Abstract
Calcium-dependent protein kinases (CPKs), the best-characterized calcium sensors in plants, regulate many aspects of plant growth and development as well as plant adaptation to biotic and abiotic stresses. However, how CPKs regulate the antioxidant defense system remains largely unknown. We previously found that impaired function of OsCPK12 leads to oxidative stress in rice, with more H2O2, lower catalase (CAT) activity, and lower yield. Here, we explored the roles of OsCPK12 in oxidative stress tolerance in rice. Our results show that OsCPK12 interacts with and phosphorylates OsCATA and OsCATC at Ser11. Knockout of either OsCATA or OsCATC leads to an oxidative stress phenotype accompanied by higher accumulation of H2O2. Overexpression of the phosphomimetic proteins OsCATAS11D and OsCATCS11D in oscpk12-cr reduced the level of H2O2 accumulation. Moreover, OsCATAS11D and OsCATCS11D showed enhanced catalase activity in vivo and in vitro. OsCPK12-overexpressing plants exhibited higher CAT activity as well as higher tolerance to oxidative stress. Our findings demonstrate that OsCPK12 affects CAT enzyme activity by phosphorylating OsCATA and OsCATC at Ser11 to regulate H2O2 homeostasis, thereby mediating oxidative stress tolerance in rice.
Key words: OsCPK12, OsCATs, oxidative tolerance, Oryza sativa L
This study reports that the calcium-dependent protein kinase OsCPK12 phosphorylates OsCATA and OsCATC at Ser11 to increase their activity and maintain H2O2 homeostasis. Overexpression of OsCPK12 can increase catalase activity and improve oxidative stress tolerance in rice.
Introduction
Calcium-dependent protein kinases (CPKs) are one of the largest families of serine/threonine protein kinases in plants. CPKs consist of an N-terminal variable domain (VNTD/V), a Ser/Thr protein kinase domain (KD/K), an autoinhibitory junction region (JD/L), and a calmodulin-like (C) domain (Cam-LD/R) that includes four EF-hand motifs. CPKs are activated via conformational changes triggered by binding of Ca2+ to the C domain (Yip Delormel and Boudsocq, 2019; Yang et al., 2022). As key actors in plant signaling that transduce calcium signals into physiological responses, CPKs are crucial for both sensors and effectors, inducing downstream effects by phosphorylating target proteins to respond to nutrient deficiency, drought, salt, and cold (Alves et al., 2021).
Phosphorylation is an important posttranslational modification performed by many CPKs. CPKs participate in biotic and abiotic responses by phosphorylating target proteins, including ion channels, transcription factors (TFs), and metabolic enzymes (Ormancey et al., 2017). During prolonged salt stress, AtCPK3 phosphorylates TPK1 at Ser42 and provides the basis for its interaction with 14-3-3 proteins, thus enabling sustained potassium efflux to balance cytosolic potassium homeostasis and increase salt tolerance (Latz et al., 2013). AtCPK13, a typical Ca2+-insensitive CPK, specifically phosphorylates and inhibits the Arabidopsis K+ channel proteins KAT2 and KAT1 to reduce stomatal opening, likely in response to cold and oxidative stress (Ronzier et al., 2014). Arabidopsis CPK3 and CPK13 phosphorylate HsfB2a to modulate its activity and regulate defense-related transcription for defense response via the herbivory-induced signaling network (Kanchiswamy et al., 2010). Arabidopsis CPK6 phosphorylates the abscisic acid (ABA)-responsive element-binding factors ABF3 at Ser126 and ABI5 at Ser16, Ser41, Ser138, Ser145, Ser418, and Thr156 to positively regulate ABA signaling and drought tolerance (Zhang et al., 2020). Arabidopsis CPK5/CPK6 and MPK3/MPK6 differentially phosphorylate and cooperatively activate WRKY33 to induce camalexin production, thereby regulating Arabidopsis immunity upon pathogen infection (Zhou et al., 2020). Hypoxia stress stimulates Arabidopsis CPK12 to relocate from the cytoplasm to the nucleus, where it interacts with and phosphorylates group VII ethylene-responsive ERF-VII TFs (RAP2.12, RAP2.3, and RAP2.2, but not HRE1 or HRE2) to modulate their abundance, thereby potentiating plant hypoxia sensing (Fan et al., 2023).
OsCPK14 interacts with and phosphorylates OsDi19-4 at Ser134 to modulate the expression of ABA-responsive genes such as OsNAC18 and OsASPG1 and to positively regulate the ABA response in rice (Wang et al., 2016). OsCPK21 phosphorylates Os14-3-3 (OsGF14e) at Tyr138 to facilitate the response to ABA and salt stress in rice (Chen et al., 2017). The rapeseed CDPKs BnaCPK5/6/11 phosphorylate the WRKY TF BnaWSR1 to regulate salicylic acid accumulation and leaf senescence (Cui et al., 2020). ZmCDPK7 interacts with and phosphorylates the small heat shock protein sHSP17.4 at Ser44 and the respiratory burst oxidase homolog RBOHB at Ser99, and it upregulates their expression to increase thermotolerance in maize (Zhao et al., 2021).
Catalase (CAT), a strong antioxidant enzyme that catalyzes the decomposition of H2O2 to water and oxygen, functions directly in plant responses to biotic and abiotic stresses (Mhamdi et al., 2010). CAT shows a weak affinity for H2O2, which can be activated by phosphorylation by some protein kinases. For example, c-Abl and Arg are families of mammalian non-receptor tyrosine kinases that bind and phosphorylate CAT at Tyr231 and Tyr386 to activate catalase when cells are treated with 0.25–1.0 mM H2O2 (Cao et al., 2003). Protein kinase Cδ (PKCδ)-mediated phosphorylation of Ser167 leads to structural changes within CAT, promoting formation of the tetrameric structure required for maximal activity (Rafikov et al., 2014). Arabidopsis CPK8 interacts with CATALASE3 (CAT3), phosphorylating it at Ser261 to regulate its activity in response to drought stress (Zou et al., 2015). STRK1 phosphorylates and activates CatC, thereby regulating H2O2 homeostasis and improving salt tolerance in rice (Zhou et al., 2018). Although many lines of evidence show that CAT is associated with photorespiration and cell death, our understanding of whether and how CAT activity is regulated by phosphorylation during these processes remains limited.
OsCPK12 functions in signal transduction pathways, the low-nitrogen stress response, and salt stress processes (Asano et al., 2010, 2012; Xing et al., 2018; Wang et al., 2019). Abiotic stresses such as drought and salt stress often lead to oxidative stress, resulting in reactive oxygen species accumulation in plants, which in turn affects various biological pathways. Our previous studies found that impaired function of OsCPK12 leads to oxidative stress in rice, which is accompanied by H2O2 accumulation, decreased CAT activity, and decreased yield (Wang et al., 2019). We also found that overexpression of OsCPK12 in rice improved tolerance to oxidative stress. However, how OsCPK12 regulates the antioxidant defense system remains largely unknown. Here, we demonstrate that OsCPK12 interacts with and phosphorylates OsCATA and OsCATC at Ser11 to increase their activity and regulate H2O2. Our study reveals the regulatory mechanism by which OsCPK12 improves plant oxidative stress tolerance.
Results
Overexpression of OsCPK12 increases CAT activity and reduces H2O2 accumulation in rice
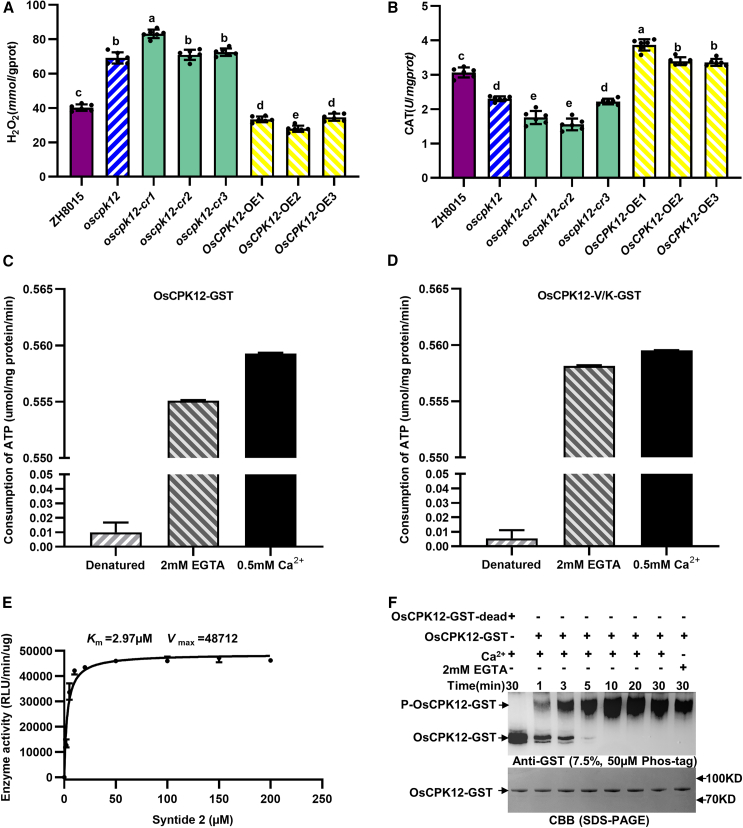
CPKs are involved not only in signaling pathways of plant growth and development but also in the response to biotic and abiotic stresses (Boudsocq and Sheen, 2013; Schulz et al., 2013). To investigate the biological function of OsCPK12 in rice, the oscpk12 mutant was identified from a population of mutant lines of the cultivar Zhonghui 8015 (ZH8015) mutagenized by ethyl methane sulfonate treatment (Wang et al., 2019). Previously, we verified that impaired function of OsCPK12 leads to early senescence in rice (Wang et al., 2019). Here, we compared the CAT activities and H2O2 levels between ZH8015 and oscpk12 using leaves at different stages. There was no significant difference in H2O2 content between wild-type and mutant leaves of 20-day-old seedlings; however, there was significantly more H2O2 in leaves of oscpk12 than in those in ZH8015 from 40 days onward (Supplemental Figure S1A). CAT activity was lower in oscpk12 than in ZH8015 at different stages (Supplemental Figure S1B). CAT activities and H2O2 levels of ZH8015, oscpk12, OsCPK12 overexpression (OsCPK12-OE), and OsCPK12 knockout (oscpk12-cr) lines were compared to further characterize the role of OsCPK12 in rice. H2O2 levels were much higher and CAT activities were much lower in leaves of oscpk12 and oscpk12-cr plants at the heading stage (Figure 1A). By contrast, OsCPK12-OE plants exhibited significantly higher CAT activity and lower H2O2 accumulation (Figure 1B). These results indicate that overexpression of OsCPK12 increased CAT activity and reduced H2O2 accumulation in rice.
Figure 1.
H2O2 content and CAT activity in ZH8015, oscpk12, oscpk12-cr, and OsCPK12-OE lines, and protein properties of OsCPK12 and truncated OsCPK12.
(A) H2O2 content in leaves of ZH8015, oscpk12, oscpk12-cr, and OsCPK12-OE lines. Data are presented as mean ± SD (n = 6). Different letters above the bars indicate significant differences (P < 0.05). P values were determined by one-way ANOVA followed by Tukey’s multiple comparison test.
(B) Enzymatic activities of CAT in leaves of ZH8015, oscpk12, oscpk12-cr, and OsCPK12-OE lines. Data are presented as mean ± SD (n = 6). Different letters above the bars indicate significant differences (P < 0.05). P values were determined by one-way ANOVA followed by Tukey’s multiple comparison test.
(C) Kinase activity of full-length OsCPK12. Recombinant OsCPK12 protein was purified, and in vitro kinase activity was determined using syntide 2 as the substrate in the absence (2 mM EGTA) or presence of 0.5 mM Ca2+. Denatured OsCPK12 was included as a negative control. Luminescence values (relative light units, RLU) were measured with a GloMax-Multi detection system. The luminescence signal is correlated with the amount of ATP present and is therefore inversely correlated with the amount of kinase activity. The amount of ATP consumed is positively correlated with kinase activity. Data are presented as mean ± SD (n = 3).
(D) Kinase activity of truncated OsCPK12. Recombinant truncated OsCPK12 protein was purified, and in vitro kinase activity was determined using syntide 2 as the substrate in the absence (2 mM EGTA) or presence of 0.5 mM Ca2+. Denatured truncated OsCPK12 was included as a negative control. Luminescence values (RLU) were determined with a GloMax-Multi detection system. The luminescence signal is correlated with the amount of ATP present and is therefore inversely correlated with the amount of kinase activity. The amount of ATP consumed is positively correlated with kinase activity. Data are presented as mean ± SD (n = 3).
(E)Km values of OsCPK12 toward the substrate syntide 2. RLU values represent the decrease in the sample relative to the control and are proportional to kinase activity. Data are presented as mean ± SD (n = 3).
(F) Auto-phosphorylation of OsCPK12 over 30 min, with or without Ca2+. Denatured OsCPK12 was included as a negative control. Protein phosphorylation detected by the immunoblots was probed with anti-GST after Phos-Tag SDS–PAGE (top), and protein loading was indicated by Coomassie brilliant blue (CBB) staining of a standard SDS–PAGE gel (bottom).
Expression of OsCPK12, subcellular localization, and protein properties of OsCPK12
Previous studies have shown that OsCPK12 is expressed in all organs but is highly expressed in green tissues such as leaves, stems, and sheaths (Wang et al., 2019). To determine transcript levels of OsCPK12 throughout the reproductive period, we analyzed the expression of OsCPK12 in leaves of ZH8015 and oscpk12 using qRT–PCR at different developmental stages. OsCPK12 expression was higher at 20 days after sowing and decreased after 30 days post sowing (Supplemental Figure S1C). OsCPK12 expression was clearly lower in oscpk12, and trends in OsCPK12 expression at different stages were roughly similar in ZH8015 and oscpk12 (Supplemental Figure S1C). These results indicate that OsCPK12 was expressed throughout the growth period, but its expression was highest at 20 days after sowing.
CPKs consist of a V domain at the N-terminal end, a Ser/Thr protein kinase K domain, an autoinhibitory junction L domain, and an R domain with four EF-hand motifs (Yip Delormel and Boudsocq, 2019; Yang et al., 2022). To verify the subcellular localization of OsCPK12 and its different domains, we first fused the full coding sequence (CDS) of OsCPK12 to the N terminus of green fluorescent protein (GFP) driven by the CaMV35S promoter and transiently expressed it in rice protoplasts. GFP signal was detected throughout the cell and merged perfectly with the mCherry signal (Supplemental Figure S2A–S2C). The GFP signal of full-length OsCPK12 also merged with the nuclear marker protein Ghd7-mCherry (Supplemental Figure S2D and S2E), the membrane marker protein Lazy1-mCherry (Supplemental Figure S2F and S2G), and the endoplasmic reticulum (ER) marker mCherry-HDEL (Supplemental Figure S2H).
N-myristoylation is a protein acylation process that is absolutely specific to the N-terminal amino acid glycine. NMT (The MYR Predictor: https://mendel.imp.ac.at/myristate/SUPLpredictor.htm) and Myristoylator (https://web.expasy.org/myristoylator/) predicted that OsCPK12 could be myristoylated at glycine2, a co-translational modification involved in membrane targeting (Boudsocq and Sheen, 2013). On the basis of this prediction, we next examined the subcellular localization of the truncated N-terminal V domain, which contained the predicted myristoylated site. As expected, the GFP signal of OsCPK12-V-GFP was distributed only in the membrane (Supplemental Figure S2I and S2J). To determine whether the V domain of OsCPK12 was necessary for membrane localization of full-length OsCPK12, the truncated N-terminal V domain and K domain of OsCPK12 and only the K domain of OsCPK12 were fused with GFP to form OsCPK12-V/K-GFP and OsCPK12-K-GFP. We found that the GFP signals of OsCPK12-V/K-GFP and OsCPK12-K-GFP were similar to those of full-length OsCPK12 (Supplemental Figures S2K, S2L, S2M, and S2N). These results demonstrated that OsCPK12 is localized in the membrane, cytoplasm, and nucleus and that the N-terminal V domain of OsCPK12 is sufficient for its membrane positioning.
To determine whether OsCPK12 is a functional protein kinase, we performed an in vitro enzyme activity assay. Previous studies have shown that Ca2+-dependency can be uncoupled from kinase activity by deliberately removing the L and R domains, often resulting in CPKs that are uninhibited by Ca2+ (Yip Delormel and Boudsocq, 2019). Full-length OsCPK12 and OsCPK12-V/K lacking the CAD domain fused with a glutathione S-transferase (GST) were expressed, purified, and confirmed by Coomassie brilliant blue (CBB) staining, and protein concentration was measured using a bicinchoninic acid (BCA) protein assay kit. The protein supernatant was used in an in vitro kinase activity assay and autophosphorylation assay. The amount of ATP consumed is positively correlated with kinase activity. Our results showed that ATP consumption was lower upon incubation of OsCPK12-GST with 2 mM ethylene glycol-bis(-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA; an extracellular calcium chelator that removes free Ca2+ from the reaction solution) than upon incubation with 0.5 mM Ca2+ (Figure 1C); similar results were obtained with OsCPK12-V/K-GST (Figure 1D). OsCPK12 activity toward syntide 2 was also analyzed. The Km value of OsCPK12 was 2.97 μM when syntide 2 was used as a substrate (Figure 1E). These findings indicated that the kinase activity of OsCPK12 and OsCPK12-V/K was enhanced after Ca2+ addition.
We next separated OsCPK12 protein incubated with Ca2+ or 2 mM EGTA for 1 to 30 min in Phos-Tag gels to detect whether OsCPK12 was auto-phosphorylated. The phosphorylated band increased in thickness with increasing reaction time (Figure 1F), indicating that OsCPK12 has autophosphorylation activity in vitro. We also detected a phosphorylated band when OsCPK12 was incubated with 2 mM EGTA (Figure 1F). Therefore, OsCPK12 can be auto-phosphorylated with or without Ca2+ in vitro.
OsCPK12 interacts with OsCATA, OsCATB, and OsCATC in vitro and in vivo
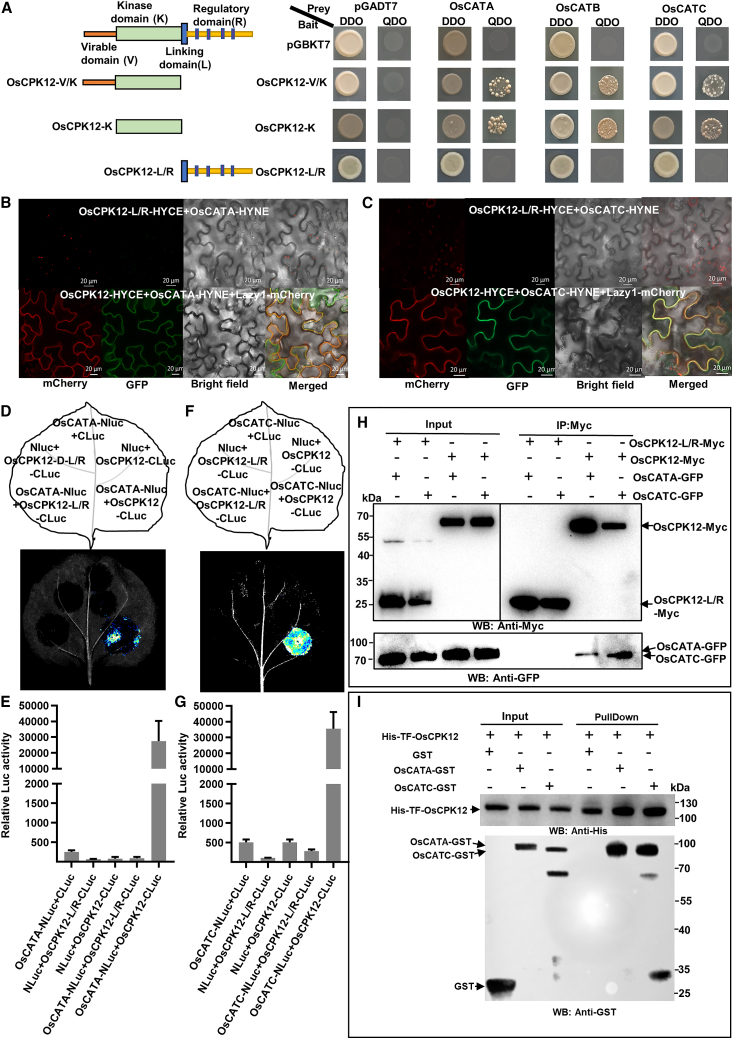
To explore the molecular mechanism by which OsCPK12 overexpression improves the antioxidant capacity of rice, we performed a yeast two-hybrid (Y2H) screen to identify OsCPK12-interacting proteins. OsCPK12-V/K was selected and fused to the Gal4DNA-binding domain of a bait vector (BD). Several interacting clones were isolated and identified as corresponding to the CAT domain-containing protein OsCATC. OsCATC is one of three genes in the rice CAT family (OsCATA, OsCATB, and OsCATC). Full-length OsCATA, OsCATB, and OsCATC were then introduced into the Gal4 activation domain of the prey vector to construct AD-OsCATA, AD-OsCATB, and AD-OsCATC. Y2H results indicated that OsCPK12 interacted with all three CATs, and OsCPK12-K containing only the kinase domain was sufficient for this interaction (Figure 2A).
Figure 2.
OsCPK12 interacts with OsCATA and OsCATC.
(A) Y2H assay of OsCPK12 interaction with OsCATA and OsCATC. DDO, double-dropout supplements (SD/-Leu-Trp); QDO, quadruple-dropout supplements (SD/-Ade-His-Leu-Trp).
(B and C) Bimolecular fluorescence complementation (BiFC) assays showing that OsCPK12 interacts with OsCATA and OsCATC. The indicated constructs were transiently expressed in N. benthamiana by agroinfiltration, and fluorescence was observed 4 days after infiltration (DAI) using a laser scanning confocal microscope (ZEISS 750). OsCPK12-L/R-HYCE was used as a negative control. Lazy1-mCherry was used as a cell membrane marker. Scale bars, 20 μm.
(D) OsCPK12 interacts with OsCATA as indicated by LCI assay. OsCPK12-CLuc and NLuc-OsCATA were transiently expressed in N. benthamiana by coinfiltration. NLuc and CLuc were the negative controls. Luminescence was monitored with a low-light, cooled, CCD imaging apparatus at 2 DAI.
(E) Quantification of LUC activity in the leaves shown in (D). Data are presented as mean ± SD (n = 3).
(F) OsCPK12 interacts with OsCATC as indicated by LCI assay. OsCPK12-CLuc and NLuc-OsCATC were transiently expressed in N. benthamiana by coinfiltration. NLuc and CLuc were the negative controls. Luminescence was monitored with a low-light, cooled, harge coupled deviceCCD imaging apparatus at 2 DAI.
(G) Quantification of LUC activity in the leaves shown in (F). Data are presented as mean ± SD (n = 3).
(H) Coimmunoprecipitation assay for interaction of OsCPK12 with OsCATA and OsCATC. The total protein extracts from rice protoplasts transfected with OsCPK12-Myc/OsCPK12-L/R-Myc, OsCATA-GFP, and OsCATC-GFP were immunoprecipitated with anti-Myc Sepharose beads. The proteins from crude lysates (left, input) and immunoprecipitated proteins (right) were detected with anti-GFP and anti-Myc antibodies.
(I) Pull-down assay for interaction of OsCPK12 with OsCATA and OsCATC. His-TF-OsCPK12, OsCATA-GST, OsCATC-GST, and GST were expressed and purified from E. coli and used in His pull-down assays. GST was used as a control. Washed His resin was immunoblotted with anti-GST and anti-His antibodies to detect the immobilized proteins and the pulled-down proteins, respectively. Input proteins were detected by immunoblotting with the indicated antibodies.
To further confirm the interaction between OsCPK12 and the OsCATs, we performed bimolecular fluorescence complementation (BiFC) assays. First, we analyzed the subcellular localization of the CATs. Previous findings suggested that OsCATA was found mainly in the cytoplasm, whereas OsCATB and OsCATC were found mainly in the peroxisome (Ye et al., 2011; Zhang et al., 2016). Consistent with previous research, our results showed that OsCATA was mainly localized in the cytoplasm (Supplemental Figure S3A and S3D) and that OsCATB and OsCATC were mainly localized in the peroxisome (Supplemental Figures S3B, S3E, S3C, and S3F). However, OsCATA-GFP, OsCATB-GFP, and OsCATC-GFP fusion proteins also showed clear nuclear localization (Supplemental Figure S3G–S3I), and all showed weak but clear plasma membrane localization when transiently expressed in rice protoplasts and Nicotiana benthamiana (Supplemental Figure S3J–S3L). To further verify the subcellular localization of OsCATA, OsCATB, and OsCATC, we fused GFP to their N-terminal ends to produce GFP-OsCATA, GFP-OsCATB, and GFP-OsCATC fusion proteins. These fusion proteins produced the same fluorescence signals as OsCATA-GFP, OsCATB-GFP, and OsCATC-GFP (Supplemental Figure S3M–S3O).
To detect the nuclear localization of the OsCATs, we performed immunoblot analysis using total proteins and nuclear proteins extracted from protoplasts of ZH8015 transiently transformed with OsCATA-GFP, OsCATB-GFP, and OsCATC-GFP. All three fusion proteins were present in total proteins and nuclear proteins, confirming their nuclear localization (Supplemental Figure S4). BiFC assays showed strong yellow fluorescent protein signals in cells coexpressing OsCPK12-HYCE and OsCATs-HYNE; these signals merged perfectly with the Lazy1-mCherry signal. However, no signal was detected in cells coexpressing OsCPK12-HYCE and GST-HYNE under the same conditions, indicating that OsCPK12 specifically interacted with CATs and that the OsCPK12/CAT complexes were localized to the plasma membrane (Figure 2B and 2C, Supplemental Figure S5A and S5B). Previous studies have reported that most CPKs are mainly or partially membrane anchored. Membrane association is typically related to a predicted N-terminal myristoylation site or to additional reversible processes such as palmitoylation, protein interactions, or phosphorylation of a polybasic domain (Meinnel and Giglione, 2008). We predicted that OsCPK12 targets the membrane to participate in signal transduction in response to stress stimulation.
We next carried out a luciferase complementation imaging (LCI) assay. N. benthamiana leaves coexpressing CATs-NLuc and CLuc-OsCPK12 showed strong fluorescence: the relative luciferase activities were nearly 30 000 for CATA-NLuc and CLuc-OsCPK12, 80 000 for CATB-NLuc and CLuc-OsCPK12l, and 40 000 for CATC-NLuc and CLuc-OsCPK12. By contrast, coexpression of the control combinations produced only background levels of LUC activity (Figure 2D, 2E, 2F, 2G, and Supplemental Figure S5C and S5D). Because OsCATA and OsCATC were found to be phosphorylated by OsCPK12 in subsequent work (Supplemental Figure S6), the interactions of OsCPK12 with OsCATA and OsCATC were further confirmed by a coimmunoprecipitation (coIP) assay. OsCATA-GFP and OsCATC-GFP fusion protein were coimmunoprecipitated with OsCPK12-Myc but not with OsCPK12-L/R-Myc (Figure 2H). We also used OsCPK12-Myc to coimmunoprecipitate OsCATA-Ha, OsCATC-Ha and CLuc-Ha, but CLuc-Ha was not coimmunoprecipitated by OsCPK12-Myc (Supplemental Figure S5E). Purified recombinant His-TF-OsCPK12 and GST/OsCATA-GST/OsCATC-GST were subjected to a pull-down assay. As expected, OsCATA-GST and OsCATC-GST were pulled down by His-TF-OsCPK12, but GST was not (Figure 2I). Taken together, these results indicate that OsCPK12 interacts with the tested OsCATs.
OsCPK12 phosphorylates OsCATA and OsCATC
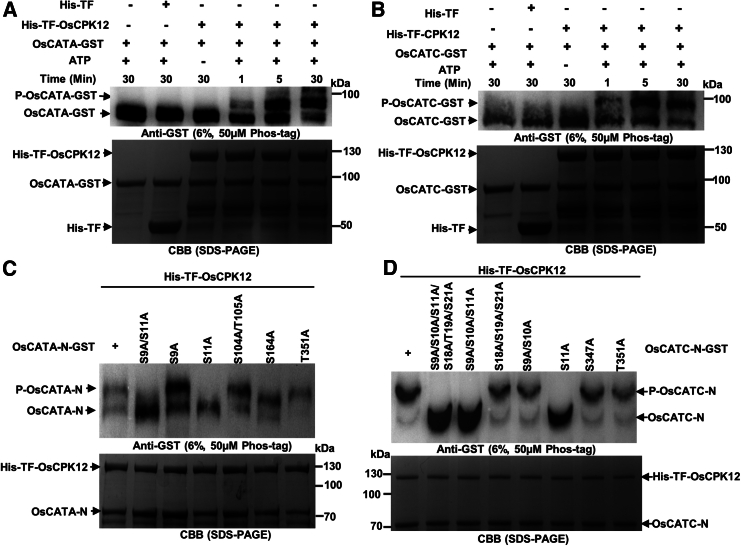
To test whether OsCPK12 phosphorylated CATs in rice protoplasts, we performed an in vivo phosphorylation assay. As shown in Supplemental Figure S6, higher bands of OsCATA-myc and OsCATC-myc were detected in ZH8015 using Phos-Tag immunoblots. There were no phosphorylation differences in OsCATB-myc. These results suggest that OsCPK12 phosphorylates OsCATA and OsCATC in vivo. To further investigate whether OsCATA and OsCATC were phosphorylated by OsCPK12, we performed an in vitro phosphorylation assay using recombinant OsCATA-GST and OsCATC-GST and His-TF-OsCPK12 or His-TF-OsCPK12-Dead (denatured His-TF-OsCPK12). As shown in Figure 3A and 3B, when OsCATA-GST and OsCATC-GST were incubated with His-TF-OsCPK12, two bands were detected on the Phos-Tag immunoblots, and the higher-mobility band increased in thickness as the reaction time increased. However, when OsCATA-GST and OsCATC-GST were incubated with denatured OsCPK12, no differential bands appeared (Supplemental Figure S7A), suggesting that OsCATA-GST and OsCATC-GST were phosphorylated in vitro in the presence of OsCPK12. To better distinguish the phosphorylated and non-phosphorylated bands, we also used truncated OsCATA (1–403 amino acids [aa]) (OsCATA-N-GST), OsCATC (1–403 aa) (OsCATC-N-GST), OsCATA (404–493 aa) (OsCATA-C-GST), and OsCATC (404–493 aa) (OsCATC-C-GST) for an in vitro phosphorylation assay. We observed that OsCPK12 phosphorylated OsCATA-N-GST and OsCATC-N-GST (Supplemental Figure S7C and S7D) but not OsCATA-C-GST or OsCATC-C-GST (Supplemental Figure S7E and S7F). Furthermore, OsCATA-N-GST and OsCATC-N-GST were not phosphorylated by His-TF-OsCPK12-Dead (Supplemental Figure S7B). These results indicate that OsCATA and OsCATC are candidate substrates of OsCPK12 and that OsCPK12 phosphorylates them at the N terminus.
Figure 3.
OsCPK12 phosphorylates OsCATA and OsCATC at Ser11 in vitro.
(A) OsCPK12 phosphorylates OsCATA in vitro. The input proteins OsCATA and OsCPK12 were detected by CBB staining. Phosphorylation activity was detected by immunoblot analysis using Phos-Tag SDS–PAGE.
(B) OsCPK12 phosphorylates OsCATC in vitro. The input proteins OsCATC and OsCPK12 were detected by CBB staining. Phosphorylation activity was detected by immunoblot analysis using Phos-Tag SDS–PAGE.
(C) OsCPK12 phosphorylates OsCATA at Ser11 in vitro. Protein phosphorylation was detected using immunoblots probed with anti-GST antibody after Phos-Tag SDS–PAGE (top), and protein loading was indicated by CBB staining in a standard SDS–PAGE gel (bottom).
(D) OsCPK12 phosphorylates OsCATC at Ser11 in vitro. Protein phosphorylation was detected using immunoblots probed with anti-GST antibody after Phos-Tag SDS–PAGE (top), and protein loading was indicated by CBB staining in a standard SDS–PAGE gel (bottom).
To identify the phosphorylation sites in OsCATA and OsCATC, we cut out bands from gels containing phosphorylated OsCATA and OsCATC (GST-OsCATA and GST-OsCATC incubated with His-TF-OsCPK12) and OsCATA-CK and OsCATC-CK (OsCATA-GST and OsCATC-GST incubated with His-TF). We extracted proteins from the bands and searched for their phosphorylation sites using liquid chromatography–tandem mass spectrometry. Seven phosphorylation sites (Ser10, Ser11, Ser104, Thr105, Ser164, Thr105, and Ser437) were detected in the OsCATA sample incubated with His-TF-OsCPK12, and 10 phosphorylation sites (Ser9, Ser10, Ser11, Ser18, Thr19, Ser21, Ser347, Thr351, Thr412, and Thr414) were detected in the OsCATC sample incubated with His-TF-OsCPK12. No phosphorylation sites were detected in the OsCATA-CK and OsCATC-CK samples incubated with His-TF (Supplemental Table 1). These results show that OsCPK12 phosphorylates OsCATA and OsCATC at the N terminus and that OsCATA and OsCATC have six (Ser10, Ser11, Ser104, Thr105, Ser164, and Thr351) and eight (Ser9, Ser10, Ser11, Ser18, Thr19, Ser21, Ser347, and Thr351) putative phosphorylation sites, respectively.
To validate specific phosphorylation sites of OsCATA and OsCATC, we replaced these Ser (S) and Thr (T) residues with Ala (A) to mimic nonphosphorylation. Results of the in vitro phosphorylation assay showed that OsCATA-NS10A-GST, OsCATA-NS104A/S105A-GST, OsCATA-NS164A-GST, and OsCATA-NT351A-GST showed a clear phosphorylation band similar to that of OsCPK12-mediated OsCATA-N (Figure 3C). However, this phosphorylation band was not detected in OsCATA-NS10A/S11A-GST or OsCATA-NS11A-GST. Therefore, Ser11 of OsCATA is the major phosphorylated residue recognized by OsCPK12. In vitro phosphorylation assays between OsCATC and OsCPK12 showed almost no phosphorylation of OsCATC-NS9A/S10A/S11A/S18A/T19A/S21A-GST, OsCATC-NS9A/S10A/S11A-GST, or OsCATC-NS11A-GST; the other single point mutant proteins OsCATC-NS18A/T19A/S21A-GST, OsCATC-NS9A/S10A-GST, OsCATC-NS347A-GST, and OsCATC-NT351A-GST showed similar OsCPK12-mediated phosphorylation (Figure 3D). These results suggest that Ser11 of OsCATC is the major OsCPK12 phosphorylation site and indicate that OsCPK12 may phosphorylate OsCATA and OsCATC at Ser11.
Knockout of OsCATA and OsCATC leads to decreased CAT activity and greater H2O2 accumulation
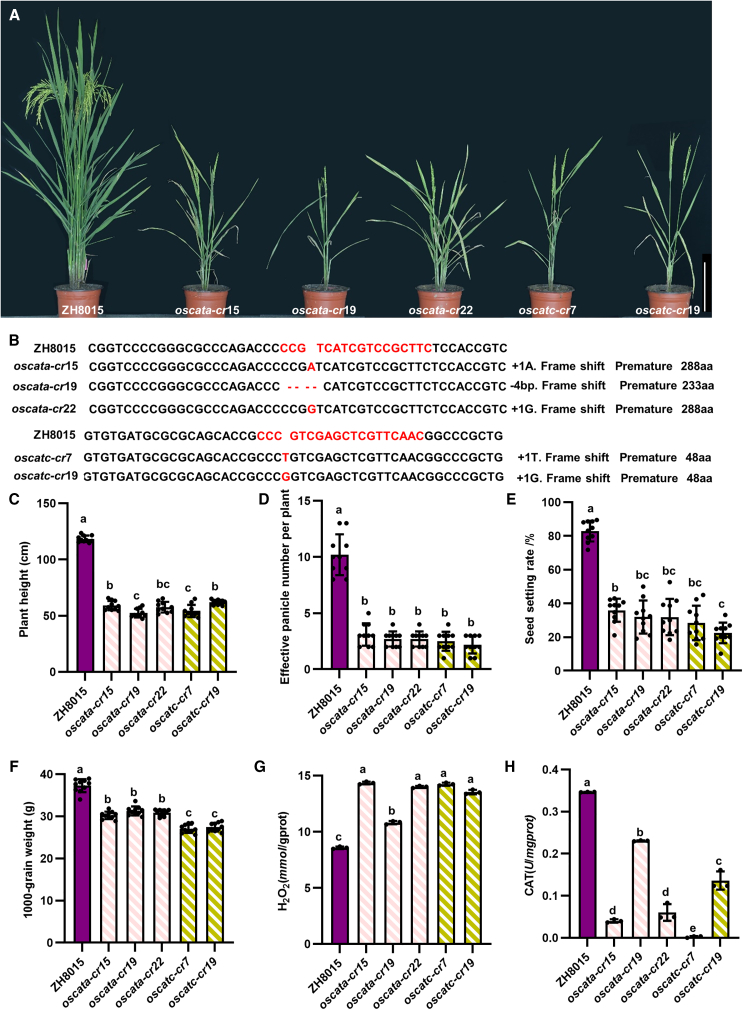
We generated various knockout lines of OsCATA (oscata-cr) and OsCATC (oscatc-cr) in the ZH8015 background by CRISPR–Cas9 to investigate their functions in rice. In total, we generated three independent oscata-cr lines and two independent oscatc-cr lines (Figure 4A and 4B). All oscata-cr lines and oscatc-cr lines showed a clear leaf-bleaching and cell-death phenotype from the three-leaf-stage (Supplemental Figure S8A, S8B, S8D, and S8E) to maturity (Figure 4A). noe1, a loss-of-function mutation of OsCATC, also exhibits leaf cell death with H2O2 overaccumulation (Lin et al., 2012).
Figure 4.
Phenotypes of oscata-cr and oscatc-cr lines.
(A) Phenotypes of oscata-cr and oscatc-cr lines. Bar, 20 cm.
(B) Genotypes of oscata-cr and oscatc-cr lines.
(C) Plant height of ZH8015, oscata-cr, and oscatc-cr lines.
(D) Effective panicle number per plant of ZH8015, oscata-cr, and oscatc-cr lines.
(E) Seed-setting rate of ZH8015, oscata-cr, and oscatc-cr lines.
(F) Thousand-grain weight of ZH8015, oscata-cr, and oscatc-cr lines. (C–F) Data are presented as mean ± SD (n = 10). Different letters above the bars indicate significant differences (P < 0.05). P values were determined by one-way ANOVA followed by Tukey’s multiple comparison test.
(G) H2O2 contents in leaves of ZH8015, oscata-cr, and oscatc-cr plants.
(H) CAT activity in leaves of ZH8015, oscata-cr, and oscatc-cr plants.
(G and H) Data are presented as mean ± SD (n = 3). Different letters above the bars indicate significant differences (P < 0.05). P values were determined by one-way ANOVA followed by Tukey’s multiple comparison test.
Plant height was significantly lower in oscata-cr lines and oscatc-cr lines than in wild-type ZH8015 (Figure 4C), as was effective panicle number per plant (Figure 4D). Seed-setting rate of the oscata-cr and oscatc-cr lines was significantly decreased to 20.20%–30.49%, which was only one-quarter that of wild-type ZH8015 (Figure 4E). Thousand-grain weights were also significantly lower in oscata-cr and oscatc-cr lines than in ZH8015 (Figure 4F). We measured H2O2 content and CAT activity in leaves of oscata-cr and oscatc-cr plants at the heading stage. H2O2 content was higher (Figure 4G) and CAT activity was lower (Figure 4H) in leaves of oscata-cr and oscatc-cr plants than in those of ZH8015, consistent with the phenotype of noe1. 3,3-diaminobenzidine (DAB) staining also revealed higher H2O2 accumulation in leaves of the oscata-cr and oscatc-cr lines at the seedling stage (Supplemental Figure S8C and S8F). These results indicate that knockout of OsCATA and OsCATC can lead to accumulation of H2O2 and decreased CAT activity in rice, seriously affecting its growth and development and causing a significant decrease in yield.
Phosphorylation of OsCATA and OsCATC increases their activity
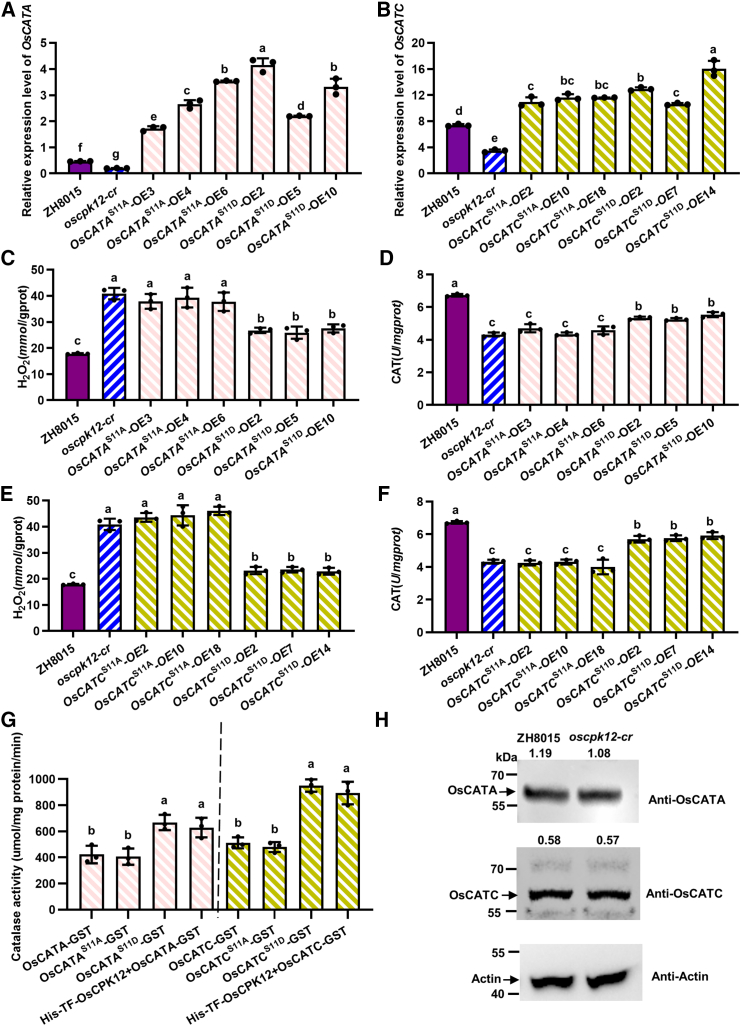
To study the significance of Ser11 phosphorylation of OsCATA and OsCATC in planta, the Ser11 residues of OsCATA and OsCATC were mutated to Ala (A) and Asp (D) to mimic non-phosphorylated proteins (OsCATAS11A and OsCATCS11A) and phosphorylated proteins (OsCATAS11D and OsCATCS11D), respectively. OsCATAS11A, OsCATCS11A, OsCATAS11D, and OsCATCS11D were overexpressed in oscpk12-cr plants to produce OsCATAS11A-OE, OsCATCS11A-OE, OsCATAS11D-OE, and OsCATCS11D-OE (Figure 5A and 5B). We measured CAT activities and H2O2 concentrations in overexpression plants and oscpk12-cr plants under normal growth conditions. Compared with leaves of OsCATAS11A-OE, those of oscpk12-cr/OsCATAS11D-OE and oscpk12-cr/OsCATCS11D-OE had significantly enhanced CAT activity and lower H2O2 accumulation (Figure 5C–5F). By contrast, the CAT activities and H2O2 concentrations of oscpk12-cr/OsCATAS11A-OE and oscpk12-cr/OsCATCS11A-OE were similar to those of oscpk12-cr plants (Figure 5C–5F). Thus, OsCATAS11D-OE and OsCATCS11D-OE can rescue the CAT activity of oscpk12-cr plants to some extent, but OsCATAS11A-OE and OsCATCS11A-OE cannot. Together, these results demonstrate that OsCATAS11D and OsCATCS11D exhibit higher CAT activity in planta.
Figure 5.
Phosphorylation of OsCATA and OsCATC at Ser11 activates their CAT activity in vivo and in vitro, and immunoblot analysis of OsCATA and OsCATC in the ZH8015 and oscpk12-cr backgrounds.
(A) Relative expression level of OsCATA in leaves of transgenic seedlings (T2) overexpressing OsCATAS11A and OsCATAS11D in the oscpk12-cr background.
(B) Relative expression levels of OsCATC in leaves of transgenic seedlings (T2) overexpressing OsCATCS11A and OsCATCS11D in the oscpk12-cr background.
(C) H2O2 content in transgenic seedlings (T2) overexpressing OsCATAS11A and OsCATAS11D in the oscpk12-cr background. ZH8015 (wild type [WT]) and oscpk12-cr seedlings were used as controls.
(D) H2O2 content in transgenic seedlings (T2) overexpressing OsCATCS11A and OsCATCS11D in the oscpk12-cr background. ZH8015 (WT) and oscpk12-cr seedlings were used as controls.
(E) CAT activity in transgenic seedlings (T2) overexpressing OsCATAS11A and OsCATAS11D in the oscpk12-cr background. ZH8015 (WT) and oscpk12-cr seedlings were used as controls.
(F) CAT activity in transgenic seedlings (T2) overexpressing OsCATCS11A and OsCATCS11D in the oscpk12-cr background. ZH8015 (WT) and oscpk12-cr seedlings were used as controls.
(G) Phosphorylation of OsCATA and OsCATC at Ser11 activates their CAT activity in vitro. OsCATAS11A-GST and OsCATCS11A-GST, recombinant OsCATA and OsCATC with Ser11 mutated to Ala. OsCATAS11D-GST and OsCATCS11D-GST, recombinant OsCATA and OsCATC with Ser11 mutated to Asp. OsCATA-GST, OsCATAS11A-GST, OsCATAS11D-GST, OsCATC-GST, OsCATCS11A-GST, OsCATCS11D-GST, and His-TF-OsCPK12 were purified for CAT activity assays in vitro. (A–G) Data are presented as mean ± SD (n = 3). Different letters above the bars indicate significant differences (P < 0.05). P values were determined by one-way ANOVA followed by Tukey’s multiple comparison test.
(H) Immunoblot analysis of OsCATA, OsCATC, and actin in the ZH8015 and oscpk12-cr backgrounds. Total proteins were extracted from leaves of ZH8015 and oscpk12-cr. Antisera against OsCATA, OsCATC, and β-actin (anti-OsCATA, anti-OsCATA, and anti-actin) were used in blotting. The data above the figure represent the protein intensity ratios of OsCATA/actin and OsCATC/actin.
To demonstrate that phosphorylation of OsCATA and OsCATC by OsCPK12 can indeed increase their catalase activity, we also performed a CAT activity assay in vitro using purified OsCATA, OsCATAS11A, OsCATAS11D, OsCATC, OsCATCS11A, and OsCATCS11D (Supplemental Figure S9). OsCATAS11D and OsCATCS11D exhibited higher CAT activity than wild-type OsCATA and OsCATC, OsCATAS11A, and OsCATCS11A (Figure 5G). CAT activities of OsCATA and OsCATC were next analyzed in the presence of OsCPK12. As expected, OsCPK12 significantly enhanced CAT activity of OsCATA and OsCATC (Figure 5G). The protein stability of OsCATB and OsCATC can also change depending on their partners. For example, APIP6 promotes the degradation of OsCATC to attenuate the efficiency of OsCATC-mediated H2O2 scavenging (You et al., 2022); rice ENHANCED DISEASE SUSCEPTIBILITY 1 (OsEDS1) interacts with and stabilizes OsCATC to improve OsCATC-mediated H2O2 scavenging ability (Liao et al., 2023); and SEMI-ROLLED LEAF 10 (SRL10) stabilizes OsCATB and enhances CAT activity to regulate leaf morphology and thermotolerance in rice (Wang et al., 2023a; 2023b). We therefore speculated that CPK12 might also alter CAT activity by changing CAT protein stability. However, protein levels of OsCATA and OsCATC were unaltered in the mutant oscpk12-cr background compared with wild-type ZH8015 (Figure 5H). We also observed similar subcellular localizations of OsCATA and OsCATC in protoplasts of ZH8015 and oscpk12-cr. These results suggest that OsCPK12-mediated phosphorylation does not regulate protein stability or subcellular localization of OsCATA and OsCATC. Rather, phosphorylation of OsCATA and OsCTAC at Ser11 by OsCPK12 enhances their CAT activities.
OsCPK12 positively regulates oxidative stress tolerance in rice
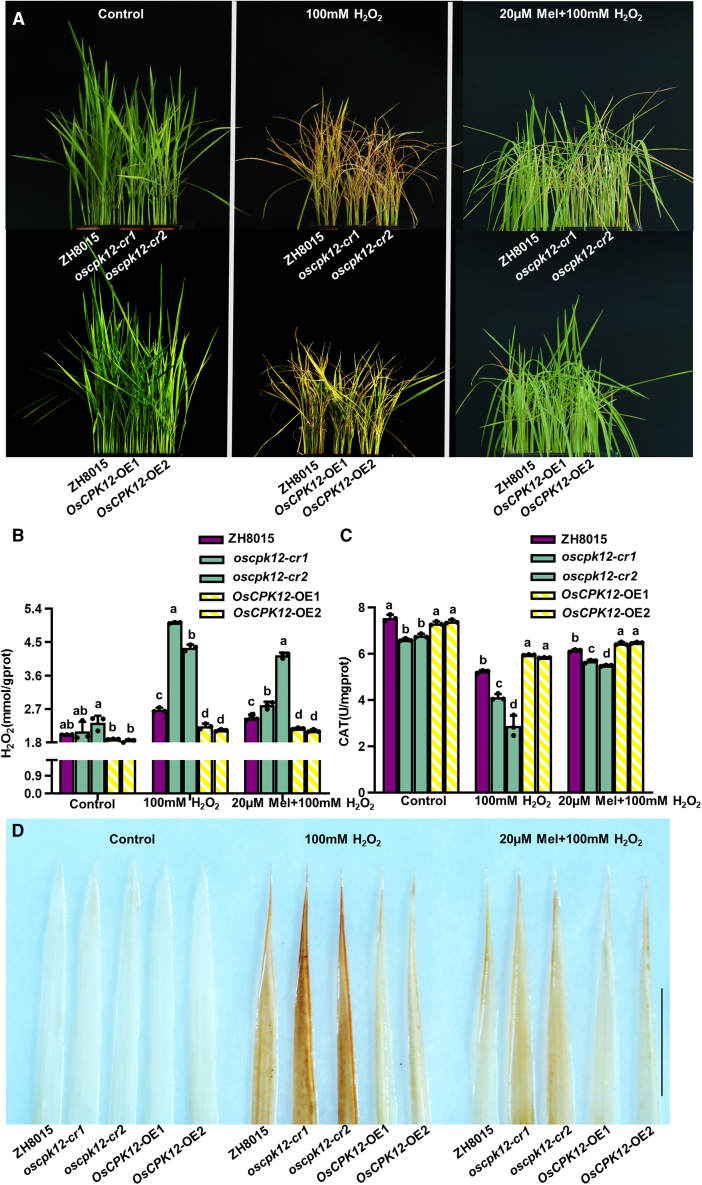
To investigate the response of OsCPK12 to oxidative stress, three-leaf-stage seedlings of the wild type, OsCPK12-overexpressing (OsCPK12-OE) transgenic plants, and knockout lines (oscpk12-cr) were treated with 100 mM H2O2 or 100 mM H2O2 and 20 μM melatonin. After 6 days of treatment, chlorophyll content, H2O2 content, and CAT activity were determined. Under normal conditions, there was no significant difference in growth among the OsCPK12-OE lines, oscpk12-cr lines, and wild-type seedlings. But after 6 days of H2O2 treatment, severe yellowing, leaf rolling, and dryness were observed in the oscpk12-cr lines (Figure 6A), and the leaves of ZH8015 appeared chlorotic, with curling and dryness. The leaves of OsCPK12-OE also appeared chlorotic with curling, but both effects were weaker than those in ZH8015. OsCPK12-OE plants had higher contents of Chla, Chlb, and Car than wild-type plants, whereas oscpk12-cr plants exhibited the opposite results (Supplemental Figure S10A and S10B).
Figure 6.
OsCPK12 promotes oxidative stress tolerance in rice.
Ten-day-old seedlings were transplanted into medium supplemented with 100 mM H2O2 or 100 mM H2O2 and 20 μM melatonin for 6 days.
(A) Phenotypic comparison of rice plants subjected to H2O2 stress and H2O2 stress + melatonin.
(B) H2O2 contents in leaves of rice plants subjected to H2O2 stress and H2O2 stress + melatonin. Data are presented as mean ± SD (n = 3). Data were analyzed using Student’s t-test; ∗P < 0.05, ∗∗P < 0.01.
(C) CAT activity in leaves of rice plants subjected to H2O2 stress and H2O2 stress + melatonin. Data are presented as mean ± SD (n = 3). Data were analyzed using Student’s t-test; ∗P < 0.05, ∗∗P < 0.01.
(D) DAB staining of H2O2 in leaves from plants in the unstressed, H2O2 stressed, and H2O2 stressed + melatonin treatments.
Lower H2O2 accumulation was observed in OsCPK12-OE leaves, whereas higher H2O2 accumulation was observed in oscpk12-cr leaves (Figure 6B). CAT activity was lower in oscpk12-cr leaves than in those of ZH8015 and OsCPK12-OE (Figure 6C). These results showed that overexpression of OsCPK12 in rice can increase oxidative stress tolerance. Melatonin is essential for maintaining the redox balance of plant cells, decreasing reactive oxygen species, and regulating biotic and abiotic stress tolerance (Hardeland, 2016; Hong et al., 2018). Addition of 20 μM melatonin significantly ameliorated the leaf-senescence phenotype caused by 100 mM H2O2 treatment and reduced H2O2 accumulation in leaves compared with 100 mM H2O2 treatment alone (Figure 6A and 6B). Accumulation of H2O2 in oscpk12-cr leaves was also assessed by DAB staining (Figure 6D). Together, these results indicated that overexpression of OsCPK12 can increase oxidative stress tolerance in rice.
Discussion
OsCPK12 is a weak Ca2+-dependent protein kinase that harbors a typical myristoylation site and is localized in the membrane, cytoplasm, and nucleus.
OsCPK12 functions in multiple signal transduction pathways; it participates in the low-nitrogen stress response, regulates leaf senescence, positively regulates salt tolerance, and negatively modulates blast resistance (Asano et al., 2010, 2012; Xing et al., 2018; Wang et al., 2019). The oscpk12 mutant usually shows leaf yellowing at about 60 days after sowing (Wang et al., 2019). Here, CAT activities and H2O2 levels differed significantly between ZH8015 and oscpk12 from 40 days onward (Supplemental Figure S1A and S1B); however, the expression level of OsCPK12 was higher at 20 days after sowing (Supplemental Figure S1C). We speculate that a specific time may be required for physiological differences to occur in plants.
CPKs are key components in plant signaling that transduce calcium signals into physiological responses and display highly variable calcium dependence in their activities. An increasing number of CDPK enzymes with low or no detectable Ca2+-dependent changes in in vitro kinase activity have been identified, despite the conserved modular structure within the CDPK family and the deduced model of Ca2+-dependent CDPK activation (Boudsocq et al., 2012). Some CPKs require an increase in cytosolic [Ca2+], whereas others are active at basal [Ca2+] and/or contain degenerate EF hands (Yip Delormel and Boudsocq, 2019; Loranger et al., 2021), pointing to involvement of additional regulatory mechanisms such as phosphorylation or binding partners. Our study indicates that the kinase activity of recombinant OsCPK12 was enhanced after Ca2+ addition (Figure 1C and 1D). Previous research has shown that Ca2+-dependency can be uncoupled from kinase activity through deliberate removal of the CAD domain, often resulting in deregulated/uninhibited CPKs (Liese and Romeis, 2013; Yip Delormel and Boudsocq, 2019; Durian et al., 2020). However, some truncated variants are totally inactive, such as TgCDPK1 and OsCPK17 (Ingram et al., 2015; Almadanim et al., 2018), or show lower activity than the calcium-activated full-length kinase (Asai et al., 2013; Dubiella et al., 2013). In our study, OsCPK12-V/K (OsCPK12 without the CAD domain) was not a constitutively active kinase (Figure 2A and 2B). The Km value can be used to determine the affinity between a substrate and an enzyme, and the Km value of a kinase often differs among different substrates. The Km value of OsCPK12 was 2.97 μM when syntide 2 was used as a substrate (Figure 1E). Km values of OsCPK12 were 57.99 and 135.3 μM using OsCATA (1–40 aa) and OsCATC (1–40 aa) peptides, respectively (Supplemental Figure S11A and S11B), indicating that the reaction rate of OsCPK12 is more sensitive to OsCATA (1–40 aa) peptides.
Subcellular localization is also essential for CPK function. CPKs exhibit different subcellular localizations, including the cytosol, nucleus, plasma membrane, ER, peroxisome, mitochondrial outer membrane, and oil bodies (Harper et al., 2004). In our study, OsCPK12 was mainly localized to the plasma membrane but was also located in the cytoplasm, nucleus, and ER (Supplemental Figure S2A–S2G). Xing et al. (2018) showed that OsCPK12 was only located in the cell membrane. The discrepancy between our results and those of Xing et al. may be due to differences in the protein expression level of OsCPK12-GFP. It has been suggested that an N-myristoylation site may be responsible for the membrane association of CPKs. Arabidopsis CPK4 and CPK11, lacking any acylation motifs, were soluble, whereas 11 other CPKs (CPK2, CPK5, CPK25, CPK3, CPK9, CPK7, CPK8, CPK10, CPK13, CPK30, and CPK32) with a predicted myristoylation site were mainly or partially associated with membranes (Boudsocq et al., 2012). NMT and Myristoylator predicted that OsCPK12 could be myristoylated at glycine2. In our study, OsCPK12-V-GFP localized in the plasma membrane (Supplemental Figure S2I and S2J). Unexpectedly, the subcellular localizations of OsCPK12-V/K and OsCPK12-K were similar to that of full-length OsCPK12. This may have occurred because membrane association is related not only to a predicted N-terminal myristoylation site but also to an additional reversible process such as palmitoylation, protein interactions, or regulation of a polybasic domain by phosphorylation (Meinnel and Giglione, 2008).
CDPKs can also be translocated in response to stress stimuli. Mesembryanthemum crysallinum CDPK1 (McCDPK1) is mainly associated with the plasma membrane in unstressed cells but shows greater nuclear localization for interaction with CSP1 under salt stress (Patharkar and Cushman, 2000). AtCPK10/30/32 translocate to the nucleus in response to nitrate (Liu et al., 2017). Cytosol-to-nucleus translocation of AtCPK5 is required for its synergistic activation of defense genes with the TFs WRKY8, 28, or 48 upon ETI activation (Gao and He, 2013). AtCPK12 is rapidly activated and phosphorylated at the Ser-186 residue during hypoxia, and phosphorylated CPK12 shuttles from the cytoplasm to the nucleus, where it phosphorylates and enhances the stability of ERF-VII TFs (Fan et al., 2023). We predict that OsCPK12 targets the membrane by allowing itself to be a membrane protein or a regulator of related membrane target proteins to participate in signal transduction in response to stress stimulation. Whether the function of OsCPK12 is affected by its different localization patterns requires further study with a genetic complementation assay.
CAT isozymes in rice synergistically regulate plant development
Plant catalases are divided into three classes (Willekens et al., 1995). AtCAT1 is mainly expressed in reproductive tissues and seeds, whereas AtCAT2 is strongly expressed in photosynthetic tissues, and AtCAT3 is expressed constitutively, especially in roots and young leaves (Du et al., 2008). AtCAT1, AtCAT2, and AtCAT3 belong to class III, class I, and class II, respectively. Three CAT homologs have been identified in rice: OsCATA, OsCATB, and OsCATC. OsCATC belongs to class I, OsCATA to class II, and OsCATB to class III (Mhamdi et al., 2012).
OsCATA is expressed in all organs and shows the highest expression in leaves; OsCATB is predominantly expressed in roots; and OsCATC is mainly expressed in leaves (Zhang et al., 2016). Our results showed that OsCATA and OsCATC were highly expressed in green tissues such as leaves, stems, and sheaths (Supplemental Figures S12A and S12C), consistent with the expression pattern of OsCPK12, whereas OsCATB was highly expressed in stems, roots, and panicles (Supplemental Figure S12B). The function of CATs is related not only to their temporal and spatial expression patterns but also to their subcellular localization. Previous studies have shown that OsCATA is localized mainly in the cytoplasm, whereas OsCATB and OsCATC are localized mainly in peroxisomes (Zhang et al., 2016; Gao et al., 2021; You et al., 2022; Liao et al., 2023). We also detected GFP fluorescence signals of OsCATA, OsCATB, and OsCATC in the nucleus and cell membrane (Supplemental Figures S2 and S3). Previous studies in rice have shown that OsCATA, OsCATB, and OsCATC interact with STRK1 on the membrane (Zhou et al., 2018). CAT3 is localized in the peroxisome, cytoplasm, and cell membrane in Arabidopsis and interacts with CPK8 on the cell membrane. Therefore, CATs can interact with some kinases on the cell membrane and become activated (Zou et al., 2015; Zhou et al., 2018). OsCATB-GFP has also been detected at the PM on the basis of weak fluorescence signals (Gao et al., 2021), and OsCATC has been found in punctate cellular compartments (Chen et al., 2023). Our results confirmed that OsCATs were indeed localized on the cell membrane but that OsCATA, OsCATB, and OsCATC were also localized in the nucleus.
Different CAT isozymes may participate synergistically in plant growth and development. Analysis of the CAT isozyme spectrum in Arabidopsis leaves showed that photorespiration was mainly related to CAT2, and a CAT3 band was also gradually obvious. In addition, some hybrid CAT holoenzyme bands were seen during growth and senescence of Arabidopsis, indicating that the CAT holoenzyme in Arabidopsis leaves involved not only CAT2 but also CAT3 (Zimmermann et al., 2006). The CAT holoenzyme in Arabidopsis leaves was in the form of a ∼220-kDa tetramer that was mainly encoded by CAT2 and CAT3 (Li et al., 2015). The predominant form of CAT in rice leaves is a homomeric complex of OsCATC isoforms (Zhang et al., 2016). Recent research also shows that OsCATA, OsCATB, and OsCATC may participate in the AvrPiz-t–ROD1–CATs–APIP6/RIP1 hierarchical regulatory immunity network, indicating that rice catalases may function as homomeric or heteromeric complexes (You et al., 2022). Our study demonstrated that OsCATA, OsCATB, and OsCATC interacted with each other in a Ser11 phosphorylation-independent manner, and lack of Ser11 phosphorylation on OsCATA and OsCATC did not affect their interaction with OsCPK12 (Supplemental Figures S13 and S14).
noe1, a loss-of-function mutation of OsCATC, also exhibits leaf cell death with H2O2 overaccumulation (Lin et al., 2012). In our study, oscata-cr and oscatc-cr showed similar photorespiration-deficit phenotypes (Figure 4 and Supplemental Figure S8), whereas oscatb-cr plants showed no apparent premature-senescence phenotype. Numerous studies have demonstrated that CAT not only functions as a key H2O2 scavenging enzyme but also plays an important role in leaf morphology. Mutation of Arabidopsis CAT2 results in hyponastic leaves because of changes in auxin levels (Gao et al., 2014), and loss of OsCATB function in rice results in abaxially rolled flag leaves (Wang et al., 2023a; 2023b). We therefore speculate that OsCATB is mainly involved in development of rice leaf morphology, whereas OsCATA and OsCATC may function directly in the photorespiratory process of rice.
OsCPK12 interacts with and phosphorylates OsCATA and OsCATC to regulate H2O2 homeostasis
H2O2 serves as a main transmitter of redox signals and functions directly in various biological processes such as senescence. Concentrations of H2O2 are normally maintained at low levels in plants to avoid cytotoxic effects. H2O2 concentration in the peroxisome may be as low as 10 μM (Foyer and Noctor, 2016) and is around 253 μM in rice leaves under normal conditions (Zhang et al., 2016). Required and appropriate levels of H2O2 are governed by CAT, ascorbate peroxidase, glutathione peroxidase, and phenol peroxidase (Asada, 1999; Dat et al., 2000). CAT, a major scavenger of H2O2, degrades H2O2 into oxygen and water and is primarily located in peroxisomes (Mhamdi et al., 2012). CATs have a very fast turnover rate but an extraordinarily low affinity for H2O2 compared with enzymes such as ascorbate peroxidase and peroxiredoxin (Foyer et al., 2009; Mhamdi et al., 2012). Apparent Km values of CAT for H2O2 reportedly range from 38 to 600 mM in various organisms (Switala and Loewen, 2002). Plants thus need mechanisms to potentiate CAT with higher scavenging efficiency for the low concentrations of H2O2 they normally maintain.
Numerous studies have demonstrated that phosphorylation of CAT may increase its activity and that different phosphorylation sites are likely to be associated with different stimuli (Rafikov et al., 2014; Zou et al., 2015; Zhou et al., 2018). For example, Endothelin-1 stimulates CAT activity through protein kinase Cδ (PKCδ)-mediated serine 167 phosphorylation (Rafikov et al., 2014). CPK8 phosphorylates the Ser261 of CAT3 to regulate CAT activity in Arabidopsis (Zou et al., 2015). STRK1 activates OsCatC by phosphorylation at Tyr210 in rice (Zhou et al., 2018). In our study, CAT activities were higher in leaves of OsCPK12-OE plants than in those of ZH8015, oscpk12, and oscpk12-cr plants (Figure 1D). OsCPK12 phosphorylates both OsCATA and OsCATC, mainly at Ser11. The Ser11 of OsCATA/OsCATC was not conserved in OsCATB (Supplemental Figure S12D), which has a Gly11 at this position. Furthermore, overexpression of continuously phosphorylated OsCATAS11D-OE and OsCATCS11D-OE increased CAT activity and reduced H2O2 content, but that of unphosphorylated OsCATAS11A-OE and OsCATCS11A-OE in oscpk12-cr plants did not. In vitro, OsCATAS11D, OsCATCS11D, and pre-phosphorylated OsCATA and OsCATC mediated by OsCPK12 all exhibited higher CAT activity than wild-type OsCATA and OsCATC, OsCATAS11A, and OsCATCS11A (Figure 5G). Our results showed that phosphorylation of OsCATA and OsCATC at Ser11 increased their CAT activity, thereby improving oxidative stress tolerance in rice.
Previous studies have shown that most wild-type CAT is present as a mixture of monomers and dimers, but S167D CAT is primarily tetrameric, as required for maximal activity, indicating that phosphorylation of specific sites can affect the activity of CAT by affecting its multimeric structure (Rafikov et al., 2014). Thus, how phosphorylation of OsCATA and OsCATC at Ser11 affects their CAT activity requires further study. Whether OsCPK12 affects the multimeric forms of OsCATA and OsCATC by phosphorylating Ser11 to increase their CAT activity also remains to be determined.
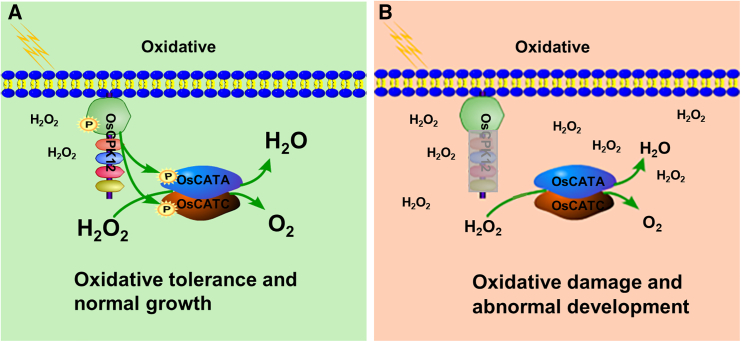
As illustrated in our model in Figure 7, phosphorylation of OsCPK12 in wild-type plants increased OsCATA and OsCATC activities via phosphorylation, mainly at Ser11, thereby inhibiting the accumulation of H2O2. However, loss of function of OsCPK12 in mutant plants prevents phosphorylation of OsCATA and OsCATC, and non-phosphorylated OsCATA and OsCATC have lower CAT activity. Low CAT enzyme activity causes accumulation of H2O2, which results in a state of oxidative stress in the plant. In summary, OsCPK12 phosphorylates OsCATA and OsCATC at Ser11 to increase their activity, thus maintaining H2O2 homeostasis, and overexpression of OsCPK12 improves tolerance to oxidative stress in rice.
Figure 7.
A proposed working model for the role of OsCPK12 in regulation of oxidative stress tolerance in rice.
In WT plants, phosphorylation of OsCPK12 increases OsCATA and OsCATC activities via phosphorylation, mainly at Ser11, thereby inhibiting accumulation of H2O2. However, loss of function of OsCPK12 in mutant plants prevents phosphorylation of OsCATA and OsCATC, and non-phosphorylated OsCATA and OsCATC have lower CAT activity. Low CAT enzyme activity causes accumulation of H2O2, which results in a state of oxidative stress in the plant. In summary, OsCPK12 phosphorylates OsCATA and OsCATC at Ser11 to increase their activity and maintain H2O2 homeostasis, and overexpression of OsCPK12 improves tolerance to oxidative stress in rice. The green background represents the WT, and the pink background represents the mutant; P, phosphate group; the larger the font, the stronger the enzyme activity.
Methods
Plant materials and growth conditions
Wild-type rice (Oryza sativa Zhonghui 8015 [ZH8015]) and OsCPK12-knockout (oscpk12-cr) and OsCPK12-OE transgenic plants were used to construct transgenic lines for functional analysis and investigation of the H2O2 stress response. For H2O2 treatment, 10-day-old seedlings (24 plants per genotype) were transferred to a hydroponic culture solution containing 100 mM H2O2 or 100 mM H2O2 and 20 μM melatonin. After 6 days, the leaves were stained with DAB. Chlorophyll content, CAT activity, and H2O2 content were measured after 6 days of treatment. All plants were grown in a greenhouse with white fluorescent light at a photon flux density of 300–350 mmol m−2 s−1 and a 16-h light, 30°C/8-h dark, 25°C cycle. Relative humidity in the greenhouse was maintained at 60%–70%.
To study the significance of Ser11 phosphorylation of OsCATA and OsCATC in planta, the CDSs of mutated OsCATA (OsCATAS11A), OsCATC (OsCATCS11A), OsCATA (OsCATAS11D), and OsCATC (OsCATCS11D) were generated by site-directed mutagenesis and subcloned into the pCAMBIA2300 vector (primers are listed in Supplemental Table 2). The mutated sequences were overexpressed in the oscpk12-cr line via Agrobacterium-mediated transformation as described previously (Toki et al., 2006). The seedlings (T2) regenerated from the positive transgenic calli were used for measurement of CAT activity and H2O2 content. ZH8015 and oscpk12-cr seedlings were used as controls.
Physiological measurements
Chlorophyll was extracted from fully expanded rice leaves. In brief, 100 mg of leaf tissue was submerged in 10 ml of 80% acetone for 24 h in darkness. Absorbance of the extract was measured at 470, 645, and 663 nm using a DU800 visible spectrophotometer (Beckman, CA, USA). All experiments were repeated with three biological replicates, and mean values of the three repeats are presented.
Fresh leaves were wiped clean and accurately weighed to within 0.1 g. One milliliter of PBS was added, and the samples were ground into homogenates under ice-bath conditions. After centrifugation at 3500 rpm for 10 min, the supernatants were used for subsequent experiments. CAT activity in vivo and contents of soluble protein (SP) and H2O2 were determined using commercial assay kits from Nanjing Jiancheng Bioengineering Research Institute (SP, A045-2; CAT, A007-1-1; H2O2, A064-1-1). Specific experimental steps were performed according to the manufacturer’s instructions.
CAT activities of purified OsCATA-GST, OsCATAS11A-GST, OsCATAS11D-GST, OsCATC-GST, OsCATCS11A-GST, and OsCATCS11D-GST were measured using a UV spectrophotometer in a reaction mixture containing 50 mM KH2PO4 (pH 7.5) and 10 mM H2O2 at 30°C. H2O2 consumption was detected as a decrease in absorbance at 240 nm, and CAT activity was calculated using a molar extinction coefficient for H2O2 of 39.4 mol−1 cm−1 (Weydert and Cullen, 2010). One unit represented the amount of enzyme that catalyzed the decomposition of 1 mM H2O2 per minute at 30°C. To determine the effect of OsCPK12 on OsCATA and OsCATC activity, purified His-TF-OsCPK12 proteins were mixed with OsCATA and OsCATC for 1 h at 30°C, and CAT activity was determined. All experiments were repeated with three biological replicates, and mean values of the three repeats are presented.
RNA isolation and qRT–PCR
Total RNA was extracted from leaves, stems, sheaths, roots, and panicles of ZH8015. RNA was treated with DNase (Promega, Madison, WI) and reverse transcribed using the ImProm-II Reverse Transcription System (Promega). Quantitative real-time PCR was performed on a LightCycler 480 instrument (Roche Applied Science, Mannheim, Germany) using GoTaq qPCR Master Mix (Promega). The reaction was performed in a 96-well plastic plate (Roche). Real-time PCR data were collected using the following cycling conditions: 5 min of initial denaturation at 95°C, followed by 45 cycles of 10 s at 95°C, 10 s at 58°C, and 15 s at 72°C. Cycling threshold (Ct) values were normalized using the UBQ10 reference gene.
Subcellular localization assay
To detect the subcellular localization of OsCPK12, OsCATA, OsCATB, and OsCATC, their full-length CDSs were fused to the N terminus of PYBA-1132-GFP (Yan et al., 2012). To observe the subcellular localization of different OsCPK12 domains, truncated derivatives of OsCPK12 were ligated into the GFP vector pYBA1132-EGFP, resulting in OsCPK12-V-GFP, OsCPK12-K-GFP, and OsCPK12-V/K-GFP. OsLazy1 encodes a fusion protein that indicates the nucleus and plasma membrane (Li et al., 2019), and OsLazy1 was fused with mCherry to generate mCherry-LA1 as described in Wang et al. (2023a), 2023b. OsGhd7 has been demonstrated to be a nuclear-localized protein (Xue et al., 2008), and it was fused with mCherry to generate OsGhd7-mCherry as described in Tu et al. (2020) and Wang et al. (2023a), 2023b. HDEL is an ER-localized signal peptide that was fused to mCherry, resulting in pCAMBIA1300-35S-ER-mCherry-HDEL (Agrisera: AS10-683-25). Lysine–serine–arginine–methionine (KSRM) is a polypeptide necessary and sufficient for targeting of transiently expressed proteins to peroxisomes (Trelease et al., 1996); it was fused with mCherry to generate KSRM-mCherry. The recombinant proteins were transiently coexpressed with empty mCherry (used as a cytoplasmic and nuclear marker) or OsGhd7-mCherry (used as a nuclear marker), Lazy1-mCherry (used as a plasma membrane and nuclear marker), mCherry-HDEL (used as an ER marker), and KSRM-mCherry (used as a peroxisomal marker) in rice protoplasts. Fluorescence was observed with a laser scanning confocal microscope (ZEISS 750) 14–24 h after transfection of rice protoplasts. Primers used to generate the constructs are listed in Supplemental Table 2.
Y2H assay
Y2H analysis was performed using the Matchmaker Gold Yeast Two-Hybrid System (Clontech) according to the manufacturer’s instructions. Truncated OsCPK12 was cloned into the pGBKT7 vector, and the resulting construct was transformed into the yeast strain Y2HGold. The transformed Y2HGold yeast strain was fused with the Y187 yeast strain containing a rice cDNA library. Medium lacking Leu, Trp, His, and Ade was used for selection. Positive clones were selected for sequencing. The interactions between OsCPK12 and CATs were verified by Y2H assays. The coding regions of OsCATA, OsCATB, and OsCATC were cloned into the pGADT7 prey plasmid and cotransformed into the yeast strain Y2HGold with OsCPK12 as bait. Primers used for Y2H are listed in Supplemental Table 2.
BiFC assay
To generate BiFC vectors, the full-length CDS of OsCPK12 was amplified via PCR and cloned into the pSPYNE-35S-pUC-SPYNE vector, resulting in CPK12-YNE, and the full-length cDNA sequences of OsCATA, OsCATB, and OsCATC were separately amplified via PCR and cloned into the pSPYNE-35S-pUC-SPYCE vector to produce OsCATA-YCE, OsCATB-YCE, and OsCATC-YCE. The constructs were transiently expressed in N. benthamiana by agroinfiltration. Four days after infiltration (DAI), fluorescence was observed using a laser scanning confocal microscope (ZEISS 750). Using an argon laser, GFP was excited at 488 nm and RFP at 552 nm. Primers used to generate the BiFC constructs are listed in Supplemental Table 2.
LCI assay
For the LCI assay, the coding region of OsCPK12 was cloned into pCAMBIA1300-CLuc, resulting in OsCPK12-CLuc, and the coding regions of OsCATA, OsCATB, and OsCATC were cloned into pCAMBIA1300-NLuc to produce OsCATA-NLuc, OsCATB-NLuc, and OsCATC-NLuc. The LCI assay was performed as described previously (Chen et al., 2008). Primers used to generate the LCI constructs are listed in Supplemental Table 2.
CoIP assay
For coIP assays, the full-length CDSs of OsCPK12, OsCATA, OsCATB, and OsCATC were separately amplified via PCR and fused with sequences encoding a GFP tag and an Myc tag driven by the 35S promoter. The constructs were coexpressed in rice protoplasts as described previously (He et al., 2018). Protoplasts were transfected and incubated for 14–20 h. Total proteins were extracted with NB buffer (50 mM Tris–MES [pH 8.0], 0.5 M sucrose, 1 mM MgCl2, 10 mM EDTA, and Roche plant protease inhibitor cocktail) and then immunoprecipitated with anti-MYC magnetic beads (Bimake, catalog no. B26301) according to the manufacturer’s instructions. Immunoprecipitated proteins were separated via SDS–PAGE (4%–25% gel) and analyzed by immunoblotting with anti-hemagglutinin (TransGen; HT301) or anti-Myc antibodies (TransGen; HT101). After incubation with a secondary antibody (HuaAn; HA1006) for 1.5 h, the immunoblot signal was visualized with Super ECL (Coolaber; SL1350). Primers used to generate the coIP constructs are listed in Supplemental Table 2.
Expression and purification of recombinant proteins
The pCold TF vector (Takara) is a fusion cold shock expression vector that expresses a 48-kDa trigger factor chaperone as a soluble tag. The pCold TF vector also contains a His-tag sequence, and we therefore named it the His-TF vector. His-TF vectors containing full-length CDSs of OsCPK12 (His-TF-OsCPK12), OsCATA (His-TF-OsCATA), and OsCATC (His-TF-OsCATC) were constructed. GST fusions with full-length CDSs of OsCPK12 (OsCPK12-GST), OsCPK12-V/K (OsCPK12-V/K-GST), OsCATA (OsCATA-GST), OsCATC (OsCATC-GST), OsCATA-N (OsCATA-N-GST), OsCATC-N (OsCATC-N-GST), OsCATA-C (OsCATA-C-GST), and OsCATC-C (OsCATC-C-GST) were also constructed for pull-down and phosphorylation assays. Mutant isomers of OsCATA-N-GST and OsCATC-N-GST, including OsCATAS9A/S11A-N-GST, OsCATAS9A-N-GST, OsCATAS11A-N-GST, OsCATAS104A/T105A-N-GST, OsCATAS164-N-GST, OsCATAT351A-N-GST, OsCATCS9A/S10A/S11A/S18A/T19A/S21A-N-GST, OsCATCS9A/S10A/S11A-N-GST, OsCATCS18A/T19A/S21A-N-GST, OsCATCS9A/S10A-N-GST, OsCATCS11A-N-GST, OsCATCS347A-N-GST, and OsCATCT351-N-GST, were constructed for validation of specific phosphorylation sites. The recombinants were transformed into Escherichia coli BL21, and protein expression was induced using 0.1 mM isopropyl β-D-thiogalactoside at 15°C for 12 h. The cells were collected and analyzed by SDS–PAGE, together with untreated control cells. The induced cells were disrupted by applying ultrasound, the mixture was centrifuged for 30 min at 10 000 rpm, and the GST-tagged proteins and His-tagged proteins in the supernatant were purified using BeaverBeads GSH (catalog no. 70601-100, Xi Yan Technology) and the His-tag protein Purification Kit (catalog no. P2226, Beyotime Biotechnology), respectively. Protein concentration was measured using an NCM BCA protein assay kit (catalog no. WB6501 New Cell and Molecular Biotech). Primers used for expressed protein constructs are listed in Supplemental Table 2.
Pull-down assay
For the in vitro GST pull-down assay, roughly 2 μg of His-TF-OsCPK12 was mixed with 4 μg of GST, 2 μg of OsCATA-GST, or 2 μg of OsCATC-GST. The mixed supernatants were incubated with 25 μl of BeyoGold His-tag resin (catalog no. P2226 Beyotime Biotechnology) at 4°C for 4 h, then washed five times with wash buffer. Finally, the protein was eluted on resin with 40 μl of eluent. His-TF-OsCPK12 was detected with an anti-His antibody (A00186-100). GST, OsCATA-GST, and OsCATC-GST were detected with an anti-GST antibody (TransGen; HT801).
Kinase activity assays and in vitro phosphorylation assay
Enzyme activity assays with recombinant full-length OsCPK12 and truncated OsCPK12 were performed using a Kinase-Glo Luminescent Kinase Assay kit (catalog no. #V6071; Promega Biotechnology). Proteins used for kinase activity assays were washed three times (10 min each) with a solution containing 2 mM EGTA to remove calcium; washed three to five times (10 min each) with a solution that lacked 2 mM EGTA; and finally eluted with the eluent to obtain purified protein. Enzymatic buffer (30 mM Tris–HCl [pH 8.0], 100 mM NaCl, 0.5 mM DTT, 20 mM MgCl2, 50 μM syntide 2, and 100 μM ATP) and 0.5 mM Ca2+ or 2 mM EGTA (as an extracellular calcium ion chelator) were added to a 200-μl centrifuge tube. Approximately 2 μg of recombinant protein was added to start the reaction, which was incubated for 0.5 h at 30°C. Denatured OsCPK12 was included as a negative control. The reactions were then boiled at 100°C for 1 min, and 50 μl of reaction mixture was transferred to a white 96-well reaction plate, where it was incubated with 50 μl Kinase-Glo reagent for 20 min at room temperature. Luminescence values (relative light units [RLU]) were then recorded on a GloMax-Multi detection system (Promega) as specified in the instructions of the Kinase-Glo Luminescent Kinase Assay kit. The luminescence signal was correlated with the amount of ATP present and was therefore inversely correlated with the amount of kinase activity. The amount of ATP consumed was positively correlated with kinase activity. OsCATA (1–40 aa) and OsCATC (1–40 aa) peptides containing the Ser11 site were synthesized by GenScript Biotechnology. For determination of OsCPK12 Km values, different concentrations of syntide 2 (0–200 μM), OsCATA(1–40 aa) (0–1000 μM), and OsCATA(1–40 aa) (0–1000 μM) were used in the reactions.
Approximately 2 μg of kinase (His-TF and His-TF-OsCPK12) and 1 μg of substrate (OsCATA-GST/OsCATA-N-GST and OsCATC-GST/OsCATC-N-GST) were used. Proteins were combined with 1× kinase buffer (100 mM Tris–HCl [pH 8.0], 5 mM DTT, 5 mM EGTA, 5 mM MgCl2, and 100 μM ATP) in a total volume of 30 μl for the in vitro phosphorylation assay. The reactions were incubated at 30°C for 30 min and stopped by adding 5× loading buffer and boiling for 5 min. An aliquot of each assay was resolved by SDS–PAGE, and the gels were stained and dried. Protein phosphorylation was detected by a 6% 50 μM Phos-Tag acrylamide gel (Phos-tag AAL-107; Wako) assay, and protein loading was indicated by CBB staining.
Statistical analysis
Data were compared using Student’s t-test or one-way analysis of variance (ANOVA); P values were generated using GraphPad Prism 8.0.
The measured RLU value was positively correlated with the amount of ATP present and thus inversely correlated with kinase activity. Curve fitting and determination of the Ca2+ K0.5 (concentration required for half-maximal activity) of OsCPK12 were performed using a dose-response-stimulation (variable slope) model in GraphPad Prism software.
ImageJ was used for gray-scale analysis of protein bands in the western blots.
Funding
This project was supported by grants from the National Natural Science Foundation of China (#32100224 and #31961143016); the Science and Technology Program of Zhejiang Province, China (2022R51009); the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2013-CNRRI); the Earmarked Fund for the China Agricultural Research System (CARS-01); and the Zhejiang Provincial Natural Science Foundation of China (grant no. LY23C130003).No conflict of interest is declared.
Author contributions
Conceptualization, B.F.W., Q.E.L., S.H.C., and L.Y.C; methodology, B.F.W. and Q.E.L.; investigation, B.F.W. and P.X.; writing – original draft, B.F.W., Q.E.L., and P.X.; writing – review & editing, B.F.W. and Q.E.L.; funding acquisition, L.Y.C., S.H.C., P.Y., and B.F.W.; resources, Y.X.Z., X.D.Z., W.X.W., D.B.C., J.L.F., Y.B.H., X.H.S., and L.P.S.
Acknowledgments
No conflict of interest is declared.
Published: December 21, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Shihua Cheng, Email: chengshihua@caas.cn.
Qunen Liu, Email: liuqunen@caas.cn.
Liyong Cao, Email: caoliyong@caas.cn.
Supplemental information
References
- Almadanim M.C., Gonçalves N.M., Rosa M.T.G., Alexandre B.M., Cordeiro A.M., Rodrigues M., Saibo N.J.M., Soares C.M., Romão C.V., Oliveira M.M., et al. The rice cold-responsive calcium-dependent protein kinase OsCPK17 is regulated by alternative splicing and post-translational modifications. Biochim. Biophys. Acta Mol. Cell Res. 2018;1865:231–246. doi: 10.1016/j.bbamcr.2017.10.010. [DOI] [PubMed] [Google Scholar]
- Alves H.L.S., Matiolli C.C., Soares R.C., Almadanim M.C., Oliveira M.M., Abreu I.A. Carbon/nitrogen metabolism and stress response networks-calcium-dependent protein kinases as the missing link? J. Exp. Bot. 2021;72:4190–4201. doi: 10.1093/jxb/erab136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K. THE WATER-WATER CYCLE IN CHLOROPLASTS: Scavenging of Active Oxygens and Dissipation of Excess Photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:601–639. doi: 10.1146/annurev.arplant.50.1.601. [DOI] [PubMed] [Google Scholar]
- Asai S., Ichikawa T., Nomura H., Kobayashi M., Kamiyoshihara Y., Mori H., Kadota Y., Zipfel C., Jones J.D.G., Yoshioka H. The variable domain of a plant calcium-dependent protein kinase (CDPK) confers subcellular localization and substrate recognition for NADPH oxidase. J. Biol. Chem. 2013;288:14332–14340. doi: 10.1074/jbc.M112.448910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T., Hayashi N., Kobayashi M., Aoki N., Miyao A., Mitsuhara I., Ichikawa H., Komatsu S., Hirochika H., Kikuchi S., et al. A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 2012;69:26–36. doi: 10.1111/j.1365-313X.2011.04766.x. [DOI] [PubMed] [Google Scholar]
- Asano T., Wakayama M., Aoki N., Komatsu S., Ichikawa H., Hirochika H., Ohsugi R. Overexpression of a calcium-dependent protein kinase gene enhances growth of rice under low-nitrogen conditions. Plant Biotechnol. 2010;27:369–373. [Google Scholar]
- Boudsocq M., Droillard M.J., Regad L., Laurière C. Characterization of Arabidopsis calcium-dependent protein kinases: activated or not by calcium? Biochem. J. 2012;447:291–299. doi: 10.1042/BJ20112072. [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Sheen J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013;18:30–40. doi: 10.1016/j.tplants.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C., Leng Y., Kufe D. Catalase activity is regulated by c-Abl and Arg in the oxidative stress response. J. Biol. Chem. 2003;278:29667–29675. doi: 10.1074/jbc.M301292200. [DOI] [PubMed] [Google Scholar]
- Chen H., Lin Q., Li Z., Chu J., Dong H., Mei Q., Xuan Y. Calcineurin B-like interacting protein kinase 31 confers resistance to sheath blight via modulation of ROS homeostasis in rice. Mol. Plant Pathol. 2023;24:221–231. doi: 10.1111/mpp.13291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008;146:368–376. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhou X., Chang S., Chu Z., Wang H., Han S., Wang Y. Calcium-dependent protein kinase 21 phosphorylates 14-3-3 proteins in response to ABA signaling and salt stress in rice. Biochem. Biophys. Res. Commun. 2017;493:1450–1456. doi: 10.1016/j.bbrc.2017.09.166. [DOI] [PubMed] [Google Scholar]
- Cui X., Zhao P., Liang W., Cheng Q., Mu B., Niu F., Yan J., Liu C., Xie H., Kav N.N.V., et al. A Rapeseed WRKY Transcription Factor Phosphorylated by CPK Modulates Cell Death and Leaf Senescence by Regulating the Expression of ROS and SA-Synthesis-Related Genes. J. Agric. Food Chem. 2020;68:7348–7359. doi: 10.1021/acs.jafc.0c02500. [DOI] [PubMed] [Google Scholar]
- Dat J., Vandenabeele S., Vranová E., Van Montagu M., Inzé D., Van Breusegem F. Dual action of the active oxygen species during plant stress responses. Cell. Mol. Life Sci. 2000;57:779–795. doi: 10.1007/s000180050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y.Y., Wang P.C., Chen J., Song C.P. Comprehensive functional analysis of the catalase gene family in Arabidopsis thaliana. J. Integr. Plant Biol. 2008;50:1318–1326. doi: 10.1111/j.1744-7909.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- Dubiella U., Seybold H., Durian G., Komander E., Lassig R., Witte C.P., Schulze W.X., Romeis T. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA. 2013;110:8744–8749. doi: 10.1073/pnas.1221294110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durian G., Sedaghatmehr M., Matallana-Ramirez L.P., Schilling S.M., Schaepe S., Guerra T., Herde M., Witte C.P., Mueller-Roeber B., Schulze W.X., et al. Calcium-Dependent Protein Kinase CPK1 Controls Cell Death by In Vivo Phosphorylation of Senescence Master Regulator ORE1. Plant Cell. 2020;32:1610–1625. doi: 10.1105/tpc.19.00810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan B., Liao K., Wang L.N., Shi L.L., Zhang Y., Xu L.J., Zhou Y., Li J.F., Chen Y.Q., Chen Q.F., et al. Calcium-dependent activation of CPK12 facilitates its cytoplasm-to-nucleus translocation to potentiate plant hypoxia sensing by phosphorylating ERF-VII transcription factors. Mol. Plant. 2023;16:979–998. doi: 10.1016/j.molp.2023.04.002. [DOI] [PubMed] [Google Scholar]
- Foyer C.H., Bloom A.J., Queval G., Noctor G. Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Annu. Rev. Plant Biol. 2009;60:455–484. doi: 10.1146/annurev.arplant.043008.091948. [DOI] [PubMed] [Google Scholar]
- Foyer C.H., Noctor G. Stress-triggered redox signalling: what's in pROSpect? Plant Cell Environ. 2016;39:951–964. doi: 10.1111/pce.12621. [DOI] [PubMed] [Google Scholar]
- Gao X., He P. Nuclear dynamics of Arabidopsis calcium-dependent protein kinases in effector-triggered immunity. Plant Signal. Behav. 2013;8 doi: 10.4161/psb.23868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Yuan H.M., Hu Y.Q., Li J., Lu Y.T. Mutation of Arabidopsis CATALASE2 results in hyponastic leaves by changes of auxin levels. Plant Cell Environ. 2014;37:175–188. doi: 10.1111/pce.12144. [DOI] [PubMed] [Google Scholar]
- Gao M., He Y., Yin X., Zhong X., Yan B., Wu Y., Chen J., Li X., Zhai K., Huang Y., et al. Ca2+ sensor-mediated ROS scavenging suppresses rice immunity and is exploited by a fungal effector. Cell. 2021;184:5391–5404.e17. doi: 10.1016/j.cell.2021.09.009. [DOI] [PubMed] [Google Scholar]
- Hardeland R. Melatonin in Plants-Diversity of Levels and Multiplicity of Functions. Front. Plant Sci. 2016;7:198. doi: 10.3389/fpls.2016.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J.F., Breton G., Harmon A. Decoding Ca(2+) signals through plant protein kinases. Annu. Rev. Plant Biol. 2004;55:263–288. doi: 10.1146/annurev.arplant.55.031903.141627. [DOI] [PubMed] [Google Scholar]
- He F., Zhang F., Sun W., Ning Y., Wang G.L. A Versatile Vector Toolkit for Functional Analysis of Rice Genes. Rice. 2018;11:27. doi: 10.1186/s12284-018-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Zhang Y., Sinumporn S., Yu N., Zhan X., Shen X., Chen D., Yu P., Wu W., Liu Q., et al. Premature leaf senescence 3, encoding a methyltransferase, is required for melatonin biosynthesis in rice. Plant J. 2018;95:877–891. doi: 10.1111/tpj.13995. [DOI] [PubMed] [Google Scholar]
- Ingram J.R., Knockenhauer K.E., Markus B.M., Mandelbaum J., Ramek A., Shan Y., Shaw D.E., Schwartz T.U., Ploegh H.L., Lourido S. Allosteric activation of apicomplexan calcium-dependent protein kinases. Proc. Natl. Acad. Sci. USA. 2015;112:4975–4984. doi: 10.1073/pnas.1505914112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchiswamy C.N., Takahashi H., Quadro S., Maffei M.E., Bossi S., Bertea C., Zebelo S.A., Muroi A., Ishihama N., Yoshioka H., et al. Regulation of Arabidopsis defense responses against Spodoptera littoralis by CPK-mediated calcium signaling. BMC Plant Biol. 2010;10:97. doi: 10.1186/1471-2229-10-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latz A., Mehlmer N., Zapf S., Mueller T.D., Wurzinger B., Pfister B., Csaszar E., Hedrich R., Teige M., Becker D. Salt stress triggers phosphorylation of the Arabidopsis vacuolar K+ channel TPK1 by calcium-dependent protein kinases (CDPKs) Mol. Plant. 2013;6:1274–1289. doi: 10.1093/mp/sss158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Liu J., Wang G., Cha J.Y., Li G., Chen S., Li Z., Guo J., Zhang C., Yang Y., et al. A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell. 2015;27:908–925. doi: 10.1105/tpc.114.135095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Liang Y., Yuan Y., Wang L., Meng X., Xiong G., Zhou J., Cai Y., Han N., Hua L., et al. OsBRXL4 Regulates Shoot Gravitropism and Rice Tiller Angle through Affecting LAZY1 Nuclear Localization. Mol. Plant. 2019;12:1143–1156. doi: 10.1016/j.molp.2019.05.014. [DOI] [PubMed] [Google Scholar]
- Liao M., Ma Z., Kang Y., Zhang B., Gao X., Yu F., Yang P., Ke Y. ENHANCED DISEASE SUSCEPTIBILITY 1 promotes hydrogen peroxide scavenging to enhance rice thermotolerance. Plant Physiol. 2023;192:3106–3119. doi: 10.1093/plphys/kiad257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liese A., Romeis T. Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK) Biochim. Biophys. Acta. 2013;1833:1582–1589. doi: 10.1016/j.bbamcr.2012.10.024. [DOI] [PubMed] [Google Scholar]
- Lin A., Wang Y., Tang J., Xue P., Li C., Liu L., Hu B., Yang F., Loake G.J., Chu C. Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol. 2012;158:451–464. doi: 10.1104/pp.111.184531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K.H., Niu Y., Konishi M., Wu Y., Du H., Sun Chung H., Li L., Boudsocq M., McCormack M., Maekawa S., et al. Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature. 2017;545:311–316. doi: 10.1038/nature22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loranger M.E.W., Huffaker A., Monaghan J. Truncated variants of Ca2+-dependent protein kinases: a conserved regulatory mechanism? Trends Plant Sci. 2021;26:1002–1005. doi: 10.1016/j.tplants.2021.07.002. [DOI] [PubMed] [Google Scholar]
- Meinnel T., Giglione C. Protein lipidation meets proteomics. Front. Biosci. 2008;13:6326–6340. doi: 10.2741/3157. [DOI] [PubMed] [Google Scholar]
- Mhamdi A., Noctor G., Baker A. Plant catalases: peroxisomal redox guardians. Arch. Biochem. Biophys. 2012;525:181–194. doi: 10.1016/j.abb.2012.04.015. [DOI] [PubMed] [Google Scholar]
- Mhamdi A., Queval G., Chaouch S., Vanderauwera S., Van Breusegem F., Noctor G. Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 2010;61:4197–4220. doi: 10.1093/jxb/erq282. [DOI] [PubMed] [Google Scholar]
- Ormancey M., Thuleau P., Mazars C., Cotelle V. CDPKs and 14-3-3 Proteins: Emerging Duo in Signaling. Trends Plant Sci. 2017;22:263–272. doi: 10.1016/j.tplants.2016.11.007. [DOI] [PubMed] [Google Scholar]
- Patharkar O.R., Cushman J.C. A stress-induced calcium-dependent protein kinase from Mesembryanthemum crystallinum phosphorylates a two-component pseudo-response regulator. Plant J. 2000;24:679–691. doi: 10.1046/j.1365-313x.2000.00912.x. [DOI] [PubMed] [Google Scholar]
- Rafikov R., Kumar S., Aggarwal S., Hou Y., Kangath A., Pardo D., Fineman J.R., Black S.M. Endothelin-1 stimulates catalase activity through the PKCδ-mediated phosphorylation of serine 167. Free Radic. Biol. Med. 2014;67:255–264. doi: 10.1016/j.freeradbiomed.2013.10.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronzier E., Corratgé-Faillie C., Sanchez F., Prado K., Brière C., Leonhardt N., Thibaud J.B., Xiong T.C. CPK13, a noncanonical Ca2+-dependent protein kinase, specifically inhibits KAT2 and KAT1 shaker K+ channels and reduces stomatal opening. Plant Physiol. 2014;166:314–326. doi: 10.1104/pp.114.240226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz P., Herde M., Romeis T. Calcium-dependent protein kinases: hubs in plant stress signaling and development. Plant Physiol. 2013;163:523–530. doi: 10.1104/pp.113.222539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Switala J., Loewen P.C. Diversity of properties among catalases. Arch. Biochem. Biophys. 2002;401:145–154. doi: 10.1016/S0003-9861(02)00049-8. [DOI] [PubMed] [Google Scholar]
- Toki S., Hara N., Ono K., et al. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006;47:969–976. doi: 10.1111/j.1365-313X.2006.02836.x. [DOI] [PubMed] [Google Scholar]
- Trelease R.N., Lee M.S., Banjoko A., Dunkelmann J. C-terminal polypeptides are necessary and sufficient for in vivo targeting of transiently-expressed proteins to peroxisomes in suspension-cultured plant cells. Protoplasma. 1996;195:156–167. [Google Scholar]
- Tu R., Wang H., Liu Q., Wang D., Zhou X., Xu P., Zhang Y., Wu W., Chen D., Cao L., et al. Characterization and genetic analysis of the oshpl3 rice lesion mimic mutant showing spontaneous cell death and enhanced bacterial blight resistance. Plant Physiol. Biochem. 2020;154:94–104. doi: 10.1016/j.plaphy.2020.05.001. [DOI] [PubMed] [Google Scholar]
- Wang B., Zhang Y., Bi Z., Liu Q., Xu T., Yu N., Cao Y., Zhu A., Wu W., Zhan X., et al. Impaired Function of the Calcium-Dependent Protein Kinase, OsCPK12, Leads to Early Senescence in Rice (Oryza sativa L.) Front. Plant Sci. 2019;10:52. doi: 10.3389/fpls.2019.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Xu J., Wang L., Zhou M., Nian J., Chen M., Lu X., Liu X., Wang Z., Cen J., et al. SEMI-ROLLED LEAF 10 stabilizes catalase isozyme B to regulate leaf morphology and thermotolerance in rice (Oryza sativa L.) Plant Biotechnol. J. 2023;21:819–838. doi: 10.1111/pbi.13999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Tu R., Ruan Z., Chen C., Peng Z., Zhou X., Sun L., Hong Y., Chen D., Liu Q., et al. Photoperiod and gravistimulation-associated Tiller Angle Control 1 modulates dynamic changes in rice plant architecture. TAG. Theoretical and applied genetics. Theor. Appl. Genet. 2023;136:160. doi: 10.1007/s00122-023-04404-z. [DOI] [PubMed] [Google Scholar]
- Wang L., Yu C., Xu S., Zhu Y., Huang W. OsDi19-4 acts downstream of OsCDPK14 to positively regulate ABA response in rice. Plant Cell Environ. 2016;39:2740–2753. doi: 10.1111/pce.12829. [DOI] [PubMed] [Google Scholar]
- Weydert C.J., Cullen J.J. Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nat. Protoc. 2010;5:51–66. doi: 10.1038/nprot.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willekens H., Inzé D., Van Montagu M., van Camp W. Catalases in plants. Mol. Breed. 1995;1:207–228. [Google Scholar]
- Xing Y., Guo S., Chen X., Du D., Liu M., Xiao Y., Zhang T., Zhu M., Zhang Y., Sang X., et al. Nitrogen Metabolism is Affected in the Nitrogen-Deficient Rice Mutant esl4 with a Calcium-Dependent Protein Kinase Gene Mutation. Plant Cell Physiol. 2018;59:2512–2525. doi: 10.1093/pcp/pcy169. [DOI] [PubMed] [Google Scholar]
- Xue W., Xing Y., Weng X., Zhao Y., Tang W., Wang L., Zhou H., Yu S., Xu C., Li X., et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- Yang X., Chen Z., Yin X., Wang Y., Yang Y., Yang Y. Genome-Wide Survey Indicates Diverse Physiological Roles of Dendrobium officinale Calcium-Dependent Protein Kinase Genes. Int. J. Mol. Sci. 2022;23:1298. doi: 10.3390/ijms23031298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X.H., Wang H., Ye Y.Y., Zeng G., Ma R.C., Mi F.G., Yao L. pYBA100: An Ease-of-use Binary Vector with LoxP-FRT Recombinase Site for Plant Transformation. Mol. Plant Breed. 2012;10:371–379. [Google Scholar]
- Ye N., Zhu G., Liu Y., Li Y., Zhang J. ABA controls H₂O₂ accumulation through the induction of OsCATB in rice leaves under water stress. Plant Cell Physiol. 2011;52:689–698. doi: 10.1093/pcp/pcr028. [DOI] [PubMed] [Google Scholar]
- Yip Delormel T., Boudsocq M. Properties and functions of calcium-dependent protein kinases and their relatives in Arabidopsis thaliana. New Phytol. 2019;224:585–604. doi: 10.1111/nph.16088. [DOI] [PubMed] [Google Scholar]
- You X., Zhang F., Liu Z., Wang M., Xu X., He F., Wang D., Wang R., Wang Y., Wang G., et al. Rice catalase OsCATC is degraded by E3 ligase APIP6 to negatively regulate immunity. Plant Physiol. 2022;190:1095–1099. doi: 10.1093/plphys/kiac317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Liu D., Yang B., Liu W.Z., Mu B., Song H., Chen B., Li Y., Ren D., Deng H., et al. Arabidopsis CPK6 positively regulates ABA signaling and drought tolerance through phosphorylating ABA-responsive element-binding factors. J. Exp. Bot. 2020;71:188–203. doi: 10.1093/jxb/erz432. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Xu Y., Xie Z., Li X., He Z.H., Peng X.X. Association-Dissociation of Glycolate Oxidase with Catalase in Rice: A Potential Switch to Modulate Intracellular H2O2 Levels. Mol. Plant. 2016;9:737–748. doi: 10.1016/j.molp.2016.02.002. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Du H., Wang Y., Wang H., Yang S., Li C., Chen N., Yang H., Zhang Y., Zhu Y., et al. The calcium-dependent protein kinase ZmCDPK7 functions in heat-stress tolerance in maize. J. Integr. Plant Biol. 2021;63:510–527. doi: 10.1111/jipb.13056. [DOI] [PubMed] [Google Scholar]
- Zhou J., Wang X., He Y., Sang T., Wang P., Dai S., Zhang S., Meng X. Differential Phosphorylation of the Transcription Factor WRKY33 by the Protein Kinases CPK5/CPK6 and MPK3/MPK6 Cooperatively Regulates Camalexin Biosynthesis in Arabidopsis. Plant Cell. 2020;32:2621–2638. doi: 10.1105/tpc.19.00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.B., Liu C., Tang D.Y., Yan L., Wang D., Yang Y.Z., Gui J.S., Zhao X.Y., Li L.G., Tang X.D., et al. The Receptor-Like Cytoplasmic Kinase STRK1 Phosphorylates and Activates CatC, Thereby Regulating H2O2 Homeostasis and Improving Salt Tolerance in Rice. Plant Cell. 2018;30:1100–1118. doi: 10.1105/tpc.17.01000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Heinlein C., Orendi G., Zentgraf U. Senescence-specific regulation of catalases in Arabidopsis thaliana (L.) Heynh. Plant Cell Environ. 2006;29:1049–1060. doi: 10.1111/j.1365-3040.2005.01459.x. [DOI] [PubMed] [Google Scholar]
- Zou J.J., Li X.D., Ratnasekera D., Wang C., Liu W.X., Song L.F., Zhang W.Z., Wu W.H. Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 Function in Abscisic Acid-Mediated Signaling and H2O2 Homeostasis in Stomatal Guard Cells under Drought Stress. Plant Cell. 2015;27:1445–1460. doi: 10.1105/tpc.15.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.