Summary
Large-scale biorepositories and databases are essential to generate equitable, effective, and sustainable advances in cancer prevention, early detection, cancer therapy, cancer care, and surveillance. The Mutographs project has created a large genomic dataset and biorepository of over 7,800 cancer cases from 30 countries across five continents with extensive demographic, lifestyle, environmental, and clinical information. Whole-genome sequencing is being finalized for over 4,000 cases, with the primary goal of understanding the causes of cancer at eight anatomic sites. Genomic, exposure, and clinical data will be publicly available through the International Cancer Genome Consortium Accelerating Research in Genomic Oncology platform. The Mutographs sample and metadata biorepository constitutes a legacy resource for new projects and collaborations aiming to increase our current research efforts in cancer genomic epidemiology globally.
Graphical abstract
Perdomo et al. present the Mutographs biorepository, a new research project that is helping scientists to understand the causes and development of cancer around the world. This information can be used to develop new ways to prevent cancer and to reduce the number of people who die from it.
Introduction
Large-scale biomedical databases and resources help to promote advances in cancer research applications in diverse areas including cancer prevention, early detection, therapy, cancer care, and surveillance. The need to diversify the current knowledge on cancer genomics around the world requires the development and sustainability of large cancer biorepositories in different geographical regions with complete epidemiological data including demographics, lists of relevant environmental exposures, and detailed clinical information.
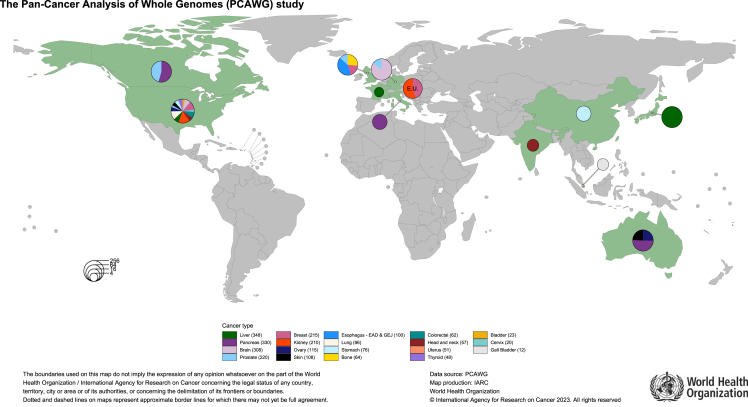
Whole-genome sequencing (WGS) of tumor-normal matched pairs is a powerful method to determine the diversity and complexity of somatic and germline mutations for both understanding the etiology and revealing diagnosis and treatment opportunities in patients with cancer. Despite the large amounts of publicly available WGS data, generated from patients with cancer as part of the global PanCancer Analysis of Whole Genomes (PCAWG) project1 (n = 3,109) of the International Cancer Genome Consortium (ICGC), the Hartwig Foundation (n = 5,520),2 and, more recently, from the Genomics England’s 100,000 Genomes Project (n = 12,222),3 these efforts have focused almost exclusively on patients from Europe, North America, and Australia, with a limited representation of cancers from Asian and African countries (Figure 1). Inclusion of more diverse populations of patients from other geographical regions in cancer genomic studies still lags behind,4 and emerging findings from comparative studies of population diversity in cancer genomics5,6 have established the necessity to expand diverse genetic and epidemiological data.
Figure 1.
Geographical distribution of Pan-Cancer Analysis of Whole Genomes (PCAWG) study
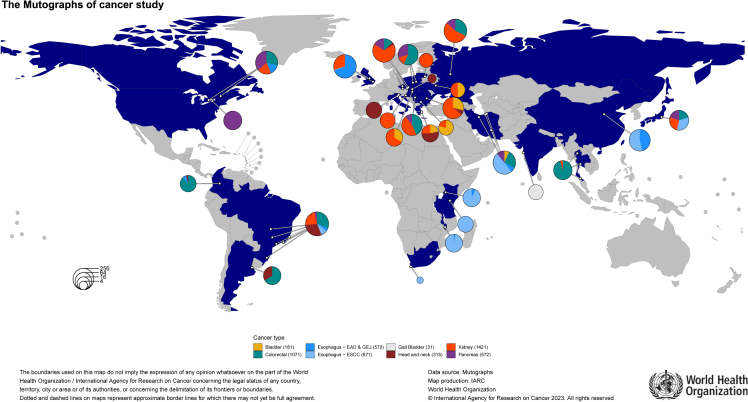
The Mutographs project is a Cancer Grand Challenges partnership funded by Cancer Research UK (CRUK: C98/A24032) with the primary objective of elucidating the causes of major global geographical and temporal differences in cancer incidence through mutational signature analysis. The project is generating mutational signatures and additional genomic descriptive analyses from WGS of thousands of paired (tumor/blood) samples from patients with cancer diagnosed with selected cancer types in 30 countries across 5 continents (Figure 2). The Mutographs study also consolidates an international research network working on cancer genomic epidemiology and exemplifies how genomic studies in cancer can promote scientific inclusion and equity through international collaboration.
Figure 2.
Geographical distribution of Mutographs cancer studies with complete whole-genome sequencing
Mutographs cases correspond to those that passed the quality controls for eligibility, tissue and blood processing, and sequencing. White circles represent cities where cases were recruited: Argentina: Buenos Aires; Brazil: Barretos, Goiania, Porto Alegre, Rio de Janeiro, Sao Paulo, and Vitoria; Bulgaria: Sofia; Canada: Montreal and Toronto; China: Shanxi; Colombia: Bogotá; Croatia: Zagreb; Czech Republic: Brno, Ceske Budejovice, Olomouc, and Prague; Greece: Athens; Hungary: Budapest; India: Mumbai; Iran: Gonbad, Gorgan, Kerman, and Tehran; Italy: Aviano and Padova; Japan: Tokyo; Kenya: Eldoret; Lithuania: Vilnius; Malawi: Blantyre; Poland: Lodz and Warsaw; Romania: Bucharest; Russia: Moscow; Serbia: Belgrade; Slovakia: Banska Bystrica; South Africa: Cape Town; Tanzania: Moshi; Thailand: Bangkok, Chiang Mai, and Hat Yai; Ukraine: Kiev; UK: Cambridge and Leeds; and US: Rochester.
Genomic epidemiology approaches for identifying new causes of cancer globally
Differences in cancer incidence between populations cannot be uniquely attributable to endogenous mutagenic processes. For instance, evidence from migrant studies7 and recent time trends8,9,10,11 show that genetic susceptibility cannot sufficiently explain these differences, indicating that lifestyle and environmental factors should also be responsible.
Traditional epidemiological studies mostly based on large retrospective case-control analysis have exhausted the possibilities of finding or confirming new potential causes of cancer because data are outdated or non-existent for current and/or relevant exposures.12,13 The more recently available large prospective, population-based cohorts and cohort consortia have improved the resolution of cancer etiological findings by improving study quality for rare exposures, linking data across sources such as electronic health records, tumor biobanks, cancer registries, geospatial data, and mobile data, among others, and independently validating previous findings.14 However, these consortia are far from being representative of many geographical areas and will fail to identify unknown exposures possibly relevant for certain regions. The inclusion of state-of-the-art genomic studies complementary to well-defined epidemiological study designs and extensive data collection redefines the new era of genomic epidemiology studies in cancer research and can help to identify unknown causes of cancer worldwide.15,16
Mutographs rationale: Elucidating global differences in cancer incidence using genomic epidemiology
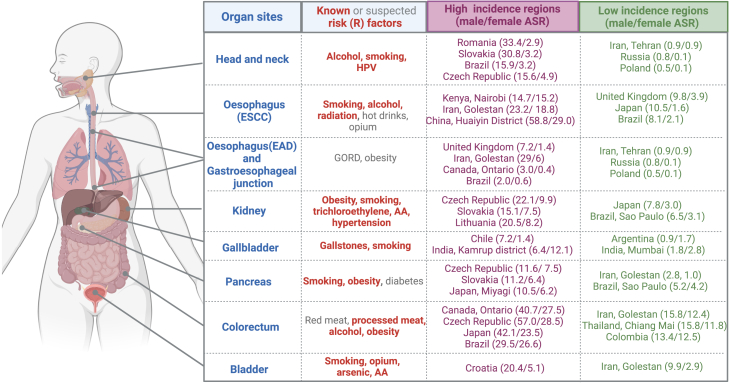
The Mutographs project focused on investigating the causes of large international differences in incidence and mortality for several cancers that are still poorly understood. Five initial cancer types, esophageal squamous cell carcinoma (ESCC), renal cell carcinoma (RCC), colorectal cancer (CRC), pancreatic ductal adenocarcinoma (PDAC), and gastroesophageal junction adenocarcinoma (GEJ), including both esophageal adenocarcinoma (EAD) and adenocarcinomas of gastric cardia, were selected based on the following criteria: (1) cancers with the highest differences in incidence across geographical regions, (2) cancers accounting for more than 10% of new cases and about 20% of deaths, and (3) cancers for which prevalence of known risk factors (i.e., smoking, alcohol, and obesity) do not fully explain the large geographical and temporal differences across regions (Figure 3).
Figure 3.
Summary of cancer subsites included in the Mutographs biorepository by known and suspected risk factors and differences of incidence by regions
Age-standardized rates (ASRs) retrieved from Cancer Incidence in Five Continents, Vol. XI.17 AA, aristolochic acid; HPV, human papillomavirus. Created with BioRender.com.
For instance, CRC, RCC, and PDAC cancer cases show a similar distribution across geographical regions, being most common in Central Europe (particularly Czech Republic), North America, and East Asia (especially Japan and South Korea) and relatively rare in Africa and certain parts of Asia.18 The known common risk factors for these three cancers are obesity19 and tobacco smoking, although their effects are modest (∼50% increased risk). Other suspected risk factors include dietary components such as animal protein, processed meat,20 and alcohol consumption for CRC21; exposure to trichloroethylene,22 aristolochic acid,23,24 and per- and polyfluoroalkyl substances for RCC25,26; and clinical conditions such a hypertension and diabetes for RCC27,28 and PDAC,29 respectively.
Esophageal cancer, in particular ESCC, is an example of a cancer with differences in incidence within regions or countries. High ESCC rates are found in localized populations, including northeastern Iran, north and central China,30 parts of Africa, and southern Brazil.31 Established risk factors include tobacco, opium,32 and alcohol,21 but population attributable fractions vary between regions. Additionally, there is strongly suggestive evidence for a role of the consumption of very hot beverages32 (tea/coffee/porridge in Africa, tea in Iran, maté in the south of Brazil) and a nutritionally deficient diet and exposure to polycyclic aromatic hydrocarbons from diverse sources, such as indoor biomass combustion.33
There has been a rapid increase of EAD in recent decades more evident among men and in specific populations,34 particularly in Western Europe, North America, Australia, and in the Golestan province of Iran. The reasons for this particular geographical distribution are still unknown.35
Three additional cancer types were subsequently integrated in the study based on marked regional incidence differences and/or specific exposures of interest: (1) head and neck cancer (HNC), including cases identified in high- and intermediate-incidence countries in Europe36 and South America37 and oversampled for cases without reported tobacco and/or alcohol consumption; (2) urinary bladder cancer (UBC), with a selection of cases diagnosed in the Kerman province in Iran with and without documented consumption of opium,38 recently classified as a carcinogenic substance (group I)39; and (3) gallbladder cancer (GBC), a cancer with the highest incidence rates in countries in South America and India and for which causes are poorly understood. Cases were selected from different regions in India where GBC is more common in women in the north, northeastern, and east (e.g., in the Kamrup urban district, incidence of GBC is 6.4 per 100,000 for men and 12.1 per 100,000 for women) compared to the southern part of India (in Mumbai, incidence is 1.8 per 100,000 for men and 2.8 per 100,000 in women)40 (Figures 2 and 3).
The eight cancer sites included in the Mutographs study are also covered in the PCAWG study. However, Mutographs provides a larger number of cases for these cancer sites (4,397 cases already sequenced in Mutographs vs. 794 in PCAWG) and a broader geographic representation with 17 countries not explored in PCAWG from additional regions in eastern Europe, South America, South and East Asia, and Eastern Africa (Figure 2).
Building up a global biorepository: Considering the local perspective and experience in global multicenter studies
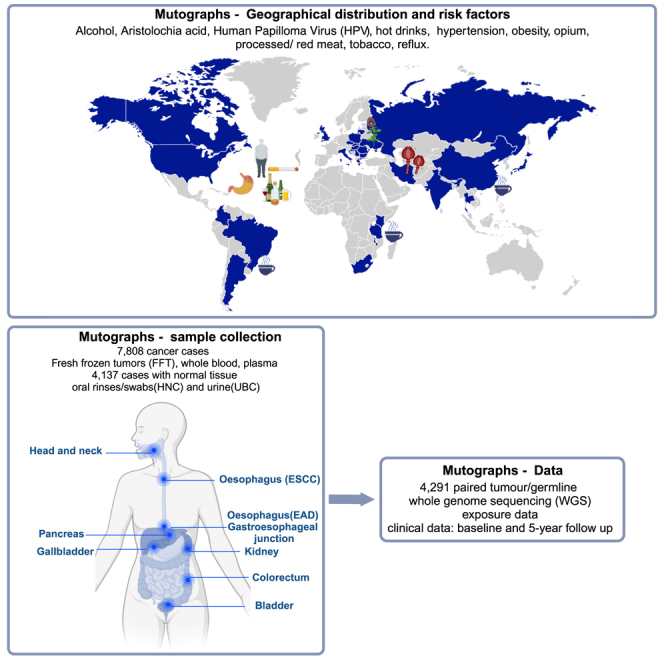
The International Agency for Research on Cancer (IARC/WHO) promotes global collaboration in cancer research through the coordination of research across countries and organizations41 and convening multidisciplinary expertise. With this long-standing track record and extensive international studies and network of partners, the Mutographs study was in the position to bring together existing studies as well as initiate de novo studies. Collaborating centers were selected among academic institutions, university hospitals, national cancer institutions, and private and public hospitals with experience in patient recruitment and sample and data acquisition for representative samples of patients with cancer. Through this international network of 50 institutional collaborators in 30 countries, between 2018 and 2022, the Mutographs team harmonized biospecimen and epidemiologic data on 7,808 cancer cases. Of the 7,808 cases, 3,023 (39%) were newly recruited, and 4,785 (61%) were based upon existing metadata and selected biorepositories.
A unified protocol for case selection and prospective recruitment with extended exposure data
Sample/data collection and inclusion criteria
Participant centers were included in the Mutographs project under two different scenarios or a combination of both.
Scenario 1: ongoing or retrospective studies and biorepository collections not being previously sequenced/analyzed, published, or included in other international genomic initiatives such as PCAWG.1 31 centers provided patients that fulfilled the required criteria for inclusion (as described below).
Scenario 2: prospective collection of newly diagnosed patients. 19 centers prospectively recruited a sample of representative patients per cancer site. Patients were excluded if they had any condition that could interfere with their ability to provide informed consent or if there were no means of obtaining adequate tissues as per the protocol requirements. Ethical approvals were first obtained from each local research ethics committee and federal ethics committee when applicable, as well as from the IARC ethics committee.
Dedicated standard operating procedures (SOPs) were designed by IARC/WHO following guidelines from the ICGC to harmonize exposure, lifestyle, pathological, and clinical information from all cases to be included in the Mutographs project. The inclusion criteria for patients were that they were at least 18 years of age; had a confirmed diagnosis of primary tumors from the list of cancer sites eligible for the study; had no prior treatment; had an availability of fresh frozen tumor and, if possible, non-tumor fresh frozen tissue (FFT) and blood samples; had an availability of core epidemiological and clinical data (retrospectively or prospectively collected); and had ethics approval and consent for genetic studies and data sharing. For all patients prospectively included, after informed consent was obtained, anthropometric measures were taken, together with relevant information regarding medical and familial history. Blood samples were drawn, and a 30 min questionnaire was administered by a trained interviewer to collect complementary lifestyle and environmental information following the Mutographs SOPs.
Large sample collection
Fresh frozen tumor tissue and blood were collected from all cases as the most suitable samples for genomic studies. Non-tumor adjacent tissue was available for 53% of the cases. Collection of oral rinses and urine was also included for a subset of the HNC and UBC cohorts, respectively. Blood collection in EDTA tubes consisted in 10 mL preserved as whole blood. When feasible, blood samples were immediately processed into buffy coat, plasma, and red blood cells, followed by storage at −80°C. These collections resulted in more than 85,000 biological samples currently stored at the IARC/WHO BioBank (https://ibb.iarc.fr). Tumor and non-tumor tissues were collected before any treatment and while avoiding routine care disruption. Unless surgery was the first line of treatment, mucosal biopsies were collected. Tumor and non-tumor tissues were snap frozen in liquid nitrogen or promptly preserved in RNAlater (RNAprotect Tissue Tubes, QIAGEN). Laboratory information management systems at local recruiting centers were used to keep track of samples collected and processed before being transferred to the IARC/WHO BioBank.
Detailed environmental exposure and clinical questionnaire: Diversifying local and regional exposure data
The focus on evaluation of exposures associated with recognized and plausible risk factors makes this biorepository a valuable source of detailed and comparable epidemiological metadata from patients with cancer diagnosed with the same tumor site but from diverse geographical regions. We compared the available exposure information among patients from cancer sites included in both PCAWG and Mutographs to estimate the extent of the project metadata. PCAWG includes information on tobacco smoking and alcohol consumption for three cancer sites. No additional exposure information was publicly accessible (Table 1). Data for those two exposures are partially available. History of tobacco smoking consumption is available for 30% of patients with PDAC from Canada, 20% of patients diagnosed with oral cavity (HNC) in India, and 80% of patients with EAD from the UK. History of alcohol consumption is absent in more than 70% of these cancer sites in PCAWG. In contrast, tobacco and alcohol consumption is documented in 97% and 81% of the Mutographs patients for the three cancer sites (Table 1). Mutographs has retrieved information on environmental exposures and risk factors for up to 90% of patients from all eight cancer sites using the following methodology.
Table 1.
Distribution of PCAWG and Mutographs cancer cases with smoking and alcohol consumption information
| Characteristic | Esophagus |
Head and neck (HNC) |
Pancreas (PDAC) |
|||
|---|---|---|---|---|---|---|
| Mutographs (GEJ) | PCAWG (EAD) | Mutographs | PCAWG | Mutographs | PCAWG | |
| Total | 570 | 100 | 315 | 57 | 572 | 330 |
| Countries | Brazil, China (Shanxi), Iran, Japan, Kenya, Malawi, Tanzania, UK | UK | Argentina, Brazil, Colombia, Czech Republic, Greece, Italy, Romania, Slovakia | India, US | Brazil, Canada, Czech Republic, Iran, Poland, Russia, Serbia, UK, US | Australia, Canada |
| Sex assigned at birth | ||||||
| Female (%) | 104 (18) | 14 (14) | 72 (23) | 10 (18) | 291 (51) | 152 (46) |
| Male (%) | 466 (82) | 86 (86) | 243 (77) | 47 (82) | 281 (49) | 176 (53) |
| Unknown (%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (0.6) |
| Age at diagnosis (years), median (IQR) | 67 (60.0, 73.0) | 70 (62.5, 76.0) | 59 (50.0, 68.0) | 53 (42.0, 62.0) | 66 (59.1, 72.0) | 65 (56.0, 73.0) |
| Unknown (%) | 13.0 (2.3) | 1.0 (1.0) | 0.0 (0.0) | 0.0 (0.0) | 6.0 (1.0) | 1.0 (0.3) |
| Tobacco status (%) | ||||||
| Current smoker | 159 (28) | 20 (20) | 152 (48) | 8 (14) | 122 (21) | 5 (1.5) |
| Ever smoker | 2 (0.4) | 0 (0) | 0 (0) | 0 (0) | 24 (4.2) | 0 (0) |
| Ex-smoker | 190 (33) | 48 (48) | 93 (30) | 0 (0) | 145 (25) | 29 (8.8) |
| Never | 171 (30) | 20 (20) | 70 (22) | 5 (8.8) | 280 (49) | 61 (18) |
| Unknown | 48 (8.4) | 12 (12) | 0 (0) | 44 (77) | 1 (0.2) | 235 (71) |
| Alcohol status (%) | ||||||
| Current drinker | 107 (19) | 0 (0) | 122 (39) | 0 (0) | 98 (17) | 0 (0) |
| Ever drinker | 46 (8.1) | 27 (27) | 21 (6.7) | 6 (11) | 166 (29) | 30 (9.1) |
| Ex-drinker | 20 (3.5) | 0 (0) | 99 (31) | 0 (0) | 28 (4.9) | 0 (0) |
| Never | 126 (22) | 0 (0) | 73 (23) | 7 (12) | 251 (44) | 11 (3.3) |
| Unknown | 271 (48) | 73 (73) | 0 (0) | 44 (77) | 29 (5.1) | 289 (88) |
EAD, esophageal adenocarcinoma; GEJ, gastroesophageal junction adenocarcinoma; HNC, head and neck cancer; PDAC, pancreatic ductal adenocarcinoma; PCWAG, the global PanCancer Analysis of Whole Genomes.
Data from prospectively recruited patients were collected using a centralized database developed in the REDCap platform,42 and IARC/WHO harmonized all retrospective data received as previously described.43,44 The following core epidemiological and clinical data were required from all participants in the study: (1) demographic details (age, sex, ethnic origin, city and place of residence, and educational status); (2) history of tobacco use, including frequency and intensity; (3) history of alcohol consumption, including frequency and intensity; (4) anthropometric data (height, weight at diagnosis); (5) medical history of diabetes, hypertension, and acid reflux/heartburn for PDAC, RCC, and GEJ, respectively; and (6) consumption of hot drinks (for ESCC) or red and processed meat (for CRC). In addition to the data collected under the core variables mentioned above, all prospectively and partially retrospectively recruited patients provided information on oral health, physical activity, occupational exposures, and family history of cancer. We also collected information on the following regional and/or population-specific exposures.
-
(1)
Opium consumption including route of administration, frequency, and intensity for patients with cancer recruited in Iran.
-
(2)
Consumption of traditional South American maté including frequency, temperature, and intensity for patients from and/or recruited in the south of Brazil and in Argentina.
-
(3)
Residential history and consumption of herbal remedies as possible sources of aristolochic acid exposure for patients with RCC from Romania, Serbia, Bulgaria, Croatia, Hungary, Greece, and Ukraine.
Clinical follow-up information up to 3 years after cancer diagnosis was retrieved, if possible, from clinical charts from retrospective collections, and additional information for up to 5 years is being collected from the prospectively recruited patients. IARC/WHO harmonized all retrospective data. All data were de-identified locally through the use of a dedicated alpha-numerical identifier system before being transferred to IARC/WHO central database.
Centralized expert pathology review
Diagnostic pathology departments from participating centers provided diagnostic details on morphology and histology of patients through standard abstract forms, together with a representative hematoxylin and eosin (H&E)-stained slide of formalin-fixed, paraffin-embedded (FFPE) tumor tissues whenever possible. For all patients, to reconfirm the original histology, IARC/WHO centralized the entire pathology workflow on FFT tumors and coordinated their digital pathology examination included in the study, as well as FFPE sections when available, via a web-based report completed by a dedicated expert panel for each cancer site. High-resolution images of FFT tumors were randomly assigned to panel members, all blind to the original diagnosis. In addition to diagnosis and confirmation of tumor type, the percentage of viable cellular elements (tumor, inflammatory, and other non-tumor cells) and necrosis were recorded. 17% of randomly selected H&E slides underwent two independent pathology evaluations. A minimum of 50% viable tumor cells were required for eligibility to WGS. The percentage of processed cases with less than 50% tumor content was 18% for UBC, 20% for RCC, 31% for CRC, 36% for GBC, 43% for HNC, 45% for ESCC, and 49% for GEJ. Tumor enrichment procedures were applied, when possible, by laser capture microdissection (LCM) of the unwanted non-tumoral area. Approximately half of the GEJ cases and 96% of PDAC cases underwent LCM to enrich tumor cellularity.
Extraction of DNA and quantification from tumor and paired blood was centrally conducted at IARC/WHO43 and is stored for subsequent analyses.
Out of the 7,808 recruited cancer cases, 4,400 were successfully processed at IARC/WHO, passed the pathological quality control metrics, and were sent to the Wellcome Sanger Institute for paired WGS and primary mutational signature analyses. 655 cases are yet to be processed and will be evaluated through our pathology pipeline and stored in the study biorepository for future studies.
Sequencing analyses and current publications
The data sequencing pipeline has been developed and validated by the Wellcome Sanger Institute as previously described.43 WGS (150 bp paired end) is performed on the Illumina NovaSeq 6000 platform with target coverage of 40× for tumors and 20× for matched non-tumor tissues. Cases are excluded if coverage is below 30× for tumors or 15× for non-tumor tissue.
The data analysis workflow focuses on 4 areas: (1) the characterization of the tumor genome for each sample with specific data generated on driver genes, copy-number profiles, evaluation of tumor mutation burden, structural rearrangements, and other cancer-specific information such as the presence of viral and/or bacterial sequences for specific cancer sites; (2) the extraction and attribution of mutational signatures based on base substitutions (single and double), insertions/deletions, copy-number variants, and chromosomal rearrangements; (3) analyses highlighting possible contributions of germline variants and ancestry distribution45 to the mutational signature profiles and associated exposures; and lastly, (4) associations between somatic genomic profiles and epidemiological data focused mainly on recognized and plausible risk factors. In 2021, the analysis on 552 patients with ESCC from eight countries with varying incidence rates was completed and showed a high prevalence of APOBEC signatures in all cases, as well as specific mutation signatures linked to opium and alcohol consumption, and homologous DNA repair deficiency.43 Analysis of 962 patients with clear cell RCC is ongoing, and preliminary results are shedding light on the contribution of environmental causes on the high risk of this cancer in Central Europe and Japan.44 Most of the sequencing and analysis efforts are now focused on cases of HNC, CRC, PDAC, and GEJ. By early 2023, we completed the sequencing of 2,777 matched-normal cancer genomes, and these samples are undergoing bioinformatics cancer genomics analysis. Data from these cases have been released to the Mutographs teams for subsequent combined analysis.
Data repository and sharing
A general description of the Mutographs project is available on the project website: https://www.mutographs.org. WGS data and patient metadata after analyses are being deposited and made publicly available via the European Genome Phenome Archive (EGA), currently associated with studies EGAS00001003542 and EGAS00001002725. All algorithms and codes used for genomic and epidemiological analysis and figures are publicly available with repositories noted in the respective publications.43,44
In addition, Mutographs is one of the participating programs of the ICGC Accelerating Research in Genomic Oncology (ARGO).46 Therefore, all genomic, exposure, and clinical data will be publicly available through the ICGC ARGO data platform after agreement of the participating centers. An online catalog of the Mutographs biorepository is under development and will allow the broader research community to propose additional projects and/or analyses beyond the scope of Mutographs.
Ongoing and future initiatives for the Mutographs biorepository
The study of the mutational signatures operative in the genomes of patients with cancer around the world will generate a comprehensive catalog of the mutational processes that cause human cancer. An increasing number of signatures of different mutation classes are being reported, and correlations are being drawn to various exposures and/or endogenous factors. Experimental validation of signatures linked to specific exposures in human sample collections adds valuable information to establish causality.47 Therefore, it is fundamental to gain a mechanistic understanding of how mutational signatures arise through experimental exploration.
To completely understand the etiology of cancer and to apply this knowledge to cancer prevention,15 analysis of non-cancer tissues from patients with cancer and patients with benign or preneoplastic conditions48 can provide insight into background mutational processes in healthy cells49 and into the effects of suspected mutagens and exposures prior to the development of symptomatic lesions and, eventually, the diagnosis of cancer.50,51,52 PROMINENT (CRUK: CGCATF-2021/100007, NIH: 1OT2CA278681-01) is a recently awarded Cancer Grand Challenges project aiming to detect and characterize mutagenic and promoting exposures before cancer develops using human tissue, mouse, and organoid models. Combined multiomics approaches are being used to understand the distributions of mutations and early neoplastic clones in non-tumor tissues from patients with cancer included in the Mutographs biorepository. Further investigation of mutagenic processes in non-tumor tissues using non-invasive sources of tissue (i.e., blood, urine, and nasopharyngeal, buccal, cervical, and anal swabs) will allow us to easily identify and monitor carcinogenic exposures and ultimately determine how these influence clinical and epidemiological patterns of cancer development. The resources created by Mutographs should also allow for subsequent studies addressing poorly understood cancer-related questions and shed light on the current and future challenges in cancer research.
Beyond Mutographs: Expanding genomic epidemiological repositories and inclusive research collaborations
The Mutographs study is an example of novel genomic initiatives needed to expand our understanding of causes and processes related to cancer onset on a global scale. Some of the key aspects that contributed to the successful creation of such a large-scale cancer biorepository and that we suggest should be applied to similar initiatives in the future include the following.
Establishing long-lasting research collaborations and standardized protocols: a fundamental pillar in any international collaborative project involves complying with a detailed and unique recruitment protocol for patient selection and sample collection while adapting to the specific local or regional context for an effective implementation of the study protocol. Such a balance was achieved in Mutographs by close communication and follow up with the different institutions throughout the duration of the project. The Mutographs study team investigated the local research needs in terms of available personnel, minimum infrastructure requirements, and institutional procedures for patient approach, routes of diagnosis, and treatment. Questionnaires were revised to include relevant exposures, adapt the questions’ wording, and avoid possible sensitive questions. Center-specific adjustments were included in the protocols if necessary. Research agreements as well as material and data transfer agreements were better established after close discussions with collaborators to comply with legal and data protection requirements in each country, the research institutions, and the funders. Essentially, publication policies and ownership of data and materials must be clearly stated to guarantee ethical research conduct and consistent use of the resulting data.
Mitigating difficulties and adapting: the COVID-19 pandemic had a profound impact on the recruitment of patients with cancer between 2020 and 2022. However, centers continuously adapted their protocols to maintain patient inclusion rates and completeness of interview data and gained experience in new strategies for patient approach. For instance, the enrollment phase of patient identification, initial interview, information about the protocol, signature of informed written consent, lifestyle questionnaires, and some clinical information was conducted exclusively via telephone or video calls, and documents were provided electronically. These are strategies that were successfully piloted under these restrictive conditions and will continue to be used in other research protocols to facilitate patient recruitment and follow up.
Investing in large, geographically and population-diverse biorepositories: in the current largest international genomic consortium, PCAWG,1 the overall ancestry distribution was heavily weighted toward donors of European descent (77% of total) followed by East Asians (16%), as expected for large contributions from European, North American, and Australian projects. Initial admixture analysis for four cancer sites in Mutographs shows that the percentages of donors of European, African, East Asian, and mixed-descent ancestry are, respectively, 25%, 30%, 32%, and 13% for ESCC; 90%, 1%, 5%, and 4% for RCC; 78%, 1%, 14%, and 7% for CRC; and 72%, 3%, 1%, and 24% for HNC. This reflects the geographical regions included based on incidence rates and risk exposures as previously discussed. Financial support is always required for participation in these large genomic epidemiology studies and should be allocated based on the contribution and needs of each collaborator. Funding bodies and cancer research agencies should envision additional financial and research capacity investment in large genomic epidemiology studies supporting participation of populations systematically underrepresented in this field.
Limitations of Mutographs and opportunities for future studies
There are cancer types not included in Mutographs that are highly relevant to understanding the etiology of cancer and should be considered in additional genomic epidemiology studies. For instance, high-incident cancers such as lung cancer adenocarcinoma, particularly prevalent in women, non-smokers, and Asian populations,53 as well as less-incident, rare but frequently aggressive cancer types, for which many of the causes are still unknown. In addition, closer attention should be paid to the integration of genomics studies evaluating new exposures emerging as possible cancer risk factors, including air pollution,54 vaping,55 and opioid use.56
The knowledge generated by new large and geographically diverse biorepositories such as Mutographs have the potential to reveal previously unknown risk factors and ultimately establish causality, specifically by linking putative risk factors to specific genomic features. This, in turn, can guide the tailoring of prevention strategies and aid in the global reduction of the burden of cancer.
Acknowledgments
This work was delivered as part of the Mutographs team supported by the Cancer Grand Challenges partnership funded by Cancer Research UK (C98/A24032). Work at the Wellcome Sanger Institute was also supported by the Wellcome Trust (grants 206194 and 220540/Z/20/A), and work at the IARC/WHO was supported by regular budget funding, the NIH/NCI (grant number R21CA191965), and grant 2018/1795 from the Wereld Kanker Onderzoek Fonds (WKOF) as part of the World Cancer Research Fund International grant program. This work was supported in part by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, NCI, NIH. The head and neck cancer collection received funding from the European Union's Horizon 2020 research and innovation programme under grant no. 825771 and the São Paulo Research Foundation, FAPESP 2018/26297-3. Work at the Masaryk Memorial Cancer Institute, Brno, Czech Republic, was supported by MH CZ - DRO (MMCI, 00209805). The Porto Alegre center in Brazil received support from Hospital de Clínicas de Porto Alegre and Fundação Médica do Rio Grande do Sul. We are grateful for the support provided by the IARC General Services, including the Laboratory Services and Biobank team led by Z. Kozlakidis, the Section of Support to Research overseen by T. Landesz under IARC’s regular budget funding, and the staff of DNA pipelines at the Wellcome Sanger Institute under the C98/A24032 grant. Farid Azmoudeh-Ardalan, Mojgan Asgari, Sophie Ferlicot, Hiva Saffar, Jean-Yves Scoazec, Stefano Serra, and Masoud Sotoudeh supported the pathological evaluation of samples. Laura Torrens Fontanals, Sergey Senkin, and Wellington Oliveira Dos Santos supported the admixture analysis. We are thankful for the work of all other collaborators in the Mutographs project who participated in the recruitment of patients in all centers. The authors would also like to thank all the patients and their families involved in these studies. We remember and celebrate Dr. Gloria Petersen for her passion for science, her inclusive intellect, and her kind and generous spirit.
Where authors are identified as personnel of the IARC/WHO, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy, or views of the IARC/WHO.
Author contributions
Conceptualization, S.P., B.A.-A., A.C.d.C., A.F.-I., and P.B.; funding acquisition, M.R.S., P.B., and L.B.A.; methodology, S.P., B.A.-A., A.C.d.C., A.F.-I., E.C., G.S., and P.B.; project administration, A.S. and L.H.; data curation, V.G., T. Cattiaux, and H.R.; resources, P.C., C. Carreira, R.C.C.P., A.N., S.O.A., P.A.-P., C. Canova, T. Chitapanaru, R.C., M.P.C., J.C.d.O., C.D., E.F., L. Ferri, R.F., L. Foretova, S.G., A.M.G., I.H., A.H., V.J., S.J., R.K., L.P.K., T.K., P.L., J.L., R.M., D. Mates, V.M., D. Menya, S. Mhatre, B.T.M., A.d.M., P.N., M.O., K.P., J.P., M.P.P., S.R., L.M.R.B., R.M.R., L.F.R.P., P.A.R.-U., S. Sangkhathat, S. Sangrajrang, T.S., E.S., B.S., C.V., J.R.V.d.P., N.S.V., M.V., J.Y., D.Z., K.Z., G.S., E.C., J.W., S.F., C.L., and S. Moody; supervision, S.P., B.A.-A., A.C.d.D., A.F.-I., and P.B.; visualization, S.P. and T. Cattiaux; writing – original draft, S.P., B.A.-A., A.C.d.D, A.F.-I., and P.B.; writing – review & editing, all authors.
Declaration of interests
M.R.S. is founder of, consultant to, and stockholder in Quotient Therapeutics. L.B.A. is a compensated consultant and has equity interest in io9, LLC, and Genome Insight. His spouse is an employee of Biotheranostics, Inc. L.B.A. is also an inventor of a US patent 10,776,718 for source identification by non-negative matrix factorization. L.B.A. declares US provisional applications with serial numbers 63/289,601; 63/269,033; and 63/483,237. L.B.A. also declares US provisional applications with serial numbers 63/366,392; 63/367,846; 63/412,835; and 63/492,348.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xgen.2024.100500.
Supplemental information
References
- 1.ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Abascal F., Abeshouse A., Aburatani H., Adams D.J., Agrawal N., Ahn K.S., Ahn S.-M., Aikata H., Akbani R., et al. Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Priestley P., Baber J., Lolkema M.P., Steeghs N., de Bruijn E., Shale C., Duyvesteyn K., Haidari S., van Hoeck A., Onstenk W., et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature. 2019;575:210–216. doi: 10.1038/s41586-019-1689-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Degasperi A., Zou X., Amarante T.D., Martinez-Martinez A., Koh G.C.C., Dias J.M.L., Heskin L., Chmelova L., Rinaldi G., Wang V.Y.W., et al. Substitution mutational signatures in whole-genome-sequenced cancers in the UK population. Science. 2022;376 doi: 10.1126/science.abl9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fatumo S., Chikowore T., Choudhury A., Ayub M., Martin A.R., Kuchenbaecker K. A roadmap to increase diversity in genomic studies. Nat. Med. 2022;28:243–250. doi: 10.1038/s41591-021-01672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wojcik G.L., Graff M., Nishimura K.K., Tao R., Haessler J., Gignoux C.R., Highland H.M., Patel Y.M., Sorokin E.P., Avery C.L., et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature. 2019;570:514–518. doi: 10.1038/s41586-019-1310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spratt D.E., Chan T., Waldron L., Speers C., Feng F.Y., Ogunwobi O.O., Osborne J.R. Racial/Ethnic Disparities in Genomic Sequencing. JAMA Oncol. 2016;2:1070–1074. doi: 10.1001/jamaoncol.2016.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parkin D.M. IARC Sci Publ; 1993. Studies of Cancer in Migrant Populations; pp. 1–10. [PubMed] [Google Scholar]
- 8.Piñeros M., Laversanne M., Barrios E., Cancela M.d.C., de Vries E., Pardo C., Bray F. An updated profile of the cancer burden, patterns and trends in Latin America and the Caribbean. Lancet Reg. Health. Am. 2022;13 doi: 10.1016/j.lana.2022.100294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan E., Soerjomataram I., Rumgay H., Coleman H.G., Thrift A.P., Vignat J., Laversanne M., Ferlay J., Arnold M. The Global Landscape of Esophageal Squamous Cell Carcinoma and Esophageal Adenocarcinoma Incidence and Mortality in 2020 and Projections to 2040: New Estimates From GLOBOCAN 2020. Gastroenterology. 2022;163:649–658.e2. doi: 10.1053/j.gastro.2022.05.054. [DOI] [PubMed] [Google Scholar]
- 10.Znaor A., Lortet-Tieulent J., Laversanne M., Jemal A., Bray F. International variations and trends in renal cell carcinoma incidence and mortality. Eur. Urol. 2015;67:519–530. doi: 10.1016/j.eururo.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Arnold M., Abnet C.C., Neale R.E., Vignat J., Giovannucci E.L., McGlynn K.A., Bray F. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology. 2020;159:335–349.e15. doi: 10.1053/j.gastro.2020.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taubes G. Epidemiology faces its limits. Science. 1995;269:164–169. doi: 10.1126/science.7618077. [DOI] [PubMed] [Google Scholar]
- 13.Peto J. Cancer epidemiology in the last century and the next decade. Nature. 2001;411:390–395. doi: 10.1038/35077256. [DOI] [PubMed] [Google Scholar]
- 14.McCullough L.E., Maliniak M.L., Amin A.B., Baker J.M., Baliashvili D., Barberio J., Barrera C.M., Brown C.A., Collin L.J., Freedman A.A., et al. Epidemiology beyond its limits. Sci. Adv. 2022;8:eabn3328. doi: 10.1126/sciadv.abn3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brennan P., Davey-Smith G. Identifying Novel Causes of Cancers to Enhance Cancer Prevention: New Strategies Are Needed. J. Natl. Cancer Inst. 2022;114:353–360. doi: 10.1093/jnci/djab204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogino S., Nowak J.A., Hamada T., Milner D.A., Jr., Nishihara R. Insights into Pathogenic Interactions Among Environment, Host, and Tumor at the Crossroads of Molecular Pathology and Epidemiology. Annu. Rev. Pathol. 2019;14:83–103. doi: 10.1146/annurev-pathmechdis-012418-012818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bray F., Colombet M., Mery L., Piñeros M., Znaor A., R Z., Ferlay J e. XI. International Agency for Research on Cancer; Lyon: 2017. (Cancer Incidence in Five Continents). (electronic version) [Google Scholar]
- 18.Bray F., Colombet M., Mery L., Piñeros M., Znaor A., Zanetti R., Ferlay J. XI. International Agency for Research on Cancer; Lyon: 2017. http://ci5.iarc.fr (Cancer Incidence in Five Continents). (electronic version) Available from: accessed on 22 March 2018. [Google Scholar]
- 19.Arnold M., Pandeya N., Byrnes G., Renehan P.A.G., Stevens G.A., Ezzati P.M., Ferlay J., Miranda J.J., Romieu I., Dikshit R., et al. Global burden of cancer attributable to high body-mass index in 2012: a population-based study. Lancet Oncol. 2015;16:36–46. doi: 10.1016/S1470-2045(14)71123-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouvard V., Loomis D., Guyton K.Z., Grosse Y., Ghissassi F.E., Benbrahim-Tallaa L., Guha N., Mattock H., Straif K., International Agency for Research on Cancer Monograph Working Group Carcinogenicity of consumption of red and processed meat. Lancet Oncol. 2015;16:1599–1600. doi: 10.1016/S1470-2045(15)00444-1. [DOI] [PubMed] [Google Scholar]
- 21.Rumgay H., Shield K., Charvat H., Ferrari P., Sornpaisarn B., Obot I., Islami F., Lemmens V.E.P.P., Rehm J., Soerjomataram I. Global burden of cancer in 2020 attributable to alcohol consumption: a population-based study. Lancet Oncol. 2021;22:1071–1080. doi: 10.1016/S1470-2045(21)00279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.IARC Monographs Vol 130 group Carcinogenicity of 1,1,1-trichloroethane and four other industrial chemicals. Lancet Oncol. 2021;22:1661–1662. doi: 10.1016/S1470-2045(21)00659-8. [DOI] [PubMed] [Google Scholar]
- 23.Das S., Thakur S., Korenjak M., Sidorenko V.S., Chung F.F.L., Zavadil J. Aristolochic acid-associated cancers: a public health risk in need of global action. Nat. Rev. Cancer. 2022;22:576–591. doi: 10.1038/s41568-022-00494-x. [DOI] [PubMed] [Google Scholar]
- 24.Grosse Y., Baan R., Straif K., Secretan B., El Ghissassi F., Bouvard V., Benbrahim-Tallaa L., Guha N., Galichet L., Cogliano V., WHO International Agency for Research on Cancer Monograph Working Group A review of human carcinogens--Part A: pharmaceuticals. Lancet Oncol. 2009;10:13–14. doi: 10.1016/s1470-2045(08)70286-9. [DOI] [PubMed] [Google Scholar]
- 25.Scelo G., Riazalhosseini Y., Greger L., Letourneau L., Gonzàlez-Porta M., Wozniak M.B., Bourgey M., Harnden P., Egevad L., Jackson S.M., et al. Variation in genomic landscape of clear cell renal cell carcinoma across Europe. Nat. Commun. 2014;5:5135. doi: 10.1038/ncomms6135. [DOI] [PubMed] [Google Scholar]
- 26.Shearer J.J., Callahan C.L., Calafat A.M., Huang W.Y., Jones R.R., Sabbisetti V.S., Freedman N.D., Sampson J.N., Silverman D.T., Purdue M.P., Hofmann J.N. Serum Concentrations of Per- and Polyfluoroalkyl Substances and Risk of Renal Cell Carcinoma. J. Natl. Cancer Inst. 2021;113:580–587. doi: 10.1093/jnci/djaa143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alcala K., Mariosa D., Smith-Byrne K., Nasrollahzadeh Nesheli D., Carreras-Torres R., Ardanaz Aicua E., Bondonno N.P., Bonet C., Brunström M., Bueno-de-Mesquita B., et al. The relationship between blood pressure and risk of renal cell carcinoma. Int. J. Epidemiol. 2022;51:1317–1327. doi: 10.1093/ije/dyac042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weikert S., Boeing H., Pischon T., Weikert C., Olsen A., Tjonneland A., Overvad K., Becker N., Linseisen J., Trichopoulou A., et al. Blood pressure and risk of renal cell carcinoma in the European prospective investigation into cancer and nutrition. Am. J. Epidemiol. 2008;167:438–446. doi: 10.1093/aje/kwm321. [DOI] [PubMed] [Google Scholar]
- 29.Ben Q., Xu M., Ning X., Liu J., Hong S., Huang W., Zhang H., Li Z. Diabetes mellitus and risk of pancreatic cancer: A meta-analysis of cohort studies. Eur. J. Cancer. 2011;47:1928–1937. doi: 10.1016/j.ejca.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Ke L. Mortality and incidence trends from esophagus cancer in selected geographic areas of China circa 1970-90. Int. J. Cancer. 2002;102:271–274. doi: 10.1002/ijc.10706. [DOI] [PubMed] [Google Scholar]
- 31.Abnet C.C., Arnold M., Wei W.Q. Epidemiology of Esophageal Squamous Cell Carcinoma. Gastroenterology. 2018;154:360–373. doi: 10.1053/j.gastro.2017.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheikh M., Poustchi H., Pourshams A., Etemadi A., Islami F., Khoshnia M., Gharavi A., Hashemian M., Roshandel G., Khademi H., et al. Individual and Combined Effects of Environmental Risk Factors for Esophageal Cancer Based on Results From the Golestan Cohort Study. Gastroenterology. 2019;156:1416–1427. doi: 10.1053/j.gastro.2018.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mwachiro M.M., Pritchett N., Calafat A.M., Parker R.K., Lando J.O., Murphy G., Chepkwony R., Burgert S.L., Abnet C.C., Topazian M.D., et al. Indoor wood combustion, carcinogenic exposure and esophageal cancer in southwest Kenya. Environ. Int. 2021;152 doi: 10.1016/j.envint.2021.106485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li M., Park J.Y., Sheikh M., Kayamba V., Rumgay H., Jenab M., Narh C.T., Abedi-Ardekani B., Morgan E., de Martel C., et al. Population-based investigation of common and deviating patterns of gastric cancer and oesophageal cancer incidence across populations and time. Gut. 2023;72:846–854. doi: 10.1136/gutjnl-2022-328233. [DOI] [PubMed] [Google Scholar]
- 35.Ryan A.M., Duong M., Healy L., Ryan S.A., Parekh N., Reynolds J.V., Power D.G. Obesity, metabolic syndrome and esophageal adenocarcinoma: epidemiology, etiology and new targets. Cancer Epidemiol. 2011;35:309–319. doi: 10.1016/j.canep.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Macfarlane T.V., Macfarlane G.J., Oliver R.J., Benhamou S., Bouchardy C., Ahrens W., Pohlabeln H., Lagiou P., Lagiou A., Castellsague X., et al. The aetiology of upper aerodigestive tract cancers among young adults in Europe: the ARCAGE study. Cancer Causes Control. 2010;21:2213–2221. doi: 10.1007/s10552-010-9641-3. [DOI] [PubMed] [Google Scholar]
- 37.Abrahão R., Perdomo S., Pinto L.F.R., Nascimento de Carvalho F., Dias F.L., de Podestá J.R.V., Ventorin von Zeidler S., Marinho de Abreu P., Vilensky M., Giglio R.E., et al. Predictors of Survival After Head and Neck Squamous Cell Carcinoma in South America: The InterCHANGE Study. JCO Glob. Oncol. 2020;6:486–499. doi: 10.1200/GO.20.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hadji M., Rashidian H., Marzban M., Naghibzadeh-Tahami A., Gholipour M., Mohebbi E., Safari-Faramani R., Seyyedsalehi M.S., Hosseini B., Bakhshi M., et al. Opium use and risk of bladder cancer: a multi-centre case-referent study in Iran. Int. J. Epidemiol. 2022;51:830–838. doi: 10.1093/ije/dyac031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.IARC Monographs Vol 126 group Carcinogenicity of opium consumption. Lancet Oncol. 2020;21:1407–1408. doi: 10.1016/S1470-2045(20)30611-2. [DOI] [PubMed] [Google Scholar]
- 40.Matsuda T., Marugame T. International comparisons of cumulative risk of gallbladder cancer and other biliary tract cancer, from Cancer Incidence in Five Continents Vol. VIII. Jpn. J. Clin. Oncol. 2007;37:74–75. doi: 10.1093/jjco/hyl158. [DOI] [PubMed] [Google Scholar]
- 41.Maurice J. IARC celebrates 50 years of cancer research. Lancet. 2016;387:2367–2368. doi: 10.1016/S0140-6736(16)30784-X. [DOI] [PubMed] [Google Scholar]
- 42.Harris P.A., Taylor R., Thielke R., Payne J., Gonzalez N., Conde J.G. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moody S., Senkin S., Islam S.M.A., Wang J., Nasrollahzadeh D., Cortez Cardoso Penha R., Fitzgerald S., Bergstrom E.N., Atkins J., He Y., et al. Mutational signatures in esophageal squamous cell carcinoma from eight countries with varying incidence. Nat. Genet. 2021;53:1553–1563. doi: 10.1038/s41588-021-00928-6. [DOI] [PubMed] [Google Scholar]
- 44.Senkin S., Moody S., Díaz-Gay M., Abedi-Ardekani B., Cattiaux T., Ferreiro-Iglesias A., Wang J., Fitzgerald S., Kazachkova M., Vangara R., et al. Geographic variation of mutagenic exposures in kidney cancer genomes. medRxiv. 2023;888 doi: 10.1101/2023.06.20.23291538. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.International Cancer Genome Consortium, ICGC ARGO 2022. http://platform.icgc-argo.org/
- 47.Koh G., Degasperi A., Zou X., Momen S., Nik-Zainal S. Mutational signatures: emerging concepts, caveats and clinical applications. Nat. Rev. Cancer. 2021;21:619–637. doi: 10.1038/s41568-021-00377-7. [DOI] [PubMed] [Google Scholar]
- 48.Mustjoki S., Young N.S. Somatic Mutations in "Benign" Disease. N. Engl. J. Med. 2021;384:2039–2052. doi: 10.1056/NEJMra2101920. [DOI] [PubMed] [Google Scholar]
- 49.Balmain A. The critical roles of somatic mutations and environmental tumor-promoting agents in cancer risk. Nat. Genet. 2020;52:1139–1143. doi: 10.1038/s41588-020-00727-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kakiuchi N., Ogawa S. Clonal expansion in non-cancer tissues. Nat. Rev. Cancer. 2021;21:239–256. doi: 10.1038/s41568-021-00335-3. [DOI] [PubMed] [Google Scholar]
- 51.Wijewardhane N., Dressler L., Ciccarelli F.D. Normal Somatic Mutations in Cancer Transformation. Cancer Cell. 2021;39:125–129. doi: 10.1016/j.ccell.2020.11.002. [DOI] [PubMed] [Google Scholar]
- 52.Colom B., Herms A., Hall M.W.J., Dentro S.C., King C., Sood R.K., Alcolea M.P., Piedrafita G., Fernandez-Antoran D., Ong S.H., et al. Mutant clones in normal epithelium outcompete and eliminate emerging tumours. Nature. 2021;598:510–514. doi: 10.1038/s41586-021-03965-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y., Vaccarella S., Morgan E., Li M., Etxeberria J., Chokunonga E., Manraj S.S., Kamate B., Omonisi A., Bray F. Global variations in lung cancer incidence by histological subtype in 2020: a population-based study. Lancet Oncol. 2023;24:1206–1218. doi: 10.1016/S1470-2045(23)00444-8. [DOI] [PubMed] [Google Scholar]
- 54.Hill W., Lim E.L., Weeden C.E., Lee C., Augustine M., Chen K., Kuan F.-C., Marongiu F., Evans E.J., Moore D.A., et al. Lung adenocarcinoma promotion by air pollutants. Nature. 2023;616:159–167. doi: 10.1038/s41586-023-05874-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goniewicz M.L., Smith D.M., Edwards K.C., Blount B.C., Caldwell K.L., Feng J., Wang L., Christensen C., Ambrose B., Borek N., et al. Comparison of Nicotine and Toxicant Exposure in Users of Electronic Cigarettes and Combustible Cigarettes. JAMA Netw. Open. 2018;1 doi: 10.1001/jamanetworkopen.2018.5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sheikh M., Brennan P., Mariosa D., Robbins H.A. Opioid medications: an emerging cancer risk factor? Br. J. Anaesth. 2023;130:e401–e403. doi: 10.1016/j.bja.2022.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.