Abstract
Pigmented rice stands out for its nutritional value and is gaining more and more attention. Wild rice, domesticated red rice, and weedy rice all have a red pericarp and a comprehensive genetic background in terms of the red-pericarp phenotype. We performed population genetic analyses using 5104 worldwide rice accessions, including 2794 accessions with red or black pericarps, 85 of which were newly sequenced in this study. The results suggested an evolutionary trajectory of red landraces originating from wild rice, and the split times of cultivated red and white rice populations were estimated to be within the past 3500 years. Cultivated red rice was found to feralize to weedy rice, and weedy rice could be further re-domesticated to cultivated red rice. A genome-wide association study based on the 2794 accessions with pigmented pericarps revealed several new candidate genes associated with the red-pericarp trait for further functional characterization. Our results provide genomic evidence for the origin of pigmented rice and a valuable genomic resource for genetic investigation and breeding of pigmented rice.
Key words: red rice, population genomics, pericarp color, re-domestication, divergence time
This study reports population genomic analyses of 5104 worldwide rice accessions (including 2794 accessions with red or black pericarps) revealing an admixed evolutionary relationship of red rice with wild rice and white rice. The split times of cultivated red and white rice populations have been estimated to be within the most recent 3500 years. The results also provide evidence for re-domestication of weedy rice into cultivated red rice, and suggest candidate genes underlying the red-pericarp trait.
Introduction
Rice has been domesticated for thousands of years and is now grown worldwide as a staple food. Rice produces grains with red, brown, purple, or black pericarps. Pigmented rice, which is rich in phenolic compounds like anthocyanins and proanthocyanidins, plays a role in biological functions such as inhibition of oxidative stress, alleviation of hyperlipidemia, and prevention of obesity (Chu et al., 2019; Yu et al., 2021). With the improvement in societal living standards, growing attention has gradually been focused on red rice because of its benefits to human health.
Red pigmentation of rice grains is attributed to two domestication-related genes, Rc and Rd (Sweeney et al., 2006; Sweeney et al., 2007). Multiple alleles of the Rc gene, which encodes a bHLH protein, have been reported. The recessive rc allele contains a 14-bp deletion in exon 6 of Rc and is a domesticated loss-of-function allele of Rc (Sweeney et al., 2006). Rcr and Rc-g contain single G-base deletions in distinct locations, both occurring upstream of the 14-bp deletion in the rc allele, which restore the reading frame of the gene and thus contribute to the red-pericarp phenotype (Brooks et al., 2008; Lee et al., 2009). The Rc-s allele is characterized by a C-to-A transversion within the Rc gene, which results in premature termination of the Rc protein and a white pericarp (Sweeney et al., 2007; Furukawa et al., 2007). Rd encodes dihydroflavonol-4-reductase, which catalyzes the conversion of dihydroflavonols into leucoanthocyanidins. Rd by itself does not generate any pigment, but it regulates accumulation of proanthocyanidins in the pericarp (Sweeney et al., 2006; Furukawa et al., 2007). The Rc-rd genotype results in brown rice grains, whereas both rc-rd and rc-Rd genotypes produce white rice grains (Furukawa et al., 2007).
The evolutionary trajectory of rice has attracted significant attention in the last 10 years (e.g., Huang et al., 2012b; Civáň et al., 2015; Choi et al., 2017; Jing et al., 2023; Wu et al., 2023). Specific efforts have been undertaken to unravel the domestication trajectory of red rice (Sweeney et al., 2007; Gross and Olsen, 2010). It has been demonstrated that the 14-bp deletion in Rc arose from Geng-japonica (GJ) rice and was later introgressed into Xian-indica (XI) and aus subpopulations. On the other hand, the C-A mutation (Rc-s) originated in the aus group and did not propagate widely during the domestication process (Sweeney et al., 2007). In the USA, the term “red rice” is also used to describe weedy rice, which is found in rice fields worldwide (Gross et al., 2010; Thurber et al., 2010; Wu et al., 2022). The major-effect loci (qSD7-1 and qPC7) that overlap with the Rc locus in weedy rice have been studied in several population genomic studies (Li et al., 2017; Qiu et al., 2017). Reversion of Rc to a functional state in weedy rice has been attributed to spontaneous back mutations (Sweeney et al., 2007; Cui et al., 2016; Sun et al., 2018), gene flow from wild rice (Song et al., 2014; Wedger et al., 2019; Wu et al., 2023), or standing variation from older landraces that lack the domestication allele (Qiu et al., 2020; Wu et al., 2022).
It is noteworthy that black grain color is not observed in wild rice (O. rufipogon), and nearly all wild rice exhibits a red pericarp (Sweeney et al., 2007). Wild plants underwent a pre-domestication cultivation phase, for which certain domestication traits are documented in archaeobotanical records, reflecting varying levels of domestication (Gross and Olsen, 2010). During the initial stage of rice domestication, farmers paid more attention to reducing seed shattering and increasing seed size (Purugganan and Fuller, 2009). Attention shifted toward enhancement and diversification of other traits, including selection for grain color (Gross and Olsen, 2010). Notably, functional changes in genes like rc (loss of function) and Kala4 (gain of function) occurred within a relatively short time frame (Oikawa et al., 2015). The specifics of how, when, and where red and white rice were established during the long domestication process remain elusive.
Although red rice has received less attention owing to shifts in dietary preferences (Ahuja et al., 2007), its significance as a crucial link between the pre- and post-domestication periods is indisputable. Wild rice, domesticated rice, and de-domesticated rice can all exhibit red pericarps. Consequently, red rice encompasses a broad spectrum of rice genetic backgrounds and provides a comprehensive resource for studying the history of rice domestication and genetic improvement. In this study, we sequenced dozens of cultivated red rice accessions from China and compiled additional available genomic data from red and black rice worldwide to form a genomic dataset for 5104 accessions (termed the 5k genomes). The dataset consists of 4315 cultivated, 230 wild, and 559 weedy rice accessions. The genomic data were used in population genomic analyses to investigate the evolutionary history of red rice. To the best of our knowledge, this is the largest collection used to date for studying the population genetics of red rice. Our results provide genomic evidence for the evolutionary trajectory of red rice and a valuable genomic resource for future research and breeding of pigmented rice.
Results
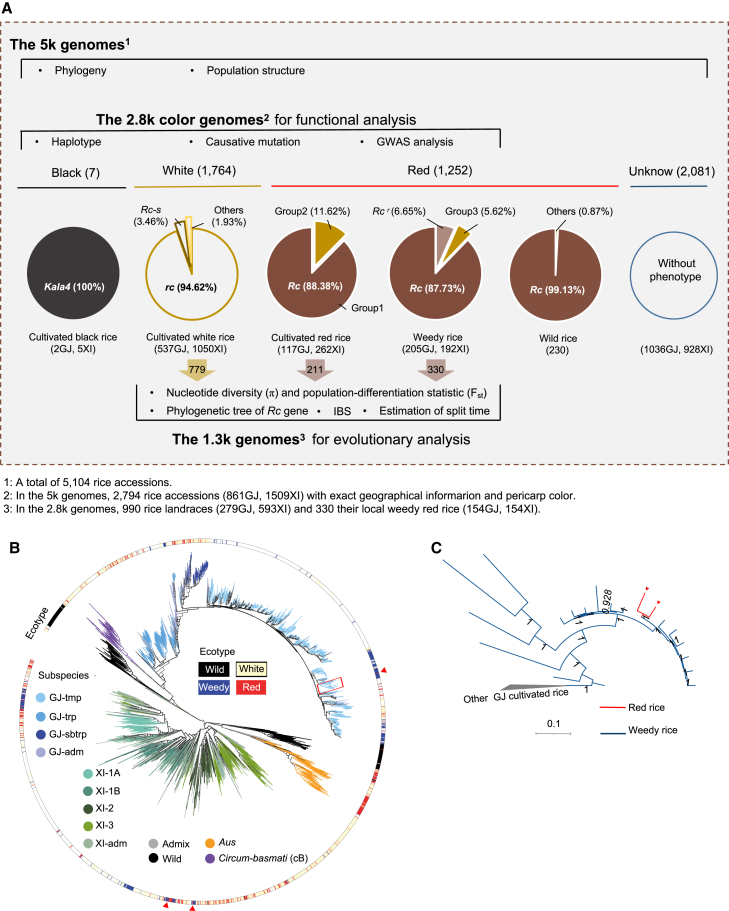
The 5k genomes used in this study
We collected 85 red rice accessions from various regions across China and sequenced them at an average depth of ∼15-fold. We also obtained high-depth whole-genome sequencing data for 5019 rice accessions from previous studies (Wang et al., 2018; Xia et al., 2019; Li et al., 2020; Qiu et al., 2020; Zheng et al., 2021). Together, the genomes of 5104 worldwide rice accessions (hereafter referred to as “the 5k genomes”) were integrated and analyzed (Figure 1A; Supplemental Figure 1; Supplementary Table 1). The 5k genomes include 2437 XI, 1897 GJ, 229 aus, 90 circum-basmati (cB), 221 admixed, and 230 wild (O. rufipogon) accessions (Figure 1A; Supplemental Figure 1). Among them, 2794 (53.30%) accessions (hereafter referred to as “the 2.8k color genomes”) were classified into black (purple), red (brown), and white groups on the basis of pericarp color (Figure 1A). Alternatively, the 2.8k color genomes could be separated into 542 cultivated red rice, seven cultivated black rice, 481 weedy rice (red pericarp), and 1764 cultivated white rice collected from 117 different countries. In terms of subspecies, the 2.8k color genomes contain 1509 XI, 861 GJ, 202 aus, 72 cB, and 150 admixed accessions. From the 2.8k color genomes, 990 landraces and 330 local weedy rice were selected as “the 1.3k genomes” dataset for evolutionary analysis (Figure 1A). The 1.3k genomes consist of 747 XI, 433 GJ, 80 aus, 26 cB, and 34 admixed accessions.
Figure 1.
Overview of the genomic data and workflow used in this study.
(A) Workflow used in this study. Among the 5k genomes, 2794 accessions with colored pericarps and geographical information (the 2.8k color genomes) were used for functional analysis, and 1320 rice accessions (the 1.3k genomes), including landraces and local weedy rice, selected from the 2.8k color genomes were used for evolutionary analysis. The accessions were classified into red, white, and black groups according to their available pericarp color. On the basis of genotypes of two functional genes (Rc and Kala4), the pigmented rice accessions (O. sativa) were further defined as black rice with the Kala4 genotype or red rice with the Rc/kala4 genotype. Most wild rice accessions (O. rufipogon) have red pericarps and the Rc/kala4 genotype.
(B) Phylogenetic tree of the 5k genomes inferred from whole-genome SNPs. Different subspecies are indicated by colored lines, and the outermost circle represents the ecotypes of the accessions.
(C) Detailed phylogenetic tree of the rice accessions in the red rectangle in (B), showing the relationship between a cultivated red rice (represented by the red lines) and weedy red rice. The cultivated red rice is surrounded by weedy red rice (blue lines). Other cultivated rice is represented by a gray cluster.
Phylogeny, population structure, and diversification of the rice populations
We first individually mapped the clean paired-end reads (average 10-fold coverage) of the 5k genomes to the Nipponbare reference genome (IRGSP-1.0). A total of ∼46 million single nucleotide polymorphisms (SNPs) and ∼5.6 million small insertions/deletions (InDels) were identified. A maximum-likelihood phylogenetic tree based on 4 251 459 high-quality synonymous SNPs clearly showed divergence of subspecies from wild groups (Figure 1B). The phylogenetic tree revealed a mosaic structure of red, black, white, and weedy rice, although specific branches were dominated by single rice types. For example, the aus group was clustered into a single clade and closely nested with wild rice (Figure 1B), with 65.6% of the accessions being cultivated red rice collected from South Asia (Supplementary Table 2). Cultivated red and white rice were scattered across the whole tree, and this population structure in the phylogenetic tree was consistent with the results of principal component analysis and ADMIXTURE analysis (Supplemental Figure 2). As expected, weedy rice clustered with both GJ and XI rice in the branches of the phylogenetic tree (Figure 1B). Some cultivated red rice accessions were located within a clade of weedy rice with high bootstrap support (Figure 1C and Supplemental Figure 3), implying that weedy rice may have been re-domesticated into cultivated red rice. On the other hand, within the XI subspecies, several clusters exhibited a mix of weedy and cultivated red rice. Weedy rice was embedded within the cultivated red rice clusters (Figure 1B). Further investigation of derived allele frequency in each range of minor allele frequency in wild rice revealed similar distribution patterns of derived allele frequencies between cultivated red rice (C) and weedy rice (W) but significantly different patterns in wild rice (x axis) (Supplemental Figure 4). These results suggested that weedy rice was likely to have been de-domesticated from cultivated red rice.
Given that landrace genomes contain limited introgression from cultivated white rice, we used the 1.3k genomes, which consist of landraces and local weedy rice, to investigate the domestication history of red rice. We used variants within the causative gene responsible for the red pericarp, i.e., Rc, to construct a phylogenetic tree. The 990 landraces, 330 weedy rice, and 180 wild rice were clearly separated according to different alleles of Rc (Figure 2A). Apparently, some red rice were at the positions closest to wild rice. Nearly all aus accessions clustered together and nested within wild ancestors, indicating that wild rice was the direct donor of the Rc gene in the aus rice subgroup (Figure 2A). Several weedy rice accessions were embedded within a group of cultivated red rice but far from wild rice (Figure 2A), suggesting that weedy rice might have been de-domesticated from red rice, retaining the Rc gene.
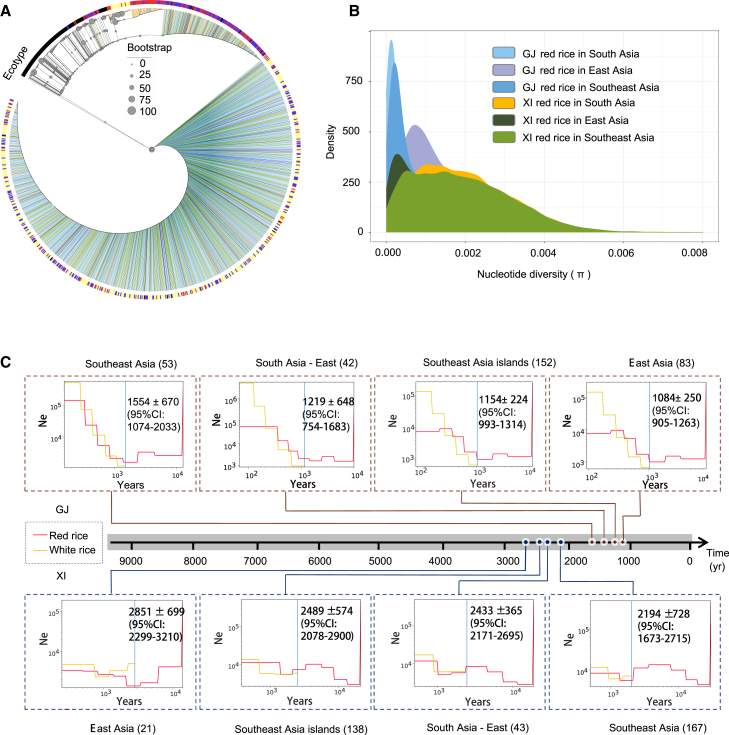
Figure 2.
Genetic diversity and differentiation of red rice.
(A) Phylogenetic tree of the 1.3k genomes based on SNPs within the Rc gene. The colored lines indicate different subspecies, and the outermost circle represents the ecotypes of the accessions; the color representation is the same as that in Figure 1B.
(B) Distribution of nucleotide diversity density of GJ and XI red rice in different regions. Subspecies and regions are indicated by different colors.
(C) The estimated split time of red rice and white rice. Top: differentiation time of GJ red and white landraces in different regions inferred by SMC++. Bottom: differentiation time of XI red and white landraces in different regions inferred by SMC++.
Nucleotide diversity (π) was estimated for different rice groups to compare their genetic diversity (Figure 2B). A density plot showed that GJ red landraces in East Asia (average π = 1.22 × 10−3) and XI red landraces in South Asia (average π = 1.92 × 10−3) were the most diverse (Figure 2B). Red rice had the highest polymorphism in areas of rice origin (i.e., GJ in East Asia and XI in South Asia; Huang et al., 2012b; Civáň and Brown, 2018), indicating a high likelihood of its domestication from wild rice. We also calculated identity by state (IBS) between the red/white landraces and wild rice derived from the same region (Supplemental Figure 5). In all regions, cultivated red landraces exhibited closer genetic proximity to wild rice than to cultivated white landraces (Supplemental Figure 5A), implying that red landraces have greater genetic introgression from wild rice and red landraces are more likely to originate from wild rice.
The split time of red and white rice landraces
To provide a temporal context of differentiation time between red rice and white rice, we used SMC++ (Terhorst et al., 2017) to estimate divergence time between the two groups based on their landraces. SMC++ combines the simplicity of sequentially Markovian coalescent methods and the scalability of site frequency spectrum methods, and its robustness has been demonstrated previously (Zhao et al., 2023). Our results showed that GJ red rice and white landraces from different regions (including East Asia, South Asia, and Southeast Asia) differentiated into different groups within the recent ∼2200 yr BP (years before present). The differentiation time of XI red landraces and white landraces in different regions was estimated to be within ∼3500 yr BP (Figure 2C and Supplemental Figure 6). These time estimates are consistent with descriptions in ancient Chinese books such as “Xin Tang Shu” and “Song Shi,” which relate that south China had white rice production and entered the era of intensive white-rice farming in the Tang dynasty (about 1500 yr BP), especially in the Taihu lake basin (Li et al., 2021). Our results indicate that the rc mutation (e.g., white rice) has been present in the rice population for a long time but was ultimately fixed at a much later stage.
Identification of candidate alleles/genes for rice red pericarp
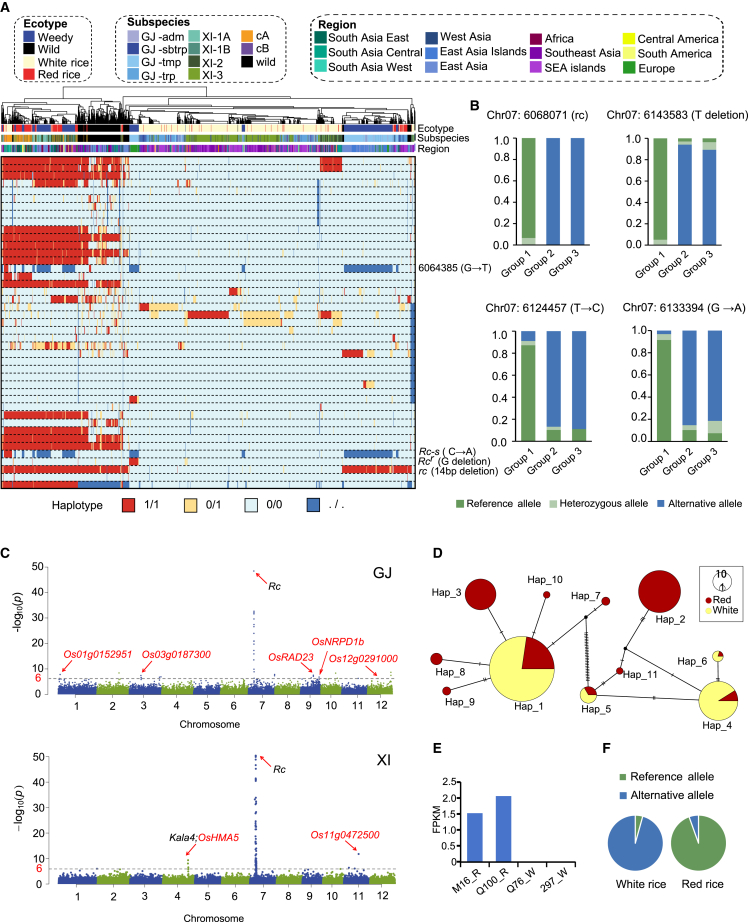
On the basis of their Rc and Kala4 (contributing to black pericarp) genotypes, pigmented rice accessions (O. sativa) were further categorized as either black rice with Kala4 or red rice with Rc/kala4. All wild rice accessions showed a red pericarp with the Rc/kala4 genotype. However, a small subset of rice accessions (84/5104, 1.7%) displayed a red pericarp that could not be explained by Rc or Kala4 (Figure 1A). As expected, the majority of cultivated white rice accessions (94.6%) contained the rc gene. In addition, 62 (3.5%) accessions harbored the Rc-s allele, which contributes to a white pericarp owing to premature termination of the Rc protein caused by a C-A mutation. Intriguingly, 34 (1.9%) accessions with white pericarps harbored the Rc gene, and the underlying genetic mechanism for this phenomenon remains to be discovered. We did not find the Rc-g allele in any of the accessions, presumably as a result of filtering by a relatively high missing rate of 0.2. To gain insight into the mutation distribution within the Rc gene, we constructed a haplotype plot of the Rc gene for the 2.8k color genomes and categorized them based on ecotypes, regions, and subspecies (Figure 3A). The haplotype plot indicated that all wild accessions harbored the Rc allele, confirming its status as the ancestral allele contributing to the wild-type red-pericarp phenotype. Furthermore, absence of proanthocyanin accumulation in some white rice from the aus group was due to the presence of the loss-of-function Rc-s allele (with a C-A transversion). In these aus white rice, a G-T transition (r2 = 0.51) was identified 3633-bp upstream of the C-A mutation in the Rc-s allele. The simultaneous deletion of these two mutations was observed in red rice across all regions except Africa (Figure 3A), perhaps reflecting human selection during the domestication process.
Figure 3.
Identification of new candidate genes/alleles for red pericarp in rice.
(A) SNP- and indel-based haplotype map of the Rc gene. The ecotype, subspecies, and source region of each accession are indicated by colored lines under the clustered tree, and important mutations, including rc, Rc-s, and Rcr, are marked at their positions. The genotypes (ref, alt, het, and missing) of each variant are color-coded, using the IRGSP-1.0 genome as the reference.
(B) Three possible sequence variants related to the contradiction between phenotype and genotype, including a deletion (T) and two single-base mutations (T to C and GnullA). All three variants had a higher rate of the reference allele in Group1 and a higher rate of the alternative allele in Group2 and Group3. Three groups were defined as Group#1 and Group#2, cultivated red rice with the Rc or rc allele, respectively, and Group#3, weedy red rice with the rc allele and without the Rcr gene.
(C) Manhattan plots for red pericarp color in the GJ (top) and XI (bottom) rice groups. The −log10(P) values from a genome-wide scan are plotted against position on each of 12 chromosomes. The horizontal dashed lines indicate the genome-wide significance thresholds (P = 2.9 × 10−7 for GJ and P = 3.0 × 10−7 for XI, equivalent to about 6.5 after the logarithm).
(D) Haplotype network of Os01g00152951 in the GJ group.
(E) Transcriptomic patterns of Os01g0152951 in the two red rice accessions presented as number of fragments per kilobase of exon model per million mapped reads (FPFM).
(F)Os04g0556000 in the XI group has a significantly different allele frequency between the red rice and white rice groups.
The majority of the 542 cultivated red rice accessions (88.4%, termed “Group1”) harbored the Rc gene, whereas 63 accessions (11.6%) harbored the rc allele but still exhibited a red pericarp (termed “Group2”). Among the 481 weedy rice accessions, the red-pericarp phenotype in most (87.7%) could be attributed to the presence of the Rc gene. Thirty-two accessions (6.7%) harbored the rc and Rcr allele, in which a G-base deletion in rc restores Rc functionality, resulting in a red pericarp. In addition, 27 accessions (5.6%, termed “Group3”) contained the rc gene but maintained a red pericarp, with the causative mutation(s) for their red-pericarp phenotype remaining unknown (Figure 1A). To further explore the genomic pattern behind the phenotypes and look for variants that contribute to a red pericarp, we focused on rice accessions that exhibited a contradiction between phenotype and genotype at the Rc locus (i.e., Group1, Group2, Group3) (Figure 1A). By comparing allele frequencies of SNPs and indels within the 100-kb flanking regions of the Rc gene among the three groups, we identified three variations as candidates that could potentially contribute to explaining the contradiction. These included a T deletion 74 266 bp downstream of the Rc gene, a G-A mutation 64 077 bp downstream, and a T-C mutation 55 140 bp downstream. The T deletion (chr07:6143583) may affect the function of a transposase-encoding gene (Os07t0212400-01). More than 95% of accessions in Group1 harbored the reference allele (C), whereas over 94% and 89% of accessions in Group2 and Group3, respectively, harbored the alternative allele (T) (Figure 3B). The G-A mutation (chr07:6133394) is an upstream variation in a gene encoding an mRNA-binding protein. More than 91% of accessions in Group1 harbored the reference allele (G), whereas more than 85% and 81% of accessions in Group2 and Group3 harbored the alternative allele (A) (Figure 3B). For the T-C mutation (chr07:6124457), over 87% of accessions in Group1 harbored the reference allele (T), and more than 86% and 88% of accessions in Group2 and Group3 harbored the alternative allele (C) (Figure 3B).
To explore new genes underlying the red-pericarp phenotype, we performed a genome-wide association study (GWAS) for the GJ and XI populations (Figure 3C). Seventeen associated loci (P = 2.9 × 10−7) were identified in the GJ population, and four associated loci were identified in the XI population (P = 3.0 × 10−7) (Supplementary Table 3). Among these loci, one in the GJ group and two in the XI group were associated with known genes involved in proanthocyanin synthesis and other pathways, such as Rc (Sweeney et al., 2006) and Kala4 (Oikawa et al., 2015) (Figure 3C), as supported by previous studies on mutants or recombinant populations. Several new loci were found to be associated with red pericarps in both GJ and XI subspecies (Figure 3C). For example, an association signal (P = 2.18 × 10−8) was found upstream of Os01g0152951 (annotated as a hypothetical conserved gene) in the GJ population at chr01:2781894. Notably, this locus harbored different dominant haplotypes in the red rice and white rice groups. Haplotype network analysis revealed that 75.7% of the GJ red rice predominantly contained haplotypes hap#2 and hap#3, whereas hap#1 and hap#4 accounted for 79.0% and 90.2% of GJ white rice, respectively (Figure 3D). To better verify the candidate genes, we downloaded RNA-seq data from a previous study of two red rice and two white rice accessions (Zainal-Abidin et al., 2020). We found a significant difference in the expression level of Os01g0152951 between the two phenotypes (Figure 3E). Further sequence analysis indicated that Os01g0152951 has been lost in the two white rice (Q76_W and 297_W) (Supplemental Figure 7). Another association signal (P = 1.49 × 10−9) was found upstream of OsHMA5 (Os04g0556000) in the XI population. This gene neighbors Kala4 and has previously been implicated in xylem loading of copper in rice (Deng et al., 2013). By comparing the allele frequency of each SNP in OsHMA5 between the red and white rice groups, we found significant difference in allele frequency at chr04:27834266 between red rice (T: 0.95; A: 0.05) and white rice (T: 0.04; A: 0.96) (Figure 3F). The linkage disequilibrium of every single SNP in Kala4 was calculated, and very low linkage (average r2 = 0.019) was found, suggesting that OsHMA5 is another promising candidate gene for the red-pericarp phenotype.
Discussion
A red pericarp is an inherent trait of wild rice (Roy and Shil, 2020), and loss of proanthocyanins in the rice pericarp is a domestication-related trait (Xia et al., 2021). Red rice, found across wild, domesticated, and de-domesticated rice, represents the ancient form of rice and preserves rich genomic imprints inherited from its ancestors. Its existence is associated with geographic and cultural contexts, particularly for the red rice landraces. Therefore, red rice maintains an intact genetic background of rice history. The broad spectrum of accessions collected in this study was useful for revealing the domestication history of red rice. Moreover, the population studies carried out here used a balanced number of pigmented and white rice accessions, enhancing the power for illuminating the evolutionary history of rice.
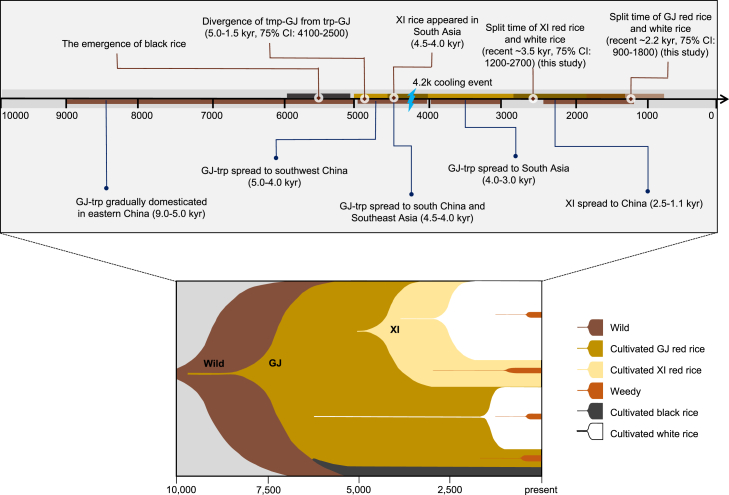
We proposed multiple origins of red rice on the basis of the genome-wide phylogeny results, the phylogenetic tree of the Rc gene, and other genetic evidence. We found that red landraces in the aus group originated from wild rice, whereas some red rice accessions were re-domesticated from weedy red rice. We found that both cultivated white and red rice could be de-domesticated into weedy rice. Although the utilization of cultivated black rice in this study was limited, the phylogenetic tree seems to support previous findings (Supplemental Figure 8) (Oikawa et al., 2015); i.e., the sequence rearrangement in the promoter of Kala4 that contributes to the black pericarp phenotype first occurred in tropical GJ and then spread to XI and subsequently to temperate GJ through natural crossing and artificial selection for the black-pericarp trait (Oikawa et al., 2015).
A considerable amount of archaeological and genomic evidence has been proposed to infer the origin of rice (Molina et al., 2011; Huang et al., 2012b; Gross and Zhao, 2014; Civáň et al., 2015; Choi et al., 2017; Choi and Purugganan, 2018; Wang et al., 2018; Zheng et al., 2021; Wu et al., 2023). The evolutionary trajectory of single domestication with multiple origins (i.e., GJ was first domesticated from wild rice, and XI originated from proto-GJ by introgression to local wild rice) has been well documented on the basis of genomic data (Huang et al., 2012b; Choi et al., 2017; Choi and Purugganan, 2018; Wu et al., 2023). The history of rice dispersal has also been reconstructed using whole-genome sequences (Gutaker et al., 2020). In brief, originating around 9000 years ago in the Yangtze Valley, rice diversified into temperate and tropical GJ types after the global cooling event about 4200 yr BP (Figure 4). It was since ∼2500 yr BP that tropical GJ rice reached Southeast Asia and diversified rapidly. GJ rice spread to South Asia by ∼4000 yr BP and led to introgression of domestication alleles into proto-XI or local O. nivara populations, leading to the emergence of XI rice (Gutaker et al., 2020). Crop plants acquire characteristics distinct from those of their wild progenitors through a process of stepwise selection, involving collective and progressive changes in several important traits. It has been reported that during the initial period of domestication, humans continued to gather wild rice, leading to slow emergence of the non-shattering trait. Because of the ongoing gene flow into proto-domestications, fixation of the trait took about 2000 years (Purugganan and Fuller, 2009). It is conceivable that rice tends to retain the characteristic red pericarp of wild rice for a long time, until completion of the initial domestication process. Meanwhile, red rice exhibits greater adaptability to local environments (Qi et al., 2022; Gu et al., 2005), which may have encouraged ancient farmers to plant red rice in order to gather more grains.
Figure 4.
A schematic evolutionary trajectory of pigmented rice.
The top panel shows the timeline of rice domestication, including evidence from previous archaeological and genomic studies (Huang et al., 2012b; Gross and Zhao, 2014; Oikawa et al., 2015; Gutaker et al., 2020; this study). The bottom panel shows a wild–red–white model of rice evolution based on the pericarp colors. Ancient wild rice experienced slow domestication and gradually produced GJ rice, most of which had a red pericarp. Over time, the proportion of white-pericarp rice increased, and this was accompanied by the emergence of weedy rice de-domesticated from both red rice and white rice. XI rice was domesticated later and experienced an evolutionary process similar to that of GJ rice.
In addition to human innovation and crop adaptation under the climatic and political conditions of the time (Zhang et al., 2015; Pei et al., 2019), people’s preference for certain crops changed agricultural systems (Xhauflair et al., 2017; Overton and Taylor, 2018), as did the red–white shift in rice. At present, white rice varieties dominate in rice production. Our estimates suggest that the red–white shift in rice production occurred about 3500 years ago (Figures 2C and 4 and Supplemental Figure 6). The widespread cultivation of white rice might be attributed to the fact that it is easier to detect insects and eliminate pathogens on a light background. In addition, less energy and time required for cooking also contributed to its popularity (Sweeney et al., 2006). The rapid and extensive adoption of white rice in cultivation may also be attributed, in part, to the recessive nature of the rc mutation. Seeds with a white pericarp will faithfully produce offspring with white pericarps (Sweeney et al., 2006). It is conceivable that the favored rc allele would not have traveled far beyond its origin if not for selection by humans who picked out the white grain as a preferred food and traded it as a commodity. Some red landraces seem likely to have served as the intermediate ancestral progenitor of cultivated white rice, having undergone extensive artificial selection and improvement over time. For example, some cultivated white rice accessions were surrounded by red rice accessions in the phylogenetic tree, suggesting their potential origin from red rice (Figure 1B).
Taken together, our results led us to propose a wild–red–white model of rice evolution based on pericarp colors (the lower panel of Figure 4). We believe that rice with a red pericarp existed during the initial domestication period of rice and gradually turned to rice with a white pericarp through a process of diversification. Red rice production predominated for most of the approximately 10 000-year evolutionary history of rice until roughly three thousand years ago, when white rice began gaining in popularity and subsequently predominated in both XI and GJ rice. As suggested by previous work and this study, cultivated white and red rice may feralize to weedy red rice (Qiu et al., 2020; Wu et al., 2022). The red–white shift happened in diverse regions and different historical periods. Therefore, since its domestication from wild rice, rice has entered into a red–white evolutionary cycle and keeps transitioning from one state (e.g., red) to another (e.g., white).
GWAS has been used to study pericarp color for a long time. Larger sample size can greatly enhance the power of GWAS (Huang et al., 2010). The substantial increase in sample size of this study compared with previous studies (Huang et al., 2010, 2012b; Wang et al., 2016; Rana et al., 2022; Yang et al., 2022) enables more comprehensive detection of associated loci. In general, the genetic architecture of the coloration traits exhibited remarkable similarity between the two subspecies, suggesting that visible traits are more susceptible to the influence of introgression between subspecies (Huang et al., 2012a). In summary, our results suggest that in addition to the significant effect of the Rc gene, effects of multiple other loci may also contribute to variation in pericarp pigmentation. These newly identified loci are attractive candidates for follow-up investigations that could expand our understanding of the genetic basis of proanthocyanidin synthesis and accumulation. More functional genomics studies are needed to further validate the effects of these genes and identify their functional variants.
Methods
Sampling of rice accessions
To capture the comprehensive genetic diversity of all rice ecotypes, 5104 rice accessions were used in this study (Supplementary Table 2), including 5019 accessions from published work (Zheng et al., 2021; Wang et al., 2018; Xia et al., 2019; Li et al., 2020; Qiu et al., 2020) and 85 newly sequenced red rice accessions. The newly sequenced accessions were collected from major rice production regions of China.
DNA isolation and genome sequencing
Genomic DNA was extracted from young leaves of each of the 85 red rice accessions using the standard cetyltrimethylammonium bromide–based protocol (Edwards et al., 1991), and 1.5 μg of DNA per sample was used as input for sequencing library construction using the TruSeq Nano DNA HT sample preparation kit (Illumina USA). The libraries were sequenced on the Illumina HiSeq 4000 platform (150-bp, paired-end reads) to generate about 1.21 Tb of raw sequence.
Sequence quality checking and filtering
To eliminate reads with artificial bias (i.e., low-quality paired reads, which primarily resulted from base-calling duplicates and adaptor contamination), raw paired reads from the 85 red rice accessions were first cleaned using NGSQC-toolkit (Patel and Jain, 2012) with default parameters. About 1.21 Tb of high-quality genomic data were retained.
Sequence alignment, variant calling, and annotation
We downloaded raw sequencing data for 5019 rice accessions from each of the cited publications and performed SNP calling from the raw short reads using the same pipeline. First, high-quality reads were mapped to the O. sativa reference genome (IRGSP-1.0) by Bowtie 2 (Langmead and Salzberg, 2012) with the default command. Variants were detected (based on the default parameters) and filtered (those with QUAL ≥ 30, DP ≥ 10, QD ≥ 2, and MQ ≥ 20 were retained) using GATK software (v3.7) (Mckenna et al., 2010). Next, all variants from different projects, including 3K-RG (Wang et al., 2018), one thousand accessions from China (Li et al., 2020), 185 accessions of wild rice (Zheng et al., 2021), a weedy rice gene pool (Qiu et al., 2020), and the 85 newly sequenced red rice accessions, were integrated into one file in VCF format. The following potential low-quality variants were removed: (1) missing rate >0.2 and (2) minor allele frequency ≤0.05. After filtering, ∼4 million high-quality SNPs and ∼0.5 million indels were retained and annotated with SnpEff (v3.6) (Cingolani et al., 2014) to profile their potential effects on predicted amino acid sequences and gene functions.
Population phylogenetic analysis
To reduce marker redundancy, we randomly picked one SNP from every 10 consecutive SNPs and constructed a SNP dataset consisting of 425 145 sites evenly distributed across the rice genome. Phylogenetic trees were constructed using FastTreeMP (Price et al., 2009) with 1000 bootstrap replicates. iTOL (http://itol.embl.de/) was used to visualize the trees.
Population structure analysis
Structure analysis was performed using ADMIXTURE v1.3.0 software (Alexander et al., 2009) with K values from 5 to 15 to estimate the standard error of parameters. Principal component analysis was performed using PLINK v1.9 (Chang et al., 2015) with the command “--pca 20.” Genetic distances were estimated by IBS using PLINK v1.9 both among and within rice subgroups and ecotypes. The distance matrix was imported into R v3.4.1 to plot a boxplot using R scripts.
Fluctuation of effective population size over time
To better understand the historical demographics of red rice compared with white rice, we tried to reconstruct past effective population size using the SMC++ method (Terhorst et al., 2017). Because SMC++ has sufficient power to deal with situations with different coverages and sample sizes, the reconstruction was carried out independently for varying numbers of subgroups of red rice and white rice from different regions around the world. Because sequences of functional genes and regulatory elements do not satisfy neutral selection assumptions and will strongly effect estimation of split time, we masked protein-coding sequences and their 3-kb flanking sequences in the follow-up analyses. Fifteen samples from different regions were randomly selected at a time, and partitioned VCF files were transferred into smc haploblock files, then further divided into 12 chromosomes. The files were used in the ESTIMATE function of SMC++ with a mutation rate of 6.5 × 10−9 to estimate past effective population sizes. Results were scaled in time using an estimated generation time of 1 year and plotted on a smooth linear timescale in R. These demographics were used to calculate split times between red rice and white rice in different regions with the SPLIT function.
Population genetic analysis
The genome of every subpopulation and subgroup was scanned with a 100-kb window size and a 10-kb step size. Population parameters were estimated for each window with VCFtools (Danecek et al., 2011). Nucleotide diversity (π) was calculated with the parameters “--window-pi 100000 --window-pi-step 10000.” The average π value in a 100-kb window was taken as the genetic diversity. The population differentiation index (FST) was measured with the settings “--fst-window-size 100000 --fst-window-step 10000.” The minor allele frequency was calculated using PLINK v1.9 with parameters “--noweb --freq” for XI red rice, XI weedy rice, and wild rice, which are all neighboring in the phylogeny. The distribution of the derived allele frequency was calculated in weedy rice (W) and cultivated red rice (C) in each range of minor allele frequency (x axis) in wild rice.
Genome-wide association study
GWAS was performed using a compressed MLM model that could effectively reduce false positives. The equation of the compressed MLM model is y = Xα + Pβ + Kμ + e, in which y is the phenotype, X is the genotype, P is the population structure matrix (Q matrix), and K is the kinship matrix. The P matrix was built from the top five principal components for population structure correction. The K matrix was built from the matrix of simple matching coefficients. The filtered VCF files were converted to tped format using PLINK v1.9 and entered into the software Efficient Mixed Model Association eXpedited (EMMAX; Kang et al., 2010) for GWAS. The significant P-value threshold was determined using the Bonferroni correction method. The Manhattan and QQ plots for GWAS were generated using the R package qqman. To verify the candidate genes, RNA sequences generated by Zainal-Abidin et al. (2020) (accession number PRJEB34340) from two red and two white rice accessions were downloaded and used to compare expression differences of the candidate genes. The two white accessions used in the RNA-seq experiment have the same mutation or allele as that in the candidate genes identified in this study, whereas the two red rice accessions have the wild-type genotype.
Data and code availability
The raw sequencing data are available at the China National Genomics Data Center (https://ngdc.cncb.ac.cn) under BioProject accession number PRJCA017658.
Funding
This work was supported by the Department of Science and Technology of Zhejiang Province (2022C02032 and 2020C02002), the National Natural Science Foundation of China (31971865), and CIC-MCP to L.F.
Author contributions
L.F. conceptualized and designed the experiments. L.X. performed most of the experiments under the supervision of L.F. and the guidance of D.W. Y.F., C.Y., and X.W. helped to collect the samples for sequencing. L.F., D.W., and Q.-H.Z. revised the manuscript.
Acknowledgments
We thank three anonymous reviewers for valuable suggestions. No conflict of interest is declared.
Published: December 6, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Supplemental information
References
- Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks S.A., Yan W., Jackson A.K., Deren C.W. A natural mutation in rc reverts white-rice-pericarp to red and results in a new, dominant, wild-type allele: Rc-g. Theor. Appl. Genet. 2008;117:575–580. doi: 10.1007/s00122-008-0801-8. [DOI] [PubMed] [Google Scholar]
- Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation plink: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.Y., Platts A.E., Fuller D.Q., Hsing Y.I., Wing R.A., Purugganan M.D. The rice paradox: multiple origins but single domestication in Asian rice. Mol. Biol. Evol. 2017;msx49:msx049. doi: 10.1093/molbev/msx049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.Y., Purugganan M.D. Multiple origin but single domestication led to Oryza sativa. G3 (Bethesda). 2018;8:797–803. doi: 10.1534/g3.117.300334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu M.J., Du Y.M., Liu X.M., Yan N., Wang F.Z., Zhang Z.F. Extraction of proanthocyanidins from chinese wild rice (zizania latifolia) and analyses of structural composition and potential bioactivities of different fractions. Molecules. 2019;24:1681. doi: 10.3390/molecules24091681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, snpeff. Fly. 2014;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civáň P., Craig H., Cox C.J., Brown T.A. Three geographically separate domestications of asian rice. Nat. Plants. 2015;1:15164. doi: 10.1038/nplants.2015.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civáň P., Brown T.A. Role of genetic introgression during the evolution of cultivated rice (Oryza sativa L.) BMC Evol. Biol. 2018;18:57. doi: 10.1186/s12862-018-1180-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Song B.K., Li L.F., Li Y.L., Huang Z., Caicedo A.L., Jia Y., Olsen K.M. Little white lies: pericarp color provides insights into the origins and evolution of southeast Asian weedy rice. G3 (Bethesda). 2016;6:4105–4114. doi: 10.1534/g3.116.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., et al. The variant call format and vcftools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng F., Yamaji N., Xia J., Ma J.F. A member of the heavy metal p-type ATPase OsHMA5 is involved in xylem loading of copper in rice. Plant Physiol. 2013;163:1353–1362. doi: 10.1104/pp.113.226225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K., Johnstone C., Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T., Maekawa M., Oki T., Suda I., Iida S., Shimada H., Takamure I., Kadowaki K.i. The rc and rd genes are involved in proanthocyanidin synthesis in rice pericarp. Plant J. 2007;49:91–102. doi: 10.1111/j.1365-313X.2006.02958.x. [DOI] [PubMed] [Google Scholar]
- Gross B.L., Olsen K.M. Genetic perspectives on crop domestication. Trends Plant Sci. 2010;15:529–537. doi: 10.1016/j.tplants.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross B.L., Reagon M., Hsu S.-C., Caicedo A.L., Jia Y., Olsen K.M. Seeing red: the origin of grain pigmentation in US weedy rice. Mol. Ecol. 2010;19:3380–3393. doi: 10.1111/j.1365-294X.2010.04707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross B.L., Zhao Z. Archaeological and genetic insights into the origins of domesticated rice. Proc. Natl. Acad. Sci. USA. 2014;111:6190–6197. doi: 10.1073/pnas.1308942110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutaker R.M., Groen S.C., Bellis E.S., Choi J.Y., Pires I.S., Bocinsky R.K., Slayton E.R., Wilkins O., Castillo C.C., Negrão S., et al. Genomic history and ecology of the geographic spread of rice. Nat. Plants. 2020;6:492–502. doi: 10.1038/s41477-020-0659-6. [DOI] [PubMed] [Google Scholar]
- Gu X.Y., Kianian S.F., Hareland G.A., Hoffer B.L., Foley M.E. Genetic analysis of adaptive syndromes interrelated with seed dormancy in weedy rice (Oryza sativa) Theor. Appl. Genet. 2005;110:1108–1118. doi: 10.1007/s00122-005-1939-2. [DOI] [PubMed] [Google Scholar]
- Huang X., Wei X., Sang T., Zhao Q., Feng Q., Zhao Y., Li C., Zhu C., Lu T., Zhang Z., et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010;42:961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- Huang X., Zhao Y., Wei X., Li C., Wang A., Zhao Q., Li W., Guo Y., Deng L., Zhu C., et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 2012;44:32–39. doi: 10.1038/ng.1018. [DOI] [PubMed] [Google Scholar]
- Huang X., Kurata N., Wei X., Wang Z.X., Wang A., Zhao Q., Zhao Y., Liu K., Lu H., Li W., et al. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012;490:497–501. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing C.Y., Zhang F.M., Wang X.H., Wang M.X., Zhou L., Cai Z., Han J.D., Geng M.F., Yu W.H., Jiao Z.H., et al. Multiple domestications of asian rice. Nat. Plants. 2023;9:1221–1235. doi: 10.1038/s41477-023-01476-z. [DOI] [PubMed] [Google Scholar]
- Kang H., Sul J., Service S., et al. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with bowtie2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Lupotto E., Powell W. G-string slippage turns white rice red. Genome. 2009;52:490–493. doi: 10.1139/g09-025. [DOI] [PubMed] [Google Scholar]
- Li L.F., Li Y.L., Jia Y., Caicedo A.L., Olsen K.M. Signatures of adaptation in the weedy rice genome. Nat. Genet. 2017;49:811–814. doi: 10.1038/ng.3825. [DOI] [PubMed] [Google Scholar]
- Li H., Liu Z., James N., Li X., Hu Z., Shi H., Sun L., Lu Y., Jia X. Agricultural transformations and their influential factors revealed by archaeobotanical evidence in Holocene Jiangsu Province, eastern China. Front. Earth Sci. 2021;9 [Google Scholar]
- Li X., Chen Z., Zhang G., Lu H., Qin P., Qi M., Yu Y., Jiao B., Zhao X., Gao Q., et al. Analysis of genetic architecture and favorable allele usage of agronomic traits in a large collection of Chinese rice accessions. Sci. China Life Sci. 2020;63:1688–1702. doi: 10.1007/s11427-019-1682-6. [DOI] [PubMed] [Google Scholar]
- Molina J., Sikora M., Garud N., Flowers J.M., Rubinstein S., Reynolds A., Huang P., Jackson S., Schaal B.A., Bustamante C.D., et al. Molecular evidence for a single evolutionary origin of domesticated rice. Proc. Natl. Acad. Sci. USA. 2011;108:8351–8356. doi: 10.1073/pnas.1104686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The genome analysis toolkit: a map reduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overton N.J., Taylor B. Humans in the environment: plants, animals and landscapes in mesolithic britain and ireland. J. World Prehist. 2018;31:385–402. [Google Scholar]
- Oikawa T., Maeda H., Oguchi T., Yamaguchi T., Tanabe N., Ebana K., Yano M., Ebitani T., Izawa T. The birth of a black rice gene and its local spread by introgression. Plant Cell. 2015;27:2401–2414. doi: 10.1105/tpc.15.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R.K., Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Q., Lee H.F., Zhang D.D., Fei J. Climate change, state capacity and nomad-agriculturalist conflicts in Chinese history. Quat. Int. 2019;508:36–42. [Google Scholar]
- Price M.N., Dehal P.S., Arkin A.P. Fasttree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan M.D., Fuller D.Q. The nature of selection during plant domestication. Nature. 2009;457:843–848. doi: 10.1038/nature07895. [DOI] [PubMed] [Google Scholar]
- Qi Q., Chu M., Yu X., Xie Y., Li Y., Du Y., Liu X., Zhang Z., Shi J., Yan N. Anthocyanins and proanthocyanidins: chemical structures, food sources, bioactivities, and product development. Food Rev Int ahead-of-print. 2022;39:4581–4609. [Google Scholar]
- Qiu J., Zhou Y., Mao L., Ye C., Wang W., Zhang J., Yu Y., Fu F., Wang Y., Qian F., et al. Genomic variation associated with local adaptation of weedy rice during de-domestication. Nat. Commun. 2017;8:15323. doi: 10.1038/ncomms15323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J., Jia L., Wu D., Weng X., Chen L., Sun J., Chen M., Mao L., Jiang B., Ye C., et al. Diverse genetic mechanisms underlie worldwide convergent rice feralization. Genome Biol. 2020;21:70. doi: 10.1186/s13059-020-01980-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana N., Kumawat S., Singh U.M., Singh V.K., Deshmukh R., Sharma T.R., Sonah H. Identification of genomic loci governing pericarp color through GWAS in rice (Oryza sativa L.) Indian J. Genet. Plant Breed. 2022;82:1–6. [Google Scholar]
- Roy S.C., Shil P. Assessment of genetic heritability in rice breeding lines based on morphological traits and caryopsis ultrastructure. Sci. Rep. 2020;10:7830. doi: 10.1038/s41598-020-63976-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B.K., Chuah T.S., Tam S.M., Olsen K.M. Malaysian weedy rice shows its true stripes: wild Oryza and elite rice cultivars shape agricultural weed evolution in Southeast Asia. Mol. Ecol. 2014;23:5003–5017. doi: 10.1111/mec.12922. [DOI] [PubMed] [Google Scholar]
- Sun X., Zhang Z., Chen C., Wu W., Ren N., Jiang C., Yu J., Zhao Y., Zheng X., Yang Q., et al. The C–S–A gene system regulates hull pigmentation and reveals evolution of anthocyanin biosynthesis pathway in rice. J. Exp. Bot. 2018;69:1485–1498. doi: 10.1093/jxb/ery001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney M.T., Thomson M.J., Cho Y.G., et al. Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genet. 2007;3:e133. doi: 10.1371/journal.pgen.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney M.T., Thomson M.J., Pfeil B.E., McCouch S. Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell. 2006;18:283–294. doi: 10.1105/tpc.105.038430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhorst J., Kamm J.A., Song Y.S. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat. Genet. 2017;49:303–309. doi: 10.1038/ng.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber C.S., Reagon M., Gross B.L., Olsen K.M., Jia Y., Caicedo A.L. Molecular evolution of shattering loci in U.S. weedy rice. Mol. Ecol. 2010;19:3271–3284. doi: 10.1111/j.1365-294X.2010.04708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja U., Ahuja S.C., Chaudhary N., Thakrar R. Asian Agri-History; 2007. Red Rices-Past, Present, and Future. [Google Scholar]
- Wang H., Xu X., Vieira F.G., Xiao Y., Li Z., Wang J., Nielsen R., Chu C. The power of inbreeding: NGS-based GWAS of rice reveals convergent evolution during rice domestication. Mol. Plant. 2016;9:975–985. doi: 10.1016/j.molp.2016.04.018. [DOI] [PubMed] [Google Scholar]
- Wang W., Mauleon R., Hu Z., Chebotarov D., Tai S., Wu Z., Li M., Zheng T., Fuentes R.R., Zhang F., et al. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature. 2018;557:43–49. doi: 10.1038/s41586-018-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedger M.J., Pusadee T., Wongtamee A., Olsen K.M. Discordant patterns of introgression suggest historical gene flow into Thai weedy rice from domesticated and wild relatives. J. Hered. 2019;110:601–609. doi: 10.1093/jhered/esz030. [DOI] [PubMed] [Google Scholar]
- Wu D., Qiu J., Sun J., Song B.K., Olsen K.M., Fan L. Weedy rice, a hidden gold mine in the paddy field. Mol. Plant. 2022;15:566–568. doi: 10.1016/j.molp.2022.01.008. [DOI] [PubMed] [Google Scholar]
- Wu D., Xie L., Sun Y., Huang Y., Jia L., Dong C., Shen E., Ye C.Y., Qian Q., Fan L. A syntelog-based pan-genome provides insights into rice domestication and de-domestication. Genome Biol. 2023;24:179. doi: 10.1186/s13059-023-03017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D., Zhou H., Wang Y., Li P., Fu P., Wu B., He Y. How rice organs are colored: the genetic basis of anthocyanin biosynthesis in rice. Crop J. 2021;9:598–608. [Google Scholar]
- Xia H., Luo Z., Xiong J., Ma X., Lou Q., Wei H., Qiu J., Yang H., Liu G., Fan L., et al. Bi-directional selection in upland rice leads to its adaptive differentiation from lowland rice in drought resistance and productivity. Mol. Plant. 2019;12:170–184. doi: 10.1016/j.molp.2018.12.011. [DOI] [PubMed] [Google Scholar]
- Xhauflair H., Revel N., Vitales T.J., Callado J.R., Tandang D., Gaillard C., Forestier H., Dizon E., Pawlik A. What plants might potentially have been used in the forests of prehistoric southeast Asia? An insight from the resources used nowadays by local communities in the forested highlands of Palawan island. Quat. Int. 2017;448:169–189. [Google Scholar]
- Yang W., Chen L., Zhao J., Wang J., Li W., Yang T., Dong J., Ma Y., Zhou L., Chen J., et al. Genome-wide association study of pericarp color in rice using different germplasm and phenotyping methods reveals different genetic architectures. Front. Plant Sci. 2022;13:841191. doi: 10.3389/fpls.2022.841191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Yang T., Qi Q., Du Y., Shi J., Liu X., Liu Y., Zhang H., Zhang Z., Yan N. Comparison of the contents of phenolic compounds including flavonoids and antioxidant activity of rice (Oryza sativa) and Chinese wild rice (Zizania latifolia) Food Chem. 2021;344 doi: 10.1016/j.foodchem.2020.128600. [DOI] [PubMed] [Google Scholar]
- Zainal-Abidin R.A., Zainal Z., Mohamed-Hussein Z.A., Abu-Bakar N., Ab Razak M.S.F., Simoh S., Sew Y.S. RNA-seq data from whole rice grains of pigmented and non-pigmented Malaysian rice varieties. Data Brief. 2020;30 doi: 10.1016/j.dib.2020.105432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.D., Pei Q., Lee H.F., Zhang J., Chang C.Q., Li B., Li J., Zhang X. The Pulse of imperial China: a quantitative analysis of long-term geopolitical and climatic cycles. Global Ecol. Biogeogr. 2015;24:87–96. [Google Scholar]
- Zhao X., Guo Y., Kang L., Yin C., Bi A., Xu D., Zhang Z., Zhang J., Yang X., Xu J., et al. Population genomics unravels the Holocene history of bread wheat and its relatives. Nat. Plants. 2023;9:403–419. doi: 10.1038/s41477-023-01367-3. [DOI] [PubMed] [Google Scholar]
- Zheng X., Pang H., Wang J., Yao X., Song Y., Li F., Lou D., Ge J., Zhao Z., Qiao W., et al. Genomic signatures of domestication and adaptation during geographical expansions of rice cultivation. Plant Biotechnol. J. 2021;20:16–18. doi: 10.1111/pbi.13730. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data are available at the China National Genomics Data Center (https://ngdc.cncb.ac.cn) under BioProject accession number PRJCA017658.