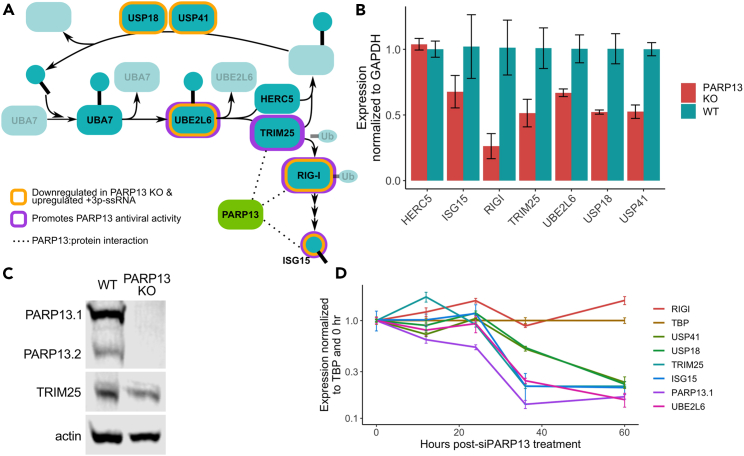
Figure 4.
Genes associated with ISGylation are dysregulated in PARP13 KO cells
(A) A schematic of the ISGylation pathway that includes ISG15, a ubiquitin-like protein; UBA7, the E1; UBE2L6, the E2; HERC5 and TRIM25, E3 ligases; and USP18 and USP41, which remove ISG15 from conjugated proteins. RIG-I, which is ubiquitinated by TRIM25, activates expression of ISG15. Genes that have been shown to promote PARP13 antiviral activity are highlighted in purple. Genes identified by the RNA-seq data to be down-regulated in PARP13 KO cells in control condition and upregulated upon mock viral infection in WT cells are highlighted in gold. Proteins shown to bind PARP13 are connected by dotted lines.
(B) qPCR of ISGylation genes in untreated WT and PARP13 KO HEK293T cells (n = 3). UBA7 expression was too low and beyond the limit of detection. Error bars are +/− 1sd.
(C) Western blot of PARP13 and TRIM25 expression in WT and PARP13 KO HEK293T cell lysates, with beta-actin as a loading control. Also Figure S2B.
(D) PARP13 KD time course in WT HEK293T cells assessing the expression of ISGylation genes shown in (B) to have reduced expression in PARP13 KO cells. Error bars are +/− 1sd.