Abstract
Antibiotics and sedatives are used in freshwater fish culture and transportation, and residue in freshwater fish pose potential risks to human health. Therefore, a throughput method was developed to detect antibiotic and sedative residues in fish, simultaneously quantifying 68 antibiotics and 9 sedatives in freshwater fish using a modified QuEChERS extraction method and UPLC-MS/MS. Matrix-matched calibrations demonstrated good correlation coefficients (R2 > 0.995), with a recovery range of 66.2–118.5%. The intra-day and inter-day relative standard deviation (RSD) were below 9.7% and 12.8%, respectively. The limits of detection (LOD) and quantification (LOQ) were 0.08–1.46 μg/kg and 0.25–4.86 μg/kg, respectively. 68.8% of analytes had weak matrix effects, and 13.0% had moderate matrix effects. In addition, diazepam and many types of antibiotics were detected in30 freshwater fish. The validation parameters were in agreement with the acceptable criteria of the Codex guidelines. The method was effective in analyzing antibiotic and sedative residues in freshwater fish.
Keywords: Antibiotic, Sedative, Freshwater fish, UPLC-MS/MS, QuEChERS
Highlights
-
•
A UPLC-MS/MS method was developed to determine 68 antibiotics and 9 sedatives in fish.
-
•
The modified QuEChERS method was optimized.
-
•
Many antibiotics, especially enrofloxacin, and diazepam were detected in freshwater fish.
1. Introduction
With the increasing demand for aquatic products, aquaculture has developed rapidly in recent years. The breeding area and density of aquatic products, especially freshwater fish, have increased steadily (Chen, Li, Liu, Li, & Yang, 2020). Antibiotics are often used to treat and prevent bacterial infection to improve survival rates and reduce loss during aquaculture and circulation (Liu, Steele, & Meng, 2017). Furthermore, sedatives are employed to attenuate freshwater fish activity, slow down the metabolic processes to enhance their growth rate, and prevent stress and weight loss (Trushenski et al., 2013). However, excessive use of antibiotics and sedatives results in drug residue in freshwater fish, which may lead to potential human health risks by dietary consumption (Hua, Yao, Lin, Li, & Yang, 2022). Sedatives and antibiotics can cause acute and chronic toxicological hazards to human health and may lead to antibiotic resistance. The concern for antibiotic and sedative residues in freshwater fish has attracted widespread attention. Therefore, developing a simple and efficient analytical method for monitoring these drugs is significant for ensuring food safety.
Several authors have reported methods to detect antibiotic or sedative residues in food, including quinolones and tetracyclines (Grande-Martinez, Moreno-Gonzalez, Arrebola-Liebanas, Garrido-Frenich, & Garcia-Campana, 2018; Guidi et al., 2018), cephalosporins (Kim, Park, Kang, Cho, & Oh, 2020), nitroimidazoles (Chen, Delmas, Hurtaud-Pessel, & Verdon, 2020), macrolides (Dickson, 2014), sulfonamides (Bortolotte, Daniel, de Campos Braga, & Reyes, 2019), and sedatives (Hong et al., 2022; Moon, Nam, Muambo, & Oh, 2023). Several studies have proposed analytical methods focusing only on a few classes of antimicrobials or sedatives. However, methods capable of simultaneously detecting antibiotics and sedatives in freshwater fish have not been reported previously.
The different physicochemical traits of antibiotics and sedatives pose a challenge in their simultaneous detection in freshwater fish. Benzodiazepine sedatives typically exhibit weak basic properties, and their physicochemical properties, such as pKa and log Kow values, vary greatly. When exposed to acid, alkali, and heat, benzodiazepine sedatives are easily hydrolyzed and become ineffective (Carter, Williams, Martin, Kamaludeen, & Kookana, 2018). However, antibiotics possess a complex structure, often containing acidic or basic functional groups, and exhibit different physicochemical properties and polarities (Min & Lingxin, 2020). Antibiotic and sedative residues are present in food at low concentrations, and fish is rich in complex matrices such as protein lipids, phospholipids, and lipoproteins. Therefore, the accuracy and sensitivity of the method are highly dependent on the sample preparation (Reinholds, Pugajeva, Perkons, & Bartkevics, 2016). Various extraction methods have been reported, most commonly involving solid-phase extraction (SPE) cartridges and liquid-liquid extraction (Chen, Ying, & Deng, 2019). Currently, the QuEChERS (quick, easy, cheap, effective, rugged, and safe) method has been widely used in food samples pretreatment. (Bang Ye, Huang, & Lin, 2022; Yang, Lin, Liu, & Lin, 2022). The QuEChERS method has several advantages over traditional approaches, including quick sample processing, requiring less solvent, and effective analysis in complex matrices (Petrarca, Braga, Reyes, & Bragotto, 2022). Recent studies have shown that combining QuEChERS and UPLC-MS/MS has a greater advantage in detecting multi-drug residues (Lee et al., 2020).
Considering the ever-increasing demand for environmentally friendly and cost-effective analytical methods, this study aimed to develop a simple and sensitive analytical method to simultaneously detect antibiotics and sedatives in freshwater fish based on a modified QuEChERS sample preparation and a UPLC–MS/MS approach. The established detection method could contribute to monitoring antibiotic and sedative residues in freshwater fish.
2. Materials and methods
2.1. Materials and reagents
Ninety-five standards, including 9 sedatives, 9 tetracyclines, 5 nitroimidazoles, 18 quinolones, 23 sulfonamides, 9 macrolides, 4 β-lactams, and 18 isotope-internal standards were used in this study. All the standards were of high purity grade (> 95%) and were purchased from Dr. Ehrenstorfer (Augsburg, Germany), Sigma-Aldrich (St. Louis, MO, USA), and Anpel (Shanghai, China). The 77 antibiotic standards and 18 isotope-internal standards are detailed in Table 1. HPLC-grade methanol was purchased from Merck (Darmstadt, Germany). LC/MS grade formic acid and HPLC-grade acetonitrile were purchased from Thermo Fisher Scientific (Fair Lawn, NJ, USA). N-propyl-ethylenediamine (PSA) adsorbent was purchased from Dikma Technologies Inc. (Beijing, China). C18 adsorbent was obtained from Supelco (Bellefonte, PA, USA). Na2EDTA, NaCl, NaOH, and Na2SO4 were purchased from Aladdin Bio-Chem Company (Shanghai, China). QuEChERS extraction salt packet (4 g anhydrous Na2SO4, 1 g NaCl, and 1 g NaCl-4 g anhydrous Na2SO4) were purchased from Jin Yang Filter Material Company (Hebei, China). Ultra-pure water (18.2 MΩcm) was obtained using Millipore direct-Q ultra-pure water system (Millipore, MA, USA).
Table 1.
Detailed information about the retention times and MRM parameters of 95 compounds.
| Class | Compound | Formula | Retention time |
Precursor Ion | Product Ion | Cone Voltage |
Collision |
Internal standard |
|---|---|---|---|---|---|---|---|---|
| Energy | ||||||||
| (min) | (V) | (eV) | ||||||
| Quinolones | Enoxacin | C15H17FN4O3 | 5.24 | 321.4 | 232.0*/303.1 | 40 | 30/35 | Enrofloxacin-d5 |
| Orbifloxacin | C19H20F3N3O3 | 6.85 | 396.0 | 352.0*/295.0 | 30 | 17/24 | Enrofloxacin-d5 | |
| Sparfloxacin | C19H22F2N4O3 | 7.77 | 393.2 | 349.1*/292.1 | 37 | 20/24 | ||
| Fleroxacin | C17H18F3N3O3 | 4.63 | 370.1 | 326.1*/269.1 | 34 | 19/25 | Enrofloxacin-d5 | |
| Norfloxacin | C16H18FN3O3 | 5.47 | 320.2 | 302.2*/276.2 | 10 | 16/16 | Norfloxacin-d5 | |
| Ciprofloxacin | C17H18FN3O3 | 5.85 | 332.2 | 314.1*/288.2 | 10 | 18/17 | Ciprofloxacin-d8 | |
| Pefloxacin | C17H20FN3O3 | 5.21 | 334.2 | 316.2*/290.2 | 10 | 20/16 | Enrofloxacin-d5 | |
| Lomefloxacin | C17H19F2N3O3 | 6.52 | 352.2 | 265.2*/308.2 | 10 | 20/15 | Oflaxacin-d3 | |
| Danofloxacin | C19H20FN3O3 | 6.32 | 358.2 | 314.1*/96.0 | 32 | 20/25 | Oflaxacin-d3 | |
| Enrofloxacin | C19H22FN3O3 | 6.25 | 360.2 | 316.1*/245.0 | 32 | 20/22 | Enrofloxacin-d5 | |
| Oflaxacin | C18H20FN3O4 | 5.20 | 362.9 | 261.1*/318.2 | 10 | 25/17 | Oflaxacin-d3 | |
| Sarafloxacin | C20H17F2N3O3 | 7.25 | 386.2 | 342.1*/299.1 | 45 | 18/27 | Sarafloxacin-d8 | |
| Difloxacin | C21H19F2N3O3 | 6.87 | 400.2 | 299.0*/356.1 | 37 | 27/21 | Difloxacin-d5 | |
| Oxolinic acid | C13H11NO5 | 8.19 | 262.0 | 244.1*/216.0 | 30 | 19/30 | Oxolinic acid‑d5 | |
| Flumequine | C14H12FNO3 | 8.85 | 262.1 | 244.1*/220.0 | 10 | 15/30 | Oxolinic acid‑d5 | |
| Marbofloxacin | C17H19FN4O4 | 4.43 | 363.2 | 320.2*/345.2 | 20 | 13/18 | Oflaxacin-d3 | |
| Nalidixic acid | C12H12N2O3 | 8.73 | 233.2 | 215.4*/187.1 | 20 | 14/23 | ||
| Cinoxacin | C12H10N2O5 | 7.95 | 263.2 | 217.1*/245.1 | 20 | 20/14 | Oxolinic acid‑d5 | |
| Tetracyclines | 4-Epioxytetracycline | C22H24N2O9 | 5.18 | 461.3 | 426.0*/444.0 | 20 | 19/16 | Tetracycline-d6 |
| Oxytetracycline | C22H24N2O9 | 5.81 | 461.3 | 426.0*/444.0 | 20 | 19/16 | Tetracycline-d6 | |
| Chlortetracycline | C22H23ClN2O8 | 7.91 | 479.1 | 444.1*/154.0 | 20 | 20/26 | Tetracycline-d6 | |
| Demeclocycline | C21H21ClN2O8 | 7.01 | 465.1 | 448.1*430.1 | 20 | 15/20 | Tetracycline-d6 | |
| 4-Epichlortetracycline | C22H23ClN2O8 | 7.31 | 479.1 | 444.1*/98.0 | 20 | 20/42 | Tetracycline-d6 | |
| Tetracycline | C22H24N2O8 | 5.54 | 445.2 | 410.1*/154 | 22 | 19/26 | Tetracycline-d6 | |
| 4-Epitetracycline | C22H24N2O8 | 4.33 | 445.0 | 410.1*/428.0 | 20 | 19/16 | Tetracycline-d6 | |
| Doxycycline | C22H24N2O8 | 8.42 | 445.2 | 428.0*/154.0 | 20 | 20/28 | Doxycycline-d3 | |
| Methacycline | C22H22N2O8 | 8.27 | 443.0 | 426.1*/154.0 | 20 | 15/27 | Tetracycline-d6 | |
| Sulfonamides | sulfaclozine | C10H9ClN4O2S | 7.84 | 285.0 | 156.0*/92.0 | 20 | 15/28 | Sulfamethoxazole-d4 |
| sulfaphenazole | C15H14N4O2S | 7.85 | 315.0 | 156*/108 | 20 | 18/25 | Sulfamethoxazole-d4 | |
| trimethoprim | C14H18N4O3 | 4.69 | 291.3 | 123.0*/230.0 | 40 | 30/15 | ||
| sulfaguanidine | C7H10N4O2S | 1.07 | 215.0 | 156.0*/92.0 | 20 | 13/22 | Sulfapyridine-d4 | |
| Sulfapyridine | C11H11N3O2S | 3.21 | 250.0 | 156.0*/184.0 | 30 | 16/16 | Sulfapyridine-d4 | |
| Sulfadiazine | C10H10N4O2S | 2.46 | 251.0 | 156.0*/92.0 | 30 | 15/27 | Sulfapyridine-d4 | |
| Sulfamethoxazole | C10H11N3O3S | 6.10 | 254.0 | 156.0*/108.0 | 30 | 16/16 | Sulfamethoxazole-d4 | |
| Sulfathiazole | C9H9N3O2S2 | 2.90 | 256.0 | 156.0*/92.0 | 30 | 15/25 | Sulfapyridine-d4 | |
| Sulfamerazine | C11H12N4O2S | 3.55 | 265.1 | 156.0*/92.0 | 30 | 19/30 | Sulfapyridine-d4 | |
| Sulfisoxazole | C11H13N3O3S | 4.65 | 268.0 | 156.0*/92.0 | 30 | 13/28 | Sulfapyridine-d4 | |
| Sulfamoxole | C11H13N3O3S | 6.97 | 268.1 | 156.0*/108 | 30 | 15/23 | Sulfamethoxazole-d4 | |
| Sulfamethizole | C9H10N4O2S2 | 4.72 | 271.0 | 156.0*/92.0 | 30 | 15/30 | Sulfamethoxazole-d4 | |
| Sulfabenzamine | C13H12N2O3S | 7.46 | 277.1 | 156.0*/92.0 | 20 | 15/25 | Sulfamethoxazole-d4 | |
| Sulfisomidine | C12H14N4O2S | 2.51 | 279.1 | 124.0*/186.0 | 35 | 20/17 | Sulfamethoxazole-d4 | |
| Sulfamethazine | C12H14N4O2S | 4.85 | 279.1 | 186.0*/124.0 | 30 | 16/19 | Sulfadimidine-d3 | |
| Sulfamethoxypyridazine | C11H12N4O3S | 4.48 | 281.0 | 156.0*/108.0 | 20 | 15/24 | Sulfamethoxazole-d4 | |
| Sulfamonomethoxine | C11H12N4O3S | 6.21 | 281.0 | 92.0*/156.0 | 30 | 22/15 | Sulfamethoxazole-d4 | |
| Sulfametoxydiazine | C11H12N4O3S | 5.25 | 281.0 | 156.0*/108.1 | 30 | 15/26 | Sulfapyridine-d4 | |
| Sulfacholrpyridazine | C10H9ClN4O2S | 5.73 | 285.1 | 156.0*/92.0 | 30 | 15/28 | Sulfamethoxazole-d4 | |
| Sulfaquinoxaline | C14H12N4O2S | 8.24 | 301.1 | 156.0*/108.0 | 30 | 16/20 | Sulfamethoxazole-d4 | |
| Sulfadoxine | C12H14N4O4S | 6.89 | 311.1 | 156.0*/92.0.0 | 10 | 17/25 | Sulfamethoxazole-d4 | |
| Sulfadimethoxine | C12H14N4O4S | 8.09 | 311.1 | 156.0*/108.0 | 30 | 18/30 | Sulfamethoxazole-d4 | |
| Sulfacetamide | C8H10N2O3S | 1.94 | 215.2 | 156.0*/91.8 | 20 | 13/22 | Sulfapyridine-d4 | |
| Macrolides | Oleandomycin | C35H61NO12 | 8.65 | 688.4 | 158.1*/544.4 | 39 | 25/15 | Erythromycin-13Cd3 |
| Erythromycin | C37H67NO13 | 8.91 | 734.5 | 158.1*/ 576.5 | 30 | 30/20 | Erythromycin-13Cd3 | |
| Clarithromycin | C38H69NO13 | 9.16 | 748.5 | 158.1*/590.5 | 30 | 25/17 | Erythromycin-13Cd3 | |
| Azithromycin | C38H72N2O12 | 8.21 | 749.5 | 591.5*/158.1 | 30 | 25/40 | Erythromycin-13Cd3 | |
| kitasamycin | C39H65NO14 | 8.87 | 772.4 | 109.4*/174 | 20 | 35/30 | Erythromycin-13Cd3 | |
| Josamycin | C42H69NO15 | 9.08 | 828.5 | 109.0*/174.2 | 40 | 40/35 | Erythromycin-13Cd3 | |
| Spiramycin | C43H74N2O14 | 8.06 | 422.2 | 174.1*/101.0 | 30 | 20/25 | Erythromycin-13Cd3 | |
| Tilmicosin | C46H80N2O13 | 8.38 | 869.5 | 174.1*/696.5 | 25 | 45/30 | ||
| Tylosin | C46H77NO17 | 8.83 | 916.5 | 174.1* /772.2 | 50 | 40/29 | Erythromycin-13Cd3 | |
| β-lactams | Dicloxacillin | C19H17Cl2N3O5S | 9.31 | 471.0 | 160.0*/ 311.0 | 20 | 15/13 | |
| Piperacillin | C23H27N5O7S | 8.71 | 518.3 | 143.1*/160.1 | 20 | 15/10 | ||
| Flucloxacillin | C21H25N5O8S2 | 9.17 | 540.1 | 296.1*/253.1 | 20 | 18/28 | ||
| Cefathiamidine | C19H28N4O6S2 | 7.75 | 473.2 | 201.0*/ 275.1 | 20 | 20/18 | ||
| nitroimidazoles | Ronidazole | C6H8N4O4 | 2.48 | 201.0 | 140*/55 | 20.0 | 11/20 | Ronidazole-d3 |
| Ornidazole | C7H10ClN3O3 | 5.40 | 220.0 | 128*/82 | 20.0 | 15/27 | Ronidazole-d3 | |
| Tinidazole | C8H13N3O4S | 3.50 | 248.0 | 121*/128 | 20.0 | 16/20 | Ronidazole-d3 | |
| Dimetridazole | C5H7N3O2 | 2.66 | 142.0 | 96*/81 | 30.0 | 14/22 | Dimetridazole-d3 | |
| Metronidazole | C6H9N3O3 | 2.30 | 172.0 | 128*/82 | 20.0 | 13/22 | Metronidazole-d4 | |
| sedatives | Midazolam | C18H13ClFN3 | 8.54 | 326.1 | 243.9*/291.0 | 20 | 16/10 | Oxazepam-d5 |
| Alprazolam | C17H13ClN4 | 9.22 | 309.1 | 281.0*/274.1 | 20 | 26/26 | Oxazepam-d5 | |
| Clonazepam | C15H10ClN3O3 | 9.01 | 316.0 | 213.8*/240.9 | 20 | 30/25 | Oxazepam-d5 | |
| Diazepam | C16H13ClN2O | 9.52 | 285.1 | 193.1*/ 154.0 | 30 | 28/26 | Diazepam-d5 | |
| Lorazepam | C15H10Cl2N2O2 | 5.25 | 321.1 | 275.0*/302.9 | 20 | 22/14 | Oxazepam-d5 | |
| Triazolam | C17H12Cl2N4 | 9.18 | 343.1 | 308.0*/314.9 | 20 | 28/25 | Oxazepam-d5 | |
| Oxazepam | C15H11ClN2O2 | 9.24 | 287.0 | 241.1*/269.1 | 30 | 24/15 | Oxazepam-d5 | |
| Nitrazepam | C15H11N3O3 | 8.99 | 282.1 | 236.2*/254.0 | 20 | 24/19 | Oxazepam-d5 | |
| Estazolam | C16H11ClN4 | 9.12 | 295.1 | 267.0*/ 192.0 | 20 | 20/13 | Oxazepam-d5 | |
| internal standard | Sulfamethoxazole-d4 | C10H7D4N3O3S | 6.07 | 258.0 | 112.0 | 25 | 25 | |
| Sulfapyridine-d4 | C11H11N3O2S | 3.17 | 253.3 | 160.1 | 30 | 15 | ||
| Sulfadimidine-d3 | C12H11D3N4O2S | 4.85 | 285.0 | 186.0 | 20 | 17 | ||
| Tetracycline-d6 | C22H18D6N2O8 | 5.49 | 451.0 | 416.0 | 30 | 20 | ||
| Doxycycline-d3 | C22H21D3N2O8 | 8.41 | 448.1 | 431.1 | 20 | 17 | ||
| Norfloxacin-d5 | C16H13D5FN3O3 | 5.44 | 325.1 | 281.1 | 30 | 18 | ||
| Ciprofloxacin-d8 | C17H10D8FN3O3 | 5.80 | 340.2 | 235.3 | 20 | 40 | ||
| Oflaxacin-d3 | C18H17D3FN3O4 | 5.18 | 365.2 | 321.2 | 10 | 17 | ||
| Enrofloxacin-d5 | C19H17D5FN3O3 | 6.22 | 365.2 | 347.2 | 20 | 22 | ||
| Sarafloxacin-d8 | C20H9D8F2N3O4 | 7.21 | 394.1 | 376.2 | 10 | 20 | ||
| Oxolinic acid‑d5 | C13H6D5NO5 | 8.19 | 267.1 | 249.1 | 10 | 16 | ||
| Difloxacin-d5 | C21H16D3F2N3O3 | 6.86 | 405.1 | 361.2 | 10 | 18 | ||
| Erythromycin-13Cd3 | 13CC36D3H64NO13 | 8.91 | 738.4 | 162.1 | 20 | 30 | ||
| Metronidazole-d4 | C6H5D4N3O3 | 2.26 | 176.0 | 128.0 | 20 | 13 | ||
| Dimetridazole-d3 | C5H4D3N3O2 | 2.62 | 145.0 | 99.0 | 20 | 16 | ||
| Ronidazole-d3 | C6D3H5N4O4 | 2.46 | 204.0 | 143.0 | 20 | 12 | ||
| Diazepam-d5 | C16H8ClD5N2O | 9.51 | 290.1 | 198.1 | 30 | 30 | ||
| Oxazepam-d5 | C15H6ClD5N2O2 | 9.22 | 292.1 | 246.0 | 20 | 20 |
2.2. Preparation of standard solutions
Individual standard stock solutions were prepared at a concentration of 100 μg/mL. β-Lactam standards were dissolved in acetonitrile/water (50:50, v/v), while quinolone standards were dissolved in methanol (add 1% NaOH), and the other standards were dissolved in methanol. The standard stock solutions were diluted with methanol/water (50:50, v/v) to obtain the mixed working standards solution, yielding a concentration of 1.0 μg/mL for β-lactams, tetracyclines, and quinolones, 0.5 μg/mL for sedatives, macrolides, and sulfonamides, and 0.1 μg/mL for nitroimidazoles. The standard stock solutions were stored at −20 °C, and working solutions were stored at 4 °C.
2.3. Freshwater fish sample preparation
The freshwater fish samples were cleaned with deionized water. After removing the fish bones, the fish muscle and skin were homogenized using a blender, and then stored at −20°C until extraction. The modified QuEChERS method was used for sample extraction. Briefly, 2 g of homogenized sample was weighed into 50 mL centrifuge tubes and spiked with 40 μL mix-internal standards. 10 mL of 80% acetonitrile containing 0.1% acetic acid and 50 mg Na2EDTA were added to the sample and shaken for 5 min. Subsequently, 1 g NaCl was added for liquid–partitioning. Ultrasonic extraction was performed for 10 min at 4 °C and centrifugation was conducted at 9000 rpm for 5 min. Next, 6 mL of the supernatant was transferred into a 15 mL centrifuge tube containing 200 mg C18 for cleanup. After the mixture was shaken for 5 min and centrifugation at 9000 rpm for 5 min. The supernatant (5 mL) was transferred and evaporated to dryness under nitrogen at 40 °C. Finally, the residue was dissolved with 1 mL of 10% methanol containing 0.1% formic acid and filtered through a 0.22 μm filter before UPLC-MS/MS analysis.
2.4. UPLC-MS/MS analysis
The target compounds were analyzed by a Waters Acquity I-Class UPLC system connected to a Xevo TQ-XS tandem quadrupole mass spectrometer (Waters, Milford, MA, USA). The chromatographic separation was carried out with an ACQUITY UPLC BEH C18 column (100 mm × 2.1 mm, 1.7 μm, Waters, Dublin, Ireland). The mobile phase consisted of two eluents, namely aqueous 0.1% formic acid as solvent A and methanol as solvent B. The gradient elution was performed as follows:0–6.0 min, 10%B-25%B; 6.0–9.0 min, 25%B-90%B; 9.0–10.0 min, 90%B-100%B; 10.0–12.0 min, 100%B; 12.0–12.1 min, 10%B; 12.1–14.5 min 10%B. The flow rate was set to 0.3 mL/min and the column temperature was maintained at 40 °C, with an injection volume of 2 μL. Multiple-reaction monitoring (MRM) mode with positive electrospray ionization (ESI+) was utilized for mass spectrometry analysis. The optimized parameters for the ESI+ mode ionization source were performed as follows: capillary voltage 3.0 kV; cone gas flow 150 L/Hr; source temperature 150 °C; nebulizer gas flow 7.0 bar; desolvation gas flow rate 1000 L/h; collision gas flow rate 0.15 mL/min; desolvation temperature 500 °C.
2.5. Method validation
To evaluate the feasibility of the developed method,validation was performed by assessing the limit of detection (LOD), the limit of quantitation (LOQ), linearity, accuracy, precision, and selectivity based on the criteria of the Codex guideline (Commission, 1993).
The seven-point matrix-matched calibration curve was utilized to evaluate the linearity and quantification of 68 antibiotics and 9 sedatives. The peak area (y) was plotted against the concentration (x, μg/L) to construct calibration curves for compounds without isotope-internal standards (IS). The analyte/IS peak area ratio was calculated as the y-axis value (ordinate) and the analyte/IS concentration ratio as the x-axis value (abscissa) to construct calibration curves for compounds with IS. A weighting factor (1/x) was applied to improve precision at the lowest calibration points. The LOD and LOQ were determined by spiking blank samples with low concentration standards and calculating the signal-to-noise ratio (S/N) of 3:1 and 10:1, respectively. Accuracy was assessed as percent recovery, which was obtained by spiking blank fish samples with the standard at three levels (n = 6). The levels of nitroimidazoles were 0.2, 1, and 2 μg/kg; the levels of quinolones, tetracyclines, and β-lactams were 2, 10, and 20 μg/kg; the levels of all other compounds were 1, 5, and 10 μg/kg. Each level contained six replicates. Precision was evaluated by repeatability (intra-day precision) and reproducibility (inter-day precision). The three concentration levels of spiked samples were determined in six replicates on the same day to assess the intra-day precision, and the experiment was repeated over three consecutive days to evaluate inter-day precision. The method's precision was calculated as the relative standard deviation (RSD) of six replicate spiked samples at each concentration. The specificity of the method was evaluated by comparing the chromatograms of blank fish samples with spiked fish samples and examining any interfering peaks (S/N > 3) occurring at the retention time (±2.5% deviation) of the target analytes.
3. Results and discussion
3.1. Optimization of the mass spectrometric detection and liquid chromatographic separation conditions
To obtain the maximal abundance of precursor ions and product ions, the parameters for precursor and product ions, collision energy, and cleavage voltage were optimized by directly injecting 100 μg/L individual standard solutions into the mass spectrometer using a syringe pump at 10 μL/min injection speed.
The primary mass spectrometry Q1 scan was performed in positive ion mode to determine the parent ions. The secondary mass spectrometry scan was performed to select two characteristic fragment product ions as qualitative and quantitative. Detailed information on the retention times and MRM parameters for the 95 compounds was listed in Table 1.
Methanol or acetonitrile are commonly used organic solvents as mobile phases in the LC–MS/MS method. Acetonitrile, as a mobile phase, has greater elution capacity than methanol(Yang et al., 2022). Different mobile phases were compared for chromatographic separation based on previous works and literature (Barbieri et al., 2019; Khaled, Singh, & Pawliszyn, 2019; Yu et al., 2022), including the four organic phases acetonitrile, methanol, acetonitrile-methanol (2:3, V/V), and acetonitrile-methanol (5:5, V/V). Compared to other organic phases, methanol provided superior separation in sulfonamides isomers (sulfameter, sulfamethoxypyridazine, and sulfamonomethoxine) and tetracyclines isomers (4-Epioxytetracycline and oxytetracycline) due to its relatively weak elution strength. According to previous reports(Bortolotte, Daniel, & Reyes, 2021; Hong et al., 2022; Kim et al., 2020; Voronov, Falev, Ul'yanovskii, & Kosyakov, 2023), different reagents were tested for the aqueous mobile phase, including 0.1% formic acid-5 mM ammonium acetate, 0.1%formic acid, and 5 mM ammonium acetate. The results revealed that the compound was more easily ionized and showed higher sensitivity in positive ionization mode when 0.1% formic acid was added to the aqueous mobile phase. Methanol and aqueous 0.1% formic acid were selected as the mobile phase compositions. The extracted ion chromatograms for the 95 compounds in the spiked sample are illustrated in Supplementary Fig. 1. All target compounds were accurately separated within 14.5 min in optimal gradient conditions.
3.2. Optimization of the sample preparation process
Spiked fish samples were used to evaluate the extraction solvent and clean-up conditions to determine the optimal QuEChERS parameters. According to previous research reports, acidified acetonitrile has commonly been used as an extraction agent for antibiotics from animal tissues due to its ability to precipitate proteins and eliminate interference from the matrix (Kim et al., 2020). However, several β-lactams degraded when using (0.2%–1%) formic acid in acetonitrile as an extractant (Turnipseed et al., 2017).
Subsequently, the extraction effect of 0.1% formic acid was evaluated in three extraction solvents (acetonitrile, 90% acetonitrile, and 80% acetonitrile). The distribution of the recoveries obtained by using three extraction solvents is shown in Fig. 1. Higher sample recovery was achieved with 80% acetonitrile as the extraction solvent compared to the other two solvents.These findings may be attributed to adding a certain amount of water, which improved the extraction of different polar compounds. However, the sample proteins coagulated into clusters during pure acetonitrile extraction, hindering compound extraction.
Fig. 1.
Distribution of the recoveries obtained by using three extraction solvents (acetonitrile, 90% acetonitrile, and 80% acetonitrile).
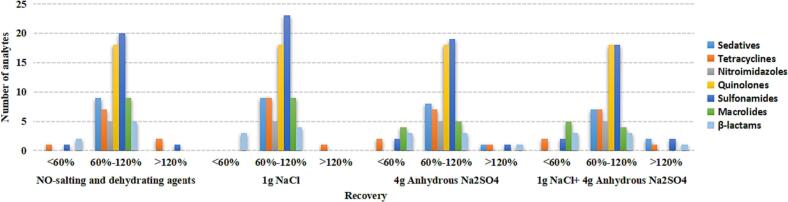
Salting-out and dehydrating agents were used to remove water and induce organic-aqueous phase separation in the QuEChERS method. NaCl, anhydrous Na2SO4, and anhydrous MgSO4 are commonly used salting and dehydrating agents. Nevertheless, a previous study reported that MgSO4 could affect the extraction due to water absorption, agglomeration, and heat generation (Wang, Tian, Ai, & Liang, 2023). Therefore, the salting-out and dehydrating effects of anhydrous Na2SO4 and NaCl were evaluated in the extraction process. We compared the effects of commonly used QuEChERS extraction salt packet (4 g anhydrous Na2SO4, 1 g NaCl, and 1 g NaCl-4 g anhydrous Na2SO4) and no salting-out and dehydrating agents on the extraction recovery. The results showed that adding 1 g NaCl as a salting-out agent exhibited good recovery for the tested compounds compared to other agents. When 4 g anhydrous Na2SO4 was added, the recovery of some macrolide antibiotics decreased (Fig. 2).
Fig. 2.
Distribution of the recoveries obtained by using four salting-out and dehydrating agents (NO-salting and dehydrating agents, 1 g NaCl, 4 g anhydrous Na2SO4, and 1 g NaCl-4 g anhydrous Na2SO4).
To reduce the matrix effect and increase recovery, the appropriate sorbent should be selected based on the physicochemical properties of the analyte. Based on previous literature reports, GCB (graphitized carbon black) is not utilized as a purification adsorbent for antibiotics due to its adsorption effect on antibiotics with benzene ring structure(Desmarchelier et al., 2018; Zhang et al., 2019). Instead, PSA and C18 are commonly employed as adsorbents for antibiotics. 5 to 10 mL of supernatant was purified using 50–200 mg of PSA or C18 adsorbent (Guo et al., 2022; Wang et al., 2023; Yang et al., 2022). Hence, this study evaluated the cleanup effect of C18 and PSA, including four kinds of purification sorbents(50 mg PSA,50 mg C18,100 mg C18, and 200 mg C18)(Fig. 3). After adding 50 mg PSA, the recoveries of β-lactam and tetracycline antibiotics showed decline. This effect was also observed in other studies, which might be attributed to the alkaline character of PSA causing low recoveries of acid analytes. (Arias et al., 2018; Guo et al., 2022). The low recoveries of the β-lactams can be explained by the interaction with PSA (Arias et al., 2018; Jung, Kim, Nam, Seo, & Yoo, 2022). The results demonstrated that good extraction recoveries were achieved with 200 mg C18 sorbent.
Fig. 3.
Distribution of the recoveries obtained by using three cleanup sorbents (50 mg PSA, 50 mg C18, 100 mg C18, 200 mg C18).
However, three β-lactams (nafcillin, penicillin G, and amoxicillin) and one tetracycline compound (minocycline) did not meet the recovery requirements (60–120%) and were removed. Finally, 68 antibiotics and 9 sedatives were successfully analyzed in freshwater fish using this method.
3.3. Method validation results
3.3.1. Linearity, LOD, and LOQ
All analytes showed good linearity, with correlation coefficients (R2) ≥ 0. 995. The ranges of the LODs and LOQs were 0.08–1.46 μg/kg and 0.25–4.86 μg/kg, respectively. The method was sensitive enough to analyze the target compounds at concentrations below the MRLs (maximum residue limits). The results of the linear range, linear equation, correlation coefficient, LOQ, LOD and MRLs were summarized in Supplementary Table 1.
3.3.2. Accuracy, precision, and specificity
The recoveries of all compounds at the three fortified levels ranged from 66.2% to 117.1% at low concentrations, from 67.9% to 118.5% at middle concentrations, and from 70.1% to 115.5% at high concentrations (Supplementary Table 1).
The intra-day precision ranged between 1.9 and 9.7%, and the inter-day value ranged between 4.1 and 12.8% (Supplementary Table 1). Moreover, the acceptable precision and accuracy indicated the reliability of the method for analyzing residues in fish.
The results of specificity showed no interference peaks around the retention times for all target compounds in blank fish samples, indicating that the method had satisfactory specificity.
3.4. Matrix effect
The matrix effect (ME), caused by the co-elution of matrix components, affects the ionization efficiency of the compound. Therefore, the ME should be evaluated to ensure accurate compound quantification. The ME was calculated as follows: ME (%) = (B/A-1) × 100, where A and B represent the slope of the standard solvent calibration and matrix-matched calibration, respectively (Khaled et al., 2019). ME < −50% or ME >50%, indicates a strong matrix inhibition or enhancement effect. In addition, −50% ≤ ME < −20% suggests a moderate matrix inhibition effect, and 50% ≥ ME >20% indicates a moderate matrix enhancement effect, while −20% ≤ ME≤20% indicates a weak matrix inhibition or enhancement effect. As shown in Fig. 4, most of the compounds showed a matrix suppression effect. 68.8% of compounds exhibited weak matrix effects, whereas 13.0% showed moderate matrix effects. In contrast, orbifloxacin, cinoxacin, sulfaguanidine, and tylosin showed strong matrix effects, ranging from −67.4 to 55.2%.
Fig. 4.
Evaluation of the matrix effects of 77 analytes in freshwater fish.
The proposed method efficiently reduces the matrix effect by utilizing isotopic-internal standards, optimizing chromatographic separation and sample processing. However, this extensive multi-residue approach did not obtain all isotopic-internal standards of each analyte. Therefore, the quantification method applied matrix-matched calibration to compensate for matrix effects.
3.5. Analysis of freshwater fish samples
To evaluate the adaptability of the developed method, freshwater fish (n = 30) were collected from supermarkets, food markets, and farmer's markets in the southeast of China from January 2023 to May 2023. The quality control procedures included sample blanks, solvent blanks, and spiking samples (n = 3),and results met the recovery criteria of 60–120%, RSD ≤ 20%, and no antibiotics detected in the blanks. The positive samples were confirmed by comparing retention times (±2.5% deviation) and relative abundance ion ratio with the standards.
Among the quinolone antibiotics, enrofloxacin was detected in seventeen samples (56.7%), with concentrations ranging from 0.69 to 17.9 μg/kg. Ciprofloxacin was detected in five samples(16.7%), with concentrations ranging from 0.39 to 1.33 μg/kg. Sarafloxacin was detected in one sample (3.3%) at a concentration of 2.46 μg/kg. Among the tetracycline antibiotics, doxycycline was detected in one sample (3.3%), with a concentration of 9.72 μg/kg. Furthermore, among the sedatives, diazepam was detected in two samples (6.7%), with concentrations ranging from 0.47 to 0.83 μg/kg. Among the sulfonamide antibiotics, trimethoprim was detected in three samples (10.0%), ranging from 0.62 to 7.29 μg/kg. Among the macrolide antibiotics, azithromycin was detected in six samples (20.0%), ranging from 0.77 to 116.0 μg/kg. β-lactams and nitroimidazole antibiotics were not detected.
3.6. Comparison with previously reported methods
Compared to previously published methods, this method is the first to simultaneously determine multiple antibiotic and sedative residues in freshwater fish. The study utilized a QuEChERS extraction procedure and isotopic-internal standards to develop a simple and quick quantification method with acceptable accuracy and precision (Table 2).
Table 2.
Comparison of previously published methods with the proposed method for the detection of multiple antibiotics and sedatives in freshwater fish.
| Compound | Total compound | Extraction method | Instrument method | LOD (μg/kg) | LOD (μg/kg) | Recovery(%) | Ref. |
|---|---|---|---|---|---|---|---|
| 10sedatives | 10 | QuEChERS | LC-MS/MS | 0.003–0.19 | 0.01–0.58 | 85–120.0 | Hong et al., 2022 |
| 5 tetracyclines | 5 | QuEChERS | UPLC–MS/MS | 0.5–1.2 | 1.7–4.4 | 80.1–105.0 | Grande-Martinez et al., 2018 |
| 2 β-lactams, 8 quinolones, 2 macrolides, 6 sulfonamides, 4 tetracyclines, 3 amphenicols | 25 | QuEChERS | UPLC–MS/MS | 0.1–3.0 | 0.30–10.0 | 71.2–119.6 | Bortolotte et al., 2019 |
| 4 sulfonamides, 11 quinolones, 4 tetracyclines, 2 malachite green | 21 | QuEChERS | UPLC–MS/MS | 0.1–0.8 | 0.2–2.5 | 67.17–109.2 | Chen, Wei, & Cao, 2019 |
| 18sulfonamides | 18 | on–line SPE | UPLC–MS/MS | 0.00146–0.0155 | 0.00490–0.0516 | 71.5–102.0 | Li, Wang, Xu, & Chakraborty, 2020 |
| 5 tetracyclines, 4 sulfonamides, 9 quinolones, 1dapson | 19 | SPE | UPLC-QTOF-MS | 2.22–15.00 | 6.67–45.46 | 93.8–107.5 | Vardali, Samanidou, & Kotzamanis, 2018 |
| 11 β-lactams, 9 quinolones, 6 sulfonamides, 7 macrolides, 4 tetracyclines, 5 synthetic antibiotics | 42 | SPE | UPLC–MS/MS | 0.1–3.0 | 0.3–9.0 | 99.0–112.0 | Reinholds et al., 2016 |
| 16 sulfonamides, 2 tetracyclines, 11 quinolones, 7 nitroimidazoles, 3 amphenicols, 5 steroids, 3 stilbenes | 47 | LLE | UPLC–MS/MS | 0.1–5.0 | 0.33–16.7 | 60.0–148.0 | Gibbs et al., 2018 |
| 4 β-lactams, 6 quinolones, 5 sulfonamides, 2 macrolides, 4 tetracyclines, 3 amideols | 24 | LLE | UPLC–Q-Exactive-Orbitrap-MS | 1.00 | 2.00 | 91.1–105.6 | Chiesa et al., 2018 |
| 9 sedatives, 9 tetracyclines, 5nitroimidazoles, 18quinolones, 23 sulfonamides,9 macrolides, 4 β-lactams | 79 | QuEChERS | UPLC–MS/MS | 0.08–1.46 | 0.25–4.86 | 65.2–118.5 | the present method |
* QuEChERS (quick, easy, cheap, effective, rugged, and safe) extraction; SPE, solid-phase extraction; LLE, liquid-liquid extraction.
4. Conclusions
A simple, fast, and sensitive method was developed to quantify 68 antibiotics and 9 sedatives in freshwater fish samples. Sample extraction was easily performed by a modified QuEChERS method, effectively removing interferences, and saving time. The method applies isotopic-internal standard quantification, making stable results with good accuracy and precision. Many antibiotics, especially enrofloxacin (found in 56.7% of freshwater fish samples) and diazepam were detected in freshwater fish. Therefore, the proposed QuEChERS-UPLC-MS/MS method is appropriate for simultaneously monitoring multi-class antibiotic and sedative residues in freshwater fish samples.
Funding
This work was supported by the fund of the Fujian Health Innovation Project (No.2022CXA035), the Natural Science Foundation of Fujian Province (No.2020J01092) and Construction of Fujian Provincial Scientific and Technological Innovation Platform (No.2019Y2001).
CRediT authorship contribution statement
Yan Yang: Writing – review & editing, Writing – original draft, Supervision, Software. Xin Li: Data curation. Jian Lin: Validation. Rong Bao: Methodology.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2024.101268.
Appendix A. Supplementary data
Supplementary material
Data availability
Data will be made available on request.
References
- Arias J.L.O., Schneider A., Batista-Andrade J.A., Vieira A.A., Caldas S.S., Primel E.G. Chitosan from shrimp shells: A renewable sorbent applied to the clean-up step of the QuEChERS method in order to determine multi-residues of veterinary drugs in different types of milk. Food Chemistry. 2018;240:1243–1253. doi: 10.1016/j.foodchem.2017.08.041. [DOI] [PubMed] [Google Scholar]
- Bang Ye S., Huang Y., Lin D.Y. QuEChERS sample pre-processing with UPLC-MS/MS: A method for detecting 19 quinolone-based veterinary drugs in goat’s milk. Food Chemistry. 2022;373(Pt B) doi: 10.1016/j.foodchem.2021.131466. [DOI] [PubMed] [Google Scholar]
- Barbieri M.V., Postigo C., Guillem-Argiles N., Monllor-Alcaraz L.S., Simionato J.I., Stella E.…López de Alda M. Analysis of 52 pesticides in fresh fish muscle by QuEChERS extraction followed by LC-MS/MS determination. Science of the Total Environment. 2019;653:958–967. doi: 10.1016/j.scitotenv.2018.10.289. [DOI] [PubMed] [Google Scholar]
- Bortolotte A.R., Daniel D., de Campos Braga P.A., Reyes F.G.R. A simple and high-throughput method for multiresidue and multiclass quantitation of antimicrobials in pangasius (Pangasionodon hypophthalmus) fillet by liquid chromatography coupled with tandem mass spectrometry. Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences. 2019;1124:17–25. doi: 10.1016/j.jchromb.2019.05.034. [DOI] [PubMed] [Google Scholar]
- Bortolotte A.R., Daniel D., Reyes F.G.R. Occurrence of antimicrobial residues in tilapia (Oreochromis niloticus) fillets produced in Brazil and available at the retail market. Food Research International. 2021;140 doi: 10.1016/j.foodres.2020.109865. [DOI] [PubMed] [Google Scholar]
- Carter L.J., Williams M., Martin S., Kamaludeen S.P.B., Kookana R.S. Sorption, plant uptake and metabolism of benzodiazepines. Science of the Total Environment. 2018;628-629:18–25. doi: 10.1016/j.scitotenv.2018.01.337. [DOI] [PubMed] [Google Scholar]
- Chen D., Delmas J.M., Hurtaud-Pessel D., Verdon E. Development of a multi-class method to determine nitroimidazoles, nitrofurans, pharmacologically active dyes and chloramphenicol in aquaculture products by liquid chromatography-tandem mass spectrometry. Food Chemistry. 2020;311 doi: 10.1016/j.foodchem.2019.125924. [DOI] [PubMed] [Google Scholar]
- Chen J., Wei Z., Cao X.-Y. QuEChERS Pretreatment Combined with Ultra-performance Liquid Chromatography–Tandem Mass Spectrometry for the Determination of Four Veterinary Drug Residues in Marine Products. Food Analytical Methods. 2019;12(5):1055–1066. doi: 10.1007/s12161-018-01431-1. [DOI] [Google Scholar]
- Chen J., Ying G.G., Deng W.J. Antibiotic residues in food: Extraction, analysis, and human health concerns. Journal of Agricultural and Food Chemistry. 2019;67(27):7569–7586. doi: 10.1021/acs.jafc.9b01334. [DOI] [PubMed] [Google Scholar]
- Chen L., Li H., Liu Y., Li Y., Yang Z. Occurrence and human health risks of twenty-eight common antibiotics in wild freshwater products from the Xiangjiang River and comparison with the farmed samples from local markets. Food Additives & Contaminants. Part A, Chemistry, Analysis, Control, Exposure & Risk Assessment. 2020;37(5):770–782. doi: 10.1080/19440049.2020.1730987. [DOI] [PubMed] [Google Scholar]
- Chiesa L., Panseri S., Pasquale E., Malandra R., Pavlovic R., Arioli F. Validated multiclass targeted determination of antibiotics in fish with high performance liquid chromatography-benchtop quadrupole orbitrap hybrid mass spectrometry. Food Chemistry. 2018;258:222–230. doi: 10.1016/j.foodchem.2018.03.072. [DOI] [PubMed] [Google Scholar]
- Commission C.A. Vol. 16. CAC/GL; 1993. Codex guidelines for the establishment of a regulatory programme for control of veterinary drug residues in foods. Part III attributes of analytical methods for residue of veterinary drugs in foods; p. 41. [Google Scholar]
- Desmarchelier A., Fan K., Minh Tien M., Savoy M.C., Tarres A., Fuger D.…Mottier P. Determination of 105 antibiotic, anti-inflammatory, antiparasitic agents and tranquilizers by LC-MS/MS based on an acidic QuEChERS-like extraction. Food Additives & Contaminants. Part A, Chemistry, Analysis, Control, Exposure & Risk Assessment. 2018;35(4):646–660. doi: 10.1080/19440049.2018.1429677. [DOI] [PubMed] [Google Scholar]
- Dickson L.C. Performance characterization of a quantitative liquid chromatography-tandem mass spectrometric method for 12 macrolide and lincosamide antibiotics in salmon, shrimp and tilapia. Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences. 2014;967:203–210. doi: 10.1016/j.jchromb.2014.07.031. [DOI] [PubMed] [Google Scholar]
- Gibbs R.S., Murray S.L., Watson L.V., Nielsen B.P., Potter R.A., Murphy C.J. Development and validation of a hybrid screening and quantitative method for the analysis of eight classes of therapeutants in aquaculture products by liquid chromatography-tandem mass spectrometry. Journal of Agricultural and Food Chemistry. 2018;66(20):4997–5008. doi: 10.1021/acs.jafc.7b05357. [DOI] [PubMed] [Google Scholar]
- Grande-Martinez A., Moreno-Gonzalez D., Arrebola-Liebanas F.J., Garrido-Frenich A., Garcia-Campana A.M. Optimization of a modified QuEChERS method for the determination of tetracyclines in fish muscle by UHPLC-MS/MS. Journal of Pharmaceutical and Biomedical Analysis. 2018;155:27–32. doi: 10.1016/j.jpba.2018.03.029. [DOI] [PubMed] [Google Scholar]
- Guidi L.R., Santos F.A., Ribeiro A., Fernandes C., Silva L.H.M., Gloria M.B.A. Quinolones and tetracyclines in aquaculture fish by a simple and rapid LC-MS/MS method. Food Chemistry. 2018;245:1232–1238. doi: 10.1016/j.foodchem.2017.11.094. [DOI] [PubMed] [Google Scholar]
- Guo X., Tian H., Yang F., Fan S., Zhang J., Ma J.…Zhang Y. Rapid determination of 103 common veterinary drug residues in milk and dairy products by ultra performance liquid chromatography tandem mass spectrometry. Frontiers in Nutrition. 2022;9 doi: 10.3389/fnut.2022.879518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Kwon N., Kang H.S., Jang E., Kim H., Han E. Development of an analytical method for detection of anesthetics and sedatives in fish. Journal of AOAC International. 2022;105(3):774–783. doi: 10.1093/jaoacint/qsab155. [DOI] [PubMed] [Google Scholar]
- Hua Y., Yao Q., Lin J., Li X., Yang Y. Comprehensive survey and health risk assessment of antibiotic residues in freshwater fish in Southeast China. Journal of Food Composition and Analysis. 2022;114 doi: 10.1016/j.jfca.2022.104821. [DOI] [Google Scholar]
- Jung Y.S., Kim D.B., Nam T.G., Seo D., Yoo M. Identification and quantification of multi-class veterinary drugs and their metabolites in beef using LC-MS/MS. Food Chemistry. 2022;382 doi: 10.1016/j.foodchem.2022.132313. [DOI] [PubMed] [Google Scholar]
- Khaled A., Singh V., Pawliszyn J. Comparison of solid-phase microextraction to solvent extraction and QuEChERS for quantitative analysis of veterinary drug residues in chicken and beef matrices. Journal of Agricultural and Food Chemistry. 2019;67(46):12663–12669. doi: 10.1021/acs.jafc.9b01570. [DOI] [PubMed] [Google Scholar]
- Kim J., Park H., Kang H.S., Cho B.H., Oh J.H. Comparison of sample preparation and determination of 60 veterinary drug residues in flatfish using liquid chromatography-tandem mass spectrometry. Molecules. 2020;25(5) doi: 10.3390/molecules25051206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Min A.Y., Han J.H., Yang Y.J., Kim H., Shin D. Development and validation of LC-MS/MS method with QuEChERS clean-up for detecting cannabinoids in foods and dietary supplements. Food Additives & Contaminants. Part A, Chemistry, Analysis, Control, Exposure & Risk Assessment. 2020;37(9):1413–1424. doi: 10.1080/19440049.2020.1769200. [DOI] [PubMed] [Google Scholar]
- Li T., Wang C., Xu Z., Chakraborty A. A coupled method of on-line solid phase extraction with the UHPLC–MS/MS for detection of sulfonamides antibiotics residues in aquaculture. Chemosphere. 2020;254 doi: 10.1016/j.chemosphere.2020.126765. [DOI] [PubMed] [Google Scholar]
- Liu X., Steele J.C., Meng X.Z. Usage, residue, and human health risk of antibiotics in Chinese aquaculture: A review. Environmental Pollution. 2017;223:161–169. doi: 10.1016/j.envpol.2017.01.003. [DOI] [PubMed] [Google Scholar]
- Min L., Lingxin C. Advances in sample pretreatment techniques for analysis of antibiotics in the coastal environment. Chinese Journal of Chromatography. 2020 doi: 10.3724/sp.j.1123.2019.07005. [DOI] [PubMed] [Google Scholar]
- Moon H., Nam A.J., Muambo K.E., Oh J.E. Simultaneous multi-residue analytical method for anesthetics and sedatives in seafood samples by LC-ESI/MSMS. Food Chemistry. 2023;404(Pt B) doi: 10.1016/j.foodchem.2022.134157. [DOI] [PubMed] [Google Scholar]
- Petrarca M.H., Braga P.A.C., Reyes F.G.R., Bragotto A.P.A. Exploring miniaturized sample preparation approaches combined with LC-QToF-MS for the analysis of sulfonamide antibiotic residues in meat- and/or egg-based baby foods. Food Chemistry. 2022;366 doi: 10.1016/j.foodchem.2021.130587. [DOI] [PubMed] [Google Scholar]
- Reinholds I., Pugajeva I., Perkons I., Bartkevics V. The application of phospholipid removal columns and ultra-high performance liquid chromatography-tandem quadrupole mass spectrometry for quantification of multi-class antibiotics in aquaculture samples. Journal of Pharmaceutical and Biomedical Analysis. 2016;128:126–131. doi: 10.1016/j.jpba.2016.05.002. [DOI] [PubMed] [Google Scholar]
- Trushenski J.T., Bowker J.D., Cooke S.J., Erdahl D., Bell T., MacMillan J.R.…Sharon S. Issues regarding the use of sedatives in fisheries and the need for immediate-release options. Transactions of the American Fisheries Society. 2013;142(1):156–170. doi: 10.1080/00028487.2012.732651. [DOI] [Google Scholar]
- Turnipseed S.B., Storey J.M., Lohne J.J., Andersen W.C., Burger R., Johnson A.S., Madson M.R. Wide-scope screening method for multiclass veterinary drug residues in fish, shrimp, and eel using liquid chromatography-quadrupole high-resolution mass spectrometry. Journal of Agricultural and Food Chemistry. 2017;65(34):7252–7267. doi: 10.1021/acs.jafc.6b04717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardali S.C., Samanidou V.F., Kotzamanis Y.P. Development and validation of an ultra performance liquid chromatography-quadrupole time of flight-mass spectrometry (in MS(E) mode) method for the quantitative determination of 20 antimicrobial residues in edible muscle tissue of European sea bass. Journal of Chromatography. A. 2018;1575:40–48. doi: 10.1016/j.chroma.2018.09.017. [DOI] [PubMed] [Google Scholar]
- Voronov I.S., Falev D.I., Ul’yanovskii N.V., Kosyakov D.S. Suspect screening and semi-quantification of macrolide antibiotics in municipal wastewater by high-performance liquid chromatography—Precursor ion scan tandem mass spectrometry. Chemosensors. 2023;11(1) doi: 10.3390/chemosensors11010044. [DOI] [Google Scholar]
- Wang H., Tian H., Ai L.F., Liang S.X. Screening and quantification of 146 veterinary drug residues in beef and chicken using QuEChERS combined with high performance liquid chromatography-quadrupole orbitrap mass spectrometry. Food Chemistry. 2023;408 doi: 10.1016/j.foodchem.2022.135207. [DOI] [PubMed] [Google Scholar]
- Yang Y., Lin G., Liu L., Lin T. Rapid determination of multi-antibiotic residues in honey based on modified QuEChERS method coupled with UPLC-MS/MS. Food Chemistry. 2022;374 doi: 10.1016/j.foodchem.2021.131733. [DOI] [PubMed] [Google Scholar]
- Yu X., Wu X., Xie Y., Tong K., Wang M., Li J.…Chen H. Development and validation of a method for determination of 43 antimicrobial drugs in Western-style pork products by UPLC-MS/MS with the aid of experimental design. Molecules. 2022;27(23) doi: 10.3390/molecules27238283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Deng Y., Zheng J., Zhang Y., Yang L., Liao C.…Luo A. The application of the QuEChERS methodology in the determination of antibiotics in food: A review. TrAC Trends in Analytical Chemistry. 2019;118:517–537. doi: 10.1016/j.trac.2019.06.012. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.