Abstract
Extracellular vesicles (EVs) have emerged as significant players in intercellular communication. They carry crucial biological information, and their uptake induces changes in the biological functioning and phenotypes of the recipient cell. Thus, there has been a great deal of interest in understanding their roles in the pathobiology of benign diseases and cancer. Moreover, EVs carry the molecular signatures of the donor cells, and therefore, their utility in biomarker development is being explored. Investigations are also underway to exploit their natural property of cargo transfer from one cell to another to develop efficient, nontoxic, and nonimmunogenic drug delivery systems. EVs originate through endosomal pathways, membrane-budding, or membrane-blebbing during apoptosis. These EV subtypes are usually expected to follow a specific size and surface marker distribution reflective of their origin; however, variations are often reported, especially under pathobiological conditions. Therefore, they are categorized mainly based on their size distribution as small, medium, and large EVs. Dynamic Light Scattering (DLS) is frequently used to measure the size distribution of nanoscale particles in a solution. Moreover, it also provides data on other biophysical properties such as polydispersity, aggregation, solubility, viscosity, and stability. This chapter describes the methods for determining the size distribution and integrity of EVs using DLS along with some constraints associated with the practical use of the technology.
Keywords: Dynamic light scattering, Extracellular vesicles, Size distribution, Integrity, Particle aggregation
1. Introduction
Extracellular vesicles (EVs) are a heterogeneous group of bilayered membranous structures released by most of the living cells into the extracellular spaces [1, 2]. Their shedding was initially thought to be a process of selective elimination of unwanted biomaterial from the cells. However, it is now well-established that EVs play an essential role in intercellular communication. EVs carry important biomolecules such as nucleic acids, proteins, and lipids from the donor cell and efficiently transfer them to the recipient cells [1, 3, 4]. Either EVs are generated inside the cells via the endosomal pathway and then released into the extracellular milieu or they simply pinch off the plasma membrane. EVs of endosomal origin are generally smaller in size (30–150 nm) and referred to as “exosomes,” whereas those originating directly from the plasma membrane are larger (100–1000 nm) and are referred to as “microvesicles” [2, 5]. However, this size distribution is not strict, and significant overlap is reported, particularly under pathobiological conditions [1, 2, 6]. The third subtype of EVs, named “apoptotic bodies,” is generated by membrane blebbing that occurs during programed cell death or apoptosis, and they are generally the largest in size distribution (1000–5000 nm) [2, 7]. The role of apoptotic bodies in intercellular communication is not appreciated, and these are usually engulfed by the neighboring phagocytic cells [8].
Recognition of EV’s role in intercellular communication has given a significant impetus to the research focused on understanding their functions and achieving clinical exploitation. Several lines of evidence suggest that EVs play a crucial role in cancer progression and metastasis [9, 10], immune response regulation [11, 12], inflammatory reactions [13], tissue regeneration [14], and modulation of therapeutic responses [3, 5]. The fact that they carry biological information from the donor cells has attracted much attention for exploring their role in biomarker development for disease diagnosis, prognosis, and monitoring of disease progression and therapeutic responses [5, 15-17]. Being a natural carrier of biomolecules that efficiently transfer them from one cell to another, the utility of EVs in drug delivery is also being explored [18, 19]. Thus, EV research is not only essential to understand biological functioning but also critical to open new vistas for their clinical exploitation as biomarkers, therapeutic targets, and targeted drug delivery systems to modulate cell behavior.
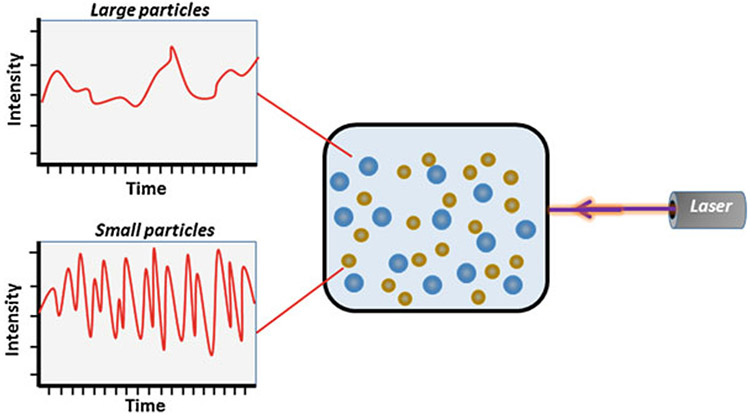
Under any circumstance, EVs can be composed of diverse subtypes differing in size, composition, and biogenic properties. The characterization of EV subtypes is complex. Although different marker proteins are ascribed for different subtypes based on their origin, none of the markers are specific. Therefore, the international society of extracellular vesicles (ISEV) recommends that EVs are characterized based on their size range [20]. Thus, we require a technology that could distinguish between subtypes based on size and provide data on other biological characteristics. Although direct imaging approaches such as Transmission Electron Microscopy (TEM), Atomic Force Microscopy (AFM), and Super-Resolution Microscopy can give accurate information on the size of EV, these are cumbersome and provide data on a small sample portion [21]. Hence, the use of Dynamic Light Scattering (DLS) in the characterization of EVs has received considerable attention [1]. DLS, also known as Photon Correlation Spectroscopy (PCS) or Quasi Elastic Light Scattering (QELS), is frequently used to determine the size distribution of nanometer-scaled particles in a solution. It also provides data on other biophysical properties of biomolecules such as polydispersity index, aggregation, solubility, viscosity, and stability [22]. DLS uses a monochromatic beam of laser light to measure the Brownian motion of particles in a colloidal suspension or polymers in solution and records the temporal fluctuation of scattered light correlated with a hydrodynamic diameter of particles [22] (Fig. 1).
Fig. 1.
Representation of dynamic light scattering principle
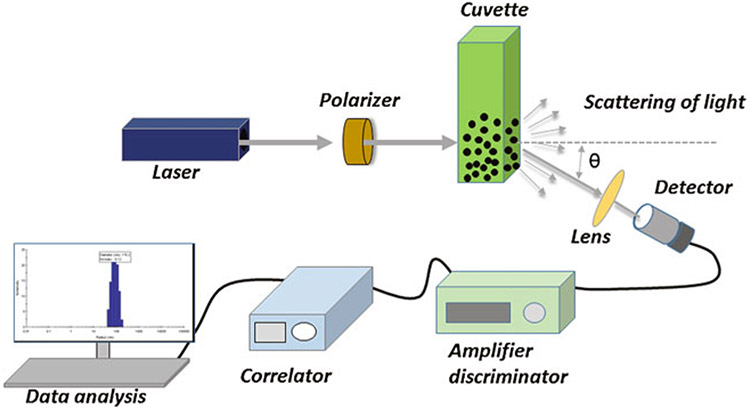
The basic setup of dynamic light scattering involves a single-frequency laser beam that passes through a polarizer to the sample placed in a cuvette holder. Incident light gets scattered due to the Brownian motion of the particles and passes through a single-photon detector (SPD), resulting in the emission of electronic current pulses. An amplifier within the SPD converts these current pulses into voltage pulses, which are then processed by a discriminator that reduces noise by normalizing with a reference threshold. The processed final output voltage pulses corresponding to a photon detection are analyzed by the digital correlator that extrapolates the most precise estimate of the diffusion coefficient () and particle’s hydrodynamic radius () (Fig. 2) by using the following Stokes-Einstein equation:
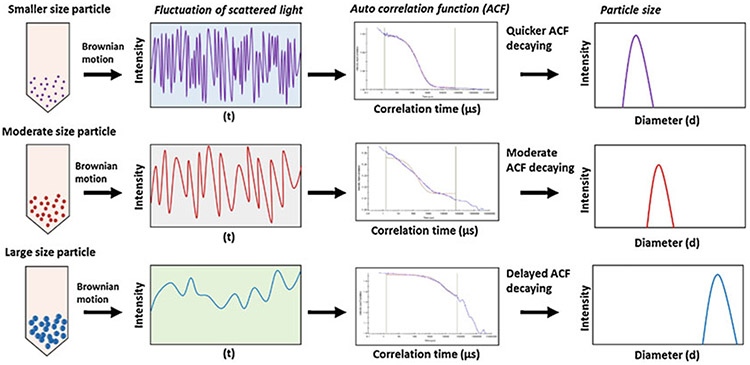
where is the translational diffusion coefficient [m2/s] as speed of the particles, is the Boltzmann constant [m2kg/Ks2], is the temperature, is the viscosity [Pa s], and is the hydrodynamic radius [m]. Besides, a heterogeneous population in a sample represented by the polydispersity index can be calculated by mathematical extrapolation of the autocorrelation function [23, 24]. Noteworthy aspects of this technology are that it requires a small quantity of samples, can provide data in a wide size range (1 nm to 6 μm), and is easy to use. A schematic is presented to describe how the variations in Brownian motion of particles in a wide size range are processed to collect data on size distribution (Fig. 3). In the following sections, we describe the requirement of materials and methodology employed for determining the size and integrity of EVs by DLS technology while also listing some of the technology limitations.
Fig. 2.
Schematic diagram of the basic setup of dynamic light scattering showing laser equipped with a polarizer, lens, measurement cuvette, photomultiplier or detector, amplifier discriminator, correlator, and computer for data handling
Fig. 3.
Schematic flow of dynamic light scattering of small-, moderate-, and large-sized particles. Particles exhibit variations in their Brownian motion depending on their size. These differences in the Brownian motion of particles affect their autocorrelation function decay, resulting in the differences in the size (hydrodynamic diameter) of particles
2. Materials
2.1. Sources of Extracellular Vesicles
Body Fluids:
EVs can be isolated from a variety of biological fluids such as blood, urine, saliva, milk, semen, bile juice, ascites, cystic, bronchoalveolar lavage, and spinal fluids.
Cell Culture Media:
Cells grown in the laboratory also shed EVs into the culture media, which can be used as a source.
2.2. Cell Culture
Following materials are needed to isolate EVs from the cell culture supernatant:
Cell line(s).
EV-free, Heat-inactivated Fetal Bovine Serum.
Optimal Cell culture media [Dulbecco’s Modified Eagle Medium (DMEM), Roswell Park Memorial Institute medium (MEM), etc.].
Tissue culture hood.
CO2 incubator set at 37 °C and 5% CO2.
Phosphate-buffered saline (PBS).
T75 culture flasks.
Serological pipette.
Trypsin-ethylenediaminetetraacetic acid (0.05%).
15 mL conical tubes.
Penicillin Streptomycin solution.
Trypan Blue.
Hemocytometer.
Bright-field Microscope.
2.3. Centrifuges
Ultracentrifugation separates EVs from the solution based on their density, size, and shape, with larger and denser particles sedimenting out first (see Note 1). Density gradient centrifugation is commonly used to separate different organelles such as mitochondria, peroxisomes, and endosomes into distinct density gradient layers of sucrose, iohexol, or iodixanol. The following material is required to perform the ultracentrifugation:
Ultracentrifuge.
Ultraclear centrifuge tubes (25 × 89 mm).
Ultraclear centrifuge tubes (13 × 51 mm).
Benchtop centrifuge.
Weighing balance.
2.4. Protease Inhibitor Cocktail
Protease inhibitor cocktail (100 ×) from Thermo Scientific™ that contains aprotinin (80 μM); bestatin (5 mM); E64 (1.5 mM); AEBSF (100 mM); leupeptin (2.0 mM); pepstatin A (1.0 mM) and EDTA (500 mM).
2.5. Commercial Kits for EV Isolation
Several kits for the isolation of EVs are available commercially based on different principles like precipitation, gel-filtration, affinity purification, etc. (see Note 2). Some of these kits can be used to isolate EVs are listed below:
Capturem™ Extracellular Vesicle Isolation Kit from Takara Bio USA, Inc.
Total Exosomes Isolation kit from Invitrogen.
PureExo isolation kit from 101Bio.
MagCapture™ Exosome Isolation Kit from Wako Life Sciences.
qEV size-exclusion column was from iZON sciences.
2.6. Dynamic Light Scattering (DLS)
DLS measures the fluctuations in scattered light intensity due to the Brownian motion of the particle present in the solution. DLS instrument is sold by many companies such as Malvern Panalytical, Beckman Coulter Inc., Wyatt Technology Corporation. The basic DLS analyzer detects the average size of the particles in a wide range. However, advanced instruments such as DelsaMax PRO light scattering analyzer from Beckman Coulter are well-equipped to determine the complete particle size distribution. In DLS, samples can be injected manually or automatically. The analyzer also comes with a temperature control option, which helps to perform temperature-controlled studies. DelsaMax PRO analyzer supports the temperature range of 4–70 °C, and the temperature controller is designed to change the temperature at a rate of 1 °C/min.
EV sample.
DelsaMax Pro (see Note 3).
Deionized water.
Ultrasonic bath sonicator.
Plastic and Quartz cuvette.
3. Methods
3.1. Isolation of Extracellular Vesicles from Conditioned Media
Collect the conditioned media in polypropylene conical centrifuge tubes (15/50 mL) from subconfluent cells grown in dishes in the tissue culture facility.
Spin the samples at a speed of 300 × g for 10 min at 4 °C to remove the cell debris (see Note 4).
Subject debris-free media to centrifugation at 2000 × g for 30 min at 4 °C in polypropylene conical centrifuge tubes to collect large-size EVs.
Transfer supernatants into a polycarbonate thick-walled tube (designed to withstand high speed) and spin at 16,500 × g for 30 min at 4 °C to pellet the moderate-size EVs.
Transfer the supernatants in the fresh thick-walled tube and spin at 120,000 × g for 2 h to pellet the small-size EVs at 4 °C.
Alternatively, isolate EVs using commercially available kits listed above (Subheading 2.5) as per the manufacturer’s suggested protocols [25].
Resuspend the pellets of EVs (obtained from different methods) in cold phosphate-buffered saline (PBS).
Add Protease inhibitor cocktail (100×) to the samples to make a final dilution to 1× (see Note 5).
Aliquot and store at 4 °C, −20 °C, or −80 °C depending on the experimental requirement.
The size and integrity of isolated EVs are determined by using DLS, as described below.
3.2. Size Distribution and Integrity of Extracellular Vesicles by DelsaMax PRO
Switch on DelsaMax PRO light scattering analyzer and computer attached to it.
Allow the analyzer to warm up for about 30 min after turning it on.
Keep EV samples in ultrasonic baths for a couple of seconds (see Note 6).
Dilute the EV samples into a 1:1000 ratio (1 μL sample:999 μL deionized water).
Keep diluted samples (~1000 μL) in a plastic cuvette and place it in the analyzer.
Once the analyzer stabilizes, start the DelsaMax analysis software on the computer.
Select Tools > Instruments from the DelsaMax analysis software menus.
Select File > New from the software menus.
In the experiment window, set parameters (select preset size).
Next, connect the software with the analyzer and begin the analysis.
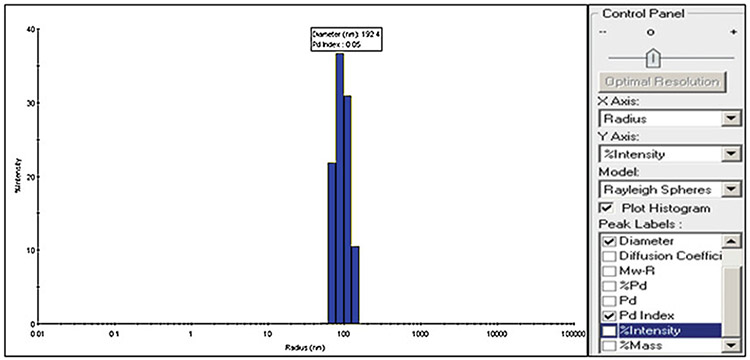
To collect desired data, select diameter, radius, Pd index, and % intensity from the control panel (Fig. 4).
Apply the same procedure to collect data from different subfractions of EVs (Large, moderate, and small).
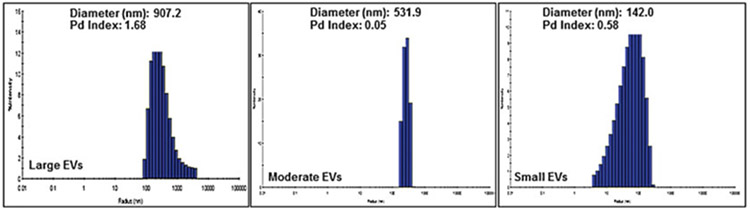
Collect data on the size distribution and polydispersity index (Fig. 5).
For the analysis of zeta potential, place the samples in a quartz cuvette.
Make sure that the sample amount does not exceed 45 μL while measuring zeta potential.
Select phase analysis light scatter from the software.
Collect data on zeta potential.
Fig. 4.
Image profile of extracellular vesicles acquired through DelsaMaxPro
Fig. 5.
Dynamic Light Scattering distribution shows peaks characteristic of large, moderate, and small extracellular vesicles
4. Notes
The ultracentrifugation method is a time-consuming process and not suitable for small sample amounts [25]. Furthermore, high speeds can affect the integrity of EVs by inducing aggregation, breakage, and coprecipitation of soluble proteins present in the biofluids. Therefore, commercially available kits can be used to address these limitations [25, 26].
Commercially available kits are easy and quick and often do not require large volumes of samples. Moreover, specialized equipment such as ultracentrifuge is also not required. However, these kits cannot be used to isolate different subtypes of EVs [25]. Therefore, isolation methods should be wisely selected based on the downstream requirements of the experimental studies.
DLS analysis is easy and quick, does not require additional chemicals, and works great in homogenous samples [27]. However, one of the limitations of DLS is the analysis of heterogeneous mixtures. The results produced by DLS remain skewed toward larger particle sizes in a heterogeneous mixture of widesize-ranged particles in the suspension. It is attributed to the fact that the intensity of scattered light is proportional to the sixth power of particle diameter, making smaller particles harder to detect.
Low-speed centrifugation helps to remove dead cell particles and other contaminants, which might interfere in the isolation process and also the integrity of EVs.
EV pellets can be stored in PBS containing a cocktail of protease inhibitors for a longer duration without losing their integrity.
EVs tend to form aggregates that interfere with DLS analysis. Therefore, to overcome this problem, mild sonication to the samples is helpful that breaks the aggregates.
Acknowledgments
The authors would like to acknowledge the funding from NIH/NCI [R01CA224306, U01CA185490 (to APS) and R01CA204801, R01CA231925 (to SS)] and USA MCI (to APS and SS).
References
- 1.Patton MC, Zubair H, Khan MA, Singh S, Singh AP (2020) Hypoxia alters the release and size distribution of extracellular vesicles in pancreatic cancer cells to support their adaptive survival. J Cell Biochem 121(1):828–839. 10.1002/jcb.29328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathieu M, Martin-Jaular L, Lavieu G, Thery C (2019) Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol 21(1):9–17. 10.1038/s41556-018-0250-9 [DOI] [PubMed] [Google Scholar]
- 3.Patel GK, Patton MC, Singh S, Khushman M, Singh AP (2016) Pancreatic cancer exosomes: shedding off for a meaningful journey. Pancreat Disord Ther 6(2):e148. 10.4172/2165-7092.1000e148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW, Simpson RJ (2018) Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat Rev Clin Oncol 15(10):617–638. 10.1038/s41571-018-0036-9 [DOI] [PubMed] [Google Scholar]
- 5.Patel GK, Khan MA, Bhardwaj A, Srivastava SK, Zubair H, Patton MC et al. (2017) Exosomes confer chemoresistance to pancreatic cancer cells by promoting ROS detoxification and miR-155-mediated suppression of key gemcitabine-metabolising enzyme, DCK. Br J Cancer 116(5):609–619. 10.1038/bjc.2017.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menck K, Sonmezer C, Worst TS, Schulz M, Dihazi gH, Streit F, et al. (2017) Neutral sphingomyelinases control extracellular vesicles budding from the plasma membrane. J Extracell Vesicles 6(1):1378056. 10.1080/20013078.2017.1378056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greening DW, Simpson RJ (2018) Understanding extracellular vesicle diversity - current status. Exp Rev Proteom 15(11):887–910. 10.1080/14789450.2018.1537788 [DOI] [PubMed] [Google Scholar]
- 8.Hochreiter-Hufford A, Ravichandran KS (2013) Clearing the dead: apoptotic cell sensing, recognition, engulfment, and digestion. Cold Spring Harb Perspect Biol 5(1):a008748. 10.1101/cshperspect.a008748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siveen KS, Raza A, Ahmed EI, Khan AQ, Prabhu KS, Kuttikrishnan S et al. (2019) The role of extracellular vesicles as modulators of the tumor microenvironment, metastasis and drug resistance in colorectal cancer. Cancers (Basel) 11(6):746. 10.3390/cancers11060746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao H, Achreja A, Iessi E, Logozzi M, Mizzoni D, Di Raimo R et al. (2018) The key role of extracellular vesicles in the metastatic process. Biochim Biophys Acta Rev Cancer 1869(1):64–77. 10.1016/j.bbcan.2017.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thery C, Ostrowski M, Segura E (2009) Membrane vesicles as conveyors of immune responses. Nat Rev Immunol 9(8):581–593. 10.1038/nri2567 [DOI] [PubMed] [Google Scholar]
- 12.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ et al. (1996) B lymphocytes secrete antigen-presenting vesicles. J Exp Med 183(3):1161–1172. 10.1084/jem.183.3.1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Console L, Scalise M, Indiveri C (2019) Exosomes in inflammation and role as biomarkers. Clin Chim Acta 488:165–171. 10.1016/j.cca.2018.11.009 [DOI] [PubMed] [Google Scholar]
- 14.Taverna S, Pucci M, Alessandro R (2017) Extracellular vesicles: small bricks for tissue repair/regeneration. Ann Transl Med 5(4):83. 10.21037/atm.2017.01.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong BS, Cho JH, Kim H, Choi EJ, Rho S, Kim J et al. (2009) Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genomics 10: 556. 10.1186/1471-2164-10-556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pang B, Zhu Y, Ni J, Thompson J, Malouf D, Bucci J et al. (2020) Extracellular vesicles: the next generation of biomarkers for liquid biopsy-based prostate cancer diagnosis. Theranostics 10(5):2309–2326. 10.7150/thno.39486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whiteside TL (2017) Extracellular vesicles isolation and their biomarker potential: are we ready for testing? Ann Transl Med 5(3):54. 10.21037/atm.2017.01.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garofalo M, Villa A, Rizzi N, Kuryk L, Rinner B, Cerullo V et al. (2019) Extracellular vesicles enhance the targeted delivery of immunogenic oncolytic adenovirus and paclitaxel in immunocompetent mice. J Control Release 294:165–175. 10.1016/j.jconrel.2018.12.022 [DOI] [PubMed] [Google Scholar]
- 19.Kanchanapally R, Deshmukh SK, Chavva SR, Tyagi N, Srivastava SK, Patel GK et al. (2019) Drug-loaded exosomal preparations from different cell types exhibit distinctive loading capability, yield, and antitumor efficacies: a comparative analysis. Int J Nanomedicine 14:531–541. 10.2147/IJN.S191313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R et al. (2018) Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 7(1):1535750. 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chuo ST, Chien JC, Lai CP (2018) Imaging extracellular vesicles: current and emerging methods. J Biomed Sci 25(1):91. 10.1186/s12929-018-0494-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim J, Yeap SP, Che HX, Low SC (2013) Characterization of magnetic nanoparticle by dynamic light scattering. Nanoscale Res Lett 8(1):381. 10.1186/1556-276X-8-381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhattacharjee S (2016) DLS and zeta potential - what they are and what they are not? J Control Release 235:337–351. 10.1016/j.jconrel.2016.06.017 [DOI] [PubMed] [Google Scholar]
- 24.Hassan PA, Rana S, Verma G (2015) Making sense of Brownian motion: colloid characterization by dynamic light scattering. Langmuir 31(1):3–12. 10.1021/la501789z [DOI] [PubMed] [Google Scholar]
- 25.Patel GK, Khan MA, Zubair H, Srivastava SK, Khushman M, Singh S et al. (2019) Comparative analysis of exosome isolation methods using culture supernatant for optimum yield, purity and downstream applications. Sci Rep 9(1):5335. 10.1038/s41598-019-41800-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helwa I, Cai J, Drewry MD, Zimmerman A, Dinkins MB, Khaled ML et al. (2017) A comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents. PLoS One 12(1):e0170628. 10.1371/journal.pone.0170628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szatanek R, Baj-Krzyworzeka M, Zimoch J, Lekka M, Siedlar M, Baran J (2017) The methods of choice for extracellular vesicles (EVs) characterization. Int J Mol Sci 18(6):1153. 10.3390/ijms18061153 [DOI] [PMC free article] [PubMed] [Google Scholar]