Figure 1.
PARP13 expression affects expression of many constitutive transcripts and is required for transcriptomic upregulation of innate immune response
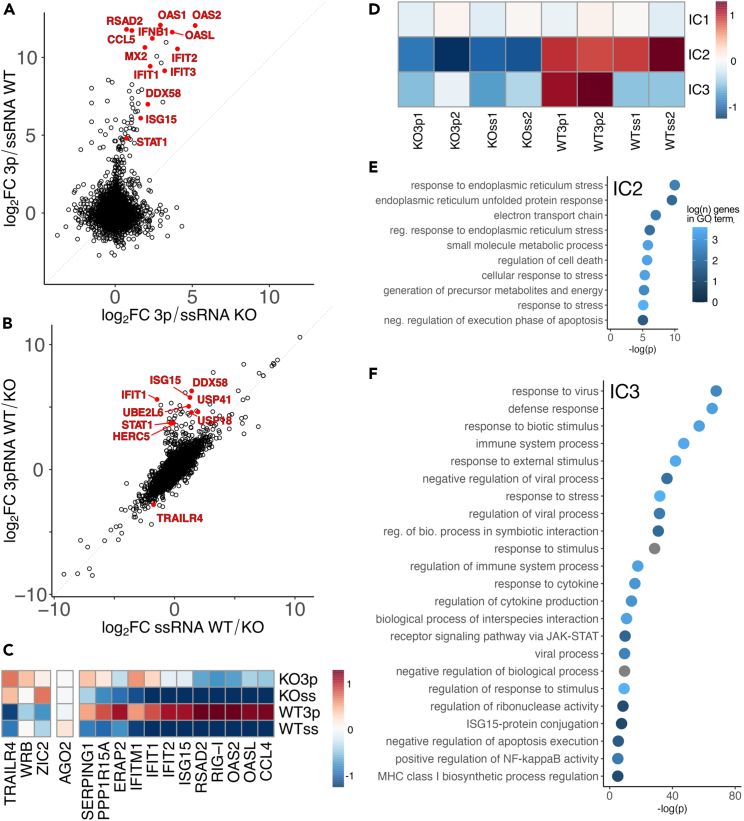
(A) Log2 fold change expression data 24 h after treatment with +3p-RNA vs. +ssRNA in HEK293T WT and PARP13 KO cells (n = 2). A selection of innate immune response genes is highlighted in red.
(B) Log2 fold change expression data from HEK293T WT vs. PARP13 KO cell 24 h after treatment with +3p-RNA or +ssRNA (n = 2).
(C) Scaled average expression relative to WT + ssRNA measured by qPCR in WT HEK293T and PARP13 KO cells and normalized to GAPDH or TBP expression (n ≥ 3). Complete replicate information Figure S1B.
(D) IC pattern weights across RNA-seq samples. The color scale represents values of the linear mixing matrix centered at 0 where columns of S contain the independent components, A is a linear mixing matrix, and data matrix X is considered to be a linear combination of non-Gaussian components such that X = SA.
(E) Top GO terms that are enriched among genes that contribute to IC2, shown in (D).
(F) Top GO terms that are enriched among genes that contribute to IC3, shown in (D).