Abstract
The three-dimensional structures of the Fab fragment of a neutralizing antibody raised against a foot-and-mouth disease virus (FMDV) of serotype C1, alone and complexed to an antigenic peptide representing the major antigenic site A (G-H loop of VP1), have been determined. As previously seen in a complex of the same antigen with another antibody which recognizes a different epitope within antigenic site A, the receptor recognition motif Arg-Gly-Asp and some residues from an adjacent helix participate directly in the interaction with the complementarity-determining regions of the antibody. Remarkably, the structures of the two antibodies become more similar upon binding the peptide, and both undergo considerable induced fit to accommodate the peptide with a similar array of interactions. Furthermore, the pattern of reactivities of five additional antibodies with versions of the antigenic peptide bearing amino acid replacements suggests a similar pattern of interaction of antibodies raised against widely different antigens of serotype C. The results reinforce the occurrence of a defined antigenic structure at this mobile, exposed antigenic site and imply that intratypic antigenic variation of FMDV of serotype C is due to subtle structural differences that affect antibody recognition while preserving a functional structure for the receptor binding site.
Foot-and-mouth disease virus (FMDV) is an important animal pathogen of the genus Aphthovirus of the Picornaviridae family (45). It causes an economically important disease of cattle and other cloven-hooved animals, and although the disease has been reasonably controlled in the developed world, it is enzootic in many countries of Africa, Asia, and South America. In the last few years, outbreaks have been recorded in Italy, Greece, and several Eastern European countries. Understanding of protective immune responses against FMDV is important for the development of safer and more effective vaccines (5, 36). One of the major antigenic sites of FMDV is located at the G-H loop of capsid protein VP1 (1, 24, 25, 49). In serotype C FMDV, antigenic site A involves a cluster of essentially continuous epitopes located within residues 136 to 150 of VP1. Site A includes the highly conserved triplet Arg141-Gly142-Asp143 (RGD) (Fig. 1A), which is involved in recognition of an integrin receptor (3, 12, 29). The function of this antigenic loop in antibody and host cell recognition has been faithfully mimicked with synthetic peptides (12, 17, 30, 33, 35, 44). Despite extensive overlap, most site A epitopes in FMDV of serotype C were distinguishable by immunochemical methods, suggesting multiple ways of antibody recognition of this antigenic site. Even though the G-H loop of VP1 appears to be disordered in crystals of native FMDV particles, a structure could be defined in chemically reduced FMDV O1BFS particles (26), as well as in a complex between an antigenic peptide and the Fab fragment of a neutralizing antibody raised against the virus (52). This structure revealed that the RGD triplet participates directly in the interaction with antibody SD6 (52, 53). Cryoelectron microscopy (18) and biochemical (51) studies have shown that SD6 is an effective neutralizer that binds monovalently to particles without causing aggregation of virions. Antibody SD6 neutralizes by blocking attachment of virus particles to cells (51). A dual participation of capsid amino acids in receptor recognition and antibody binding has recently been observed also for poliovirus (15) and rhinovirus (47). However, in addition to monoclonal antibody (MAb) SD6, other MAbs neutralize C-S8c1 by binding to distinct epitopes within the G-H loop of VP1, as evidenced by the isolation and sequencing of MAb-resistant mutants and by the distinct reactivities of the MAbs with variant synthetic peptides (32, 33; for a review, see reference 30). However, no structural information on the interaction of these antibodies with site A is available. Although the reactivities with MAbs of eight synthetic peptides that included replacements at the RGD triplet suggested an important influence of these residues in the interactions (41), their direct participation in antibody recognition is based only on structural studies with SD6 (52, 53).
FIG. 1.
(A) Amino acid sequence of the peptide antigen A15 (VP1 residues 136 to 150 of C-S8c1). The Arg-Gly-Asp motif is underlined. (B and C) Alignment of the amino acid sequences of the light (B) and heavy (C) chains of the variable regions of antibodies 4C4 and SD6. Residues that differ between the two antibodies are in boldface. The positions of the CDRs are indicated by horizontal lines above the sequences.
In the present report we describe the three-dimensional structure of the Fab fragment of site A-specific, neutralizing MAb 4C4 alone and in a complex with antigenic peptide A15, representing site A of C-S8c1 (Fig. 1A). MAb 4C4 is a neutralizing antibody raised against FMDV C1 Brescia It/64 (7), and it defines an epitope which is distinct from that defined by MAb SD6 (32). In addition we have quantitated the interaction of other site A-specific MAbs with substituted synthetic peptides representing variant forms of site A. The results suggest common features in the modes of interaction of different antibodies with antigenic site A, in particular the direct participation of Asp143, a residue which belongs to the receptor recognition triplet RGD.
These observations have a number of implications for understanding the immunodominance of this antigenic loop of FMDV and the mechanisms of escape of the virus from neutralization in connection with the dynamics of RNA virus evolution. The results also provide relevant information for vaccine design.
MATERIALS AND METHODS
Virus and antibodies.
C-S8c1 is a biological clone derived from isolate C1 Sta Pau Sp/70, as described previously (48). C1 Sta Pau was isolated from a diseased swine in Santa Pau, Girona, Spain, in 1970. C1 Brescia It/64 was isolated from cattle in Brescia, Italy, in 1964 and adapted to cell culture (7, 38).
Site A-specific MAbs SD6, 4C4, 7JDI, 7CA11, 6D11, 7FC12, and 5A2 have been previously described (32, 34). They were raised against the following FMDV antigens: MAb SD6 against C-S8c1 (34); MAbs 4C4, 6D11, and 5A2 against C1 Brescia It/64 (7); and MAbs 7JD1, 7CA11, and 7FC12 against C3 Indaial Br/71 (31). Neutralizing MAb 4C4 reacts with both C1 Brescia It/64 and C-S8c1 in enzyme-linked immunosorbent assays (ELISA), with isolated VP1 in Western blots, and with synthetic peptides representing antigenic site A of FMDV C-S8c1 (32). The VP1 G-H loop of FMDV C1 Brescia It/64 is identical to that of C-S8c1 except that it has T instead of A at position 140 and A instead of T at position 149 (27, 38).
Purification and sequencing of the Fab fragment of MAb 4C4.
MAb 4C4 was purified from ascitic fluid by protein A-Sepharose (Pharmacia) affinity chromatography and ammonium sulfate fractionation; its purity was >90% as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The Fab fragment was obtained by digestion with soluble papain in phosphate-buffered saline (PBS)–0.8 mM EDTA–4.2 mM Cys for 4 h at 37°C with an immunoglobulin G-to-enzyme ratio of 104:1 (wt/wt). The reaction was stopped by addition of iodoacetamide (Sigma) at 6 mM (final concentration). The solution was then dialyzed against PBS, and the protein was concentrated by ammonium sulfate precipitation to 85% saturation. The Fab moiety was purified by protein A-Sepharose chromatography followed by concentration in a Centricon-30 (Amicon) filter and Sephadex G-200 chromatography. The purification was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and by analytical isoelectric focusing. Two different isoforms were found and were separated by fast protein liquid chromatography chromatofocusing in a MonoP column (Pharmacia).
The amino acid sequences of the variable regions of the 4C4 Fab fragment were deduced from the corresponding mRNA sequences. The mRNAs encoding the variable region of the 4C4 κ light chain and the variable-constant CHI hinge region of the 4C4 γ2a heavy chain were reverse transcribed and amplified by PCR. The oligonucleotide 5′ primers were those described by Coloma et al. (9); the 3′ primers corresponded to segments within the constant regions of the BALBc murine κ light chain and γ2a heavy chain, respectively. The nucleotide sequences of the amplified DNAs were determined by using the Femtomol sequencing system (Promega) and were used to deduce the corresponding amino acid sequences (Fig. 1B and C).
Synthetic peptides.
A total of 240 analogs of peptide A15 (Fig. 1A) were prepared by systematic single-residue replacement at every position with 16 of the genetically coded amino acids; for synthetic simplicity, Cys, Met, and Trp were not used. Peptides were synthesized by solid-phase procedures and analyzed as previously described (35). Each peptide included at least 80% of the target sequence, as determined by analytical high-pressure liquid chromatography. The identities of the peptides were confirmed by amino acid analysis and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF). Peptides were dissolved in PBS and adjusted to neutral pH when necessary, and soluble-peptide concentrations were determined by amino acid analysis (35).
Immunochemical procedures.
A competition ELISA was used to quantitate the reactivity of MAbs with substituted synthetic peptides (41). ELISA plates were coated with 5 pmol of peptide A15 (Fig. 1A) coupled to keyhole limpet hemocyanin overnight at 4°C. After extensive washing of the wells with PBS, mixtures of a nonsaturating amount of MAb and increasing amounts (1, 5, 25, 125, and 625 pmol) of the synthetic peptide (preincubated for 2 h at room temperature) were added to each well. Incubation of the plates was for 1 h at room temperature. After extensive washing with 0.05% Tween 20–0.1% bovine serum albumin in PBS, the enzymatic reaction was carried out with o-phenylenediamine as a substrate. Absorbance was read at 492 nm; background values obtained without MAb and synthetic peptide (A492 < 0.05) were subtracted. A relative IC50 (concentration of peptide which causes 50% of inhibition of binding of a nonsaturating amount of MAb to antigen) was determined for each variant peptide by dividing its IC50 by the IC50 of the homologous (A15) peptide.
Crystallization and data collection.
Crystals of the 4C4 Fab fragment, both isolated and complexed with the 15-amino-acid peptide containing the sequence of antigenic site A of FMDV (residues 136 to 150 of VP1 [Fig. 1A]), were obtained by the hanging-drop vapor diffusion technique at room temperature. For the isolated Fab, crystals of about 0.7 by 0.3 by 0.05 mm3 grew at pH 7.5 with 0.1 M Tris-HCl as the buffer and 0.4 M MgCl2 and 18% polyethylene glycol 4000 as precipitants. The Fab concentration was 7 mg/ml. The crystals were orthorhombic, with space group P21212 and unit cell parameters a = 115.9 Å, b = 183.6 Å, and c = 42.7 Å and containing two Fab molecules per asymmetric unit, which would correspond to a specific volume of 2.25 Å3/Da and to an approximate volume solvent content of 45%. The data were collected at 100 K by means of cryocrystallographic techniques with 30% glycerol as a cryoprotectant. A total of 125 images were recorded on a Mar Research Imaging Plate on a Rigaku rotating anode. The intensities were evaluated and internally scaled by using programs Denzo and Scalepack (42). The data were 96% complete at 3.0-Å resolution, giving an internal agreement factor Rsymm of 10% and an I/ς in the last resolution shell of 8.5. Crystals of the complex (0.6 by 0.15 by 0.05 mm3) were obtained at pH 8.5 with Tris-HCl as the buffer and 0.4 M LiCl and 18% polyethylene glycol 4000 as precipitants. The Fab concentration was 7 mg/ml and the peptide concentration was 1.5 mg/ml. The crystals were orthorhombic, with space group P212121 and unit cell parameters a = 48.6 Å, b = 68.7 Å, and c = 155.9 Å and with one complex molecule per asymmetric unit, which corresponds to 2.6 Å3/Da and a volume solvent content of 52%. The X-ray data were collected at room temperature with a Mar Research Imaging Plate in a Rigaku rotating anode generator and were reduced with the Denzo package. Data were 95% complete at 3.2-Å resolution (Rsymm = 8.9%), and the I/ς in the last resolution shell was 5.8.
Structure solution and refinement.
Both structures were determined by molecular replacement with the AMoRe Package (39). The structure of the isolated Fab fragment was solved and refined first, and then the final model obtained was used to solve the Fab-peptide complex structure. The starting model was taken from the structure of the SD6 Fab fragment (53). The correctly oriented and positioned models were subjected to rigid-body refinement with the X-PLOR program (6). After some cycles of rigid-body refinement treating the variable and constant modules as independent bodies, the resulting R value was 35% in the resolution range 15 to 3.5 Å. 2Fo-Fc and Fo-Fc electron density maps were computed after omitting the noncommon residues or side chains between SD6 and 4C4 and were examined by using the graphic program FRODO (21). Most of the truncated residues were visible in this difference map. After manual fitting of residues to the density, the positional refinement was started with the program X-PLOR. The model was improved by iterative cycles of rebuilding and refinement. The final model contained 434 protein residues and no solvent molecules and was refined to Rcryst and Rfree values of 0.198 and 0.266, respectively, for 11,599 reflections with F > 2ςF for data between 8 and 3 Å. The root mean square deviations from ideality of bonds and angles were 0.011 Å and 2.5°. The final model was then used to solve the structure of the 4C4-peptide complex. After rigid-body refinement, the Rcryst and Rfree values were 0.372 and 0.374, respectively. Examination of the 2Fo-Fc and Fo-Fc electron density maps calculated at this stage clearly indicated the presence of the oligopeptide occupying the antigen binding site. Some conformational rearrangements in the complementarity-determining regions (CDRs) of the antibody, in particular in CDRH3, were also visible. The final model for the 4C4-peptide complex structure was obtained by iterative cycles of model building, using program O (22) and X-PLOR refinement, including a bulk solvent correction. The refined model contained 434 Fab residues, 11 of the 15 peptide residues, and no solvent molecules. This model was refined to Rcryst and Rfree values of 0.20 and 0.278, respectively, for 7,900 reflections with F > 2ςF in the resolution shell 8 to 3.2 Å. The root mean square deviation for bond lengths is 0.008 Å, and that for bond angles 2.1°.
Coordinates of both unliganded and complexed 4C4 structures will be deposited with the Brookhaven Protein Data Bank and are available directly from the authors on request until they have been processed and released.
RESULTS
Overall structure of a complex between antigenic peptide A15 and the Fab fragment of MAb 4C4.
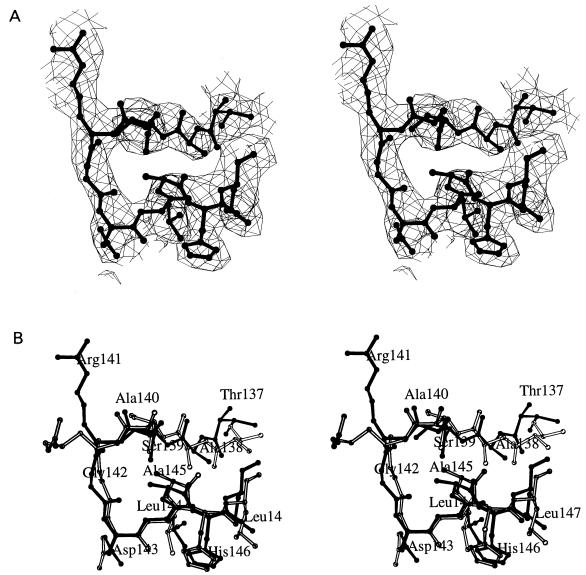
Crystals of the Fab fragment of MAb 4C4 unbound and in a complex with synthetic peptide A15 were obtained and analyzed by X-ray crystallography as detailed in Materials and Methods. The quality of the final electron density maps allowed the positioning with confidence of most residues and side chains for the unbound and complexed 4C4 Fab fragment. In both structures the Fab fragment comprised a total of 434 residues (218 residues from the light chain and 216 from the heavy chain). The electron density corresponding to the peptide was clear for 11 of the 15 residues involved, including the cell recognition RGD motif (Fig. 2A). The terminal Tyr136 and Thr148 to Thr150 could not be positioned in the electron density map. The 11 peptide residues complexed to 4C4 Fab displayed a compact folded conformation (Fig. 2) stabilized by nine intrapeptide hydrogen bonds and a buried hydrophobic surface area of 143.4 Å2 (the solvent radius used was 1.7 Å). The RGD triplet appeared in an open-turn conformation followed by a short helical segment involving residues Asp143 to Leu147. The N-terminal residues (Thr137 to Ala140) were in an extended conformation (Fig. 2).
FIG. 2.
Stereoviews of the antigenic peptide A15 complexed with antibody. (A) Fo-Fc omit map of the peptide in the complex with 4C4 Fab at 3.2-Å resolution. The final peptide model (thick lines) is shown for clarity. The N-terminal residue Tyr136 and the C-terminal residues Thr148 to Thr150 have not been included in the model. There is some extra density in the N-terminal region that would correspond to the Tyr136 main chain. (B) Superimposition of the A15 peptide conformations found in the 4C4-A15 (filled lines) and SD6-A15 (empty lines) complexes. Only residues Thr137 to Leu147, which have been positioned in the two peptide models, are shown.
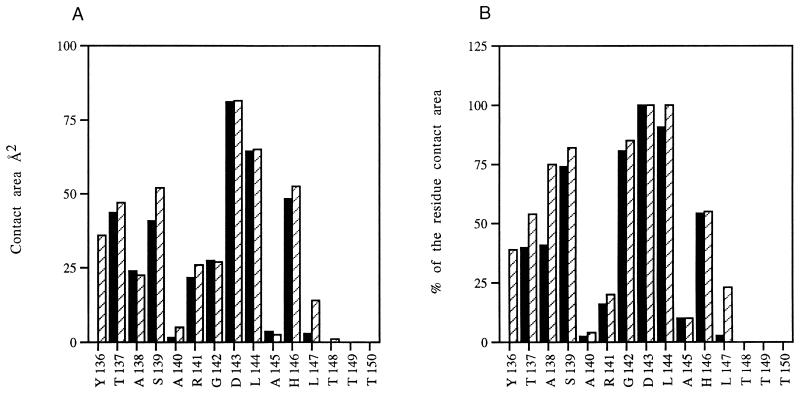
The antigenic peptide interacts with each of the CDRs of the Fab through a total of 11 hydrogen bonds (Table 1 and Fig. 3). Asp143 participates with the largest contact area and with 100% of its molecular surface in the interaction with the antibody (Fig. 4). It is important to emphasize that this Asp143 belongs to the highly conserved RGD receptor recognition site (3, 12, 29). In addition, Thr137, Ser138, and His146 participate in a number of polar interactions (Table 1 and Fig. 3), while residue Leu144 dominates the hydrophobic interactions (Fig. 4).
TABLE 1.
Hydrogen bonds between the antigenic peptide A15 and the Fab fragment in complexes between A15 and 4C4 or SD6a
| MAb | Bond interaction site in:
|
Location | Distance (Å) | |
|---|---|---|---|---|
| FMDV C-S8c1 peptide | MAb Fab fragment | |||
| 4C4 | Thr137 N | Arg54 Nη2 | L2 | 2.6 |
| Thr137 O | Asp104 N | H3 | 3.2 | |
| Thr137 Oγ1 | Asp34 Nδ2 | L1 | 2.9 | |
| Ser139 Oγ | Asn96 Oδ1 | L3 | 3.4 | |
| Asp143 O | Tyr59 Oη | H2 | 3.4 | |
| Asp143 Oδ1 | Arg9 Nη2 | H3 | 2.7 | |
| Asp143 Oδ2 | Arg99 Nη1 | H3 | 2.8 | |
| Asp143 Oδ2 | Thr50 Oγ1 | H2 | 3.0 | |
| His146 Nδ1 | Tyr59 Oη | H2 | 2.6 | |
| His146 Nδ2 | Thr33 Oγ | H1 | 3.2 | |
| His146 Nδ2 | Ser52 Oγ | H2 | 2.8 | |
| SD6 | Tyr136 Oη | Glu100 Oɛ2 | H3 | 2.6 |
| Tyr136 Oη | Gly103 N | H3 | 3.2 | |
| Thr137 O | Asp104 N | H3 | 2.7 | |
| Ser139 Oγ | Ser31 Oγ | L1 | 3.2 | |
| Ser139 Oγ | Asn96 Oδ1 | L3 | 3.3 | |
| Arg141 O | Glu97 N | L3 | 2.6 | |
| Arg141 Nɛ | Glu97 Oɛ1 | L3 | 3.1 | |
| Asp143 O | Tyr59 Oη | H2 | 3.4 | |
| Asp143 Oδ1 | Arg99 Nη2 | H3 | 3.3 | |
| Asp143 Oδ2 | Arg99 Nη1 | H3 | 3.2 | |
| Asp143 Oδ2 | Thr50 Oγ1 | H2 | 2.6 | |
| His146 Nδ1 | Tyr59 Oη | H2 | 2.7 | |
| His146 Nδ2 | Ser52 Oγ | H2 | 3.2 | |
| His146 Nδ2 | Ser53 Oγ | H2 | 2.8 | |
Procedures for structure determination are detailed in Materials and Methods. The amino acid sequences of antigenic peptide A15 and of the variable regions of MAbs 4C4 and SD6 are given in Fig. 1.
FIG. 3.
Stereoview of the 4C4 Fab-peptide interactions. The peptide residues are shown as thick lines, and the Fab residues in direct contact with the peptide is shown as thinner lines. Broken lines indicate hydrogen bonds between Fab and peptide residues.
FIG. 4.
Contact area expressed in square angstroms (A) and as percentage of the total residue surface (B) for the FMDV C-S8c1 peptide in the 4C4-A15 (▪) and SD6-A15 (▨) complexes.
Comparison between the structures of two site A-specific antibodies and their complexes with antigenic peptide A15.
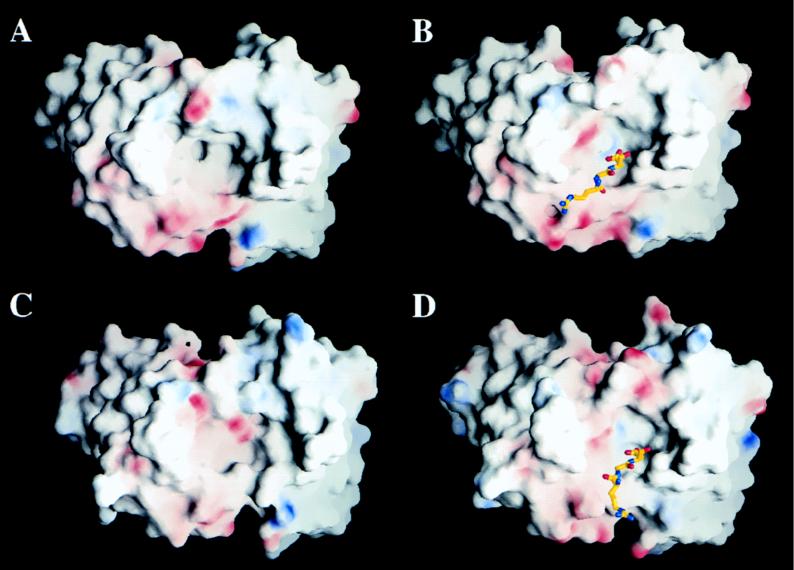
The structure of the peptide A15 complexed with the Fab fragment of 4C4 allowed a detailed comparison with the previously determined structure of the same antigenic peptide complexed with the Fab fragment of SD6 (52). MAb SD6 defines an immunochemically distinct site A epitope with different degrees of conservation among FMDV isolates (32). The variable regions of the light and heavy chains of SD6 and 4C4 show an average of 88% amino acid sequence identity, with variations clustered mainly at CDR1 and CDR3 of the heavy chain (Fig. 1B and C). Accordingly, the main structural differences between the unbound antibodies are located in the CDRs of the heavy chains and, in particular, in CDRH3 (Fig. 5). These structural differences, together with the divergence in amino acid sequence, lead to two very distinct paratopes differing both in shape and in charge distribution (Fig. 6A and C).
FIG. 5.
Stereodiagrams of the CDRH3 regions for Fab 4C4 (main chain tracings depicted as dark ribbons) and Fab SD6 (clear ribbons). (A and B) The two CDRH3 regions are shown superimposed as found in the unbound Fab fragments (A) and in the Fab fragments complexed with peptide A15 (B). The amino acid side chains are represented as filled and empty balls and sticks for 4C4 and SD6, respectively. The N-terminal (ArgH98) and C-terminal (PheH107) amino acids of the CDRH3 loop are labeled. (C and D) Conformational rearrangements of the CDRH3 loop of 4C4 (C) and SD6 (D) upon binding to antigen. In each of the two diagrams, the unbound CDRH3 loops are depicted as clear ribbons and the bound loops are depicted as dark ribbons. The side chains of ArgH99 and AspH104 which are responsible for the formation of the pocket that fits the antigenic residue Asp143 in both complexes are drawn as balls and sticks (see text and Fig. 6). The backbone tracing of the peptide antigen is also shown (thin ribbon), and the Arg141-Gly142-Asp143 recognition motif is highlighted.
FIG. 6.
Views of the paratope surface in the unbound (A) and bound (B) 4C4 structures and comparison with the unbound (C) and bound (D) SD6 structures. The electrostatic potentials on these surface were calculated and drawn with GRASP (40). Positively and negatively charged regions are represented with blue and red, respectively. The RGD triplet inside the binding pocket of the complexes is also shown. In both complexes the redistribution of charges to form a pocket to accommodate Asp143 upon peptide binding is apparent.
The A15 antigen has very similar conformations in the SD6 and 4C4 complexes (Fig. 2B). The root mean square deviation between the main chain peptide atoms of residues Thr137 to Leu147 in the two complexes is only 0.5 Å, a value close to the experimental error for crystal structures determined at about 3.0-Å resolution. The high structural similarity extends to the disposition of side chains, with the sole exception of Arg141 (Fig. 2B). This residue displays weak electron density in the two peptide structures, probably due to high mobility or multiple conformations.
In the two peptide complexes the antibodies show large modifications when compared with the unbound structures (Fig. 5). The rearrangements induced by the complex formation create, in both SD6 and 4C4, a pocket that tightly fits the A15 peptide structure. The concave paratopes acquire similar shapes and a similar distributions of charged and polar groups in the two complexes, resulting in almost identical interactions with the peptide antigen (Fig. 6). The only significant difference is observed for Tyr136, a residue which was in direct contact with antibody SD6 and has not been located in the structure of the 4C4 complex (Table 1). The important antigen residue Asp143 reproduces very similar specific interactions with SD6 and 4C4. In the complex with 4C4, ArgH99 of the Fab fragment neutralizes the negative charge of the aspartate residue. ArgH99 is in turn held in place by interactions with the Fab residue AspH104, following a pattern similar to that observed in the SD6 complex (52). It is remarkable that the two Fab fragments are closer in structure in the complexes than in the unbound state. Furthermore, the residual differences in conformation that can still be identified serve to compensate for the differences in sequence, helping to produce the same types of peptide-Fab interactions in the two complexes.
Pattern of reactivity of different site A-specific antibodies with substituted antigenic peptides.
Since antibodies 4C4 and SD6 interact with peptide A15 in very similar fashions (Fig. 3 and 4), it was interesting to extend to other site A-specific antibodies an analysis of the peptide residues required for antibody recognition. To this aim, the effect of single amino acid replacements in antigenic peptide A15 on the interaction with MAbs SD6, 4C4, 5A2, 6D11, 7JD1, 7FC12, and 7CA11 was analyzed by competitive ELISA (Fig. 7). The results indicate several common features among all antibodies in their requirements for positive binding to peptide A15. In particular, the intolerance of Gly142, Asp143, and Leu144 to all or most replacements tested suggests that these residues must play some central role in binding to the different antibodies analyzed. Other residues exert effects which are specific for one or a group of antibodies. As an example, the contribution of Ala138 to antibody binding was considerable for the interaction with MAb SD6 and undetectable for that with MAb 7FC12. Also, His146, a residue found repeatedly replaced in MAb-resistant mutants of C-S8c1 (28, 32, 33), participates in the interaction with all antibodies tested, but some replacements at this site have strong effects with regard to binding to some antibodies and negligible effects on binding to other antibodies (Fig. 7). The results suggest that epitope specificity in site A of FMDV of serotype C is achieved by subtle structural alterations involving residues located around the Arg-Gly-Asp triplet.
FIG. 7.
Reactivities of substituted antigenic peptide A15 with site A-specific MAbs. Competitive ELISA were performed as described in Materials and Methods. In each panel the MAb used is indicated at the top. The residues of peptide A15 are listed in the left column, and the amino acids used for single-site substitution are listed in the top row. Relative IC50s are indicated as black (IC50 > 100), heavily striped (IC50 = 30 to 100), lightly striped (IC50 = 5 to 30), or empty (IC50 < 5) boxes. The last two columns indicate in symbolic and numerical forms the average IC50s for all tested substitutions of a given residue. Primary data for IC50 determinations will be provided upon request. The origin of each MAb and the procedure used for synthesis of replaced peptides are given in Materials and Methods.
DISCUSSION
Implications of the striking similarities of two antigen-antibody complexes.
Neutralizing MAbs SD6 and 4C4 were raised against two related but different C1 FMDVs in two different laboratories (7, 34). Despite the two antibodies defining two distinguishable epitopes within antigenic site A (32), a synthetic peptide representing this site acquired a very similar compact structure in the two complexes (Fig. 2). The high similarity of the two loop structures clearly suggests that they represent a predominant, biologically relevant conformation. Upon binding to antigen, both antibodies underwent considerable structural rearrangements to attain a similar pattern of interactions with the peptide. Remarkably, Asp143, which is part of the integrin receptor recognition triplet RGD (3, 12, 29), plays a critical role in recognition of the two antibodies. However, residues Leu144 and His146 also play an important role in the interaction (Table 1 and Fig. 4). In fact, a weak reactivity was observed between MAb 4C4 and a synthetic peptide spanning residues 144 to 156 of the G-H loop of VP1 (32). Recently, a number of MAb SD6-resistant mutants of multiply passaged C-S8c1 have been isolated and characterized (28). Interestingly, some of these mutants included substitutions at the RGD triplet and replicated normally in BHK-21 cells (28). A mutant including the substitution Asp143→Gly as the only replacement in antigenic site A failed to react, or reacted very weakly, with site A-specific MAbs (46), in agreement with the results of reactivity of these MAbs with substituted synthetic peptides (Fig. 7). These observations fully support the critical role of Asp143 in the interaction of FMDV of serotype C with MAbs directed to antigenic site A.
The immunodominance of the exposed antigenic site A (1, 25) may be reflected in the evoking of antibodies which share structural features and mode of binding to antigen. The structural similarity among antibodies could be favored by restrictions in the B-cell receptor repertoire in the process of development of B- and T-cell tolerance to self antigens (8, 13). In the present case, the self antigen would be the RGD triplet acting as a widespread cell recognition motif in a conformation similar to that found in FMDV (19, 20, 23, 26, 52). Elimination of self-reactive cells could limit the repertoire of antiviral antibodies directed to structures which include an RGD motif. The possibility of restricted modes of antigen-antibody recognition was reinforced by the study of the effects of single amino acid replacements on the interaction of antigenic peptide A15 with additional antibodies. Despite the different antibodies having been raised against different serotype C viruses, common features in the interaction with antigen are evident (Fig. 7). The immunodominance of antigenic site A extended also to the generation of neutralizing antibodies by divergent site A variants of FMDV C-S8c1 (4). In contrast to the critical role of the RGD motif of FMDV of serotype C in the interaction with antibodies, mutants of FMDV of serotype A12 lacking the RGD motif reacted with MAbs directed to site A (29, 37). This difference illustrates the complexity of the mode of interaction of aphthoviruses with antibodies and suggests that multiple molecular mechanisms may underlie antibody escape even within the same picornavirus genus.
The structural disorder of the exposed loop that constitutes site A (1, 26) can be interpreted by two nonexclusive models: (i) the G-H loop is able to move as a rigid body relative to the capsid, as some evidence suggests (18, 43), and (ii) the loop is intrinsically flexible and, like a peptide in solution, samples different conformations, each recognized by a different antibody (16, 32). Indirect evidence of this conformational sampling was provided by the observation that fusion of the G-H loop of FMDV C-S8c1 to different regions of β-galactosidase led to different effects on the binding affinity of different anti-FMDV antibodies, including some tested in the present work (2). However, the facts that the antigen structures in the complexes with two different MAbs are very similar to each other and closely related to the one found in the reduced forms of FMDVs O1BFS and O1K (24, 26) suggest that the conformational sampling may be limited to a narrow panel of structures. Furthermore, recent examination of a cyclic peptide representing site A by proton two-dimensional nuclear magnetic resonance spectroscopy has also provided evidence of a noncanonical turn at the RGD and of a nascent helix in the C-terminal part of this antigen free in solution (14). Therefore, several lines of evidence suggest that site A may undergo mostly hinge movements that preserve the internal loop structure.
Subtle structural variations underlie intratypic antigenic diversity of FMDV.
The different epitopes previously defined on antigenic site A of FMDV of serotype C (32) showed distinct degrees of conservation during the natural evolution of the virus in the field. Some epitopes have been conserved from the earliest FMDV of type C analyzed, GGC/1926, to isolates from the last decade, while other epitopes are strictly isolate specific (27, 31). The conserved RGD triplet and some neighboring residues involved in cell receptor recognition were critical for binding to each antibody tested. In contrast, substitutions within the hypervariable regions (positions 138 to 140 and 148 to 150) flanking the conserved residues (Fig. 1A) often affected recognition by only one or a few antibodies (27, 30, 32, 41). As a consequence, two different mechanisms of antigenic diversification of site A in the field were distinguished: one involved accumulation of noncritical substitutions at the hypervariable segments, and the other resulted from fixation of single, critical replacements in the central segment (27). The results reported here point to the interesting conclusion that intratypic antigenic variation of FMDV type C must generally involve subtle structural modifications which affect antigen-antibody recognition. In some cases, the two available antigen-antibody complexes offer an interpretation of the differential effects of amino acid substitutions on the binding of antigen to antibodies 4C4 and SD6. For example, both Ala138 and Ser139 show a higher percentage of residue contact area with SD6 than with 4C4 (Fig. 4B). The reactivities of the two MAbs with substituted A15 peptides show that replacements of Ala138 and Ser139 have, on average, a more pronounced effect on the interaction with SD6 than on that with 4C4 (Fig. 7). However, a quantitative assessment of these and other effects of amino acid substitutions on binding of antigen to antibodies will require appropriate modeling studies based on the structural information now available. Such studies are now in progress, and they may also contribute to defining those residues within the contact or structural epitopes which constitute the functional epitopes (30, 54) and thus participate in antigenic variation.
In a recent series of vaccination experiments, it was observed that FMDV escape variants were selected with high frequency in cattle immunized with synthetic peptides which included site A sequences (50). The structural and functional studies reported here suggest that it may be possible to reduce the frequency of escape mutants by immunization with cocktails of variant peptides. Such mixtures should be designed with careful consideration of the requirements of the virus to maintain a functional VP1 G-H loop.
The population dynamics of RNA viruses imply the frequent generation of mutations despite many of them being lethal (10, 11). Perhaps the need to preserve a functional open-turn conformation in the RGD motif has dictated that antibodies directed to site A will often belong to a class which will recognize the essential, most protruding residues. This, in turn, required a mechanism for evasion of neutralizing antibodies which involved only subtle structural modifications in order to preserve the conformation of the receptor recognition site. FMDV exists because it found the molecular mechanisms to fulfill such a compromise.
ACKNOWLEDGMENTS
We thank M. del Val for pointing out to us the possibility of B-cell repertoire restrictions and C. Escarmís and L. Menéndez-Arias for valuable suggestions.
Work at Centre de Investigació i Desenvolupament in Barcelona was supported by grants PB92-0707 and PB 95-0218 from DGICYT, that in Madrid was supported by grant PB 94-0034-C02-01 from DGICYT and Fundación Ramón Areces, and that at Universitat de Barcelona was supported by grants PB94-0845 and PB95-1131 from DGICYT. The Barcelona groups are part of CERBA from Generalitat de Catalunya.
REFERENCES
- 1.Acharya R, Fry E, Stuart D, Fox G, Rowlands D J, Brown F. The three dimensional structure of foot-and-mouth disease virus at 2.9 Å resolution. Nature. 1989;337:709–716. doi: 10.1038/337709a0. [DOI] [PubMed] [Google Scholar]
- 2.Benito A, Mateu M G, Villaverde A. Improved mimicry of a foot-and-mouth disease virus antigenic site by a viral peptide displayed on β-galactosidase surface. Bio/Technology. 1995;13:801–804. doi: 10.1038/nbt0895-801. [DOI] [PubMed] [Google Scholar]
- 3.Berinstein A, Roivainen M, Hovi T, Mason P W, Baxt B. Antibodies to the vitronectin receptor (integrin αv β3) inhibit binding and infection of foot-and-mouth disease virus to cultured cells. J Virol. 1995;69:2664–2666. doi: 10.1128/jvi.69.4.2664-2666.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borrego B, Camarero J A, Mateu M G, Domingo E. A highly divergent antigenic site of foot-and-mouth disease virus retains its immunodominance. Viral Immunol. 1995;8:11–18. doi: 10.1089/vim.1995.8.11. [DOI] [PubMed] [Google Scholar]
- 5.Brown F. New approaches to vaccination against foot-and-mouth disease. Vaccine. 1992;10:1022–1026. doi: 10.1016/0264-410x(92)90111-v. [DOI] [PubMed] [Google Scholar]
- 6.Brünger A T. XPLOR manual, version 3.0. New Haven, Conn: Yale University; 1992. [Google Scholar]
- 7.Capucci L, Brocchi E, De Simone F, Panina G F. Report of a session of the research group of the standing technical committee of the European Commission for the Control of Foot-and-Mouth Disease. Brescia, Italy: Food and Agriculture Organization, United Nations; 1984. Characterisation of monoclonal antibodies against foot-and-mouth disease virus; pp. 32–39. [Google Scholar]
- 8.Casanova J-L, Maryanski J L. Antigen-selected T-cell receptor diversity and self-nonself homology. Immunol Today. 1993;14:391–394. doi: 10.1016/0167-5699(93)90140-G. [DOI] [PubMed] [Google Scholar]
- 9.Coloma M J, Hastings A, Wims L A, Morrison S L. Novel vectors for the expression of antibody molecules using variable regions generated by polymerase chain reaction. J Immunol Methods. 1992;152:89–104. doi: 10.1016/0022-1759(92)90092-8. [DOI] [PubMed] [Google Scholar]
- 10.Domingo E, Holland J J. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:141–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 11.Domingo E, Mateu M G, Escarmís C, Martínez-Salas E, Andreu D, Giralt E, Verdaguer N, Fita I. Molecular evolution of aphthovirus. Virus Genes. 1996;11:125–135. doi: 10.1007/BF01728659. [DOI] [PubMed] [Google Scholar]
- 12.Fox G, Parry N, Barnett P V, McGinn B, Rowlands D J, Brown F. The cell attachment site on foot-and-mouth disease virus includes the amino acid sequence RGD (argine-glycine-aspartic acid) J Gen Virol. 1989;70:625–637. doi: 10.1099/0022-1317-70-3-625. [DOI] [PubMed] [Google Scholar]
- 13.Goodnow C C, Cyster J G, Hartley S B, Bell S E, Cooke M P, Healy J I, Akkaraju S, Rathmell J C, Pogue S L, Shokat K P. Self-tolerance checkpoints in B lymphocyte development. Adv Immunol. 1995;59:279–367. doi: 10.1016/s0065-2776(08)60633-1. [DOI] [PubMed] [Google Scholar]
- 14.Haack T, Camarero J A, Roig X, Mateu M G, Domingo E, Andreu D, Giralt E. A cyclic disulfide peptide reproduces in solution the main structural features of a native antigenic site of foot-and-mouth disease virus. Int J Biol Macromol. 1997;20:209–219. doi: 10.1016/s0141-8130(97)01163-x. [DOI] [PubMed] [Google Scholar]
- 15.Harber J, Bernhardt G, Lu H-H, Sgro J-Y, Wimmer E. Canyon rim residues, including antigenic determinants, modulate serotype-specific binding of polioviruses to mutants of the poliovirus receptor. Virology. 1995;214:559–570. doi: 10.1006/viro.1995.0067. [DOI] [PubMed] [Google Scholar]
- 16.Harrison S C. Finding the receptors. Nature. 1989;338:305. doi: 10.1038/338205a0. [DOI] [PubMed] [Google Scholar]
- 17.Hernández J, Valero M L, Andreu D, Domingo E, Mateu M G. Antibody and host cell recognition of foot-and-mouth disease virus (serotype C) cleaved at the Arg-Gly-Asp (RGD) motif: a structural interpretation. J Gen Virol. 1996;77:257–264. doi: 10.1099/0022-1317-77-2-257. [DOI] [PubMed] [Google Scholar]
- 18.Hewat E A, Verdaguer N, Fita I, Blakemore W, Brookes S, King A, Newman J, Domingo E, Mateu M G, Stuart D. Structure of the complex of an Fab fragment of a neutralizing antibody with foot-and-mouth disease virus: positioning of a highly mobile antigenic loop. EMBO J. 1997;16:1492–1500. doi: 10.1093/emboj/16.7.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hynes R O. Integrins: versatility modulation and signalling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 20.Jones E Y, Harlos K, Bottomley M J, Robinson R C, Driscoll P C, Edwards R M, Clements J M, Dudgeon T J, Stuart D I. Crystal structure of an integrin-binding fragment of vascular cell adhesion molecule-1 at 1.8 Å resolution. Nature. 1995;373:539–544. doi: 10.1038/373539a0. [DOI] [PubMed] [Google Scholar]
- 21.Jones T A. Interactive computer graphics: FRODO. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- 22.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 23.Krezel A M, Wagner G, Seymour-Ulmer J, Lazarus R A. Structure of the RGD protein decorsin: conserved motif and distinct function in leech proteins that affect blood clotting. Science. 1994;264:1944–1947. doi: 10.1126/science.8009227. [DOI] [PubMed] [Google Scholar]
- 24.Lea S, Abu-Ghazaleh R, Blakemore W, Curry S, Fry E, Jackson T, King A, Logan D, Newman J, Stuart D. Structural comparison of two strains of foot-and-mouth disease virus subtype O1 and a laboratory antigenic variant, G67. Structure. 1995;3:571–580. doi: 10.1016/s0969-2126(01)00191-5. [DOI] [PubMed] [Google Scholar]
- 25.Lea S, Hernández J, Blakemore W, Brocchi E, Curry S, Domingo E, Fry E, Abu-Ghazaleh R, King A, Newman J, Stuart D, Mateu M G. The structure and antigenicity of a type C foot-and-mouth disease virus. Structure. 1994;2:123–139. doi: 10.1016/s0969-2126(00)00014-9. [DOI] [PubMed] [Google Scholar]
- 26.Logan D, Abu-Ghazaleh R, Blakemore W, Curry S, Jackson T, King A, Lea S, Lewis R, Newman J, Parry N, Rowlands D, Stuart D, Fry E. Structure of a major immunogenic site of foot-and-mouth disease virus. Nature. 1993;362:566–568. doi: 10.1038/362566a0. [DOI] [PubMed] [Google Scholar]
- 27.Martínez M A, Hernández J, Piccone M E, Palma E L, Domingo E, Knowles N, Mateu M G. Two mechanisms of antigenic diversification of foot-and-mouth disease virus. Virology. 1991;184:695–706. doi: 10.1016/0042-6822(91)90439-i. [DOI] [PubMed] [Google Scholar]
- 28.Martínez M A, Verdaguer N, Mateu M G, Domingo E. Evolution subverting essentiality: dispensability of the cell attachment Arg-Gly-Asp motif in multiply passaged foot-and-mouth disease virus. Proc Natl Acad Sci USA. 1997;94:6798–6802. doi: 10.1073/pnas.94.13.6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mason P W, Rieder E, Baxt B. RGD sequence of foot-and-mouth disease virus is essential for infecting cells via the natural receptor but can be bypassed an antibody-dependent enhancement pathway. Proc Natl Acad Sci USA. 1994;91:1932–1936. doi: 10.1073/pnas.91.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mateu M G. Antibody recognition of picornaviruses and escape from neutralization: a structural view. Virus Res. 1995;38:1–24. doi: 10.1016/0168-1702(95)00048-u. [DOI] [PubMed] [Google Scholar]
- 31.Mateu M G, Da Silva J L, Rocha E, De Brum D L, Alonso A, Enjuanes L, Domingo E, Barahona H. Extensive antigenic heterogeneity of foot-and-mouth disease virus of serotype C. Virology. 1988;166:113–124. doi: 10.1016/0042-6822(88)90060-8. [DOI] [PubMed] [Google Scholar]
- 32.Mateu M G, Martínez M A, Capucci L, Andreu D, Giralt E, Sobrino F, Brocchi E, Domingo E. A single amino acid substitution affects multiple overlapping epitopes in the major antigenic site of foot-and-mouth disease virus of serotype C. J Gen Virol. 1990;71:629–637. doi: 10.1099/0022-1317-71-3-629. [DOI] [PubMed] [Google Scholar]
- 33.Mateu M G, Martínez M A, Rocha E, Andreu D, Parejo J, Giralt E, Sobrino F, Domingo E. Implications of a quasispecies genome structure: effect of frequent, naturally occurring amino acid substitutions on the antigenicity of foot-and-mouth disease virus. Proc Natl Acad Sci USA. 1989;86:5883–5887. doi: 10.1073/pnas.86.15.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mateu M G, Rocha E, Vicente O, Vayreda F, Navalpotro C, Andreu D, Pedroso E, Giralt E, Enjuanes L, Domingo E. Reactivity with monoclonal antibodies of viruses from an episode of foot-and-mouth disease. Virus Res. 1987;8:261–274. doi: 10.1016/0168-1702(87)90020-7. [DOI] [PubMed] [Google Scholar]
- 35.Mateu M G, Valero M L, Andreu D, Domingo E. Systematic replacement of amino acid residues within an Arg-Gly-Asp-containing loop of foot-and-mouth disease virus: effect on cell recognition. J Biol Chem. 1996;271:12814–12819. doi: 10.1074/jbc.271.22.12814. [DOI] [PubMed] [Google Scholar]
- 36.McCullough K C, De Simone F, Brocchi E, Capucci L, Crowther J R, Kihm U. Protective immune response against foot-and-mouth disease. J Virol. 1992;66:1835–1840. doi: 10.1128/jvi.66.4.1835-1840.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenna T S C, Lubroth J, Rieder E, Baxt B, Mason P W. Receptor binding-site-deleted foot-and-mouth disease (FMD) virus protects cattle from FMD. J Virol. 1995;69:5787–5790. doi: 10.1128/jvi.69.9.5787-5790.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer R F, Pacciarini M, Hilyard E J, Ferrari S, Vakharia V N, Donini G, Brocchi E, Molitor T W. Genetic variation of foot-and-mouth disease virus from field outbreaks to laboratory isolation. Virus Res. 1994;32:299–312. doi: 10.1016/0168-1702(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 39.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. 1994;A50:157–163. [Google Scholar]
- 40.Nicholls A, Sharp K, Honig B. GRASP—graphical representation and analysis of structural properties of proteins. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 41.Novella I S, Borrego B, Mateu M G, Domingo E, Giralt E, Andreu D. Use of substituted and tandem-repeated peptides to probe the relevance of the highly conserved RGD tripeptide in the immune response against foot-and-mouth disease virus. FEBS Lett. 1993;330:253–259. doi: 10.1016/0014-5793(93)80883-v. [DOI] [PubMed] [Google Scholar]
- 42.Otwinowki Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1996;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 43.Parry N, Fox G, Rowlands D, Brown F, Fry E, Acharya R, Logan D, Stuart D. Structural and serological evidence for a novel mechanism of antigenic variation in foot-and-mouth disease virus. Nature. 1990;347:569–572. doi: 10.1038/347569a0. [DOI] [PubMed] [Google Scholar]
- 44.Rowlands D J, Clarke B E, Carroll A R, Brown F, Nicholson B H, Bittle J L, Houghten R A, Lerner R A. Chemical basis of antigenic variation in foot-and-mouth disease virus. Nature. 1983;306:694–697. doi: 10.1038/306694a0. [DOI] [PubMed] [Google Scholar]
- 45.Rueckert R R. Picornaviridae: the viruses and their replication. In: Fields B N, et al., editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 609–654. [Google Scholar]
- 46.Ruiz-Jarabo, C. M., N. Sevilla, and E. Domingo. 1997. Unpublished results.
- 47.Smith T J, Chase E S, Schmidt T J, Olson N H, Baker T S. Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon. Nature. 1996;383:350–354. doi: 10.1038/383350a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sobrino F, Dávila M, Ortín J, Domingo E. Multiple genetic variants arise in the course of replication of foot-and-mouth disease virus in cell culture. Virology. 1983;128:310–318. doi: 10.1016/0042-6822(83)90258-1. [DOI] [PubMed] [Google Scholar]
- 49.Strohmaier K, Franze R, Adam K H. Location and characterization of the antigenic portion of the FMDV immunizing protein. J Gen Virol. 1982;59:295–306. doi: 10.1099/0022-1317-59-2-295. [DOI] [PubMed] [Google Scholar]
- 50.Taboga O, Tami C, Carrillo E, Núñez J I, Rodríguez A, Saiz J C, Blanco E, Valero M-L, Roig X, Camarero J A, Andreu D, Mateu M G, Giralt E, Domingo E, Sobrino F, Palma E L. A large-scale evaluation of peptide vaccines against foot-and-mouth disease: lack of solid protection in cattle and isolation of escape mutants. J Virol. 1997;71:2606–2614. doi: 10.1128/jvi.71.4.2606-2614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verdaguer N, Fita I, Domingo E, Mateu M G. Efficient neutralization of foot-and-mouth disease virus by monovalent antibody binding. J Virol. 1997;71:9813–9816. doi: 10.1128/jvi.71.12.9813-9816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verdaguer N, Mateu M G, Andreu D, Giralt E, Domingo E, Fita I. Structure of the major antigen loop of foot-and-mouth disease virus complexed with a neutralizing antibody: direct involvement of the Arg-Gly-Asp motif in the interaction. EMBO J. 1995;14:1690–1696. doi: 10.1002/j.1460-2075.1995.tb07158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verdaguer N, Mateu M G, Bravo J, Domingo E, Fita I. Induced pocket to accommodate the cell attachment Arg-Gly-Asp motif to a neutralizing antibody against foot-and-mouth disease virus. J Mol Biol. 1996;256:364–376. doi: 10.1006/jmbi.1996.0092. [DOI] [PubMed] [Google Scholar]
- 54.Webster D M, Henry A H, Rees A. Antibody-antigen interactions. Curr Opin Struct Biol. 1994;4:123–129. doi: 10.1016/0959-440x(94)90267-4. [DOI] [PubMed] [Google Scholar]