Abstract
Malaria remains a major public health problem worldwide, including in Southeast Asia. Chemotherapeutic agents such as chloroquine (CQ) are effective, but problems with drug resistance and toxicity have necessitated a continuous search for new effective antimalarial agents. Here we report on a virtual screening of ∼300 diarylpentanoids and derivatives, in search of potential Plasmodium falciparum lactate dehydrogenase (PfLDH) inhibitors with acceptable drug-like properties. Several molecules with binding affinities comparable to CQ were chosen for in vitro validation of antimalarial efficacy. Among them, MS33A, MS33C and MS34C are the most promising against CQ-sensitive (3D7) with EC50 values of 1.6, 2.5 and 3.1 μM, respectively. Meanwhile, MS87 (EC50 of 1.85 μM) shown the most active against the CQ-resistant Gombak A strain, and MS33A and MS33C the most effective P. knowlesi inhibitors (EC50 of 3.6 and 5.1 μM, respectively). The in vitro cytotoxicity of selected diarylpentanoids (MS33A, MS33C, MS34C and MS87) was tested on Vero mammalian cells to evaluate parasite selectivity (SI), showing moderate to low cytotoxicity (CC50 > 82 μM). In addition, MS87 exhibited a high SI and the lowest resistance index (RI), suggesting that MS87 may exert effective parasite inhibition with low resistance potential in the CQ-resistant P. falciparum strain. Furthermore, the in vivo toxicity of the molecules on early embryonic development, the cardiovascular system, heart rate, motor activity and apoptosis were assessed in a zebrafish animal model. The overall results indicate the preliminary potential of diarylpentanoids, which need further investigation for their development as new antimalarial agents.
Keywords: Diarylpentanoids, Molecular docking, Antimalarial activity, PfLDH inhibitors, Zebrafish
Abbreviations
- P. falciparum
Plasmodium falciparum
- PfLDH
P. falciparum lactate dehydrogenase
- LDH
lactate dehydrogenase
- CQ
Chloroquine
- PAINS
Pan assay interference compounds
- ADMET
absorption, distribution, metabolism, excretion, and toxicity
1. Introduction
Malaria is a serious parasitic disease caused by Plasmodium spp. According to the World Malaria Report (2022), there are an estimated 247 million malaria cases worldwide in 84 malaria-endemic countries (including Southeast Asia; cases 10% and deaths 3%) and 619,000 deaths until 2021. Most of this statistic comes from the African region, which accounts for about 95% of global cases, with an estimated 234 million cases in 2021. At the same time, Southeast Asia contributed approximately 2% of global malaria cases, with India accounting for 79% of cases in this region [1]. The five identified Plasmodium of malaria-causal species in humans are Plasmodium falciparum, P. vivax, P. malaria, P. ovale and P. knowlesi [2], with P. falciparum and P. vivax mainly responsible for >95% of malaria cases worldwide. In particular, a significant increase in the number of P. knowlesi infections has been recorded in the Southeast Asia region, especially in Malaysia. Although no indigenous non-zoonotic malaria cases or deaths were reported from 2018 to 2021, a total of 17,125 P. knowlesi cases and 48 deaths have been reported since 2017, with 3575 cases and 13 deaths in 2021, in Malaysia alone, and thus remains a prominent public health burden [1,[3], [4], [5]]. Plasmodium infections in humans show a wide range of signs and symptoms, ranging from asymptomatic to severe malaria. Severe malaria, classified according to World Health Organization (WHO) criteria, includes impaired consciousness, hypoglycaemia, jaundice, acute respiratory distress, and severe anaemia [6].
Since then, several types of first-line antimalarials with different mechanisms of action have been developed and used for treatment and chemoprophylaxis. The best-known antimalarials often recommended by WHO are quinine derivatives such as chloroquine (CQ), antifolate combination drugs [e.g., sulfadoxine-pyrimethamine (SP) and sulfalene-pyrimethamine (metakelfin)] and artemisinin-based combination therapies (ACTs). However, the emergence and rapid spread of multidrug-resistant malaria parasites, is leading to an increase in malaria incidence, epidemics and a high number of corresponding morbidity and mortality cases [7]. As current antimalarials become increasingly ineffective, in addition to their toxicity issue [8], there is an urgent need to develop new drugs to treat and control the disease, if possible, with newer and effective alternative mechanisms of action against drug-resistant and drug-sensitive Plasmodium.
As technologies become more advanced and efficient, more discoveries are anticipated since chemoprospecting activities can be carried out faster, with higher throughput and with greater promise of success. To date, our research efforts have led to the construction of a compound library of approximately 300 synthetic diarylpentanoid analogues and derivatives (Fig. 1), mainly targeting inflammation and inflammation-related diseases. The compounds are simple but diverse in structure and have good stability and drug-like properties, making them potential candidates for the discovery of new hit molecules against malaria. The origin of diarylpentanoids, also known as monocarbonyl curcuminoids, monoketone curcuminoids and monocurcuminoids, is due to the clinical limitations of curcumin, including its poor stability and bioavailability. In response, researchers have undertaken the synthesis of various monocarbonyl analogues of curcumin in which the β-diketone moiety has been omitted to improve their biological properties [9]. These analogues have shown significant potential as anti-cancer, anti-inflammatory, anti-parasitic and neuroprotective agents [[10], [11], [12]] and have thus paved the way for the development of new therapeutics with diverse pharmacological activities.
Fig. 1.
Collection of diarylpentanoid analogues and derivatives categorized into differemt series: Series 1 (3-hydroxy-1,5-diphenylpenta-2,4-dien-1-ones), series 2 (3-benzylidene-2-hydroxycyclohexenyl)phenylmethanones), series 3 (2-styryl-4H-chromen-4-ones), series 4 (1,5-diphenylpenta-1,4-dien-3-ones), series 5 (2,5-di(benzylidene)cyclopentan-1-ones), series 6 (2,6-di(benzylidene)cyclohexan-1-ones), series 7 (1,5-diphenylpenta-2,4-dien-1-ones), series 8 (1-(3-phenyl-5-styryl-4,5-dihydro-1H-pyrazol-1-yl)ethanones), series 9 (chalcones), series 10 (N-(3-(5-phenylpenta-2,4-dienoyl)phenyl)benzenesulfonamides), series 11 (5-phenyl-3-styryl-1H-pyrazoles), and series 12 ((2,3-dihydro-1H-xanthen-4-yl)phenylmethanones).
Previously, Viira et al. (2016) reported several diarylpentanoids that showed remarkable antimalarial activity in vitro against the 3D7 strain of P. falciparum, with IC50 values between 30 and 240 nM in the SYBR green I fluorescence assay [13]. Subsequently, the extensive antimalarial research on curcuminoids was extended to the in vivo model, in which [14] found a significant suppression of 56.8% and 40.5% of parasites with encapsulated curcumin and free curcumin (dosage of 5 mg/kg), respectively, four days after treatment in mice. In addition, the diarylpentanoids have shown low toxicity both in vitro and in vivo. This includes lower cytotoxicity and genotoxicity observed in cell-based models [15] and minimal effects on weight gain in mice, as well as favorable histopathological evaluations and safety profiles in humans [16]. These results suggest a promising safety profile that underscores the potential for further development of diarylpentanoids as therapeutic agents.
Following these results, we herein report the virtual screening of in-house diarylpentanoids (molecular docking, druglikeness and ADMET prediction), synthesis and in vitro lactate dehydrogenase (LDH)-inhibitory activity against CQ-sensitive strain of P. falciparum 3D7, CQ-resistant strains of P. falciparum Gombak A and P. knowlesi A1H1. The PfLDH assay evaluates the inhibition of the parasite during its asexual stage within the intraerythrocytic stage. It is commonly used to measure parasite viability in drug susceptibility testing, as PfLDH activity is closely associated with the presence of parasites in the bloodstream [17,18]. Therefore, this assay serves as an indicator of parasite viability in providing us a preliminary insight into the potential antimalarial efficacy of our compounds. The selected bioactive compounds were also evaluated for their in vitro cytotoxicity against Vero cells and in vivo toxicity in the zebrafish embryo model to further explore the therapeutic potential and clinical relevance of diarylpentanoids, especially as antimalarial agents.
2. Results and discussion
2.1. Molecular docking, and in silico predictions of drug-likeness and ADMET properties
In silico study is a computational method used in drug discovery and development to predict drug-like molecules and targets using bioinformatics tools [19]. It involves the use of molecular docking approach, to predict potential binding interactions, activities, and affinities of drugs (or also called ligands) to macromolecules with a view to rationally selecting compounds for validation of in vitro activity with a view to developing potential drug candidates [[19], [20], [21]]. In silico drug design is cost-effective and play an important role in all phases of drug development, from preclinical discovery to late-stage clinical development [22]. Consequently, molecular docking was applied in this work to investigate the binding interactions and affinities of approximately 300 diarylpentanoid analogues and derivatives from our in-house library with the target protein P. falciparum lactate dehydrogenase (PfLDH, PDB ID: 1CET).
PfLDH is an important enzyme in the glycolytic metabolic pathway of the malaria parasite that is highly expressed during the blood stage and directly correlates with parasitaemia, making it a potential therapeutic target for antimalarials [23,24]]. Crystal structures of PfLDH have been previously established [25], with the active site of PfLDH located in a cleft between the NADH-binding domain and the substrate-binding domain. These active site loop of PfLDH contains a five-amino acid insertion that is unique to the malarial form of the enzyme and is absent in human LDH. Site-directed mutagenesis studies have shown that these additional amino acids are essential for inhibition of the enzyme and represent a potential new target for new antimalarial drugs [26]. Moreover, the crystal structure of the PfLDH-CQ complex protein shows that CQ occupies a similar position as the cofactor adenyl ring in the NADH-binding pocket of the enzyme [24]. To this end, the co-crystallised CQ was separated from the protein in the 1CET crystal structure and re-docked to PfLDH. An RMSD of 2.384 Å was found, and the RMSD <3.000 Å was considered legitimate [27], indicating the reproducibility of the experimentally observed binding mode for the PfLDH inhibitors. The grid box was therefore approved, and consequently can be used for docking diarylpentanoids to the same active site. The full docking results with the predicted binding energy are shown in Table S1 (in Supplementary Materials).
Although molecular docking is valuable for predicting the binding affinity of molecules, discrepancies in reliability have been reported when it comes to correlating scoring functions with experimental validation [[28], [29], [30]]. Consequently, the selection of compounds for antimalarial in vitro assessment in this study is not only based on docking results, i.e. highest binding affinity. Instead, additional criteria are crucial for the protocol selection of compounds in this work. (1) Physiological relevance influences the behaviour of a compound in biological systems, with a closer binding affinity to a standard drug indicating a better suitability for therapeutic use [31]. (2) Minimizing side effects is of critical importance. Excessively high binding affinities can lead to unexpected side effects or off-target interactions, emphasizing the need for binding affinities that are closer to control [32]. (3) Consistency in the dose-response relationship is key to predicting biological effects. Extremely high binding affinities can disrupt typical dose-response relationships and favour compounds with moderately low binding affinities [33]. Finally, (4) specificity and selectivity of drugs play a role. Higher binding affinity does not guarantee better specificity, so selecting compounds closer to control affinities increases specificity and reduces off-target effects [34].
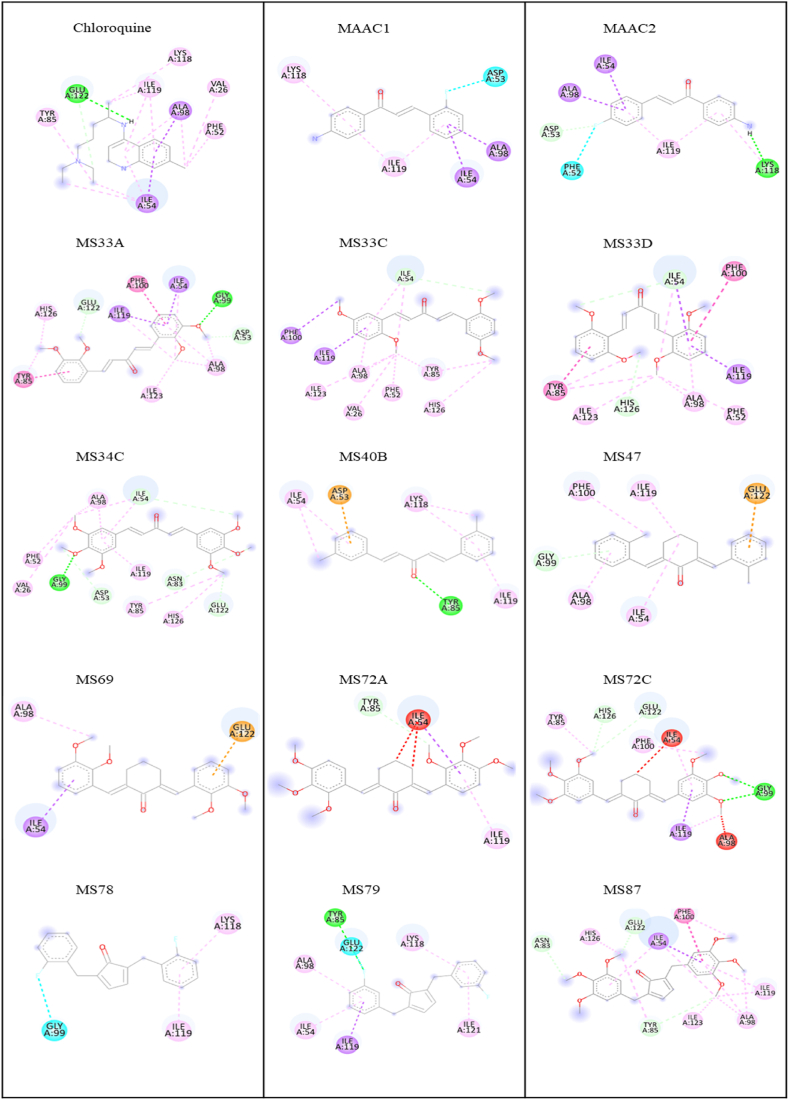
As shown in Table 1, several compounds (MAAC1, MAAC2, MS33A, MS33C, MS33D, MS34C, MS40B, MS47, MS78, MS79, and MS87; the structures are shown in Fig. 2) with binding affinity values between −5.2 and −7.2 kcal/mol, comparable to CQ (−6.0 kcal/mol), were reasonably selected for in vitro validation of antimalarial activity. In contrast, compounds 122, 123, and 124 with the highest binding affinity (−8.0 to −8.2 kcal/mol) are not readily available at present, and starting materials for synthesis are also limited, so they were not included in the study. Although compounds with the highest predicted binding affinity are preferred by default because there are more interactions with the target protein, the compounds with the lowest binding affinity (readily available samples of MS72C, MS72A, and MS69; structures in Fig. 2) were also considered for later comparison by in vitro biological evaluation. All chemical interactions and the amino acid residues involved are listed and shown in Table 1 and Fig. 3, respectively. As previously reported, an aromatic ring of CQ is hydrophobically bound to Ile54, Ala98, and Ile119 via a van der Waals interaction. In addition, the 4-NH group of CQ forms a hydrogen bond with Glu22 [25]. The above amino acid residues are critical for the orientation and binding of CQ in the binding pocket of PfLDH to exert its therapeutic effect against malaria, reproduced from our molecular docking studies, and are shown in Fig. 3.
Table 1.
Binding affinity and important interactions of selected compounds with the active site residues of PfLDH.
| Compound | Binding Affinity (kcal/mol) | Protein Ligand Interactions |
Compound | Binding Affinity (kcal/mol) | Protein Ligand Interactions |
||||
|---|---|---|---|---|---|---|---|---|---|
| aReceptor | Interaction | Bond distance (Å) | aReceptor | Interaction | Bond distance (Å) | ||||
| MAAC1 | −6.7 | Ile54 | -sigma | 3.78 | MS47 | −6.7 | Ile54 | Alkyl | 4.79 |
| Ala98 | -sigma | 3.53 | Ala98 | -alkyl | 4.08 | ||||
| Ile119 | -alkyl | 4.83 | Ile119 | Alkyl | 3.61 | ||||
| MAAC2 | −6.8 | Ile54 | -sigma | 3.77 | Glu122 | -anion | 4.04 | ||
| Ala98 | -sigma | 3.53 | MS69 | −4.0 | Ile54 | -sigma | 3.76 | ||
| Ile119 | -alkyl | 4.81 | Ala98 | Alkyl | 3.80 | ||||
| MS33A | −5.8 | Ile54 | -sigma | 3.93 | Glu122 | -anion | 3.14 | ||
| Ala98 | -alkyl | 3.86 | MS72A | −0.7 | Ile119 | -alkyl | 4.69 | ||
| Ile119 | -sigma | 3.62 | MS72C | −0.1 | Ile54 | -alkyl | 4.90 | ||
| Glu122 | C–H bond | 3.53 | Ile119 | Alkyl | 3.88 | ||||
| MS33C | −5.6 | Ile54 | C–H bond | 3.76 | Glu122 | C–H bond | 3.76 | ||
| Ala98 | -alkyl | 4.71 | MS78 | −7.2 | Ile119 | -alkyl | 4.34 | ||
| Ile119 | -sigma | 3.83 | MS79 | −6.8 | Ile54 | -alkyl | 4.37 | ||
| MS33D | −5.5 | Ile54 | C–H bond | 3.61 | Ala98 | -alkyl | 5.08 | ||
| Ala98 | -alkyl | 5.00 | Ile119 | -sigma | 3.70 | ||||
| Ile119 | -sigma | 3.78 | Glu122 | Halogen (F) | 3.40 | ||||
| MS34C | −5.8 | Ile54 | C–H bond | 3.77 | MS87 | −5.2 | Ile54 | -sigma | 3.65 |
| Ala98 | -alkyl | 4.15 | Ala98 | -alkyl | 5.20 | ||||
| Ile119 | -alkyl | 4.62 | Ile119 | Alkyl | 4.50 | ||||
| Glu122 | C–H bond | 3.75 | Glu122 | C–H bond | 3.63 | ||||
| MS40B | −5.9 | Ile54 | Alkyl | 5.26 | CQ | −6.0 | Ile54 | -sigma | 3.59 |
| Ile119 | -alkyl | 4.61 | Ala98 | -alkyl | 4.05 | ||||
| Ile119 | -alkyl | 4.81 | |||||||
| Glu122 | H-bond | 2.31 | |||||||
Important amino acid residues responsible for the orientation and binding of CQ in the active site of PfLDH.
Fig. 2.
The selected diarylpentanoid analogues and derivatives with the highest and lowest binding energy against PfLDH (PDB ID: 1CET) from molecular docking studies. MAAC1 [(4-Amino-phenyl)-3-(2-fluoro-phenyl)-propenone; −6.7 kcal/mol], MAAC2 [(4-amino-phenyl)-3-(4-fluoro-phenyl)-propenone; −6.8 kcal/mol], MS33A [1,5-bis(2,3-dimethoxyphenyl)penta-1,4-dien-3-one; −5.8 kcal/mol],MS33C [1,5-bis(2,5-dimethoxyphenyl)penta-1,4-dien-3-one; −5.6 kcal/mol], MS33D [1,5-bis(2,6-dimethoxyphenyl)penta-1,4-dien-3-one; −5.5 kcal/mol], MS34C [1,5-bis(3,4,5-trimethoxyphenyl)penta-1,4-dien-3-one; −5.8 kcal/mol], MS40B [1,5-bis(3-chlorophenyl)penta-1,4-dien-3-one; −5.9 kcal/mol], MS47 [2,6-bis(2-chlorobenzylidene)cyclohexanone; −6.7 kcal/mol], MS69 [2,6-bis(2,3-dimethoxybenzylidene)cyclohexanone; −4.0 kcal/mol], MS72A [2,6-bis(2,3,4-trimethoxy-benzylidene)cyclohexanone; −0.7 kcal/mol], MS72C [2,6-bis(3,4,5-trimethoxybenzylidene)cyclohexanone; −0.1 kcal/mol], MS78 [2,5-bis(2-fluorobenzylidene)cyclopentanone; −7.2 kcal/mol], MS79 [2,5-bis(3-fluorobenzylidene)cyclopentanone; −6.8 kcal/mol], MS87 [2,5-bis(3,4,5-trimethoxybenzylidene)cyclopentanone; −5.2 kcal/mol].
Fig. 3.
Binding interactions of selected diarylpentanoids with the active site of PfLDH (Green: H-bond; Light green: C–H bond; Pink: alkyl and π-alkyl; Purple: π-sigma; Orange: π-anion; Cyan: halogen bond)
A complete summary of the chemical interactions between selected molecules and amino acid residues within the active binding site of the PfLDH protein can be found in Table S2 (in Supplementary Material). Based on the molecular docking results (Table 1), all compounds interacted hydrophobically (via π-alkyl, π-sigma, or alkyl) with the key PfLDH amino acid residue Ile119, comparable to Ahmed et al. (2017) [35]. Another essential amino acid residue Glu122 formed a hydrogen bond with CQ. Conversely, the same Glu122 formed weaker C–H bonds with MS34C, MS33A, MS87, and MS72; a halogen bond with MS79 and an electrostatic π-anion interaction with MS69 and MS47 were observed, suggesting lower antimalarial properties compared with CQ. All diarylpentanoids also showed hydrophobic interactions with crucial Ile54 and Ala98 residues, except for compounds MS72A, MS72C, MS78, and MS40B. As with the alkyl interactions of CQ with Val26 and Phe52, the same interactions were detected with MS34C, MS33C, and MS33D. Different forms of hydrophobic interaction with Tyr85 were also spotted with several compounds but MAAC1, MAAC2, MS47, MS69, and MS78. Since the binding of diarylpentanoids and CQ in the cavity pocket of the PfLDH protein is almost identical, we concluded that some of the selected molecules might show comparable PfLDH inhibition to CQ when tested in an in vitro assay. However, it is suspected that the selected diarylpentanoids might have a different mechanism of action than CQ, as they belong to different classes of compound.
The selected compounds were then evaluated for their physicochemical properties (Table 2), including Lipinski's rule of 5 (RO5), aggregators, pan-assay interference compounds (PAINS), and absorption, distribution, metabolism, excretion, and toxicity (ADMET) predictions (Table 3). In silico filtering of drug properties is one of the most important computational methods for predicting drug properties of compounds before they are synthesized and validated by in vitro assays, to reduce failure rates in drug discovery process [36].
Table 2.
Physicochemical properties, PAINS, and aggregation of diarylpentanoid analogues and derivatives.
| Compound | MW (g/mol) | HBD | HBA | TPSA | NRB | RO5 violation | LogP | LogS | PAINS | PAINS‐associated group | Aggregator probability | Oral bioavailability |
Fsp3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VEBER | EGAN | |||||||||||||
| MAAC1 | 241.26 | 2 | 2 | 43.09 | 3 | 0 | 3.003 | −4.480 | No | – | 0.225 | Good | Good | 0.105 |
| MAAC2 | 241.26 | 2 | 2 | 43.09 | 3 | 0 | 2.880 | −4.244 | No | – | 0.085 | Good | Good | 0.105 |
| MS33A | 354.40 | 0 | 5 | 53.99 | 8 | 0 | 3.437 | −5.638 | No | – | 0.12 | Good | Good | 0.0 |
| MS33C | 354.40 | 0 | 5 | 53.99 | 8 | 0 | 3.859 | −6.292 | No | – | 0.138 | Good | Good | 0.15 |
| MS33D | 354.40 | 0 | 5 | 53.99 | 8 | 0 | 3.688 | −6.198 | No | – | 0.103 | Good | Good | 0.0 |
| MS34C | 414.45 | 0 | 7 | 72.45 | 10 | 0 | 2.922 | −5.110 | No | – | 0.07 | Good | Good | 0.346 |
| MS40B | 303.18 | 0 | 1 | 17.07 | 4 | 0 | 4.882 | −6.630 | No | – | 0.208 | Good | Good | 0.346 |
| MS47 | 343.25 | 0 | 1 | 17.07 | 2 | 0 | 5.437 | −6.759 | Intermediate | ene_one_ene A (57) | 0.329 | Good | Good | 0.292 |
| MS69 | 394.46 | 0 | 5 | 53.99 | 6 | 0 | 3.949 | −5.747 | Intermediate | ene_one_ene A (57) | 0.241 | Good | Good | 0.32 |
| MS72A | 454.51 | 0 | 7 | 72.45 | 8 | 0 | 3.464 | −5.563 | Intermediate | ene_one_ene A (57) | 0.135 | Good | Good | 0.19 |
| MA72C | 454.51 | 0 | 7 | 72.45 | 8 | 0 | 3.493 | −5.463 | Intermediate | ene_one_ene A (57) | 0.160 | Good | Good | 0.19 |
| MS78 | 296.31 | 0 | 1 | 17.07 | 2 | 0 | 4.532 | −6.073 | Intermediate | ene_one_ene A (57) | 0.295 | Good | Good | 0.19 |
| MS79 | 296.31 | 0 | 1 | 17.07 | 2 | 0 | 4.485 | −6.209 | Intermediate | ene_one_ene A (57) | 0.192 | Good | Good | 0.261 |
| MS87 | 440.49 | 0 | 7 | 72.45 | 8 | 0 | 3.275 | −5.197 | Intermediate | ene_one_ene A (57) | 0.167 | Good | Good | 0.0 |
| CQ | 319.87 | 1 | 3 | 31.39 | 7 | 0 | 4.511 | −3.902 | No | – | 0.039 | Good | Good | 0.5 |
Table 3.
Antimalarial activity of selected diarylpentanoid analogues and derivatives against P. falciparum CQ-sensitive (3D7), P. falciparum CQ-resistant (Gombak A) and P. knowlesi A1H1.
| Compound | P. falciparum 3D7 (CQ-sensitive) EC50 (μM) ± SD | P. falciparum Gombak A (CQ-resistant) EC50 (μM) ± SD | P. knowlesi PKA1H1 EC50 (μM) ± SD | Vero cell (Normal cell) CC50 (μM) ± SD | Selectivity Index (SI) |
Resistance Index (RI) on P. falciparum strains |
||
|---|---|---|---|---|---|---|---|---|
| P. falciparum 3D7 (CQ-sensitive) | P. falciparum Gombak A (CQ-resistant | P. knowlesi PKA1H1 | P. falciparum 3D7 (CQ-sensitive) | |||||
| MAAC1 | 12.0 ± 0.7 | >200 | 17.4 ± 0.8 | 157.0 ± 0.9 | 13.1 | NC | 9.0 | NC |
| MAAC2 | 39.3 ± 5.1 | >200 | 13.4 ± 0.7 | NDa | NDa | NDa | NDa | NDa |
| MS33A | 3.1 ± 0.2 | 87.5 ± 0.9 | 3.6 ± 0.2 | 82.6 ± 1.7 | 26.6 | 1.0 | 22.9 | 28.2 |
| MS33C | 2.5 ± 0.4 | 56.2 ± 1.3 | 5.1 ± 0.2 | 163.1 ± 1.8 | 65.2 | 2.9 | 32.0 | 22.5 |
| MS33D | 21.7 ± 0.8 | 74.4 ± 2.4 | 23.7 ± 2.6 | 225.3 ± 3.0 | 10.4 | 3.0 | 9.5 | 3.4 |
| MS34C | 1.6 ± 0.2 | 8.8 ± 0.9 | 11.8 ± 1.1 | 134.4 ± 3.2 | 84.0 | 15.4 | 11.4 | 5.5 |
| MS40B | 13.7 ± 0.8 | 45.6 ± 1.3 | 16.6 ± 0.5 | 164.0 ± 2.1 | 12.0 | 3.6 | 9.9 | 3.3 |
| MS47 | 16.1 ± 0.5 | 51.4 ± 2.7 | 21.7 ± 1.2 | 170.6 ± 1.7 | 10.6 | 3.3 | 7.9 | 3.2 |
| MS69 | 785.8 ± 4.2 | 28.7 ± 2.0 | 324.1 ± 5.9 | ND | ND | ND | ND | ND |
| MS72A | 530.0 ± 3.6 | >200 | 507.5 ± 1.3 | ND | ND | ND | ND | ND |
| MS72C | 515.1 ± 4.9 | >200 | 430.1 ± 2.3 | ND | ND | ND | ND | ND |
| MS78 | 689.1 ± 4.2 | >200 | 354.8 ± 7.1 | ND | ND | ND | ND | ND |
| MS79 | 329.3 ± 2.6 | >200 | 194.6 ± 2.7 | ND | ND | ND | ND | ND |
| MS87 | 10.8 ± 1.3 | 1.85 ± 2.8 | 11.4 ± 0.9 | 564.1 ± 0.4 | 52.2 | 304.9 | 49.5 | 0.17 |
| CQ | 0.0017 ± 0.2 | 5.56 ± 3.2 | 0.0018 ± 0.1 | 491.7 ± 2.4 | >2000 | 89.0 | >2000 | >2000 |
ND = Not determine (EC50 > 200 μM).
NDa = Not determine due to limited sample.
NC = Not calculated.
SI > 10 (Acceptable value); SI > 50 (Ideal value) RI < 100 (High resistance); RI < 10 (Intermediate resistance); RI < 1 (Low resistance).
According to RO5, an orally active drug-like compound should have no more than one violation of the following standards: hydrogen bond donor (HBD) ≤ 5, hydrogen bond acceptor (HBA) ≤ 10, molecular weight (MW) < 500 Da, and an octanol-water partition coefficient (LogP) ≤ 5 [37]. In addition to assessing their oral bioavailability, all molecules were screened according to the rules of Veber [good oral bioavailability if total surface area (TPSA) ≤ 140 Å2 and no more than 10 rotatable bonds (NRB ≤10)] and Egan (good oral bioavailability if −1 ≤ log P ≤ 5.8 and TPSA ≤130 Å2) [38,39]. Table 2 shows that all fourteen selected compounds meet these RO5 requirements and are predicted to have good bioavailability, satisfying both Veber and Egan rules. Moreover, LogP (lipid solubility) and LogS (water solubility) are important parameters in in silico filtration of drugs as they are related to the pharmacokinetic behavior of drugs, i.e., the dissolution, absorption, distribution, and transport of the drug in the human body [40,41]. Usually, the optimal value for LogP is 0 < LogP < 3, while the optimal value for logS is ≥ −4. Unfortunately, all diarylpentanoid analogues and derivatives have a logP > 3 and a suboptimal logS value, except MS34C, suggesting that the compounds may have poor water solubility.
We also projected the selected molecules to potential pan-assay interference compounds (PAINS) and aggregator predictors via the FAFDrugs4 and ChemAGG web servers, respectively. Both these PAINS and aggregators can interact non-specifically with lipid bilayers, altering their physicochemical properties, and forming colloidal aggregates that can interfere with the screening process and lead to false positive results [42,43]. The result showed that several molecules (MS47, MS69, MS72A, MS72C, MS78, MS79, and MS87) were considered as PAINS intermediates possessing ene_one_ene A (57) fragments with low aggregation probability (probability less than 1). However, since the total substitution of the mentioned PAINS fragments was below the rejection threshold set by the public filter (1 for each compound), the desired molecules were classified as low-risk compounds. Note that a compound is classified as a PAINS molecule if it contains a high-risk structural warning and/or exceeds the threshold for the number of undesirable substructures in the same compound [44,45]. On the other hand, Fsp3 is a measure of carbon bond saturation (sp3-hybridised carbon) and it is said that increasing Fsp3 improves the physicochemical properties of drug candidates such as logP, solubility, and ADMET profile [46]. Compounds with higher Fsp3 are more likely to progress from the discovery phase to clinical trials and eventually lead to marketed drugs [47]. It should be noted that the average Fsp3 value for 1179 approved drugs is 0.47 [48], while the highest Fsp3 value (0.346) was found for MS34C and MS40B, indicating their potential for development as drugs.
Prediction of ADMET properties is also important in the early stages of drug discovery and development to improve the efficiency of drug candidates. A summary of ADMET results and a detailed discussion are provided in Table S3 and Section S2 (Supplementary Material).
Given the promising chemical interactions that mimic CQ in the active site of PfLDH and the predicted good physicochemical properties that conform to RO5, Veber, and Egan rules for bioavailability, we concluded that it was worthwhile to validate the selected molecules for in vitro biological evaluation. We even hypothesize that some molecules could lead to good Plasmodium inhibition, as they showed similar chemical interactions within the PfLDH receptor and comparable binding affinity as CQ.
2.2. Synthesis
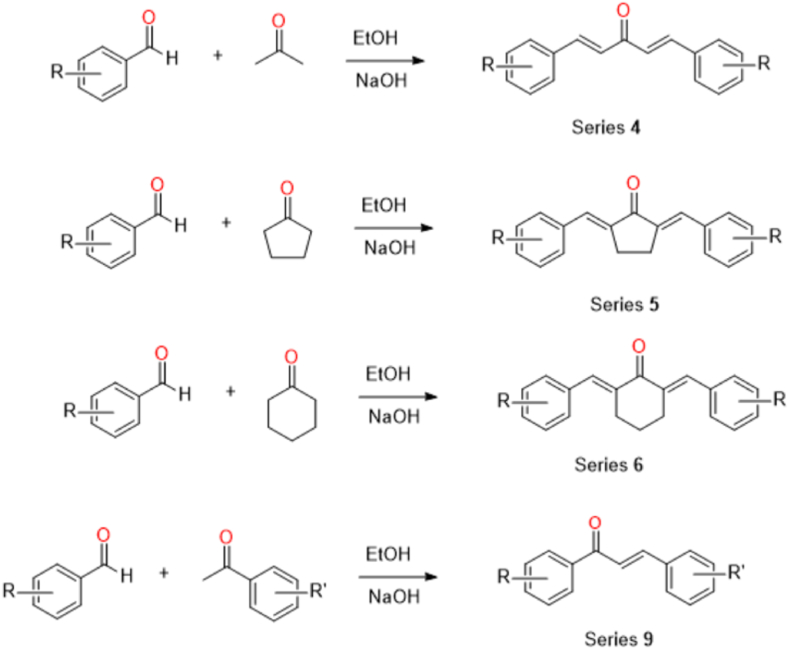
All the selected compounds (of series 4, 5, 6, and 9) listed in Table 1 have been previously described synthetically [49,50]. In general, substituted benzaldehydes and acetone (series 4), cyclopentanone (series 5), cyclohexanone (series 6) or substituted acetophenones (series 9) were mixed via a base-catalysed Claisen-Schmidt condensation reaction (Scheme 1) under ethanolic conditions. The yielded crude products were either purified by column chromatography (hexane: ethyl acetate) or recrystallized with methanol to give a pure compound, which was then structurally characterised by spectroscopic methods (see Figs. S1–S18 in the Supplementary Material), and subsequently investigated by in vitro antimalarial assays. It is important to emphasise that although these diarylpentanoids are derived from the curcuminoid structure, their stability is incomparable. Several studies have shown that diarylpentanoids have better stability under different physiological buffer conditions [51], improved chemical and metabolic stability in terms of bioavailability and increased intestinal permeability and water solubility [52].
Scheme 1.
Synthesis routes for the preparation of predicted bioactive antimalarial agents (of series 4, 5, 6 and 9) against PfLDH.
2.3. In vitro antiplasmodial activity and cytotoxic (vero cells) activities
The in vitro parasite inhibitory activity of all selected diarylpentanoid analogues and derivatives was determined against CQ-sensitive strains of P. falciparum 3D7, CQ-resistant strain of P. falciparum Gombak A and P. knowlesi A1H1, using CQ as the standard reference. The effective concentration of a drug that produces a half-maximal response (EC50) against different Plasmodium strains is listed in Table 3. The antimalarial activity of the selected compounds was categorized based on ranges, i.e., potent activity with EC50 < 1 μM, active with 1 < EC50 < 20 μM, moderate with 21 < EC50 < 100 μM, weak with 100 < EC50 < 200 μM, and inactive EC50 > 200 μM [53,54].
Among the selected diarylpentanoids, seven compounds (MAAC1, MS33A, MS33C, MS34C, MS40B, MS47 and MS87) showed active antimalarial activity against CQ-sensitive P. falciparum 3D7, two compounds (MAAC2 and MS33D) had moderate parasite inhibitory activity, while five other compounds were inactive with EC50 > 200 μM. A detailed examination of the bioactivity data revealed probable structure-activity relationships (SARs) between the assayed diarylpentanoid compounds. Interestingly, three molecules displayed significant dose-dependent inhibition of Plasmodium at EC50 < 5 μM (MS33A, MS33C, and MS34C; Supplementary Fig. S20), with MS34C registering as the most potent compound in the series with an EC50 value of 1.6 ± 0.2 μM (see Fig. S19 in Supplementary Material for other molecules). This significant activity might be contributed by the less bulky and flexible structure (C5 linker with acetone) associated with di- or tri-methoxylphenyl groups. It could also be inferred that the more electron donating groups (-OCH3) present in the aromatic ring, the better the electron density (since hydrophobic interactions are important) and antimalarial properties. This could be seen from the activity of MS34C > MS33C > MS33A. For the activity comparison, CQ showed the most potent antimalarial activity (EC50 value of 1.7 nM) against P. falciparum 3D7 strain, as expected, in parallel with its role as a reference antimalarial agent.
Predictably, the selected MS69, MS72A, and MS72C with the lowest binding affinity (−0.1 to −4.0 kcal/mol) to PfLDH exhibited the lowest activity with EC50 values > 500 μM. This significant loss of antimalarial activity could be due to the bulky structure consisting of a cyclohexanone C5 linker. This seems to indicate that the cyclohexanone fragment probably limits the flexibility of the compounds and does not properly fit into the active binding sites of the PfLDH protein and therefore does not significantly interact with the enzyme and inhibit its activity, which is essential for the antimalarial effect. On the other hand, although researchers typically correlate experimental EC50 values with binding affinity (derived from a molecular docking study), in most cases it is impossible to obtain a 1:1 correlation between these values. It is unlikely that this was the case for the compounds with the highest binding affinity MS78 and MS79, which caused a decrease in antimalarial activity with an EC50 of >500 μM. The most likely rationale is that this lower activity is due to the bulky and less flexible structure of the cyclopentanone linker (reducing the flexibility of the compound) in conjunction with the most electronegative atom (-F), which increases the electrophilicity of the molecules and thus diminishes the targeted activity.
Consequently, all selected molecules were also evaluated for their in vitro antiplasmodial activity against CQ-resistant Gombak A. Two compounds (MS87 and MS34C) showed considerable activity with EC50 values ranging from 1.85 to 8.75 μM, while other compounds exhibited moderate to poor Plasmodium inhibitory activity (EC50 > 25 μM). In contrast to previous SAR, MS87 with cyclopentanone-containing C5 linker showed better activity (EC50 1.85 μM) than other straight-chain C5 linker molecules when compared to their activity against the CQ-sensitive P. falciparum strain 3D7 (see Fig. S20 in Supplementary Material).
To assess the selectivity of the parasites, the in vitro cytotoxicity of the selected compounds (EC50 < 100 μM) was tested against mammalian Vero cells, demonstrating moderate to no cytotoxic activity (82.6 μM < CC50 < 564.1 μM, see Fig. S22 in Supplementary Material). It is worth noting that the compounds were classified as highly toxic to normal mammalian cells when the obtained CC50 value is < 10 μM, according to the cytotoxic threshold of Burger and Fiebig [55,56]. In addition, to understand the efficacy and potential of diarylpentanoids as safer and selective drugs, the selectivity indices (SI) were determined. The SI calculation for the CQ-sensitive 3D7 strain inhibition revealed that the tested molecules had SI values ranging from 10.6 to 84.0, although these values are much lower than the SI of CQ (>2000). Nevertheless, this is an interesting result, as SI > 10 is considered the minimum acceptable value and SI > 50 is an ideal value for the profile of the target candidate, since common antimalarial drugs should penetrate at least three membranes (the membrane of the red blood cell, the membrane of the parasitophore vacuole, and the plasma membrane of the parasite) after entering the bloodstream in order to exert their effect [57]. Additionally, the ideal drug would be cytotoxic only at very high concentrations and have antiviral activity at very low concentrations, thus yielding a high SI value and thereby able to eliminate the target virus at concentrations well below its cytotoxic concentration [58]. Meanwhile, inhibition of the P. falciparum strain Gombak A resulted in a high SI value of 304.9 for MS87 only, indicating that MS87 acts in parallel with strong inhibitory effects on the CQ-resistant strain.
Another remarkable observation was that the resistance index (RI) of the compounds was much lower (up to 28.2) than that of CQ (RI > 2000), proving that this class of compounds is potentially effective against both CQ-sensitive and CQ-resistant P. falciparum strains, while MS87 (RI = 0.17) showed better antiplasmodial activity against CQ-resistant than CQ-sensitive strains (RI < 1), indicating that this bioactive molecule may induce lower resistance. This is in line with the RI definition of Nzila & Mwai (2010), according to which an RI of 100 and 10 signifies a higher and intermediate level of resistance, respectively. Shalini et al. (2020), on the other hand, concluded that an RI < 0.7 is considered potentially low resistance to CQ [[59], [60]]. Relatively, a low RI indicates a promising antimalarial because it is equipotent regardless of strain susceptibility, which in turn indicates its effectiveness in combating drug resistance [61].
In addition to the increase in prevalence of P. knowlesi in Malaysia, we also tested all selected diarylpentanoids for their activity against the P. knowlesi PKA1H1 strain. A similar trend to the inhibition of the CQ-sensitive 3D7 strain was observed, with the compounds with the highest and lowest binding affinities showing lowest inhibition of the parasites (EC50 > 194 μM). Interestingly, two molecules (MS33A and MS33C) showed the strongest inhibition against the PKA1H1 strain with EC50 values of 3.6 and 5.1 μM, respectively. While the other compounds showed moderate antiplasmodial activity (11.4 < EC50 < 23.7 μM; see Fig. S21 in Supplementary Material). Thus, it can be concluded that the dimethoxylphenyl moiety with aliphatic C5 linker contributes significantly to the antimalarial properties against P. knowlesi compared to other chemical scaffolds.
2.4. In vivo toxicity test using zebrafish animal model
Clinical studies in human volunteers and in vivo mammalian models (e.g., rodents) have already demonstrated their toxicological reliability in identifying potential adverse effects of chemicals, drugs, and environmental pollutants on human health and help establish safe exposure levels for these substances. Although these models offer many advantages, they have suffered from cost, time, ethical, and animal welfare issues, as well as human endpoints in animal testing facilities. Until recently, all research using in vivo models had to follow the guidelines of standard ethics committees in which the “3 Rs” principle (Replacement, Refinement, and Reduction) must be fully implemented, and most importantly, the U.S. Environmental Protection Agency (EPA) has announced that it will eliminate the use of mammals in chemical testing by 2035 [62]. In addition, under new legislation, U.S. Food and Drug Administration (FDA) no longer must require animal testing before human drug trials [63]. Consistent with these principles and mandate, zebrafish (Danio rerio) have emerged as a promising alternative model for in vivo toxicology studies because they are genetically analogous to humans, have rapid external development and high reproductive rates, are transparent, and can be easily genetically modified [64]. According to the EU Directive 2010/63/EU on the protection of animals used for scientific purposes, early life-stages of zebrafish up to the stage when they are capable of independent feeding 5 dpf (days post fertilization) are not protected as animals. This makes it a valuable and efficient tool with a good dose-to-toxicity response, which is beneficial for drug discovery.
CQ and artemisinin are both pharmacotherapies commonly used as first-line treatments for malaria in certain regions of the world. However, their use is commonly associated with resistance and detrimental effects, including cardiotoxicity [65] and ototoxicity [66] with CQ administration, and severe haemolytic anaemia has been reported following parenteral artemisinin treatment [67], although the underlying cellular and molecular mechanisms for both drugs remain unclear. Therefore, we tested the toxic effects of selected significant compounds (MS33A, MS33C, and MS34C with EC50 < 10 μM; MS87 was not included due to problems with solubility in egg water) on early development of zebrafish embryos 24 h post fertilisation (hpf), cardiovascular system at 30 hpf, heart rate at 72 hpf, and cell death/apoptosis at 96 hpf. We also tested motor behaviour by measuring spontaneous tail coiling (STC) movements in diarylpentanoids-treated zebrafish embryos at 30 hpf.
First, we examined the effects of all selected compounds, including MS33A, MS33C and MS34C, on the early development of zebrafish embryos. At 24 hpf, embryos treated with MS33C and MS34C up to a high concentration of 200 μM showed no significant effect on early embryonic development, whereas the majority of MS33A-exposed embryos exhibited gastrulation defects at 100 μM, but not at 50 μM and lower concentrations (Fig. S23 in Supplementary Material). Gastrulation is the process of cell migration and differentiation that leads to the formation of the three germ layers of complex multicellular organisms and is the prelude to the full establishment of the body plan and organogenesis [68]. Defects in gastrulation, especially early gastrulation, can lead to severe developmental defects and affect the proper formation of organs, including the heart and nervous system.
To evaluate the cardiotoxicity of bioactive diarylpentanoids, we studied the effects of MS33A, MS33C and MS34C on the heart rate of treated larvae. The zebrafish heart is one of the first organs to develop and function and is therefore considered a primary target for developmental toxicity. The strategy for early detection of adverse cardiac symptoms in drug candidates is critical, as cardiotoxicity remains the most common reason for drug development failure [69]. Considering the cardiotoxic effects of the antimalarial drug CQ, the heart beats of zebrafish at 72 hpf in the control and treatment groups were counted 2 h after treatment. The results in Fig. 4A and B shows no obvious abnormalities in the heart rates of the compounds (MS33C and MS34C, up to 200 μM) compared to the control, confirming their non-cardiotoxic potential. However, zebrafish embryos treated with MS33A at concentrations of 50 and 25 μM showed a decrease in heartbeats, although they were relatively normal at a concentration of 12.5 μM, indicating possible cardiotoxicity (Fig. 4C).
Fig. 4.
Effects of MS33C, MS34C and MS33A treatments on heart rate of zebrafish larvae at 72 hpf. (A) MS33C-treated larvae exhibited normal heart beats up to a high concentration of 200 μM. (B) MS34C- treated larvae displayed normal heart beats up to a high concentration of 200 μM. (C) MS33A-treated larvae showed a decrease in heart beats from 25 μM to 50 μM compared to control 0.1% DMSO-treated larvae. n = 10 larvae for each concentration of tested compounds. The graphs shown are representative of three independent experiments. Mean values were expressed as bar graphs; ns indicates not significant; ****p < 0.0001.
Subsequently, the chemically induced toxic effects of MS33A, MS33C and MS34C were assessed up to 96 h and the LC50 of the compounds were determined in triplicate following OECD Guideline 236 [Fish Embryo Acute Toxicity Test (FET)] [70]. The respective LC50 was determined as 100 μM for MS33A and <200 μM for both MS33C and MS34C. The LC50 is considered a standardized approach for comparing the acute toxicity of the tested chemicals, which includes short-term chemical exposure to zebrafish and helps establish safe exposure levels to define the range of concentration limits for further detailed testing.
Furthermore, most embryos were able to survive at a concentration of 25 μM without showing toxic effects or abnormalities after exposure to each chemical for up to 72 h. There were no detectable morphological changes in embryos treated with MS33C and MS34C, i.e., body size, yolk expansion, pigment cell development, eye size, somite formation, and blood circulation of treated larvae compared to control larvae, while embryos treated with MS33A resulted in heart edema at 50 μM but not at 25 μM, as shown in Fig. 5A.
Fig. 5.
Effects of MS33C, MS34C and MS33A treatments on cardiovascular development. (A) At 72 hpf, chemical treatment with MS33C and MS34C up to 200 μM had no effect on normal heart development. However, zebrafish larvae treated with MS33A showed heart edema at 50 μM but not at 25 μM. The heart region is indicated by arrow. Anterior is to the left, lateral view. n = 5 larvae for each concentration. (B) Examination of blood vessel development in living embryos using a transgenic zebrafish line, Tg(kdrl:egfp), at 30 hpf. Growing intersegmental vessels (ISVs, arrows) in the trunk region were labelled by fluorescent GFP expression under control of the endothelial cell-specific kdrl gene promoter. MS33C and MS34C treatment had no effect on normal vessel development up to 200 μM. However, MS33A affected ISVs development at 50 μM but not at 25 μM. n = 3 embryos for each concentration. Scale bar = 200 μm.
To understand the defects in heart rate and heart edema in MS33A-treated zebrafish larvae, we introduced a transgenic zebrafish line, Tg(kdrl:egfp), in which blood vessel development can be monitored at the cellular level in the living animal by expression of green fluorescent protein (GFP) under control of the vascular endothelial cell-specific kdrl gene promoter. As shown in the cardiotoxicity assays, a similar observation was made in the development of blood vessels after MS33A treatment (Fig. 5B), with MS33C- and MS34C-exposed groups showing no significant difference in the development of intersegmental vessels (ISVs) of transgenic Tg(kdrl:egfp) zebrafish. However, embryos treated with MS33A show inhibitory effects on ISV development in the trunk region after treatment with a concentration of 50 μM, whereas ISVs at lower concentrations are similar to control larvae. This suggests that MS33A may have an anti-angiogenic activity that affects blood circulation and heart rate, eventually leading to heart edema. These cardiotoxic effects of MS33A are comparable to those of CQ, in contrast to the safer compounds MS33C and MS34C.
It should be emphasized that the early growth of the circulatory system of the zebrafish composes a strong resemblance to other vertebrates [71]. The vascular system of the zebrafish is very well organized and provides an opportunity to analyse the effects of toxic compounds at the cellular and molecular levels. In addition, studying the effects of toxic compounds on angiogenesis/blood vessel development in zebrafish may provide a useful model for studying cardiotoxic properties.
It is relatively recent that the STC test in zebrafish embryos is considered a powerful tool for screening developmental neurotoxicity (DNT) induced by chemical compounds [72]. STC in zebrafish involves the side-to-side contraction of the trunk muscles and is considered the first motor behaviour of zebrafish embryos. This first motor activity begins at 17 hpf, peaks at 20 hpf, and gradually decreases after 26 hpf [73]. To identify the potential neurotoxic effects of MS33A, MS33C and MS34C, we analysed STC activity in zebrafish embryos by exposing them to different concentrations of the tested compounds (50 μM, 25 μM, 12.5 μM, and 6.25 μM), compared to the control with 0.1% DMSO. After video recording, the average number of STC movements in a minute was counted in zebrafish embryos at 24 hpf for the different concentrations of each compound. No significant change was observed in the mean STC activity of the embryos treated with MS33A, MS33C, or MS34C compared with the control, indicating that these diarylpentanoid analogues do not have significant neurotoxic effects (refer Fig. S24 in Supplementary Material).
Moreover, zebrafish is an alternative, powerful vertebrate model that has been used extensively to study apoptotic cell death during normal development and under cellular stress conditions. Apoptosis is known as programmed cell death and has been morphologically classified into distinguishable patterns in dying cells, such as plasma membrane blebbing, cell shrinkage, fragmentation, and condensation of the nucleus. Over the past two decades, a detailed picture of the molecular basis of cell-intrinsic and cell-extrinsic apoptosis signalling pathways in zebrafish has emerged. Recent technological advances have enabled the labelling and visualisation of apoptotic cells in this zebrafish in vivo model system [73,74]. including the use of the efficient and vital fluorescent dye acridine orange (AO) in animal studies. To further verify the relative toxicity of the selected compounds, we analysed the apoptotic cells (stained with AO) in zebrafish larvae after treatment. Zebrafish embryos were treated at developmental stage 6 hpf (with concentrations of 25 μM, 12.5 μM, 6.25 μM) and incubated until 96 hpf. As shown in Fig. S25 (in Supplementary Material), no negative effect on apoptosis was observed compared to diarylpentanoids-treated larvae and control.
It is important to note that the evaluation of the toxicity of diarylpentanoids in the zebrafish in vivo model has the highest translatability to toxicity in humans. This is since 70% of human genes have a zebrafish homologue and 82% of human genes associated with disease have a zebrafish homologue, and thus, zebrafish technology can be considered a useful pre-filter to select the safest lead candidates early in the drug discovery process [75]. Nevertheless, possible damage caused by these compounds to the mitochondria, microtubules and DNA of Plasmodium strains should be further investigated in future research. Additionally, we predicted the mechanism of action of the antimalarials (based on the results of in vitro antimalarials and in vivo toxicity) using molecular docking, as described in Section S6 in Supplementary Material.
3. Conclusion
The present study describes an integration of in silico molecular docking studies, in vitro bioassessment and cytotoxicity and in vivo toxicity profiling using zebrafish for the discovery of new antimalarials. This work supports the correlation between high binding affinities derived from molecular docking results and favorable in vitro activity. However, given the observed discrepancies in the reliability of correlation of scoring functions with experimental validation, additional selection criteria: physiological relevance, minimization of side effects, dose-response consistency and specificity/selectivity were introduced to guide the selection of compounds for in vitro antimalarial assessment. Consequently, selected readily available molecules with similar chemical interactions and binding affinity comparable to CQ (−6.0 kcal/mol) were chosen for in vitro validation of antimalarial efficacy. Compounds MS33A, MS33C, and MS34C are the most promising against CQ-sensitive (3D7) with EC50 values of 1.6, 2.5, and 3.1 μM, MS87 (EC50 of 1.85 μM) as the most active against CQ-resistant Gombak A strain, and MS33A and MS33C as the most potent P. knowlesi inhibitors (EC50 of 3.6 and 5.1 μM). In addition, the compounds (with EC50 < 100 μM of PfLDH inhibition) were also evaluated against the Vero mammalian cell line, confirming that these compounds exhibited moderate to low cytotoxicity. Interestingly, MS87 exhibited a high SI and a good RI of 304.9 and 0.17, respectively, suggesting that MS87 acts in parallel with potent inhibitory effects with potentially lower resistance in the CQ-resistant P. falciparum strain. In addition, the same active molecules were molecularly docked to various glycolytic enzymes of P. falciparum and to haemoglobin. It was predicted that the selected diarylpentanoids (excluding MS34C), have a high potential to inhibit PfDHFR-TS as their probable mechanism of action. Furthermore, no toxic effects on normal embryonic development, blood vessel formation, heart rate, cell death/apoptosis, and motor behaviour were observed when these active diarylpentanoids (MS33C and MS34C) were tested for in vivo toxicity in zebrafish animal models, with the exception of MS33A, which has potential cardiotoxicity. Overall, the preliminary results suggest the potential of diarylpentanoids for further development of new antimalarial agents. However, in-depth pharmacological efficacy studies involving in vivo experiments, metabolic stability, and pharmacokinetic studies remain to be conducted to determine the likely mechanism of action. Nevertheless, it can be concluded that virtual screening still holds promise for filtering out new hit compounds from a large chemical library for drug discovery, especially when integrated and validated with biological experiments and preclinical toxicity tests.
4. Experimental
4.1. In silico studies
4.1.1. Molecular docking
Molecular docking was initially performed to investigate potential chemical interactions and binding affinities between the common drug CQ and diarylpentanoids at the PfLDH receptor to predict their antimalarial properties. The crystal structure of PfLDH (PDB ID: 1CET) was first retrieved from the RCSB Protein Data Bank (https://www.rcsb.org/). Consequently, the protein was prepared using AutoDock Tools version 1.5.6 by adding polar hydrogen and assigning charges to PfLDH and saved as a .pdbqt file. Discovery Studio Visualizer v19.1.0.18.287 was then used to remove all ligands, water, small compounds, and residues and saved in .pdb format. The docking input files were created with the following parameters for the search grid box: Center x: 36,472, Center y: 16,000, Center z: 19,806, Size x: 14, Size y: 14, Size z: 14, and Spacing: 1 before being saved in dock.txt format. Accordingly, the General Amber Force Field (GAFF) was applied to optimize the entire in-house library of diarylpentanoid analogues and derivatives (∼300 compounds) and CQ (compounds were defined as ligands), which were then saved as a .pdb file. The AutoDock tool version 1.5.6 was used afterwards to add all hydrogen atoms, define them as ligands, and save the results in the .pdbqt file. Docking programming was done with automatic docking software (AutoDock Vina) and Discovery Studio Visualizer v19.1.0.18,287 was used to determine the binding affinities and interactions formed between the enzyme and the ligands [76].
4.1.2. Physicochemical properties and ADMET prediction
The druglikeness of a molecule can be predicted from its physiochemical properties. First, all 2D chemical structures of diarylpentanoid analogues and derivatives (∼300 compounds) were drawn using ChemDraw Ultra 12.0 to create the Simplified Molecular Input Line Entry System (SMILES). The SMILES were then translated into Symyx Spatial Data File (SDF) format using OpenBabel 3.1.1 according to the recommendations of the FAF-Drugs4 Bank Formatter before being imported as an input file into the FAF-Drugs4 Web Server. Meanwhile, to identify potential aggregators, the identical SDF file containing the desired chemicals was loaded into the ChemAGG Web Server (https://admet.scbdd.com/ChemAGG/index/). The aggregator identification results were displayed in scores: Aggregator (1) and non-aggregator (0). Finally, ADMET profiles could be created by adding the current SMILES to SwissADME (https://admet.scbdd.com/ChemAGG/index/) [76].
4.2. Synthesis
4.2.1. Material and equipment
All chemicals and reagents were purchased from Merck and Sigma Aldrich, while the organic solvents used for spectroscopic measurements were of spectroscopic grade. All chemicals, reagents, and solvents were used directly for the synthesis without prior purification. The progress of the reaction was monitored by thin-layer chromatography using aluminium TLC sheets of Merck silica gel 60 f254 (layer thickness 0.2 mm). After completion of the reaction, the resulting mixtures were first extracted with suitable organic solvents, dried over anhydrous MgSO4, and concentrated in vacuo. This was followed by purification by recrystallization (series 4–6) or gravitational column chromatography (series 9) using Merck silica gel 60 PF254 no. 7734 (mesh size 70–230) and Merck silica gel 60 PF254 no. 9835 (mesh size 230–400). 1H and 13C-NMR spectra were measured in acetone-d6 using a Varian 500 MHz NMR spectrometer (Varian Inc., California, USA). Chemical shifts were expressed in δ (ppm) values relative to the standard tetramethylsilane (TMS), while melting points were determined using a Fisher-Johns melting point apparatus. Meanwhile, HRMS analysis was performed using a Thermo Scientific Dionex Ultimate 3000 Series RS pump coupled to a Thermo Scientific Dionex Ultimate 3000 Series TCC-3000RS column and a Thermo Fisher Scientific Ultimate 3000 Series WPS-3000RS autosampler controlled by Chromeleon 7.2 software (Thermo Fisher Scientific, Waltham, MA and Dionex Softron GMbH Part of Thermo Fisher Scientific, Germany). Separations were performed using an ACQUITY UPLC® BEH C18 analytical column (2.1 mm × 100 mm; particle size, 1.7 μm) (Waters, Milford, MA, USA) equipped with a Van Guard BEH C18 precolumn (2.1 mm × 5 mm; particle size, 1.7 m) (Waters, Milford, MA, USA) maintained at 40 °C. The mobile phase consisted of solutions A (0.1% v/v formic acid in water) and B (0.1% v/v formic acid in acetonitrile solution). A gradient program was used for elution, initially containing 5% solution B for 2 min, followed by 5–95% solution B from 2 to 15 min, 95% solution B from 15 to 18 min, and 5% solution B from 18.5 to 20 min. The mobile phase was added at a flow rate of 0.3 mL per minute and an injection volume of 1 μL. HRAMS data were acquired using a Thermo Scientific Q-Exactive Orbitrap mass spectrometer controlled by Xcalibur 3.0.63 software (Thermo Fisher Scientific, Waltham, MA) operating at a resolution of 140,000 in full-scan and 70,000 in MS/MS-scan mode. The main parameters of the heat electrospray ionization (HESI) source were optimized as follows: spray voltage, 4.0 kV; capillary temperature, 320 °C, sheath gas flow rate 35; auxiliary gas flow rate 102; heater temperature, 350 °C; S-lens RF level 55.
4.2.2. General synthetic procedure
A mixture of 2 mmol (2 equiv.) of substituted benzaldehydes and 1 mmol (1 equiv.) of appropriate acetone [series 4 (MS33A, MS33C, MS33D, MS34C, MS40B)], cyclopentanone [series 5 (MS78, MS79, MS87)], or cyclohexanone [series 6 (MS47, MS69, MS72A, MS72C)] was dissolved in 10 ml of ethanol and stirred in an ice bath to prepare the selected known diarylpentanoid analogues. To this solution, 10 ml of a 40% NaOH solution was then added dropwise over several minutes and then stirred at room temperature for about 10 h. The reaction was neutralized with a diluted HCl solution to quench the reaction, forming a precipitate that was consequently filtered and dried in vacuo. The crude product was recrystallized from MeOH–water mixture to give a pure compound whose structure was confirmed by spectroscopic methods [49]. It is worth noting that the only available spectroscopic data attached for the published and known compounds in series 4–6 are high performance liquid chromatography (HPLC) profiles with 89–99% purity (see Figs. S1–S12 in Supplementary Material) and 1H-NMR spectrum.
Meanwhile, MAAC1 and MAAC2 (series 9) were newly synthesized by a one-pot Claisen-Schmidt condensation reaction. In this reaction, equimolar 4-aminoacetophenones and fluorobenzaldehydes were mixed and dissolved with 1 ml methanol in a round bottom flask and stirred for 5 min. Then 1 ml of 6 M NaOH was added dropwise to the resulting mixture solution and stirred continuously overnight. After 24 h, the crude products were quenched to pH 3 with ice and 1 M HCl to induce precipitation of the target molecules before filtration. In further steps, the products were purified by recrystallization and/or column chromatography and then structurally characterized by spectroscopic methods [50]. All required spectroscopic data for the newly synthesized MAAC1 and MAAC 2 (of series 9) are attached for reference (Figs. S13–S18 in Supplementary Material).
1,5-Bis(2,3-dimethoxyphenyl)penta-1,4-dien-3-one (MS33A). Following the general procedure stated, 2,3-dimethoxylbenzaldehyde (0.3323 g, 2 mmol), reacted with acetone (0.0581 g, 1 mmol) and was purified through recrystallization with MeOH–water mixture given the product as pale-yellow solid; mp 106–108 °C; HPLC purity 89.47%; 1H NMR (500 MHz, acetone-d6) δ in ppm: 8.07 (d, J = 16.0 Hz, 2H, 2 x COCH=CH), 7.41 (t, J = 4.5 Hz, 2H, 2 x Ar-CH), 7.31 (d, J = 16.0 Hz, 2H, 2 x COCH=CH), 7.14 (d, J = 4.5 Hz, 4H, 2 x Ar-CH), 3.91 (s, 6H, 2 x -OCH3), 3.89 (s, 6H, 2 x -OCH3); 13C NMR (125 MHz, acetone-d6) δ in ppm: 188.6 (C=O), 153.3 (2 x Ar-COCH3), 152.6 (2 x COCH=CH), 149.3 (2 x Ar-COCH3), 141.5 (2 x Ar–C), 123.3 (2 x COCH=CH), 121.9 (2 x Ar-CH), 120.8 (2 x Ar-CH), 114.5 (2 x Ar-CH), 61.7 (2 x -OCH3), 56.1 (2 x -OCH3); MS (EI): m/z found for C21H22O5 is 354 [M]+ [49].
1,5-Bis(2,5-dimethoxyphenyl)penta-1,4-dien-3-one (MS33C). Following the general procedure stated, 2,5-dimethoxylbenzaldehyde (0.3323 g, 2 mmol), reacted with acetone (0.0581 g, 1 mmol) and was purified through recrystallization with MeOH–water mixture given the product as pale-yellow solid; mp105–106 °C; HPLC purity 91.28%; 1H NMR (500 MHz, acetone-d6) δ in ppm: 7.98 (d, J = 16.0 Hz, 2H, 2 x COCH=CH), 6.88 (d, J = 16.0 Hz, 2H, 2 x COCH=CH), 6.84 (d, J = 8.0 Hz, 4H, 2 x Ar-CH), 6.82 (s, 2H, 2 x Ar-CH), 3.96 (s, 6H, 2 x -OCH3), 3.85 (s, 6H, 2 x -OCH3). 13C NMR (125 MHz, acetone-d6) δ in ppm: 188.4 (C=O), 152.8 (2 x Ar-COCH3),152.6 (2 x COCH=CH), 151.5 (2 x Ar-COCH3), 123.3 (2 x COCH=CH), 115.9 (2 x Ar–C),115.2 (2 x Ar-CH), 114.5 (2 x Ar-CH), 111.5 (2 x Ar-CH), 56.2 (2 x -OCH3), 55.8 (2 x -OCH3); MS (EI): m/z found for C21H22O5 is 354 [M]+ [49].
1,5-Bis(2,6-dimethoxyphenyl)penta-1,4-dien-3-one (MS33D). Following the general procedure stated, 2,6-dimethoxylbenzaldehyde (0.3323 g, 2 mmol), reacted with acetone (0.0581 g, 10 mmol) and was purified through recrystallization with MeOH–water mixture given the product as pale-yellow solid; mp153–155 °C; HPLC purity 93.31%; 1H NMR (500 MHz, acetone-d6) δ in ppm: 8.17 (d, J = 16.0 Hz, 2H, 2 x COCH=CH), 7.59 (d, J = 16.0 Hz, 2H, 2 x COCH=CH), 7.36 (t, J = 8.5 Hz, 2H, 2 x Ar-CH), 6.75 (d, J = 8.5 Hz, 4H, 4 x Ar-CH), 3.90 (s, 12H, 4 x -OCH3); 13C NMR (125 MHz, acetone-d6) δ in ppm: 188.6 (C=O), 158.6 (4 x Ar-COCH3), 152.6 (2 x COCH=CH), 129.9 (2 x Ar-CH) 123.3 (2 x COCH=CH), 114.7 (2 x Ar-CH), 106.5 (4 x Ar-CH), 56.2 (4 x -OCH3); MS (EI): m/z found for C21H22O5 is 354 [M]+ [49].
1,5-Bis(3,4,5-trimethoxyphenyl)penta-1,4-dien-3-one (MS34C). Following the general procedure stated, 3,4,5-trimethoxylbenzaldehyde (0.3924 g, 2 mmol), reacted with acetone (0.0581 g, 1 mmol) and was purified through recrystallization with MeOH–water mixture given the product as pale-yellow solid; mp 105–107 °C; HPLC purity 90.01%; 1H NMR (500 MHz, acetone-d6) δ in ppm: 7.71 (d, J = 16.0 Hz, 2H, 2 x COCH=CH), 7.24 (d, J = 16.0 Hz, 2H, 2 x COCH=CH), 7.09 (s, 4H, 2 x Ar-CH), 3.92 (s, 12H, 4 x -OCH3), 3.80 (s, 6H, 2 x -OCH3); 13C NMR (125 MHz, acetone-d6) δ in ppm: 188.6 (C=O), 153.0 (4 x Ar-COCH3), 142.2 (COCH=CH)138.4 (2 x Ar-COCH3),126.4 (2 x Ar-CH), 123.3 (COCH=CH), 103.8 (2 x Ar-CH) 60.8 (2 x -OCH3), 56.1 (4 x -OCH3); MS (EI): m/z found for C23H26O7 is 414 [M]+ [49].
1,5-Bis(3-chlorophenyl)penta-1,4-dien-3-one (MS40B). Following the general procedure stated, 3-chlorobenzaldehyde (0.2811 g, 2 mmol), reacted with acetone (0.0581 g, 1 mmol) and was purified through recrystallization with MeOH–water mixture given the product as pale-yellow solid; mp123–126 °C; HPLC purity 89.38%; 1H NMR (500 MHz, acetone-d6) δ in ppm: 7.83 (s, 2H, 2 x Ar-CH), 7.78 (d, J = 16.0 Hz, 2H, COCH=CH), 7.73 (d, J = 7.0 Hz, 2H, 2 x Ar-CH), 7.54 (m, 4H, 4 x Ar-CH), 7.42 (d, J = 16.0 Hz, 2H, COCH=CH); 13C NMR (125 MHz, acetone-d6) δ in ppm: 188.6 (C=O), 142.2 (COCH=CH), 136.6 (2 x Ar–C), 134.2 (2 x Ar-C-Cl), 130.0 (2 x Ar-CH), 128.0 (2 x Ar-CH), 126.6 (2 x Ar-CH), 126.4 (2 x Ar-CH), 123.3 (COCH=CH); MS (EI): m/z found for C17H13Cl2O is 303 [M]+ [49].
2,6-Bis(2-chlorobenzylidene)cyclohexanone (MS47). Following the general procedure stated, 2-chlorobenzaldehyde (0.2811 g, 2 mmol), reacted with acetone (0.0581 g, 1 mmol) and was purified through recrystallization with MeOH–water mixture given the product as pale-yellow solid; mp 90–92 °C; HPLC purity 92.54%; 1H NMR (500 MHz, acetone-d6) δ in ppm: 7.85 (s, 2H, 2 x COC=CH), 7.56 (m, 4H, 4 x Ar-CH), 7.45 (m, 4H, 4 x Ar-CH), 2.87 (t, J = 6.5 Hz, 4H, 2 x COCCH2CH2), 1.78 (quintet, J = 6.5 Hz, 2H, COCCH2CH2); 13C NMR (125 MHz, acetone-d6) δ in ppm: 188.6 (C=O), 142.2 (2 x COC=CH), 136.6 (2 x Ar-CC=CH), 134.2 (2 x Ar-C-Cl), 130.0 (2 x Ar-CH), 128.0 (2 x Ar-CH), 126.6 (2 x Ar-CH), 126.4 (2 x Ar-CH), 123.3 (2 x COCH=CH); MS (EI): m/z found for C20H16Cl2O is 343 [M]+ [49].
2,6-Bis(2,3-dimethoxybenzylidene)cyclohexanone (MS69). Following the general procedure stated, 2,3-dimethoxylbenzaldehyde (0.3323 g, 2 mmol), reacted with cyclohexanone (0.0982 g, 1 mmol) and was purified through recrystallization with MeOH–water mixture given the product as pale-yellow solid; mp107–109 °C; HPLC purity 91.18%; 1H NMR (500 MHz, acetone-d6) δ in ppm: 7.90 (s, 2H, COC=CH), 7.12 (m, 4H, 4 x Ar-CH), 7.03 (dd, J = 1.5 Hz, 7.5 Hz, 2H, 2 x Ar-CH), 3.90 (s, 6H, 2 x -OCH3), 3.81 (s, 6H, 2 x -OCH3), 2.88 (m, 4H, 2 x COCCH2CH2), 1.76 (quintet, J = 6.0 Hz, 2H, 2 x COCCH2CH2); 13C NMR (125 MHz, acetone-d6) δ in ppm: 190.4 (C=O), 154.8 (2 x Ar-C-OCH3), 153.3 (2 x Ar-C-OCH3), 145.5 (2 x COC=CH), 137.1 (2 x COC=CH), 121.9 (2 x Ar-CH), 120.8 (2 x Ar-CH), 115.9 (2 x Ar-C-CHCO), 114.5 (2 x Ar-CH), 61.7 (2 x -OCH3), 26.1 (COCCH2CH2), 25.1 (COCCH2CH2); MS (EI): m/z found for C24H26O5 is 394 [M]+ [49].
2,6-Bis(2,3,4-trimethoxybenzylidene)cyclohexanone (MS72A). Following the general procedure stated, 2,3,4-trimethoxylbenzaldehyde (0.3924 g, 2 mmol), reacted with cyclohexanone (0.0982 g, 1 mmol) and was purified through recrystallization with MeOH–water mixture given the product as pale-yellow solid; mp 163–165 °C; HPLC purity 94.69%; 1H NMR (500 MHz, acetone-d6) δ in ppm: 7.89 (s, 2H, COC=CH), 7.20 (d, J = 8.5 Hz, 2H, 2 x Ar-CH), 6.88 (d, J = 8.5 Hz, 2H, 2 x Ar-CH), 3.91 (s, 6H, 2 x -OCH3), 3.87 (s, 6H, 2 x -OCH3), 3.84 (s, 6H, 2 x -OCH3), 2.90 (t, J = 6.5 Hz, 4H, COCH2CH2), 1.77 (quintet, J = 6.5 Hz, 2H, COCH2CH2); 13C NMR (125 MHz, acetone-d6) δ in ppm: 190.4 (C=O), 156.0 (2 x Ar-C-OCH3), 149.1 (2 x Ar-C-OCH3), 145.5 (2 x COC=CH), 143.3 (2 x Ar-C-OCH3), 137.1 (2 x COC=CH), 122.7 (2 x Ar-CH), 108.2 (2 x Ar-C-CH), 104.0 (2 x Ar-CH), 61.7 (2 x -OCH3), 60.8 (2 x -OCH3), 56.1 (2 x -OCH3), 26.1 (COCCH2CH2), 25.1 (COCCH2CH2); MS (EI): m/z found for C26H30O7 is 454 [M]+ [49].
2,6-Bis(3,4,5-trimethoxybenzylidene)cyclohexanone (MS72C). Following the general procedure stated, 3,4,5-trimethoxylbenzaldehyde (0.3924 g, 2 mmol), reacted with cyclohexanone (0.0982 g, 1 mmol) and was purified through recrystallization with MeOH–water mixture given the product as pale-yellow solid; mp 147–149 °C; HPLC purity 94.16%; 1H NMR (500 MHz, acetone-d6) δ in ppm: 7.64 (s, 2H, COC=CH), 6.86 (s, 4H, 4 x Ar-CH), 3.90 (s, 12H, 4 x -OCH3), 3.79 (s, 6H, 4 x -OCH3), 2.82 (t, J = 6.5 Hz, 4H, COCH2CH2), 1.77 (quintet, J = 6.5 Hz, 2H, COCH2CH2); 13C NMR (125 MHz, acetone-d6) δ in ppm: 190.4 (C=O), 153.0 (4 x Ar-C-OCH3), 138.4 (2 x Ar-C-OCH3), 137.1 (2 x COC=CH), 132.2 (2 x COC=CH), 125.0 (2 x Ar-C-CH), 107.4 (2 x Ar-CH), 60.8 (2 x -OCH3), 56.1 (4 x -OCH3), 26.1 (COCCH2CH2), 25.1 (COCCH2CH2); MS (EI): m/z found for C26H30O7 is 454 [M]+ [49].
2,5-Bis(2-fluorobenzylidene)cyclopentanone (MS78). Following the general procedure stated, 2-fluorobenzaldehyde (0.2482 g, 2 mmol), reacted with cyclopentanone (0.0841 g, 1 mmol) and was purified through recrystallization with MeOH–water mixture given the product as pale-yellow solid; mp 180–182 °C; HPLC purity 98.81%; 1H NMR (500 MHz, acetone-d6) δ in ppm: 7.82 (t, J = 8.0 Hz, 2H, 2 x Ar-CH), 7.71 (s, 2H, COC=CH), 7.52 (m, 2H, 2 x Ar-CH), 7.36 (t, J = 8.0 Hz, 2H, 2 x Ar-CH), 7.27 (t, J = 10 Hz, 2H, 2 x Ar-CH), 3.17 (s, 4H, 2 x COC–CH2); 13C NMR (125 MHz, acetone-d6) δ in ppm: 196.6 (C=O), 161.3 (Ar-CF), 146.1 (2 x COC=CH2), 143.6 (2 x COC=CH2), 129.5 (2 x Ar-CH), 128.0 (2 x Ar-CH), 124.2 (2 x Ar-CH), 123.1 (2 x Ar–C), 115.4 (2 x Ar-CH), 29.4 (2 x COC–CH2); MS (EI): m/z found for C19H14F2O is 296 [M]+ [49].
2,5-Bis(3-fluorobenzylidene)cyclopentanone (MS79). Following the general procedure stated, 3-fluorobenzaldehyde (0.2482 g, 2 mmol), reacted with cyclopentanone (0.0841 g, 1 mmol) and was purified through recrystallization with MeOH–water mixture given the product as pale-yellow solid; mp 215–217 °C; HPLC purity 98.08%; 1H NMR (500 MHz, acetone-d6) δ in ppm: 7.57 (s, 2H), 7.54 (d, 2H, J = 8.5 Hz), 7.48 (d, 4H, J = 8.5 Hz), 7.24 (m, 2H), 3.22 (s, 4H); 13C NMR (125 MHz, acetone-d6) δ in ppm: 196.6 (C=O), 162.8 (2 x Ar-CF), 143.6 (2 x COC=CH), 136.8 (2 x Ar–C), 132.8 (2 x COC=CH), 130.2 (2 x Ar-CH), 124.1 (2 x Ar-CH), 114.7 (2 x Ar-CH), 113.9 (2 x Ar-CH), 35.4 (COC–CH2), 29.4 (COC–CH2); MS (EI): m/z found for C19H14F2O is 296 [M]+ [49].
2,5-Bis(3,4,5-trimethoxybenzylidene)cyclopentanone (MS87). Following the general procedure stated, 3,4,5-trimethoxylbenzaldehyde (0.3924 g, 2 mmol), reacted with cyclopentanone (0.0841 g, 1 mmol) and was purified through recrystallization with MeOH–water mixture given the product as pale-yellow solid; mp 195–197 °C; HPLC purity 90.32%; 1H NMR (500 MHz, acetone-d6) δ in ppm: 7.42 (s, 2H, 2 x COC=CH), 7.01 (s, 4H, 4 x Ar-CH), 3.93 (s, 12H, 4 x -OCH3), 3.81 (s, 6H, 2 x -OCH3), 3.22 (s, 4H, 2 x COC–CH2); 13C NMR (125 MHz, acetone-d6) δ in ppm: 196.6 (C=O), 153.0 (4 x Ar-C-OCH3), 143.6 (2 x CO–C=CH), 138.4 (2 x Ar-C-OCH3), 132.8 (2 x CO–C=CH), 125.0 (2 x Ar–C), 107.4 (2 x Ar-CH), 60.8 (2 x -OCH3), 56.1 (4 x -OCH3), 29.4 (COC–CH2); MS (EI): m/z found for C25H29O7 is 440 [M]+ [49].
(4-Amino-phenyl)-3-(2-fluoro-phenyl)-propenone (MAAC1, new compound). Following the general procedure stated, 4-aminoacetophenone (0.1352 g, 1 mmol), reacted with 2-fluorobenzaldehyde (0.1241 g, 1 mmol) and was purified through recrystallization with ethanol given the product as dark yellow solid; mp 142–144 °C; 1H NMR (500 MHz, acetone-d6): δ in ppm: 7.96 (d, J = 8.5 Hz, 2H, 2 x Ar-CH), 7.95 (d, J = 16.0 Hz, 1H, COCH=CH), 7.89 (dd, J = 8.5 Hz, 1H, Ar-CH), 7.87 (d, J = 15.5 Hz, 1H, COCH=CH), 7.47 (dd, J = 7.5 Hz, 1H, Ar-CH), 7.25 (td, J = 8.5, 2.5 Hz, 2H, 2 x Ar-CH), 6.76 (d, J = 8.5 Hz, 2H, 2 x Ar-CH), 5.61 (s, 2H, NH2); 13C NMR (125 MHz, acetone-d6) δ in ppm: 185.9 (C=O), 162.2 (C–F), 153.6 (C–NH2), 133.3 (COCH=CH), 131.6 (Ar–C), 131.0 (2 x Ar-CH), 128.9 (Ar-CH), 124.6 (Ar–C), 123.3 (COCH=CH), 116.0 (Ar-CH), 115.8 (Ar-CH), 113.1 (2 x Ar-CH); MS (EI): m/z found for C15H13FNO is 241 [M]+; HRMS (ESI): m/z calcd for C15H12FNO+H+: 242.0981 [M+H]+; found: 242.0969.
(4-Amino-phenyl)-3-(4-fluoro-phenyl)-propenone (MAAC2, new compound). Following the general procedure stated, 4-aminoacetophenone (0.1352 g, 1 mmol), reacted with 4-fluorobenzaldehyde (0.1241 g, 1 mmol) and was purified through recrystallization with ethanol given the product as pale-yellow solid; mp 146–148 °C; 1H NMR (500 MHz, acetone-d6) δ in ppm: 8.11 (d, J = 9.0 Hz, 2H, 2 x Ar-CH), 8.10 (d, J = 14.5 Hz, 2H, 2 x CH=CH), 8.10 (d, J = 9.0 Hz, 2H, 2 x Ar-CH), 7.29 (d, J = 9.0 Hz, 2H, 2 x Ar-CH), 7.27 (t, J = 8.5 Hz, 2H, 2 x Ar-CH), 7.27 (s, 2H, NH2); 13C NMR (125 MHz, acetone-d6) δ in ppm: 185.7 (C=O), 162.3 (C–F), 153.9 (C–NH2), 133.5 (COCH=CH), 131.4 (Ar–C), 131.0 (2 x Ar-CH), 128.7 (2 x Ar-CH), 124.6 (Ar–C), 123.3 (COCH=CH), 115.7 (2 x Ar-CH), 113.2 (2 x Ar-CH); MS (EI): m/z found for C15H13FNO is 241 [M]+; HRMS (ESI): m/z calcd for C15H12FNO+H+: 242.0981 [M+H]+; found: 242.0969.
4.3. In vitro pharmacological studies
4.3.1. Antimalarial assessment against Plasmodium falciparum strains and P. knowlesi A1H1
Plasmodium parasites were cultured according to the methods of Trager and Jensen (1976) [77] to determine the anti-malarial activity of compounds against CQ-sensitive P. falciparum (3D7) strain and CQ-resistant strain of P. falciparum (Gombak A). Meanwhile, P. knowlesi A1H1 was contributed by Robert W. Moon, London School of Hygiene and Tropical Medicine, London, UK, and method for culture adaptation into human blood was referred to Moon et al. (2013) [78]. Parasites were obtained from the Malaria Culture Laboratory of Universiti Malaya, Malaysia and maintained in continuous culture in fresh human group O+ (Rhesus-positive) erythrocytes suspended in 2% hematocrit in RPMI 1640 containing 10% AlbumaX I, glucose (3 g/L), hypoxanthine (45 μg/L), and gentamicin (50 μg/L). Whereas 10% horse serum was used as a substitute for AlbumaX I in P. knowlesi culture. Plasmodium lactate dehydrogenase (pLDH) assays were performed with slight modifications as described by Makler et al. (1993) [79]. Parasites were plated in the asynchronous phase at 2% hematocrit and 2% parasitemia in 100 μL of the test compounds, using CQ as the reference antimalarial drug, while the unparasitized erythrocytes without the test compounds and the parasitized erythrocytes without the test compounds were used as blanks and controls, respectively, for the assay. The corresponding concentration used for CQ and the test diarylpentanoids in this study ranged from 0.00001 μM to 100 μM and 0.002 μM–2000 μM. The plates were placed in an incubator containing 5% CO2 and incubated at 37 °C for 48 h for P. falciparum, and 24 h for P. knowlesi. After incubation, the plates were subjected to three 30-min freeze-thaw cycles to lyse and resuspend the erythrocytes in culture. Malstat reagent (100 μl) and NBT/PES solution (25 μL) were then added to each well of a new 96-well microtiter plate. Consequently, the culture (25 μL) was added to the corresponding well of the reagent plate to initiate the PfLDH reaction. The colour development of the plate was monitored calorimetrically at 650 nm after 1 h of incubation in the dark. The data obtained were analysed using nonlinear regression via GraphPad Prism software to obtain EC50 values. EC50 values of the tested compounds were derived from three independent experiments.
4.3.2. Cytotoxicity against vero cell
Cytotoxicity is the toxicity caused by the action of chemotherapeutic agents on living cells. One of the parameters used to determine the cytotoxicity of the selected compounds is the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assay. A cell culture was prepared at a concentration of 7 × 103 cells/mL and plated on 96-well plates (100 μL/well). Diluted series of the tested compounds (MS33A, MS33C, MS34C, MS87 and CQ) were added to each well at the indicated concentrations (1600, 800, 400, 200, 100, 50 and 25 μM) and incubated for 72 h. At the end of incubation, MTT solution was added to the cells and incubation continued for an additional 3 h. After solubilization of the purple formazan crystals with DMSO, the optical density (OD) of the assayed diarylpentanoids was measured using an ELISA reader at a wavelength of 570 nm. Cytotoxicity was determined as the cytotoxic concentration causing 50% inhibition of cell growth (CC50 value). After determining the percentage of cell viability, graphs of the percentage cell viability against each concentration were generated using Graph Pad Prism software. CC50 values of the tested compounds were calculated from three independent experiments [80].
4.4. In vivo toxicity test using zebrafish animal model
4.4.1. Zebrafish husbandry and embryo collection
A line of wild-type zebrafish (Danio rerio) kept in our animal facility was reared under specific environmental conditions. All in vivo experiments were conducted with the approval of the Institutional Animal Care and Use Committee (IACUC) of Chungnam National University (Approval number: 202012A-CNU-170). Zebrafish were maintained in 2-L aquaria at 25–27 °C on a 10:14 h dark-light cycle and fed brine shrimp (Artemia) five times daily. The aquarium water system was maintained by a constant circulating pump. Fish were kept in a ratio of 3 males to 3 females in a single tank to provide a stress-free environment for initiation of the reproductive cycle. Fertilized eggs were collected after 24 h and incubated in an egg buffer solution at 28 °C. Only embryos that had reached the early gastrulation stage (6 hpf) with intact chorionic membranes were selected, while unfertilized or dead eggs were discarded [73].
4.4.2. Acute toxicity, LC50, STC and cardiotoxicity testing on zebrafish embryos
The acute toxicity assay for fish embryos was conducted in accordance with Test Guideline (TG) 236 of the Organization for Economic Co-operation and Development (OECD) [70]. To test the acute toxicity on early embryonic development, ten fertilized zebrafish embryos were collected at an early developmental stage of 6 hpf, placed on the 24-well plate and treated with 1 ml of the different selected diarylpentanoid concentrations (200 μM, 100 μM, 50 μM, 25 μM, 12.5 μM and 6.25 μM) and the control (0.1% DMSO). First, the selected MS33A, MS33C, and MS34C with the strongest malarial potential (EC50 < 10 μM) were dissolved in DMSO to prepare an 80 mM stock solution, and then serially diluted into the embryo water at an initial concentration of 200 μM. The plates were then placed in the incubator to maintain 28 °C and dark conditions. At each 24-h interval (4 endpoints were taken-24, 48, 72, and 96 h), the number of dead embryos was counted, embryo development was monitored, and test solutions were replaced with freshly prepared test chemicals for each concentration. Signs of a dead embryo include coagulation of the embryo, lack of somite formation, non-detachment of the tail, and/or lack of heartbeat. All these characteristics can be easily detected by light microscope. In addition, the LC50 of MS33A, MS33C and MS34C was determined at different time intervals. Consequently, zebrafish embryos were anesthetized with tricaine (Sigma-Aldrich), dechorionated, and embedded in 3.5% methylcellulose gel for microscopic imaging. The exposed embryos were visualized with a Nikon AZ100 microscope (Nikon, Tokyo, Japan), digital images were captured with a Nikon Digital Sight DS-Fil1 digital camera (Nikon) and processed with NIS-Elements F 3.0 (Nikon) [73].
The possible neurotoxicity of diarylpentanoids could be investigated using the STC movements of zebrafish. At developmental stage 22 hpf, 10 embryos were collected for each test concentration (50 μM, 25 μM, 12.5 μM, and 6.25 μM) and incubated for 2 h before STC movements were recorded, selecting only embryos without morphological malformations. The tested embryos were first habituated for 2 min, and subsequently, the number of STC movements within 1 min was manually counted under a standard dissecting microscope. A complete STC movement was represented by a whole-body contraction that brings the tail tip to the head and includes two alternating lateral (left-right) flexions of the whole body [73].
Meanwhile, the cardiotoxicity of the selected compounds was examined at 48 hpf. Ten embryos were collected per test concentration (25 μM, 12.5 μM and 6.25 μM) and incubated for 2 h before calculating the number of heart beats of all exposed zebrafish embryos for 1 min. Counting was performed under a standard dissecting microscope.
4.4.3. Evaluation of adverse effect on the formation of blood vessel
The transgenic zebrafish line Tg(kdrl:egfp) was treated with 0.1% DMSO (as a control) and with different exposure concentrations of MS33A (25 and 50 μM), MS33C and MS34C (200 μM), in which the tested concentration being chosen depending on teratogenicity and LC50 experiments. Treatment with the selected diarylpentanoids began at the developmental stage of 10 hpf and was examined at 30 hpf, when normal blood vessel formation was observed in control zebrafish. Subsequently, the treated embryos were dechorionated and embedded in 3% methylcellulose on a glass slide for bioimaging, and live images were acquired using a CELENA® S Digital Imaging System (Logos Biosystems, Anyang, Korea) [73].
4.4.4. Evaluation of apoptosis
Cell death in zebrafish larvae treated with diarylpentanoids was examined using the fluorescent stain acridine orange (AO), which is commonly used as a marker of apoptotic cells in zebrafish. In each assay, ten zebrafish embryos were placed in egg water containing 4 μg/mL AO (Sigma) for 20 min. The live zebrafish embryos were then washed five times with the egg buffer water for 5 min each, anesthetized with tricaine (Sigma), and embedded in 3% methylcellulose gel before being observed by stereomicroscopy and fluorescence microscopy as previously described [73].
4.4.5. Statistical analysis
Statistical analyses were performed using SPSS (SPSS v. 25.0). One-way analysis of variance (ANOVA) was used to determine the effect of each treatment group on the control. Data were presented as mean ± standard deviation (SD) using GraphPad Prism (Graphpad Software, USA). Data were significantly different when p ≤ 0.05. To ensure that these values were independent, data were analysed per well to avoid any interaction bias between embryos [73].
Data availability
Data will be made available on request.
Funding
This work was supported by the Korea Environment Industry & Technology Institute (KEITI) under the Core Technology Development Project for Environmental Diseases Prevention and Management (RE2021003310003) for the experiments with zebrafish in vivo.
CRediT authorship contribution statement
Amirah Hani Ramli: Writing – original draft, Investigation, Formal analysis. Puspanjali Swain: Writing – original draft, Investigation, Formal analysis. Muhammad Syafiq Akmal Mohd Fahmi: Investigation. Faridah Abas: Resources, Conceptualization. Sze Wei Leong: Resources. Bimo Ario Tejo: Resources, Conceptualization. Khozirah Shaari: Resources. Amatul Hamizah Ali: Writing – original draft, Investigation, Formal analysis. Hani Kartini Agustar: Validation, Methodology, Data curation, Conceptualization. Rusdam Awang: Methodology, Investigation, Formal analysis, Data curation. Yee Ling Ng: Resources. Yee Ling Lau: Resources. Mohammad Aidiel Md Razali: Investigation, Formal analysis. Siti Nurulhuda Mastuki: Investigation, Formal analysis. Norazlan Mohmad Misnan: Resources, Investigation, Formal analysis, Data curation. Siti Munirah Mohd Faudzi: Writing – review & editing, Visualization, Validation, Supervision, Resources, Project administration, Methodology, Data curation, Conceptualization. Cheol-Hee Kim: Validation, Supervision, Project administration, Methodology, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The author also thanks Prof. Dr. Faridah Abas for the collection of diarylpentanoids analogues, Prof. Dr. Khozirah Shaari and Assoc. Prof. Dr. Bimo Ario Tejo for the financial support of the in vitro antimalarial and cytotoxicity assays.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e27462.
Contributor Information
Amirah Hani Ramli, Email: hanidoux98@gmail.com.
Puspanjali Swain, Email: puspanjaliswain20@gmail.com.
Muhammad Syafiq Akmal Mohd Fahmi, Email: syafiqakmal1@gmail.com.
Faridah Abas, Email: faridah_abas@upm.edu.my.
Sze Wei Leong, Email: leongszewei@um.edu.my.
Bimo Ario Tejo, Email: bimo.tejo@upm.edu.my.
Khozirah Shaari, Email: khozirah@yahoo.com.my.
Amatul Hamizah Ali, Email: amatulhamizahali@yahoo.com.my.
Hani Kartini Agustar, Email: hani_ag@ukm.edu.my.
Rusdam Awang, Email: rusdam_awang@upm.edu.my.
Yee Ling Ng, Email: yeeling0726@gmail.com.
Yee Ling Lau, Email: lauyeeling@um.edu.my.
Mohammad Aidiel Md Razali, Email: aidiel980429@gmail.com.
Siti Nurulhuda Mastuki, Email: ctnurulhuda@upm.edu.my.
Norazlan Mohmad Misnan, Email: norazlan.misnan@moh.gov.my.
Siti Munirah Mohd Faudzi, Email: sitimunirah@upm.edu.my.
Cheol-Hee Kim, Email: zebrakim@cnu.ac.kr.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.WHO . vols. 1–293. World Health Organization; Geneva: 2022. https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2022 (World Malaria Report). [Google Scholar]
- 2.Singh B., Daneshvar C. Human infections and detection of Plasmodium knowlesi. Clin. Microbiol. Rev. 2013;26:165–184. doi: 10.1128/CMR.00079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper D.J., Rajahram G.S., William T., Jelip J., Mohammad R., Benedict J., Alaza D.A., Malacova E., Yeo T.W., Grigg M.J., Anstey N.M., Barber B.E. Plasmodium knowlesi Malaria in Sabah, Malaysia, 2015-2017: ongoing increase in incidence despite near-elimination of the human-only Plasmodium species. Clin. Infect. Dis. 2020;70:361–367. doi: 10.1093/cid/ciz237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hussin N., Lim Y.A.L., Goh P.P., William T., Jelip J., Mudin R.N. Updates on malaria incidence and profile in Malaysia from, 2013 to 2017. Malar. J. 2020;19:55–69. doi: 10.1186/s12936-020-3135-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goh X.T., Lim Y.A.L., Vythilingam I., Chew C.H., Lee P.C., Ngui R., Tan T.C., Yap N.J., Nissapatorn V., Chua K.H. Increased detection of plasmodium knowlesi in sandakan division, sabah as revealed by PlasmoNex. Malar. J. 2013;12:264–273. doi: 10.1186/1475-2875-12-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization . third ed. 2015. Guidelines for the Treatment of Malaria.https://www.who.int/malaria/publications/atoz/9789241549127/en/ [Google Scholar]
- 7.Bloland P.B. vols. 1–27. World Health Organization; Geneva: 2001. https://apps.who.int/iris/handle/10665/66847 (Drug Resistance in Malaria). [Google Scholar]
- 8.Wani W.A., Jameel E., Baig U., Mumtazuddin S., Hun L.T. Ferroquine and its derivatives: new generation of antimalarial agents. Eur. J. Med. Chem. 2015;101:534–551. doi: 10.1016/j.ejmech.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang W., Qiu C., Li S., Zhou L., Hu M., Chen X., Yu B., Hong Y., Liu Z., Xia Q. Crystal structure of 2, 5-bis ((E)-2-(trifluoromethyl) benzylidene) cyclopentan-1-one, C21H14F6O. Z. Kristallogr. - New Cryst. Struct. 2017;232:921–922. doi: 10.1515/ncrs-2017-0080. [DOI] [Google Scholar]
- 10.Baldwin P.R., Reeves A.Z., Powell K.R., Napier R.J., Swimm A.I., Sun A., Giesler K., Bommarius B., Shinnick T.M., Snyder J.P., Liotta D.C. Monocarbonyl analogs of curcumin inhibit growth of antibiotic sensitive and resistant strains of Mycobacterium tuberculosis. Eur. J. Med. Chem. 2015;92:693–699. doi: 10.1016/j.ejmech.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohd Faudzi S.M., Leong S.W., Auwal F.A., Abas F., Wai L.K., Ahmad S., Tham C.L., Shaari K., Lajis N.H., Yamin B.M. In silico studies, nitric oxide, and cholinesterases inhibition activities of pyrazole and pyrazoline analogs of diarylpentanoids. Arch. Pharm. 2021;354 doi: 10.1002/ardp.202000161. [DOI] [PubMed] [Google Scholar]
- 12.Hussain H., Ahmad S., Shah S.W.A., Ghias M., Ullah A., Rahman S.U., Kamal Z., Khan F.A., Khan N.M., Muhammad J., Almehmadi M. Neuroprotective potential of synthetic mono-carbonyl curcumin analogs assessed by molecular docking studies. Molecules. 2021;26:7168. doi: 10.3390/molecules26237168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Viira B., Gendron T., Lanfranchi D.A., Cojean S., Horvarth D., Marcou G., Varnek A., Maes L., Maran U., Loiseau P.M., Davioud-Charvet E. In silico mining for antimalarial structure-activity knowledge and discovery of novel antimalarial curcuminoids. Molecules. 2016;21:853. doi: 10.3390/molecules21070853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Busari Z.A., Dauda K.A., Morenikeji O.A., Afolayan F., Oyeyemi O.T., Meena J., Sahu D., Panda A.K. Antiplasmodial activity and toxicological assessment of curcumin PLGA-encapsulated nanoparticles. Front. Pharmacol. 2017;8:622. doi: 10.3389/fphar.2017.00622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clariano M., Marques V., Vaz J., Awam S., Afonso M.B., Jesus Perry M., Rodrigues C.M. Monocarbonyl analogs of curcumin with potential to treat colorectal cancer. Chem. Biodivers. 2023;20 doi: 10.1002/cbdv.202300222. [DOI] [PubMed] [Google Scholar]
- 16.Shetty D., Kim Y.J., Shim H., Snyder J.P. Eliminating the heart from the curcumin molecule: monocarbonyl curcumin mimics (MACs) Molecules. 2014;20:249–292. doi: 10.3390/molecules20010249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maji A.K. Drug susceptibility testing methods of antimalarial agents. Tropenmed. Parasitol. 2018;8:70. doi: 10.4103/2229-5070.248695. 10.4103%2F2229-5070.248695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fokou P.V.T., Tali B.M.T., Dize D., Mbouna C.D.J., Ngansop C.A.N., Keumoe R., Tchokouaha L.R.Y., Tchouankeu J.C., Escudie F., Duffy J., Boyom F.F. Implementation and continued validation of the malaria Plasmodium falciparum lactate dehydrogenase-based colorimetric assay for use in antiplasmodial drug screening. Anal. Biochem. 2022;648 doi: 10.1016/j.ab.2022.114669. [DOI] [PubMed] [Google Scholar]
- 19.Baspinar Y. In: The Chemistry and Bioactive Components of Turmeric. Gopi S., thomas S., Kunnumakkara A.B., Aggarwal B.B., Amalraj A., editors. The Royal Society of Chemistry; London: 2020. Molecular docking studies of curcumin; pp. 239–248. [DOI] [Google Scholar]
- 20.Rodrigues R.P., Andrade S.F., Mantoani S.P., Eifler-Lima V.L., Silva V.B., Kawano D.F. Using free computational resources to illustrate the drug design process in an undergraduate medicinal chemistry course. J. Chem. Educ. 2015;92:827–835. doi: 10.1021/ed500195d. [DOI] [Google Scholar]
- 21.Rajkhowa S., Deka R.C. In: Methods and Algorithms for Molecular Docking-Based Drug Design and Discovery. Dastmalchi S., Hamzeh-Mivehroud M., Sokouti B., editors. IGI Global; Pennysylvania: 2017. Protein-ligand docking methodologies and its application in drug discovery; pp. 196–219. (IGI Global. In Oncology: Breakthroughs in Research and Practice). 891-914. [Google Scholar]
- 22.Wadood A., Ahmed N., Shah L., Ahmad A., Hassan H., Shams S. In-silico drug design: an approach which revolutionarised the drug discovery process. OA Drug. Des. Deliv. 2013;1:3. doi: 10.13172/2054-4057-1-1-1119. [DOI] [Google Scholar]
- 23.Salim N.O., Fuad F.A.A., Khairuddin F., Seman W.M.K.W., Jonet M.A. Purifying and characterizing bacterially expressed soluble lactate dehydrogenase from plasmodium knowlesi for the development of anti-malarial drugs. Molecules. 2021;26:6625. doi: 10.3390/molecules26216625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J., Kim T.I., Lê H.G., Yoo W.G., Kang J.M., Ahn S.K., Myint M.K., Lin K., Kim T.S., Na B.K. Genetic diversity of Plasmodium vivax and Plasmodium falciparum lactate dehydrogenases in Myanmar isolates. Malar. J. 2020;19:60. doi: 10.1186/s12936-020-3134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Read J.A., Wilkinson K.W., Tranter R., Sessions R.B., Brady R.L. Chloroquine binds in the cofactor binding site of Plasmodium falciparum lactate dehydrogenase. J. Biol. Chem. 1999;274:10213–10218. doi: 10.1074/jbc.274.15.10213. [DOI] [PubMed] [Google Scholar]
- 26.Turgut‐Balik D., Sadak D., Celik V. Analysis of active site loop amino acids of lactate dehydrogenase from Plasmodium vivax by site‐directed mutagenesis studies. Drug Dev. Res. 2006;67:175–180. doi: 10.1002/ddr.20089. [DOI] [Google Scholar]
- 27.Ramírez D., Caballero J. Is it reliable to take the molecular docking top scoring position as the best solution without considering available structural data? Molecules. 2018;23:1038. doi: 10.3390/molecules23051038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim S., Loo J., Alshagga M., Alshawsh M., Ong C., Pan Y. Protein-ligand identification and in vitro inhibitory effects of cathine on 11 major human drug metabolizing cytochrome p450s. Int. J. Toxicol. 2022;41:355–366. doi: 10.1177/10915818221103790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Araújo J., Bastos R., Santos G., Alves M., Figueiredo K., Sousa L., Rocha J. Molecular docking and evaluation of antileishmania activity of a ruthenium complex with epiisopiloturine and nitric oxide. J. Biosci. Med. 2020;8:42–53. doi: 10.4236/jbm.2020.85005. [DOI] [Google Scholar]
- 30.Ong C.H., Tham C.L., Harith H.H., Firdaus N., Israf D.A. Drug repurposing of clinically approved drugs to target epithelial-mesenchymal transition using molecular docking approach. Malays. J. Med. Health Sci. 2023;19:15–23. doi: 10.47836/mjmhs19.5.4. [DOI] [Google Scholar]
- 31.Cruz-Aguado J.A., Penner G. Determination of ochratoxin A with a DNA aptamer. J. Agric. Food Chem. 2008;56:10456–10461. doi: 10.1021/jf801957h. [DOI] [PubMed] [Google Scholar]
- 32.Kenakin T. The mass action equation in pharmacology. Br. J. Clin. Pharmacol. 2016;81:41–51. doi: 10.1111/bcp.12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klekota J., Brauner E., Roth F.P., Schreiber S.L. Using high-throughput screening data to discriminate compounds with single-target effects from those with side effects. J. Chem. Inf. Model. 2006;46:1549–1562. doi: 10.1021/ci050495h. [DOI] [PubMed] [Google Scholar]
- 34.Pisabarro M., Serrano L. Rational design of specific high-affinity peptide ligands for the Abl-SH3 domain. Biochemistry. 1996;35:10634–10640. doi: 10.1021/bi960203t. [DOI] [PubMed] [Google Scholar]
- 35.Ahmed N., Anwar S., Thet Htar T. Docking based 3D-QSAR study of tricyclic guanidine analogues of batzelladine K as anti-malarial agents. Front. Chem. 2017;5:36. doi: 10.3389/fchem.2017.00036. 10.3389/fchem.2017.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ani R.S., Anand B., Deepa O.S. In silico prediction tool for drug-likeness of compounds based on ligand based screening. Int. J. Res. Pharm. Sci. 2020;11:6273–6281. doi: 10.26452/ijrps.v11i4.3310. [DOI] [Google Scholar]
- 37.Tijjani H., Olatunde A., Adegunloye A.P., Ishola A.A. In: Coronavirus Drug Discovery. Egbuna C., editor. Elsevier; Amsterdem: 2022. In silico insight into the interaction of 4-aminoquinolines with selected SARS-CoV-2 structural and nonstructural proteins; pp. 313–333. [Google Scholar]
- 38.Veber D.F., Johnson S.R., Cheng H.Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 39.Vázquez-Vuelvas O.F., Enríquez-Figueroa R.A., García-Ortega H., Flores-Alamo M., Pineda-Contreras A. Crystal structure of the chalcone (E)-3-(furan-2-yl)-1-phenylprop-2-en-1-one. Acta Crystallogr. E. 2015;71:161–164. doi: 10.1107/S205698901500047X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan C., Peng Y., Deng C., Gong D., Cao A. Boosting n-octanol/water partition coefficients prediction with an improved gene expression programming method. J. Phys.: Conf. Ser. 2020;1486 doi: 10.1088/1742-6596/1486/4/042042. [DOI] [Google Scholar]
- 41.Schelenz T., Schäfer W. Zur physikalisch‐chemischen charakterisierung von 5‐Amino‐1‐aryl‐1H‐tetrazolen: charakterisierung von wasserlöslichkeiten und ihre beziehungen zu octan‐1‐ol/wasser‐verteilungskoeffizienten. J. Prakt. Chem. 2000;342:91–95. doi: 10.1002/(SICI)1521-3897(200001)342:1<91::AID-PRAC91>3.0.CO;2-F. [DOI] [Google Scholar]
- 42.Magalhães P.R., Reis P.B., Vila-Viçosa D., Machuqueiro M., Victor B.L. Optimization of an in silico protocol using probe permeabilities to identify membrane pan-assay interference compounds. J. Chem. Inf. Model. 2022;62:3034. doi: 10.1021/acs.jcim.2c00372. 2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z.-Y., Yang Z.-J., Dong J., Wang L.-L., Zhang L.-X., Ding J.-J., Ding X.-Q., Lu A.-P., Hou T.-J., Cao D.-S. Structural analysis and identification of colloidal aggregators in drug discovery. J. Chem. Inf. Model. 2019;59:3714–3726. doi: 10.1021/acs.jcim.9b00541. [DOI] [PubMed] [Google Scholar]
- 44.Lagorce D., Bouslama L., Becot J., Miteva M.A., Villoutreix B.O. FAF-Drugs4: free ADME-tox filtering computations for chemical biology and early stages drug discovery. Bioinformatics. 2017;33:3658. doi: 10.1093/bioinformatics/btx491. [DOI] [PubMed] [Google Scholar]
- 45.Baell J.B., Holloway G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 46.Für C.S., Riszter G., Gerencsér J., Szigetvári Á., Dékány M., Hazai L., L, Keglevich G., Bölcskei H. Synthesis of spiro [cycloalkane-pyridazinones] with high Fsp3 character. Lett. Drug Des. Discov. 2020;17:731–744. doi: 10.2174/1570180816666190710130119. [DOI] [Google Scholar]
- 47.Wei W., Cherukupalli S., Jing L., Liu X., Zhan P. Fsp3: a new parameter for drug-likeness. Drug Discov. Today. 2020;25:1839–1845. doi: 10.1016/j.drudis.2020.07.017. [DOI] [PubMed] [Google Scholar]
- 48.Bhuvan Kumar N.N., Mukhina O.A., Kutateladze A.G. Photoassisted synthesis of enantiopure alkaloid mimics possessing unprecedented polyheterocyclic cores. J. Am. Chem. Soc. 2013;135:9608–9611. doi: 10.1021/ja4042109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hosoya T., Nakata A., Yamasaki F., Abas F., Shaari K., Lajis N.H., Morita H. Curcumin-like diarylpentanoid analogues as melanogenesis inhibitors. J. Nat. Med. 2012;66:166–176. doi: 10.1007/s11418-011-0568-0. [DOI] [PubMed] [Google Scholar]
- 50.Mohd Faudzi S.M., Abdullah M.A., Abdull Manap M.R., Ismail A.Z., Rullah K., Mohd Aluwi M.F.F., Ramli A.N.M., Abas F., Lajis N.H. Inhibition of nitric oxide and prostaglandin E2 production by pyrrolylated-chalcones: synthesis, biological activity, crystal structure analysis, and molecular docking studies. Bioorg. Chem. 2020;94 doi: 10.1016/j.bioorg.2019.103376. [DOI] [PubMed] [Google Scholar]
- 51.Mohd Faudzi S.M., Leong S.W., Abas F., Mohd Aluwi M.F.F., Rullah K., Lam K.W., Abdul Bahari M.N., Ahmad S., Tham C.L., Shaari K., Lajis N.H. Synthesis, biological evaluation, QSAR studies of diarylpentanoid analogues as potential nitric oxide inhibitors. Med. Chem. Comm. 2015;6:1069–1080. doi: 10.1039/C4MD00541D. [DOI] [Google Scholar]
- 52.Pantiora P., Furlan V., Matiadis D., Mavroidi B., Perperopoulou F., Papageorgiou A.C., Sagnou M., Bren U., Pelecanou M., Labrou N.E. Monocarbonyl curcumin analogues as potent inhibitors against human glutathione transferase P1-1. Antioxidants. 2023;12 doi: 10.3390/antiox12010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dolabela M.F.N., Oliveira S.G., Nascimento J.M., Peres J.M., Wagner H., Póvoa M.M., de Oliveira A.B. In vitro antiplasmodial activity of extract and constituents from Esenbeckia febrifuga, a plant traditionally used to treat malaria in the Brazilian Amazon. Phytomedicine. 2008;15:367–372. doi: 10.1016/j.phymed.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 54.Basco L.K., Marquet F., Makler M.M., Lebras J. Plasmodium falciparum and Plasmodium vivax: lactate-dehydrogenase activity and its application for in vitro drug susceptibility assay. Exp. Parasitol. 1995;80:260–271. doi: 10.1006/expr.1995.1032. [DOI] [PubMed] [Google Scholar]
- 55.Burger A.M., Fiebig H.H. In: Handbook of Anticancer Pharmacokinetics and Pharmacodynamics. Figg W.D., McLeod H.L., editors. Springer; Berlin: 2014. Preclinical screening for new anticancer agents; pp. 23–38. [Google Scholar]
- 56.Wiji Prasetyaningrum P., Bahtiar A., Hayun H. Synthesis and cytotoxicity evaluation of novel asymmetrical mono-carbonyl analogs of curcumin (AMACs) against Vero, HeLa, and MCF7 cell lines. Sci. Pharm. 2018;86:25. doi: 10.3390/scipharm86020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oleinikov A.V. Malaria parasite Plasmodium falciparum proteins on the surface of infected erythrocytes as targets for novel drug discovery. Biochemistry. 2022;87:S192–S202. doi: 10.1134/S0006297922140152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pritchett J.C., Naesens L., Montoya J. In: Human Herpesviruses HHV-6A, HHV-6B & HHV-7. third ed. Flamand L., Lautenschlager I., Krueger G.R.F., Ablashi D.V., editors. Elsevier; Amsterdam: 2014. Treating HHV-6 infections: the laboratory efficacy and clinical use of anti-HHV-6 agents; pp. 311–331. [Google Scholar]
- 59.Nzila A., Mwai L. In vitro selection of Plasmodium falciparum drug-resistant parasite lines. J. Antimicrob. Chemother. 2010;65:390–398. doi: 10.1093/jac/dkp449. https://doi:10.1093/jac/dkp449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shalini Kumar S., Gendrot M., Fonta I., Mosnier J., Cele N., Awolade P., Singh P., Pradines B., Kumar V. Amide tethered 4-aminoquinoline-naphthalimide hybrids: a new class of possible dual function antiplasmodials. ACS Med. Chem. Lett. 2020;11:2544–2552. doi: 10.1021/acsmedchemlett.0c00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pal K., Raza M.K., Legac J., Rahman A., Manzoor S., Bhattacharjee S., Rosenthal P.J., Hoda N. Identification, in-vitro antiplasmodial assessment and docking studies of series of tetrahydrobenzothieno[2,3-d]pyrimidine-acetamide molecular hybrids as potential antimalarial agents. Eur. J. Med. Chem. 2023;248 doi: 10.1016/j.ejmech.2022.115055. [DOI] [PubMed] [Google Scholar]
- 62.Grimm D. EPA plan to end animal testing splits scientists. Science. 2019;365:1231. doi: 10.1126/science.365.6459.1231. [DOI] [PubMed] [Google Scholar]
- 63.FDA no longer needs to require animal tests before human drug trials. https://www.science.org/content/article/fda-no-longer-needs-require-animal-tests-human-drug-trials, (accessed 11 June 2023).
- 64.Howe K., Clark M.D., Torroja C.F., Torrance J., Berthelot C., Muffato M. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blignaut M., Espach Y., van Vuuren M., Dhanabalan K., Huisamen B. Revisiting the cardiotoxic effect of chloroquine. Cardiovasc. Drugs Ther. 2019;33:1–11. doi: 10.1007/s10557-018-06847-9. [DOI] [PubMed] [Google Scholar]
- 66.Davis S.N., Wu P., Camci E.D., Simon J.A., Rubel E.W., Raible D.W. Chloroquine kills hair cells in zebrafish lateral line and murine cochlear cultures: implications for ototoxicity. Hear. Res. 2020;395 doi: 10.1016/j.heares.2020.108019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang R., Yan S., Zhu X., Li C.Y., Liu Z., Xiong J.W. Antimalarial drug artemisinin depletes erythrocytes by activating apoptotic pathways in zebrafish. Exp. Hematol. 2015;43 doi: 10.1016/j.exphem.2014.11.012. 331-41.e8. [DOI] [PubMed] [Google Scholar]
- 68.Bruce A.E.E., Heisenberg C.P. Mechanisms of zebrafish epiboly: a current view. Curr. Top. Dev. Biol. 2020;136:319–341. doi: 10.1016/bs.ctdb.2019.07.001. [DOI] [PubMed] [Google Scholar]
- 69.Maciag M., Wnorowski A., Mierzejewska M., Plazinska A. Pharmacological assessment of zebrafish-based cardiotoxicity models. Biomed. Pharmacother. 2022;148 doi: 10.1016/j.biopha.2022.112695. [DOI] [PubMed] [Google Scholar]
- 70.OECD . OECD Publishing; Paris: 2013. Test No. 236: Fish Embryo Acute Toxicity (FET) Test, OECD Guidelines for the Testing of Chemicals, Section 2. [Google Scholar]
- 71.Gore A.V., Monzo K., Cha Y.R., Pan W., Weinstein B.M. Vascular development in the zebrafish. Cold Spring Harb. Perspect. Med. 2012;2 doi: 10.1101/cshperspect.a006684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zindler F., Beedgen F., Brandt D., Steiner M., Stengel D., Baumann L., Braunbeck T. Analysis of tail coiling activity of zebrafish (Danio rerio) embryos allows for the differentiation of neurotoxicants with different modes of action. Ecotoxicol. Environ. Saf. 2019;186 doi: 10.1016/j.ecoenv.2019.109754. [DOI] [PubMed] [Google Scholar]
- 73.Mat Zian N.F.A., Swain P., Mohd Faudzi S.M., Zakaria N., Wan Ibrahim W.N., Abu Bakar N., Shaari K., Stanslas J., Choi T.I., Kim C.H. Mapping molecular networks within Clitoria ternatea Linn. against LPS-induced neuroinflammation in microglial cells, with molecular docking and in vivo toxicity assessment in zebrafish. Pharmaceuticals. 2022;15:467. doi: 10.3390/ph15040467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eimon P.M., Ashkenazi A., A The zebrafish as a model organism for the study of apoptosis. Apoptosis. 2010;15:331–349. doi: 10.1007/s10495-009-0432-9. [DOI] [PubMed] [Google Scholar]
- 75.Cassar S., Adatto I., Freeman J.L., Gamse J.T., Iturria I., Lawrence C., Muriana A., Peterson R.T., Van Cruchten S., Zon L.I. Use of zebrafish in drug discovery toxicology. Chem. Res. Toxicol. 2020;33:95–118. doi: 10.1021/acs.chemrestox.9b00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mohd Fahmi M.S.A., Swain P., Ramli A.H., Wan Ibrahim W.N., Saleh Hodin N.A., Abu Bakar N., Tan Y.S., Mohd Faudzi S.M., Kim C.H. In silico studies, X-ray diffraction analysis and biological investigation of fluorinated pyrrolylated-chalcones in zebrafish epilepsy models. Heliyon. 2023;9 doi: 10.1016/j.heliyon.2023.e13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trager W., Jensen J.B. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 78.Moon R.W., Hall J., Rangkuti F., Ho Y.S., Almond N., Mitchell G.H., Pain A., Holder A.A., Blackman M.J. Adaptation of the genetically tractable malaria pathogen Plasmodium knowlesi to continuous culture in human erythrocytes. Proc. Natl. Acad. Sci. USA. 2013;110:531–536. doi: 10.1073/pnas.1216457110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Makler M., Ries J., Williams J., Bancroft J., Piper R., Gibbins B., Hinrichs D. Parasite lactate dehydrogenase as an assay for Plasmodium falciparum drug sensitivity. Am. J. Trop. Med. Hyg. 1993;48:739–741. doi: 10.4269/ajtmh.1993.48.739. [DOI] [PubMed] [Google Scholar]
- 80.Leong S.W., Chia S.L., Abas F., Yusoff K. Synthesis and in-vitro anti-cancer evaluations of multi-methoxylated asymmetrical diarylpentanoids as intrinsic apoptosis inducer against colorectal cancer. Bioorg. Med. Chem. Lett. 2020;30 doi: 10.1016/j.bmcl.2020.127065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.