Abstract
2-Bromo-5,6-dichloro-1-β-d-ribofuranosyl benzimidazole (BDCRB) is a member of a new class of benzimidazole ribonucleosides which inhibit human cytomegalovirus (HCMV) late in the replication cycle without inhibiting viral DNA synthesis. We show here that polygenomic concatemeric HCMV DNA does not mature to unit genome length in the presence of BDCRB. To discover the locus of action, virus resistant to BDCRB was selected by serial passage in the presence of the compound. Genetic mapping experiments with BDCRB-resistant virus demonstrated that the resistance phenotype mapped to one amino acid (Asp344Glu; low resistance) or two amino acids (Asp344Glu and Ala355Thr; high resistance) within the product of exon 2 of the HCMV UL89 open reading frame. The HCMV UL89 open reading frame and its homologs are among the most conserved open reading frames in the herpesviruses, and their products have sequence similarities to a known ATP-dependent endonuclease from the double-stranded DNA bacteriophage T4. These findings strongly suggest that BDCRB prevents viral DNA maturation by interacting with a UL89 gene product and that the UL89 open reading frame may encode an endonucleolytic subunit of the putative HCMV terminase. Further, since mammalian cell DNA replication does not involve a DNA maturation step, compounds which inhibit viral DNA maturation should be selective and safe.
Primary infection with human cytomegalovirus (HCMV) in a person with a normal immune system is generally asymptomatic; however, infrequently, an immunocompetent individual may present with a self-limiting, mild mononucleosis syndrome (12). In contrast, in the immunocompromised population (AIDS patients, organ transplant recipients, and neonates), HCMV can cause systemic disease with consequences including blindness (due to CMV retinitis), pneumonitis, and gastroenteritis, etc. (12). The clinically available drug therapies for treating HCMV disease include the viral DNA polymerase inhibitors ganciclovir (Cytovene), foscarnet sodium (Foscavir), and cidofovir (Vistide). These all have low oral bioavailability and/or dose-related toxicities (26, 32) which limit their usefulness, particularly in AIDS patients on antiretroviral therapies. Additionally, resistance to ganciclovir and foscarnet sodium in this patient group has been correlated with clinical failure (20). Thus, there is a clear need for an orally bioavailable, effective, nontoxic anti-HCMV agent to treat the immunocompromised patient population.
As part of our studies to find new inhibitors of HCMV, we found that 2-bromo-5,6-dichloro-1-β-d-ribofuranosyl benzimidazole (BDCRB) and its 2-chloro analog (TCRB) were active against HCMV at noncytotoxic concentrations (43). These compounds are analogs of 5,6-dichloro-1-β-d-ribofuranosyl benzimidazole (DRB), a molecule shown to have activity against some viruses (likely by inhibiting transcription by cellular RNA polymerase II [36, 46]). As demonstrated by us previously, the addition of 2-position halogens to DRB to generate BDCRB and TCRB lowered cell toxicity and increased anti-HCMV activity considerably (43). These compounds did not have activity against other herpesviruses (e.g., herpes simplex virus [HSV] and varicella-zoster virus [VZV]). In addition, BDCRB and TCRB do not inhibit HCMV DNA synthesis at concentrations which completely prevent production of infectious virus (19). Finally, electron microscopy of HCMV-infected cells demonstrated that TCRB allowed capsids to form in the nucleus and that most of the capsids accumulated at an apparent common stage, with an apparent internal scaffolding ring and lacking obvious internal DNA (19).
As a follow-up to the demonstrations that these compounds allow apparent abortive capsid assembly and do not block viral DNA synthesis, we biochemically characterized the state of selected viral mRNAs, general viral protein syntheses, and viral DNA maturation following treatment with BDCRB. We report herein that these compounds do not inhibit transcription of measured mRNAs or general viral protein synthesis; rather, they block maturation of polygenomic precursor HCMV DNA to unit genome length. Since there are numerous points at which intervention can prevent viral DNA maturation, we used DNA from laboratory-derived BDCRB-resistant HCMV to genetically map the BDCRB resistance phenotype. BDCRB resistance mapped within exon 2 of the HCMV UL89 open reading frame (ORF), strongly suggesting that UL89 is a molecular target for BDCRB viral inhibition.
MATERIALS AND METHODS
Chemicals.
BDCRB was synthesized in the laboratories of L. B. Townsend as described previously (43). Ganciclovir was synthesized at the Wellcome Research Laboratories.
Cell and viral culture and antiviral sensitivity measurements.
Human embryonic lung fibroblasts (MRC-5 cells) were from Whittaker Scientific (Walkersville, Md.), and HCMV strain AD169 was obtained from the American Type Culture Collection (Rockville, Md.). Monolayer cultures were grown at 37°C in an atmosphere of 5% CO2 in Eagle’s minimal essential medium (MEM; GIBCO-BRL) with Earle’s salts and containing antibiotics and supplemented with fetal calf serum (HyClone Laboratories, Logan, Utah). The concentration of serum used for growth of cells was 8% (vol/vol).
For viral infection, monolayers were grown to confluence. The medium was removed, and virus was added and suspended in the minimum volume of MEM with the serum concentration reduced to 2%. After incubation with gentle rocking for 90 min, the virus suspension was removed and replaced with MEM containing 2% serum.
Antiviral susceptibilities were determined by plaque reduction assays (8).
Measurement of HCMV DNA synthesis.
MRC-5 cells were inoculated into 24-well plates at ∼5 × 104 cells/well, incubated to confluence (∼1.1 × 105 cells/well), and infected at a multiplicity of infection (MOI) of 2. At various times after infection, duplicate wells were harvested by aspirating the medium from the wells, adding 0.2 ml of lysis buffer (0.1 M Tris-HCl [pH 8], 50 mM EDTA, 0.2% sodium dodecyl sulfate [SDS], 0.1 mg of proteinase K per ml), and incubating the wells for 10 min at 37°C. The cell lysate was incubated for 1 h at 55°C and extracted with 600 μl of TE (10 mM Tris-HCl, 1 mM EDTA [pH 7.0])-saturated phenol-chloroform (1:1). To 200 μl of the aqueous layer was added 25 μl of 3 N NaOH. After incubation at 95°C for 15 min, the samples were made to a final molarity of 1.5 M ammonium acetate, 0.15 M ammonium H2 phosphate, 5 mM EDTA (pH 6.5) and aliquots were blotted onto GIBCO-BRL supported nitrocellulose (catalog no. 1465MH) membranes under vacuum and washed with the buffer described above The samples were cross-linked to the membrane with a Stratalinker 1800 UV oven (Stratagene, La Jolla, Calif.) on the Autolink setting. Standards of purified AD169 DNA, 1 to 250 ng, were also blotted. HCMV DNA was then measured by quantitative hybridization as described below.
Quantitative hybridization of nucleic acids.
The hybridization probe was prepared from suitable cosmids labelled with [α-32P]dCTP by use of random primers and T7 DNA polymerase (Pharmacia) after restriction digestion to linearize the molecules. For measuring total HCMV DNA, a 1:1 mixture of cosmids pC7S31 and pCS37 was used as probe. The HCMV DNA pieces were unique long DNA fragments from approximately 0.22 to 0.39 map unit (cosmid pC7S31) and 0.46 to 0.63 map unit (cosmid pC7S37). The plasmids used for measuring RNA levels are described in “Measurements of viral transcription and translation” below. A PhosphorImager and its associated ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.) was used to quantitate HCMV DNA levels. Prehybridization of the membranes was carried out in a mixture of 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), Ficoll, polyvinylpyrrolidine, and bovine serum albumin, each at 0.2%, 0.5% SDS, and 50 μg of salmon sperm DNA per ml at 45°C for 2 h. The probe was denatured by the addition of 0.1 volume of 1 N NaOH and incubation at 37°C for 5 min, neutralized by the addition of 6× SSPE, and filtered through a sterile Costar 8110 μStar filter (pore size, 0.22 μm). The prehybridization solution was replaced with hybridization solution (6× SSPE, 0.5% SDS, 50 μg of salmon sperm DNA per ml) containing 1 × 106 to 2 × 106 cpm of probe per ml. Hybridization was performed for 16 h at 65°C. The membranes were then washed as follows: with 6× SSPE–0.5% SDS at room temperature twice for 15 min; with 1× SSPE–0.5% SDS at 45 to 65°C twice for 15 min; with 0.1× SSPE–0.5% SDS at 65°C for 30 min to 1 h. The membranes were blotted dry and wrapped in Saran Wrap for quantitation with a PhosphorImager.
Measurements of viral transcription and translation.
MRC-5 cells were infected at an MOI of 1. In experiments to measure immediate-early gene transcription, cycloheximide (100 μg/ml) was added 1 h prior to infection, and the cells were harvested at 24 h postinfection (hpi). Late transcripts were measured in infected cultures without cycloheximide and sampled at 72 hpi. The cells were harvested by scraping and washing with phosphate-buffered saline. Poly(A) RNA was isolated from the pelleted cells with a FastTrack kit (Invitrogen Corp., San Diego, Calif.), suspended in 0.1 M NaCl–0.01 M Na citrate–3% (vol/vol) formaldehyde, heated at 65°C for 15 min, and then blotted onto Nytran membranes (Schleicher & Schuell, Inc., Keene, N.H.). After being baked under vacuum at 80°C for 30 min, the RNA was measured by quantitative hybridization as described above.
For detection of immediate-early transcripts, the probe was prepared from plasmid pHD101SV1 (16), which carries HCMV DNA encompassing UL118 to UL128. Two different plasmids were used for detection of late transcripts, i.e., p1818, encoding the UL86 154-kDa major capsid protein, and pp71, which encodes the UL83 65-kDa matrix protein.
For measurement of the synthesis of viral proteins, MRC-5 monolayers were infected as described above. At 48 and 72 hpi, [35S]methionine (10 μM, 10 μCi/ml) was added in GIBCO-BRL Select-Amine MEM. Twenty-four hours later, the cells were harvested as described above and lysed with Nonidet P-490 to prepare nuclei (4). The nuclei were lysed in SDS loading buffer and run on SDS-polyacrylamide gels (4). The radioactivity on the dried gels was measured with a PhosphorImager and ImageQuant software.
Selection of BDCRB-resistant virus.
The selection of three BDCRB-resistant HCMV strains was initiated by infecting MRC-5 monolayers with AD169 at an MOI of 0.5. The infection was allowed to proceed in the presence of BDCRB at a concentration starting at 1 μM and increasing finally to 10 μM. Three populations of BDCRB-resistant virus were independently derived and termed 1038rA, -B, and -C. They were tested for BDCRB resistance by a plaque reduction assay, and isolates from each of the populations were plaque-purified three times. Final drug susceptibility testing by plaque reduction assay, genetic mapping, and DNA sequencing was performed on plaque-purified strains termed 1038rA 5-2-3, 1038rB 8-3-3, and 1038rC 4-3-2.
HCMV DNA analysis by contour-clamped homogeneous electric field (CHEF) gel electrophoresis.
MRC-5 cell monolayers were infected with virus strain AD169 at an MOI of 1. Intact viral DNA was prepared at various times postinfection by embedding cells in agarose blocks and incubating them at 45°C for 48 h in a mixture of 0.45 M EDTA (pH 9.0), 1% sarcosyl sodium, and 0.5 mg of proteinase K per ml. The agarose plugs containing intact DNA were stored in 10 mM Tris-HCl–50 mM EDTA (pH 7.5) at 4°C. Parameters for resolving the high-molecular-weight viral DNA were selected by entering DNA of the desired size range for separation (10 to 500 kb) into a ChefMapper gel apparatus (Bio-Rad, Mellville, N.Y.). The parameters were as follows: 1% agarose–0.5× Tris-borate-EDTA buffer, a 14°C run temperature, a 120° pulse angle (−60° to +60°), a voltage gradient of 6 V/cm, and a pulse time from 0.47 s with a linear ramp to 44.69 s over 20.18 h. DNA was stained with 1 μg of ethidium bromide per ml for 30 min, UV irradiated in a Stratalinker 1800 at an energy setting of 1500, soaked in transfer buffer (0.4 N NaOH, 1.5 M NaCl) for 15 min, and then transferred to Nytran filters (Schleicher & Schuell) by blotting under pressure for 12 to 16 h (Posiblot apparatus; Stratagene). The filter was hybridized with a 32P-radiolabelled DNA probe made with cosmids pC7S31 and pC7S37, and viral DNA levels were quantitated as described in “Quantitative hybridization of nucleic acids” above.
Vector construction, PCR amplification, and sequence analysis.
High-molecular-weight (HMW) BDCRB-resistant HCMV DNA was prepared from cell-free 1038rB 8-3-3 virus by standard extraction methods (41). The resistant viral DNA was partially digested with Sau3A and ligated into the complementary BamHI site of the SuperCos cosmid (Stratagene). The ligation mixture was then packaged into lambda particles with a Gigapack II packaging extract as described in the manufacturer’s instructions (Stratagene). The GigaPack II packaging extract was used to transfect Escherichia coli NM544 as described in the Stratagene instructions. Overlapping cosmid clones which covered the HCMV genome were identified and characterized by hybridization analysis with a defined library of HCMV DNA fragments and with restriction endonuclease mapping. For hybridization analysis, the cosmid DNA was isolated by a standard alkaline lysis method (4) and bound to nitrocellulose filters under basic conditions essentially as described by Schleicher & Schuell. The DNA was diluted into 3 N NaOH–10 mM Tris–1 mM EDTA, heated at 65°C for 90 min, and applied to Nytran filters with a BioDot apparatus (Bio-Rad, Hercules, Calif.). The filters were hybridized with 32P-radiolabelled HCMV fragments isolated from previously characterized cosmid libraries and subjected to autoradiography with X-ray film by standard methods (4). To verify that the cosmids indeed had the expected DNA content, they were digested with restriction enzymes and separated by agarose gel electrophoresis, and the resulting ethidium bromide staining pattern was compared with the expected pattern based on the published sequence of strain AD169 (13). The corresponding restriction digest of the AD169 laboratory strain was run for restriction pattern confirmation. To subclone a 6.2-kb fragment which transferred resistance to BDCRB, cosmid pCU32 was digested with EcoRI and HindIII and the 6.2-kb fragment was isolated with Gene Clean (Bio 101, La Jolla, Calif.) and ligated into the vector pBR322 digested with EcoRI and HindIII.
PCR amplifications used to generate DNA fragments for transfections and for sequencing were performed with the primer sets termed 1038r-1 (AGT CCC TTT TCT GTC TCC TCT CTA CA) plus 1038r-2 (CCA TGA TGA CGG CGA TGA TAA) to produce fragment PCR-12 and 1038r-3 (CGC TTC TTA CGA CTA ACC TTG A) plus 1038r-4 (TCG GAT GCT CTG TGT AGA GAG G) to produce fragment PCR-34. PCR amplifications were performed on a Perkin-Elmer/Roche (Branchburg, N.J.) model 9600 thermal cycler with the following parameters in Perkin-Elmer GeneAmp PCR buffer with 1.5 mM MgCl2: 1 cycle of 94°C for 3.5 min and then 30 cycles of 94°C for 1 min, 58°C for 2 min, and 72°C for 3 min. We used ∼10 to 100 ng of DNA for the amplification of cosmids. Viral DNA for amplifications was generated by diluting a cell-free virion supernatant with a titer of about 106 PFU/ml with an equal volume of 1× PCR buffer plus nonionic detergents and proteinase K (50 mM KCl, 10 mM Tris-HCl [pH 8.3], 1.5 mM MgCl2, 0.45% Nonidet P-40, 0.45% Tween 20, 50 μg of proteinase K per ml). The mixture was incubated at 55° for 60 min and then at 95°C for 15 min to inactivate the proteinase K. Five microliters of this lysate was used for amplification in a 50-μl PCR mixture. Gel-filtration-grade oligonucleotides used for PCR and sequencing were purchased from Midland Certified Reagent Company (Midland, Tex.). Sequenase with 7-deaza-dGTP and [α-35S]dATP was used for sequencing plasmid pUB1 and PCR products as recommended by the manufacturer (United States Biochemicals, Cleveland, Ohio).
Genetic mapping.
Transfections for genetic mapping (marker transfer) were performed with CaPO4 as described previously (40). Infectious HCMV AD169 DNA was isolated as reported before (41) and coprecipitated with a 3- to 12-fold molar excess of the DNA fragments from the resistant viral DNA cloned into cosmids, isolated DNA fragments, or PCR products. The cosmid DNA used in the transfections was isolated by standard alkaline lysis methods (4) and digested with EcoRI prior to coprecipitation with the infectious AD169 DNA. In general, a higher molar excess of fragments gave a greater percentage of recombinants but fewer plaques following transfection (data not shown). Transfected fragments were (i) entire cosmids digested with EcoRI, (ii) isolated DNA restriction enzyme subfragments from cosmid pCU32, (iii) restriction enzyme-digested fragments from plasmid pUB1, or (iv) DNA pieces amplified by PCR from cosmid pCU32 or from cell-free BDCRB-resistant virus. The DNA subfragments used in the transfections were isolated following gel electrophoresis with Gene Clean and sterilized by ethanol precipitation. Transfected cultures were incubated in the absence of BDCRB until the maximum cytopathic effect was achieved, at which time the progeny virus was harvested and stored at −135°C. For plating efficiencies, 10,000 to 40,000 virions were plated on MRC-5 monolayers under agarose in the presence or absence of 2 μM BDCRB. Plating efficiencies were calculated as the percentage of infectious virus resistant to BDCRB. Amino acid sequence alignments with the HCMV UL89 ORF product were performed with the Compare, Gap, or BestFit programs in the GCG program suite (23). Motif searches were performed as described previously (21).
RESULTS
Effect of BDCRB on viral DNA synthesis, transcription, and translation.
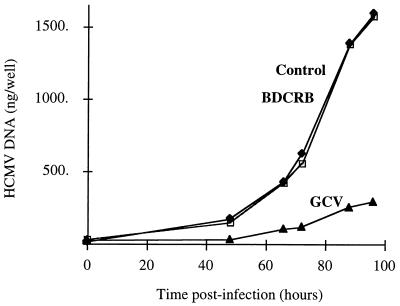
Previous studies by Drach et al. (19) established that HCMV DNA synthesis continued at concentrations of TCRB and BDCRB that inhibited viral replication. The data in Fig. 1 confirm this observation and extend it to show that synthesis of HCMV DNA was approximately exponential until near the end of the infection cycle at about 100 hpi. Ganciclovir added at approximately the nominal plaque reduction 50% inhibitory concentration (IC50) inhibited DNA synthesis as expected, reducing the yield approximately eightfold at the end of a single infection cycle. However, BDCRB at 2.5 times the nominal IC50 had no detectable effect on DNA synthesis (Fig. 1).
FIG. 1.
Synthesis of HCMV DNA in the presence of BDCRB and ganciclovir (GCV). MRC-5 cell monolayers were infected with HCMV strain AD169 at an MOI of 1.5. The infection was allowed to proceed in the presence of BDCRB (1.5 μM) or GCV (5 μM). At different times postinfection, cell monolayers were washed and collected for viral DNA quantification in a dot blot format with 32P-radiolabelled HCMV-specific probes.
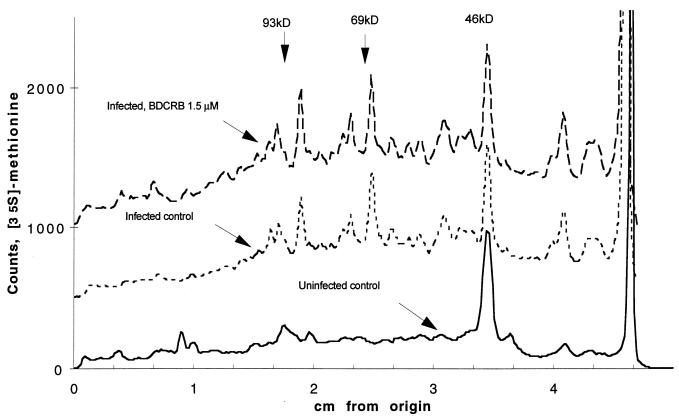
Since DRB has been reported to inhibit the synthesis of specific mRNAs (46), it was possible that the close homolog BDCRB also inhibited mRNA synthesis and could thereby inhibit the synthesis of HCMV proteins required for viral DNA packaging. We therefore measured the levels of representative early and late mRNAs (immediate-early mRNA, UL86 154-kDa capsid mRNA, and UL83 65-kDa matrix protein mRNA). No decreases in the levels of these mRNAs were observed in the presence of 25 μM BDCRB (data not shown). We also measured overall viral protein synthesis by radiolabelling infected cell cultures with [35S]methionine in the absence or presence of 1.5 μM BDCRB. The radiolabelled protein profiles on 7.5% acrylamide gels were identical with and without BDCRB in infected cell cultures at 48 and 72 hpi (the 72-hpi profile is shown in Fig. 2).
FIG. 2.
PhosphorImager scans of methionine-labelled proteins separated by SDS-polyacrylamide gel electrophoresis. Nuclear lysates were prepared at 72 hpi from HCMV-infected MRC-5 cells with or without 1.5 μM BDCRB or from control uninfected MRC-5 cells. Baselines for infected control samples and infected samples with 1.5 μM BDCRB were set arbitrarily at 500 and 1,000 counts, respectively, for ease of comparing the scans.
Inhibition of processing of DNA concatemers.
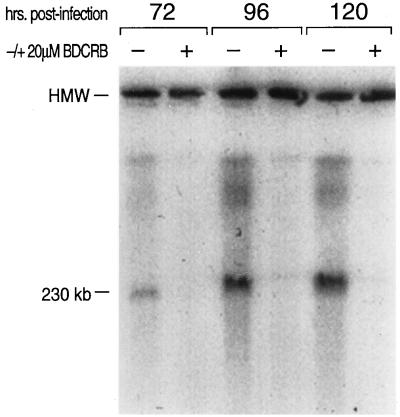
Because neither DNA, RNA, nor protein synthesis was obviously inhibited by BDCRB, we characterized the state of the viral DNA by CHEF gel electrophoresis (14) (a form of pulsed-field gel electrophoresis). Pulsed-field gel electrophoresis analysis has been used extensively to separate intact HMW DNA molecules (28), including genomes of other herpesviruses (22, 48). Cells infected with HCMV strain AD169 were sampled at various times postinfection, and the replicating viral DNA was examined by CHEF electrophoresis and Southern analysis with HCMV-specific DNA probes. In samples taken at 24 to 48 hpi, most of the replicating DNA (HMW DNA) remained in the sample wells during electrophoresis (data not shown), as would be expected for the concatemeric replicative intermediate. The HMW DNA also did not leave the wells under separation conditions under which 5.7-Mb chromosomal DNA molecules of Schizoaccharomyces pombe did enter the gel (data not shown). CHEF electrophoresis and Southern analysis demonstrated that between 72 and 120 hpi, the HMW DNA was converted to a form which migrated with a mobility expected for linear HCMV genomic DNA (Fig. 3). BDCRB at 20 μM strongly inhibited this conversion, indicating that it might be inhibiting viral precursor DNA processing.
FIG. 3.
Inhibition of HCMV DNA concatemer processing by BDCRB. HCMV-infected cells were grown with and without BDCRB added immediately following infection. HMW DNA was separated by CHEF gel electrophoresis and located in the gel by Southern analysis with 32P-radiolabelled HCMV DNA fragments. The image was captured by exposure of the filter to a PhosphorImager screen and analyzed with ImageQuant software. The band at the top of the gel, labelled HMW, is the well where most of the DNA remained. The 230-kb band is HCMV DNA migrating at the position expected for genome-length DNA. Concatemeric phage lambda genomic DNA was run on the same gel to serve as molecular size standards.
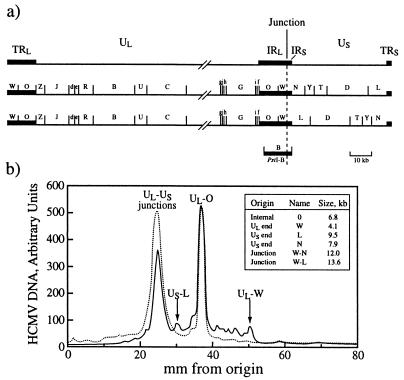
The endless nature of the HMW DNA which accumulated with BDCRB treatment was confirmed by analysis of the genomic termini. Because HCMV polygenomic precursor DNA normally lacks terminal ends (29), we treated infected cells with BDCRB and characterized the proportions of genomic termini relative to internal unique long-unique short (UL-US) junctions. The UL and US regions invert around the a sequence during DNA replication to generate equimolar amounts of four DNA isomers (29). The EcoRI restriction digest of HCMV genomic DNA gives two possible patterns when unit-length genomes are probed with the PstI B fragment which spans the UL-US junction and thereby identifies internal UL-US junctions as well as termini (Fig. 4a). DNA from infected cells at 72 hpi, or from mature HCMV virions, was digested with EcoRI, and the fragments were separated by electrophoresis and blotted onto nylon filters. The filters were probed with the HCMV DNA Pst1 B fragment shown in Fig. 4a. The concentration of genomic ends (US-L and UL-W) in replicating HCMV DNA is clearly reduced to almost zero at 72 hpi in the presence of BDCRB (Fig. 4b), as expected if the DNA were in a head-to-tail concatemeric form. This reduction in the concentration of genomic ends was accompanied by a corresponding increase in the concentration of the UL-US junction fragments (Fig. 4b). Table 1 shows the data for a series of similar experiments generated over time following infection. Because of the relatively low abundance of the end fragments, their quantitation by integration of the PhosphorImager scans after background subtraction is prone to some error. Nevertheless, the data clearly show that as infection progressed, the end fragment UL-W reappeared in the absence, but not in the presence, of BDCRB, as expected from the pulsed-field gel observations.
FIG. 4.
Restriction digestion analysis of end and junction sequences of HCMV DNA from intact virions and from infected cells. MRC-5 cell monolayers were infected at an MOI of 1.5 with AD169, and the infection was allowed to proceed in the absence or presence of 25 μM BDCRB for 72 h. Viral DNA was digested with EcoRI, separated by size on an agarose gel, transferred to a nylon filter, and then hybridized with a probe which hybridized only with the region which contained the junctions and ends. (a) The HCMV genome is shown above the two DNA isomers which are distinguished when an EcoRI digest of genomic DNA is probed with the PstI B fragment. Abbreviations: TRL and IRL, terminal and internal UL inverted repeats, respectively; IRS and TRS, terminal and internal US inverted repeats, respectively. The location of the PstI B fragment used as the probe is shown below the two isomers. It is important to note that the PstI B fragment also hybridizes with the genomic terminal repeats. (b) The DNA fragment profile for EcoRI-digested viral DNA probed with radiolabelled PstI fragment B is shown. The inset box contains the origins, names, and sizes of ends and junction regions shown on the DNA profile. —, without BDCRB; - - - , with 25 μM BDCRB added. The positions of the quantified US and UL ends as they separated under these gel conditions are designated US-L and UL-W, respectively. All data are normalized to the intensity of the UL EcoRI O fragment, which, as an internal repeat fragment, always occurs at a frequency of twice per genome.
TABLE 1.
Ratios of UL-US junction and end fragment UL-W concentration to internal fragment UL-O concentration in the presence or absence of BDCRB during synthesis and maturation of HCMV DNAa
| Fragment | Ratio of fragment to UL-O fragment
|
||||||
|---|---|---|---|---|---|---|---|
| AD169 virion DNAb | 72 h
|
96 h
|
120 h
|
||||
| − | + | − | + | − | + | ||
| Junction | 1.05 | 1.21 | 1.18 | 1.32 | 1.22 | 0.97 | 1.27 |
| UL-O | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| UL-W | 0.12 | 0 | 0 | 0.02 | 0 | 0.14 | 0.015 |
Ratios were calculated from Southern analyses run as described in the legend to Fig. 4 in either the presence (+) or absence (−) of 25 μM BDCRB. All values were normalized to the amount of the EcoRI O fragment generated. Samples were collected at the postinfection times shown.
AD169 virion DNA was isolated and digested in side-by-side experiments, and fragment ratios were calculated as a control for linear DNA.
Selection and characterization of resistant virus.
The packaging and processing of concatemeric DNA involve multiple steps, inhibition of any one of which could result in the observed block in viral DNA maturation. To more closely define the process inhibited by BDCRB, we genetically mapped the location of mutations which conferred BDCRB resistance.
Three independently derived BDCRB-resistant HCMV strains were generated by passaging the wild-type strain, AD169, in the presence of BDCRB concentrations increasing from 1 to 10 μM. The IC50 values for BDCRB-resistant isolates were determined by plaque reduction analysis (8) and ranged from ∼10-fold (1038rA and 1038rC) to ∼30-fold (1038rB) over that of the parent strain, AD169. Strains 1038rA, 1038rB, and 1038rC were plaque-purified three times, and IC50s of BDCRB and TCRB were determined by a plaque reduction assay (Table 2). Strain 1038rB 8-3-3 was also resistant to the 2-chloro analog of BDCRB (TCRB), indicating a common mechanism of action for these closely related analogs (Table 2).
TABLE 2.
Inhibition of HCMV strains by benzimidazole ribosides
| Virus straina | IC50 (μM)b of:
|
|
|---|---|---|
| BDCRB | TCRB | |
| 1038rA | 6 | NDc |
| 1038rB | 20 | ND |
| 1038rC | 6 | ND |
| 1038rA 5-2-3 | 6 ± 0.6 | ND |
| 1038rB 8-3-3 | 18 ± 5 | >50 |
| 1038rC 4-3-2 | 6 | ND |
| AD169 | 0.6 ± 0.1 | 2.8 |
1038rA, -B, and -C are three independently isolated strains. 1038rA 5-2-3, 1038rB 8-3-3, and 1038rC 4-3-2 are the original isolate strains (1038rA, 1038rB, and 1038rC, respectively) plaque purified three times.
Results are from a single plaque reduction assay if no standard error is given; otherwise, the results are from at least three assays with the IC50 given as a mean ± standard error.
ND, not determined.
DNA maturation characterization.
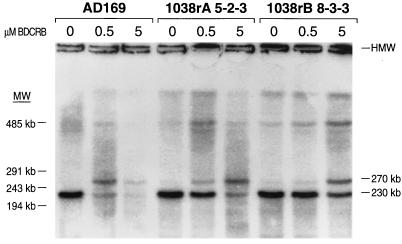
The effects of BDCRB on HCMV DNA maturation in cells infected with BDCRB-sensitive or -resistant virus were compared by CHEF electrophoresis and Southern analysis. All samples were taken at 96 hpi. The image of the blotted and hybridized gel is shown in Fig. 5. Processing of the concatemeric DNA to monomer size by the plaque-purified BDCRB-resistant viruses 1038rA 5-2-3 and 1038rB 8-3-3 was much less sensitive to inhibition by BDCRB than that by the wild-type parental virus, AD169. At partially inhibitory concentrations of BDCRB, a form of HCMV DNA which migrated at the position expected for a linear 270-kb DNA molecule, in addition to the 230-kb monomer-genome-size molecule, was observed. This novel 270-kb band is strongest at 0.5 μM BDCRB for the wild-type strain, AD169, and at 5 μM BDCRB for both of the mutant viruses. The 270-kb band form was not obvious in experiments without BDCRB, indicating that if this band is a DNA maturation intermediate, it is transient. It should be noted that DNA of a nonlinear form may electrophorese at unexpected apparent molecular weights. Experiments are in progress to determine the DNA content and structure of this molecular form.
FIG. 5.
Analysis of HCMV DNA maturation by CHEF electrophoresis. The band visualized at the top of the gel (labelled HMW) is DNA which did not leave the loading well. The 230-kb bands at the bottom of the gel migrate at the position expected for viral DNA molecules of monomer genome size, based on lambda genome concatemeric molecular size markers (MW) loaded on the same gel.
Genetic mapping of the BDCRB resistance gene.
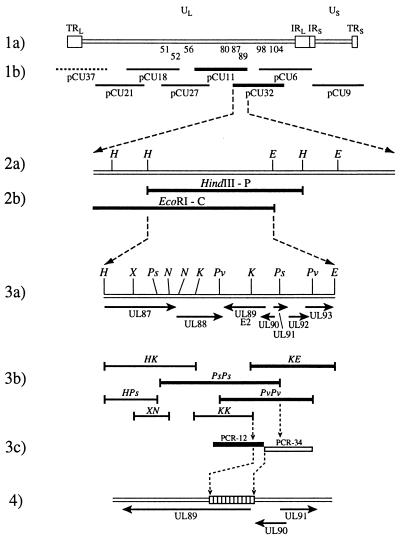
To identify the viral gene responsible for the BDCRB resistance phenotype, discrete DNA fragments were used for recombination into the wild-type AD169 (BDCRB-sensitive) genome. We first generated a cosmid library (shown schematically in Fig. 6, line 1b) with DNA from the most resistant plaque-purified HCMV strain, 1038rB 8-3-3. Two of the seven cosmids tested with marker transfer, pCU11 and pCU32, were able to transfer BDCRB resistance to the laboratory HCMV strain AD169 as shown by the percentage of progeny resistant to BDCRB (i.e., plating efficiency) in these experiments (Table 3, experiment 1). Additional marker transfers with HindIII or EcoRI restriction fragments (Fig. 6, line 2b) further localized resistance to within a 6,320-bp HindIII P-EcoRI C fragment overlap (Fig. 6, line 3a). Other fragments from within the pCU11-pCU32 overlap were tested but did not transfer resistance. Marker transfers with restriction fragments (Fig. 6, line 3b) defined the resistance phenotype to within the overlap of three fragments (KE, PsPs, and PvPv [Fig. 6, line 3b]) as shown by plating efficiencies in Table 3, experiment 2. Finally, the viral resistance phenotype was defined to within a 394-bp region with two PCR amplification products (PCR-12 and PCR-34) (Fig. 6, lines 3c and 4; plating efficiencies shown in Table 3, experiment 3). The plaque-purified recombinants generated in this way demonstrated a drug resistance phenotype similar to that of the parental virus strains (data not shown).
FIG. 6.
Schematic representation of defined DNA fragments from BDCRB-resistant virus used in marker transfer experiments. (Line 1a) Genome structure of HCMV strain AD169. Boxes marked TRL and TRS are terminal repeat elements, and boxes marked IRL and IRS are internal repeat elements. Directly beneath the UL region are indicated the approximate locations of ORFs (by number). (Line 1b) Schematic representation showing location and overlap of cosmids from a library generated from 1038rB 8-3-3 DNA. Thick bars represent cosmids which transferred resistance, solid lines represent cosmids which did not transfer resistance, and a dashed line represents a cosmid which was not tested. (Line 2a) EcoRI and HindIII restriction sites within the pCU11-pCU32 overlap shown in line 1b. (Line 2b) Schematic colinear representation of two fragments isolated from pCU11 (HindIII P and EcoRI C) which transferred resistance. (Line 3a) Schematic representation of the 6,320-bp pCU11-pCU32 overlap fragment, with restriction sites indicated by vertical lines. Directly below this line is a schematic colinear representation in which the horizontal arrows indicate the lengths and orientations of the indicated ORFs. (Line 3b) Schematic colinear representation of restriction fragments isolated from the 6,320-bp fragment used in marker transfers. The fragments are named according to the restriction sites at their ends. Thick bars represent fragments which transferred resistance in marker transfer experiments, while thin bars represent fragments which were unable to transfer resistance. (Line 3c) Schematic representation of PCR products amplified from BDCRB-resistant virus DNA and used for genetic transfers and sequencing. A solid bar represents the fragment that transferred BDCRB resistance. (Line 4) Schematic representation of the 394-bp region which conferred resistance in marker transfer experiments. Below line 4 is shown a schematic colinear representation in which the horizontal arrows indicate the lengths and orientations of the indicated ORFs. Restriction endonuclease abbreviations: E, EcoRI; H, HindIII; K, KpnI; N, NotI; Ps, PstI; Pv, PvuI; X, XbaI.
TABLE 3.
Plating efficiencies for genetic mapping experiments
| Expta | Transfected DNAb | Plating efficiency (%)c |
|---|---|---|
| 1 | None | <0.02 |
| pCU11 | 2.7 | |
| pCU27 | <0.02 | |
| pCU32 | 0.41 | |
| 2 | None | <0.01 |
| KE | 0.14 | |
| PsPs | 0.12 | |
| PvPv | 0.54 | |
| 3 | None | <0.01 |
| PCR-B12 | 0.10 | |
| PCR-B34 | <0.01 | |
| 4 | None | <0.01 |
| PCR-A12d | 0.34 | |
| PCR-C12d | 0.36 |
Experiments 1 through 3 were run with cosmids or different DNA fragments from strain 1038rB 8-3-3. Four other cosmids tested in experiment 1 which are not shown in this table had plating efficiencies of <0.02%. Similarly, restriction fragments tested in experiment 2 which are not shown had plating efficiencies of <0.01%. Experiment 4 was a mapping experiment run with PCR fragments from 1038rA 5-2-3 or 1038rC 4-3-2.
None, negative control in which AD169 was precipitated without additional fragments. Italicized designations represent restriction sites at the ends of the fragments and correspond to abbreviations defined in the legend to Fig. 6.
Plating efficiency is the percentage of plated marker transfer progeny which formed plaques in the presence of 2 μM BDCRB.
PCR amplification products from 1038rA 5-2-3 and 1038rC 4-3-2 (i.e., PCR-A12 and PCR-C12, respectively) were generated with PCR primers as described in Materials and Methods and transfected into cells.
To demonstrate that the lower level of resistance found for 1038rA 5-2-3 and 1038rC 4-3-2 was also due to a mutation(s) giving rise to an amino acid substitution(s) in this region, PCR amplification products were generated from the three-times-plaque-purified resistant viruses 1038rA 5-2-3 and 1038rC 4-3-2 and used for sequencing (see below) and in marker transfer experiments. Plating efficiencies (Table 3, experiment 4) indicated that BDCRB resistance also resided within the 362-amino-acid region of the product of UL89 exon 2 in both 1038rA 5-2-3 and 1038rC 4-3-2.
Sequencing the BDCRB resistance-conferring region of UL89.
We sequenced the region which transferred BDCRB resistance by amplifying by PCR that region and directly sequencing the PCR product. The strains sequenced included the parental strain, AD169, used for selection of the resistant viral strains, the original resistant viruses (1038rA 5-2-3, 1038rB 8-3-3, and 1038rC 4-3-2), and resistant recombinant progeny which had been plaque purified three times. As shown in Table 4, 1038rA 5-2-3 and 1038rC 4-3-2 (and their BDCRB-resistant recombinant progeny) had a single conservative Asp344Glu (D344E) amino acid substitution which was necessary and sufficient to produce BDCRB resistance at an IC50 of 6 μM. In these two viruses, the D344E amino acid substitution was due to a single C-to-G nucleotide change at the wobble position. No other amino acid substitutions were observed. 1038rB 8-3-3, with the highest level of resistance, had two amino acid substitutions, i.e., D344E, due to a single C-to-A nucleotide change at the wobble position, and Ala355Thr (A355T), due to a G-to-A nucleotide substitution at the first position of the codon. One resistant progeny virus recovered (rBREK 2-1-4) carried only the A355T mutation, which most likely arose as the result of a DNA repair process or from partial recombinational transfer. In a plaque reduction assay, BDCRB had an IC50 towards strain rBREK of 4 μM.
TABLE 4.
Sequence comparison of the product of the UL89 exon 2 region carrying BDCRB resistance with ORF products of other herpesviruses
| Virusa | Amino acidsb |
|---|---|
| 1038rB 8-3-3 | E T |
| 1038rA 5-2-3 | E |
| 1038rC 4-3-2 | E |
| rBREK 2-1-4 | T |
| HCMV AD169 | TNTTSDSTCF LTRLNNAPFD MLNVVSYVCE |
| EBV | VNSADQATSF LYKLKDAQER LLNVVSYVCQ |
| HSV-1 | TNTGKASTSF LYNLRGAADE LLNVVTYICD |
| VZV | TNTGKASTSF LYNLRGSSDQ LLNVVTYVCD |
| HHV-6-U | TNSGNHSTSF LMKLNNSPFE MLSVVSYVCE |
| HVS | VNSGDRATSF LFNLKNASEK MLNIVNYICP |
| EHV-1 | TNTGKASTSF LYNLKGAADD LLNVVTYICD |
| BHV-1 | TNTGKASTSF LYNLKGASDG LLNVVTYICN |
| HHV-7 | TNSGNHSTSF LTKLSNSPFE MLTVVSYVCE |
| CONc | tNtgkasTsF LynLkga.d. lLnvVtYvC. |
Abbreviations: HHV-6-U, human herpesvirus 6 (Uganda); HVS, herpesvirus saimiri; EHV-1, equine herpesvirus type 1; BHV-1, bovine herpesvirus type 1; HHV-7, human herpesvirus 7.
The amino acid substitutions at positions 344 and 355 in the sequences of UL89 exon 2 products of 1038rB 8-3-3, 1038rA 5-2-3, 1038rC 4-3-2, and rBREK 2-1-4 are shown aligned above the HCMV AD169 sequence, and the corresponding sequences of other herpesvirus homologs are shown below it.
Consensus sequence for the alignment of HCMV AD169 with the other herpesvirus homologs shown in this table. Capital letters indicate amino acids that are completely conserved, lowercase letters indicate amino acids that are predominantly conserved, and periods indicate limited conservation.
The sequence of the UL89 amino acid region which transferred resistance is shown with four other herpesvirus sequences aligned as described by Davison (17), i.e., those of HSV type 1 (HSV-1), VZV, Epstein-Barr virus (EBV), and human herpesvirus 6, and with the sequences of four additional herpesviruses in Table 4. The amino acid sequence similarities in this short stretch are very clear, and a potential consensus sequence is given (Table 4). However, of the amino acid substitutions found in the sequences of the resistant viruses, only the A (alanine) at amino acid position 355 is present in some other herpesviruses while the D (aspartic acid) at position 344 is not conserved, although the EBV and herpesvirus saimiri homologs have the acidic residue D one amino acid removed. Finally, although it has been shown previously that the T4 bacteriophage gp17 DNA cleavage subunit of the terminase (9) has overall similarities to these homologs (17), there were no similarities to gp17 within this defined region.
DISCUSSION
The herpesviruses and many other double-stranded DNA (dsDNA) viruses replicate their genomes through the formation of polygenomic concatemeric intermediates. This provides the virus with a partial solution to the problem of replication of the ends of the linear viral genome (44) but at the same time makes it necessary that the concatemeric DNA be packaged and cut into monomeric genomes to produce infectious viral particles. Eukaryotic cells, in contrast to these viruses, solve the end replication problem by the synthesis of telomeres at the chromosome ends (10). The formation and processing of DNA concatemers have no known counterpart in mammalian cells, and thus inhibition of concatemer processing should have no effect on the host cells of the virus. Concatemer processing provides a novel and potentially selective target for inhibition of herpesviruses, as was pointed out by Hammerschmidt and Sugden (24).
We show here that the benzimidazole riboside BDCRB inhibits growth of HCMV by inhibiting this essential process of DNA maturation. Cleavage of concatemers to monomeric genomes is inhibited without inhibition of the synthesis of viral DNA, mRNA, or proteins. BDCRB and TCRB are the first antiviral agents to exploit this target.
The processes of herpesvirus DNA maturation and packaging into capsids are complex and not well understood. It is known that the polygenomic viral DNA is packaged into preformed capsids in unit genome lengths with specific nucleotide sequences at the termini (29) and that at least six HSV ORFs (UL6, UL15, UL25, UL28, UL32, and UL33) (1–3, 33, 37, 38, 42, 45) are involved, although the proteins encoded by these ORFs have no defined biochemical function. Each has a close homolog in HCMV (encoded by UL104, UL89, UL77, UL56, UL52, and UL51, respectively), about which even less is known. It is possible that inhibition by BDCRB of the function of any one of these proteins, or of capsid assembly or maturation, could produce the observed inhibition of DNA cleavage.
In this report, we show by marker transfer and sequence comparison that resistance to BDCRB in three separately selected HCMV strains was conferred by one or two nucleotide substitutions within the UL89 ORF. Thus, we infer that a protein encoded by this gene is a molecular target of BDCRB.
UL89 of HCMV is known only as a potential ORF encoding a 674-amino-acid protein (13). However, this gene is very highly conserved across the herpesvirus family. For example, HSV-1 UL15 is 41% identical and 62% similar, while VZV 42/45 is 38% identical and 61% similar, to the HCMV UL89 ORF based on our analysis with the GCG GAP program. Another similarity is that UL89 and its homologs are among the relatively few herpesvirus ORFs which contain spliced genes (15, 17, 18).
Although there is currently no information on transcription or translation of the UL89 ORF in infected cells, HSV1 UL15 has been studied (5–7, 15, 18, 33, 47). The gene product is essential; in a strain of HSV-1 with a temperature-sensitive mutation in UL15, a shift to the nonpermissive temperature prevented generation of infectious virus but did not block DNA synthesis or capsid assembly (33). This UL15 temperature-sensitive phenotype is similar to that seen following treatment of HCMV-infected cells with benzimidazole ribonucleosides (19), and it is likely that UL15 of HSV and UL89 of HCMV have not only sequence homology but a common essential function.
The DNA maturation and packaging processes of many dsDNA bacteriophages (e.g., lambda, T3, T7, and T4) have been relatively well characterized (reviewed in reference 9). The bacteriophage precursor DNA is cleaved by a terminase activity which requires two viral proteins in a large-subunit-plus-small-subunit structure of various stoichiometries. The cleavage of precursor DNA during maturation is a function of the large subunit. Davison (17) identified significant amino acid sequence similarities between protein gp17, the endonucleolytic terminase component of bacteriophage T4, and five herpesvirus UL89 homologs. He proposed that this gene family, including UL15 of HSV and UL89 of HCMV, encodes a DNA terminase.
In addition to this sequence homology, many similarities suggest that the dsDNA bacteriophage may indeed provide good models for DNA maturation and packaging in the herpesviruses. In both, a HMW polygenomic precursor dsDNA is synthesized, matured, and packaged into preformed capsids (9, 29). An assembly protein is found in the preformed capsids of dsDNA bacteriophage (9, 27) and of herpesviruses (25, 34), and in both cases, it is proteolytically removed to make room for the viral DNA. In addition, the DNA packaging process itself may be similar for both as evidenced by the similar organization of DNA inside the capsids (11).
On the other hand, maturation of HCMV DNA to unit genome size requires a sequence-specific cleavage in the region known as the a sequence (30, 39), but cleavage by the T4 gp17 terminase is not sequence specific (9). There is no obvious homology between the sequence-specific terminases of bacteriophages T3, T7, and lambda and the proteins of the UL89 family.
The terminase complexes have a variety of activities, e.g., DNA binding, prohead (capsid) binding, ATPase activity, and endonucleolytic cleavage of the precursor viral DNA. We searched the UL89 protein for amino acid motifs which might be consistent with terminase activities and which might allow us to understand the significance of the amino acid substitutions which conferred BDCRB resistance. It had been previously reported that the product of the UL89 ORF and other herpesvirus protein homologs contain a motif (located at amino acids GKT219/DE310) which may indicate the ability to bind ATP (17). We used the GCG Motifs program (23) to find that the UL89 protein sequence also contains a nearly consensus ATP-binding P-loop motif (compare amino acids AYDYFGKT457 of the UL89 protein with the P-loop consensus sequence AFDYFGKT) such as is found in thymidine kinase and helicases (35). Both of these motifs are somewhat distant (at least in the linear sense) from the UL89 gene mutations which confer BDCRB resistance. Possibly, protein folding allows the mutations in UL89 to modify the binding pocket for one of these putative ATP binding sites. In addition, it is important to note that two or more ATP-binding motifs are often found in the large endonucleolytic subunit of the bacteriophage terminase (9, 31).
The occurrence of these motifs, together with the homology with the T4 gp17 protein and the blockage of concatemer cleavage resulting from its inhibition by BDCRB, is consistent with previous suggestions that the gene family represented by UL89 of HCMV encodes a DNA terminase (17), possibly the putative large endonucleolytic subunit. This is, however, strictly the most attractive model and remains to be proven.
Since the UL89 family is a highly conserved herpesvirus gene (7, 13), it might be expected that a compound active as an inhibitor against one could be active against other herpesviruses and thus be a potential broad-spectrum antiherpesvirus agent. It is therefore surprising that neither BDCRB nor TCRB inhibits HSV or VZV replication (19). All three of the mutant HCMV gene products isolated here had a common D344E amino acid substitution and, in one case, an additional A344T mutation which elevated the resistance level. The limited number of mutations may imply that it is difficult to generate resistance without causing a lethal defect in a well-conserved and essential protein product. The HSV and VZV homologs do not have an acidic amino acid at the position corresponding to the mutated amino acid, Asp-344, and this may explain the lack of activity against these viruses.
Nevertheless, the herpesvirus terminases remain a unique target for potent and potentially very selective antiviral agents, with the potential for broad-spectrum activity if compounds binding to more conserved regions of the protein can be found.
In addition, these novel compounds present a unique opportunity to characterize further the process of herpesvirus DNA maturation and packaging and to characterize the function of the HCMV UL89 ORF.
ACKNOWLEDGMENTS
Plasmids pHD101SV1 and pHD713SV2 were provided by Michelle Davis. We thank Jennie Jacobsen for sequence alignments of UL89 homologs and Michael Agostino for homology and motif searches.
REFERENCES
- 1.Addison C, Rixon F J, Palfreyman J W, O’Hara M, Preston V G. Characterisation of a herpes simplex virus type 1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology. 1984;138:246–259. doi: 10.1016/0042-6822(84)90349-0. [DOI] [PubMed] [Google Scholar]
- 2.Addison C, Rixon F J, Preston V G. Herpes simplex virus type 1 UL28 gene product is important for the formation of mature capsids. J Gen Virol. 1990;71:2377–2384. doi: 10.1099/0022-1317-71-10-2377. [DOI] [PubMed] [Google Scholar]
- 3.al-Kobaisi M F, Rixon F J, McDougall I, Preston V G. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology. 1991;180:380–388. doi: 10.1016/0042-6822(91)90043-b. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 5.Baines J D, Cunningham C, Nalwanga D, Davison A. The UL15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the UL15 gene product. J Virol. 1997;71:2666–2673. doi: 10.1128/jvi.71.4.2666-2673.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baines J D, Poon A P W, Rovnak J, Roizman B. The herpes simplex virus 1 UL15 gene encodes two proteins and is required for cleavage of genomic viral DNA. J Virol. 1994;68:8118–8124. doi: 10.1128/jvi.68.12.8118-8124.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baines J D, Roizman B. The cDNA of UL15, a highly conserved herpes simplex virus 1 gene, effectively replaces the two exons of the wild-type virus. J Virol. 1992;66:5621–5626. doi: 10.1128/jvi.66.9.5621-5626.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biron K K, Elion G B. In vitro susceptibility of varicella-zoster virus to acyclovir. Antimicrob Agents Chemother. 1980;18:443–447. doi: 10.1128/aac.18.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Black L W. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 10.Blackburn E H, Szostak J W. The molecular structure of centromeres and telomeres. Annu Rev Biochem. 1984;53:163–194. doi: 10.1146/annurev.bi.53.070184.001115. [DOI] [PubMed] [Google Scholar]
- 11.Booy F P, Newcomb W W, Trus B L, Brown J C, Baker T S, Steven A C. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Britt W J, Alford C A, editors. Cytomegalovirus. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. [Google Scholar]
- 13.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C A I, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrel B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 14.Chu G, Vollrath D, Davis R W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986;234:1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- 15.Costa R H, Draper K G, Kelly T J, Wagner E K. An unusual spliced herpes simplex virus type 1 transcript with sequence homology to Epstein-Barr virus DNA. J Virol. 1985;54:317–328. doi: 10.1128/jvi.54.2.317-328.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis M G, Kenney S C, Kamine J, Pagano J S, Huang E S. Immediate-early gene region of human cytomegalovirus trans-activates the promoter of human immunodeficiency virus. Proc Natl Acad Sci USA. 1987;84:8642–8646. doi: 10.1073/pnas.84.23.8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davison A J. Channel catfish virus: a new type of herpesvirus. Virology. 1992;186:9–14. doi: 10.1016/0042-6822(92)90056-u. [DOI] [PubMed] [Google Scholar]
- 18.Dolan A, Arbuckle M, McGeoch D J. Sequence analysis of the splice junction in the transcript of herpes simplex virus type 1 gene UL15. Virus Res. 1991;20:97–104. doi: 10.1016/0168-1702(91)90064-3. [DOI] [PubMed] [Google Scholar]
- 19.Drach J C, Townsend L B, Biron K K, Kern E R. Program and abstracts of the 32nd Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1992. Selective inhibition of human cytomegalovirus by polyhalogenated benzimidazole ribonucleosides, a new class of antivirals; p. 402. [Google Scholar]
- 20.Field A K, Biron K K. “The end of innocence” revisited: resistance of herpesviruses to antiviral drugs. Clin Microbiol Rev. 1994;7:1–13. doi: 10.1128/cmr.7.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuchs R. Predicting protein function: a versatile tool for the Apple Macintosh. Comput Appl Biosci. 1994;10:171–178. doi: 10.1093/bioinformatics/10.2.171. [DOI] [PubMed] [Google Scholar]
- 22.Garber D A, Beverley S M, Coen D M. Demonstration of circularization of herpes simplex virus DNA following infection using pulsed field gel electrophoresis. Virology. 1993;197:459–462. doi: 10.1006/viro.1993.1612. [DOI] [PubMed] [Google Scholar]
- 23.Genetics Computer Group. Program manual for the GCG package. 7th ed. Genetics Computer Group, University of Wisconsin, Madison; 1991. [Google Scholar]
- 24.Hammerschmidt W, Sugden B. DNA replication of herpesviruses during the lytic phase of their life-cycles. Mol Biol Med. 1990;7:45–57. [PubMed] [Google Scholar]
- 25.Irmiere A, Gibson W. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology. 1983;130:118–133. doi: 10.1016/0042-6822(83)90122-8. [DOI] [PubMed] [Google Scholar]
- 26.Jacobson M A. Current management of cytomegalovirus disease in patients with AIDS. AIDS Res Hum Retroviruses. 1994;10:917–923. doi: 10.1089/aid.1994.10.917. [DOI] [PubMed] [Google Scholar]
- 27.King J, Casjens S. Catalytic head assembling protein in virus morphogenesis. Nature. 1974;251:112–119. doi: 10.1038/251112a0. [DOI] [PubMed] [Google Scholar]
- 28.Lai E, Birren B W, Clark S M, Simon M I, Hood L. Pulsed field gel electrophoresis. Biotechniques. 1989;7:34–42. [PubMed] [Google Scholar]
- 29.Mocarski E S. Cytomegaloviruses and their replication. In: Fields B N, editor. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2447–2492. [Google Scholar]
- 30.Mocarski E S, Liu A C, Spaete R R. Structure and variability of the a sequence in the genome of human cytomegalovirus (Towne strain) J Gen Virol. 1987;68:2223–2230. doi: 10.1099/0022-1317-68-8-2223. [DOI] [PubMed] [Google Scholar]
- 31.Morita M, Tasaka M, Fujisawa H. Analysis of functional domains of the packaging proteins of bacteriophage T3 by site-directed mutagenesis. J Mol Biol. 1994;235:248–259. doi: 10.1016/s0022-2836(05)80031-2. [DOI] [PubMed] [Google Scholar]
- 32.Polis M A, Spooner K M, Baird B F, Manischewitz J F, Jaffe H S, Fisher P E, Falloon J, Davey R T, Jr, Kovacs J A, Walker R E, Whitcup S M, Nussenblatt R B, Lane H C, Masur H. Anticytomegaloviral activity and safety of cidofovir in patients with human immunodeficiency virus infection and cytomegalovirus viruria. Antimicrob Agents Chemother. 1995;39:882–886. doi: 10.1128/aac.39.4.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poon A P W, Roizman B. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J Virol. 1993;67:4497–4503. doi: 10.1128/jvi.67.8.4497-4503.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robson L, Gibson W. Primate cytomegalovirus assembly protein: genome location and nucleotide sequence. J Virol. 1989;63:669–676. doi: 10.1128/jvi.63.2.669-676.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saraste M, Sibbald P R, Wittinghofer A. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 36.Sehgal P B, Tamm I. Benzimidazoles and their nucleosides. Antibiot Chemother. 1980;27:93–138. doi: 10.1159/000385391. [DOI] [PubMed] [Google Scholar]
- 37.Sherman G, Bachenheimer S L. Characterization of intranuclear capsids made by ts morphogenic mutants of HSV-1. Virology. 1988;163:471–480. doi: 10.1016/0042-6822(88)90288-7. [DOI] [PubMed] [Google Scholar]
- 38.Sherman G, Gottlieb J, Challberg M D. The UL8 subunit of the herpes simplex virus helicase-primase complex is required for efficient primer utilization. J Virol. 1992;66:4884–4892. doi: 10.1128/jvi.66.8.4884-4892.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spaete R, Mocarski E S. The a sequence of the cytomegalovirus genome functions as a cleavage/packaging signal for herpes simplex virus defective genomes. J Virol. 1985;54:817–824. doi: 10.1128/jvi.54.3.817-824.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sullivan V, Biron K K, Talarico C, Stanat S C, Davis M, Pozzi L M, Coen D M. A point mutation in the human cytomegalovirus DNA polymerase gene confers resistance to ganciclovir and phosphonylmethoxyalkyl derivatives. Antimicrob Agents Chemother. 1993;37:19–25. doi: 10.1128/aac.37.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sullivan V, Talarico C L, Stanat S C, Davis M, Coen D M, Biron K K. A protein kinase homologue controls phosphorylation of ganciclovir in human cytomegalovirus-infected cells. Nature. 1992;358:162–164. doi: 10.1038/358162a0. [DOI] [PubMed] [Google Scholar]
- 42.Tengelsen L A, Pederson N E, Shaver P R, Wathen M W, Homa F L. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J Virol. 1993;67:3470–3480. doi: 10.1128/jvi.67.6.3470-3480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Townsend L B, Devivar R V, Turk S R, Nassiri M R, Drach J C. Design, synthesis, and antiviral activity of certain 2,5,6-trihalo-1-(beta-d-ribofuranosyl)benzimidazoles. J Med Chem. 1995;38:4098–4105. doi: 10.1021/jm00020a025. [DOI] [PubMed] [Google Scholar]
- 44.Watson J D. Origin of concatemeric T7 DNA. Nature New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 45.Weller S K, Carmichael E P, Aschman D P, Goldstein D J, Schaffer P A. Genetic and phenotypic characterization of mutants in four essential genes that map to the left half of HSV-1 UL DNA. Virology. 1987;161:198–210. doi: 10.1016/0042-6822(87)90186-3. [DOI] [PubMed] [Google Scholar]
- 46.Yankulov K, Yamashita K, Roy R, Egly J M, Bentley D L. The transcriptional elongation inhibitor 5,6-dichloro-1-beta-d-ribofuranosylbenzimidazole inhibits transcription factor IIH-associated protein kinase. J Biol Chem. 1995;270:23922–23925. doi: 10.1074/jbc.270.41.23922. [DOI] [PubMed] [Google Scholar]
- 47.Yu D, Sheaffer A K, Tenney D J, Weller S K. Characterization of ICP6::lacZ insertion mutants of the UL15 gene of herpes simplex virus type 1 reveals the translation of two proteins. J Virol. 1997;71:2656–2665. doi: 10.1128/jvi.71.4.2656-2665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X, Efstathiou S, Simmons A. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: implications for viral DNA amplification strategies. Virology. 1994;202:530–539. doi: 10.1006/viro.1994.1375. [DOI] [PubMed] [Google Scholar]