Summary
We designed a massively parallel reporter assay (MPRA) in an Epstein-Barr virus transformed B cell line to directly characterize the potential for histone post-translational modifications, i.e., histone quantitative trait loci (hQTLs), expression QTLs (eQTLs), and variants on systemic lupus erythematosus (SLE) and autoimmune (AI) disease risk haplotypes to modulate regulatory activity in an allele-dependent manner. Our study demonstrates that hQTLs, as a group, are more likely to modulate regulatory activity in an MPRA compared with other variant classes tested, including a set of eQTLs previously shown to interact with hQTLs and tested AI risk variants. In addition, we nominate 17 variants (including 11 previously unreported) as putative causal variants for SLE and another 14 for various other AI diseases, prioritizing these variants for future functional studies in primary and immortalized B cells. Thus, we uncover important insights into the mechanistic relationships among genotype, epigenetics, and gene expression in SLE and AI disease phenotypes.
Keywords: SLE, lupus, histone quantitative trait loci, quantitative trait loci, QTL, massively parallel reporter assay, MPRA, risk variants, functional mechanisms
A massively parallel reporter assay (MPRA) characterizes the potential for histone post-translational modifications (i.e., histone quantitative trait loci [hQTLs]) to modulate regulatory activity and uncovers important insights into the mechanistic relationships among genotype, epigenetics, and gene expression in SLE and autoimmune disease phenotypes.
Introduction
Genetic variations in the regulatory non-coding genome are a significant contributor to autoimmune (AI [MIM: 109100]) disease susceptibility and progression.1 Approximately 90% of AI disease-associated variants are non-coding, with ∼60% mapping to immune cell enhancers and other types of cis-regulatory elements (cREs; e.g., enhancers, promoters, and CTCF-occupied elements [silencers and insulators] that modulate the cell type and context-specific expression of nearby and/or distant genes).2,3 The activity of a cRE is, in turn, influenced by complex interactions between histone modifications (e.g., acetylation, methylation, phosphorylation) that influence chromatin structure, transcription factor (TF) accessibility, genetic variation, and the TFs that co-localize at the cRE leading to disrupted immune homeostasis.4
Our laboratory previously integrated epigenetic and genotypic data from systemic lupus erythematosus (SLE [MIM: 152700]) patient-derived Epstein-Barr virus (EBV)-transformed B cells to assess the degree to which genetic variants in non-coding regions of the genome influence H3K4me1 and H3K27ac histone modifications, i.e., histone quantitative trait loci (hQTLs).5 H3K4me1 and H3K27ac hQTLs were found to be enriched on AI disease risk haplotypes. hQTLs also disproportionately influenced gene expression variability compared with non-hQTL variants; however, the direct regulatory potential of hQTLs remained unclear.
In this study, we designed a massively parallel reporter assay (MPRA) to systematically and directly explore the regulatory potential attributed to these hQTL variants.5 MPRA leverages a vector containing a reporter gene (typically green fluorescent protein [GFP]), a promoter, and thousands of barcoded DNA sequences that carry selected variants. A change in the reporter gene expression is indicative of regulatory activity of the sequence, as well as the functional allelic effects of variants carried on the sequence.6,7,8 Since some of the identified hQTLs were shown to modulate the effect of expression QTLs (eQTLs) positioned within the same chromatin looping network in EBV B cells, our MPRA design also evaluated the regulatory potential of the eQTLs shown to interact with hQTLs. Last, a subset of SLE and AI disease index SNPs were also evaluated. Determining how non-coding genetic variants, especially hQTLs and eQTLs, alter the activity of regulatory elements and influence gene expression is vital to understanding how such intricate regulatory mechanisms contribute to complex traits and human disease.
Material and methods
Study population
All experiments were approved by the Institutional Review Board at the Oklahoma Medical Research Foundation (OMRF) and proper informed consent was obtained prior to study initiation. The EBV-transformed B cell line used in the MPRA was generated from a non-Hispanic, White, 55-year-old female with SLE enrolled in the Lupus Family Registry and Repository9 and provided by OMRF’s Arthritis and Clinical Immunology Biorepository Core (https://aci-cores.omrf.org/biorepository/). Race and ethnicity were self-reported using a form with fixed categories but confirmed by genetic similarity (via principal-components analysis) to other self-reported non-Hispanic White individuals.
Massively parallel reporter assay
Variant selection and oligo generation
A 67,035-oligo library (32,481 variants) (Agilent Technologies) was designed to test hQTLs previously identified in 25 lymphoblastoid cell lines (LCLs) from European-American individuals with SLE,5 proxies (r2 ≥ 0.8) of selected hQTLs, published SLE or AI disease index SNPs (p ≤ 5E−08), proxies of SLE and AI disease index SNPs, marginal eQTLs identified in 358 CEU, FIN, GBR, and TSI gEUVADIS10 LCLs and eQTLs found previously to interact with hQTLs,5 proxies of interacting eQTLs, and additional SNPs located on AI haplotypes that included a hQTL. Original hQTL discovery utilized the combined haplotype test,11 and was based upon allele-specific ChIP-seq read mapping in heterozygous individuals.5 Original discovery of eQTLs found to interact with hQTLs5 was performed with the Matrix eQTL software package with expression quantifications standardized to the normal distribution. Testable eQTL interactions were required to have variants with minor allele frequency (MAF) ≥0.05 and transcripts with coefficient of variation ≥0.15 across gEUVADIS RNA-seq samples. Variant proxies were evaluated to test whether variants that modulate histones or expression are more likely causal than variants with similar genetic profiles not known to modulate these phenotypes. Location controls and random controls were also included as potential negative controls. Location controls met the following criteria as previously described6: (1) MAF ≥5%; (2) location within 150-1,000 bp of an hQTL SNP; (3) low LD with hQTL SNP (r2 ≤ 0.25); and (4) no significant eQTL signal in public eQTL databases. LD and MAF information were determined using the 1000 Genomes Project CEU, FIN, GBR, and TSI reference genomes.12,13 Random control SNPs were selected randomly across the genome and their MAF was matched to the overall MAF distribution of the other non-control SNPs. Oligos were generated using 150 base pair (bp) of hg19 genomic sequence flanking the reference and alternate alleles of each selected variant (74 bp 5′ and 75 bp 3′ of the allele of interest) with 15 bp adapters added to each end (5′ ACTGGCCGCTTGACG [150bp oligo] CACTGCGGCTCCTGC 3′) (Table S1).
Plasmid library construction
MPRA was performed as described previously6,14 with minor modifications. First, oligo-barcode libraries were constructed by 28X parallel PCR reactions to add 20-bp random barcodes to the synthesized 180-bp oligos. MpraΔorf libraries were assembled using Gibson Assemble Master Mix (NEB E2611L). The GFP amplicon containing a minimal promoter, GFP open reading frame, and partial 3′UTR was amplified from the pGL4.23:minP GFP plasmid and inserted into purified mpraΔorf plasmids by Gibson Assembly. Constructed mpra:gfp libraries were transformed into NEB 10-β E. coli (NEBC3020K) by electroporation and expanded in 5 L LB media supplemented with 100 μg/mL carbenicillin at 37°C, shaking for 16 h. The mpra:gfp plasmid libraries were purified using the Qiagen Plasmid Plus Giga Kit.
MPRA library transfections
EBV B cells were cultured in RPMI medium supplemented with 15% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine at 37°C, 5% CO2. Cells were seeded at 5 × 105 cells/mL 36 h before transfection. Cells were collected and split into six transfections with 100 million cells and 100 μg mpra:gfp plasmid library per replicate. Transfection was performed with the Neon transfection system in 100-μL tips containing 10 million cells per tip with three pulses of 1200 V and 20 ms each. After transfection, cells were cultured with RPMI supplemented with 15% FBS and without antibiotics for 24 h. Cells were then collected and lysed in RLT buffer (Qiagen Midi RNeasy 75144) by passing through 18-gauge needles. Cell lysate was stored in −80°C until RNA purification.
MPRA library complexity validation
The fragment containing the oligo-barcode combination was amplified from mpraΔorf plasmids and attached to Illumina sequencing adapters with the Illumina TruSeq Universal Adapter and unique P7 index primers. Libraries were sequenced using 2 × 150 PE reads on the Illumina NovaSeq platform.
Sequencing library preparations
Total cell lysis was thawed on ice and lysed again by passing 5–10 times through 18-gauge needles. GFP mRNA extraction, pull-down, and cDNA synthesis was performed as previously described.6,14 Plasmid libraries and cDNA samples were amplified, and Illumina sequencing adaptors were added using the Illumina TruSeq Universal Adapter and TruSeq_Index primer (NEB E7335S). Libraries were sequenced using the Illumina NovaSeq targeting 400 million reads per sample.
Promoter capture HiC
Leukoreduction chambers were obtained from the Oklahoma Blood Institute. Primary B cells were isolated with negative magnetic bead selection (StemCell 19054). Cells were seeded at 3 × 106 cells/mL in RPMI medium supplemented with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine. After 1 h incubation at 37°C, 5% CO2, cells were treated with 5 μg/mL R837 (TLR7 agonist), 1 μg/mL CD40 ligand, and 3 μg/mL IgG/M for 48 h to induce SLE-like inflammation responses. HiC libraries were generated with a pool of 5 million B cells from 10 donors following the Hi-C 3.0 protocol.15 Capture enrichment was performed using the Arima Human Promoter Panel kit (Arima A510008 and A302010) following the manufacturer’s instructions. Libraries were sequenced using the Illumina NovaSeq PE150 targeting 200 million reads per sample. Data were processed following the Arima Genomics pipeline (https://github.com/ArimaGenomics/CHiC) and interactions were viewed on the WashU Epigenome Browser (http://epigenomegateway.wustl.edu/browser/).
Data analysis
Oligo-barcode associations and barcode counting
Paired oligo-barcode associations were determined in the four sequenced mpraΔorf plasmid control libraries using analysis scripts (e.g., MPRAmatch.wdl) and the pipeline designed by Dr. Ryan Tewhey (https://github.com/tewhey-lab/MPRA_oligo_barcode_pipeline).6 Pairs with alignment score error rates of greater than 5% were discarded. Only barcodes that uniquely mapped to one oligo were used for downstream analysis. Oligo-barcode pairs from the four libraries were then merged together for a total of 66,949 (99.87%) oligos captured and 145 M total oligo-barcode pairs in the initial oligo-barcode pools. Barcode read counting for each oligo-barcode pair was determined in each of the four plasmid control replicates and six transfected EBV B cell line replicates using the MPRAcount.wdl script. Reads were totaled across all barcodes associated with each oligo.
Expression modulating sequence and allele-specific expression modulating variant identification
Expression modulating sequence (emSeq) and allele-specific expression modulating variant (emVar) discovery utilized Dr Ryan Tewhey’s MPRA count analysis pipeline (https://github.com/tewhey-lab/MPRAmodel). Oligo counts were normalized by DESeq216 and modeled as a negative binomial distribution to obtain estimates of variance in oligo counts across all samples. EmSeqs—defined as sequences with regulatory activity that modulate GFP reporter gene expression—were determined for the plasmid controls and library replicates and tested for significant expression differences in GFP reporter gene expression using a Wald’s test.6 A fold change (FC) difference of 1.5 between the plasmid controls and EBV library replicates and a false discovery rate (FDR) <0.05 were required for significance. EmSeqs were then assessed for allele-specific transactivation potential (emVars) by comparing log2 ratios of the reference vs. alternate alleles using a Student’s t test.6 An FC = 1.25 between the two alleles and an FDR <0.05 were required for significance. For the 320 multiallelic variants, the reference allele was compared with each alternate allele separately.
TF motif enrichment analysis
The findMotifs.pl program within HOMER17 was utilized to evaluate all emSeqs and hQTL emSeqs for known TF motif enrichment. For the emSeq analysis, FASTA sequences of identified emSeq oligos were used as target sequences and FASTA sequences for all remaining oligos were used as background sequences; for the hQTL emSeq analysis, target FASTA sequences of hQTL emSeq oligos were compared with FASTA sequences of non-hQTL emSeq oligos as the background.
Annotations
Prior to downstream analysis, variant positions were converted to hg38 using the UCSC LiftOver tool (https://genome.ucsc.edu/cgi-bin/hgLiftOver). Human ENCODE CRE information (encodeCcreCombined.bb)3 was downloaded from the UCSC genome browser (http://hgdownload.soe.ucsc.edu), converted to a bed file using the bigBedToBed tool, and evaluated for overlaps with MPRA variant locations for annotation. SLE and AI index SNP identification was initially obtained from the NHGRI-EBI GWAS catalog18 in 2018; annotations were later updated using the NHGRI-EBI GWAS catalog v1.0.2 download on March 27, 2023. Published index SNPs (p < 5E−8) from the following AI diseases were identified: ankylosing spondylitis (AS [MIM: 106300]), autoimmune thyroid disorders (AITD [MIM: 608173]), which include Graves’ disease (GRD [MIM: 275000]) or Hashimoto’s thyroiditis (HT [MIM: 140300]), celiac disease [MIM: 212750], inflammatory bowel disease (IBD [MIM: 266600], which includes Crohn disease [CD] and ulcerative colitis [UC]), Kawasaki disease (KD [MIM: 611775]), multiple sclerosis (MS [MIM: 126200]), myasthenia gravis (MG [MIM: 254200]), myositis [MIM: 160750], primary biliary cirrhosis (PBC [MIM: 109720]), psoriasis [MIM: 177900], rheumatoid arthritis (RA [MIM: 180300]), sarcoidosis [MIM: 181000], Sjögren’s disease (SjD [MIM: 270150]), systemic sclerosis (scleroderma, SS [MIM: 181750]), type 1 diabetes mellitus (T1D [MIM: 222100]), and vitiligo [MIM: 606579]. Mapped and nearest gene annotations were obtained from Ensembl’s variant effect predictor tool (https://useast.ensembl.org/Homo_sapiens/Tools/VEP; release 109, February 2023).19
Results
hQTLs demonstrate strong regulatory activity and are enriched for interferon regulatory factor TFs
A total of 66,865 (99.87%) oligos were recovered from the plasmid control and EBV B cell line libraries with sufficient oligos having >10 barcodes per oligo (98.9% and 91.3% in the plasmid controls and EBV B cell replicates, respectively) and >20 mean reads count per oligo (98% and 96.2% in the plasmid controls and EBV B cell replicates, respectively) (Figures S1A–S1D). Experimental replicates of each library produced strong reproducibility of the normalized read counts, and the EBV B cell samples produced more normalized reads per oligo, on average, than the plasmid controls (Figures S1E and S1F). Oligos with <20 associated barcodes (n = 1,475) and <20 average reads in the plasmid controls (n = 569) were removed from the downstream analysis, resulting in a total of 64,821 oligos with an average coverage of 1,271 barcodes/oligo and 31,665 total variants for analysis.
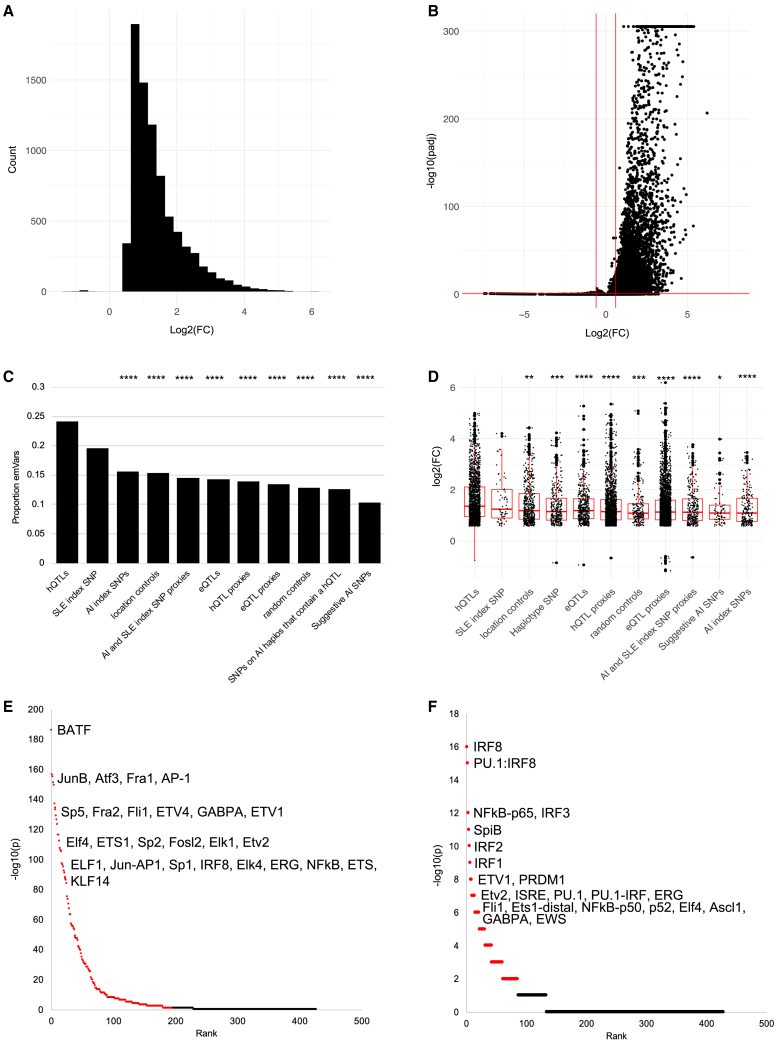
Our post-QC dataset included 4,039 hQTL variants; 6,810 hQTL proxies (r2 > 0.8); 161 SLE index SNPs (p ≤ 5E−8); 802 AI index SNPs (p ≤ 5E−8); 1,198 SLE and AI index SNP proxies; 2,001 eQTLs determined to interact with hQTLs; 11,793 interacting eQTL proxies; 1,526 proxies of a hQTL located on an AI haplotype (“haplotype SNPs”); 1,968 location controls; 975 random controls; and 425 additional AI index SNPs with p ≤ 1E−6 (“suggestive AI SNPs”). A total of eight and 15 hQTLs were also SLE and AI index SNPs, respectively. We found that 7,911 oligos (emSeqs) collectively carrying 4,780 (15.1%) tested variants demonstrated regulatory activity of the GFP reporter gene (Figure 1A; Table S2). The vast majority of emSeqs (n = 7,898; 99.8%) exhibited higher regulatory activity in the EBV B libraries compared with controls; this was not unexpected, however, since we used a construct with a low basal activity promoter making it easier to detect inducible effects6 (Figure 1B). When evaluating the different types of variants, hQTLs produced the highest proportion of emSeqs (24%, n = 976), followed by published SLE index SNPs (20%, n = 32) (Figure 1C). While there was no significant difference in emSeq proportions between the hQTLs and SLE index SNPs, the proportion of hQTL emSeqs was significantly higher than every other variant type tested including AI disease index SNPs (15%, n = 25, χ2 = 28.02, p < 0.00001) and eQTLs (14%, n = 286, χ2 = 78.89, p < 0.00001). hQTLs also displayed the strongest effects on regulatory activity compared with other tested variant types, with mean and median EBV B/control FC = 3.06 (log2(FC) = 1.61) and 2.60 (log2(FC) = 1.36), respectively, demonstrating that many sequences harboring hQTLs are capable of inducing significantly stronger regulatory activity than sequences harboring eQTLs (mean and median FC = 2.60 and 2.27, respectively, tmean = 5.43, p < 0.0001) or other variants in strong LD with them (Figure 1D).
Figure 1.
Properties of emSeqs in the MPRA
(A) Histogram distribution of emSeq regulatory activity (log2(FC)) in six EBV B cell replicates compared with four plasmid controls. Positive values represent increased regulatory activity and negative values represent decreased activity in EBV B cells relative to plasmid controls. Oligo count is plotted on the y axis.
(B) Volcano plot of emSeq effect sizes (−log10(padj) from DESeq2) in EBV B cells relative to controls. Horizontal red line represents padj ≤ 0.05; vertical red lines (log2(FC) ± 0.58) represent a 1.5 FC difference between the EBV B replicates and plasmid controls.
(C) Proportion of emSeqs within each variant type. Significant differences between the proportion of emSeqs within hQTLs and the other variant types are shown: ∗∗∗∗chi-square p < 0.0001.
(D) Boxplots of emSeq effect sizes (log2(FC)) for each variant type. The x axis is sorted in descending order by mean log2(FC) of the variant types. Significant differences in the means of hQTL effect sizes compared with the other variant types are shown: ∗t test p < 0.05; ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p <0.0001.
(E and F) Significant TFs enriched in all emSeqs (E) and hQTL emSeqs (F). TF rank and HOMER −log10(p) are plotted. FDR≤0.05 effects are highlighted in red. Top TFs are indicated.
A significantly higher proportion of emSeqs were located in ENCODE cREs than non-emSeqs (84% vs. 80%; χ2 = 64.9; p < 0.00001) (Figure S2A). EmSeqs were most commonly found within ENCODE’s enhancer-like signatures (ELS) characterized by high DNase and H3K27ac activity and low H3K4m3 activity,3 as well as more frequently located >2kB of an annotated TSS (distal ELS, dELS, 61.3%) than within 2kB of an annotated TSS (promoter ELS, pELS, 13%). We also found that 91% of hQTL emSeqs were located in a cRE (70% in a dELS), which was not unexpected since they were originally identified as H3K27ac and H3K4me1 hQTLs5 and is consistent with their suggestive role in modulating regulatory activity (Figures S2B and S2C). While a high proportion of SLE index emSeqs were also found in cREs (90%, n = 57), only 50% were located in dELS, and had the highest proportion of variants located in DNase-H3K4me3 (6%) and CTCF-only (6%) cREs compared with the other variants tested, suggesting that a proportion of SLE index SNPs likely exhibit different regulatory functions than hQTLs or the other tested variants typed (Figures S2D–S2G).
When emSeqs were tested for enrichment of TF motifs compared with the 57,358 sequences that did not exhibit regulatory activity, an enrichment of 196 TFs was observed with most being members of the C2H2-zinc finger (ZF; n = 32), basic leucine zipper (bZIP; n = 27), basic-helix-loop-helix (bHLH; n = 25), or homeobox (n = 25) and erythroblast transformation specific (ETS; n = 25) families. The most strongly enriched TFs among emSeqs, however, belonged to the bZIP and ETS families (Figure 1E; Table S3). The strongest bZIP family TFs included BATF [MIM: 612476] (p = 1E−186; 11.25% of emSeqs included the motif vs. 3.16% in non-emSeqs), JUNB [MIM: 165161] (p = 1E−156; 9.28% vs. 2.55%), ATF3 [MIM: 603148] (p = 1E−155; 10.52% vs. 3.23%), and others. Strong ETS family TFs included FLIi1 [MIM: 193067] (p = 1E−133; 13.85% in emSeqs vs. 5.68% in non-emSeqs), ETV4 [MIM: 600711] (p = 1E−129; 13.24% vs. 5.37%), GABPA [MIM: 600609] (p = 1E−126; 11.57% vs. 4.37%), and others (Figure 1E; Table S3). When we narrowed the analysis to specifically identify TFs enriched within hQTL emSeqs compared with non-hQTL emSeqs, we observed enrichment of 87 TFs, with strongest enrichment among TF members of the interferon regulatory factor (IRF) family (e.g., IRF8 [MIM: 601565], IRF3 [MIM: 603734], IRF2 [MIM: 147576], IRF1 MIM: 147575], etc.), indicating that these hQTLs are potentially important in the regulation of interferon signaling responses in B cells (Figure 1F; Table S4).
Strong EmVars are dominated by hQTLs
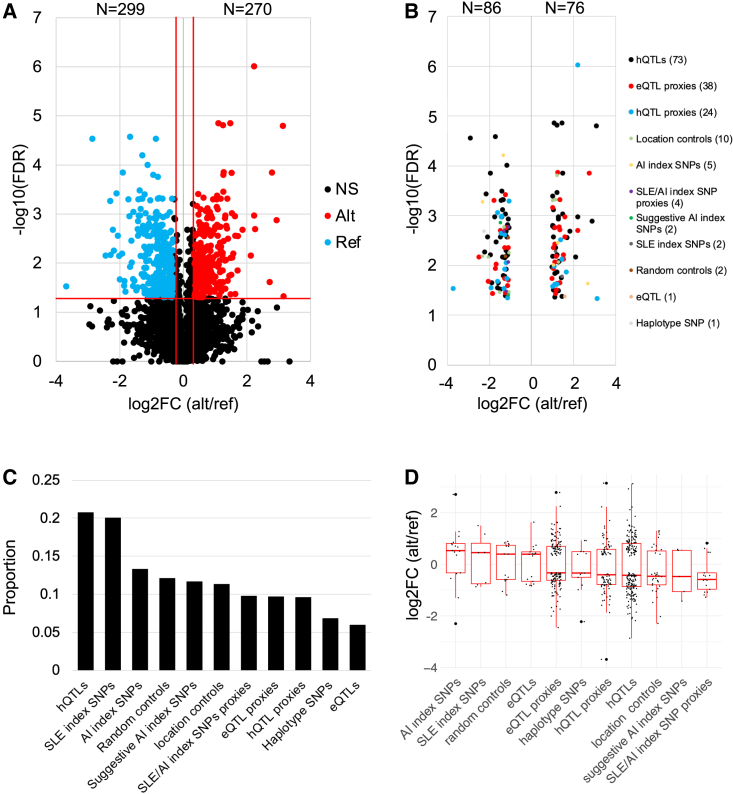
Of the variants that exhibited at least one emSeq, 4,765 (99.7%) variants had both alleles represented to detect significant allele-specific differential regulatory effects (emVars). A total of 567 variants (11.9%; 572 alleles due to multiallelic variants) were identified as emVars, with 28.5% (n = 162) boasting strong allele-specific differences in regulatory activity (FC > 2; log2(FC) > 1 or < −1). Within these 162 strong emVars, the majority were hQTLs (45%, n = 73), followed by eQTL proxies (n = 38, 23%), and hQTL proxies (n = 15, 24%) (Figures 2A and 2B; Table S5). A total of 299 variants (302 alleles due to multiallelic variants) displayed significant increases in activation of regulatory activity with the reference allele and 270 with the alternate allele (Figure 2A). When looking at the different types of variants evaluated, hQTLs and published SLE index SNPs produced the highest proportion of emVars (21% [201/971] and 20% [7/35], respectively) (Figure 2C). While random SNPs only produced a significantly higher proportion of emVars than the eQTLs (p = 0.03), the proportion observed within this variant type was higher than unexpected. After evaluating their locations with regard to other regulatory information, we found that 5.4% and 18.21% were located within enhancers and in long non-coding RNAs, which was significantly higher than that of eQTLs and the different proxy groups, which may account for the similar proportion of emSeq effects observed in the random controls compared with several other variant types. When comparing effects driven by the alternate or reference allele, the alternate allele more frequently demonstrated higher regulatory activity (median log2(FC) > 0) for AI and SLE index SNPs and eQTLs, while the reference allele more often demonstrated significantly higher regulatory activity (median log2(FC) < 0) in the other variant types (Figure 2D).
Figure 2.
Strong emVars are dominated by hQTLs
(A) Volcano plot of emVar effect sizes (log2(FC)) relative to the alternate/reference alleles. Horizontal red line represents q ≤ 0.05; vertical red lines (log2(FC) = ±0.32) represent a 1.25-FC difference between the alternate/reference alleles. Red dots indicate alternate alleles that demonstrate significantly higher regulatory activity than the reference allele; blue dots indicate reference alleles that have significantly higher activity.
(B) emVars (n = 162) with effect sizes >2.0, colored by variant type, demonstrating the high number of hQTLs (45%, black dots) with strong effects. The log2(FC) of alternate/reference allele is plotted on the x axis; y axis is the −log10(FDR).
(C) Proportions of emVars within each type.
(D) The log2(FC) of alternate/reference allele effect size plotted by variant type, sorted by descending median log2FC alternate/reference.
EmVars identify candidate causal variants for SLE and AI disease risk haplotypes
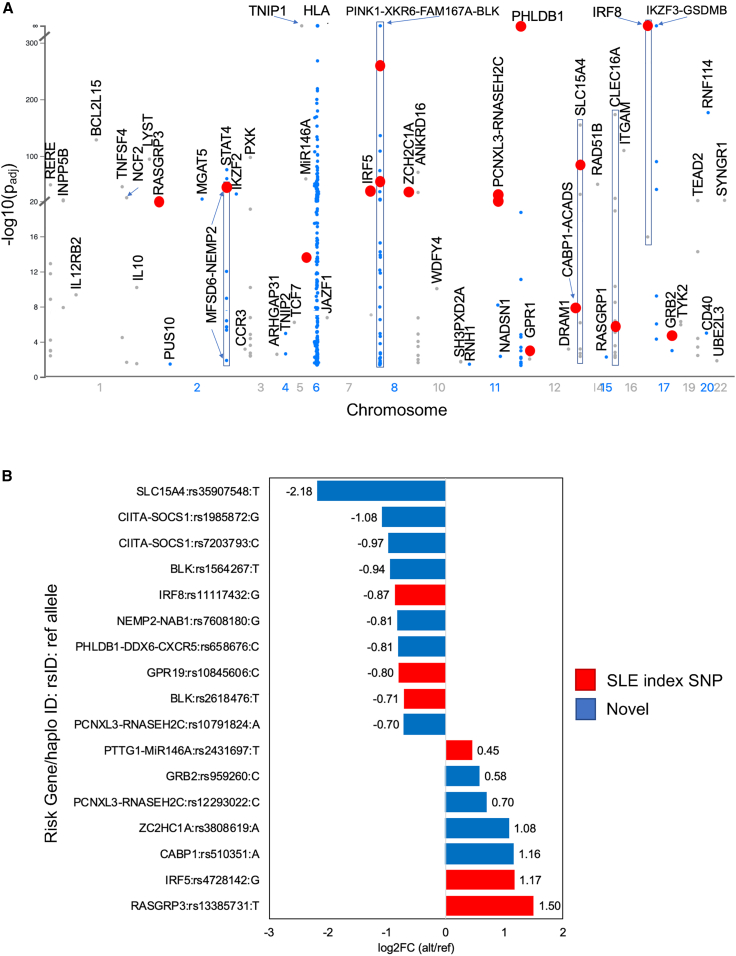
We next focused on effects located on SLE risk haplotypes to identify putative causal variants for disease. In addition to the selected 161 SLE index SNPs that passed QC, an additional 2,446 of the tested variants were located on SLE risk haplotypes (D’>0.8 to an SLE index SNP) (Table S2, column AI). A total of 381 (15%) variants were emSeqs (35 SLE index SNPs and 346 SLE haplotype SNPs) located on 50 SLE risk haplotypes; 208 of which were within the HLA region (Figure 3A). A total of 35 (9%) of these (17 outside of the HLA region) demonstrated significant differential regulatory activity between the two alleles and are, thus, putative causal variant candidates for SLE risk haplotypes. Six of the 17 non-HLA emVars were SLE index SNPs (Table 1; Figures 3B, S3A–S3F, and S4A–S4F; Table S5) and the remaining 11 are previously unreported putative causal variant candidates of SLE (Table 1; Figures 3B, 4, 5, S3G–S3K, and S4G–S4K; Table S5). We also identified 14 AI index emVars that we now nominate as putative causal variants for RA, T1D, MS, UC, CD, KD, PBC, GD, and vitiligo (Tables 1 and S5; Figure S5). We focus discussion on several previously unreported SLE candidate causal variants identified below.
Figure 3.
EmSeq and emVar variants on SLE risk haplotypes
(A) Manhattan plot of significant emSeqs (−log10(padj) as determined by DESeq2 plotted on SLE risk haplotypes). Only the allele with the highest regulatory activity is plotted for each variant. SLE risk gene/haplotype is indicated. Red dots represent emVars.
(B) Effect sizes of emVars on SLE risk haplotypes. The risk gene/haplotype and reference allele are provided for each variant. Positive effects indicate the alternate allele significantly increases regulatory activity over the reference allele; negative effects indicate the reference allele significantly increases activity over the alternate allele. Candidate causal SLE index SNPs are indicated in red. Previously unreported candidate causal variants on SLE haplotypes are indicated in blue.
Table 1.
Non-HLA candidate causal variants for SLE and AI disease risk genes/haplotypes
| Reported Gene/Haplotype | SNP | Chr | Pos (Mbp) | Typea | Reference | Alt | Ref log2FC | Alt log2FC | AE log2FC | −log10 FDR | Index SNP diseaseb | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SLE risk haplotype effects | RASGRP3 | rs13385731 | 2 | 33.48 | A | T | C | 0.22 | 1.72 | 1.50 | 1.77 | SLE |
| NEMP2-NAB1 | rs7608180 | 2 | 190.58 | D | G | T | 1.67 | 0.86 | −0.81 | 2.63 | novel | |
| PTTG1-miR146A | rs2431697 | 5 | 160.45 | A | T | C | 0.25 | 0.70 | 0.45 | 1.55 | SLE | |
| IRF5 | rs4728142 | 7 | 128.93 | A | G | A | 0.62 | 1.79 | 1.17 | 2.27 | SLE; MS; RA; UC |
|
| BLK | rs1564267 | 8 | 11.48 | I | T | C | 1.44 | 0.49 | −0.94 | 2.63 | novel | |
| BLK | rs2618476 | 8 | 11.50 | A & C | T | C | 1.27 | 0.56 | −0.71 | 2.03 | SLE | |
| PKIA-ZC2HC1A | rs3808619 | 8 | 78.67 | C | A | C | 0.39 | 1.47 | 1.08 | 3.10 | novel | |
| RNASEH2C-OVOL1 | rs12293022 | 11 | 65.76 | E | C | T | 0.57 | 1.27 | 0.70 | 1.44 | novel | |
| RNASEH2C-OVOL1 | rs10791824 | 11 | 65.79 | C | A | G | 1.99 | 1.29 | −0.70 | 1.42 | novel | |
| PHLDB1-DDX6-CXCR5 | rs658676 | 11 | 118.70 | F | C | T | 2.23 | 1.43 | −0.81 | 3.31 | novel | |
| GPR19 | rs10845606 | 12 | 12.68 | A | C | A | 0.68 | −0.12 | −0.80 | 1.64 | SLE | |
| CABP1-SPPL3 | rs510351 | 12 | 120.75 | E | A | G | −0.02 | 1.14 | 1.16 | 2.74 | novel | |
| SLC15A4 | rs35907548 | 12 | 128.80 | C | T | C | 3.70 | 1.51 | −2.18 | 2.26 | novel | |
| CIITA-SOCS1 | rs1985872 | 12 | 11.05 | G | G | C | 2.44 | 1.36 | −1.08 | 2.05 | novel | |
| CIITA-SOCS1 | rs7203793 | 16 | 11.09 | G | C | G | 0.99 | 0.02 | −0.97 | 1.57 | novel | |
| IRF8 | rs11117432 | 16 | 85.99 | A & C | G | A | 3.50 | 2.63 | −0.87 | 4.54 | SLE; PBC | |
| GRB2 | rs959260 | 17 | 75.37 | H | C | T | 0.17 | 0.75 | 0.58 | 1.96 | novel | |
| AI disease index SNPs | C5orf3 | rs26232 | 5 | 103.26 | B | C | T | 3.24 | 1.94 | −1.30 | 3.37 | RA |
| SLC22A23 | rs17309827 | 6 | 3.43 | B | T | G | 2.43 | 3.23 | 0.81 | 1.62 | CD | |
| IKZF1 | rs62447205 | 7 | 50.40 | B | A | G | 1.53 | 2.80 | 1.27 | 1.63 | T1D | |
| PVT1 | rs2019960 | 8 | 128.18 | B | T | C | 0.79 | 1.32 | 0.53 | 1.13 | MS | |
| IL2RA | rs12251307 | 10 | 6.08 | B | C | T | 2.76 | 0.46 | −2.30 | 2.44 | T1D | |
| GLYAT | rs11229555 | 11 | 58.64 | B | G | T | 1.79 | 1.46 | −0.33 | 1.88 | UC | |
| FADS1-FADS2 | rs968567 | 11 | 61.83 | B | C | T | 0.45 | 3.16 | 2.71 | 1.62 | RA; T1D | |
| ERBB3 | rs11171739 | 12 | 56.08 | B | C | T | 0.73 | 1.24 | 0.51 | 1.14 | T1D | |
| DLEU1 | rs9591325 | 13 | 50.24 | B | T | C | 2.32 | 2.80 | 0.47 | 2.16 | MS | |
| STAT3 | rs9891119 | 17 | 42.36 | B | A | C | 0.74 | 0.20 | −0.55 | 1.72 | CD; MS | |
| CD4 | rs4813003 | 20 | 46.13 | B | C | A,T | 0.0002 | 0.85 | 0.85 | 1.72 | KD | |
| CYCSP42 | rs2823286 | 21 | 15.45 | B | G | A | 1.12 | 1.47 | 0.36 | 1.21 | CD; UC | |
| UBE2L3 | rs2256609 | 22 | 21.57 | B | A | G | −0.09 | 0.67 | 0.76 | 1.52 | CD | |
| C1QTNF6 | rs229527 | 22 | 37.19 | B | C | A,G | 1.10 | 1.77 | 0.67 | 1.72 | GD; Vit |
The allele that demonstrates the allelic effect for each emVar is in italics. aType: A = SLE index SNP; B = AI index SNP; C = hQTL; D = hQTL proxy (r2 > 0.8); E = eQTL proxy (r2 > 0.8); F = haplotype SNP; G = proxy of SLE/AI index SNP (r2 > 0.8); H = suggestive AI index SNP (p < 1E−6); I = location control. bIndex SNP disease: CD = Crohn disease; GD = Graves’ disease; KD = Kawasaki disease; MS = multiple sclerosis; PBC = primary biliary cirrhosis; RA = rheumatoid arthritis; SLE = systemic lupus erythematosus; T1D = type 1 diabetes; UC = ulcerative colitis; Vit = vitiligo. Novel = variant not currently reported as an SLE index SNP (p < 5E−08) in the GWAS catalog.
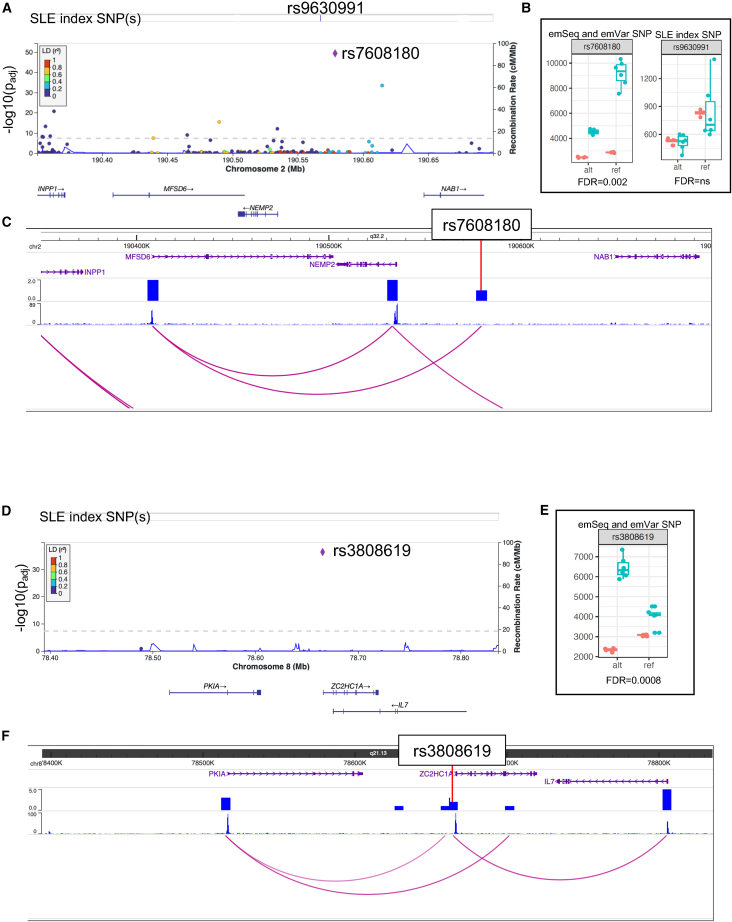
Figure 4.
Candidate variant identification on the NEMP2-NAB1 and PKIA-ZC2HC1A SLE risk haplotypes
(A and D) LocusZoom plots demonstrate emSeq effects on the haplotypes of (A) NEMP2-NAB1 and (D) PKIA-ZC2HC1A. Evaluated index SNPs are indicated. Variants evaluated, their genomic location, and genes in the region are presented on the x axis. The previously unreported emVar is represented as a purple diamond. LD values between each variant and the emVar are colored based on their r2 value (see LD key).
(B and E) Boxplots of the counts for each allele (alt and ref) in the EBV B (green) and plasmid control (orange) replicates. emVar FDR q values are given.
(C and F) Screenshot from the WashU Epigenome Browser for each haplotype region. Gene positions are provided on the top, followed by density HiC bed tracks, HiC bigwig tracks, and the interactions between gene promoters and the HiC data. The emVar position is provided.
Figure 5.
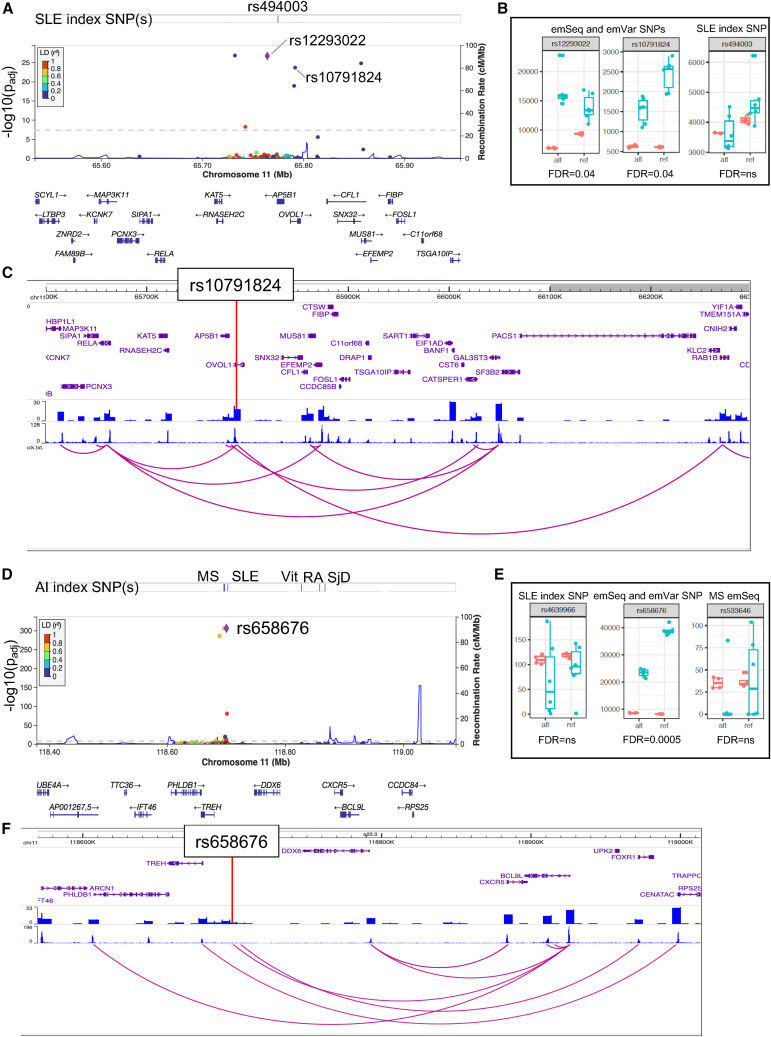
Candidate variant identification on the RNASEH2C-OVOL1 and PHLDB1-DDX6-CXCR5 SLE risk haplotypes
(A and D) LocusZoom plots demonstrate emSeq effects on the haplotypes of (A) RNASEH2C-OVOL1 and (D) PHLDB1-DDX6-CXCR5. Evaluated index SNPs are indicated. Variants evaluated, their genomic location, and genes in the region are presented on the x axis. The previously unreported emVar is represented as a purple diamond. LD values between each variant and the emVar are colored based on their r2 value (see LD key).
(B and E) Boxplots of the counts for each allele (alt and ref) in the EBV B (green) and plasmid control (orange) replicates. emVar FDR q values are given.
(C and F) Screenshot from the WashU Epigenome Browser for each haplotype region. Gene positions are provided on the top, followed by density HiC bed tracks, HiC bigwig tracks, and the interactions between gene promoters and the HiC data. The emVar position is provided.
There is currently one reported SLE index SNP in the region between NEMP2 [MIM: 616497] and NAB1 [MIM: 600800] on chromosome 2: rs9630991.20 We evaluated this variant along with 153 other variants on the haplotype and, while multiple variants were shown to be emSeqs (Figure 4A; Table S2), only rs7608180, a variant 11,264 bp downstream of rs9630991 (D’ = 0.95, r2 = 0.35), was shown to be an emVar with the reference G allele (FC = 0.57, FDR q = 0.002) (Figures 3B and 4B; Table S5). Our promoter capture HiC data collected in primary B cells demonstrates that, while the region containing rs7608180 lies between NEMP2 and NAB1, it interacts with the promoter of the upstream major facilitator superfamily domain containing 6 (MFSD6 [MIM: 613476]) gene, a gene predicted to enable MHC class I protein binding and receptor activity,21 thus prioritizing MFSD6 as a possible SLE risk locus in the region (Figure 4C).
Association between SLE and rs4739134 has been reported between the region of PKIA [MIM: 606059] and ZC2HC1A on chromosome 8.22 We, unfortunately, did not include this variant in our study. We did, however, evaluate an hQTL variant (rs3808619) that is strongly correlated with rs4739134 (D’ = 0.99, r2 = 0.98). rs3808619, located ∼600 bp downstream of ZC2HC1A, was a strong emSeq (Figure 4D; Table S2) and emVar, with the alternate C allele producing a significant allelic effect (FC = 2.11, FDR q = 0.0008) (Figures 3B and 4E; Table S5). Our promoter capture HiC data show an interaction between rs3808619 and the downstream interleukin 7 (IL7 [MIM: 146660]) gene promoter (Figure 4F). IL7 is a cytokine important for B and T cell development and has been associated with SLE nephritis.23,24 Taken together, we nominate the hQTL rs3808619 as a putative causal variant for SLE with potential regulatory action on IL7.
Two index SNPs (rs494003 and rs10896045) are reported for the SLE risk haplotype spanning RNASEH2C [MIM: 610330] to OVOL1 [MIM: 602313] on chromosome 1125,26; only rs494003 was evaluated in our study and failed to modulate regulatory activity (Figure 5A; Table S2). Other SNPs in the region were emSeqs, but only two SNPs were discovered to be emVars (rs12293022 and rs10791824 [an hQTL]). Rs12293022 demonstrated significantly increased regulatory activity with the alternate T allele (FC = 1.62, FDR q = 0.036), while the reference A allele of hQTL rs10791824 produced increased regulatory activity relative to the alternate G allele (FC = 0.61, FDR q = 0.038) (Figures 3B and 5B; Table S5). Our promoter capture HiC data show that the region containing rs10791824 interacts with the promoters of multiple genes besides OVOL1, including upstream genes RELA [MIM: 164014] and AP5B1 [MIM: 614367], and downstream gene RAB1B [MIM: 612565] (Figure 5C).
The region spanning PHLDB1 [MIM: 612834], DDX6 [MIM: 600326], and CXCR5 [MIM: 601613] on chromosome 11 has been associated with multiple AI diseases, including three associations with SLE (rs4639966, rs480958, and rs4936441).26,27 We evaluated 126 variants in this region including rs4639966, as well as index SNPs in the region for MS (rs533646),28 vitiligo (rs638893),29 RA (rs10790268),30,31 and SjD (rs7119038).32 None of the tested index SNPs were emSeqs (Figure 5D; Table S2). However, rs658676, a variant in strong LD with MS index SNP rs533646 (D’ = 0.962, r2 = 0.888), produced a significant emSeq and was the only tested variant in the region that produced an emVar (FC = 0.57, FDR q = 0.0005 with the reference C allele) (Figures 3B and 5E; Table S5). Our promoter capture HiC data show that the region containing rs658676 interacts with the promoter of B cell lymphoma 9-like protein (BCL9L [MIM: 609004]), a gene just downstream and overlapping a portion of CXCR5 (Figure 5F).
The region on chromosome 16 that spans CIITA [MIM: 600005], CLEC16A [MIM: 611303], and SOCS1 [MIM: 603597] is also associated with multiple AI diseases (SLE, celiac disease, MS, T1D, CD, psoriasis, and PBC).22,26,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50 We evaluated 169 variants in the region including two of the five SLE index SNPs (rs9652601 and rs7200786) and index SNPs for MS (rs2286974, rs7200786, rs12927355), T1D (rs12708716, rs12927355, rs741172, rs2903692), PBC (rs413024, rs1646019), psoriasis (rs367569), and CD (rs423674). Only two index SNPs modulated regulatory activity (PBC: rs413024 and CD: rs423674), but neither produced an emVar (Tables S2 and S5; Figure S3G). Instead, we identified emVars with two other variants in CLEC16A, rs1985872 and rs7203793 (D’ = 0.89, r2 = 0.64), both with the reference alleles displaying significantly higher regulatory activity than the alternate alleles (Table S5; Figure S4G), prioritizing these variants as candidate causal variants for SLE and multiple other AI diseases.
Discussion
In this study, we used an MPRA to evaluate the regulatory activity of previously identified hQTLs as well as other variant types implicated in SLE and AI disease, eQTLs found to interact with hQTLs, and various SNP proxies. We discovered that of 24% (976/4039) of the tested hQTL variants are positioned in regulatory sequences (emSeqs) and that 20.7% exhibited significant allelic effects (emVars), suggesting that they are putative causal variants. In contrast, only 9.6% (90/942) of the tested variants in strong LD (r2 > 0.8) with hQTLs (hQTL proxies) were emVars, demonstrating that a priori knowledge of hQTLs can facilitate the identification of putative causal variants in the context of strong LD using standard statistical methods. Further, tested hQTL emSeqs exhibited significantly stronger effects on regulatory activity than the eQTL emSeqs and the other types of variants tested. Notably, only 6% of tested eQTLs were found to be emVars, confirming a previous report that only a low proportion of eQTLs overall are likely to be causal.51
Using this approach, we also identified and nominate 17 potential non-HLA causal variants in established but incompletely characterized SLE risk haplotypes: RASGRP3, PTTG1-miR146A, IRF5, BLK, GPR19, IRF8, NEMP2-NAB1, BLK, PKIA-ZC2HC1A, PCNXL3-RNASEH2C, PHLDB1-CXCR5, CABP1, SLC15A4, CIITA-SOCS1, and GRB2. For several of these regions, this approach successfully narrowed the >150 evaluated variants in strong LD to one or two variants per region. We also identified allele-dependent regulation for 14 additional published non-HLA AI index SNPs for loci associated with MS, RA, PBC, CD, T1D, KD, UC, GD and vitiligo, prioritizing these variants for future functional studies in B cell lines and B cells.
Our study is the largest MPRA study of SLE to date, testing 161 SLE index SNPs, 2,446 SNPs on SLE risk haplotypes, and 29,058 additional variants for both functional potential and allelic effects. In 2021, Lu et al.14 evaluated 91 known SLE index SNPs and 2,990 variants in strong LD, identifying 51 emVars. We expanded upon the study by Lu et al. by testing an additional 113 SLE index SNPs, 2,391 SNPs in LD with SLE index SNPs, and 777 published index SNPs in other AI diseases (Figures S6 and S6B; Tables S2 and S5). When comparing the regulatory determinations for the 328 oligo sequences (166 variants) evaluated in both studies (Figure S6C), 204 oligo sequences (62%) produced concordant regulatory effects: 51 were emSeqs and 153 sequences did not modulate regulatory activity. For the 124 oligo sequences (67 variants) that exhibited discordant effects, 14 emSeqs identified in our study and 110 identified by Lu et al. were not confirmed by the other study (Table S2). Our study appears to be more conservative in its determinations despite using the same significance and FC thresholds to call emSeqs and emVar variants. Therefore, we feel confident in the effects that we identify but note that we have likely missed others.
The limitations of this study include the use of EBV B cells because variants that only function in non-B cell types or require specific conditions would not have been detected by our MPRA strategy. Further, because of the minimal promoter used in the MPRA construct, the observed effects were limited largely to inducible rather than suppressive effects on regulatory activity. Last, the primary goal of our study was to evaluate the functional implications of previously identified hQTLs, and, therefore, the MPRA was not designed to test every variant in LD with SLE or other AI index SNPs or every eQTL positioned in each risk locus. Therefore, while we successfully identified multiple likely causal effects within SLE and AI disease risk haplotypes, there are many others that we have not evaluated. Additionally, selected index SNPs were identified using the GWAS catalog; since risk variants not identified by GWAS or the ImmunoChip arrays (e.g., candidate gene studies or fine mapping studies) are not included in the catalog, they were not considered for this report.
In summary, our study expands our understanding of hQTLs, demonstrating that our tested hQTLs likely regulate aspects of the innate and adaptive immune responses through interactions with IRF TFs. Further, many of the identified hQTL emSeqs and emVars are located in dELS cREs that likely function as causal variants for complex trait phenotypes. In addition, we nominate 31 causal variants for SLE and AI diseases. Thus, we uncover important insights into the mechanistic relationships between genotype, epigenetics, and regulatory activity in SLE and AI disease phenotypes.
Data and code availability
-
•
The codes utilized during this study were developed by Dr. Ryan Tewhey and are available with full documentation and examples at https://github.com/tewhey-lab/MPRA_oligo_barcode_pipeline and https://github.com/tewhey-lab/MPRAmodel.
-
•
All sequencing data that support the findings of this study are available at NCBI Gene Expression Omnibus (GEO): GSE254502. The Hi-C promoter capture data supporting the current study have not yet been deposited in a public repository because they are part of a bigger project that will be deposited at a later time. All other relevant data are available from the corresponding author upon request.
Web resources
-
(1)
https://aci-cores.omrf.org/biorepository/- OMRF’s Arthritis and Clinical Immunology Biorepository Core
-
(2)
http://epigenomegateway.wustl.edu/browser/- WashU Epigenome Browser
-
(3)
http://hgdownload.soe.ucsc.edu - the UCSC genome browser downloads page
-
(4)
https://genome.ucsc.edu/cgi-bin/hgLiftOver - UCSC LiftOver tool
-
(5)
https://github.com/ArimaGenomics/CHiC - Arima Genomics pipeline
-
(6)
https://github.com/tewhey-lab/MPRA_oligo_barcode_pipeline and https://github.com/tewhey-lab/MPRAmodel – Dr. Ryan Tewhey’s MPRA analyses pipelines
-
(7)
http://omim.org – Online Mendelian Inheritance in Man
-
(8)
https://omrf.org/research-faculty/core-facilities/next-generation-sequencing/- OMRF’s Clinical Genomics Center
-
(9)
https://useast.ensembl.org/Homo_sapiens/Tools/VEP - Ensembl’s variant effect predictor tool
Acknowledgments
We would like to thank Dr. Ryan Tewhey for providing the pGL4:23:ΔxbaΔluc and pGL4.23:minP GFP plasmids. We would also like to thank Drs. Ryan Tewhey (The Jackson Laboratory), Leah Kottyan and Xioaming Lu (Cincinnati Children’s Hospital Medical Center), and Kaiyu Jiang (University of Buffalo) for their expert advice regarding different aspects of their MPRA protocols. All sequencing experiments were conducted by the Clinical Genomics Center at the OMRF (https://omrf.org/research-faculty/core-facilities/next-generation-sequencing/). Research reported in this publication was supported by the Presbyterian Health Foundation (Oklahoma City, OK), and the National Institute of Arthritis and Musculoskeletal and Skin Diseases, the National Institute of Allergy and Infectious Diseases, and the National Institute of General Medical Sciences under award numbers R01 AR073606, R01 AI156724, P20 RR020143, P30 AR053483, and P30 GM103510. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or other funding agencies.
Author contributions
Conceptualization: P.M.G. Study design: P.M.G., Y.F., J.A.K. Data curation: J.A.K., R.C.P. Formal analysis: J.A.K. Investigation: Y.F., J.G., S.P. Project Administration: P.M.G. Visualization: J.A.K. Writing – original draft: Y.F., J.A.K., K.L.T., P.M.G. Writing – reviewing and editing: P.M.G., Y.F., J.A.K., J.G., R.C.P., K.L.T., K.G., D.A.M., S.P.
Declaration of interests
The authors declare no competing interests.
Footnotes
It can be found online at https://doi.org/10.1016/j.xhgg.2024.100279.
Supplemental information
References
- 1.Zhang F., Lupski J.R. Non-coding genetic variants in human disease. Hum. Mol. Genet. 2015;24:R102–R110. doi: 10.1093/hmg/ddv259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farh K.K.-H., Marson A., Zhu J., Kleinewietfeld M., Housley W.J., Beik S., Shoresh N., Whitton H., Ryan R.J.H., Shishkin A.A., et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ENCODE Project Consortium. Moore J.E., Purcaro M.J., Pratt H.E., Epstein C.B., Shoresh N., Adrian J., Kawli T., Davis C.A., Dobin A., et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020;583:699–710. doi: 10.1038/s41586-020-2493-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L., Xue G., Liu J., Li Q., Wang Y. Revealing transcription factor and histone modification co-localization and dynamics across cell lines by integrating ChIP-seq and RNA-seq data. BMC Genom. 2018;19:914. doi: 10.1186/s12864-018-5278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pelikan R.C., Kelly J.A., Fu Y., Lareau C.A., Tessneer K.L., Wiley G.B., Wiley M.M., Glenn S.B., Harley J.B., Guthridge J.M., et al. Enhancer histone-QTLs are enriched on autoimmune risk haplotypes and influence gene expression within chromatin networks. Nat. Commun. 2018;9:2905. doi: 10.1038/s41467-018-05328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tewhey R., Kotliar D., Park D.S., Liu B., Winnicki S., Reilly S.K., Andersen K.G., Mikkelsen T.S., Lander E.S., Schaffner S.F., Sabeti P.C. Direct Identification of Hundreds of Expression-Modulating Variants using a Multiplexed Reporter Assay. Cell. 2016;165:1519–1529. doi: 10.1016/j.cell.2016.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melnikov A., Zhang X., Rogov P., Wang L., Mikkelsen T.S. Massively parallel reporter assays in cultured mammalian cells. J. Vis. Exp. 2014;17:51719. doi: 10.3791/51719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patwardhan R.P., Hiatt J.B., Witten D.M., Kim M.J., Smith R.P., May D., Lee C., Andrie J.M., Lee S.-I., Cooper G.M., et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat. Biotechnol. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasmussen A., Sevier S., Kelly J.A., Glenn S.B., Aberle T., Cooney C.M., Grether A., James E., Ning J., Tesiram J., et al. The lupus family registry and repository. Rheumatology. 2011;50:47–59. doi: 10.1093/rheumatology/keq302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lappalainen T., Sammeth M., Friedländer M.R., 't Hoen P.A.C., Monlong J., Rivas M.A., Gonzàlez-Porta M., Kurbatova N., Griebel T., Ferreira P.G., et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McVicker G., van de Geijn B., Degner J.F., Cain C.E., Banovich N.E., Raj A., Lewellen N., Myrthil M., Gilad Y., Pritchard J.K. Identification of genetic variants that affect histone modifications in human cells. Science. 2013;342:747–749. doi: 10.1126/science.1242429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Via M., Gignoux C., Burchard E.G. The 1000 Genomes Project: new opportunities for research and social challenges. Genome Med. 2010;2:3. doi: 10.1186/gm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delaneau O., Marchini J., 1000 Genomes Project Consortium Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat. Commun. 2014;5:3934. doi: 10.1038/ncomms4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu X., Chen X., Forney C., Donmez O., Miller D., Parameswaran S., Hong T., Huang Y., Pujato M., Cazares T., et al. Global discovery of lupus genetic risk variant allelic enhancer activity. Nat. Commun. 2021;12:1611. doi: 10.1038/s41467-021-21854-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akgol Oksuz B., Yang L., Abraham S., Venev S.V., Krietenstein N., Parsi K.M., Ozadam H., Oomen M.E., Nand A., Mao H., et al. Systematic evaluation of chromosome conformation capture assays. Nat. Methods. 2021;18:1046–1055. doi: 10.1038/s41592-021-01248-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sollis E., Mosaku A., Abid A., Buniello A., Cerezo M., Gil L., Groza T., Güneş O., Hall P., Hayhurst J., et al. The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource. Nucleic Acids Res. 2023;51:D977–D985. doi: 10.1093/nar/gkac1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cunningham F., Allen J.E., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Austine-Orimoloye O., Azov A.G., Barnes I., Bennett R., et al. Ensembl 2022. Nucleic Acids Res. 2022;50:D988–D995. doi: 10.1093/nar/gkab1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y.-F., Zhang Y., Lin Z., Zhang H., Wang T.-Y., Cao Y., Morris D.L., Sheng Y., Yin X., Zhong S.-L., et al. Identification of 38 novel loci for systemic lupus erythematosus and genetic heterogeneity between ancestral groups. Nat. Commun. 2021;12:772. doi: 10.1038/s41467-021-21049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alliance of Genome Resources Consortium Harmonizing model organism data in the alliance of genome resources. Genetics. 2022;220 doi: 10.1093/genetics/iyac022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langefeld C.D., Ainsworth H.C., Cunninghame Graham D.S., Kelly J.A., Comeau M.E., Marion M.C., Howard T.D., Ramos P.S., Croker J.A., Morris D.L., et al. Transancestral mapping and genetic load in systemic lupus erythematosus. Nat. Commun. 2017;8 doi: 10.1038/ncomms16021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanley S., Mok C.C., Vanarsa K., Habazi D., Li J., Pedroza C., Saxena R., Mohan C. Identification of Low-Abundance Urinary Biomarkers in Lupus Nephritis Using Electrochemiluminescence Immunoassays. Arthritis Rheumatol. 2019;71:744–755. doi: 10.1002/art.40813. [DOI] [PubMed] [Google Scholar]
- 24.Lauwerys B.R., Husson S.N., Maudoux A.L., Badot V., Houssiau F.A. sIL7R concentrations in the serum reflect disease activity in the lupus kidney. Lupus Sci. Med. 2014;1 doi: 10.1136/lupus-2014-000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris D.L., Sheng Y., Zhang Y., Wang Y.-F., Zhu Z., Tombleson P., Chen L., Cunninghame Graham D.S., Bentham J., Roberts A.L., et al. Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nat. Genet. 2016;48:940–946. doi: 10.1038/ng.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin X., Kim K., Suetsugu H., Bang S.-Y., Wen L., Koido M., Ha E., Liu L., Sakamoto Y., Jo S., et al. Meta-analysis of 208370 East Asians identifies 113 susceptibility loci for systemic lupus erythematosus. Ann. Rheum. Dis. 2021;80:632–640. doi: 10.1136/annrheumdis-2020-219209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han J.-W., Zheng H.-F., Cui Y., Sun L.-D., Ye D.-Q., Hu Z., Xu J.-H., Cai Z.-M., Huang W., Zhao G.-P., et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 28.International Multiple Sclerosis Genetics Consortium IMSGC. Beecham A.H., Patsopoulos N.A., Xifara D.K., Davis M.F., Kemppinen A., Cotsapas C., Shah T.S., Spencer C., Booth D., et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat. Genet. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang X.-F., Zhang Z., Hu D.-Y., Xu A.-E., Zhou H.-S., Sun L.-D., Gao M., Gao T.-W., Gao X.-H., Chen H.-D., et al. Association analyses identify three susceptibility Loci for vitiligo in the Chinese Han population. J. Invest. Dermatol. 2013;133:403–410. doi: 10.1038/jid.2012.320. [DOI] [PubMed] [Google Scholar]
- 30.Laufer V.A., Tiwari H.K., Reynolds R.J., Danila M.I., Wang J., Edberg J.C., Kimberly R.P., Kottyan L.C., Harley J.B., Mikuls T.R., et al. Genetic influences on susceptibility to rheumatoid arthritis in African-Americans. Hum. Mol. Genet. 2019;28:858–874. doi: 10.1093/hmg/ddy395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lessard C.J., Li H., Adrianto I., Ice J.A., Rasmussen A., Grundahl K.M., Kelly J.A., Dozmorov M.G., Miceli-Richard C., Bowman S., et al. Variants at multiple loci implicated in both innate and adaptive immune responses are associated with Sjögren’s syndrome. Nat. Genet. 2013;45:1284–1292. doi: 10.1038/ng.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bentham J., Morris D.L., Graham D.S.C., Pinder C.L., Tombleson P., Behrens T.W., Martín J., Fairfax B.P., Knight J.C., Chen L., et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 2015;47:1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trynka G., Hunt K.A., Bockett N.A., Romanos J., Mistry V., Szperl A., Bakker S.F., Bardella M.T., Bhaw-Rosun L., Castillejo G., et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Genet. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.International Multiple Sclerosis Genetics Consortium Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science. 2019;365 doi: 10.1126/science.aav7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.International International Multiple Sclerosis Genetics Consortium, Wellcome Trust Case Control Consortium 2. Sawcer S., Hellenthal G., Pirinen M., Spencer C.C.A., Patsopoulos N.A., Moutsianas L., Dilthey A., Su Z., et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cooper J.D., Smyth D.J., Smiles A.M., Plagnol V., Walker N.M., Allen J.E., Downes K., Barrett J.C., Healy B.C., Mychaleckyj J.C., et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat. Genet. 2008;40:1399–1401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plagnol V., Howson J.M.M., Smyth D.J., Walker N., Hafler J.P., Wallace C., Stevens H., Jackson L., Simmonds M.J., et al. Type 1 Diabetes Genetics Consortium, , . Type 1 Diabetes Genetics Consortium Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrett J.C., Clayton D.G., Concannon P., Akolkar B., Cooper J.D., Erlich H.A., Julier C., Morahan G., Nerup J., Nierras C., et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Todd J.A., Walker N.M., Cooper J.D., Smyth D.J., Downes K., Plagnol V., Bailey R., Nejentsev S., Field S.F., Payne F., et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Onengut-Gumuscu S., Chen W.-M., Burren O., Cooper N.J., Quinlan A.R., Mychaleckyj J.C., Farber E., Bonnie J.K., Szpak M., Schofield E., et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat. Genet. 2015;47:381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Márquez A., Kerick M., Zhernakova A., Gutierrez-Achury J., Chen W.-M., Onengut-Gumuscu S., González-Álvaro I., Rodriguez-Rodriguez L., Rios-Fernández R., González-Gay M.A., et al. Meta-analysis of Immunochip data of four autoimmune diseases reveals novel single-disease and cross-phenotype associations. Genome Med. 2018;10:97. doi: 10.1186/s13073-018-0604-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andlauer T.F.M., Buck D., Antony G., Bayas A., Bechmann L., Berthele A., Chan A., Gasperi C., Gold R., Graetz C., et al. Novel multiple sclerosis susceptibility loci implicated in epigenetic regulation. Sci. Adv. 2016;2 doi: 10.1126/sciadv.1501678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hakonarson H., Grant S.F.A., Bradfield J.P., Marchand L., Kim C.E., Glessner J.T., Grabs R., Casalunovo T., Taback S.P., Frackelton E.C., et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448:591–594. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- 45.Ellinghaus D., Ellinghaus E., Nair R.P., Stuart P.E., Esko T., Metspalu A., Debrus S., Raelson J.V., Tejasvi T., Belouchi M., et al. Combined analysis of genome-wide association studies for Crohn disease and psoriasis identifies seven shared susceptibility loci. Am. J. Hum. Genet. 2012;90:636–647. doi: 10.1016/j.ajhg.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Juran B.D., Hirschfield G.M., Invernizzi P., Atkinson E.J., Li Y., Xie G., Kosoy R., Ransom M., Sun Y., Bianchi I., et al. Immunochip analyses identify a novel risk locus for primary biliary cirrhosis at 13q14, multiple independent associations at four established risk loci and epistasis between 1p31 and 7q32 risk variants. Hum. Mol. Genet. 2012;21:5209–5221. doi: 10.1093/hmg/dds359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsoi L.C., Spain S.L., Knight J., Ellinghaus E., Stuart P.E., Capon F., Ding J., Li Y., Tejasvi T., Gudjonsson J.E., et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet. 2012;44:1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J.Z., van Sommeren S., Huang H., Ng S.C., Alberts R., Takahashi A., Ripke S., Lee J.C., Jostins L., Shah T., et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu J.Z., Almarri M.A., Gaffney D.J., Mells G.F., Jostins L., Cordell H.J., Ducker S.J., Day D.B., Heneghan M.A., Neuberger J.M., et al. Dense fine-mapping study identifies new susceptibility loci for primary biliary cirrhosis. Nat. Genet. 2012;44:1137–1141. doi: 10.1038/ng.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dubois P.C.A., Trynka G., Franke L., Hunt K.A., Romanos J., Curtotti A., Zhernakova A., Heap G.A.R., Adány R., Aromaa A., et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abell N.S., DeGorter M.K., Gloudemans M.J., Greenwald E., Smith K.S., He Z., Montgomery S.B. Multiple causal variants underlie genetic associations in humans. Science. 2022;375:1247–1254. doi: 10.1126/science.abj5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The codes utilized during this study were developed by Dr. Ryan Tewhey and are available with full documentation and examples at https://github.com/tewhey-lab/MPRA_oligo_barcode_pipeline and https://github.com/tewhey-lab/MPRAmodel.
-
•
All sequencing data that support the findings of this study are available at NCBI Gene Expression Omnibus (GEO): GSE254502. The Hi-C promoter capture data supporting the current study have not yet been deposited in a public repository because they are part of a bigger project that will be deposited at a later time. All other relevant data are available from the corresponding author upon request.