Abstract
Primary macrophages are infected by macrophage (M)-tropic but not T-cell line (T)-tropic human immunodeficiency virus type 1 (HIV-1) strains, and CCR5 and CXCR-4 are the principal cofactors utilized for CD4-mediated entry by M-tropic and T-tropic isolates, respectively. Macrophages from individuals homozygous for an inactivating mutation of CCR5 are resistant to prototype M-tropic strains that depend on CCR5 but are permissive for a dual-tropic isolate, 89.6, that can use both CCR5 and CXCR-4, as well as CCR2b, CCR3, and CCR8. Here we show that 89.6 entry into CCR5-deficient macrophages is blocked by an anti-CXCR-4 antibody and by the CXCR-4-specific chemokine SDF but not by the ligands to CCR2b or CCR3. Reverse transcription-PCR demonstrated expression of CXCR-4 but not CCR3 or CCR8 in macrophages, while CCR2b was variable. Macrophage surface expression of CXCR-4 was confirmed by immunofluorescence staining and flow cytometry. Thus, CXCR-4 is expressed by primary macrophages and functions as a cofactor for entry by dual-tropic but not T-tropic HIV-1 isolates, and macrophage resistance to T-tropic strains does not result from a lack of the T-tropic entry cofactor CXCR-4. Since CXCR-4 on macrophages can be used by some but not other isolates, these results indicate that HIV-1 strains differ in how they utilize chemokine receptors as cofactors for entry and that the ability of a chemokine receptor to mediate HIV-1 entry differs, depending on the cell type in which it is expressed.
Macrophage (M)-tropic human immunodeficiency virus type 1 (HIV-1) strains infect primary macrophages and lymphocytes but not CD4+ transformed cell lines, while T-cell line (T)-tropic HIV-1 strains infect lymphocytes and cell lines but not macrophages. The chemokine receptors CCR5 and CXCR-4 are the principal cofactors that enable entry by M-tropic and T-tropic strains, respectively, when introduced along with CD4 into otherwise nonpermissive cells (8, 17, 19, 20, 22). Certain dual-tropic strains that infect macrophages, lymphocytes, and transformed cell lines can utilize both CXCR-4 and CCR5 (19, 46). Target cell tropism is largely determined at the level of virus entry and is encoded mainly by the HIV-1 env gene (4, 34, 43). The reciprocal patterns of cofactor use by M-tropic and T-tropic strains suggest a simple model for the cellular determinants of tropism in which CCR5 would be expressed by macrophages, CXCR-4 would be expressed by transformed cell lines, and both would be expressed by lymphocytes. Whether this is accurate, however, remains to be determined.
Several molecules in addition to CCR5 and CXCR-4 also support entry by more restricted subsets of HIV-1 isolates. These include CCR3, CCR2b, and a growing list of known or putative chemokine receptors, such as CCR8 (also known as chemR1), the cytomegalovirus receptor US28, and others (8, 15, 19, 38, 40). Most have been identified in heterologous transfection-based systems, however, and defining their role in the infection of native target cells is critical for understanding HIV-1 pathogenesis and developing therapeutic agents targeted at cofactor-mediated viral entry.
Recently, a mutant allele of the CCR5 gene (ccr5Δ32) was identified that encodes a truncated protein which is not expressed on the cell surface and cannot support HIV entry (16, 28, 42). Individuals homozygous for ccr5Δ32 are resistant to HIV-1 infection, and lymphocytes and macrophages from these individuals are resistant to infection with M-tropic HIV-1 isolates (14, 16, 28, 39, 42). Although resistance in vivo is incomplete (1), this nevertheless shows that CCR5 is the principal entry cofactor used in primary macrophages and lymphocytes by prototype M-tropic strains and confirms the critical role of CCR5-dependent M-tropic strains in person-to-person HIV-1 transmission. The presence of this naturally occurring CCR5 knockout model also offers the opportunity to examine pathways other than CCR5 that may be utilized by certain viruses for entry into primary target cells.
HIV-1 89.6 is a dual-tropic primary isolate that infects both macrophages and some transformed cell lines (9) and can use both CCR5 and CXCR-4 as cofactors for entry, as well as CCR3, CCR2b, and CCR8 (19, 39, 40). While primary macrophages derived from ccr5Δ32-homozygous individuals are resistant to infection with most M-tropic prototype HIV-1 isolates, we recently found that these cells were permissive for strain 89.6 (39). We have demonstrated 89.6 replication in CCR5-deficient macrophages from each of six ccr5Δ32-homozygous donors tested (data not shown). To address mechanisms of entry into macrophages in addition to CCR5, and to better understand how different HIV-1 strains utilize entry cofactors in primary cells, we sought to identify the pathway used by 89.6 for entry into CCR5-deficient macrophages.
CCR5-independent, CD4-dependent infection of primary macrophages.
To be sure that infection of CCR5-deficient macrophages by strain 89.6 reflected a CD4-mediated entry pathway and not a distinct mechanism independent of CD4, we tested whether infection would be inhibited by blocking CD4 (Fig. 1). The CCR5 genotypes of blood donors were determined by PCR (39), and macrophages were isolated from peripheral blood mononuclear cells by a stringent two-step selective adherence procedure and maintained in culture as previously described (11, 39). After 1 week in culture, the macrophages were infected overnight with equal amounts of cell-free virus stocks based on p24 antigen content, washed, and sampled periodically by enzyme-linked immunosorbent assay (Dupont, Wilmington, Del.) for p24 antigen production. Cells infected with 89.6 were incubated for 1 h prior to and throughout the infection with an anti-CD4 monoclonal antibody (MAb), no. 19 (21), or with a control MAb, B33.1 (10).
FIG. 1.
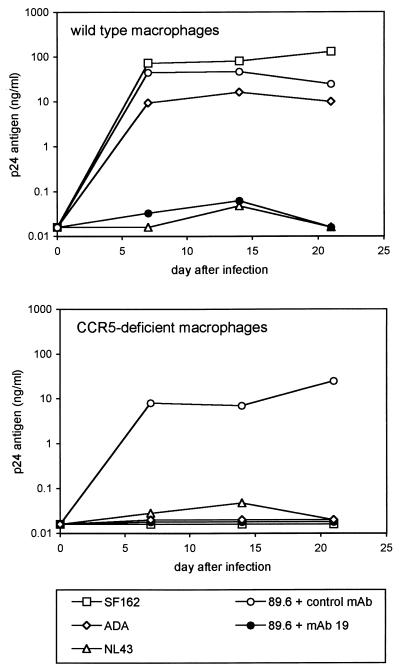
HIV-1 replication in normal and CCR5-deficient macrophages. Monocytes were isolated from individuals homozygous for the wild-type CCR5 allele or ccr5Δ32, plated at 2 × 105 per well in 48-well plastic tissue culture plates, and allowed to differentiate into macrophages in vitro. After 7 days in culture they were infected overnight with 20 ng of p24 antigen of M-tropic (SF162 or ADA), T-tropic (NL43), or dual-tropic (89.6) strains. Cultures were then washed, and the supernatant was sampled periodically for p24 antigen. Cells infected with 89.6 were incubated for 1 h prior to and during infection with the anti-CD4 MAb no. 19 or the control MAb B33.1 (each at 10 μg/ml), which was then maintained in the medium throughout the experiment.
As shown in Fig. 1, 89.6 infection of both normal and CCR5-deficient macrophages was blocked by the anti-CD4 MAb but not by the control MAb. Thus, infection of primary macrophages in the absence of functional CCR5 requires CD4 and does not result from a fundamentally different, CD4-independent entry mechanism. As expected, the prototype M-tropic strain SF162 infected wild-type but not CCR5-deficient macrophages (Fig. 1).
In addition to CCR5 and CXCR-4, 89.6 can also utilize the chemokine receptors CCR3, CCR2b, and CCR8 (also known as chemR1) for entry (19, 25, 40). We showed previously that macrophages lacking functional CCR5 were not permissive for the M-tropic strain YU-2, which can use both CCR3 and CCR5, suggesting that CCR3 was not involved in CCR5-independent entry (8, 39). Here we tested the M-tropic strain ADA, which can use both CCR8 and CCR3 in addition to CCR5 (8, 40). ADA also failed to replicate in macrophages lacking functional CCR5 (Fig. 1). These results suggest that 89.6 infection of CCR5-deficient macrophages is unlikely to result from utilization of either CCR3 or CCR8.
No replication by the T-tropic strain NL43 (Fig. 1) or several other T-tropic isolates (data not shown) was seen in either wild-type or ccr5Δ32-homozygous macrophages. Since T-tropic strains replicate efficiently in lymphocytes even in the absence of CCR5 (14, 28, 39), this confirmed that our macrophage cultures were not significantly contaminated with T cells and that 89.6 replication did not result from infection of contaminating lymphocytes. The lack of NL43 replication in CCR5-deficient macrophages also indicates that the absence of functional CCR5 in macrophages does not lead to enhanced permissiveness for T-tropic HIV-1 strains.
Detection of chemokine receptor-specific RNA in macrophages.
To determine which chemokine receptors used by 89.6 might be involved in entry independent of CCR5, we carried out reverse transcription-PCR (RT-PCR) on RNA from purified monocyte-derived macrophages with primers that detect CCR5, CXCR-4, CCR3, CCR2b, and CCR8 (chemR1), as well as CD4 (Fig. 2). RNA was extracted from 7-day-old macrophage cultures with TRIZOL-LS (GIBCO-BRL, Grand Island, N.Y.) and incubated with RNase-free DNase I (30 min at 37°C in the presence of 5 mM MgCl2) to eliminate any residual genomic DNA, and DNase was inactivated by the addition of EDTA (5 mM) and heating (65°C for 10 min). rTth polymerase (Perkin-Elmer, Emeryville, Calif.) was used for both reverse transcription and PCR amplification as directed by the manufacturer, using specific antisense primers for reverse transcription and 45 cycles of PCR amplification. Sense (S) and antisense (A) primers used were as follows: CCR5-S (5′-CGTCTCTCCCAGGAATCATCTTTAC-3′) and CCR5-A (5′-TTGGTCCAACCTGTTAGAGCTACTG-3′), which yield a 356-bp CCR5 product; CXCR4-S (5′-GAACTTCCTATGCAAGGCAGTCC-3′) and CXCR4-A (5′-CCATGATGTGCTGAAACTGGAAC-3′), which amplify a 304-bp CXCR-4 product; CD4-S (5′-GCAATTGCTAGTGTTCGGATTGA-3′) and CD4-A (5′-GTCAGCTTTTCAACTGTAAAGGCG-3′), which yield a 347-bp CD4 product; CCR2-S (5′-GCGGAATCTTCTTCATCATCCTC-3′) and CCR2-A (5′-CCTCTTCTTCTCGTTTCGACACC-3′), which yield a 338-bp CCR2b product; CCR3-S (5′-AGCTGGAGGCATTTCCACACTC-3′) and CCR3-A (5′-TTCATGCAGCAGTGGGAGTAGG-3′), which yield a 311-bp CCR3 product; and CCR8-S (5′-TCCATGCCGTGTATGCCC-3′) and CCR8-A (5′-CCACGTTGAATGGGACCC-3′), which yield a 363-bp CCR8 product. Specificity of the primers was confirmed with plasmid DNA, DNA extracts of human and nonhuman cells, and RNA extracts of cells expressing specific chemokine receptors (17), and the identity of RT-PCR products was also verified in selected experiments by Southern blotting with specific internal oligonucleotide probes (data not shown).
FIG. 2.
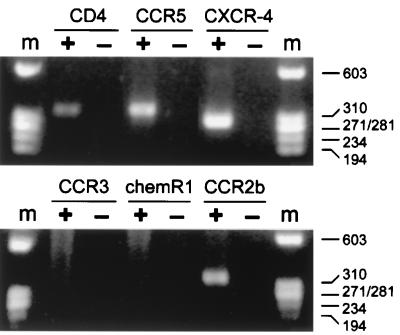
RT-PCR detection of chemokine receptor expression in macrophages. RNA was extracted from 7-day-old macrophage cultures and subjected to reverse transcription and PCR amplification for CCR5, CXCR-4, CCR3, CCR8 (chemR1), and CCR2b, as well as CD4. Products were separated on agarose gels and stained with ethidium bromide. Macrophages from a CCR5 wild-type-homozygous donor are shown, and the same pattern was seen with cells from ccr5Δ32-homozygous donors. Amplification was done following reverse transcription with (+) or without (−) reverse transcription enzyme present. m, molecular size standards.
As expected, RT-PCR signals for CD4 and CCR5 were detected in macrophages (Fig. 2). Surprisingly, we also saw a strong signal for CXCR-4. This was consistent among macrophages from multiple wild-type- and ccr5Δ32-homozygous donors. We confirmed that this signal represented RNA and not cellular DNA contamination, since no band was seen if reverse transcription was omitted prior to PCR (Fig. 2). In addition, the amplified band was verified as CXCR-4 by Southern blotting with a CXCR-4-specific internal oligonucleotide probe (data not shown).
In contrast to CXCR-4, neither CCR3 nor CCR8 (chemR1) yielded an RT-PCR signal in macrophages. Based on serial dilutions of plasmid DNA, the primer pairs can detect 20 cDNA molecules for CCR8 and 2,000 cDNA molecules for CCR3 (data not shown). When amplification for CCR2b was done, no signal was seen in most macrophage cultures (data not shown), but it did give a positive band in approximately one-third of the donors tested (Fig. 2). Based on serial dilutions of plasmid, the CCR2b primers can detect 200 molecules of cDNA (data not shown). This occasional detection of CCR2b suggests either that there is significant donor-to-donor variability in CCR2b expression or that the level of expression under these culture conditions is low and just at the threshold of detection. Thus, CXCR-4 is the only cofactor used by 89.6, other than CCR5, that was uniformly detected in these macrophages by RT-PCR. There were no differences in patterns of cofactor expression detected by RT-PCR when macrophages from homozygous wild-type and ccr5Δ32 donors were compared (data not shown).
CXCR-4 is present on the surface of primary monocyte-derived macrophages.
Expression of CXCR-4 RNA by primary macrophages was somewhat unexpected, since these cells are resistant to infection by CXCR-4-dependent T-tropic HIV-1 isolates. Therefore, to determine whether CXCR-4 protein was present on the cell surface, we carried out immunofluorescence staining with the anti-CXCR-4 MAb 12G5 (21). One-week-old cultured macrophages were detached with EDTA (1 mM for 5 min) and gentle scraping, suspended in staining buffer (SB) (phosphate-buffered saline with 1 mg of bovine serum albumin/ml and 0.02% sodium azide), and incubated for 30 min with 5% rat serum and 5% rabbit serum. They were then stained for 30 min with murine MAbs diluted in SB, washed, and incubated for 30 min with fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G (Biosource, Camarillo, Calif.) diluted 1:200 in SB supplemented with 50% fetal bovine serum. The cells were washed again, fixed with 4% paraformaldehyde, and analyzed by flow cytometry. All incubations were carried out at 4°C. The MAbs used were as follows: 12G5 (6 μg/ml) to detect CXCR-4, no. 19 (10 μg/ml) to detect CD4, OKT3 (diluted 1:10) to stain T lymphocytes, and OKM1 (diluted 1:10) to stain macrophages. A major histocompatibility complex class I MAb was used as a positive control (w6/32; 1:10 dilution of hybridoma supernatant), and an irrelevant MAb directed against HIV-1 gp120 (D47; 10 μg/ml) served as a negative control that was isotype matched with 12G5 and no. 19. OKT3 and OKM1 were obtained from Ortho Diagnostic Systems (Raritan, N.J.); 12G5, no. 19, and w6/32 (21, 35) were provided by J. Hoxie (University of Pennsylvania); and D47 (5) was provided by R. Doms (University of Pennsylvania). In parallel we examined the CD4+ T-cell line SUP-T1, which is highly permissive for CXCR-4-dependent strains. As shown in Fig. 3, staining with OKM1 and OKT3 demonstrated highly purified macrophages that were devoid of contaminating T cells. This level of purity also indicates that our detection of CXCR-4 in the macrophage cultures by RT-PCR was unlikely to be the result of lymphocyte contamination.
FIG. 3.
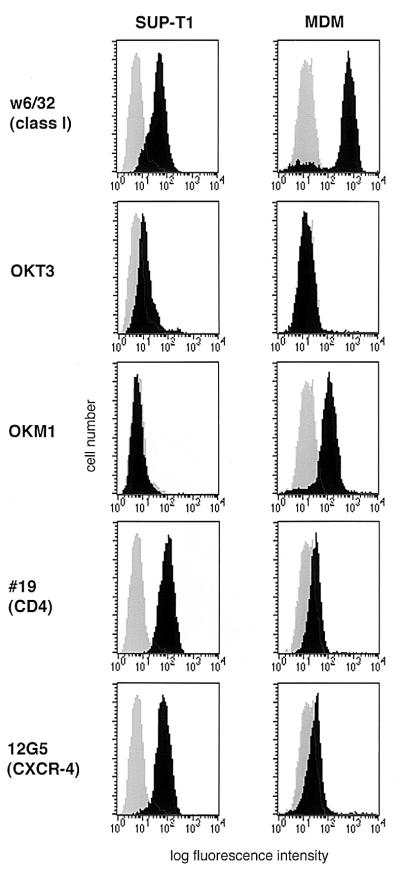
Macrophage surface expression of CXCR-4 by flow cytometry. Seven-day-old cultures of monocyte-derived macrophages (MDM) were detached and stained with MAbs for major histocompatibility complex class I as a positive control (w6/32), a T-cell marker (OKT3), a macrophage marker (OKM1), CD4 (#19), and CXCR-4 (12G5). Cells were then analyzed by flow cytometry with a minimum of 104 cells. Specific MAb profiles are indicated by the black histogram, and the negative control antibody is shown as the shaded histogram in each graph. Macrophages from a CCR5 wild-type-homozygous donor are shown and are representative of cells from six different donors. The same patterns were seen with cells from ccr5Δ32-homozygous donors. The SUP-T1 cell line was stained in parallel.
CXCR-4 staining of macrophages with MAb 12G5 revealed a population with a single peak that was clearly positive compared with the control MAb (Fig. 3). This was confirmed with macrophages from six donors and verified with two isotype-matched negative control MAbs (data not shown). Background fluorescence after staining with the negative control antibody was generally higher in macrophages than in SUP-T1 cells, as described previously (10). Nevertheless, the mean channel fluorescence for 12G5 in macrophages from six donors was consistently twice that of the negative control MAb (mean ± standard error of the mean, 28.3 ± 3.8 for 12G5 compared with 14.4 ± 2.3 for the control MAb). This contrasts with SUP-T1 cells, for which the 12G5 mean channel fluorescence was considerably higher than that of the negative control MAb (118 ± 24 for 12G5 versus 6.99 ± 1.02 for the control). With a stringent cutoff to determine positive staining (a fluorescence level greater than 99% of the cells stained with the isotype-matched negative control MAb), (11.3 ± 2.6)% of macrophages were positive for CXCR-4 (range, 5.3 to 23.3%) compared to (85.3 ± 5.3)% of SUP-T1 cells. Thus, fluorescence-activated cell sorter analysis revealed CXCR-4 on the surface of primary macrophages, although the level of expression was low compared to that of transformed cell lines. The same pattern of staining for CXCR-4, CD4, and other markers was seen with macrophages derived from CCR5 wild-type- and ccr5Δ32-homozygous individuals (data not shown).
Entry into macrophages lacking CCR5 is inhibited by blocking CXCR-4.
Because these studies revealed both CXCR-4 gene expression and protein on the surface of macrophages but no CCR3 or CCR8 and inconsistent CCR2b expression, we next examined whether CCR5-independent entry into macrophages could be inhibited by agents directed against CXCR-4 or other potential cofactors. Entry was determined by PCR detection of viral DNA with primers directed at early reverse transcription products. One-week-old cultures of macrophages were infected as described above with the M-tropic strain SF162, the T-tropic strain NL43, and the dual-tropic isolate 89.6, using DNase-treated (50 U/ml for 30 min at room temperature) cell-free virus stocks. To test for blocking, cells were incubated for 1 h prior to and throughout the infection with the anti-CXCR-4 MAb 12G5, the anti-CD4 MAb no. 19, or the control MAb B33.1 (all at 10 μg/ml) or with specific chemokines (Peprotech, Rocky Hill, N.J.). The chemokines MCP-1, MCP-3, and eotaxin were used at 1 μg/ml and SDF was used at 2.5 μg/ml (2, 24, 33). Two days after infection the cells were lysed and amplified with primers that detect conserved regions of the HIV-1 long terminal repeat (LTR), followed by Southern blotting. The LTR primers, PCR amplification conditions, and Southern blotting protocol have been described previously (19).
In agreement with the replication data shown in Fig. 1, SF162 entered wild-type macrophages but not CCR5-deficient macrophages while 89.6 entered both normal and CCR5-deficient macrophages (Fig. 4). The T-tropic strain NL43 failed to generate viral DNA in either wild-type or CCR5-deficient macrophages (Fig. 4), which indicates that NL43 is blocked in macrophages at an early stage of infection, consistent with entry. This also confirms that the absence of functional CCR5 in macrophages does not result in enhanced entry by T-tropic isolates. Entry by both 89.6 and SF162 was blocked by the anti-CD4 antibody but not by a control antibody (Fig. 4), which is also consistent with the infection experiments and shows that the PCR signals reflect actual infection and not DNA carryover in the virus inoculum. In agreement with this, no band was seen if virus stocks were heat inactivated before infection (data not shown).
FIG. 4.
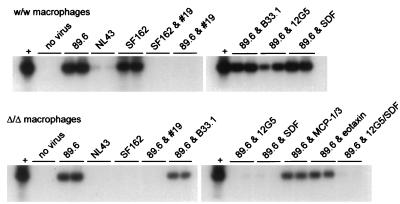
Inhibition of HIV-1 entry into normal and CCR5-deficient macrophages. Macrophages were isolated from individuals homozygous for the wild-type CCR5 allele (w/w) or the ccr5Δ32 allele (Δ/Δ). After 7 days in culture they were infected with 20 ng of p24 antigen of the dual-tropic isolate 89.6, the T-tropic strain NL43, or the M-tropic strain SF162. The indicated wells were incubated for 1 h prior to and throughout the infection with the anti-CD4 MAb no. 19 (#19), the anti-CXCR-4 MAb 12G5, or the control antibody B33.1 or with SDF, MCP-1 and MCP-3, or eotaxin. Three days after infection the cells were lysed and subjected to PCR amplification with primers that detect conserved regions of the HIV-1 LTR, followed by Southern blotting. Infections were done in duplicate and amplified in independent PCR reactions, both of which are shown. Amplification with β-globin primers showed a positive signal in all wells (data not shown). HIV plasmid DNA was used as a positive control (+) for amplification. Data are representative of replicate experiments with cells from three ccr5Δ32-homozygous donors.
We then determined whether CCR5-independent entry into macrophages would be inhibited by agents that target specific chemokine receptors. MAb 12G5 has been shown to block CXCR-4-mediated entry of some HIV-1 and HIV-2 strains, including 89.6 (21, 47). SDF, the chemokine ligand for CXCR-4, also blocks CXCR-4-mediated entry by 89.6 and other isolates (2, 33). We also examined MCP-1 and MCP-3, which are the ligands for CCR2b, and eotaxin, which is the ligand for CCR3 and can block CCR3-mediated infection (24). As shown in Fig. 4, entry of 89.6 into CCR5-deficient macrophages was inhibited by both 12G5 and SDF. Thus, specific targeting of CXCR-4 prevented CCR5-independent infection of macrophages by strain 89.6, and this indicates that CXCR-4 is the cofactor utilized by 89.6 for CD4-mediated entry into these cells. In contrast, no evidence was seen to suggest a role for CCR2 or CCR3, since neither MCP-1 and MCP-3 nor eotaxin blocked infection (Fig. 4). As expected, neither SDF nor 12G5 blocked infection of macrophages expressing wild-type CCR5, indicating that the inhibition seen in ccr5Δ32-homozygous macrophages did not result from a nonspecific effect of the chemokine or antibody on macrophages (Fig. 4).
Significance of CXCR-4-mediated infection of macrophages.
The discovery that distinct entry cofactors are used by M-tropic and T-tropic HIV-1 variants initially suggested that tropism could be easily explained by cell-specific distribution of CXCR-4 on lymphocytes and transformed cell lines but not macrophages and of CCR5 on primary macrophages and lymphocytes but not transformed cell lines. In this report, however, we show that tropism patterns are not explained by this simple paradigm, since the inability of T-tropic strains to enter and infect primary macrophages does not result from an absence of CXCR-4 on these target cells. In addition, because T-tropic strains use CXCR-4 on transformed cells but not on macrophages, our data also indicate that the ability of a cofactor to support HIV-1 entry can vary markedly, depending on the cellular context in which it is expressed. Furthermore, since CXCR-4 expressed on macrophages supports entry of 89.6 but not T-tropic isolates, these results show that HIV-1 strains differ in the way they utilize this cofactor.
The reason(s) that macrophage CXCR-4 supports entry of 89.6 but not T-tropic strains may offer important clues about the function of these molecules or their interaction with other components of the cell surface. One straightforward explanation might relate to levels of expression in different cell types, combined with strain differences in the efficiency of receptor and coreceptor utilization. Macrophages express low levels of CD4 compared to those of other CD4-positive cell types, and our data show that levels of CXCR-4 immunofluorescence are also low compared with those of T-cell lines. Recently, CD4 levels were found to have distinct effects on entry by different HIV-1 isolates, such that T-cell line-adapted T-tropic strains could utilize CXCR-4 in the presence of low CD4 levels while primary T-tropic isolates required higher CD4 levels (27). It is possible that in the presence of low CD4 levels on macrophages, low levels of CXCR-4 can be used by a dual-tropic strain like 89.6 but not by T-tropic strains. A related explanation we initially considered was that a stoichiometric interaction between CD4 and the chemokine receptor might be needed for entry. Since CD4 levels in macrophages are low, the presence of CCR5 might leave insufficient CD4 available to associate with the small amount of CXCR-4 present. However, our finding that T-tropic strains also failed to enter macrophages from ccr5Δ32-homozygous donors argues against this possibility.
Alternatively, CXCR-4 may be expressed in macrophages in a form different from that in lymphocytes and cell lines, resulting in utilization by some but not other isolates. Studies with chimeric chemokine receptors have shown that individual HIV-1 strains interact differently with cofactors and that isolates rely on distinct although somewhat overlapping domains of the cofactors (30, 36, 37, 41). It is possible, therefore, that on the surface of primary macrophages there are molecules associated with CXCR-4 that interfere with its ability to function as a cofactor for T-tropic but not dual-tropic strains or that differences between macrophages and other cells in glycosylation or other posttranslational modifications of CXCR-4 might affect its ability to mediate entry by some but not other strains. Supporting this possibility, the anti-CXCR-4 MAb 12G5 is able to block infection by some HIV-1 strains in a manner that is both strain specific and dependent on the cell in which the cofactor is expressed (31).
While there clearly exists a general association between tropism and cofactor selectivity, these results show that cofactor utilization in heterologous cells does not necessarily predict use in primary cell targets. Others have also identified exceptions where cofactor selectivity does not predict tropism (7, 18). Similarly, although the finding of CXCR-4 expression by primary macrophages contrasts with the host range of CXCR-4-dependent T-tropic HIV-1 strains, it is consistent with several previous reports of CXCR-4 expression in monocytes or macrophages (3, 29, 31) and calcium currents induced by SDF (33). Our study extends those observations to show that CXCR-4 is expressed on macrophages in a form that is functional as a cofactor for entry for some HIV-1 isolates.
In addition to CCR5 and CXCR-4, some HIV-1 isolates can use a growing list of other known or putative chemokine receptors for entry in heterologous systems. A critical question is which of them are involved in infection of primary cells that are relevant in vivo. CCR3 can be used by many M-tropic strains and mediates infection of microglia (24). In agreement with other reports (12), we found no evidence for CCR3 expression or cofactor function in monocyte-derived macrophages. Similarly, strain 89.6 can use CCR8 (40) but we found no evidence for expression or entry cofactor function in these cultures. In contrast, CCR2b is well described in macrophages (13, 44), yet we found variable and inconsistent expression and no evidence that it was involved in CCR5-independent macrophage infection by an isolate that uses CCR2b in transfection-based systems. A likely explanation is that expression levels vary under different culture conditions and in these cultures there were extremely low levels that were just at or below the threshold of detection by RT-PCR and were insufficient to support entry. It is also possible that CCR2b utilization in heterologous systems may not predict entry function in primary target cells, similar to our findings involving T-tropic isolates and CXCR-4 in macrophages.
T-tropic strains were blocked in our macrophage cultures at an early stage of infection, prior to the generation of initial reverse transcription products. This points to a defect in entry, which is similar to results reported by most other laboratories and is supported by direct evidence that env genes derived from M-tropic but not T-tropic strains can mediate fusion with macrophage cell membranes (4, 34). However, HIV-1 host cell tropism is relative, rather than absolute, and low-level replication in macrophages by T-tropic strains may occasionally be seen (11). In addition, some groups have reported fusion and/or entry by T-tropic strains in macrophages despite their failure to replicate (26, 45). Our finding that CXCR-4 is present on macrophages may provide a means of reconciling these results. Since there are clearly steps subsequent to entry at which replication of even CCR5-dependent HIV-1 and simian immunodeficiency virus strains may be restricted in macrophages (6, 23, 32), there may be conditions under which CXCR-4 does mediate entry by T-tropic strains into macrophages, but downstream events then lead to restricted replication.
Acknowledgments
We thank J. Hoxie, R. Smyth, D. Kolson, S. Isaacs, and G. Besson for valuable discussions and critical reading of the manuscript. We also thank the blood donors who generously provided cells; J. Hoxie, R. Doms, R. Horuk, and J. Hesselgesser for MAbs and other reagents; and C. Woods for M-CSF.
This work was supported by grants AI-35502, NS-27405, and HL-58004 from the National Institutes of Health.
REFERENCES
- 1.Biti R, Ffrench R, Young J, Bennetts B, Stewart G. HIV-1 infection in an individual homozygous for the CCR5 deletion allele. Nat Med. 1997;3:252–253. doi: 10.1038/nm0397-252. [DOI] [PubMed] [Google Scholar]
- 2.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 3.Bleul C C, Wu L J, Hoxie J A, Springer T A, Mackay C R. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broder C C, Berger E A. Fusogenic selectivity of the envelope glycoprotein is a major determinant of human immunodeficiency virus type 1 tropism for CD4+ T-cell lines vs primary macrophages. Proc Natl Acad Sci USA. 1995;92:9004–9008. doi: 10.1073/pnas.92.19.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broder C C, Earl P L, Long D, Abedon S T, Moss B, Doms R W. Antigenic implications of human immunodeficiency virus type 1 envelope quaternary structure: oligomer-specific and -sensitive monoclonal antibodies. Proc Natl Acad Sci USA. 1994;91:11699–11703. doi: 10.1073/pnas.91.24.11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chackerian B, Long E M, Luciw P A, Overbaugh J. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J Virol. 1997;71:3932–3939. doi: 10.1128/jvi.71.5.3932-3939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng-Mayer C, Liu R, Landau N R, Stamatatos L. Macrophage tropism of human immunodeficiency virus type 1 and utilization of the CC-CKR5 coreceptor. J Virol. 1997;71:1657–1661. doi: 10.1128/jvi.71.2.1657-1661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choe H, Farzan M, Sun Y, Sullivan N, Rollins R, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 9.Collman R, Balliet J W, Gregory S A, Friedman H, Kolson D L, Nathanson N, Srinivasan A. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J Virol. 1992;66:7517–7521. doi: 10.1128/jvi.66.12.7517-7521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collman R, Godfrey B, Cutilli J, Rhodes A, Hassan N F, Sweet R, Douglas S D, Friedman H, Nathanson N, Gonzalez-Scarano F. Macrophage-tropic strains of human immunodeficiency virus type 1 utilize the CD4 receptor. J Virol. 1990;64:4468–4476. doi: 10.1128/jvi.64.9.4468-4476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collman R, Hassan N F, Walker R, Godfrey B, Cutilli J, Hastings J C, Friedman H, Douglas S D, Nathanson N. Infection of monocyte-derived macrophages with human immunodeficiency virus type 1 (HIV-1). Monocyte-tropic and lymphocyte-tropic strains of HIV-1 show distinctive patterns of replication in a panel of cell types. J Exp Med. 1989;170:1149–1163. doi: 10.1084/jem.170.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Combadiere C, Ahuja S K, Murphy P M. Cloning and functional expression of a human eosinophil CC chemokine receptor. J Biol Chem. 1995;270:16491–16494. doi: 10.1074/jbc.270.28.16491. [DOI] [PubMed] [Google Scholar]
- 13.Combadiere C, Ahuja S K, Van Damme J, Tiffany H L, Gao J L, Murphy P M. Monocyte chemoattractant protein-3 is a functional ligand for CC chemokine receptors 1 and 2B. J Biol Chem. 1995;270:29671–29675. doi: 10.1074/jbc.270.50.29671. [DOI] [PubMed] [Google Scholar]
- 14.Connor R I, Paxton W A, Sheridan K E, Koup R A. Macrophages and CD4+ T lymphocytes from two multiply exposed, uninfected individuals resist infection with primary non-syncytium-inducing isolates of human immunodeficiency virus type 1. J Virol. 1996;70:8758–8764. doi: 10.1128/jvi.70.12.8758-8764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O’Brien S. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CCR5 structural gene. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 17.Deng H, Liu R, Ellmeir W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 18.Dittmar M T, McKnight A, Simmons G, Clapham P R, Weiss R A. HIV-1 tropism and co-receptor use. Nature. 1997;385:495–496. doi: 10.1038/385495a0. [DOI] [PubMed] [Google Scholar]
- 19.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3 and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 20.Dragic T, Litwin V, Allaway G, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 21.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 22.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–876. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 23.Fouchier R A M, Brouwer M, Kootstra N A, Huisman H G, Schuitemaker H. HIV-1 macrophage tropism is determined at multiple levels of the viral replication cycle. J Clin Invest. 1994;94:1806–1814. doi: 10.1172/JCI117529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He J L, Chen Y Z, Farzan M, Choe H Y, Ohagen A, Gartner S, Busciglio J, Yang X Y, Hofmann W, Newman W, Mackay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 25.Horuk, R., J. Hesselgesser, Y. Zhou, D. Faulds, D. Taub, J. Rucker, B. J. Doranz, and R. W. Doms. The CC chemokine I309 is a functional ligand for ChemR1/CCR8 and inhibits ChemR1/CCR8 dependent infection by diverse HIV-1 strains. J. Biol. Chem., in press. [DOI] [PubMed]
- 26.Huang Z-B, Potash M J, Simm M, Shahabuddin M, Chao W, Gendelman H E, Eden E, Volsky D J. Infection of macrophages with lymphotropic human immunodeficiency virus type 1 can be arrested after viral DNA synthesis. J Virol. 1993;67:6893–6896. doi: 10.1128/jvi.67.11.6893-6896.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozak S L, Platt E J, Madani N, Ferro F E, Jr, Peden K, Kabat D. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 29.Loetscher M, Geiser T, O’Reilly T, Zwahlen R, Baggiolini M, Moser B. Cloning of a human seven-transmembrane domain receptor, LESTR, that is highly expressed in leukocytes. J Biol Chem. 1994;269:232–237. [PubMed] [Google Scholar]
- 30.Lu Z H, Berson J F, Chen Y H, Turner J D, Zhang T Y, Sharron M, Jenks M H, Wang Z X, Kim J, Rucker J, Hoxie J A, Peiper S C, Doms R W. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc Natl Acad Sci USA. 1997;94:6426–6431. doi: 10.1073/pnas.94.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKnight Á, Wilkinson D, Simmons G, Talbot S, Picard L, Ahuja M, Marsh M, Hoxie J A, Clapham P R. Inhibition of human immunodeficiency virus fusion by a monoclonal antibody to a coreceptor (CXCR4) is both cell type and virus strain dependent. J Virol. 1997;71:1692–1696. doi: 10.1128/jvi.71.2.1692-1696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mori K, Ringler D J, Desrosiers R C. Restricted replication of simian immunodeficiency virus strain 239 in macrophages is determined by env but is not due to restricted entry. J Virol. 1993;67:2807–2814. doi: 10.1128/jvi.67.5.2807-2814.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J L, Arenzana-Seisdedos F, Schwartz O, Heard J M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 34.O’Brien W A, Koyanagi Y, Namazie A, Zhao J-Q, Diagne A, Idler K, Zack J A, Chen I S Y. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 35.Parham P, Barnstable C J, Bodmer W F. Use of a monoclonal antibody (W6/32) in structural studies of HLA-A,B,C antigens. J Immunol. 1979;123:342–349. [PubMed] [Google Scholar]
- 36.Picard L, Simmons G, Power C A, Meyer A, Weiss R A, Clapham P R. Multiple extracellular domains of CCR-5 contribute to human immunodeficiency virus type 1 entry and fusion. J Virol. 1997;71:5003–5011. doi: 10.1128/jvi.71.7.5003-5011.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Picard L, Wilkinson D A, McKnight A, Gray P W, Hoxie J A, Clapham P R, Weiss R A. Role of the amino-terminal extracellular domain of CXCR-4 in human immunodeficiency virus type 1 entry. Virology. 1997;231:105–111. doi: 10.1006/viro.1997.8506. [DOI] [PubMed] [Google Scholar]
- 38.Pleskoff O, Tréboute C, Brelot A, Heveker N, Seman M, Alizon M. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science. 1997;276:1874–1878. doi: 10.1126/science.276.5320.1874. [DOI] [PubMed] [Google Scholar]
- 39.Rana S, Besson G, Cook D G, Rucker J, Smyth R J, Yi Y, Turner J D, Guo H-H, Du J-G, Peiper S C, Lavi E, Samson M, Libert F, Liesnard C, Vassart G, Doms R W, Parmentier M, Collman R G. Role of CCR5 in infection of primary macrophages and lymphocytes by macrophage-tropic strains of human immunodeficiency virus: resistance to patient-derived and prototype isolates resulting from the Δccr5 mutation. J Virol. 1997;71:3219–3227. doi: 10.1128/jvi.71.4.3219-3227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Margulies B, Collman R G, Doranz B J, Parmentier M, Doms R W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rucker J, Samson M, Doranz B J, Libert F, Berson J F, Yi Y, Smyth R J, Collman R G, Broder C C, Vassart G, Doms R W, Parmentier M. Regions in the β-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 42.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cogniaux J, Forceille C, Muyldermans G, Verhofstede C, Bortonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection of Caucasian individuals bearing mutant alleles of the CCR5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 43.Shioda T, Levy J A, Cheng-Mayer C. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 44.Sica A, Saccani A, Borsatti A, Power C A, Wells T N, Luini W, Sozzani S, Mantovani A. Bacterial lipopolysaccharide rapidly inhibits expression of C-C chemokine receptors in human monocytes. J Exp Med. 1997;185:969–974. doi: 10.1084/jem.185.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simmons G, McKnight A, Takeuchi Y, Hoshino H, Clapham P R. Cell-to-cell fusion, but not virus entry in macrophages by T-cell line tropic HIV-1 strains: a V3 loop-determined restriction. Virology. 1995;209:696–700. doi: 10.1006/viro.1995.1307. [DOI] [PubMed] [Google Scholar]
- 46.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strizki J M, Turner J D, Collman R G, Hoxie J, González-Scarano F. A monoclonal antibody (12G5) directed against CXCR-4 inhibits infection with the dual-tropic human immunodeficiency virus type 1 isolate HIV-189.6 but not the T-tropic isolate HIV-1HxB. J Virol. 1997;71:5678–5683. doi: 10.1128/jvi.71.7.5678-5683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]