Abstract
Purpose
This study aimed to investigate the diagnostic and prognostic values of neuropilin-1 (NRP-1) in triple-negative breast cancer (TNBC) and analyze its immune function in the tumor microenvironment.
Methods
Based on The Cancer Genome Atlas (TCGA), Gene Expression Omnibus, Genotype Tissue Expression, Immune Cell Abundance Identifier (ImmuCellAI), Reactome, and Genomics of Drug Sensitivity in Cancer databases, the cancer tissues from 50 patients with TNBC and corresponding adjacent noncancerous tissues from 10 patients (tissue microarrays were purchased from Shanghai Xinchao Biotechnology Co., Ltd.) were collected for validation. Bioinformatics combined with immunohistochemistry was used to analyze the relationship among NRP-1 expression, prognosis, tumor immune cell infiltration, immune genes, and drug resistance so as to investigate the role of NRP-1 in the development of TNBC.
Results
A significant difference in NRP-1 gene expression was found between the cancerous and noncancerous tissues (p-value < 0.05); NRP-1 expression was high in carcinoma. No significant correlation was found between NRP-1 protein expression levels and each stage in the TCGA database. Prognostic expression survival analysis showed that the survival probability of patients with high NRP-1 expression was significantly lower than that of patients with low NRP-1 expression (p-value < 0.05), suggesting that the gene might be a pro-oncogene. The data from 50 clinical samples also confirmed that the NRP-1 expression was significantly higher in triple-negative breast cancer (TNBC) tissues than in adjacent noncancerous tissues. The NRP-1 expression significantly correlated with the tumor diameter and pathological grade (p-value < 0.05), but not with age, stage, and ki67 (p-value > 0.05). The Kaplan–Meier survival curves suggested that the median overall survival was significantly shorter in patients with high NRP-1 expression than in those with low NRP-1 expression (13.6 months vs 15.2 months, p-value < 0.05). The 300 genes most significantly positively associated with this gene were selected for Gene Ontology (including Biological Process, Molecular Function, and Cellular Component groups) and Kyoto Encyclopedia of Genes and Genomics enrichment analysis. The findings showed that NRP-1 was involved in immune regulation in TNBC. In addition, the NRP-1 expression in TNBC positively correlated with a variety of immune cells and checkpoints.
Conclusion
NRP-1 can be used as a potential biomarker and therapeutic target in TNBC.
Keywords: Biomarkers, Immune infiltrates, NRP-1, Therapeutic targets, Triple-negative breast cancer, tumor purity
1. Introduction
Breast cancer is the leading cause of death from malignant tumors in women worldwide, with high morbidity and mortality [1]. Immunohistochemistry showed that triple-negative breast cancer (TNBC), with negative estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 (HER-2) as its special subtype [2], accounted for about 15%–20% of breast cancers [3]. Traditional treatment is ineffective because of its biological characteristics such as high invasiveness, strong recurrence, and easy distant metastasis [4,5]. Therefore, new TNBC treatment targets need to be urgently explored.
Neuropilin-1 (NRP-1) is a non-tyrosine kinase transmembrane glycoprotein receptor [6], collapsin/semaphorin family receptor, a class of neuropilins, and a co-receptor for vascular endothelial growth factor, which plays an important role in neurodevelopment, axonal guidance, angiogenesis, tumor proliferation, and metastasis [7]. A previous study found that the NRP-1 expression was significantly higher in non-small-cell lung cancer (NSCLC) tissues than in the adjacent normal tissues [8], and high NRP-1 expression was associated with poor clinicopathological features and prognosis of NSCLC [9]. However, in breast cancer, NRP-1 is promising as a serological diagnostic marker for detecting early-stage breast cancer [10,11]. However, the expression, prognosis, and related immune function of NRP-1 in TNBC are still unclear. This study aimed to further investigate whether NRP-1 was involved in the metastasis, immune infiltration, related signaling pathways, and drug efficacy of TNBC by combining a variety of bioinformatics methods with immunohistochemistry, thus providing a new theoretical basis and potential biological target for the immunotherapy of TNBC to benefit more patients.
2. Materials and methods
2.1. The Cancer Genome Atlas database
The Cancer Genome Atlas (TCGA) (https://portal.gdc.com), the Tumor Genome Atlas Project, is by far the largest cancer gene information database in the world. The expression values (mean) of NRP-1 in tumor tissues from 33 cancers were downloaded and presented in order from low to high. The expression values of NRP-1 in each stage of TNBC were downloaded from the TCGA database. The mutation data for TNBC were downloaded from the Genomic Data Commons (GDC) TCGA cohort in the University of Cingifornia Sisha Cruz (UCSC) XENA.
2.2. Genotype Tissue Expression database
The Genotype Tissue Expression (GTEx) database is a tissue-specific database of gene expression and regulation derived from samples of multiple tissues and organs of the human body. The expression and distribution of NRP-1 in normal human tissues were obtained from this database.
2.3. Gene Expression Omnibus database
GSE202203 was found in the Gene Expression Omnibus (GEO) database, and differential expression analysis of the genes was performed using the Transcripts Per Million(TPM)values of the NRP-1 gene. Associated boxplots were made using the ggplot 2 package and ggpubr package in R language, where Tumor represented TNBC tissue and Normal represented normal tissue. GEO's GSE202203 comprised 431 samples, 333 carcinomas, and 98 adjacent carcinomas. Prognostic analysis was performed using the R language survminer package and survival package to generate the best cutoff value. The value in the high-expression group was higher than the best cutoff, and the value in the low-expression group was lower than the best cutoff, and finally the survival curve was obtained.
2.4. Gene Ontology versus Kyoto Encyclopedia of Genes and Genomics enrichment analysis and Gene Set Enrichment Analysis
First, the correlation analysis of the NRP-1 gene with all genes was performed, and 300 genes with the most significant positive correlation with the NRP-1 gene were selected for Gene Ontology (GO) [including Biological Process (BP), Molecular Function (MF), and Cellular Component (CC) groups] and Kyoto Encyclopedia of Genes and Genomics (KEGG) enrichment analysis to predict the functional pathway of this gene. The GO analysis is a powerful bioinformatics tool to identify biological processes, cellular adequacy, and molecular functions associated with NRP-1. The mechanism of action of NRP-1 was investigated using Gene Set Enrichment Analysis (GSEA), and the R package used for GO, KEGG, and GSEA was “clusterprofiler.”
2.5. Gene Set Variation Analysis
Gene Set Variation Analysis (GSVA), called GSVA Gene Set, is a pathway-level difference analysis. The GSVA Gene Set used in this study was the MSigDB database MSigDB databasev7.1, updated in March 2020.
2.6. Cibersort algorithm for immune infiltration
Cibersort is a single-cell-type analysis method. Currently, Cibersort is used to measure the proportion of tumor-infiltrating immune cells in TNBC in published articles [12] and the Immune Cell Abundance Identifier (ImmuCellAI) database. Then, we further analyzed the association of NRP-1 with immune cell subsets. A p-value <0.05 indicated a statistically significant difference.
2.7. Genomics of Drug Sensitivity in Cancer database
The data analysis of the relationship between genes and half-maximal inhibitory concentration (IC50) of drugs using Genomics of Drug Sensitivity in Cancer 2 (GDSC2) (https://www.cancerrxgene.org/) and Spearman method was adopted to plot the IC50 difference of each drug in the high– and low–gene expression groups and the correlation between genes and IC50, respectively.
2.8. Analytical steps
We analyzed the following to explore the situation of NRP-1 in triple-negative breast cancer: “Expression of NRP-1 in TNBC,” as described in Section 3.1. the “Correlation analysis of NRP-1 with tumor diameter and pathological grade in patients with TNBC,” as described in Section 3.2, which illustrates the correlation between NRP-1 and the main clinical parameters; the “Elevated NRP-1 expression suggested a poor prognosis in patients,' discussed in Section 3.3, describes the relationship between NRP-1 expression and prognosis; NRP-1 highly expressed genes and related signaling pathways in TNBC, as described in Sections 3.4, 3.5. Further, the immune microenvironment is an essential factor in the occurrence and development of tumors; therefore, the correlation between NRP-1 and immune infiltration was also investigated, and described in Section 3.6. Finally, we explored the potential for drugs to be effective in patients with NRP-1-expressing TNBC.
2.9. Immunohistochemical staining
Cancer tissue chips of 40 patients with TNBC and corresponding adjacent tissue chips of 10 patients were purchased from Shanghai Xinchao Biotechnology Co., Ltd. All patients were female, with a median age of 53 (28–84) years. None of the patients received radiotherapy, chemotherapy, or other tumor-specific treatment before surgery. According to the World Health Organization classification of breast tumors, 22 cases were classified as G2 and 18 cases as G3. Tumor node metastasis (TNM) clinical staging: 4 cases in stage 0, 25 cases in stage I–II, and 11 cases in stage III–IV. For the detection of NRP-1 antibody, 5 spots (4 cancer tissues and 1 corresponding paracancerous tissue) were not included in the statistical analysis due to exfoliation and the absence of cancer tissue. The tissue chip was placed in the oven; the temperature was adjusted to 63 °C, and the wax was baked for 1 h. Dewaxing was done in an automated stainer for 15 min per tank with two cylinders of xylene, 7 min per tank with two cylinders of 100% alcohol, 5 min per tank with one cylinder of 90% alcohol, 5 min per tank with one cylinder of 80% alcohol, and 5 min per tank with one cylinder of 70% alcohol. The slides were placed in the Target Retrieval Instrument, and retrieval was initiated after selecting the program. After repair, the slides were placed in distilled water at room temperature and allowed to cool naturally for more than 10 min. For primary antibody incubation, the slides were rinsed with phosphate-buffered saline (PBS); diluted primary antibody working solution was added to the slides, which were refrigerated at 4 °C overnight. For secondary antibody incubation, the slides were removed from the refrigerator, warmed at room temperature for 45 min, and washed with PBS. They were then placed in the DAKO, and the appropriate procedures to run the blocking were selected following the Autostainer Link 48 Instructions; secondary antibody binding and DAB chromogenic procedures were performed. Then, the hematoxylin staining was performed for 1 min. The tissues were then immersed in 0.25% alcohol hydrochloric acid (400 mL of 70% alcohol + 1 mL of concentrated hydrochloric acid) for about 10 s and rinsed with tap water for 5 min. The staining results showed that the NRP-1 protein was mainly expressed in the cytoplasm, and CD274 was expressed in the cell membrane and cytoplasm. A brown or brownish-yellow staining at the corresponding site of the cells determined positive staining. The staining intensity was scored according to the depth of cell staining, which was recorded as 0 points (negative), 0.5 points (0.5 +), 1 point (1 +), 2 points (2 +), and 3 points (3 +). According to the percentage of positive cells in the total number of tumor cells, the staining positive rate score was recorded as 0%–100%. The overall immunohistochemical score was the product of the “staining intensity score” and the “staining positivity” and ranged from 0% to 300%. For cytoplasmic NRP-1, the total score was ≤160% for the low-expression group and >160% for the high-expression group; for cytoplasmic CD274, the total score was ≤15% for the low-expression group and >15% for the high-expression group.
2.10. Statistical analysis
Statistical analysis was performed using the R software package (4.1.1). The Prism 8 software was used for data analysis. Wilcoxon rank sum test was used to analyze the data of the two groups. The chi-square test or Fisher exact test was used for the statistical analysis of enumeration data. The Kaplan–Meier method was used to draw survival curves for overall survival (OS).
3. Results
3.1. Expression of NRP-1 in TNBC
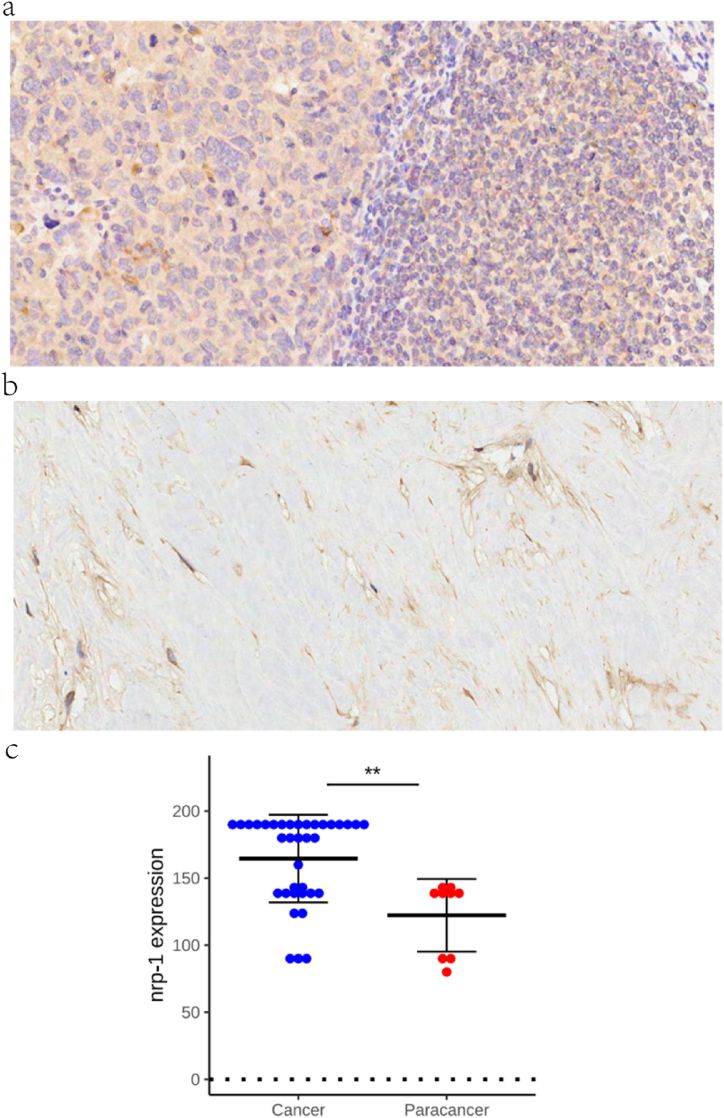
The expression levels of NRP-1 in 33 tumors were first analyzed using the TCGA database to investigate the expression of NRP-1 in TNBC. The findings revealed that the NRP-1 expression was higher in TNBC tissues (Fig. 1a). In the TCGA and GTEx databases, no significant difference was found in the NRP-1 expression between tumor and normal tissues in Adrenocortical Carcinoma (ACC), Bladder Urothelial Carcinoma, Kidney Renal Papillary Cell Carcinoma (KIRP), Pheochromocytoma and Paraganglioma (PCPG), Prostate Adenocarcinoma(PRAD), and Sarcoma(SARC), but a significant difference was found in TNBC (Fig. 1b). The expression was further detected using the GEO database. NRP-1 showed high expression in cancer tissues and adjacent normal tissues in differential expression analysis, with p-value <0.001, and the results similarly showed high expression in cancer, as shown in the box plot in Fig. 1c. The validation of NRP-1 expression in TNBC by immunohistochemistry revealed that the positive rate of NRP-1 in cancer tissues and adjacent noncancerous tissues in TNBC was 80% and 20%, respectively (Fig. 2a and b). The expression level of NRP-1 protein was significantly higher in cancer tissues than in adjacent noncancerous tissues (p-value < 0.001) (Fig. 2c). In addition, according to the GTEx database, NRP-1 was significantly highly expressed in adipose tissue, thyroid, breast, and lungs (Fig. 3). These findings indicated that the NRP-1 expression was upregulated in TNBC, suggesting that NRP-1 played an important regulatory role in the development of cancer.
Fig. 1.
(a) Expression of NRP-1 in cancer tissues in the TCGA database (b) Pan-cancer expression analysis of NRP-1 in 33 tumors (c) Differential expression of NRP-1 in TNBC tissues and normal tissues in the GEO database.
Fig. 2.
(a) High expression of NRP-1 in TNBC tissues ( × 200) (b) Low expression of NRP-1 in TNBC tissues ( × 200) (c) NRP-1 protein expression levels in cancer and adjacent noncancerous tissues.
Fig. 3.
Expression of NRP-1 in human normal tissue and organs in the GTEx database.
3.2. Correlation analysis of NRP-1 with tumor diameter and pathological grade in patients with TNBC
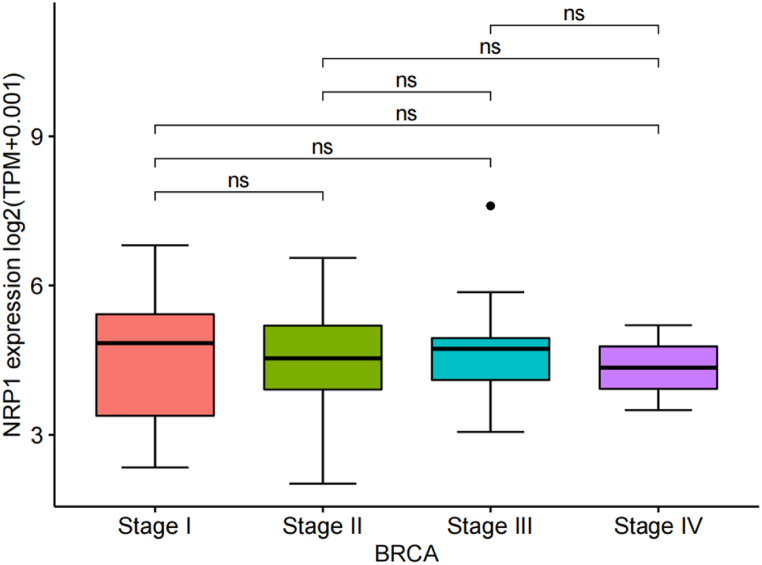
As shown in Fig. 4, no significant correlation was found between NRP-1 protein expression levels and each tumor node metastasis (TNM) stage of TNBC in the TCGA database. In Table 1, the tissue experimental results of 50 patients showed that the expression of NRP-1 was significantly correlated with tumor diameter (x2 = 6.218,p-value = 0.013) and pathological grade (x2 = 4.392,p-value = 0.036), but not with age (x2 = 0.008,p-value = 0.927), TNM stage (x2 = 1.506,p-value = 0.471), and ki67 (x2 = 2.676,p-value = 0.102). This is also consistent with our bioinformatics results.
Fig. 4.
Correlation between NRP-1 protein expression level and stages of TNBC.
Table 1.
Relationship between NRP-1 expression and clinicopathologic parameters in patients with TNBC.
| Clinicopathologic parameters | NRP-1 |
Total | x2 | p-value | ||
|---|---|---|---|---|---|---|
| Low expression | High expression | |||||
| Age (year) | ≤60 | 11 | 17 | 28 | 0.008 | 0.927 |
| >60 | 3 | 5 | 8 | |||
| Pathologic grade | I/II | 8 | 5 | 13 | 4.392 | 0.036 |
| III | 6 | 17 | 23 | |||
| Tumor diameter (cm) | ≤2 | 9 | 5 | 14 | 6.218 | 0.013 |
| >2 | 5 | 17 | 22 | |||
| T stage | T1/T2 | 11 | 15 | 26 | 1.407 | 0.495 |
| T3/T4 | 3 | 5 | 8 | |||
| Tis | 0 | 2 | 2 | |||
| N stage | N0 | 8 | 13 | 21 | 0.013 | 0.908 |
| N1/N2/N3 | 6 | 9 | 15 | |||
| TNM stage | I/II | 10 | 13 | 23 | 1.506 | 0.471 |
| III/IV | 4 | 7 | 11 | |||
| 0 | 0 | 2 | 2 | |||
| Ki67 | ≤0.3 | 9 | 8 | 17 | 2.676 | 0.102 |
| >0.3 | 5 | 14 | 19 | |||
T, tumor; N, node; M, metastasis; TNM stage, tumor node metastasis stage.
3.3. Elevated NRP-1 expression suggested a poor prognosis in patients
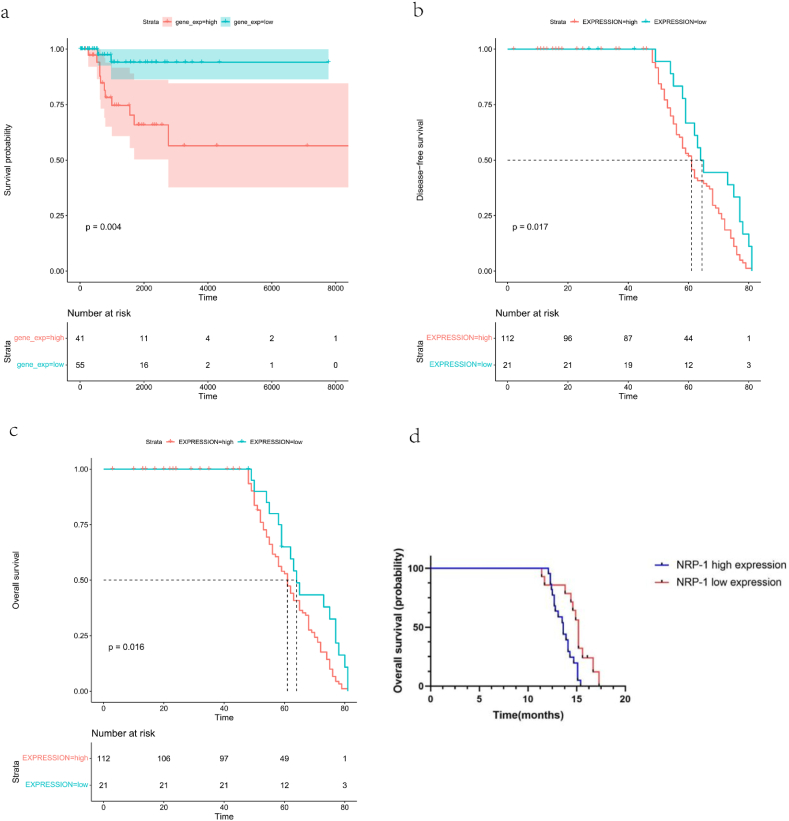
In the TCGA database, the survival probability was significantly lower in patients in the high-expression group than in those in the low-expression group, as shown in Fig. 5a. The p-value calculated using log-rank (Mantel–Cox) was less than 0.05, and the difference was extremely significant. Therefore, the prognosis of patients in the high–NRP-1 expression group was poor, suggesting that this gene might be a pro-oncogene. In the GEO database, the disease-free survival (Fig. 5b) and OS (Fig. 5c) were analyzed, and the p-value was calculated to be 0.0062 and 0.017 for NRP-1, respectively. This suggested that patients with high expression of NRP-1 had significantly shorter survival than those with low expression. The data from 50 clinical samples were similarly confirmed (Fig. 5d). The Kaplan–Meier survival curves suggested that the median OS was significantly shorter in patients with high NRP-1 expression than in those with low NRP-1 expression (13.6 months vs 15.2 months, p-value < 0.05).
Fig. 5.
(a) Survival probability of patients with high and low expression of NRP-1 in TNBC (b) Disease-free Survival in patients with high and low NRP-1 expression in TNBC (c) Overall Survival in patients with high and low NRP-1 expression in TNBC (d) Clinical data validated overall survival in patients with high and low NRP-1 expression in TNBC.
3.4. Pathway analysis of GO and KEGG with NRP-1 and its co-expressed genes in TCGA TNBC
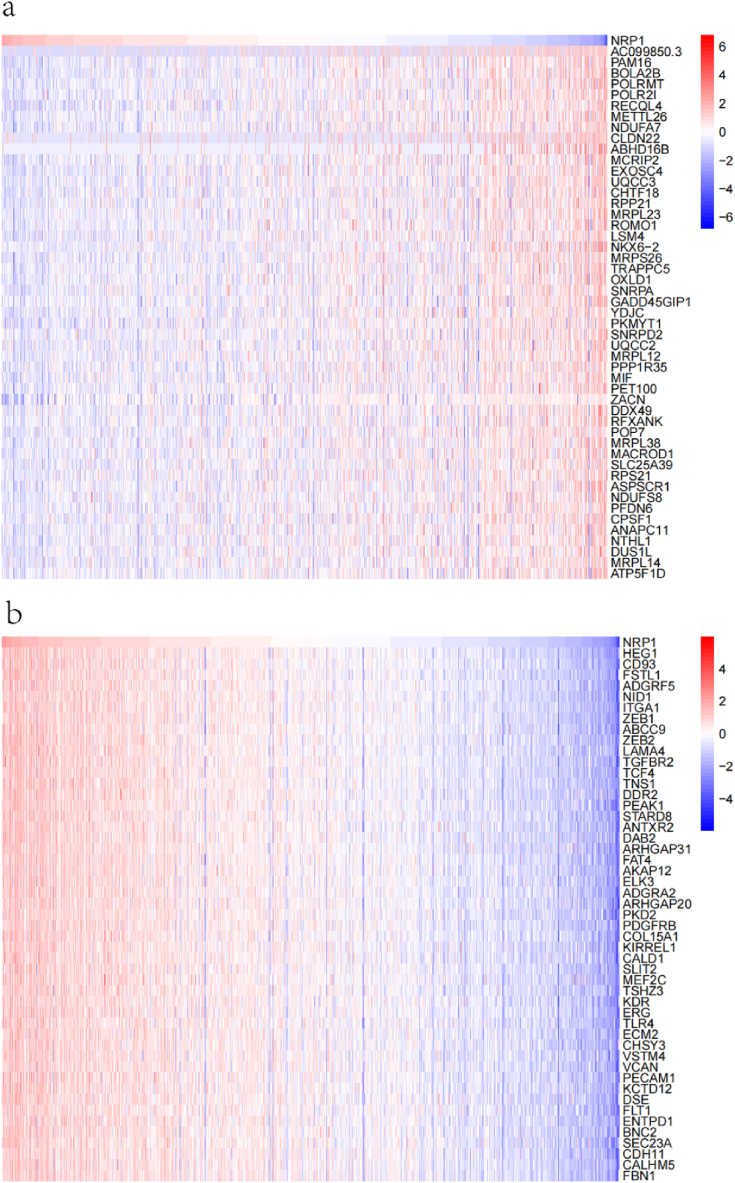
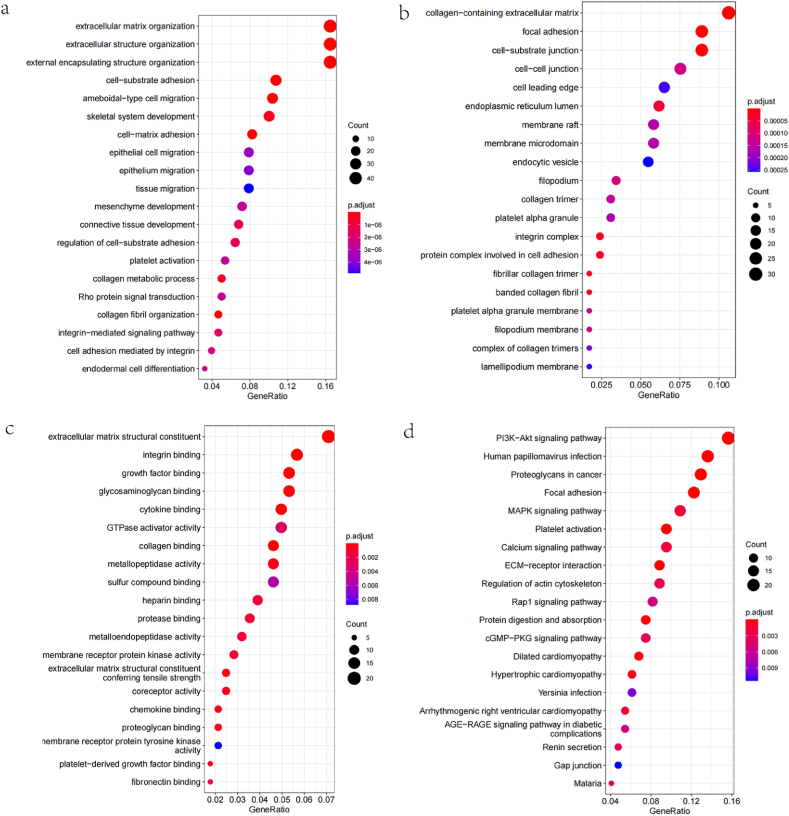
The data mining techniques in the TCGA database were used to identify genes having positive and negative correlations with NRP-1, and the top 50 genes positively and negatively correlated with NRP-1 were obtained; the results are shown in correlation heat maps in Fig. 6a and b. GO and KEGG enrichment analyses were then performed on 300 genes positively correlated with NRP-1 to investigate the associated pathways and biological functions of NRP-1. The GO analysis showed that NRP-1 was enriched in major biological processes (extracellular matrix organization, extracellular structure organization, and external encapsulating structure organization), cellular components (focal adhesion, cell−substrate), and molecular functions (actin binding) to varying degrees (Fig. 7a–7c). The KEGG pathway analysis showed that the PI3K−Akt pathway was enriched in NRP-1-co-expressed genes (Fig. 7d).
Fig. 6.
(a) Negative heat map of NRP-1 in TNBC (b) Positive heat map of NRP-1 in TNBC.
Fig. 7.
(a) GO-BP bubble drawing (b) GO-CC bubble drawing (c) GO-MF bubble drawing (d) KEGG bubble map.
3.5. GSEA and GSVA were performed to identify NRP-1-related signaling pathways
GSEA analysis was performed to further investigate the molecular mechanism of NRP-1 in TNBC. In GO terms (Fig. 8a), GSEA revealed multiple sets of functional genes, including angiogenesis, cytokine production, cell morphogenesis, protein phosphorylation, cell activation, system development, mitogen-activated protein kinase (MAPK) cascade, lyticole, ossification, and ameboidal-type cell migration. However, in KEGG terms (Fig. 8b), the top 20 signaling pathways affected by NRP-1 mainly included the cGMP−PKG signaling pathway, PI3K−Akt signaling pathway, MAPK signaling pathway, platelet activation, ECM−receptor interaction, focal adhesion, Rap 1 signaling pathway, Ras signaling pathway, and human cytomegalovirus infection. In addition, in the Reactome (Fig. 8c) terminology, NRP-1 was shown to be closely related to extracellular matrix organization, Platelet activation, Signaling and aggregation, GTPase cycle, Response to elevated platelet cytosolic Ca2+, Anti−inflammatory response favoring Leishmania parasite infection, Vesicle−mediated transport, GPCR downstream signaling, Adaptive immune system. In TNBC, the median number of genes was used to divide the samples into two groups with high and low expression for the pathway-level difference analysis (Fig. 8d). Fifteen pathways with the most significant positive and negative correlations with TNBC were taken for correlation mapping, and the abscissa showed correlation coefficients, with yellow indicating positive correlation and blue indicating negative correlation. The results showed that PID_LYMPH_ANGIOGENESIS_PATHWAY, GO VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY, and GO RESPONSE TO HEPATOCYTE GROWTH FACTOR were positively correlated pathways (Fig. 8e), and the corresponding GO RNA BINDING, MODY HIPPOCAMPUS GROWTH NATAL, and GO MITOCHONDRIAL PROTON TRANSPORTING ATP PRESYNTHASE COMPLEX COUPLING FACTOR FO pathways were listed as negatively correlated pathways. These results strongly suggested that NRP-1 was involved in the development of TNBC.
Fig. 8.
(a) GSEA-GO wave plot (b) GSEA-KEGG wave plot (c) GSEA-Reactome wave plot (d) GSVA analysis volcano plot (e) GSVA correlation plot.
3.6. Correlation analysis between NRP-1 and immune infiltration
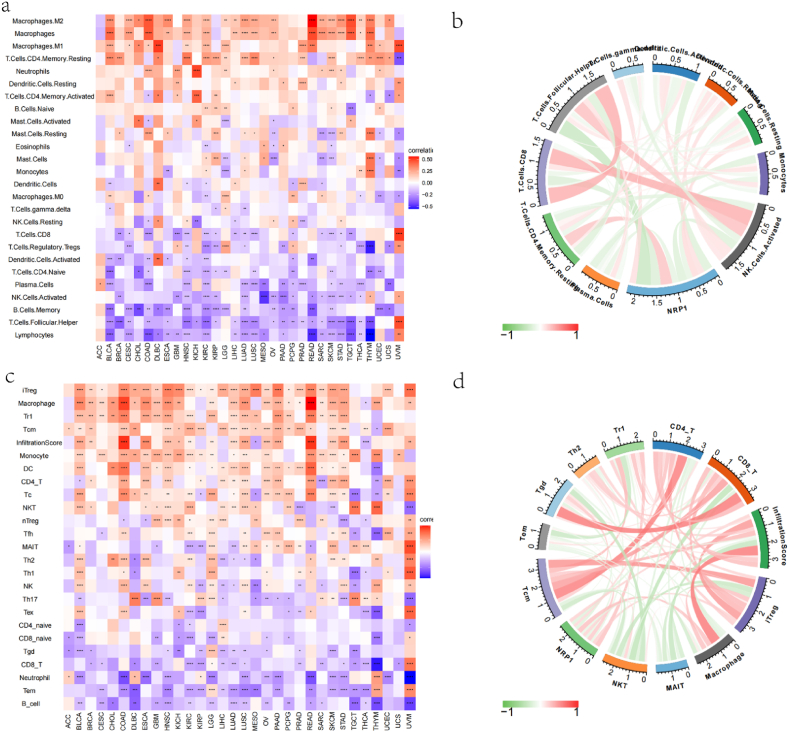
The association of the NRP-1 gene with 26 immune cell infiltrates (Fig. 9a and b), including B.Cells.Memory, B.Cells.Naïve, Dendritic.Cells, Dendritic.Cells.Activated, Dendritic.Cells.Resting, Eosinophils, Lymphocytes, Macrophages, Macrophages.M0, Macrophages.M1,Macrophages.M2,Mast.Cells, Mast.Cells.Activated, Mast.Cells.Resting, Monocytes, Neutrophils, NK.Cells.Activated, NK.Cells.Resting, Plasma.Cells, T.Cells.CD4.Memory.Activated, T.Cells.CD4.Memory.Resting, T.Cells.CD4.Naïve, T.Cells.CD8,T.Cells.Follicular.Helper,T.Cells.Gamma.Delta, andT.Cells.Regulatory.Tregs, was investigated to explore the relationship between the two. The results showed that the expression level of NRP-1 significantly correlated with that of Dendritic.Cells.Activated (r = −0.23, p-value = 0.0072), Mast.Cells.Resting (r = 0.17, p-value = 0.0435), NK.Cells.Activated (r = −0.22, p-value = 0.0084), T.Cells.CD4.Memory.Resting (r = 0.3, p-value = 4e−04), T.Cells.CD8 (r = −0.25, p-value = 0.0033), and T.Cells.Follicular.Helper (r = −0.38, p-value < 0.001). Similarly, in the ImmuCellAI database (Fig. 9c and d), among the 24 immune cell infiltrates, the expression level of NRP-1 significantly correlated with CD4T (r = 0.2, p-value = 0.0202), CD8T (r = −0.21, p-value = 0.0145), InfiltrationScore (r = 0.23, p-value = 0.0077), iTreg (r = 0.23, p-value = 0.0074), Macrophage (r = 0.32, p-value = 1e−04), NKT (r = 0.22, p-value = 0.0102), Tcm (r = 0.27, p-value = 0.0014), Tgd (r = −0.2, p-value = 0.0176), and Tr1 (r = 0.31, p-value = 2e−04).
Fig. 9.
(a) Heat map showing the association of NRP-1 with 26 immune cell infiltrates (b) Circle plot showing the correlation of NRP-1 with 26 immune cell infiltrates (c) Heat map showing the association of NRP-1 with 24 immune cell infiltrates (d) Circle plot showing the correlation of NRP-1 with 24 immune cell infiltrates.
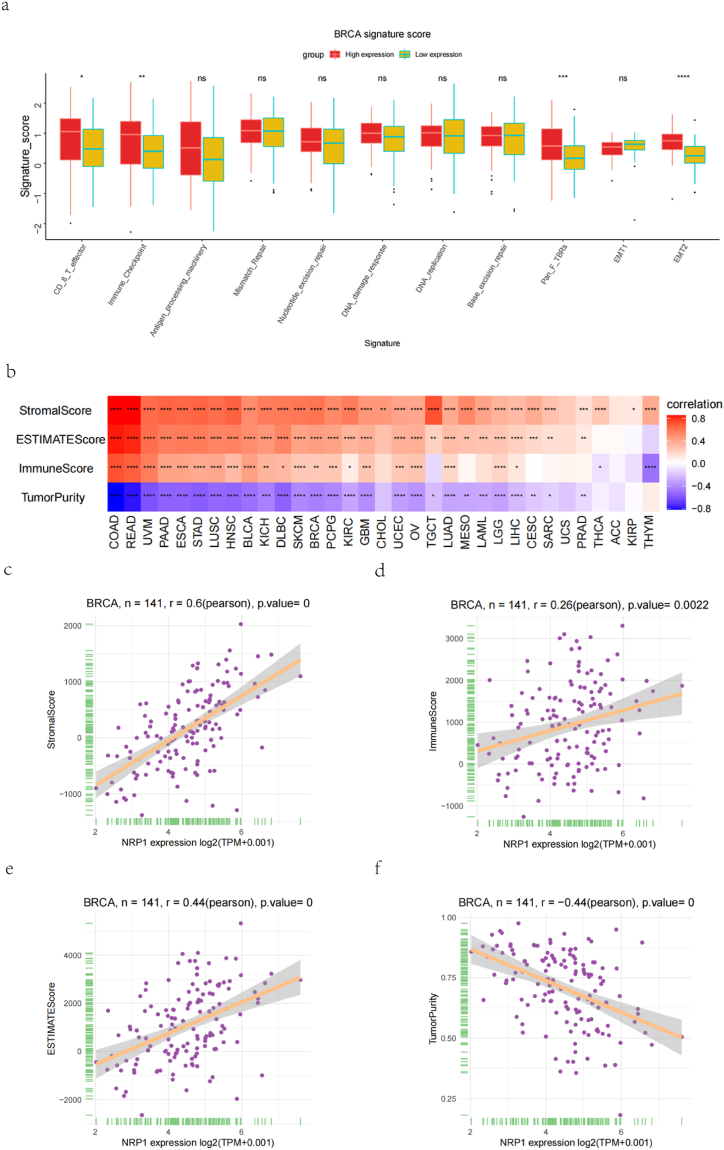
In TNBC, a box plot (Fig. 10a) was used to show the fraction of pathways that differed in the high- and low-expression groups of genes to further evaluate the impact of NRP-1 on the tumor microenvironment, and a heat plot was used to show the correlation between genes and pathway scores (Fig. 10b). The scores of EMT2 and PanFTBRs pathways were high in different expression groups of genes, demonstrating that NRP-1 was closely related to these pathways. The StromalScore positively correlated with the NRP-1 expression (n = 141, r = 0.6, p-value < 0.001) (Fig. 10c); the ImmuneScore positively correlated with the NRP-1 expression (n = 141, r = 0.26, p-value = 0.0022) (Fig. 10d); the ESTIMATEScore positively correlated with the NRP-1 expression (n = 141, r = 0.44, p-value < 0.001) (Fig. 10e); and the TumorPurity negatively correlated with the NRP-1 expression (n = 141, r = − 0.44, p-value < 0.001) (Fig. 10f). Further investigation of the correlation between NRP-1 and immune genes in TNBC revealed that the expression level of NRP-1 positively correlated with that of the immune-activating genes ENTPD1 and CXCL12 (Fig. 11a) and significantly positively correlated with that of the immunosuppressive gene KDR (Fig. 11b) in TNBC. Further, the expression level of NRP-1 positively correlated with that of the chemokine receptor CX3CR1 (Fig. 11c), significantly positively correlated with that of chemokine CXCL12 (Fig. 11d), and positively correlated with that of iron death ZEB1 (Fig. 11e).
Fig. 10.
(a) Correlation between high and low NRP-1 expression levels and pathway score (b) Heat map of correlation between NRP-1 gene and pathway fraction (c) StromalScore correlated with NRP-1 expression (d) ImmuneScore correlated with NRP-1 expression (e) ESTIMATEScore correlated with NRP-1 expression (f) TumorPurity correlated with NRP-1 expression.
Fig. 11.
(a) Association of NRP-1 with immune-activating genes (b) Association of NRP-1 with immunosuppressive genes (c) Association of NRP-1 with chemokine receptors (d) NRP-1 association with chemokines (e) Association of NRP-1 with iron death genes.
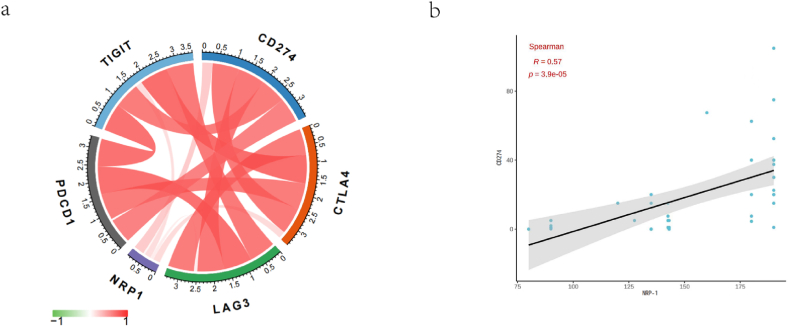
The expression level of NRP-1 positively correlated with that of TIGIT, CD274, CTLA4, and LAG3, and weakly correlated with that of PDCD1 (Fig. 12a). Immunohistochemistry was performed in 50 patients with TNBC, and we similarly verified that the expression level of NRP-1 positively correlated with that of CD274 (r = 0.57, p-value < 0.001) (Fig. 12b). These findings further supported that NRP-1 played an essential role in immune escape in the microenvironment of TNBC.
Fig. 12.
(a) NRP-1 associated with immune checkpoints (b) NRP-1 associated with CD274.
3.7. Drug resistance analysis between NRP-1 and IC50
The relationship between genes and drug IC50 was analyzed using data from GDSC2 (https://www.cancerrxgene.org/), by including 198 drugs to assess the relationship between NRP-1 and drug efficacy versus resistance. The findings revealed that the higher the NRP-1, the lower the IC50, the higher the target affinity, and the better the efficacy. Dasatinib and trametinib (p-value < 0.05) (Fig. 13a) may be effective drugs against NRP-1 and are expected to improve the life cycle of patients with TNBC. The analysis of IC50 values (Fig. 13b) for high and low expression levels of NRP-1 in patients with TNBC for the two drugs showed no significant difference for trametinib, while the expression levels were significantly different and more effective for dasatinib.
Fig. 13.
(a) Correlation between NRP-1 and drug IC50(b) IC50 values for high and low NRP-1 expression.
4. Discussion
TNBC, as a special type of breast cancer with relatively unique clinicopathology, has a poor prognosis due to its own biological characteristics and limited treatment modalities [13,14]. In the present study, we found that NRP-1 was highly expressed in TNBC tissues compared to normal and paracancerous tissues. In addition, patients with high NRP-1 expression had significantly poorer survival probability, disease-free survival, and OS. These findings suggested that NRP-1, as a pro-oncogene, might promote the development of TNBC [15,16]. We obtained similar conclusions in further clinical data validation. No significant correlation was found between NRP-1 expression levels in cancer tissues and tumor stage, whereas a significant correlation was found between high NRP-1 expression and tumor diameter and pathological grade. These results confirmed that NRP-1, as an independent biomarker, might promote the development of precision medicine.
Some previous studies found [17,18] that tumor purity in colon cancer was an independent risk factor affecting patient prognosis, and low tumor purity was associated with a poor prognosis and more immune-related proteins [programmed death ligand 1(PD-1), PD-L1, CTLA-4, LAG-3]. However, higher tumor purity was related to better sensitivity to immune efficacy in gynecological tumors. This coincided with our belief that advanced TNBC usually had low tumor purity due to the high expression of pathological-grade NRP-1, resulting in high StromalScore and ImmuneScore in nontumor cells such as immune cells, stromal cells, and stromal cells, thus affecting the patient's life cycle. This might be related to the fact that low tumor purity was more likely to lead to high mutation rates in critical pathways and changes in the microenvironment.
In recent years, targeted therapy against immune checkpoints has become a hot spot in cancer therapy [19], and immune checkpoint inhibitors are considered to effectively prolong the life cycle of patients and alleviate disease progression [20]. However, immune-related adverse reactions and drug resistance reactions are usually induced due to immune microenvironment and tumor heterogeneity [21]. In this study, NRP-1 positively correlated with the degree of immune cell infiltration in most TNBCs, while all four immune checkpoints also positively correlated with the expression of NRP-1. Further pan-cancer analysis revealed that NRP-1 expression positively correlated with the degree of immune cell infiltration in many types of tumor tissues. These results suggested that NRP-1 might regulate tumor immunity through multiple immune cell populations. The analysis of NRP1 further confirmed that the high and low expression of the NRP-1 gene was affected by CD8Teffector and ImmuneCheckpoint, with significant differences in expression. Therefore, these results suggested that using immune checkpoint inhibitors associated with NRP-1 might significantly improve the efficacy of TNBC. The Vignali [22]team also created a transgenic mouse model demonstrating that memory CD8 + T cell subsets from patients with advanced cancer had higher levels of NRP-1, but CD8 + T cell NRP-1 deletion combined with immune checkpoint blockade therapy improved tumor clearance. Based on these findings, we concluded that NRP-1, as a critical checkpoint, existed in the immune microenvironment of TNBC [23]. In addition, GO and KEGG analysis results suggested that NRP-1 was associated with cell activation, phylogeny, and angiogenesis, and acted mainly through the PI3K−Akt signaling pathway. Further, GSEA achieved similar conclusions. These results suggested that NRP-1 not only played a role in normal embryonic development and angiogenesis but also participated in key immune signaling pathways.
This study had some limitations. First, the bioinformatics analysis data retrieved from public databases and the clinical samples used for validation need to be confirmed by further prospective studies. Second, the clinical sample size used for experimental validation in this study was small. In addition, although NRP-1 was found to be closely associated with immune cell infiltration in different tumors, the specific mechanism was not clear, and further mechanistic studies are needed to explore it. In summary, this study used bioinformatics methods to verify the value of NRP-1 in the diagnosis and prognosis of TNBC, combined with further verification of clinical samples, making the conclusion more persuasive. In this study, we found that NRP-1 might be involved in the development and progression of TNBC as well as in the corresponding immune regulation. Therefore, NRP-1 could be used as a potential diagnostic and prognostic biomarker and therapeutic target to provide new ideas for treating breast cancer.
Ethics statement
This study was reviewed and approved by [the Ethics Committee of Shanghai Xinchao Biotechnology Co., Ltd.], with the approval number: [No.: YB M-05-02]. All participants/patients (or their proxies/legal guardians) provided informed consent to participate in the study. All participants/patients (or their proxies/legal guardians) provided informed consent for the publication of their anonymised case details and images.
Funding statement
No grant numbers apply.
Data availability statement
Data associated with our study had not been deposited in publicly available repositories. Data will be made available on request.
CRediT authorship contribution statement
Xiao Ma: Writing – review & editing, Writing – original draft, Conceptualization. Haonan Liu: Data curation. Congcong Shi: Methodology. Yang Zhao: Validation, Supervision. Hongmei Wang: Software, Methodology. Zhengxiang Han: Project administration, Conceptualization.
Declaration of competing interest
This study did not receive any financial or non-financial assistance from any third party. In the past 3 years, no financial interests or relationships related to the subject matter but not relevant to this article have been identified. All authors declare that there are no patent or copyright issues related to the work presented in the manuscript. The authors have no conflicts of interest.
Acknowledgments
None.
References
- 1.Azamjah N., Soltan-Zadeh Y., Zayeri F. Global trend of breast cancer mortality rate: a 25-year study. Asian Pac. J. Cancer Prev. APJCP. 2019;20(7):2015–2020. doi: 10.31557/APJCP.2019.20.7.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang T., et al. Changes of EGFR and SMC4 expressions in triple-negative breast cancer and their early diagnostic value. Gland Surg. 2021;10(3):1118–1124. doi: 10.21037/gs-21-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong S., Alahari S.K. Combination treatment of bicalutamide and curcumin has a strong therapeutic effect on androgen receptor-positive triple-negative breast cancers. Anti Cancer Drugs. 2020;31(4):359–367. doi: 10.1097/CAD.0000000000000880. [DOI] [PubMed] [Google Scholar]
- 4.Yao M., et al. Research on correlations of miR-585 expression with progression and prognosis of triple-negative breast cancer. Clin. Exp. Med. 2022;22(2):201–207. doi: 10.1007/s10238-021-00704-0. [DOI] [PubMed] [Google Scholar]
- 5.Borri F., Granaglia A. Seminars in Cancer Biology. Elsevier; 2021. Pathology of triple negative breast cancer. [DOI] [PubMed] [Google Scholar]
- 6.Issitt T., et al. Neuropilin-1 controls endothelial homeostasis by regulating mitochondrial function and iron-dependent oxidative stress. iScience. 2019;11:205–+. doi: 10.1016/j.isci.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaddis D.E., et al. Neuropilin-1 expression on CD4 T cells is atherogenic and facilitates T cell migration to the aorta in atherosclerosis. J. Immunol. 2019;203(12):3237–3246. doi: 10.4049/jimmunol.1900245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding Z., et al. Neuropilin 1 modulates TGF-beta 1-induced epithelial-mesenchymal transition in non-small cell lung cancer. Int. J. Oncol. 2020;56(2):531–543. doi: 10.3892/ijo.2019.4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding Z., et al. The regulation of Neuropilin 1 expression by miR-338-3p promotes non-small cell lung cancer via changes in EGFR signaling. Mol. Carcinog. 2019;58(6):1019–1032. doi: 10.1002/mc.22990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rachner T.D., et al. Soluble Neuropilin-1 is an independent marker of poor prognosis in early breast cancer. J. Cancer Res. Clin. Oncol. 2021;147(8):2233–2238. doi: 10.1007/s00432-021-03635-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seifi-Alan M., et al. Neuropilin-1 expression is associated with lymph node metastasis in breast cancer tissues. Cancer Manag. Res. 2018;10:1969–1974. doi: 10.2147/CMAR.S169533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorsson V., et al. The immune landscape of cancer. Immunity. 2018;48(4):812–830 e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim C., et al. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell. 2018;173(4):879–893 e13. doi: 10.1016/j.cell.2018.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howard F.M., Olopade O.I. Epidemiology of triple-negative breast cancer: a review. Cancer J. 2021;27(1):8–16. doi: 10.1097/PPO.0000000000000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J., et al. The miR-124-3p/neuropilin-1 Axis contributes to the proliferation and metastasis of triple-negative breast cancer cells and Co-activates the TGF-beta pathway. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.654672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naik A., et al. Neuropilin-1 associated molecules in the blood distinguish poor prognosis breast cancer: a cross-sectional study. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-03280-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong Z., Zhang J., Guo W. Tumor purity as a prognosis and immunotherapy relevant feature in gastric cancer. Cancer Med. 2020;9(23):9052–9063. doi: 10.1002/cam4.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng Y., et al. Tumor purity as a prognosis and immunotherapy relevant feature in cervical cancer. Aging. 2021;13(22) doi: 10.18632/aging.203714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B., Chan H.L., Chen P.J.C.m.c. Immune checkpoint inhibitors: basics and challenges. Curr. Med. Chem. 2019;26(17):3009–3025. doi: 10.2174/0929867324666170804143706. [DOI] [PubMed] [Google Scholar]
- 20.Darvin P., et al. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp. Mol. Med. 2018;50(12):1–11. doi: 10.1038/s12276-018-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramos-Casals M., et al. Immune-related adverse events of checkpoint inhibitors. Nat. Rev. Dis. Prim. 2020;6(1):1–21. doi: 10.1038/s41572-020-0160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C., et al. Neuropilin-1 is a T cell memory checkpoint limiting long-term antitumor immunity. Nat. Immunol. 2020;21(9):1010–1021. doi: 10.1038/s41590-020-0733-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chuckran C.A., et al. Neuropilin-1: a checkpoint target with unique implications for cancer immunology and immunotherapy. J Immunother Cancer. 2020;8(2) doi: 10.1136/jitc-2020-000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data associated with our study had not been deposited in publicly available repositories. Data will be made available on request.