Abstract
The shoot apical meristem (SAM) is responsible for overall shoot growth by generating all aboveground structures. Recent research has revealed that the SAM displays an autonomous heat stress (HS) memory of a previous non-lethal HS event. Considering the importance of the SAM for plant growth, it is essential to determine how its thermomemory is mechanistically controlled. Here, we report that HEAT SHOCK TRANSCRIPTION FACTOR A7b (HSFA7b) plays a crucial role in this process in Arabidopsis, as the absence of functional HSFA7b results in the temporal suppression of SAM activity after thermopriming. We found that HSFA7b directly regulates ethylene response at the SAM by binding to the promoter of the key ethylene signaling gene ETHYLENE-INSENSITIVE 3 to establish thermotolerance. Moreover, we demonstrated that HSFA7b regulates the expression of ETHYLENE OVERPRODUCER 1 (ETO1) and ETO1-LIKE 1, both of which encode ethylene biosynthesis repressors, thereby ensuring ethylene homeostasis at the SAM. Taken together, these results reveal a crucial and tissue-specific role for HSFA7b in thermomemory at the Arabidopsis SAM.
Key words: ethylene response, heat stress adaptation, shoot apical meristem, SAM, thermopriming, thermomemory
The shoot apical meristem displays an autonomous heat stress memory of a previous non-lethal heat stress event. This study reports that Arabidopsis transcription factor HSFA7b controls ethylene homeostasis at the shoot apical meristem in response to thermopriming by directly controlling the expression of ethylene biosynthesis and signaling genes, thus playing a tissue-specific role in thermomemory.
Introduction
As sessile organisms, plants are frequently exposed to unpredictable and often life-threatening environmental challenges such as heat stress (HS); however, evolution has established response strategies for coping with these variable conditions to ensure survival. In particular, plants employ a stress memory mechanism that enables them to “memorize” exposure to a first, moderate, and non-lethal stress (called priming), during which information about the past stress is stored (memory phase) to facilitate a faster or stronger response to the next, potentially more threatening stress signal (triggering) (Hilker et al., 2016). The initial phase of an HS response (priming and memory) involves a complex interplay between transcription factors (TFs) of the HEAT SHOCK FACTOR (HSF) family and HS memory genes (Lämke et al., 2016; Liu et al., 2019). HSFs bind to heat shock elements (HSEs, with a conserved 5′-nGAAnnTTCn-3′ sequence) in promoters of HS-inducible genes, including those encoding HEAT SHOCK PROTEIN (HSP) chaperones (Nover et al., 2001; Park and Seo, 2015), to confer thermotolerance. Although components of the HS memory machinery have been characterized (Balazadeh, 2022), our understanding of the mechanisms that underlie HS memory is limited. To date, the molecular framework of HS memory has mainly been studied in whole Arabidopsis thaliana (Arabidopsis) seedlings (Stief et al., 2014; Sedaghatmehr et al., 2016; Friedrich et al., 2021). However, it has recently been shown that the Arabidopsis shoot apical meristem (SAM) directly senses changes in temperature and displays a strong transcriptional HS memory for genes involved in protein folding, primary carbohydrate metabolism, and meristem maintenance (Olas et al., 2021a). Importantly, the SAM engages an HS response and memory network that differs in many aspects from that of whole seedlings: the expressional timing of HS memory genes and the types of genes that generate thermomemory, including HSFs, differ among organs, providing strong evidence for tissue- or organ-specific heat memories.
The SAM is a highly organized assortment of cells required for proper and continuous growth of aboveground plant organs (Groß-Hardt and Laux, 2003). It harbors stem cells whose descendants form shoot structures like leaves, flowers, and their derivatives (seeds and fruits) (Uchida and Torii, 2019). The balance between stem cell loss and renewal enables plants to form new organs throughout their lifespan (Uchida and Torii, 2019), supporting the plant’s high developmental plasticity during the stress period. Changes in SAM homeostasis, including modifications of phytohormone levels, alter meristem activity. Although various hormones, including abscisic acid and ethylene, have been linked to stress acclimation (Ruonala et al., 2006), surprisingly little is currently known about how the SAM’s hormonal status is affected and transcriptionally controlled by stress. Considering the key importance of the SAM for plant growth, we expect that specific molecular mechanisms have evolved to enable the SAM to respond appropriately to environmental challenges. This is particularly relevant for seedlings that have not yet established axillary meristems; severe damage of the SAM by abiotic stress will lead to termination of shoot growth and, hence, death of the individual, precluding inheritance of genetic information to the next generation.
Here, we report that HSFA7a and HSFA7b are involved in controlling HS memory at the SAM. We show that the absence of functional HSFA7a and HSFA7b proteins leads to temporary suppression of SAM activity after thermopriming. Furthermore, we find that HSFA7b regulates the ethylene response during HS by directly regulating the expression of ethylene biosynthesis and signaling genes through binding to HSEs in their promoter regions. Furthermore, by studying the most downstream components of the ethylene signaling pathway, we demonstrate that a functional ethylene response at the SAM requires HSFA7b. Taken together, our data show that HSFA7b controls ethylene homeostasis during thermopriming, enabling plants to establish thermotolerance.
Results
HSFA7a and HSFA7b display transcriptional heat stress memory at the SAM
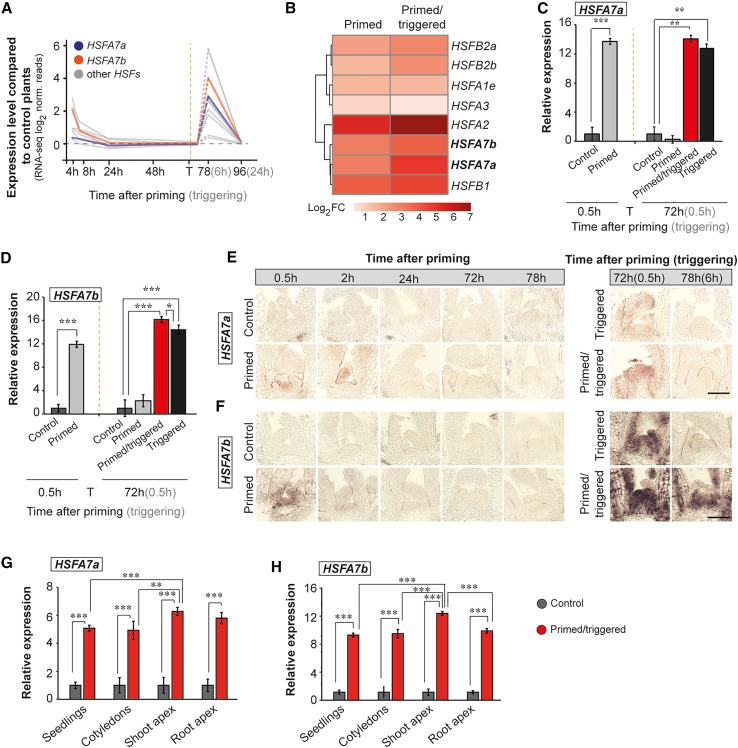
Transcriptome analysis of shoot apices from wild-type Arabidopsis (Col-0) seedlings (Olas et al., 2021a) revealed that expression of HSFA7a and HSFA7b was induced at 4 h after priming HS and was hyper-induced in response to a more severe triggering HS (6 h after triggering), suggesting the existence of transcriptional HS memory (Balazadeh, 2022) (Figure 1A). Because the expression of both HSFs was not analyzed directly after HS treatments, we subjected Col-0 plants to a previously established thermomemory assay (Supplemental Figure 1) (Sedaghatmehr et al., 2016; Olas et al., 2021a) and performed RNA-sequencing (RNA-seq) of shoot apices from Col-0 wild-type plants at 0.5 h after priming and 0.5 h after triggering to confirm the thermomemory pattern of HSFA7a and HSFA7b expression at the SAM (Figure 1B). Transcriptome analysis of HSF genes revealed that both HSFA7a and HSFA7b are induced at the SAM after priming and show hyper-induction upon triggering HS (Figure 1B). We confirmed the transcriptional induction of both genes by qRT–PCR (Figure 1C and 1D). To gain information on HSFA7a and HSFA7b expression at the SAM at a higher spatial resolution, we performed RNA in situ hybridization (Figure 1E and 1F). We did not detect HSFA7a or HSFA7b expression at the Col-0 SAM under control conditions (C plants). However, both genes were rapidly induced at the SAM of primed (P) plants directly after the priming HS, and expression was hyper-induced at the SAM of primed/triggered (PT) plants after triggering compared with P and triggered (T) plants, confirming that HSFA7a and HSFA7b are transcriptional HS memory genes. Our data also confirmed higher expression of HSFA7b than HSFA7a at the SAM after heat treatment. Next, we tested expression of both HSFs in whole seedlings, cotyledons, and shoot and root apices of PT plants at 0.5 h after triggering (Figure 1G and 1H). Transcripts of HSFA7a and HSFA7b were upregulated in all organs, with the highest expression detected at the SAM. This suggests that HSFA7a and HSFA7b serve as integral components of HS memory throughout the entire plant. As HSFA7a and HSFA7b share 59% amino acid sequence similarity (Supplemental Figure 2A), we tested their capacity for interaction by yeast two-hybrid assays. Supplemental Figure 2B shows that both TFs may interact to form a heterodimer likely involved in regulation of target genes.
Figure 1.
HSFA7a and HSFA7b act as HS memory genes at the shoot apical meristem (SAM).
(A) Expression profiles of HEAT SHOCK TRANSCRIPTION FACTOR (HSF) genes at the SAM of Col-0 plants at 4, 8, 24, 48, 78, and 96 h after priming (6 and 24 h after triggering) normalized to those of untreated control plants. Blue, HSAF7a; orange, HSAF7b; gray, other HSFs.
(B) Heatmap showing the log2 fold change (log2 FC) in expression of HSFs at the SAM of Col-0 wild-type plants at 0.5 h after priming and 0.5 h after triggering.
(C and D) Expression level of HSFA7a(C) and HSFA7b(D) at the SAM of Col-0 plants after priming and triggering treatments obtained by qRT–PCR.
(E and F) RNA in situ hybridization using HSFA7a(E) and HSFA7b(F) as probes on longitudinal sections through the meristems of control, primed, primed/triggered (PT), and triggered plants. Scale bars, 100 μm.
(G and H) Tissue-specific expression of HSFA7a(G) and HSFA7b(H) in 8-day-old control and PT plants at 0.5 h after triggering treatment analyzed by qRT–PCR. Error bars indicate SD (n = 3). Asterisks indicate statistically significant differences (Student’s t-test: ∗P ≤ 0.05; ∗∗P ≤ 0.01; ∗∗∗P ≤ 0.001) compared with control conditions. In (A), (C), and (D), the vertical dashed line represents the time point of the triggering (T) treatment. Time is given in hours (h) after priming (black color) and triggering (gray color) treatments.
HSFA7b is crucial for maintaining thermomemory
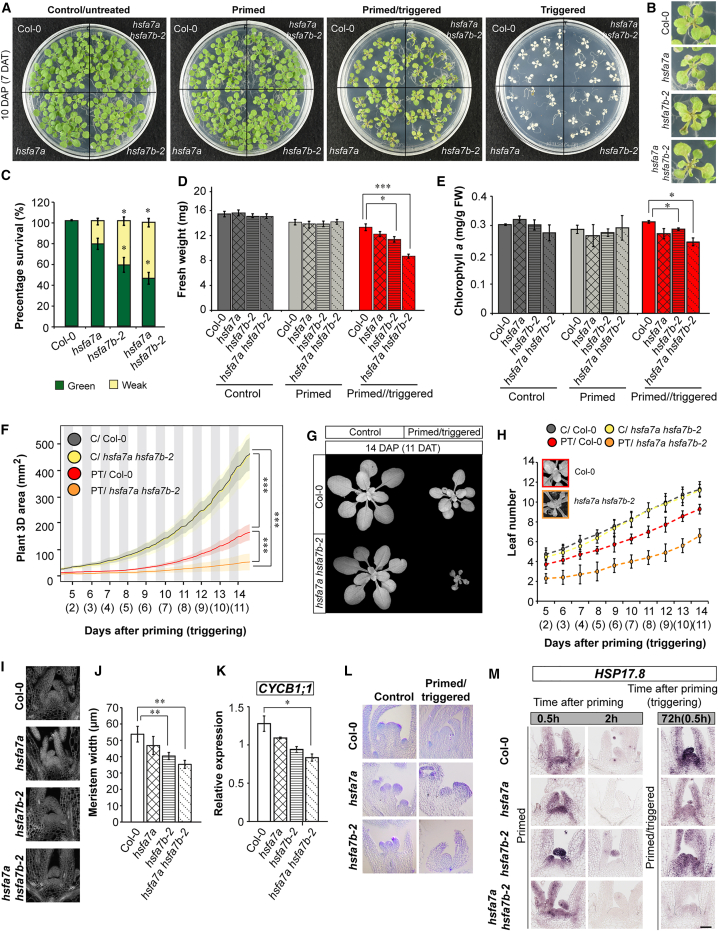
To test whether HSFA7a/b support HS memory, we subjected homozygous hsfa7a, two independent hsfa7b T-DNA insertion lines (hsfa7b-1 and hsfa7b-2), and hsfa7a hsfa7b-2 double mutants to the thermopriming assay (Figure 2A and 2B, Supplemental Figure 3). When Col-0 seedlings were subjected to a triggering HS in the absence of a prior priming stimulus, cotyledons bleached and no new leaves formed, even after an extended cultivation period, as reported previously (Sedaghatmehr et al., 2016; Olas et al., 2021a). By contrast, Col-0 PT seedlings established new leaves at the SAM after the triggering HS, and shoot development progressed (Sedaghatmehr et al., 2016; Olas et al., 2021a) (Figure 2A and 2B). Importantly, although growth of hsfa7a, hsfa7b-1, hsfa7b-2, hsfa7a hsfa7b-2, and Col-0 plants was similar in control and primed conditions, we observed impaired growth recovery, reduced survival, decreased biomass, and lower chlorophyll content after triggering HS in previously primed hsfa7a, hsfa7b-1, hsfa7b-2, and hsfa7a hsfa7b-2 plants compared with Col-0. These changes were most pronounced in hsfa7a hsfa7b-2 seedlings, clearly demonstrating defective HS memory in the double mutant (Figure 2A–2E, Supplemental Figure 4A and 4B). Of particular importance, however, is that both basal and acquired thermotolerance were unaltered in hsfa7a, hsfa7b-2, and hsfa7a hsfa7b-2 seedlings compared with the Col-0 wild type (Supplemental Figure 4C and 4D), providing evidence for a specific involvement of HSFA7a and HSFA7b in thermomemory rather than a general response to HS. Our conclusion that HSFA7b and HSFA7a are required for functional thermomemory was substantiated by high-resolution 3D imaging of dynamic plant growth (Apelt et al., 2015): the PT hsfa7a hsfa7b-2 mutant produced significantly smaller rosettes than Col-0 by the end of the imaging period (i.e., 11 days after triggering, DAT), together with a lower relative rosette expansion growth rate (Figure 2F, 2G, and Supplemental Figure 4E). Interestingly, we observed that the growth restriction of PT hsfa7a hsfa7b-2 mutants compared with Col-0 wild-type plants became more pronounced when the plants were grown in soil instead of Murashige and Skoog (MS) medium (Supplemental Figure 5). We also tested whether thermopriming induced cell death 24 h after triggering HS by performing Trypan Blue staining of cotyledons (Supplemental Figure 6A). Although only 29% of Col-0 PT plants displayed lesions, 44%, 43%, and 75% of hsfa7a and hsfa7b-2 single and hsfa7a hsfa7b-2 double mutants, respectively, showed dead cells (Supplemental Figure 6B), demonstrating that thermopriming triggers cell death in the absence of HSFA7a/b. In summary, the combined observations demonstrate that HSFA7a and HSFA7b are required for maintenance of thermomemory.
Figure 2.
HSFA7a and HSFA7b are required for thermomemory at the SAM.
(A and B) Growth recovery phenotypes of the control (C), primed (P), primed/triggered (PT), and triggered (T) Col-0 wild-type and the hsfa7a, hsfa7b-2, and hsfa7a hsfa7b-2 mutant plants grown on medium under a long-day (16-h light/8-h dark) photoperiod at 7 days after triggering (DAT; 10 days after priming [DAP]). Note, no new leaf formation was observed in the PT hsfa7a hsfa7b-2 double mutant (B) at that time.
(C) Percentage survival of seedlings in different phenotype classes analyzed at 7 DAT. “Green” indicates seedlings in which shoot regeneration continued and almost the entire plant was green; “weak” indicates seedlings in which shoot regeneration was weak and plants were mostly pale.
(D) Fresh weight of C, P, and PT Col-0, hsfa7a, hsfa7b-2, and hsfa7a hsfa7b-2 mutant plants analyzed at 7 DAT.
(E) Chlorophyll a content analyzed in C, P, and PT Col-0 and mutant plants at 10 DAT.
(F) Three-dimensional total plant area of C and PT Col-0 and hsfa7a hsfa7b-2 double-mutant plants grown in soil under a neutral photoperiod (neutral day, 12-h light/12-h dark) measured over time (n ≥ 6 for each condition). Lines and shaded areas represent mean and SD, respectively.
(G) Growth phenotype of C and PT Col-0 and hsfa7a hsfa7b-2 plants analyzed in (F) at 14 DAP (11 DAT).
(H) Leaf numbers produced by C and PT Col-0 and hsfa7a hsfa7b-2 double-mutant plants. The data were calculated from the plants analyzed in (F). Note that plants analyzed in (F) to (H) were transferred to soil 1 day after thermopriming and grown under neutral-day conditions.
(I) Calcofluor White–stained sections through meristems of PT Col-0, hsfa7a, hsfa7b-2, and hsfa7a hsfa7b-2 plants at 6 h after triggering.
(J) Meristem width of PT Col-0, hsfa7a, hsfa7b-2, and hsfa7a hsfa7b-2 plants at 6 h after triggering.
(K) Expression level of CYCLIN B1;1 (CYCB1;1) at the shoot apex of PT Col-0, hsfa7a, hsfa7b-2, and hsfa7a hsfa7b-2 at 0.5 h after triggering.
(L) Toluidine Blue–stained sections through meristems of 14-day-old C and PT Col-0, hsfa7a, and hsfa7b-2 mutant plants.
(M) RNA in situ hybridization using HEAT SHOCK PROTEIN 17.8 (HSP17.8) as a probe on longitudinal sections through meristems of P and PT Col-0, hsfa7a, hsfa7b-2, and hsfa7a hsfa7b-2 plants. Error bars in (C) to (E), (H), (J), and (K) represent SD (n = 3). Asterisks indicate statistically significant differences (Student’s t-test: ∗P ≤ 0.05; ∗∗P ≤ 0.01, ∗∗∗P ≤ 0.001) compared with Col-0 under the same conditions.
Meristem activity is more severely affected in the absence of functional HSFA7b
Next, to test whether meristem activity is altered in hsfa7 mutants compared with Col-0 wild-type plants in response to thermopriming, we analyzed the leaf initiation rate (LIR)—the read-out of vegetative meristem activity based on monitoring of new leaf emergence (Figure 2H). Analysis of LIR of double-mutant and Col-0 plants revealed that thermopriming affects new leaf appearance significantly more strongly in PT hsfa7a hsfa7b-2 than in PT Col-0 plants, whereas LIR did not differ between wild-type and mutant plants under control conditions, suggesting that HS alters meristem activity more severely in the absence of both HSFs and further indicating that HSFA7a and HSFA7b are essential components of SAM thermomemory. Therefore, we next tested whether the altered growth phenotype of hsfa7a hsfa7b-2 seedlings may be due to abnormalities at the SAM (Figure 2). As shown in Figure 2I and 2J, meristem size was significantly reduced in PT hsfa7b-2 single and hsfa7a hsfa7b-2 double mutants compared with PT Col-0 plants. Furthermore, qRT–PCR expression analysis of the cell cycle marker gene CYCLIN B1;1 (Olas et al., 2021b) at the shoot apex of C, P, and PT Col-0 and hsfa7a hsfa7b-2 mutant plants revealed that cell proliferation is significantly affected in the double mutant after triggering HS (Figure 2K; Supplemental Figure 6C and 6D), suggesting that temporal inhibition of meristem activity occurs in response to thermopriming. Because meristem function was more severely affected in the hsfa7b-2 mutant than in hsfa7a, we analyzed meristem development of 14-day-old (6 DAT) C and PT Col-0, hsfa7a, and hsfa7b-2 plants by Toluidine Blue staining (Figure 2L). Although meristems of Col-0, hsfa7a, and hsfa7b-2 plants were at the reproductive stage under control conditions, differences in SAM development became evident in PT plants. We observed that the meristems of PT Col-0 and hsfa7b-2 mutants remained in a vegetative state compared with control Col-0 and hsfa7b-2 plants. By contrast, no difference was observed between controls and PT hsfa7a mutants, suggesting that the temporal inhibition of meristem activity in response to thermopriming in the double mutant is due mainly to the lack of functional HSFA7b.
Last, we analyzed whether decreased meristem activity in hsfa7 mutants due to lack of functional HSFA7a and/or HSFA7b affects thermopriming capacity of the SAM. We analyzed expression of the HS memory marker HEAT SHOCK PROTEIN 17.8 (HSP17.8) (Olas et al., 2021a) by RNA in situ hybridization (Figure 2M). Expression of HSP17.8 was similar at the SAM of Col-0, hsfa7a, hsfa7b-2, and hsfa7a hsfa7b-2 plants after priming HS. By contrast, after triggering, expression of HSP17.8 in the SAM was reduced in both hsfa7 single-gene mutants and virtually absent in the hsfa7a hsfa7b-2 double mutant, demonstrating that both HSFs are important for establishing thermomemory at the SAM.
Transcriptome analysis of hsfa7a and hsfa7b-2 shoot apices
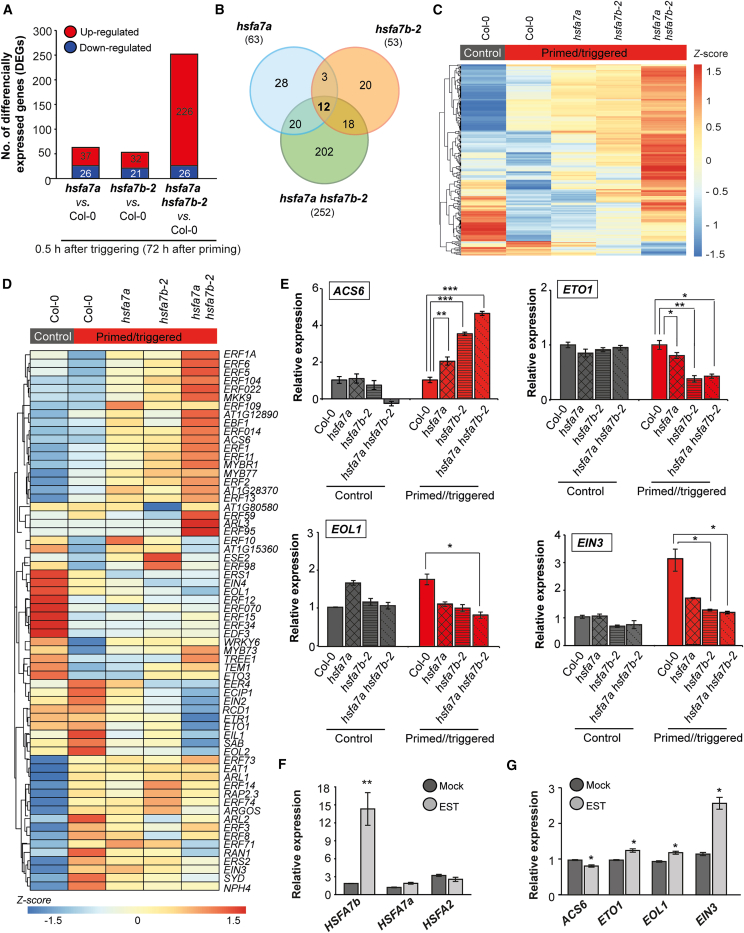
To determine the molecular basis of the phenotypic changes in the hsfa7a/b mutants, we performed RNA-seq of shoot apices from C, P, and PT Col-0 wild-type, hsfa7a, hsfa7b-2, and hsfa7a hsfa7b-2 plants at 0.5 h after priming and 0.5 h after triggering treatments (Supplemental Figure 7; Supplemental Data 1). In response to priming and triggering, several hundred genes were significantly differentially expressed between hsfa7a/b mutants and Col-0. To identify high-confidence genes, we used a fold-change (FC) criterion for gene expression of |FC| > 1.5 (Supplemental Table 1). Moreover, because hsfa7a/b mutants are impaired in HS memory but not in basal or acquired thermotolerance, we focused on differentially expressed genes (DEGs) observed after the triggering HS (Figure 3A). First, we generated a Venn diagram of the DEGs identified in hsfa7a, hsfa7b-2, and hsfa7a hsfa7b-2 (Figure 3B). The intersection of differentially expressed transcripts shared among all three mutants after triggering treatment included only 12 genes. In particular, 28 DEGs (of 63) were specific to hsfa7a, 20 (of 53) to hsfa7b-2, and 202 (of 252) to hsfa7a hsfa7b-2 after triggering HS. Next, because thermomemory was most strongly affected in the double mutant, we aimed to identify DEGs specific for the PT hsfa7a hsfa7b-2 double mutant (Supplemental Data 2). We generated a heatmap of the 202 genes (Figure 3C) affected only in hsfa7a hsfa7b-2 shoot apices after triggering HS. Clearly, the expression profiles of hsfa7a hsfa7b-2–specific genes differed strongly from those of Col-0 and hsfa7a and hsfa7b-2 single mutants, suggesting that the HSFA7a–HSFA7b complex controls a different set of genes in response to HS than do the individual HSFA7a/b proteins. Furthermore, several ethylene-related genes, including the ethylene biosynthesis–related genes 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE 6 (ACS6), ETHYLENE OVERPRODUCER 1 (ETO1), and ETO1-LIKE 1 (EOL1) and the ethylene signaling–related genes ETHYLENE-INSENSITIVE 2 and 3 (EIN2 and EIN3), EIN3-BINDING F BOX PROTEIN 1 (EBF1), and ETHYLENE-INSENSITIVE-LIKE 1 (EIL1), were differentially expressed between Col-0 and hsfa7a hsfa7b-2, indicating that the response to ethylene is affected by thermopriming at the SAM.
Figure 3.
Thermopriming affects expression of ethylene response genes at the shoot apical meristem (SAM) in an HSFA7b-dependent manner.
(A) Total number of differentially expressed genes (DEGs; false discovery rate <0.05 and |FC| > 1.5) at the shoot apices of primed and primed/triggered (PT) hsfa7a, hsfa7b-2, and hsfa7a hsfa7b-2 mutants compared with PT Col-0 wild-type plants, with numbers of up- (red) and downregulated (blue) genes.
(B) Venn diagrams of overlapping DEGs at 0.5 h after triggering in hsfa7a/b single and hsfa7a hsfa7b-2 double mutants compared with control Col-0 plants.
(C) Heatmap of 202 DEGs identified for the PT hsfa7a hsfa7b-2 double mutant and their relative expression levels (Z-score normalized).
(D) Heatmap depicting relative expression (Z-score normalized) of genes from the GO-term category “ethylene biosynthesis and response” in shoot apices of the Col-0 control and PT Col-0, hsfa7a, hsfa7b-2, and hsfa7a hsfa7b-2.
(E) Expression levels of selected ethylene-related genes analyzed by qRT–PCR at the SAM of control and PT Col-0, hsfa7a/b, and hsfa7a hsfa7b-2 plants at 0.5 h after triggering.
(F and G) Expression levels of (F)HEAT SHOCK TRANSCRIPTION FACTOR A7a (HSFA7a), HSFA7b, HSFA2, and (G) ethylene-related genes in β-estradiol (EST)- and mock-treated HSFA7b-IOE plants. Error bars represent SD (n = 3). Asterisks indicate statistically significant differences (Student’s t-test: ∗P ≤ 0.05, ∗∗P ≤ 0.01, and ∗∗∗P ≤ 0.001) compared with Col-0 under the same conditions (E) and compared with the mock treatment (F and G).
Thermopriming affects expression of ethylene response genes in an HSFA7b-dependent manner
The finding that expression of ethylene-related genes is affected by triggering HS in PT hsfa7a hsfa7b-2 plants prompted us to investigate whether ethylene response plays a role in HS memory at the SAM. Figure 3D shows that expression of ethylene biosynthesis and signaling genes clearly differed among the hsfa7a/b single mutants, the double mutants, and Col-0. The most pronounced changes were observed in hsfa7b-2 and hsfa7a hsfa7b-2, suggesting that expression of ethylene-related genes mostly requires HSFA7b, whereas HSFA7a has a weaker effect. We confirmed this by testing the expression of ACS6, ETO1, EIN2, EIN3, EBF1, and EIL1 in meristems of C and PT plants at 0.5 h after triggering (Figure 3E, Supplemental Figure 8). Notably, expression of ACS6 was upregulated at the SAM of PT hsfa7b-2 and hsfa7a hsfa7b-2 plants compared with untreated and PT Col-0 plants, suggesting that the lack of functional HSFA7b protein affects ethylene levels. Furthermore, expression of all tested genes involved in ethylene signaling was downregulated at the SAM of PT hsfa7b-2 and hsfa7a hsfa7b-2 plants compared with untreated and PT Col-0 plants, suggesting that ethylene signaling requires functional HSFA7b protein. Reduced expression of these genes after triggering HS was also observed in hsfa7a, although the effect was much weaker than that observed in hsfa7b-2 single and double mutants, indicating a more important effect of HSFA7b. To validate the role of HSFA7b in ethylene signaling, we generated a β-estradiol (EST)-inducible HSFA7b overexpression line (HSFA7b-IOE; Supplemental Figure 9A) and performed RNA-seq after EST induction and identified 1219 up- and 548 downregulated genes, respectively, compared with the mock treatment (Supplemental Figure 9B). A clustered heatmap of ethylene-related genes revealed their altered expression in EST-induced HSFA7b-IOE seedlings (Supplemental Figure 9C). The changes in expression of HSFA7b, ACS6, ETO1, EIN2, EBF1, EIN3, and EIL1 in HSFA7b-IOE seedlings were confirmed by qRT–PCR (Figure 3F, G, and Supplemental Figure 9D).
Collectively, our data suggest that HSFA7b, with a minor effect from HSFA7a, regulates expression of ethylene biosynthesis and signaling genes, and therefore ethylene response, at the SAM during thermomemory.
HSFA7b directly regulates ethylene biosynthesis and signaling
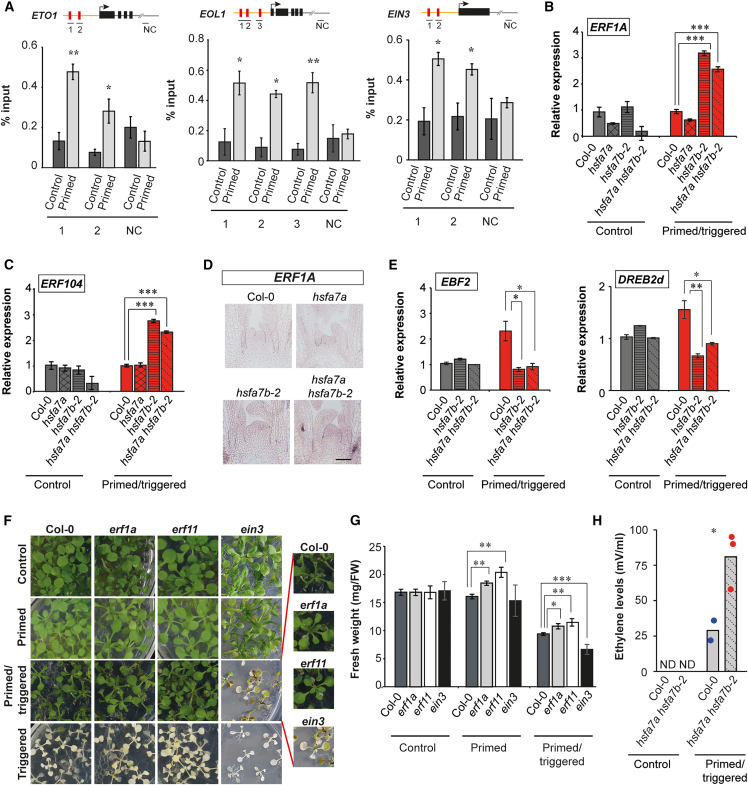
The results above support a model in which HSFA7b controls ethylene response at the SAM during thermopriming by regulating expression of ethylene signaling genes as well as ethylene biosynthesis genes. Analysis of the promoters of several ethylene-related genes revealed the presence of putative HSEs, suggesting that HSFs directly regulate their expression. Therefore, to confirm the transcriptional regulation of ethylene-related genes by HSFA7b, we generated a pHSF7b:HSFA7b-GFP transgenic line (Supplemental Figure 10) and performed chromatin immunoprecipitation (ChIP)–qPCR analysis on selected target genes involved in controlling ethylene biosynthesis (ETO1 and EOL1) and ethylene sensing and signaling (EIN3) at 0.5 h after priming HS and in C seedlings (Figure 4A). Binding of HSFA7b was highly enriched at the EOL1, ETO1, and EIN3 promoters after priming HS, demonstrating direct regulation of these genes, and thus the ethylene response, during thermopriming.
Figure 4.
HSFA7b controls ethylene response at the shoot apical meristem (SAM) during thermomemory.
(A) Binding of HSFA7b to the promoter regions of ETHYLENE OVERPRODUCER 1 (ETO1), ETO1-LIKE 1 (EOL1), and ETHYLENE-INSENSITIVE 3 (EIN3) under primed conditions compared with control, determined by ChIP–qPCR. The y axis represents the relative enrichment compared with the input (in %), and the x axis represents the genomic regions depicted in the schematics. The schematic representation of each gene depicts the regions analyzed by ChIP–qPCR (arrow, TSS; yellow line, promoter; black boxes, exons; red boxes, heat shock elements; gray line, 3′ UTR; NC; negative control).
(B and C) Expression level of ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 (ERF1A) and ETHYLENE RESPONSE FACTOR 104 (ERF104) genes analyzed by qRT–PCR at the SAM of control (C) and primed/triggered (PT) Col-0, hsfa7a/b, and hsfa7a hsfa7b-2 plants at 0.5 h after triggering.
(D) RNA in situ hybridization using an ERF1A-specific probe on longitudinal sections through meristems of PT Col-0, hsfa7a/b, and hsfa7a hsfa7b-2 plants. Scale bar, 100 μm.
(E) Expression of EIN3-BINDING F BOX PROTEIN 2 (EBF2) and DREB2d at the SAM of C and PT Col-0, hsfa7b-2, and hsfa7a hsfa7b-2 mutant plants at 0.5 h after triggering.
(F) Phenotypes of C, primed (P), PT, and triggered (T) Col-0, erf1a, erf11, and ein3 mutant plants. Images were taken 10 days after triggering.
(G) Fresh weights of C, P, and PT Col-0, erf1a, erf11, and ein3 mutant plants. n = 20.
(H) Ethylene levels in C and PT Col-0 and hsfa7a hsfa7b-2 mutant plants at 7 days after triggering. Bars show mean values, and colored dots represent individual replicates. Note that ethylene levels were not detected (ND) in C Col-0 and hsfa7a hsfa7b-2 plants, and in one PT Col-0 replicate. The experiment was performed once using three biological replicates. Asterisks indicate statistically significant differences compared with the control (Student’s t-test: ∗P ≤ 0.05, ∗∗P ≤ 0.01, and ∗∗∗P ≤ 0.001).
To confirm that the ethylene response is altered in the absence of HSFA7b at the SAM, we analyzed the expression of downstream transcriptional regulatory components of the ethylene response, i.e., ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1A (ERF1A), ETHYLENE RESPONSIVE FACTOR 104 (ERF104), and ERF11 by qRT–PCR (Figure 4B, C, and Supplemental Figure 11A). Expression of ERFs was higher at the shoot apex of PT hsfa7b-2 and hsfa7a hsfa7b-2 plants compared with that of untreated and PT Col-0 plants, demonstrating that the ethylene response is affected in the absence of functional HSFA7b. We also performed RNA in situ hybridization for ERF1A (Figure 4D). After triggering HS, ERF1A was weakly expressed at the SAM of PT Col-0 plants, but its expression increased in hsfa7b-2 and hsfa7a hsfa7b-2 mutants, compared with Col-0 and hsfa7a, further indicating that HSFA7b is involved in control of the ethylene response at the SAM in response to thermopriming. Next, because HSFA7b physically binds to HSEs of the EIN3 promoter, and given that EIN3 is the master regulator of the transcriptional control of ethylene-related genes (Potuschak et al., 2003), we tested whether the absence of functional HSFA7b protein affects expression of EIN3 direct targets (Cheng et al., 2013) (Supplemental Figure 11B). To this end, we generated a heatmap that illustrates the differential expression of ethylene-related direct targets of EIN3 in C and PT Col-0 and hsfa7b-2 plants. This revealed substantial differences in the expression of these genes between Col-0 and mutant plants, suggesting that they are targets of the HSFA7b–EIN3 transcriptional cascade. To confirm this observation, we analyzed the expression of two selected genes, EBF2 and DREB2d, by qRT–PCR (Figure 4E). Expression of both genes was upregulated at the SAM of PT Col-0 plants compared with C seedlings (Col-0) at 0.5 h after triggering HS. Interestingly, their transcriptional activation in response to triggering HS was abolished in PT hsfa7b-2 single and hsfa7a hsfa7b-2 double mutants, confirming that the activation of some of EIN3 direct targets during HS requires functional HSFA7b protein.
Next, to investigate whether changes in the ethylene response affect thermomemory, we subjected the ethylene signaling and response mutants erf1a, erf11, and ein3 to the thermomemory assay (Figure 4F). The erf1a and erf11 mutants exhibited a better recovery after priming plus triggering HS (PT) than Col-0 (Figure 4F and 4G). Conversely, the PT ein3 mutant exhibited a weaker growth recovery (Figure 4F and 4G), confirming that components of the ethylene response pathway are functional elements of HS memory.
The finding that HSFA7b directly regulates expression of ETO1 and EOL1 genes in response to thermopriming and the fact that proteins encoded by these genes negatively regulate ethylene biosynthesis through the inhibition of ACS enzymes (Yoshida et al., 2005) suggest that ethylene levels may accumulate in Col-0 plants after triggering HS. This also indicates that HSFA7b plays a role in regulating these levels. Consequently, we measured ethylene levels in C and PT Col-0 and hsfa7a hsfa7b-2 plants at 7 days after triggering and observed an increased ethylene level in PT Col-0 plants (Figure 4H). Importantly, the ethylene level was higher in the PT hsfa7a hsfa7b-2 double mutant than in Col-0, consistent with our observation that HSFA7b modulates ethylene biosynthesis during thermopriming.
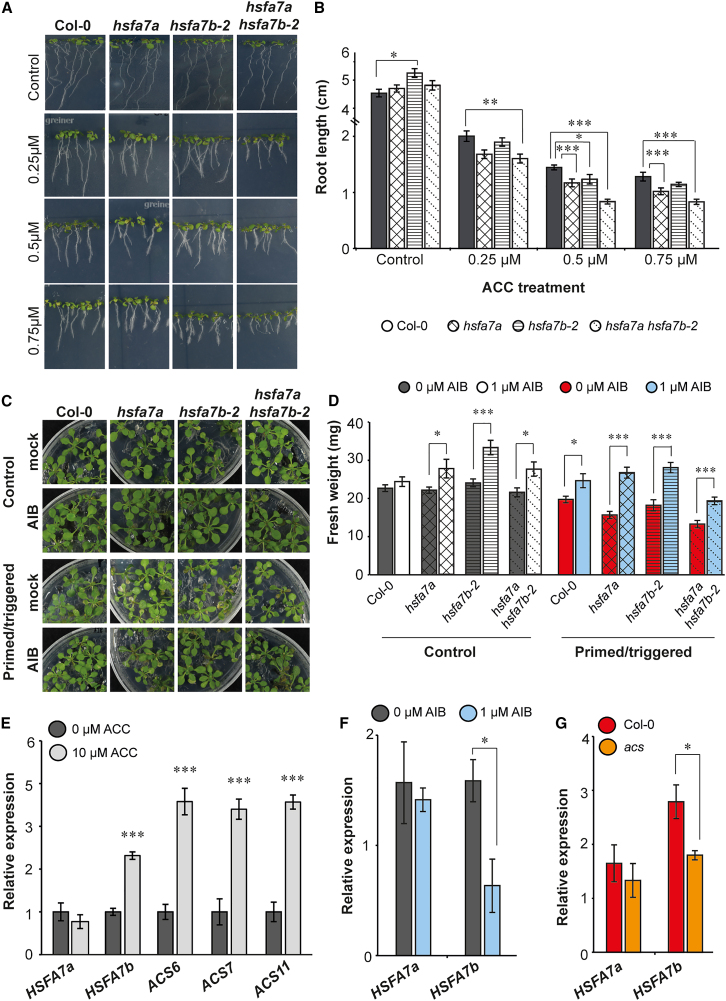
This result prompted us to test whether changes in ethylene level alter molecular and/or morphological phenotypes of hsfa7b-2 and hsfa7a hsfa7b-2 mutants. To this end, we grew Col-0, hsfa7a, hsfa7b-2, and hsfa7a hsfa7b-2 seedlings in the presence or absence of the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) (Figure 5A). Without ACC, all plants grew normally, and with ACC, root growth was reduced in all plants in a dose-dependent manner (Figure 5A and 5B). All mutants showed a stronger response to ACC treatment than did Col-0, and the greatest effect was found in hsfa7a hsfa7b-2, suggesting that lack of HSFA7a/b increases the sensitivity of plants to ethylene (Figure 5A and 5B). Given that ACC also serves as a signaling molecule, we grew Col-0 and hsfa7a/b mutant plants on medium supplemented with the ethylene biosynthesis inhibitor α-aminoisobutyric acid (AIB), which blocks ACC oxidase, the final enzyme in the ethylene biosynthesis pathway (Merritt et al., 2001). We then performed the thermopriming assay to determine how ethylene itself influences the thermopriming capacity of Arabidopsis plants. We observed a notable improvement in the recovery of the PT hsfa7a and hsfa7b-2 single and double mutants compared with PT plants grown without AIB (Figure 5C and 5D). This finding indicates that the inhibitor effectively counteracts the negative effects of ethylene on mutant growth. Furthermore, our results demonstrated that by blocking ethylene biosynthesis, we significantly enhanced the growth and recovery of PT Col-0 plants (Figure 5C and 5D), providing evidence that ethylene indeed contributes to the reduced growth observed in PT plants in response to triggering HS. Next, we tested whether expression of HSFA7a and HSFA7b is affected by ACC or AIB treatments (Figure 5E and 5F). We found that expression of HSFA7b, but not HSFA7a, is induced at the shoot apex in response to ACC treatment (Figure 5E), whereas its expression is significantly suppressed when AIB is present in the medium (Figure 5F). This result suggests that ethylene modulates HSFA7b expression at the SAM. To confirm that transcriptional induction of HSFA7b in response to thermopriming requires ethylene biosynthesis, we analyzed expression of HSFA7b and HSFA7a in PT Col-0 and octuple acs mutant plants (Figure 5G). We found that expression of HSFA7b, but not HSFA7a, is significantly compromised in the acs mutant compared with Col-0 after triggering HS, indicating that ACC/ethylene positively influences HSFA7b transcription in response to HS. Taken together, our data demonstrate that HSFA7b controls ethylene homeostasis in response to thermopriming at the SAM of Arabidopsis (Figure 6), suggesting a tissue-specific role of HSFA7b.
Figure 5.
Ethylene content modulates expression of HSFA7b.
(A) Col-0 and hsfa7a and hsfa7b-2 single- and double-mutant seedlings grown on 0.5 MS medium supplemented with or without 0.25, 0.5, and 0.75 μM of the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC). Images were taken at 7 days after germination.
(B) Root length of Col-0 and mutant plants grown on increasing concentrations of ACC.
(C) Shoot phenotypes of control (C) and primed/triggered (PT) Col-0, hsfa7a, hsfa7b-2, and hsfa7a hsfa7b-2 seedlings grown on 0.5 MS medium supplemented with or without 1 μM of the ethylene inhibitor α-aminoisobutyric acid (AIB). Images were taken at 7 days after triggering (DAT).
(D) Fresh weight of C and PT Col-0 and mutant plants grown on media supplemented with or without AIB at 7 DAT.
(E) Expression level of HEAT SHOCK TRANSCRIPTION FACTOR A7a (HSFA7a), HSFA7b, and 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE 6, 7, and 11 (ACS6, ACS7, ACS11) genes at the SAM of plants grown on medium with or without ACC.
(F) Transcript levels of HSFA7a and HSFA7b in plants grown on medium with or without the inhibitor AIB.
(G) Expression of HSFA7a and HSFA7b at the SAM of PT Col-0 and acs mutant plants at 0.5 h after triggering. Error bars represent SD (n = 3). Asterisks indicate statistically significant differences compared with the control (Student’s t-test: ∗P ≤ 0.05, ∗∗P ≤ 0.01, and ∗∗∗P ≤ 0.001).
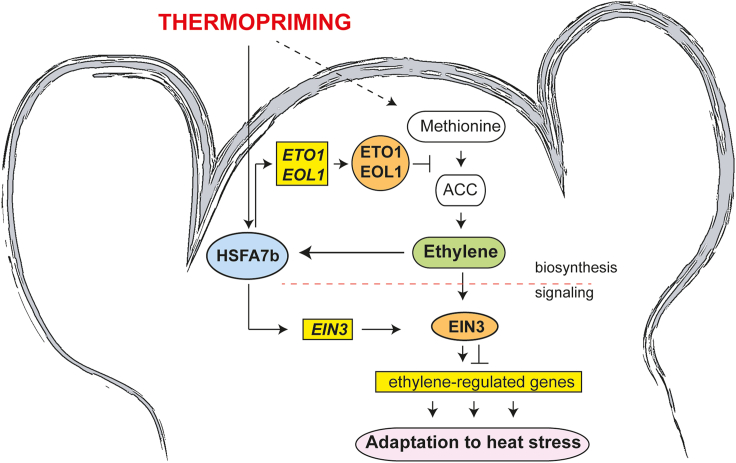
Figure 6.
Schematic model showing regulation of the ethylene response by HSFA7b at the shoot apex during thermomemory.
HSFA7b controls the ethylene response by binding to HSEs in the promoter regions of negative regulators of ethylene biosynthesis genes, including ETHYLENE OVERPRODUCER 1 (ETO1) and ETO1-LIKE 1 (EOL1). The direct transcriptional activation of ETO1 and EOL1 by HSFA7b leads to downregulated transcription of 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE genes at the SAM, most likely resulting in inhibition of ethylene production. The increased ethylene content during thermopriming modulates the expression of HSFA7b. Furthermore, HSFA7b directly regulates expression of the ethylene signaling gene ETHYLENE-INSENSITIVE 3 (EIN3) to activate a transcriptional HS cascade and establish thermotolerance at the SAM. Yellow rectangular boxes indicate genes, blue ovals indicate proteins, and pink rectangles with rounded ends indicate cellular processes. Solid lines, direct interactions; dashed lines, indirect interactions.
Discussion
The global temperature increase poses a serious threat to agriculture. Recent research suggests that climate change triggers mismatches between above- and belowground plant phenology and that shoots and roots respond differently to environmental input (Liu et al., 2022). It has therefore become imperative to study tissue-specific responses to HS in order to improve crop thermotolerance.
Here, we show that SAM-expressed HSFA7a and HSFA7b generate HS memory in Arabidopsis seedlings: hsfa7b-2 and hsfa7a hsfa7b-2 mutants have reduced thermomemory compared with wild-type plants, whereas their basal and acquired thermotolerance are unaffected. The HS memory of the hsfa7a hsfa7b-2 SAM is strongly impaired, as evidenced by reduced expression of the memory gene HSP17.8 after triggering HS. The main role of molecular chaperone HSPs is to protect plants from the devastating effects imposed by the stressor (here, HS) (Wang et al., 2004).
A key finding reported here is that HSFA7b controls ethylene response at the SAM during thermopriming, whereas HSFA7a has a weaker effect on the expression of ethylene-related genes. For example, the ethylene response genes ERF1A and ERF11 are strongly upregulated only at the SAM of hsfa7b-2 and hsfa7a hsfa7b-2 mutants after triggering HS. ERF11 negatively regulates abiotic stress tolerance in plants (Young et al., 2004; Habben et al., 2014). Accordingly, erf11 seedlings display better growth recovery than Col-0 after priming/triggering HS.
Direct regulation of the ethylene response at the SAM by HSFA7b was supported by ChIP–qPCR analyses, which revealed direct interaction with EIN3, a key transcriptional regulator of ethylene signaling. Previous research has shown that EIN3 binds to the ERF95 and ERF97 promoters to mediate thermotolerance (Huang et al., 2021). Thus, HSFA7b controls HS memory by affecting ethylene signaling through binding to the EIN3 promoter to activate an ERF transcriptional cascade at the SAM. In agreement with this scenario, our analysis revealed that expression of EIN3 is strongly reduced at the SAM of hsfa7b-2 single and hsfa7a/b double mutants. Furthermore, transcripts of EIN3 direct targets (Cheng et al., 2013) are strongly affected at the SAM of PT hsfa7b-2 mutants compared with PT Col-0 plants, indicating that activation of these genes during HS requires functional HSFA7b protein. Furthermore, we found that the ein3 mutant is impaired in thermomemory, confirming that EIN3 is an important component of HS memory at the SAM. Interestingly, previous research demonstrated that EIN3 is specifically expressed in the organizing center of the SAM (Zeng et al., 2021). Furthermore, EIN3 overexpression plants showed increased expression of WUS and CLV3, whereas ein3eil2eil3 mutants showed reduced SAM size and lower WUS and CLV3 expression (Zeng et al., 2021), demonstrating that endogenous ethylene enriched at the SAM is crucial for stem cell function. Our results support a model in which HSFA7b controls EIN3 at the SAM to maintain stem cell integrity during HS.
Importantly, our data show that HSFA7b also regulates ethylene response by controlling ethylene biosynthesis. ChIP–qPCR and transcriptome analysis demonstrated that HSFA7b directly controls the expression of ETO1 and EOL1, whose products negatively regulate ethylene biosynthesis by inhibiting ACS enzymes, thereby controlling ethylene production (Yoshida et al., 2005). In fact, our data show that ethylene production is increased in the absence of HSFA7b after PT treatment, as revealed by the reduced expression of ETO1 and EOL1 and the corresponding increase in expression of the ethylene biosynthesis gene ACS6 after PT in hsfa7a hsfa7b-2 plants compared with Col-0. The reduced growth recovery after PT in the hsfa7a hsfa7b-2 double mutant and the hypersensitivity to ACC treatment are attributable to increased ethylene levels in the mutant. Consistent with this notion, Col-0 and hsfa7a/b mutants exhibit improved growth recovery after triggering HS when grown in the presence of the ethylene inhibitor AIB. Interestingly, we found that expression of HSFA7b, but not HSFA7a, is mediated by ethylene during HS, as evidenced by reduced expression of HSFA7b in plants treated with AIB or in PT acs mutants. Previous studies have reported an accumulation of ethylene after HS (Ketsa et al., 1999; Hays et al., 2007; Wu and Yang, 2019). Increased ethylene production under HS is correlated with accelerated leaf senescence (Iqbal et al., 2017). In accordance with this observation, we found that chlorophyll breakdown, senescence activation, and programmed cell death occur more rapidly in the double mutant than in the Col-0 wild type after triggering HS. Furthermore, a previous study showed that ethylene mediates various growth and developmental processes in plants, including flowering time and fruit development, under non-stress conditions mainly through crosstalk with other phytohormone signaling pathways (Iqbal et al., 2017). Increased physiological and morphological abnormalities observed in hsfa7b-2 mutants compared with Col-0 wild-type plants were linked to temporal changes in meristem activity. Furthermore, the reduced expression of cell cycle marker genes and smaller meristem size in hsfa7b-2 mutants after triggering HS support the previous finding that ethylene negatively regulates cell proliferation at the SAM (Street et al., 2015).
Overall, our study suggests that ethylene regulation via HSFA7b at the SAM functions through canonical ethylene signaling and biosynthesis pathways (Figure 6).
Methods
Plant material and growth conditions
Arabidopsis Col-0 seedlings were grown on 0.5 MS medium with 1% sucrose under long-day (16-h light/8-h dark) conditions at 22°C with 160 μmol m−2 s−1 of photosynthetically active radiation in a controlled growth chamber (Fitotron SCG 120, Weiss Technik, Loughborough, UK). For 3D growth analysis (see below) plants were grown under neutral-day conditions (12-h light/12-h dark). Thermomemory, basal thermotolerance, and acquisition of thermotolerance were assayed as described previously (Stief et al., 2014; Olas et al., 2021a). In brief, 5-day-old seedlings were subjected to the priming treatment (1.5 h at 37°C, 1.5 h at 22°C, 45 min at 44°C) at 6 h after dawn, followed by a recovery/memory phase at 22°C for 3 days and a triggering stimulus (1.5 h at 44°C) at 9 h after dawn. The hsfa7a (SALK_080138), hsfa7b-1 (GK-498E08), hsfa7b-2 (SALK_152004), acs (acs8 acs11 acs2-1 acs4-1 acs5-2 acs6-1 acs7-1 acs9-1), erf1a (SALK_036267), erf11 (SALK_116053), and ein3 (N_8052) mutants were reported previously (Charng et al., 2007; Tsuchisaka et al., 2009; Dubois et al., 2015) and were obtained from the Nottingham Arabidopsis Stock Centre collection. Homozygous lines were confirmed by PCR, and the primer sequences for genotyping are listed in Supplemental Table 2. The hsfa7a hsfa7b-2 double mutant was generated by crossing the homozygous hsfa7a and hsfa7b-2 single mutants. To generate the HSFA7b complementation line (pHSFA7b:HSFA7b-GFP), a modified pGreen0229 plant transformation vector containing a C-terminal GFP tag was used (Hellens et al., 2000). The HSFA7b open reading frame (without the stop codon) and its 2-kb promoter region were amplified by PCR from Arabidopsis Col-0 leaf genomic DNA. To generate the EST-inducible HSFA7b overexpression line (HSFA7b-IOE), the HSFA7b coding sequence was cloned into the pER8 vector (Zuo et al., 2000) downstream of the EST-inducible promoter.
Determination of plant size, RER, and fresh weight
Plants were classified into two phenotypic classes based on their recovery phenotype at 7 days after priming/triggering treatment. Seedlings in which shoot regeneration continued and the shoot remained green were classified as “green,” and seedlings with weak shoot regeneration and a mostly pale appearance were classified as “weak.” Fresh weights of rosettes from C, P, and PT Col-0 and mutant plants were measured 7 days after the triggering treatment.
Plant 3D rosette area and relative expansion growth rate (RER) of Col-0 wild-type (n = 10 for C; n = 6 for PT) and hsfa7a hsfa7b-2 mutant plants (n = 12 for C; n = 6 for PT) were analyzed using an established 3D camera-based imaging system (Apelt et al., 2015). Plants were transferred to soil 1 day after triggering and continuously imaged in a growth chamber (model E-36L; Percival Scientific) for several days as described previously (Olas et al., 2021a).
Chlorophyll measurement
For chlorophyll measurement, whole rosettes were harvested and analyzed as described previously (Olas et al., 2021b). Assays were performed in 96-well plates, and absorbances at 645 and 665 nm were determined using a Synergy microplate reader (Bio-Tek). For all assays, two technical replicates were measured per biological replicate.
RNA in situ hybridization
For RNA in situ hybridization experiments, meristems of plants grown under long-day conditions were harvested into freshly prepared formaldehyde/acetic acid/alcohol (FAA) fixative solution after different time points of the thermomemory assay. The samples were then transferred to embedding cassettes and fixed overnight using an automated tissue processor (Leica ASP200S, Wetzlar, Germany). After fixation, the samples were embedded in paraffin wax using an embedding system (HistoCore Arcadia, Leica, Wetzlar, Deutschland). Longitudinal sections through the apices (8-μm thickness) were prepared using a rotary microtome (Leica RM2255). Slides were stored until use for RNA in situ hybridization.
RNA in situ hybridization was performed as described previously (Olas et al., 2019). To compare changes in gene expression, slides containing longitudinal sections through meristems of Col-0 and mutant plants were processed at the same time to eliminate variations in the duration of probe hybridization and signal development before imaging.
Trypan Blue, Toluidine Blue, and Calcofluor White staining
Trypan Blue staining was performed 24 h after triggering treatment as reported previously (Fernández-Bautista et al., 2016). In brief, seedlings of Col-0 and mutant plants were incubated in 0.4 mg/ml Trypan Blue, dissolved in phenol/glycerol/lactic acid/water/ethanol (1:2:1:1:8) at room temperature for 30 min, and then de-stained by washing three times in 90% EtOH.
For Toluidine Blue and Calcofluor White staining, the sections were incubated in Histo-Clear and processed through an ethanol series. The slides were stained with 0.01% Toluidine Blue/sodium borate solution or Calcofluor White solution, respectively.
qRT–PCR and RNA-seq
Shoot and root apices, cotyledons, and whole rosettes of Arabidopsis plants were collected during and after thermopriming treatment to study gene expression. Total RNA was isolated using the TRIzol method (Ambion/Life Technologies, Darmstadt, Germany). DNA digestion and cDNA synthesis were performed using the Turbo DNA-free DNase I kit (Ambion/Life Technologies, Darmstadt, Germany) and the RevertAid H minus reverse transcriptase kit (ThermoFisher Scientific, Darmstadt, Germany), respectively. The qRT–PCR measurements were performed using SYBR Green-PCR Master Mix (Applied Biosystems/Life Technologies, Darmstadt, Germany). Relative expression of each gene was analyzed using the comparative cycle threshold method (Livak and Schmittgen, 2001) with TUBULIN2 (TUB2) as the reference gene. Primer sequences are listed in Supplemental Table 2.
For RNA-seq, shoot apices of Col-0, hsfa7a, hsfa7b-2, and hsfa7a hsfa7b-2 were dissected in three biological replicates at 0.5 h after the priming treatment and 0.5 h after the triggering treatment. In addition, mock- and EST-treated HSFA7b-IOE seedlings were harvested for transcriptome analysis (n = 3). Total RNA was isolated using the mirVana RNA isolation kit (Ambion/Life Technologies, Darmstadt, Germany) according to the manufacturer’s protocol. Library preparation and sequencing to generate paired-end reads (2 × 100 base pairs) were performed by BGI Tech Solutions Co., Ltd (Hong Kong, China).
STAR (version 2.5.2b) (Dobin et al., 2013) was used to align the reads to the Arabidopsis reference genome (Arabidopsis thaliana, TAIR10). The aligned reads were quantified using HTSeq (version 0.9.1) (Anders et al., 2015). For details of the libraries, read numbers, and alignments, see Supplemental Data 1. Genome annotations were obtained from Araport11, RepTAS, and miRbase (Liu et al., 2012; Cheng et al., 2017; Kozomara et al., 2019). Differential gene expression analysis was performed with DESeq2 (1.20.0) (Love et al., 2014) using criteria of false discovery rate <0.05 and |FC| > 1.5 for DEGs. Detailed pairwise comparisons for DEGs are provided in Supplemental Data 2. Heatmaps were constructed from normalized expression values generated by applying variance stabilizing transformation using DESeq2. Gene Ontology (GO) analysis was performed using PANTHER 16.0 (Gene List Analysis, http://pantherdb.org/). Genes involved in ethylene response were manually selected from The Arabidopsis Information Resource website (TAIR).
Yeast two-hybrid assay
For bait construction, the coding sequence of HSFA7b was cloned into the pDEST32 vector (containing the GAL4 binding domain) using GATEWAY cloning. The positive clone was transformed into the yeast strain pJ684α. For prey construction, the coding sequence of the functional HSFA7b was cloned into the pDEST22 vector (containing the GAL4 activation domain) using GATEWAY cloning. The positive clone was transformed into the yeast strain YM4271a. The prey with the HSFA7a protein was obtained from a TF library containing approximately 1200 Arabidopsis TFs established in vector pDEST222 in yeast strain YM4271a. To test for interactions, yeast cells were mated in the following combinations: BD-HSFA7b–AD-HSFA7b and BD-HSFA7b–AD-HSFA7a. The empty GAL4 BD bait vector (pDEST32) and GAL4 AD prey vector (pDEST22) combination was used as the negative control. After mating for 3 days, the cells were checked for positive interactions by plating on the following selection media: SD-Leu-Trp (mating control) and SD-Leu-Trp-His + 3AT (3-amino-1,2,4-triazole), where HIS3 is the reporter gene and 3AT is a competitive inhibitor of the histidine biosynthesis enzyme encoded by HIS3.
ChIP–qPCR
Five-day-old control and primed pHSFA7b:HSFA7b-GFP seedlings were used for ChIP–qPCR experiments as reported previously (Olas et al., 2021a). In brief, 1 g fresh weight of seedlings was harvested within 0.5 h after the priming treatment. Crosslinking was performed by vacuum infiltration (approximately −950 mbars) for 20 min, and ChIP–qPCR was performed using the Diagenode Universal Plant kit (Diagenode, Seraing, Belgium) according to the manufacturer’s protocol. The sheared chromatin was immunoprecipitated using anti-GFP antibody (Abcam 290), with no-antibody as a control. Three biological replicates were used for each ChIP reaction.
The promoter regions (1-kb upstream of the TSS) of putative HSFA7b target genes were analyzed for the presence of HSEs using the online tool Plant Promoter Analysis Navigator 3.0 (Plant PAN 3.0) (Chow et al., 2019). Primers amplifying 100- to 200-base pair regions were used for qPCR with immunoprecipitated chromatin as the template. Primer sequences are listed in Supplemental Table 2.
Estradiol, ACC, and AIB treatments
For β-estradiol treatment, 10-day-old HSFA7b-IOE seedlings were incubated for 16 h with 10 μM β-estradiol or ethanol (0.1%, v/v; mock treatment). After incubation, seedlings were harvested for transcriptome analysis.
To check the sensitivity of Col-0 and hsfa7a/b mutants to ACC, seedlings were grown on 0.5 MS medium supplemented with or without different concentrations of ACC (0.25, 0.5, and 0.75 μM). Images were taken 7 days after germination.
To examine the impact of ethylene on thermomemory, Col-0 and hsfa7a and hsfa7b-2 single- and double-mutant seeds were sown on 0.5 MS medium supplemented with an ethylene inhibitor, 1 μM α-aminoisobutyric acid (AIB). Seeds grown on 0.5 MS medium without AIB were treated as controls. The thermomemory assay was performed as described previously. Images were taken 7 days after triggering.
To check the expression of HSFA7a and HSFA7b in response to ACC and AIB treatments, Col-0 seedlings were grown on 0.5 MS medium with or without 10 μM ACC or 1 μM AIB for 10 days.
Ethylene measurement
Ethylene levels were determined using a GC-2025 capillary gas chromatograph (Shimadzu, Germany) in both control and PT Col-0 and hasfa7a hsfa7b-2 double-mutant plants. Plants were grown on plates containing 0.5 MS medium and subjected to the thermopriming assay. Prior to the triggering treatment, the plate covers were replaced with lids containing a small hole sealed with tape to enable sampling of the plate headspace. Seven days after triggering, 1 ml of headspace sample was collected with a GC-2025 injection needle and injected into the GC-2025 SPL injection unit preset at 200°C and 93.3 kPa. A Shimadzu SH-Rt-Alumina BOND/KCl column was used to separate the gases. Data were collected for 1.75 min with an FID detector set at 200°C. Under these conditions, ethylene was detected at a retention time of 1.53 min. Peak analysis was performed with LabSolutions software (Shimadzu), including the i-PeakFinder algorithm that automatically detected and quantified heights of the ethylene peaks. The thermopriming experiment was performed once with three independent biological replicates.
Statistical analysis
Statistical significance was calculated using Student’s t-test: ∗P ≤ 0.05; ∗∗P ≤ 0.01; ∗∗∗P ≤ 0.001.
Data and code availability
Sequencing data are available at the NCBI Sequence Read Archive (SRA) under BioProject ID PRJNA877651.
AGI codes
HSFA7a, AT3G51910; HSFA7b, AT3G63350; ERF1A, AT4G17500; EIN3, AT3G20770; TUB2, AT5G62690; ETO1, AT3G51770. Additional AGI codes are provided in the supplementary tables.
Funding
B.M.-R. thanks the DFG for funding Collaborative Research Centre 973 “Priming and Memory of Organismic Responses to Stress” (www.sfb973.de), the European Union’s Horizon 2020 Research and Innovation Programme for funding project PlantaSYST (SGA-CSA No. 739582 under FPA No. 664620), the European Regional Development Fund for funding project BG05M2OP001-1.003-001-C01 through the Bulgarian “Science and Education for Smart Growth” Operational Programme, and the MPI-MP and the University of Potsdam for financial support. B.M.-R. and S.J. thank the International Max Planck Research School “Primary Metabolism and Plant Growth” (IMPRS-PMPG) for support. J.J.O. thanks the DFG (OL 767/1-1) and Leibniz Institute of Vegetable and Ornamental Crops for funding.
Author contributions
J.J.O. and B.M.-R. conceived the study and designed the experiments. S.J. conducted experiments with help from J.J.O. F.A. performed RNA-seq analysis with contributions from A.K. F.A. and D.B. measured plant 3D growth. I.F.A. measured ethylene levels. M.G.A. measured chlorophyll content. F.F. generated the pHSFA7b::HSFA7b:GFP transgenic line. C.G. provided infrastructure for I.F.A. and advice for set-up of the ethylene assay. J.J.O. wrote the manuscript with contributions from S.J. and B.M.-R. All authors read and commented on the manuscript before submission.
Acknowledgments
We thank Eike Kamann for cloning and technical assistance and Sarah Richard and Philip Cieslak for general lab work. The authors declare no competing interests.
Published: November 2, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Bernd Mueller-Roeber, Email: bmr@uni-potsdam.de.
Justyna J. Olas, Email: olas@igzev.de.
Supplemental information
References
- Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apelt F., Breuer D., Nikoloski Z., Stitt M., Kragler F. Phytotyping4D: a light-field imaging system for non-invasive and accurate monitoring of spatio-temporal plant growth. Plant J. 2015;82:693–706. doi: 10.1111/tpj.12833. [DOI] [PubMed] [Google Scholar]
- Balazadeh S. A ‘hot’cocktail: The multiple layers of thermomemory in plants. Curr. Opin. Plant Biol. 2022;65 doi: 10.1016/j.pbi.2021.102147. [DOI] [PubMed] [Google Scholar]
- Charng Y.-y., Liu H.-c., Liu N.-y., Chi W.-t., Wang C.-n., Chang S.-h., Wang T.-t. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 2007;143:251–262. doi: 10.1104/pp.106.091322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.Y., Krishnakumar V., Chan A.P., Thibaud-Nissen F., Schobel S., Town C.D. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017;89:789–804. doi: 10.1111/tpj.13415. [DOI] [PubMed] [Google Scholar]
- Cheng M.-C., Liao P.-M., Kuo W.-W., Lin T.-P. The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 2013;162:1566–1582. doi: 10.1104/pp.113.221911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C.-N., Lee T.-Y., Hung Y.-C., Li G.-Z., Tseng K.-C., Liu Y.-H., Kuo P.-L., Zheng H.-Q., Chang W.-C. PlantPAN3. 0: a new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 2019;47:D1155–D1163. doi: 10.1093/nar/gky1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M., Van den Broeck L., Claeys H., Van Vlierberghe K., Matsui M., Inzé D. The ETHYLENE RESPONSE FACTORs ERF6 and ERF11 antagonistically regulate mannitol-induced growth inhibition in Arabidopsis. Plant Physiol. 2015;169:166–179. doi: 10.1104/pp.15.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Bautista N., Domínguez-Núñez J., Moreno M.M., Berrocal-Lobo M. Plant tissue trypan blue staining during phytopathogen infection. Bio-protocol. 2016;6:e2078. [Google Scholar]
- Friedrich T., Oberkofler V., Trindade I., Altmann S., Brzezinka K., Lämke J., Gorka M., Kappel C., Sokolowska E., Skirycz A., et al. Heteromeric HSFA2/HSFA3 complexes drive transcriptional memory after heat stress in Arabidopsis. Nat. Commun. 2021;12:3426. doi: 10.1038/s41467-021-23786-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groß-Hardt R., Laux T. Stem cell regulation in the shoot meristem. J. Cell Sci. 2003;116:1659–1666. doi: 10.1242/jcs.00406. [DOI] [PubMed] [Google Scholar]
- Habben J.E., Bao X., Bate N.J., DeBruin J.L., Dolan D., Hasegawa D., Helentjaris T.G., Lafitte R.H., Lovan N., Mo H., et al. Transgenic alteration of ethylene biosynthesis increases grain yield in maize under field drought-stress conditions. Plant Biotechnol. J. 2014;12:685–693. doi: 10.1111/pbi.12172. [DOI] [PubMed] [Google Scholar]
- Hays D.B., Do J.H., Mason R.E., Morgan G., Finlayson S.A. Heat stress induced ethylene production in developing wheat grains induces kernel abortion and increased maturation in a susceptible cultivar. Plant Sci. 2007;172:1113–1123. [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- Hilker M., Schwachtje J., Baier M., Balazadeh S., Bäurle I., Geiselhardt S., Hincha D.K., Kunze R., Mueller-Roeber B., Rillig M.C., et al. Priming and memory of stress responses in organisms lacking a nervous system. Biol. Rev. 2016;91:1118–1133. doi: 10.1111/brv.12215. [DOI] [PubMed] [Google Scholar]
- Huang J., Zhao X., Bürger M., Wang Y., Chory J. Two interacting ethylene response factors regulate heat stress response. Plant Cell. 2021;33:338–357. doi: 10.1093/plcell/koaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal N., Khan N.A., Ferrante A., Trivellini A., Francini A., Khan M.I.R. Ethylene role in plant growth, development and senescence: interaction with other phytohormones. Front. Plant Sci. 2017;8:475. doi: 10.3389/fpls.2017.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketsa S., Chidtragool S., Klein J., Lurie S. Ethylene synthesis in mango fruit following heat treatment. Postharvest Biol. Technol. 1999;15:65–72. [Google Scholar]
- Kozomara A., Birgaoanu M., Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lämke J., Brzezinka K., Altmann S., Bäurle I. A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. EMBO J. 2016;35:162–175. doi: 10.15252/embj.201592593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wang H., Li N., Shao J., Zhou X., van Groenigen K.J., Thakur M.P. Phenological mismatches between above-and belowground plant responses to climate warming. Nat. Clim. Change. 2022;12:97–102. [Google Scholar]
- Liu J., Feng L., Gu X., Deng X., Qiu Q., Li Q., Zhang Y., Wang M., Deng Y., Wang E., et al. An H3K27me3 demethylase-HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res. 2019;29:379–390. doi: 10.1038/s41422-019-0145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Jung C., Xu J., Wang H., Deng S., Bernad L., Arenas-Huertero C., Chua N.-H. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell. 2012;24:4333–4345. doi: 10.1105/tpc.112.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt F., Kemper A., Tallman G. Inhibitors of ethylene synthesis inhibit auxin-induced stomatal opening in epidermis detached from leaves of Vicia faba L. Plant Cell Physiol. 2001;42:223–230. doi: 10.1093/pcp/pce030. [DOI] [PubMed] [Google Scholar]
- Nover L., Bharti K., Döring P., Mishra S.K., Ganguli A., Scharf K.-D. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell stress & chaperones. 2001;6:177–189. doi: 10.1379/1466-1268(2001)006<0177:aathst>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olas J.J., Apelt F., Annunziata M.G., John S., Richard S.I., Gupta S., Kragler F., Balazadeh S., Mueller-Roeber B. Primary carbohydrate metabolism genes participate in heat-stress memory at the shoot apical meristem of Arabidopsis thaliana. Mol. Plant. 2021;14:1508–1524. doi: 10.1016/j.molp.2021.05.024. [DOI] [PubMed] [Google Scholar]
- Olas J.J., Apelt F., Watanabe M., Hoefgen R., Wahl V. Developmental stage-specific metabolite signatures in Arabidopsis thaliana under optimal and mild nitrogen limitation. Plant Sci. 2021;303 doi: 10.1016/j.plantsci.2020.110746. [DOI] [PubMed] [Google Scholar]
- Olas J.J., Van Dingenen J., Abel C., Działo M.A., Feil R., Krapp A., Schlereth A., Wahl V. Nitrate acts at the Arabidopsis thaliana shoot apical meristem to regulate flowering time. New Phytol. 2019;223:814–827. doi: 10.1111/nph.15812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.-J., Seo Y.-S. Heat shock proteins: a review of the molecular chaperones for plant immunity. Plant Pathol. J. 2015;31:323–333. doi: 10.5423/PPJ.RW.08.2015.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T., Lechner E., Parmentier Y., Yanagisawa S., Grava S., Koncz C., Genschik P. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell. 2003;115:679–689. doi: 10.1016/s0092-8674(03)00968-1. [DOI] [PubMed] [Google Scholar]
- Ruonala R., Rinne P.L.H., Baghour M., Moritz T., Tuominen H., Kangasjärvi J. Transitions in the functioning of the shoot apical meristem in birch (Betula pendula) involve ethylene. Plant J. 2006;46:628–640. doi: 10.1111/j.1365-313X.2006.02722.x. [DOI] [PubMed] [Google Scholar]
- Sedaghatmehr M., Mueller-Roeber B., Balazadeh S. The plastid metalloprotease FtsH6 and small heat shock protein HSP21 jointly regulate thermomemory in Arabidopsis. Nat. Commun. 2016;7:12439. doi: 10.1038/ncomms12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stief A., Altmann S., Hoffmann K., Pant B.D., Scheible W.-R., Bäurle I. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell. 2014;26:1792–1807. doi: 10.1105/tpc.114.123851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street I.H., Aman S., Zubo Y., Ramzan A., Wang X., Shakeel S.N., Kieber J.J., Schaller G.E. Ethylene inhibits cell proliferation of the Arabidopsis root meristem. Plant Physiol. 2015;169:338–350. doi: 10.1104/pp.15.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A., Yu G., Jin H., Alonso J.M., Ecker J.R., Zhang X., Gao S., Theologis A. A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics. 2009;183:979–1003. doi: 10.1534/genetics.109.107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N., Torii K.U. Stem cells within the shoot apical meristem: identity, arrangement and communication. Cell. Mol. Life Sci. 2019;76:1067–1080. doi: 10.1007/s00018-018-2980-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Vinocur B., Shoseyov O., Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Wu Y.-S., Yang C.-Y. Ethylene-mediated signaling confers thermotolerance and regulates transcript levels of heat shock factors in rice seedlings under heat stress. Bot. Stud. 2019;60 doi: 10.1186/s40529-019-0272-z. 23-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Nagata M., Saito K., Wang K.L.C., Ecker J.R. Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol. 2005;5 doi: 10.1186/1471-2229-5-14. 14-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young T.E., Meeley R.B., Gallie D.R. ACC synthase expression regulates leaf performance and drought tolerance in maize. Plant J. 2004;40:813–825. doi: 10.1111/j.1365-313X.2004.02255.x. [DOI] [PubMed] [Google Scholar]
- Zeng J., Li X., Ge Q., Dong Z., Luo L., Tian Z., Zhao Z. Endogenous stress-related signal directs shoot stem cell fate in Arabidopsis thaliana. Nat. Plants. 2021;7:1276–1287. doi: 10.1038/s41477-021-00985-z. [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000;24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are available at the NCBI Sequence Read Archive (SRA) under BioProject ID PRJNA877651.
AGI codes
HSFA7a, AT3G51910; HSFA7b, AT3G63350; ERF1A, AT4G17500; EIN3, AT3G20770; TUB2, AT5G62690; ETO1, AT3G51770. Additional AGI codes are provided in the supplementary tables.