Abstract
While most glial cell types in the central nervous system (CNS) arise from neuroectodermal progenitors, some, like microglia, are mesodermally derived. To understand mesodermal glia development and function, we investigated C. elegans GLR glia, which envelop the brain neuropil and separate it from the circulatory system cavity. Transcriptome analysis shows that GLR glia combine astrocytic and endothelial characteristics, which are relegated to separate cell types in vertebrates. Combined fate acquisition is orchestrated by LET-381/FoxF, a fate-specification/maintenance transcription factor also expressed in glia and endothelia of other animals. Among LET-381/FoxF targets, the UNC-30/Pitx2 transcription factor controls GLR glia morphology and represses alternative mesodermal fates. LET-381 and UNC-30 co-expression in naive cells is sufficient for GLR glia gene expression. GLR glia inactivation by ablation or let-381 mutation disrupts locomotory behavior and promotes salt-induced paralysis, suggesting brain-neuropil activity dysregulation. Our studies uncover mechanisms of mesodermal glia development and show that like neuronal differentiation, glia differentiation requires autoregulatory terminal selector genes that define and maintain the glial fate.
Keywords: Glia Development, let-381, Locomotory Behavior, Terminal Selector, unc-30
Subject terms: Chromatin, Transcription & Genomics; Development; Neuroscience
Synopsis
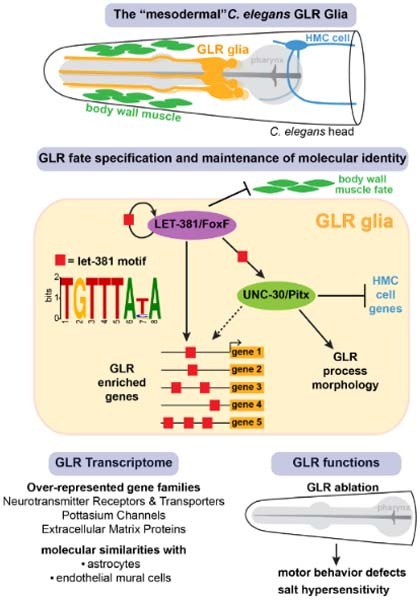
Mesodermal glia have crucial functions in the nervous system, although the mechanisms regulating their development remain elusive. Here, transcriptomic and mutational analyses identify a gene regulatory network required for specification and maintenance of mesodermally derived GLR glia in C. elegans.
In let-381/FoxF null mutants, GLR glia are not specified and some GLR lineages adopt sister muscle lineage fates instead.
LET-381/FoxF acts as an autoregulatory terminal selector to maintain GLR gene expression via a common cis-regulatory motif.
UNC-30/Pitx2 represses head mesodermal cell fate in GLR glia and regulates GLR morphology and gene expression.
GLR glia ablation results in severe motor behavior defects and salt hypersensitivity.
GLR glia that envelop the C. elegans central nervous system show a mixed astrocyte/endothelial cell identity and are specified by two co-operating transcription factors.
Introduction
Glia are abundant and diverse cellular components of most, if not all, nervous systems, and are anatomically positioned to affect every aspect of signal transduction and processing in the brain. Glia dynamically regulate neuronal activity in response to presynaptic cues, provide insulation around axons and at synapses, and supply trophic support for neuron survival (Allen and Lyons, 2018). Most glia, including astrocytes and myelinating glia, are derived from neuroectodermal precursors (Kastriti and Adameyko, 2017; Rowitch and Kriegstein, 2010). By contrast, other glia, such as microglia, which are born in the yolk sac and migrate into the CNS, arise from mesodermal progenitors (Ginhoux et al, 2010; Ginhoux et al, 2013; Ginhoux and Prinz, 2015). Transcription factors regulating the development of some neuroectodermal glia are known (Hochstim et al, 2008; Rowitch and Kriegstein, 2010; Wegner, 2020); however, less is understood about the control of mesodermal glia differentiation. Furthermore, only a few factors sufficient to confer specification of certain glia subtypes in naive cellular settings are known (Canals et al, 2018).
The nematode C. elegans has been instrumental in uncovering basic principles of glia development and function (Shaham, 2015a). The nervous system of the adult C. elegans hermaphrodite contains 56 glial cells. 50 of these derive from the AB blast cell lineage, which also produces neurons and epithelial cells. Six GLR glia derive from the MS blastomere, which primarily generates body wall and pharyngeal muscle (Fig. 1A) (Sulston et al, 1983). Thus, as in vertebrates, C. elegans possesses glia of both neuroectodermal and mesodermal origin. Some genes affecting C. elegans neuroepithelial glia development have been characterized (Mizeracka et al, 2021; Shaham, 2015b; Wallace Sean et al, 2016; Zhang et al, 2020); however, virtually nothing is known about GLR glia development and functions. GLR glia extend intricate, non-overlapping sheet-like processes that ensheath the inner aspect of the C. elegans brain neuropil (the nerve ring; Fig. 1B) and that are adjacent to neuromuscular synapses (White et al, 1986). At the nerve ring, GLR glia are electrically coupled to the RME motoneurons through gap junctions (White et al, 1986), and uptake extracellular GABA (Gendrel et al, 2016). More anteriorly, GLR glia extend thin processes that fasciculate with sensory neuron dendrites. GLR glia also physically separate the nerve ring from the pseudocoelomic body cavity which surrounds the pharynx and acts as a rudimentary circulatory system. The proximity to synapses and to the circulatory system, the association with GABA, and the ability to phagocytose injured neurons (Altun and Hall, 2016; Nass et al, 2002; White et al, 1986) make comparisons between GLR glia and astrocytes tempting.
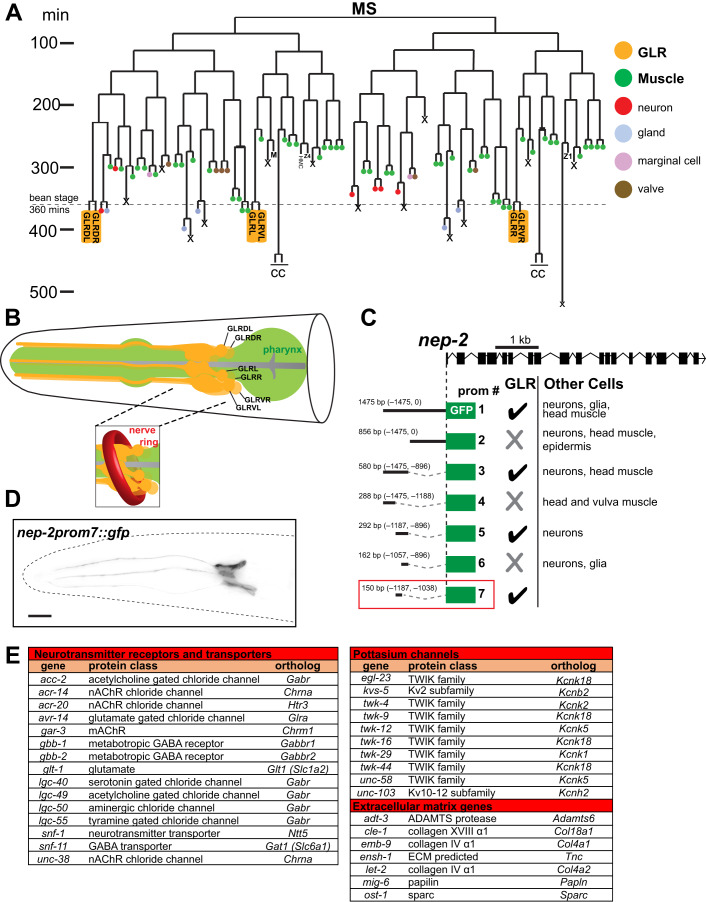
Figure 1. Generation of a GLR-specific driver to study the expression profile of the mesodermal GLR glia.
(A) GLR glia (yellow boxes) derive from the lineage of the blast cell MS. This lineage produces mainly body wall muscle and pharyngeal muscle cells (green). GLR glia (yellow) are born at around the embryonic bean stage (360 min of embryonic development). The HMC cell and coelomocytes (CC) also derive from the MS lineage. Schematic adapted from (Sulston et al, 1983). (B) Schematic representation of the GLR glia (yellow). Pharynx is shown in green. The inset shows how C. elegans Nerve Ring (red) wraps around the sheet-like GLR glia processes. Schematic redrawn and modified from (Altun and Hall, 2016). (C) Cis-regulatory dissection analysis for the gene nep-2 resulted in isolation of a GLR glia-specific driver, prom7 (red box). (D) Fluorescence image of an L4 C. elegans showing expression of nep-2prom7::gfp specifically in GLR glia. Anterior is left, dorsal is up, and scale bar is 10 μm. (E) Genes from three families (neurotransmitter receptors and transporters, potassium channels and extracellular matrix genes) are overrepresented among GLR-enriched genes.
Here, we describe the transcriptome of adult C. elegans GLR glia, uncovering similarities with both astrocytes and endothelial cells. We use the transcriptome to develop a molecular toolkit for labeling and manipulating GLR glia, which we use to identify and characterize two master regulators of GLR glia development. We show that LET-381, the C. elegans ortholog of the forkhead transcription factor FoxF, promotes GLR glia fate specification and maintenance and that UNC-30/Pitx2 transcription factor controls GLR glia morphology and represses the acquisition of an alternative mesodermal fate. Importantly, the expression of both transcription factors in a naive cell is sufficient to promote GLR glia gene expression. Through genetic studies, we order let-381, unc-30, and other genes into a pathway for GLR glia development. Finally, we show that let-381 autoregulation-deficient mutants, as well as animals in which GLR glia are genetically ablated, exhibit specific defects in locomotory behavior and are hypersensitive to salt, suggesting important roles for these glia in coordinating neuronal activity.
Results
GLR glia gene expression reveals similarities with astrocytes and endothelial cells
Although GLR glia reporters have been previously described, none are exclusively expressed in these cells (Krause et al, 1994; Ringstad et al, 2009; Ringstad and Horvitz, 2008; Warren et al, 2001; Yamada et al, 2010). To identify drivers allowing specific genetic manipulation and marking of GLR glia, we performed promoter dissection studies of known GLR glia-expressed genes (Appendix Fig. S1A–E), isolating a 150 bp cis-regulatory sequence from the gene nep-2 that, when fused to gfp, promotes expression only in GLR glia (Fig. 1C,D). Expression of this reporter is first detected in first-stage (L1) larvae and is maintained through adulthood.
To identify genes regulating GLR glia development and functions, we generated a stable transgenic C. elegans strain expressing nuclear YFP using the nep-2 regulatory sequence (Appendix Fig. S1F). nep-2prom7::nls::yfp expressing cells were isolated from dissociated L4 larvae using fluorescence-activated cell sorting (FACS), and lysed to isolate mRNA. Following mRNA amplification and RNA-seq, we compared transcript abundances between GLR glia (YFP-positive) and all other cells (YFP-negative). We identified 886 genes with enriched GLR glia expression (P < 0.05, log2-fold enrichment >1, Dataset EV1) out of 13,794 genes with any GLR glia expression (>50 reads, Dataset EV1; Appendix Fig. S1G). To validate this list, we confirmed the expression of 39 GLR glia-enriched genes using transgenic and endogenous gfp reporters (green/blue highlights in Dataset EV1). In addition, all previously known GLR glia genes show strong enrichment in our analysis.
Using PANTHER gene ontology over-representation analysis (Mi et al, 2019; Thomas et al, 2022), we find that genes involved in synaptic transmission, including neurotransmitter receptors and transporters, potassium channels, and genes encoding extracellular matrix proteins, are overrepresented among GLR glia-enriched genes (Fig. 1E). Genes from these transcript classes as well as other GLR-enriched genes are also overrepresented in murine astrocytes (e.g., snf-11/Gat1, gbb-1/Gabbr1, gbb-2/Gabbr2, glt-1/Glt1, ensh-1/Tnc, pll-1/Plcd4) (Batiuk et al, 2020; Yang and Jackson, 2019; Zhang et al, 2014), suggesting similarities between GLR glia and astrocyte transcriptomes. Genes encoding ion, amino acid, and neurotransmitter transporters, as well as extracellular matrix proteins are enriched also in endothelial cells of the blood–brain barrier (Munji et al, 2019), and other GLR-enriched genes, including let-381/Foxf, dep-1/Ptprb, tag-68/Smad6, T16A9.4/Ece1, gei-1/Dlc-1, slcf-2/Slc2a1, unc-115/Ablim1, mrp-2/Abcc6, are also enriched in endothelial cells of the central nervous system (Batiuk et al, 2020; Munji et al, 2019; Zhang et al, 2014). In vertebrates, astrocyte endfeet are found in proximity to endothelial blood vessels, and in C. elegans, GLR glia separate the circulatory cavity from the nerve ring. It is intriguing to speculate that to conserve cell numbers, C. elegans may have merged astrocytic and endothelial functions into the GLR glia cell type. Such functional and anatomic compression has also been observed in the C. elegans motor circuit (Gao et al, 2018).
let-381/FoxF is required early to specify GLR glia fate
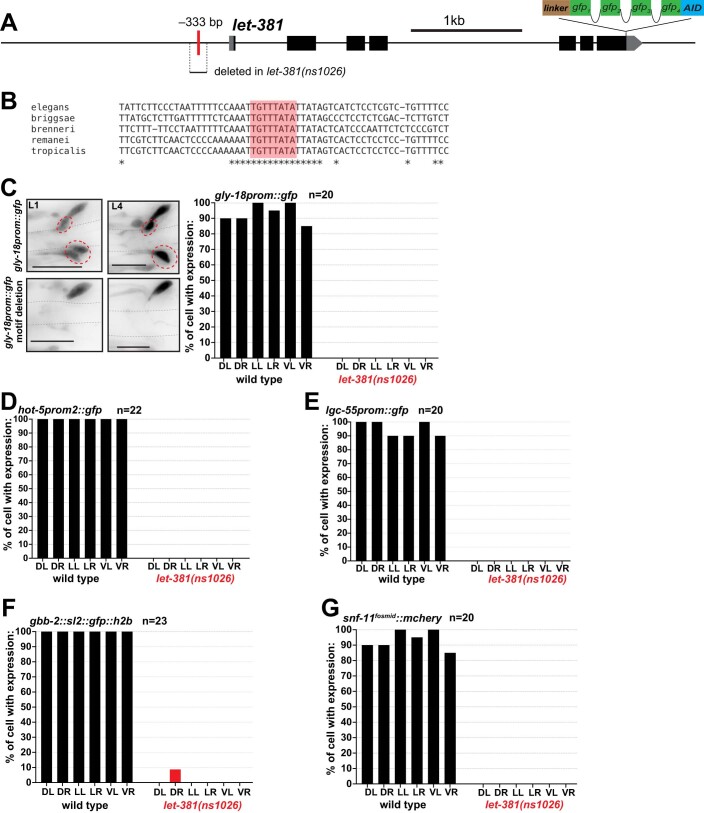
To understand how the unusual fate merger of GLR glia arises, we sought to identify transcription factors that control GLR glia fate specification and differentiation (Table EV1). Transcripts encoding LET-381, the sole C. elegans ortholog of the Forkhead domain transcription factor FOXF (Amin et al, 2010), are highly enriched in GLR glia. Recent studies suggest that FoxF genes are expressed in phagocytic glia and independently in endothelial mural cells (Reyahi et al, 2015; Scimone et al, 2018), raising the possibility that LET-381 could govern a combined glia/endothelial gene expression pattern in GLR glia. To follow let-381 expression in developing animals, we used CRISPR/Cas9 to insert gfp coding sequences into the endogenous let-381 locus (Fig. 2A). Transgenic homozygotes display nuclear GFP fluorescence in likely GLR glia precursors (pre-bean; Fig. 2B), as previously identified by lineaging of a let-381 transcriptional reporter (Murray et al, 2012), and in GLR glia until adulthood (Fig. 2B). Expression is also detected in the head mesodermal cell (HMC) and in coelomocytes (Fig. 2B; Appendix Fig. S2A), cell types also generated by the MS lineage (Fig. 1A). Animals carrying the gfp reporter transgene do not exhibit the lethality associated with loss of let-381 (see below), suggesting that let-381 gene function is retained.
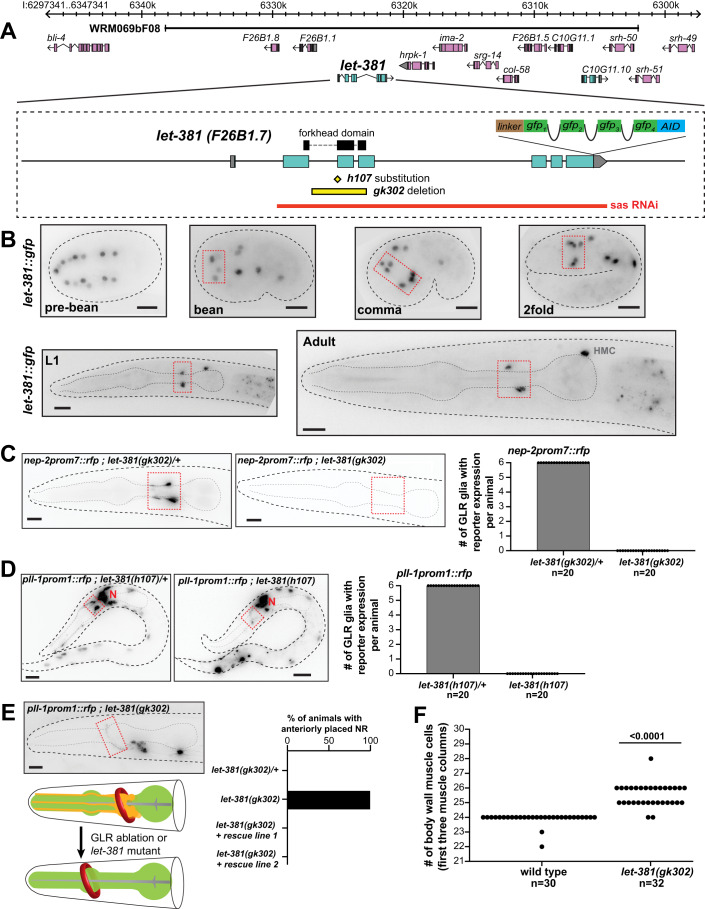
Figure 2. let-381/FoxF is required for GLR glia fate specification.
(A) let-381 genomic locus showing mutant alleles, reporters, fosmid genomic clones and RNAi sequences used in this study. (B) Expression of the endogenous let-381::gfp reporter at different stages during development. Dashed red boxes outline expression in GLR glia. (C) Absence of nep-2prom7::rfp expression in GLR glia (dashed red box) in homozygous let-381(gk302) mutants (right) as opposed to heterozygous animals (left). Quantification (number of GLR glia with nep-2prom7::rfp expression) is shown in the bar graph on the right. (D) Absence of pll-1prom1::rfp expression in GLR glia (dashed red box) in homozygous let-381(h107) mutants (right) as opposed to heterozygous animals (left). Quantification (number of GLR glia with pll-1prom1::rfp expression) is shown in the bar graph. Red “N” denotes pll-1prom1::rfp expression in neurons. (E) Similar to GLR glia-ablated animals (schematic), let-381(gk302) homozygous mutants lacking GLR glia exhibit anteriorly displaced nerve ring (NR). Dashed red box outlines a neuronal axon of the NR. Red circle in schematic indicates the Nerve Ring and pharynx is shown in green. Quantification is shown in the bar graph. Transgenic animals carrying the fosmid genomic clone WRM069bF08 with wild-type let-381 (rescue lines 1 and 2) display normal NR position. (F) Number of body wall muscle cells in the first three muscle columns of head and neck in wild type and let-381(gk302) mutants. Data information: unpaired t test was used for statistical analysis in (F). Anterior is left, dorsal is up and scale bars are 10 μm for all animal images. Source data are available online for this figure.
To determine whether let-381 promotes GLR glia fate specification, we introduced transgenic and endogenous GLR glia reporters into animals homozygous for the previously identified let-381 null alleles gk302 and h107. gk302 contains a deletion spanning LET-381 DNA binding domain encoding sequences; and h107 is a G to A substitution at a splice acceptor site (Fig. 2A). Animals homozygous for either allele undergo late-embryonic/early-larval arrest, with a few gk302 animals surviving to become sterile adults (sterility may reflect improper sex muscle specification in let-381 mutants (Amin et al, 2010)). Homozygous mutant gk302 or h107 animals fail to express five different GLR glia reporters that we tested (nep-2prom7::gfp, pll-1prom1::rfp, gly-18prom::gfp, hlh-1::gfp, unc-30::gfp) (Fig. 2C,D; Appendix Fig. S2B), suggesting that GLR glia are not generated in these mutants. Consistent with this, laser ablation of GLR glia precursors was previously shown to cause anterior displacement of the nerve ring (Shah et al, 2017) and we find a similar defect in let-381 mutants (Fig. 2E). In addition, the GLR glia sister lineage produces head body wall muscle cells (Fig. 1A), and we observe extra muscle cells in the heads of let-381(gk302) mutants (Fig. 2F). All let-381(gk302) mutant defects are rescued by a transgene containing the wild-type let-381 locus (let-381 fosmid WRM069bF08; Fig. 2A,E; Appendix Fig. S2C,D). Taken together, our results suggest that LET-381 is required for the specification of GLR glia, and in its absence, some GLR lineages adopt sister muscle lineage fates instead.
let-381/FoxF is continuously and cell-autonomously required to maintain GLR glia gene expression
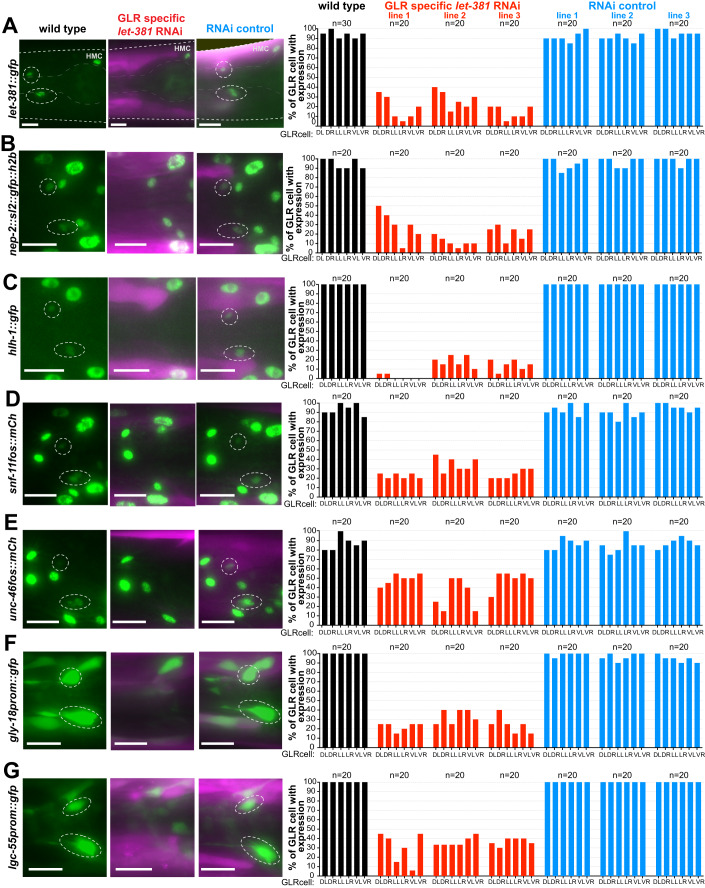
Although LET-381 is required early to specify GLR glia, its expression during larval development and in adults suggests it may have additional later roles. To test this idea, we knocked down let-381 in GLR glia of let-381::gfp animals by RNAi, using transgenic constructs co-transcribing sense and antisense let-381 sequences (Esposito et al, 2007) from the postembryonic nep-2prom7 promoter. These animals, also homozygous for the RNAi-sensitizing allele eri-1(mg366), downregulate GFP expression specifically in GLR glia, but not in other let-381 expressing cells, confirming RNAi specificity and efficacy (Fig. 3A). Importantly, let-381 RNAi transgenes downregulate expression of GLR glia reporters for nep-2/neprilysin, hlh-1/MyoD/Myf, snf-11/GAT GABA transporter, unc-46/LAMP-like, gly-18/N-acetylglucosaminyl transferase, and lgc-55/tyramine receptor (Fig. 3B–G). Expression of unc-30/Pitx2 is not affected by let-381 RNAi (see below), allowing us to visualize the cells and determine that they are still generated. Indeed, let-381 RNAi animals neither display an anteriorly displaced nerve ring nor have extra head muscle cells (Appendix Fig. S3A,B). Thus, LET-381 is required post-embryonically to maintain GLR glia gene expression, and this function is distinct from its role in generating GLR glia.
Figure 3. Postembryonic let-381 knockdown affects GLR glia gene expression.
(A–G) gfp or mCherry-based reporter expression (green) in GLR glia of endogenously tagged (A) let-381, (B) nep-2, and (C) hlh-1 and transgenic (D) snf-11 (E) unc-46, (F) gly-18, and (G) lgc-55 in wild-type (left column), GLR-specific postembryonic let-381 RNAi (middle column) and RNAi control animals (right column). Fluorescence images of L4 animals are shown. GFP expression in GLR glia (dashed white circles) is downregulated in the let-381 RNAi animals but not affected in RNAi control. RNAi and control lines carry a co-injection marker expressed in body wall muscle (magenta). Quantification is shown in bar graphs on the right. Each bar represents % of expression in each of the six GLR glia (DL, DR, LL, LR, VL, VR) in the three different backgrounds (wild type = black, GLR-specific RNAi = red, RNAi control = blue). Three independent extrachromosomal lines were scored for the RNAi and RNAi controls. Data information: anterior is left, dorsal is up, and scale bars are 10 μm for all animal images. Source data are available online for this figure.
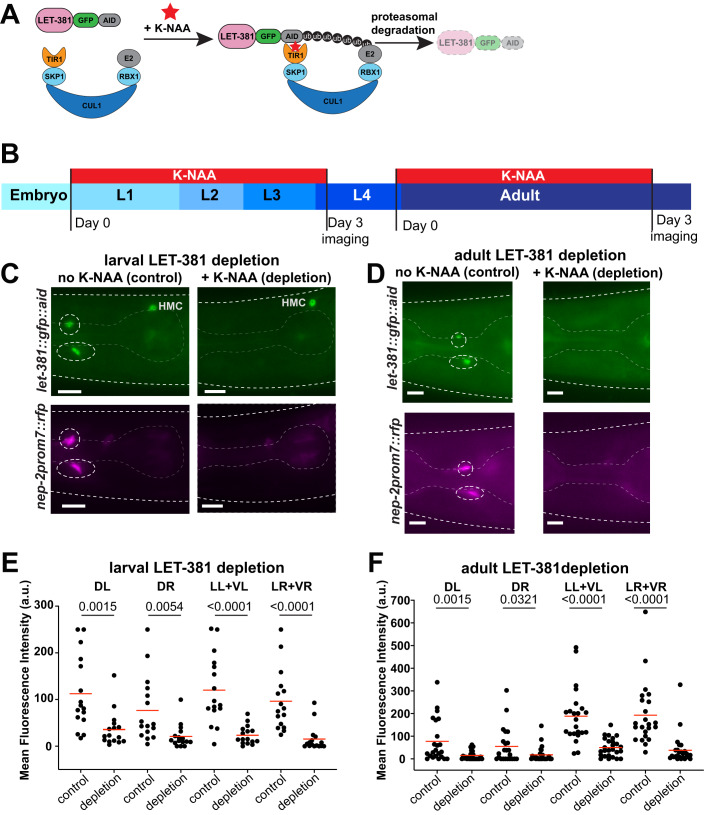
To probe dynamic functions of let-381, we used CRISPR/Cas9 to insert sequences encoding an auxin-inducible degron (AID) (Zhang et al, 2015) into the let-381 genomic locus (Fig. 2A). In the presence of GLR glia-expressed TIR1 and a synthetic auxin analog (K-NAA) (Martinez et al, 2020), AID-tagged LET-381 protein is degraded within 2 hours specifically in GLR glia (Fig. 4A,C, upper left panel; Appendix Fig. S4). A three-day exposure to K-NAA starting either at the L1 or late-L4/young-adult stages downregulates nep-2prom7::rfp reporter expression (Fig. 4B–F). Thus, let-381 functions cell-autonomously and is continuously required to maintain GLR glia gene expression, even in adults.
Figure 4. Acute larval and adult LET-381 depletion results in loss of GLR gene expression.
(A) Schematic representation of auxin-induced degradation (Zhang et al, 2015) of LET-381. TIR1 is transgenically provided and expressed specifically in GLR glia by the nep-2prom7 promoter. (B) Timeline of the auxin-inducible depletion experiment. Synchronized populations of L1 or late-L4/Young-adult animals were placed on plates containing the auxin analog K-NAA and imaged after a three-day exposure to K-NAA. Fluorescence intensities of gene expression in GLR glia were compared to age-matched animals grown on control plates without K-NAA. (C, D) Fluorescence images showing the result of (C) larval and (D) adult depletion of LET-381 using auxin-inducible degradation. Animals grown on control plates without K-NAA (left panels) show expression of let-381::gfp (green) and nep-2prom7::rfp (magenta) in GLR glia (dashed white circles). Age-matched animals grown on K-NAA-containing plates (right panels) show depletion of endogenous let-381::gfp expression specifically in the GLR glia; as shown expression in HMC remains unaffected. As a result, expression of the GLR-specific nep-2prom7::rfp reporter is downregulated in GLR glia. (E, F) Quantification (mean fluorescence intensity in cell bodies) of (E) larval and (F) adult LET-381 depletion on expression of nep-2prom7::rfp reporter in each GLR glia. Cell bodies of Lateral and Ventral GLR glia are too close to be clearly distinguished, thus they were grouped (LL + VL, LR + VR) for quantification purposes for this experiment. Red lines in dot plots indicate averages. Data information: unpaired t test used for statistical analysis in (E, F). n = 16 for control and depletion in (E), n = 23 for control and n = 26 for depletion in (F). a.u. = arbitrary units. Anterior is left, dorsal is up and scale bars are 10 μm for all animal images. Source data are available online for this figure.
LET-381 binding motifs are required and sufficient for GLR glia gene expression
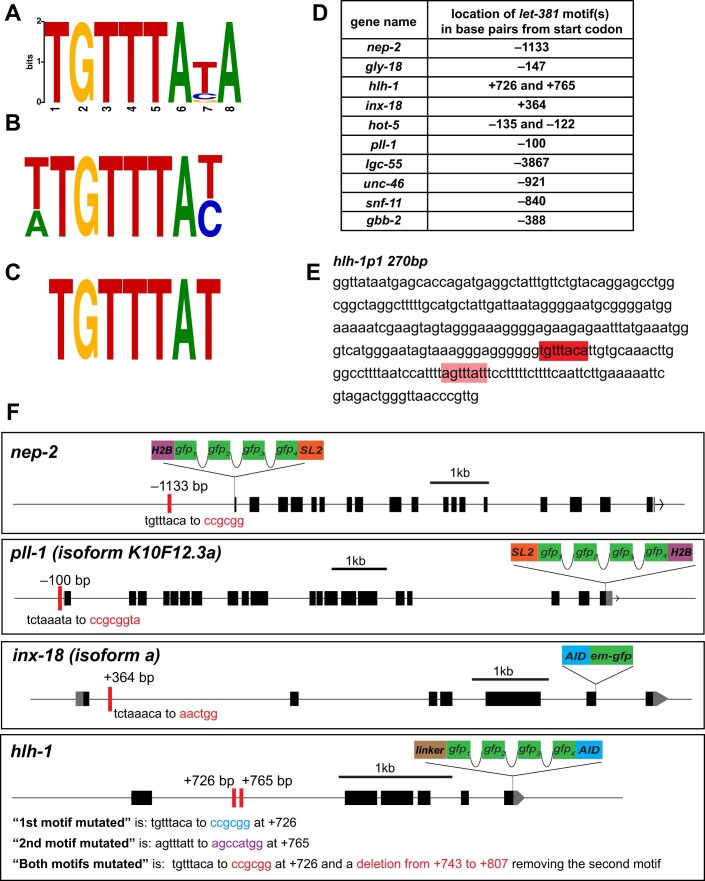
The expression of LET-381 in GLR glia throughout its life suggests it may function as a terminal selector, directly co-regulating expression of terminal differentiation gene batteries via a shared cis-regulatory motif (Hobert, 2011). To test this idea, we used sequences identified in our promoter dissection studies, ranging in size from 125 to 1022 bp, as input for the motif discovery tool MEME (Bailey and Elkan, 1994). This analysis identified a TGTTTA(C/T/G)A sequence common to all sequence inputs (Fig. EV1A). Remarkably, this sequence is highly similar to a previously identified FoxF binding sequence in mice (Peterson et al, 1997) and to a C. elegans LET-381 binding sequence identified through protein binding microarrays (Narasimhan et al, 2015) (Fig. EV1B,C). Notably, we find this motif in regulatory regions of all genes whose expression in GLR glia is downregulated by let-381 knockdown (Fig. EV1D). We refer to the TGTTTA(C/T/G)A sequence as the let-381 motif.
Figure EV1. let-381 motifs are required for endogenous GLR gene expression and let-381 autoregulation in GLR glia.
(A) let-381 motif identified in this study. (B) Motif of the let-381 ortholog foxf from (Peterson et al, 1997). (C) let-381 motif from (Narasimhan et al, 2015). Similarities between the three motifs are apparent. (D) Locations (distances from start codons) of let-381 motifs of genes whose expression in GLR is downregulated in let-381 mutants (either GLR-specific let-381 RNAi and/or the let-381 autoregulatory allele). (E) Minimal promoter hlh-1prom1 was one of the promoters used in MEME to identify common motifs present in GLR glia genes. The let-381 motif identified by MEME is highlighted in dark red. A let-381 motif with slightly altered sequence (light red) was identified manually later and is required, together with the first motif, to control hlh-1 expression in GLR glia. (F) Schematics showing details on endogenous gfp-based tags, location of let-381 motifs and their mutation for nep-2, pll-1, hlh-1 and inx-18 genes. Red bars represent let-381 motifs. Distance from ATG is indicated above each motif. Nucleotide changes for each motif mutation is shown below the motifs.
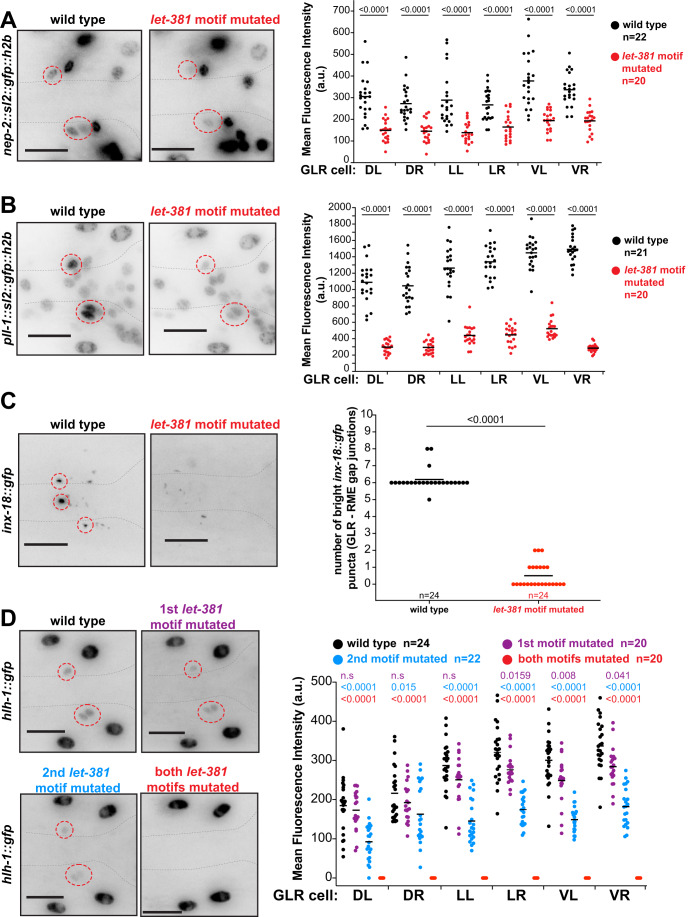
To assess how the motif affects gene expression, we used CRISPR/Cas9 to mutate it in different genomic locations (Fig. EV1F). As shown in Fig. 5, mutating let-381 motifs in upstream regulatory regions of the nep-2 and pll-1 genes, fused endogenously to gfp, significantly reduces gfp expression (Fig. 5A,B). Animals homozygous for an endogenous inx-18::gfp insertion allele we generated localize GFP in bright puncta marking the gap junctions between the GLR glia and the RME motoneurons. Mutagenesis of the inx-18 let-381 motif eliminates these bright puncta (Fig. 5C). Finally, disrupting either of two motifs in the gene hlh-1 only slightly reduces endogenous hlh-1::gfp expression; however, disrupting both together completely abolishes expression (Fig. 5D), suggesting that in some contexts, LET-381 binds multiple sites in the same gene.
Figure 5. let-381 motifs are required for endogenous gene expression in GLR glia.
(A–D) Endogenous expression of (A) nep-2, (B) pll-1, (C) hlh-1, and (D) inx-18 in wild-type and let-381 motif-mutated animals (details on endogenous gfp reporters and molecular identity of let-381 motif mutations are shown in Fig. EV1F). Animal images are on the left. Dashed circles outline expression in GLR glia. Quantifications are shown in the dot plots on the right. Data information: Mean fluorescence expression intensity for each GLR glia cell for (A, B, D) and number of bright gap-junction puncta for (C) is compared between the wild-type and let-381-motif-mutated backgrounds. Black lines indicate averages. Unpaired t test used for statistical analysis. a.u. = arbitrary units. Anterior is left, dorsal is up, and scale bars are 10 μm for all animal images. Source data are available online for this figure.
To further test the idea that let-381 is a terminal selector gene, we identified motifs in 15 GLR glia-enriched genes whose expression was not previously verified. We then generated animals transgenic for sequences surrounding let-381 motifs from each gene fused to gfp coding sequences (Table EV2). We found that let-381 motif-containing regions (ranging in size from 149 to 254 bp) from 14/19 genes are sufficient to drive GFP expression in GLR glia. Our experiments, therefore, support the idea that let-381 is a terminal selector gene, controlling the coordinate expression of genes expressed in differentiated GLR glia.
LET-381 regulates its own expression to maintain GLR glia identity
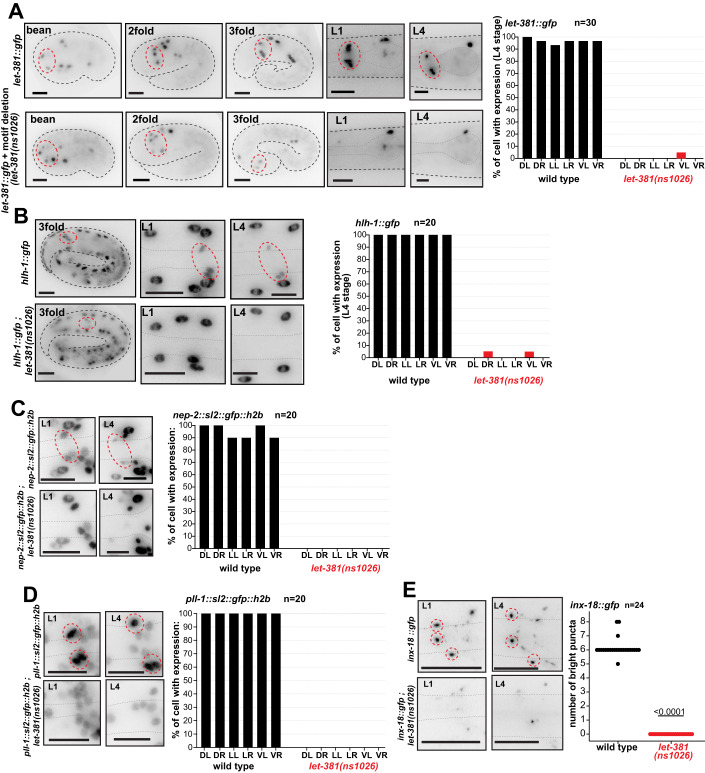
Given the requirement for let-381 in maintaining GLR glia gene expression even in adults, we wondered how let-381 expression itself is maintained. We identified a conserved let-381 motif upstream of the let-381 first exon (Fig. EV2A,B). We wondered whether, through positive feedback, this element could account for sustained let-381 expression through adulthood, and used CRISPR/Cas9 to delete this motif. In mutant animals, let-381(ns1026), endogenous let-381::gfp expression is observed in GLR glia of bean-stage embryos at levels similar to wild type. However, expression gradually wanes and is completely lost in L1 larva and older animals (Fig. 6A). Thus, let-381 is required to maintain its own expression through an autoregulatory let-381 motif. Expression of nine downstream GLR glia genes (but not unc-30, see below) is also gradually lost by the L1 stage (Figs. 6B–E and EV2C–G), further supporting the notion that LET-381 is required for GLR glia fate maintenance. Of note, let-381 autoregulation mutants exhibit neither an anteriorly displaced nerve ring nor extra head muscles, consistent with the RNAi and AID knockdown results.
Figure EV2. GLR gene expression is lost in let-381 autoregulatory mutant animals.
(A) Schematic showing the location of the let-381 motif (red bar) in the let-381 promoter region and region deleted in the let-381(ns1026) mutation. (B) Conservation of the let-381 autoregulatory motif sequence (red box) is shown among five nematode species. Asterisks indicate conserved nucleotides. (C–G) Effect of let-381 autoregulatory motif deletion, let-381(ns1026), on expression of (C) gly-18, (D) hot-5, (E) lgc-55, (F) gbb-2, and (G) snf-11 in GLR glia. Bar graphs show quantifications of gene expression at the L4 stage. For (C) animal images showing gene expression at L1 and L4 stages in wild-type and mutant backgrounds are shown on the left. Dashed red circles outline expression in GLR glia. Data information: Anterior is left, dorsal is up and scale bars are 10 μm for all animal images.
Figure 6. let-381 positively regulates its own expression.
(A) Endogenous let-381::gfp expression in different development stages in two different backgrounds: wild type (top), let-381 autoregulatory motif deletion (bottom). Animal images are shown on the left and quantification of expression in the L4 stage is shown on the right: percentage of each GLR cell expressing let-381::gfp in wild-type and autoregulatory motif deletion backgrounds. Red dashed circles outline GLR glia. (B–E) Effect of autoregulatory motif deletion on endogenous (B) hlh-1, (C) nep-2, (D) pll-1, and (E) inx-18 gfp reporter expression in GLR glia at different developmental stages. Quantification is shown on the right of each animal image panel for L4 animals. Red dashed circles outline GLR glia. Data information: unpaired t test used for statistical analysis in (E). Anterior is left, dorsal is up, and scale bars are 10 μm for all animal images. Source data are available online for this figure.
unc-30/Pitx2, a let-381/FoxF target, controls GLR glia gene expression and shape
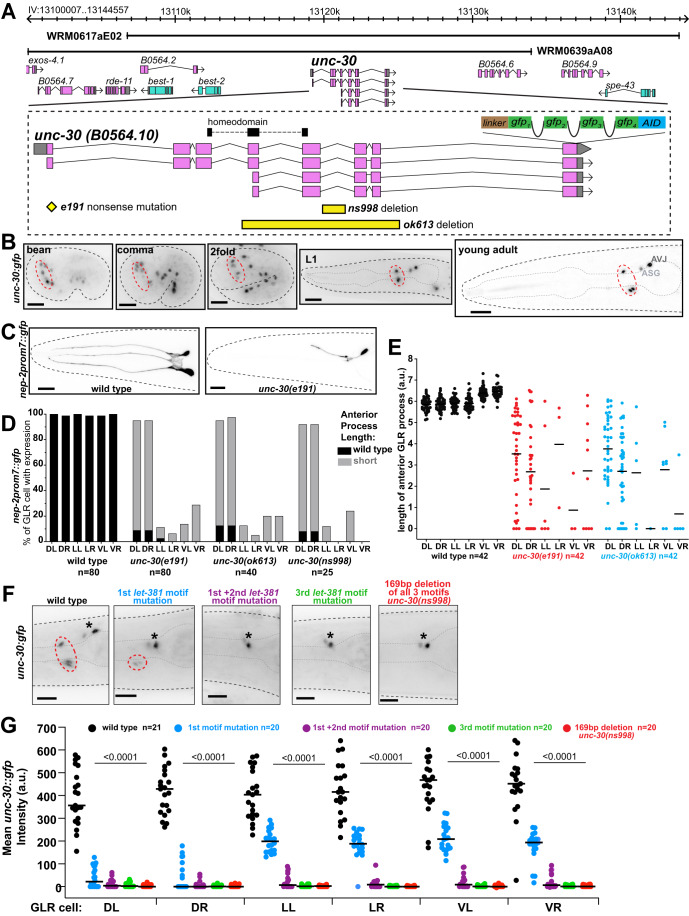
Our transcriptome studies revealed that transcripts encoding the transcription factor UNC-30/Pitx2 are also significantly enriched in GLR glia (Table EV1). UNC-30 was previously identified as a terminal selector of GABAergic identity in ventral cord neurons (Cinar et al, 2005; Eastman et al, 1999; Jin et al, 1994), and a recent study showed that GLR glia are GABA-positive by immunostaining and express the GABA-related genes snf-11/GAT, gta-1/GABA-T, and unc-46/LAMP (Gendrel et al, 2016). We found that animals carrying an endogenous unc-30::gfp reporter we generated (Fig. 7A) express GFP in GLR glia starting at the embryonic bean stage and through to adulthood (Fig. 7B). We also observed unc-30::gfp expression in the ASG, AVJ, DD, VD, and PVP neuron classes, all derived from the AB lineage (Fig. 7B; Appendix Fig. S5A).
Figure 7. unc-30 acts downstream of let-381 to control GLR glia gene expression and the length of GLR anterior process.
(A) unc-30 genomic locus showing mutant alleles, reporters, fosmid genomic clones used in this study. (B) Expression of the endogenous unc-30::gfp reporter at different stages during development. Dashed red circles outline GLR glia. (C) nep-2prom7::gfp expression in a wild-type L4 (left) and a unc-30(e191) null mutant L4 animal (right). GFP expression is lost in the lateral and ventral GLR glia in unc-30(e191). The anterior process of the dorsal GLR glia still expressing GFP is shorter than that of a wild-type animal. (D) Quantification of percentage of each GLR glia cell with nep-2prom7::gfp expression (L4 stage) for different unc-30 mutant backgrounds. (E) Quantification of the length of GLR anterior processes (L4 stage) for different unc-30 mutant backgrounds. (F) Images of L4 animals showing differences in endogenous unc-30::gfp expression upon mutation of the let-381 motifs present in the fifth intron of unc-30. Dashed red circles outline GLR glia and black asterisks denote expression in the ASG and AVJ head neurons. (G) Quantification of unc-30::gfp fluorescence intensity for each GLR glia cell; black lines in bar graphs indicate averages. Data information: unpaired t test used for statistical analysis in (G). a.u. = arbitrary units. Anterior is left, dorsal is up, and scale bars are 10 μm for all animal images. Source data are available online for this figure.
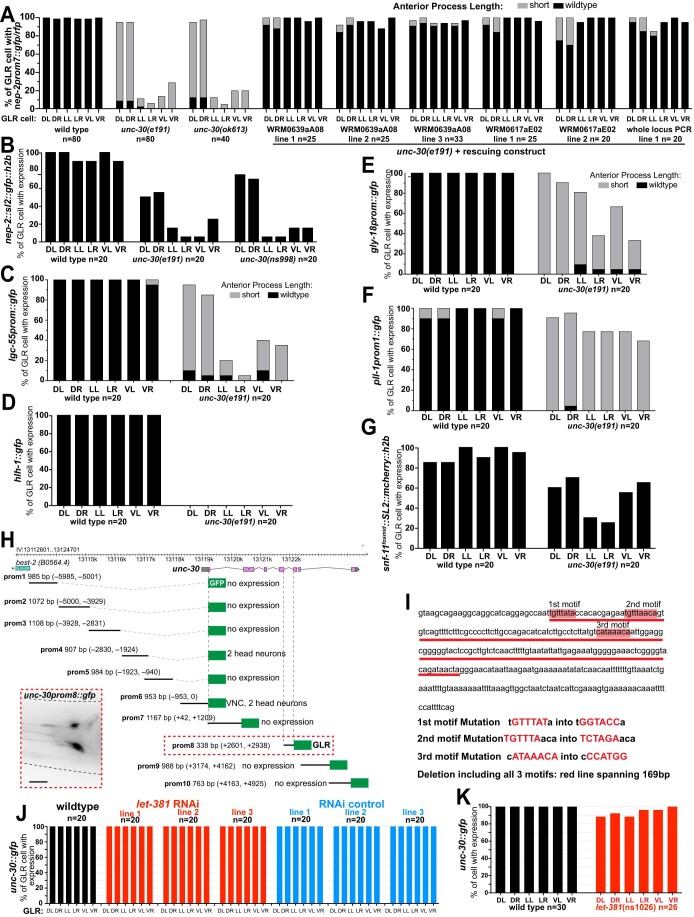
To determine whether unc-30 controls GLR glia gene expression, we crossed gene reporters into unc-30(e191) mutants, harboring an early nonsense mutation (Fig. 7A). Endogenous hlh-1::gfp expression is completely abolished in these animals, and expression of endogenous nep-2::gfp, transgenic nep-2prom7::gfp, or lgc-55prom::gfp is also lost, but mainly in lateral and ventral GLR glia (Figs. 7C,D and EV3B–D). Expression of transgenic gly-18prom::gfp, pll-1prom1::gfp and snf-11fosmid::SL2::mCherry:H2B reporters is reduced but not abolished (Fig. EV3E–G). In most animals in which reporter expression is not extinguished, GLR glia anterior processes are shortened (Fig. 7C,E). Similar findings are observed with the unc-30(ok613) deletion mutant (Fig. 7A,C,E), and all GLR glia defects are rescued with transgenes carrying the wild-type unc-30 locus (Fig. EV3A). The segregation of these unstable extrachromosomal rescuing transgenes to MS-lineage but not AB-lineage cells is sufficient to rescue GLR defects of unc-30(e191) mutants, suggesting that UNC-30 functions cell-autonomously in GLR glia (Appendix Fig. 5B–D). Thus, UNC-30 regulates GLR glia gene expression and also controls GLR glia morphology. Gene expression of dorsal GLR glia appears largely unaffected in unc-30 mutants. This suggests that UNC-30 has a more restricted effect on GLR gene expression than LET-381, and that other, yet unidentified transcription factors, may act as LET-381 cofactors to control gene expression in the dorsal GLR.
Figure EV3. Effect of unc-30 on GLR gene expression.
(A) Transgenic constructs containing different fosmid clones (WRM) or PCR amplicons carrying wild-type copies of UNC-30 can rescue the effect of unc-30(e191) on GLR gene expression and anterior process length. (B-G) Effect of unc-30 mutation on expression of different genes in GLR glia. Expression of (E) gly-18, (F) pll-1 and (G) snf-11 is affected at a lesser extent compared to (B) nep-2, (C) lgc-55 and (D) hlh-1. (H) Cis-regulatory dissection analysis of unc-30. The fifth intron (prom8) of unc-30 is sufficient to drive expression in GLR glia. (I) Three let-381 motifs are found in the fifth intron of unc-30 (red boxes). Details on let-381 motif mutation alleles are shown below the DNA sequence. (J, K) Endogenous unc-30::gfp expression is not affected by postembryonic let-381 knockdown either (J) by GLR-specific RNAi or (K) in the GLR-specific let-381 autoregulatory motif deletion allele let-381(ns1026). Data information: Anterior is left, dorsal is up and scale bars are 10 μm for all animal images.
How might let-381 and unc-30 interact? We found that the fifth intron of unc-30 contains three conserved let-381 motifs located within an 88 bp sequence (Fig. EV3H,I). Sequences derived from this intron are sufficient to promote GFP expression in GLR glia (Fig. EV3H). Furthermore, mutating the upstream let-381 motif alone substantially reduces endogenous unc-30::gfp expression, and simultaneous mutation of the two upstream motifs, a mutation of the downstream motif alone, or a 169 bp deletion removing all three motifs all specifically abolish unc-30::gfp expression in GLR glia but not in other unc-30 expressing cells (Figs. 7F,G and EV3I). Remarkably, the unc-30(ns998) 169 bp deletion allele, which abolishes unc-30::gfp expression only in GLR glia, phenocopies the effect of unc-30 null alleles on GLR glia gene expression and anterior process length (Figs. 7D and EV3B), supporting the notion that unc-30 functions cell-autonomously in GLR glia. We found no effects of unc-30 loss on the expression of let-381; however, let-381 expressing cells are often displaced along the dorsoventral and left-right axes in unc-30 mutants (Fig. 8A,B). Together, these results support the conclusion that unc-30 acts cell-autonomously to control GLR glia gene expression and anterior process length and that let-381 controls unc-30 expression in GLR glia. Previous studies suggest that UNC-30 may regulate its own expression (Cinar et al, 2005; Hobert, 2011), suggesting that following initial LET-381 binding, unc-30 may become independent of let-381. This notion is supported by our findings that unc-30::gfp expression in GLR glia is unaffected when let-381 is knocked down post-embryonically (Fig. EV3J,K).
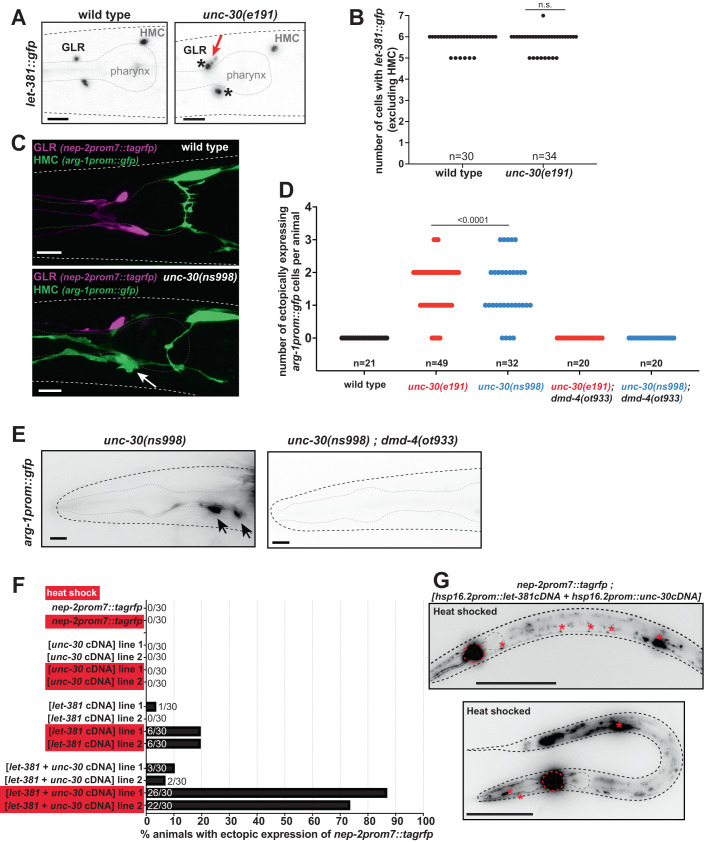
Figure 8. unc-30 represses HMC gene expression in GLR glia.
(A) Images showing let-381::gfp expression in wild type (left) and unc-30(e191) mutants (right). Expression is observed in GLR glia anterior to the pharynx bulb and the HMC above and posterior to the pharynx bulb. In wild-type background animals, GLR glia have a small sesame-like nucleus shape. In contrast in unc-30 mutants, some GLR glia nuclei appear larger and more round (black asterisks), reminiscent to the nucleus of the HMC cell. In addition, GLR glia are often mispositioned along the dorsoventral or left-right axis in unc-30 mutants. Red arrow points to dorsally mispositioned cells. (B) Number of let-381::gfp expressing cells is unaffected in unc-30(e191) null mutants. (C) Expression of GLR-specific nep-2prom7::tagrfp (magenta) and HMC-specific arg-1prom::gfp (green) in wild-type and unc-30(ns998) mutant backgrounds. Fluorescence images of L4 animals are shown. In the GLR-specific unc-30(ns998) mutant background, GLR glia lose GLR-specific RFP expression and ectopically express HCM-specific GFP instead (white arrow). (D) Quantification of ectopic expression of the HMC-specific arg-1prom::gfp in GLR glia in unc-30(e191) and unc-30(ns998) mutants. (E) Ectopic arg-1prom::gfp expression in unc-30 mutants is not observed in the dmd-4(ot933) mutant background. Black arrows point to ectopic arg-1prom::gfp expression. (F) Percentage of animals displaying ectopic expression of the GLR glia-specific reporter nep-2prom7::rfp, upon heat-shock-induced misexpression of let-381 and unc-30. Heat-shocked animals (red boxes) are compared to age-matched non-heat-shocked controls. (G) Images showing animals with ectopic nep-2prom7::rfp expression after heath shock-induced misexpression of LET-381 and UNC-30. Red dashed circles outline the expression GLR glia. Red asterisks point to expression in ventral nerve cord motor neurons and stomatointestinal muscle (upper panel) and body wall and pharynx muscle (bottom panel). Data information: unpaired t test used for statistical analysis in (B, D). Anterior is left, dorsal is up, and scale bars are 10 μm for (A, C, E). Scale bars are 100 μm for (G). Source data are available online for this figure.
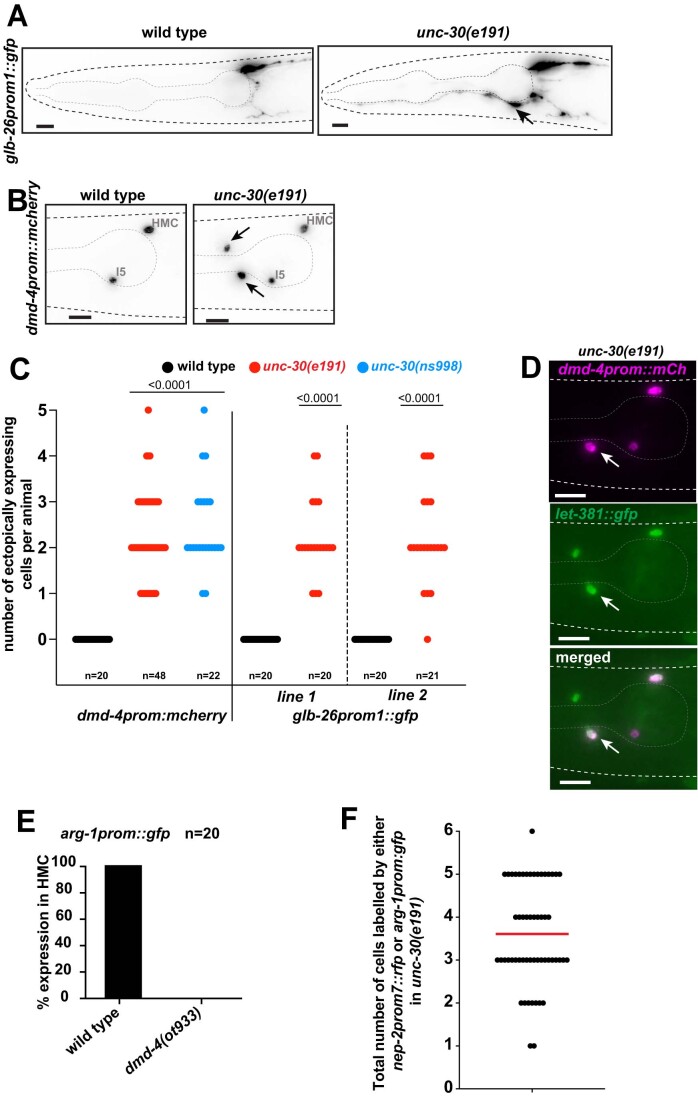
unc-30/Pitx2 represses expression of HMC genes in GLR glia
Wild-type GLR glia have small, sesame-seed-shaped nuclei. We noticed that in unc-30 mutants, GLR glia nuclei are larger and rounder, resembling the nucleus of the head mesodermal cell (HMC), another let-381-expressing cell (Fig. 8A). To determine whether unc-30 loss results in GLR glia acquiring additional HMC characteristics, we generated a nep-2prom7::tagrfp GLR glia reporter strain also expressing arg-1prom::gfp, an HMC reporter. We found that in unc-30(e191) mutants, some GLR glia that lose RFP expression now ectopically express the HMC reporter. unc-30(ns998) mutants exhibit similar defects (Fig. 8C,D). Likewise, two additional HMC reporters, glb-26prom::gfp and dmd-4::his24::mCherry, are also mis-expressed in GLR glia of unc-30 mutants (Fig. EV4A–C). While HMC-converted GLR glia retain let-381::gfp expression (Fig. EV4D), we never observe GLR glia expressing a mix of GLR glia-specific and HMC-specific reporters. Finally, we found that ectopic expression of the HMC reporter arg-1prom::gfp requires the DMRT transcription factor DMD-4 (Figs. 8D,E and EV4E), normally required for arg-1prom::gfp expression in the HMC (Bayer et al, 2020). We conclude that UNC-30 acts in GLR glia to repress dmd-4-dependent HMC-specific gene expression. This model succinctly explains the differential effect of unc-30 loss on the various reporters we tested (Fig. EV4B–G): snf-11, gly-18 and pll-1 reporters are normally expressed in both GLR glia and HMC and are therefore less affected by an unc-30 mutation than nep-2, lgc-55, or hlh-1, which are expressed in GLR glia but not in HMC.
Figure EV4. unc-30 represses HMC gene expression in GLR glia.
(A) Fluorescence images showing expression of glb-26prom::gfp in wild type and unc-30(e191) mutants. (B) Fluorescence images showing expression of dmd-4prom::mCherry in wild type and unc-30(e191) mutants. Arrows point to ectopic expression in unc-30(e191) mutants. (C) Quantification of ectopic expression of the two HMC reporters shown in (A) and (B) in unc-30 mutant backgrounds. (D) Cells ectopically expressing (white arrow) the HMC reporter dmd-4prom::mCherry (magenta) always co-express let-381::gfp (green). (E) Expression of arg-1prom::gfp is lost in HMC in dmd-4(ot933) mutants. (F) Total number of GLR glia cells expressing either the GLR glia-specific nep-2prom7::rfp or the HMC-specific arg-1prom::gfp in unc-30(e191) mutants. Data information: unpaired t test used for statistical analysis in (C). Anterior is left, dorsal is up and scale bars are 10 μm for all animal images.
Does unc-30 control of GLR glia-specific gene expression require inhibition of HMC gene expression? We found that even though the total number of cells expressing let-381::gfp is unaltered between wild type and unc-30 mutants (6 total cells—excluding HMC; Fig. 8B), the number of cells expressing either a GLR glia or an ectopic HMC reporter rarely adds up to 6 (Fig. EV4F), indicating that there are GLR glia that lose GLR glia gene expression without acquiring HMC fate. Thus, it is likely that unc-30 mediates GLR glia-specific gene expression independently of HMC gene expression suppression.
let-381 and unc-30 are together sufficient to induce GLR glia gene expression
The broader effect of let-381 loss on GLR glia gene expression suggests that expression of this gene in naive cells should drive the GLR glia expression program in these cells. Surprisingly, we found that this is not the case (Fig. 8F). Indeed, broad inducible misexpression of let-381 cDNA using a heat-shock promoter results in minimal misexpression of the nep-2prom7::tagrfp GLR glia reporter. We found a similar result using unc-30 cDNA alone. However, misexpression of both let-381 and unc-30 results in highly penetrant misexpression of nep-2prom7::tagrfp in many cells, including body wall muscle, pharyngeal muscle, stomatointestinal muscle and ventral nerve cord motoneurons (Fig. 8F,G). Thus, let-381 is not sufficient to induce GLR glia fate and must cooperate with unc-30.
GLR glia-defective animals display locomotion abnormalities
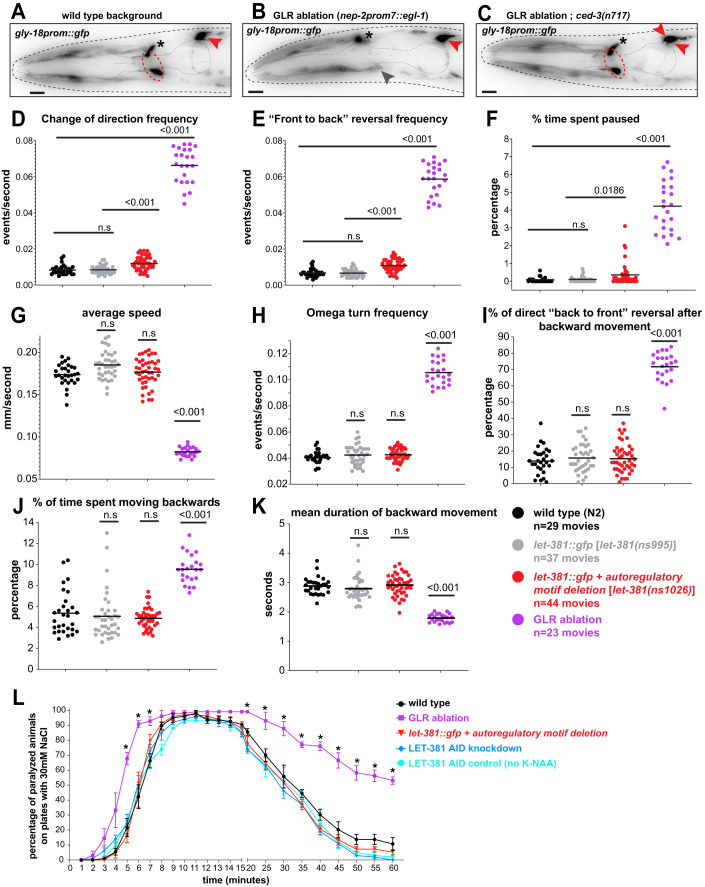
Our development of tools to manipulate gene expression and function specifically in GLR glia allowed us to interrogate the functions of these cells. To do so, we generated a nep-2prom7::egl-1 transgenic line driving postembryonic expression of EGL-1, a pro-apoptotic caspase activator, only in GLR glia. We observed GLR glia loss, rescued by the ced-3(n717) caspase mutation, confirming that GLR glia in this strain die by apoptosis (Fig. 9A–C). We next recorded movies and analyzed the locomotion of GLR glia-ablated animals freely moving on an agar plate without food (Katz et al, 2018; Katz et al, 2019). Remarkably, we found that while GLR glia-ablated animals can move, they exhibit severe defects in all locomotion parameters we analyzed, including reduced locomotion rates, increased turning frequency, increased reversal rates, and increased pausing (Fig. 9D–K; Movies EV1 and EV2). These defects are similar to those observed following ablation of CEPsh glia, C. elegans astrocytes that line the outer aspect of the nerve ring and regulate synaptic function (Katz et al, 2018; Katz et al, 2019). In addition, while wild-type animals exposed to high-salt concentrations are initially paralyzed and then recover, GLR glia-ablated animals paralyze more quickly and recover at a much slower rate (Fig. 9L), suggesting that GLR glia may play an important role in the control of solute permeability and ionic balance in the nerve ring. Thus, GLR glia are important for coordinated locomotion and nerve ring function.
Figure 9. GLR glia-defective animals display locomotion abnormalities and hypersensitivity at high-salt concentrations.
(A–C) Images of L4 animals showing expression of gly-18prom::gfp reporter in (A) wild-type, (B) GLR glia ablation and (C) GLR glia ablation; ced-3(n717) backgrounds. GLR glia are outlined by red dashed circles. Black asterisks denote expression in a neuronal cell just above the dorsal GLR glia and red arrowheads point to HMC cells. In the GLR glia-ablation background, the nerve ring is anteriorly displaced as noted by a head muscle arm (gray arrowhead) penetrating the nerve ring at the anterior pharynx bulb. The HMC sister cell that normally dies by programmed apoptotic cell death in a wild-type animal, survives and also expresses gly-18prom::gfp in the ced-3(n7171) mutant background in (C). (D–K) Locomotion parameters of foraging wild-type (black), control (gray), GLR glia-defective (red), and GLR glia-ablated (purple) animals were analyzed using automated tracking (Katz et al, 2018; Katz et al, 2019). (D) change of direction frequency, (E) “front to back” reversal frequency, (F) % time spent paused, (G) average speed, (H) omega turn frequency, (I)% direct “back to front” reversal after backward movement, (J) % time spent moving backwards, (K) mean duration of backward movement. Black lines in dot plots indicate averages. (L) GLR glia-ablated animals paralyze at a significantly higher rate than wild-type animals and animals in which LET-381 is downregulated [let-381(ns1026) and LET-381 AID knockdown] when exposed to 300 mM NaCl. GLR glia-ablated animals also recover motility at a significantly lower rate. Data information: unpaired t test was used for statistical analysis in (D–L), *<0.05 in (L). For (L), 4 replicates were performed per genotype, n = 20–25 for replicate, error bars indicate standard error of the mean. Anterior is left, dorsal is up, and scale bars are 10 μm for all animal images. Source data are available online for this figure.
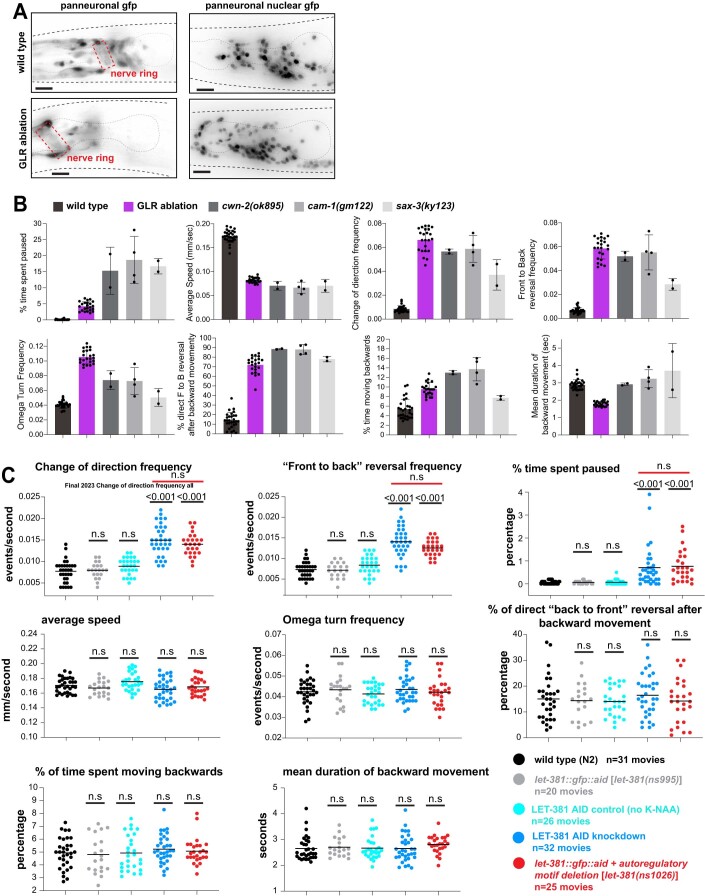
We noted that, like animals whose GLR glia progenitors are laser ablated during embryogenesis (Shah et al, 2017), nep-2prom7::egl-1 ablated animals have anteriorly displaced and sometimes defasciculated nerve rings (Fig. EV5A). To determine whether the abnormal locomotion we observed is a consequence of these structural defects, we examined the behavior of axon-guidance mutant strains that also possess anteriorly displaced nerve rings (Kennerdell et al, 2009; Zallen et al, 1999). Indeed, these mutants also exhibit defects in all locomotion parameters tested. However, the magnitude of these defects is not always the same as in GLR glia-ablated animals (Fig. EV5B). Thus, while some of the locomotion defects may be attributed to structural defects in the nerve ring, it appears that animals lacking GLR glia may be additionally compromised. To uncouple the physical positioning of the nerve ring from other GLR glia functions in C. elegans locomotion, we examined movement of animals carrying the let-381 autoregulatory motif mutation, which does not result in nerve ring displacement. This strain, let-381(ns1026), was generated from the let-381(ns995) strain, containing an insertion of gfp into the let-381 locus. As in GLR glia-ablated animals, we find specific defects in let-381(ns1026) mutants: animals change direction more frequently than let-381(ns995) controls (Fig. 9D), display a higher frequency of reversals (Fig. 9E), and tend to pause more often (Fig. 9F). Other behaviors defective in ablated animals are unaltered (Fig. 9G–K). Auxin-dependent LET-381::AID downregulation late in development (24 hour auxin exposure of L4 animals) does not disturb nerve ring positioning and affects locomotion to the same extent as the let-381(ns1026) autoregulatory mutation (Fig. EV5C). Unlike GLR ablation, neither let-381 autoregulatory mutant nor the let-381::AID knockdown result in hypersensitivity to high salt. We can only speculate that as opposed to GLR ablation, the GLR glia processes are still physically present in these mutants, and may therefore constitute enough of a physical barrier around the nerve ring, to protect it from the effects of sudden salt concentration shifts. Taken together, our observations support the conclusion that GLR glia regulate C. elegans locomotory behavior not only by ensuring nerve ring positioning but perhaps in non-structural ways as well, revealing a previously uncharacterized function for these cells.
Figure EV5. Locomotion defects of GLR-ablated animals could partially be due to anteriorly displaced nerve ring.
(A) In wild-type animals (top row), axons of the nerve ring (dashed red box) are located between the two pharyngeal bulbs. In GLR-ablated animals (bottom row) the nerve ring is anteriorly displaced, located on top of the anterior pharynx bulb. As evidenced in the images on the right, not only the axonal projections, but also neuronal cell bodies (panneuronal nuclear gfp) are anteriorly displaced. Panneuronal gfp = unc-119prom::gfp, panneuronal nuclear gfp = rab-3prom1::nls::yfp. (B) cwn-2(ok895), cam-1(gm122), sax-3(ky123) mutants with anteriorly displaced nerve rings exhibit locomotion defects to the same direction, although of different magnitude as the GLR glia-ablated animals. (C) Auxin (K-NAA) dependent LET-381::AID knockdown results in similar defects in the same locomotion parameters as the let-381(ns1026) autoregulatory mutation. Genotypes are: wild-type N2 (black), let-381(ns995) control (gray), LET-381::AID knockdown [let-381(ns995);nsIs879 (nep-2prom7::TIR1)] exposed to K-NAA auxin (dark blue), LET-381::AID control [let-381(ns995) ; nsIs879 (nep-2prom7::TIR1)] not exposed to K-NAA auxin (light blue) and let-381(ns1026) autoregulatory mutation (red). Data information: in (B) wild type n = 29 movies, GLR ablation n = 23 movies, cwn-2(ok895) n = 2 movies, cam-1(gm122) n = 4 movies, sax-3(ky123) n = 2 movies. Bar height indicates average (center of error bars) and error bars show standard deviation in (B). Unpaired t test used for statistical analysis in (C); controls (gray and light blue) were compared to wild type (black). LET-38::AID knockdown (dark blue) was compared to its control group (light blue) and let-381(ns1026) was compared to its control (gray). No statistically significant differences were observed between the LET-381::AID knockdown and let-381(ns1026) as indicated by the red line on the top of the three upper diagrams. Anterior is left, dorsal is up and scale bars are 10 μm for all animal images.
Discussion
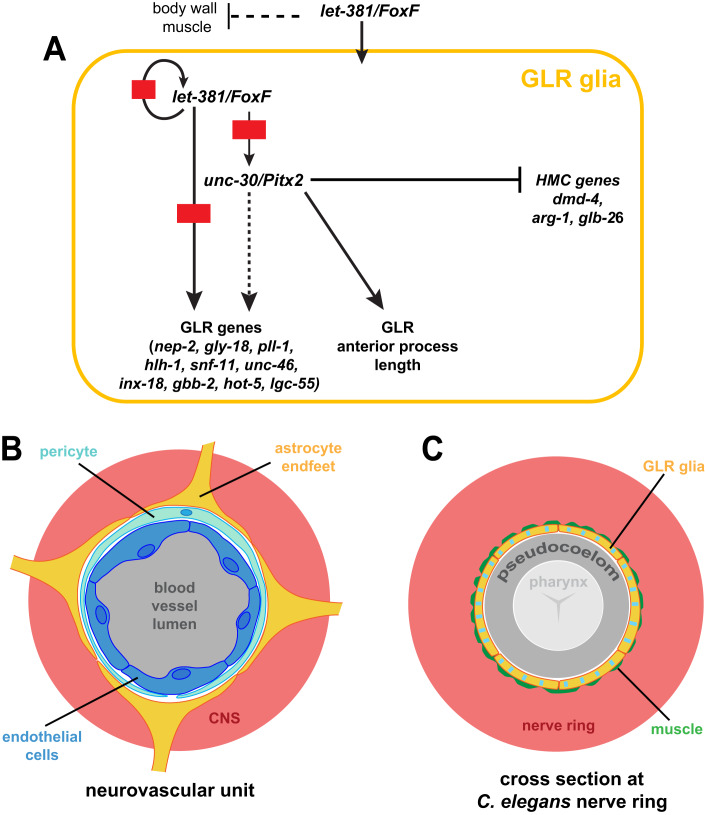
We describe here a gene regulatory network for the specification and maintenance of C. elegans GLR glia (Fig. 10A). Early in development, let-381/FoxF is required to specify GLR glia fate and to suppress sister-lineage body wall-muscle gene expression. It does so, in part, by promoting the expression of dozens of GLR glia-enriched genes, all of which possess cis-regulatory LET-381 binding motifs. Such a motif upstream of let-381 ensures that once turned on, the gene remains continuously expressed, as are its targets. Among these targets is unc-30/Pitx2, required for the expression of some let-381-dependent genes and for generating GLR glia anterior processes of appropriate length. unc-30 also independently prevents expression in GLR glia of genes and traits of the HMC, a non-contractile MS-lineage-derived cell (Choi et al, 2023). Together, LET-381 and UNC-30 can bestow GLR glia gene expression onto naive cells that do not normally express these transcription regulators.
Figure 10. Regulatory network for the specification and identity maintenance of the C. elegans GLR glia.
(A) Schematic of the regulatory network identified in this study controlling fate specification and differentiation of GLR glia. (B) At the neurovascular unit, endothelial cells (dark blue), pericytes (light blue), and astrocytic endfeet (yellow) form the Blood–Brain Barrier isolating the central nervous system (CNS—red) from blood circulation (gray). (C) Similarly, the GLR glia sheet-like processes (yellow with blue stripes to show a mixed astrocytic-endothelial/mural fate) isolate the C. elegans nerve ring (red) from the pseudocoelom (gray). A thin layer of head muscle arms (green) penetrates the C. elegans nerve ring and therefore GLR flat processes are in close proximity to head neuromuscular junctions (White et al, 1986). The pseudocoelom is shown larger than its actual volume.
While transcriptional control of glial cell fate is generally not well understood, neuronal fate specification by terminal selector transcription factors has been studied extensively in C. elegans, Drosophila, chordates, and mice (Hobert and Kratsios, 2019). These master regulators, often acting with a specific cofactor (Hobert, 2011; Hobert and Kratsios, 2019), establish and maintain neuron identity by co-regulating a wide variety of neuronal features, including gene expression and synaptic connectivity. Terminal selectors also ensure neuron identity by repressing alternative neuronal fates (Feng et al, 2020; Remesal et al, 2020). Here we show that similar fate control is exercised in C. elegans GLR glia by let-381/FoxF and its cofactor unc-30/Pitx2. It is therefore plausible that such mechanisms also specify the fates of glial cells in other settings, and may account for regional differences between glia in the vertebrate brain.
The formation of intestinal muscle from visceral mesoderm using FoxF transcription factors is conserved among Drosophila (Zaffran et al, 2001), planaria (Scimone et al, 2018), Xenopus (Mahlapuu et al, 2001) and the mouse (Ormestad et al, 2006; Tseng et al, 2004). In C. elegans, let-381/FoxF is not expressed in the 4 intestine-associated muscles. Nonetheless, it is expressed in GLR glia, the head mesodermal cell (HMC), and the endocytic coelomocytes, all derived from the mesoderm-like lineage of the blast cell MS. Similar to our findings in GLR glia, let-381/FoxF functions with ceh-34/Six2 in coelomocytes (Amin et al, 2010). Intriguingly, in planaria, FoxF1 promotes specification of phagocytic cells, including phagocytic glia (Scimone et al, 2018). These findings suggest that FoxF transcription factors are key cell-fate regulators of non-contractile mesodermally derived cells, including mesodermal glia. Indeed, single-cell sequencing of mouse CNS cells reveals FoxF1/2 expression in blood vessel endothelial and mural cells (Hupe et al, 2017; Saunders et al, 2018; Vanlandewijck et al, 2018; Zhang et al, 2014), with FoxF2 driving pericyte differentiation and maintenance of the blood–brain barrier (Reyahi et al, 2015). While FoxF genes are expressed only at low levels in microglia (Vanlandewijck et al, 2018), it remains possible that these genes direct early microglia differentiation.
Intriguingly, cerebellar astrocytes, which often ensheath blood vessels, also express FoxF1 (Kalinichenko et al, 2003), raising the possibility that FoxF genes play a broader role in specifying the entire neurovascular unit. Supporting this idea, we show here that C. elegans GLR glia merge astrocyte and mural cell molecular and anatomical characteristics (Fig. 10B,C). In phylogenetically older species, such as sturgeons (subclass: Chondrostei), astrocytes are thought to be the main components of the blood–brain barrier (Bundgaard and Abbott, 2008). Might GLR glia, by analogy then, form a barrier that isolates the C. elegans nerve ring from the pseudocoelom? Unlike endothelial cells, GLR glia do not adhere to each other to form a tight seal, and gaps between the cells are evident on electron micrographs (White et al, 1986). Furthermore, neuronal cell bodies, which reside outside the nerve ring, are not ensheathed and could have direct access to materials within the pseudocoelom. Nonetheless, roles for GLR glia in blocking diffusion of synaptically released factors into the pseudocoelom, which can then have systemic effects, are plausible, as are more general roles in regulating the extracellular environment of the nerve ring. Indeed, our behavior studies of GLR glia-ablated animals and GLR glia mutants reveal disruptions in the coordination of locomotion, suggesting effects on neuronal signaling within the nerve ring. GLR glia are electrically coupled with the RME head motoneurons and head muscle and thus motor defects of GLR mutants could be due to disruption of synchronized activity of this gap-junction network. Alternatively, GLR glia mediated changes in neurotransmitter or neuropeptide signaling of an underlying motor network could account for the observed defects; indeed, some GABA, glutamate, acetylcholine, tyramine and putative neuropeptide receptors are enriched in GLR glia. GLR glia could also be important for regulation of potassium homeostasis in the extra-synaptic space, as supported by the over-representation of TWIK potassium channels among GLR-enriched genes, therefore having a broader role on neuronal excitability and synchronization. While nerve ring positioning is not affected in let-381(ns1026) mutants, microscopic structural defects may exist in individual neurons. Dissecting the effects of GLR glia mutants on neuronal development and activity would therefore be crucial to understand GLR glia roles in regulation of motor behavior. The similarities in locomotory defects between CEPsh astrocyte-ablated animals and GLR glia-ablated animals suggest that both glial types affect a similar set of underlying processes. CEPsh glia wrap around the outside aspect of the nerve ring, are not in direct contact with the pseudocoelom, and are not enriched for endothelial genes, further supporting the idea that GLR glia have merged astrocyte and endothelial functions.
Methods
Caenorhabditis elegans strains and handling
Animals were grown on nematode growth media (NGM) plates seeded with E. coli (OP50) bacteria as a food source unless otherwise mentioned. Strains were maintained by standard methods (Brenner, 1974). The wild-type is strain N2, C. elegans variety Bristol RRID:WB-STRAIN:WBStrain00000001. A complete list of strains generated and used in this study is listed below (Table 1). A few of the strains were previously published, and/or obtained from the Caenorhabditis Genetic Center (CGC), or the TransgeneOme project.
Table 1.
List of strains used and generated in this study.
| Strain name | Genotype | Comment | Reference |
|---|---|---|---|
| N2 | wild type | CGC | |
| VC706 | let-381(gk302) I/hT2 (I;III) | (C. elegans Deletion Mutant Consortium 2012) | |
| KR429 | dpy-5(e61) let-381(h107) unc-13(e450) I; sDp2 (I;f) | (Howell et al, 1987) | |
| CB845 | unc-30(e191) IV | (Brenner, 1974) | |
| VC295 | unc-30(ok613) IV | (C. elegans Deletion Mutant Consortium 2012) | |
| GR1373 | eri-1(mg366) IV | (Kennedy et al, 2004) | |
| CX3198 | sax-3(ky123) X | (Zallen et al, 1999) | |
| VC636 | cwn-2(ok895) IV | (C. elegans Deletion Mutant Consortium 2012) | |
| NG2615 | cam-1(gm122) II | (Forrester et al, 1999) | |
| DE60 | dnIs13 [gly-18prom::gfp + unc-119( + )] I l; unc-119(e2498) III | (Warren et al, 2001) | |
| MT20492 | lin-15B&lin-15A(n765) X l; nIs471 [lgc-55prom::gfp + lin-15(+)] | (Ringstad et al, 2009) | |
| OH13025 | otIs567 [unc-46(fosmid)::SL2::H2B::mCHOPTI) + pha-1(+)] | (Gendrel et al, 2016) | |
| OH13027 | otIs569 [snf-11(fosmid)::SL2::H2B::mChopti + pha-1(+)] | (Gendrel et al, 2016) | |
| PD4443 | ccIs4443 [arg-1prom::gfp + dpy-20( + )] IV | (Kostas and Fire, 2002) | |
| SD1633 | ccIs4251 [(pSAK2) myo-3p::GFP::LacZ::NLS + (pSAK4) myo-3p::mitochondrial GFP + dpy-20( + )] I; stIs10539 [dmd-4p::HIS-24::mCherry + unc-119(+)] | (Liu et al, 2009) | |
| OH16770 | ccIs4443 [arg-1prom::gfp + dpy-20( + )] IV; him-5 (e1490) V ; dmd-4(ot933) X | (Bayer et al, 2020) | |
| OP185 | unc-119 (ed3) III; wgIs185 [fkh-2-1::TY1::EGFP::3xFLAG(92C12) + unc-119(+)] | (Sarov et al, 2006) | |
| BC10849 | dpy-5(e907) I; sIs10707 [sre-6prom::GFP + dpy-5(+)] | (McKay et al, 2003) | |
| OP447 | unc-119(tm4063) III; wgIs447 [tag-68::TY1::EGFP::3xFLAG + unc-119(+)] | (Sarov et al, 2006) | |
| OH9545 | otIs287 [rab-3prom1::2xnsl::yfp, rol-6(su1006)] IV | (Stefanakis et al, 2015) | |
| OH441 | otIs45 [unc-119prom::gfp] | (Altun-Gultekin et al, 2001) | |
| CF4587 | muIs253 [eft-3prom::sfGFP1-10::unc-54 3’UTR + Cbr-unc-199( + )] II ; unc-119(ed3) III | (Goudeau et al, 2021) | |
| JN332 | peEx3332 [nep-2prom(S)::Venus, rol-6(su1006)] | (Yamada et al, 2010) | |
| OH15262 | otIs669 (NeuroPAL) | (Yemini et al, 2021) | |
| OS11153 | nsEx5558 [nep-2prom1::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11157 | nsEx5562 [nep-2prom2::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11158 | nsEx5563 [nep-2prom3::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11162 | nsEx5567 [nep-2prom4::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11163 | nsEx5568 [nep-2prom5::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11165 | nsEx5570 [nep-2prom6::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11166 | nsEx5571 [nep-2prom7::GFP, pha-1(+)] line 1; pha-1 (e2123) III ; otIs356 [rab-3prom1::NLS-TagRFP] V | This study | |
| OS11176 | nsEx5581 [lgc-55prom1::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11177 | nsEx5582 [lgc-55prom2::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11178 | nsEx5583 [lgc-55prom3::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11180 | nsEx5585 [lgc-255prom4::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11181 | nsEx5586 [lgc-55prom5::GFP, pha-1(+) line 1; pha-1 (e2123) III | This study | |
| OS11182 | nsEx5587 [lgc-55prom6::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11183 | nsEx5588 [lgc-55prom7::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11017 | nsEx5824 [egl-6prom1::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11232 | nsEx5825 [egl-6prom2::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11234 | nsEx5827 [egl-6prom3::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11236 | nsEx5829 [egl-6prom4::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11202 | nsEx5591 [gly-18prom1::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11203 | nsEx5592[gly-18prom2::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11204 | nsEx5593 [gly-18prom3::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11205 | nsEx5594 [gly-18prom4::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11206 | nsEx5595 [gly-18prom5::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11208 | nsEx5597 [gly-18prom6::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11209 | nsEx5598 [gly-18prom7::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11211 | nsEx5809 [gly-18prom8::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11212 | nsEx5810 [gly-18prom9::GFP, pha-1(+)] line 1; pha-1 (e2123) III ; otIs356 [rab-3prom1::NLS-TagRFP] V | This study | |
| OS11168 | nsEx5573 [hlh-1prom1::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11171 | nsEx5576 [hlh-1prom2::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11172 | nsEx5577 [hlh-1prom3 cloned in pSM-GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11959 | nsEx6055 [inx-18prom1::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11960 | nsEx6056 [inx-18prom2::GFP, pha-1(+)] line 1; pha-1 (e2123) | This study | |
| OS11962 | nsEx6058 [inx-18prom3::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS12795 | nxEx6276 [inx-18prom4::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS12797 | nxEx6278 [inx-18prom5::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS12798 | nxEx6279 [inx-18prom6::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS12800 | nxEx6281 [inx-18prom7::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS12801 | nxEx6282 [inx-18prom8::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS12802 | nxEx6283 [inx-18prom9::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS12819 | nsEx6288 [inx-18prom10::gfp, pha-1 + ] line 1; pha-1(e2123) III | This study | |
| OS12821 | nsEx6290 [inx-18prom11::gfp, pha-1 + ] line 1; pha-1(e2123) III | This study | |
| OS14565 | nsEx7189 [unc-30prom1::gfp + pha-1 + ] line 1; pha-1(e2123) III | This study | |
| OS14566 | nsEx7190 [unc-30prom2::gfp + pha-1 + ] line 1; pha-1(e2123) III | This study | |
| OS14567 | nsEx7191 [unc-30prom3::gfp + pha-1 + ] line 1; pha-1(e2123) III | This study | |
| OS14568 | nsEx7192 [unc-30prom4::gfp + pha-1 + ] line 1; pha-1(e2123) III | This study | |
| OS14569 | nsEx7193 [unc-30prom5::gfp + pha-1 + ] line 1; pha-1(e2123) III | This study | |
| OS14570 | nsEx7194 [unc-30prom6::gfp + pha-1 + ] line 1; pha-1(e2123) III | This study | |
| OS14571 | nsEx7195 [unc-30prom7::gfp + pha-1 + ] line 1; pha-1(e2123) III | This study | |
| OS13296 | nsEx6463 [unc-30prom8::gfp + pha-1 + ] line 1; pha-1(e2123) III | This study | |
| OS14572 | nsEx7196 [unc-30prom9::gfp + pha-1 + ] line 1; pha-1(e2123) III | This study | |
| OS14573 | nsEx7197 [unc-30prom10::gfp + pha-1 + ] line 1; pha-1(e2123) III | This study | |
| OS14094 | nsEx6871 [F41G4.8prom1::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14173 | nsEx6921 [F41G4.8prom2::gfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::tagrf] V | This study | |
| OS14164 | nsEx6914 [twk-4prom1::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14158 | nsEx6908 [twk-4prom2::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14098 | nsEx6875 [twk-4prom3::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14157 | nsEx6907 [ocr-1prom1::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14273 | nsEx6966 [ocr-1prom2::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14097 | nsEx6874 [ocr-1prom3::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14099 | nsEx6876 [twk-9prom1::gfp +pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14172 | nsEx6920 [mig-6prom1::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14156 | nsEx6906 [mig-6prom2::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14174 | nsEx6922 [mig-6prom3::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14161 | nsEx6911 [mig-6prom4::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14175 | nsEx6923 [mig-6prom5::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14198 | nsEx6941 [mig-17prom1::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14188 | nsEx6931 [mig-17prom2::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14176 | nsEx6924 [let-2prom1::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14159 | nsEx6909 [let-2prom2::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14160 | nsEx6910 [let-2prom3::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14187 | nsEx6930 [let-2prom4::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14197 | nsEx6940 [let-2prom5::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14276 | nsEx6969 [acc-2prom1::gfp + pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS14268 | nsEx6961 [oac-7prom1::gfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14307 | nsEx6973 [kvs-5prom1::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14203 | nsEx6942 [kvs-5prom2::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14272 | nsEx6965 [lbp-1prom1::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14279 | nsEx6972 [lbp-1prom2::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14308 | nsEx6974 [adt-3prom1::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14343 | nsEx7015 [R03E9.2prom1::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14455 | nsEx7118 [R03E9.2prom2::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14345 | nsEx7017 [R03E9.2prom3::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14346 | nsEx7018 [R03E9.2prom4::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14398 | nsEx7067 [F49B2.6prom1::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14399 | nsEx7068 [F49B2.6prom2::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14395 | nsEx7064 [nta-1prom1::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14575 | nsEx7199 [nta-1prom2::rfp + pha-1 + ] line 1; pha-1 (e2123)III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14400 | nsEx7069 [T14B4.9prom1::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14458 | nsEx7121 [T14B4.9prom2::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14483 | nsEx7138 [T14B4.9prom3::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14459 | nsEx7122 [tbc-12prom1::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14574 | nsEx7198 [tbc-12prom2::rfp + pha-1 + ] line 1; pha-1 (e2123) III ; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14482 | nsEx7137 [tbc-12prom3::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14484 | nsEx7139 [tbc-12prom4::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14454 | nsEx7117 [pgp-4prom1::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14457 | nsEx7120 [pgp-4prom2::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14205 | nsEx6944 [F19B10.3prom1::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14576 | nsEx7200 [haf-7prom1::rfp + pha-1 + ] line 1; pha-1 (e2123); nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS14456 | nsEx7119 [haf-7prom 2::rfp + pha-1 + ] line 1; pha-1 (e2123) III; nsls758 [nep-2prom7::nls::yfp] V | This study | |
| OS13444 | nsEx6539 [glb-26prom1::gfp + pha-1 + ] line 1; pha-1(e2123) III | This study | |
| OS13445 | nsEx6540 [glb-26prom1::gfp + pha-1 + ] line 2; pha-1(e2123) III | This study | |
| OS12140 | nsEx6132 [WRM0641dF02 gly-18::sl2::snl::yfp::h2b, pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS12142 | nsEx6134 [WRM065dE01 gpx-8::sl2::snl::yfp::h2b, pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS12145 | nsEx6137[F28H7.2prom1::gfp, pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS12147 | nsEx6139 [T28A11.3prom1::gfp, pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS12149 | nsEx6141[ZC317.2prom1::gfp, pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS12151 | nsEx6143 [F35D2.3prom1::gfp, pha-1 + ] line 1; pha-1 (e2123) III | This study | |
| OS12789 | nsEx6078 [F13D12.10prom1::GFP, pha-1(+)] line 1; pha-1 (e2123) III; nsIs700 V | This study | |
| OS11967 | nsEx6063 [mnp-1prom3::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11219 | nsEx5817 [ncam-1prom2::GFP, pha-1(+)] line 1; pha-1 (e2123) III | This study | |
| OS11956 | nsEx6052 [unc-54prom3::GFP, pha-1(+)] line 1; pha-1 (e2123) | This study | |
| OS12370 | nsEx6202 [WRM061BG12 best-22 TY1::EGFP::3xFLAG(92C12) + pha-1(+)] line 1; pha-1 (e2123) III |
Tagged fosmid generated by (Sarov et al, 2006). Transgenic line generated in this study. |
|
| OS14023 | nsEx6821 [WRM069bF08 (let-381 fosmid) + myo-3p::mCherry] line 1; let-381(gk302) I; nsIs746 [nep-2prom7::gfp] V | This study | |
| OS14024 | nsEx6822 [WRM069bF08 (let-381 fosmid) + myo-3p::mCherry] line 2; let-381(gk302) I; nsIs746 [nep-2prom7::gfp] V | This study | |
| OS12240 | nsEx6151 [WRM0639aA08 (unc-30 fosmid) + unc-122prom::gfp)] line 1; unc-30 (e191) IV; nsIs700 [nep-2prom7::tagrfp] V | This study | |
| OS12241 | nsEx6152 [WRM0639aA08 (unc-30 fosmid) + unc-122prom::gfp)] line 2; unc-30 (e191) IV; nsIs700 [nep-2prom7::tagrfp] V | This study | |
| OS12242 | nsEx6153 [WRM0639aA08 (unc-30 fosmid) + unc-122prom::gfp)] line 3; unc-30 (e191) IV; nsIs700 [nep-2prom7::tagrfp] V; | This study | |
| OS12246 | nsEx6157 [WRM0617aE02 (unc-30 fosmid) + unc-122prom::rfp] line 1; unc-30 (e191) IV; nsIs746 [nep-2prom7::gfp] V | This study | |
| OS12247 | nsEx6158 [WRM0617aE02 (unc-30 fosmid) + unc-122prom::rfp] line 2; unc-30 (e191) IV; nsIs746 [nep-2prom7::gfp] V | This study | |
| OS12244 | nsEx6155 [unc-30 whole locus PCR + unc-122prom::gfp] line 1; unc-30 (e191) IV; nsIs700 [nep-2prom7::tagrfp] V | This study | |
| OS13665 | nsEx6668 [nep-2p7::let-381 sas RNAi + myo-3::mCherry] line 1; let-381(ns995) I; eri-1(mg366) IV | This study | |
| OS13666 | nsEx6669 [nep-2p7::let-381 sas RNAi + myo-3::mCherry] line 2; let-381(ns995) I; eri-1(mg366) IV | This study | |
| OS13667 | nsEx6670 [nep-2p7::let-381 sas RNAi + myo-3::mCherry] line 3; let-381(ns995) I; eri-1(mg366) IV | This study | |
| OS13668 | nsEx6671 [nep-2p7 (sas RNAi control) + myo-3::mCherry] line 1; let-381(ns995) I; eri-1(mg366) IV | This study | |
| OS13820 | nsEx6667 [nep-2p7 (sas RNAi control) + myo-3::mCherry] line 2 ; nep-2(syb4689) II; eri-1(mg366) IV | This study | |
| OS13672 | nsEx6675 [nep-2p7 (sas RNAi control) + myo-3::mCherry] line 3; ccIs4443 [arg-1prom::gfp + dpy-20(+)] eri-1(mg366) IV | This study | |
| OS13435 | nsEx6530 [hsp-16,2prom::let-381 cDNA, hsp-16,2prom::unc-30 cDNA, pha-1 + ] line 1; pha-1(e2123) III; nsIs700 [nep-2prom7::tagrfp] V | This study | |
| OS13436 | nsEx6531 [hsp-16,2prom::let-381 cDNA, hsp-16,2prom::unc-30 cDNA, pha-1 + ] line 2; pha-1(e2123) III; nsIs700 [nep-2prom7::tagrfp] V | This study | |
| OS13438 | nsEx6533 [hsp-16,2prom::let-381 cDNA, pha-1 + ] line 1; pha-1(e2123) III; nsIs700 [nep-2prom7::tagrfp] V | This study | |
| OS13439 | nsEx6534 [hsp-16,2prom::let-381 cDNA, pha-1 + ] line 2; pha-1(e2123) III; nsIs700 [nep-2prom7::tagrfp] V | This study | |
| OS13440 | nsEx6535 [hsp-16,2prom::unc-30 cDNA, pha-1 + ] line 1; pha-1(e2123) III; nsIs700 [nep-2prom7::tagrfp] V | This study | |
| OS13441 | nsEx6536 [hsp-16,2prom::unc-30 cDNA, pha-1 + ] line 2; pha-1(e2123) III; nsIs700 [nep-2prom7::tagrfp] V | This study | |
| OS11484 | nsIs700 [nep-2prom7::tagrfp] V | This study | |
| OS11703 | nsIs746 [nep-2prom7::gfp] V | This study | |
| OS11715 | nsIs758 [nep-2prom7::nls::yfp] V | This study | |
| OS12099 | nsIs831 [pll-1prom1::tagrfp] X | This study | |
| OS12103 | nsIs835 [hot-5prom2::gfp] X | This study | |
| OS12767 | nsIs879 [nep-2p7::TIR1::mCardinal + unc-122prom::mCherry] | This study | |
| OS12156 | nsIs854 [nep-2prom7::egl-1, myo-3mCherry]; nsIs831 [pll-1prom1::tagrfp] X | This study | |
| OS12700 | unc-30(ns959[unc-30::gfp:::degron(AID)]) IV | This study | |
| OS13288 | let-381(ns995[let-381::gfp::aid]) I | This study | |
| PHX4689 | nep-2(syb4689[gfp::h2b::sl2::nep-2]) II | This study | |
| PHX3025 | hlh-1(syb3025[hlh-1::gfp::aid]) II | This study | |
| PHX2879 | inx-18(syb2879[inx-18::aid::emgfp]) IV | This study | |
| PHX5792 | pll-1(syb5792[pll-1::sl2::gfp::h2b]) III | This study | |
| PHX5704 | gbb-1(syb5704[gbb-1::sl2::gfp::h2b]) X | This study | |
| PHX5759 | gbb-2(syb5759[gbb-2::sl2::gfp::h2b]) IV | This study | |
| OS13744 | nep-2(ns1012[*syb4689]) II | ns1012 = let-381 motif mutation | This study |
| OS14214 | pll-1(ns1040[*syb5792]) III | ns1040 = let-381 motif mutation | This study |
| OS13748 | hlh-1(ns1016[*syb3025]) II | ns1016 = 1st let-381 motif mutation | This study |
| OS13842 | hlh-1(ns1027[*syb3025]) II | ns1027 = 2nd let-381 motif mutation | This study |
| OS13747 | hlh-1(ns1015[*syb3025]) II | ns1015 = 1st + 2nd motif mutation | This study |
| OS13742 | inx-18(ns1010[*syb2879]) | ns1010 = let-381 motif mutation | This study |
| OS13838 | let-381(ns1026[*ns995]) I | ns1026 = let-381 motif deletion | This study |
| OS13308 | unc-30(ns1000[*ns959]) IV | ns1000 = 1st let-381 motif mutation | This study |
| OS13307 | unc-30(ns999[*ns959]) IV | ns999 = 1st + 2nd let-381 motifs mutation | This study |
| OS13309 | unc-30(ns1001[*ns959]) IV | ns1001 = 3rd let-381 motif mutation | This study |
| OS13306 | unc-30(ns998[*ns959]) | ns998 = three let-381 motifs deletion | This study |
CRISPR/Cas9 genome editing
CRISPR/Cas9 genome editing was performed using Cas9, tracrRNAs, and crRNAs from IDT, as described in (Dokshin et al, 2018).
Generation of deletion alleles was performed by use of two crRNAs and a ssODN donor as follows:
-
unc-30(ns998[*ns959]) [deletion of all three let-381 motifs from the 5th intron of unc-30]:
crRNAs (tctcgtgtggtataaacaat, actcggggtacagataacta) ssODN (gtcaggtaagcagaaggcaggcatcaggagttaattgggaacataattaagaatgaaaaaatatatcaaca)
-
let-381(ns1026[*ns995]) [deletion of let-381 autoregulatory motif]:
crRNAs (tggttgaagagacatacatc, ttatggatggaaaacagacg) ssODN (tcatcatacttttccctctatcttctcaaccagatctgttttccatccataagccaccaccccattctgc). CRISPR/Cas9 generated indel: deletion from –481 to –340 (housing the tgtttata let-381 motif) and random insertion of a 34 bp sequence cttatcttctcaatcttctcaaccagatgtgttg.
-
hlh-1(ns1015[*syb3025]) [mutation of the 1st let-381 motif at +726 from tgtttaca to ccgcgg and a deletion from +743 to +807 containing the 2nd let-381 motif at +765)
crRNAs (gtgtttacattgtgcaaact, tcttgaaaaattcgtagact) ssODN (atgggaatagtaaagggaggggggtgccgcggttgtgcaaactgggttaacccgttgtaaacataaatcgctaataggaa)
let-381 motif substitutions were performed by a single crRNA and a ssODN donor containing the desired mutations.
-
nep-2(ns1012[*syb4689]) [substitution of let-381 motif at –1133 tgtttaca to ccgcgg]
crRNA (caattgaggaacactgggcg) ssODN (cattccgattcccacttggcactgtgccaagttgcgcccagtgttcctcaattgccgcggacagcggctccggggggc)
-
pll-1(ns1040[*syb5792]) [substitution of let-381 motif at –100 from tctaaata to ccgcggta]
crRNA (acattttggcgtcgacggcg) ssODN (tttctagtagtagcaacagctcacaagacattttgaagcgccgtcgacgccaaaaccgcggtagaaaagaagaaaaaggaaaaaaactggaaacgg)
-
hlh-1(ns1016[*syb3025]) [substitution of the 1st let-381 motif +726 from tgtttaca to ccgcgg]
crRNA (gtgtttacattgtgcaaact) ssODN (atgggaatagtaaagggaggggggtgccgcggttgtgcaataagccttttaatccattttagtttatttcctttttcttt)
-
hlh-1(ns1027[*syb3025]) [substitution of the 2nd let-381 motif at +765 from agtttatt to agccatgg]
crRNA (gtgtttacattgtgcaaact) ssODN (aggggagaagagaatttatgaaatgggtcatgggaatagtaaagggaggggggtgtttacattgtgcaaacttttgccttttaatccattttagccatggtcctttttcttttcaattcttgaaaaattcgtagactg)
-
inx-18(ns1010[*syb2879]) [substitution of the let-381 motif at +364 from tctaaaca to aactgg]
crRNA (ggtcatttctcataggaaga) ssODN (aaacttcttgacatttttggtcatttctcataggaagacttgatttccatggaaacatttttgggcggcggcgggct)
-
unc-30(ns1000[*ns959]) [substitution of the 1st let-381 motif]
crRNA (tctcgtgtggtataaacaat) ssODN (cagtcaggtaagcagaaggcaggcatcaggagaaaattgtttataccacacgagaatctagaacagtgtcagttttctttcgccccttcttg)
-
unc-30(ns999[*ns959]) [substitution of the 1st + 2nd let-381 motifs]
crRNA (tctcgtgtggtataaacaat) ssODN (cagtcaggtaagcagaaggcaggcatcaggagaaaattggtaccaccacacgagaatctagaacagtgtcagttttctttcgccccttcttg)
unc-30(ns1001[*ns959]) [substitution of the 3rd let-381 motif]
crRNA (cctcttatgtcataaacaattgg) ssODN (ctttcgccccttcttgccagacatcatcttgaatcttatgtcccatggattggaggcgggggtactccgcttgtctcaac)
Generation of the let-381 and unc-30 knock-in reporters was performed by a single crRNA and a dsDNA asymmetric hybrid-donor containing a linker::gfp::aid cassette for C-terminal tagging (see sequence below) and 120 bp flanking arms with homology to target sequence.
let-381(ns995[let-381::gfp::aid]) crRNA (gctattccacaagatttat)
-
unc-30(ns959[unc-30::gfp::aid]) crRNA (aagtggtccactgtactgac)
Cassette sequence:
GGAGCATCGGGAGCCTCAGGAGCATCGATGAGTAAAGGAGAAGAACTTTTCACTGGAGTTGTCCCAATTCTTGTTGAATTAGATGGTGATGTTAATGGGCACAAATTTTCTGTCAGTGGAGAGGGTGAAGGTGATGCAACATACGGAAAACTTACCCTTAAATTTATTTGCACTACTGGAAAACTACCTGTTCCATGGgtaagtttaaacatatatatactaactaaccctgattatttaaattttcagCCAACACTTGTCACTACTTTCTgTTATGGTGTTCAATGCTTcTCgAGATACCCAGATCATATGAAACgGCATGACTTTTTCAAGAGTGCCATGCCCGAAGGTTATGTACAGGAAAGAACTATATTTTTCAAAGATGACGGGAACTACAAGACACgtaagtttaaacagttcggtactaactaaccatacatatttaaattttcagGTGCTGAAGTCAAGTTTGAAGGTGATACCCTTGTTAATAGAATCGAGTTAAAAGGTATTGATTTTAAAGAAGATGGAAACATTCTTGGACACAAATTGGAATACAACTATAACTCACACAATGTATACATCATGGCAGACAAACAAAAGAATGGAATCAAAGTTgtaagtttaaacatgattttactaactaactaatctgatttaaattttcagAACTTCAAAATTAGACACAACATTGAAGATGGAAGCGTTCAACTAGCAGACCATTATCAACAAAATACTCCAATTGGCGATGGCCCTGTCCTTTTACCAGACAACCATTACCTGTCCACACAATCTGCCCTTTCGAAAGATCCCAACGAAAAGAGAGACCACATGGTCCTTCTTGAGTTTGTAACAGCTGCTGGGATTACACATGGCATGGATGAACTATACAAACCTAAAGATCCAGCCAAACCTCCGGCCAAGGCACAAGTTGTGGGATGGCCACCGGTGAGATCATACCGGAAGAACGTGATGGTTTCCTGCCAAAAATCAAGCGGTGGCCCGGAGGCGGCGGCGTTCGTGAAG
bold = linker
non-bold, non-italicized = gfp with introns
bold + italicized = auxin-inducible degron
Generation of the fkh-9 knock-in reporter was performed as previously described in (Goudeau et al, 2021). Cas9, tracrRNA, crRNA and ssODN carrying the 2x(sfGFP11) tag were injected, as described in (Dokshin et al, 2018), into strain CF4587 which ubiquitously expresses sfGFP(1-10). crRNA (ccagtcttttcttcttcaat), ssODN (ctttctccaatcaaggccgagagccagtcttttcttcttcaaggaggaggatccggtgattccggcggcgttgacgtactcgtggaggaccatgtggtcacgtcctcctcccgtgaccacatggtcctccacgagtacgtcaacgccgccggaatcacctagacaccgattctgaactgattgaattagtcgcagttacg)
The following knock-in reporter alleles were generated by SUNY Biotech:
hlh-1(syb3025[hlh-1::gfp::aid]) C-terminal tagging with linker::gfp::aid.
inx-18(syb2879[inx-18::aid::emgfp]) tagged with aid::emgfp at position +6629 bp from inx-18 start codon.
nep-2(syb4689[gfp::h2b::sl2::nep-2]) N-terminal tagging with gfp::h2b::sl2.
pll-1(syb5792[pll-1::sl2::gfp::h2b]), gbb-1(syb5704[gbb-1::sl2::gfp::h2b]), gbb-2(syb5759[gbb-2::sl2::gfp::h2b]) C-terminal tagging with sl2::gfp::h2b.
Generation of transgenic reporters and rescuing constructs and neuron identification
All reporter gene fusions for the cis-regulatory analysis and the minimal promoters containing let-381 motifs were generated by a PCR fusion approach (Hobert, 2002). Genomic promoter fragments were fused to gfp or tagrfp followed by the unc-54 3’ UTR. Promoters were initially amplified with primers A (forward) and B (reverse) and gfp or tagrfp was amplified by primers C (forward) and D (reverse). For the fusion step, amplification was done using primers A* and D* as previously described (Hobert, 2002). PCR fusion DNA fragments were injected as simple extrachromosomal arrays in a pha-1(e2123) mutant background strain in the following concentrations: promoter fusion (50 ng/μL), pha-1 rescuing plasmid (pBX) (50 ng/μL). Promoter size and distance from the start codon for each promoter is shown in Fig. 1C for nep-2, Appendix Fig. S1A–E for lgc-55, egl-6, gly-18, hlh-1, inx-18, respectively, and Fig. EV3H for unc-30. Standard 20 bp oligos were used to amplify these promoters from wild-type (N2) genomic DNA. Oligos are listed in Appendix Table S1.
The minimal GLR glia-specific nep-2prom7 (amplification primers: cccattccgattcccacttg, cttaaaatatgatgaggtcg) was subcloned by Gibson cloning into the TIR1 coding plasmid pLZ31 (Zhang et al, 2015). nep-2p7::TIR1::unc-54 3’UTR was amplified by PCR and injected at 50 ng/μL with unc-122::mCherry (50 ng/μL) as co-injection marker. Similarly, nep-2prom7 was subcloned by Gibson cloning into the egl-1 expressing plasmid pTB28 (Bacaj et al, 2008). nep-2prom7::egl-1::unc-54 3’UTR was amplified by PCR and injected at 50 ng/μL with myo-3prom::mCherry (50 ng/μL) as co-injection marker.
Rescuing let-381 fosmids were injected as simple extrachromosomal arrays at 50 ng/μL with a myo-3prom::mCherry co-injection marker (25 ng/μL) and pBluescript (25 ng/μL) as filler DNA to reach a minimum DNA injection concertation of 100 ng/μL.
Rescuing unc-30 fosmids were linearized using NotI (NEB R3189S) and injected as complex extrachromosomal arrays in the following concentrations: linearized fosmid (30 ng/μL), linearized unc-122prom::gfp (5 ng/μL) as co-injection marker, sonicated E. coli (strain OP50) genomic DNA (100 ng/μL).
Rescuing “unc-30 whole locus”, including a 2.8kB promoter, unc-30 gene with introns and 3’UTR, was PCR amplified from genomic DNA (primers ctgttgctcgaaaacttccg, gattaggtagaaggtagaga) and injected as simple extrachromosomal arrays at 50 ng/μL with unc-122prom::gfp (40 ng/μL) as co-injection marker.
Integrated transgenic constructs (nsIs700, nsIs746, nsIs758, nsIs831, nsIs835, nsIs879, nsIs854) were generated by exposure to 33.4 μg/mL trioxsalen (Sigma T6137) and UV irradiation using a Stratagene Stratalinker UV 2400 Crosslinker (360 μJ/cm2 × 100) as previously described (Kage-Nakadai et al, 2014).
Fosmid reporters were generated as previously described (Tursun et al, 2009). More specifically, gly-18 and gpx-8 fosmids (WRM0641dF02 and WRM065dE01, respectively) were tagged at the C-terminus of the target genes with SL2::YFP::H2B amplified from pBALU23 (Stefanakis et al, 2015). Fosmid reporters were linearized using NotI and injected in the pha-1(e2123) mutant background strain as complex extrachromosomal arrays in the following concentrations: linearized fosmid (30 ng/μL), linearized pBX (pha-1 rescuing plasmid) (2.5 ng/μL), sonicated E.coli (strain OP50) genomic DNA 100 ng/μL.
Colocalization with the NeuroPAL transgene otIs669 as previously described (Yemini et al, 2021), was used to identify the neurons expressing unc-30::gfp.
Temporally controlled, GLR glia-specific LET-381 protein degradation
Exposure of C. elegans to a synthetic auxin analog K-NAA results in ubiquitination and subsequent proteasomal degradation of auxin-inducible degron (AID)-tagged proteins (Martinez et al, 2020). This occurs only in the presence of transgenically provided TIR1, the substrate recognition component of the E3 ubiquitin ligase complex. Strains carrying let-381::gfp::aid [let-381(ns995)] were crossed into a strain expressing TIR1 specifically in GLR glia (nsIs879 [nep-2prom7::TIR1]). K-NAA, 1-napthaleneacetic Acid Potassium Salt (PhytoTech Labs #N610), was dissolved in sterile H2O to prepare a 200 mM stock solution. OP50-seeded NGM plates were coated with K-NAA for a final concentration of 4 mM. Synchronized populations of L1 or late-L4/young-adult animals were transferred on K-NAA plates and grown at 20 °C for the duration of the experiment. Age-matched animals placed on OP50-seeded NGM plates coated with sterile H2O, were used as controls.
Cell-specific RNAi
A let-381 fragment, spanning from +421 to +1578 from the let-381 start codon (Fig. 2A), was amplified by PCR and fused in the sense and antisense orientation under the GLR glia-specific promoter nep-2prom7 (method described in (Esposito et al, 2007)). These sense and antisense let-381 fragments were injected at 40 ng/μL each, together with 25 ng/μL of the co-injection marker myo-3prom::mCherry driving expression in body wall muscle. Animals injected with unfused nep-2prom7 (40 ng/μL) together with the co-injection marker myo-3prom::mCherry (25 ng/μL) were used as controls. Three independent extrachromosomal arrays were scored for each genotype. GLR glia are refractory to RNAi; thus, RNAi-sensitized background strains carrying eri-1(mg366) (Kennedy et al, 2004) were used for these experiments.
Mosaic analysis
unc-30(e191) ; nsIs746 [nep-2prom7::gfp] animals were injected with extrachromosomal arrays consisting of NotI digested unc-30 fosmid WRM0617aE02 (30 ng/μL), rab-3prom1::2xnls::tagrfp (panneuronal::rfp) plasmid (35 ng/μL), unc-122:rfp (coelomocyte::rfp) plasmid (35 ng/μL) and sonicated E. coli (strain OP50) gDNA (80 ng/μL). Once extrachromosomal lines were established, mosaic animals carrying the rescuing array in the MS lineage or the AB lineage were picked based on RFP expression under a dissecting fluorescence microscope. Rescue of the unc-30(e191) mutant phenotype on GLR gene expression (nsIs746) was then assessed by imaging of mosaic animals on a confocal microscope.
Heat-shock-induced misexpression
One step RT-PCR (Invitrogen #12594025) was used to isolate let-381 and unc-30 cDNAs (primers for let-381: atggaatgctcaacag, ctagcaatccgataaatc, primers for unc-30: atggatgacaatacggccac, ctaaagtggtccactgtact), which were subsequently cloned under the heat-shock inducible promoter hsp-16.2 by Gibson cloning. These constructs were injected at 50 ng/μL to generate transgenic lines carrying hsp-16.2::let-381 cDNA (nsEx6533, nsEx6534), or hsp-16.2::unc-30 cDNA (nsEx6535, nsEx6536) or both (nsEx6530, nsEx6531) and crossed into a strain expressing RFP specifically in GLR glia (nsIs700 [nep-2prom7::tagrfp]). Twenty-five 1-day adult animals (P0) from each genotype were heat-shocked by incubating parafilm plates in a 32 °C water bath for 2.5 h. Heat-shocked P0s were kept at 25 °C and allowed to lay progeny (F1s) for 15 h before being removed from the plate. F1 progeny were grown at 25 °C for another 24 h and then scored for ectopic expression of the GLR glia-specific reporter. Age-matched, non-heat-shocked animals were used as controls.
High-salt motility assays
One day adult animals (24 h after L4 stage, blinded for each genotype) were placed on plates without food, containing 300 mM NaCl and scored for their motility over a period of 60 min: every 1 min for the first 15 min and every five minutes after that. Animals were score for loss of motility (paralysis) and recovery from paralysis. Twenty to twenty-five animals were tested for each genotype in four replicate experiments.
Analysis of animal locomotion
Locomotion of wild-type, GLR glia-ablated (nsIs854), GLR glia-defective let-381(ns1026), and control let-381(ns995) animals were recorded and analyzed as described in detail in (Katz et al, 2018; Katz et al, 2019). Briefly, 20–40 1-day adult animals were picked to an unseeded plate, washed three times with M9 buffer, and transferred to a 6 cm plate containing 4 ml of NGM-agar without food, with a high-osmolarity barrier (4 M fructose) at the periphery of the plate to prevent wandering of animals off the plate. Animals were allowed to habituate for 20 min, then the plate was placed under a camera and locomotion was recorded for 30 min at two frames per second and thereafter analyzed as previously described (Katz et al, 2018; Katz et al, 2019). Several plates were recorded and analyzed for each genotype (29 for wild-type, 37 for control, and 44 for GLR glia-defective animals) over the span of three days. For motor behavior assays with auxin-dependent LET-381::AID downregulation, synchronized let-381(ns995); nsIs879 L4 larvae were grown on OP50-seeded, 4 mM K-NAA plates for 24 h prior to motor behavior recordings. Locomotion assay plates also contained 4 mM of K-NAA. Age-matched animals of the same genotype, never exposed to K-NAA were used as control.
FACS-based GLR glia glia cell isolation
For isolation of the GLR glia, synchronized L1 larvae expressing nuclear-localized YFP in the GLR glia cells (OS11715) were grown on thirty 10-cm plates seeded with HB101 E. coli bacteria. After 48 h at 20 °C, L4 larvae animals were washed off plates and subsequently washed ten times with M9 to remove excess bacteria. Each wash consisted of a brief (10 s, 1300 rpm) centrifugation, such that most animals were pelleted, but bacteria stayed in suspension. Animals were then dissociated using SDS-DTT (0.25% SDS; 200 mM DTT; 20 mM HEPES, pH 8.0; 3% sucrose) and Pronase E (15 mg/ml) as previously described (Katz et al, 2019). We used 2:1 ratio of SDS-DTT to a volume of packed animals pellet, followed by 4 min incubation on ice. After five washes with ice-cold egg buffer (1.18 M NaCl; 480 mM KCl; 20 mM CaCl2; 20 mM MgCl2; 250 mM HEPES, pH 7.3), 4:1 ratio of room temperature Pronase E was added to the packed animals pellet and animals were then incubated rotating at 20 °C for 5 min, followed by 12 min of gentle homogenization (2 mL Dounce homogenizer, pestle clearance 0.0005–0.0025 inches, DWK Life Sciences). After three washes with ice-cold egg buffer to remove Pronase E, cells were filtered through a 5 μM filter to remove undigested animal fragments and immediately sorted by FACS.
GLR glia cell sorting was done using a BD FACS Aria sorter equipped with a 488-nm laser (Rockefeller University Flow Cytometry Resource Center), with egg buffer as the sheath buffer to preserve cell viability. Dead cell exclusion was carried out using DAPI, while DRAQ5 was used to distinguish nucleated cells from non-nucleated cell fragments. Gates for size and granularity were adjusted to exclude cell aggregates and debris. Gates for fluorescence were established using wild-type (N2) nonfluorescent animals. 500,000–800,000 YFP-positive events were sorted per replicate (4 replicates total), representing 0.3–0.7% of total events (after scatter exclusion), which is roughly the expected labeled-cell frequency in the animal (~0.6%). YFP-negative events from the same gates of size and granularity, representing all other cell types, were also sorted for comparison. Cells were sorted directly into TRIzol LS (Thermo Fisher Scientific) at a ratio 3:1 (TRIzol to cell volume).
RNAi extraction and sequencing
RNA was extracted from the sorted cells following the TRIzol LS protocol guidelines, until the isopropanol precipitation step, then RNA was re-suspended in extraction buffer of an RNA isolation kit (PicoPure, Arcturus) and isolation continued according to the manufacturer’s guideline. This two-step purification protocol helps obtain RNA of high quality when starting with samples of large volumes and resulted in a yield of around 10–80 ng per replica with RNA integrity number (RIN) ≥8, as measured by a Bioanalyzer (Agilent). All subsequent steps were performed by the Rockefeller University Genomics Resource Center. Briefly, mRNA amplification and cDNA preparation were performed using the SMARTer mRNA amplification kit (Takara #634940). Labeled samples were sequenced using an Illumina NextSeq 500 sequencer using 75 base pair single read protocols.
RNA-seq quality assessment and differential expression analysis
Fastq files were generated with CASAVA v1.8.2 (Illumina), and examined using the FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) application for sequence quality. Reads were aligned to customized build genome that combine C. elegans WS262 genome release (https://downloads.wormbase.org/releases/WS262/species/c_elegans/PRJNA13758/) and the “nep-2prom7::nls::::yfp::unc-54 3’UTR” transgene using the STAR v2.3 aligner with parameters (Dobin et al, 2012) (--out- FilterMultimapNmax 10 --outFilterMultimapScoreRange 1). Mapping rate was >72% with >62 million uniquely mapped reads. The alignment results were evaluated through RNA-SeQC v1.17 to make sure all samples had a consistent alignment rate and no obvious 5′ or 3′ bias (DeLuca et al, 2012). Aligned reads were summarized through featureCounts (Liao et al, 2013) with gene models from Ensemble (Caenorhabditis_elegans. WBcel235.77.gtf) at gene level unstranded: specifically, the uniquely mapped reads (NH “tag” in bam file) that overlapped with an exon (feature) by at least 1 bp on either strand were counted, and then the counts of all exons annotated to an Ensemble gene (meta-features) were summed into a single number. rRNA genes, mitochondrial genes, and genes with length <40 bp were excluded from downstream analysis.
The experiment was done with four independent replicates. DESeq2 was applied to normalize count matrix and to perform differential gene expression analysis, comparing RNA counts derived from the GLR glia cells (YFP-positive) to RNA counts that were derived from all other C. elegans cells, using the negative binomial distribution (Love et al, 2014). Raw and processed RNA-seq data have been deposited at Gene Expression Omnibus with accession number GEO: GSE234746.
Microscopy
Animals were anesthetized using 100 mM NaN3 (sodium azide) and mounted on 5% agarose pads on glass slides. Z-stack images (each ~7 μm thick) were acquired using either a Zeiss confocal microscope LSM990 (images in Figs. 7C and 8D; Appendix Fig. S5D,E) or a Zeiss compound microscope Axio Imager M2 (all other images) using MicroManager software (version 1.4.22) (Edelstein et al, 2010). ImageJ (Schneider et al, 2012) was used to produce maximum projections of z-stack images (2–30 slices) presented in the Figures. Figures were prepared by using Adobe Illustrator.
Quantification and statistical analysis
All microscopy fluorescence quantifications were done in ImageJ (Schneider et al, 2012). Mutant and control animals were imaged during the same imaging session with all acquisition parameters maintained constant between the two groups. The fluorescence intensity of gene expression in GLR glia and the HMC cell (Figs. 4E, F, 5A, B, D, and 7G) was measured in the plane with the strongest signal within the z-stack in a region drawn around the GLR glia nucleus (for nuclear reporters) or cell body (for cytoplasmic reporters). A single circular region in an adjacent area was used to measure background intensity for each animal; this value was then subtracted from the fluorescence intensity of reporter expression for each GLR glia/HMC cell. Quantification of the length of the GLR glia anterior process (Fig. 7E) was performed in maximum intensity projections. A line was drawn along the anterior process for each GLR glia cell and its length was measured and normalized to the length of the pharynx for each animal. Quantification of number of puncta for inx-18::gfp CRISPR knock-in reporter (Figs. 5C and 6E), quantification of number or percentage of GLR glia or HMC with reporter expression (Figs. 2C,D, 3A–G, 6A–E, 7D, 8B, D, EV2C–G, EV3A-G,J–K, and EV4C,E,F; Appendix Fig. S2B,C), quantification of animals with anteriorly displaced nerve ring (Fig. 2E) and quantification of number of head muscle cells (Fig. 2F; Appendix Figs. S2D and S3A) were performed by manual counting using ImageJ.
Prism (GraphPad) was used for statistical analysis as described in Figure legends. Unpaired, two-sided Students’ t test was used to determine the statistical significance between the two groups. n.s. denotes not statistically significant (P value > 0.05).
Supplementary information
Acknowledgements
The authors thank Oliver Hobert and Yuichi Lino for strains; The Rockefeller University Flow Cytometry and Genomics Resource Centers for technical support; Menachem Katz and members of the Shaham lab for experimental advice, comments, and discussion. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported in part by funds from a Leon Levy Foundation Fellowship to NS and by National Institutes of Health grant R35NS105094 to SS.
Expanded view
Author contributions
Nikolaos Stefanakis: Conceptualization; Formal analysis; Funding acquisition; Investigation; Visualization; Methodology; Writing—original draft; Writing—review and editing. Jessica Jiang: Investigation. Yupu Liang: Software; Formal analysis. Shai Shaham: Conceptualization; Funding acquisition; Writing—original draft; Project administration; Writing—review and editing.
Data availability
Raw and processed RNA-seq data have been deposited at Gene Expression Omnibus with accession number GEO: GSE234746. This paper does not report any original code. All newly generated critical strains will be available at the Caenorhabditis Genetics Center (CGC). Any additional information required to reanalyze the data reported and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author.
Disclosure and competing interests statement
The authors declare no competing interests.
Supplementary information
Expanded view data, supplementary information, appendices are available for this paper at 10.1038/s44318-024-00049-w.
References
- Allen NJ, Lyons DA. Glia as architects of central nervous system formation and function. Science. 2018;362:181–185. doi: 10.1126/science.aat0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun ZF, Hall DH (2016) Handbook of C. elegans anatomy. WormAtlas
- Altun-Gultekin Z, Andachi Y, Tsalik EL, Pilgrim D, Kohara Y, Hobert O. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development. 2001;128:1951–1969. doi: 10.1242/dev.128.11.1951. [DOI] [PubMed] [Google Scholar]
- Amin NM, Shi H, Liu J. The FoxF/FoxC factor LET-381 directly regulates both cell fate specification and cell differentiation in C. elegans mesoderm development. Development. 2010;137:1451–1460. doi: 10.1242/dev.048496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacaj T, Tevlin M, Lu Y, Shaham S. Glia are essential for sensory organ function in C. elegans. Science. 2008;322:744–747. doi: 10.1126/science.1163074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Batiuk MY, Martirosyan A, Wahis J, de Vin F, Marneffe C, Kusserow C, Koeppen J, Viana JF, Oliveira JF, Voet T, et al. Identification of region-specific astrocyte subtypes at single cell resolution. Nat Commun. 2020;11:1220. doi: 10.1038/s41467-019-14198-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer EA, Stecky RC, Neal L, Katsamba PS, Ahlsen G, Balaji V, Hoppe T, Shapiro L, Oren-Suissa M, Hobert O. Ubiquitin-dependent regulation of a conserved DMRT protein controls sexually dimorphic synaptic connectivity and behavior. eLife. 2020;9:e59614. doi: 10.7554/eLife.59614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundgaard M, Abbott NJ. All vertebrates started out with a glial blood-brain barrier 4-500 million years ago. Glia. 2008;56:699–708. doi: 10.1002/glia.20642. [DOI] [PubMed] [Google Scholar]
- Canals I, Ginisty A, Quist E, Timmerman R, Fritze J, Miskinyte G, Monni E, Hansen MG, Hidalgo I, Bryder D, et al. Rapid and efficient induction of functional astrocytes from human pluripotent stem cells. Nat Methods. 2018;15:693–696. doi: 10.1038/s41592-018-0103-2. [DOI] [PubMed] [Google Scholar]
- C. elegans Deletion Mutant Consortium (2012) large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 2:1415–1425 [DOI] [PMC free article] [PubMed]
- Choi U, Hu M, Zhang Q. The head mesodermal cell couples FMRFamide neuropeptide signaling with rhythmic muscle contraction in C. elegans. Nat Commun. 2023;14:4218. doi: 10.1038/s41467-023-39955-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinar H, Keles S, Jin Y. Expression profiling of GABAergic motor neurons in Caenorhabditis elegans. Curr Biol. 2005;15:340–346. doi: 10.1016/j.cub.2005.02.025. [DOI] [PubMed] [Google Scholar]
- DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire MD, Williams C, Reich M, Winckler W, Getz G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2012;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokshin GA, Ghanta KS, Piscopo KM, Mello CC. Robust genome editing with short single-stranded and long, partially single-stranded DNA donors in Caenorhabditis elegans. Genetics. 2018;210:781–787. doi: 10.1534/genetics.118.301532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman C, Horvitz HR, Jin Y. Coordinated transcriptional regulation of the unc-25 glutamic acid decarboxylase and the unc-47 GABA vesicular transporter by the Caenorhabditis elegans UNC-30 homeodomain protein. J Neurosci. 1999;19:6225–6234. doi: 10.1523/JNEUROSCI.19-15-06225.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N (2010) Computer control of microscopes using µManager. Curr Protoc Mol Biol Chapter 14:Unit14.20 10.1002/0471142727.mb1420s92 [DOI] [PMC free article] [PubMed]
- Esposito G, Di Schiavi E, Bergamasco C, Bazzicalupo P. Efficient and cell specific knock-down of gene function in targeted C. elegans neurons. Gene. 2007;395:170–176. doi: 10.1016/j.gene.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Feng W, Li Y, Dao P, Aburas J, Islam P, Elbaz B, Kolarzyk A, Brown AE, Kratsios P. A terminal selector prevents a Hox transcriptional switch to safeguard motor neuron identity throughout life. eLife. 2020;9:e50065. doi: 10.7554/eLife.50065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester WC, Dell M, Perens E, Garriga G. A C. elegans Ror receptor tyrosine kinase regulates cell motility and asymmetric cell division. Nature. 1999;400:881–885. doi: 10.1038/23722. [DOI] [PubMed] [Google Scholar]
- Gao S, Guan SA, Fouad AD, Meng J, Kawano T, Huang Y-C, Li Y, Alcaire S, Hung W, Lu Y, et al. Excitatory motor neurons are local oscillators for backward locomotion. eLife. 2018;7:e29915. doi: 10.7554/eLife.29915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel M, Atlas EG, Hobert O. A cellular and regulatory map of the GABAergic nervous system of C. elegans. eLife. 2016;5:e17686. doi: 10.7554/eLife.17686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Lim S, Hoeffel G, Low D, Huber T. Origin and differentiation of microglia. Front Cell Neurosci. 2013;7:45. doi: 10.3389/fncel.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Prinz M. Origin of microglia: current concepts and past controversies. Cold Spring Harb Perspect Biol. 2015;7:a020537. doi: 10.1101/cshperspect.a020537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudeau J, Sharp CS, Paw J, Savy L, Leonetti MD, York AG, Updike DL, Kenyon C, Ingaramo M. Split-wrmScarlet and split-sfGFP: tools for faster, easier fluorescent labeling of endogenous proteins in Caenorhabditis elegans. Genetics. 2021;217:iyab014. doi: 10.1093/genetics/iyab014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Hobert O. Regulation of terminal differentiation programs in the nervous system. Annu Rev Cell Dev Biol. 2011;27:681–696. doi: 10.1146/annurev-cellbio-092910-154226. [DOI] [PubMed] [Google Scholar]
- Hobert O, Kratsios P. Neuronal identity control by terminal selectors in worms, flies, and chordates. Curr Opin Neurobiol. 2019;56:97–105. doi: 10.1016/j.conb.2018.12.006. [DOI] [PubMed] [Google Scholar]
- Hochstim C, Deneen B, Lukaszewicz A, Zhou Q, Anderson DJ. Identification of positionally distinct astrocyte subtypes whose identities are specified by a homeodomain code. Cell. 2008;133:510–522. doi: 10.1016/j.cell.2008.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell AM, Gilmour SG, Mancebo RA, Rose AM. Genetic analysis of a large autosomal region in Caenorhabditis elegans by the use of a free duplication. Genet Res. 1987;49:207–213. [Google Scholar]
- Hupe M, Li MX, Kneitz S, Davydova D, Yokota C, Kele J, Hot B, Stenman JM, Gessler M. Gene expression profiles of brain endothelial cells during embryonic development at bulk and single-cell levels. Sci Signal. 2017;10:eaag2476. doi: 10.1126/scisignal.aag2476. [DOI] [PubMed] [Google Scholar]
- Jin Y, Hoskins R, Horvitz HR. Control of type-D GABAergic neuron differentiation by C. elegans UNC-30 homeodomain protein. Nature. 1994;372:780–783. doi: 10.1038/372780a0. [DOI] [PubMed] [Google Scholar]
- Kage-Nakadai E, Imae R, Yoshina S, Mitani S. Methods for single/low-copy integration by ultraviolet and trimethylpsoralen treatment in Caenorhabditis elegans. Methods. 2014;68:397–402. doi: 10.1016/j.ymeth.2014.02.036. [DOI] [PubMed] [Google Scholar]
- Kalinichenko VV, Gusarova GA, Shin B, Costa RH. The forkhead box F1 transcription factor is expressed in brain and head mesenchyme during mouse embryonic development. Gene Expr Patterns. 2003;3:153–158. doi: 10.1016/s1567-133x(03)00010-3. [DOI] [PubMed] [Google Scholar]
- Kastriti ME, Adameyko I. Specification, plasticity and evolutionary origin of peripheral glial cells. Curr Opin Neurobiol. 2017;47:196–202. doi: 10.1016/j.conb.2017.11.004. [DOI] [PubMed] [Google Scholar]
- Katz M, Corson F, Iwanir S, Biron D, Shaham S. Glia modulate a neuronal circuit for locomotion suppression during sleep in C. elegans. Cell Rep. 2018;22:2575–2583. doi: 10.1016/j.celrep.2018.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz M, Corson F, Keil W, Singhal A, Bae A, Lu Y, Liang Y, Shaham S. Glutamate spillover in C. elegans triggers repetitive behavior through presynaptic activation of MGL-2/mGluR5. Nat Commun. 2019;10:1882. doi: 10.1038/s41467-019-09581-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Fetter RD, Bargmann CI. Wnt-Ror signaling to SIA and SIB neurons directs anterior axon guidance and nerve ring placement in C. elegans. Development. 2009;136:3801–3810. doi: 10.1242/dev.038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostas SA, Fire A. The T-box factor MLS-1 acts as a molecular switch during specification of nonstriated muscle in C. elegans. Genes Dev. 2002;16:257–269. doi: 10.1101/gad.923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Harrison SW, Xu S-Q, Chen L, Fire A. Elements regulating cell-and stage-specific expression of the C. elegans MyoD family homolog hlh-1. Dev Biol. 1994;166:133–148. doi: 10.1006/dbio.1994.1302. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liu X, Long F, Peng H, Aerni SJ, Jiang M, Sánchez-Blanco A, Murray JI, Preston E, Mericle B, Batzoglou S, et al. Analysis of cell fate from single-cell gene expression profiles in C. elegans. Cell. 2009;139:623–633. doi: 10.1016/j.cell.2009.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlapuu M, Ormestad M, Enerback S, Carlsson P. The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development. 2001;128:155–166. doi: 10.1242/dev.128.2.155. [DOI] [PubMed] [Google Scholar]
- Martinez MAQ, Kinney BA, Medwig-Kinney TN, Ashley G, Ragle JM, Johnson L, Aguilera J, Hammell CM, Ward JD, Matus DQ. Rapid degradation of Caenorhabditis elegans proteins at single-cell resolution with a synthetic auxin. G3. 2020;10:267–280. doi: 10.1534/g3.119.400781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay SJ, Johnsen R, Khattra J, Asano J, Baillie DL, Chan S, Dube N, Fang L, Goszczynski B, Ha E, et al. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb Symp Quant Biol. 2003;68:159–169. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Huang X, Ebert D, Mills C, Guo X, Thomas PD. Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0) Nat Protoc. 2019;14:703–721. doi: 10.1038/s41596-019-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizeracka K, Rogers JM, Rumley JD, Shaham S, Bulyk ML, Murray JI, Heiman MG. Lineage-specific control of convergent differentiation by a Forkhead repressor. Development. 2021;148:dev199493. doi: 10.1242/dev.199493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munji RN, Soung AL, Weiner GA, Sohet F, Semple BD, Trivedi A, Gimlin K, Kotoda M, Korai M, Aydin S, et al. Profiling the mouse brain endothelial transcriptome in health and disease models reveals a core blood–brain barrier dysfunction module. Nat Neurosci. 2019;22:1892–1902. doi: 10.1038/s41593-019-0497-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JI, Boyle TJ, Preston E, Vafeados D, Mericle B, Weisdepp P, Zhao Z, Bao Z, Boeck M, Waterston RH. Multidimensional regulation of gene expression in the C. elegans embryo. Genome Res. 2012;22:1282–1294. doi: 10.1101/gr.131920.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan K, Lambert SA, Yang AW, Riddell J, Mnaimneh S, Zheng H, Albu M, Najafabadi HS, Reece-Hoyes JS, Fuxman Bass JI, et al. Mapping and analysis of Caenorhabditis elegans transcription factor sequence specificities. eLife. 2015;4:e06967. doi: 10.7554/eLife.06967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nass R, Hall DH, Miller DM, 3rd, Blakely RD. Neurotoxin-induced degeneration of dopamine neurons in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2002;99:3264–3269. doi: 10.1073/pnas.042497999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormestad M, Astorga J, Landgren H, Wang T, Johansson BR, Miura N, Carlsson P. Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development. 2006;133:833–843. doi: 10.1242/dev.02252. [DOI] [PubMed] [Google Scholar]
- Peterson RS, Lim L, Ye H, Zhou H, Overdier DG, Costa RH. The winged helix transcriptional activator HFH-8 is expressed in the mesoderm of the primitive streak stage of mouse embryos and its cellular derivatives. Mech Dev. 1997;69:53–69. doi: 10.1016/s0925-4773(97)00153-6. [DOI] [PubMed] [Google Scholar]
- Remesal L, Roger-Baynat I, Chirivella L, Maicas M, Brocal-Ruiz R, Perez-Villalba A, Cucarella C, Casado M, Flames N. PBX1 acts as terminal selector for olfactory bulb dopaminergic neurons. Development. 2020;147:dev186841. doi: 10.1242/dev.186841. [DOI] [PubMed] [Google Scholar]
- Reyahi A, Nik AM, Ghiami M, Gritli-Linde A, Ponten F, Johansson BR, Carlsson P. Foxf2 is required for brain pericyte differentiation and development and maintenance of the blood-brain barrier. Dev Cell. 2015;34:19–32. doi: 10.1016/j.devcel.2015.05.008. [DOI] [PubMed] [Google Scholar]
- Ringstad N, Abe N, Horvitz HR. Ligand-gated chloride channels are receptors for biogenic amines in C. elegans. Science. 2009;325:96–100. doi: 10.1126/science.1169243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringstad N, Horvitz HR. FMRFamide neuropeptides and acetylcholine synergistically inhibit egg-laying by C. elegans. Nat Neurosci. 2008;11:1168–1176. doi: 10.1038/nn.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468:214–222. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- Sarov M, Schneider S, Pozniakovski A, Roguev A, Ernst S, Zhang Y, Hyman AA, Stewart AF. A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nat Methods. 2006;3:839–844. doi: 10.1038/nmeth933. [DOI] [PubMed] [Google Scholar]
- Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, et al. Molecular diversity and specializations among the cells of the adult mouse brain. Cell. 2018;174:1015–1030.e1016. doi: 10.1016/j.cell.2018.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Wurtzel O, Malecek K, Fincher CT, Oderberg IM, Kravarik KM, Reddien PW. foxF-1 controls specification of non-body wall muscle and phagocytic cells in planarians. Curr Biol. 2018;28:3787–3801.e3786. doi: 10.1016/j.cub.2018.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PK, Santella A, Jacobo A, Siletti K, Hudspeth AJ, Bao Z. An in toto approach to dissecting cellular interactions in complex tissues. Dev Cell. 2017;43:530–540.e534. doi: 10.1016/j.devcel.2017.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham S. Glial development and function in the nervous system of Caenorhabditis elegans. Cold Spring Harb Perspect Biol. 2015;7:a020578. doi: 10.1101/cshperspect.a020578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham S. Glial development and function in the nervous system of Caenorhabditis elegans. Cold Spring Harb Perspect. 2015;8:4. doi: 10.1101/cshperspect.a020578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanakis N, Carrera I, Hobert O. Regulatory logic of pan-neuronal gene expression in C. elegans. Neuron. 2015;87:733–750. doi: 10.1016/j.neuron.2015.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Thomas PD, Ebert D, Muruganujan A, Mushayahama T, Albou LP, Mi H. PANTHER: making genome-scale phylogenetics accessible to all. Protein Sci. 2022;31:8–22. doi: 10.1002/pro.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng HT, Shah R, Jamrich M. Function and regulation of FoxF1 during Xenopus gut development. Development. 2004;131:3637–3647. doi: 10.1242/dev.01234. [DOI] [PubMed] [Google Scholar]
- Tursun B, Cochella L, Carrera I, Hobert O. A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS ONE. 2009;4:e4625. doi: 10.1371/journal.pone.0004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlandewijck M, He L, Mäe MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Laviña B, Gouveia L, et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018;554:475–480. doi: 10.1038/nature25739. [DOI] [PubMed] [Google Scholar]
- Wallace Sean W, Singhvi A, Liang Y, Lu Y, Shaham S. PROS-1/Prospero is a major regulator of the glia-specific secretome controlling sensory-neuron shape and function in C. elegans. Cell Rep. 2016;15:550–562. doi: 10.1016/j.celrep.2016.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CE, Krizus A, Dennis JW. Complementary expression patterns of six nonessential Caenorhabditis elegans core 2/I N-acetylglucosaminyltransferase homologues. Glycobiology. 2001;11:979–988. doi: 10.1093/glycob/11.11.979. [DOI] [PubMed] [Google Scholar]
- Wegner M (2020) Chapter 34—Specification of oligodendrocytes. In: Rubenstein J, Rakic P, Chen B, Kwan KY (eds) Patterning and cell type specification in the developing CNS and PNS, 2nd edn. Academic Press, pp 847–866
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Yamada K, Hirotsu T, Matsuki M, Butcher RA, Tomioka M, Ishihara T, Clardy J, Kunitomo H, Iino Y. Olfactory plasticity is regulated by pheromonal signaling in Caenorhabditis elegans. Science. 2010;329:1647–1650. doi: 10.1126/science.1192020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Jackson R. Astrocyte identity: evolutionary perspectives on astrocyte functions and heterogeneity. Curr Opin Neurobiol. 2019;56:40–46. doi: 10.1016/j.conb.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yemini E, Lin A, Nejatbakhsh A, Varol E, Sun R, Mena GE, Samuel ADT, Paninski L, Venkatachalam V, Hobert O. NeuroPAL: a multicolor atlas for whole-brain neuronal identification in C. elegans. Cell. 2021;184:272–288.e211. doi: 10.1016/j.cell.2020.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffran S, Kuchler A, Lee HH, Frasch M. biniou (FoxF), a central component in a regulatory network controlling visceral mesoderm development and midgut morphogenesis in Drosophila. Genes Dev. 2001;15:2900–2915. doi: 10.1101/gad.917101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen JA, Kirch SA, Bargmann CI. Genes required for axon pathfinding and extension in the C. elegans nerve ring. Development. 1999;126:3679–3692. doi: 10.1242/dev.126.16.3679. [DOI] [PubMed] [Google Scholar]
- Zhang A, Noma K, Yan D. Regulation of gliogenesis by lin-32/Atoh1 in Caenorhabditis elegans. G3 Genes|Genomes|Genetics. 2020;10:3271–3278. doi: 10.1534/g3.120.401547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ward JD, Cheng Z, Dernburg AF. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development. 2015;142:4374–4384. doi: 10.1242/dev.129635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed RNA-seq data have been deposited at Gene Expression Omnibus with accession number GEO: GSE234746. This paper does not report any original code. All newly generated critical strains will be available at the Caenorhabditis Genetics Center (CGC). Any additional information required to reanalyze the data reported and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author.