Abstract
Background
Caffeic acid (CA) is a naturally occurring phenolic compound with diverse pharmacologic properties. CA plays a crucial role in hemostasis by increasing platelet count. However, the mechanism by which CA regulates platelets to promote hemostasis remains unclear.
Objectives
We aim to identify the potential target pathways and genes by which CA regulates platelets to promote hemostasis.
Methods
We performed RNA sequencing (RNA-seq) analysis of mouse platelet pools in both the CA-gavaged group and phosphate-buffered saline–gavaged group.
Results
The 12,934 expressed transcripts had been annotated after platelet RNA-seq. Compared with the phosphate-buffered saline group, 987 differentially expressed genes (DEGs) were identified, of which 466 were downregulated and 521 were upregulated in CA group. Gene Ontology, Kyoto Encyclopedia of Genes and Genomes, and Reactome gene set enrichment analysis demonstrated that upregulated DEGs were enriched in the pathways of hemostasis, platelet activation, signaling, aggregation, and degranulation. Moreover, Kyoto Encyclopedia of Genes and Genomes and Reactome gene set enrichment analysis revealed that 5 of the 25 cosignificantly upregulated DEGs were essential in CA-mediated platelet regulation to promote hemostasis.
Conclusion
Our findings of platelet RNA-seq analysis demonstrate that CA regulates the gene expression of hemostasis and platelet activation–related pathways to increase platelet count and promote hemostasis. It will also provide reference molecular resources for future research on the function and mechanism by which CA regulates platelets to promote hemostasis.
Keywords: caffeic acid, differentially expressed genes, gene set enrichment analysis, hemostasis, platelet, RNA-seq
Graphical abstract
Essentials
-
•
Caffeic acid (CA) plays an important role in the process of hemostasis.
-
•
Platelet RNA sequencing reveals CA affects hemostasis and platelet activation–related pathways.
-
•
Platelet RNA sequencing reveals the 5 potential target genes of CA affecting hemostasis.
-
•
CA regulates hemostasis and platelet activation-related pathways and genes to promote hemostasis.
1. Introduction
Caffeic acid (CA), a chemical structure of 3,4-dihydroxycinnamic acid (Figure 1A), is a naturally occurring phenolic compound widely found in numerous fruits, vegetables, and herbal medicines [1]. CA has various pharmacologic activities, including antioxidant [[2], [3], [4]], antibacterial [[4], [5], [6]], antiinflammatory [7,8], anticancer [4,9,10], and neuroprotective [11,12] properties. Currently, the mechanisms underlying these beneficial properties of CA are still not fully understood. CA plays a crucial role in hemostasis by increasing platelet count [[13], [14], [15], [16]], especially for treating thrombocytopenia caused by various causes [17]. However, the specific mechanism by which CA regulates platelets to promote hemostasis remains unclear.
Figure 1.
Overview of the study protocol and associated metrics. (A) The chemical structure and appearance of caffeic acid (CA). (B) CA administration was performed by mouse gavage experiments. Red boxes indicate mice in the CA group that were gavaged 18 mg CA/kg daily for 7 days, and green boxes indicate mice in the phosphate-buffered saline (PBS) group that were gavaged the same volume of PBS for 7 days. (C) The program overview was as follows: identify the target pathways and genes in platelets that are regulated by CA to promote hemostasis. (D) The first 3 drops of blood were discarded, and the next 3 drops of blood were collected and diluted with acid-citrate dextrose anticoagulant for platelet count. (E) The mouse tail bleeding time assay. (F) The cartridge membrane with collagen and adenosine diphosphate-coated membrane (C/ADP) was selected for platelet function assays. Red dots indicate mice in the CA group, and green dots indicate mice in the PBS group. Error bars indicate the SEM, and significant differences (P < .05) between the CA and PBS groups are indicated with an asterisk (∗). (G) Fluorescein isothiocyanate-labeled CD62P or phycoerythrin-labeled JON/A (GPIIbIIIa, integrin αIIbβ3) were used to evaluate platelet activation indicators. Red indicates the 3 platelet pools in the CA group, and green indicates the 3 platelet pools in the PBS group. DEGs, differentially expressed genes; GO, Gene Ontology; GSEA, gene set enrichment analysis (GSEA); KEGG, Kyoto Encyclopedia of Genes and Genomes; qPCR, quantitative polymerase chain reaction.
Platelets are anucleated and simplified cells produced by the cytoplasmic lysis of megakaryocytes [18]. Platelets also play a crucial role in hemostasis and are key regulators of thrombosis, inflammation, and immunity [[18], [19], [20], [21], [22]]. However, anucleated platelets internally contain megakaryocyte-derived mRNA and all components required for mRNA processing and translation [[23], [24], [25]], whose RNA can be exchanged with other cells [26]. We hypothesize that CA regulates the gene expression of platelet-related pathways to increase platelet count and promote hemostasis.
RNA sequencing (RNA-seq) has a high signal-to-noise ratio and wide application, making it an essential experimental method for analyzing differentially expressed genes (DEGs) at the transcriptional level [27]. Blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics can be achieved using RNA-seq analysis of tumor-educated platelets [28]. RNA-seq analysis of venous blood platelets can be used to identify the DEGs in diabetic kidney disease [29]. A prothrombotic profile was revealed using RNA-seq analysis of reticulated platelets [30]. Several DEGs were found by performing genome-wide RNA-seq analysis of human and mouse platelet transcriptomes [31]. These reasons above allowed our study to identify the potential target pathways and genes by which CA regulates platelets to promote hemostasis by performing RNA-seq analysis of mouse platelet pools in CA-gavaged group and phosphate-buffered saline (PBS)-gavaged group (CA vs PBS group).
2. Methods
2.1. Reagents
CA (purity 99.99%) was provided by Dezhou Deyao Pharmaceutical Co, Ltd. Its appearance is a pale yellow, nearly water-insoluble powder with a slightly sour taste (Figure 1A). Using dimethyl sulfoxide or ethanol can improve the solubility of CA [32] but introduces new variables to the experiment. Therefore, we formulated 3-mg/mL CA and PBS suspension for gavage experiments in this study. PBS was obtained from Gibco (Thermo Fisher Scientific).
2.2. Animals
Specific pathogen-free C57BL/6 male and female mice, aged 6 to 8 weeks and weighing 23 ± 2 g, were purchased from the laboratory animal center of Huazhong University of Science and Technology (HUST). All animal experiments have followed Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines (https://arriveguidelines.org/). Also, all experimental procedures were approved by the local animal ethics committee and conducted following the Code of Practice for the Care and Use of Animals for Scientific Purposes described by the Medical Research Committee of Tongji Medical College, HUST. At the end of the experiment, all animals were killed by anesthetic overdose.
2.3. Mouse gavage experiment
We performed 2 batches of mouse gavage experiments. One batch was used for mouse platelet pools RNA-seq analysis; the other batch was used for quantitative polymerase chain reaction (qPCR) validation, mouse tail bleeding time, and platelet functional assays (Figure 1C). The mice were randomly divided into different groups (treatment group and control group) of 6 female and 6 male mice in 2 cages each. After 2-3 days of normal feeding, the mice in treatment group were gavaged 18 mg CA/kg daily for 7 days. Similarly, the mice in control group were gavaged the same volume of PBS for 7 days (Figure 1B).
2.4. Mouse tail bleeding time assays
The mouse tail bleeding time assay was carried out according to the method of Liu et al. [33]. The tail bleeding time was monitored using a stop clock; each mouse was only monitored for 20 minutes, and over 20 minutes was calculated as 20 minutes.
2.5. Blood collection and platelet count
Standardized blood collection procedures are usually required to avoid mild platelet activation in vitro [34,35]. Chloral hydrate (250 mg/kg) was used for mouse anesthesia. As much whole blood was collected as possible from each mouse to isolate and purify platelets. It was inconvenient to collect a large amount of whole blood by orbital vein puncture and cardiac puncture methods, which could easily lead to platelet activation and affect the accuracy of subsequent platelet RNA-seq data. Therefore, we used the blood collection method (carotid artery cannulation, ADInstruments) that caused the least platelet activation. For each mouse, the first 3 drops of blood were discarded, and the next 3 drops were collected and diluted with acid-citrate dextrose (ACD) anticoagulant for platelet count (KT6200VET, Genrui Biotech) before collecting approximately 1.5 mL ACD anticoagulant whole blood for subsequent experiments.
2.6. Platelet functional assays
Platelet function assays were performed using the platelet function analyzer (PFA)-200 (Siemens Healthcare). The PFA is a popular platelet function and primary hemostasis screening instrument with test results reported as closure time (CT) [36,37]. The cartridge membrane with collagen and adenosine diphosphate-coated membrane (C/ADP; Siemens Healthcare, Cat# B4170-21) was selected for platelet function assays.
2.7. Platelet isolation and purification
During the process of mouse platelet isolation and purification, the addition of prostaglandin E1 could reduce platelet aggregation and activation [38]. Thus, according to the platelet isolation and purification process described by Rowley et al. [31], prostaglandin E1 (300 μM) was applied in the relevant solutions, and Anti-Ter-119 MicorBeads (Miltenyi Biotec, Cat# 130-049-901) and CD45 MicorBeads (Miltenyi Biotec, Cat# 130-052-301) were used to remove residual red blood cells and white blood cells in platelets, respectively. Briefly, 3 platelet pools (2 female and 2 male mice isolated platelets per pool) were harvested as 3 samples (CA1, CA2, and CA3) in CA group; the other 3 samples (PBS1, PBS2, and PBS3) were harvested in PBS group.
2.8. Platelet activation analysis
BD FACSVia Flow cytometry (BD Biosciences) was used to detect platelet activation in 6 platelet pools during carotid artery cannulation and platelet isolation. Allophycocyanin-labeled CD61 (integrin β3, GPIIIa) antibody (BioLegend, Cat# 104316) was used to label all platelets. Fluorescein isothiocyanate-labeled CD62P (P-selectin, Selp) antibody (BD Biosciences, Cat# 553744) and phycoerythrin-labeled JON/A (GPIIbIIIa, integrin αIIbβ3) antibody (Emfret Analytics, Cat# M023-2) were used to label activated platelets.
2.9. RNA extraction, library construction, and sequencing
Trizol reagent (Invitrogen, Cat# 15596026CN) was used to extract the total RNA. Agilent 2100 Bioanalyzer (Agilent Technologies) was used to detect the quality and concentration of each RNA sample. In the first batch of mouse gavage experiments, 6 platelet pool samples (CA1, CA2, CA3, PBS1, PBS2, and PBS3) and 500 ng total RNA per sample were used for RNA library construction (Figure 1D). Next, the 6 RNA libraries were sequenced by Novogene (China) using the Novaseq 6000 (Illumina) with a sequencing read length of 150 bp paired-end reads (PE150).
2.10. RNA reverse transcription and quantitative polymerase chain reaction validation
In the second batch of mouse gavage experiments, using the same method, the next 6 platelet pool samples (CA4, CA5, CA6, PBS4, PBS5, and PBS6) were obtained for qPCR validation. RNA (500 ng per sample) was used for reverse transcription. The HiScript III RT SuperMix for qPCR (+gDNA wiper) Kit (Vazyme Biotech, Cat# R323-01) was selected for the synthesis of full-length cDNA. Using the Primer Premier 5 software (Premier Biosoft), a pair of specific primers was designed for each DEG (Table 1) and sent to Genecreate (China) for synthesis. The qPCR validation was performed using the ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech, Cat# Q711) and CFX96 Deep Well Real-Time System (BioRad). The qPCR validation data were processed using the 2–ΔΔCt method [39] after normalization to the housekeeping gene Actb.
Table 1.
Specific primer sequences of 7 differentially expressed genes and housekeeping gene Actb used for quantitative polymerase chain reaction validation.
| Gene name | Forward primer sequence (5' to 3') | Reverse primer sequence (5' to 3') |
|---|---|---|
| Tln1 | TAACCGTGAGGGTGCTGAAA | CGGCTTGTGCCAACTTCTC |
| Flna | CCCAGGATTTTCACCCAGAC | AGAGGAGCCTTCCCAGCAT |
| Vwf | TTCCAGCCTGCGGACATT | TCCTTTTGCTACGGTGAGACA |
| Thbs1 | GCCCCAACCTTCCCAACT | GCGGTCTCCCACATCATCTC |
| Itga6 | GCTTCCTCGTTTGGCTATGAT | TTAATGTAGACGTAAACTGCACCC |
| Ptpn11 | GAATGACTTCTGGCGGATGG | GGGCGGCACTTTCTTTGA |
| Itgb3 | CGGGAAAATCCGCTCTAAA | AGCTCACCGTGTCTCCAATCT |
| Actba | CAACTGGGACGACATGGA | CCATCACAATGCCTGTGG |
The housekeeping gene was used for quantitative polymerase chain reaction validation.
2.11. RNA sequencing analysis
The original image files obtained from RNA-seq were converted using CASAVA base recognition to obtain sequence data stored in the fastq format, known as raw reads. For quality control, FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to obtain clean reads, and the clean reads were aligned to the reference mouse genome (GRCm38) from Ensemble (http://ftp.ensembl.org/pub/release-102/fasta/mus_musculus/dna/Mus_musculus.GRCm38.dna_sm.primary_assembly.fa.gz) for gene annotation using Hisat2 (v2.0.5), and featureCounts (1.5.0-p3) was used to normalize and calculate the expression of each gene with the fragments per kilobase million method. DESeq2 (v1.20.0) was used to analyze the DEGs in CA vs PBS group. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Reactome gene set enrichment analysis (GSEA) of DEGs were implemented using the clusterProfiler R package (v3.8.1), and normalized enrichment score (NES) of each GSEA data set was implemented using the local version of GSEA tool (https://www.gsea-msigdb.org/gsea/index.jsp).
2.12. Statistical analysis
Statistical analysis was done using R (3.4.1). Benjamini and Hochberg correction was performed for multiple comparison corrections [40]. When performing DEG analysis with DESeq2 (v1.20.0), a P value ≤ .05 and | log2-fold change in CA vs PBS group | > 0 were set as the threshold for significant differential expression. If not specifically stated, all of the data obtained in this study were shown as mean ± SEM of n (n ≥ 3) independent experiments. Differences between normally distributed values of 2 groups were analyzed by an unpaired Student’s t-test, with P value < .05 considered statistically significant. All charts were drawn using Adobe Illustrator (v18.1.1.0) and GraphPad Prism 5 (GraphPad Software), some of which were drawn in the style of Dario et al. [30].
3. Results
3.1. Caffeic acid increases mouse platelet count
To detect changes in platelets in both groups, we discarded the first 3 drops of blood and collected the next 3 drops of blood, which were diluted with ACD anticoagulant from each mouse for platelet count (Figure 1D). Compared with the PBS group, the platelet count was significantly increased in the CA group (P = .009). It was indicated that CA had the effect of increasing mouse platelet count.
3.2. Influence of caffeic acid on mouse tail bleeding time and platelet function
To find out whether CA affects hemostasis in mice, we detected the mouse tail bleeding time after the second batch of mouse gavage experiments (Figure 1E). We also used the PFA-200 to determine the CT of platelet-blocking C/ADP pores (Figure 1F). Compared with the PBS group, the tail bleeding time was shortened without significance in CA group (P = .096), but the CT of the C/ADP was significantly shortened in CA group (P = .048). The results showed that CA had the effect of promoting hemostasis in mice.
3.3. Quality control of samples and RNA sequencing data meet requirements
The platelet activation ratio (Q1 + Q2 + Q3) of each sample was <6%, indicating that no significant platelet activation was detected throughout the platelet isolation and purification process (Figure 1G). The results demonstrated that the total RNA concentration of each sample was >10 ng/μL and the RNA integrity numbers >5 (Table 2), which were consistent with the RNA library construction requirements. As shown in Table 2, the 6 RNA libraries generated an average of 46.84 million reads each, both the Q20 and Q30 values were >90%, the percentage of unidentified bases <0.02%, the GC contents were about 50%, the total mapping rates >80%, and the multiple mapping rates <15%, indicating that the data quality met the requirements of RNA-seq analysis.
Table 2.
The quality indicators of 6 platelet pool samples.
| Sample name | CA1 | CA2 | CA3 | PBS1 | PBS2 | PBS3 |
|---|---|---|---|---|---|---|
| RNA concentrationa | 119 pg/μL | 73 pg/μL | 183 pg/μL | 232 pg/μL | 123 pg/μL | 242 pg/μL |
| RIN valuea | 8.1 | 8.2 | 6.0 | 7.3 | 7.5 | 7.2 |
| Raw readsb | 47,768,682 | 47,994,714 | 45,691,272 | 47,923,640 | 45,550,924 | 46,829,908 |
| Raw basesb | 7.17 G | 7.20 G | 6.85 G | 7.19 G | 6.83 G | 7.02 G |
| Error rateb, % | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 |
| Q20b, % | 97.36 | 97.68 | 97.45 | 97.56 | 97.31 | 97.47 |
| Q30b, % | 93.19 | 93.78 | 93.47 | 93.61 | 93.11 | 93.39 |
| GCb, % | 50.44 | 49.79 | 49.52 | 49.37 | 48.24 | 48.70 |
| Total mapc, % | 95.05 | 93.16 | 92.95 | 96.21 | 83.91 | 96.02 |
| Multimapc, % | 11.58 | 6.17 | 6.06 | 7.50 | 5.48 | 6.65 |
| Exonc, % | 96.25 | 95.36 | 95.73 | 96.09 | 95.94 | 96.11 |
| Intronc, % | 2.14 | 2.51 | 2.40 | 2.21 | 2.37 | 2.24 |
| Intergenicc, % | 1.61 | 2.13 | 1.86 | 1.70 | 1.69 | 1.65 |
CA, caffeic acid; PBS, phosphate-buffered saline, RIN, RNA integrity number.
The indicators of RNA quality and each RNA sample were diluted 100- to 200-fold for measurement.
The quality indicators of sequencing reads.
The reference sequence alignment results.
3.4. Differentially expressed gene analysis and gene ontology gene set enrichment analysis revealed that the upregulation of differentially expressed genes was related to the hemostasis regulation of caffeic acid
The systematic clustering and DEG analysis were performed on the RNA-seq data of 6 samples in CA vs PBS group. The 12,934 expressed transcripts were annotated (Figure 2A, each dot representing a transcript, Supplementary Table S1), of which 82.15% encode proteins, 7.79% pseudogenes, 2.76% antisense RNA, 3.74% lincRNA, and 3.56% others (Figure 2B). Compared with PBS group, we identified 987 DEGs, of which 466 were downregulated and 521 were upregulated in CA group (Figure 2C, Supplementary Figure S1, and Table S1).
Figure 2.
Differentially expressed genes (DEGs) in platelet pool RNA sequencing data of mice in the caffeic acid (CA) vs phosphate-buffered saline (PBS) group. (A) M-vs-A (MA) plot showing the log2 gene expression distribution plotted against log2-fold change for each gene. Red dots indicate significantly upregulated genes, gray dots indicate nondifferentially expressed genes, and green dots indicate significantly downregulated genes in the CA group. (B) Pie chart illustrating the biotypes of expressed transcripts. (C) Heatmap of DEGs using a negative binominal linear model provided by DESeq2. Each row in the heatmap indicates the log-ratio intensity of data of 1 DEG across 6 samples. The expression level is represented as a gradient from low (green) to high (red). (D–F) Gene Ontology (GO) analysis of DEGs. The bar length represents the fold enrichment. Red bars indicate terms enriched in the CA group, and green bars indicate terms enriched in the PBS group. The color range is proportional to the log10-fold P value. The top 5 enriched terms are shown for GO domains: (D) biological process, (E) cellular component, and (F) molecular function.
GO is a structured, controlled vocabulary for systematically analyzing gene function at the molecular and cellular levels [41]. We performed GO GSEA on the 466 downregulated DEGs in CA group (Supplementary Table S2). The top 5 biological processes in GO GSEA were aerobic respiration, electron transport chain, mitochondrial adenosine triphosphate synthesis coupled electron transport, mitochondrial electron transport, and adenosine triphosphate synthesis coupled electron transport (Figure 2D). The top 5 cellular components in GO GSEA were the mitochondrial membrane, respiratory chain, respiratory chain complex, mitochondrial respiratory chain, and mitochondrial protein complex (Figure 2E). The top 5 molecular functions in GO GSEA were the electron transfer activity, cytochrome-c oxidase activity, heme-copper terminal oxidase activity, oxidoreductase activity, and proton transmembrane transporter activity (Figure 2F). Similarly, the 521 upregulated DEGs were also analyzed in the CA group (Supplementary Table S2). The top 5 biological processes in GO GSEA were cell-substrate adhesion, regulation of blood coagulation and hemostasis, actin cytoskeleton organization, blood coagulation, and hemostasis and coagulation (Figure 2D). The top 5 cellular components in GO GSEA were the actin cytoskeleton, cell-cell junction, side of the membrane, ruffle, and adherens junction (Figure 2E). The top 5 molecular functions in GO GSEA were the cell adhesion molecule binding, actin binding, cadherin binding, integrin binding, and actin filament binding (Figure 2F). The results showed that the 466 downregulated DEGs were related to the aerobic respiration metabolism regulation of CA, and the 521 upregulated DEGs were related to the hemostasis regulation of CA.
3.5. Kyoto Encyclopedia of Genes and Genomes and Reactome gene set enrichment analysis screened out 25 upregulated differentially expressed genes that were related to the hemostasis regulation of caffeic acid
KEGG is a collection of databases for the biological interpretation of RNA-seq-generated large-scale datasets [42]. Next, we performed KEGG GSEA on 521 upregulated DEGs in CA group (Supplementary Table S3). We discovered that the top 5 pathways with KEGG GSEA were platelet activation (log10-fold P = 3.75; NES = 1.78), proteoglycans in cancer (log10-fold P = 3.59; NES = 1.63), extracellular matrix -receptor interaction (log10-fold P = 3.39; NES = 1.58), regulation of actin cytoskeleton (log10-fold P = 3.15; NES = 1.80), and focal adhesion (log10-fold P = 3.14; NES = 1.55) (Figure 3A). In these significantly enriched KEGG pathways, there were 44 significantly upregulated DEGs that may be related to the hemostasis regulation of CA (Supplementary Table S5).
Figure 3.
Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome gene set enrichment analysis (GSEA) of platelet pool RNA sequencing upregulated differentially expressed genes (DEGs) in the CA group. (A) KEGG and Reactome GSEA of upregulated DEGs revealed enrichment of categories involved in platelet activation and hemostasis in the CA group. Bars indicate the normalized enrichment score. The color range is proportional to the log10-fold P value. (B) Heatmap with leading edge genes identified by KEGG and Reactome GSEA. Each row in the heatmap indicates the relative expression of 1 upregulated DEG across 6 samples. The expression level is represented as a gradient from low (green) to high (red).
The Reactome database (https://reactome.org) is an extended version of the ordered network of molecular transformations—the classical metabolic map providing molecular details of signal transduction, transport, DNA replication, metabolism, and other cellular processes [43]. Similarly, we performed Reactome GSEA on the 521 DEGs upregulated in CA group (Supplementary Table S4). The top 6 pathways with Reactome GSEA were closely correlated with hemostasis (log10-fold P = 6.79; NES = 1.47), platelet activation, signaling and aggregation (log10-fold P = 6.30; NES = 1.72), platelet degranulation (log10-fold P = 4.86; NES = 1.61), response to elevated platelet cytosolic Ca2+ (log10-fold P = 4.61; NES = 1.59), cell-cell communication (log10-fold P = 3.70; NES = 1.52), and integrin cell surface interactions (log10-fold P = 2.86; NES = 1.65) (Figure 3A). We also found that there were 57 significantly upregulated DEGs that may be related to the hemostasis regulation of CA in these pathways (Supplementary Table S5).
After performing KEGG and Reactome GSEA of upregulated DEGs in CA group, the main targeted pathways of CA affecting hemostasis were concentrated, including the pathways of hemostasis, platelet activation, signaling, aggregation, and degranulation (Figure 3A). The 25 cosignificantly upregulated DEGs may be related to hemostatic regulation of CA (Figure 3B, Table 3), of which 16 DEGs (Tln1, Flna, Vwf, Thbs1, Ptpn11, Itgb3, Vcl, Gp1ba, Cd36, Itpr2, Actn1, Mapk14, Timp3, Gnaq, Syk, and Lyn) were distributed in the main targeted pathways. Through further comparative analysis, 5 (Tln1, Flna, Vwf, Thbs1, and Ptpn11) (P < .001) of these 16 upregulated DEGs were most likely to be the targeted genes of CA affecting hemostasis.
Table 3.
The expression levels of 25 significantly upregulated differentially expressed genes using the Kyoto Encyclopedia of Genes and Genomes, Reactome gene set enrichment analysis, and the expression levels of corresponding genes in mouse platelet transcriptome datasets by Rowley et al. [31].
| Gene name | Gene description | CA (mean RPKM) | PBS (mean RPKM) | Log2-fold change (CA vs PBS) | P value | Rowley et al. [31] (mean RPKM) |
|---|---|---|---|---|---|---|
| Tln1 | Talin 1 | 17676.74 | 10116.43 | 0.805198 | 6.3 × 10−8 | 760.5494 |
| Flna | Filamin, α | 23650.1 | 12946.33 | 0.869359 | 7.5 × 10−8 | 841.1512 |
| Vwf | Von Willebrand factor | 31425.06 | 16533.91 | 0.926569 | 4.0 × 10−6 | 873.7162 |
| Thbs1 | Thrombospondin 1 | 28320.78 | 15961.16 | 0.827318 | 4.5 × 10−5 | 732.041 |
| Itga6 | Integrin αVI | 25338.74 | 14544.83 | 0.800862 | 6.0 × 10−5 | 726.1951 |
| Ptpn11 | Protein tyrosine phosphatase, nonreceptor type 11 | 10434.69 | 6376.263 | 0.710537 | 7.7 × 10−4 | 297.0055 |
| Itgb3 | Integrin β3 | 35609.47 | 23886.92 | 0.576014 | 1.7 × 10−3 | 1060.948 |
| Vcl | Vinculin | 1668.783 | 792.2697 | 1.074242 | 1.7 × 10−3 | 88.84931 |
| Gp1ba | Glycoprotein 1b, α polypeptide | 5448.669 | 3302.963 | 0.722379 | 2.1 × 10−3 | 246.8136 |
| Cd36 | CD36 molecule | 702.9394 | 144.5864 | 2.279118 | 3.7 × 10−3 | 0.407926 |
| Hspg2 | Perlecan (heparan sulfate proteoglycan 2) | 63.95594 | 0.887052 | 6.100749 | 4.3 × 10−3 | 0.042957 |
| Itpr2 | Inositol 1,4,5-triphosphate receptor 2 | 3826.323 | 2419.123 | 0.661846 | 4.6 × 10−3 | 55.99499 |
| Actn1 | Actinin, α 1 | 3653.581 | 2264.198 | 0.690777 | 9.5 × 10−3 | 312.9807 |
| Mapk14 | Mitogen-activated protein kinase 14 | 2044.594 | 1328.725 | 0.621744 | 1.2 × 10−2 | 81.7842 |
| Gpc1 | Glypican 1 | 29.51418 | 0 | 7.24639 | 1.2 × 10−2 | 0 |
| Parvb | Parvin, β | 42200.59 | 31068.58 | 0.441844 | 1.4 × 10−2 | 1624.163 |
| Kdr | Kinase insert domain protein receptor | 219.0124 | 32.74483 | 2.73728 | 1.5 × 10−2 | 2.871863 |
| Itgb2 | Integrin β2 | 11345 | 7986.086 | 0.506569 | 1.5 × 10−2 | 626.1656 |
| Timp3 | Tissue inhibitor of metalloproteinase 3 | 124.8425 | 21.59947 | 2.539816 | 1.7 × 10−2 | 0.940079 |
| Prkacb | Protein kinase, cAMP-dependent, catalytic, β | 320.203 | 138.7467 | 1.208162 | 2.5 × 10−2 | 8.240311 |
| Gnaq | Guanine nucleotide–binding protein, α q polypeptide | 674.3222 | 399.9358 | 0.753103 | 2.6 × 10−2 | 21.2075 |
| Syk | Spleen tyrosine kinase | 2104.623 | 1547.619 | 0.443477 | 3.9 × 10−2 | 59.61091 |
| Itga2 | Integrin αII | 1883.193 | 1307.344 | 0.526695 | 4.2× 10−2 | 100.8845 |
| Lyn | LYN proto-oncogene, Src family tyrosine kinase | 6537.115 | 4528.37 | 0.529748 | 4.5 × 10−2 | 232.4545 |
| Iqgap1 | IQ motif containing GTPase activating protein 1 | 155.3689 | 32.21947 | 2.267254 | 1.2 × 10−2 | 2.411546 |
CA, caffeic acid; PBS, phosphate-buffered saline; RPKM, reads per kilobase per million mapped reads.
3.6. Quantitative polymerase chain reaction validation confirmed the reliability and accuracy of the RNA sequencing data
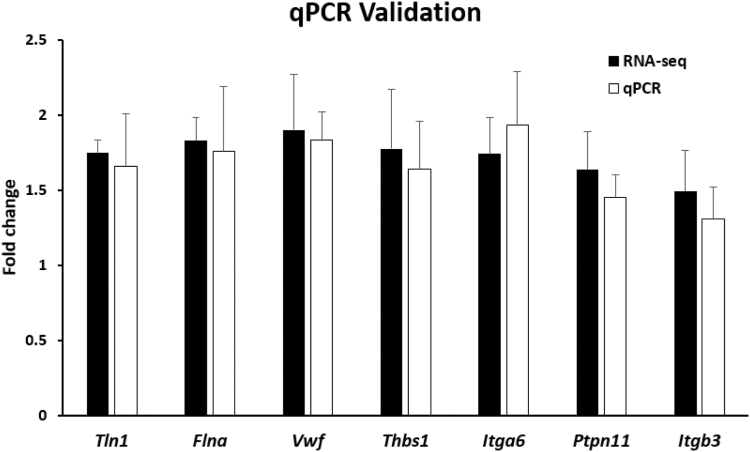
To verify the reliability and accuracy of the RNA-seq data, we selected the top 7 upregulated DEGs (Tln1, Flna, Vwf, Thbs1, Igta6, Ptpn11, and Itgb3) ranked in descending order of P value in Table 3 for further qPCR validation. As shown in Figure 4, for these 7 upregulated DEGs, the fold change (CA vs PBS group, the second batch) identified using qPCR was similar to the fold change (CA vs PBS group, the first batch) obtained using RNA-seq analysis, although the fold change (CA vs PBS group) of these 2 batches were not always entirely consistent. Nevertheless, the results of qPCR validation confirmed the reliability and accuracy of our RNA-seq data. In addition, as shown in Supplementary Figure S2, we selected the 25 genes in Table 3 as an example, and found the relative expression level (vs Actb) in PBS group’s RNA-seq was similar to the relative expression level (vs Actb) in Rowley et al.’s [31] mouse platelet transcriptomes, which also indirectly reflected the reliability and accuracy of our RNA-seq data.
Figure 4.
Quantitative polymerase chain reaction (qPCR) validation of the expression levels of differentially expressed genes identified through RNA sequencing (RNA-seq). Seven differentially expressed genes were randomly selected for qPCR validation and compared with data obtained using RNA-seq. Black bars represent the RNA-seq results, and white bars represent the qPCR validation results. Data are shown in fold change for the caffeic acid vs phosphate-buffered saline group. Error bars indicate the SEM with 3 samples, and significant differences (P < .05) between qPCR and RNA-seq are indicated with an asterisk (∗).
4. Discussion
For the first time, RNA-seq analysis was performed on platelet pools from mice gavaged with CA and platelet pools from mice gavaged with PBS (Figure 1B, C). The main findings of the present study are that the significant gene enrichment of hemostasis and platelet activation–related pathways were detected in the platelet pools from mice gavaged with CA, and the main target pathways (hemostasis, platelet activation, signaling, aggregation, and degranulation) and the 5 (Tln1, Flna, Vwf, Thbs1, and Ptpn11) potential target genes of CA affecting hemostasis were screened out.
GO GSEA results showed that 987 DEGs were related to the aerobic respiration metabolism regulation and the hemostasis regulation of CA. The antioxidant activities of CA had been reported; Gülçin [2] studied the antioxidant properties of CA through various in vitro antioxidant assays. Khan et al. [3] summarized and described the inhibitory mechanism against oxidative stress of CA. In addition, 466 downregulated DEGs from the GO GSEA results (Figure 2D–F) supported that the in vivo antioxidant properties of CA were likely to be achieved by downregulating the expression of genes related to the aerobic respiration process (Supplementary Tables S2–S4). Correspondingly, 521 upregulated DEGs from the GO GSEA results (Figure 2D–F) supported that CA was involved in regulating coagulation and hemostasis, which coincides with our previous speculation. Studies on CA in increasing platelet count had been reported. Du et al. [13] discovered that CA could promote the recovery of platelet count in irradiated mice, Zhang et al. [14] found that CA could also increase the platelet count of cytarabine-induced thrombocytopenia mice, and Qin et al. [17] revealed that CA had a good effect on increasing the platelet count of primary immune thrombocytopenia. These findings may be related to the upregulated expression of potential target genes Flna and Ptpn11. FLNA is a key determinant of platelet structural integrity and shape change, and platelet count is correlated with the Flna level [44,45]. PTPN11 is a major regulator of megakaryocyte development, platelet production, and function; platelet counts were reduced in Ptpn11 knockout mice [46]. Our study showed that the platelet counts of mice in the CA group were significantly higher than those in the PBS group (Figure 1D), but the platelet count range in the PBS group (444∼699 × 109/L) was lower than that of Li et al. [47] normal 6 weeks mice (466 ∼ 818 × 109/L), which was related to the detection after we diluted whole blood with ACD.
In addition, studies on CA in promoting hemostasis had also been reported. Zhu et al. [15] reported that CA could shorten thrombin time and prothrombin time, activated partial thromboplastin time in leukopenia and thrombocytopenia model mice, increase fibrinogen content and enhance hemostasis and coagulation function. Lin et al. [16] also reported that CA shortened prothrombin time and activated partial thromboplastin time at medium and low concentrations compared with the control group. These may be related to the upregulated expression of potential target genes Tln1, Vwf, and Thbs1. TLN1 is crucial for hemostasis, platelet aggregation, and adhesion. Tln1 knockout mice have prolonged tail bleeding time [48], and endothelial cells-specific deletion of talin1 causes vascular hemorrhage and death [49]. Thbs1-deficient platelets show increased bleeding times and diminished thrombosis in vivo, and at sites of vascular injury, activated platelets release Thbs1, which acts to suppress inhibitory cAMP signaling and promote hemostasis [50]. Subsequently, the result of the tail bleeding time and platelet functional assays in the second batch of mice supported the idea that CA promotes hemostasis (Figure 1E, F), although the tail bleeding time was shortened without significant difference in the CA vs PBS group, which may be related to sample size. Moreover, the PFA-200 measurement is very sensitive to variation in platelet count and von Willebrand factor (VWF) levels [36,37] and the plasma VWF is a major determinant of hemostasis, but platelet VWF also contributes to platelet adhesion and plays a role in local hemostasis [51]. The shortened CT may be due to increased platelet count and platelet VWF levels, but we cannot rule out the effect of increased plasma VWF levels after CA treatment, and the 5 potential target genes of CA affecting hemostasis will be focused, especially Vwf.
The regulation of CA on platelet activation may have 2 effects. First, Zhu et al. [52] reported that CA could increase the expression level of CD62P (Selp) on the surface of mouse platelet membranes and activate platelets (flow cytometry) after 100 mg/kg or 200 mg/kg CA continuous gavaging for 10 days. Our platelet RNA-seq analysis demonstrated that upregulated DEGs were enriched in the related pathway of platelet activation. Although the fold change of Selp expression level (CA vs PBS group) was not significantly increased after 18 mg/kg CA continuous gavaging for 7 days (flow cytometry). On the other side, Park [53] found that mouse platelets could inhibit the expression of Selp under CA incubation (nonflow cytometry). Lu et al. [54] confirmed that 25-100 μM CA could inhibit ADP-induced platelet P-selectin (Selp) expression in vitro (flow cytometry). Nam et al. [55] found that 500 μM CA could also inhibit thrombin-induced platelet aggregation and P-selectin (Selp) expression in vitro, but 125 μM or 250 μM CA could not (flow cytometry). These studies indicated whether flow cytometry can detect changes of Selp expression, which may depend on the concentration of CA and the duration of continuous administration. Although flow cytometry is a more straightforward method for platelet activation assay, the role of CA in promoting platelet activation still needs to be further validated with new methods.
We acknowledge that this study has some limitations. We reviewed the literature related to the field of CA, and most of the studies in vitro and in vivo confirmed that CA has platelet-inhibiting activity under the concentration conditions mentioned in the corresponding literature [[53], [54], [55]], which is worthy of affirmation. RNA-seq analysis in this study showed that 521 upregulated DEGs were related to platelet activation pathway under the condition of 18 mg/kg CA gavage dose (Figure 3A), but no obvious thrombosis was observed in mice (figure not shown). This contradicts the previously confirmed platelet-inhibiting activity of CA. This may be related to the concentration of CA, the mode of administration, the duration of continuous administration, the method of blood collection and platelet isolation in the later stage, and the detection object and the detection technology. At the same time, we have also noticed that some researchers have observed that caffeic acid at certain concentrations has antioxidant [2,3], hemostasis, and elevated platelets [[13], [14], [15], [16], [17]]. RNA-seq analysis in this study showed that under the condition of gavage CA concentration, upregulated DEGs related to the hemostatic pathway (Figure 2D), downregulated DEGs related to the aerobic respiratory pathway (Figure 2D), platelet count was increased (Figure 1C), and the CT and tail bleeding time were shortened (Figure 1E, F). These conclusions support the phenomena observed by these researchers. This also prompts us to think deeply about whether CA has platelet inhibitory activity within a certain concentration range and antioxidant, hemostatic, and platelet elevation effects in another concentration range, which needs to be further studied.
In conclusion, CA regulates the gene expression of hemostasis and platelet activation–related pathways to increase platelet count and promote hemostasis.
Acknowledgments
We would like to thank Dr Li Guo from the University of Utah for providing technical guidance for taking blood from the carotid artery of mice and isolating and purifying platelets, Drs Chenggong Li and Wenjing Luo from Union Hospital, Tongji Medical College, HUST, for their technical guidance on the operation of the cytometer, and Mr Bing Wang from the laboratory animal center of HUST for his valuable suggestions on the convenience of mouse breeding.
Funding
The study was supported by National Key Research and Development Program of China (No. 2022YFC2304600).
Author contributions
J.D., Y.H., L.X., and S.L. designed the research and wrote the manuscript. L.X., Y.D., Y.C., and Z.C. conducted experiments. J.D., L.X., and Y.D. analyzed and mapped RNA-seq results. J.D., Y.H., L.X., S.L., and F.F. interpreted the results and reviewed the paper.
Relationship Disclosure
None of the authors have any disclosures relevant to this paper.
Data availability
The RNA-seq raw data of mouse platelet pools in both groups were deposited at the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/) under project number PRJNA838396. Additional details are found in the Supplemental Materials.
Footnotes
Handling Editor: Dr Henri Spronk
The online version contains supplementary material available at https://doi.org/10.1016/j.rpth.2024.102349.
Supplementary material
References
- 1.Clifford M.N. Chlorogenic acids and other cinnamates – nature, occurrence and dietary burden. J Sci Food Agric. 1999;79:362–372. [Google Scholar]
- 2.Gülçin I. Antioxidant activity of caffeic acid (3,4-dihydroxycinnamic acid) Toxicology. 2006;217:213–220. doi: 10.1016/j.tox.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Khan F.A., Maalik A., Murtaza G. Inhibitory mechanism against oxidative stress of caffeic acid. J Food Drug Anal. 2016;24:695–702. doi: 10.1016/j.jfda.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mude H., Maroju P.A., Balapure A., Ganesan R., Ray Dutta J. Water-soluble caffeic acid-dopamine acid-base complex exhibits enhanced bactericidal, antioxidant, and anticancer properties. Food Chem. 2022;374 doi: 10.1016/j.foodchem.2021.131830. [DOI] [PubMed] [Google Scholar]
- 5.Kępa M., Miklasińska-Majdanik M., Wojtyczka R.D., Idzik D., Korzeniowski K., Smoleń-Dzirba J., et al. Antimicrobial potential of caffeic acid against Staphylococcus aureus clinical strains. Biomed Res Int. 2018;2018 doi: 10.1155/2018/7413504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinho E., Soares G., Henriques M. Evaluation of antibacterial activity of caffeic acid encapsulated by β-cyclodextrins. J Microencapsul. 2015;32:804–810. doi: 10.3109/02652048.2015.1094531. [DOI] [PubMed] [Google Scholar]
- 7.Kassim M., Achoui M., Mustafa M.R., Mohd M.A., Yusoff K.M. Ellagic acid, phenolic acids, and flavonoids in Malaysian honey extracts demonstrate in vitro anti-inflammatory activity. Nutr Res. 2010;30:650–659. doi: 10.1016/j.nutres.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 8.Kim S.R., Jung Y.R., Kim D.H., An H.J., Kim M.K., Kim N.D., et al. Caffeic acid regulates LPS-induced NF-κB activation through NIK/IKK and c-Src/ERK signaling pathways in endothelial cells. Arch Pharm Res. 2014;37:539–547. doi: 10.1007/s12272-013-0211-6. [DOI] [PubMed] [Google Scholar]
- 9.Gomes C.A., da Cruz T.G., Andrade J.L., Milhazes N., Borges F., Marques M.P. Anticancer activity of phenolic acids of natural or synthetic origin: a structure-activity study. J Med Chem. 2003;46:5395–5401. doi: 10.1021/jm030956v. [DOI] [PubMed] [Google Scholar]
- 10.Yang G., Fu Y., Malakhova M., Kurinov I., Zhu F., Yao K., et al. Caffeic acid directly targets ERK1/2 to attenuate solar UV-induced skin carcinogenesis. Cancer Prev Res (Phila) 2014;7:1056–1066. doi: 10.1158/1940-6207.CAPR-14-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Socała K., Szopa A., Serefko A., Poleszak E., Wlaź P. Neuroprotective effects of coffee bioactive compounds: a review. Int J Mol Sci. 2020;22:107. doi: 10.3390/ijms22010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim I.H., Yan B.C., Park J.H., Yeun G.H., Yim Y., Ahn J.H., et al. Neuroprotection of a novel synthetic caffeic acid-syringic acid hybrid compound against experimentally induced transient cerebral ischemic damage. Planta Med. 2013;79:313–321. doi: 10.1055/s-0032-1328211. [DOI] [PubMed] [Google Scholar]
- 13.Du Z., Jia H., Liu J., Yang L., Jiang X., Cheng F. Preliminary study on hematopoietic reconstruction effects of caffeic acid in lethally irradiated mice. Chin J Radiol Health. 2015;24:449–452. [Google Scholar]
- 14.Zhang Q., Hu Z.L., Wang F.W., Mou Y.L., Yu R.J. Discussion on the effect of caffeic acid tablets on cytarabine-induced thrombocytopenia in mice [abstract] The 9th National Member Congress of Chinese Pharmacological Society and National Conference of Pharmacology. 2007 [Google Scholar]
- 15.Zhu X.L., Mou J.X., Ren H.Y., Lu Y., Yang J.L., Li N., et al. Effects of caffeic acid on coagulation function in leucopenia and thrombocytopenia mice. Pharmacology and Clinics of Chinese Materia Medica. 2014;30:46–48. [Google Scholar]
- 16.Lin Z.C., Fang Y.J., Huang A.Y., Chen L.Y., Guo S.H., Chen J.W. Chemical constituents from Sedum aizoon and their hemostatic activity. Pharm Biol. 2014;52:1429–1434. doi: 10.3109/13880209.2014.895019. [DOI] [PubMed] [Google Scholar]
- 17.Qin P., Wei Y., Hou M., Zhao C., Shen Z. A multicenter clinical trial of caffeic acid tablet in treatment of 103 primary immune thrombocytopenia patients. Zhonghua Xue Ye Xue Za Zhi. 2015;36:103–106. doi: 10.3760/cma.j.issn.0253-2727.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leslie M. Cell biology. Beyond clotting: the powers of platelets. Science. 2010;328:562–564. doi: 10.1126/science.328.5978.562. [DOI] [PubMed] [Google Scholar]
- 19.Koupenova M., Clancy L., Corkrey H.A., Freedman J.E. Circulating platelets as mediators of immunity, inflammation, and thrombosis. Circ Res. 2018;122:337–351. doi: 10.1161/CIRCRESAHA.117.310795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maouia A., Rebetz J., Kapur R., Semple J.W. The immune nature of platelets revisited. Transfus Med Rev. 2020;34:209–220. doi: 10.1016/j.tmrv.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jurk K., Kehrel B.E. The role of platelets in haemostasis, thrombosis, immune defense and inflammation. Dtsch Med Wochenschr. 2008;133:1130–1135. doi: 10.1055/s-2008-1077228. [DOI] [PubMed] [Google Scholar]
- 22.Smyth S.S., McEver R.P., Weyrich A.S., Morrell C.N., Hoffman M.R., Arepally G.M., et al. Platelet functions beyond hemostasis. J Thromb Haemost. 2009;7:1759–1766. doi: 10.1111/j.1538-7836.2009.03586.x. [DOI] [PubMed] [Google Scholar]
- 23.Clancy L., Beaulieu L.M., Tanriverdi K., Freedman J.E. The role of RNA uptake in platelet heterogeneity. Thromb Haemost. 2017;117:948–961. doi: 10.1160/TH16-11-0873. [DOI] [PubMed] [Google Scholar]
- 24.Thon J.N., Devine D.V. Translation of glycoprotein IIIa in stored blood platelets. Transfusion. 2007;47:2260–2270. doi: 10.1111/j.1537-2995.2007.01455.x. [DOI] [PubMed] [Google Scholar]
- 25.Mills E.W., Green R., Ingolia N.T. Slowed decay of mRNAs enhances platelet specific translation. Blood. 2017;129:e38–e48. doi: 10.1182/blood-2016-08-736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Risitano A., Beaulieu L.M., Vitseva O., Freedman J.E. Platelets and platelet-like particles mediate intercellular RNA transfer. Blood. 2012;119:6288–6295. doi: 10.1182/blood-2011-12-396440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tarazona S., García-Alcalde F., Dopazo J., Ferrer A., Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011;21:2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Best M.G., Sol N., Kooi I., Tannous J., Westerman B.A., Rustenburg F., et al. RNA-seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer cell. 2015;28:666–676. doi: 10.1016/j.ccell.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang B.L., Yang X.H., Jin H.M., Zhan X.L. Identification of differentially expressed genes in diabetic kidney disease by RNA-seq analysis of venous blood platelets. FEBS Open Bio. 2021;11:2095–2109. doi: 10.1002/2211-5463.13199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bongiovanni D., Santamaria G., Klug M., Santovito D., Felicetta A., Hristov M., et al. Transcriptome analysis of reticulated platelets reveals a prothrombotic profile. Thromb Haemost. 2019;119:1795–1806. doi: 10.1055/s-0039-1695009. [DOI] [PubMed] [Google Scholar]
- 31.Rowley J.W., Oler A.J., Tolley N.D., Hunter B.N., Low E.N., Nix D.A., et al. Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes. Blood. 2011;118:e101–e111. doi: 10.1182/blood-2011-03-339705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamia N., Miriana K., Hélène G.G., Sophie F. Effect of dimethylsulfoxide, ethanol, α- and β-cyclodextrins and their association on the solubility of natural bioactive compounds. J Mol Liq. 2020;310 [Google Scholar]
- 33.Liu Y., Jennings N.L., Dart A.M., Du X.J. Standardizing a simpler, more sensitive and accurate tail bleeding assay in mice. World J Exp Med. 2012;2:30–36. doi: 10.5493/wjem.v2.i2.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu K.K. Platelet activation mechanisms and markers in arterial thrombosis. J Intern Med. 1996;239:17–34. doi: 10.1046/j.1365-2796.1996.331661000.x. [DOI] [PubMed] [Google Scholar]
- 35.Michelson A.D. Flow cytometry: a clinical test of platelet function. Blood. 1996;87:4925–4936. [PubMed] [Google Scholar]
- 36.Emmanuel J.F., Soma M., Ronny V., Kent C., Geoffrey K., Sarah J., et al. Harmonizing platelet function analyzer testing and reporting in a large laboratory network. Int J Lab Hematol. 2022;44:934–944. doi: 10.1111/ijlh.13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emmanuel J., Favaloro R.B. External quality assessment/proficiency testing and internal quality control for the PFA-100 and PFA-200: an update. Semin Thromb Hemost. 2014;40:239–253. doi: 10.1055/s-0034-1365844. [DOI] [PubMed] [Google Scholar]
- 38.Hawker R.J., Turner V.S., Mitchell S.G. Use of prostaglandin E1 during preparation of platelet concentrates. Transfus Med. 1996;6:249–254. doi: 10.1111/j.1365-3148.1996.tb00076.x. [DOI] [PubMed] [Google Scholar]
- 39.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 41.Consortium G.O. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosa J.P., Raslova H., Bryckaert M. Filamin A: key actor in platelet biology. Blood. 2019;134:1279–1288. doi: 10.1182/blood.2019000014. [DOI] [PubMed] [Google Scholar]
- 45.Jurak Begonja A., Hoffmeister K.M., Hartwig J.H., Falet H. FlnA-null megakaryocytes prematurely release large and fragile platelets that circulate poorly. Blood. 2011;118:2285–2295. doi: 10.1182/blood-2011-04-348482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mazharian A., Mori J., Wang Y.J., Heising S., Neel B.G., Watson S.P., et al. Megakaryocyte-specific deletion of the protein-tyrosine phosphatases Shp1 and Shp2 causes abnormal megakaryocyte development, platelet production, and function. Blood. 2013;121:4205–4220. doi: 10.1182/blood-2012-08-449272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li G.L., Liu K., Li X.C., Kuang S.S., Wang G. Determination and comparison of blood physiological and biochemical indexes between SPF BALB/c mice and C57BL/6 mice in different physiological stages and genders. Guangdong Agricultural Sciences. 2019;46:102–109. [Google Scholar]
- 48.Petrich B.G., Marchese P., Ruggeri Z.M., Spiess S., Weichert R.A., Ye F., et al. Talin is required for integrin-mediated platelet function in hemostasis and thrombosis. J Exp Med. 2007;204:3103–3111. doi: 10.1084/jem.20071800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pulous F.E., Carnevale J.C., Al-Yafeai Z., Pearson B.H., Hamilton J.A.G., Henry C.J., et al. Talin-dependent integrin activation is required for endothelial proliferation and postnatal angiogenesis. Angiogenesis. 2021;24:177–190. doi: 10.1007/s10456-020-09756-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aburima A., Berger M., Spurgeon B.E.J., Webb B.A., Wraith K.S., Febbraio M., et al. Thrombospondin-1 promotes hemostasis through modulation of cAMP signaling in blood platelets. Blood. 2021;137:678–689. doi: 10.1182/blood.2020005382. [DOI] [PubMed] [Google Scholar]
- 51.Kanaji S., Fahs S.A., Shi Q., Haberichter S.L., Montgomery R.R. Contribution of platelet vs. endothelial VWF to platelet adhesion and hemostasis. J Thromb Haemost. 2012;10:1646–1652. doi: 10.1111/j.1538-7836.2012.04797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu X.L., Pan X.Z., Ren H.Y., Yang J.L., Li X.J. Effects of caffeic acid on expression of glucoproteins on the platelet membrane surface in leucopenia and thrombocytopenia mice. Pharmacology and Clinics of Chinese Materia Medica. 2013;29:24–26. [Google Scholar]
- 53.Park J.B. 5-Caffeoylquinic acid and caffeic acid orally administered suppress P-selectin expression on mouse platelets. J Nutr Biochem. 2009;20:800–805. doi: 10.1016/j.jnutbio.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 54.Lu Y., Li Q., Liu Y.Y., Sun K., Fan J.Y., Wang C.S., et al. Inhibitory effect of caffeic acid on ADP-induced thrombus formation and platelet activation involves mitogen-activated protein kinases. Sci Rep. 2015;5 doi: 10.1038/srep13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nam G.S., Park H.J., Nam K.S. The antithrombotic effect of caffeic acid is associated with a cAMP-dependent pathway and clot retraction in human platelets. Thromb Res. 2020;195:87–94. doi: 10.1016/j.thromres.2020.07.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq raw data of mouse platelet pools in both groups were deposited at the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/) under project number PRJNA838396. Additional details are found in the Supplemental Materials.