werck-Reichhart D, Renault H. Evolutionary interplay between sister cytochrome P450 genes shapes plasticity in plant metabolism. Nature Communications. 2016;7:13026. doi: 10.1038/ncomms13026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Lynch M, Katju V. The altered evolutionary trajectories of gene duplicates. Trends in Genetics. 2004;20:544–549. doi: 10.1016/j.tig.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Ma WJ, Pannell JR. Sex determination: separate sexes are a double turnoff in melons. Current Biology. 2016;26:R171–R174. doi: 10.1016/j.cub.2015.12.026. [DOI] [PubMed] [Google Scholar]
- Ma L, Wang Q, Mu J, Fu A, Wen C, Zhao X, Gao L, Li J, Shi K, Wang Y, Zhang X, Zhang X, Fei Z, Grierson D, Zuo J. The genome and transcriptome analysis of snake gourd provide insights into its evolution and fruit development and ripening. Horticulture Research. 2020;7:199. doi: 10.1038/s41438-020-00423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Hultin-Rosenberg L, Axelsson E, Ellegren H. Rapid evolution of female-biased, but not male-biased, genes expressed in the avian brain. Molecular Biology and Evolution. 2007;24:2698–2706. doi: 10.1093/molbev/msm208. [DOI] [PubMed] [Google Scholar]
- Mank JE. Sex chromosomes and the evolution of sexual dimorphism: lessons from the genome. The American Naturalist. 2009;173:141–150. doi: 10.1086/595754. [DOI] [PubMed] [Google Scholar]
- Mank JE. The transcriptional architecture of phenotypic dimorphism. Nature Ecology & Evolution. 2017;1:6. doi: 10.1038/s41559-016-0006. [DOI] [PubMed] [Google Scholar]
- Mank JE. Sex-specific morphs: the genetics and evolution of intra-sexual variation. Nature Reviews. Genetics. 2023;24:44–52. doi: 10.1038/s41576-022-00524-2. [DOI] [PubMed] [Google Scholar]
- Mao XZ, Cai T, Olyarchuk JG, Wei LP. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- Marinić M, Lynch VJ. Relaxed constraint and functional divergence of the progesterone receptor (PGR) in the human stem-lineage. PLOS Genetics. 2020;16:e1008666. doi: 10.1371/journal.pgen.1008666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP. Towards a more nuanced understanding of the relationship between sex-biased gene expression and rates of protein-coding sequence evolution. Molecular Biology and Evolution. 2011;28:1893–1900. doi: 10.1093/molbev/msr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP, Connallon T. The faster-X effect: integrating theory and data. Trends in Genetics. 2013;29:537–544. doi: 10.1016/j.tig.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, Bendahmane A, Renner SS. Sex chromosomes in land plants. Annual Review of Plant Biology. 2011;62:485–514. doi: 10.1146/annurev-arplant-042110-103914. [DOI] [PubMed] [Google Scholar]
- Moore JC, Pannell JR. Sexual selection in plants. Current Biology. 2011;21:R176–R182. doi: 10.1016/j.cub.2010.12.035. [DOI] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Research. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle LC, Wu M, Gibson MJS. Reproductive proteins evolve faster than non-reproductive proteins among Solanum species. Frontiers in Plant Science. 2021;12:635990. doi: 10.3389/fpls.2021.635990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat F, Mbengue N, Winge SB, Trefzer T, Leushkin E, Sepp M, Cardoso-Moreira M, Schmidt J, Schneider C, Mößinger K, Brüning T, Lamanna F, Belles MR, Conrad C, Kondova I, Bontrop R, Behr R, Khaitovich P, Pääbo S, Marques-Bonet T, Grützner F, Almstrup K, Schierup MH, Kaessmann H. The molecular evolution of spermatogenesis across mammals. Nature. 2023;613:308–316. doi: 10.1038/s41586-022-05547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Weaver S, Smith MD, Wertheim JO, Murrell S, Aylward A, Eren K, Pollner T, Martin DP, Smith DM, Scheffler K, Kosakovsky Pond SL. Gene-wide identification of episodic selection. Molecular Biology and Evolution. 2015;32:1365–1371. doi: 10.1093/molbev/msv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschietti JP, Wengier DL. How many receptor-like kinases are required to operate a pollen tube. Current Opinion in Plant Biology. 2018;41:73–82. doi: 10.1016/j.pbi.2017.09.008. [DOI] [PubMed] [Google Scholar]
- Muyle A. How different is the evolution of sex-biased gene expression between plants and animals? A commentary on: “Sexual dimorphism and rapid turnover in gene expression in pre-reproductive seedlings of A dioecious herb.”. Annals of Botany. 2019;123:iv–v. doi: 10.1093/aob/mcz081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi S, Godfrey AK, Hughes JF, Goodheart ML, Mitchell RN, Page DC. Conservation, acquisition, and functional impact of sex-biased gene expression in mammals. Science. 2019;365:eaaw7317. doi: 10.1126/science.aaw7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DH, Rogers TF, Dean R, Wright AE. How to identify sex chromosomes and their turnover. Molecular Ecology. 2019;28:4709–4724. doi: 10.1111/mec.15245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa F, Windbichler N, Waterhouse RM, Cagnetti A, D’Amato R, Persampieri T, Lawniczak MKN, Nolan T, Papathanos PA. Rapid evolution of female-biased genes among four species of Anopheles malaria mosquitoes. Genome Research. 2017;27:1536–1548. doi: 10.1101/gr.217216.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch J, Ellegren H. The evolutionary causes and consequences of sex-biased gene expression. Nature Reviews. Genetics. 2013;14:83–87. doi: 10.1038/nrg3376. [DOI] [PubMed] [Google Scholar]
- Parvathy ST, Udayasuriyan V, Bhadana V. Codon usage bias. Molecular Biology Reports. 2022;49:539–565. doi: 10.1007/s11033-021-06749-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pröschel M, Zhang Z, Parsch J. Widespread adaptive evolution of Drosophila genes with sex-biased expression. Genetics. 2006;174:893–900. doi: 10.1534/genetics.106.058008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Wu Y, Zhu Z, Chen R, Zhang L. Biosynthesis and regulation of diterpenoids in medicinal plants. Chinese Journal of Natural Medicines. 2022;20:761–772. doi: 10.1016/S1875-5364(22)60214-0. [DOI] [PubMed] [Google Scholar]
- Renner SS. The relative and absolute frequencies of angiosperm sexual systems: dioecy, monoecy, gynodioecy, and an updated online database. American Journal of Botany. 2014;101:1588–1596. doi: 10.3732/ajb.1400196. [DOI] [PubMed] [Google Scholar]
- Rowe L, Chenoweth SF, Agrawal AF. The genomics of sexual conflict. The American Naturalist. 2018;192:274–286. doi: 10.1086/698198. [DOI] [PubMed] [Google Scholar]
- Sanderson BJ, Wang L, Tiffin P, Wu ZQ, Olson MS. Sex-biased gene expression in flowers, but not leaves, reveals secondary sexual dimorphism in Populus balsamifera. The New Phytologist. 2019;221:527–539. doi: 10.1111/nph.15421. [DOI] [PubMed] [Google Scholar]
- Sandler G, Beaudry FEG, Barrett SCH, Wright SI. The effects of haploid selection on Y chromosome evolution in two closely related dioecious plants. Evolution Letters. 2018;2:368–377. doi: 10.1002/evl3.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer H, Renner SS. A three-genome phylogeny of Momordica (Cucurbitaceae) suggests seven returns from dioecy to monoecy and recent long-distance dispersal to Asia. Molecular Phylogenetics and Evolution. 2010;54:553–560. doi: 10.1016/j.ympev.2009.08.006. [DOI] [PubMed] [Google Scholar]
- Schaefer H, Renner SS. Phylogenetic relationships in the order Cucurbitales and a new classification of the gourd family (Cucurbitaceae) TAXON. 2011;60:122–138. doi: 10.1002/tax.601011. [DOI] [Google Scholar]
- Scharf KD, Berberich T, Ebersberger I, Nover L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2012;1819:104–119. doi: 10.1016/j.bbagrm.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Scharmann M, Rebelo AG, Pannell JR. High rates of evolution preceded shifts to sex-biased gene expression in Leucadendron, the most sexually dimorphic angiosperms. eLife. 2021;10:e67485. doi: 10.7554/eLife.67485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader L, Pan H, Bollazzi M, Schiøtt M, Larabee FJ, Bi X, Deng Y, Zhang G, Boomsma JJ, Rabeling C. Relaxed selection underlies genome erosion in socially parasitic ant species. Nature Communications. 2021;12:2918. doi: 10.1038/s41467-021-23178-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppey M, Manni M, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness. Methods in Molecular Biology. 2019;1962:227–245. doi: 10.1007/978-1-4939-9173-0_14. [DOI] [PubMed] [Google Scholar]
- Shiryev SA, Papadopoulos JS, Schäffer AA, Agarwala R. Improved BLAST searches using longer words for protein seeding. Bioinformatics. 2007;23:2949–2951. doi: 10.1093/bioinformatics/btm479. [DOI] [PubMed] [Google Scholar]
- Singh A, Agrawal AF, Parsch J. Two forms of sexual dimorphism in gene expression in Drosophila melanogaster : their coincidence and evolutionary genetics. Molecular Biology and Evolution. 2023;40:msad091. doi: 10.1093/molbev/msad091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skogsmyr I, Lankinen A. Sexual selection: an evolutionary force in plants? Biological Reviews of the Cambridge Philosophical Society. 2002;77:537–562. doi: 10.1017/s1464793102005973. [DOI] [PubMed] [Google Scholar]
- Smith MD, Wertheim JO, Weaver S, Murrell B, Scheffler K, Kosakovsky Pond SL. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Molecular Biology and Evolution. 2015;32:1342–1353. doi: 10.1093/molbev/msv022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson AG, Bertin RI. In: Pollination Biology. Real L, editor. New York: Academic Press; 1983. Male competition, female choice, and sexual selection in plants; pp. 109–151. [Google Scholar]
- Teo ZWN, Song SY, Wang YQ, Liu J, Yu H. New insights into the regulation of inflorescence architecture. Trends in Plant Science. 2014;19:158–165. doi: 10.1016/j.tplants.2013.11.001. [DOI] [PubMed] [Google Scholar]
- Tosto NM, Beasley ER, Wong BBM, Mank JE, Flanagan SP. The roles of sexual selection and sexual conflict in shaping patterns of genome and transcriptome variation. Nature Ecology & Evolution. 2023;7:981–993. doi: 10.1038/s41559-023-02019-7. [DOI] [PubMed] [Google Scholar]
- Toubiana W, Armisén D, Dechaud C, Arbore R, Khila A. Impact of male trait exaggeration on sex-biased gene expression and genome architecture in a water strider. BMC Biology. 2021;19:89. doi: 10.1186/s12915-021-01021-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan MM, Wang Q, Webster FX, Kiemle D, Hong YJ, Tantillo DJ, Coates RM, Wray AT, Askew W, O’Donnell C, Tokuhisa JG, Tholl D. Formation of the unusual semivolatile diterpene rhizathalene by the Arabidopsis class I terpene synthase TPS08 in the root stele is involved in defense against belowground herbivory. The Plant Cell. 2013;25:1108–1125. doi: 10.1105/tpc.112.100057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltsos P. Not all sex-biased genes are the same. The New Phytologist. 2019;221:10–11. doi: 10.1111/nph.15531. [DOI] [PubMed] [Google Scholar]
- Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K. RELAX: detecting relaxed selection in a phylogenetic framework. Molecular Biology and Evolution. 2015;32:820–832. doi: 10.1093/molbev/msu400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle CA, Kulkarni A, Extavour CG. Evolutionary dynamics of sex-biased genes expressed in cricket brains and gonads. Journal of Evolutionary Biology. 2021;34:1188–1211. doi: 10.1111/jeb.13889. [DOI] [PubMed] [Google Scholar]
- Williams JH, Reese JB. Evolution of development of pollen performance. Current Topics in Developmental Biology. 2019;131:299–336. doi: 10.1016/bs.ctdb.2018.11.012. [DOI] [PubMed] [Google Scholar]
- Wright F. The “effective number of codons” used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- Wright AE, Harrison PW, Zimmer F, Montgomery SH, Pointer MA, Mank JE. Variation in promiscuity and sexual selection drives avian rate of Faster-Z evolution. Molecular Ecology. 2015;24:1218–1235. doi: 10.1111/mec.13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Raven P, Hong D. Flora of China. Beijing: Science Press; 2011. [Google Scholar]
- Wu H, Zhao G, Gong H, Li J, Luo C, He X, Luo S, Zheng X, Liu X, Guo J, Chen J, Luo J. A high-quality sponge gourd (Luffa cylindrica) genome. Horticulture Research. 2020;7:128. doi: 10.1038/s41438-020-00350-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH, Wong WSW, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Molecular Biology and Evolution. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yue T, Guo Y, Qi X, Zheng W, Zhang H, Wang B, Liu K, Zhou B, Zeng X, He Y, Su B. Sex-biased regulatory changes in the placenta of native highlanders contribute to adaptive fetal development. bioRxiv. 2023 doi: 10.1101/2023.05.24.542081. [DOI] [PMC free article] [PubMed]
- Zemp N, Tavares R, Muyle A, Charlesworth D, Marais GAB, Widmer A. Evolution of sex-biased gene expression in a dioecious plant. Nature Plants. 2016;2:16168. doi: 10.1038/nplants.2016.168. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Hambuch TM, Parsch J. Molecular evolution of sex-biased genes in Drosophila. Molecular Biology and Evolution. 2004;21:2130–2139. doi: 10.1093/molbev/msh223. [DOI] [PubMed] [Google Scholar]
- Zhang JZ, Nielsen R, Yang ZH. Evaluation of an Improved branch-site likelihood method for detecting positive selection at the molecular level. Molecular Biology and Evolution. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Parsch J. Positive correlation between evolutionary rate and recombination rate in Drosophila genes with male-biased expression. Molecular Biology and Evolution. 2005;22:1945–1947. doi: 10.1093/molbev/msi189. [DOI] [PubMed] [Google Scholar]
- Zhang LB, Simmons MP, Kocyan A, Renner SS. Phylogeny of the Cucurbitales based on DNA sequences of nine loci from three genomes: implications for morphological and sexual system evolution. Molecular Phylogenetics and Evolution. 2006;39:305–322. doi: 10.1016/j.ympev.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sturgill D, Parisi M, Kumar S, Oliver B. Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature. 2007;450:233–237. doi: 10.1038/nature06323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Li ZW, Ma B, Hou QC, Wan XY. Phylogeny and functions of LOB domain proteins in plants. International Journal of Molecular Sciences. 2020;21:2278. doi: 10.3390/ijms21072278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MM, Zhang SQ. Mitogen-activated protein kinase cascades in plant signaling. Journal of Integrative Plant Biology. 2022;64:301–341. doi: 10.1111/jipb.13215. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wu S, Bai Y, Sun H, Jiao C, Guo S, Zhao K, Blanca J, Zhang Z, Huang S, Xu Y, Weng Y, Mazourek M, K Reddy U, Ando K, McCreight JD, Schaffer AA, Burger J, Tadmor Y, Katzir N, Tang X, Liu Y, Giovannoni JJ, Ling K-S, Wechter WP, Levi A, Garcia-Mas J, Grumet R, Fei Z. Cucurbit genomics database (CuGenDB): a central portal for comparative and functional genomics of cucurbit crops. Nucleic Acids Research. 2019;47:D1128–D1136. doi: 10.1093/nar/gky944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X-M, Zhao P, Wang W, Zou J, Cheng T, Peng X, Sun M. A comprehensive, genome-wide analysis of autophagy-related genes identified in tobacco suggests A central role of autophagy in plant response to various environmental cues. DNA Research. 2015;22:245–257. doi: 10.1093/dnares/dsv012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Zhao P, Sun M-X, Bozhkov P. Autophagy in sexual plant reproduction: new insights. Journal of Experimental Botany. 2021;72:7658–7667. doi: 10.1093/jxb/erab366. [DOI] [PubMed] [Google Scholar]